Abstract
Autophagy is a catabolic process used by eukaryotic cells for the degradation and recycling of cytosolic proteins and excess or defective organelles. In yeast, autophagy is primarily a response to nutrient limitation, whereas in higher eukaryotes it also plays a role in developmental processes. Due to its essentially unlimited degradative capacity, it is critical that regulatory mechanisms are in place to modulate the timing and magnitude of the autophagic response. One set of proteins that seems to function in this regard includes a complex that contains the Atg1 kinase. Aside from Atg1, the proteins in this complex participate primarily in either nonspecific autophagy or specific types of autophagy, including the cytoplasm to vacuole targeting pathway, which operates under vegetative growth conditions, and peroxisome degradation. Accordingly, these proteins are prime candidates for factors that regulate the conversion between these pathways, including the change in size of the sequestering vesicle, the most obvious morphological difference. The atg17Δ mutant forms a reduced number of small autophagosomes. As a result, it is defective in peroxisome degradation and is partially defective for autophagy. Atg17 interacts with both Atg1 and Atg13, via two coiled-coil domains, and these interactions facilitate its inclusion in the Atg1 complex.
INTRODUCTION
Cells must be able to respond to changes in their environment. These changes may be dictated in a random manner as the result of varying nutrient conditions or they may be part of a precise program that is part of a developmental plan, and the response may be either biosynthetic or catabolic. In recent years, increased emphasis has been placed upon the degradative pathways that allow cells to respond to stress or to specific hormonal cues. One of the primary pathways that play a role in cellular remodeling through degradation is autophagy (Klionsky, 2004; Levine and Klionsky, 2004). Autophagy presumably occurs in all eukaryotic cells and has been documented from yeast to humans (Reggiori and Klionsky, 2002; Wang and Klionsky, 2003). It involves the sequestration of cytoplasm within double-membrane cytosolic vesicles (Klionsky and Ohsumi, 1999; Kim and Klionsky, 2000; Klionsky and Emr, 2000). The completed vesicles subsequently fuse with endosomes and/or lysosomes, allowing the breakdown and recycling of the cargo.
The protein components that form the autophagic machinery were first identified through molecular genetic studies in various yeasts, including Saccharomyces cerevisiae, Pichia pastoris, and Hansenula polymorpha (Tsukada and Ohsumi, 1993; Thumm et al., 1994; Harding et al., 1995; Titorenko et al., 1995; Tuttle and Dunn, 1995). Homologues of most of the corresponding genes, termed ATG (Klionsky et al., 2003), have been identified in higher eukaryotes, and recent studies have demonstrated the importance of autophagy in human health and disease (Cuervo, 2004; Shintani and Klionsky, 2004a). Autophagy is generally thought to occur in a nonspecific manner; however, studies in yeast have provided clear examples of specific types of autophagy. For example, when methylotrophic yeast are shifted from conditions where peroxisomes are essential to ones where they are superfluous, these organelles are rapidly and specifically degraded (Veenhuis et al., 1978; Tuttle et al., 1993). Even cytosolic proteins may be targets for specific degradation (Onodera and Ohsumi, 2004). S. cerevisiae presents an unusual situation in that there is not only specific and nonspecific autophagy but also a biosynthetic route, the cytoplasm to vacuole targeting (Cvt) pathway, that uses the autophagic machinery. However, there are various differences between the Cvt and autophagy pathways both in terms of when they operate and the structural features of the sequestering vesicles. The Cvt pathway may be constitutive, but it is of primary importance during growth in rich medium. Cvt vesicles are ∼140–160 nm in diameter and exclude bulk cytosol (Baba et al., 1997); the main cargo of the Cvt pathway are the vacuolar hydrolases aminopeptidase I (Ape1) and α-mannosidase (Ams1) (Klionsky et al., 1992; Hutchins and Klionsky, 2001). In contrast, autophagy operates at a basal level under vegetative conditions and is induced by starvation. Autophagosomes are 300–900 nm in diameter and include bulk cytosol and entire organelles (Baba et al., 1994). Presumably some factors regulate the switch between the two pathways and also determine the type of vesicle that is formed. Thus, S. cerevisiae provides a unique system in which to characterize the regulatory pathways that modulate the autophagic response.
Most of the Atg proteins are used by both the Cvt and autophagy pathways; however, a subset of these proteins are relatively specific for one or the other pathway, and these components are likely candidates for factors that control the conversion between them. A central protein required for both pathways is the serine/threonine kinase Atg1, which may form a complex that includes the specific components (Figure 1). Atg1 has been shown to interact with Atg11, Atg13, and Atg17 (Funakoshi et al., 1997; Kamada et al., 2000; Kim et al., 2001b). Atg13 also binds Vac8 (Scott et al., 2000), whereas Atg17 interacts with Atg24, a PX domain protein that binds Atg20 (Nice et al., 2002). The Atg11, Atg20, Atg24 and Vac8 proteins are reported to be relatively specific for the Cvt pathway, whereas Atg17 is considered to be relatively specific for autophagy. Atg13 has been shown to be involved in modulating the kinase activity of Atg1 and an atg13Δ mutant is suppressed by overexpression of Atg1 (Funakoshi et al., 1997; Kamada et al., 2000); however, the role of Atg1 kinase activity itself is not known. Atg11 plays a role in cargo packaging and transport in the Cvt pathway (Shintani et al., 2002). In contrast, the functions of most of the remaining proteins are not well characterized. A preliminary analysis of Atg17 suggests that it also affects Atg1 kinase activity (Kamada et al., 2000), but little else is known about this protein. In this report, we have investigated the role of Atg17 in the Cvt, pexophagy and autophagy pathways. Atg17 interacts with both Atg1 and Atg13, and the interaction with Atg1 may be dependent upon Atg13. We found that atg17Δ represents a unique class of mutants that is normal for the Cvt pathway, partially defective for autophagy, and blocked in pexophagy. Atg17 seems to modulate the size of the autophagic response because the atg17Δ mutant has fewer, and smaller, autophagosomes.
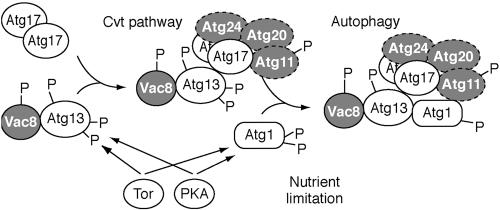
Figure 1.
Schematic model depicting assembly of the Atg1 kinase complex. Atg17 first interacts with the Atg13-Vac8 proteins. Atg13 and Atg1 may be phosphorylated by Tor and/or PKA (Budovskaya et al., 2004; Schmelzle et al., 2004) or by downstream effectors. The highly phosphorylated form of Atg13 has a lower affinity for Atg1 (Kamada et al., 2000) and this form of the complex may promote the Cvt pathway. Partial dephosphorylation of Atg13 and Atg1 allows the assembly of the putative holo-complex that triggers autophagy. Atg17 is depicted as interacting with Atg1 via Atg13, but the interaction may be direct (see text). Factors that have previously been characterized as being relatively specific for the Cvt pathway are depicted in gray. The timing of assembly of the Atg11, Atg20, and Atg24 proteins is not known as denoted by dotted lines. See text for additional details.
MATERIALS AND METHODS
Strains and Media
The S. cerevisiae strains used in this study are listed in Table 1. Tagging of proteins by integration of genes at the corresponding chromosomal loci and gene deletions were performed by a PCR-based procedure (Longtine et al., 1998). For gene disruption, the entire coding region was replaced with the Escherichia coli kanr, S. cerevisiae TRP1, Schizosaccharomyces pombe HIS5, Kluyveromyces lactis URA3, S. kluyveri HIS3, or the S. cerevisiae TRP1, LEU2, or URA3 gene by using PCR primers containing ∼40 bases of identity to the regions flanking the open reading frame (ORF). For integration, the PCR-amplified product of green fluorescent protein (GFP) at the 3′ end of the ATG1 and ATG17 chromosomal loci, or yellow fluorescent protein (YFP) at the 3′ end of the ATG9 chromosomal locus were used to generate strains expressing fusion proteins under the control of the native promoters. The template for integration was pFA6a-GFP-His3MX (oligonucleotide sequences available upon request). Western blotting, PCR, or both verified putative gene knockout and tagged strains. The functionality of tagged constructs was confirmed by examining the processing of precursor Ape1 and/or the processing of GFP-Atg8.
Table 1.
Yeast strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| atg17Δ | BY4742 atg17Δ::KAN | ResGen/Invitrogen (Carlsbad, CA) |
| BY4742 | MATα his3Δ leu2Δ lys2Δ ura3Δ | ResGen/Invitrogen |
| CWY233 | SEY6210 atg13Δ::KAN | This study |
| CWY239 | SEY6210 atg17Δ::KAN | This study |
| CWY241 | SEY6210 ATG17-GFP::HIS5 S.p. | This study |
| CWY242 | CWY241 atg1Δ::URA3 | This study |
| CWY263 | CWY241 atg13Δ::KAN | This study |
| CWY270 | PJ69-4A atg17Δ::KAN | This study |
| CWY276 | PJ69-4A vac8Δ::KAN | This study |
| CWY277 | PJ69-4A atg13Δ::KAN | This study |
| CWY278 | TN124 vac8Δ::KAN | This study |
| CWY279 | TN124 atg17Δ::KAN | This study |
| CWY321 | CWY241 vac8Δ::KAN | This study |
| CWY332 | SEY6210 vac8Δ::KAN atg17Δ::URA3 | This study |
| CWY333 | SEY6210 atg20Δ::HIS5 S.p. atg17Δ::URA3 | This study |
| D3Y112 | TN124 atg20Δ::TRP1 | Nice et al. (2002) |
| DKY6901 | PJ69-4A atg1Δ::KAN | This study |
| FRY136 | SEY6210 ATG9-YFP::HIS3 S.k. | This study |
| FRY143 | SEY6210 vps4Δ::TRP1 pep4Δ::LEU2 | This study |
| HAY572 | TN124 atg1Δ::URA3 | Abeliovich et al. (2003) |
| HCY31 | FRY143 atg17Δ::KAN | This study |
| HCY32 | SEY6210 ATG1-GFP::HIS5 S.p. atg13Δ::TRP1 | This study |
| HCY35 | FRY143 vac8Δ::URA3 K.l. | This study |
| HCY36 | FRY143 atg17Δ::KAN vac8Δ::URA3 K.l. | This study |
| HCY40 | SEY6210 ATG1-GFP::HIS5 S.p. atg17Δ::TRP1 | This study |
| HCY42 | SEY6210 ATG1-GFP::HIS5 S.p. vac8Δ::URA3 | This study |
| HCY43 | TN124 vac8Δ::KAN atg17Δ::TRP1 | This study |
| JLY4 | KTY89 atg1Δ::URA3 | This study |
| KTY89 | FRY136 atg17Δ::KAN | Reggiori et al. (2004a) |
| PJ69-4A | MATa leu2-3,112 trp1-Δ901 ura3-52 his3-Δ200 gal4Δ gal80Δ LYS2::GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ | James et al. (1996) |
| PSY143 | SEY6210 ATG1-GFP::HIS3 S.k. | Reggiori et al. (2004a) |
| SEY6210 | MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 GAL | Robinson et al. (1988) |
| TN124 | MATa leu2-3,112 trp1 ura3-52 pho8::pho8Δ60 pho13Δ::LEU2 | Noda et al. (1995) |
| WHY001 | SEY6210 atg1Δ::HIS5 S.p. | Shintani et al. (2002) |
| YTS178 | SEY6210 vac8Δ::HIS5 S.p. | This study |
Yeast were grown in YPD medium (1% yeast extract, 2% peptone, and 2% glucose) or synthetic minimal medium (SD; 0.67% yeast nitrogen base, 2% glucose, and auxotrophic amino acids and vitamins as required). Starvation medium was SD-N (0.17% yeast nitrogen base without ammonium sulfate or amino acids containing 2% glucose). Peroxisomes were induced by incubation in synthetic glycerol (0.67% YNB without amino acids, pH 5.5, 3% glycerol, and 0.1% glucose) and oleic acid (YTO; 0.67% YNB, 0.1% Tween 40, and 0.1% oleic acid) media as described previously (Hutchins et al., 1999).
Plasmids
Plasmids expressing protein A (pRSCu416-CuProtA) and GFP-ATG8 [pGFP-Aut7(414)] have been described previously (Kim et al., 2002; Abeliovich et al., 2003). For the deletion of Atg17 domains, the full-length gene or truncated ORFs were PCR amplified and ligated into the ClaI/SalI sites of pRSCu416-CuProtA (Kim et al., 2002) for affinity isolation analysis and the ClaI/PstI sites of the pGBDU-C1 vector for yeast two-hybrid analysis. For analysis of the localization of Atg17 lacking coiled-coil domains, N-terminal GFP fusion proteins were generated; for fusion of GFP at the N terminus, the full-length ATG17 gene or truncated ORFs were PCR amplified and ligated into the ClaI/SalI sites of pRS416-CuGFP (Kim et al., 2001a). To create the constructs for the Pho8Δ60 autophagy assay the full-length ATG17 gene or truncated ORFs were excised from pGFP-ATG17(416), or truncated versions, and inserted into the ClaI/SalI sites of pRS426-CuGFP (Kim et al., 2001a).
Protein Extraction and Immunoblot Analysis
S. cerevisiae strains were generally grown at 30°C to early mid-log phase in YPD or SD-N medium, treated with 10% trichloroacetic acid (TCA), and washed twice with acetone. The dry cell pellet was then resuspended in MURB buffer (50 mM Na2HPO4, 25 mM MES, pH 7.0, 1% SDS, 3 M urea, 0.5% 2-mercaptoethanol, 1 mM NaN3, and 0.05% bromophenol blue) and disrupted by vortex with an equal volume of acid-washed glass beads, for 5 min. The samples were incubated at 70°C for 10 min, unlysed cells were removed by centrifugation, and then aliquots (A600 = 0.2) were resolved by SDS-PAGE. After Western blotting, membranes were probed with appropriate antiserum or antibodies.
Antisera against Ape1 (Klionsky et al., 1992), Fox3 (Hutchins et al., 1999), and Atg1 (Abeliovich et al., 2003) have been described previously. To generate antiserum against Atg17 and Atg13, DNA segments corresponding to amino acid residues 150–417 encoded by the ATG17 ORF and residues 1–350 and 519–738 encoded by the ATG13 ORF were amplified by PCR and fused to a maltose-binding protein tag. The resulting plasmids were then transformed into the Escherichia coli BL21 strain. Expression of the fusion protein was induced in cells at log phase by adding isopropyl-1-thio-β-d-galactopyranoside (final concentration of 0.5 mM) for 3 h. A crude cell lysate was prepared and purified by column chromatography as described by the suppliers of the vector fusion system; the pMAL protein fusion plasmid and purification system was from New England Biolabs (Beverly, MA). The resulting purified antigens were used to generate antiserum in New Zealand White rabbits using standard procedures (Harlow and Lane, 1999).
Protein A Affinity Isolation Analysis
For examining protein interactions with coiled-coil domain-deleted Atg17, the pRS416-CuProtA plasmids expressing Atg17ΔCC1, ΔCC3, or ΔCC4 were transformed into atg17Δ (CWY239) cells. The transformed cells were grown to mid-log phase and grown in SMD lacking the amino acids needed to maintain plasmid selection. Twenty-five A600 equivalents of yeast cells were harvested and resuspended in 1 ml of immunoprecipitation (IP) buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.5% Tween 20, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitor cocktail; Roche Diagnostics, Indianapolis, IN). A half volume (500 μl) of acid-washed glass beads was added, and the cells were lysed by vortex at 4°C for 5 min. Cell lysates were subsequently incubated with 20 μl of IgG-Sepharose beads (Amersham Biosciences, Piscataway, NJ) at 4°C for 3–6 h. The beads were then washed in IP lysis buffer six times and eluted with SDS-PAGE sample buffer. The proteins associated with Atg17 were affinity isolated with IgG-Sepharose beads, resolved by SDS-PAGE, and analyzed by immunoblot with antisera against Atg1 or Atg13.
Alkaline Phosphatase and Pexophagy Assays
The alkaline phosphatase (ALP) assay has been described previously (Noda et al., 1995; Abeliovich et al., 2003). The analysis of Fox3 (thiolase) degradation was performed as described previously (Hutchins et al., 1999).
Yeast Two-Hybrid Assays
Yeast two-hybrid assays were done as described previously (James et al., 1996). The PCR product of full-length ATG17, carrying 5′ ClaI and 3′ PstI sites was cloned into pGAD-C1 and pGBDU-C1. Plasmids were transformed into strain PJ69-4A, and interactions were assayed by streaking colonies on SD plates lacking either histidine or adenine and testing for growth. For the mutated versions of Atg17, the partially deleted ORFs were PCR amplified and ligated into the pGBDU-C1 vector.
Transport of GFP-Atg8 to the Vacuole
The strains harboring the GFP-ATG8 [pGFP-Aut7(414)] plasmid were grown in SMD medium lacking auxotrophic amino acids at 30°C to A600 = 1.0 and were cultured in SD-N medium for 4 h to induce starvation conditions. At various time points, 1 ml of culture was harvested and used to prepare a protein extract. Protein extracts equivalent to A600 = 0.2 U of yeast cells were subjected to SDS-PAGE and probed with anti-GFP monoclonal antibody (Covance Research Products, Berkeley, CA).
Microscopy
For fluorescent microscopy, cells were grown to mid-log phase in YPD or SD medium lacking the appropriate selective nutrients and stained with 0.8 μM FM 4-64 (Molecular Probes, Eugene, OR) for 15–20 min. Cells were then washed in YPD or SD and resuspended in the same medium for a further 0.5–1 h before viewing. The cells were viewed using a DeltaVision Spectris microscope (Applied Precision, Issaquah, WA) fitted with differential interference contrast optics and Olympus camera IX-HLSH100 with softWoRx software (Applied Precision). Electron microscopy was performed as described previously (Kaiser and Schekman, 1990).
RESULTS
Atg17 Is Required for Pexophagy and Efficient Autophagy but Not the Cvt Pathway
Autophagy, the cytoplasm to vacuole targeting pathway, and the degradation of peroxisomes by pexophagy show extensive overlap with regard to the components of the protein machinery that drive each process (Klionsky et al., 2003). The precursor form of the resident vacuolar hydrolase aminopeptidase I (prApe1) is transported to the vacuole via the Cvt pathway or autophagy depending on the nutrient conditions, whereas excess peroxisomes are degraded when cells are shifted from conditions where they are essential (such as growth on oleic acid or methanol) to rich media (reviewed in Klionsky, 2004). Some atg mutants are blocked in all of these pathways, whereas others are specifically defective in a subset. For example, atg18Δ cells are defective in all three pathways, whereas atg21Δ cells that lack a homologous gene are only defective for the Cvt pathway but not for nitrogen starvation-induced autophagy or pexophagy (Stromhaug et al., 2004). In contrast, the atg20Δ and atg24Δ strains are defective in both the Cvt pathway and pexophagy but not autophagy (Nice et al., 2002). In particular, many of the proteins that interact with the Atg1 kinase seem to be relatively specific for the Cvt pathway or autophagy (Kamada et al., 2000; Kim et al., 2001b). The atg17Δ mutant is another example of a specific mutant, but in this case a preliminary analysis indicated that the cells are defective for autophagy but not the Cvt pathway (Kamada et al., 2000). We decided to extend the analysis of the role of Atg17 in autophagy-related pathways.
We first examined the role of Atg17 in the Cvt pathway by monitoring the processing of prApe1 under rich and starvation conditions; mutants that are defective in the Cvt pathway but that are not completely defective for autophagy are able to process some or all of the prApe1 that accumulates under vegetative conditions when the cells are shifted to starvation conditions (Scott et al., 2000; Kim et al., 2001b). For controls, we used the atg1Δ strain that is completely defective for both the Cvt and Atg pathways and the vac8Δ strain; this latter mutant is blocked in the Cvt pathway but shows complete reversal of prApe1 accumulation when shifted to starvation conditions due to the induction of autophagy. As expected, atg1Δ cells accumulated only the precursor form, whereas wild-type cells maintained only the mature form of Ape1 under either nutrient condition (Figure 2A). In addition, the vac8Δ mutant displayed an intermediate phenotype and was able to mature the accumulated precursor Ape1 after autophagic induction. In contrast, the atg17Δ mutant accumulated only the mature form of Ape1 under either condition, suggesting that Atg17 is not needed for the Cvt pathway (Figure 2A), in agreement with the previous study (Kamada et al., 2000).
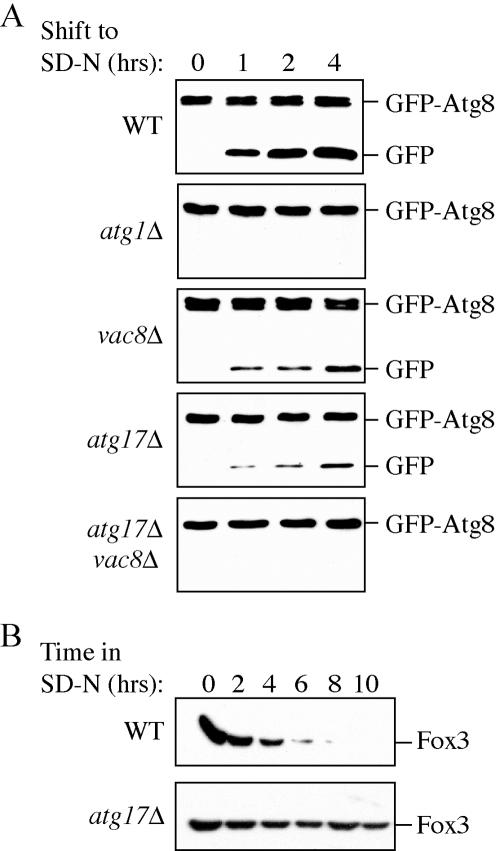
Figure 2.
The atg17Δ mutant is partially defective in autophagy. (A) The atg17Δ mutant is not defective in the Cvt pathway. Wild-type (SEY6210), atg1Δ (WHY001), vac8Δ (YTS178), atg17Δ (CWY239), atg17Δ vac8Δ (CWY332), and atg17Δ atg20Δ (CWY333) strains were grown in SMD and shifted to SD-N for the indicated times to induce autophagy. atg17Δ cells were not defective for processing of prApe1 by the Cvt or autophagy pathways. The atg17Δ mutation in combination with vac8Δ, but not atg20Δ, was blocked in the autophagic delivery of prApe1. (B) Pho8Δ60, a marker for nonspecific autophagy, indicates an autophagy defect in the atg17Δ strain. The wild-type (TN124), atg1Δ (HAY572), atg20Δ (D3Y112), vac8Δ (CWY278), atg17Δ (CWY279), and atg17Δ vac8Δ (HCY43) strains were grown in YPD and shifted to SD-N for 4 h. Samples were collected and protein extracts assayed for ALP activity. The results represent the mean and SD of three experiments.
The observation that atg17Δ cells were able to process prApe1 under starvation conditions, however, was unexpected because the atg17Δ mutant is reported to be defective in autophagy (Kamada et al., 2000). This result suggested one of two possibilities: either atg17Δ cells were not defective for autophagy, or prApe1 was imported to the vacuole through the Cvt pathway under vegetative conditions. To distinguish between these possibilities, we constructed double mutant strains. The vac8Δ mutant is characterized as being relatively specific for the Cvt pathway, although it does not display normal autophagy (Scott et al., 2000). The atg17Δ vac8Δ double mutant was not able to process prApe1 under either condition (Figure 2A), suggesting that vacuolar import of prApe1 in the atg17Δ strain under starvation conditions was due to the Cvt pathway, and conversely, reversal of prApe1 accumulation under starvation conditions in the vac8Δ mutant was due to autophagy. We decided to extend this analysis by examining an additional double mutant strain, atg17Δ atg20Δ. As noted above, the atg20Δ mutant is defective for the Cvt pathway but not autophagy (Nice et al., 2002). Surprisingly, the double mutant cells were able to process prApe1 under starvation conditions, at a level similar to that seen with the atg20Δ single mutant (Figure 2A; Nice et al., 2002). This result suggested that the atg17Δ cells are not completely defective for autophagy, because import in the atg17Δ atg20Δ double mutant that took place during starvation could not occur through the Cvt pathway. The prApe1-processing defect observed in the atg17Δ vac8Δ double mutant strain under starvation conditions was likely due to a cumulative or synthetic defect.
Because of our results with the double mutants, we next examined the role of Atg17 in autophagy by using the nonspecific autophagy marker Pho8Δ60 (Noda et al., 1995); the truncated form of ALP, Pho8, can only be delivered to the vacuole through autophagy. Once delivered to the vacuole, Pho8Δ60 is processed to the mature, active form. Thus, measuring Pho8Δ60 activity under starvation conditions provides a quantitative analysis of autophagy. Wild-type cells grown in rich medium displayed a basal or background level of Pho8Δ60-derived ALP activity (Figure 2B). After autophagy was induced by starvation for 4 h, the level of activity increased substantially. In contrast, the atg1Δ mutant did not show an increase in Pho8Δ60 activity after starvation (Figure 2B). As expected, the Cvt pathway-specific atg20Δ cells showed almost wild-type levels of autophagy after starvation. In contrast, the vac8Δ mutant was severely compromised for autophagy based on Pho8Δ60 activity even though these cells were able to process prApe1 completely when shifted to starvation conditions in agreement with previous studies (Figure 2, A and B; Scott et al., 2000). The atg17Δ mutant was essentially completely blocked for uptake of Pho8Δ60 (Figure 2B) in agreement with the previous analysis by Kamada et al. (2000). The atg17Δ mutant in combination with vac8Δ was similarly blocked for Pho8Δ60 activity in agreement with the inability of this strain to process prApe1 in starvation conditions (Figure 2A).
Thus, the atg17Δ mutant alone and in some combinations was at least partially competent for autophagy based on maturation of prApe1 but seemed to be defective for autophagy based on activation of Pho8Δ60. The activation of latent ALP activity from Pho8Δ60 is currently the most common method used to examine nonspecific autophagy. Because of the difference between the two sets of data described above, we decided to examine the viability of the atg17Δ strain in starvation conditions. Wild-type cells are relatively resistant to starvation, whereas autophagy mutants are starvation sensitive (Tsukada and Ohsumi, 1993). For example, atg1Δ mutant cells die within ∼3 to 4 d after a shift from nutrient-rich to nitrogen-depleted medium. The atg17Δ mutant displayed an intermediate phenotype; atg17Δ cells died within ∼6 d (our unpublished data). This is similar to the result seen with the atg8Δ mutant (Abeliovich et al., 2000), which is only partially blocked in autophagy, suggesting that atg17Δ cells may have only a partial autophagy defect.
We decided to further analyze the requirement for Atg17 in autophagy through a biochemical approach. Atg8 is an ubiquitin-like protein that is conjugated to phosphatidylethanolamine and is the only Atg protein that seems to remain associated with the autophagosome membrane (Kirisako et al., 1999; Huang et al., 2000; Ichimura et al., 2000; Kirisako et al., 2000). GFP-Atg8 can be used to monitor delivery of vesicles to the vacuole by fluorescence microscopy (Kim et al., 2001a). On delivery, the GFP moiety is proteolytically removed; as a result, the appearance of free GFP can serve as a biochemical assay to monitor vesicle delivery (Shintani and Klionsky, 2004b). Single and double mutant strains were transformed with a plasmid encoding GFP-Atg8, grown in rich medium to mid-log phase, and shifted to starvation medium to induce autophagy. The formation of free GFP was detected by immunoblot by using anti-GFP antibody at various time points.
In wild-type cells, the amount of free GFP increased during the course of starvation, whereas the atg1Δ negative control accumulated only the full-length GFP-Atg8 hybrid (Figure 3A). In the vac8Δ mutant that is partially defective for autophagy (Figure 2, A and B), a reduced level of free GFP accumulated. The atg17Δ mutant showed an increase in free GFP similar to the vac8Δ strain (Figure 3A), indicating that Atg17 is not absolutely required for autophagy but that the level of autophagy in the atg17Δ mutant was reduced relative to a wild-type strain. To rule out the possibility that the atg17Δ mutation showed reduced levels of free GFP due to decreased expression of the hybrid protein, we analyzed the expression level of GFP-Atg8 in the atg17Δ mutant; the expression level was similar to that of wild-type cells (our unpublished data). Finally, atg17Δ vac8Δ double mutants showed no free GFP. These results suggest that the atg17Δ mutant has an intermediate autophagy defect, whereas the atg17Δ vac8Δ double mutant shows a greater synthetic defect.
Figure 3.
Autophagy and pexophagy occur at a reduced level in atg17Δ mutant cells. (A) Wild-type (SEY6210), atg1Δ (WHY001), vac8Δ (YTS178), atg17Δ (CWY239), and atg17Δ vac8Δ (CWY332) strains expressing GFP-Atg8 from pGFP-Aut7(414) were grown in SMD lacking auxotrophic amino acids and shifted to SD-N medium. At the indicated times, aliquots were removed, the proteins precipitated with TCA, and resolved by SDS-PAGE. Full-length GFP-Atg8 and free GFP were detected by immunoblot by using anti-GFP antibodies as described in Materials and Methods. The band running just below full-length GFP-Atg8 is a cross-contaminant. (B) atg17Δ cells are deficient in peroxisome degradation. Wild-type (BY4742) and atg17Δ cells were shifted from oleic acid-containing medium to SD-N, and pexophagy was monitored by Western blot by using an antibody against peroxisomal thiolase (Fox3) as described in Materials and Methods.
Finally, we examined the degradation of peroxisomes by pexophagy. Peroxisomes were induced to proliferate by growth on oleic acid as the sole carbon source as described in Materials and Methods. Cells were then shifted to nitrogen starvation medium to cause the excess peroxisomes to be transported to the vacuole and degraded. Following the peroxisome matrix protein Fox3 allowed us to monitor this process. The level of Fox3 decreased rapidly and was essentially undetectable after 8 h when wild-type cells were shifted from oleic acid to SD-N medium (Figure 3B). In contrast, the level of Fox3 remained largely unchanged relative to the wild-type strain over the time course examined in the atg17Δ mutant (Figure 3B), indicating that Atg17 is needed for efficient pexophagy, another specific type of autophagy. The atg17Δ strain did display some reduction in the level of Fox3, particularly at much later time points (our unpublished data). Similar results were seen for the vac8Δ mutant (our unpublished data). Together, these data suggest that Atg17 is defective for bulk autophagy and pexophagy but that the atg17Δ mutant retains some limited capacity for specific autophagic import, in particular uptake of prApe1 in the form of a Cvt complex.
Atg17 Is Required for Formation of Normal-sized Autophagosomes
The data for prApe1 maturation, Pho8Δ60 activity, GFP-Atg8 processing, and pexophagy in the atg17Δ mutant under starvation conditions made it unclear as to the requirement for Atg17 in autophagy. Accordingly, we decided to obtain morphological data to resolve this issue. Much of the subcellular vesicular traffic within the yeast cell terminates at the vacuole. For endocytosis or the vacuolar protein sorting (Vps) pathways, single-membrane transport vesicles fuse with the limiting membrane of the vacuole to release their cargo within the vacuole lumen. In contrast, delivery of cargo through the Cvt/Atg pathways and the multivesicular body (Mvb) pathway (Katzmann, 2004) use transient double-membrane vesicles or compartments; after fusion of the vesicle outer membrane with the vacuole, a single membrane vesicle is released within the vacuole lumen and is subsequently degraded. The breakdown of these vesicles is blocked in a pep4Δ mutant that is defective in the activity of vacuolar proteinase A, allowing the accumulated vesicles to be observed by electron microscopy. To eliminate confusion due to the presence of vesicles derived from the Mvb pathway, we relied on the vps4Δ mutation, which blocks this pathway but not autophagy (Babst et al., 1998; Reggiori et al., 2004b). Cells harboring the pep4Δ vps4Δ double mutation were grown in YPD, shifted to SD-N for 4 h to induce autophagy, and examined by electron microscopy to allow the detection of autophagic bodies, the single-membrane intravacuolar vesicles that result from fusion of the autophagosome with the vacuole (Takeshige et al., 1992), as described in Materials and Methods.
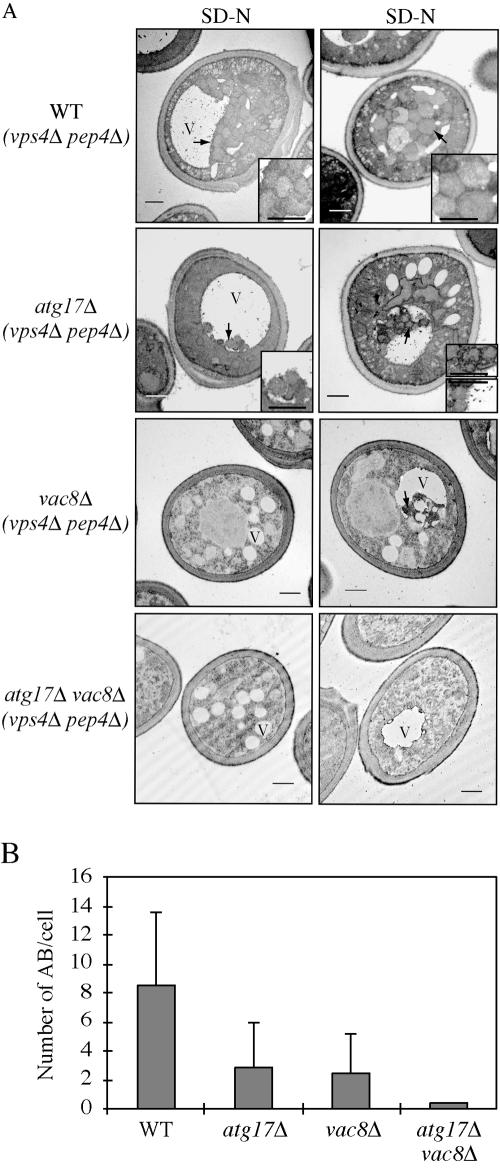
The wild-type (pep4Δ vps4Δ mutant) vacuole was filled with a large number of autophagic bodies that accumulated after starvation (Figure 4). In contrast, the atg17Δ strain (also harboring pep4Δ vps4Δ mutations) accumulated a reduced number of autophagic bodies, showing approximately a 60% reduction. Furthermore, the size of the autophagic bodies in the atg17Δ mutant also was substantially reduced; however, they were clearly larger than Cvt bodies that are ∼150 nm in diameter (Baba et al., 1997). We extended this analysis by examining two additional strains, vac8Δ and atg17Δ vac8Δ, that displayed autophagy defects based on the Pho8Δ60 assay (Figure 2B). The vac8Δ and atg17Δ vac8Δ mutants showed two phenotypes. Approximately 60–70% of the cells had multiple fragmented vacuoles due to the vac8Δ mutation (Figure 4A, left); the remaining cells had one prominent vacuole along with additional smaller vacuolar compartments (Figure 4A, right). In the vac8Δ mutant, both the large and fragmented vacuoles had autophagic bodies, but they were smaller and reduced in number relative to the wild-type strain (Figure 4, A and B). The atg17Δ vac8Δ double mutant lacked any detectable autophagic bodies. These results are in agreement with the Pho8Δ60 and GFP-Atg8 data (Figures 2B and 3A) that showed that the vac8Δ and atg17Δ vac8Δ mutants had blocks in bulk autophagy and autophagosome delivery to the vacuole, with the latter strain showing a more severe defect. Overall, these results suggest that Atg17 is involved in regulating the size and number of autophagosomes—that is the magnitude of the autophagic response—during starvation but that the atg17Δ mutant is not completely defective for autophagy.
Figure 4.
(A) The atg17Δ mutant generates fewer and smaller autophagosomes. The wild-type (FRY143; vps4Δ pep4Δ), atg17Δ (HCY31; vps4Δ pep4Δ), vac8Δ (HCY35; vps4Δ pep4Δ), and atg17Δ vac8Δ (HCY36; vps4Δ pep4Δ) strains were grown to mid-log stage in YPD and transferred to SD-N medium for 4 h. Cells were fixed with permanganate and examined by electron microscopy as described in Materials and Methods. Arrows mark the locations of autophagic bodies. The bars in the main images and insets (2× magnification) represent 0.5 μm. V, vacuole. (B) Quantification of autophagic body accumulation. Fifty sections for each strain were scored for autophagic body (AB) accumulation.
Atg17 Interacts with Atg1 and Atg13
Atg17 was identified by yeast two-hybrid screening as an interacting partner of Atg1 and is known to be required for Atg1 kinase activity similar to Atg13 (Kamada et al., 2000). In addition, Atg13 is known to interact with Vac8 by yeast two-hybrid and coimmunoprecipitation analyses (Scott et al., 2000). To analyze the interaction among these proteins in a putative Atg1 complex, we performed yeast two-hybrid analyses using Atg1, Atg13, Atg17, and Vac8 hybrid proteins. Transformants carrying various combinations of two-hybrid plasmids were tested for growth on histidine- or adenine-deficient plates. We observed interactions between Atg13 and Atg17, between Atg1 and Atg17, between Atg13 and Vac8 (Figure 5), and self-interactions with Atg17 (our unpublished data). Furthermore, none of the interactions we detected were dependent upon the presence of the remaining Atg proteins or Vac8. For example, Atg1 interacted with Atg17, and this interaction did not depend upon the presence of Atg13. These results suggest that the interactions between Atg1, Atg13, Atg17, and Vac8 do not significantly or absolutely depend upon other proteins in this putative complex.
Figure 5.
Yeast two-hybrid analysis among Atg1 complex components. The wild-type (PJ69–4A), atg1Δ (DKY6901), atg13Δ (CWY277), atg17Δ (CWY270), or the vac8Δ (CWY276) strains were transformed with the empty AD or BD vectors or with the vectors containing full-length Atg1, Atg13, Atg17, or Vac8 as indicated. After selection on plates lacking uracil and leucine, the transformants were patched onto plates additionally lacking either adenine, or histidine and containing 10 mM 3-aminotriazole. The plates were grown for 5 d at 30°C.
One of the drawbacks of the two-hybrid method for detecting protein–protein interactions is that it does not necessarily represent physiological conditions because the interactions must take place within the nucleus. To examine the physiological interaction of Atg17 with Atg1 and Atg13, we performed an affinity isolation analysis by using cells that contain a chimeric form of Atg17 fused to protein A. The extracts from cells expressing protein A-Atg17 (PA-Atg17) were incubated with IgG-Sepharose to isolate the protein A chimera and its interacting proteins. After eluting proteins from the Sepharose beads, interacting proteins were detected by immunoblot as described in Materials and Methods. As a control, we used cells expressing protein A alone.
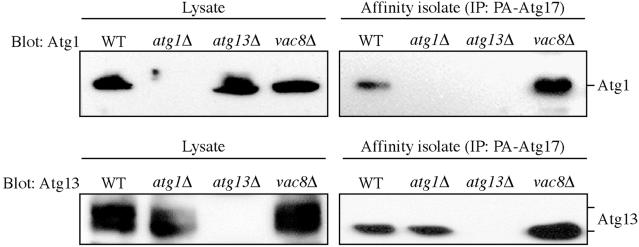
Atg1 and Atg13 were detected by immunoblot as interacting proteins after affinity isolation of PA-Atg17 from cell lysates (Figure 6). These proteins were not found in the affinity isolate from control cells expressing protein A alone (our unpublished data) or from the corresponding deletion mutants (Figure 6). These results suggest that Atg17 specifically interacts with both Atg1 and Atg13, indicating that these proteins form part of an Atg1 complex. Atg17 seemed to interact preferentially with a less phosphorylated form of Atg13, and partially dephosphorylated Atg13 is reported to have a higher affinity for Atg1 (Kamada et al., 2000); however, it is possible that dephosphorylation occurred nonspecifically during the course of the affinity isolation. Furthermore, the interaction between Atg17 and Atg1 was no longer detected in the atg13Δ mutant (Figure 6), indicating that Atg13 is required for the interaction between Atg17 and Atg1. In contrast, the Atg17–Atg13 interaction was not dependent on the presence of Atg1. The absence of Vac8 did not interfere with the ability of Atg17 to interact with either Atg1 or Atg13 (Figure 6). There is a discrepancy between these data and the results with the two-hybrid analysis where Atg17 and Atg1 showed an interaction in the atg13Δ strain (Figure 5). This difference may reflect the different sensitivity and/or stringency of the respective assays.
Figure 6.
Atg17 interacts with Atg13 and Atg1. The wild-type (SEY6210), atg1Δ (WHY001), atg13Δ (CWY233), and vac8Δ (YTS178) strains expressing protein A-tagged Atg17 or the empty vector were grown in SMD plus casamino acids (0.5%). Protein extracts (lysate) were prepared and incubated with IgG-Sepharose beads as described in MATERIALS AND METHODS. The resulting immunocomplexes (affinity isolate) were resolved by SDS-PAGE and analyzed by Western blot by using serum directed against Atg1 or Atg13. No bands were detected from samples prepared from the strains carrying the empty protein A vector.
According to the above-mentioned results (Figures 5 and 6), Atg13 directly interacts with Atg17, and the interaction is not altered in the atg1Δ mutant. To gain further information about the nature of the Atg13–Atg17 intermediate complex, we next decided to map the domain(s) of Atg13 that is involved in interacting with Atg17 and vice versa. Accordingly, we performed yeast two-hybrid analyses by using various constructs containing different fragments or different truncates of Atg13 or Atg17. The C terminus of Atg13, encompassing the region between residues 521 and 738, is known to be necessary for the interaction with Vac8 (Scott et al., 2000; Figure 7A). We found that the domain between amino acids 280 and 520 is required for the interaction with Atg17 (Figure 7A). Part of this domain overlaps with the binding site for Atg1 (Kamada et al., 2000). Next, we examined the interacting domain(s) in Atg17. This protein contains five predicted coiled-coil domains within its 417 amino acids (Figure 7B). Because a coiled-coil motif often mediates a protein–protein interaction, we constructed a series of coiled-coil deleted mutants and performed a yeast two-hybrid analysis. In particular, the Atg17ΔCC1, ΔCC2, ΔCC3, ΔCC4, and ΔCC5 proteins were deleted for amino acids 1–90 (relying on the methionine residue at position 91 for the new start codon), 129–177, 186–236, 266–312, and 335–417, respectively (Figure 7B). We found that the Atg17ΔCC2, ΔCC4, and ΔCC5 proteins supported growth on plates lacking histidine when the two-hybrid strain was transformed with the corresponding Atg17 two-hybrid plasmid and a two-hybrid plasmid encoding Atg13 or Atg1 (Figure 7B). In contrast, the Atg17ΔCC1 or ΔCC3 plasmids did not support growth.
Figure 7.
Regions in the coiled-coil domains of Atg17 are needed for interaction with its binding partners. (A) Schematic representation of Atg13, including the location of domains that bind Atg1, Atg17, and Vac8. Regions of Atg13 present in the activation domain (AD) two-hybrid protein constructs are depicted. The ability of the corresponding proteins to interact with the full-length Atg17 and Vac8 or an N-terminally truncated Atg1 binding domain two-hybrid protein is indicated on the right. The data for interactions between Atg1 and Atg13 are from Kamada et al. (2000) and for Atg13 and Vac8 are from Scott et al. (2000). ND, not determined. (B) A schematic representation of Atg17, indicating the location of predicted coiled-coil domains, is shown at the top. Regions of Atg17 deleted from the binding domain (BD) two-hybrid protein construct are depicted. The ability of the corresponding proteins to interact with the full-length Atg1 or Atg13 activation domain two-hybrid protein is indicated on the right. “+” means that the strain containing the two plasmids was able to grow on plates lacking histidine, and “–” indicates an inability to grow after 5 d. A deletion of the first or third coiled-coil domain in Atg17 blocked interaction with both Atg13 and Atg1.
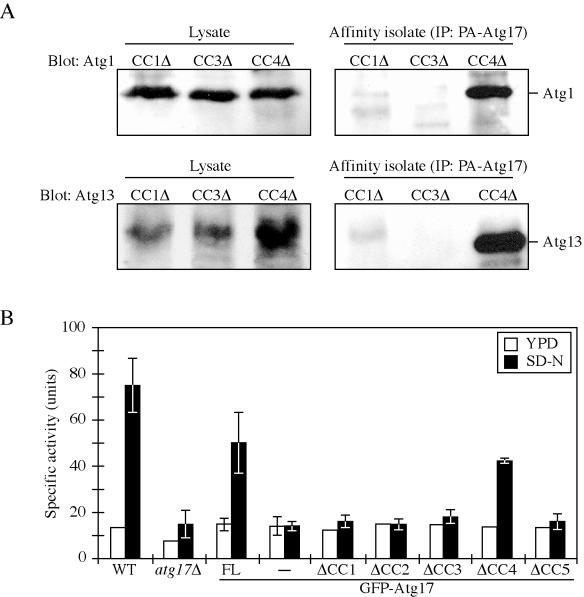
To extend the analysis of protein interactions, we performed affinity isolation in cells transformed with plasmids expressing Atg17ΔCC1, ΔCC3, or ΔCC4 fused to protein A. The presence of Atg1 or Atg13 was determined by Western blot after affinity purification of the protein A-tagged constructs. We found that Atg1 and Atg13 were efficiently recovered from cells expressing Atg17ΔCC4 (Figure 8A). In contrast, there was essentially no recovery of either protein from cells where the only copy of Atg17 lacked either CC1 or CC3. These data confirm the results from the two-hybrid analysis, suggesting that the first and third coiled-coil regions of Atg17 mediate the interactions with Atg1 and Atg13.
Figure 8.
The first and third Atg17 coiled-coil domains are required for interaction with Atg1 and Atg13 and for autophagic function. (A) Affinity isolation analysis. The atg17Δ (CWY239) strains expressing the indicated protein A-tagged coiled-coil domain-deleted Atg17 proteins were grown in SMD lacking auxotrophic amino acids. Protein extracts (lysate) were prepared and incubated with IgG-Sepharose beads as described in Materials and Methods. The resulting immunocomplexes (affinity isolate) were resolved by SDS-PAGE and analyzed by Western blot by using serum directed against Atg1 or Atg13. (B) Pho8Δ60 assay. The wild-type (TN124) and atg17Δ (CWY279) strain or the atg17Δ strain expressing full-length Atg17 or coiled-coil domain-deleted Atg17 in pRS426-CuGFP plasmids were grown in SMD lacking auxotrophic amino acids and shifted to SD-N for 4 h. The atg17Δ strain transformed with the pRS426-CuGFP empty plasmid was used as a negative control. Samples were collected and protein extracts assayed for ALP activity as described in Materials and Methods. The results represent the mean and SD of three experiments.
We next used the Pho8Δ60 assay to determine whether the coiled-coil domains of Atg17 played a functional role in autophagy. The atg17Δ mutant was severely blocked for Pho8Δ60 activity under starvation conditions, relative to the wild-type strain (Figures 2B and 8B). An N-terminal GFP fusion to full-length Atg17 restored the Pho8Δ60 activity of the atg17Δ strain to ∼67% that of the wild-type strain, indicating that this construct retained substantial Atg17 function (Figure 8B). The Atg17ΔCC1, ΔCC2, ΔCC3, and ΔCC5 proteins were not able to restore autophagic capacity to the atg17Δ strain and displayed essentially background levels of Pho8Δ60 activity. In contrast, the strain expressing Atg17ΔCC4 recovered ∼57% of the wild-type activity. These data suggest that the interactions mediated via the CC1 and CC3 regions of Atg17 are important for autophagy. Although the second and fifth coiled-coil domains of Atg17 are not needed for interaction with Atg1 or Atg13 (Figure 7B), these parts of the protein may bind other Atg proteins or may affect the structure of Atg17 so that the respective deletion constructs also lose autophagic activity.
The Atg1–Atg13 Complex Is Required for Normal Localization of Atg17
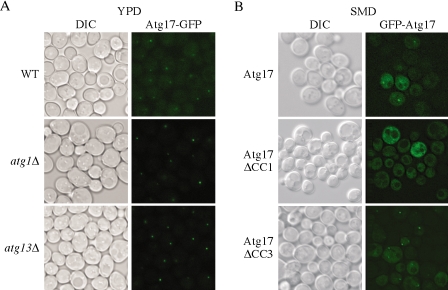
Based on the above-mentioned data, we concluded that the direct interaction between Atg17 and Atg13, and the possibly indirect interaction between Atg17 and Atg1, are important for autophagy. Next, we examined whether the proteins that interact with Atg17 influence its localization in vivo. To visualize Atg17, we generated an Atg17-GFP hybrid by integration at the 3′ end of the ATG17 chromosomal locus. The resulting Atg17-GFP protein was functional (our unpublished data). Cells expressing Atg17-GFP were grown to mid-log phase and imaged by fluorescence microscopy. In wild-type cells, Atg17-GFP displayed a prominent punctate dot and also a partial cytosolic diffuse pattern (Figure 9A). The punctate structure was located in a perivacuolar region and represents the pre-autophagosomal structure (PAS; Suzuki et al., 2001; Kim et al., 2002). We then analyzed the localization of Atg17-GFP in atg1Δ or atg13Δ mutants. In either mutant strain Atg17-GFP displayed a very bright single punctate dot located on the PAS, without any or with very limited diffuse cytosolic staining (Figure 9A), and the same phenotype was seen in an atg1Δ atg13Δ double mutant (our unpublished data). These observations suggest that Atg17 may exist in two pools, one cytosolic and one located at the PAS, and that Atg1 and Atg13 are involved in disassembling Atg17 from the PAS.
Figure 9.
Atg17 localization depends on the first coiled-coil domain. Cells from (A) wild-type (CWY241), atg1Δ (CWY242), and atg13Δ (CWY263) strains were grown in YPD. The cells were examined by fluorescence microscopy as described in Materials and Methods. (B) For localization of Atg17 mutants, the atg17Δ (CWY239) strain expressing full length Atg17 or coiled-coil domain deleted Atg17 on pRS416-CuGFP plasmids was grown in SMD lacking auxotrophic amino acids and examined as described in A. DIC, differential interference contrast.
We extended this analysis by examining whether the loss of the corresponding Atg17 interaction domains affected the localization of Atg17. To observe localization of Atg17 mutants lacking the coiled-coil domains, we constructed single-copy GFP fusion plasmids expressing truncated versions of Atg17 and examined the cells by fluorescence microscopy. In wild-type cells, plasmid-encoded GFP-Atg17 showed a single punctate dot and diffuse cytosolic staining similar to chromosomally tagged Atg17-GFP (Figure 9, A and B). When we analyzed the localization of GFP-Atg17 mutants, we found that GFP-Atg17ΔCC2, ΔCC3, and ΔCC4 showed a staining pattern similar to the full-length GFP-Atg17 (Figure 9B; our unpublished data). In contrast, GFP-Atg17ΔCC1 displayed a unique localization pattern; all of the protein was diffuse in the cytosol and did not show a punctate dot (Figure 9B). These data suggest that the first coiled-coil region of Atg17 is involved in localization of the protein to the PAS, possibly through its interaction with Atg1 and Atg13. We cannot conclude, however, that the interaction of Atg17 with Atg1 and/or Atg13 is the only factor determining its localization; GFP-Atg17ΔCC3 displayed essentially wild-type localization even though this mutant is not able to interact with Atg1 or Atg13 (Figures 7B and 8A). Furthermore, Atg17-GFP displayed a single, bright punctate dot at the PAS in the atg1Δ and atg13Δ mutant strains. Together, these data suggest that the presence of Atg17CC1 is able to compensate for the absence of the CC3 domain, but not vice versa, and possibly that another component aside from Atg1 and Atg13 also may bind the first coiled-coil region and play a role in determining Atg17 localization. Alternatively, it is possible that Atg17ΔCC1 is not folded correctly, although the protein is stable.
Additionally, we decided to examine the localization of Atg17 in cells lacking Vac8, because Vac8 interacts with Atg13, but its function in the Cvt and Atg pathways is not known. The Atg17–GFP pattern in the vac8Δ strain was intermediate between that of the wild-type and atg1Δ strains under vegetative conditions but looked similar to that seen in wild-type cells in starvation conditions (our unpublished data). In contrast, the Atg17-GFP pattern seen in atg20Δ or atg24Δ cells looked essentially the same as wild type (our unpublished data). These data suggest that Cvt-specific components that interact as part of the Atg1 complex, including Vac8, Atg20, and Atg24, are not significantly involved in the normal localization of Atg17 under starvation conditions, whereas Vac8 may have some effect on localization during vegetative growth.
Finally, we examined the role of Atg17 in the localization of two other Atg proteins, Atg1 and Atg9. Atg1-GFP in wild-type cells showed a punctate dot and a diffuse cytosolic pattern similar to that seen with Atg17 (Figure 10A). The Atg1 pattern was similar in the atg17Δ (Figure 10A), atg13Δ, and vac8Δ mutant strains (our unpublished data). Atg9 and Atg23 are unusual among the Atg proteins in that they display multiple punctate dots in a wild-type strain (Noda et al., 2000; Tucker et al., 2003; Figure 10B). This localization is dependent upon Atg1 and Atg13; in the corresponding mutants, Atg9 is restricted to the PAS (Reggiori et al., 2004a; Figure 10B). Because Atg17 is part of the Atg1 complex, we examined the effect of deleting ATG17 on the localization of Atg9. In growing conditions, Atg9-YFP was partly restricted to the PAS in the atg17Δ mutant, although the intensity of the punctate signal was reduced and additional dots could occasionally be seen (Figure 10B); that is, the phenotype was intermediate between the wild-type and atg1Δ strains. This result is essentially in agreement with a previous analysis of Atg9 localization (Reggiori et al., 2004a). We then extended the analysis by examining the transport of Atg9 after knocking out ATG1 (TAKA assay). In an atg1Δ atg17Δ double mutant, Atg9-YFP was completely restricted to the PAS, indicating that Atg17 temporally acts at the same stage or after Atg1 in the retrieval process that allows cycling of Atg9.
Figure 10.
Localization of Atg1 and Atg9 in the atg17Δ mutant. (A) Wild-type (PSY143), atg17Δ (HCY032), atg13Δ (HCY040), and vac8Δ (HCY042) cells expressing Atg1-GFP were grown in YPD and shifted to SD-N for 2 h before microscopy. Atg1 showed both a punctate and diffuse cytosolic localization. The data for localization of Atg1-GFP in atg13Δ or vac8Δ were essentially the same as those shown for wild-type and atg17Δ. (B) Atg9 cycling is defective in cells lacking Atg17. Integrated Atg9-YFP was expressed in wild-type (FRY136), atg1Δ (FRY138), atg17Δ (KTY89), and atg1Δ atg17Δ (JLY4) cells. The cells were grown in YPD or appropriate selective media to mid-log phase and FM 4-64 was added to the culture medium for 15 min, after which the cells were pelleted and resuspended in YPD or SMD, and further incubated for 30 min, followed by imaging by fluorescence microscopy. In the merged panels Atg9-YFP is shown in green to facilitate visualization relative to the FM 4-64 dye. DIC, differential interference contrast.
DISCUSSION
Atg17 is part of a putative multiprotein complex, including Atg1 kinase. In addition to Atg17, most of the other proteins in the putative complex have been characterized as being relatively specific for either autophagy or the Cvt pathway (Figure 1; Kamada et al., 2000; Scott et al., 2000; Kim et al., 2001b; Nice et al., 2002), leading to the hypothesis that this complex plays a role in regulating the conversion between these two pathways; however, the assignment of a particular mutant as “specific” has been somewhat arbitrary. For example, vac8Δ cells are completely blocked for import of prApe1 under vegetative but not starvation conditions (Scott et al., 2000). Thus, the Vac8 protein has been classified as specific for the Cvt pathway; however, the vac8Δ mutant displays a substantial defect in autophagy based on the Pho8Δ60 assay (Scott et al., 2000; Figure 2B). Similarly, in the present report, we find based on electron microscopy data and analysis of GFP-Atg8 processing that the vac8Δ mutant displays a significant autophagic defect (Figures 3 and 4). One conclusion from these observations is that it is insufficient to rely on one particular assay to reliably characterize an atg mutant as being specific to autophagy versus the Cvt pathway.
The initial characterization of the atg17Δ mutant suggested that it was specific for autophagy; however, this conclusion was based largely on the fact that prApe1 is matured in the atg17Δ mutant under vegetative conditions, whereas the strain is defective for autophagy as determined by the inability to activate Pho8Δ60 (Kamada et al., 2000); maturation of prApe1 was not determined under starvation conditions. Partly for the reasons described above, we decided to undertake a more detailed analysis of the role of Atg17. Although we found that atg17Δ cells are normal for processing of prApe1 in vegetative conditions, the same was true for cells incubated in SD-N to induce autophagy (Figure 2A). Nonetheless, in agreement with the data of Kamada et al. (2000), we found essentially a complete block in uptake of Pho8Δ60 (Figure 2B). These two sets of data are somewhat contradictory with regard to the requirement of Atg17 for autophagy; however, prApe1 is delivered to the vacuole by a specific autophagic mechanism that uses a receptor (Scott et al., 2001; Shintani et al., 2002), whereas Pho8Δ60 is a nonspecific marker for bulk autophagy. To resolve this discrepancy, we undertook three additional assays to monitor autophagy. We found that the atg17Δ mutant displayed an intermediate starvation-sensitivity phenotype relative to the atg1Δ strain (our unpublished data), showed limited vacuolar import of the autophagosome marker GFP-Atg8 (Figure 3A), and was essentially defective for pexophagy (Figure 3B).
Our conclusion from these various assays was that the atg17Δ mutant was severely, but not fully, defective for autophagy. To fully understand the nature of this phenotype, we carried out an analysis of atg17Δ cells by using electron microscopy (Figure 4). We used a pep4Δ vps4Δ background to allow accumulation of autophagic bodies within the vacuole lumen and to eliminate the presence of Mvb-derived vesicles. We found that the atg17Δ mutant accumulated autophagic bodies that were smaller, and fewer in number compared with those seen in the wild-type strain. Thus, Atg17 is not absolutely required for autophagy but seems to play an important role in determining the magnitude of the autophagic response. The electron microscopy data provide an explanation for the results with prApe1, Pho8Δ60 and peroxisome degradation. The reduction in autophagosome capacity apparently causes a severe block in uptake of Pho8Δ60. In contrast, prApe1 is still imported with essentially wild-type kinetics due to its use of a specific receptor. Even though pexophagy is a specific process, the smaller autophagosomes are apparently unable to sequester an organelle of this size (Figure 3B); peroxisomes from oleate-grown cells are typically in the range of 0.2–0.4 μm in diameter (Thieringer et al., 1991). Because Pho8Δ60 is a soluble cytosolic protein, it seems surprising that a low level is not taken up inside the aberrant autophagosomes that are produced in the atg17Δ strain. Pho8Δ60 uptake, however, is not efficient even in wild-type cells, and the level of uptake by atg17Δ cells may be below the level of detection. This possibility is supported by the observation that GFP is liberated from GFP-Atg8 in the atg17Δ mutant, albeit at a lower level relative to the wild-type strain, similar to the vac8Δ mutant (Figure 3A). Atg8 is a component of the autophagosome; hence, any autophagosome that fuses with the vacuole will deliver some Atg8 into the lumen. Thus, cleavage of GFP from GFP-Atg8 is a very sensitive marker for autophagy.
An atg17Δ vac8Δ double mutant resulted in a complete block in uptake of prApe1 (Figure 2A) or processing of GFP-Atg8 (Figure 3A). Similarly, we observed by electron microscopy that the atg17Δ vac8Δ double mutant was completely unable to form autophagic bodies and presumably autophagosomes. One explanation for these data is that Vac8 and Atg17 have a similar function; in the absence of either individual protein there is sufficient activity to carry out autophagy. Indeed, both the vac8Δ and atg17Δ mutants are only partially defective in autophagosome formation (Figure 4). Although the vac8Δ strain was previously reported to produce normal autophagosomes (Scott et al., 2000), in our present analysis the vac8Δ mutant generated smaller and fewer autophagic bodies relative to the wild-type strain (Figure 4), in agreement with the autophagic defect detected by the Pho8Δ60 and GFP-Atg8 assays (Figures 2B and 3A). We do not consider the explanation of similar functions for Vac8 and Atg17 likely, however, because the vac8Δ mutant is completely defective for import of prApe1 under vegetative conditions, whereas atg17Δ cells are not blocked in prApe1 import. Finally, Vac8 interacts with several proteins and plays a role in a range of processes, including organelle inheritance (Wang et al., 1998) and piecemeal microautophagy of the nucleus (Roberts et al., 2003), suggesting that it has different functions than Atg17. Because the vac8Δ mutant displayed a severe, although not complete, defect in autophagy based on uptake of Pho8Δ60 (Figure 2B), we propose that the two mutations have a synthetic defect. A synthetic defect also would fit with the fact that these two proteins are part of a common complex. Atg13 modulates Atg1 kinase activity and Atg17 may have a similar role, via Atg13. The function of Vac8 in the Cvt and autophagy pathways is not known, but its interaction with Atg13 may allow it to also modulate Atg1 kinase activity.
To gain further information about the interaction among the components of the Atg1 kinase complex, we mapped interacting domains in Atg13 and Atg17 (Figure 7). The domains in Atg13 that interact with Atg1 and Vac8 have been mapped previously (Kamada et al., 2000; Scott et al., 2000). We found that the Atg17-interacting domain overlapped partially with that of Atg1 but was separate from that of Vac8. A finer degree of analysis would probably separate the Atg1- and Atg17-binding domains; however, we chose to focus on Atg17. Two of the five predicted coiled-coil domains of Atg17 were needed for interaction with Atg1 and Atg13 (Figures 1, 7B, and 8A). It is possible that the interaction between Atg17 and Atg1 is mediated through Atg13; however, we obtained conflicting data on this point. Two-hybrid assays indicated a direct interaction between Atg17 and Atg1 (Figure 5), whereas affinity isolation suggested that these two proteins did not interact in the absence of Atg13 (Figure 6). These differences may reflect the greater stringency of the affinity isolation, or a higher sensitivity of the two-hybrid assay. Although the two-hybrid assay also is better suited to detect transient interactions, it is limited by the use of hybrid proteins that must interact in a nonphysiological context. We extended the analysis of protein interactions by carrying out affinity isolation with the mutated Atg17 proteins and were able to confirm that the first and third coiled-coils of Atg17 are necessary for binding Atg1 and Atg13 (Figure 8A). During the course of these studies, Ohsumi and colleagues published an analysis of Atg17 (Kabeya et al., 2005). They found that a mutation of cysteine at position 24, corresponding to a site within the first coiledcoil domain, eliminated the interaction between Atg17, Atg13, and Atg1, further supporting our results; however, they were unable to determine whether the interaction between Atg1 and Atg17 was direct or mediated through Atg13.
When the level of autophagy was examined with Atg17 mutants lacking predicted coiled-coil domains, the Atg17ΔCC2 mutant showed low Pho8Δ60 activity after starvation, similar to the result seen with the Atg17ΔCC1 or ΔCC3 mutants (Figure 8B). Although Atg17ΔCC2 is not involved in the interaction with Atg1 and Atg13, we have not determined whether the Atg17CC2 domain is involved in interactions with other proteins that might be important for autophagy. Similarly, the Atg17ΔCC5 mutant may be defective for Pho8Δ60 activity due to the loss of other binding sites that do not involve Atg1 or Atg13. Alternatively, this large deletion may affect the folding of the protein resulting in a loss of function, although the mutant protein was stable.
When we analyzed the localization of GFP-Atg17 mutants, we found that GFP-Atg17ΔCC1 displayed a diffuse cytosolic signal and did not show a punctate dot, a localization pattern different from that seen for Atg17-GFP in the atg1Δ and atg13Δ mutants (Figure 9). As part of our analysis, we generated a set of smaller deletions within the coiled-coil regions in a preliminary study to more finely map the Atg17 interaction sites. We found that smaller deletions within the first coiled-coil region resulted in mutant GFP-Atg17 proteins that displayed single, bright punctate dots and that were also defective in interacting with Atg13 (our unpublished data). These data suggest that a specific region(s) within the first coiled-coil domain of Atg17 may be involved in its localization to the PAS, and that this region(s) might be separate from those that interact with Atg1 and Atg13.
The role of Atg1 kinase activity is not clear, and there have been conflicting reports suggesting that higher kinase activity plays a more important role in the autophagy (Kamada et al., 2000) or Cvt (Abeliovich et al., 2003) pathways. In the present analysis, we found that the localization of Atg17 was dependent on Atg1 kinase activity particularly in rich conditions (Figure 8A; our unpublished data). Atg17 localization was more severely affected in starvation conditions in the atg1Δ strain than in the presence of the kinase dead Atg1K54A mutant (our unpublished data). This finding is in agreement with previous studies that suggest that Atg1 may have a structural role in autophagy (Abeliovich et al., 2003). We suggest that higher kinase activity is more important for the Cvt pathway but that Atg1 kinase activity is needed for both the Cvt and autophagy pathways (Abeliovich et al., 2003; Reggiori et al., 2004a) in agreement with the recent study by Kabeya et al. (2005).
Finally, it may be informative to note that ATG17 only has homologues in yeast, whereas ATG13 and VAC8 do not have homologues in higher eukaryotes outside of plants. Autophagosomes in higher eukaryotes seem to be of a uniform size before and after starvation (Dunn, 1990). Thus, regulation of sequestering vesicle size seems to be peculiar to yeast. We propose that the direct interaction between Atg17 and Atg13, and the direct or indirect interaction with Atg1 is critical for regulating autophagosome size. Although the Atg1 complex is proposed to be involved in controlling the conversion between the autophagy and Cvt pathways, this regulation may not take place at the stage of autophagic induction. We have shown that Atg1 and Atg13 participate at a later stage, retrieval of Atg9 from the PAS (Reggiori et al., 2004a). An Atg1-dependent defect in Atg9 retrieval that is mediated through Atg17 could provide one explanation for the smaller and fewer autophagosomes seen in the atg17Δ mutant. That is, the absence of Atg17 may partially interfere with Atg9 cycling (Figure 10), resulting in a reduced supply of membrane to the sequestering vesicles. In contrast, atg1Δ or atg13Δ strains are completely defective in this step and are unable to form vesicles of any size. Additional studies concerning the functions of the components of the Atg1 complex should provide further insight into the mechanism of vesicle size regulation.
Acknowledgments
We thank members of the Klionsky laboratory for helpful discussions. This work was supported by Public Health Service Grant GM53396 from the National Institutes of Health (to D.J.K.). F. R. was supported by a Swiss National Foundation Fellowship for advanced researchers.
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E04–10–0894) on May 18, 2005.
References
- Abeliovich, H., Dunn, W. A., Jr., Kim, J., and Klionsky, D. J. (2000). Dissection of autophagosome biogenesis into distinct nucleation and expansion steps. J. Cell Biol. 151, 1025–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abeliovich, H., Zhang, C., Dunn, W. A., Jr., Shokat, K. M., and Klionsky, D. J. (2003). Chemical genetic analysis of Apg1 reveals a non-kinase role in the induction of autophagy. Mol. Biol. Cell 14, 477–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba, M., Osumi, M., Scott, S. V., Klionsky, D. J., and Ohsumi, Y. (1997). Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J. Cell Biol. 139, 1687–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba, M., Takeshige, K., Baba, N., and Ohsumi, Y. (1994). Ultrastructural analysis of the autophagic process in yeast: detection of autophagosomes and their characterization. J. Cell Biol. 124, 903–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst, M., Wendland, B., Estepa, E. J., and Emr, S. D. (1998). The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 17, 2982–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budovskaya, Y. V., Stephan, J. S., Reggiori, F., Klionsky, D. J., and Herman, P. K. (2004). The Ras/cAMP-dependent protein kinase signaling pathway regulates an early step of the autophagy process in Saccharomyces cerevisiae. J. Biol. Chem. 279, 20663–20671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo, A. M. (2004). Autophagy: in sickness and in health. Trends Cell Biol. 14, 70–77. [DOI] [PubMed] [Google Scholar]
- Dunn, W. A., Jr. (1990). Studies on the mechanisms of autophagy: formation of the autophagic vacuole. J. Cell Biol. 110, 1923–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funakoshi, T., Matsuura, A., Noda, T., and Ohsumi, Y. (1997). Analyses of APG13 gene involved in autophagy in yeast, Saccharomyces cerevisiae. Gene 192, 207–213. [DOI] [PubMed] [Google Scholar]
- Harding, T. M., Morano, K. A., Scott, S. V., and Klionsky, D. J. (1995). Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J. Cell Biol. 131, 591–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow, E., and Lane, D. (1999). Using Antibodies: A Laboratory Manual, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Huang, W.-P., Scott, S. V., Kim, J., and Klionsky, D. J. (2000). The itinerary of a vesicle component, Aut7p/Cvt5p, terminates in the yeast vacuole via the autophagy/Cvt pathways. J. Biol. Chem. 275, 5845–5851. [DOI] [PubMed] [Google Scholar]
- Hutchins, M. U., and Klionsky, D. J. (2001). Vacuolar localization of oligomeric α-mannosidase requires the cytoplasm to vacuole targeting and autophagy pathway components in Saccharomyces cerevisiae. J. Biol. Chem. 276, 20491–20498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins, M. U., Veenhuis, M., and Klionsky, D. J. (1999). Peroxisome degradation in Saccharomyces cerevisiae is dependent on machinery of macroautophagy and the Cvt pathway. J. Cell Sci. 112, 4079–4087. [DOI] [PubMed] [Google Scholar]
- Ichimura, Y., et al. (2000). A ubiquitin-like system mediates protein lipidation. Nature 408, 488–492. [DOI] [PubMed] [Google Scholar]
- James, P., Halladay, J., and Craig, E. A. (1996). Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya, Y., Kamada, Y., Baba, M., Takikawa, H., Sasaki, M., and Ohsumi, Y. (2005). Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol. Biol. Cell 16, 2544–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser, C. A., and Schekman, R. (1990). Distinct sets of SEC genes govern transport vesicle formation and fusion early in the secretory pathway. Cell 61, 723–733. [DOI] [PubMed] [Google Scholar]
- Kamada, Y., Funakoshi, T., Shintani, T., Nagano, K., Ohsumi, M., and Ohsumi, Y. (2000). Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 150, 1507–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann, D. J. (2004). Ubiquitin-Mediated Vacuolar Sorting and Degradation, Georgetown, TX: Landes Bioscience.
- Kim, J., Huang, W.-P., and Klionsky, D. J. (2001a). Membrane recruitment of Aut7p in the autophagy and cytoplasm to vacuole targeting pathways requires Aut1p, Aut2p, and the autophagy conjugation complex. J. Cell Biol. 152, 51–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J., Huang, W.-P., Stromhaug, P. E., and Klionsky, D .J. (2002). Convergence of multiple autophagy and cytoplasm to vacuole targeting components to a perivacuolar membrane compartment prior to de novo vesicle formation. J. Biol. Chem. 277, 763–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J., Kamada, Y., Stromhaug, P. E., Guan, J., Hefner-Gravink, A., Baba, M., Scott, S. V., Ohsumi, Y., Dunn, W. A., Jr., and Klionsky, D. J. (2001b). Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J. Cell Biol. 153, 381–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J., and Klionsky, D. J. (2000). Autophagy, cytoplasm-to-vacuole targeting pathway, and pexophagy in yeast and mammalian cells. Annu. Rev. Biochem. 69, 303–342. [DOI] [PubMed] [Google Scholar]
- Kirisako, T., Baba, M., Ishihara, N., Miyazawa, K., Ohsumi, M., Yoshimori, T., Noda, T., and Ohsumi, Y. (1999). Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 147, 435–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako, T., Ichimura, Y., Okada, H., Kabeya, Y., Mizushima, N., Yoshimori, T., Ohsumi, M., Takao, T., Noda, T., and Ohsumi, Y. (2000). The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J. Cell Biol. 151, 263–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky, D. J. (2004). Autophagy, Georgetown, TX: Landes Bioscience.
- Klionsky, D. J., et al. (2003). A unified nomenclature for yeast autophagy-related genes. Dev. Cell 5, 539–545. [DOI] [PubMed] [Google Scholar]
- Klionsky, D. J., Cueva, R., and Yaver, D. S. (1992). Aminopeptidase I of Saccharomyces cerevisiae is localized to the vacuole independent of the secretory pathway. J. Cell Biol. 119, 287–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky, D. J., and Emr, S. D. (2000). Autophagy as a regulated pathway of cellular degradation. Science 290, 1717–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky, D. J., and Ohsumi, Y. (1999). Vacuolar import of proteins and organelles from the cytoplasm. Annu. Rev. Cell Dev. Biol. 15, 1–32. [DOI] [PubMed] [Google Scholar]
- Levine, B., and Klionsky, D. J. (2004). Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell 6, 463–477. [DOI] [PubMed] [Google Scholar]
- Longtine, M. S., McKenzie, A., III, Demarini, D. J., Shah, N. G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J. R. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961. [DOI] [PubMed] [Google Scholar]
- Nice, D. C., Sato, T. K., Stromhaug, P. E., Emr, S. D., and Klionsky, D. J. (2002). Cooperative binding of the cytoplasm to vacuole targeting pathway proteins, Cvt13 and Cvt20, to phosphatidylinositol 3-phosphate at the pre-autophagosomal structure is required for selective autophagy. J. Biol. Chem. 277, 30198–30207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda, T., Kim, J., Huang, W.-P., Baba, M., Tokunaga, C., Ohsumi, Y., and Klionsky, D. J. (2000). Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J. Cell Biol. 148, 465–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda, T., Matsuura, A., Wada, Y., and Ohsumi, Y. (1995). Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 210, 126–132. [DOI] [PubMed] [Google Scholar]
- Onodera, J., and Ohsumi, Y. (2004). Ald6p is a preferred target for autophagy in yeast, Saccharomyces cerevisiae. J. Biol. Chem. 279, 16071–16076. [DOI] [PubMed] [Google Scholar]
- Reggiori, F., and Klionsky, D. J. (2002). Autophagy in the eukaryotic cell. Eukaryot. Cell 1, 11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori, F., Tucker, K. A., Stromhaug, P. E., and Klionsky, D. J. (2004a). The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev. Cell 6, 79–90. [DOI] [PubMed] [Google Scholar]
- Reggiori, F., Wang, C.-W., Nair, U., Shintani, T., Abeliovich, H., and Klionsky, D. J. (2004b). Early stages of the secretory pathway, but not endosomes, are required for Cvt vesicle and autophagosome assembly in Saccharomyces cerevisiae. Mol. Biol. Cell 15, 2189–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, P., Moshitch-Moshkovitz, S., Kvam, E., O'Toole, E., Winey, M., and Goldfarb, D. S. (2003). Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol. Biol. Cell 14, 129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, J. S., Klionsky, D. J., Banta, L. M., and Emr, S. D. (1988). Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell. Biol. 8, 4936–4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelzle, T., Beck, T., Martin, D. E., and Hall, M. N. (2004). Activation of the RAS/cyclic AMP pathway suppresses a TOR deficiency in yeast. Mol. Cell. Biol. 24, 338–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, S. V., Guan, J., Hutchins, M. U., Kim, J., and Klionsky, D. J. (2001). Cvt19 is a receptor for the cytoplasm-to-vacuole targeting pathway. Mol. Cell 7, 1131–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, S. V., Nice, D. C., III, Nau, J. J., Weisman, L. S., Kamada, Y., Keizer-Gunnink, I., Funakoshi, T., Veenhuis, M., Ohsumi, Y., and Klionsky, D. J. (2000). Apg13p and Vac8p are part of a complex of phosphoproteins that are required for cytoplasm to vacuole targeting. J. Biol. Chem. 275, 25840–25849. [DOI] [PubMed] [Google Scholar]
- Shintani, T., Huang, W.-P., Stromhaug, P. E., and Klionsky, D. J. (2002). Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev. Cell 3, 825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani, T., and Klionsky, D. J. (2004a). Autophagy in health and disease: a double-edged sword. Science 306, 990–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani, T., and Klionsky, D. J. (2004b). Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J. Biol. Chem. 279, 29889–29894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stromhaug, P. E., Reggiori, F., Guan, J., Wang, C.-W., and Klionsky, D. J. (2004). Atg21 is a phosphoinositide binding protein required for efficient lipidation and localization of Atg8 during uptake of aminopeptidase I by selective autophagy. Mol. Biol. Cell 15, 3553–3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, K., Kirisako, T., Kamada, Y., Mizushima, N., Noda, T., and Ohsumi, Y. (2001). The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 20, 5971–5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshige, K., Baba, M., Tsuboi, S., Noda, T., and Ohsumi, Y. (1992). Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J. Cell Biol. 119, 301–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thieringer, R., Shio, H., Han, Y. S., Cohen, G., and Lazarow, P. B. (1991). Peroxisomes in Saccharomyces cerevisiae: immunofluorescence analysis and import of catalase A into isolated peroxisomes. Mol. Cell. Biol. 11, 510–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thumm, M., Egner, R., Koch, B., Schlumpberger, M., Straub, M., Veenhuis, M., and Wolf, D. H. (1994). Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 349, 275–280. [DOI] [PubMed] [Google Scholar]
- Titorenko, V. I., Keizer, I., Harder, W., and Veenhuis, M. (1995). Isolation and characterization of mutants impaired in the selective degradation of peroxisomes in the yeast Hansenula polymorpha. J. Bacteriol. 177, 357–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada, M., and Ohsumi, Y. (1993). Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 333, 169–174. [DOI] [PubMed] [Google Scholar]
- Tucker, K. A., Reggiori, F., Dunn, W. A., Jr., and Klionsky, D. J. (2003). Atg23 is essential for the cytoplasm to vacuole targeting pathway and efficient autophagy but not pexophagy. J. Biol. Chem. 278, 48445–48452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuttle, D. L., and Dunn, W. A., Jr. (1995). Divergent modes of autophagy in the methylotrophic yeast Pichia pastoris. J. Cell Sci. 108, 25–35. [DOI] [PubMed] [Google Scholar]
- Tuttle, D. L., Lewin, A. S., and Dunn, W. A., Jr. (1993). Selective autophagy of peroxisomes in methylotrophic yeasts. Eur. J. Cell Biol. 60, 283–290. [PubMed] [Google Scholar]
- Veenhuis, M., Zwart, K., and Harder, W. (1978). Degradation of peroxisomes after transfer of methanol-grown Hansenula polymorpha into glucose-containing media. FEMS Micro. Lett. 3, 21–28. [Google Scholar]
- Wang, C.-W., and Klionsky, D. J. (2003). The molecular mechanism of autophagy. Mol. Med. 9, 65–76. [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. X., Catlett, N. L., and Weisman, L. S. (1998). Vac8p, a vacuolar protein with armadillo repeats, functions in both vacuole inheritance and protein targeting from the cytoplasm to vacuole. J. Cell Biol. 140, 1063–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]