Abstract
The ability of Candida albicans to switch cellular morphologies is crucial for its ability to cause infection. Because the cell cycle machinery participates in Saccharomyces cerevisiae filamentous growth, we characterized in detail the two C. albicans B-type cyclins, CLB2 and CLB4, to better understand the molecular mechanisms that underlie the C. albicans morphogenic switch. Both Clb2p and Clb4p levels are cell cycle regulated, peaking at G2/M and declining before mitotic exit. On hyphal induction, the accumulation of the G1 cyclin Cln1p was prolonged, whereas the accumulation of both Clb proteins was delayed when compared with yeast form cells, indicating that CLB2 and CLB4 are differentially regulated in the two morphologies and that the dynamics of cyclin appearance differs between yeast and hyphal forms of growth. Clb2p-depleted cells were inviable and arrested with hyper-elongated projections containing two nuclei, suggesting that Clb2p is not required for entry into mitosis. Unlike Clb2p-depleted cells, Clb4p-depleted cells were viable and formed constitutive pseudohyphae. Clb proteins lacking destruction box domains blocked cell cycle progression resulting in the formation of long projections, indicating that both Clb2p and Clb4p must be degraded before mitotic exit. In addition, overexpression of either B-type cyclin reduced the extent of filamentous growth. Taken together, these data indicate that Clb2p and Clb4p regulate C. albicans morphogenesis by negatively regulating polarized growth.
INTRODUCTION
Candida albicans, an important human pathogen, is multimorphic, forming yeast, pseudohyphae, and true hyphae depending on the growth conditions. Mutations that lock the organism in a particular morphology reduce virulence, leading to the suggestion that the ability to switch between the morphologies is important for virulence (Mitchell, 1998; Gow et al., 2002). A widely accepted model suggests that yeast form cells are essential for efficient dissemination through the body, whereas the filamentous forms are required for tissue invasion (Ernst, 2000). Understanding how C. albicans regulates and executes these morphogenic transitions is vital to understanding the pathogenesis of C. albicans infections.
Analogous to the growth of S. cerevisiae, C. albicans yeast cells are round to ovoid shaped, grow by budding, and readily separate at sites of septation. Pseudohyphal cells also grow by budding and display distinct constrictions at septa, although they are more elongated and do not readily separate. In contrast to both yeast and pseudohyphal cells, true hyphal cells form long thin tubes with parallel cell walls that lack obvious septal constrictions.
Cellular differences between the three morphologies include extent of polarization, the ability to separate after mitosis, placement of the septin ring, and movement of the nuclei (Sudbery et al., 2004). Actin cables and patches play a central role in cell morphology by marking sites of cell wall expansion and thus affecting the extent of cell polarization in the different morphological forms (Anderson and Soll, 1986). Cell separation is also unique in the three morphologies. Yeast form daughter cells readily separate from their mothers, whereas pseudohyphal cells remain attached, forming long chains of cells. Cells within the true hyphal tubes do not separate (Mitchell and Soll, 1979; Sudbery, 2001). Septin ring placement occurs at the mother-bud neck of yeast and pseudohyphal cells and persists through mitosis. In contrast, a more diffuse septin band appears at the mother-germ tube neck of hyphal cells during early germ tube evagination. This basal septin band disappears and a stable septin ring forms, usually within the tube, ∼10–15 μm from the mothergerm tube junction (Sudbery, 2001).
In S. cerevisiae, the connection between cell cycle progression and filamentous growth has been well established (Rua et al., 2001). Pseudohyphal cells undergo longer periods of polarized growth and an extended G2 phase compared with yeast form cells. Regulation of this extended G2 phase is thought to occur through the cyclin-dependent kinase, Cdc28p (Kron et al., 1994; Ahn et al., 2001) and its association with both G1 and mitotic cyclins. The G1 cyclins, Cln1p and Cln2p, both positively regulate pseudohyphal growth: overexpression of either CLN causes elongated growth (Oehlen et al., 1998; Loeb et al., 1999a; Madhani et al., 1999). In contrast, the mitotic cyclin Clb2p negatively regulates pseudohyphal growth: clb2Δ strains exhibit elongated bud growth and a delay in G2/M phase (Ahn et al., 1999). Effectors of Cdk-cyclin activity, including the Swe1p kinase, and the transcriptional repressors Xbp1p and Whi3p, also affect the degree of filamentation in S. cerevisiae cells (Mosch and Fink, 1997; Edgington et al., 1999; Gari et al., 2001; Miled et al., 2001).
Evidence for cell cycle–regulated filamentous growth in C. albicans also exists, but has been somewhat controversial. Soll et al. (1985) quantitatively demonstrated a commitment point after which a budding cell, when challenged with high temperature and high pH medium, cannot form hyphal projections until the next cell cycle. This observation was recently challenged by Liu and colleagues (Hazan et al., 2002), who reported that yeast form cells exposed to serum at high temperature can induce hyphal-like growth throughout the cell cycle. The difference may be due to the conditions used in the two studies; serum is a very potent inducer, whereas pH is a weaker inducer of hyphal growth. Another difference is the semantic definition of hyphal cells: those induced with serum later in the cell cycle formed “bottle nose” shaped cells that had constrictions at the mother-bud neck and thus do not conform to the definition of a true hyphal cell (septum within a narrow, parallel-walled tube).
We previously demonstrated the importance of Fkh2p, a potential cell cycle regulatory transcription factor, in C. albicans morphogenesis and virulence (Bensen et al., 2002). Fkh2p affects the transcription of multiple transcripts, including a putative B-type cyclin (previously termed CYB99 and now named CLB4). Furthermore, S. cerevisiae Fkh2p regulates a group of cell cycle–regulated transcripts including CLB2, the major mitotic cyclin. To determine the role of mitotic cyclins in C. albicans morphogenesis, we characterized the two C. albicans B-type cyclins, CLB2 and CLB4. Our results indicate that CLB2 is an essential cyclin, whereas CLB4 is not essential, that both cyclins affect morphogenesis and thus are candidates for regulating hyphal growth, and that both B-cyclins are negative regulators of polarized growth, although deleting them causes very different morphologies.
MATERIALS AND METHODS
Media and Growth Conditions
Rich media (YPAD), synthetic complete media (SDC), and synthetic minimal medium lacking specific nutrients have been described previously (Sherman, 1991). Hyphal growth was induced with YPAD + 10% bovine calf serum at 37°C (Sigma, St. Louis, MO). Ura– auxotrophs were selected on SDC containing 0.1% 5-fluoroorotic acid (5-FOA) and 80 mg/l uridine. All media contained 80 mg/l uridine except when Ura+ prototrophs were being selected.
Plasmids
To place CLB2 under transcriptional control of the MET3 promoter, the 5′-end of CLB2 was amplified by PCR from genomic DNA isolated from strain SC5314 (Gillum et al., 1984) using primer 635 engineered to include a BamHI site (underlined) (5′-GGCGGATCCATGCCACAAGTCACTAAAACT-3′) and primer 636 engineered to include a PstI site (underlined) (5′-GGGCCTGCAGATTTGAACCACTTCAACAGA-3′). The PCR product was digested with BamHI and PstI and ligated into pBluescript II SK+ (Invitrogen, Carlsbad, CA) digested with BamHI and PstI, generating pBSK-CLB2–5′-2. The BamHI/PstI fragment from pBSK-CLB2–5′-2 was subcloned into pCaDIS (Care et al., 1999) digested with BamHI and PstI to generate pDIS-CLB2–5′. To place CLB4 under transcriptional control of the MET3 promoter, the 5′-end of CLB4 was amplified by PCR from genomic DNA isolated from strain SC5314 using primers 637 engineered to include a BamHI site (5′-GGCGGATCCATGCGATCTTATAAATCATCC-3′) and 638 engineered to include a PstI site (5′-GGGCCTGCAGCTGTTTTCTTTATCTTCTTCGTC-3′). The PCR product was digested with BamHI and PstI and ligated into pCaDIS digested with BamHI and PstI generating pDIS-CLB4–5′. The PCR derived CLB2 and CLB4 fragments in pDIS-CLB2–5′ and pDIS-CLB4–5′ were sequenced and found to match the genomic sequences of orf6.7127 and orf6.8006, respectively, at the Stanford Genome Technology Center (http://www-sequence.stanford.edu/group/candida/index.html).
To HA tag CLB2, the last 1100 nucleotides of CLB2 were amplified by PCR from genomic DNA isolated from strain BWP17 using primer CLB2-FLAG5 (5′-GGCCCTGCAGGATGCCACAAGTCACTAAAACTAATAATG-3′) and primer CLB2–3HA engineered to include a EcoRV site (5′-GCTTGATATCCTCTTCTGCTTCTGCTACCAC-3′). The PCR product was ligated into pBluescript II SK+ (Invitrogen), generating pBSK-CLB2HA. The SalI/EcoRV fragment (1.1 kb) from pBSK-CLB2HA was subcloned into pCaHA (a gift from Peter Sudbery) digested with SalI and EcoRV, generating the plasmid pCaHA-CLB2. To place CLB2HA under transcriptional control of the ACT1 promoter, a two-step strategy was followed. First, the CLB2 open reading frame (ORF) was amplified by PCR from genomic DNA isolated from strain BWP17 using primer CLB2-FLAG5 engineered to include a Sse8387I site (5′-GGCCCTGCAGGATGCCACAAGTCACTAAAACTAATAATG-3′) and primer CLB2-FLAG3 engineered to include a SphI site (5′-GCCGCATGCCTCTTCTGCTTCTGCTACCACTATTTC-3′). The PCR product was cloned at the EcoRV site of pBluescript II SK+ generating pBSK-CLB2. The Sse8387I/SphI fragment from pBSK-CLB2 was subcloned into pFLAG-Act1 (Umeyama et al., 2002) digested with Sse8387I and SphI, generating pFLAGAct1-CLB2. Second, the CLB2HA fragment present in pCaHA-CLB2 was amplified using the primer pCaHA-5 (5′-AGGGAACAAAATCTGGG-3′) and primer pCaHA-3′ engineered to include a SphI site (5′-ACACATGCATGCTTAACCGGCATAGTCTGG-3′) and cloned into pGEM vector, generating pGEM-CLB2HA. Finally, the SalI/SphI fragment of CLB2 presented in pFLAGAct1-CLB2 was substituted by the SalI/SphI fragment of CLB2HA presented in pGEM-CLB2HA, generating pFLAGAct1-CLB2HA. To HA tag CLB4, the last 1100 nucleotides of CLB4 were amplified by PCR from genomic DNA isolated from strain BWP17 using primer CLB4Flag5 (5′-GGCCCTGCAGGATGCGATCTTATAAATCATCCATAACGG-3′) and primer CLB4–3HA engineered to include a EcoRV site (5′-GCTTGATATCACTCTGGGACATTATGTGACG-3′). The PCR product was ligated into pBluescript II SK+ (Invitrogen), generating pBSK-CLB4HA. The EcoRV fragment (1.1 kb) from pBSK-CLB4HA was subcloned into pCaHA digested with EcoRV to generate pCaHA-CLB4. To place CLB4HA under transcriptional control of the ACT1 promoter, a two-step strategy was followed. First, the CLB4 ORF was amplified by PCR from genomic DNA isolated from strain BWP17 using primer CLB4-FLAG5 engineered to include a Sse8387I site (5′-GGCCCTGCAGGATGCGATCTTATAAATCATCCATAACGG-3′) and primer CLB4-FLAG3 engineered to include a XhoI site (5′-GGCCTCGAGACTCTGGGACATTATGTGACGAAAATATTC-3′). The PCR product was cloned at the EcoRV site of pBluescript II SK+ generating pBSK-CLB4. The Sse8387I/XhoI fragment from pBSK-CLB4 was subcloned into pFLAG-Act1 digested with Sse8387I and XhoI, generating pFLAGAct1-CLB4. Second, the CLB4HA fragment present in pCaHA-CLB4 was amplified using the primer pCaHA-5 (5′-AGGGAACAAAATCTGGG-3′) and primer pCaHA-3′ engineered to include a SphI site (5′-ACACATGCATGCTTAACCGGCATAGTCTGG-3′). The PCR product was digested with SphI, and the generated 750-base pairs fragment was used to substitute the SphI fragment of CLB4 presented in pFLAGAct1-CLB4 generating pFLAGAct1-CLB4HA. Sequence of all cloned fragments in pCaHA-CLB2, pFLAGAct1-CLB2HA, pCaHA-CLB4, and pFLAGAct1-CLB4HA were confirmed by DNA sequencing.
To generate pMG2093, a HindIII-BglII fragment containing 13 myc epitopes from plasmid pFA6a-13myc-TRP1 (Longtine et al., 1998) was ligated into pGFP-HIS1 (Gerami-Nejad et al., 2001) digested with HindIII and BglII.
Strains
C. albicans strains used in this study are shown in Table 1. All strains were checked for correct genome integration by PCR and/or Southern analysis (unpublished data). Disruption strains were constructed using a previously described PCR-mediated gene disruption system (Wilson et al., 1999).
Table 1.
Yeast strains used in this study
| Strain | Geneotype | Source |
|---|---|---|
| SC5314 | Prototrophic | Gillum et al. (1984) |
| BWP17 | ura3Δ::λimm434/ura3Δ:λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG | Wilson et al. (1999) |
| YJB5221 | BWP17 clb2::HIS1/CLB2 | This study |
| YJB5919 | BWP17 clb2::HIS1/PMET3-CLB2:URA3::clb2 | This study |
| YJB6144 | BWP17 clb4::ARG4/CLB4 | This study |
| YJB6145 | BWP17 clb4::ARG4/CLB4 | This study |
| YJB6146 | BWP17 clb4::ARG4/CLB4 | This study |
| YJB6147 | ura3Δ::λimm434/ura3Δ:λimm434 HIS1::his1::hisG/his1::hisG arg4::hisG/arg4::hisG | This study |
| YJB6284 | ura3Δ::λimm434/ura3Δ::λimm434 HIS1::his1::hisG/his1::hisG ARG4:URA3::arg4::hisG/arg4::hisG | Bensen et al. (2002) |
| YJB6407 | BWP17 clb4::ARG4/PMET3-CLB4:URA3::clb4 | This study |
| YJB6409 | BWP17 clb4::ARG4/PMET3-CLB4:URA3::clb4 | This study |
| YJB6667 | BWP17 clb4::ARG4/clb4::HIS1 | This study |
| YJB6854 | ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG ARG4:URA3::arg4::hisG/arg4::hisG clb4::ARG4/clb4::HIS1 | This study |
| YJB7421 | BWP17 clb4::ARG4/PMET3-CLB4:URA3::clb4 TUB1-YFP:HIS1/TUB1 | This study |
| YJB7783 | ura3Δ::λimm434/ura3Δ:λimm434 HIS1::his1::hisG/his1::hisG ARG4::arg4::hisG/arg4::hisG | This study |
| YJB7820 | ura3Δ::λimm434/ura3Δ:λimm434 his1::hisG/his1::hisG ARG4::arg4::hisG/arg4::hisG clb2::HIS1//PMET3-CLB2:URA3::clb2 | This study |
| YJB7824 | ura3Δ::λimm434/ura3Δ:λimm434 HIS1::his1::hisG/his1::hisG arg4::hisG/arg4::hisG clb4::ARG4/PMET3-CLB4:URA3::clb4 | This study |
| YJB7878 | ura3Δ::λimm434/ura3Δ:λimm434 HIS1::his1::hisG/his1::hisG ARG4::arg4::hisG/arg4::hisG PMET3:URA3::RP10/RP10 | This study |
| YJB8333 | YJB7783 PMET3-clb2Δdb:URA3/CLB2 | This study |
| YJB8339 | YJB7783 PMET3-clb4Δdb:URA3/CLB4 | This study |
| YJB8365 | BWP17 clb2::ARG4/CLB2 | This study |
| YJB8366 | BWP17 clb2::ARG4/CLB2 | This study |
| YJB8447 | BWP17 clb2::ARG4/PMET3-CLB2:URA3::clb2 | This study |
| YJB8450 | BWP17 clb2::ARG4/PMET3-CLB2:URA3::clb2 | This study |
| YJB8606 | BWP17 clb2::ARG4/PMET3-CLB2:URA3::clb2 TUB1-GFP:HIS1/TUB1 | This study |
| YJB8744 | BWP17 PMET3-CLB2-Myc:HIS1:URA3::clb2/clb2::ARG4 | This study |
| YJB8761 | BWP17 PMET3-CLB2:URA3/CLB2 | This study |
| YJB8763 | BWP17 PMET3-clb2Δdb:URA3/CLB2 | This study |
| YJB8765 | BWP17 PMET3-CLB4:URA3/CLB4 | This study |
| YJB8768 | BWP17 PMET3-clb4Δdb:URA3/CLB4 | This study |
| YJB8784 | BWP17 PMET3-CLB2-Myc:HIS1:URA3/CLB2 | This study |
| YJB8785 | BWP17 PMET3-clb2Δdb-Myc:HIS1:URA3/CLB2 | This study |
| YJB8787 | BWP17 PMET3-CLB4-Myc:HIS1:URA3/CLB4 | This study |
| YJB8795 | BWP17 PMET3-CLB4-Myc:HIS1:URA3::clb4/clb4::ARG4 | This study |
| YJB8796 | BWP17 PMET3-clb4Δdb-Myc:HIS1:URA3/CLB4 | This study |
| YJB6983 | BWP17 clb2::UAU1/CLB2 | This study |
| YJB6985 | BWP17 clb4::UAU1/CLB4 | This study |
| JCB64 | BWP17 CLB2/CLB2-HA:URA3 | This study |
| JCB77 | BWP17 CLB4/CLB4-HA:URA3 | This study |
| JCB126 | BWP17 RP10::pFLAGAct1-CLB2HA::URA3 | This study |
| JCB103 | BWP17 RP10::pFLAGAct-CLB4HA::URA3 | This study |
| JCB157 | BWP17 clb4::HIS1//CLB4-HA:URA3 | This study |
| JCB158 | BWP17 CLB2/clb2:HIS1 | This study |
| JCB169 | BWP17 clb2::HIS1/CLB2-HA:URA3 | This study |
clb2::HIS1 and clb2::ARG4 PCR products were amplified using primers 597 (5′-CATAGTAATGCCACAAGTCACTAAAACTAATAATGAAAATGAGTTTAGACTTACTAGATCAAAAGGTTTTCCCAGTCACGACGTT-3′) and 598 (5′-GCATTGTAAAGATAAGAACCTAGATCCAATAGTCATCAAAACTTTACTCTTCTGCTTCTGCTACCTGTGGAATTGTGAGCGGATA-3′) with plasmids pGEM-HIS1 or pRS-ARG4ΔSpeI, respectively (Wilson et al., 1999). The clb2::HIS1 product was transformed into BWP17 (Wilson et al., 1999) to generate the heterozygous strain YJB5221. The clb2::ARG4 product was transformed into BWP17 to generate heterozygous strains YJB8365 and YJB8366. CLB2 was placed under control of the MET3 promoter by transforming YJB5221, YJB8365, and YJB8366 with pDIS-CLB2–5′ linearized with ClaI generating YJB5919, YJB8447, and YJB8450, respectively. YJB5919 was made prototrophic by transforming it with pRS-ARG4ΔSpeI digested with EcoRI. Multiple CLB4 heterozygotes were constructed by transforming BWP17 with a clb4::ARG4 PCR product amplified using the primers 722 (5′-GGTTCACAAATTATGCGATCTTATAAATCATCCATAACGGATGAAAATGAGTTGACAAAACAAAGACTTAGTTTTCCCAGTCACGACGTT-3′) and 723 (5′-TTTGGTTAGATTGAATAAAATGTAAAGTACCGCATTGTTGGAAACTTGTCTTTAGATCAACTCTGGGACATTGTGGAATTGTGAGCGGATA-3′) with pRS-ARG4ΔSpeI as DNA template generating strains YJB6144, YJB6145, and YJB6146. The MET3 promoter was inserted 5′ to CLB4 by transforming YJB6144 and YJB6145 with pDIS-CLB4–5′ digested with EcoRV generating YJB6407 and 6409, respectively. A homozygous CLB4 deletion strain was constructed by transforming YJB6146 with a clb4::HIS1 PCR product (amplified using primers 722 and 723 with pGEM-HIS1 as template) generating YJB6667. YJB6407 and YJB6667 were made prototrophic by transformation with NotI-digested pRS-ARG-URA-BN generating YJB7824 and YJB6854, respectively.
Tub1p was tagged at the C-terminus with green fluorescent protein (GFP) or yellow fluorescent protein (YFP) by PCR-mediated gene modification (Gerami-Nejad et al., 2001) using primers 650 (5′-TTGGCTGCTTTAGAGAGAGATTATATTGAAGTTGGTACTGATTCTTTCCCTGAAGAAGAAGAAGAATATGGTGGTGGTTCTAAAGGTGAAGAATTATT-3′) and 719 (5′-ATAATGAAAAACCCAGACCTTTGTAATTAAAAAAATTTAAACATTAGCAACAAAGTAAGAACACGATCAAGAATTCCGGAATATTTATGAGAAAC-3′) with pGFP-HIS1 or pYFP-HIS1 (Gerami-Nejad et al., 2001) as the template. The TUB1-GFP:HIS1 fragment was transformed into YJB8450, generating YJB8606. The TUB1-YFP:HIS1 fragment was transformed into YJB6409 generating, YJB7421.
The destruction box of CLB2 was removed by transforming a PMET3-clb2Δdb:URA3 PCR fragment, amplified with primers 1247 (5′-CTAAGTAGTACATCTGCAAATACTACATCTACAGTCAAAAATAGGTGAGAGCAACCACAAGAAGAGAAATTCTAGAAGGACCACCTTTGATTG-3′) and 1522 (5′-TTTGTAGAGGGGCATTGTCTCCACCCAATGGTTTTCTTTTATTTGTTGGTTTGGCATTTGTTATGTATTGCATTTTAATAAACGCGGATCC-3′) with pURA3-PMET3-GFP (Gerami-Nejad et al., 2004) as template, into strains YJB7783 and BWP17, generating YJB8333 and YJB8763, respectively. This places the MET3 promoter with an ATG codon adjacent to the codon encoding amino acid 76 of CLB2. The destruction box of CLB4 was removed by transforming a PMET3-clb4Δdb:URA3 PCR fragment, amplified with primers 1249 (5′-GGATTTCCAACTATTTGCTATCAAAGGTAAATAACTACATTCGTTTAAATTGTGTTTTGCGGTCCACTCGTCTAGAAGGACCACCTTTGATTG-3′) and 1523 (5′-TTGAGCTGTTTCTGTGATGATTGGTGTTTGTTTGTGTTAGTTTCACTCTTGGGTGATTTTTATTCTCCTGCATTTTAATAAACGCGGATCC-3′) with pURA3-PMET3-GFP as template, into strains YJB7783 and BWP17, generating YJB8339 and YJB8768, respectively. This places the MET3 promoter with an ATG codon adjacent to the codon encoding amino acid 53 of CLB4.
BWP17 was sequentially transformed with NruI-linearized pGEM-HIS1 to generate the His+ prototroph YJB6147 followed by transformation with pRS-ARG4ΔSpeI digested with EcoRI generating the His+ Arg+ prototroph YJB7783. A MET3 promoter control strain was generated by transforming BglII-linearized pCaEXP (Care et al., 1999) into YJB7783 forming YJB7878. This strain has the MET3 promoter integrated near the RP10 locus but does not impart transcriptional activity to an ORF.
CLB2 and CLB4 were disrupted with the UAU1 cassette by amplifying a clb2::UAU1 fragment using primers 597 and 598 and a clb4:UAU1 fragment using primers 722 and 723 with the plasmid pBME101 (Enloe et al., 2000) as DNA template. The clb2::UAU1 and clb4::UAU1 fragments were transformed independently into BWP17 to generate JBY6983 and JBY6985, respectively.
CLB2 and CLB4 were placed under control of the MET3 promoter by transforming BWP17 with PMET3-CYB1:URA3 (amplified with primers 1525 (5′-CTTGATGCTGTACTTTTGATCTAGTAAGTCTAAACTCATTTTCATTATTAGTTTTAGTGACTTGTGGCATTTTAATAAACGCGGATCC-3′ and 1247 using plasmid pURA3-PMET3-GFP as template) and PMET3-CLB4:URA3 (amplified with primers 1524 (5′-TACTTTTGGCTCTAAGTCTTTGTTTTGTCAACTCATTTTCATCCGTTATGGATGATTTATAAGATCGCATTTTAATAAACGCGGATCC-3′ and 1249 using plasmid pURA3-PMET3-GFP as template) fragments generating strains YJB8761 and YJB8765, respectively.
To Myc tag CLB2, YJB8447, YJB8761, and YJB8763 were transformed with a CLB2-Myc:HIS1 fragment amplified from primers 1441 (5′-GCACATAACTCATGTTCATCTTCTTTCATTTCCTCATTTATGCATTGTAAAGATAAGAACCTAGATCCAAGAATTCCGGAATATTTATGAGAAAC-3′) and 1017 (5′-ATCGACCCATAGGCTAACATTAGAAGATGATGACGAAGAAGAAGAAATAGTGGTAGCAGAAGCAGAAGAGGGTGGTGGTCGGATCCCCGGGTTAATTAA-3′) with plasmid pMG2093 generating YJB8744, YJB8784, and YJB8785, respectively. To Myc tag CLB4, YJB6407, YJB8765, and YJB8768 were transformed with a CLB4-Myc:HIS1 fragment amplified from primers 1442 (5′-AGTACTTAACCAATGCATAGAATTAATGATTTTGGTTAGATTGAATAAAATGTAAAGTACCGCATTGTTGGAATTCCGGAATATTTATGAGAAAC-3′) and 1019 (5′-CAAGGAAAGAAGGTATAGAAAAAGTTCACTTTTTGTTCAAGAATATTTTCGTCACATAATGTCCCAGAGTGGTGGTGGTCGGATCCCCGGGTTAATTAA-3′) with plasmid p2093 generating YJB8795, YJB8787 and YJB8796, respectively.
The CLB2 gene was HA tagged by transforming BWP17 with pCaHA-CLB2 linearized with ClaI generating strain JCB64. The CLB4 gene was HA tagged by transforming BWP17 strain with pCaHA-CLB4 partially digested with SacI, generating strain JCB77. CLB2HA was placed under transcriptional control of ACT1 promoter by transforming BWP17 with pFLAGAct1-CLB2HA linearized with StuI to target the integration of the plasmid into the RP10 locus, generating strain JCB126. CLB4HA was placed under transcriptional control of the ACT1 promoter by transforming BWP17 with pFLAGAct1-CLB4HA linearized with StuI to target the integration of the plasmid into the RP10 locus, generating strain JCB103.
To demonstrate HA-tagged cyclins were functional, BWP17 was transformed with a clb2::HIS1 fragment amplified from primers S1CLB2 (5′-ACGACACAATAAATTTTAATTCATTAATCAACCAACGAACCAGCCAAACCAAAATTAATTCACATTTATACTCACTGTTTGTCATTTTCATCTCATAGTAGAAGCTTCGTACGCTGCAGGTC-3′) and S2CLB2 (5′-CAAATTATAGGGTAATGCACATAACTCATGTTCATCTTCTTTCATTTCCTCATTTATGCATTGTAAAGATAAGAACCTAGATCCAATAGTCATCAAAACTTCTGATATCATCGATGAATTCGAG-3′) with plasmid pFA-HIS1 (Gola et al., 2003) generating strain JCB158. The remaining CLB2 allele in JCB158 was HA tagged by transforming JCB158 with pCaHA-CLB2 linearized with ClaI generating strain JCB169. JCB77 was transformed with a clb4::HIS1 fragment amplified from primers S1CLB4 (5′-ATAGAAATAGAGTCTGTATTCAGTCGATTGATTTTGGTGTTTTTATTTTTATCTTTTTTTTTGAACAACACGCTCATTTATTCCAGAGGTTCACAACATTGAAGCTTCGTACGCTGCAGGTC-3′) and S2CLB4 (5′-AGCTTCAACAAATCAGTACTTAACCAATGCATAGAATTAATGATTTTGGTTAGATTGAATAAAATGTAAAGTACCGCATTGTTGGAAACTTGTCTTTAGATCTGATATCATCGATGAATTCGAG-3′) with plasmid pFA-HIS1 generating strain JCB157.
Cell Synchronization
To isolate unbudded G1 cells, strains were grown at 30°C in YPD to OD600 = 1.5, sonicated, and then loaded into the separation chamber of a JE-5.0 elutriation system (Beckman Coulter, Fullerton, CA) maintained at 4200 rpm and a flow rate of 32 ml/min. After loading, fresh YPD medium was used to recover the cells. To collect the cells from the chamber, the rate was gradually reduced to 3800 rpm and the outflow was gradually increased. Unbudded cells were collected, concentrated by centrifugation, and then released into fresh prewarmed medium. Samples were taken every 15 min and protein extracts were made.
Protein Extract Preparation and Western Blotting
Figure 1 and Figure 7: Total protein extracts were prepared from 1.6 × 108 frozen cells. Cells were resuspended in 20 μl of RIPA buffer (10 mM sodium phosphate, 1% Triton X-100, 0.1% SDS, 10 mM EDTA, 150 mM NaCl, pH 7) and 200 μl of glass beads (0.4 mm) were added. Cells were broken for 20 s in a Hybaid Ribolyser (Hybaid, Teddington, Middlesex, United Kingdom), and the crude extract was recovered by washing with 200 μl of RIPA. Soluble extracts were obtained by centrifugation of total extracts at 12,000 rpm during 10 min at 4°C. For Western blots, 30 μg of protein extract was run on a 8% SDS-polyacrylamide gel, transferred to Hybond-P (Amersham Biosciences, Piscataway, NJ) membrane and probed with anti-HA (12CA5, 1:500), anti-PSTAIRE (Santa Cruz Biotechnology, Santa Cruz, CA, 1:3000), and anti-MYC (9E10, 1:500) antibodies. Secondary antibodies conjugated to horseradish peroxidase were diluted 1:15.000. Immunoblots were developed using the Supersignal West Pico kit (Pierce Biotechnology, Rockford, IL). Quantification of Westerns was performed by densitometric analysis.
Figure 1.
Clb2p and Clb4p accumulation is delayed in hyphal cells. G1 cells in logarithmic growing cultures of strains JCB64 (Clb2-HAp), JCB77 (Clb4-HAp), and YJB8228 (Cln1-MYCp) were isolated by elutriation (see Materials and Methods) and released into YPD at 30°C (yeast form growth) or YPD + 5% fetal calf serum (FCS) at 37°C (hyphal growth). At 15-min intervals after release, cells were collected and assayed by Western blotting for cyclin levels (B). Cdc28p (anti-PSTAIRE) was used as a loading control. (A) The amount of each cyclin relative to Cdc28p was plotted against time for yeast form (black line) and hyphal growth conditions (red line). Buds and germ tubes were scored as indicated in legend.
Figure 7.
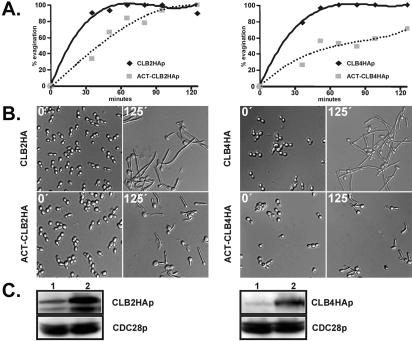
Clb2p and Clb4p inhibit filamentous growth. Overnight cultures of strains JCB64 (PCLB2-CLB2-HA), JCB126 (PACT1-CBL2-HA), JCB77 (PCLB4-CLB4-HA), and JCB103 (PACT1-CLB4-HA) were diluted in YPD medium, grown at 30°C for 3 h, collected by centrifugation, and resuspended in prewarmed YPD + 10% FCS at 37°C to a final cell density of 0.3 OD600. At the indicated times, cells were removed, photographed (B), and scored for % evagination (A; cells with germ tubes/total cells). (C) Western blot showing protein levels of cyclins at time 0. (C, left) lane 1, JCB64 (PCLB2-CLB2-HA); lane 2, JCB126 (PACT1-CBL2-HA). (C, right) Lane 1, JCB77 (PCLB4-CLB4-HA); lane 2, JCB103 (PACT1-CLB4-HA). Cdc28p, detected with anti-PSTAIRE, is shown as a loading control.
Figure 2, Figure 5, and Figure 8: Total protein extracts were prepared by lysing cells with 0.2 g glass beads (0.5 mm) and 50 μl 2% SDS with vigorous vortexing for 90 s in 13 × 100-mm glass tubes. Tubes were heated to 100°C for 3 min followed by the addition of 0.25 ml Laemmli sample buffer (Laemmli, 1970) and returned to 100°C for 1 min. Lysates were transferred to fresh microfuge tubes and centrifuged at 16,000 × g for 10 min. Supernatants were boiled for 3 min before separation by SDS-PAGE. Proteins were transferred to a PVDF membrane, blocked for 1 h with 4% skimmed milk in TBST (20 mM Tris, pH 7.6, 137 mM NaCl, 0.1% Tween-20), and incubated overnight at 4°C with anti-PSTAIRE antibody (Santa Cruz Biotechnology) diluted 1:10,000 or anti-Myc antibody (9E10) diluted 1:500. Secondary antibodies conjugated to horseradish peroxidase were diluted 1:10.000. Immunoblots were developed using the Supersignal West Pico kit (Pierce Biotechnology). Quantification of Westerns was performed by densitometric analysis.
Figure 2.
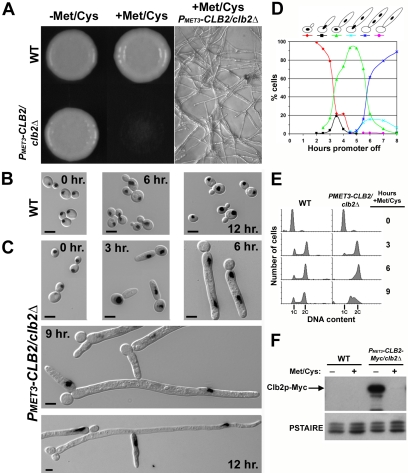
Clb2p-depleted cells display elongated filaments. (A) Stationary phase cultures of strains YJB7878 (WT) and YJB7820 (PMET3-CLB2/clb2Δ) grown in SDC –Met –Cys (promoter on) were spotted on SDC –Met –Cys agar or SDC +Met +Cys agar (promoter off) and allowed to grow at 30°C for 3 d. The right panel is a magnified region of the SDC +Met +Cys plate, where PMET3-CLB2/clb2Δ. (B) YJB7878 (WT) and (C) YJB7820 (PMET3-CLB2/clb2Δ) were grown to stationary in SDC –Met –Cys conditions and released into SDC +Met +Cys. Aliquots were taken at the indicated times after release, fixed in ethanol, and stained with DAPI to visualize nuclei. Shown are merged images of DIC and DAPI micrographs. Bar, 5 μm. (D) Nuclear migration in Clb2p-depleted cells. A saturated culture of YJB7820 (PMET3-CLB2/clb2Δ) grown in SDC –Met –Cys (promoter on) was released into SDC +Met +Cys (promoter off) at 30°C. Aliquots were removed at the indicated times, fixed in ethanol, and stained with Sytox Green (see Materials and Methods). Sytox Green stained nuclear material in Clb2p-depleted cells was scored as follows: small-budded with one stained spot (nucleus) in the mother (red), elongated bud tube with one spot at the mother-bud neck (black), one spot in mother and one spot in the germ tube (green), one spot at the mother-tube neck and one spot within the tube (turquoise), two spots within the tube (blue), one spot only, located in the tube (purple). (E) Flow cytometric analysis of Clb2p-depleted cells. Sytox Green–stained Clb2p-depleted cells from (A) and YJB7878 (WT) were analyzed for DNA content by flow cytometry. 2C and 4C DNA content is shown below each histogram. (F) Clb2p-Myc protein levels of strains YJB7878 (WT) and YJB8744 (PMET3-CLB2-Myc/clb2Δ) grown for 6 h in SDC +Met +Cys (promoter off) or SDC –Met –Cys (promoter on). Bars, 5 μm.
Figure 5.

clb4Δ/clb4Δ and Clb4p-depleted cells display constitutive pseudohyphal growth. (A) YJB6284 (WT) and (B and D) YJB6854 (clb4Δ/clb4Δ) cells were grown to logarithmic phase in SDC, fixed in ethanol, and labeled with (A and B) DAPI or (D) Calcofluor White. (C) YJB7824 (PMET3-CLB4/clb4Δ) was grown to saturation in SDC –Met –Cys (promoter on) and released into SDC +Met +Cys (promoter off) at 30°C. Cells were removed at the indicated times, fixed with ethanol, and stained with DAPI. (A–C) Merged images of DIC and inverse fluorescent micrographs. (F) Clp4p-Myc protein levels of strains YJB7878 (WT) and YJB8795 (PMET3-CLB4-Myc/clb4Δ) grown for 6 h in SDC +Met +Cys (promoter off) or SDC –Met –Cys (promoter on). Bars, 5 μm.
Figure 8.
Clb2p and Clb4p must be degraded to exit mitosis. Saturated cultures of (A) YJB8333 (PMET3-clb2Δdb/CLB2) and (B) YJB8339 (PMET3-clb4Δdb/CLB4) grown in SDC +Met +Cys (promoter off) were diluted into SDC –Met –Cys (promoter on) and incubated at 30°C. At the indicated times, cells were removed and stained with Sytox Green as described in Figure 3A (A and B) or Calcofluor White (C). Shown are merged images of DIC and inverted fluorescence micrographs (A and B) or DIC (C, left) and fluorescence micrographs (C, right). Cells in A and B were scored for nuclear content as described in Figure 3A and graphed (A, and B, right). (D) Western blot of cell lysates from stratins YJB7878 (WT), YJB8784 (PMET3-CLB-Myc/CLB2), YJB8785 (PMET3-clb2Δdb-Myc/CLB2), YJB8787 (PMET3-CLB4-Myc/CLB4), and YJB8796 (PMET3-clb4Δdb-Myc/CLB4) grown overnight to saturation in SDC +Met +Cys (promoter off) and diluted into SDC –Met –Cys (promoter on) for 6 h at 30°C. Blots were probed with anti-Myc and anti-PSTAIRE antibodies. Bars, 5 μm.
Morphological Observations
Calcofluor White (Sigma) staining was performed by incubating formaldehyde- or ethanol-fixed cells in 0.01 mg/ml Calcofluor for 30 min in phosphate-buffered saline (PBS) at room temperature and washing with PBS. Nuclei were visualized by incubating fixed cells in PBS containing 0.1 μg/ml 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) for at least 30 min at room temperature. For Alexa Fluor 568 phalloidin (Molecular Probes, Eugene, OR) staining of actin, cells were fixed for 1 h in 3.7% formaldehyde, washed with 50 mM potassium phosphate buffer, pH 6.6 (PK buffer), and resuspended in PK buffer containing 0.1% Triton X-100 for 30 min. Cells were washed twice with PBS and incubated in a 1:10 dilution of Alexa Fluor 568 phalloidin (200 U/ml). Differential interference contrast (DIC) and epifluorescence microscopy were performed with a Nikon Eclipse E600 photomicroscope (Melville, NY) equipped with a standard UV filter set, an Endow GFP bandpass emission filter set (Chroma Technology, Brattleboro, VT) and YFP filter set 41028 (Chroma Technology). Digital images were collected with a DVC-1310 digital camera (Digital Video Camera, Austin, TX), captured using C-View V2.1 software (Digital Video Camera), and processed with ImageJ (National Institutes of Health, Bethesda, MD) and Adobe Photoshop (Adobe Systems, San Jose, CA).
Flow Cytometry
Approximately 1 × 107 cells were fixed in 70% ethanol overnight at 4°C. Cells were washed twice with wash solution A (50 mM Tris-HCl, pH 8.0, 5 mM EDTA) followed by treatment with 2 mg/ml RNase (Sigma) in wash solution A for 16 h at 37°C. RNase-treated cells were resuspended in a 5 mg/ml pepsin solution in 55 mM HCl for 1 h at 37°C. Protease-treated cells were washed twice in wash solution B (50 mM Tris-HCl, pH 7.5, 5 mM EDTA) followed by staining in 1 μM Sytox Green (Molecular Probes) in wash solution B for at least 1 h at 4°C. Cells were analyzed on a FACSCalibur (BD Biosciences, San Jose, CA) using CellQuest Pro software (BD Biosciences) to capture data.
RESULTS
The Candida albicans Genome Encodes Two B-type Cyclins
A search for ORFs encoding B-type cyclins in the C. albicans genome identified the previously described CYB1 gene (orf6.7127, orf19.1446; Damagnez and Cottarel, 1996) and a novel open reading frame, orf19.7186 (orf6.8006), which has been referred to as CYB99 (http://www-sequence.stanford.edu/group/candida/index.html), CYB4 (Bensen et al., 2002), and CYB2 (Zheng and Wang, 2004). Orf19.7186 encodes a 486 amino acid protein with a predicted molecular weight of 57.26 kDa, a conserved cyclin box (aa 264–295), and a putative destruction box (aa 44–52). Orf19.7186 is most similar to S. cerevisiae Clb3p and Clb4p, with a slightly higher overall similarity to Clb3p but a higher similarity to Clb4p within a conserved internal 124-base pairs sequence (Clb3p: aa203–327, Clb4p: aa242–366, orf19.7186: aa266–390; unpublished data, Table 2). Cyb1p most closely resembles S. cerevisiae Clb1p and Clb2p with a slightly higher similarity to Clb2p. Our genome search did not uncover a Clb5p/Clb6p homolog. C. albicans cyctochrome b genes have also been termed CYB in the literature, resulting in confusion when referring to these very different gene products. Thus, with the permission of Dr. Guillaume Cottarel, we have renamed the CYB1 gene, CLB2 and we named orf19.7186 CLB4. When necessary, we use ScCLB2 and CaCLB2 to distinguish between the S. cerevisiae and C. albicans genes.
Table 2.
Amino acid percent identity/similarity of C. albicans B-type cyclins to S. cerevisiae B-type cyclins
| CaClb4p | ScClb1p | ScClb2p | ScClb3p | ScClb4p | ScClb5p | ScClb6p | |
|---|---|---|---|---|---|---|---|
| CaClb2p | 32/43 | 39/50 | 42/52 | 31/42 | 35/46 | 38/45 | 38/48 |
| CaClb4p | 35/45 | 33/44 | 44/57 | 43/54 | 33/43 | 32/45 |
Values were obtained using the GAP program from the Wisconsin Package, version 10.2 (Genetics ComputerGroup, Madison, WI).
Clb2p and Clb4p Levels Are Cell Cycle Regulated
To determine if C. albicans Clb2p and Clb4p levels oscillate during the cell cycle, we monitored the endogenous levels of HA-tagged Clb2p, Clb4p, and Cln1p in cultures that were synchronized by elutriation and released into medium supporting yeast-form growth. HA-tagged cyclins were functional as determined by the ability of a lone HA-tagged Clb2p or Clb4p to support wild-type growth (unpublished data). Clb2p-HA and Clb4p-HA were both absent from the starting cultures, indicating that G1 cells do not accumulate significant amounts of these B-cyclins (Figure 1). Clb2p-HA levels increased rapidly, peaking at 60–75 min after synchronous release, which correlates with the time just before G2/M as determined by budding index. A decrease in Clb2p-HA levels correlated with an increase in binucleate cells, with the trough occurring at 120–135 min after release, indicating that Clb2p was degraded after mitosis. Like Clb2p-HA levels, Clb4p-HA levels rapidly increased, peaking 90 min after release and correlating with G2/M phase. The trough of Clb4p-HA also occurred at 135 min after release, correlating with the end of mitosis. C. albicans Cln1p-myc levels increased just after the release of the elutriated cells, peaking 45 min after release and returning to low levels at 90 min as G2/M cells were accumulating. This is consistent with the previous characterization of C. albicans CLN1, in which CLN1 mRNA was found to peak at G1/S (Loeb et al., 1999b). The accumulation of Clb proteins before and during G2/M phase and their degradation at the end of mitosis is consistent with the properties of mitotic cyclins in other organisms (Ito, 2000; Irniger, 2002).
Clb2p and Clb4p Accumulation Is Delayed in Hyphal Cells
Cyclin-HA protein profiles for cells growing as true hyphae were determined by releasing unbudded G1 cells, which had been isolated by elutriation, into hyphal-inducing medium. When compared with Clb protein profiles of yeast form cells, both Clb2p-HA and Clb4p-HA accumulated and peaked at later time points in the hyphal cells (Figure 1). Furthermore, Cln1p-myc levels appeared earlier and persisted longer in hyphal cells than in yeast form cells (Figure 1) before dropping at the time of the first mitosis in hyphal cells. As in yeast form cells, Clbp-HA levels peaked at the time that Cln1p-HA levels were at a minimum. Taken together, these data suggest that Cln1p is stabilized during germ tube formation, whereas Clb2p and Clb4p accumulation is delayed in germ tubes relative to their levels in yeast-form cells. It should be noted that this delay occurs despite the fact that hyphal cells are induced at a higher temperature (37°C) than the yeast-form cells (30°C). A similar stabilization of Cln1p was seen in S. cerevisiae pseudohyphae (Vallier et al., 1994).
Clb2p-depleted Cells Grow Highly Elongated Filaments
The six B-type cyclins in S. cerevisiae have highly redundant functions (Andrews and Measday, 1998). The lack of B-type cyclin gene redundancy in C. albicans suggested that CLB2 and/or CLB4 might be essential genes. To test this hypothesis, we used the UAU1 (ura3-ARG4-ura3) single transformation gene function test (Enloe et al., 2000). In this procedure, one copy of a gene is disrupted with the UAU1 cassette, which encodes ARG5 flanked by two incomplete copies of URA3. Growth on 5-FOA in the absence of arginine selects for strains that underwent homozygosis of the UAU1 cassette, followed by recombination between the URA3 copies on one the UAU1 cassettes to yield a functional URA3 gene. For nonessential genes disrupted by UAU1, homozygosis results in the loss of the remaining wild-type copy of the gene. For essential genes, homozygosis of the UAU1-disrupted gene requires a gene triplication for survival. Thus, the presence of a wild-type copy of the gene in every Arg+ Ura+ isolate from a strain carrying a UAU1-disrupted gene copy strongly suggests that the gene is essential. Consistent with the idea that CLB2 is an essential gene, we found a wild-type copy of CLB2 in all of the 70 Arg+ Ura+ derivatives of a clb2::UAU1 strain.
To determine the fate of cells depleted of Clb2p, we generated a strain in which the only copy of CLB2 was under control of the inducible-MET3 promoter (PMET3-CLB2/clb2Δ). When grown under conditions that induce expression from the MET3 promoter (absence of methionine and cysteine), the PMET3-CLB2/clb2Δ strain grew round, smooth colonies and formed normal yeast-form cells indistinguishable from wild-type cells (Figure 2A; unpublished data), indicating that there was sufficient CLB2 in the strains to produce wild-type growth. In contrast, under conditions that repressed expression from the MET3 promoter (presence of methionine and cysteine), the PMET3-CLB2/clb2Δ strain did not form colonies. Close inspection of the plates revealed that the Clb2p-depleted cells formed highly elongated cellular structures that eventually stopped growing (Figure 2A). Western blot analysis of protein lysates from a PMET3-CLB2-myc/clb2Δ stain indicated that very little, if any, Clb2p was produced under repressing (presence of methionine and cysteine) conditions (Figure 2F). Furthermore, the wild-type growth phenotype of the PMET3-CLB2-myc/clb2Δ strain grown in SDC –Met –Cys indicated that the epitope-tagged Clb2p was functional (unpublished data). These data are consistent with the idea that CLB2 is an essential gene.
The terminal phenotype of Clb2p-depleted cells was determined by analyzing cell cycle progression of the PMET3-CLB2/clb2Δ strain after the MET3 promoter was repressed. After overnight growth in PMET3-CLB2–inducing conditions (SDC –Met/Cys), the PMET3-CLB2/clb2Δ strain was released into PMET3-CLB2–repressing conditions (SDC +Met/Cys) and cells were collected and fixed at several times after promoter shut off. Wild-type cells grew normally in SDC +Met/Cys, indicating that the repressing conditions did not affect morphogenesis (Figure 2B). After 3 h, Clb2p-depleted cells had formed hyperpolarized buds (Figure 2C). This hyperpolarization continued until extremely long filaments were seen (Figure 2C, 12 h.). At later time points, highly polarized branches emanated both from round mother cells and from hyperpolarized buds. After 24 h, the Clb2p-depleted cells were highly vacuolated and stopped growing (unpublished data). When Clb2p was reexpressed in cells that had been depleted for Clb2p for 3, 6, 9, and 12 h, budded cells emerged from the elongated cells. The timing of new bud emergence exhibited some delay in cells depleted for longer periods of time, but budding yeast cells did emerge. Therefore, the phenotypes seen in Clb2p-depleted cells are not due to cell death but rather cell cycle arrest. The extreme polarized growth of Clb2p-depleted cells suggests that Clb2p is required for isotropic (and consequently yeast-form) growth.
Clb2p-depleted Cells Arrest in Late Anaphase
DAPI staining revealed that Clb2p-depleted cells could undergo only a single nuclear division (Figure 2C). Even after 12 h, very few cells contained more than two nuclei although they remained alive: return of these cells to medium lacking methionine and cysteine eventually yielded budding yeast cells (unpublished data). A more quantitative assessment of nuclear division was carried out with Clb2p-depleted cells in which nuclear DNA was stained with Sytox Green and scored for nuclear content (Figure 2D). Between 3–5 h after Clb2p depletion, nuclear division occurred, resulting in one nucleus in the round mother cell and one in the elongated bud. After division, the nucleus in the mother cell migrated into the bud/germ tube, such that the mother cell lacked any nuclear-staining material and the bud/germ tube was binucleate. Flow cytometric analysis indicated that these cells accumulate with a total of 2C DNA content (Figure 2E), implying that each individual stained nucleus in the binucleate cells contains 1C DNA content. At 24 h, nuclear staining was very weak and diffuse, suggesting the breakdown of nuclear material. The lack of >2C DNA content suggests that these nuclei had arrested and did not undergo additional rounds of DNA replication. Although we cannot rule out the possibility that a small amount of Clb2p persists at the start of this experiment, the above results suggest that 1) Clb2p is not essential for entry into a first round of mitosis and 2) that progression of the cell cycle is blocked by the depletion of Clb2p following this first round of mitosis.
To better characterize the cell cycle-arrest point of Clb2p-depleted cells, we visualized spindles using a Tub1p-GFP fusion protein. A PMET3-CLB2/clb2Δ TUB1-GFP strain was grown in the presence of methionine and cysteine to deplete Clb2p, as described above, and Tub1p-GFP was visualized in live cells. In the vast majority of cells, an elongated spindle connected two spindle pole bodies (Figure 3A). The lack of spindle breakdown seen in Clb2p-depleted cells suggests that they are arrested/delayed in late anaphase/early telophase and thus are unable to exit mitosis.
Figure 3.
(A) Clb2p-depleted cells arrest with long spindles. A stationary culture of YJB8606 (PMET3-CLB2/clb2Δ TUB1-GFP) grown in SDC –Met –Cys (promoter on) was released into SDC +Met +Cys (promoter off) at 30°C. At the indicated times, fluorescent micrographs were taken of unfixed cells. Arrows indicate long spindles. (B) Clb2p-depleted cells do not form septa. A stationary culture of YJB7820 (PMET3-CLB2/clb2Δ) grown in SDC –Met –Cys was released into SDC +Met +Cys at 30°C. After 9 h, cells were removed, fixed with ethanol and stained with Calcofluor White to visualize septa formation. Arrowhead highlights diffuse staining at filament branch point. (C) Actin is hyperpolarized in Clb2p-depleted cells. YJB7820 (PMET3-CLB2/clb2Δ) was grown as in B. Aliquots were removed at 3 and 6 h after release into SDC +Met +Cys, fixed with formaldehyde, and stained with Alexa Fluor 568 phalloidin (see Materials and Methods). Paired images of DIC (top) and inverted fluorescent micrographs (bottom) are shown. Bars, 5 μm.
Clb2p-depleted cells rarely formed septa, as detected by calcofluor staining (Figure 3B). Although staining was seen in the initial mother-tube neck, a septum did not form in the elongated daughter cell, indicating that the filament was one continuous cellular compartment. Later migration of the mother nucleus through the mother-tube neck into the tube is consistent with the idea that cytokinesis does not occur in Clb2p-depleted cells. At filament branch points, irregular, diffuse calcofluor staining was observed occasionally (Figure 3B, arrowhead).
Alexa phalloidin staining of Clb2p-depleted cells revealed that actin patches localized to the growing tips of the elongated buds and remained concentrated at the tips after prolonged Clb2p depletion (Figure 3C). No obvious actin ring was seen at the mother bud neck. These data suggest that Clb2p is required for depolarization of actin cortical patches in the growing bud and is consistent with the continued hyphal-like apical cell wall expansion and hyperpolarized growth of Clb2p-depleted cells. Thus, cells depleted for Clb2p are able to enter, but are unable to exit, the first mitotic division, do not form true septa, and grow as hyperpolarized, highly elongated cells with a hyphal-like morphology.
Cells Lacking Clb4p Display Constitutive Pseudohyphal Growth
We next focused our attention on the other C. albicans B-cyclin, Clb4p. We first asked if CLB4 is an essential gene using the UAU1 test (Enloe et al., 2000). In contrast to CLB2, 7 of 30 Arg+ Ura+ clb4::UAU1 isolates did not contain a wild-type CLB4 allele, indicating that CLB4 is not essential. Thus, we constructed a homozygous clb4Δ/clb4Δ strain in addition to a strain in which the only copy of CLB4 was under the control of the MET3 promoter (PMET3-CLB4/clb4Δ). PMET3-CLB4/clb4Δ cells, when grown on solid medium in the absence of methionine and cysteine (promoter on), grew into round colonies indistinguishable from colonies of wild-type cells (Figure 4). However, colonies formed from PMET3-CLB4/clb4Δ cells grown on solid medium containing methionine and cysteine (promoter off) were irregularly shaped and had a wrinkled appearance (Figure 4). When grown on solid medium that normally supports yeast form growth (SDC), clb4Δ/clb4Δ cells formed wrinkled colonies indistinguishable from the irregularly shaped colonies of Clb4p-depleted (pMET3-CLB4 promoter off) cells (Figure 4).
Figure 4.
clb4Δ/clb4Δ and Clb4p-depleted cells produce wrinkled colonies. YJB7878 (WT) and YJB7824 (PMET3-CLB4/clb4Δ) were grown for 3 d on SDC –Met –Cys agar (promoter on) and SDC +Met +Cys agar (promoter off). YJB6284 (WT) and YJB6854 (clb4Δ/clb4Δ) were grown for 3 d on SDC agar.
Individual clb4Δ/clb4Δ cells resembled pseudohyphae; they were elongated and formed chains that did not separate readily (Figure 5B). Clb4p-depleted cells exhibited an identical pseudohyphal-like growth phenotype, indicating that the morphology of the clb4Δ/clb4Δ cells was due to the absence of CLB4 (Figure 5C). Nuclear division appeared normal: each clb4Δ/clb4Δ pseudohyphal cell contained nuclear material (Figure 5, B and C). This indicates that, like Clb2p, Clb4p is not essential for the onset of mitosis. Furthermore, septum formation, detected by calcofluor staining between mother and daughter clb4Δ/clb4Δ cells, occurred in the absence of Clb4p (Figure 5D). No detectable Clb4p was detected in a Western blot of protein lysates from a PMET3-CLB4-myc/clb4Δ strain grown under repressing (presence of methionine and cysteine) conditions (Figure 5E). When grown under expressing conditions (absence of methionine and cysteine), PMET3-CLB4-myc/clb4Δ cells grew normally indicating that that epitope tagged Clb4p was functional (unpublished data). Thus, in contrast to Clb2p, Clb4p is not necessary for mitotic exit or cytokinesis.
Alexa-phalloidin staining revealed actin patches organized primarily at the apical end of elongated Clb4p-depleted cells, with some delocalized patches along the cortex (Figure 6). The concentration of tip patches was less pronounced in Clb4p-depleted cells than in Clb2p-depleted cells. Actin was also seen at sites of septation, suggesting that the actin contractile ring forms in the absence of Clb4p. Together, these data indicate that Clb4p negatively regulates pseudohyphal growth, yet is not essential for the depolarization of actin cortical patches or for cell cycle progression.
Figure 6.
Actin localizes to sites of polarized growth in Clb4p-depleted cells. YJB7824 (PMET3-CLB4/clb4Δ) was grown to stationary phase in SDC –Met –Cys (promoter on) and released into SDC +Met +Cys at 30°C. Cells were removed at the indicated times and stained with Alexa Fluor 568 phalloidin as in Figure 5C. DIC (top) and inverted fluorescent micrographs (bottom) are shown. Bars, 5 μm.
Excess Clb2p or Clb4p Inhibit Filamentous Growth
The phenotypic analysis of both C. albicans B-type cyclins discussed above indicates that Clb2p and Clb4p negatively regulate filamentous growth under yeast growth conditions. To further test this hypothesis, we induced hyphal growth in strains overexpressing either CLB2 or CLB4 from the ACT1 promoter. Relative to their endogenous promoters, CLB2 and CLB4 were expressed at 3- and 4.6-fold higher levels from the ACT1 promoter, respectively (Figure 7). CLB2- and CLB4-overexpressing strains formed hyphae less efficiently than strains expressing CLB2 and CLB4 at endogenous levels (Figure 7). In the overexpressing strains, fewer germ tubes formed, and those germ tubes that did form were shorter. The CLB4-overexperssing strain reduced the serum response to a greater extent than the CLB2-overexpressing strain. These data support the hypothesis that Clb2p and Clb4p inhibit filamentous growth.
Clb2p and Clb4p Must Be Degraded to Exit Mitosis
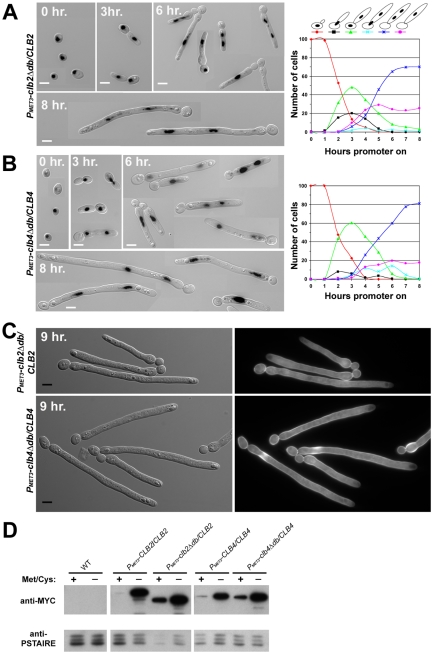
In S. cerevisiae, completion of mitosis requires the ubiquitin-dependent degradation of mitotic cyclins (Morgan, 1999). Required for this degradation is the destruction box (db) motif, a sequence found near the N-terminus of B-type cyclins. In S. cerevisiae, stabilization of Clb2p by removal of the db results in cell cycle arrest at a late stage of mitosis (Surana et al., 1993). To determine the effect of mitotic cyclin stabilization on C. albicans cell cycle control, we produced N-terminal truncations of Clb2p (Clb2pΔdb) and Clb4p (Clb4pΔdb) lacking destruction box motifs by integrating the MET3 promoter into CLB2 and into CLB4 immediately downstream of the destruction box domains in the two CLB genes. To verify that the truncated Clb proteins were being expressed, these strains were tagged with a Myc epitope. Myc-tagged copies of Clb2pΔdb and Clb4pΔdb were both present in cells when expressed from the MET3 promoter (Figure 8D). Even when the MET3 promoter was repressed, cells containing PMET3-clbΔdb-Myc showed a significant increase in protein when compared with cells containing PMET3-CLB-Myc. This is likely due to the stabilization of the small amount of Clbp transcribed under MET3-repressing conditions. Cells expressing either Clb2pΔdb in the presence of wild-type Clb2p or Clb4pΔdb in the presence of wild-type Clb4p grew with highly elongated buds that could not support further growth (Figure 8, unpublished data). Thus, both destruction box mutants exhibited a dominant phenotype that led to eventual cell death.
Like Clb2p-depleted cells, some clbΔdb expressing strains were able to undergo a single nuclear division. However, unlike Clb2p-depleted cells, which had very few cells with a single nucleus after 8 h, a significant number of clbΔdb cells contained a single nucleus in the elongated bud (Figure 8, A and B). The presence of a single nucleus after 8 h was slightly more evident in clb2Δdb cells (26%; n = 214) than in clb4Δdb cells (18%; n = 218). At later time points, clb2Δdb strains were highly branched, whereas clb4Δdb strains exhibited much less branching (Figure 8). For example, at 14 h after induction of expression, 41% (n = 315) of clb2Δdb cells displayed two or more branches emanating from the initial germ tube, whereas <1% (n = 292) of clb4Δdb cells had more than one branch. Furthermore, calcofluor staining revealed that the elongated tubes did not contain septa, indicating that cells were not proceeding through the cell cycle to cytokinesis (Figure 8C). Together, these data indicate that stabilization of either mitotic cyclin causes cell cycle arrest accompanied by polarized growth and suggests that nuclear division is inefficient in the presence of stabilized mitotic cyclins.
Clbp-depleted Cells Respond to Serum
In C. albicans, the ability to form true hyphal cells is associated with the ability to cause infection. To determine if Clb-deficient cells can respond to the signals that induce formation of true hyphae, we exposed Clb2p- and Clb4p-depleted cells to serum, a strong hyphal inducer. Wild-type cells responded normally to these hyphal-inducing conditions (Figure 9A). When stationary PMET3-CLB2/clb2Δ cells were released into serum-containing medium in the presence of methionine and cysteine to repress the MET3 promoter, germ tube-like structures were seen emanating from round, yeast cells (Figure 9B). The average width of these tubes (1.20 ± 0.15 μm, n = 17) was similar to those formed from wild-type cells (1.23 ± 0.12 μm, n = 24). A distinct septum was apparent in Clb2p-depleted cells within the length of the germ tube. This is in clear contrast to what is seen in Clb2p-depleted cells grown in yeast-form conditions, where a septal-band is seen at the mother-bud neck but not in the elongated bud (compare Figure 9B with 3B). These data suggest that the control of septation is different in hyphal cells than in yeast cells. In hyphal cells, the requirement for Clb2p in septin formation appears to be bypassed.
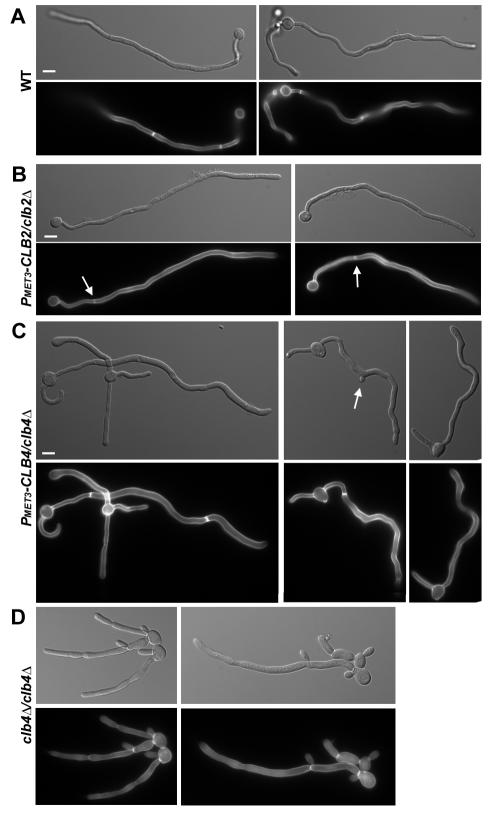
Figure 9.
Clb2p- and Clb4p-depleted cells respond to hyphal inducing conditions. (A) Saturated cultures of YJB7878 (WT), (B) YJB7820 (PMET3-CLB2/clb2Δ), and (C) YJB7824 (PMET3-CLB4/clb4Δ) grown in SDC –Met –Cys (promoter on) were diluted into SDC +Met +Cys + 10% serum (promoter off) and incubated at 37°C for 6.5 h to induce hyphal formation. A saturated culture of (D) YJB6854 (clb4Δ/clb4Δ) grown in YPAD was diluted into YPAD + 10% serum and incubated at 37°C for 6 h. All cells were fixed with formaldehyde and stained with Calcofluor White to visualize septa. Bars, 5 μm.
Stationary PMET3-CLB4/clb4Δ cells incubated in serum-containing medium in the presence of methionine and cysteine (promoter off) also formed long projections (Figure 9C). In contrast to the Clb2p-depleted germ tubes, Clb4p-depleted germ tubes were significantly wider (1.63 ± 0.25 μm, n = 24) than wild-type tubes. Furthermore, the tube formed after the first septum was wider (2.03 ± 0.28 μm, n = 24) than the initial tube, suggesting that there is not only a defect in the initial polarization of a germ tube, but also in the maintenance of hyphal growth in cells lacking Clb4p. When clb4Δ/clb4Δ cells were exposed to serum-containing media (Figure 9D), the initial elongations formed were significantly wider than wild-type germ tubes (2.26 ± 0.48 μm, n = 10 vs. 1.29 ± 0.15 μm, n = 10) and were also wider than the Clb4p-depleted cells exposed to serum (2.26 ± 0.48 μm, n = 10 and 2.11 ± 0.33 μm, n = 10, for cells before and after the first septum, respectively). Thus, clb4Δ/clb4Δ cells have a stronger defect with respect to serum induction than do Clb4p-depleted cells, suggesting that a small amount of Clb4p is present in the Clb4p-depleted cells. This is consistent with what has been seen for other genes expressed from the MET3 promoter (Bassilana et al., 2003). Even though cells deficient for Clb4p (both Clb4p-depeleted and clb4Δ/Δ cells) displayed defects in both the extent of hyperpolarization and septum placement in response to serum, they remained capable of responding to the polarized growth signal in serum: cells were more polarized when grown in serum than in serum-free medium. This indicates that serum has components that can induce polarized growth in the absence of the cell cycle changes that occur in true hyphae.
DISCUSSION
C. albicans Clb2p and Clb4p Are Mitotic Cyclins
The data presented here indicate that the protein products of the CLB2 and CLB4 genes are mitotic cyclins. The levels of both proteins increase as the number of cells entering G2/M phase rise and they decrease as cells exit mitosis. Consistent with previous CLN1 mRNA profiling, Cln1p levels increase during G1 and decrease with the accumulation of G2/M cells and of Clbp levels (Loeb et al., 1999b). This is similar to the S. cerevisiae paradigm where Clb1–4p inhibit transcription of CLN1/2, suggesting that a similar regulatory network may occur in C. albicans (Koch et al., 1996).
The C. albicans genome sequence (http://www-sequence.stanford.edu/group/candida/index.html) does not include an obvious homolog to S. cerevisiae CLB5/6 (Zheng and Wang, 2004). We do not know if CaClb2/4p can perform the S phase functions performed by ScClb5/6p.
CLB2 Is Essential and Required for Mitotic Exit
CLB2 is essential for vegetative growth. Clb2p-depleted cells formed highly elongated filaments with polarized actin at their tips, suggesting that, as in S. cerevisiae, Clb2p is required for the switch from apical to isotropic cell wall expansion. DNA replication and nuclear division in these cells suggests that Clb2p-depleted cells arrest in a late anaphase/early telophase stage of the cell cycle and that Clb2p is required for mitotic exit. Clb2p could fulfill this mitotic exit function through direct activation of the mitotic exit network or as an indirect consequence of Cdc28p/Clb2p inactivity, for example by activating a checkpoint or by inactivating another cyclin/CDK activity. In S. cerevisiae, a misoriented spindle results in the activation of a checkpoint blocking APC/CCdc20p and therefore mitotic exit (Irniger, 2002). A similar checkpoint could be triggered in Clb2p-depleted C. albicans cells by elongated spindles that do not cross a mother-bud neck, because no septum was formed in these cells.
Clb2p does not appear to be required for entry into S-phase or for the onset of mitosis. Flow cytometric data and morphological analysis suggest that DNA replication occurs in Clb2p-depleted cells with little or no delay. Furthermore, divided nuclear material was connected by a long spindle, indicating that anaphase can initiate in the absence of Clb2p. In contrast, in S. cerevisiae, a PGAL1-CLB1 clb2Δ clb3Δ strain arrests before nuclear division with 2C DNA content and a short spindle, indicating a preanaphase block (Richardson et al., 1992) and S. pombe requires the Cdc13 B-type cyclin to enter mitosis (Hayles et al., 1994). However, we cannot rule out the possibility that a small amount of CLB2 is expressed in the MET3 “promoter off” conditions used for our experiments and that this small amount of Clb2p is sufficient to allow progression through early anaphase. Another possibility is that Clb2p cell cycle functions are masked by Clb4p redundancy.
Clb4p Negatively Regulates Pseudohyphal Growth
Unlike CLB2, CLB4 is not an essential gene. Both the homozygote (clb4Δ/clb4Δ) and the pMET3-regulated strains grew as pseudohyphal-like cells under conditions supporting yeast-form growth in wild-type cells. As with Clb2p-depleted cells, cells lacking Clb4p were able to enter S-phase, suggesting that neither B-type cyclin is absolutely required for the initiation of DNA replication. Unlike Clb2p, Clb4p is not required for mitotic exit, because cells continued to grow and divide without Clb4p. Therefore, Clb2p is the only B-type cyclin essential for growth and for mitotic exit in C. albicans.
Like fkh2Δ/fkh2Δ cells, cells lacking Clb4p formed constitutive pseudohyphae and these pseudohyphae were more elongated when exposed to strong hyphal induction conditions than when grown under conditions favoring pseudohyphal growth (Bensen et al., 2002). Thus, these cells respond to signals in serum that promote polarized growth, despite the fact that their cell cycle defects cause them to have the cell cycle features (constricted septa, wider cell; Sudbery et al., 2004) of pseudohyphae rather than true hyphae. This indicates that serum induction of hyphal growth involves separable functions: polarized growth and altered cell cycle features such as the timing and position of septin ring formation.
The polarized growth of cells lacking either of the B-cyclins and the reduced polarization of cells containing excess B-cyclins is consistent with the idea that B-cyclins are negative regulators of polarized growth in C. albicans, because they are in other organisms. However, the clb-Δdb mutants, which arrested in the presence of undegradable Clbs, also produced highly polarized cells. Other types of cell cycle delays also result in a polarized growth response in C. albicans; for example, treatment with hydroxyurea or nocodazole (Bai et al., 2002; Hazan et al., 2002; Bachewich et al., 2003) or depletion of Cdc5p (Bachewich et al., 2003). Interestingly, even when cells arrest in response to a lack of the Cln3, they initially grow isotropically and then elongate (Bachewich and Whiteway, 2005; Chapa y Lazo et al., 2005). We propose that a default response of C. albicans cells to many types of cell cycle delay or arrest is to continue growing in a polarized manner, often resulting in a morphology resembling long pseudohyphal-like cells.
Mitotic Exit Requires Clb2p and Clb4p Degradation
Stabilization of either mitotic cyclin, by removal of their destruction box motif, resulted in the accumulation of the majority of cells at a late anaphase/early telophase stage of the cell cycle, indicating that, like S. cerevisiae mitotic cyclins (Surana et al., 1993), C. albicans mitotic cyclins must be degraded before mitotic exit can occur. A significant minority of cells (∼20%) accumulated with a single nucleus, suggesting arrest before anaphase. In vertebrate cells, defects in cyclin B degradation has been reported to cause a in blockage or delay in metaphase (Stemmann et al., 2001; Chang et al., 2003) or telophase (Murray et al., 1989).
Mitotic Cyclins as Key Regulators of Filamentous Growth
An important result revealed by these studies is that the expression pattern of both a G1 cyclin and two B-type cyclins is altered in true hyphae. When compared with yeast-form cells, Cln1p was found to accumulate earlier and to persist for longer times in hyphal-induced cells, despite the fact that the hyphal cells are grown at a higher temperature (37°C) than the yeast-form cells (30°C). Furthermore, accumulation of both Clb2p and Clb4p was delayed in hyphal-induced cells relative to their accumulation in yeast-form cells. These data suggest that the initial germ tube undergoes an extended G1 phase during which Cln1p is stabilized (Figure 1B). The delay seen in Clb2p and Clb4p accumulation could be required for the hyperpolarization of hyphal cells as well as for other hyphal cell characteristics.
These results lead to a conclusion that is very different from those reached by Liu and coworkers (Hazan et al., 2002), who suggested that the cell cycle is not altered by hyphal induction. One explanation for this difference may be the difference in elutriation methods used to synchronize cells in the two studies. Hazen et al. (2002) elutriated cells at 4°C, which delayed yeast-form bud formation (buds appeared at 100 min). We elutriated cells at room temperature, such that cells rapidly reentered the cell cycle (buds appeared at 30 min). Therefore, the delay induced by stressing the cells at 4°C may have masked the cell cycle delay that occurs during hyphal induction. In addition, the cell cycle delay in hyphal cells may have been further masked by the differences in growth temperature for hyphal (37°C) and yeast (30°C) cells.
The delay in Clb2p and Clb4p accumulation upon hyphal induction could be physiologically relevant, because the expression of either of these CLB genes under the actin promoter gave rise to shorter and wider germ tubes (Figure 6). This suggests that regulation of the B-cyclins is important at the onset of germ tube formation, perhaps to ensure that hyphal cells grow in a hyper-polarized manner. Consistent with this idea, in S. cerevisiae the G1 cyclins appear to be stabilized (Barral et al., 1995), and thus B-cyclin expression is presumably delayed, in pseudohyphal cells (Kron et al., 1994; Kron and Gow, 1995).
Hyphal growth differs from yeast and pseudohyphal growth in several ways. An important characteristic of hyphal growth is the hyperpolarization of the actin cytoskeleton and of new cell wall deposition. Recent work from Wang and coworkers revealed the presence of a G1 cyclin (Hgc1p) with an important role in hyphal morphogenesis (Zheng and Wang, 2004). Constitutive overexpression of Hgc1p does not induce hyphal growth in the absence of serum, suggesting that Hgc1p contributes to, but is not sufficient for, hyperpolarized growth. We propose that the hyperpolarized growth of hyphae is accomplished by a balance between the G1 cyclins (prolonged accumulation of Cln1, presence of Hgc1) and the delayed appearance of the B-cyclins (Clb2p and Clb4p), which negatively regulate polarized growth.
Acknowledgments
We thank Zosia Piotroswska for UAU1 analysis, Maryam Gerami-Nejad for help with strain and plasmid construction, Mark McClellan for technical assistance and Cheryl Gale for critical review of the manuscript. We thank Peter Sudbery and Yoshimasa Uehara for providing plasmids. We acknowledge the assistance of the Flow Cytometry Core Facility of the University of Minnesota Cancer Center, a comprehensive cancer center designated by the National Cancer Institute, supported in part by P30 CA77598. This work was supported by the National Institutes of Heath (NIH) AI/DE14666 to J.B. and grants from the Ministerio de Ciencia y Tecnología (PM99–0182 and BMC2003–05758) to J.C.-B. E.B. was supported by training funds AI T32-AI 07421 and a postdoctoral fellowship (AI10647) from the NIH. J.A.C.-B. is recipient of a fellowship from the Consejería de Educación, Ciencia y Tecnología de la Junta de Extremadura.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–12–1081) on May 11, 2005.
References
- Ahn, S. H., Acurio, A., and Kron, S. J. (1999). Regulation of G2/M progression by the STE mitogen-activated protein kinase pathway in budding yeast filamentous growth. Mol. Biol. Cell 10, 3301–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn, S. H., Tobe, B. T., Gerald, J. N., Anderson, S. L., Acurio, A., and Kron, S. J. (2001). Enhanced cell polarity in mutants of the budding yeast cyclin-dependent kinase Cdc28p. Mol. Biol. Cell 12, 3589–3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, J. M., and Soll, D. R. (1986). Differences in actin localization during bud and hypha formation in the yeast Candida albicans. J. Gen. Microbiol. 132, 2035–2047. [DOI] [PubMed] [Google Scholar]
- Andrews, B., and Measday, V. (1998). The cyclin family of budding yeast: abundant use of a good idea. Trends Genet. 14, 66–72. [DOI] [PubMed] [Google Scholar]
- Bachewich, C., Thomas, D. Y., and Whiteway, M. (2003). Depletion of a polo-like kinase in Candida albicans activates cyclase-dependent hyphal-like growth. Mol. Biol. Cell 14, 2163–2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachewich, C., and Whiteway, M. (2005). Cyclin Cln3p links G1 progression to hyphal and pseudohyphal development in Candida albicans. Eukaryot. Cell 4, 95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai, C., Ramanan, N., Wang, Y. M., and Wang, Y. (2002). Spindle assembly checkpoint component CaMad2p is indispensable for Candida albicans survival and virulence in mice. Mol. Microbiol. 45, 31–44. [DOI] [PubMed] [Google Scholar]
- Barral, Y., Jentsch, S., and Mann, C. (1995). G1 cyclin turnover and nutrient uptake are controlled by a common pathway in yeast. Genes Dev. 9, 399–409. [DOI] [PubMed] [Google Scholar]
- Bassilana, M., Blyth, J., and Arkowitz, R. A. (2003). Cdc24, the GDP-GTP exchange factor for Cdc42, is required for invasive hyphal growth of Candida albicans. Eukaryot. Cell 2, 9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensen, E. S., Filler, S. G., and Berman, J. (2002). A forkhead transcription factor is important for true hyphal as well as yeast morphogenesis in Candida albicans. Eukaryot. Cell 1, 77–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Care, R. S., Trevethick, J., Binley, K. M., and Sudbery, P. E. (1999). The MET3 promoter: a new tool for Candida albicans molecular genetics. Mol. Microbiol. 34, 792–798. [DOI] [PubMed] [Google Scholar]
- Chang, D. C., Xu, N., and Luo, K. Q. (2003). Degradation of cyclin B is required for the onset of anaphase in mammalian cells. J. Biol. Chem. 278, 37865–37873. [DOI] [PubMed] [Google Scholar]
- Chapa y Lazo, B., Bates, S., and Sudbery, P. (2005). The G1 cyclin Cln3 regulates morphogenesis in Candida albicans. Eukaryot. Cell 4, 90–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damagnez, V., and Cottarel, G. (1996). Candida albicans CDK1 and CYB1, cDNA homologues of the cdc2/CDC28 and cdc13/CLB1/CLB2 cell cycle control genes. Gene 172, 137–141. [DOI] [PubMed] [Google Scholar]
- Edgington, N. P., Blacketer, M. J., Bierwagen, T. A., and Myers, A. M. (1999). Control of Saccharomyces cerevisiae filamentous growth by cyclin-dependent kinase Cdc28. Mol. Cell. Biol. 19, 1369–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enloe, B., Diamond, A., and Mitchell, A. P. (2000). A single-transformation gene function test in diploid Candida albicans. J. Bacteriol. 182, 5730–5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst, J. F. (2000). Transcription factors in Candida albicans—environmental control of morphogenesis. Microbiol. UK 146, 1763–1774. [DOI] [PubMed] [Google Scholar]
- Gari, E., Volpe, T., Wang, H., Gallego, C., Futcher, B., and Aldea, M. (2001). Whi3 binds the mRNA of the G1 cyclin CLN3 to modulate cell fate in budding yeast. Genes Dev. 15, 2803–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerami-Nejad, M., Berman, J., and Gale, C. A. (2001). Cassettes for PCR-mediated construction of green, yellow and cyan fluorescent protein fusions in Candida albicans. Yeast 18, 859–864. [DOI] [PubMed] [Google Scholar]
- Gerami-Nejad, M., Hausauer, D., McClellan, M., Berman, J., and Gale, C. (2004). Cassettes for the PCR-mediated construction of regulatable alleles in Candida albicans. Yeast 21, 429–436. [DOI] [PubMed] [Google Scholar]
- Gillum, A. M., Tsay, E. Y., and Kirsch, D. R. (1984). Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 198, 179–182. [DOI] [PubMed] [Google Scholar]
- Gola, S., Martin, R., Walther, A., Dunkler, A., and Wendland, J. (2003). New modules for PCR-based gene targeting in Candida albicans: rapid and efficient gene targeting using 100 bp of flanking homology region. Yeast 20, 1339–1347. [DOI] [PubMed] [Google Scholar]
- Gow, N., Brown, A., and Odds, F. (2002). Fungal morphogenesis and host invasion. Curr. Opin. Microbiol. 5, 366. [DOI] [PubMed] [Google Scholar]
- Hayles, J., Fisher, D., Woollard, A., and Nurse, P. (1994). Temporal order of S phase and mitosis in fission yeast is determined by the state of the p34cdc2-mitotic B cyclin complex. Cell 78, 813–822. [DOI] [PubMed] [Google Scholar]
- Hazan, I., Sepulveda-Becerra, M., and Liu, H. (2002). Hyphal elongation is regulated independently of cell cycle in Candida albicans. Mol. Biol. Cell 13, 134–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- http://www-sequence.stanford.edu/group/candida/index.html. Stanford C. albicans sequencing.
- Irniger, S. (2002). Cyclin destruction in mitosis: a crucial task of Cdc20. FEBS Lett. 532, 7–11. [DOI] [PubMed] [Google Scholar]
- Ito, M. (2000). Factors controlling cyclin B expression. Plant Mol. Biol. 43, 677–690. [DOI] [PubMed] [Google Scholar]
- Koch, C., Schleiffer, A., Ammerer, G., and Nasmyth, K. (1996). Switching transcription on and off during the yeast cell cycle: Cln/Cdc28 kinases activate bound transcription factor SBF (Swi4/Swi6) at start, whereas Clb/Cdc28 kinases displace it from the promoter in G2. Genes Dev. 10, 129–141. [DOI] [PubMed] [Google Scholar]
- Kron, S. J., and Gow, N.A.R. (1995). Budding yeast morphogenesis: signaling, cytoskeleton and cell cycle. Curr. Opin. Cell Biol. 7, 845–855. [DOI] [PubMed] [Google Scholar]
- Kron, S. J., Styles, C. A., and Fink, G. R. (1994). Symmetric cell division in pseudohyphae of the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 5, 1003–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Loeb, J. D., Kerentseva, T. A., Pan, T., Sepulveda-Becerra, M., and Liu, H. (1999a). Saccharomyces cerevisiae G1 cyclins are differentially involved in invasive and pseudohyphal growth independent of the filamentation mitogen-activated protein kinase pathway. Genetics 153, 1535–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb, J. D., Sepulveda-Becerra, M., Hazan, I., and Liu, H. (1999b). A G1 cyclin is necessary for maintenance of filamentous growth in Candida albicans. Mol. Cell. Biol. 19, 4019–4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M. S., McKenzie, A., 3rd, Demarini, D. J., Shah, N. G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J. R. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961. [DOI] [PubMed] [Google Scholar]
- Madhani, H. D., Galitski, T., Lander, E. S., and Fink, G. R. (1999). Effectors of a developmental mitogen-activated protein kinase cascade revealed by expression signatures of signaling mutants. Proc. Natl. Acad. Sci. USA 96, 12530–12535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miled, C., Mann, C., and Faye, G. (2001). Xbp1-mediated repression of CLB gene expression contributes to the modifications of yeast cell morphology and cell cycle seen during nitrogen-limited growth. Mol. Cell. Biol. 21, 3714–3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, L. H., and Soll, D. R. (1979). Temporal and spatial differences in septation during synchronous mycelium and bud formation by Candida albicans. Exper. Mycol. 3, 298–309. [Google Scholar]
- Mitchell, T. G. (1998). Medical mycological research and training: needs and opportunities. ASM News 64, 17–23. [Google Scholar]
- Morgan, D. O. (1999). Regulation of the APC and the exit from mitosis. Nat. Cell Biol. 1, E47–E53. [DOI] [PubMed] [Google Scholar]
- Mosch, H. U., and Fink, G. R. (1997). Dissection of filamentous growth by transposon mutagenesis in Saccharomyces cerevisiae. Genetics 145, 671–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray, A. W., Solomon, M. J., and Kirschner, M. W. (1989). The role of cyclin synthesis and degradation in the control of maturation promoting factor activity. Nature 339, 280–286. [DOI] [PubMed] [Google Scholar]
- Oehlen, L. J., Jeoung, D. I., and Cross, F. R. (1998). Cyclin-specific START events and the G1-phase specificity of arrest by mating factor in budding yeast. Mol. Gen. Genet. 258, 183–198. [DOI] [PubMed] [Google Scholar]
- Richardson, H., Lew, D. J., Henze, M., Sugimoto, K., and Reed, S. I. (1992). Cyclin-B homologs in Saccharomyces cerevisiae function in S phase and in G2. Genes Dev. 6, 2021–2034. [DOI] [PubMed] [Google Scholar]
- Rua, D., Tobe, B. T., and Kron, S. J. (2001). Cell cycle control of yeast filamentous growth. Curr. Opin. Microbiol. 4, 720–727. [DOI] [PubMed] [Google Scholar]
- Sherman, F. (1991). Getting started with yeast. In: Methods Enzymology: Guide to Yeast Genetics and Molecular Biology, Vol. 194, ed. C. Guthrie and G. R. Fink, San Diego: Academic Press, 3–20. [Google Scholar]
- Soll, D. R., Herman, M. A., and Staebell, M. A. (1985). The involvement of cell wall expansion in the two modes of mycelium formation of Candida albicans. J. Gen. Microbiol. 131, 2367–2375. [DOI] [PubMed] [Google Scholar]
- Stemmann, O., Zou, H., Gerber, S.A., Gygi, S. P., and Kirschner, M. W. (2001). Dual inhibition of sister chromatid separation at metaphase. Cell 107, 715–726. [DOI] [PubMed] [Google Scholar]
- Sudbery, P., Gow, N., and Berman, J. (2004). The distinct morphogenic states of Candida albicans. Trends Microbiol. 12, 317–324. [DOI] [PubMed] [Google Scholar]
- Sudbery, P. E. (2001). The germ tubes of Candida albicans hyphae and pseudohyphae show different patterns of septin ring localization. Mol. Microbiol. 41, 19–31. [DOI] [PubMed] [Google Scholar]
- Surana, U., Amon, A., Dowzer, C., McGrew, J., Byers, B., and Nasmyth, K. (1993). Destruction of the CDC28/CLB mitotic kinase is not required for the metaphase to anaphase transition in budding yeast. EMBO J. 12, 1969–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeyama, T., Nagai, Y., Niimi, M., and Uehara, Y. (2002). Construction of FLAG tagging vectors for Candida albicans. Yeast 19, 611–618. [DOI] [PubMed] [Google Scholar]
- Vallier, L. G., Coons, D., Bisson, L. F., and Carlson, M. (1994). Altered regulatory responses to glucose are associated with a glucose transport defect in grr1 mutants of Saccharomyces cerevisiae. Genetics 136, 1279–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, R. B., Davis, D., and Mitchell, A. P. (1999). Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181, 1868–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, X., and Wang, Y. (2004). Hgc1, a novel hypha-specific G1 cyclin-related protein regulates Candida albicans hyphal morphogenesis. EMBO J. 23, 1845–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]