Abstract
Spatially distinct populations of microfilaments, characterized by different tropomyosin (Tm) isoforms, are present within a neuron. To investigate the impact of altered tropomyosin isoform expression on neuronal morphogenesis, embryonic cortical neurons from transgenic mice expressing the isoforms Tm3 and Tm5NM1, under the control of the β-actin promoter, were cultured in vitro. Exogenously expressed Tm isoforms sorted to different subcellular compartments with Tm5NM1 enriched in filopodia and growth cones, whereas the Tm3 was more broadly localized. The Tm5NM1 neurons displayed significantly enlarged growth cones accompanied by an increase in the number of dendrites and axonal branching. In contrast, Tm3 neurons displayed inhibition of neurite outgrowth. Recruitment of Tm5a and myosin IIB was observed in the peripheral region of a significant number of Tm5NM1 growth cones. We propose that enrichment of myosin IIB increases filament stability, leading to the enlarged growth cones. Our observations support a role for different tropomyosin isoforms in regulating interactions with myosin and thereby regulating morphology in specific intracellular compartments.
INTRODUCTION
The actin cytoskeleton plays an essential role in the structural changes that initially establish neuronal shape and subsequently contribute to the morphological differentiation of neurons. Actin filaments have been implicated in the initial sprouting of neurites, whereas microtubules strengthen and support the new extensions (Smith, 1988, 1994).
Tropomyosin (Tm) isoforms, integral components of actin microfilaments, form coiled-coil head-to-tail dimers that bind along the major groove of actin polymers (Phillips et al., 1979). Tms are derived from four highly conserved genes known as the αTmfast, βTm, γTm (Tm5NM), and δTm genes that, via alternative splicing, give rise to >40 isoforms (Lees-Miller and Helfman, 1991; Dufour et al., 1998; Cooley and Bergtrom, 2001). Despite the well understood function of Tms in muscle where, together with the troponin complex, they regulate contraction in a calcium-dependent manner, little is known about their role in nonmuscle cells. In vitro studies have implicated Tms in the stabilization of the actin cytoskeleton by protecting actin filaments from the severing action of gelsolin (Ishikawa et al., 1989) and the depolymerizing action of ADF/cofilin (Bernstein and Bamburg, 1982). Gene transfection studies have demonstrated that tropomyosin isoforms can regulate the organization of actin filaments in transformed cells (Prasad et al., 1993; Boyd et al., 1995; Gimona et al., 1996) and the neuroepithelial cell line B35 (Bryce et al., 2003).
In neurons, a strict repertoire of Tm isoforms is known to be expressed. These include TmBr3, Tm5a and Tm5b from the αTmfast gene; multiple products from the γTm gene; and Tm4 from the δTm gene. Most interestingly, Tm isoforms have been previously shown to be spatially and temporally regulated, identifying distinct subcellular compartments. The Tm5a and Tm5b isoforms are enriched in the growth cones of freshly plated primary cortical neurons (Schevzov et al., 1997). In contrast, a product from the γTm gene is absent from the growth cone but enriched in the axon hillock and proximal region of axons and dendrites (Weinberger et al., 1996; Schevzov et al., 1997). Finally, as reported by Had et al., (1994), Tm4 is enriched in the growth cone, whereas TmBr3 is absent. Together, these studies suggest that the observed specific expression and spatial segregation of Tm isoforms in neurons may lead to the formation of actin filaments with specific functions (Gunning et al., 1998).
The aim of the present study was to perturb the normal repertoire of Tm isoforms and assess their effects on neuronal morphogenesis. Consequently, embryonic primary cortical neurons cultured from transgenic mice overexpressing Tm3 and the human homologue of Tm5NM1 were investigated. Overexpression of these tropomyosin isoforms had a profound and differential effect on neuronal morphology and differentiation. Embryonic cortical neurons overexpressing Tm5NM1 displayed significantly enlarged growth cones with Tm5a, and myosin IIB enriched in the periphery. In addition, an increase in the number of dendrites and axonal branching was observed in the Tm5NM1 neurons. In contrast, inappropriate expression of the high-molecular-weight tropomyosin isoform (Tm3) results in an initial inhibition of neurite outgrowth followed by a significant decrease in the number and length of dendrites. Hence we conclude that the expression of Tm isoforms is informative for the establishment of neuronal structure.
MATERIALS AND METHODS
Cell Culture
Primary cortical neurons were prepared as described previously (Schevzov et al., 1997). In brief, brains from embryonic day 14.5 stage mouse embryos were removed and transferred to a culture dish containing phosphate-buffered saline (PBS) calcium-magnesium free. The cerebral cortices were freed from the meninges placed in a fresh tube and treated for 15 min with 0.25% trypsin, 0.15% DNaseI (Roche, Sydney, Australia) at 37°C. In a sterile hood, the tissue was gently triturated by passing through a glass Pasteur pipette. The tissue pieces were allowed to settle for 2 min, and the cell suspension was plated onto poly-l-lysine (1 mg/ml in PBS; Sigma-Aldrich, Castle Hill, Australia)–coated eight-well chamber slides or 100-mm dishes. Cells were cultured in Neurobasal media containing 2 mM l-glutamine and B27 supplements (Invitrogen, Melbourne, Australia) at 37°C in a humidified atmosphere with 5% CO2. The stable transfection of tropomyosin isoforms into the B35 neuroepithelial cell line has been described in detail in Bryce et al. (2003). In brief, the B35 cells (Schubert et al., 1974) were maintained in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (FBS) (Invitrogen) and 2 mM l-glutamine (Invitrogen) at 37°C in a humidified atmosphere with 5% CO2. Stably transfected Tm3 and Tm5NM1 B35 clonal cell lines were generated using Lipofectamine 2000 according to manufacturer's instructions (Invitrogen). Cells were differentiated by the addition of 1 mM dibutyryl cAMP (Sigma-Aldrich, Sydney, Australia).
Generation and Screening of Transgenic Animals
Transgenic animals carrying the human Tm5NM1 cDNA were generated and screened as described previously (Bryce et al., 2003). The transgenic animals carrying the rat Tm3 cDNA were generated and screened in the same manner as the Tm5NM1 transgenic animals. The human Tm5NM1 differs from the mouse at amino acid 4, isoleucine in the human, and serine in the mouse. The rat Tm3 is identical to the mouse.
Immunofluorescence Staining
Cells for immunofluorescence staining were fixed with 4% paraformaldehyde in PBS for 15 min, rinsed three times with PBS, and then permeabilized with chilled methanol for 15 min. The neurons were then incubated in blocking solution/wash solution (2% FBS in PBS) for 30 min and then incubated with the primary antibody diluted in blocking solution for 1 h at room temperature. The cells were rinsed three times with wash solution and then incubated with the secondary antibody for 1 h. Cells were finally rinsed three times in PBS before mounting a coverslip onto the chamber slides with 1,2-diazabicyclo (2.2.2) octane (Sigma-Aldrich, Castle Hill). Slides were examined with a Leica TCSP2 laser scanning confocal microscope by using a 63× objective. Neurons were optically sectioned in the x-y plane and serial sections of 0.5-μm thickness were taken with eight scan averaging. Images were processed using Adobe Photoshop 5.0 (Adobe Systems, Mountain View, CA). The primary rabbit antibodies used were WSα/9d antiserum (Weinberger et al., 1996) at 1:1000, WS5/9d (Weinberger et al., 1996) at 1:200, and MHCIIB (Rochlin et al., 1995) at 1:250 (a kind gift of Robert Adelstein, National Institutes of Health, Bethesda, MD). The mouse monoclonal antibodies used were LC1 (Warren et al., 1995) at 1:200 (a kind gift of Jim Lin, University of Iowa, Iowa City, IA), C4 total actin (Lessard, 1988) at 1:500 (a kind gift of Jim Lessard, Cincinnati Children's Hospital Medical Center, Cincinnati, OH), β-actin (clone AC-74) at 1:1000 (Sigma-Aldrich, Castle Hill), TM311 (Sigma-Aldrich, Castle Hill) at 1:500; CG3 (IgM class) at 1:250 (a kind gift of Jim Lin), and SMI 310 phosphorylated neurofilament H at 1:500 (Sternberger Monoclonals, Baltimore, MD). The primary sheep antibody used was γ/9d at 1:100 (Percival et al., 2004). The secondary antibodies used at 1:1000 were Alexa Fluor488 donkey anti-mouse IgG (H+L) conjugate and Cy3 donkey antirabbit IgG (H+L) (Jackson ImmunoResearch Laboratories, West Grove, PA).
Gel Electrophoresis and Immunoblotting Blots
Primary cortical neurons grown on poly-l-lysine (1 mg/ml, Sigma-Aldrich, Castle Hill)–coated 100-mm plastic dishes were washed with PBS, lysed in SDS solubilization buffer (10 mM Tris, pH 7.6, 2% SDS, 2 mM dithiothreitol), and the extracts were heated at 95°C for 3 min. The proteins were precipitated as described by Wessel and Flugge (1984), resolubilized in SDS solubilization buffer, and the protein concentration was determined by using a BCA protein assay kit (Pierce Chemical, Rockford IL). Before electrophoresis, protein samples were solubilized and boiled for 5 min in 2× sample buffer (1× buffer contains 0.125 M Tris, pH 6.8, 0.5% SDS, 5% glycerol, 5% 2-mercaptoethanol, 0.005% bromphenol blue). SDS-PAGE was performed according to Laemmli (1970) on 12.5% acrylamide and 0.1% bis-acrylamide. Proteins were transferred to Immobilon-P polyvinylidene difluoride (Millipore, Billerica, MA) for 2 h at 80 V according to Towbin et al. (1979). A 5% low fat skim milk in Tris-buffered saline (TBS) (100 mM Tris HCl, pH 7.5, 150 mM NaCl) solution was used to block nonspecific binding on the blot. Primary and secondary (anti-rabbit, -sheep, and -mouse immunoglobulin-conjugated horseradish peroxidase; Amersham Biosciences UK, Little Chalfont, Buckinghamshire, United Kingdom) antibodies were incubated for 1 h each, and 3 × 10-min washes with TBS with 0.05% Tween 20 were carried out after each antibody incubation. Blots were developed with the Western lighting chemiluminescence reagent (PerkinElmer Life and Analytical Sciences, Boston, MA) and exposed to Fuji x-ray film (Eastman Kodak, Rochester, NY). Quantitation was performed using an Amersham Biosciences (Sunnyvale, CA) densitometer as described previously (Bryce et al., 2003).
Morphological Analysis of Neurons
Neurons were plated on chamber slides as described above, cultured for 5 d, and immunofluorescence stained with an actin antibody (C4) and the phosphorylated neurofilament H antibody to confirm that the longest neurite was the axon. Fluorescence images were taken using a Spot II cooled charge-coupled device digital camera (Diagnostic Instruments, Sterling Heights, MI) mounted on an Olympus Bx50 microscope. For neurite length determinations, the neurites were traced and the morphometry analysis was performed using Image-Pro Plus version 4.0 (Media Cybernetics, Silver Spring, MD). A branch was defined as an extension from the axon and was >10 μm in length. The length of the axon or dendrite was defined as the distance from the soma to the tip of the process. Surface area measurement of the growth cones was performed by tracing the perimeter of the growth cone and the proximal region of the growth cone was delineated as the point where it first splayed from the axon. An average of 50 neurons from each group was selected from a number of chamber slides from two independent cultures.
To determine the fluorescence intensity of the MHCIIB staining in the growth cones, cells were cultured and images taken as described above. The intensity of fluorescence was measured in the leading edge and central region of the growth cone by using the color sample tool function witha3by3 image pixel average in Adobe Photoshop 5.5 (Adobe Systems). The percentage of growth cones with a leading edge to central region intensity ratio of >1 was calculated from an average of 80 growth cones from two independent experiments. All statistical analysis was performed using nonparametric analysis of variance (ANOVA) test (Kruskal-Wallis) on the SPSS program (SPSS, Chicago, IL). All error bars indicate SD.
RESULTS
Expression of Tm Transgenes in Mouse Brain and Primary Neurons
Transgenic mice were generated with gene constructs that placed either the rat Tm3 or human Tm5NM1 coding regions under the control of the human β-actin promoter (Qin and Gunning, 1997; Bryce et al., 2003). Tm3, derived from the αTm gene (Figure 1A), was chosen as a suitable nonneuronal Tm because we had previously demonstrated that primary cortical neurons lack expression of high-molecular-weight tropomyosin isoforms such as Tm3 (Schevzov et al., 1997; Hughes et al., 2003). In contrast, Tm5NM1, derived from the γTm gene (Figure 1A), was chosen as an appropriate neuronal Tm isoform because its expression has been previously demonstrated to correlate with neurite outgrowth (Weinberger et al., 1996). We reasoned that if the choice of Tm isoforms is informative for neuronal morphogenesis, then these two isoforms should differ in their impact on this process.
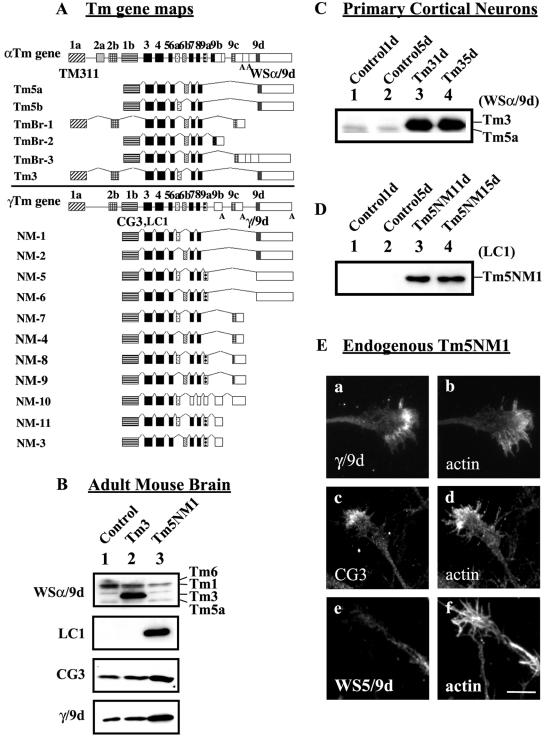
Figure 1.
Organization of Tm genes, their expression in transgenic mice, and subcellular localization in neurons. (A) Schematic representation of the organization of the α, and γ mammalian Tm genes and the respective neuronal isoforms. Exons, shaded boxes, are numbered 1–9; unshaded boxes correspond to 3′ untranslated sequences; and introns are shown as lines. The black shaded exons are common to all genes. The name of the Tm antibody is indicated in bold below the exon where the epitope is found. (B, C, and D) Equal loading (10 μg) of total cellular protein was isolated and electrophoresed on a 12.5% low-bis SDS-PAGE gel. (B) Immunoblots of adult mouse brain. Lane 1, control brain; lane 2, Tm3 transgenic; and lane 3, Tm5NM1 were probed with WSα/9d to detect Tm3, LC1 to detect the exogenous Tm5NM1, CG3 to detect all products from the γTm gene, and γ/9d to detect Tm5NM1 and Tm5NM2. (C) Immunoblot of Tm3 primary cortical neurons. Lane 1, control 1 d old; lane 2, control 5 d; lane 3, Tm3 1 d; and lane 4, Tm3 5 d, probed with the WSα/9d antibody. (D) Immunoblot of Tm5NM1 primary cortical neurons. Lane 1, control 1 d old; lane 2, control 5 d; lane 3, Tm5NM1 1 d; and lane 4, Tm5NM1 5 d, probed with the LC1 antibody. (E) Endogenous Tm5NM1 sorts to the peripheral region of the growth cone unlike Tm5NM2. Control primary cortical neurons cultured for 5 d were double immunofluorescence stained with γ/9d (a), actin (b), CG3 (c), actin (d), WS5/9d (e), and actin (f). Bar, 10 μm.
The expression of these transgenes was confirmed by Western blot. Adult mouse brain protein lysate from control or homozygous Tm3 and Tm5NM1 transgenic mice was reacted with the WSα/9d antibody, which detects products from the α and β Tm genes (including Tm3) but not from the γ or δ Tm genes (Figure 1B) (Schevzov et al., 1997). The Tm3 transgenic mouse brain shows high level of Tm3 in comparison with no detectable Tm3 in control and Tm5NM1 transgenic brains (Figure 1B). The exogenously expressed Tm5NM1 was monitored with the LC1 antibody that specifically identifies the human Tm5NM1 but not the endogenous mouse isoform based on the single amino acid difference between them (Warren et al., 1995). No detectable human Tm5NM1 is identified in either control or Tm3 brains, but a strong signal is present in the Tm5NM1 lane (Figure 1B). Furthermore, the LC1 antibody detects no other Tm proteins. The CG3 antibody raised against exon 1b of the γTm gene detects all nonmuscle products from the γTm gene, including the exogenous Tm5NM1 (Figure 1B). A two-fold increase in the 30-kDa band is seen in the Tm5NM1 transgenic brain sample, indicating that the level of total Tm5NM1 has at least doubled in this sample. In contrast, no significant changes in the total output from the γTm gene are seen in the Tm3 brain sample (Figure 1B). The γ/9d antibody, which detects exon 9d of the γTm gene (Figure 1A), similarly shows a significant increase (2-fold) in the Tm5NM1 brain sample corresponding to the endogenous product plus the exogenous Tm5NM1 (Figure 1B). This confirms that the Tm5NM1 level is at least twice that in control animals.
Embryonic primary cortical neurons isolated from control or heterozygous 14.5ED transgenic embryos were cultured for 1 and 5 d. Tm3 cortical neurons show substantial levels of Tm3 (Figure 1C, lanes 3 and 4, respectively). This demonstrates that neurons are capable of accumulating considerable levels of a nonneuronal tropomyosin. A significant level of the exogenous Tm5NM1 is detected in the Tm5NM1 cortical neurons with the LC1 antibody (Figure 1D, lanes 3 and 4). The level of expression of the exogenous proteins, both Tm3 and Tm5NM1, in the primary cortical neurons, remained unaltered after 5 d in culture.
Primary Neurons Differentially Sort Tm3 and Tm5NM1
The endogenous Tm5NM1 isoform localizes to the peripheral region of the growth cone. Control primary cortical neurons were double immunostained with the γ/9d antibody (recognizes Tm5NM1 and Tm5NM2) and actin. Figure 1E shows staining of both neurite and the peripheral region of the growth cone with γ/9d (Figure 1E, a). The majority of growth cones (80%, n = 50 in two separate cultures) showed this pattern of staining. The CG3 antibody also displays this pattern of staining (Figure 1E, c). Previous staining with the antibody WS5/9d, which specifically shows the location of Tm5NM2 (Percival et al., 2004), reacts only with the shafts of neurites (Figure 1E, e) (Hannan et al., 1995; Schevzov et al., 1997). This therefore suggests that Tm5NM1, but not Tm5NM2, is located in the growth cone (Figure 1E, a).
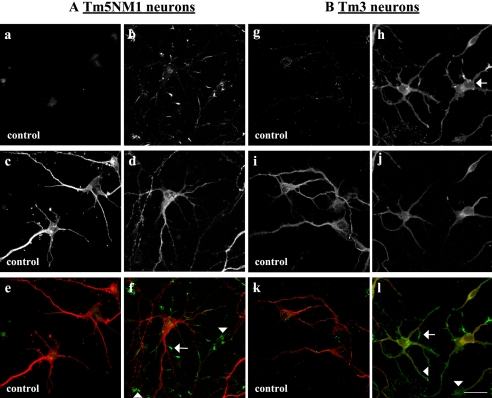
The exogenous Tm5NM1 isoform localizes to the tips of neurites and filopodia. Both control and Tm5NM1 transgenic primary neurons were stained with WS5/9d to identify neuronal processes (Hannan et al., 1995) and the Tm5NM1-specific antibody LC1. Figure 2 shows that control primary cortical neurons labeled with WS5/9d (Figure 2, c and e) display no background labeling with the LC1 antibody (Figure 2a). In contrast, LC1 intensely stains the growth tips of neurons (Figure 2, b and f, arrow and arrowheads) and only poorly stains the processes and cell bodies (Figure 2b). We therefore conclude that the exogenous Tm5NM1 protein sorts to the growth cone of the neuron as does the endogenous Tm5NM1 protein and confirm that the γ/9d product in the axon shaft is Tm5NM2. In addition, this shows that the Tm5NM1 protein alone contains information sufficient to precisely target it to the growth tips of neurites.
Figure 2.
Subcellular localization of the exogenous Tm5NM1 and Tm3. Primary cortical neurons were cultured in vitro for 5 d and double immunofluorescence stained with the LC1 antibody to visualize the exogenous Tm5NM1 (green) or TM311 antibody to visualize the exogenous Tm3 (green) and WS5/9d (red), which tracks with the endogenous Tm5NM2. (A) The exogenous Tm5NM1 (b and f) was enriched in the filopodia, (f, arrow), and the peripheral regions of growth cones (f, arrowheads). In contrast control neurons showed no such staining (a, c, and e). (e) Merged image of a (green) and c (red). (f) Merged image of b (green) and d (red). (B) The exogenous Tm3 protein was preferentially enriched in the filopodia (l, arrow), the peripheral region of the growth cone (l, arrowhead), and the cell body (h, arrow). In contrast, control neurons showed no such staining (g and k). (k) Merged image of g (green) and i (red). (l) Merged image of h (green) and j (red). Bar, 40 μm.
In contrast, Tm3 is much more broadly distributed throughout primary neurons. Staining with the TM311 antibody, which detects exon1a of the αTm gene (Figure 1A), confirmed that primary cortical neurons from control mice do not accumulate significant levels of high-molecular-weight tropomyosins, including Tm3 (Figure 2g cf. h). The TM311 antibody revealed widely distributed staining of Tm3 in the Tm3 cortical neurons (Figure 2, h and l). Cell bodies (Figure 2h, arrow), processes, growth cones (Figure 2l, arrowhead), and filopodia (Figure 2l, arrow) were all stained for Tm3. Comparison with Tm5NM1 allows us to conclude that the proteins Tm3 and Tm5NM1 contain different sorting information because the two transgenes differ only in the tropomyosin coding region.
Differential Impact of Tropomyosin Isoforms on Neuronal Morphogenesis
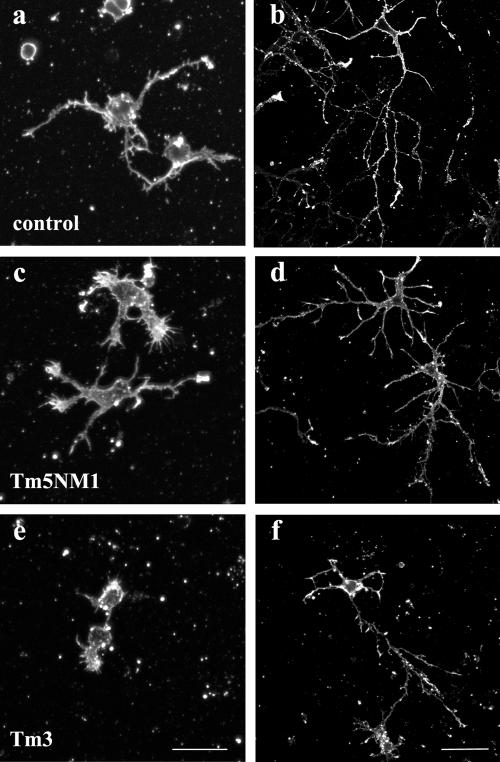
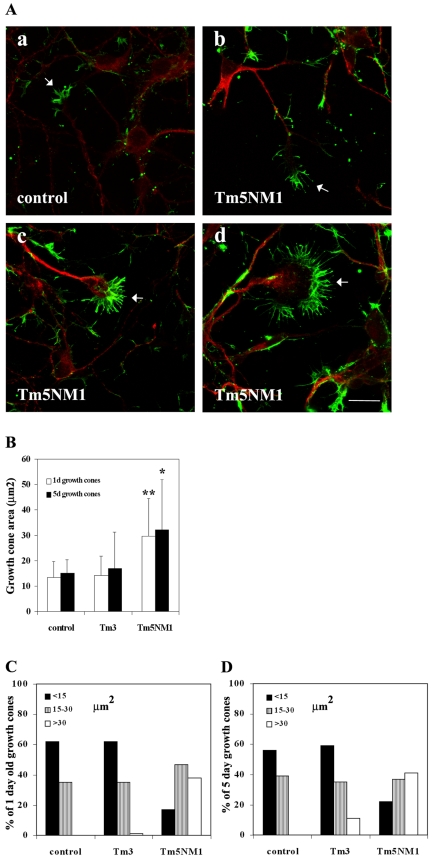
Expression of Tm3 and Tm5NM1 had profound and differential effects on neuronal morphogenesis. Embryonic cortical neurons were cultured in vitro for 1 and 5 d, and their overall morphology was analyzed by immunofluorescence staining with an actin antibody. After 1 d in culture, both control and Tm5NM1 neurons (Figure 3, a and c, respectively) extended neurites, whereas attenuation of neurite outgrowth was observed in the Tm3 neurons to the point where neurite outgrowth was barely detectable (Figure 3e). Despite such poor initial outgrowth, after 5 d in culture the Tm3 neurons were able to extend substantial neurites (Figure 3f). After 5 d, a significant increase in axonal branching is the predominate phenotype observed in the Tm5NM1 neurons (Figure 3d) compared with control or Tm3 neurons (Figure 3, b and f, respectively).
Figure 3.
Differential impact of tropomyosin isoforms on neuronal morphogenesis. Primary cortical neurons were cultured in vitro for 1 and 5 d and immunofluorescence stained with an actin antibody. After 1 d in culture, both control (a) and Tm5NM1 (c) neurons extend neurites, whereas attenuation of neurite outgrowth is observed in the Tm3 neurons (e). After 5 d, a significant increase in axonal branching is the predominate phenotype observed in the Tm5NM1 neurons (d) compared with control (b) or Tm3 neurons (f). Bar, 20 μm (a, c, and e) and 40 μm (b, d, and f).
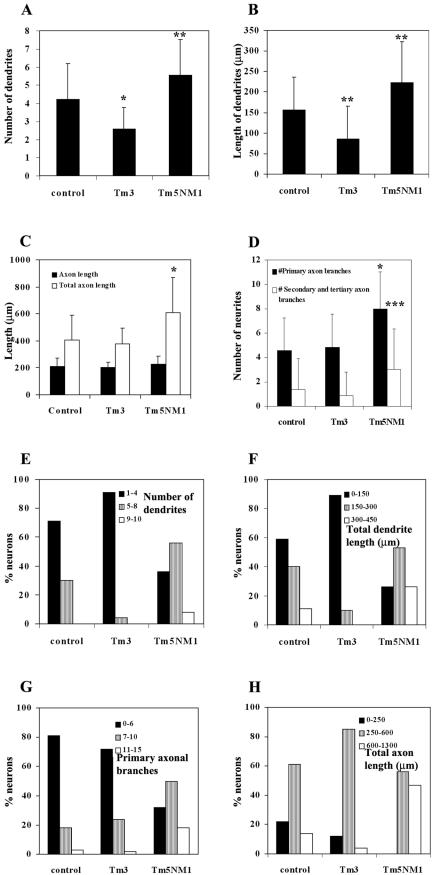
The morphogenesis of these cells was quantitated by measuring the number and length of dendrites, the length of the axon excluding or including axonal branches, and the number of primary/secondary axonal branches. We found a significant decrease in the number and the total length of the dendrites in the Tm3 neurons (Figure 4A, p < 0.001, and B, p <0.005, respectively). In contrast, the number and length of the dendrites per cell were significantly increased in the Tm5NM1 neurons (Figure 4A, p < 0.005, and B, p < 0.005, respectively). There was no significant difference in the length of the axon in 5-d-old Tm3 or Tm5NM1 neurons compared with controls (Figure 4C, black columns). However, when the lengths of both primary and secondary axonal branches were included in the measurements, a significant increase was seen in the Tm5NM1 neurons (Figure 4C, white columns, p < 0.001). The significant increase in the length of the axon, including the branches seen in the Tm5NM1 neurons is due to the significant increase in the number of both primary and secondary/tertiary axonal branches (Figure 4D, p < 0.001 and p < 0.01, respectively). In conclusion, elevated levels of Tm3 initially have a dramatic impact on neurite outgrowth, but with time in culture the predominant feature of these neurons is a significant decrease in the number and length of dendrites. In contrast, Tm5NM1 promotes an overall increase in the growth of neurites, including dendrites and axonal branching.
Figure 4.
Tm3 impacts on dendrites and Tm5NM1 affects both dendrites and axonal branching. All measurements were performed on 5-d-old neurons. A dendrite or branch was defined as a process that was >10 μm in length. The axon was the longest process, confirmed by the presence of phosphorylated neurofilament H. An average of 50 neurons from each group was selected from a number of chamber slides from two independent cultures. A nonparametric ANOVA test was chosen because the data did not display a Gaussian distribution. Asterisks indicate *p < 0.001, **p < 0.005, and ***p < 0.01. Note that the Tm3 neurons showed a significant decrease in the number of dendrites (A) and total length of dendrites (B). In contrast, an increase in the number and length of dendrites is seen in the Tm5NM1 (A and B). The Tm5NM1 neurons also showed a significant increase in the total axon length (C) due to an increase in the number of axonal branches (D). The data were also plotted as a % of neurons with a particular range of dendrites (E), dendrite length (F), number of 1° axonal branches (G), and total axon length (H). This shows that the Tm5NM1 neurons have a wider range of morphologies.
The impact of these tropomyosins on number and length of dendrites and axon length and branching was further analyzed as shown in Figure 4, E–H. Each of the parameters was broken into three groups corresponding to small, medium, and large. We then measured the distribution of neurons in each culture between these three groups. We reasoned that if the impact of Tm3 and Tm5NM1 was restricted to their effects on a small number of neurons, then analysis would reveal the presence of an extreme population of altered neurons in these cultures. The results show a systematic shift across all groups for dendrite number (Figure 4E) and length (Figure 4F), indicating progressive effects of Tm3 and Tm5NM1 on most neurons. The same result also is observed for the impact of Tm5NM1 on axon length (Figure 4H) and branching (Figure 4G)
In addition to the observed impact on branching seen in the Tm5NM1 neurons, we also observed a significant increase in the size of the growth cones (Figure 5B, p < 0.001, for 1- and p < 0.005 for 5-d-old growth cones). Immunofluorescence staining with β-actin demonstrated significantly larger growth cones with long filopodia in the Tm5NM1 neurons compared with control (Figure 5A, a cf. b, c, and d, arrows) or Tm3 neurons (Figure 8k). Quantitation of growth cone area was performed on 1- and 5-d-old neurons that were stained with a total actin antibody. The data also were plotted as the percentage of growth cones having a particular range of size. In both the control and Tm3 neurons, we found that the size of the majority of their 1- and 5-d-old growth cones were less than 15 μm2. There were few if any growth cones with surface areas >30 μm2 seen in the control or Tm3 neurons. In contrast, Tm5NM1 neurons displayed a systematic shift to larger growth cone sizes (Figure 5, C and D). This suggests that overexpression of Tm5NM1 promotes an increased surface area for most growth cones.
Figure 5.
Overexpression of Tm5NM1 results in enlarged growth cones. (A) Primary cortical neurons were cultured for 5 d and double immunofluorescence stained with the WS5/9d (red) antibody and β-actin (green). The merged images are depicted. Control neurons (a) and Tm5NM1 neurons (c, d, and e). Note the enlarged growth cones (arrows) and long filopodia in the Tm5NM1 neurons. Bar, 20 μm. (B) The surface areas of 1- and 5-d-old growth cones were determined by using Image-Pro Plus version 4.0 of neurons stained with a total actin antibody, C4. An average of 50 growth cones from each group was selected from a number of chamber slides from two independent cultures. A nonparametric ANOVA test was chosen because the data did not display a Gaussian distribution. Asterisks indicate *p < 0.001 and **p < 0.005. The data also were plotted as a percentage of neurons with a particular range of growth cone size for the 1- (C) and the 5 (D)-d-old neurons.
Figure 8.
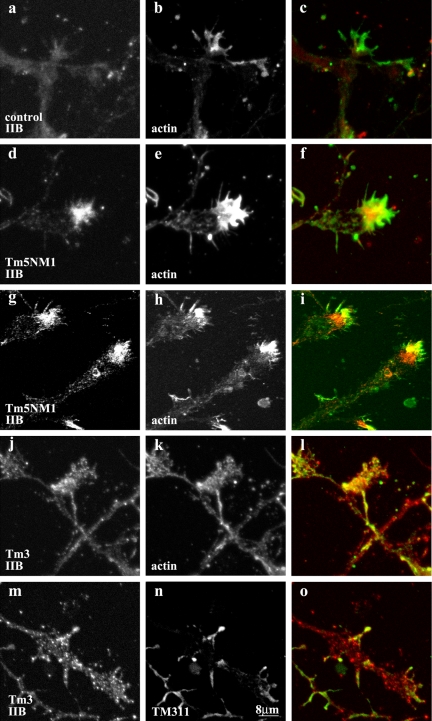
Recruitment of MHCIIB by Tm5NM1 to the growth cone. Primary cortical neurons cultured for 5 d were double immunofluorescence stained with MHCIIB (a, d, g, j, and m) and actin (b, e, and k) or LC1 (h) or TM311 to detect Tm3 (n). (c) Merged image of a (red) and b (green). (f) Merged image of d (red) and e (green). (i) Merged imaged of g (red) and h (green). (l) Merged image of j (red) and K (green). (o) Merged image of m (red) and n (green). Note the enrichment of MHCIIB in the peripheral region of the Tm5NM1 growth cones (d and g) where actin (e) and the exogenous Tm5NM1 (h) also are enriched. In contrast, such enrichment of MHCIIB was not observed in control (a) or Tm3 growth cones (j and m). Bar, 8 μm.
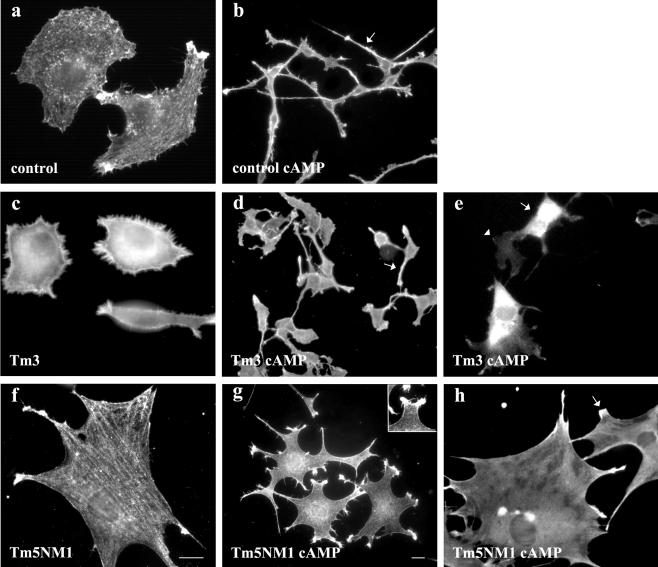
To confirm the differential impact of Tm3 and Tm5NM1 on neuronal morphogenesis, we examined the impact of these tropomyosins in the B35 neuroepithelial cell line. Transfection of these gene constructs into B35 cells allowed us to isolate clones that stably express Tm3 or Tm5NM1 (Bryce et al., 2003). Exposure of B35 cells to 1 mM dibutyryl cAMP induces morphological differentiation involving the elaboration of neurites. Figure 6b shows the impact of 48-h exposure to dibutyryl cAMP on control cells. The cell body is greatly reduced in size, and the cells elaborate multiple processes (Figure 6, a cf. b). The Tm3-transfected cells, in contrast, display very poor process outgrowth in response to dibutyryl cAMP (Figure 6, c and d). The average length of neurites from the control B35 cells was 115 ± 6 μm, whereas the Tm3 B35 cells displayed neurites which were only 44 ± 2.5 μm in length (n = 20 cells, p < 0.05). This is very similar to the primary cortical neurons expressing Tm3 (Figure 3e) and suggests that elevated levels of Tm3 interferes with the mechanism(s) responsible for neurite outgrowth. The Tm5NM1 transfectants display a remarkably different phenotypic response to dibutyryl cAMP (Figure 6, f and g). The cells begin to elaborate what seems to be the start of many processes reminiscent of the increased branching phenotype that is observed in the Tm5NM1 cortical neurons (Figure 6g cf. Figure 3d). Due to the nature of the processes extended by the Tm5NM1 transfectants, after cAMP induction, it was difficult to measure their length. Although cells do show some discrete neurites, for the majority of them the point at which the neurite first splays from the body of the cells was difficult to assess and hence an accurate measure of process length could not be made. Furthermore, the lamellipodia-like structures seen in the B35 cells after dibutyryl cAMP (Figure 6g, inset) also resemble the lamellipodia at the tips of neurites in 1-d-old cultures of the Tm5NM1 cortical neurons (Figure 3c). In addition we also observed that the subcellular localization of the exogenous Tm3 and Tm5NM1 proteins in the morphologically differentiated B35 cells resembled that seen in the primary cortical neurons. Hence exogenous Tm3 was found diffusely localized to the cell body, whereas the exogenous Tm5NM1 was enriched in the very tips of the cells (Figure 6, e and h, respectively). These results suggest that the impact of Tm3 and Tm5NM1 on neuronal morphogenesis also can be seen in a model cell line.
Figure 6.
Attenuation of neurite outgrowth also is seen in Tm3 stably transfected B35 neuroepithelial cells. B35 cells were cultured in the absence (a, c, and f) and 48 h presence of dibutyryl cAMP (b, d, e, g, and h). Control B35 cells immunofluorescence stained with TM311 antibody (a) and β-actin (b), Tm3 B35 cells immunofluorescence stained with TM311 (c and e) and β-actin (d), and Tm5NM1 B35 cells immunofluorescence stained with LC1 (f and h) and β-actin (g). Elevated levels of Tm3 results in a significant decrease in the length of neurites after dibutyryl cAMP stimulation. Inset in g shows the lamellipodia like structure. Bar, 10 μm in f corresponds to a, c, e, f, and h. Bar, 10 μm in g corresponds to b, d, and g.
Recruitment of Tm5a and Myosin IIB by Tm5NM1 to the Growth Cone
Alterations in the expression of actin and tropomyosin by in vitro transfection studies in different cell types have previously been shown to change the expression of other members of these gene families (Schevzov et al., 1993; Bryce et al., 2003). Consequently, we hypothesized that the observed increase in growth cone size seen in Tm5NM1 neurons could be due to changes in the expression and/or subcellular localization of other cytoskeletal proteins.
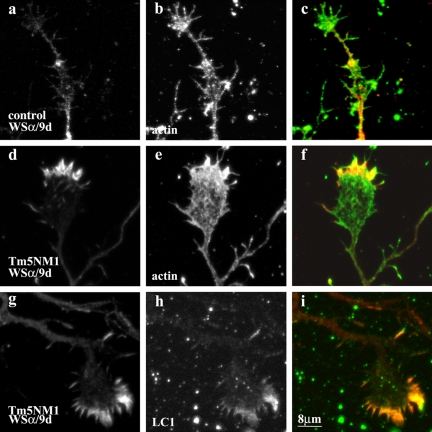
In control embryonic cortical neurons, the subcellular localization of Tm5a/5b has previously been shown to be initially present along the neurites and the growth cone in 24-h-old neurons (Schevzov et al., 1997). However, by 48 h, Tm5a/5b becomes restricted to the neurite and diminished in the growth cone. More recently, protein gels that resolve Tm5a and Tm5b indicate that these cultures express predominantly Tm5a (our unpublished data). In Figure 7, we show that in 5-d-old control neurons Tm5a is predominantly present along the neurite and highly diminished in the growth cone (Figure 7, a and b). A total of 75% of growth cones displayed this staining pattern (n = 128 in two separate cultures). Conversely, in 5-d-old Tm5NM1 neurons, the predominant localization of Tm5a is in the peripheral region of the growth cone (Figure 7, d and g). We observed that 78% of Tm5NM1 growth cones showed enriched levels of Tm5a (n = 77 in two separated cultures). Tm5a and Tm5b can form heterodimers with Tm5NM1 (Temm-Grove et al., 1996). We therefore performed double labeling to test whether the staining of these proteins is coincident. Double immunofluorescence staining clearly demonstrated that Tm5a colocalizes with the Tm5NM1 protein as detected with the LC1 antibody (Figure 7, g, h, and merge i). This is consistent with the formation of heterodimers. Similarly in the Tm5NM1 stable transfected B35, Tm5a also was recruited to stress fibers detected with the LC1 antibody (Bryce et al., 2003). There were, however, a minority of Tm5NM1 growth cones that displayed enriched staining of LC1 but lacked Tm5a staining.
Figure 7.
Recruitment of Tm5a by Tm5NM1 to the growth cone in vitro. Primary cortical neurons were cultured for 5 d and double immunofluorescence stained with WSα/9d that detects Tm5a (a, d, and g) and actin (b and e) or LC1 (h). (c) Merged image of a (red) and b (green). (f) Merged image of d (red) and e (green). (i) Merged image of g (red) and h (green). Note the enrichment of Tm5a in the peripheral region of the Tm5NM1 growth cone (d and g) where actin (e) and the exogenous Tm5NM1 (h) also are enriched. In control neurons, Tm5a (a) is highly diminished in 5-d-old growth cones although actin is present (b). Bar, 8 μm.
Expression of the Tm5NM1 but not the Tm3 protein was found to recruit myosin IIB to certain growth cones of these neurons. Initial visualization of myosin IIB in control neurons shows it uniformly distributed throughout the neurite and central region of the growth cone but largely excluded from the leading edge and filopodia (Figure 8, a and b). In contrast, in the Tm5NM1 neurons myosin IIB was found enriched at the leading edge of the growth cone and in filopodia (Figure 8, d and e). This enrichment of myosin IIB seems related to the distribution of Tm5NM1 because costaining reveals some degree of colocalization of myosin IIB with Tm5NM1 (Figure 8, g and h). However, this overlap occurs only at the leading edge, which indicates that the enrichment of myosin IIB is not completely accounted for by physical linkage of myosin IIB to Tm5NM1-containing filaments. The enrichment of myosin IIB in the growth cone of Tm3 neurons was not apparent in the growth cones and not at the leading edge (Figure 8, j and k) where the exogenous Tm3 protein is found (Figure 8, m and n).
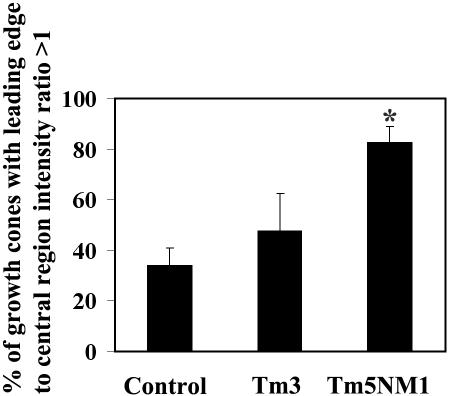
To further confirm the above-mentioned findings, quantitation of the myosin IIB staining was determined by measuring the fluorescence intensity at the leading edge and central region of the growth cone. The percentage of growth cones with a leading edge to central region intensity ratio of >1 was calculated and shown in Figure 9. More than 80% of Tm5NM1 neurons show enrichment of myosin IIB at the leading edge, which is significantly greater and more than double that observed for control neurons. In contrast, Tm3 neurons were not significantly different from controls. We therefore conclude that tropomyosin isoforms can promote reorganization of a myosin motor in an isoform-specific manner.
Figure 9.
Quantitative analysis of MHCIIB localization to the growth cone periphery. Primary cortical neurons cultured for 5 d were double immunofluorescence stained with MHCIIB and actin. The intensity of fluorescence was measured in the leading edge (confirmed by the actin staining) and central region of the growth. The percentage of growth cones with a leading edge to central region intensity ratio of >1 was calculated from an average of 80 growth cones from two independent experiments. Asterisk indicates *p < 0.001.
DISCUSSION
The diversity of actin filament function requires mechanisms that can both regulate spatially separated pools in the same cell and generate filaments with distinct biochemical characteristics. The creation of >40 Tm isoforms and their spatial segregation in a single cell provides an attractive mechanism to meet this challenge. In this article, we have provided evidence that Tms can regulate both quantitative and qualitative properties of neuronal morphology. This supports a model of actin filament function in which both the size of a specific Tm-bound filament population and the function of different filament populations are regulated by the segregation of Tm isoforms.
Regulation of Tropomyosin Sorting by Protein Sequence
The difference in location of Tm3 and Tm5NM1 in primary neurons provides a strong argument that the protein sequences alone can direct specific targeting of tropomyosins. Previous work has demonstrated specific targeting of tropomyosin mRNAs in neurons that parallel, but do not precisely correspond to, the location of the proteins (Hannan et al., 1995, 1998). The targeting of the mRNAs may therefore play a role in expediting targeting of tropomyosin isoforms rather than absolutely specifying the site of accumulation of specific tropomyosins.
The specificity of targeting of Tm5NM1 contrasted with the much more diffuse distribution of Tm3. Indeed, the exogenous Tm5NM1 seemed to precisely mirror the location of the corresponding endogenous product to the growth cone and filopodia. In contrast, the wide distribution of the Tm3 may reflect that of Tm5a/5b (Schevzov et al., 1997). These isoforms are derived from the αTm gene (Figure 1A, map of Tm genes) and carry the same carboxyl terminal exon 9d, although they differ at their amino terminus. The differential sorting of Tm isoforms may therefore be attributed to protein sequence differences between α and γ genes.
It is possible that sorting of tropomyosin isoforms may be influenced by the local organization of the actin filaments. In the growth cone, actin filaments are known to be organized into different structures, depending on their localization. A polarized bundled array of F-actin is found in the filopodia, whereas within the lamellipodia, F-actin adopts a meshwork-like array (Lewis and Bridgman, 1992). Dynamic comet-like arrays, similar to those found in the tails of bacterial and viral pathogens, termed intrapodia emanate from the transition region into the peripheral region of the growth cone (Dent and Kalil, 2001). Finally, within the transition region, actin can adopt an arc-like structure that has kinetics distinct from lamellipodial and filopodial actin (Schaefer et al., 2002). It is this region that seems to be sparse for γTm isoforms.
Sorting of tropomyosins does not seem to use simple geographical signals. Tm5NM1 is sorted in neurons to the very growth tip of processes. One might therefore predict that it would be similarly sorted to areas of membrane remodeling in all cells. In contrast, in epithelial (Temm-Grove et al., 1998), neuroepithelial (Bryce et al., 2003), and fibroblast cells (Percival et al., 2004), Tm5NM1 is sorted to stress fibers. This is not consistent with a universal geographical destination. The similarity that does exist between all Tm5NM1 destinations is the organization of actin into bundles of tension-bearing microfilaments. This supports the possibility that it is the organization of actin that influences the site of accumulation of specific isoforms.
Qualitative and Quantitative Impact of Tropomyosin on Neuronal Morphology
Differentiation and morphogenesis in the brain and skeletal muscle are accompanied by extensive changes in Tm isoform expression (Gunning et al., 1990; Stamm et al., 1993; Weinberger et al., 1993, 1996; Dufour et al., 1998). Although consistent with a direct role for tropomyosin in morphogenesis, no experiments have addressed the specificity of tropomyosin isoform function in this process. Do Tm levels regulate morphogenesis quantitatively or qualitatively and is this isoform specific? Our results support an isoform-specific role and indicate that individual isoforms carry both quantitative and qualitative information.
The impact of Tm5NM1 expression on growth cone size provides a clear demonstration that isoform sorting can be used to regulate the size of a specific intracellular compartment. Previous studies that have manipulated actin and tropomyosin isoform expression in myogenic and neuroepithelial cells have led to global changes in cell dimension (Schevzov et al., 1992; von Arx et al., 1995; Mounier et al., 1997; Bryce et al., 2003). In each case, the manipulated gene products have altered the organization of actin filaments throughout the cell. The only exception has been the impact of γ-actin on cell membrane morphology in two studies (Schevzov et al., 1992; von Arx et al., 1995). In Tm5NM1 expression in neurons, the protein is specifically sorted to growth cones, and it is this compartment alone that shows an increase in size. The organization of actin is not changed, only dimension seems to be altered. The morphology of the growth cone also seems to be qualitatively normal but quantitatively increased. The sorting of isoforms to a specific compartment therefore provides a mechanism to control dimension independent of shape.
The differential impact of Tm5NM1 and Tm3 indicates that Tm isoforms carry different structural information at the level of cell shape. This is entirely consistent with previous studies that have shown that Tm isoforms differ in their ability to restore cell shape to transformed cells (Gimona et al., 1996) and in their impact on neuroepithelial cells (Bryce et al., 2003; Figure 6). In addition, the original demonstration that β and γ-actin carry different structural information (Schevzov et al., 1992) may be accounted for by their differential impact on Tm isoform levels and organization (Schevzov et al., 1993). In all cases, the alterations in cell shape are consistent with changes in the organization of actin filaments. These studies are therefore all consistent with a direct role for Tm isoforms in regulating cell structure, and the current study demonstrates that Tm isoform choice can regulate the overall shape of primary neurons. The morphological outcomes that we have observed in the Tm-overexpressing cortical neurons in culture seem also to impact on brain structure in vivo. We have preliminary data to indicate progressive neurodegeneration in the cerebellum (Meaney, Jeffrey, Schevzov, Weinberger, Gunning, unpublished data). This is currently the subject of an intensive and separate study.
Mechanism of Tm Regulation of Neurogenesis
The impact of Tm5NM1 on growth cone size seems likely to act, at least in part, via its recruitment of MHCIIB. Bridgman et al. (2001) have previously demonstrated that myosin IIB is essential for growth cone motility and morphology. Antisense studies and chromophore-assisted laser inactivation of myosin IIB in neurons in vitro, show a significant reduction in neurite length (Wylie et al., 1998; Diefenbach et al., 2002). In addition, neurons isolated from the myosin IIB knockout mice (Tullio et al., 2001) displayed a narrow growth cone phenotype in vitro (Bridgman et al., 2001). This suggests that MHCIIB is required for growth cone spreading. Our results are also consistent with a model in which the level of MHCIIB in the growth cone determines spreading. In our case, elevated levels of Tm5NM1 in the growth cone promote recruitment of MHCIIB and increased size. Similarly, we have found that elevated expression of Tm5NM1 in B35 cells leads to increased cell spreading (Bryce et al., 2003). In this case, incorporation of Tm5NM1 into stress fibers is accompanied by recruitment of MHCIIA and increased myosin activity. This therefore provides a mechanism by which a structural protein can regulate the contractility of microfilaments in an isoform-specific manner.
It is intriguing that the overexpression of Tm3 impacted on dendritic but not axonal growth. Similarly, in a Drosophila study (Li and Gao, 2003) loss-of-function mutation in tropomyosin II impacted on the dendritic field. The Rho GTPase family of molecules has been shown to be involved in determining neuronal morphology. We know that Cdc42 and Rac are positive regulators of neurite outgrowth, whereas Rho inhibits neurite extension (Luo, 2000). Interestingly, unique effects of each Rho GTPase in different neuronal compartments have been reported. Both in Drosophila and mice, constitutively active Rac causes a selective defect on axonal outgrowth without notably affecting dendritic growth. In contract, Cdc42 mutations affect both axon and dendrites (Luo et al., 1994, 1996). We therefore postulate that the distinct molecular composition of microfilaments in different neuronal compartments may regulate not only local cell structure but also its ability to respond to different signals.
Acknowledgments
We thank Dr. Julie Martyn for the critical reading of the manuscript. This work was supported by the Australian National Health and Medical Research Council (NHMRC) grants to P. G. P. G. is a Principal Research Fellow of the NHMRC.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–10–0951) on May 11, 2005.
Abbreviations used: Tm, tropomyosin.
References
- Bernstein, B. W., and Bamburg, J. R. (1982). Tropomyosin binding to F-actin protects the F-actin from disassembly by brain actin-depolymerizing factor (ADF). Cell Motil. 2, 1–8. [DOI] [PubMed] [Google Scholar]
- Boyd, J., Risinger, J. I., Wiseman, R. W., Merrick, B. A., Selkirk, J. K., and Barrett, J. C. (1995). Regulation of microfilament organization and anchorage-independent growth by tropomyosin 1. Proc. Natl. Acad. Sci. USA 92, 11534–11538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridgman, P. C., Dave, S., Asnes, C. F., Tullio, A. N., and Adelstein, R. S. (2001). Myosin IIB is required for growth cone motility. J. Neurosci. 21, 6159–6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryce, N. S., et al. (2003). Specification of actin filament function and molecular composition by tropomyosin isoforms. Mol. Biol. Cell 14, 1002–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooley, B. C., and Bergtrom, G. (2001). Multiple combinations of alternatively spliced exons in rat tropomyosin-α gene mRNA: evidence for 20 new isoforms in adult tissues and cultured cells. Arch. Biochem. Biophys. 390, 71–77. [DOI] [PubMed] [Google Scholar]
- Dent, E. W., and Kalil, K. (2001). Axon branching requires interactions between dynamic microtubules and actin filaments. J. Neurosci. 21, 9757–9769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diefenbach, T. J., Latham, V. M., Yimlamai, D., Liu, C. A., Herman, I. M., and Jay, D. G. (2002). Myosin 1c and myosin IIB serve opposing roles in lamellipodial dynamics of the neuronal growth cone. J. Cell Biol. 30, 1207–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour, C., Weinberger, R. P., Schevzov, G., Jeffrey, P. L., and Gunning, P. (1998). Splicing of two internal and four carboxyl-terminal alternative exons in nonmuscle tropomyosin 5 pre-mRNA is independently regulated during development. J. Biol. Chem. 273, 18547–18555. [DOI] [PubMed] [Google Scholar]
- Gimona, M., Kazzaz, J. A., and Helfman, D. M. (1996). Forced expression of tropomyosin 2 or 3 in v-Ki-ras-transformed fibroblasts results in distinct phenotypic effects. Proc. Natl. Acad. Sci. USA 93, 9618–9623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunning, P., Gordon, M., Wade, R., Gahlmann, R., Lin, C. S., and Hardeman, E. (1990). Differential control of tropomyosin mRNA levels during myogenesis suggests the existence of an isoform competition-autoregulatory compensation control mechanism. Dev. Biol. 138, 443–453. [DOI] [PubMed] [Google Scholar]
- Gunning, P., Hardeman, E., Jeffrey, P., and Weinberger, R. (1998). Creating intracellular structural domains: spatial segregation of actin and tropomyosin isoforms in neurons. Bioessays 20, 892–900. [DOI] [PubMed] [Google Scholar]
- Had, L., Faivre-Sarrailh, C., Legrand, C., Mery, J., Brugidou, J., and Rabie, A. (1994). Tropomyosin isoforms in rat neurons: the different developmental profiles and distributions of TM-4 and TMBr-3 are consistent with different functions. J. Cell Sci. 107, 2961–2973. [DOI] [PubMed] [Google Scholar]
- Hannan, A. J., Gunning, P., Jeffrey, P. L., and Weinberger, R. P. (1998). Structural compartments within neurons: developmentally regulated organization of microfilament isoform mRNA and protein. Mol. Cell. Neurosci. 11, 289–304. [DOI] [PubMed] [Google Scholar]
- Hannan, A. J., Schevzov, G., Gunning, P., Jeffrey, P. L., and Weinberger, R. P. (1995). Intracellular localization of tropomyosin mRNA and protein is associated with development of neuronal polarity. Mol. Cell. Neurosci. 6, 397–412. [DOI] [PubMed] [Google Scholar]
- Hughes, J. A., Cooke-Yarborough, C. M., Chadwick, N. C., Schevzov, G., Arbuckle, S. M., Gunning, P., and Weinberger, R. P. (2003). High-molecular-weight tropomyosins localize to the contractile rings of dividing CNS cells but are absent from malignant pediatric and adult CNS tumors. Glia 42, 25–35. [DOI] [PubMed] [Google Scholar]
- Ishikawa, R., Yamashiro, S., and Matsumura, F. (1989). Differential modulation of actin-severing activity of gelsolin by multiple isoforms of cultured rat cell tropomyosin. Potentiation of protective ability of tropomyosins by 83-kDa nonmuscle caldesmon. J. Biol. Chem. 264, 7490–7497. [PubMed] [Google Scholar]
- Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Lees-Miller, J. P., and Helfman, D. M. (1991). The molecular basis for tropomyosin isoform diversity. Bioessays 13, 429–437. [DOI] [PubMed] [Google Scholar]
- Lessard, J. L. (1988). Two monoclonal antibodies to actin: one muscle selective and one generally reactive. Cell Motil. Cytoskeleton 10, 349–362. [DOI] [PubMed] [Google Scholar]
- Lewis, A. K., and Bridgman, P. C. (1992). Nerve growth cone lamellipodia contain two populations of actin filaments that differ in organization and polarity. J. Cell Biol. 119, 1219–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W., and Gao, F. B. (2003). Actin filament-stabilizing protein tropomyosin regulates the size of dendritic fields. J. Neurosci. 23, 6171–6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, L. (2000). Rho GTPases in neuronal morphogenesis. Nat. Rev. Neurosci. 1, 173–180. [DOI] [PubMed] [Google Scholar]
- Luo, L., Hensch, T. K., Ackerman, L., Barbel, S., Jan, L. Y., and Jan, Y. N. (1996). Differential effects of the Rac GTPase on Purkinje cell axons and dendritic trunks and spines. Nature 379, 837–840. [DOI] [PubMed] [Google Scholar]
- Luo, L., Liao, Y. J., Jan, L. Y., and Jan, Y. N. (1994). Distinct morphogenetic functions of similar small GTPases: Drosophila Drac1 is involved in axonal outgrowth and myoblast fusion. Genes Dev. 8, 1787–1802. [DOI] [PubMed] [Google Scholar]
- Mounier, N., Perriard, J. C., Gabbiani, G., and Chaponnier, C. (1997). Transfected muscle and non-muscle actins are differentially sorted by cultured smooth muscle and non-muscle cells. J. Cell Sci. 110, 839–846. [DOI] [PubMed] [Google Scholar]
- Percival, J. M. Hughes, J. A., Brown, D. L., Schevzov, G., Heimann, K., Vrhovski, B., Bryce, N., Stow, J. L., and Gunning, P. W. (2004). Targeting of a tropomyosin isoform to short microfilaments associated with the Golgi complex. Mol. Biol. Cell 15, 268–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips, G. N., et al. (1979). Crystal structure and molecular interactions of tropomyosin. Nature 278, 413–417. [DOI] [PubMed] [Google Scholar]
- Prasad, G. L., Fuldner, R. A., and Cooper, H. L. (1993). Expression of transduced tropomyosin 1 cDNA suppresses neoplastic growth of cells transformed by the ras oncogene. Proc. Natl. Acad. Sci. USA 90, 7039–7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, H., and Gunning, P. (1997). The 3′-end of the human β-actin gene enhances activity of the β-actin expression vector system: construction of improved vectors. J. Biochem. Biophys. Methods 36, 63–72. [DOI] [PubMed] [Google Scholar]
- Rochlin, M. W., Itoh, K., Adelstein, R. S., and Bridgman, P. C. (1995). Localization of myosin II A and B isoforms in cultured neurons. J. Cell Sci. 108, 3661–3670. [DOI] [PubMed] [Google Scholar]
- Schaefer, A. W., Kabir, N., and Forscher, P. (2002). Filopodia and actin arcs guide the assembly and transport of two populations of microtubules with unique dynamic parameters in neuronal growth cones. J. Cell Biol. 158, 139–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schevzov, G., Gunning, P., Jeffrey, P. L., Temm-Grove, C., Helfman, D. M., Lin, J. J., and Weinberger, R. P. (1997). Tropomyosin localization reveals distinct populations of microfilaments in neurites and growth cones. Mol. Cell. Neurosci. 8, 439–454. [DOI] [PubMed] [Google Scholar]
- Schevzov, G., Lloyd, C., and Gunning, P. (1992). High level expression of transfected β- and γ-actin genes differentially impacts on myoblast cytoarchitecture. J. Cell Biol. 117, 775–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schevzov, G., Lloyd, C., Hailstones, D., and Gunning, P. (1993). Differential regulation of tropomyosin isoform organization and gene expression in response to altered actin gene expression. J. Cell Biol. 121, 811–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert, D., Heinemann, S., Carlisle, W., Tarikas, H., Kimes, B., Patrick, J., Steinbach, J. H., Culp, W., and Brandt, B. L. (1974). Clonal cell lines from the rat central nervous system. Nature 249, 224–227. [DOI] [PubMed] [Google Scholar]
- Smith, C. L. (1994). The initiation of neurite outgrowth by sympathetic neurons grown in vitro does not depend on assembly of microtubules. J. Cell Biol. 127, 1407–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, S. J. (1988). Neuronal cytochmechanics: the actin-based motility of growth cones. Science 242, 708–715. [DOI] [PubMed] [Google Scholar]
- Stamm, S., Casper, D., Lees-Miller, J. P., and Helfman, D. M. (1993). Brain-specific tropomyosins TMBr-1 and TMBr-3 have distinct patterns of expression during development and in adult brain. Proc. Natl. Acad. Sci. USA 90, 9857–9861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temm-Grove, C. J., Guo, W., and Helfman, D. M. (1996). Low molecular weight rat fibroblast tropomyosin 5 (TM-5): cDNA cloning, actin-binding, localization, and coiled-coil interactions. Cell Motil. Cytoskeleton 33, 223–240. [DOI] [PubMed] [Google Scholar]
- Temm-Grove, C. J., Jockusch, B. M., Weinberger, R. P., Schevzov, G., and Helfman, D. M. (1998). Distinct localizations of tropomyosin isoforms in LLC-PK1 epithelial cells suggests specialized function at cell-cell adhesions. Cell Motil. Cytoskeleton 40, 393–407. [DOI] [PubMed] [Google Scholar]
- Towbin, H., Staehelin, T., and Gordon, J. (1979). Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76, 4350–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullio, A. N., Bridgman, P. C., Tresser, N. J., Chan, C. C., Conti, M. A., Adelstein, R. S., and Hara, Y. (2001). Structural abnormalities develop in the brain after ablation of the gene encoding nonmuscle myosin II-B heavy chain. J. Comp. Neurol. 433, 62–74. [DOI] [PubMed] [Google Scholar]
- von Arx, P., Bantle, S., Soldati, T., and Perriard, J. C. (1995). Dominant negative effect of cytoplasmic actin isoproteins on cardiomyocyte cytoarchitecture and function. J. Cell Biol. 131, 1759–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren, K. S., Lin, J. L., McDermott, J. P., and Lin, J. J. (1995). Forced expression of chimeric human fibroblast tropomyosin mutants affects cytokinesis. J. Cell Biol. 129, 697–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger, R. P., Henke, R. C., Tolhurst, O., Jeffrey, P. L., and Gunning, P. (1993). Induction of neuron-specific tropomyosin mRNAs by nerve growth factor is dependent on morphological differentiation. J. Cell Biol. 120, 205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger, R., Schevzov, G., Jeffrey, P., Gordon, K., Hill, M., and Gunning, P. (1996). The molecular composition of neuronal microfilaments is spatially and temporally regulated. J. Neurosci. 16, 238–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel, D., and Flugge, U. I. (1984). A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138, 141–143. [DOI] [PubMed] [Google Scholar]
- Wylie, S. R., Wu, P. J., Patel, H., and Chantler, P. D. (1998). A conventional myosin motor drives neurite outgrowth. Proc. Natl. Acad. Sci. USA 95, 12967–12972. [DOI] [PMC free article] [PubMed] [Google Scholar]