Abstract
Nucleostemin is a p53-interactive cell cycle progression factor that shuttles between the nucleolus and nucleoplasm, but it has no known involvement in ribosome synthesis. We found the dynamic properties of nucleostemin differed strikingly from fibrillarin (a protein directly involved in rRNA processing) both in response to rRNA transcription inhibition and in the schedule of reentry into daughter nuclei and the nucleolus during late telophase/early G1. Furthermore, nucleostemin was excluded from the nucleolar domains in which ribosomes are born—the fibrillar centers and dense fibrillar component. Instead it was concentrated in rRNA-deficient sites within the nucleolar granular component. This finding suggests that the nucleolus may be more subcompartmentalized than previously thought. In support of this concept, electron spectroscopic imaging studies of the nitrogen and phosphorus distribution in the nucleolar granular component revealed regions that are very rich in protein and yet devoid of nucleic acid. Together, these results suggest that the ultrastructural texture of the nucleolar granular component represents not only ribosomal particles but also RNA-free zones populated by proteins or protein complexes that likely serve other functions.
INTRODUCTION
The nucleolus is a specialized domain of the nucleus that was established as the site of ribosome synthesis four decades ago (Vincent and Miller, 1966). However, it has recently become apparent that the nucleolus has other functions as well (Pederson, 1998a,b; Olson et al., 2000, 2003; Pederson and Politz, 2000; Leung and Lamond, 2003). In support of this new concept, proteomic analysis of purified nucleoli has revealed the presence in nucleoli of many proteins with no known or obvious relationship to ribosome synthesis (Andersen et al., 2002; Scherl et al., 2002). A key question that therefore has arisen is how these newly recognized functions and proteins are spatially localized in relation to the well defined intranucleolar sites of ribosome synthesis.
One newly suggested function of the nucleolus is a role in the assembly of the signal recognition particle (Jacobson and Pederson, 1998; Ciufo and Brown, 2000; Pederson and Politz, 2000; Politz et al., 2000, 2002; Grosshans et al., 2001; Alavian et al., 2004; Sommerville et al., 2005). However, signal recognition particle (SRP) RNA does not spatially overlap substantially with 28S rRNA in the mammalian cell nucleolus, nor is it present in the regions in which rRNA transcription or initial processing take place (Politz et al., 2002). It thus seems that there might be subdomains within the nucleolus that are devoid of nascent ribosomes and within which certain other macromolecules that are unrelated to ribosome synthesis reside.
In the present study, we tested the hypothesis that the nucleolar landscape contains domains that are not devoted to ribosome synthesis. We investigated the intranucleolar localization of a protein that has no known relationship to ribosome synthesis or other RNA biosynthesis or metabolism: nucleostemin, a p53-interacting protein that is expressed in stem cells and tumor cells (Tsai and McKay, 2002, 2005; Liu et al., 2004; Misteli, 2005). Because nucleostemin's role in regulating p53 and cell cycle progression is thought to occur in the nucleoplasm, its transient nucleolar residence is most unlikely to be linked to the ribosome synthesis pathway. We therefore reasoned that nucleostemin might define novel subnucleolar domains occupied by proteins that were related to other functions.
Our results indeed reveal a distinctive intranucleolar localization of nucleostemin as well as other unusual properties with respect to its dynamic behavior that contrast strikingly with nucleolar proteins that are involved in ribosome synthesis. Furthermore, we present electron spectroscopic imaging results that define protein-rich, RNA-deficient regions within the granular component of the nucleolus, likely to represent the sites at which ribosome nonrelated nucleolar components reside.
MATERIALS AND METHODS
Cell Culture
Mouse 3T3 cells were propagated in DMEM with 10% newborn calf serum. The rat L6 myoblast cell line was grown in DMEM with 10% fetal bovine serum (FBS), and rat normal rat kidney (NRK) fibroblasts were propagated in F-12K medium with 10% FBS. Murine embryonic stem cells (designated AB2.2) were derived from the inner cell mass of E3.5 blastocyst stage mouse embryos (strain 129SvBrd) and maintained in DMEM with 15% FBS and leukemia inhibitory factor (500 U/ml; Chemicon International, Temecula, CA). The experiments reported here were carried out with cells that were at 65–75% confluence, except for murine embryo-derived stem cells, which were examined at lower cell density. In some experiments, cells were treated with actinomycin D at a final concentration of 0.1 μg/ml for 2 or 4 h. In these cases, control cells were exposed for an equal amount of time to 0.001% (vol/vol) ethanol (although we observed no differences between the results obtained with 0.001% ethanol-treated cells and cells not exposed to ethanol).
Immunocytochemistry and In Situ Hybridization
For immunostaining, cells were fixed and permeabilized as detailed previously (Politz et al., 2002). Nucleostemin was detected with a chicken polyclonal immunoglobulin Y antibody raised against a peptide corresponding to amino acids 522–538 of murine nucleostemin (dilution of 1:500; Tsai and McKay, 2002) followed by fluorescein-conjugated rabbit anti-chicken IgY (dilution of 1:100; Promega, Madison, WI) or a Cy3-conjugated F(ab′)2 fragment of donkey anti-chicken IgY (dilution of 1:200; Jackson ImmunoResearch Laboratories, West Grove, PA). No differences were observed in the nucleostemin immunostaining patterns obtained with these two secondary antibodies. Nucleolar fibrillar centers were detected by transfecting cells with a plasmid encoding a green fluorescent fusion protein of the rDNA-specific upstream binding factor (Chen and Huang, 2001). The dense fibrillar component of the nucleolus was detected with a murine monoclonal antibody (mAb) to mouse fibrillarin (Reimer et al., 1987), followed by rhodamine-coupled goat anti-mouse IgG. In situ nucleic acid hybridization to detect 28S rRNA or signal recognition particle RNA was carried out as described previously (Politz et al., 2002), except that oligos 2 and 4 were used to detect 28S rRNA (Politz et al., 2002). These two rat 28S rRNA probes and the rat SRP RNA probes used in this study cross-react with mouse 28S rRNA and SRP RNA, respectively. Combined immunostaining followed by in situ hybridization, microscopy, and image processing were performed as described previously (Politz et al., 2002), except that three-dimensional image stacks were deconvolved using exhaustive photon reassignment (Carrington et al., 1995). All two-dimensional (2D) images were scaled (using MetaMorph; Universal Imaging, Downingtown, PA) to exclude background signal (defined by the signal level observed after treatment with secondary antibody alone).
Electron Spectroscopic Imaging
Human neuroblastoma SK-N-SH cells were fixed with 2% formaldehyde at room temperature for 5 min and postfixed with 2% glutaraldehyde at room temperature for 15 min. Cells were dehydrated in steps with increasing concentrations of ethanol and embedded in Quetol resin (Ren et al., 2003). Sections of 70-nm thickness were cut with an ultramicrotome, picked up onto electron microscopy grids, and coated with a carbon film of 3-nm thickness to stabilize the specimens in the electron beam.
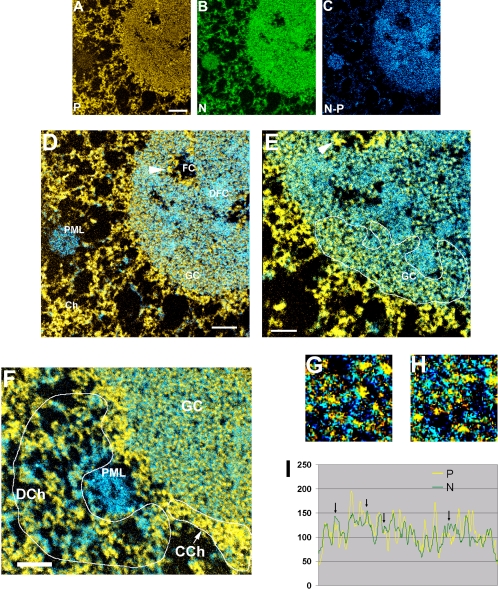
Electron spectroscopic imaging was carried out as described previously (Eskiw et al., 2003; Dellaire et al., 2004), by using a Tecnai 20 transmission electron microscope equipped with an imaging filter (Gatan). The microscope was operated with an accelerating voltage of 200 kV and with an energy-selecting slit aperture of 20 eV. Images were collected with a 12-bit cooled charge-coupled device detector. Phosphorus maps were calculated by the division of a postedge image collected at 155 eV by a preedge image collected at 120 eV. Similarly, nitrogen maps were formed with preedge and postedge images collected at 385 and 415 eV, respectively. Phosphorus (red) and nitrogen (green) maps (Figure 7, A and B) were computationally colored to distinguish protein-based from nucleic acid-based structures. Subtraction of nitrogen maps from phosphorus maps also was used to delineate the composition of structures in the images.
Figure 7.
Electron spectroscopic images of a nucleolus and surrounding nucleoplasm of a SK-N-SH cell nucleus. A phosphorus map (A), nitrogen map (B), and nitrogen minus phosphorus map (C) are used to distinguish protein-based structures from chromatin-based structures. An overlay of the phosphorus map onto the nitrogen minus phosphorus map is shown in D, and a region of higher magnification is shown in E. In D–H, yellow denotes the phosphorus map and blue the nitrogen minus phosphorus map. Various structures are labeled in D and F: chromatin (Ch), PML, fibrillar center (FC), dense fibrillar component (DFC), and granular component (GC). The DNA of a fibrillar center is indicated with an arrowhead. The arrowhead in D and E can be used to determine the rotation of the field in the lower and higher magnification images. The areas labeled DCh and in CCh in F refer to regions of decondensed and condensed chromatin, respectively. (G and H) Two fields of the GC shown at higher magnification and with enhanced contrast. (I) Linescan through a GC region; arrows point to N-rich peaks. Bar, 300 nm (A–D), 160 nm (E and F) (left); the full width of the high-magnification panels (G and H) corresponds to 115 nm.
ImageJ (public domain software) was used for the quantification of phosphorus and nitrogen levels from the elemental maps. Regions of interest were delineated corresponding to chromatin on the periphery of the nucleolus, DNA in the fibrillar centers, the dense fibrillar component, the granular component, and individual granules within the granular component.
RESULTS AND DISCUSSION
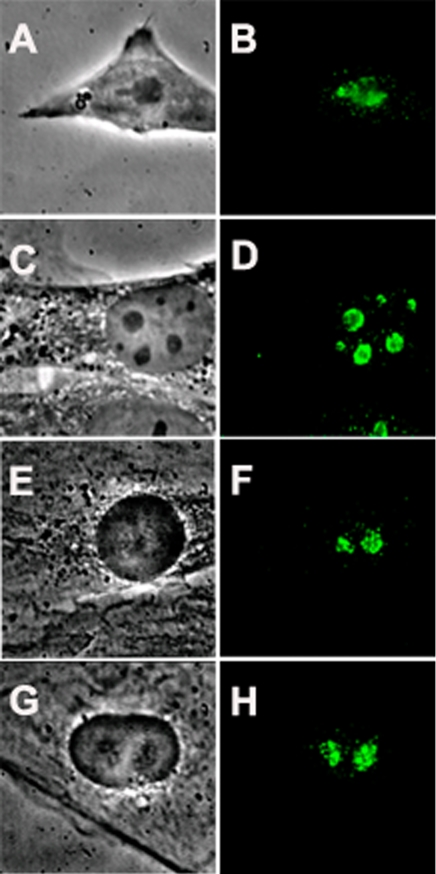
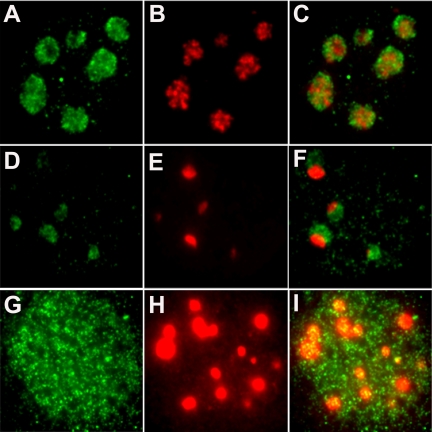
Nucleostemin was discovered in rat embryonic neural stem cells (Tsai and McKay, 2002). In the present investigation, we used a polyclonal antibody to nucleostemin that reacts with a single 60- to 70-kDa band in immunoblots (Tsai and McKay, 2002). As shown in Figure 1, A and B, we found that nucleostemin is also expressed in murine embryo-derived stem cells, with a predominantly nucleolar localization. Nucleostemin also is expressed at high levels in certain other, nontumor cell lines, e.g., Chinese hamster ovary cells and mouse 3T3 cells (Tsai and McKay, 2002; Figure 1, C and D), as well as rat myoblasts (Figure 1, E and F) and NRK cells (Figure 1, G and H). Because the relatively flat morphology of 3T3 cells and NRK cells is more favorable for immunostaining studies than the rounder shape of most stem cells, we used 3T3 and NRK cells in the present investigation.
Figure 1.
Nucleolar localization of nucleostemin. Murine and rat cells were stained with a peptide antibody raised against mouse nucleostemin (Tsai and McKay, 2002) followed by detection of the primary antibody with fluorescein-conjugated anti-chicken IgY. A, C, E, and G are phase contrast micrographs, and B, D, F, and H are the corresponding immunofluorescence images. (A and B) Murine embryonic stem cells. (C and D) Mouse 3T3 cells. (E and F) Rat L6 myoblasts. (G and H) Rat NRK cells. Each panel shows a microscope field 35 μm in width.
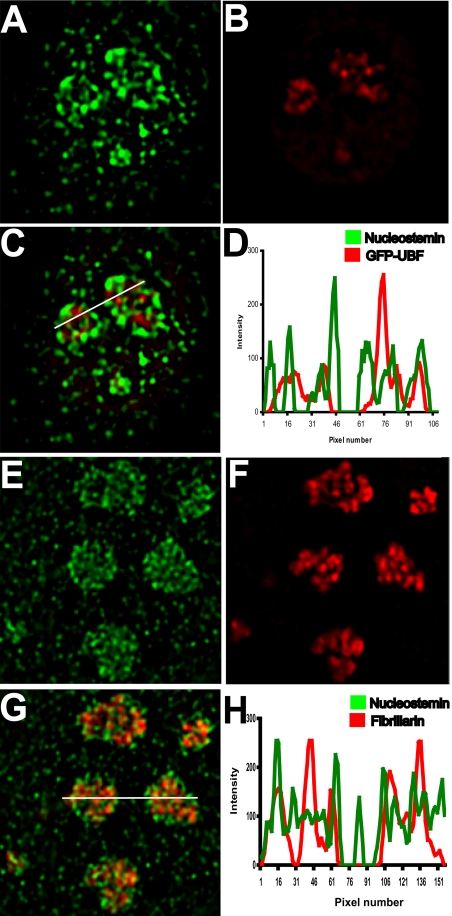
High-resolution digital imaging microscopy was used to examine the localization of nucleostemin in relation to the three classical regions of the nucleolus that have been defined by their ultrastructural appearance and by their roles in ribosome synthesis. These regions are 1) the fibrillar centers, which are the sites of the repeated rRNA genes; 2) the dense fibrillar component, which surrounds the fibrillar centers and into which the nascent rRNA extends and some of its processing occurs; and 3) the granular component, in which ribosome assembly steps are completed (Spector, 1993; Shaw and Jordan, 1995; Scheer and Hock, 1999; Koberna et al., 2003). Figure 2 shows single planes from deconvolved optical stacks of cells stained with nucleostemin antibody. Nucleostemin signal is concentrated in nucleoli and also is present at lower levels in the nucleoplasm, consistent with its reported shuttling behavior (Tsai and McKay, 2005). Nucleostemin (Figure 2A) did not concentrate in the fibrillar centers of 3T3 cells, as defined by the presence of the RNA polymerase I-specific transcription factor upstream binding factor (UBF) (Figure 2B). The merged images (Figure 2C) show little overlap between the two proteins, and a linescan through the nucleoli (Figure 2D) shows that very few of the intensity peaks coincide. Figure 2, E–H, show the results of a comparable experiment in which the localization of nucleostemin (Figure 2E) was compared with fibrillarin (Figure 2F), which is a specific marker for the dense fibrillar component. As can be seen in the merged image (Figure 2G), although there were a few regions where the two proteins overlap (small yellow foci), the intranucleolar distribution patterns of each protein were very different. These results indicate that nucleostemin does not participate in the transcription or early processing of rRNA. The localization of nucleostemin in the granular component was confirmed at the electron microscopic level by correlative analysis of the second antibody fluorescence in thin sections (Bazett-Jones, unpublished data).
Figure 2.
Nucleostemin localization in relation to fibrillar centers and the dense fibrillar component of the nucleolus. Mouse 3T3 cells were transfected with a plasmid encoding a green fluorescent protein (GFP) fusion of the rDNA-specific UBF to mark nucleolar fibrillar centers (A–D) or were immunostained with an mAb to fibrillarin to mark the dense fibrillar component of the nucleolus (E–H), followed by immunostaining for nucleostemin in both cases. A midplane from a deconvolved image stack of one nucleus is shown in each panel. (A) Nucleostemin. (B) UBF. (C) Merged image. (D) Plot showing intensity at each pixel along line drawn right to left in C (green, nucleostemin; red, UBF). (E) Nucleostemin. (F) Fibrillarin. (G) Merged image. (H) Plot showing intensity at each pixel along line drawn left to right in G (green, nucleostemin; red, fibrillarin). Each panel shows a microscope field 16 μm in width.
It can be seen in Figure 2, C and G, that a substantial fraction of nucleostemin was localized in peripheral regions of the nucleolus, in what seems to be a relatively restricted domain of the granular component. Because ribosome assembly events are thought to be taking place throughout the granular component, the relatively restricted localization pattern of nucleostemin suggests that it may not be stoichiometrically associated with nascent ribosomes.
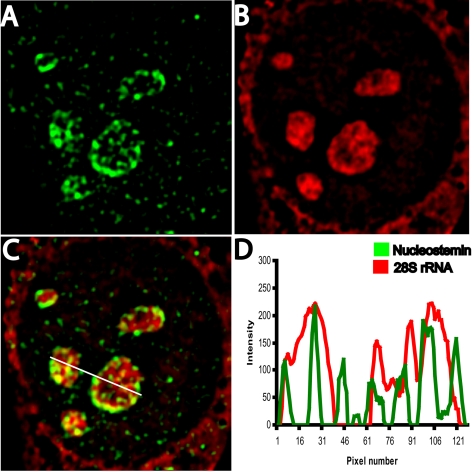
The most definitive markers for the ribosome-containing regions of the granular component of the nucleolus are the ribosomal RNAs that are present in the ribosomal subunits. We therefore performed nucleostemin immunostaining (Figure 3A) in combination with detection of 28S rRNA by in situ nucleic acid hybridization (Figure 3B). It can be seen that although some regions contained both nucleostemin and 28S rRNA (yellow in the merged image, Figure 3C; overlapping peaks in linescan shown in Figure 3D), the distribution pattern of nucleostemin (green) was nevertheless different from that of 28S rRNA (red), with high levels of nucleostemin present where 28S rRNA was present at only low concentration and vice versa. This lack of complete colocalization was especially evident with respect to the nucleostemin that occupied the outer edge of the nucleoli, but it also was observed with respect to more interiorly located nucleostemin (Figure 3, C and D). Thus, nucleostemin is not invariantly associated with 28S rRNA-containing ribosomal subunits in the granular component of the nucleolus.
Figure 3.
Localization of nucleostemin and 28S rRNA. 3T3 cells were subjected to immunostaining for nucleostemin followed by in situ hybridization for 28S rRNA (see Materials and Methods). Image stacks were captured and deconvolved, and a midplane of a representative nucleus is shown. (A) Nucleostemin. (B) 28S rRNA. (C) Merged image. (D) Plot showing intensity at each pixel along line drawn left to right in C (green, nucleostemin; red, 28S rRNA). Each panel shows a microscope field 19 μm in width.
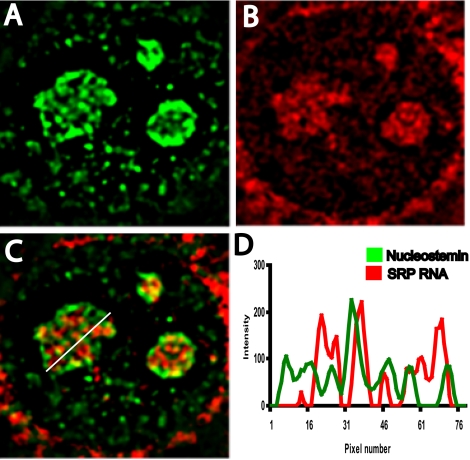
The nature of the regions of the granular component that do not contain 28S rRNA has not been defined. 18S rRNA has been observed to be colocalized with 28S rRNA in the granular component (Lazdins et al., 1997; Stavreva and McNally, personal communication of unpublished data), arguing against the possibility that the 28S rRNA-deficient regions represent separate loci that contain 18S rRNA. We recently investigated the distribution of signal recognition particle RNA within the nucleolus and found that, like nucleostemin, it is not appreciably present in fibrillar centers or the dense fibrillar component but rather primarily concentrates in regions of the granular component that are deficient in 28S rRNA (Politz et al., 2002). It was therefore of interest to investigate the intranucleolar localization of nucleostemin in comparison with SRP RNA (Figure 4) to learn more about these rRNA-deficient nucleolar regions. Would all “nonribosomal” nucleolar constituents (e.g., nucleostemin and SRP RNA) localize to the same granular component subdomains? As can be seen in the merged image (Figure 4C; linescan plot in Figure 4D), we found that nucleostemin and SRP RNA did not extensively colocalize within the nucleolus. In some cases, a region of nucleostemin overlapped with an SRP RNA-rich region, but most of the regions occupied by each entity were discrete. Therefore, because nucleostemin and 28S rRNA only partially overlapped (Figure 3C), and SRP and 28S rRNA also only partially overlap (Politz et al., 2002), it follows that there are numerous sites throughout the granular component at which only one of these three entities resides, unaccompanied by the other two. Thus, the landscape of the granular component is molecularly heterogeneous at the spatial resolution of these localization studies. Stated differently, these results rule out the possibility that various nonribosome-related molecules are confined to a common set of sites within the granular component.
Figure 4.
Localization of nucleostemin and signal recognition particle RNA. 3T3 cells were subjected to immunostaining for nucleostemin followed by in situ hybridization for signal recognition particle RNA. (A) Nucleostemin. (B) SRP RNA. (C) Merged image. (D) Plot showing intensity at each pixel along line drawn right to left in C (green, nucleostemin; red, SRP RNA). Each panel shows a microscope field 17 μm in width.
The degree of noncolocalization of the three nucleolar entities under discussion—nucleostemin, 28S rRNA, and SRP RNA—cannot be attributed merely to possible differences in their relative nucleolar abundance. Although a molecular species that is present in the nucleolus at a lower abundance would not necessarily display extensive spatial overlap with all the regions occupied by a more abundant entity, the former would be expected, at the least, to coreside with a subset of the latter if they were both confined to common sites in the nucleolus. But this is not what we observed. Rather, a considerable portion of each of the three entities is concentrated at sites in the nucleolus where neither of the other two are concentrated.
To further test nucleostemin's spatial segregation from components involved in the ribosome pathway, we investigated its behavior after actinomycin treatment. When mammalian cells are treated with low concentrations of actinomycin, the synthesis of rRNA is selectively inhibited (Perry, 1962; Roberts and Newman, 1966; Perry and Kelley, 1970). As a consequence, the nucleolus undergoes a reorganization in which the fibrillar centers, dense fibrillar component, and the granular component condense and become more spatially segregated from one another than usual (Hadjiolov, 1985). Notwithstanding these profound changes in rRNA synthesis and nucleolar organization, most of the ribosome-processing proteins that have been studied are observed to remain associated with these segregated nucleoli (although these proteins themselves spatially reorganize within the nucleoli). It was therefore of interest to examine the effects of a low concentration of actinomycin on the behavior of nucleostemin. Figure 5, A–C, show, as a control, the nucleostemin (A), fibrillarin (B), and merged (C) images for cells treated for 4 h with the same concentration of ethanol [0.001% (vol/vol)] as was present in the actinomycin experiments. Two hours after treating cells with a low concentration actinomycin (0.1 μg/ml), fibrillarin was observed to be concentrated into a single, large domain located near the edge of each nucleolus (Figure 5F, red), whereas nucleostemin retained its typical widespread distribution throughout the nucleoli (Figure 5F, green). In continuing contrast to the behavior of fibrillarin, after 4 h of actinomycin treatment nucleostemin no longer was concentrated in the nucleolus but instead was distributed throughout the nucleoplasm (Figure 5G), whereas fibrillarin was still retained in the nucleoli (Figure 5H). This highly differential behavior of nucleostemin and fibrillarin after low actinomycin treatment was observed in ∼50% of the cells in some experiments, and in nearly 100% of the cells in others. The basis of this experiment-to-experiment variation has not been explored in detail but did not seem to be related to cell density. In addition to 3T3 cells (Figure 5), a nucleolar departure of nucleostemin after low actinomycin treatment also was observed in NRK cells (our unpublished data). When the total nuclear signal was quantitated and normalized for nuclear area, there was no significant difference in the average amount of nucleostemin present in nuclei either before or after actinomycin treatment (758 ± 41 intensity units/pixel in untreated cells and 724 ± 25 intensity units/pixel in cells treated with actinomycin for 4 h). Therefore, the actinomycin effect represents a net translocation of nucleostemin to the nucleoplasm and not degradation of the protein.
Figure 5.
Contrasting responses of nucleostemin versus fibrillarin to selective inhibition of rRNA synthesis. 3T3 cells were treated with actinomycin at 0.1 μg/ml, and the intranuclear localization of nucleostemin and fibrillarin was detected by immunostaining. The panels show 2D images of single nuclei. (A–C) control cells, treated for 4 h with 0.001% (vol/vol) ethanol alone. (D–F) Cells treated with actinomycin for 2 h. (G–I) Cells treated with actinomycin for 4 h. (A, D, and G) Nucleostemin. (B, E, and H) Fibrillarin. (C, F, and I) Merged images. Nucleostemin signal in G and I is shown twice as bright as that in A and D so that the distribution is easily visualized, but the total nuclear signal in populations of cells treated with actinomycin for 4 h was not significantly different from that in populations of cells treated for 2 h (D) or not at all (A). These images are not deconvolved; therefore, signal that seems to overlap may lie above or below the plane of focus. Each panel shows a microscope field 20 μm in width.
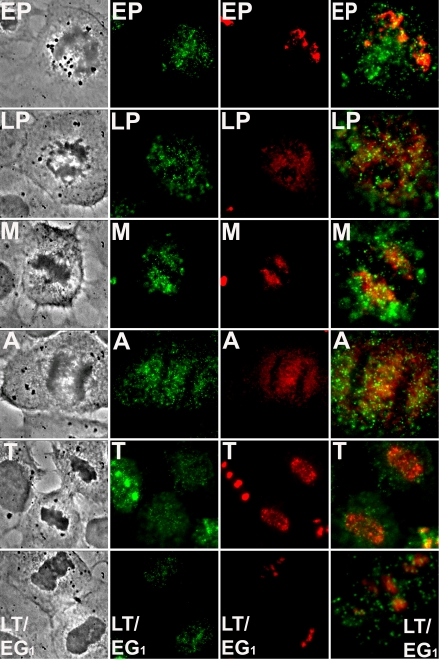
To further investigate the degree to which nucleostemin and the ribosome-related nucleolar protein fibrillarin differ with respect to their intracellular dynamics, we examined their behavior during and after mitosis. Nucleoli disassemble in late G2/prophase and begin to reform in telophase around nucleolar organizer regions with the subsequent appearance of prenucleolar bodies, followed by their coalescence into the definitive nucleoli of the early postmitotic cell (Dousett et al., 2000; Dundr et al., 2000; Leung et al., 2004). We stained cells for both nucleostemin and fibrillarin, and imaged cells that were in different stages of mitosis. Nucleostemin had already left nucleoli at early prophase (Figure 6, “EP”, green), whereas fibrillarin did not become similarly dispersed until late prophase (Figure 6, “LP”, red). After metaphase (Figure 6, “M”) and anaphase (Figure 6, “A”), fibrillarin was observed to begin concentrating within the reforming nuclei in telophase (Figure 6, “T”, red), as has been observed previously (Dousett et al., 2000). However, nucleostemin had not completely entered the nuclei at this stage (Figure 6, “T”, green). By the time cytokinesis was completed (Figure 6, “LT/EG1”), the fibrillarin had become concentrated into the nucleolus (red), whereas much of the nucleostemin was still dispersed throughout the nucleoplasm (green). These results agree with the localization behavior of nucleostemin during mitosis initially reported by Tsai and McKay (2002), and, in addition, show how this behavior contrasts with that of fibrillarin. Thus, these two nucleolar proteins of very different function also demonstrate temporally independent mitotic schedules.
Figure 6.
Differential dynamic behavior of nucleostemin and fibrillarin during postmitotic reformation of nucleoli. NRK cells were immunostained for nucleostemin and fibrillarin as described in Materials and Methods, and cells in various stages of mitosis were imaged. Cell cycle stages were identified by the criteria of chromatin or chromosome condensation or decondensation, nucleolar and nuclear envelope integrity, the extent of progression of the cleavage furrow, and the absence or presence of the midbody, all as determined by phase contrast microscopy. Each horizontal row of images consists of a phase contrast micrograph (left), nucleostemin immunostaining (left center), fibrillarin immunostaining (right center), and a merged image (right). EP, early prophase; LP, late prophase; M, metaphase; A, anaphase; T, telophase; and LT/EG1, late telophase/early G1. Each panel in the first three columns shows a microscope field 35 μm across. Panels in the last column showing the merged images are slightly enlarged (EP, 18 μm; LP, 18 μm; M, 16 μm; A, 20 μm; T, 25 μm; LT, upper cell shown, 11 μm, wide).
The differential time course of the reentry of fibrillarin and nucleostemin into nucleoli in telophase/early G1 is reminiscent of a comparable finding made recently as regards the temporally contrasting appearances of various mRNA splicing-related proteins into interchromatin granule clusters (a.k.a. speckles) within postmitotic daughter nuclei (Prasanth et al., 2003; Bubulya et al., 2004). It also is interesting to note that a previous study (Mintz and Spector, 2000) on the location of various proteins within speckles (during interphase) revealed a surprising degree of discrete, intraspeckle compartmentalization, even in these nucleoplasmic structures that are so much smaller than nucleoli. It will be interesting to see whether such similarly segregated zones of molecular composition are present in other nuclear bodies, e.g., Cajal bodies and promyelocytic leukemia (PML) nuclear bodies (Gall, 2000; Borden, 2002.)
Because these findings suggested the possibility that the landscape of the nucleolar granular component is one in which RNA-rich territories are interspersed with RNA-deficient (protein-rich) territories, we turned to the method of electron spectroscopic imaging (ESI). This technique is performed in the transmission electron microscope and is based on the principle of electron energy loss spectroscopy (Dellaire et al., 2004). In this method, some electrons that pass through the specimen lose characteristic amounts of energy by exciting or ionizing the specimen's atoms. Thus, the chemical composition of the specimen can be determined to a very high level of both elemental accuracy and spatial resolution with an electron spectrometer. In ESI, however, the electron spectrometer also acts as an imaging lens, so that element-specific maps of the specimen can be obtained. By comparing computationally colored phosphorus (Figure 7A) and nitrogen (Figure 7B) maps, structures that are nucleic acid rich can be distinguished from ones that are protein based. If the phosphorus map is subtracted from the nitrogen map, areas that contain protein structures that do not overlap with the phosphorus-rich backbone of DNA or RNA can be identified (Figure 7C). Overlaying the phosphorus map (yellow) onto the nitrogen minus phosphorus map (blue) facilitates definition of nucleic acid rich versus protein-rich structures (Figure 7, D–F). Chromatin, for example, is represented in shades of yellow, structures that are composed largely or entirely of protein, such as the core of a PML nuclear body, are represented in blue, and ribonucleoprotein structures in the nucleolus, which have intermediate phosphorus to nitrogen ratios, are shown in intermediate shades of yellow and blue (FC, DFC, and GC in Figure 7, D and F).
In addition to the qualitative information in computationally colored images, quantification of phosphorus and nitrogen levels provides additional information on the biochemical composition of subregions within the nucleolus. To obtain phosphorus and nitrogen ratios of the nucleolar domains, an internal standard was required. We chose to use regions of the most highly condensed chromatin at the periphery of the nucleolus for this purpose. This chromatin would be composed of ∼50% protein and 50% nucleic acid, based on the assumption that such chromatin is almost entirely nucleosomal, with little associated nonhistone chromosomal protein. This assumption is supported in Figure 7F. The chromatin in the region indicated by “CCh” is highly condensed and seems to be associated with little additional protein that does not overlap with the phosphorus of the DNA (few structures in the nitrogen minus phosphorus image; blue). In contrast, the chromatin in region “DCh” is less condensed and associated with a significant amount of protein, which coats or cross-links the chromatin fibers (structures colored blue in the nitrogen minus phosphorus image, Figure 7C).
The ESI results provide both confirmatory and new, higher resolution information that refines and extends the current model of nucleolar organization. First, the ESI data show that a major component of the fibrillar center is DNA (arrowhead in Figure 7, D and E). Quantification of phosphorus and nitrogen levels also reveals biochemical relationships of protein and nucleic acid composition in subnucleolar compartments. Comparisons of P and N ratios can be converted to stoichiometric relationships by using internal standards such as chromatin or ribosomes (Bazett-Jones et al., 1999). (This approach is superior to quantification from electron energy loss spectra. Reliable values for the partial cross section of scattering of these elements, required for quantification from spectra, have not been determined.) The P-to-N ratio of chromatin is 0.129 (based on nucleosomal composition), a value similar to the measured P-to-N ratio of the ribosomal gene chromatin in the fibrillar center (0.140, Table 1). The P-to-N ratio over large regions of either the dense fibrillar component (0.079) or the granular component (0.081) predicts an overall nucleic acid content of 31%. However, the P-to-N ratio of the granules themselves in the granular component (0.116) is significantly higher than that of the overall granular compartment and predicts a 45% nucleic acid content of the granules. This nucleic acid content is similar to that of mature ribosomes (54%). The difference in the P-to-N content of the granules in comparison with that of the entire granular component predicts that 14% of the granular component, corresponding to the spaces between the granules, is composed of protein and that this intergranular protein is not coresident with nucleic acid. This is further supported qualitatively by the high-magnification images (Figure 7, G and H) representing areas selected from a granular component region (Figure 7F), showing protein-based structures (blue) interspersed with the phosphorus-rich preribosomes (yellow). Linescans of the phosphorus and nitrogen maps passing through the granular component reveal quantitative differences in the distribution of the two elements (Figure 7I). The vertical arrows reveal relatively high levels of nitrogen (corresponding to predominantly protein-based structures) between the phosphorus peaks (corresponding to preribosomes). Similar ESI results were obtained in mouse 3T3 cells (our unpublished data), indicating that the existence of separate protein-rich and phosphorus-rich domains within the granular component is a general feature of at least mammalian nucleoli. We conclude that this ribosome-free domain of the granular component is populated by macromolecules that likely serve other functions.
Table 1.
Phosphorus/nitrogen ratios of nucleolar regions
| Nuclear or nucleolar region | Mean P/N (arbitrary units) | Measurement converted to theoretical value of P/N for chromatin | Measured value converted to P/N based on theoretical values for chromatin |
|---|---|---|---|
| Chromatin | 1.35 ± 0.15 | 0.129 | |
| Fibrillar center | 1.46 ± 0.15 | 0.140 | |
| Dense fibrillar component | 0.82 ± 0.23 | 0.079 | |
| Granular component | 0.84 ± 0.10 | 0.081 | |
| Granules in granular component | 1.21 ± 0.11 | 0.116 |
Chromatin has a theoretical P/N of 0.129, a nucleoprotein complex that is 50% protein/50% nucleic acid. From relating the measured P/N ratio of chromatin to the theoretical chromatin value, the measured P/N of 0.081 for the entire GC (granules and intergranules) indicates that 31% of the overall GC corresponds to nucleic acid. The granules themselves (within the GC) have a P/N of 0.116, predicting a nucleic acid content of 45%. Therefore, 14% (45-31%) of the GC is protein not associated with nucleic acid. This may be an underestimate, because the theoretical nucleic acid content of the ribosome is 54%, indicating that proteins that are not ribosomal proteins also associate or overlap with the granules, over and above the 14% (perhaps as much as 14 + 9 = 23% of the GC is protein that is not ribosomal protein). See Results and Discussion.
In summary, the results of this investigation establish that nucleostemin, a nucleolar protein with no known role in the production of ribosomes, has a distinctive intranucleolar localization, an unusual response to nucleolar segregation, and a delayed time course of nucleolar reentry after cell division. The results suggest that a substantial fraction of this protein is localized in regions of the nucleolar granular component that seem to contain very little, if any, rRNA. Electron spectroscopic analysis confirmed the existence of protein-rich, RNA-deficient regions within the granular component. Numerous ultrastructural studies of the nucleolus (using standard heavy metal stains) have revealed the granular component to contain electron-opaque foci surrounded by electron-translucent regions (Hadjiolov, 1985). The molecular nature of these interstitial regions of the granular component has never been defined. One possibility has been that this material is some sort of proteinaceous architecture that underlies the ribonucleoprotein particles that constitute the granularity of this nucleolar component. Our results with nucleostemin, a known shuttling protein, raise the alternative possibility that these electron-translucent regions of the granular component are composed of proteins transiently visiting the nucleolus, rather than a stably organized structure. Further work will be required to test this hypothesis, and it is to be noted that the two ideas are not mutually exclusive.
To paraphrase the term “plurifunctional nucleolus” that was coined previously (Pederson, 1998a), the present results indicate that the nucleolus is spatially pluralistic and strongly suggest that the nucleolus is functionally pluralistic as well. Much remains to be learned, however, about the full repertoire of molecules and functions that reside in those regions of the nucleolus where ribosome production is not taking place. Nucleostemin may only be the first of many yet to be discovered. For example, the RNA and protein components of telomerase have been reported to transiently visit the nucleolus (Pederson, 2004, and references cited therein; Zhang et al., 2004), and it is intriguing to consider the possibility that telomerase and the cell cycle-related, p53-interactive nucleostemin have similar locations when visiting the nucleolus.
Acknowledgments
We thank to Robert Tsai (Texas A&M University, College Station, TX) and Ronald McKay (National Institutes of Health, Bethesda, MD) for kindly providing nucleostemin antibody; Sui Huang (Northwestern University Feinberg School of Medicine, Chicago, IL) for the GFP-UBF plasmid; and Joan Steitz (Howard Hughes Medical Institute, Yale University School of Medicine, New Haven, CT) for fibrillarin mAb. We thank Marilyn Keeler and Stephen Jones (University of Massachusetts Medical School, Worcester, MA) for providing murine embryonic stem cells, and we gratefully acknowledge Christine Powers in the Pederson laboratory for expert technical assistance. We also thank Ren Li in the Bazett-Jones laboratory for valuable technical assistance in preparing the samples for electron microscopy and for collecting the electron spectroscopic images. This study was supported by Grant RGP0031 from the Human Frontier Scientific Program Organization (T. P.) and Grant FRN 14311 from the Canadian Institutes of Health Research (D.P.B.-J.) D.P.B.-J. is the recipient of the Canada Research Chair in Molecular and Cellular Imaging.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E05–02–0106) on April 27, 2005.
References
- Alavian, C. N., Politz, J.C.R., Lewandowski, L. B., Powers, C. M., and Pederson, T. (2004). Nuclear export of signal recognition particle RNA in mammalian cells. Biochem. Biophys. Res. Commun. 313, 351–355. [DOI] [PubMed] [Google Scholar]
- Andersen, J. S., Lyon, C. E., Fox, A. H., Leung, A.K.L., Lam, Y. W., Steen, H., Mann, M., and Lamond, A. I. (2002). Directed proteomic analysis of the human nucleolus. Curr. Biol. 12, 1–11. [DOI] [PubMed] [Google Scholar]
- Bazett-Jones, D. P., Hendzel, M. J., and Kruhlak, M. J. (1999). Stoichiometric analysis of protein- and nucleic acid-based structures in the cell nucleus. Micron 30, 151–157. [DOI] [PubMed] [Google Scholar]
- Borden, K.L.B. (2002). Pondering the promyelocytic leukemia protein (PML) puzzle: possible function for PML nuclear bodies. Mol. Cell. Biol. 22, 5259–5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubulya, P. A., Prasanth, K. V., Deerinck, T. J., Gerlich, D., Beaudouin, J., Ellisman, M. H., Ellenberg, J., and Spector, D. L. (2004). Hypophosphorylated SR splicing factors transiently localize around active nucleolar organizing regions in telophase daughter nuclei. J. Cell Biol. 167, 51–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington, W. A., Lynch, R. M., Moore, E. D., Isenberg, G., Fogarty, K. E., and Fay, F. S. (1995). Superresolution three-dimensional images of fluorescence in cells with minimal light exposure. Science 268, 1483–1487. [DOI] [PubMed] [Google Scholar]
- Chen, D., and Huang, S. (2001). Nucleolar components involved in ribosome biogenesis cycle between the nucleolus and the nucleoplasm in interphase cells. J. Cell Biol. 153, 169–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciufo, L. F., and Brown, J. D. (2000). Nuclear export of yeast signal recognition particle lacking Srp54p by the Xpo1p/Crm1p NES-dependent pathway. Curr. Biol. 10, 1256–1264. [DOI] [PubMed] [Google Scholar]
- Dellaire, G., Nissman, R., and Bazett-Jones, D. P. (2004). Correlative light and electron spectroscopic imaging of chromatin in situ. Methods Enzymol. 375, 465–478. [DOI] [PubMed] [Google Scholar]
- Dousett, T., Wang, C., Verheggen, C., Chen, D., Hernandez-Verdun, D., and Huang, S. (2000). Initiation of nucleolar assembly is independent of RNA polymerase I transcription. Mol. Biol. Cell 11, 2705–2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr, M., Misteli, T., and Olson, M.O.J. (2000). The dynamics of postmitotic reassembly of the nucleolus. J. Cell Biol. 150, 433–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskiw, C. H., Dellaire, G., Mymryk, J. S., and Bazett-Jones, D. P. (2003). Size, position and dynamic behavior of PML nuclear bodies following cell stress as a paradigm for supramolecular trafficking and assembly. J. Cell Sci. 116, 4455–4466. [DOI] [PubMed] [Google Scholar]
- Gall, J. G. (2000). Cajal bodies: the first 100 years. Annu. Rev. Cell Dev. Biol. 16, 273–300. [DOI] [PubMed] [Google Scholar]
- Grosshans, H., Deinert, K., Hurt, E., and Simos, G. (2001). Biogenesis of the signal recognition particle (SRP) involves import of SRP proteins into the nucleolus, assembly with the SRP-RNA, and Xpo1p-mediated export. J. Cell Biol. 153, 745–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjiolov, A. A. (1985). The Nucleolus and Ribosome Biogenesis, Vienna, Austria: Springer.
- Jacobson, M. R., and Pederson, T. (1998). Localization of signal recognition particle RNA in the nucleolus of mammalian cells. Proc. Natl. Acad. Sci. USA 95, 7981–7986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koberna, K., Malinsky, J., Pliss, A., Masata, M., Vecerova, J., Fialova, M., Bednar, J., and Raska, I. (2003). Ribosomal genes in focus: new transcripts label the dense fibrillar components and form clusters indicative of “Christmas trees” in situ. J. Cell Biol. 157, 743–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazdins, I. B., Delannoy, M., and Sollner-Webb, B. (1997). Analysis of nucleolar transcription and processing domains and pre-rRNA movements by in situ hybridization. Chromosoma 105, 481–495. [DOI] [PubMed] [Google Scholar]
- Leung, A.K.L., and Lamond, A. I. (2003). The dynamics of the nucleolus. Crit. Rev. Eukaryot. Gene Expr. 13, 39–54. [DOI] [PubMed] [Google Scholar]
- Leung, A.K.L., Gerlich, D., Miller, G., Lyon, C., Lam, Y. W., Lleres, D., Daigle, N., Zomerdijk, J., Ellenberg, J., and Lamond, A. I. (2004). Quantitative kinetic analysis of nucleolar breakdown and reassembly during mitosis in live cells. J. Cell Biol. 166, 787–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S. J., Cai, Z. W., Liu, Y. J., Dong, M. Y., Sun, L. Q., Hu, G. F., Wei, Y. Y., and Lao, W. D. (2004). Role of nucleostemin in growth regulation of gastric cancer, live cancer and other malignancies. World J. Gastroenterol. 10, 1246–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintz, P. J., and Spector, D. L. (2000). Compartmentalization of RNA processing factors within nuclear speckles. J. Struct. Biol. 129, 241–251. [DOI] [PubMed] [Google Scholar]
- Misteli, T. (2005). Going in GTP cycles in the nucleolus. J. Cell Biol. 168, 177–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson, M.O.J., Dundr, M., and Szebeni, A. (2000). The nucleolus: an old factory with unexpected capabilities. Trends Cell Biol. 10, 189–196. [DOI] [PubMed] [Google Scholar]
- Olson, M.O.J., Hingorani, K., and Szebeni, A. (2003). Conventional and non-conventional roles of the nucleolus. Int. Rev. Cytol. 219, 199–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson, T. (1998a). The plurifunctional nucleolus. Nucleic Acids Res. 26, 3871–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson, T. (1998b). Growth factors in the nucleolus? J. Cell Biol. 143, 279–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson, T. (2004). Can telomerase be put in its place? J. Cell Biol. 164, 637–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson, T., and Politz, J. C. (2000). The nucleolus and the four ribonucleoproteins of translation. J. Cell Biol. 148, 1091–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry, R. P. (1962). The cellular sites of ribosomal and 4S RNA. Proc. Natl. Acad. Sci. USA 48, 2179–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry, R. P., and Kelley, D. E. (1970). Inhibition of RNA synthesis by actinomycin D: characteristic dose-response of different RNA species. J. Cell. Physiol. 76, 127–139. [DOI] [PubMed] [Google Scholar]
- Politz, J. C., Yarovoi, S., Kilroy, S. M., Gowda, K., Zwieb, C., and Pederson, T. (2000). Signal recognition particle components in the nucleolus. Proc. Natl. Acad. Sci. USA 97, 55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz, J. C., Lewandowski, L. B., and Pederson, T. (2002). Signal recognition particle RNA localization within the nucleolus differs from the classical sites of ribosome synthesis. J. Cell Biol. 159, 411–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth, K. V., Sacco-Bubulya, P. A., Prasanth, S. G., and Spector, D. L. (2003). Sequential entry of components of gene expression machinery into daughter nuclei. Mol. Biol. Cell 14, 1043–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer, G., Pollard, K. M., Penning, C. A., Ochs, R. L., Lischwe, L. A. Busch, H., and Tan, E. M. (1987). mAb from a (New Zealand Black × New Zealand White) F1 mouse and some human scleroderma target a Mr 34,000 nucleolar protein of the U3 RNP particle. Arthritis Rheum. 30, 793–800. [DOI] [PubMed] [Google Scholar]
- Ren, Y., Kruhlak, M. J., and Bazett-Jones, D. P. (2003). Same serial section light and energy-filtered transmission electron microscopy. J. Histochem. Cytochem. 51, 605–612. [DOI] [PubMed] [Google Scholar]
- Roberts, W. K., and Newman, J. (1966). Use of low concentrations of actinomycin D in the study of RNA synthesis in Ehrlich ascites cells. J. Mol. Biol. 20, 63–73. [DOI] [PubMed] [Google Scholar]
- Scheer, U., and Hock, R. (1999). Structure and function of the nucleolus. Curr. Opin. Cell Biol. 11, 385–390. [DOI] [PubMed] [Google Scholar]
- Scherl, A., Coute, Y., Deon, C., Calle, K., Kindbeiter, K., Sanchez, J.-D., Greco, A., Hochstrasser, D., and Diaz, J. J. (2002). Functional proteomic analysis of human nucleolus. Mol. Biol. Cell 13, 4100–4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw, P. J., and Jordan, E. G. (1995). The nucleolus. Annu. Rev. Cell Dev. Biol. 11, 93–121. [DOI] [PubMed] [Google Scholar]
- Sommerville, J., Brumwell, C. B., Politz, J.C.R., and Pederson, T. (2005). Signal recognition particle assembly in relation to the function of amplified nucleoli of Xenopus oocytes. J. Cell Sci. 118, 1299–1307. [DOI] [PubMed] [Google Scholar]
- Spector, D. L. (1993). Macromolecular domains within the cell nucleus. Annu. Rev. Cell Biol. 9, 265–315. [DOI] [PubMed] [Google Scholar]
- Tsai, R.Y.L., and McKay, R.D.G. (2002). A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes Dev. 16, 2991–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, R.Y.L., and McKay, R.D.G. (2005). A multistep, GTP-driven mechanism controlling the dynamic shuttling of nucleostemin. J. Cell Biol. 168, 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent, W. S., and Miller, Jr., O. L. (1966). International symposium on the nucleolus. Its structure and function. Montevideo, Uruguay. J. Natl. Cancer Inst. Monogr. 23, 1–610. [Google Scholar]
- Zhang, S., Hemmerich, P., and Grosse, F. (2004). Nucleolar localization of the human telomeric repeat binding factor 2 (TRF2). J. Cell Sci. 117, 3935–3945. [DOI] [PubMed] [Google Scholar]