Abstract
Background
Lymphatic Filariasis (LF) is a neglected tropical disease affecting more than 882 million people in 44 countries of the world. A multi-epitope prophylactic/therapeutic vaccination targeting filarial defense proteins would be invaluable to achieve the current LF elimination goal.
Method
Two groups of proteins, namely Anti-oxidant (AO) and Heat shock proteins (HSPs), have been implicated in the effective survival of the filarial parasites in their hosts. Several B-cell, CTL, and T-helper epitopes were predicted from the three anti-oxidant proteins GST, GPx, and SOD. Likewise, epitopes were also predicted for HSP110, HSP90, and HSP70. Among the predicted epitopes, screening was applied to include only non-allergenic, non-toxic epitopes to construct two MEVs, PVAO and PVHSP. The epitopes for each group of proteins were connected to each other by the inclusion of suitable linkers and an adjuvant. The 3D models for PVAO and PVHSP were predicted, and validated, followed by prediction of physicochemical properties using bioinformatics tools. The binding free energy of PVAO and PVHSP with Toll like Receptors (TLR) TLR1/2, TLR4, TLR5, TLR6, and TLR9 was calculated with HawkDock. The immunogenicity of both the MEVs were assessed by Immune simulation after which codon adaptation and in-silico cloning were carried out.
Results
Conservation of the selected AOs and HSPs in other parasitic nematode species suggested that both the generated chimera could be helpful in cross-protection too. The 3D models of both MEVs contained more than 97% residues in allowed regions, as predicted by PROCHECK server. High MMGBSA and docking scores were obtained between MEVs and TLR4, TLR1/2, TLR6, and TLR9. Molecular dynamics simulation confirmed the stability of candidate vaccines in dynamic conditions present in the biological systems. The in-silico immune simulation indicated significantly high levels of IgG1, T-helper, T-cytotoxic cells, INF-γ, and IL-2 responses following immunization with PVAO and PVHSP.
Conclusion
The immunoinformatics approaches used in this study confirmed that, the designed vaccines are capable of eliciting sustained immunity against LF, however, additional in-vivo studies would be required to confirm their efficacy. Furthermore, by employing multi-epitope structures and constructing two different cocktail vaccines for LF, this study can form an important milestone in the development of future LF vaccine/s.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12879-024-10272-9.
Keywords: Filarial anti-oxidant proteins, Filarial heat shock proteins, Filarial cocktail vaccine, Multi-epitope vaccines
Background
Lymphatic filariasis (LF) is a neglected tropical disease and is a major barrier to socioeconomic development in endemic nations [1]. Filarial parasites Wuchereria bancrofti, Brugia malayi, and Brugia timori are the causative agents of Lymphatic filariasis and are transmitted by mosquito vectors of the genus Anopheles, Culex, and Aedes [2]. W. bancrofti is responsible for around 90% of filarial parasite infections, followed by B. malayi and B. timori (https://www.who.int/news-room/fact-sheets/detail/lymphatic-filariasis). Lymphatic filariasis is common in ~ 44 countries and affects around 882 million individuals [3, 4]. According to World Health Organization (WHO), LF alone accounts for approximately 2.8 million disability-adjusted life-years, making it the second most frequent cause of long-term disability after mental illness [5]. WHO has approved a combination of Ivermectin (IVM), Diethylcarbamazine (DEC), and Albendazole for Mass drug administration (MDA) for LF treatment and control. This drug combination has the ability to kill microfilariae and late embryonic stages but has little effect on adult filarial worms, because of which the adult worms remain alive for several years with in the human host [6].
In chronic patients, LF infections culminate into hydrocele, lymphedema, and ultimately elephantiasis [7]. The WHO launched the Global Program to Eliminate Lymphatic Filariasis (GPELF) in 2000, which recommended MDA to the entire population at risk of LF. Though the MDA program has considerably decreased global LF transmission, there are still questions about its long-term benefits and prevention of subsequent infections. In addition, the development of resistance to the MDA drugs is an upcoming challenge for LF elimination [8]. As a result, a more justifiable modus operandi would be, the development of an LF vaccine to prevent the occurrence or relapse of the disease [9]. No licensed LF vaccines are available till date, although several studies on vaccine candidates like BmHSP and BmALT-1 etc., have been conducted in the past [10, 11]. Recently, multi-epitope vaccines (MEVs) have emerged as a novel approach to vaccine development since they can circumvent the impact of damaging epitopes of the intact antigen [12]. Epitope-based vaccines offer additional advantages of safety, stability, and can effectively address the issues associated with conventional vaccines such as poor safety record, disease outbreak due to immunization etc [13]. The approach has already been used to design vaccines against SARS CoV-2, Mycobacterium tuberculosis [14], Schistosoma mansoni [15], and Onchocerca volvulus [16]. Earlier studies are indicative of protective immunity in LF, which is evidenced by the presence of endemic normal people living in endemic regions [17]. Cross-reacting antibodies and high levels of pro-inflammatory cytokines are both associated with potential immunity against the filarial parasites in people living in endemic areas. Therefore, developing a vaccine might be an effective way to combat the pathophysiology and to prevent new LF infections [18].
The filarial worms have highly developed enzymatic and non-enzymatic antioxidants for reducing the harmful effects of pro-oxidants, which are reactive oxygen and nitrogen species produced by the host immune cells. The filarial nematodes antioxidant system comprises of enzymes like Glutathione-S-Transferase (GST), Superoxide Dismutase (SOD), and Glutathione Peroxidase (GPx) which neutralize the hydrogen peroxide, superoxide radicals, hydroxyl ions, and other pro-oxidants generated by the hosts’ immune cells [19]. Thus, current antifilarial studies employ targeted chemo- and immunotherapeutic strategies to circumvent the filarial redox regulation system’s defenses [20]. Notably, an earlier vaccine targeting the filarial Glutathione redox system has been shown to reduce worm burden in test animals [21]. Since, the parasites’ antioxidant machinery is crucial for the survival therefore the anti-oxidant enzymes may be considered as promising vaccine candidates. Another, highly conserved class of proteins known as heat shock proteins (HSP) are well-known for their functions as molecular chaperones [22, 23]. The expression of HSPs, for either refolding damaged proteins or selecting them for degradation, is often upregulated in response to a rise in temperature in the majority of cells. HSPs expression increases quickly in response to environmental changes in temperature, pH, and pressure [24]. The filarial larvae (microfilariae L1 stage), in particular, must adapt to a substantial decrease in temperature during transmission from the homeoiotherm mammalian host to the poikilotherm arthropod vector and vice versa as different classes of HSPs are expressed when L3 is transferred from the vector to the mammalian host [25]. Therefore, in this work, the epitopic region of the crucial anti-oxidant enzymes and antigenic Heat shock proteins were predicted and two MEVs PVAO and PVHSP were constructed using in-silico approaches [26]. The constituent proteins were assessed for amino acid conservation with homologous proteins in other related parasites and humans to address the cross-protection potential of the developed vaccine candidates.
Methods
Retrieval of protein sequences and preliminary characterization
The protein sequences for W. bancrofti GST (A0A0H5S7P0, 208 aa) [27] SOD (A0A0K0IYY8, 223 aa), GPx (J9E3A6, 220 aa), HSP70 (EJW86287.1, 645 aa) [28], HSP 90 (EJW88125.1, 717 aa), and HSP110 (A0A0K0IYH1, 817 aa) were downloaded in FASTA format from the UniProt (https://www.uniprot.org/) and GenBank database (https://www.ncbi.nlm.nih.gov/genbank/). To discriminate secretory proteins from non-secretory, SignalP6 server (https://services.healthtech.dtu.dk/services/SignalP-6.0/) was used [29]. The DeepLoc 1.0 (https://services.healthtech.dtu.dk/services/DeepLoc-1.0/) server was used to predict the subcellular and cellular membrane localization of the chosen PVAO and PVHSP proteins. The DeepLoc 1.0 server uses neural networks trained on UniProt proteins combined with experimental evidences of subcellular localization [30].
Protein conservation analysis in related nematodes
A BLASTp search was conducted on the UniProt database to obtain the percentage identities of homologous relatives of the selected protein. The potential for selected proteins to provide cross-protection against additional filarial infections was examined. After performing consecutive sequence alignment of homologous proteins in related filarial worms, the protein conservation was predicted. O. volvulus, L. loa, O. ochengi, B. malayi, L. sigmodontis and W. bancrofti filarial parasites was examined and epitopes were chosen based on their significance for human and animal health.
Prediction of linear B-cell epitopes
The protein sequences in FASTA format were used for prediction of B-cell epitopes. ABCpred (https://webs.iiitd.edu.in/raghava/abcpred/ABC_submission.html) and B-cell tools of the Immune Epitope Database (IEDB) (http://tools.iedb.org/bcell/) servers were used [31]. ABCpred can predict linear B-cell epitopes using artificial neural network and the server has been trained using BciPep database and has better performed for B-cell epitope prediction. The IEDB server predicts regions that are most likely to be recognized as epitopes in a typical B-cell response. The antigen’s sequence properties and Hidden Markov Models (HMMs) were used to predict the linear B cell epitopes. Since the use of multiple servers to confirm the predicted epitopes is recommended, hence the epitopes that were predicted both by ABCpred and IEDB servers only were chosen for vaccine constructions.
Cytotoxic T lymphocytes (CTL) and helper T-cell epitope prediction
For the chosen proteins, cytotoxic-T-lymphocyte (CTL) epitopes were predicted using the freely accessible NetCTL 1.2 server (MHC supertype A1) (http://www.cbs.dtu.dk/services/NetCTL/). The technique combines the prediction of TAP transport efficiency, proteasomal C terminal cleavage, and peptide MHC class I binding [32]. Additionally, a weight matrix was used to predict TAP effectiveness. The NetMHCII 2.3 server (http://www.cbs.dtu.dk/services/NetMHCII/) has been used to predict HTL epitopes with a length of 9-mer for human alleles. The NetMHCII 2.3 server predicts peptide binding to the HLA-DR, HLA-DQ, and HLA-DP alleles [33]. MHC II epitopes were predicted based on receptor affinity and percentile scores assigned to each anticipated epitope [34]. The percentile rank is directly related to IC50 and inversely related to epitope affinity. The predicted CTL and HTL epitopes were reassessed for antigenicity by VaxiJen server. For the estimation of CTL and HTL epitopes, the threshold value was set at 0.5 following earlier researches in the field [35].
Designing of vaccine candidates
The candidate vaccine sequence was predicted by using a combination of high scoring CTLs, high affinity HTLs, and B-cell epitopes predicted by BepiPred 3.0.(https://services.healthtech.dtu.dk/services/BepiPred-3.0/) Some predicted sequences had the capacity to function as both B-cell and T-cell epitopes [36]. Using linkers GPGPG and AAY, various epitopes were connected and to increase the vaccine’s immunogenicity, a 50 S ribosomal protein L7/L12 (Locus RL7 MYCTU) adjuvant, agonist of TLR4 (Accession number P9WHE3) was added with an EAAAK linker at the N-terminal of the constructed vaccines. UniProt database was used to obtain the adjuvant’s sequence.
IFN-γ inducing epitope scanning
Interferon gamma (IFN-γ) is a cytokine that effectively responds to MHC antigens, stimulates macrophages and natural killer cells, and participates in both innate and adaptive immune responses. IFN-γ epitopes were predicted for the selected antioxidant and heat shock proteins using the IFNepitope server (http://crdd.osdd.net/raghava/ifnepitope/scan.php). The predicted method was based on a hybrid approach using the Support Vector Machine (SVM) and a motif.
Evaluation of toxicity, allergenicity, Antigenicity, and physicochemical properties of the developed vaccine candidates
The toxicity of all the epitopes were analysed by ToxinPred server (https://webs.iiitd.edu.in/raghava/toxinpred/multi_submit.php). The VaxiJen v2.0 server (https://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html) was used for the analysis of antigenicity of the whole vaccine and the ANTIGENpro server (https://scratch.proteomics.ics.uci.edu/) was used for the validation. The allergenicity of the vaccine candidates was evaluated with AllergenFP v.1.0 (http://ddg-pharmfac.net/AllergenFP/) [37] and AllerTOP v.2.0 (https://www.ddg-pharmfac.net/AllerTOP/) [38]. The solubility of the vaccine candidates were predicted against the average solubility of the Escherichia coli protein in the ProteinSol server (https://protein-sol.manchester.ac.uk/) [39]. The ExPASy-ProtParam (http://web.expasy.org/protparam/) was used for the evaluation of the physicochemical properties of the vaccine candidates [40].
Secondary and tertiary structure construction and refinement
Secondary structure of the vaccine candidates was predicted from PSIPRED 4.0 server (http://bioinf.cu.ucl.ac.uk.psipred/) [41]. For structural determination of candidate vaccines, the tertiary structures were constructed with I-TASSER server (https://zhanggroup.org/I-TASSER/) [42]. Out of the five models provided by the I-TASSER server, the model having best RMSD and TM scores was selected. The 3D model of the multi-epitope vaccine peptide was improved by a three-step method in the ModRefiner (https://zhanglab.ccmb.med.umich.edu/ModRefiner/) server. In the ModRefiner server firstly, any missing residues were added, secondly, mild relaxation was performed and lastly, a more intense relaxation was carried out for the construction and refinement of protein structure.
Tertiary structure validation
A critical step in the model-building process is detecting any probable errors in the predicted 3-D models. For a single input structure, ProSA (https://prosa.services.came.sbg.ac.at/prosa.php) determines an overall quality score; this score is then displayed alongside all previously identified protein structures [43]. The structure probably contains mistakes if the estimated score is outside the range that is typical of native proteins then the 3D molecular viewer will highlight and display such structural flaws. In addition, non-bonded atom-atom interactions were examined using the ERRAT site (http://services.mbi.ucla.edu/ERRAT/) which compares the predicted structures to trustworthy high-resolution crystallographic structures. A Ramachandran plot, which is based on the Van der Waal radius of the side chains of amino acids, was obtained using the PROCHECK server (https://saves.mbi.ucla.edu/results?job=17609&p=procheck). The Ramachandran plot, is a method for visualizing the energetically permissible and disallowed dihedral angles psi (ψ) and phi (ϕ) of an amino acid. The PROCHECK server was employed to verify protein structures using the Ramachandran plot separating the Glycine and Proline residues [44].
Prediction of discontinuous B-cell epitopes, promiscuous MHC class-I epitopes and IFN-γ inducing epitopes in candidate vaccines
Both the candidate vaccines PVAO and PVHSP were submitted in FASTA format to the IEDB server for prediction of discontinuous B-cell epitopes in the final vaccine constructs. Promiscuous MHC class-I epitopes were predicted by the ProPred1 server (http://crdd.osdd.net/raghava/propred1/) for both PVAO and PVHSP. A quantitative matrix for forecasting promiscuous MHC class-I binding peptides is used by the ProPred1 server. The ProPred1 achieves a high-level accuracy of 75% at the preset threshold of 4%. IFN-γ epitopes were predicted for the candidate vaccine peptide with the IFNepitope server (http://crdd.osdd.net/raghava/ifnepitope/scan.php).
Molecular docking analysis and interaction studies
Molecular docking was performed for the analysis of the binding affinity between the structurally refined vaccine constructs and the human TLRs as well as MHCI and MHCII. TLR1/2 (PDB ID-2Z7X) TLR4 (PDB ID-3FXI), TLR5 (PDB ID-3J0A), TLR6 (PDB ID-4OM7) TLR9 (PDB ID-5GMH) MHC-I (PDB ID-7RK7) and MHC-II (PDB ID-8JRK) were retrieved from the RCSB protein databank (https://www.rcsb.org/) and were used for docking. The molecular docking process was rendered by PatchDock (https://bioinfo3d.cs.tau.ac.il/PatchDock/php.php) and HawkDock (http://cadd.zju.edu.cn/hawkdock/) servers based on protein-protein rigid-body docking. Subsequently, lowest energy-weighted scores of the various docked complexes were obtained to identify the best ligand-receptor interaction from a group of ligand-receptor complexes. The final selected docked complex interactions were visualized by BIOVIA Discovery Studio 3.0 software and PDBsum (https://www.ebi.ac.uk/thornton-srv/databases/pdbsum/).
Calculation of free binding energies (MM/GBSA)
Binding free energy was calculated to confirm the docked data’s reliability and the bound molecules’ stability. This study utilized the HawkDock server to assess binding free energy via the Molecular Mechanics/Generalized Born Surface Area (MM/GBSA) method (http://cadd.zju.edu.cn/hawkdock/#MMGBSA_submit) [45] for the docked complexes of PVAO and PVHSP with MHC-I, MHC-II, TLR1/2, TLR4, TLR5, TLR6, and TLR9.
Disulfide engineering
To ensure the vaccination model’s stability, disulfide engineering was performed on the Design 2.0 webserver (http://cptweb.cpt.wayne.edu/DbD2/). The parameters χ3 angle were set for − 87° and + 97° and the Cα-Cβ-Sγ angle to 114.6° [46]. The introduction of novel disulfide bonds into proteins has been extensively utilized to improve protein stability, modify functional characteristics, and enable the study of protein dynamics.
Normal mode analysis
The conformational stability of the selected docked complex was analyzed through iMODS (https://imods.iqfr.csic.es/) web server [47]. NMA mobility is represented with arrows, which indicates motion dynamics. Deformability plots show the parts of the structures that are not rigid, whereas eigenvalues show the level of rigidity of these structures. The B-factor, variance, covariance map, and linking matrix of the desired structure are also provided by this server.
Molecular Dynamics Simulation of MEVs
The online server SiBioLead for Macromolecular Simulations (https://sibiolead.com/MDSIM) was used to perform molecular dynamics (MD) simulations of the protein-water complexes. The SiBioLead server force field was chosen to be the OPLS/AA. The other input parameters included the water model SPC, the box type Triclinic, the salt type NaCl, and the options to neutralize and add 0.15 M NaCl salt. The 5,000-step integrator of the steepest descent was chosen using energy minimization parameters. Equilibration and MD run parameters were chosen as NVT/NPT equilibration form, 300 K temperature, 1 (one) bar pressure, leap-frog MD integrator, 100 ns simulation period, and roughly 5,000 frames per simulation. The simulation output has been evaluated based on the average number of H-bonds in each frame over time, the root mean square deviation (RMSD) of the given structure over time, the root mean square fluctuation (RMSF) of each residue in the given structure, the radius of gyration (Rg) or structural compactness, the solvent-accessible surface area (SASA), and Gromacs energy analysis [48].
Java Codon Adaptation and In-silico cloning
The multi-epitope vaccine constructs were expressed using reverse translation and codon optimization using the Java Codon Adaptation Tool (JCat) server (http://www.prodoric.de/JCat). Codon optimization was performed to express the MEV vaccine constructs in the E. coli (strain K12) host, since the codon usage of E. coli differs from that of the native host filarial parasites [49]. Three additional parameters were introduced to escape the prokaryotic ribosome binding site, restriction enzyme cleavage sites, and rho-independent transcription termination [50]. To estimate the expression levels of proteins, the Codon Adaptation Index (CAI) and percentage GC content are included in the JCat output. CAI offers insights into codon use biases. A score of 1.0 is considered optimal, but a value of > 0.8 is considered good [51]. The GC content of a sequence should fall within a range of 30–70%, as values outside this range may negatively impact the transcriptional and translational efficiency [52]. In order to clone the optimized gene sequence of the final vaccine constructs in the E. coli pET-30a (+) vector, NdeI, and XhoI restriction sites were introduced to the N and C-terminals of the candidate vaccine sequences, respectively. Finally, this sequence was inserted into the pET-30a (+) vector with SnapGene 6.1.2 software for in-silico cloning.
Immune simulation
The C-ImmSim server (http://150.146.2.1/C-IMMSIM/index.php) was used to simulate an immunological response in a computational environment [53]. The agent-based model C-ImmSim predicts immunological interactions by using machine learning approaches and a position-specific scoring matrix (PSSM) for immune epitope prediction. Simultaneous simulation of three compartments defines three distinct anatomical regions in mammals: (i) the bone marrow, where new lymphoid and myeloid cells are produced by hematopoietic stem cells; (ii) the thymus, where naive T-cells are selected to prevent autoimmunity; and (iii) a tertiary lymphatic organ, such as a lymph node. The time intervals were set at 1, 84, and 168, where each interval represents 8 h. The first interval corresponds to the injection at time = 0. The remaining simulation settings were set to their default values [54] and three shots were administered, with a time gap of 28 days. To evaluate clonal selection, 12 doses of the injections were administered four weeks apart to mimic frequent antigen exposure typical of an endemic area. The plot also allows for the deduction of Simpson index, D, which is a measure of diversity.
Results
Selection of proteins and phylogenetic analysis
For construction of MEVs, the adult filarial antigens were selected either from W. bancrofti and B. malayi databases (Table 1). The antigens GST and GPx belonging to the family of anti-oxidant proteins are expressed in all life stages of the filarial parasites thus are crucial for the survival [55, 56]. The antigens SOD [57], HSP70 [58], HSP90 [59] and HSP110 were chosen based on their reactivities in immunological studies [60]. The homology analysis of AO and HSP proteins was confirmed by performing a BLASTp for proteins against the UniProt and GenBank database (Table 1). The cellular localization of the proteins was predicted by DeepLoc to be located either in the Cytoplasm/soluble (A0A0H5S7P0, J9E3A6, A0A0K0IYY8, EJW86287.1, EJW88125.1, A0A0K0IYH1), Lysosomes (EJW86287.1, EJW88125.1, A0A0K0IYH1), Cell membrane (A0A0K0IYY8, J9E3A6), or Nucleus (A0A0H5S7P0, EJW88125.1, A0A0K0IYH1) (Supplementary Table 1). The signal peptide was checked in all the protein sequences using SignalP 6.0 server and none of the sequences contained signal peptides. Antigenic properties of antioxidant and heat shock proteins chosen for construction of multi-epitopic vaccines were predicted by the ANTIGENpro server. The highest antigenicity was shown by HSP110 (0.915) and the lowest antigenicity by GST (0.517). The BLASTp was performed against Homo sapiens proteins to assess the safety profile of the proposed vaccine candidates. The vaccine constructs exhibited no similarity with any human protein. This shows that vaccine constructs may be entirely safe for usage, with no risk of an autoimmune response post-vaccination (Supplementary Table 2).
Table 1.
Similarity of filarial proteins with Homo sapiens
| Protein | Percent identity | |||||
|---|---|---|---|---|---|---|
| W. bancrofti | B. malayi | Loa-loa | O. volvulus | O. ochengi | Homo sapiens | |
| Superoxide dismutase | 95.19 | 100 | 91.40 | 86.10 | 86.55 | 6.20 |
| Glutathione peroxidase | 100 | 95.68 | 94.37 | NA | 88.12 | 11.2 |
| Glutathione-S-transferase | 99.04 | 100 | 84.26 | 79.81 | 80.29 | 8.0 |
| HSP110 (VDM08680.1) | 97.31 | 100 | 88.39 | NA | 88.74 | 9.0 |
| HSP90 (EJW88125.1) | 100 | 99.58 | NA | NA | NA | 4.5 |
| HSP70 (EJW86287.1) | 100 | 98.08 | 85.28 | 81.25 | 81.73 | 11.5 |
B-cell, CTL and HTL epitope prediction
In the next step, linear B-cell and T-cell epitope prediction was applied to the functional sequences of the proteins. Linear B-cell epitopes of PVAO having 7 to 20 amino acids residues were predicted with two different servers i.e. ABCpred and IDEB server. After prediction, only those epitopes were chosen which were jointly predicted by both servers (Table 2). The linear B-cell epitopes, with antigen scores higher than 0.75, in ABCpred analysis were included in the vaccine construction. Out of the three antioxidant proteins, a total of 15 CTLs (MHC supertype A1) were predicted using NetCTL1.2 server, out of which 9 were selected based on high antigenicity scores (> 0.75) as given by ANTIGENpro server. Amongst the three HSPs sequences, a total of 15 CTLs were predicted and all the 15 CTLs were included for PVHSP construction (Table 3). MHC-II epitopes having high binding affinity with HLA DR, DQ and DP (IC50 scores) were selected for construction of MEVs. All the highly antigenic (VaxiJen score > 0.5) epitopes as predicted by NetMHCII 2.3 server were included in the construction of PVAO, and PVHSP vaccine constructs (Table 4).
Table 2.
Selection of B-cell epitopes and MHC-I epitopes for PVAO construction (VaxiJen score > 0.50)
| S.N. | B-cell Epitope Sequence | Start point | Length | MHC-I Epitope Sequence | VaxiJen Score | MHC-I binding Allele |
|---|---|---|---|---|---|---|
| 1 | LRGDAGVSG | 8 | 9 | VAEVYINSY (2.2877) | 1.2984 | HLA-A1 |
| HLA-B62 | ||||||
| 2 | IIYFQQGSGGSITTI | 17 | 15 | DAGVSGIIY (0.9587) | 0.7657 | HLA-A1 |
| 3 | SGSVSGLT | 32 | 8 | |||
| 4 | PGLHGFH | 40 | 7 | |||
| 5 | VHQYGDQT | 47 | 8 | |||
| 6 | NGCTSAGDHY | 55 | 10 | |||
| 7 | NPFGKTHGG | 65 | 9 | |||
| 8 | PNDRIKHIGDLGNIVAGANG | 74 | 20 | |||
| 9 | DIKLRGPLSVIGHSLVVHA | 103 | 19 | |||
| 10 | ANTDDLGQG | 121 | 20 | |||
| 11 | TGNMREESLKTGNAGSRLAC | 130 | 8 | |||
| 12 | YEFTVKDI | 6 | MTAIKADDY (3.2387) | 1.1358 | HLA-A1 | |
| 13 | NGVDVSLEKYRGHVCLIVNV | 14 | 20 | LTNSNYTEL (0.8885) | 1.0250 | HLA-A1 |
| 14 | CKSGATDKNYRQLQEMHTRL | 35 | 20 | GKDVSLEKY (0.8132) | 0.5930 | HLA-A*0201 |
| 15 | NQFGGQEPWAEAEI | 67 | 14 | |||
| 16 | KKFVTEKYGVQFDMFSKIK | 81 | 19 | |||
| 17 | VNGSDADDLYK | 100 | 11 | |||
| 18 | FLKSRQHGTLTNNIKWNFSK | 111 | 20 | |||
| 19 | QPVKRYSPTTAPYDIEGDIM | 138 | 20 | |||
| 20 | PIRLVLVDQGIKFTD | 15 | 15 | YTKMIYQAY(2.7894) | 1.5349 | HLA-A1 |
| HLA-A26 | ||||||
| HLA-B8 | ||||||
| HLA-62 | ||||||
| 21 | DRINASDWPSMKSHFHFGQ | 30 | 19 | HTKYTKMIY (2.2179) | 1.5031 | HLA-A1 |
| HLA-A26 | ||||||
| HLA-B62 | ||||||
| 22 | LPCLYDGDHQIVQSG | 49 | 15 | FTDDRINAS (1.2726) | 1.3981 | HLA-A1 |
| 23 | AILRHLA | 64 | 7 | AYDTEKDSY (0.9831) | 1.0288 | HLA-A1 |
| 24 | RKHNLNGGNELETTHID | 71 | 17 | |||
| 25 | MFCEGVRDLHTKYTKMIYQA | 88 | 20 | |||
| 26 | ILPVELAKFE | 119 | 10 | |||
| 27 | KLLATRD | 129 | 20 | |||
| 28 | DGKNFILGEKISYVDFVLFE | 136 | 7 | |||
| 29 | CLDKFPLLKAYHQRMEDRPG | 167 | 20 |
Table 3.
Selection of B-cell epitopes and MHC-I epitopes for PVHSP constructions (VaxiJen score > 0.50)
| S.N. | B-cell Epitope sequence | Start point | Length | MHC-I Epitope sequence | VaxiJen Score | MHC-I Binding Allele |
|---|---|---|---|---|---|---|
| 1 | AFTTRGRSMGVAARQQLNTN | 40 | 20 | FSKLFYDTY (2.3138) | 1.6016 | HLA-A1 |
| HLA-A26 | ||||||
| 2 | RVRILLTNLLRESGKKAEEV | 318 | 20 | KLTVLGATY (0.8951) | 1.4512 | HLA-A1 |
| HLA-A3 | ||||||
| HLA-B58 | ||||||
| HLA-B62 | ||||||
| 3 | AAMKCAILSPAFKVRDFSVK | 378 | 20 | NTNIKNTII (0.7662) | 1.4508 | HLA-A1 |
| 4 | ETDDTKG | 520 | MSEEMNGET(0.9951) | 2.1441 | HLA-A1 | |
| 5 | NQEKEENTNAPSNDVVLP | 527 | 7 | QLEFRALLF (0.9500) | 2.0230 | HLA-A1 |
| HLA-B62 | ||||||
| 6 | PPKAEPAKEPEKGSDGQKGE | 775 | 18 | MSLIINTFY(0.9468) | 1.9252 | HKA-A1 |
| HLA-A3 | ||||||
| HLA-B58 | ||||||
| HLA-B62 | ||||||
| 7 | DDEAEEE | 222 | 7 | LMSLIINTF (0.7788) | 1.6891 | HLA-A1 |
| HLA-A24 | ||||||
| HLA-B8 | ||||||
| HLA-B58 | ||||||
| HLA-B62 | ||||||
| 8 | KKDEDKEKKE | 229 | 10 | ETALLSSGF (0.7567) | 1.5783 | HLA-A26 |
| 9 | GEIEDVGE | 239 | 8 | VKDLVVLLF (0.8568) | 1.2065 | HLA-A1 |
| HLA-A24 | ||||||
| 10 | DEEEDKKEKDKK | 247 | 12 | MKENQKQIY (0.9639) | 0.9724 | HLA-A1 |
| 11 | LAVKHFSVEGQLEFRALLFV | 306 | 20 | TRGTKITLY (0.7913) | 0.8785 | HLA-A1 |
| HLA-B27 | ||||||
| 12 | SAGGMPGG | 613 | 8 | YMTDPIDEY (1.7255) | 0.8258 | HLA-A1 |
| HLA-A26 | ||||||
| HLA-B62 | ||||||
| 13 | MPGGMPSGAPGAGSTGGGPT | 621 | 20 | YSDNQPGVL (1.7835) | 1.2493 | HLA-A1 |
| HLA-B39 | ||||||
| 14 | KSTAGDTHLGGEDFDNRMVN | 220 | 20 | YTNITRARF (0.9825) | 0.8258 | HLA-A1 |
| HLA-A26 | ||||||
| HLA-B58 | ||||||
| HLA-B62 | ||||||
| 15 | AVITVPAYFNDSQRQATKDS | 142 | 20 | SSSQASIEI (0.7980) | 0.5904 | HLA-A1 |
| HLA-B58 |
Table 4.
Selection of MHC-II epitopes for constructions of PVAO and PVHSP (VaxiJen score > 0.50)
| Name of vaccine constructs | MHC-II Epitope sequence | VaxiJen Score | MHC-II Binding Allele | No. of MHC-II binding allele |
|---|---|---|---|---|
| PVAO | KNFILGEKI | 0.6758 | DRB1_0101 | 2 |
| HLA-DPA10201-DPB1010 | ||||
| DIKLRGPLS | 2.9048 | DRB1_0101 | 2 | |
| DRB1_1602 | ||||
| PLSVIGHSL | 1.1870 | DRB1_0101 | 2 | |
| HLA-DQA10501-DQB10303 | ||||
| LSVIGHSLV | 0.6973 | DRB1_0101 | 1 | |
| PVHSP | YIKITPNKA | 0.7987 | DRB1_0101 | 11 |
| DRB1_0401 | ||||
| DRB1_0405 | ||||
| BRB1_1602 | ||||
| DRB3_0202 | ||||
| DRB5_0101 | ||||
| DRB1_0701 | ||||
| DRB1_0802 | ||||
| DRB1_1001 | ||||
| DRB1_1101 | ||||
| DRB1_1301 | ||||
| ERIMKAQAL | 0.5397 | DRB1_0101 | 1 | |
| AILSPAFKV | 1.3767 | DRB1_0101 | 1 | |
| DRB1_0801 | 4 | |||
| DRB1_1001 | ||||
| DRB1_1302 | ||||
| VRILLTNLL | 1.1471 | DRB1_0101 | 10 | |
| DRB1_0103 | ||||
| DRB1_0404 | ||||
| DRB1_405 | ||||
| DRB1_1501 | ||||
| DRB4_0101 | ||||
| DRB1_0701 | ||||
| DRB1_1302 | ||||
| HLA-DPA10301-DPB10402 | ||||
| HLA-DQA10102-DQB10501 |
Construction of multi-epitopic vaccine candidates
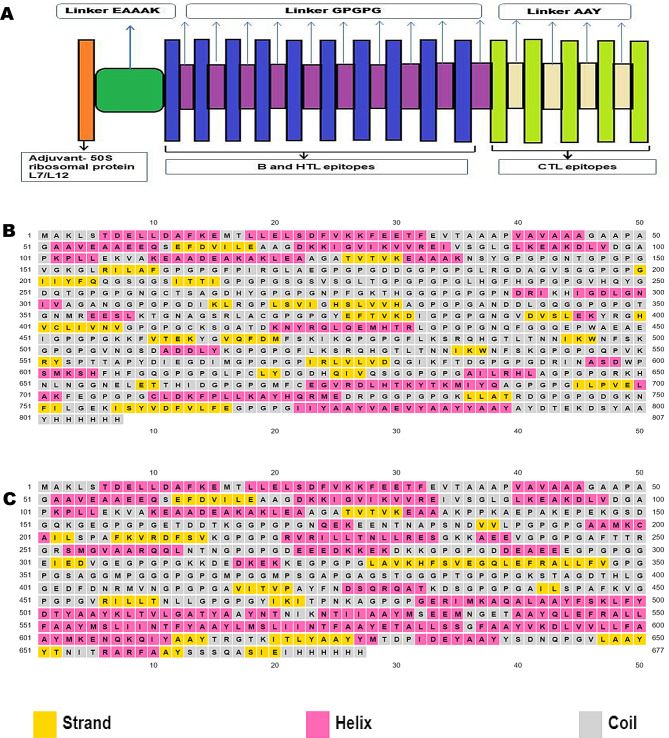
Two separate potential multi-epitope vaccines PVAO consisting of GST, GPx and SOD epitopes and PVHSP consisting of HSP70, HSP90 and HSP110 epitopes were constructed. The PVHSP and PVAO were constructed by combining highly antigenic CTLs (score > 0.5), high affinity B-cell epitopes (Threshold value = 0.50), and HTL epitopes (score > 0.75). A linker GPGPG connected HTL and B-cell epitopes and another linker AAY was used for adding CTL (MHC supertype A1) epitopes in the vaccine constructs. A 50 S ribosomal protein L7/L12, chosen as an adjuvant was added to the N-terminal of vaccine constructs through an EAAAK linker which increased the vaccine`s stability and immunogenicity (Fig. 1A).
Fig. 1.
Secondary structure of the candidate vaccines (A) Schematic diagram of polypeptide multi-epitope vaccine with adjuvant, epitopes, and linkers. The secondary structure features strand, helix, and coiled regions in (B) PVAO and (C) PVHSP, respectively
Prediction of discontinuous B-cell epitopes, promiscuous MHC class-I epitopes and IFN-γ inducing epitopes in candidate vaccine constructs
The IFN-γ epitopes of the PVAO and PVHSP vaccine constructs were predicted by IFNepitope server. Twenty three (23) IFN-γ epitopes were predicted for PVAO and eighteen (18) IFN- epitopes were predicted for PVHSP vaccine constructs (Table 5). The candidate vaccine PVAO was predicted to contain 8 and PVHSP to contain 3 discontinuous B-cell epitopes by the IEDB server (Supplementary Table 5). Promiscuous MHC class-I epitopes of HLA-A1, HLA-A2, HLA-A*0201 and HLA-A*0205 were predicted for both PVAO and PVHSP (Supplementary Table 6).
epitopes were predicted for PVHSP vaccine constructs (Table 5). The candidate vaccine PVAO was predicted to contain 8 and PVHSP to contain 3 discontinuous B-cell epitopes by the IEDB server (Supplementary Table 5). Promiscuous MHC class-I epitopes of HLA-A1, HLA-A2, HLA-A*0201 and HLA-A*0205 were predicted for both PVAO and PVHSP (Supplementary Table 6).
Table 5.
IFN-γ epitope of chimeric vaccine candidates of antioxidant and heat shock proteins of filarial parasite
| Peptide | Sequence | Method | Result | Score |
|---|---|---|---|---|
| PVAO | ||||
| EGDIMGPGPGPIRLVL | SVM | NEGATIVE | -0.27343023 | |
| YAAYYAAYAYDTEKDS | MERCI | POSITIVE | 1 | |
| VLSYKVEKAGRCDIH | SVM | NEGATIVE | -0.56939587 | |
| LGEKISYVDFVLFEGP | SVM | POSITIVE | 0.319 | |
| LQEMHTRLGPGPGNQF | SVM | NEGATIVE | -0.4640616 | |
| PGPGILPVELAKFEGP | SVM | NEGATIVE | -0.17574 | |
| PVELAKFEGPGPGCLD | SVM | NEGATIVE | -0.406873 | |
| GDRINASDWPSMKSHF | SVM | NEGATIVE | -0.13892 | |
| PGPGMFCEGVRDLHTK | SVM | NEGATIVE | -0.2640 | |
| MFSKIKGPGPGFLKSR | SVM | NEGATIVE | -0.3202906 | |
| GPGPGAILRHLAGPGP | SVM | POSITIVE | 0.32084744 | |
| QIVQSGGPGPGAILRH | SVM | NEGATIVE | -0.17945 | |
| PGCLDKFPLLKAYHQR | SVM | NEGATIVE | -0.01395 | |
| GVNGSDADDLYKGPGP | MERCI | NEGATIVE | 1 | |
| PGIIYAAYVAEVYAAY | MERCI | NEGATIVE | 4 | |
| GKLLATRDGPGPGDGK | SVM | NEGATIVE | -0.47270355 | |
| KDLVDGAPKPLLEKVA | MERCI | NEGATIVE | 1 | |
| PCLYDGDHQIVQSGGP | MERCI | POSITIVE | 1 | |
| LKTGNAGSRLACGPGP | MERCI | NEGATIVE | 2 | |
| PGRKHNLNGGNELETT | SVM | NEGATIVE | -0.575877 | |
| PGPGDGKNFILGEKIS | SVM | POSITIVE | 0.07909736 | |
| VVREIVSGLGLKEAKD | MERCI | NEGATIVE | 2 | |
| YDTEKDSYAAYHHHHH | SVM | POSITIVE | 0.82982 | |
| PV HSP | ||||
| QATKDSGPGPGAILSP | MERCI | NEGATIVE | 1 | |
| GSTGGGPTGPGPGKST | MERCI | NEGATIVE | 3 | |
| GETDDTKGGPGPGNQE | SVM | NEGATIVE | -0.59720259 | |
| TDPIDEYAAYYSDNQP | MERCI | POSITIVE | 1 | |
| GMPGGGPGPGMPGGMP | SVM | POSITIVE | 1.0220518 | |
| QKQIYAAYTRGTKITL | SVM | NEGATIVE | -0.32601724 | |
| GDTHLGGEDFDNRMVN | MERCI | POSITIVE | 2 | |
| MPGGMPSGAPGAGSTG | MERCI | NEGATIVE | 1 | |
| GDTHLGGEDFDNRMVN | MERCI | POSITIVE | 2 | |
| VEAAEEQSEFDVILEA | SVM | POSITIVE | 0.31781947 | |
| YAAYYSDNQPGVLAAY | SVM | POSITIVE | 0.11582581 | |
| GATYAAYNTNIKNTII | SVM | NEGATIVE | -0.061137575 | |
| LGLKEAKDLVDGAPKP | MERCI | NEGATIVE | 1 | |
| YMSEEMNGETAAYQLE | SVM | NEGATIVE | -0.6340472 | |
| DVILEAAGDKKIGVIK | SVM | POSITIVE | 0.5297006 | |
| RVRILLTNLLRESGKK | MERCI | POSITIVE | 1 | |
| PGPGLAVKHFSVEGQL | SVM | POSITIVE | 0.18118755 | |
| EVTAAAPVAVAAAGAA | MERCI | NEGATIVE | 1 |
Prediction of toxicity, allergenicity, antigenicity, and physiochemical properties of the vaccine candidates
All the epitopes used in this study were found to be non-toxic (Supplementary Table 7). The antigenicity of PVAO as predicted by VaxiJen v2.0 was 0.530 and by ANTIGENpro was 0.652, which showed the antigenic nature of this construct. The score of PVHSP obtained by VaxiJen was 0.879 and by ANTIGENpro the score was 0.9234, which indicated that PVHSP could be more antigenic than PVAO. Both the PVHSP and PVAO were non-allergenic as predicted by AllerTOPv2. The predicted molecular masses from ExPASy-ProtParam serve were 82.24 kDa for PVAO and 70.24 kDa for PVHSP. The PVAO and PVHSP constructs were slightly acidic in nature as per the PI values. The GRAVY scores for PVAO -0.438 and − 0.343 for PVHSP suggesting the hydrophilic nature of these molecules (Table 6). The physicochemical properties such as a number of amino acids, molecular weight, formula, total no of atoms, estimated half-life, instability index, and aliphatic index, for both the constructs were satisfactory for their further development as vaccine candidates [40].
Table 6.
VaxiJen score, ANTIGENpro score, and Physiochemical properties of the vaccine candidates PVAO and PVHSP
| Name of Vaccines | VaxiJen score | Antigen pro score | Solubility | No. of amino acids | Molecular weight (Daltons) | Theoretical isoelectric Point (PI) | Formula | Total no. of atoms | Estimate half life | Instability index | Aliphatic index | (GRAVY) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PPVAO | 0.5304 | 0.6527 | 0.9018 | 807 | 82241.26 | 6.30 |
C 3663 3663 H 5635 5635 N 1025 1025 O 1104 1104 S 17 17 |
11,444 |
30 h (mammalian reticulocytes, in vitro). > 20 h (yeast, in vivo). > 10 h (E. coli, in vivo). |
21.31 | 65.07 | -0.438 |
| PV HSP | 0.8749 | 0.9234 | 0.9018 | 677 | 70247.17 | 5.17 |
C 3126 3663 H 4892 5635 N 830 1025 O 977 1104 S 16 17 |
9841 | 30 h (mammalian reticulocytes, in vitro). >20 h (yeast, in vivo). >10 h (E. coli, in vivo). | 33.77 | 70.64 | -0.343 |
Prediction of secondary and tertiary structures
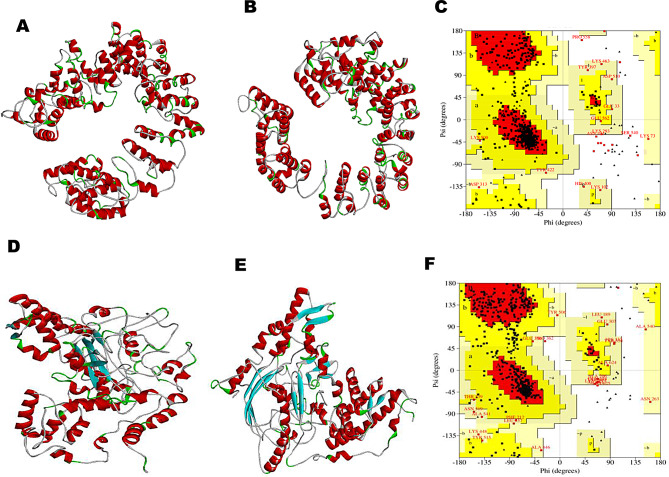
The secondary structure of PVAO and PVHSP and the helix, strand and coil region of both constructs are compared in Fig. 1B and C. The secondary structure of the peptide vaccines was predicted by PSIPRED that works on the probable sequence produced by PSI-BLAST. PSIPRED works on the secondary structure with the highest score for generating a protein prediction in terms of helix, strands, and coil. PVAO consisted of 56% helix, 0% beta-sheet, and 43% random coil, whereas, PVHSP consisted of 37% helix, 14% beta-sheet and, 48% random coil. The candidate vaccine sequences were subjected to 3D structure prediction with I-TASSER and 5 structures were predicted for each peptide vaccines (Fig. 2). The PVAO model with the highest C-score of 0.57 and PVHSP model with the highest C-score of -1.94 was chosen for further studies. Additionally, the model topology was also validated by TM score and RMSD value (Supplementary Fig. 4). A negative “Z” score was calculated by the ProSA web server (PVAO -2.61 and PVHSP -5.37) which evidenced that the predicted vaccines scored within the range typically found for native proteins (Supplementary Fig. 5). Since the estimated ERRAT values were greater than 50 (PVAO 78.907 and PVHSP 76.097) (Supplementary Fig. 5), the improved 3D structures were considered acceptable to serve as trustworthy models for additional evaluations [61]. The Ramachandran plot analysis showed 84.4%, 11.8%, 2.4%, and 1.6% residues of PVAO and 64.1%, 26.6%, 7.0%, and 2.3% residues of PVHSP in most favored, additionally allowed, generously allowed, and disallowed regions respectively (Supplementary Fig. 6). ModRefiner server was utilized to validate 3D structure in terms of highest values for parameters like RMSD and TM score. The model 1 provided by ModRefiner server for PVAO had RMSD and TM scores of 0.558 and 0.9967 respectively. Similarly for PVHSP the model 1 had RMSD and TM scores as 0.438 and 0.9977 respectively (Fig. 2B and E). In both constructs, model 1 showed the best scores, hence these were chosen for further studies. Disulfide engineering is a crucial technique for incorporating disulfide bonds into a target protein to improve its stability, increase functional properties, and facilitate the comprehension of its dynamics. The PVAO vaccine construct had 78 potential pairs for forming disulfide bonds while 60 potential disulfide sites were predicted for PVHSP vaccine constructs (Supplementary Table 8).
Fig. 2.
Protein modeling, refinement, and validation of PVAO and PVHSP vaccine constructs. (A) Tertiary structure of PVAO. (B) Refined structure of PVAO. (C) Ramachandran plot illustrating the quality of PVAO. (D) Tertiary structure of PVHSP. (E) Refined structure of PVHSP. (F) Ramachandran plot illustrating the quality of PVHSP
Molecular docking of PVAO and PVHSP and calculation of binding energy
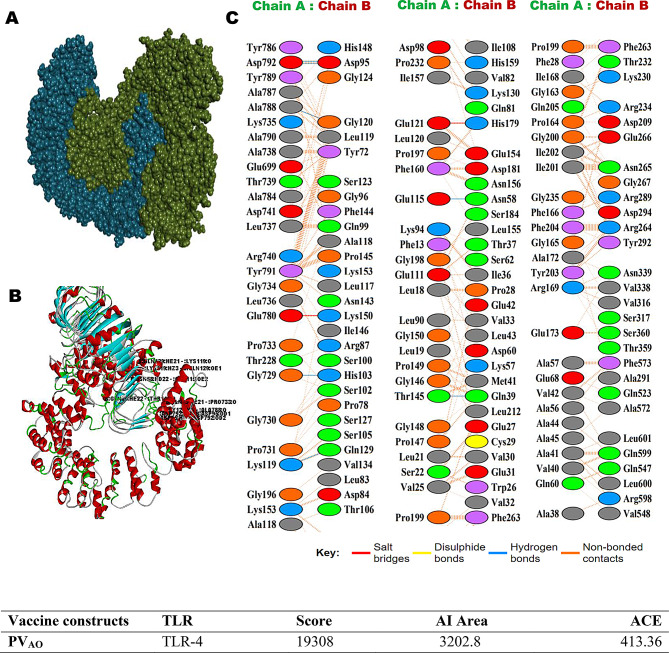
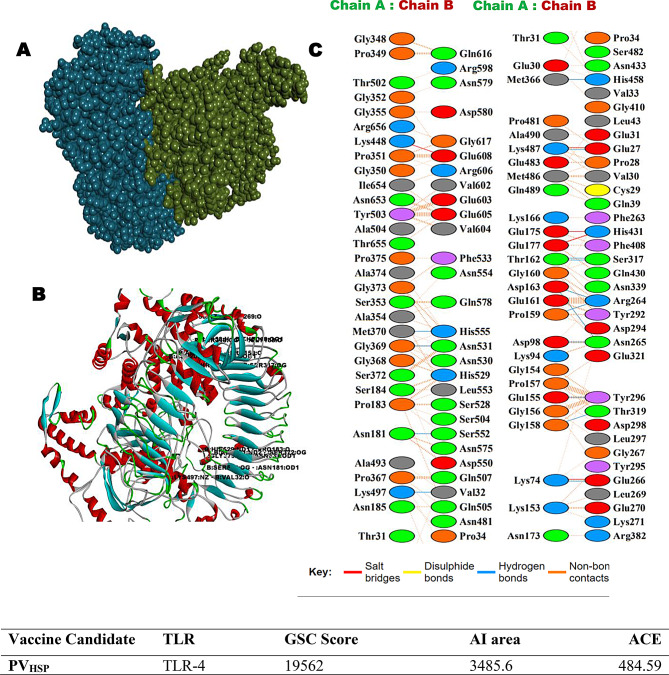
The involvement of TLR4s in lymphatic filariasis infection is well studied and TLR4 has been implicated for conferring protective immunity [62]. Hence the in-silico interactions of PVAO and PVHSP with TLR4 were examined by PatchDock server and visualized by Biovia Discovery Studio (Figs. 3 and 4). The top-scoring PVAO-TLR4 complex had a GSC score of 19,308, Area 3202.80, and ACE 413.30, whereas the PVHSP-TLR4 complex had a GSC score of 19,562, Area 3485.60, and ACE 484.59. The amino acids residues K119, D792, Q39, N58, D95, Q99, G120, Q129, L19, V42, E121, G150, G200, G734, D792, P28, V30, G96, S102, S123, K153, H179, L600 in PVAO, and K74, K487, K153, G158, T319, T162, G373, K497, R264, N265, T319, H529, N531, S552, K153, G352, S353, G369, K487, S317, H458, Q507, H555, in PVHSP formed hydrogen bonds with the TLR4 molecule (Supplementary Table 9). In summary, each vaccine candidates formed more than 20 hydrogen bonds with TLR4 molecule in the docked complex.
Fig. 3.
PVAO vaccine constructs 3D structural analysis and molecular docking (A) Diagram of docking mode of the vaccine construct (green) and TLR4 (blue) complex. (B) Docked conformation and hydrogen bond interaction maps of the vaccine construct and TLR4. (C) The interacting residues between the docked vaccine construct (Chain A) and TLR4 (Chain B)
Fig. 4.
PVHSP vaccine constructs 3D structural analysis and molecular docking (A) Diagram of docking mode of the vaccine construct (green) and TLR4 (blue) complex. (B) Docked conformation and hydrogen bond interaction maps of the vaccine construct and TLR4. (C) The interacting residues between the docked vaccine construct (Chain A) and TLR4 (Chain B)
Calculation of binding energies (MM/GBSA)
Molecular docking was also performed with HawkDock server for the analysis of the binding energy between the vaccine constructs and other TLRs such as TLR1/2, TLR4, TLR5, TLR6, and TLR9, as well as with MHC I and MHC II (Table 7). A higher docking score generally suggests a better binding affinity between the target receptors and the compounds or epitopes being tested. The MMGBSA scores obtained for TLR1/2, TLR6, TLR9, and MHC-II were in the range of -19.78 kcal/mol to -49.47 kcal/mol, which indicated that the candidate vaccines could also engage many TLRs effectively thus, leading to a stronger immune response against the filarial parasites.
Table 7.
Molecular Docking performed by HawkDock server, showing docking score and MM/GBSA energy
| Vaccine | TLRs | MHCs | ||||||
|---|---|---|---|---|---|---|---|---|
| TLR1/2 | TLR4 | TLR5 | TLR6 | TLR9 | I | II | ||
| PVAO | PDB ID | 2Z7X | 3FXI | 3J0A | 4OM7 | 5GMH | 7RK7 | 8JRK |
| Docking Score | -5336.90 | -6367.14 | -5524.64 | -3466.35 | -5781.04 | -6237.58 | -5771.49 | |
| MM/GBSA(kcal/mol) | -27.86 | -20.48 | -20.19 | -19.78 | -29.03 | -39.65 | -23.3 | |
| PVHSP | Docking Score | -9213.25 | -4359.40 | -7321.85 | -3579.20 | -6528.98 | -5401.40 | -4907.02 |
| MM/GBSA(kcal/mol) | -49.47 | -19.28 | -10.65 | -30.06 | -43.79 | -8.26 | -24.6 | |
Normal mode analysis
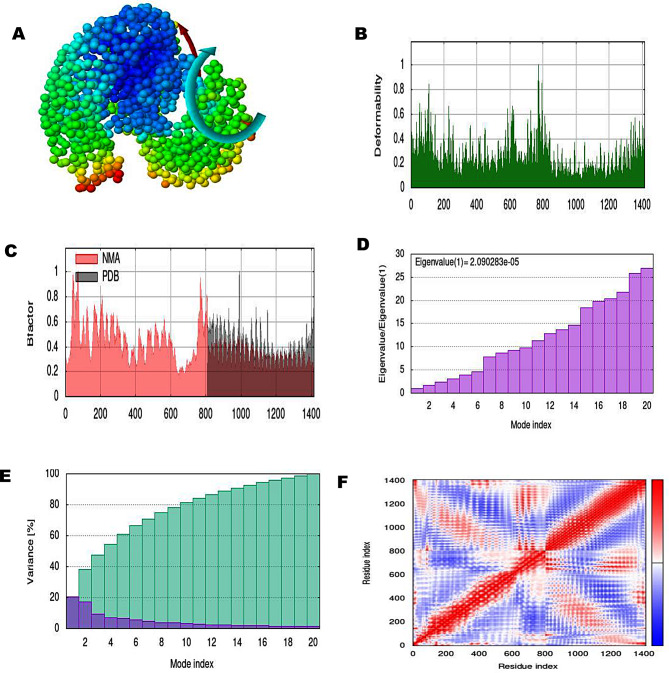
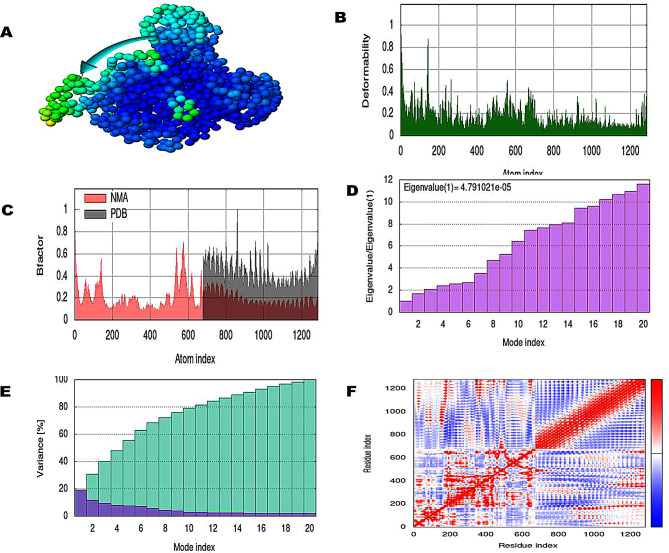
The normal mode analysis of the docked MEV-TLR4 complexes has been carried out by the iMODS server. The NMA results gave information about domain mobility towards each other as shown with the arrow (Figs. 5A and 6A). The deformability plot indicated the flexible region of the complexes in the form of peaks (Figs. 5B and 6B). The B-factor represented atomic positional changes in the flexible region of the docked complexes (Figs. 5C and 6C). The eigenvalue of the PVAO-TLR4 complex was 2.090283e-05 and eigenvalue of PVHSP-TLR4 complex was 4.79102e-05 reflecting formation of stable structures (Figs. 5D and 6D). Eigenvalue is inversely related to variance as shown in Figs. 5E and 6E where purple color represents individual and green color represents cumulative variance. Connection between residues was deducted by covariance map (Figs. 5F and 6F), where red signified correlated, white signified un-correlated and blue anti-correlated residues.
Fig. 5.
A normal mode analysis of the PVAO-TLR4 complex showing NMA mobility (affine-model arrows), deformability, B-factor, Eigenvalue, variance (purple color shows individual variance and green color shows cumulative variances), and co-variance map (red correlated, white uncorrelated, and blue anti-correlated)
Fig. 6.
Normal mode analysis of PVHSP-TLR4 complex (A) NMA mobility (affine-model arrows), (B) deformability, (C) B-factor, (D) Eigenvalue, (E) variance (purple color represents individual variance and green color indicates cumulative variances), and (F) co-variance map (red correlate, white uncorrelated, and blue anti-correlated)
Molecular dynamics simulation analysis
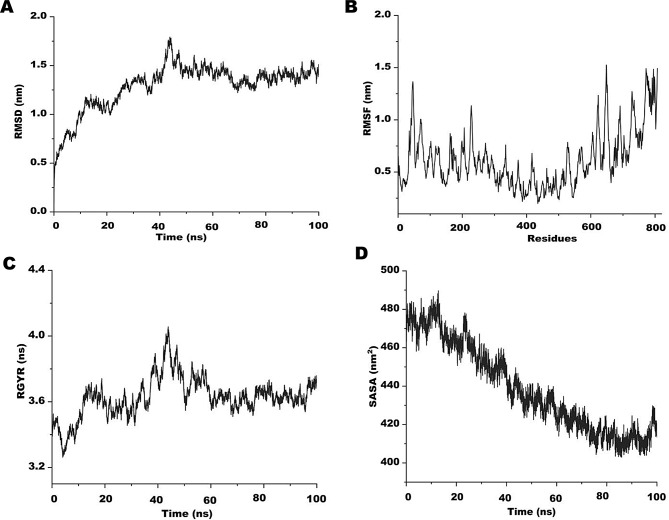
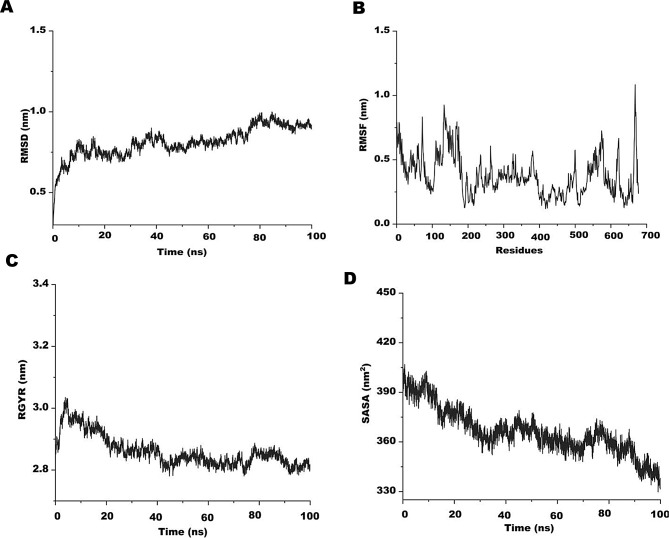
In a simulated environment that is closely analogous to real systems, MD simulation is a well-accepted technique for effectively analyzing the stability of protein complexes. Molecular dynamics (MD) simulations were carried out for 100 ns using the Linux-based molecular dynamics program of SiBioLead server to further support the stability of the vaccine constructs. The SPC water model was applied to solvate the protein in addition to the OPLS/AA force field. It is crucial to reduce the global energy of molecules, hence to stabilize the conformations, an energy minimization step was added for the first 5000 steps. In order to learn more about the construct’s Radius of Gyration (Rg), Root Mean Square Deviation (RMSD), and Root Mean Square Fluctuation (RMSF), the simulation trajectories were plotted against time. During the MD simulation, it was observed that initially, both PVAO and PVHSP systems fluctuated slightly from 2 ns to 15 ns, after which the trajectories were stable over the entire run of 100 ns. The PVAO system had a minimum RMSD value of 1.30 nm and a maximum of 1.75 nm with a total fluctuation of 0.45 nm (Fig. 7A). The RMSD value for the PVHSP vaccine over a time period of 100 ns varied between 0.80 nm and 1.09 nm (Fig. 8A). The compactness and stability was assessed by Rg during the simulation run. Both the MD trajectories showed higher values for Rg at the beginning of the run however as the simulation progressed, the vaccines attained a much steady plateaued form that continued till the completion as shown in Figs. 7B and 8B.
Fig. 7.
Molecular dynamics (MD) simulation of PVAO (A) Root mean square deviation (RMSD) plot. (B) Root mean square fluctuation (RMSF) plot (C) Radius of gyration plot. (D) Alteration in protein exposure to the solvent is shown by the complex’s Solvent Accessible Surface Area (SASA)
Fig. 8.
Molecular dynamics (MD) simulation of PVHSP (A) Root mean square deviation (RMSD) plot. (B) Root mean square fluctuation (RMSF) plot. (C) Radius of gyration plot. (D) Alteration in protein exposure to the solvent is shown by the complex’s Solvent Accessible Surface Area (SASA)
The RMSF trajectory of the two candidate vaccines were plotted against time, and was observed that the amino acid residues of PVAO from 80 to 110 (1.49 nm) was the highest and for 410 to 425 (0.71 nm) was the lowest in the entire run. In case of PVHSP, the RMSF was maximal for residues 130 to 190 (1 nm) while residues 308 to 335 (0.89 nm) showed lowest fluctuations (Figs. 7C and 8C). Similar observations were also seen in the Solvent Accessible Surface Area (SASA) analysis of both PVAO and PVHSP (Figs. 7D and 8D).
Optimization of codon for peptide vaccine and in-silico cloning
To achieve maximal expression in E-coli (strain k12), the codon optimization was performed with Java Codon Adaptation Tool (JCat). The length of the optimized PVAO codon was 2421 nucleotides and for PVHSP 2031 nucleotides. The Codon Adaptation index (CAI) for PVAO was 0.9497 and for PVHSP was 0.9553, which was much above the threshold CAI of 0.5. In the next step, recombinant plasmid pET30a (+) was designed separately for PVAO and PVHSP (Fig. 9). The in-silico integration of PVHSP in pET30a (+) expression vector between NdeI and XhoI restriction sites resulted in a product size of 7260 base-pair. Similarly, PVAO was also inserted into the pET30a (+) expression vector between the NdeI and XhoI restriction sites resulting in a product of 7650 base-pairs.
Fig. 9.
In silico cloning of the PVAO and PVHSP (A) Cloning vector pET-30a (+) has multiple restriction sites. (B) Insert of PVHSP vaccine constructs. (C) Insert of PVAO vaccine constructs (D) Final product of PVHSP (7260 bp) (E) Final product of PVAO (7650 bp)
Immune simulation
Immunological simulations were done by the C-ImmSim server, which produced results that were in line with the real immunological reactions as evidence by the generation of secondary responses too. The primary response was associated with an elevation in the IgM levels. The secondary and tertiary responses showed significant increases in the B-cell population of IgG1, IgG2 IgM, and IgG+ IgM producing cells in the host. The simulation showed a marked decrease in antigen concentration after secondary immune response, which confirms generation of immunological memory (Figs. 10 and 11). The simulation programmed 12 injections of MEVs repeated for a period of 28 days to sufficiently increase IgG1 levels and population of T-helper cells during the entire period of vaccination. Increase in the population of helper T-cell and cytotoxic T-cells are usually associated with the production of memory cells which is important for the effective clearance of antigens upon further exposure.
Fig. 10.
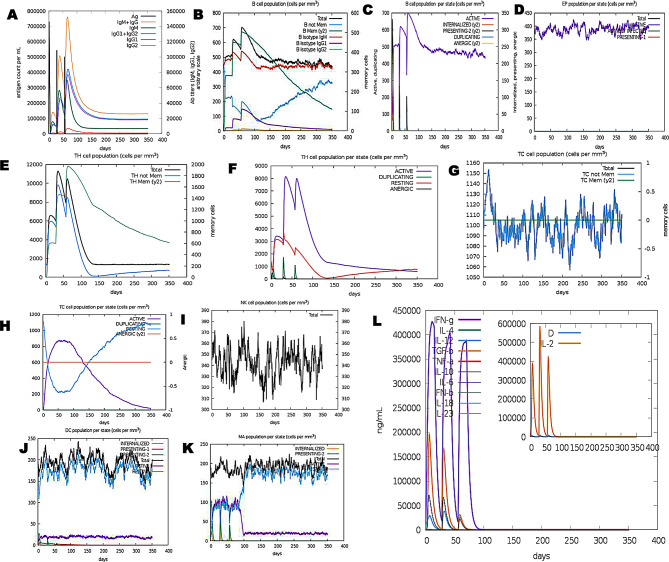
In-silico immune simulation of PVAO in the C-ImmSim server (A) The primary B cell antibody produced after antigen stimulation is IgM + IgG (yellow) and changes over time. (B) Active B cell (purple) secretion after antigen stimulation. (C) The number and proportion of helper T cell subtypes. (D) Secretion from active epithelial cells (purple) after antigen stimulation (E, F) Changes in the secretion level of helper T lymphocytes and the secretion levels of helper T lymphocytes of different memory types. (G, H) Changes in the level of CD8 + T secretion after antigen stimulation and the secretion of different types of CD8 + T cells. (I) Expression of NK cells after antigen stimulation. (J) Expression of DC cells (black) after antigen stimulation. (K) Expression in Macrophages (black) after antigen stimulation. (L) Changes in secretion levels of cytokines, mainly IFN-γ (purple) and IL-2 (yellow)
Fig. 11.
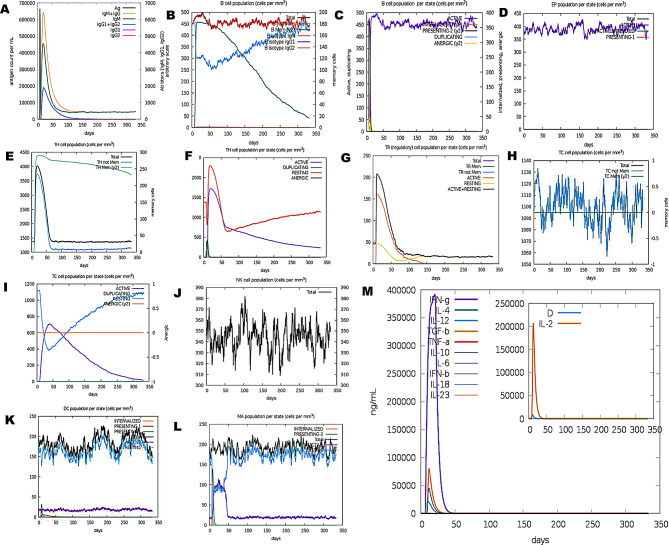
In-silico immune simulation of PVHSP in the C-ImmSim server. (A) The primary B cell antibody produced after antigen stimulation is IgM + IgG (yellow) and changes over time. (B) Active B cell (purple) secretion after antigen stimulation. (C) The number and proportion of helper T cell subtypes. (D) Secretion from active epithelial cells (purple) after antigen stimulation (E, F) Changes in the secretion level of helper T lymphocytes and the secretion levels of helper T lymphocytes of different memory types. (G) Changes in the number of regulatory T cell subtypes. (H, I) Changes in the level of CD8 + T secretion after antigen stimulation and the secretion of different types of CD8 + T cells. (J) NK cell expression after antigen stimulation. (K) Expression in Macrophages (black) after antigen stimulation. (L) Expression of DC cells (black) after antigen stimulation. (M) Changes in secretion levels of cytokines, mainly IFN-γ (purple) and IL-2 (yellow)
The administration of booster doses led to an improved immune response in terms of combined levels of IgG1 and IgG2, as well as levels of IgM and IgG1 (Supplementary Table 10). The total IgG and IgM antibody titer for both vaccines was between 780,000 per ml and 700,000 per ml following the third booster dose. The IgM titer was around 650,000 per ml for PVAO and construct vaccines, compared to 330,000 for PVHSP vaccines, respectively. Additionally, with the PVAO and PVHSP construct vaccines, the combined IgG1 and IgG2 levels were 180,000 per ml and 400,000 per ml, respectively. In addition, a significant amount of IgG1 was produced in response to all vaccine booster shots (Figs. 10 and 11). The result for Simpson Index D analysis shows that the total B-cell response induced by PVAO was higher than PVHSP although both vaccines induced sufficient humoral memory and effector responses (Fig. 10 (B) and 11 (B)). The immunological response upon vaccinations is significantly influenced by interleukins and cytokines. After vaccination, the antigen-presenting cells (APCs) process the antigen and present them to the T-cells. This mechanism activates T-cells which subsequently release cytokines including IFN-γ and IL-2, leading to B-cell and T-cell proliferation and differentiation. Subsequently, the antibodies produced by B-cells can then neutralize the pathogen against which the vaccine was created. The levels of interleukins (IL) and cytokines significantly increased after the vaccinations were given. The IFN-γ levels increased gradually and peaked at about 1,000,000 ng/ml (PVAO) and 425,000 ng/ml (PVHSP), further increased levels of cytokines such IFN-γ, IL-2, IL-10, IL-12, and TGF-β were also seen in response to the booster doses.
Discussion
Till date, LF treatment and control strategies across the globe are completely dependent upon the success of MDA programs. Neither the MDA programs are cost-effective nor the drugs are adulticidal; hence, the development of an LF vaccine proves to be the best alternative to control filarial infection. Earlier vaccine candidates like HSP70 [63] and GST [21], though successful in the laboratory, could not be translated into commercial LF vaccines. Therefore the approach has now shifted to developing epitope based vaccines. Multi-epitope vaccines comprise of overlapping epitopes that are recognized by several TCR clones and can elicit humoral, cytotoxic, and helper T-cell immune responses simultaneously. MEVs also possess the capability to produce long term immunogenicity [45, 64]. Epitope-based vaccines offer several potential benefits, such as enhanced safety and the flexibility to strategically modify epitopes to enhance their effectiveness and range [12].
The filarial parasites are long-lived organisms surviving for several years inside the human body. Owing to their robust antioxidant enzymatic system, filarial worms easily defend themselves against the reactive oxygen species generated during the oxidative burst of the hosts’ macrophages, neutrophils, and eosinophils. The anti-oxidant enzymes such as, SOD, GST, and GPx are crucial for the survival of the parasites [21, 57, 65]. In previous studies, mice vaccinated with naked DNA constructs expressing GPX, Cu/Zn cytosolic superoxide dismutase, demonstrated a significant level of protection in comparison to a control group against S. mansoni infections [66, 67]. Moreover, filarial GSTs have already been investigated as prospective LF vaccine candidates [68]. Another key player in the survival and development of filarial parasites are the HSPs. The filarial HSP’s are prominent immunogens and have been explored as potential vaccine candidates [28]. Filarial HSP70 from W. bancrofti and Brugia spp. have been investigated as an immunogenic proteins leading to the generation of high IgG4 levels in humans [58]. Therefore, in this study an attempt was made to design two MEVs comprising of filarial AOs GST, GPx, and SOD (PVAO) and HSPs 70, 90, and 110 (PVHSP) and to compare their in-silico efficacies using immuno-informatics approaches.
Additionally, it seemed imperative to create an MEV that could offer cross-protection against other filarial parasites. The minimal identity percentage that might preclude self-peptide and epitope cross-reactivity has not been documented by any investigation. Therefore, non-homology of the filarial AO and HSP sequences with the human proteome was a key consideration in selecting the antigens initially [69]. The primary sequence of candidate vaccines PVAO and PVHSP had multiple CD8+, CD4+, and B-cell epitopes that could trigger a potent immune response which is very important for vaccine development since B-cells produce antibodies and also help CD8+ and CD4+ T-cells in generating long-lasting adaptive immunity [70]. The chosen epitope sequences demonstrated their strong immunogenic potential, as evidenced by the results of the ANTIGENpro server. The induction of allergenicity rather than an immunological response by vaccination is one of the main barriers to the development of vaccines; therefore, the list of epitopes was narrowed down to those that wouldn’t be expected to trigger an allergic reaction.
For designing the multi-epitope-based vaccination, each selected B-cell, CD8+ and CD4+ epitope was connected with its neighbors using GPGPG and AAY linkers. The sequences GPGPG and AAY linkers have been seen to produce very less immunogenicity [7]. Adjuvants are crucial parts of MEVs as boosting innate immunity is also critical to improving the therapeutic efficacy of a vaccine. Additionally, a 50 S ribosomal protein L7/L12 adjuvant was combined with the first MHC-I epitopes using the EAAAK linker. The presence of the EAAAK linker, imparts a distinctive ability to auto-cleave, hence allowing for the efficient isolation of the desired protein from the fusion protein without protease treatment. Also, the linker has proven cost-effective in protein purification [71]. In the next step, the preliminary 3D structures of the candidate vaccines PVAO and PVHSP were obtained with I-TASSER server. Further improvement to obtain a high-quality 3D model, was attained following loop refinement and energy minimization of the initial model with ModRefiner. The resulting Z-score of the improved modelled structure of PVAO and PVHSP were comparable to previous studies, indicating that the predicted structure corresponded well with the X-ray crystallography values of comparable-sized proteins [72].
The refined model of candidate vaccines was next subjected to prediction for IFN-γ epitopes, discontinuous and promiscuous B-cell epitopes. PVAO and PVHSP vaccine constructs were predicted to contain IFN-γ epitopes that can elicit IFN-γ secretion in the host. Earlier IFN-γ secretions have also proven useful in controlling filarial infections in clinical cases [73]. The discontinuous B-cell epitopes play an important role in humoral immune responses by eliciting the production of antibodies. PVAO and PVHSP had several discontinuous B-cell epitopes, showing that both the candidate vaccines could induce ample quantities of antibody production. Identifying promiscuous epitopes with robust binding affinity to various HLA (HLA-A1, HLA-A2, HLA-A*0201, and HLA-A*0205 supertype) alleles is essential for eliciting protective immunity, and both candidate vaccines had several promiscuous B-cell epitopes, hence confirming for extensive population coverage. Further, to confirm the antigenicity of the designed PVAO and PVHSP vaccines, VaxiJen, and ANTIGENpro servers were employed. When compared to other LF MEVs constructed using Aspartic protease and Thioredoxin proteins, the PVHSP had the highest ANTIGENpro score (0.9234). Upon comparing the VaxiJen scores, Thioredoxin MEV demonstrated a slightly greater score of 0.9513 than PVHSP (0.8749) which confirms that the MEVs designed in this study had sufficient immunogenicity too [19, 62].
The initiation and establishment of Lymphatic filariasis disease are significantly influenced by TLR-mediated responses [74, 75]. TLRs are a subclass of pattern recognition receptors (PRRs) that are expressed on the surface of many different cell types, including immune cells (such as DCs and macrophages) and non-immune cells (like fibroblasts and epithelial cells). A wide range of molecular patterns present on the pathogen also known as Pathogen Associated Molecular Patterns (PAMPS), are recognized by TLRs [76]. Immediate response to pathogens by the host immune system via TLR recognition involves the production of pro-inflammatory cytokines and amplified co-stimulatory signal. Furthermore, the development of new vaccines is largely dependent on TLR-mediated immune activation because TLRs trigger signaling cascades that produce type 3-interferon (INF-γ), which in turn is necessary for macrophage activation and elevated expression of MHC-I and MHC-II [77]. Subsequently, the candidate vaccines were studied for immune cell receptor interactions by the HawkDock server. Both PVAO and PVHSP showed stable binding, evidenced by high docking scores and binding energies with MHC-II, TLR1/2, TLR4, TLR6, and TLR9, which strongly indicate towards the generation of long term immunity against LF through multiple TLR mediated pathways as well as cytotoxic responses.
The most important and primary innate immune response against LF infection is known to be generated by TLR4 [78]. Therefore, the molecular docking of TLR4 with PVAO and PVHSP was accomplished by the PatchDock server to obtain detailed molecular interactions. The results clearly highlighted the importance of electrostatic and Van der Waal interactions in the binding of MEVs with TLR4 [62]. The MEVs and TLR4 complex was additionally stabilized because of the formation of several H-bonds (Figs. 3 and 4, and Supplementary Table 9). These findings show that PVAO and PVHSP could stably bind to the immune receptors, suggesting that MEVs might elicit a potent immune response in-vivo too. Moreover, the normal mode analysis from iMODS showed that the PVAO and PVHSP with TLR4 complex had sufficiently high eigenvalues, which establishes the lower level of deformability. Consequently, the deformability graph also showed a potential increase in complex rigidity for the designed vaccines. Non-uniform stiffness/immobility profiles, as well as high B-factor, deformability, and mobility indices, gave much flexibility to the PVAO and PVHSP vaccines.
The PVAO and PVHSP could be expressed in the E. coli system, as supported by the estimated half-life of PVAO and PVHSP (> 10 h for both PVAO and PVHSP), the index of instability (PVAO 21.31 and PVHSP 33.77), and the aliphatic index. The candidate vaccines’ predicted solubility score also added support towards their successful production. In-silico serological analysis for immune reactivity is the initial step in verifying a multi-epitope candidate vaccines; therefore, the recombinant protein must be expressed in an appropriate host, such as E. coli expression systems [50, 79]. Codon optimization for PVAO and PVHSP were done so that the recombinant vaccine protein is abundantly expressed in high levels in E. coli (strain K12). Both the codon adaptability index and the GC content for PVAO and PVHSP was 0.9497, 0.9553, and 57.001%, 53.126%, respectively, which is suitable for expression of MEVs in bacteria. Therefore, a well-characterized expression vector pET30a (+) was used for in-silico cloning of the MEV.
The stability of the PVAO and PVHSP constructs in a dynamic environment was assessed by molecular dynamics simulation using SiBioLead services. The proposed vaccines displayed constant conventional thermodynamic behavior, with RMSDs leveling up for less than 20 ns only. The RMSD estimates molecular deviation from the specified original/reference structure, which may be used to ensure ligand-target stability/confinement as well as the validity of the MD procedure [80]. Obtaining low RMSD values that are swiftly equilibrated has been linked to the effective convergence of the simulated models, requiring no additional molecular manipulation. In general, the C-RMSF flexibility analysis tool estimates the deviations of protein residues from their reference positions, providing an evaluation of protein residues in terms of their respective dynamic behavior as represented by flexibility and fluctuation. Within the vaccine’s planned structure, the addition of lengthy helices with flexible loop connections would rationalize the early relaxation and considerable convergence into more stable compacted conformations. Broadly, Rg is responsible for the global stability of protein ternary complexes, where the mass-weighted RMSD for atom groups in regard to their respective common center of mass is specified as a stability parameter. Therefore, the observed dynamic behavior of the simulated candidate vaccines to exhibit low Rgs’ demonstrates substantial stability and compactness of PVAO and PVHSP. The results of the SASA analysis for candidate vaccines were consistent with the above preferential complex stability as the simulated both PVAO and PVHSP showed a steadier SASA tone along the mean equilibration plateau and up to the end of the simulation. As SASA is a quantitative measurement of the interaction between protein and solvent, correlating for the molecular surface area (MSA) accessible to solvent, low SASA tones indicate relative structural reduction under the stress of solvent surface charges resulting in more compact, stable conformations.
The vaccine candidates designed must be capable of eliciting a potent immunological response from the host in order to be effective. The dynamics of the human immune system in response to the candidate vaccines was monitored by in-silico immunological simulation using the C-ImmSim server. It was observed that after exposure to PVAO and PVHSP, the immune responses were strongly stimulated and the levels of antibodies (IgG, IgM) were sufficiently high. As a result, the secondary immune response related to the injections of the built vaccine was also activated after the initial immune reaction was generated by the vaccine injections. Post-vaccinations, there was a significant rise in the concentration of several immune cells, including memory B-cells, plasma B-cells, helper T-cells, cytotoxic T-cells, and various types of antibodies. The results also indicated that the population of cytotoxic T-cells and helper T-cells was increased along with an increase in the population of memory cells [19]. Additionally, low Simpson indexes were predicted for PVAO and PVHSP by the C-ImmSim server, which is indicative of multiple forms of immune responses.
In this study, multiple approaches and servers were used to assess the immunogenicity of the predicted vaccines; still, in general multi-epitope vaccines suffer from certain drawbacks. It has been seen that the epitope prediction tools fail to appropriately take into account the necessity of identifying the appropriate antigen processing sites. For instance, the composition of antigen processing pathways differs according to pro-inflammatory signals and changes among distinct cell types; thus, present prediction algorithms may be inadequate for assessing the processing efficacy of parasite antigens in an infected target cell. Further in-vitro and in-vivo experiments like expressing the candidate vaccines in bacterial systems and validating their immunogenicity by diverse immunological assays are needed to ascertain the safety and efficacy before human administration.
Conclusion
The global burden of lymphatic filarial infections warrants better chemotherapeutic and diagnostic strategies as well as, effective vaccines too. In this work, two vaccines (PVAO and PVHSP) comprising several B-cell and T-cell (HTL and CTL) epitopes were designed using immuno-informatics methods. The peptide vaccines may have preventive and therapeutic advantages since the proteins bearing these epitopes are expressed in the microfilariae and infective stages of the parasite. The in-silico findings showed that the multi-epitope-based peptide vaccines were stable in 3D architecture and may have the capacity to elicit powerful immune responses against the filarial parasites. Moreover, the vaccine model substantially supports the capabilities to induce both humoral and cell-mediated immune responses. Based on antigenicity, physiochemical properties, and molecular dynamics simulation, PVHSP turned out to be a better candidate vaccine than PVAO. Though our in-silico analysis revealed a robust immunogenic response, in-vitro and in-vivo validation for immunogenicity and host safety is still required before human administration. Also, the modified vaccine candidates were anticipated to provide cross-protection, which will allow additional Neglected tropical disease control projects focused on related filarial diseases such as onchocerciasis and loiasis to reap benefits from the initiative.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Sunil Kumar is grateful to the NBCFDC JRF Ref. No. 231620090211 Junior Research Fellowship, University Grants Commission (UGC), New Delhi. Ayushi Mishra is thankful to the Council of Scientific and Industrial Research (CSIR), India (09/013(0832)/2018-EMR-I) for providing a Senior Research Fellowship (SRF). Tripti Singh is thankful to the University Grants Commission (UGC), New Delhi for providing Junior Research Fellowship Ref. No. 211610077377. The authors are also grateful to DBT-BHU Interdisciplinary School of Life Sciences for providing laboratory space.
Author contributions
SK, AS, VK: Conceptualization, Designing, Data Curation, Study implementation and Analysis. SK, AS, AKS: Formal analysis, Writing, and editing of the manuscript. SK, AM, TS: Investigating, Methodology analysis. AS: Supervision, Project administration, and Validation. All authors read and approved the final manuscript.
Funding
No specific funding was received for this work.
Data availability
All data supporting the findings of this study are available within the paper and its Supplementary Information.
Declarations
Ethical approval
This study does not involve any human or animal subject.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hotez PJ, Molyneux DH, Fenwick A, Kumaresan J, Sachs SE, Sachs JD, Savioli L. Control of neglected tropical diseases. N Engl J Med. 2007;357(10):1018–27. [DOI] [PubMed] [Google Scholar]
- 2.Nutman TB. Insights into the pathogenesis of disease in human lymphatic filariasis. Lymphatic Res Biology. 2013;11(3):144–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cromwell EA, Schmidt CA, Kwong KT, Pigott DM, Mupfasoni D, Biswas G, Shirude S, Hill E, Donkers KM, Abdoli A. The global distribution of lymphatic filariasis, 2000–18: a geospatial analysis. Lancet Global Health. 2020;8(9):e1186–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kumar V, Mishra A, Yadav AK, Rathaur S, Singh A. Lymphatic filarial serum proteome profiling for identification and characterization of diagnostic biomarkers. PLoS ONE. 2022;17(7):e0270635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Das PK, Shenoy RK. Helminthic Diseases: Filariasis. 2017.
- 6.Kwarteng A, Ahuno ST, Akoto FO. Killing filarial nematode parasites: role of treatment options and host immune response. Infect Dis Poverty. 2016;5(1):86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gorai S, Das NC, Gupta PSS, Panda SK, Rana MK, Mukherjee S. Designing efficient multi-epitope peptide-based vaccine by targeting the antioxidant thioredoxin of bancroftian filarial parasite. Infect Genet Evol. 2022;98:105237. [DOI] [PubMed] [Google Scholar]
- 8.Collyer BS, Irvine MA, Hollingsworth TD, Bradley M, Anderson RM. Defining a prevalence level to describe the elimination of lymphatic filariasis (LF) transmission and designing monitoring & evaluating (M&E) programmes post the cessation of mass drug administration (MDA). PLoS Negl Trop Dis. 2020;14(10):e0008644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Melendez V, Turner C, Khatri V, Davis J, Chauhan N, Nagalati Sudhakar DS, Cabullos R, Carter D, Gray SA, Kalyanasundaram R. Pre-clinical development of a vaccine for human lymphatic filariasis. Front Trop Dis. 2022;3:998353. [Google Scholar]
- 10.Morris CP, Evans H, Larsen SE, Mitre E. A comprehensive, model-based review of vaccine and repeat infection trials for filariasis. Clin Microbiol Rev. 2013;26(3):381–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Samykutty A, Dakshinamoorthy G, Kalyanasundaram R. Multivalent vaccine for lymphatic filariasis. Procedia Vaccinol. 2010;3:12–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou W-Y, Shi Y, Wu C, Zhang W-J, Mao X-H, Guo G, Li H-X, Zou Q-M. Therapeutic efficacy of a multi-epitope vaccine against Helicobacter pylori infection in BALB/c mice model. Vaccine. 2009;27(36):5013–9. [DOI] [PubMed] [Google Scholar]
- 13.Bouazzaoui A, Abdellatif AA, Al-Allaf FA, Bogari NM, Al-Dehlawi S, Qari SH. Strategies for vaccination: conventional vaccine approaches versus new-generation strategies in combination with adjuvants. Pharmaceutics. 2021;13(2):140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bibi S, Ullah I, Zhu B, Adnan M, Liaqat R, Kong W-B, Niu S. In silico analysis of epitope-based vaccine candidate against tuberculosis using reverse vaccinology. Sci Rep. 2021;11(1):1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanches RC, Tiwari S, Ferreira LC, Oliveira FM, Lopes MD, Passos MJ, Maia EH, Taranto AG, Kato R, Azevedo VA. Immunoinformatics design of multi-epitope peptide-based vaccine against Schistosoma mansoni using transmembrane proteins as a target. Front Immunol. 2021;12:621706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shey RA, Ghogomu SM, Esoh KK, Nebangwa ND, Shintouo CM, Nongley NF, Asa BF, Ngale FN, Vanhamme L, Souopgui J. In-silico design of a multi-epitope vaccine candidate against onchocerciasis and related filarial diseases. Sci Rep. 2019;9(1):4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weil GJ, Curtis KC, Fischer PU, Won KY, Lammie PJ, Joseph H, Melrose WD, Brattig NW. A multicenter evaluation of a new antibody test kit for lymphatic filariasis employing recombinant Brugia malayi antigen Bm-14. Acta Trop. 2011;120:S19–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mukherjee S, Karnam A, Das M, Babu SPS, Bayry J. Wuchereria bancrofti filaria activates human dendritic cells and polarizes T helper 1 and regulatory T cells via toll-like receptor 4. Commun Biology. 2019;2(1):169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Das NC, Gorai S, Gupta PSS, Panda SK, Rana MK, Mukherjee S. Immune targeting of filarial glutaredoxin through a multi-epitope peptide-based vaccine: a reverse vaccinology approach. Int Immunopharmacol. 2024;133:112120. [DOI] [PubMed] [Google Scholar]
- 20.Mukherjee S, Joardar N, Sinha Babu SP. Redox regulatory circuits as targets for therapeutic intervention of bancroftian filariasis: biochemical, molecular, and pharmacological perspectives. Oxidative Stress Microb Dis. 2019;185–208.
- 21.Rathaur S, Yadav M, Gupta S, Anandharaman V, Reddy MV. Filarial glutathione-S-transferase: a potential vaccine candidate against lymphatic filariasis. Vaccine. 2008;26(32):4094–100. [DOI] [PubMed] [Google Scholar]
- 22.Ellis RJ. Stress proteins as molecular chaperones. In Eden WV, Young DB, editors. Stress proteins in medicine. New York: Marcel Dekker; 1996. pp. 1–26.
- 23.Ahmad F, Sharma S, Yadav S, Rathaur S. The HSP90 inhibitor 17-AAG induced calcium‐mediated apoptosis in filarial parasites. Drug Dev Res. 2022;83(8):1867–78. [DOI] [PubMed] [Google Scholar]
- 24.Lang BJ, Guerrero ME, Prince TL, Okusha Y, Bonorino C, Calderwood SK. The functions and regulation of heat shock proteins; key orchestrators of proteostasis and the heat shock response. Arch Toxicol. 2021;95(6):1943–70. [DOI] [PubMed] [Google Scholar]
- 25.Chung M, Teigen LE, Libro S, Bromley RE, Olley D, Kumar N, Sadzewicz L, Tallon LJ, Mahurkar A, Foster JM. Drug repurposing of bromodomain inhibitors as potential novel therapeutic leads for lymphatic filariasis guided by multispecies transcriptomics. Msystems. 2019;4(6). 10.1128/msystems. 00596 – 00519. [DOI] [PMC free article] [PubMed]
- 26.Ullah A, Waqas M, Aziz S, ur Rahman S, Khan S, Khalid A, Abdalla AN, Uddin J, Halim SA, Khan A. Bioinformatics and immunoinformatics approach to develop potent multi-peptide vaccine for coxsackievirus B3 capable of eliciting cellular and humoral immune response. Int J Biol Macromol. 2023;239:124320. [DOI] [PubMed] [Google Scholar]
- 27.Gupta S, Bhandari YP, Reddy MV, Harinath BC, Rathaur S. Setaria Cervi: immunoprophylactic potential of glutathione-S-transferase against filarial parasite Brugia malayi. Exp Parasitol. 2005;109(4):252–5. [DOI] [PubMed] [Google Scholar]
- 28.Ravi V, Kubofcik J, Bandopathyaya S, Geetha M, Narayanan R, Nutman T, Kaliraj P. Wuchereria bancrofti: cloning and characterization of heat shock protein 70 from the human lymphatic filarial parasite. Exp Parasitol. 2004;106(1–2):1–10. [DOI] [PubMed] [Google Scholar]
- 29.Nielsen H. Predicting secretory proteins with SignalP. Methods Mol Biol. 2017;1611:59–73. [DOI] [PubMed]
- 30.Almagro Armenteros JJ, Sønderby CK, Sønderby SK, Nielsen H, Winther O. DeepLoc: prediction of protein subcellular localization using deep learning. Bioinformatics. 2017;33(21):3387–95. [DOI] [PubMed]
- 31.Jespersen MC, Peters B, Nielsen M, Marcatili P. BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 2017;45(W1):W24–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peters B, Bulik S, Tampe R, Van Endert PM, Holzhütter HG. Identifying MHC class I epitopes by predicting the TAP transport efficiency of epitope precursors. J Immunol (Baltimore Md: 1950). 2003;171(4):1741–9. [DOI] [PubMed] [Google Scholar]
- 33.Nielsen M, Lund O. NN-align. An artificial neural network-based alignment algorithm for MHC class II peptide binding prediction. BMC Bioinformatics. 2009;10(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khatoon N, Pandey RK, Prajapati VK. Exploring Leishmania secretory proteins to design B and T cell multi-epitope subunit vaccine using immunoinformatics approach. Sci Rep. 2017;7(1):8285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kar PP, Srivastava A. Immuno-Informatics analysis to identify novel vaccine candidates and design of a Multi-epitope based vaccine candidate against Theileria parasites. Front Immunol. 2018;9:2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Das NC, Patra R, Gupta PSS, Ghosh P, Bhattacharya M, Rana MK, Mukherjee S. Designing of a novel multi-epitope peptide based vaccine against Brugia malayi: an in silico approach. Infect Genet Evol. 2021;87:104633. [DOI] [PubMed] [Google Scholar]
- 37.Dimitrov I, Bangov I, Flower DR, Doytchinova I. AllerTOP v. 2—a server for in silico prediction of allergens. J Mol Model. 2014;20:1–6. [DOI] [PubMed] [Google Scholar]
- 38.Magnan CN, Zeller M, Kayala MA, Vigil A, Randall A, Felgner PL, Baldi P. High-throughput prediction of protein antigenicity using protein microarray data. Bioinformatics. 2010;26(23):2936–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hebditch M, Carballo-Amador MA, Charonis S, Curtis R, Warwicker J. Protein–Sol: a web tool for predicting protein solubility from sequence. Bioinformatics. 2017;33(19):3098–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Williams KL, Appel RD, Hochstrasser DF. Protein identification and analysis tools in the ExPASy server. Methods Mol Biology (Clifton NJ). 1999;112:531–52. [DOI] [PubMed] [Google Scholar]
- 41.Wang S, Peng J, Ma J, Xu J. Protein secondary structure prediction using deep convolutional neural fields. Sci Rep. 2016;6(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5(4):725–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wiederstein M, Sippl MJ. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35(Web Server issue):W407–10. [DOI] [PMC free article] [PubMed]
- 44.Lovell SC, Davis IW, De Arendall PI III, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Cα geometry: ϕ, ψ and Cβ deviation. Proteins Struct Funct Bioinform. 2003;50(3):437–50. [DOI] [PubMed] [Google Scholar]
- 45.Malik M, Khan S, Ullah A, Hassan M, Haq MU, Ahmad S, Al-Harbi AI, Sanami S, Abideen SA, Irfan M et al. Proteome-wide screening of potential vaccine targets against Brucella melitensis. Vaccines 2023, 11(2). [DOI] [PMC free article] [PubMed]
- 46.Craig DB, Dombkowski AA. Disulfide by Design 2.0: a web-based tool for disulfide engineering in proteins. BMC Bioinformatics. 2013;14:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.López-Blanco JR, Aliaga JI, Quintana-Ortí ES, Chacón P. iMODS: internal coordinates normal mode analysis server. Nucleic Acids Res. 2014;42(W1):W271–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tumskiy RS, Tumskaia AV. Multistep rational molecular design and combined docking for discovery of novel classes of inhibitors of SARS-CoV-2 main protease 3CLpro. Chem Phys Lett. 2021;780:138894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grote A, Hiller K, Scheer M, Münch R, Nörtemann B, Hempel DC, Jahn D. JCat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 2005;33(suppl2):W526–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rosano GL, Ceccarelli EA. Recombinant protein expression in Escherichia coli: advances and challenges. Front Microbiol. 2014;5:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morla S, Makhija A, Kumar S. Synonymous codon usage pattern in glycoprotein gene of Rabies virus. Gene. 2016;584(1):1–6. [DOI] [PubMed] [Google Scholar]
- 52.Fu H, Liang Y, Zhong X, Pan Z, Huang L, Zhang H, Xu Y, Zhou W, Liu Z. Codon optimization with deep learning to enhance protein expression. Sci Rep. 2020;10(1):17617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rapin N, Lund O, Bernaschi M, Castiglione F. Computational immunology meets bioinformatics: the use of prediction tools for molecular binding in the simulation of the immune system. PLoS ONE. 2010;5(4):e9862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tahir ul Qamar M, Rehman A, Tusleem K, Ashfaq UA, Qasim M, Zhu X, Fatima I, Shahid F, Chen L-L. Designing of a next generation multiepitope based vaccine (MEV) against SARS-COV-2: Immunoinformatics and in silico approaches. PLoS ONE. 2020;15(12):e0244176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gupta S, Singh A, Yadav M, Singh K, Rathaur S. MALDI mass sequencing and characterization of filarialglutathione-S-transferase. Biochem Biophys Res Commun. 2007;356(2):381–5. [DOI] [PubMed] [Google Scholar]
- 56.Singh A, Rathaur S. Identification and characterization of a selenium-dependent glutathione peroxidase in Setaria Cervi. Biochem Biophys Res Commun. 2005;331(4):1069–74. [DOI] [PubMed] [Google Scholar]
- 57.Rathaur S, Sharma S, Singh RN, Henkle K, Selkirk ME. Antibody responses of Wuchereria bancrofti patients to recombinant Brugia pahangi superoxide dismutase. Indian J Exp Biol. 2001;39(1):35–40. [PubMed] [Google Scholar]
- 58.Ahmad F, Liebau E, Rathaur S. Human immune response against filarial HSP70 and its role in the diagnosis of lymphatic filariasis. Parasite Immunol. 2023;45(5):e12978. [DOI] [PubMed] [Google Scholar]
- 59.Ahmad F, Sharma S, Yadav S, Rathaur S. The HSP90 inhibitor 17-AAG induced calcium-mediated apoptosis in filarial parasites. Drug Dev Res. 2022;83(8):1867–78. [DOI] [PubMed] [Google Scholar]
- 60.Milani A, Basirnejad M, Bolhassani A. Heat-shock proteins in diagnosis and treatment: an overview of different biochemical and immunological functions. Immunotherapy. 2019;11(3):215–39. [DOI] [PubMed] [Google Scholar]
- 61.Yazdani Z, Rafiei A, Yazdani M, Valadan R. Design an efficient multi-epitope peptide vaccine candidate against SARS-CoV-2: an in silico analysis. Infect Drug Resist. 2020;13:3007–22. [DOI] [PMC free article] [PubMed]
- 62.Das NC, Sen Gupta PS, Biswal S, Patra R, Rana MK, Mukherjee S. In-silico evidences on filarial cystatin as a putative ligand of human TLR4. J Biomol Struct Dyn. 2022;40(19):8808–24. [DOI] [PubMed] [Google Scholar]
- 63.Hartmann W, Singh N, Rathaur S, Brenz Y, Liebau E, Fleischer B, Breloer M. Immunization with B rugia malayi Hsp70 protects mice against L itomosoides sigmodontis challenge infection. Parasite Immunol. 2014;36(4):141–9. [DOI] [PubMed] [Google Scholar]
- 64.Ullah A, Rehman B, Khan S, Almanaa TN, Waheed Y, Hassan M, Naz T, Ul Haq M, Muhammad R, Sanami S et al. An In Silico Multi-epitopes Vaccine Ensemble and Characterization Against Nosocomial Proteus penneri. Molecular biotechnology 2023. [DOI] [PubMed]
- 65.Singh A, Kamal S, Rathaur S. Filarial selenium glutathione peroxidase: a probable immunodiagnostic marker for lymphatic filariasis. Trans R Soc Trop Med Hyg. 2010;104(8):524–8. [DOI] [PubMed] [Google Scholar]
- 66.LoVerde PT, Carvalho-Queiroz C, Cook R. Vaccination with antioxidant enzymes confers protective immunity against challenge infection with Schistosoma mansoni. Memórias do Instituto Oswaldo Cruz. 2004;99:37–43. [DOI] [PubMed] [Google Scholar]
- 67.Dabir S, Dabir P, Goswamy K, Reddy M. Prophylactic evaluation of recombinant extracellular superoxide dismutase of Brugia malayi in jird model. Vaccine. 2008;26(29–30):3705–10. [DOI] [PubMed] [Google Scholar]
- 68.Veerapathran A, Dakshinamoorthy G, Gnanasekar M, Reddy MVR, Kalyanasundaram R. Evaluation of Wuchereria bancrofti GST as a vaccine candidate for lymphatic filariasis. PLoS Negl Trop Dis. 2009;3(6):e457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Michel-Todó L, Reche PA, Bigey P, Pinazo MJ, Gascón J, Alonso-Padilla J. In silico design of an epitope-based Vaccine Ensemble for Chagas Disease. Front Immunol. 2019;10:2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Slamanig SA, Nolte MA. The bone marrow as sanctuary for plasma cells and memory T-cells: implications for adaptive immunity and vaccinology. Cells. 2021;10(6):1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu YJ, Fan CY, Li YK. Protein purification involving a unique auto-cleavage feature of a repeated EAAAK peptide. J Chromatogr B Anal Technol Biomedical life Sci. 2009;877(31):4015–21. [DOI] [PubMed] [Google Scholar]
- 72.Bhattacharya M, Sharma AR, Patra P, Ghosh P, Sharma G, Patra BC, Lee SS, Chakraborty C. Development of epitope-based peptide vaccine against novel coronavirus 2019 (SARS‐COV‐2): Immunoinformatics approach. J Med Virol. 2020;92(6):618–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Soboslay P, Lüder C, Hoffmann W, Michaelis I, Helling G, Heuschkel C, Dreweck C, Blanke C, Pritze S, Banla M. Ivermectin-facilitated immunity in onchocerciasis; activation of parasite‐specific Th1‐type responses with subclinical Onchocerca volvulus infection. Clin Experimental Immunol. 1994;96(2):238–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Venugopal PG, Nutman TB, Semnani RT. Activation and regulation of toll-like receptors (TLRs) by helminth parasites. Immunol Res. 2009;43:252–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kanzler H, Barrat FJ, Hessel EM, Coffman RL. Therapeutic targeting of innate immunity with toll-like receptor agonists and antagonists. Nat Med. 2007;13(5):552–9. [DOI] [PubMed] [Google Scholar]
- 76.Mukherjee S, Mukherjee S, Maiti TK, Bhattacharya S, Sinha Babu SP. A novel ligand of toll-like receptor 4 from the Sheath of Wuchereria bancrofti Microfilaria induces Proinflammatory Response in macrophages. J Infect Dis. 2017;215(6):954–65. [DOI] [PubMed] [Google Scholar]
- 77.Fouzder C, Mukhuty A, Das S, Chattopadhyay D. TLR signaling on protozoan and helminthic parasite infection. Toll-like Receptors; 2019.
- 78.Duan T, Du Y, Xing C, Wang HY, Wang RF. Toll-like receptor signaling and its role in cell-mediated immunity. Front Immunol. 2022;13:812774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen R. Bacterial expression systems for recombinant protein production: E. Coli and beyond. Biotechnol Adv. 2012;30(5):1102–7. [DOI] [PubMed] [Google Scholar]
- 80.Soltan MA, Behairy MY, Abdelkader MS, Albogami S, Fayad E, Eid RA, Darwish KM, Elhady SS, Lotfy AM, Alaa Eldeen M. In silico designing of an epitope-based vaccine against common E. Coli pathotypes. Front Med. 2022;9:829467. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available within the paper and its Supplementary Information.