Abstract
Background
NDR1/HIN1-like (NHL) genes play crucial roles in Psa resistance. Kiwifruit canker, caused by Pseudomonas syringae pv. Actinidiae (Psa) infection is one of the most serious diseases affecting the kiwifruit industry. However, the key NHL has not yet been identified in kiwifruit.
Results
In this study, we conducted a genome-wide identification of NHL family in kiwifruit (Actinidia eriantha). A total of 33 AeNHLs were divided into five domain-conserved subfamilies, which were mainly assigned into phytohormones and defense responses. The expression of AeNHL genes was analyzed to identify key genes in response to Psa, and we found AeNHL17 was highly expressed upon Psa inoculation. Transgenic tobacco overexpressing AeNHL17 presented higher resistance to Psa than wild-type (WT) tobacco, implying a key role for AeNHL17 in Psa resistance. Finally, we carried out a stable genetic transformation of kiwifruit (A. chinensis), which is sensitive to Psa, and found that the overexpression of AeNHL17 increased resistance to infection. AeNHL17-silenced plants exhibited larger disease lesions than control plants.
Conclusions
Our findings revealed the function of AaNHL17 in Psa resistance, providing new data regarding the functional analysis of the NHL gene family in kiwifruit.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-024-05936-2.
Keywords: NDR1/HIN1-like gene family, Kiwifruit, Psa, AeNHL17, Disease resistance
Introduction
Kiwifruit (Actinidia spp.) is native to China and is a rich germplasm resource with a wide geographical distribution. It is a domesticated fruit plant and has become an important horticultural crop [1, 2]. However, kiwifruit bacterial canker (KBC) caused by Pseudomonas syringae pv. Actinidae (Psa) have led to devastation of the kiwifruit industry worldwide [3–5]. This disease was first reported in Japan and was later discovered in China and other kiwifruit-producing areas such as Italy, Korea, and New Zealand [6–9]. KBC is gradually becoming the primary factor limiting the kiwifruit industry development globally [10]. The currently discovered Psa lines can be divided into four types based on genomic comparison, phylogeny, and origin [11]. The Psa biovar is highly virulent, and once it systemically invades a plant, it may quickly lead to plant death [12]. Therefore, exploring the disease defense mechanisms of kiwifruit is crucial for preventing Psa infections.
Plants have evolved sophisticated regulatory networks to resist infection, such as PAMP-triggered immunity (PTI), effector-triggered immunity (ETI), hypersensitive response (HR), and systemic acquired resistance (SAR) [13, 14]. PTI and ETI are basic defense responses when plants face environmental attacks. Further pathogen resistance mediated by resistance genes, is usually associated with HR and SAR [15]. These processes require the tight coordination of multiple phytohormones, including jasmonic acid (JA), ethylene (ET) and salicylic acid (SA) [16]. Previous studies have shown that the pathogen infection response can increase the expression of the immune signal SA, which is usually consistent with the increased transcription level of pathogenesis-related (PR) genes and strengthens disease resistance [17, 18].
NDR1/HIN-like (NHL) genes are characterized by sequence and structural similarity to Non-race-specific disease resistance 1 (NDR1) of Arabidopsis thaliana and Harpin-induced 1 (HIN1) of Nicotiana tabacum [19, 20]. NDR1 is necessary for resistance genes to recognize pathogens and plays a role in plant disease resistance responses by encoding plasma membrane-localized proteins [20]. HIN1 is triggered by pathogens and plays critical roles in the plant defense pathway [21]. Most NHL proteins have two highly conserved motifs, including a conserved late embryogenesis abundant (LEA) domain [22]. The LEA domain belongs to a protein family involved in osmotic regulation, and these proteins typically participate in protecting plants from biotic and abiotic damage [23].
Previous studies have shown that NHL genes play an important role in plant-pathogen interactions and resistance to abiotic stress [22, 24]. For example, the expression of NHL3 is significantly induced by a variety Pseudomonas strains in Arabidopsis, and the overexpression of NHL3 strengthens plant resistance to Pseudomonas syringae pv. tomato DC3000 [22, 25]. The hypersensitive response of NHL10 cells to Cucumber mosaic virus (CMV) infection is controlled by the SA signaling pathway [26]. Similarly, the overexpression of StPOTHR1 can strengthen resistance against P. infestans in potatoes [27]. In pepper, transient overexpression of CaNHL4 increases plant resistance, and CaNHL4-silenced plants show significantly increased susceptibility to different pathogens [28]. Overall, these studies demonstrate that NHL family members identified from different plants play essential roles in resistance and stress responses to pathogen infection. However, the NHL gene family in kiwifruit and its function in response to biotic and abiotic stress remain largely unknown.
In the present study, we identified the NHL gene family based on whole-genome sequences of A. eriantha. The phylogenetic relationships, chromosomal localization, and other structural features of AeNHLs were analyzed systematically. Subsequently, we isolated an NHL gene AeNHL17 which is highly similar to pathogen resistance-related NHLs from other species. The function of AeNHL17 has been characterized in transgenic tobacco and kiwifruit plants. These findings provide evidence for further studies on the function of NHL in kiwifruit disease resistance and provide new genes for resistance breeding.
Results
Identification of AeNHLs in A. eriantha
To determine the NHL gene family in A. eriantha, 45 NDR1/HIN1-like sequences from A. thaliana were used as queries for BLAST search in Kiwifruit Genome Database. The BLAST search results were refined by manual selection to remove redundant sequences, and SMART was used to confirm the presence of the LEA-2 domain (PF03168). Table 1 shows that a total of 33 AeNHL genes were acquired, which were named AeNHL1 to AeNHL33 based on the reference genome localization. The CDS lengths of AeNHLs was 573 bp (AeNHL7) to 1614 bp (AeNHL13) (Table 1). The molecular weights (MW) were in the range of 20.55 to 60.23 kDa, and the isoelectric point (pI) was in the range of 7.09 (AeNHL30) to 10.43 (AeNHL21) (Table 1).
Table 1.
Characterization of identified AeNHL genes in A. eriantha
| Name | Gene ID | Chromosome location | CDS (bp) | MW (kDa) | pI |
|---|---|---|---|---|---|
| AeNHL1 | DTZ79_02g00550 | LG2:472472–473,110 | 639 | 23.79 | 9.65 |
| AeNHL2 | DTZ79_02g01580 | LG2:1280992–1,281,645 | 654 | 24.72 | 10.04 |
| AeNHL3 | DTZ79_02g07380 | LG2:7318550–7,319,164 | 615 | 22.32 | 10.23 |
| AeNHL4 | DTZ79_03g00560 | LG3:726701–727,339 | 639 | 23.66 | 9.43 |
| AeNHL5 | DTZ79_03g08330 | LG3:8554895–8,555,527 | 633 | 23.76 | 9.73 |
| AeNHL6 | DTZ79_05g01590 | LG5:2153616–2,154,368 | 753 | 28.43 | 9.86 |
| AeNHL7 | DTZ79_06g09910 | LG6:16778957–16,779,943 | 573 | 20.55 | 9.21 |
| AeNHL8 | DTZ79_07g03930 | LG7:4045388–4,047,763 | 594 | 21.32 | 8.96 |
| AeNHL9 | DTZ79_08g00870 | LG8:1128557–1,133,851 | 624 | 23.31 | 10.09 |
| AeNHL10 | DTZ79_08g00880 | LG8:1142089–1,142,784 | 696 | 26.43 | 9.51 |
| AeNHL11 | DTZ79_08g16200 | LG8:27133049–27,134,597 | 819 | 30.79 | 9.27 |
| AeNHL12 | DTZ79_11g02010 | LG11:2056415–2,059,016 | 969 | 35.30 | 9.86 |
| AeNHL13 | DTZ79_11g08930 | LG11:14856415–14,864,364 | 1614 | 59.00 | 9.16 |
| AeNHL14 | DTZ79_14g04210 | LG14:4840482–4,849,361 | 1602 | 59.10 | 10.12 |
| AeNHL15 | DTZ79_14g08860 | LG14:13459555–13,460,278 | 651 | 24.12 | 10.17 |
| AeNHL16 | DTZ79_15g07790 | LG15:10126568–10,129,976 | 867 | 32.63 | 9.87 |
| AeNHL17 | DTZ79_15g07880 | LG15:10343397–10,352,083 | 696 | 26.54 | 9.45 |
| AeNHL18 | DTZ79_15g13030 | LG15:17940403–17,945,263 | 1257 | 48.34 | 9.81 |
| AeNHL19 | DTZ79_15g13230 | LG15:18066546–18,074,706 | 1587 | 60.24 | 10.01 |
| AeNHL20 | DTZ79_16g00990 | LG16:1085317–1,086,881 | 882 | 33.20 | 8.90 |
| AeNHL21 | DTZ79_16g01530 | LG16:1612922–1,613,673 | 726 | 26.23 | 10.43 |
| AeNHL22 | DTZ79_18g06590 | LG18:14558202–14,562,670 | 861 | 32.38 | 9.84 |
| AeNHL23 | DTZ79_19g03750 | LG19:11576715–11,580,733 | 615 | 21.85 | 8.95 |
| AeNHL24 | DTZ79_19g14810 | LG19:24463556–24,465,866 | 885 | 32.74 | 9.05 |
| AeNHL25 | DTZ79_20g11920 | LG20:18639964–18,642,612 | 900 | 33.27 | 8.84 |
| AeNHL26 | DTZ79_21g06400 | LG21:7075917–7,076,969 | 957 | 34.88 | 8.81 |
| AeNHL27 | DTZ79_23g17320 | LG23:21317601–21,321,313 | 924 | 33.96 | 9.80 |
| AeNHL28 | DTZ79_25g05970 | LG25:15152300–15,155,603 | 801 | 29.17 | 9.71 |
| AeNHL29 | DTZ79_26g11040 | LG26:18413635–18,416,550 | 774 | 28.40 | 9.56 |
| AeNHL30 | DTZ79_27g07370 | LG27:8537730–8,538,593 | 864 | 31.30 | 7.09 |
| AeNHL31 | DTZ79_28g08090 | LG28:12897954–12,900,898 | 1575 | 57.89 | 9.06 |
| AeNHL32 | DTZ79_28g08100 | LG28:12904890–12,915,001 | 972 | 36.23 | 10.21 |
| AeNHL33 | DTZ79_00g01080 | LG0:1911615–1,912,520 | 627 | 23.07 | 9.30 |
Phylogenetic analysis and chromosome location of the AeNHLs
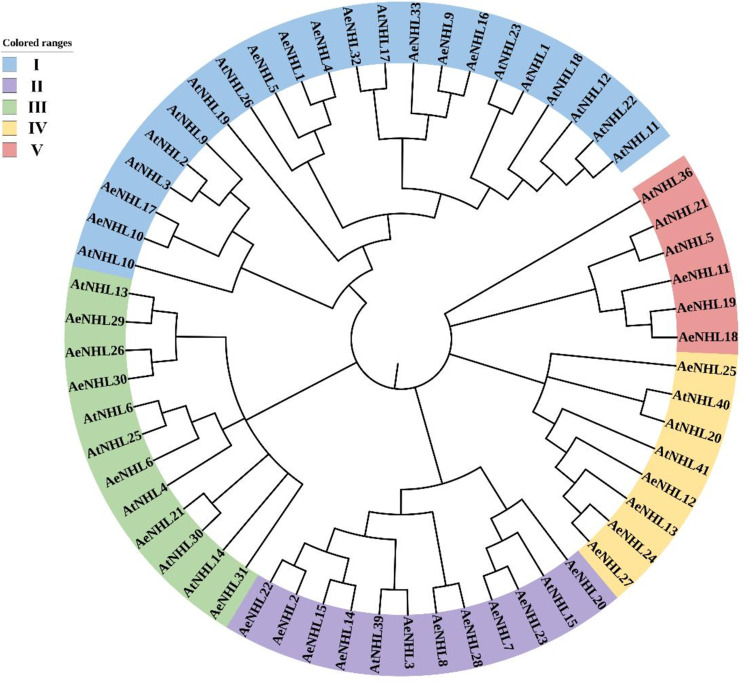
To understand the evolutionary relationship between NHL proteins, 27 AtNHL and 33 AeNHL proteins were used to perform sequence alignment and to construct an unrooted NJ tree. The total 60 NHL proteins were divided into five well-conserved subgroups (Fig. 1). Group I had the largest number and included nine AeNHL proteins, while Group V had the smallest number and contained three AeNHL proteins (Fig. 1). AeNHL17 and AeNHL10 were closer to AtNHL3. The login numbers of the NHL protein homolog sequences used in the unrooted NJ tree are listed in Supplementary Table S2.
Fig. 1.
Unrooted phylogenetic tree of NHL protein family from A. eriantha (Ae) and A. thaliana (At). MEGA 11.0.13 was used to construct the phylogenetic tree based on the NHL protein sequences, and the five distinct subgroups of NHL proteins were shown. iTOL [29] tool was used to annotate the phylogenetic tree
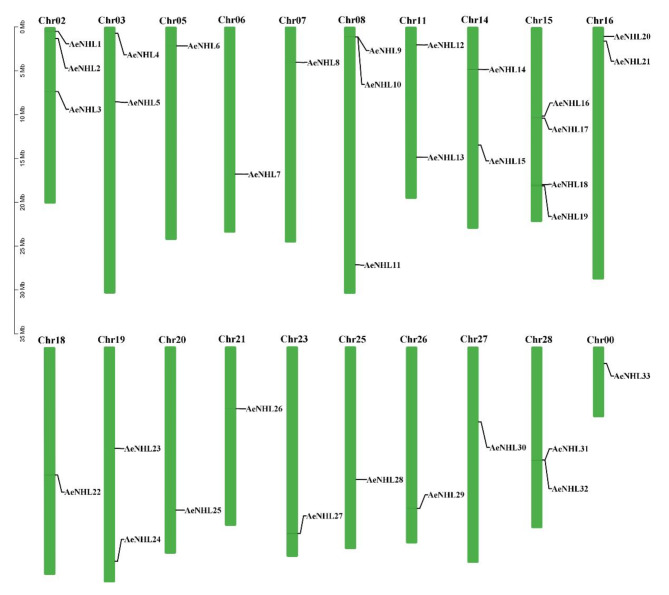
The chromosomal localization of AeNHLs was further determined using a simplified physical map, which revealed that 33 AeNHL genes were unevenly distributed across 20 chromosomes in A. eriantha genome (Fig. 2). Chromosomes (Chr) 5, 6, 7, 18, 20, 21, 23, 25, 26, 27, and 0 contained one copy each; Chr3, 11, 14, 16, 19, and 28 contained two copies each; Chr 2 and 8 contained three copies each; and Chr 15 contained four copies.
Fig. 2.
Chromosomal positions of AeNHL genes. Chromosome numbers are indicated at the top of each chromosome. Scale represents a 5 Mb chromosomal distance
Analysis of cis-elements and conserved motifs in AeNHLs
To better understand the expression and regulation of AeNHL members, their promoter sequences were extracted and regulatory elements were explored. The results showed that more than 15 regulatory elements were abundant in the promoter regions of AeNHLs, including stress and defense response elements, light responsive elements, and hormone regulation elements, including auxin (IAA), methyl jasmonate (MeJA), abscisic acid (ABA), and gibberellin (GA) (Fig. 3a). Furthermore, we discovered that some AeNHL genes contained salicylic acid (SA)-responsive elements, indicating that these members may be involved in the SA-mediated responses. These results imply that the function of AeNHL proteins in kiwifruit may be related to disease resistance.
Fig. 3.
Schematic representations of the predicted regulatory elements and the conserved motifs in the AeNHL family. (a) Schematic illustration of the predicted regulatory elements in the promoter regions of AeNHLs by PlantCare. Each regulatory element is marked with a different color. (b) Schematic representation of the conserved motifs predicted in the AeNHL proteins. MEME was used to predict the conserved motifs, and TBtools was used to display the results
MEME motif analysis indicated that AeNHL proteins have two widely distributed motifs: motifs 1 and 3. Additionally, AeNHL members in the same phylogenetic subgroup had similar and specific motif sequences. For example, AeNHL1 and AeNHL4 were located in the same subgroup as motifs 1, 2, 3, 4, and 9 (Fig. 3b). The similarity in motifs between AeNHL proteins suggests that these protein structures are conserved in specific subgroups. Collectively, our results indicated that AeNHL proteins within the same subfamily have conserved motifs and domains, implying a similar potential role in kiwifruit disease defense.
Expression profiling of AeNHLs
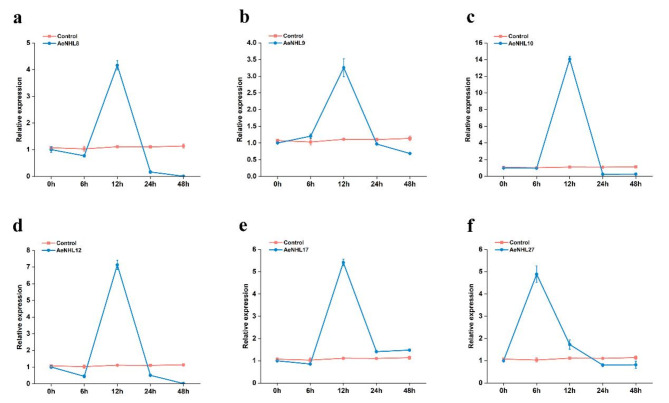
Quantitative real-time RT-PCR was performed to explore the global expression profiles of AeNHL members during Psa infection. The expression of AeNHL8, AeNHL9, AeNHL10, AeNHL12, AeNHL17 and AeNHL27 was significantly induced and rapidly increased, peaking at 12 h post-inoculation (Fig. 4). Conversely, the expression of some AeNHL genes decreased rapidly, whereas some members showed not expression (Supplementary Fig. S1). Expression levels of AeNHL9, AeNHL10, and AeNHL17 in different tissues and floral organs were also investigated.
Fig. 4.
Relative expression of the AeNHL8,AeNHL9,AeNHL10,AeNHL12,AeNHL17 and AeNHL27 genes in kiwifruit leaves after Psa infection. (a-f) Expression level of AeNHL8,AeNHL9,AeNHL10,AeNHL12,AeNHL17 and AeNHL27 at 6, 12, 24, 48 h after Psa infection. Control indicates plants without Psa infection. The values represent the means ± standard errors (SEs) of three biological replications
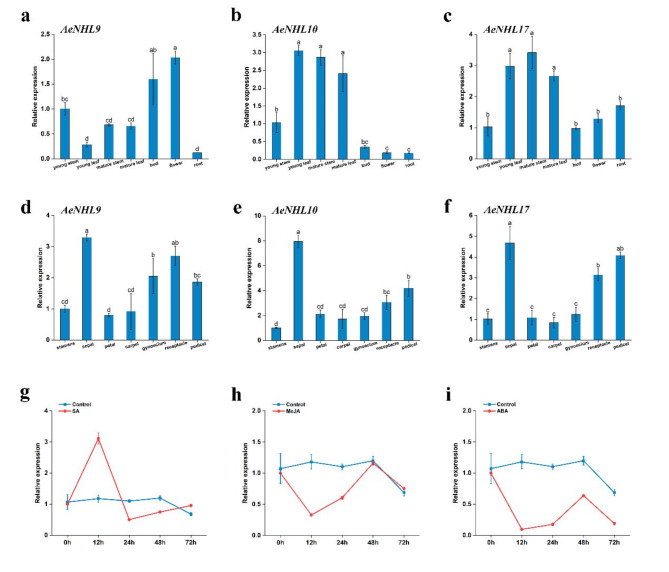
The results showed that these three genes were expressed in almost all the tissues and floral organs; however, their expression patterns differed. For instance, the expression of AeNHL9 was the highest in flowers and the lowest in roots. AeNHL17 showed the highest expression in mature stems and the lowest expression in buds (Fig. 5a-c). Among the floral organs, the highest levels of these three genes were observed in the sepals (Fig. 5d-f).
Fig. 5.
Expression patterns of NHL genes in different tissues and effect of SA, MeJA, and ABA in the expression of AeNHL17 gene. (a-f) Relative expression levels of AeNHL9, AeNHL10 and AeNHL17 genes in different tissues (young stem, young leaf, mature stem, mature leaf, bud, flower and root) and floral organ (stamens, sepal, petal, carpel, gynoecium, receptacle and pedicel). Data were presented as mean values ± SD (n = 3). Lowercase letters in the column denoted the significance level of mean differences at 0.05 (LSD method). (g-i) Expression level of AeNHL17 at 0, 12, 24, 48, 72 h after SA, MeJA, ABA treatments. Control indicates plants without above-mentioned treatments. The values represent the means ± standard errors (SEs) of three biological replications
To further determine the temporal expression levels of AeNHL17 in response to SA, MeJA, and ABA treatments, qRT-PCR was performed. As shown in Fig. 5g, AeNHL17 expression gradually increased, with a single peak 12 h after the plants were treated with SA. Upon MeJA and ABA treatment, the transcription level of AeNHL17 rapidly declined and minimum expression levels were observed at 12 h, after which it gradually increased and then peaked at 48 h (Fig. 5h-i).
Sequence analysis and subcellular localization of AeNHL17
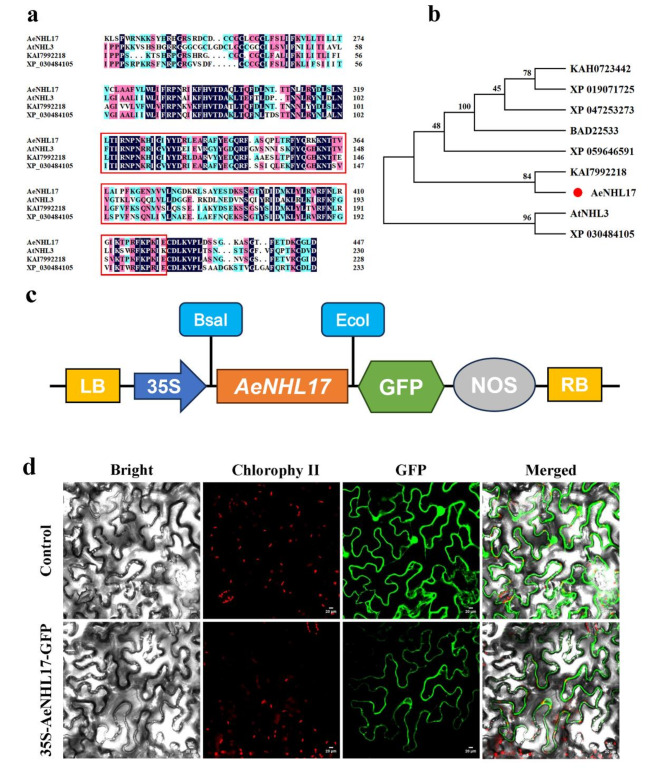
To identify the potential functions of AeNHL gene family members, we selected AeNHL17, which is highly responsive to pathogens and SA stress for further study. The full length CDS of AeNHL17 was 696 bp, encoding 231 amino acids with a pI of 9.45 and a MW of 26.5 kDa. Protein sequence and phylogenetic analysis showed that AeNHL17 had an LEA_2 domain and clustered with KAI7992218 from Camellia lanceoleosa (Fig. 6a-b).
Fig. 6.
Multiple sequence alignment, phylogenetic analysis, and subcellular localization of AeNHL17 protein. (a) Amino acid sequence alignments of AeNHL17, AtNHL3, KAI7992218 and XP_030484105. The LEA_2 domain was highlighted in red box. (b) Phylogenetic tree of AeNHL17 (indicated by a red dot) and other species proteins of Arabidopsis thaliana (AtNHL3), Camellia lanceoleosa (KAI7992218), Cornus florida (XP_059646591), Capsicum annuum (XP_047253273), Cannabis sativa (XP_030484105), Solanum tuberosum (KAH0723442), Solanum lycopersicum (XP_019071725) and Nicotiana tabacum (BAD22533). (c) Schematic representation of 35 S::AeNHL17-GFP fusion construct. (d) Subcellular localization of the control (35 S-GFP) and recombinant plasmid (35 S-AeNHL17-GFP) transiently expressed in tobacco leaves and visualized under a confocal microscope. Bar = 20 μm
To explore the subcellular localization of AeNHL17 protein, a pBWA(V)HS-AeNHL17-GFP fusion vector was constructed (Fig. 6c). The recombinant plasmid driven by CaMV 35 S promoter was transiently transformed into tobacco leaves. Two days after infiltration, confocal microscopy was used to visualize the green fluoresce signals of the cells. The AeNHL17 protein was located in the plasma membrane according to the green fluorescence signal (GFP). In contrast, the GFP signal from the control plasmid was distributed throughout the cells (Fig. 6d).
Overexpression of AeNHL17 enhanced disease resistance in transgenic tobacco and A. chinensis
To explore the function of AeNHL17, overexpression of AeNHL17 was performed through the Agrobacterium-mediated transformation method in tobacco and kiwifruit, respectively. A specific pair of primers was used to identify positive transgenic plants by using polymerase chain reaction (PCR) analysis (Figs. 7a and 8a). Under normal growth conditions, transgenic tobacco, transgenic kiwifruit, and wild-type (WT) plants did not show obvious phenotypic differences (Supplementary Fig S2).
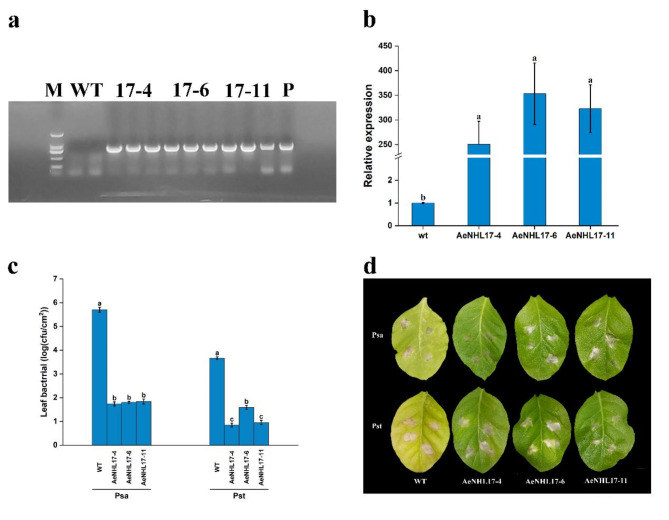
Fig. 7.
Overexpression of AeNHL17 enhances the disease resistance of tobacco. (a) Identify positive transgenic plants by using PCR analysis. (b) Relative expression of AeNHL17 in WT and transgenic tobacco lines. (c) Bacterial growth of Pseudomonas syringae pv. Actinidiae (Psa) and Pseudomonas syringae pv. Tomato DC3000 (Pst) in wild-type (WT) and transgenic lines (AeNHL17-4, AeNHL17-6, AeNHL17-11) at 3 days after inoculation. Bacterial growth is expressed as mean values of viable bacteria per square centimeter of leaf tissue. CFU: colony forming unit. (d) Disease symptom from representative leaves of WT and transgenic tobacco lines at 7 days after inoculation with Psa and Pst. Data were presented as mean values ± SD (n = 3). Lower-case letters in the column denote the significance level of mean differences at 0.05 (LSD method)
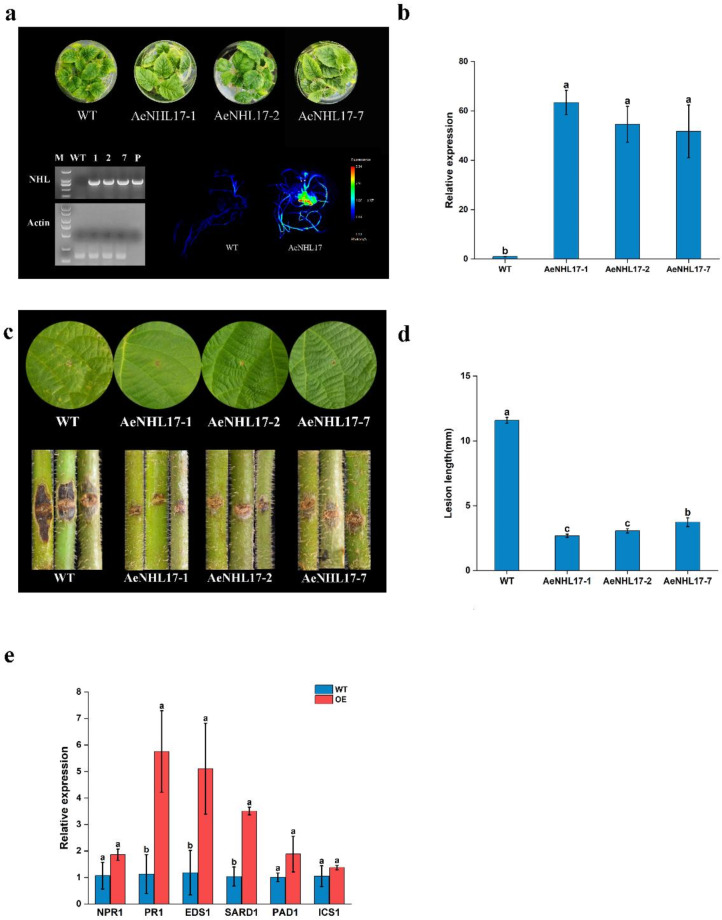
Fig. 8.
Overexpression of AeNHL17 enhances the disease resistance of kiwifruit plants. (a) PCR and fluorescence image for identification of positive transgenic kiwifruit plants based on the expression GFP reporter genes. (b) Relative expression of AeNHL17 in WT and transgenic kiwifruit lines. (c) Disease symptom of leaves and stems in WT and transgenic kiwifruit lines at 21 days after inoculation with Psa. (d) Lesion length of stems in WT and transgenic kiwifruit lines at 21 days after inoculation with Psa. (e) Effects of overexpression of AeNHL17 on the expression of the SA-responsive genes NPR1, PR1, EDS1, SARD1, PAD1 and ICS1 in transgenic lines and WT. The all results are presented from three independent experiments. Data were presented as mean values ± SD (n = 3). Lower-case letters in the column denote the significance level of mean differences at 0.05 (LSD method)
Three T2 generation plants of transgenic tobacco lines were randomly selected to evaluate whether overexpression of AeNHL17 could enhance plant resistance to pathogenic bacteria. Over-expression of AeNHL17 in tobacco was confirmed by semi-quantitative RT-PCR, and the transgenic lines showed higher expression of AeNHL17 (Fig. 7b). Transgenic and WT tobacco plants were inoculated with Psa and Pst pathogens, respectively. Three days after inoculation, compared to the transgenic lines, the bacterial numbers of both Psa and Pst in the WT increased dramatically (Fig. 7c). Seven days later, the WT leaves turned yellow and displayed necrotic lesions around the inoculation site, whereas the transgenic lines did not show any obvious changes, except for the inoculation wound (Fig. 7d).
Over-expression of AeNHL17 in kiwifruit was confirmed by semi-quantitative RT-PCR, and the transgenic lines showed GFP fluorescence and higher expression of AeNHL17 (Fig. 8a-b). For further inoculation tests, the leaves and stems of both the transgenic and WT lines were inoculated with Psa. The leaves of WT plants turned curly and exhibited yellow disease lesions compared to the transgenic lines at 21 days post-inoculation, whereas the transgenic lines did not show obvious changes, except for the inoculation wound (Fig. 8c). As shown in Fig. 8d, the inoculation area of the WT stem turned brown and the lesion length was as long as 11.83 mm, which was significantly longer than that of the transgenic lines. These results displayed that AeNHL17-overexpressed tobacco and kiwifruit plants significantly inhibited pathogen infection, suggesting that the positive function of AeNHL17 in enhances disease defense.
To further understand the influences of AeNHL17 in the SA-mediated pathways, the transcription level of some responsive genes was detected in transgenic kiwifruit lines and WT plants, such as pathogenesis-related gene 1 (PR1), phytoalexin deficient 4 (PAD4), non-expresser of pathogenesis-related gene 1 (NPR1), isochorismate synthase 1 (ICS1), SAR deficient 1 (SARD1) and enhanced disease susceptibility 1 (EDS1). The results showed that the expression levels of these genes were significantly higher in the transgenic lines than in the WT (Fig. 8e). In summary, we speculated that AeNHL17 might participate in the disease resistance response by interacting with SA signaling pathways in kiwifruit.
Silencing AeNHL17 in A. eriantha increases susceptibility to Psa
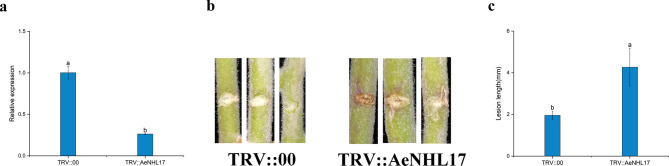
VIGS was utilized to silence AeNHL17 (TRV::AeNHL17). The transcription level of AeNHL17 was detected by qPCR analysis at 15 days after infection to evaluate the silencing efficacy of the recombinant plasmid. Compared to the control, the transcription of AeNHL17-silenced plants was dramatically reduced (Fig. 9a), demonstrating that the silencing effect of AeNHL17 was sufficient for subsequent pathogen inoculation experiments. Next, the control and silenced plants were simultaneously inoculated with Psa, and lesion length was measured 21 days after inoculation. Larger disease lesions appeared in the silenced plants than in the control plants (Fig. 9b, c), indicating that silencing AeNHL17 might increase the susceptibility of A. eriantha to Psa.
Fig. 9.
Silencing AeNHL17 in A. eriantha increases susceptibility to Psa. (a) The expression level of AeNHL17 was quantified by qPCR at 15 days after inoculation with TRV1 + TRV::AeNHL17 and TRV1 + TRV::00, respectively. (b-c) Silencing AeNHL17 increases infection of Psa. Disease symptom of stems and lesion length was measured at 21 days after inoculation. The values represent means ± standard errors (SEs) of three biological replications. The lower-case letters in the column denote the significance level of mean differences at 0.05
Identification of proteins interacting with AeNHL17
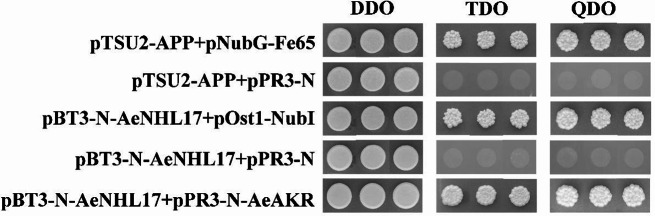
To further determine the potential proteins that interact with AeNHL17, yeast two-hybrid assays were performed. First, yeast cells co-transformed with pBT3-N-AeNHL17 and pPR3-N showed good growth on DDO, but were not produced on TDO and QDO agar plates (Fig. 10). This revealed that neither the auto-activation of AeNHL17 nor the pBT3-N-AeNHL17 bait plasmid could be used for subsequent library screening.
Fig. 10.
Yeast two-hybrid assays of AeNHL17 protein. The pBT3-N-AeNHL17 and pOST1-NubI, pPR3-N were co-transformed into yeast NMY51 strains and cultured on DDO, TDO, QDO agar plates, respectively. The CDS of AeAKR was cloned into the prey vector pPR3-N and co-transformed with the bait vector pBT3-N-AeNHL17. Yeast cells co-transformed with pTSU2-App and pNubG-Fe65, pTSU2-App and pPR3-N were used as positive and negative controls, respectively
Eleven AeNHL17-interaction proteins were identified in at least two independent screens (Supplementary Table S3). Based on functional annotations, these proteins were roughly classified into three categories: transport-related proteins, photosynthesis-related proteins and metabolic-related proteins. We selected aldo-keto reductases (AKR), which play important roles in stress defense by eliminating toxic and reactive oxygen species (ROS), as candidate proteins for point-to-point verification. pBT3-N-AeNHL17 co-transformed with pPR3-N-AeAKR grew on DDO, TDO, and QDO medium plates, whereas the negative control did not grow (Fig. 10). These results indicated that AeNHL17 interact with AeAKR.
Discussion
Bacterial cankers have long been a major problem in the kiwifruit industry. Therefore, improving kiwifruit resistance is an effective way to solve this problem. It has been demonstrated that NHL gene family is participated in the process of development and disease defense in many plants, such as tobacco, Arabidopsis, cotton, grape, and soybean [19, 22, 30–32]. However, there are no reports on NHL family members in kiwifruit, particularly their functions in regulating disease resistance. Here, the NHL gene family members of kiwifruit were identified, and their protein characteristics and gene expression were analyzed. In addition, we systematically revealed the role of AeNHL17 in regulating disease resistance.
In the present study, 33 NHL family genes in A. eriantha were identified (Table 1). Protein sequences analysis, indicated that AeNHL gene family members have a conserved LEA domain. Previous studies have documented that proteins with the LEA domain are widely distributed in higher plants and are involved in adjusting plant growth and development, and resisting environmental stresses [23]. LEA-containing proteins in soybean can regulate seed germination under cold stress via the ABA regulation pathway [33]. In tomato, the expression of some LEA genes rapidly increases under stress and phytohormone treatments [34]. NHLs are a large class of plant defense-related proteins that contain two conserved motifs, NPNKKIGIYYD and PKFYQPHKS, implying that there is also a conserved function between homologous genes [22, 35]. In this study, 15 conserved motifs in 33 AeNHL proteins were identified, which is in accordance with the number of conserved motifs in soybean and pepper [28, 33]. Two highly conserved motifs (1 and 3) were found in AeNHL gene family. These results suggest that NHL gene family members may be participate in kiwifruit defense resistance.
Phylogenetic analysis revealed that the AeNHL proteins were split into five different subgroups, and homologs with similar domains were divided into the same group, with AeNHL17 being closer to AtNHL3 (Fig. 1). Interestingly, it has been reported that the overexpression NHL3 in Arabidopsis can significantly increase transgenic plant resistance to virulent Pseudomonas syringae [36], indicating that AeNHL17 play a vital role in defense resistance in kiwifruit.
Plant hormones such as SA, ABA, GA, and MeJA play critical roles in the response to biotic and abiotic stresses, which are controlled by cis-acting regulatory elements [37]. Cis-element analysis indicated that the promoter region of AeNHLs included numerous stress- and defense-related elements and hormone induction elements (Fig. 3a), indicating that members of AeNHL gene family might be involved in the kiwifruit resistance response. Most studies have demonstrated that NHLs involve in the SA-mediated defense response pathway and other stress response [26, 38]. In kiwifruit, AeNHL25 contained four SA-responsive elements, and other members including AeNHL3, AeNHL5, AeNHL10, AeNHL11, AeNHL12, AeNHL14, AeNHL15, AeNHL16, AeNHL18, AeNHL19, AeNHL21, AeNHL24, AeNHL28 and AeNHL32, also contain SA-responsive elements (Fig. 3a). These results indicate that AeNHL genes may play a positive role in disease defense by being involved in the SA-induced pathway. The results of qPCR analysis indicated that AeNHL17 was strongly triggered after treatment with exogenous SA (Fig. 5g). However, the promoter of AeNHL17 lacks the cis-acting elements of SA, suggesting that AeNHL17 may not be directly involved in the SA signaling pathway. In addition, the experimental results showed that the expression of the SA signaling pathway genes NPR1, ICS1,PAD4, EDS1, PR1, and SARD1 increased in AeNHL17 transgenic plants, indicating that overexpression of AeNHL17 in kiwifruit might improve the transcription level of the SA pathway genes. The promoter region of AeNHL17 contained ABA- and JA-responsive elements (Fig. 3a). However, the expression of AeNHL17 decreased with JA and ABA treatments (Fig. 5h-i). The results indicated that ABA and JA might have negative regulatory roles in Psa infection, and these results were consistent with those of a previous study [39]. Therefore, we speculated that the different expression patterns of individual AeNHL genes led to differences in response to exogenous hormone treatment in this study, and the predicted regulatory elements in the promoter regions were not strongly correlated with the activated expression of particular genes.
The plasma membrane is very important for pathogen signal recognition or downstream defense signal generation [36, 40]. It has been documented that NHL proteins are localized on the plasma membrane [41]. Similarly, our results indicated that AeNHL17 was mainly localized on the plasma membrane (Fig. 6d); therefore, we hypothesized that AeNHL17 might also participate in pathogen perception and the rapid induction of defense responses. The results showed that AeNHL17 was highly induced by Psa infection (Fig. 4e). Overexpression of NHL3 enhances the resistance of Arabidopsis to Pseudomonas syringae pv. tomato DC3000 [36]. Transgenic tobacco and kiwifruit showed that the disease symptoms were significantly lower after P. syringae infection than in WT plants (Figs. 7b and 8c). In contrast, silencing AeNHL17 in kiwifruit increased susceptibility to Psa (Fig. 9b-c). The overexpression lines showed enhanced resistance, and no significant morphological or phenotypic changes were observed in the transgenic plants. Taken together, these findings indicate that AeNHL17 participates in the rapid disease resistance of plants during pathogen infection.
Under biotic and abiotic stress conditions, the expression of reactive oxygen species (ROS) increases significantly, leading to lipid peroxidation, which produces toxic compounds such as 4-hydrocynonenal (4-HNE) and malondialdehyde (MDA), causing cell damage [42, 43]. Aldo-keto reductases (AKR) are a superfamily of oxidoreductases that can reduce aldehydes and ketones to alcohol [44]. It has been reported that AKR1 could effectively eliminate toxic 4-HNE and MDA compounds and enhances stress resistance in rice [45]. Overexpression of PsAKR1 reduced the levels of methylglyoxal (MG), H2O2, and MDA, and improved chlorophyll content and plant survival under salt stress [43]. In our study, the interaction between AeNHL17 and AeAKR suggests that AKR might improve chlorophyll content and enhance membrane stability in kiwifruit under pathogenic attack. In transgenic kiwifruits, the interaction between AeNHL17 and AeAKR may enhances the ability of cells to eliminate toxic 4-HNE and MDA compounds in Psa infection. However, further research is required to elucidate the precise mechanisms underlying AKR in kiwifruit.
Conclusion
Here, we identified 33 potential NHL family members in A. eriantha for the first time using a genome-wide method, and then classified and analyzed these genes. We found that AeNHL17 plays an important role in kiwifruit disease defense. Functional analysis indicated that overexpression of AeNHL17 in kiwifruit and tobacco bestows transgenic plants with resistance to pathogens, based on the noticeable differences in disease lesion symptoms between transgenic and WT lines. Based on these results, we propose that the enhanced resistance of kiwifruit may be caused by a complex interaction involving AeNHL17, AeAKR and defense genes related to the SA signaling pathway (Fig. 11). Further functional analyses of the kiwifruit NHL genes are required to enhance our understanding of their biological roles. Our findings will contribute to the genetic improvement of kiwifruit and the development of more resistance resources.
Fig. 11.
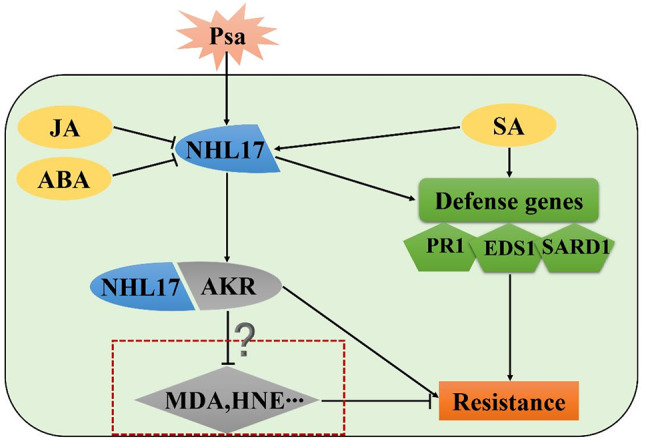
The schematic diagram of the mechanism by which overexpressed AeNHL17 enhanced the resistance of kiwifruit under Psa attacked. Both Psa and SA could induce the expression of AeNHL17, while JA and ABA exhibit negative regulatory effects. AeNHL17 interacts with AeAKR protein and AKR could eliminate MAD and HNE. In parallel, overexpression of AeNHL17 could enhance the accumulation of resistance genes expression in the SA pathway. They work together to improve the disease resistance of transgenic kiwifruit
Materials and methods
Plant materials and bacterial stains
The kiwifruit materials Zhongmi Cuixue 1 Hao (Actinidia eriantha), ‘Hongyang’ (Actinidia chinensis), and tobacco NC89 (Nicotiana tabacum) were grown in soil (perlite: vermiculite = 1:1). Kiwifruit plants were grown for 4 months and NC89 for 4 weeks at the artificial climate chamber under a 75% relative humidity, 25℃ temperature and 12 h/12 h light/dark.
Agrobacterium tumefaciens GV3101 and EHA105 were cultured in liquid LB medium with 25 mg/ml rifampicin (rif) and 50 mg/ml kanamycin (kan) for 2 d at 200 rpm 28℃ in an orbital shaker. After two days, the bacterial medium was resuspended in MS (pH 5.8) at the desired concentrations for infiltration.
Pseudomonas syringae pv. tomato DC3000 (Pst) and Pseudomonas syringae pv. actinidiae (Psa) was grown in KB medium supplemented for 1d at 23℃ in an orbital shaker at 150 rpm. After 24 h, bacterial cultures were resuspended in 10 mM magnesium chloride (MgCl2) for infiltration.
Genome-wide identification of NHL genes
To identify the members of AeNHLs, genome-wide sequences files of A. eriantha ‘White’ were downloaded from the Kiwifruit Genome Database (KGD) (https://kiwifruitgenome.org/) [46, 47]. 45 Arabidopsis NHL gene family sequences were downloaded from the TAIR database (https://www.arabidopsis.org/). By using the BLASTp program, we settled the candidate sequence with E ≤ e− 10. The theoretical isoelectric point (pI) and molecular weight (MW) were calculated using ProtParam website (https://web.expasy.org/protparam/). And Hidden Markov models (HMMs) from the SMART database (http://smart.embl.de/) were used to predict protein domain structures [48]. These protein sequences, including the LEA-2 conserved domain, were regarded as AeNHL genes of A. eriantha.
Phylogenetic analysis and chromosomal localization
Multiple sequence alignments (MAS) of AeNHL proteins were performed, and an unrooted phylogenetic tree was constructed using MEGA 11.0.13 software with the neighbor-joining (NJ) method [49]. Bootstrap replications were set to 1000. The chromosome location information of AeNHLs was extracted based on positional information from KGD [47]. All AeNHL genes were mapped to their respective loci in the kiwifruit genome and visualized using TBtools software [50].
Conserved motif and cis-element enrichment analysis
To determine the conserved motifs in the 33 AeNHL protein sequences, the online Multiple Em for Motif Elicitation (MEME) tool was used [51] (https://meme-suite.org/meme/tools/meme). The optimization parameter settings were as follows: maximum number of motifs, 15; site distribution, zero or one occurrence per sequence; and optimum width of each motif, between 6 and 50 residues. The motif structure was displayed using TBtools [50].
To explore the regulatory elements of AeNHL genes, a 2000 bp upstream sequence of the start codon (ATG) was extracted from the whole genome files using TBtools, and the sequences were run through the online PlantCare website (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). The regulatory element structure was shown by TBtools [50, 52].
Expression analysis of AeNHLs
Quantitative real-time RT-PCR was performed to examine the expression patterns of some members of AeNHL family in various tissues and floral organs. Total RNAs were extracted using an RNA Extraction Kit (Vazyme, China), and the first-strand cDNAs were synthesized using reverse transcriptase (ReverTra, TOYOBO) following the manufacturer’s instructions.
qRT-PCR was performed using the NovoStart SYBR qPCR SuperMix Plus Kit (Novoprotein, China) on a Roche Light 480 System (Roche, USA) to evaluate the transcript levels of AeNHL family members. The Primer premier 5.0 tool was used to design primers based on the CDS of each gene (Supplementary Table S1). The relative changes in gene transcription levels were calculated using the 2−ΔΔCT method, and the kiwifruit actin was used as the internal reference gene to normalize the results. All analysis were performed using four technical replicates and three biological replicates.
Cloning of the AeNHL17 cDNA
To clone the full-length cDNA of AeNHL17, specific-primers were designed based on the open-reading frame (ORF) of the A. eriantha gene DTZ71_15g07880 (Supplementary Table S1). The PCR products were purified using a GeneJET Gel Extraction Kit (Thermo Scientific, Germany) and cloned into the pCE2 TA/Blunt-Zero Cloning vector (Vazyme, China) for sequencing. A minimum four clones were sequenced.
The MW and pI of AeNHL17 were predicted using the ExPASy tool (https://web.expasy.org/protparam/). The homologous proteins from other species of AeNHL17 were retrieved from the TIAR website and NCBI database. The MEGA 11.0.13 software was used to construct an unrooted phylogenetic tree, and the DNAMAN (v9.0.1.116) tool was used to determine the MSA for AeNHL17 and other homologous proteins.
Subcellular localization of AeNHL17 protein
For subcellular localization, the ORF sequence without the stop codon of AeNHL17 was inserted into a pBWA(V)HS-GFP vector using the restriction sites BsaI and EcoRI. Plasmids harboring GFP alone were used as controls. These recombinant plasmids were transformed into tobacco leaves by infiltration with A. tumefaciens GV3101. A laser confocal microscope was used to observe the GFP green fluorescence signals.
Exogenous hormones and pathogen treatment
To investigate the effects of exogenous hormones on AeNHL17 expression, the A. eriantha grafted plants were treated with 5 mmol/L SA (Sigma, USA), 50 µmol/L ABA (Sigma, USA), or 0.1 mmol/L MeJA (Sigma, USA) at three months after grafting, respectively. The mock plants were treated with sterile water.
For Psa inoculation, an overnight fresh bacterial culture was adjusted to a final concentration of 1 × 108 cfu/L with sterile 10 mM MgCl2 (Sigma, USA) solution, and then sprayed onto the kiwifruit leaves. Each experiment was conducted in triplicates replicates, using at least three independent plants. All samples were collected at 0, 12, 24, 48, and 72 h, were frozen in liquid nitrogen immediately and stored at -80℃ for RNA isolation.
Overexpression and VIGS vectors construction
To construct a vector for the overexpression of AeNHL17, the restriction sites of XbaI and BamHI were introduced into the full-length cDNA of AeNHL17, and the fragment was cloned into the pBI121-GFP vector. For virus-induced gene silencing (VIGS), an XbaI- BamHI fragment containing 300 bp of AeNHL17 was cloned into a pTRV2 vector. All expression vector primers used throughout the experiment are provided in the Supporting Information.
Agrobacterium-mediated transformation
The vector pBI121-AeNHL17-GFP was transformed into A. tumefaciens strains EHA105 and GV3101, respectively. Tobacco genetic transformation was performed according to a previously reported Agrobacterium-mediated transformation method using GV3101 [53], and transformation of kiwifruit (A. chinensis cv. Hongyang) was performed using EHA105 based on the instructions reported by Wang [54]. The DNA of the wild-type (WT) and transgenic plants was extracted using a plant genomic DNA kit (TIANGEN, China), and positive transgenic plants were confirmed by PCR amplification using the primers 35 S-F and AeNHL17-R (Supplementary Table S1).
For the kiwifruit VIGS assays, pTRV1 was used along with pTRV2 for silencing. Agrobacterium GV3101 with pTRV1, pTRV2 empty vector or pTRV2-AeNHL17 vector was resuspended in induction medium (10 μm MES, 10 μm MgCl2, 400µ macetosyringone) and diluted to OD600 0.7, followed by incubating for 3 h at room temperature. The diluted cultures containing pTRV1 and pTRV2-AeNHL17 were then mixed in a 1:1 ratio, and a mixture of pTRV1 and pTRV2 was used as a mock control. The mixed culture was then infiltrated into the stems of A. eriantha plants.
Measurement of pathogen resistance in transgenic plants
To determine the disease resistance of AeNHL17 transgenic lines, the plants were inoculated with both Psa and Pst pathogens according to a previously described protocol with a few modifications [39, 55]. The strains were resuspended at 1 × 108 cfu/L in sterile 10 mM MgCl2 solution. Leaves of 5-week-old transgenic and non-transgenic tobacco plants were inoculated with Psa and Pst. Kiwifruit infection was carried out in 6-month-old transgenic and WT plants by leaf spraying and stem inoculation with Psa. Three days after inoculation, the tobacco leaves were collected from the treated plants, washed with sterile water, and ground with sterile MgCl2 (10 mM). The suspensions were serially diluted and plated onto KB agar plates. The number of CFU per disk leaf was calculated after 3 days incubation at 23℃. Phenotypic changes in tobacco were observed continuously after infection and photographed on the seventh day. Leaf and stem lesion sizes of kiwifruit were monitored and measured 21 days post-inoculation.
Yeast two-hybrid assays
For Y2H assays, the DUALmembrane system (Clontech, USA) was used to screen proteins interacting with AeNHL17, following a previously described protocol [56, 57]. The amplified full-length CDS of AeNHL17 was digested with SfiI enzyme and cloned into the pBT3-N vector to construct the bait. To evaluate whether pBT3-N-AeNHL17 exhibited self-activation and toxicity, pBT3-N-AeNHL17 was co-transformed with pPR3-N and pOST1-NubI respectively. The interaction between pTSU2-App and pNubG-Fe65 was used as the positive control, whereas pTSU2-App and pPR3-N were used as negative controls. They spotted onto SD/-Trp/-Leu (double dropout supplement, DDO), SD/-Trp/-Leu/-His (triple dropout supplement, TDO) and SD/-Trp/-Leu/-His/-Ade (quadruple dropout supplement, QDO) solid mediums. For library screening, the expression vector pBT3-N-AeNHL17 and library plasmids were co-transformed and plated on QDO solid mediums. pBT3-N-AeNHL17 was co-transformed with the reciprocal protein and spotted onto DDO, TDO and QDO plates for point-to-point validation. The yeast NMY51 strain was used. The primers used for Y2H are listed in Supplementary Table S1.
Statistical analysis
At least three replicates of the experiments and data were used in this study. For statistical analysis, the Software OriginPro 2023 (OriginLab Corporation, USA) was used to calculate the means and standard deviations of all data. The Dunn-Sidak test was performed to evaluate the significance level with P < 0.05 or P < 0.01.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Not applicable.
Author contributions
L.S. and X.Q. designed and supervised this project. M.Z. carried out the majority of the experiments. R.F. and M.L. performed expression analysis and data collection. R.W. and Y.L. assisted with the analysis of the results. L.S., X.Q., J.F. and J.C. provided guidance on the study. M.Z. and L.S. wrote the manuscript. All the authors reviewed and approved the ultimate manuscript.
Funding
This work was financially supported by the Major Science and Technology Projects of Henan Province (221100110400), National Key Research and Development Program (2022YFD1600700), China Agriculture Research System of MOF and MARA (CARS-26), Agricultural Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences (CAAS-ASTIP-2024-ZFRI-03), and Henan Agriculture Research System (HARS-22-09-S).
Data availability
All the experimental data are included in the main text and the supplementary data.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Lei-Ming Sun, Email: sunleiming@caas.cn.
Xiu-Juan Qi, Email: qixiujuan@caas.cn.
References
- 1.Sui L, Liu Y, Zhong C, Huang H. Geographical distribution and morphological diversity of red-fleshed kiwifruit germplasm (Actinidia chinensis Planchon) in China. Genet Resour Crop Ev. 2013;60(6):1873–83. [Google Scholar]
- 2.Huang S, Ding J, Deng D, Tang W, Sun H, Liu D, Zhang L, Niu X, Zhang X, Meng M, Yu J, Liu J, Han Y, Shi W, Zhang D, Cao S, Wei Z, Cui Y, Xia Y, Zeng H, Bao K, Lin L, Min Y, Zhang H, Miao M, Tang X, Zhu Y, Sui Y, Li G, Sun H, Yue J, Sun J, Liu F, Zhou L, Lei L, Zheng X, Liu M, Huang L, Song J, Xu C, Li J, Ye K, Zhong S, Lu B-R, He G, Xiao F, Wang H-L, Zheng H, Fei Z, Liu Y. Draft genome of the kiwifruit Actinidia chinensis. Nat Commun. 2013;4(1):2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mazzaglia A, Studholme DJ, Taratufolo MC, Cai RM, Almeida NF, Goodman T, Guttman DS, Vinatzer BA, Balestra GM. Pseudomonas syringae Pv. Actinidiae (PSA) isolates from recent bacterial canker of kiwifruit outbreaks belong to the same genetic lineage. PLoS ONE 2012, 7(5). [DOI] [PMC free article] [PubMed]
- 4.Vanneste JL. The scientific, economic, and social impacts of the New Zealand outbreak of bacterial canker of kiwifruit (Pseudomonas syringae Pv. Actinidiae). Annu Rev Phytopathol 2017: 377–99. [DOI] [PubMed]
- 5.McCann HC, Li L, Liu Y, Li D, Pan H, Zhong C, Rikkerink EHA, Templeton MD, Straub C, Colombi E, Rainey PB, Huang H. Origin and evolution of the kiwifruit canker pandemic. Genome Biol Evol. 2017;9(4):932–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scortichini M. Occurrence of Pseudomonas syringae Pv. Actinidiae on kiwifruit in Italy. Plant Pathol. 1994;43:1035–8. [Google Scholar]
- 7.Koh Y, Kim G, Jung J, Lee Y, Hur J. Outbreak of bacterial canker on Hort16A (Actinidia chinensis Planchon) caused by Pseudomonas syringae Pv. Actinidiae Korea New Zeal J Crop Hort. 2010;38:275–82. [Google Scholar]
- 8.Scortichini M, Marcelletti S, Ferrante P, Petriccione M, Firrao G. Pseudomonas syrPvgae pv. actinidiae: a re-emerging, multi-faceted, pandemic pathogen. Mol Plant Pathol. 2012;13(7):631–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Serizawa S, Ichikawa T, Takikawa Y, Tsuyumu S, Goto M. Occurrence of bacterial canker of kiwifruit in Japan description of symptoms, isolation of the pathogen and screening of bactericides. Japanese J Phytopathol. 1989;55(4):427–36. [Google Scholar]
- 10.Sawada H, Fujikawa T. Genetic diversity of Pseudomonas syringae Pv. Actinidiae, pathogen of kiwifruit bacterial canker. Plant Pathol. 2019;68(7):1235–48. [Google Scholar]
- 11.McCann HC, Rikkerink EHA, Bertels F, Fiers M, Lu A, Rees-George J, Andersen MT, Gleave AP, Haubold B, Wohlers MW, Guttman DS, Wang PW, Straub C, Vanneste J, Rainey PB, Templeton MD. Genomic analysis of the kiwifruit pathogen Pseudomonas syringae pv. actinidiae provides insight into the origins of an emergent plant disease. Plos Pathog. 2013, 9(7). [DOI] [PMC free article] [PubMed]
- 12.Donati I, Cellini A, Sangiorgio D, Vanneste JL, Scortichini M, Balestra GM, Spinelli F. Pseudomonas syringae pv. actinidiae: ecology, infection dynamics and disease epidemiology. Microb Ecol 2020, 80(1):81–102. [DOI] [PMC free article] [PubMed]
- 13.Kinkema M, Fan WH, Dong XN. Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell. 2000;12(12):2339–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones J, Dangl J. The plant immune system. Nature. 2006;444(7117):323–9. [DOI] [PubMed] [Google Scholar]
- 15.Balint-Kurti P. The plant hypersensitive response: concepts, control and consequences. Mol Plant Pathol. 2019;20(8):1163–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi H, Shen Q, Qi Y, Yan H, Nie H, Chen Y, Zhao T, Katagiri F, Tang D. BR-SIGNALING KINASE1 physically associates with FLAGELLIN SENSING2 and regulates plant innate immunity in Arabidopsis. Plant Cell. 2013;25(3):1143–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yalpani N, Silverman P, Wilson TM, Kleier DA, Raskin I. Salicylic acid is a systemic signal and an inducer of pathogenesis-related proteins in virus-infected tobacco. Plant Cell 1991, 3(8):809–18. [DOI] [PMC free article] [PubMed]
- 18.Vlot AC, Dempsey DMA, Klessig DF. Salicylic acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol. 2009;47(1):177–206. [DOI] [PubMed] [Google Scholar]
- 19.Takahashi Y, Berberich T, Yamashita K, Uehara Y, Miyazaki A, Kusano T. Identification of tobacco HIN1 and two closely related genes as spermine-responsive genes and their differential expression during the tobacco mosaic virus-induced hypersensitive response and during leaf- and flower-senescence. Plant Mol Biol. 2004;54(4):613–22. [DOI] [PubMed] [Google Scholar]
- 20.Century KS, Shapiro AD, Repetti PP, Dahlbeck D, Holub E, Staskawicz BJ. NDR1, a pathogen-induced component required for Arabidopsis disease resistance. Science. 1997;278(5345):1963–5. [DOI] [PubMed] [Google Scholar]
- 21.Peng H, Pu Y, Yang X, Wu G, Qing L, Ma L, Sun X. Overexpression of a pathogenesis-related gene NbHIN1 confers resistance to tobacco mosaic virus in Nicotiana benthamiana by potentially activating the jasmonic acid signaling pathway. Plant Sci. 2019;283:147–56. [DOI] [PubMed] [Google Scholar]
- 22.Dormann P, Gopalan S, He SY, Benning C. A gene family in Arabidopsis thaliana with sequence similarity to NDR1 and HIN1. Plant Physiol and Bioch 2000, 38(10):789–96.
- 23.Hong BS, Zong SL, Ming AS. LEA proteins in higher plants: structure, function, gene expression and regulation. Colloids Surf B. 2005;45(3):131–5. [DOI] [PubMed] [Google Scholar]
- 24.Shapiro AD, Zhang C. The role of NDR1 in avirulence gene-directed signaling and control of programmed cell death in Arabidopsis. Plant Physiol. 2001;127(3):1089–101. [PMC free article] [PubMed] [Google Scholar]
- 25.Varet A, Parker J, Tornero P, Nass N, Lee J. NHL25 and NHL3, two NDR1/HIN1-1ike genes in Arabidopsis thaliana with potential role(s) in plant defense. Mol Plant Microbe in. 2002;15(6):608–16. [DOI] [PubMed] [Google Scholar]
- 26.Zheng MS, Takahashi H, Miyazaki A, Hamamoto H, Shah J, Yamaguchi I, Kusano T. Up-regulation of Arabidopsis thaliana NHL10 in the hypersensitive response to cucumber mosaic virus infection and in senescing leaves is controlled by signalling pathways that differ in salicylate involvement. Planta. 2004, 218(5):740–750. [DOI] [PubMed]
- 27.Chen Q, Tian Z, Jiang R, Zheng X, Xie C, Liu J. StPOTHR1, a NDR1/HIN1-like gene in Solanum tuberosum, enhances resistance against Phytophthora infestans. Biochem Bioph Res Co. 2018;496(4):1155–61. [DOI] [PubMed] [Google Scholar]
- 28.Liu C, Peang H, Li X, Liu C, Lv X, Wei X, Zou A, Zhang J, Fan G, Ma G, Ma L, Sun X. Genome-wide analysis of NDR1/HIN1-like genes in pepper (Capsicum annuum L.) and functional characterization of CaNHL4 under biotic and abiotic stresses. Hortic Res. 2020;7:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Letunic I, Bork P. Interactive tree of life (iTOL) v6: recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res; 2024. [DOI] [PMC free article] [PubMed]
- 30.Chong J, Le Henanff G, Bertsch C, Walter B. Identification, expression analysis and characterization of defense and signaling genes in Vitis vinifera. Plant Physiol Bioch. 2008;46(4):469–81. [DOI] [PubMed] [Google Scholar]
- 31.Zhang X, Xue Y, Wang H, Nisa ZU, Jin X, Yu L, Liu X, Yu Y, Chen C. Genome-wide identification and characterization of NHL gene family in response to alkaline stress, ABA and MEJA treatments in wild soybean (Glycine soja). PeerJ. 2022;10:e14451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo XH, Wei F, Jian HL, Lian BY, Dang XY, Yang MQ, Fu XK, Ma L, Lu JH, Wang HT, Wei HL, Yu SX. Systematic analysis of the NDR1/HIN1-like (NHL) family in Gossypiumhirsutum reveals a role of GhNHL69 in responding to cold stress. Industrial Crops and Products 2023, 206.
- 33.Wang J, Wu R, Shangguan T, Chen G, Zheng Y, Tao X, Li S, Wang Y, Xu S. NDR1/HIN1-like genes may regulate Glycine max seed germination under chilling stress through the ABA pathway. Plant Growth Regul. 2022;98(3):613–24. [Google Scholar]
- 34.Jia C, Guo B, Wang B, Li X, Yang T, Li N, Wang J, Yu Q. The LEA gene family in tomato and its wild relatives: genome-wide identification, structural characterization, expression profiling, and role of SlLEA6 in drought stress. BMC Plant Biol. 2022;22(1):596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu L, Liu S. Genome-wide identification and phylogenetic analysis of the ERF gene family in cucumbers. Genet Mol Biol. 2011;34(4):624–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Varet A, Hause B, Hause G, Scheel D, Lee J. The Arabidopsis NHL3 gene encodes a plasma membrane protein and its overexpression correlates with increased resistance to Pseudomonas syringae Pv. Tomato DC3000. Plant Physiol. 2003;132(4):2023–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ku YS, Sintaha M, Cheung MY, Lam HM. Plant hormone signaling crosstalks between biotic and abiotic stress responses. Int J Mol Sci 2018, 19(10). [DOI] [PMC free article] [PubMed]
- 38.Bao Y, Song WM, Zhang HX. Role of Arabidopsis NHL family in ABA and stress response. Plant Signal Behav. 2016;11(5):e1180493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song Y, Sun L, Lin M, Chen J, Qi X, Hu C, Fang J. Comparative transcriptome analysis of resistant and susceptible kiwifruits in response to Pseudomonas syringae Pv. Actinidiae during early infection. PLoS ONE. 2019;14(2):e0211913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Knepper C, Savory EA, Day B. The role of NDR1 in pathogen perception and plant defense signaling. Plant Signal Behav. 2011;6(8):1114–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coppinger P, Repetti PP, Day B, Dahlbeck D, Mehlert A, Staskawicz BJ. Overexpression of the plasma membrane-localized NDR1 protein results in enhanced bacterial disease resistance in Arabidopsis thaliana. Plant J. 2004;40(2):225–37. [DOI] [PubMed] [Google Scholar]
- 42.Éva C, Zelenyánszki H, Tömösközi-Farkas R, Tamás L. Transgenic barley expressing the Arabidopsis AKR4C9 aldo-keto reductase enzyme exhibits enhanced freezing tolerance and regenerative capacity. S Afr J Bot. 2014;93:179–84. [Google Scholar]
- 43.Vemanna RS, Babitha KC, Solanki JK, Amarnatha Reddy V, Sarangi SK, Udayakumar M. Aldo-keto reductase-1 (AKR1) protect cellular enzymes from salt stress by detoxifying reactive cytotoxic compounds. Plant Physiol Bioch. 2017;113:177–86. [DOI] [PubMed] [Google Scholar]
- 44.Guan X, Yu L, Wang A. Genome-wide identification and characterization of Aldo-Keto Reductase (AKR) gene family in response to abiotic stresses in Solanum lycopersicum. Int J Mol Sci 2023, 24(2). [DOI] [PMC free article] [PubMed]
- 45.Turóczy Z, Kis P, Török K, Cserháti M, Lendvai Á, Dudits D, Horváth GV. Overproduction of a rice aldo–keto reductase increases oxidative and heat stress tolerance by malondialdehyde and methylglyoxal detoxification. Plant Mol Biol. 2011;75(4):399–412. [DOI] [PubMed] [Google Scholar]
- 46.Yue J, Liu J, Tang W, Wu YQ, Tang X, Li W, Yang Y, Wang L, Huang S, Fang C, Zhao K, Fei Z, Liu Y, Zheng Y. Kiwifruit Genome Database (KGD): a comprehensive resource for kiwifruit genomics. Hortic Res. 2020;7(1):117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang W, Sun X, Yue J, Tang X, Jiao C, Yang Y, Niu X, Miao M, Zhang D, Huang S, Shi W, Li M, Fang C, Fei Z, Liu Y. GigaScience. Chromosome-scale genome assembly of kiwifruit Actinidia eriantha with single-molecule sequencing and chromatin interaction mapping. 2019, 8(4). [DOI] [PMC free article] [PubMed]
- 48.Letunic I, Bork P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018;46(D1):D493–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tamura K, Stecher G, Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 2021;38(7):3022–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020;13(8):1194–202. [DOI] [PubMed] [Google Scholar]
- 51.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Magali L, Patrice D, Gert T, Kathleen M, Yves M, Yves VDP, Pierre R, Stephane R. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 2002(1):1. [DOI] [PMC free article] [PubMed]
- 53.Horsch RB, Fry JE, Hoffmann NL, Wallroth M, Eichholtz D, Rogers SG, Fraley RT. A simple and general method for transferring genes into plants. Science. 1985;227(4691):1229–31. [DOI] [PubMed]
- 54.Wang T, Karunairetnam S, Wu R, Wang Y-Y, Gleave AP. High efficiency transformation platforms for kiwifruit (Actinidia spp.) functional genomics. Acta Hortic. 2012;929:143–8. [Google Scholar]
- 55.Sun LM, Fang JB, Zhang M, Qi XJ, Chen JY. Molecular cloning and functional analysis of the NPR1 homolog in kiwifruit (Actinidia Eriantha). Front Plant Sci. 2020;11:551201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen Q, Wei T. Membrane and nuclear yeast two-hybrid systems. In: Plant Virology: Methods and Protocols. 2022: 93–104. [DOI] [PubMed]
- 57.Zhang H, Cheng G, Yang Z, Wang T, Xu J. Identification of sugarcane host factors interacting with the 6K2 protein of the sugarcane mosaic virus. Int J Mol Sci 2019, 20(16). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the experimental data are included in the main text and the supplementary data.