Abstract
Subsurface environments are among Earth’s largest habitats for microbial life. Yet, until recently, we lacked adequate data to accurately differentiate between globally distributed marine and terrestrial surface and subsurface microbiomes. Here, we analyzed 478 archaeal and 964 bacterial metabarcoding datasets and 147 metagenomes from diverse and widely distributed environments. Microbial diversity is similar in marine and terrestrial microbiomes at local to global scales. However, community composition greatly differs between sea and land, corroborating a phylogenetic divide that mirrors patterns in plant and animal diversity. In contrast, community composition overlaps between surface to subsurface environments supporting a diversity continuum rather than a discrete subsurface biosphere. Differences in microbial life thus seem greater between land and sea than between surface and subsurface. Diversity of terrestrial microbiomes decreases with depth, while marine subsurface diversity and phylogenetic distance to cultured isolates rivals or exceeds that of surface environments. We identify distinct microbial community compositions but similar microbial diversity for Earth’s subsurface and surface environments.
The microbiomes of Earth’s surface and subsurface environments are different in composition, but similar in diversity.
INTRODUCTION
Microbial life is pervasive in Earth’s hugely varied habitats and environments. Microorganisms have adapted to grow in acidic and alkaline springs, salterns, deserts, environments with temperature extremes greater than 122°C or lower than −20°C, and from ambient pressures up to pressures greater than those of abyssal oceanic trenches (1–4). Although surface ecosystems are thought to harbor the majority of Earth’s total biomass (5), most of Earth’s bacterial and archaeal biomass carbon is stored in subsurface ecosystems from less than 1 m to many kilometers beneath Earth’s surface. The local irregularity of boundaries between surface and subsurface ecosystems (6) due to the variability of physicochemical and biological parameters with depth supports only a loose definition of subsurface environments, which include soils, rocks, or sediments deeper than 1 to 8 m below Earth’s land surface or deeper than 0.1 to 1 m below the seafloor (mbsf) (7–12). For the “subsurface” category in this study, we use samples from aquifers, rock fracture fluids, sediments, and rock cores that originated from 0.2 to 491 m below the seafloor or 15 to 4375 m below ground.
At continental margins, especially at the mouths of large rivers, sediment thickness can exceed 10 km (13). Similarly, the habitable continental crust can exceed 15 km in thickness in the Canadian, Fennoscandian, and Siberian shields (7). Archaea, bacteria, and eukaryotic microorganisms inhabit this subsurface biosphere (12). Marine subsurface ecosystems particularly have relatively high proportions and abundances of Archaea compared to most other ecosystems (8, 14–17). Subsurface ecosystems may host more than half of all microbial cells (∼5 to 12 × 1029) on Earth (7, 18–21) despite sometimes very low cell densities, for example, in subsurface sediments of oligotrophic South Pacific Gyre as low as approximately 100 cells per cubic centimeter of sediment (22).
Continuous deposition of organic matter from the overlying ocean and concomitant burial explains the introduction and presence of most microbes in deep sediment layers beneath the ocean floor. Life in marine deep subsurface sediments often bears a resemblance to the shallow subsurface life at a given location (23–26). Terrestrial subsurface ecosystems exhibit a similar connection to the surface world, with entrainment of shallow subsurface communities into the deeper realms via recharge and movement of fluids (27). This similarity exists despite the considerable environmental differences between surface and subsurface environments (i.e., differences in pressure, light, oxygen, energy, and nutrient availabilities, as well as available pore space within which cells might exist) that generally select for distinct microbial communities. Subsurface microbes can disperse via marine and terrestrial aquifers (28, 29), fracture fluids (30), and porewaters (31). Hydrodynamic and hydrogeological processes such as eruptions of mud volcanoes (32, 33) or fluid seepage (34, 35) can reintroduce microbes from subsurface to surface environments.
Intrinsic or acquired capacities to metabolize subsurface energy, carbon, and nutrient sources and/or improved survival and dormancy strategies allow microbes to survive and live in subsurface environments. The predominant occurrence of certain microbial lineages in subsurface ecosystems suggests that some organisms are better equipped for the subsurface than others. Coping strategies for survival may play a role, yet evidence also suggests that with increasing depth, e.g., in marine sediments, microbes change their expressed extracellular peptidases to those that specialize in highly degraded detrital proteins (36). Microbes can also increase cellular lifespan by slowing genome transcription (37) or actively expressing mRNA for DNA repair enzymes (38). Among many other processes acetogenesis (39, 40), methanogenesis (41); hydrogen, methane, and sulfur oxidation (42, 43); fermentation of microbial biomass and necromass (44–46); symbiosis (47, 48); serpentinization (49); and even radiolysis (50, 51) might contribute to the subsistence of deep life. While a portion of cells persist in a dormant state (52), many organisms actively metabolize (22, 53, 54) but often with generation times of decades to centuries (24, 55–58).
While subsurface ecosystems harbor substantial biomass, diversity, and many lineages that are phylogenetically distant to cultured isolates, the degree to which the biodiversity of subsurface ecosystems directly compares with that of surface ecosystems remains unclear. Moreover, although global meta-analyses exist for either the marine (17) or the terrestrial subsurface (59), a standardized global dataset encompassing both marine and terrestrial subsurface environments was not available to date. Our study expands the insights into terrestrial subsurface diversity and composition beyond that of an earlier study that used data from two sequencing platforms (454 pyrosequencing and early Illumina) and multiple 16S ribosomal RNA (rRNA) primers, which thus allowed only a database-dependent operational taxonomic unit (OTU) approach (59). We also extend our observations beyond the bacterial domain (59) including a synthesis of archaeal communities, and we increase the total amount of samples and data, allowing a broader comparison of biomes and diverse environments.
Previous studies have compared marine and terrestrial surface environments suggesting distinct microbiomes between land and sea (60–64). Factors such as salinity, pH, and the availability of nutrients were identified as being among the major drivers of community variance (60, 62, 65–67). These works, however, focused on the principles structuring each habitat (63) on sea and land-derived bioaerosols (64) or on the ocean-land connectivity at specific sites (60–62). A standardized comparison of microbial metabolites across diverse marine and terrestrial environments is available (68), and numerous global surveys have studied either marine (69–71) or terrestrial microbiomes (72, 73). Standardized comparisons of microbial community structure between sea and land, however, have not been published to our knowledge. Datasets that can compare marine and terrestrial microbiomes on a global scale are available (67, 74, 75) yet were analyzed with a different focus. To understand the differences and commonalities in the microbiomes of global surface and subsurface environments, we here provide a comparison between surface and subsurface as well as between marine and terrestrial environments. We investigate environments that span a broad range of depths from surface environments (under the influence of relatively fresh photosynthesis-derived organic matter) to very deep and isolated environments (cutoff from photosynthetic primary production for at least centuries). For this global comparison, we analyzed 1442 globally distributed 16S rRNA gene amplicon datasets and taxonomic marker genes of 147 metagenomes. We investigate (i) whether microbial communities of marine and terrestrial biomes and of surface and subsurface environments fundamentally differ, (ii) whether subsurface environments are generally less diverse than surface ecosystems, (iii) whether subsurface environments harbor distinct clades, and (iv) whether marine and terrestrial subsurface communities share a core microbiome.
RESULTS AND DISCUSSION
The extended Census of Deep Life atlas allows comparisons of surface and subsurface biodiversity
The Census of Deep Life (CoDL) under the auspices of the Deep Carbon Observatory organized a decadal effort (2010–2020) to characterize microbial diversity and function in subsurface ecosystems worldwide, carried out by more than two dozen groups and hundreds of researchers. After the completion of the CoDL, we compiled, re-analyzed, and compared 964 bacterial and 478 archaeal 16S rRNA gene amplicon datasets from 35 individual globally distributed CoDL projects together with 15 additional non-CoDL projects that originated mainly from surface ecosystems (Fig. 1, A and B, and datasets S1 and S2). Archaeal and bacterial 16S rRNA gene amplicon sequence variants (ASVs; quasi–strain-level microbial lineages) were amplified using domain-specific primers. We obtained 31,099 unique archaeal and 377,374 unique bacterial ASVs from a total of 4.9 × 107 archaeal and 9.6 × 107 bacterial reads. On average, we found 155 archaeal and 1183 bacterial ASVs per sample (datasets S3 and S4). Analyses of 147 metagenomes from 49 globally distributed CoDL subsurface projects complement the amplicon datasets. The metagenomes were used for taxonomic analyses based on two different taxonomic markers, 16S rRNA gene sequences retrieved by phyloFlash (pF16S; before read assembly) and ribosomal protein S3 (rpS3) genes (after read assembly). We obtained a total of 1552 unique archaeal, 23,206 unique bacterial, and 1387 unique eukaryotic pF16S genes, as well as 154 different archaeal and 1601 different bacterial rpS3 genes (datasets S5 and S6).
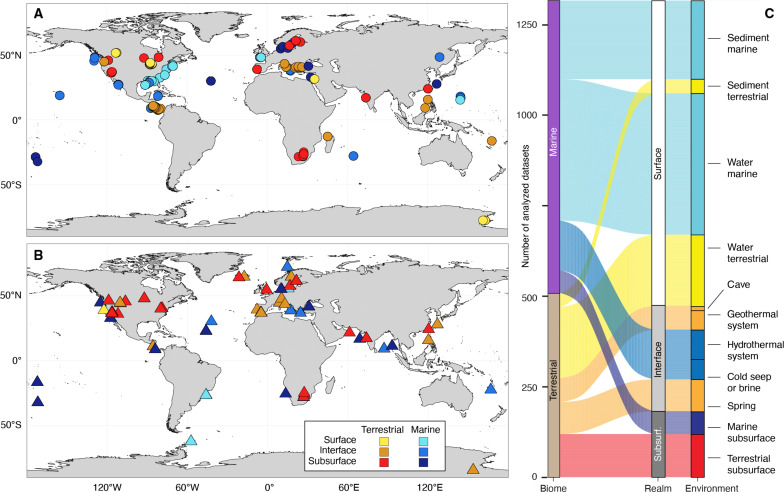
Fig. 1. Geographic location and origin of samples.
Maps of samples used for metabarcoding of 16S rRNA gene amplicon taxonomic marker genes (A) and shotgun metagenomic analyses of unassembled and de novo assembled taxonomic marker genes (B). Each symbol represents one project, which comprises multiple individual samples. Both terrestrial and marine samples may contain rock, sediment, or water samples. Further maps showing sample material, pH, and temperature are included in fig. S1. (C) Overview of sample origins derived from marine and terrestrial biomes, depth realms, and environments.
We analyzed the communities first by grouping the amplicon datasets based on the biome from which the samples originated: marine biome (nArchaea = 304 datasets, nBacteria = 503 datasets) and terrestrial biome (nArchaea = 174, nBacteria = 461). Within the marine and terrestrial biome, we grouped samples based on the depth realm they originated from: surface, interface, and subsurface.
Surface datasets (nArchaea = 183, nBacteria = 599) include water samples from oceans and lakes (at various depths in the water column) and shallow (<0.1 mbsf) sediment samples from oceans, estuaries, and lakes. Surface ecosystems represent environments under the influence of relatively fresh photosynthesis-derived organic matter. Subsurface datasets (nArchaea = 85, nBacteria = 122) originated from ecosystems that have likely been cut-off from photosynthetic primary production for decades to centuries. These ecosystems were accessed via boreholes or mines and include deep sediments, aquifers, and fracture fluids. Marine subsurface sediments were further subdivided based on depositional setting: shelf, slope, and abyssal domains. The location of each domain is defined by water depth (76–78). Shelf environments roughly correspond to water depths <200 m, except the Antarctic region where shelf area corresponds to water depths <500 m. The abyssal plain corresponds to areas of water depths >3500 m. Sediments under other water depths are referred to as slopes. Interface datasets (nArchaea = 210, nBacteria = 243) originated from environments that are subjected to influences from surface and subsurface environments and processes. Interface environments can be surface sites that are driven by energy from the subsurface, or vice versa, and included water and sediment samples from caves, hot springs, hydrothermal vents, and cold seeps. We also grouped and analyzed the samples based on the 11 major environments they originated from, as listed in Fig. 1C.
We included samples from four different types of environmental materials: water or brine (724 samples), biofilm or mat (35 samples), sediment or soil (458 samples), and rock (102 samples). Samples from different materials were dispersed relatively homogeneous across the different biomes (figs. S2 and S3) and depth realms (figs. S4 and S5) to minimize diversity effects caused by the sample material. Sediments had the highest alpha diversity, while rocks had the lowest, in both the marine and terrestrial biome. To minimize batch effects and biases, we sequenced and analyzed all surface, interface, and subsurface samples using the same archaeal or bacterial primers, the same chemistry, the same Illumina instrument, and the same bioinformatic pipeline. We minimized the potential impact of contamination by including blanks and controls and by removing notorious contaminants listed in the Supplementary Materials (79, 80). We minimized composition bias by normalizing template concentration and by standardizing workflows (81–83). Our dataset can deeply sample microbial richness and evenness, compare microbiomes at high taxonomic resolution, and study community similarities and shifts across large scales. Future work could expand our analyses to explore changes in the subsurface microbiome over time, perform long read 16S rRNA gene sequencing to achieve higher phylogenetic resolution, or use metagenomics to assess metabolic capabilities. These analyses have been done at individual sites (37, 84, 85) and promise—at a global scale—to greatly expand our understanding of the roles and nature of subsurface life.
Substantial differences exist between marine and terrestrial microbiomes
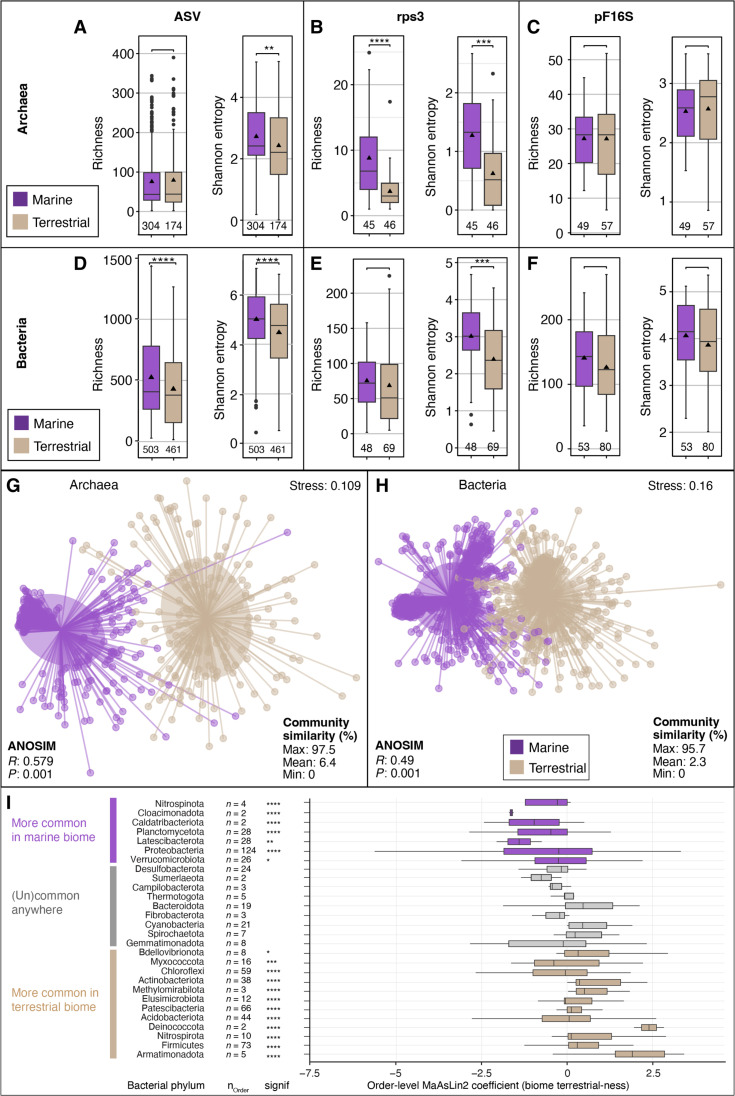
We found substantial differences in archaeal and bacterial community structure between marine and terrestrial biomes. For this analysis, we grouped all datasets solely based on whether they originated from a marine or terrestrial environment, regardless of their sample material or association with a surface, interface, or subsurface environment. Alpha diversity (community diversity per locality/sample) and beta diversity (community diversity between localities/samples) of marine and terrestrial microbiomes showed the same trends across ASV, rpS3, and pF16S-based analyses. Richness, estimated richness (Chao1), and evenness (Shannon entropy and inverse Simpson diversity) were similar or higher for archaea and bacteria in marine relative to terrestrial microbiomes (Fig. 2, A to F, and fig. S6). The high diversity in the marine biome was caused in both domains by sediments and rocks, which are more diverse in the sea than on land. Marine waters were similarly diverse as terrestrial waters for bacteria and slightly higher for archaea, whereas biofilms and microbial mats tended to be higher in diversity on land for both domains (figs. S2 and S3). In the subsampled datasets, which was done to account for the unequal sampling effort, i.e., different sequencing depth, we found, on average, 75 and 79 archaeal and 512 and 349 bacterial ASVs in marine and terrestrial samples, respectively (dataset S7). Microbial community structure and composition were overlapping but significantly different between the marine and terrestrial biomes (Fig. 2, G and H; RArc: 0.58, PArc: 0.001; RBac: 0.49, PBac: 0.001). On average, only 2 to 6% of ASVs, 2% of rpS3 genes, and 8 to 9% of pF16S genes were shared between any marine and terrestrial dataset (Fig. 2, G and H, and figs. S7 and S8). ASV-based gamma diversity (total community diversity per biome/environment) tended to be higher for archaea in the terrestrial biome and for the bacteria in the marine biome (figs. S6 and S7).
Fig. 2. Microbial diversity in marine and terrestrial biome.
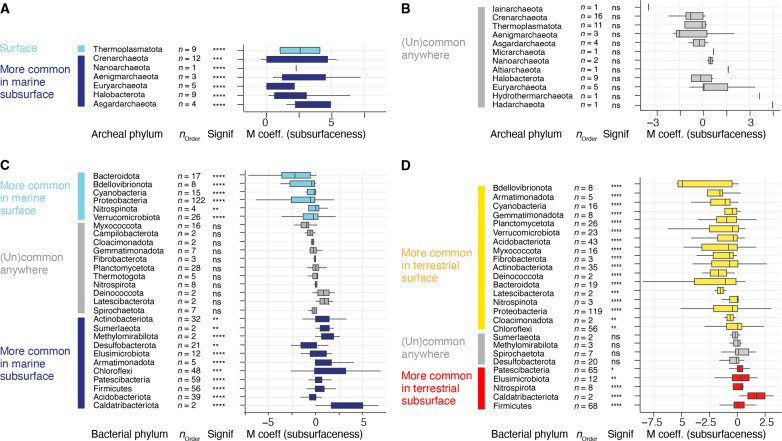
Archaeal (A to C) and bacterial (D to F) alpha diversity (per sample community richness and evenness) in marine and terrestrial biomes using 16S rRNA gene ASVs [(A) and (D)], as well as metagenome-derived ribosomal protein S3 genes [rpS3; (B) and (E)] and 16S rRNA gene sequences detected by phyloFlash (pF16S). To allow comparison, the datasets were subsampled to the same number of reads. Pairwise comparisons were performed using a Wilcoxon rank sum test. Significance: **P < 0.01, ***P < 0.001, ****P < 0.0001. The number of analyzed datasets is shown below the boxplots. Community dissimilarity between marine and terrestrial communities was shown by nonmetric multidimensional scaling ordinations of ASV-based dissimilarity matrices using 478 archaeal (G) and 964 bacterial datasets (H). Each dot represents the community structure of a dataset and is connected to the group centroid (weighted average mean of within-group distances); ellipses depict 1 SD of the centroid. Statistical testing using ANOSIM showed that the groups are overlapping but significantly different (R ∼ 0.5, P < 0.001). (I) Differential sequence abundance analyses of marine versus terrestrial bacterial phyla. The phyla are ordered from top to bottom based on increasing phylum level MaAsLin2 coefficient, i.e., likeliness of their occurrence in terrestrial-derived samples (“terrestrialness”). Boxplots summarize order level MaAsLin2 coefficients, i.e., terrestrialness, within the listed phyla. Note that due to ease of visualization, boxplots are also shown for very small number (n) of orders. Significance levels are: *P < 0.01, **P < 0.001, ***P < 0.0001, and ****P < 0.00001. Additional phyla, particularly those that lack cultured representatives, are shown in fig. S6.
A multivariate association analysis (86) of our dataset suggests that most bacterial phyla are significantly more sequence abundant and prevalent in the marine biome (fig. S9A). Phyla that are more common in marine environments include well-known Proteobacteria, Planctomycetota, and Verrucomicrobiota, but also less prominent phyla such as Marinimicrobia, Nitrospinota, Fermentibacterota, Caldatribacteriota, Latescibacterota, Aerophobota, and Gemmatimonadota (Fig. 2I and fig. S9A). In terrestrial environments, we found more commonly Firmicutes, Chloroflexi, Acidobacteriota, Actinobacteriota, Nitrospirota, and Patescibacteria, as well as Methylomirabilota, Armatimonadota, and Elusimicrobiota among others. Both the marine- and terrestrial-associated phyla comprise members of the two major monophyletic branches of the bacterial tree Gracilicutes (mostly diderm lineages) and Terrabacteria (monoderm and atypical diderm lineages), respectively (87). We also find a versatile third group of bacterial phyla that occupy both biomes at comparable relative sequence abundances (RSAs) and prevalences, also regardless of being mono- or diderms. These include Desulfobacteria, Bacteroidota, Cyanobacteria, Spirochaetota, Gemmatimonadota, Campilobacterota, and Thermotogota. Hence, despite the marked differences between sea and land at ASV, species, and genus level, there are cosmopolitan lineages but at higher phylogenetic levels (e.g., phylum and class). Most archaeal phyla were more sequence abundant and prevalent in the terrestrial biome (fig. S9B). Only Thermoplasmatota were significantly more common in marine environments, and Crenarchaeota and Asgardarchaeota were found averaged across each biome at similar RSAs and prevalence. All other archaeal phyla including Euryarchaeota, Halobacterota, Hadarchaeota, Nanoarchaeota, Aenigmarchaeota, and Altiarchaeota were more sequence abundant and prevalent in the terrestrial biome. Relative sequence abundances of archaeal and bacterial phyla (fig. S9, C and D) support the trend revealed by the association analyses; however, association analyses or differential abundance analyses can change with every sample and lineage added or removed and thus must be interpreted with caution. Robust affiliations to a certain biome may only be universally true for the lineages with highest significances, those at the very top and bottom of the graph. Some phyla contribute disproportionally to ASV-level diversity including Nitrososphaeria, Bathyarchaeia, Chloroflexi, and Planctomycetota (fig. S9, E and F), being responsible for a larger part of the ASV-level diversity than their RSA suggests.
Subsurface microbiomes can be as diverse as surface microbiomes
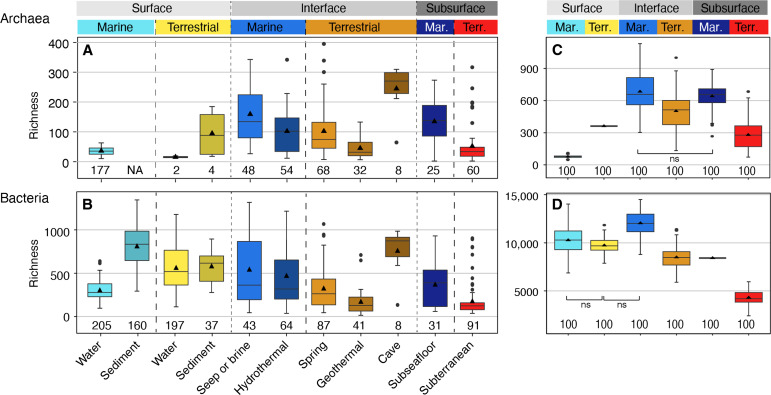
Within each biome, differences in alpha diversity between surface, interface, and subsurface environments, i.e., depth realms, were overall less pronounced than between the biomes (figs. S10 and S11). Across depth realms, communities of either biome—marine or terrestrial—shared many ASVs (figs. S12 and S13, and movies S1 and S2). Archaeal and bacterial communities in the three depth realms were distinct but with considerable overlaps. The larger differences in community structure between marine and terrestrial biomes than between depth realms indicate that the divide between microbial life on land and in the sea is more pronounced than the divide between surface and subsurface communities. We found that species richness and evenness in many subsurface environments rival those in surface environments (fig. S14). This finding was consistent across the 11 investigated environment types, where levels of microbial diversity are often comparable across the surface, interface, and subsurface (Fig. 3, A to D), i.e., being within the same order of magnitude. Archaeal alpha diversity was overall highest in samples from cold seeps and brines, caves, springs, and the deep marine subsurface (Fig. 3A). Bacterial alpha diversity was, on average, highest in the cave samples and marine sediments, followed by terrestrial water and sediment, seeps and vents. Marine subsurface bacterial diversity rivaled the diversity found in springs and marine surface water (Fig. 3B). Total archaeal diversity (gamma diversity) was significantly higher in marine interface and subsurface environments than in the marine surface, even after correcting for the different numbers of samples per group using a subsampling approach (Fig. 3C and fig. S10, A to D). Similarly, bacterial gamma diversity was highest in marine interface environments (Fig. 3D). In the terrestrial biome, archaeal gamma diversity was comparable across all depth realms (Fig. 3C), whereas bacterial diversity was highest in the surface (Fig. 3D). Overall, archaeal and bacterial diversity was only sampled exhaustively in marine waters based on species accumulation curves (fig. S15), corroborating previous findings that vast numbers of microbial species remain to be found across global biomes (11, 74, 88–90).
Fig. 3. Observed archaeal and bacterial richness across environments based on 16S rRNA gene ASVs.
Archaeal (A) and bacterial (B) richness found in the 11 studied environments (n: number of included samples). Environments are grouped on the basis of biome (marine, terrestrial) and depth realm (surface, interface, subsurface). Note that Archaeal terrestrial water and sediment samples contain too few datapoints for robust visualization using boxplots, yet the plots were retained for completeness. NA, no value available. Total archaeal (C) and bacterial richness (D) using a subsampling approach to account for different group sizes (100 iterations using 1142 archaeal or 2271 bacterial reads, respectively). Normalized gamma diversity corroborates that archaeal diversity is highest in marine interface and subsurface ecosystems even when different group sizes are considered. All pairwise comparisons were significantly different (P < 0.001) except for those indicated with ns (not significant). Shannon entropy values and a subsampling approach using 50,000 reads show the same trends (fig. S11). The diversity found in individual subsurface environments and their comparison to surface and interface environments is shown in the fig. S14.
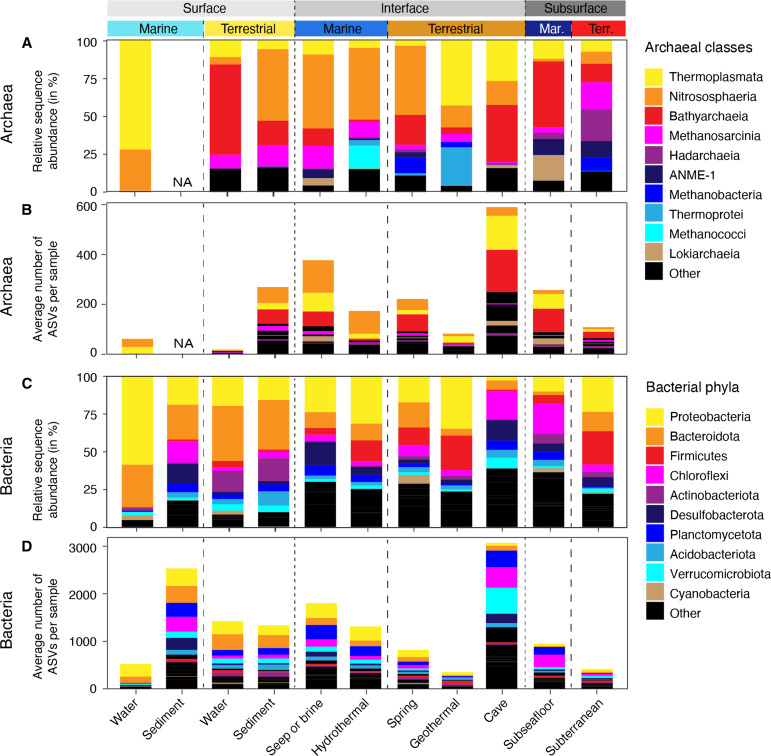
Thermoplasmata, Nitrososphaeria, and Bathyarchaeia together comprised more than half of the archaeal RSAs in any investigated environment, except the terrestrial subsurface (Fig. 4A and fig. S16). The contribution of these three classes to global archaeal ASV richness was similarly outsized (Fig. 4B), while other clades that can occur at relatively high RSAs contributed very little to overall archaeal richness, including Hadarchaea, ANME-1, Methanobacteria, Thermoprotei, and Methanococci (Fig. 4, A and B, and fig. S16). This suggests that a large part—if not most—of the global archaeal abundance and richness can be attributed to three archaeal classes Thermoplasmata, Nitrososphaeria, and Bathyarchaeia. Bacterial communities were largely dominated by Proteobacteria and Bacteroidota, except those of caves and the subseafloor where Chloroflexi played an important role (Fig. 4C). The contribution of the lineages to overall bacterial richness was often not proportional to their RSA. Proteobacteria, Bacteroidota, Firmicutes, Cyanobacteria, and Desulfobacterota contributed less, while Chloroflexi, Planctomycetota, and Verrucomicrobia contributed more richness than expected based on their sequence abundance (Fig. 4D). We found increasing archaeal richness and evenness with depth, supporting the notion that archaea are well suited for life in subsurface and subsurface-influenced, i.e., interface, environments. The importance of archaea in marine subsurface ecosystems has previously been demonstrated by studies assessing community structure using lipid analyses (8), cell abundances (16), and meta-omics (14, 91). A high diversity of archaea in the terrestrial subsurface, rivaling that of terrestrial surface ecosystems, is less well documented. This work robustly shows that average archaeal diversity in the terrestrial subsurface is often equal to and can even exceed the diversity found at terrestrial surface environments, a finding that contextualizes the high archaeal diversity described in previous studies of the terrestrial subsurface (59, 92, 93). Similarly unexpected is the high diversity of bacteria in the marine subsurface in comparison to surface ecosystems, supporting previous findings that focused on marine sediments (17). This work collectively improves our understanding of the contribution of archaea and bacteria to global microbial diversity and ecosystem function. To further investigate the global trend and corroborate the high diversity of archaeal and bacterial diversity in marine subsurface ecosystems, future studies should include datasets of additional surface environments such as soils and freshwater sediments, as those were underrepresented in our datasets.
Fig. 4. Relative sequence abundance and richness of most important lineages across environments.
Relative sequence abundance of top 10 most sequence abundant archaeal classes (A) and bacterial phyla (C) in the 11 studied environments. Contribution of most sequence abundant lineages to average number of archaeal (B) and bacterial (D) ASVs per sample (richness). Note that the most abundant clades do not necessarily show the highest richness, e.g., Planctomycetota are the seventh most sequence abundant clade [(C)] yet the third most diverse [(D)].
Interface and subsurface microbiomes are phylogenetically more diverse in marine than in terrestrial biomes
Average microbial species-level diversity was significantly higher in marine interface and subsurface environments than in terrestrial interface and subsurface environments (Fig. 3, C and D, and fig. S10). This was the case for archaea and bacteria and for all tested alpha diversity indices concerning richness, evenness, and estimated richness, as well as gamma diversity. We corroborated this ASV-based trend through comparisons of the diversity of rpS3 and pF16S genes (fig. S10), for which marine interface and subsurface microbiomes were also either significantly more diverse or statistically indistinguishable. Beta diversity between the marine and terrestrial subsurface was also high based on ASVs (fig. S12, A and B), showing that analogous to surface ecosystems marine and terrestrial subsurface environments harbor many unique microbial species and comprise fundamentally different habitats. Generally, the average community dissimilarity between terrestrial samples was greater than between marine samples. The processes that lead to lower richness and greater dissimilarity in the terrestrial subsurface cannot be explained with our dataset but could be due to fundamental properties of the environment, including high habitat heterogeneity, high disturbance, or low dispersal. The diversity trends were also supported by rpS3 and pF16S gene-based analyses (fig. S12, C and D).
Certain microbial lineages are globally abundant in the subsurface
Archaeal and bacterial communities differed between the surface and subsurface environments regarding composition, RSA and prevalence, i.e., the percentage of samples in which a lineage occurred (Figs. 5 and 6 and figs. S16 and S17). Certain archaeal and bacterial lineages were significantly more prevalent in samples from subsurface than surface ecosystems (fig. S18). In the marine biome, most archaeal phyla including Euryarchaeota and Asgardarchaeota predominantly occurred in the subsurface (Fig. 5A), except for Thermoplasmatota, which are very abundant in the ocean. Terrestrial archaeal phyla did not have a significant preferential occurrence in surface or subsurface environments (Fig. 5B). Among the bacteria, the phyla Cyanobacteria, Bdellovibrionota, Verrucomicrobiota, Planctomycetota, Bacteroidota, and Proteobacteria are statistically more common in surface ecosystems of the marine and terrestrial biome (Fig. 5, C and D, and figs. S19, S20, and S21). Firmicutes, Caldatribacteriota, Elusimicrobiota, and Patescibacteria are among those that are more likely found in the marine and terrestrial subsurface (Fig. 5, C and D, and figs. S17 and S18). In the marine subsurface, common phyla include Aerophobota, Chloroflexi, Desulfobacterota, and Methylomirabilota, while Nitrospirota was particularly common in the terrestrial subsurface. This finding supports previous reports of microbes that preferentially occur in deep subsurface environments including the sulfate-reducing firmicute Candidatus Desulforudis audaxviator (94–96), certain Spirochaeta (97), or organisms affiliating with (Cald)atribacteriota (17, 98–104). In line with previous studies, we show that Proteobacteria are ubiquitous and often dominant in marine (17) and terrestrial subsurface (59) ecosystems analogous to their well-documented abundance in surface ecosystems (Fig. 5 and figs. S19 to S21) (74).
Fig. 5. Multivariate association analyses of microbial lineages.
Analyses compare the occurrence of archaeal (A and B) bacterial (C and D) in marine [(A) and (C)] and terrestrial [(B) and (D)] surface versus subsurface realms. The phyla are ordered from top to bottom based on increasing likeliness of their occurrence in subsurface-derived samples (increasing MaAsLin2 coefficients; “subsurfaceness”). Boxplots summarize order level MaAsLin2 coefficients within the listed phyla. Note that due to ease of visualization, boxplots are even shown for very small number (n) of orders. The significance of the MaAsLin2 phylum level coefficient is shown in the column denoted “signif.” Significance levels are: not significant (ns), *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. An analysis based on all surface versus subsurface samples regardless of the biome of origin is shown in fig. S18, and additional bacterial phyla are shown in figs. S19 to S21.
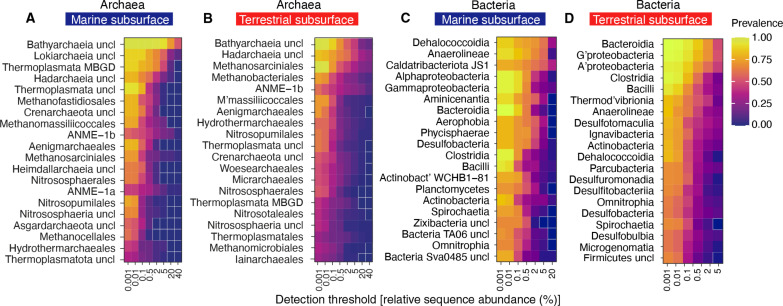
Fig. 6. Subsurface core microbiomes.
The heatmaps show potential archaeal order level core microbiomes in the marine (A) and terrestrial subsurface (B), as well as bacterial class level core microbiomes in the marine (C) and terrestrial subsurface (D). The 20 most relevant lineages, regarding their prevalence and RSAs, are shown as rows, and each column color represents the lineages prevalence at a certain RSA threshold. Uncultured/unclassified lineages are denoted by “uncl,” and the closest known phylogenetic level or the closest phylogenetic level with an isolated representative is shown. For example, Bathyarchaeia uncl are all uncultured/unclassified/order-level clades in the class Bathyarchaeia. Fields that are zero have a white outline.
We have identified the most common subsurface class-level bacteria and order-level archaea based on their RSAs and prevalence and defined four categories. The first category comprises lineages with high RSAs and a high prevalence, e.g., Bathyarchaeia in the marine subsurface or Gammaproteobacteria in the terrestrial subsurface, agreeing with findings of earlier studies (17, 59). Both lineages occur in all samples of the respective biome (prevalence = 1) at up to 2% RSA (Fig. 6, A and D). Bathyarchaeia still occur at more than half of all marine subsurface samples (prevalence > 0.5) with RSAs of >40% (Fig. 6A), underlining their role as key players in the subsurface (105). The second category comprises lineages that have a high prevalence and low RSAs, i.e., they occur at almost all sites but are mostly rare. This category comprises terrestrial subsurface Bathyarchaeia and Methanosarciniales, as well as marine Gammaproteobacteria. Bathyarchaeia and Gammaproteobacteria were shown to be abundant and diverse by 16S rRNA gene amplicon studies in the marine (17, 105, 106) and terrestrial (59, 107, 108) subsurface biome, respectively. Our work finds that both lineages are in fact abundant and/or prevalent in all studied subsurface environments, further highlighting them as two of Earth’s most widespread and sequence abundant microbial phyla. Remarkably, many other lineages also occur globally in marine and terrestrial subsurface environments at high prevalence and low RSA, including Bacilli, Clostridia, Bacteroidia, and Alphaproteobacteria (Fig. 6, C and D) and are thus truly cosmopolitan, albeit being mostly rare. Lineages of categories 1 and 2, being highly prevalent, likely comprise many generalists that have adapted to diverse habitats. Consistent with this idea, we find that, e.g., Nitrosopumilus, Methanosaeta, Pseudomonas, Bacillus, and Desulfovibrio are among the most common genera in the subsurface (figs. S22 and S23). The third category features those with low prevalence and high sequence abundance, which means that they do not occur at all sites, but when they occur, they are very sequence abundant or even dominant. This pattern fits to a specialist lifestyle in which organisms are very successful only in select habitats to which they are well adapted. This category includes marine Lokiarchaeia and ANME-1a, as well as Hadarchaeia and ANME-1b, and the latter too exhibit this distribution in both the marine and terrestrial subsurface. In the bacterial domain, examples include JS1 Caldatribacteriota, which constitute up to 20% of the community in about half of the marine subsurface samples (Fig. 6C), as well as marine Dehalococcoidia, Aminicenantia, Aerophophia, Phycisphaerae, and Desulfobacteria. The fourth category represents organisms with low prevalences and low sequence abundances. These lineages are generally rare, and their occurrence may be stochastic. Depending on an emphasis on prevalence or sequence abundance, the lineages in the first three categories, which are prevalent or abundant or both, can be interpreted as a core microbiome of the marine or terrestrial subsurface at the given phylogenetic level. Lineages that occur in our dataset widely, almost exclusively, in the marine subsurface include Lokiarchaeia, Heimdallarchaeia, Methanofastidiosales, Caldatribacteriota, Phycisphaerae, Aerophobia, and Aminicenantia. Lineages that occurred in our dataset almost exclusively in the terrestrial subsurface include Woesechaeales, Iainchaeales, Micrarchaeales, Thermodesulfovibrionia, Desulfotomaculia, and Ignavibacteria, a finding supported by previous studies (42, 92). Although there is little community overlap between subsurface environments at species level, there are lineages at higher phylogenetic levels that occur in the marine and the terrestrial subsurface biome, including Bathyarchaeia, Hadarchaeia, ANME-1b, Alphaproteobacteria, Gammaproteobacteria, Anaerolineae, Bacteroidia, Bacilli, and Clostridia, which can be considered a global subsurface core microbiome at the given phylogenetic level.
Phylogenetic distance relative to cultured isolates is high in the subsurface
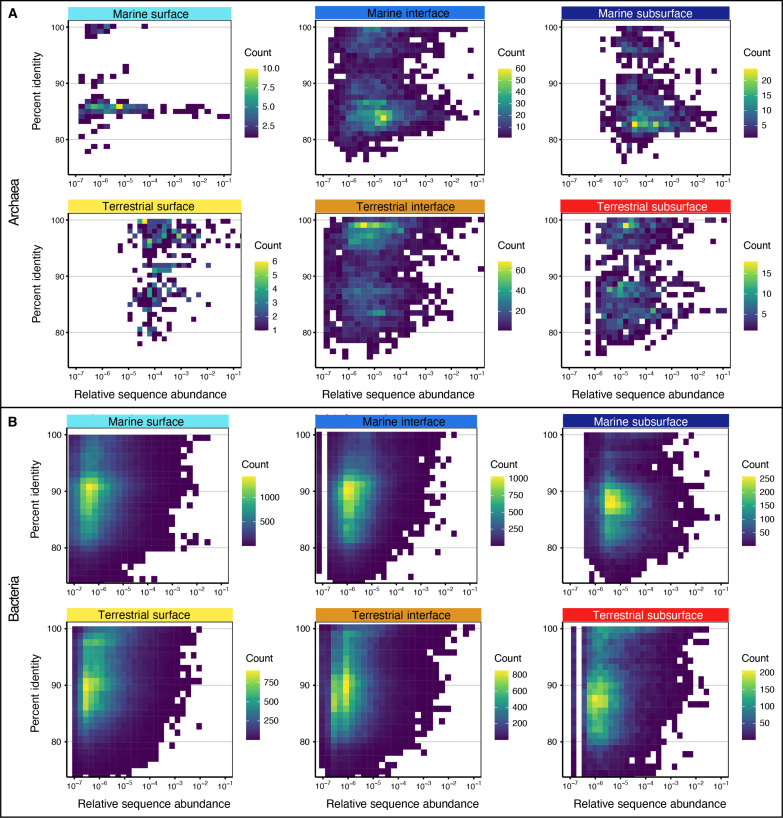
Many lineages that are abundant in Earth’s environments belong to branches in the tree of life that have few or no cultured representatives (74, 109). This is particularly true for lineages that are abundant in subsurface environments, e.g., Lokiarchaeia, Bathyarchaeia, Hadarchaeia, and Caldatribacteriota. Together, with these lineages often outsized contribution to richness (Fig. 4, B and D), it is likely that the subsurface holds a substantial degree of uncharted phylogenetic and likely metabolic diversity. We thus analyzed the phylogenetic distance of the detected lineages to their closest isolated relatives. For this, we mapped ASV against a SILVA reference database of cultivated isolates. Our analyses show that most archaeal ASVs in the marine subsurface share only 80 to 90% sequence identity to the closest isolated relative (Fig. 7A), on average 88% (fig. S24A), indicating that many uncharted family-, order-, and class-level clades can be expected (110). In the marine surface and interface and in terrestrial environments, archaeal ASVs are, on average, more closely related to a cultured isolate (95 to 97% sequence identity; Fig. 7A and fig. S24), nevertheless representing potential unseen lineages. In the case of bacteria, average sequence identity to the closest isolate is also lowest in the marine subsurface (88% on average), supporting the notion of extensive phylogenetically distant lineages in the seafloor (17). However, bacterial phylogenetic distance relative to cultured organisms is similarly high in all other environments, with most ASV being only ∼90% sequentially identical to the closest isolate (Fig. 7B), ranging, on average, from 89 to 92% (fig. S24). Notably, many of the ASVs that are phylogenetically distant from their closest cultured relative have high RSAs (Fig. 7). This is especially true for Archaea, for which several phylogenetically distant ASV had RSAs between 1 and 10% (Fig. 7A). These findings support the notion of a largely untapped reservoir of uncultivated archaeal diversity in the subsurface (111). However, our findings also highlight the sequence abundance of lineages phylogenetically distant from the next isolate in most other environments, particularly in the bacterial domain. This supports findings of a previous global study in which most microbial population genomes were distantly related to reference genomes, suggesting that uncultivated organisms dominate the diversity within most phyla and environments (74).
Fig. 7. Microbial phylogenetic distance relative to cultured organisms.
Percent identity values (PIVs) of archaeal (A) and bacterial (B) ASVs relative to their closest cultured relative for the studied biomes and realms. Each square in the density plot represents one ASV and depicts its PIV (y axis) and RSA (x axis, logarithmic). The color gradient shows how many ASVs have identical PIV and RSA. The more yellow, the more ASVs are represented by the square.
The CoDL atlas reveals global continua and great divides of microbial diversity
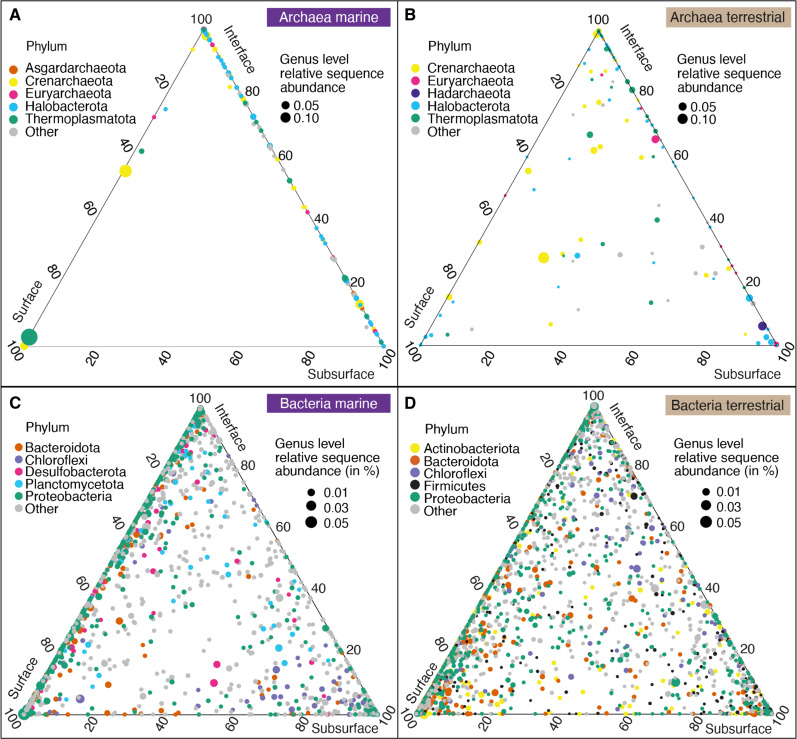
Overall, we demonstrate that communities in the marine and terrestrial subsurface can be as phylogenetically diverse as those on the surface, harboring a substantial part of Earth’s biodiversity. This finding confirms previous insights from global marine sediments (17) and the terrestrial subsurface (59) yet compares the two distinct biomes in one global analysis. Our comparison of marine and terrestrial microbiomes and depth realms shows that microbial diversity is particularly high in interface environments that are influenced by surface and subsurface processes. The communities at interface ecosystems shared community membership with those of surface and subsurface environments, which reflects the location, as well as the exchange of fluids and other materials between the depth realms. Mud volcanoes and methane seeps have been shown to connect surface and subsurface environments (32–34), as have marine crustal fluids (112, 113). Archaeal interface communities shared more genus-level clades with the subsurface (Fig. 8, A and B), whereas bacterial interface communities shared more genus-level clades with the surface (Fig. 8, C and D), suggesting that different ecological processes shape archaeal and bacterial communities at these sites.
Fig. 8. Genus-level diversity within and between depth realms.
Ternary plots show archaeal (A and B) and bacterial genus-level diversity (C and D) in the marine [(A) and (C)] and terrestrial biome [(B) and (D)]. Each circle is a genus-level clade, circle size is average RSA in the respective biome, and circle colors represent to which of the top 5 phyla the lineage belongs. The location of the circle shows the proportional average RSA in each depth realm (surface, interface and subsurface) scaled to sum 100%. For example, if a circle lies exactly in the center of the plot, then the respective lineage has an equal RSA in each of the three depths, if it lies at one of the corners (e.g., surface), then it means it occurs only in the surface, and circles that lie on the sides of the plot occur in only two of the three depths.
Despite harboring many distinct microbial lineages, the microbiomes of surface, interface, and subsurface environments showed a relatively high overlap in community composition. In previous studies of individual sites and sediment cores, it was shown that deep marine sediment layers contain communities similar to the surface seeded at the seafloor but getting more different with depth (24–26). Our standardized dataset provides further evidence at a large scale for such a diversity continuum with depth (fig. S12 and movies S1 and S2) that connects surface and subsurface communities across environments and deep time. Moreover, many of the environments we studied cannot be easily defined as surface or subsurface environment. Subsurface or subsurface-impacted ecosystems are not necessarily energy depleted and, in fact, can be relatively energy rich, such as hydrocarbon seeps (114); brines (115); hydrothermal vents (116); oil, coal, and shale deposits (43, 56, 117); serpentinizing systems (118–120); and caves (121). The subsurface is often replete with methane, hydrogen, and other energy and carbon sources (50, 122–125). An absence of oxidants, nutrients, trace metals, or extreme physical or chemical conditions, however, can shape the specific community structures and hinder microbial growth or activity in relatively energy-rich ecosystems (126, 127). Depth and oxygen content are also not always reliable proxies for subsurface conditions, e.g., in marine sediments with very low organic matter content, oxic conditions can prevail throughout the sediment column (128) as can oxygen be produced biotically or abiotically in subsurface ecosystems (129). Sediment depth and sediment age can be greatly different between locations (77), with past surface conditions or depositional environments (fig. S25) apparently being imprinted into a given subsurface sample (25, 26). Defining environments and samples by a few select parameters such as depth, oxygen content, or age is useful, but to fully understand the subsurface geobiosphere, we may need to consider large-scale gradients and continua, analogous to the concept of the critical zone in soil science (130, 131).
Our analyses further corroborate a major divide between marine and terrestrial microbiomes, showing significant differences in local and overall richness. In contrast to the studied depth realms, the differences between marine and terrestrial microbiomes were notable, with little to no overlap in community structure (Fig. 2 and fig. S6) between land- and sea-derived samples. This divide was also found on the basis of the analysis of metagenome-derived 16S rRNA and rpS3 genes (Fig. 2 and fig. S8). The taxonomic differentiation between marine and terrestrial microbial communities mirrors the differences between animal communities in these biomes (132) and is likely caused by the very different chemical and physical properties that shape these environments (133). The environmental drivers responsible for the gradients, continua, and divides remain undefined, but they likely include factors such as salinity (65, 66), energy availability (78), geological activity (134, 135), hydrologic conditions and recharge rates (136, 137), and constrictions of void space within which microbial cells might exist (127). Improvements in global scale modeling and increasing availability of data can provide the tools to describe the surface and subsurface on large scales, refine our insights into microbial provinces in the subsurface (138), and potentially define regions of similar subsurface biogeochemical regimes analogous to Longhurst provinces for the surface ocean (139).
MATERIAL AND METHODS
Dataset specifications
To investigate the microbial diversity and composition of surface, interface, and subsurface microbiomes, we used 478 archaeal and 964 bacterial 16S rRNA gene amplicon datasets sequenced at the W. M. Keck Ecological and Evolutionary Genetics Facility of the Marine Biological Laboratory (Woods Hole, MA, USA). Most archaeal and bacterial datasets from subsurface and interface environments were part of the CoDL, and most datasets from surface ecosystems were not collected during the CoDL but were shared by numerous other investigators (see Acknowledgements). All sequencing datasets were prepared using identical primers, nearly identical library preparation protocols, identical sequencing chemistries, and bioinformatic analyses to minimize batch effects and biases and ensure comparability of communities based on ASVs. We removed datasets from enrichment samples, blanks, and controls, as well as datasets that we considered as failed runs (<2000 bacterial reads or <1000 archaeal reads) and subsurface datasets that contained lineages belonging to known contaminants of subsurface samples (80). Contextual data for each sample/sequence dataset is listed in dataset S2. Datasets S8 and S9, representing the archaeal and bacterial analysis logs using the “biome” grouping as a representative workflow). We are aware that certain environments are overrepresented (e.g., marine surface), while others are underrepresented (e.g., caves) or missing (e.g., soils). However, the highlighted global trends are likely reliable due to the size and breadth of the dataset, the comparability of environments, and the robustness of the results (e.g., toward the removal of reads; fig. S11).
16S rRNA gene library preparation and amplicon sequencing
Genomic DNA was extracted by the laboratories responsible for each project (dataset S1). DNA extraction could not be standardized due to commercial and logistical obstacles and laboratory-specific scientific practices and instrumentation. It was shown that DNA extraction kits can affect diversity indices and community structure (140); however, as this work comprises datasets from more than 50 laboratories worldwide, we can very likely exclude a systematic extraction bias, i.e., most/all samples of a certain tested category were extracted with the same method, which differs from the method used for samples of other categories. Sequencing was performed at the W. M. Keck Ecological and Evolutionary Genetics Facility at MBL. The bacterial 16S rRNA gene V4-V5 variable region was amplified using the forward primer 518F (5′-CCAGCAGCYGCGGTAAN-3′) and reverse primers 926R (5′-CCGTCAATTCNTTTRAGT-3′, 5′-CCGTCAATTTCTTTGAGT-3′, and 5′-CCGTCTATTCCTTTGANT-3′) (141). The archaeal 16S rRNA gene V4-V5 variable region was amplified using the forward primers 517F (5′-GCCTAAAGCATCCGTAGC-3′, 5′-GCCTAAARCGTYCGTAGC-3′, 5′-GTCTAAAGGGTCYGTAGC-3′, 5′-GCTTAAAGNGTYCGTAGC-3′, and 5′-GTCTAAARCGYYCGTAGC-3′) and reverse primer 958R (5′-CCGGCGTTGANTCCAATT-3′) (142). Amplicons were sequenced using Illumina’s v3 600-cycle (paired-end) reagent kit on a MiSeq (Illumina Inc., San Diego, CA, USA). Reads were demultiplexed on the basis of the combination of index (CASAVA 1.8) and barcode (custom python scripts).
16S rRNA gene amplicon-based community analyses
Raw sequences were analyzed using DADA2 (143) following the DADA2 Pipeline Tutorial v1.16 (https://benjjneb.github.io/dada2/tutorial.html). Each Illumina run was analyzed separately to use run-specific error profiles, as is recommended best practice for big data (https://benjjneb.github.io/dada2/bigdata.html). Briefly, forward and reverse reads were quality-trimmed to 275 and 205 base pairs (bp), respectively, and primer sequences (17-bp forward, 21-bp reverse) were removed. Reads with more than two expected errors were discarded, and paired reads were merged. All runs were combined, and chimeric sequences were removed. Species level taxonomy was assigned with the silva_nr_v138_train_set and silva_species_assignment_v138 based on the Silva small subunit reference database SSURef v138 [release date: 16 December 2019; (144)]. After sequence quality control and the removal of enrichments, blanks, and biological and technical replicates, we removed all sequences that affiliated with known contaminants (a list is given in the Supplementary Materials). Of the samples that remained after contaminant removal, we only analyzed the datasets comprising more than 1000 archaeal or 2000 bacterial reads because samples below these thresholds were either failed runs or originally highly contaminated. After all quality control steps, we obtained 478 archaeal and 964 bacterial amplicon datasets. Archaeal datasets contained a total of 4.9 × 107 sequence reads belonging to 31,099 unique ASVs. Archaeal samples had, on average, 1.02 × 105 reads and 155 ASVs (dataset S3). Bacterial datasets comprised a total of 9.6 × 107 sequence reads belonging to 377,374 unique ASVs. Bacterial samples had, on average, 0.99 × 105 reads and 1183 unique ASVs (dataset S4). Amplicon datasets may not quantitatively represent the sampled community yet are reliable to compare community structure and make ecological interpretations (83). Alpha diversity (richness, Shannon entropy, inverse Simpson diversity, and Chao1-estimated richness) was calculated from the ASV-by-sample table using a subsampling approach to account for unequal sampling effort. We used 1142 and 2271 randomly chosen reads from each archaeal and bacterial sample, respectively, calculated 10 iterations of the respective diversity index per sample, took the average value of the 10 iterations, and visualized these averages as boxplots. Total diversity within groups of samples (gamma diversity) was calculated similarly; 1142 archaeal or 2271 bacterial reads were used to subsample the ASV table; gamma diversity (total richness and Shannon) was calculated from n samples, where n is the sample number of the smallest category; this procedure was iterated 100 times, and the values were shown as boxplots. Even when using very stringent subsampling conditions of 50,000 randomly chosen archaeal and bacterial sequences, respectively, the trends in alpha and beta diversity did not change substantially; hence, we decided to include as many samples as possible. Bray-Curtis dissimilarities (145) between all samples were calculated and used for two-dimensional nonmetric multidimensional scaling (NMDS) ordinations with 20 random starts (146). All analyses were carried out with VisuaR (https://github.com/EmilRuff/VisuaR)—a workflow based on the R statistical environment, custom R scripts and several R packages including vegan (147) and ggplot2. Exemplary analysis logs (datasets S8 and S9), the used DADA2 script (dataset S10), the VisuaR v40 script (dataset S11), and an example VisuaR user input file (dataset S12) for this study are available as the Supplementary Materials.
Gene sequence identity analyses
To determine the relatedness of detected 16S rRNA gene sequences to those of the closest isolated strain, we performed a BLASTn search of each 16S rRNA gene amplicon sequence against a database of sequences from cultured archaeal and bacterial isolates, selected to yield only the single best hit for each amplicon. The isolate database was created with makeblastdb using sequences from cultured isolates in the SILVA NR reference database v138.2 available at (www.arb-silva.de/). We included recently cultured (Cald)atribacteria (102). Only alignment lengths >350 (97% of bacterial amplicons and 62% of archaeal amplicons) were analyzed.
Metagenomic DNA sequencing, contig assembly, gene annotation, and count
Paired-end libraries (2 × 151 bp) were prepared for a subset of samples using an Illumina TruSeq DNA library preparation kit (Illumina, San Diego, CA, USA) and sequenced on the Illumina NextSeq500 platform. Raw demultiplexed reads were quality processed using bbduk.sh in BBMap v.38.71 (Bushnell B; https://sourceforge.net/projects/bbmap/) in two successive steps by first removing Illumina adapters from read termini (with options: ktrim = r minlen = 40 mink = 11 tbo tpe k = 23 hdist = 1 hdist2 = 1 ftm = 5) before quality filtering against PhiX genome to an average quality threshold of 12 (with options: maq = 12 trimq = 12 qtrim = rl maxns = 3 minlen = 40 k = 31 hdist = 1). Quality trimmed reads mapping to the human HG19 genome with more than 95% identity were discarded using bbmap.sh in BBMap (with options: local minratio = 0.9 usemodulo maxindel = 3 bwr = 0.16 bw = 12 quickmatch fast minhits = 2 qtrim = rl trimq = 12 untrim idtag printunmappedcount kfilter = 25 maxsites = 1 k = 14). Cleaned reads were used to construct de novo assemblies for each sample using metaSPAdes (v3.13.0) with default parameters (148). Open reading frames (ORFs) were predicted on assembled contigs ≥1 kb using Prodigal (v2.6.3) (setting -p meta) (149). Predicted genes were clustered at ≥95% sequence identity and ≥ 80% overlap using dedupe.sh in BBMap (with option arc = t am = t ac = t mid = 95 mlp = 80) to produce a unique gene catalog of 15,663,206 nonredundant genes. The unigene was functionally assigned to a KEGG Orthology (KO) using kofamscan (v1.3) (150). rpS3 sequences were extracted from all predicted ORFs (151) using hmmsearch (152) against custom hidden Markov models for Archaea and Bacteria (https://github.com/AJProbst/rpS3_trckr), yielding a total of 8691 rpS3 sequences. RpS3 amino acids were dereplicated at ≥99% identity and ≥80% overlap with dedupe.sh (with options: arc = t am = t ac = t mid = 99 mlp = 80) to produce 4392 rpS3 single-gene sequences, equivalent to microbial species. Taxonomic classification of rpS3 genes was performed using Kaiju (v1.7.3, with option: greedy-5 mode) (153).
For taxonomic and gene count analyses, cleaned paired-end reads were subsampled to a depth of 2 M reads per sample using reformat.sh in BBMap (with options: samplereadstarget = 2,000,000, sampleseed = 121) and then mapped back to the annotated genes using BBMap with read alignment of ≥95% for the unigene (with options: minid = 0.95 idfilter = 0.95, ambiguous = random) and ≥ 99% for rpS3 genes (with options: minid = 0.99 idfilter = 0.99, ambiguous = random). Gene count tables were normalized to gene per million (e.g., equivalent to transcript per million), following (154). Metagenomic short reads were mapped to the SILVA SSU reference database (144) to assign nearest taxonomic units, and full-length 16S/18S rRNA gene sequences were reconstructed from metagenomes using phyloFlash v3.4 (155). Metagenome-derived rpS3 and 16S rRNA genes were subsampled (datasets S3 and S4), analyzed, and visualized analogous to the amplicon-derived 16S rRNA genes using the same VisuaR community analysis workflow. Overall, metagenome based diversity trends (figs. S6 to S8 and S10 to S12) and microbial composition (fig. S17, B and C) were similar to those detected by metabarcoding.
Statistical analyses
Differences in alpha diversity metrics between conditions were tested using the Wilcoxon signed rank test (ggsignif) as implemented in ggplot2 (156). P values were corrected stringently using the Bonferroni method. We used analysis of similarity (157) as implemented in vegan to test whether community structure between conditions was significantly different, as visualized in NMDS plots. Multivariate associations were tested as implemented in the MaAsLin2 method (86).
Acknowledgments
We are indebted to all our colleagues that shared their datasets with us: W. Brazelton (U of Utah), Z. Cardon (Marine Biological Laboratory), N. Fernandez-Gonzalez (U de Valladolid), M. Saxton (Miami U), L. Maignien (U of Western Brittany), K. Meyer (U Willamette), B. Osborne (Brown U), J. Rich (U of Maine), A. Roguet, R. Newton, D. Dila and S. McLellan (U of Wisconsin Milwaukee), I. Tiago (U of Coimbra), E. Trembath-Reichert (Arizona State U), F. Trigodet (U Chicago), and L. Amaral Zettler (Royal Netherlands Institute for Sea Research). Without including outgroup datasets generously provided by these researchers, the comparison between surface and subsurface biomes would not have been possible. We are also grateful to all colleagues participating in the CoDL, who organized field work and collected and processed samples, as well as the expeditions and programs that enabled the sample collection. For further details on specific CoDL projects, results, site descriptions, funding, and publications, refer to the Supplementary Materials and dataset S1. We wish to acknowledge the support of the Sloan Foundation and the Deep Carbon Observatory. Sequencing was performed at the Marine Biological Laboratory (Woods Hole, MA, United States), and we are grateful for the assistance of S. Huse, J. Vineis, and A. Voorhis. Among CoDL principal investigators and colleagues, we especially thank P. Adam, T. Bornemann, B. Briggs, S. D’Hondt, A. Eleish, E. Gaidos, A. Hoarfrost, F. Huang, T. Kieft, R. Leon-Zayas, K. Locey, M. Osburn, L. Purkamo, K. Rogers, and B. Sherwood Lollar for helpful discussions. Above all we want to thank our friend, colleague, and mentor J. Amend for being a continuous source of inspiration, an ally of early career scientists, and a driving force in geobiology and subsurface science. This study was conceived during the Deep Carbon Observatory funded “Origins and Movement of Life” workshop, which J. Amend co-organized and cohosted in 2016.
Funding: This work was supported by the Simons Foundation (824763, to S.E.R.), the Human Frontier Science Program (RGEC34/2023, to S.E.R.), the Deep Carbon Observatory (Alfred P. Sloan Foundation G-2016-7206, to K.G.L.), the Office of Biological and Environmental Research of the US Department of Energy (DE-SC0020369, to K.G.L.), the National Science Foundation Division of Ocean Sciences (NSF OCE-2145434, to A.D.S.), the CNRS Chaires de Professeur Junior (to J.A.B.), the Agence Nationale de la Recherche (ANR23-CPJ1-0172-01, to J.A.B.), the Center for Dark Energy Biosphere Investigations (C-DEBI OCE-0939564, to J.A.H.), and the Deep Life I (2011-12-01, to J.A.H. and M.O.S.). This is C-DEBI contribution number 619.
Author contributions: Conceptualization: S.E.R., A.J.P., M.L.S., J.L., and F.C. Methodology: S.E.R., I.H.d.A., K.G.L., C.S.S., A.D.S., J.A.B., J.A.H., A.J.P., H.G.M., J.L., and F.C. Investigation: S.E.R., I.H.d.A., M.M., J.P.P., C.M., K.G.L., C.S.S., A.D.S., A.S., A.M., B.K.R., J.A.B., C.L., M.O.S., S.B.J., H.G.M., J.L., and F.C. Visualization: S.E.R. and I.H.d.A. Supervision: S.E.R., B.K.R., H.G.M., and F.C. Writing—original draft: S.E.R., I.H.d.A., K.G.L., and J.A.H. Writing—review and editing: S.E.R., I.H.d.A., K.G.L., C.S.S., A.D.S., J.A.B., M.O.S., J.A.H., A.J.P., M.L.S., J.L., and F.C.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: 16S rRNA gene amplicon datasets and metagenomic datasets are available under the NCBI umbrella Bioproject PRJNA183206, and the individual project accessions are listed in dataset S1. Contextual data are available in dataset S2. The DADA2-based workflow to process 16S rRNA gene amplicon sequences is provided in dataset S10. The R-based workflow to analyze marker gene data, VisuaR, is available on GitHub (https://github.com/EmilRuff/VisuaR) and datasets S11 and S12 (exemplary user input and workflow for archaeal ASV analysis grouped by “biome”). All other data needed to evaluate the conclusions in this paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
The PDF file includes:
Supplementary Text
Supplementary Methods
Figs. S1 to S24
Legends for datasets S1 to S12
Legends for movies S1 and S2
References
Other Supplementary Material for this manuscript includes the following:
Datasets S1 to S12
Movies S1 and S2
REFERENCES AND NOTES
- 1.Takai K., Nakamura K., Toki T., Tsunogai U., Miyazaki M., Miyazaki J., Hirayama H., Nakagawa S., Nunoura T., Horikoshi K., Cell proliferation at 122°C and isotopically heavy CH4 production by a hyperthermophilic methanogen under high-pressure cultivation. Proc. Natl. Acad. Sci. U.S.A. 105, 10949–10954 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Merino N., Aronson H. S., Bojanova D. P., Feyhl-Buska J., Wong M. L., Zhang S., Giovannelli D., Living at the extremes: Extremophiles and the limits of life in a planetary context. Front. Microbiol. 10, 780 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edgcomb V. P., Molyneaux S. J., Böer S., Wirsen C. O., Saito M., Atkins M. S., Lloyd K., Teske A., Survival and growth of two heterotrophic hydrothermal vent archaea, Pyrococcus strain GB-D and Thermococcus fumicolans, under low pH and high sulfide concentrations in combination with high temperature and pressure regimes. Extremophiles. 11, 329–342 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Ando N., Barquera B., Bartlett D. H., Boyd E., Burnim A. A., Byer A. S., Colman D., Gillilan R. E., Gruebele M., Makhatadze G., Royer C. A., Shock E., Wand A. J., Watkins M. B., The molecular basis for life in extreme environments. Annu. Rev. Biophys. 50, 343–372 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Bar-On Y. M., Phillips R., Milo R., The biomass distribution on Earth. Proc. Natl. Acad. Sci. U.S.A. 115, 6506–6511 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Teske A., Sørensen K. B., Uncultured archaea in deep marine subsurface sediments: Have we caught them all? ISME J. 2, 3–18 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Magnabosco C., Lin L.-H., Dong H., Bomberg M., Ghiorse W., Stan-Lotter H., Pedersen K., Kieft T. L., van Heerden E., Onstott T. C., The biomass and biodiversity of the continental subsurface. Nat. Geosci. 11, 707–717 (2018). [Google Scholar]
- 8.Lipp J. S., Morono Y., Inagaki F., Hinrichs K.-U., Significant contribution of Archaea to extant biomass in marine subsurface sediments. Nature 454, 991–994 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Federle T. W., Dobbins D. C., Thornton-Manning J. R., Jones D. D., Microbial biomass, activity, and community structure in subsurface soils. Groundwater 24, 365–374 (1986). [Google Scholar]
- 10.Biddle J. F., Sylvan J. B., Brazelton W. J., Tully B. J., Edwards K. J., Moyer C. L., Heidelberg J. F., Nelson W. C., Prospects for the study of evolution in the deep biosphere. Front. Microbiol. 2, 285 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whitman W. B., Coleman D. C., Wiebe W. J., Prokaryotes: The unseen majority. Proc. Natl. Acad. Sci. U.S.A. 95, 6578–6583 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beaver R. C., Neufeld J. D., Microbial ecology of the deep terrestrial subsurface. ISME J. 18, wrae091 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Straume E. O., Gaina C., Medvedev S., Hochmuth K., Gohl K., Whittaker J. M., Abdul Fattah R., Doornenbal J. C., Hopper J. R., GlobSed: Updated total sediment thickness in the world’s oceans. Geochem. Geophys. Geosyst. 20, 1756–1772 (2019). [Google Scholar]
- 14.Biddle J. F., Lipp J. S., Lever M. A., Lloyd K. G., Sørensen K. B., Anderson R., Fredricks H. F., Elvert M., Kelly T. J., Schrag D. P., Sogin M. L., Brenchley J. E., Teske A., House C. H., Hinrichs K.-U., Heterotrophic Archaea dominate sedimentary subsurface ecosystems off Peru. Proc. Natl. Acad. Sci. U.S.A. 103, 3846–3851 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie S., Lipp J. S., Wegener G., Ferdelman T. G., Hinrichs K.-U., Turnover of microbial lipids in the deep biosphere and growth of benthic archaeal populations. Proc. Natl. Acad. Sci. U.S.A. 110, 6010–6014 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lloyd K. G., May M. K., Kevorkian R. T., Steen A. D., Meta-analysis of quantification methods shows that archaea and bacteria have similar abundances in the subseafloor. Appl. Environ. Microbiol. 79, 7790–7799 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoshino T., Doi H., Uramoto G., Wörmer L., Adhikari R. R., Xiao N., Morono Y., D’Hondt S., Hinrichs K.-U., Inagaki F., Global diversity of microbial communities in marine sediment. Proc. Natl. Acad. Sci. U.S.A. 117, 27587–27597 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kallmeyer J., Pockalny R., Adhikari R. R., Smith D. C., D’Hondt S., Global distribution of microbial abundance and biomass in subseafloor sediment. Proc. Natl. Acad. Sci. 109, 16213–16216 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flemming H.-C., Wuertz S., Bacteria and archaea on Earth and their abundance in biofilms. Nat. Rev. Microbiol. 17, 247–260 (2019). [DOI] [PubMed] [Google Scholar]
- 20.McMahon S., Parnell J., Weighing the deep continental biosphere. FEMS Microbiol. Ecol. 87, 113–120 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Parkes R. J., Cragg B., Roussel E., Webster G., Weightman A., Sass H., A review of prokaryotic populations and processes in sub-seafloor sediments, including biosphere:Geosphere interactions. Mar. Geol. 352, 409–425 (2014). [Google Scholar]
- 22.Morono Y., Ito M., Hoshino T., Terada T., Hori T., Ikehara M., D’Hondt S., Inagaki F., Aerobic microbial life persists in oxic marine sediment as old as 101.5 million years. Nat. Commun. 11, 3626 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walsh E. A., Kirkpatrick J. B., Rutherford S. D., Smith D. C., Sogin M., D’Hondt S., Bacterial diversity and community composition from seasurface to subseafloor. ISME J. 10, 979–989 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Starnawski P., Bataillon T., Ettema T. J. G., Jochum L. M., Schreiber L., Chen X., Lever M. A., Polz M. F., Jørgensen B. B., Schramm A., Kjeldsen K. U., Microbial community assembly and evolution in subseafloor sediment. Proc. Natl. Acad. Sci. U.S.A. 114, 2940–2945 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirkpatrick J. B., Walsh E. A., D’Hondt S., Microbial selection and survival in subseafloor sediment. Front. Microbiol. 10, 956 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petro C., Zäncker B., Starnawski P., Jochum L. M., Ferdelman T. G., Jørgensen B. B., Røy H., Kjeldsen K. U., Schramm A., Marine deep biosphere microbial communities assemble in near-Surface sediments in Aarhus Bay. Front. Microbiol. 10, 758 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hubalek V., Wu X., Eiler A., Buck M., Heim C., Dopson M., Bertilsson S., Ionescu D., Connectivity to the surface determines diversity patterns in subsurface aquifers of the Fennoscandian shield. ISME J. 10, 2447–2458 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyer J. L., Jaekel U., Tully B. J., Glazer B. T., Wheat C. G., Lin H., Hsieh C., Cowen J. P., Hulme S. M., Girguis P. R., Huber J. A., A distinct and active bacterial community in cold oxygenated fluids circulating beneath the western flank of the Mid-Atlantic ridge. Sci. Rep. 6, 22541 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Putman L. I., Sabuda M. C., Brazelton W. J., Kubo M. D., Hoehler T. M., McCollom T. M., Cardace D., Schrenk M. O., Microbial communities in a serpentinizing aquifer are assembled through strong concurrent dispersal limitation and selection. mSystems 6, e0030021 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y., Dekas A. E., Hawkins A. J., Parada A. E., Gorbatenko O., Li K., Horne R. N., Microbial community composition in Deep-Subsurface reservoir fluids reveals natural interwell connectivity. Water Resour. Res. 56, e2019WR025916 (2020). [Google Scholar]
- 31.Scheidweiler D., Miele F., Peter H., Battin T. J., de Anna P., Trait-specific dispersal of bacteria in heterogeneous porous environments: From pore to porous medium scale. J. R. Soc. Interface 17, 20200046 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoshino T., Toki T., Ijiri A., Morono Y., Machiyama H., Ashi J., Okamura K., Inagaki F., Atribacteria from the subseafloor sedimentary biosphere disperse to the hydrosphere through submarine mud volcanoes. Front. Microbiol. 8, 1135 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruff S. E., Felden J., Gruber-Vodicka H. R., Marcon Y., Knittel K., Ramette A., Boetius A., In situ development of a methanotrophic microbiome in deep-sea sediments. ISME J. 13, 197–213 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chakraborty A., Ruff S. E., Dong X., Ellefson E. D., Li C., Brooks J. M., McBee J., Bernard B. B., Hubert C. R. J., Hydrocarbon seepage in the deep seabed links subsurface and seafloor biospheres. Proc. Natl. Acad. Sci. U.S.A. 117, 11029–11037 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gittins D. A., Desiage P.-A., Morrison N., Rattray J. E., Bhatnagar S., Chakraborty A., Zorz J., Li C., Horanszky O., Cramm M. A., Bisiach F., Bennett R., Webb J., MacDonald A., Fowler M., Campbell D. C., Hubert C. R. J., Geological processes mediate a microbial dispersal loop in the deep biosphere. Sci. Adv. 8, eabn3485 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steen A. D., Kevorkian R. T., Bird J. T., Dombrowski N., Baker B. J., Hagen S. M., Mulligan K. H., Schmidt J. M., Webber A. T., Royalty T. M., Alperin M. J., Kinetics and identities of extracellular peptidases in subsurface sediments of the white Oak River Estuary, North Carolina. Appl. Environ. Microbiol. 85, e00102-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bird J. T., Tague E. D., Zinke L., Schmidt J. M., Steen A. D., Reese B., Marshall I. P. G., Webster G., Weightman A., Castro H. F., Campagna S. R., Lloyd K. G., Uncultured microbial phyla suggest mechanisms for multi-thousand-year subsistence in Baltic sea sediments. MBio 10, e02376-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Orsi W. D., Edgcomb V. P., Christman G. D., Biddle J. F., Gene expression in the deep biosphere. Nature. 499, 205–208 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Lever M. A., Acetogenesis in the energy-starved deep biosphere—A paradox? Front. Microbiol. 2, 284 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He Y., Li M., Perumal V., Feng X., Fang J., Xie J., Sievert S. M., Wang F., Genomic and enzymatic evidence for acetogenesis among multiple lineages of the archaeal phylum Bathyarchaeota widespread in marine sediments. Nat. Microbiol. 1, 16035 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Newberry C. J., Webster G., Cragg B. A., Parkes R. J., Weightman A. J., Fry J. C., Diversity of prokaryotes and methanogenesis in deep subsurface sediments from the Nankai Trough, Ocean Drilling Program Leg 190. Environ. Microbiol. 6, 274–287 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Momper L., Casar C. P., Osburn M. R., A metagenomic view of novel microbial and metabolic diversity found within the deep terrestrial biosphere at DeMMO: A microbial observatory in South Dakota, USA. Environ. Microbiol. 25, 3719–3737 (2023). [DOI] [PubMed] [Google Scholar]
- 43.Ruff S. E., Humez P., de Angelis I. H., Diao M., Nightingale M., Cho S., Connors L., Kuloyo O. O., Seltzer A., Bowman S., Wankel S. D., McClain C. N., Mayer B., Strous M., Hydrogen and dark oxygen drive microbial productivity in diverse groundwater ecosystems. Nat. Commun. 14, 3194 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Møller M. H., Glombitza C., Lever M. A., Deng L., Morono Y., Inagaki F., Doll M., Su C., Lomstein B. A., D:L-amino acid modeling reveals fast microbial turnover of days to months in the subsurface hydrothermal sediment of Guaymas Basin. Front. Microbiol. 9, 967 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bradley J. A., Amend J. P., LaRowe D. E., Necromass as a Limited Source of Energy for Microorganisms in Marine Sediments. J. Geophys. Res. Biogeosci. 123, 577–590 (2018). [Google Scholar]
- 46.Pérez Castro S., Borton M. A., Regan K., Hrabe de Angelis I., Wrighton K. C., Teske A. P., Strous M., Ruff S. E., Degradation of biological macromolecules supports uncultured microbial populations in Guaymas Basin hydrothermal sediments. ISME J. 15, 3480–3497 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chaudhari N. M., Overholt W. A., Figueroa-Gonzalez P. A., Taubert M., Bornemann T. L. V., Probst A. J., Hölzer M., Marz M., Küsel K., The economical lifestyle of CPR bacteria in groundwater allows little preference for environmental drivers. Environ. Microbiome 16, 24 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schwank K., Bornemann T. L. V., Dombrowski N., Spang A., Banfield J. F., Probst A. J., An archaeal symbiont-host association from the deep terrestrial subsurface. ISME J. 13, 2135–2139 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schrenk M. O., Brazelton W. J., Lang S. Q., Serpentinization, Carbon, and Deep Life. Rev. Mineral. Geochemistry. 75, 575–606 (2013). [Google Scholar]
- 50.Sauvage J. F., Flinders A., Spivack A. J., Pockalny R., Dunlea A. G., Anderson C. H., Smith D. C., Murray R. W., D’Hondt S., The contribution of water radiolysis to marine sedimentary life. Nat. Commun. 12, 1297 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin L.-H., Hall J., Lippmann-Pipke J., Ward J. A., Sherwood Lollar B., DeFlaun M., Rothmel R., Moser D., Gihring T. M., Mislowack B., Onstott T. C., Radiolytic H2 in continental crust: Nuclear power for deep subsurface microbial communities. Geochem. Geophys. Geosyst. 6, Q07003 (2005). [Google Scholar]
- 52.Lennon J. T., Jones S. E., Microbial seed banks: The ecological and evolutionary implications of dormancy. Nat. Rev. Microbiol. 9, 119–130 (2011). [DOI] [PubMed] [Google Scholar]
- 53.Schippers A., Neretin L. N., Kallmeyer J., Ferdelman T. G., Cragg B. A., John Parkes R., Jørgensen B. B., Prokaryotic cells of the deep sub-seafloor biosphere identified as living bacteria. Nature 433, 861–864 (2005). [DOI] [PubMed] [Google Scholar]
- 54.Morono Y., Terada T., Nishizawa M., Ito M., Hillion F., Takahata N., Sano Y., Inagaki F., Carbon and nitrogen assimilation in deep subseafloor microbial cells. Proc. Natl. Acad. Sci. U.S.A. 108, 18295–18300 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jørgensen B. B., Deep subseafloor microbial cells on physiological standby. Proc. Natl. Acad. Sci. U.S.A. 108, 18193–18194 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trembath-Reichert E., Morono Y., Ijiri A., Hoshino T., Dawson K. S., Inagaki F., Orphan V. J., Methyl-compound use and slow growth characterize microbial life in 2-km-deep subseafloor coal and shale beds. Proc. Natl. Acad. Sci. U.S.A. 114, E9206–E9215 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Price P. B., Sowers T., Temperature dependence of metabolic rates for microbial growth, maintenance, and survival. Proc. Natl. Acad. Sci. U.S.A. 101, 4631–4636 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Braun S., Morono Y., Littmann S., Kuypers M., Aslan H., Dong M., Jørgensen B. B., Lomstein B. A., Size and carbon content of sub-seafloor microbial cells at landsort deep, Baltic Sea. Front. Microbiol. 7, 1375 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Soares A., Edwards A., An D., Bagnoud A., Bradley J., Barnhart E., Bomberg M., Budwill K., Caffrey S. M., Fields M., Gralnick J., Kadnikov V., Momper L., Osburn M., Mu A., Moreau J. W., Moser D., Purkamo L., Rassner S. M., Sheik C. S., Sherwood Lollar B., Toner B. M., Voordouw G., Wouters K., Mitchell A. C., A global perspective on bacterial diversity in the terrestrial deep subsurface. Microbiology 169, 001172 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Y., Sheng H.-F., He Y., Wu J.-Y., Jiang Y.-X., Tam N. F.-Y., Zhou H.-W., Comparison of the levels of bacterial diversity in freshwater, Intertidal wetland, and marine sediments by using millions of illumina tags. Appl. Environ. Microbiol. 78, 8264–8271 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Handler E. R., Andersen S. D. J., Gradinger R., McGovern M., Vader A., Poste A. E., Seasonality in land-ocean connectivity and local processes control sediment bacterial community structure and function in a High Arctic tidal flat. FEMS Microbiol. Ecol. 100, fiad162 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tee H. S., Waite D., Lear G., Handley K. M., Microbial river-to-sea continuum: gradients in benthic and planktonic diversity, osmoregulation and nutrient cycling. Microbiome 9, 190 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fenchel T., Whitfield M., Meadows P., Huisman J., Microbial ecology on land and sea [and discussion]. Philos. Trans. Biol. Sci. 343, 51–56 (1994). [Google Scholar]
- 64.Lang-Yona N., Flores J. M., Haviv R., Alberti A., Poulain J., Belser C., Trainic M., Gat D., Ruscheweyh H., Wincker P., Sunagawa S., Rudich Y., Koren I., Vardi A., Terrestrial and marine influence on atmospheric bacterial diversity over the north Atlantic and Pacific Oceans. Commun. Earth Environ. 3, 121 (2022). [Google Scholar]
- 65.Lozupone C. A., Knight R., Global patterns in bacterial diversity. Proc. Natl. Acad. Sci. U.S.A. 104, 11436–11440 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jurdzinski K. T., Mehrshad M., Delgado L. F., Deng Z., Bertilsson S., Andersson A. F., Large-scale phylogenomics of aquatic bacteria reveal molecular mechanisms for adaptation to salinity. Sci. Adv. 9, eadg2059 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thompson L. R., Sanders J. G., McDonald D., Amir A., Ladau J., Locey K. J., Prill R. J., Tripathi A., Gibbons S. M., Ackermann G., Navas-Molina J. A., Janssen S., Kopylova E., Vázquez-Baeza Y., González A., Morton J. T., Mirarab S., Zech Xu Z., Jiang L., Haroon M. F., Kanbar J., Zhu Q., Jin Song S., Kosciolek T., Bokulich N. A., Lefler J., Brislawn C. J., Humphrey G., Owens S. M., Hampton-Marcell J., Berg-Lyons D., McKenzie V., Fierer N., Fuhrman J. A., Clauset A., Stevens R. L., Shade A., Pollard K. S., Goodwin K. D., Jansson J. K., Gilbert J. A., Knight R., Earth Microbiome Project Consortium , A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 551, 457–463 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Voser T. M., Campbell M. D., Carroll A. R., How different are marine microbial natural products compared to their terrestrial counterparts? Nat. Prod. Rep. 39, 7–19 (2022). [DOI] [PubMed] [Google Scholar]
- 69.Sunagawa S., Coelho L. P., Chaffron S., Kultima J. R., Labadie K., Salazar G., Djahanschiri B., Zeller G., Mende D. R., Alberti A., Cornejo-Castillo F. M., Costea P. I., Cruaud C., D’Ovidio F., Engelen S., Ferrera I., Gasol J. M., Guidi L., Hildebrand F., Kokoszka F., Lepoivre C., Lima-Mendez G., Poulain J., Poulos B. T., Royo-Llonch M., Sarmento H., Vieira-Silva S., Dimier C., Picheral M., Searson S., Kandels-Lewis S., Bowler C., de Vargas C., Gorsky G., Grimsley N., Hingamp P., Iudicone D., Jaillon O., Not F., Ogata H., Pesant S., Speich S., Stemmann L., Sullivan M. B., Weissenbach J., Wincker P., Karsenti E., Raes J., Acinas S. G., Bork P., Boss E., Bowler C., Follows M., Karp-Boss L., Krzic U., Reynaud E. G., Sardet C., Sieracki M., Velayoudon D., Structure and function of the global ocean microbiome. Science 348, 1261359 (2015). [DOI] [PubMed] [Google Scholar]
- 70.Mestre M., Ruiz-González C., Logares R., Duarte C. M., Gasol J. M., Sala M. M., Sinking particles promote vertical connectivity in the ocean microbiome. Proc. Natl. Acad. Sci. U.S.A. 115, E6799–E6807 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Acinas S. G., Sánchez P., Salazar G., Cornejo-Castillo F. M., Sebastián M., Logares R., Royo-Llonch M., Paoli L., Sunagawa S., Hingamp P., Ogata H., Lima-Mendez G., Roux S., González J. M., Arrieta J. M., Alam I. S., Kamau A., Bowler C., Raes J., Pesant S., Bork P., Agustí S., Gojobori T., Vaqué D., Sullivan M. B., Pedrós-Alió C., Massana R., Duarte C. M., Gasol J. M., Deep ocean metagenomes provide insight into the metabolic architecture of bathypelagic microbial communities. Commun. Biol. 4, 604 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Delgado-Baquerizo M., Oliverio A. M., Brewer T. E., Benavent-González A., Eldridge D. J., Bardgett R. D., Maestre F. T., Singh B. K., Fierer N., A global atlas of the dominant bacteria found in soil. Science 359, 320–325 (2018). [DOI] [PubMed] [Google Scholar]
- 73.Graham E. B., Camargo A. P., Wu R., Neches R. Y., Nolan M., Paez-Espino D., Kyrpides N. C., Jansson J. K., McDermott J. E., Hofmockel K. S., Soil Virosphere Consortium , A global atlas of soil viruses reveals unexplored biodiversity and potential biogeochemical impacts. Nat. Microbiol., 1873–1883 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nayfach S., Roux S., Seshadri R., Udwary D., Varghese N., Schulz F., Wu D., Paez-Espino D., Chen I., Huntemann M., Palaniappan K., Ladau J., Mukherjee S., Reddy T. B. K., Nielsen T., Kirton E., Faria J. P., Edirisinghe J. N., Henry C. S., Jungbluth S. P., Chivian D., Dehal P., Wood-Charlson E. M., Arkin A. P., Tringe S. G., Visel A., IMG/M Data Consortium, Woyke T., Mouncey N. J., Ivanova N. N., Kyrpides N. C., Eloe-Fadrosh E. A., A genomic catalog of Earth’s microbiomes. Nat. Biotechnol. 39, 499–509 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shaffer J. P., Nothias L.-F., Thompson L. R., Sanders J. G., Salido R. A., Couvillion S. P., Brejnrod A. D., Lejzerowicz F., Haiminen N., Huang S., Lutz H. L., Zhu Q., Martino C., Morton J. T., Karthikeyan S., Nothias-Esposito M., Dührkop K., Böcker S., Kim H. W., Aksenov A. A., Bittremieux W., Minich J. J., Marotz C., Bryant M. M., Sanders K., Schwartz T., Humphrey G., Vásquez-Baeza Y., Tripathi A., Parida L., Carrieri A. P., Beck K. L., Das P., González A., McDonald D., Ladau J., Karst S. M., Albertsen M., Ackermann G., DeReus J., Thomas T., Petras D., Shade A., Stegen J., Song S. J., Metz T. O., Swafford A. D., Dorrestein P. C., Jansson J. K., Gilbert J. A., Knight R., the Earth Microbiome Project 500 (EMP500) Consortium , Standardized multi-omics of Earth’s microbiomes reveals microbial and metabolite diversity. Nat. Microbiol. 7, 2128–2150 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.A. Vion, L. Menot, Continental margins between 140m and 3500m depth (IFREMER - COMARGE, 2009) (2009); www.marineregions.org/maps.php?album=3264&pic=64937).
- 77.LaRowe D. E., Arndt S., Bradley J. A., Burwicz E., Dale A. W., Amend J. P., Organic carbon and microbial activity in marine sediments on a global scale throughout the Quaternary. Geochim. Cosmochim. Acta 286, 227–247 (2020). [Google Scholar]
- 78.Bradley J. A., Arndt S., Amend J. P., Burwicz E., Dale A. W., Egger M., LaRowe D. E., Widespread energy limitation to life in global subseafloor sediments. Sci. Adv. 6, eaba0697 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Salter S. J., Cox M. J., Turek E. M., Calus S. T., Cookson W. O., Moffatt M. F., Turner P., Parkhill J., Loman N. J., Walker A. W., Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 12, 87 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sheik C. S., Reese B. K., Twing K. I., Sylvan J. B., Grim S. L., Schrenk M. O., Sogin M. L., Colwell F. S., Identification and removal of contaminant sequences from ribosomal gene databases: Lessons from the census of deep life. Front. Microbiol. 9, 840 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kennedy K., Hall M. W., Lynch M. D. J., Moreno-Hagelsieb G., Neufeld J. D., Evaluating bias of Illumina-based bacterial 16S rRNA gene profiles. Appl. Environ. Microbiol. 80, 5717–5722 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schirmer M., Ijaz U. Z., D’Amore R., Hall N., Sloan W. T., Quince C., Insight into biases and sequencing errors for amplicon sequencing with the Illumina MiSeq platform. Nucleic Acids Res. 43, e37 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Piwosz K., Shabarova T., Pernthaler J., Posch T., Šimek K., Porcal P., Salcher M. M., Bacterial and eukaryotic small-subunit amplicon data do not provide a quantitative picture of microbial communities, but they are reliable in the context of ecological interpretations. mSphere 5, e00052-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Anderson R. E., Graham E. D., Huber J. A., Tully B. J., Microbial populations are shaped by dispersal and recombination in a low biomass subseafloor habitat. MBio, e0035422 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.J. E. Hinkle, J. P. Chanton, M. A. Moynihan, S. E. Ruff, A. Teske, Complex bacterial diversity of Guaymas Basin hydrothermal sediments revealed by synthetic long-read sequencing (LoopSeq). bioRxiv 589229 [Preprint] (2024). 10.1101/2024.04.12.589229. [DOI]
- 86.Mallick H., Rahnavard A., McIver L. J., Ma S., Zhang Y., Nguyen L. H., Tickle T. L., Weingart G., Ren B., Schwager E. H., Chatterjee S., Thompson K. N., Wilkinson J. E., Subramanian A., Lu Y., Waldron L., Paulson J. N., Franzosa E. A., Bravo H. C., Huttenhower C., Multivariable association discovery in population-scale meta-omics studies. PLOS Comput. Biol. 17, e1009442 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Coleman G. A., Davín A. A., Mahendrarajah T. A., Szánthó L. L., Spang A., Hugenholtz P., Szöllősi G. J., Williams T. A., A rooted phylogeny resolves early bacterial evolution. Science 372, eabe0511 (2021). [DOI] [PubMed] [Google Scholar]
- 88.Staley J. T., Konopka A., Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu. Rev. Microbiol. 39, 321–346 (1985). [DOI] [PubMed] [Google Scholar]
- 89.Sogin M. L., Morrison H. G., Huber J. A., Welch D. M., Huse S. M., Neal P. R., Arrieta J. M., Herndl G. J., Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc. Natl. Acad. Sci. U.S.A. 103, 12115–12120 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lloyd K. G., Steen A. D., Ladau J., Yin J., Crosby L., Phylogenetically novel uncultured microbial cells dominate earth microbiomes. mSystems 3, e00055-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vuillemin A., Wankel S. D., Coskun Ö. K., Magritsch T., Vargas S., Estes E. R., Spivack A. J., Smith D. C., Pockalny R., Murray R. W., D’Hondt S., Orsi W. D., Archaea dominate oxic subseafloor communities over multimillion-year time scales. Sci. Adv. 5, eaaw4108 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Probst A. J., Ladd B., Jarett J. K., Geller-McGrath D. E., Sieber C. M. K., Emerson J. B., Anantharaman K., Thomas B. C., Malmstrom R. R., Stieglmeier M., Klingl A., Woyke T., Ryan M. C., Banfield J. F., Differential depth distribution of microbial function and putative symbionts through sediment-hosted aquifers in the deep terrestrial subsurface. Nat. Microbiol. 3, 328–336 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.He C., Keren R., Whittaker M. L., Farag I. F., Doudna J. A., Cate J. H. D., Banfield J. F., Genome-resolved metagenomics reveals site-specific diversity of episymbiotic CPR bacteria and DPANN archaea in groundwater ecosystems. Nat. Microbiol. 6, 354–365 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chivian D., Brodie E. L., Alm E. J., Culley D. E., Dehal P. S., DeSantis T. Z., Gihring T. M., Lapidus A., Lin L.-H., Lowry S. R., Moser D. P., Richardson P. M., Southam G., Wanger G., Pratt L. M., Andersen G. L., Hazen T. C., Brockman F. J., Arkin A. P., Onstott T. C., Environmental genomics reveals a single-species Ecosystem deep within earth. Science. 322, 275–278 (2008). [DOI] [PubMed] [Google Scholar]
- 95.Labonté J. M., Field E. K., Lau M., Chivian D., Van Heerden E., Wommack K. E., Kieft T. L., Onstott T. C., Stepanauskas R., Single cell genomics indicates horizontal gene transfer and viral infections in a deep subsurface Firmicutes population. Front. Microbiol. 6, 349 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Karnachuk O. V., Frank Y. A., Lukina A. P., Kadnikov V. V., Beletsky A. V., Mardanov A. V., Ravin N. V., Domestication of previously uncultivated Candidatus Desulforudis audaxviator from a deep aquifer in Siberia sheds light on its physiology and evolution. ISME J. 13, 1947–1959 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Karnachuk O. V., Lukina A. P., Kadnikov V. V., Sherbakova V. A., Beletsky A. V., Mardanov A. V., Ravin N. V., Targeted isolation based on metagenome-assembled genomes reveals a phylogenetically distinct group of thermophilic spirochetes from deep biosphere. Environ. Microbiol. 23, 3585–3598 (2021). [DOI] [PubMed] [Google Scholar]
- 98.Carr S. A., Orcutt B. N., Mandernack K. W., Spear J. R., Abundant Atribacteria in deep marine sediment from the Adélie Basin, Antarctica. Front. Microbiol. 6, 872 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lee Y. M., Hwang K., Il Lee J., Kim M., Hwang C. Y., Noh H.-J., Choi H., Lee H. K., Chun J., Hong S. G., Shin S. C., Genomic insight into the predominance of candidate phylum Atribacteria JS1 lineage in marine sediments. Front. Microbiol. 9, 2909 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Vuillemin A., Ariztegui D., Horn F., Kallmeyer J., Orsi W. D., PASADO Science Team , Microbial community composition along a 50 000-year lacustrine sediment sequence. FEMS Microbiol. Ecol. 94, fiy029 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Vuillemin A., Vargas S., Coskun Ö. K., Pockalny R., Murray R. W., Smith D. C., D’hondt S., Orsi W. D., Atribacteria reproducing over millions of years in the atlantic abyssal subseafloor. MBio 11, e01937-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Katayama T., Nobu M. K., Kusada H., Meng X., Hosogi N., Uematsu K., Yoshioka H., Kamagata Y., Tamaki H., Isolation of a member of the candidate phylum ‘Atribacteria’ reveals a unique cell membrane structure. Nat. Commun. 11, 6381 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Moger-Reischer R. Z., Glass J. I., Wise K. S., Sun L., Bittencourt D. M. C., Lehmkuhl B. K., Schoolmaster D. R., Lynch M., Lennon J. T., Evolution of a minimal cell. Nature. 620, 122–127 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Glass J. B., Ranjan P., Kretz C. B., Nunn B. L., Johnson A. M., Xu M., McManus J., Stewart F. J., Microbial metabolism and adaptations in Atribacteria -dominated methane hydrate sediments. Environ. Microbiol. 23, 4646–4660 (2021). [DOI] [PubMed] [Google Scholar]
- 105.Kubo K., Lloyd K. G., Biddle J. F., Amann R., Teske A., Knittel K., Archaea of the Miscellaneous Crenarchaeotal Group are abundant, diverse and widespread in marine sediments. ISME J. 6, 1949–1965 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Biddle J. F., Fitz-Gibbon S., Schuster S. C., Brenchley J. E., House C. H., Metagenomic signatures of the Peru Margin subseafloor biosphere show a genetically distinct environment. Proc. Natl. Acad. Sci. U.S.A. 105, 10583–10588 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dutta A., Dutta Gupta S., Gupta A., Sarkar J., Roy S., Mukherjee A., Sar P., Exploration of deep terrestrial subsurface microbiome in late Cretaceous Deccan traps and underlying Archean basement, India. Sci. Rep. 8, 17459 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Purkamo L., Kietäväinen R., Nuppunen-Puputti M., Bomberg M., Cousins C., Ultradeep microbial communities at 4.4 km within crystalline bedrock: Implications for habitability in a planetary context. Life. 10, 2 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rinke C., Schwientek P., Sczyrba A., Ivanova N. N., Anderson I. J., Cheng J.-F., Darling A. E., Malfatti S., Swan B. K., Gies E. A., Dodsworth J. A., Hedlund B. P., Tsiamis G., Sievert S. M., Liu W.-T., Eisen J. A., Hallam S. J., Kyrpides N. C., Stepanauskas R., Rubin E. M., Hugenholtz P., Woyke T., Insights into the phylogeny and coding potential of microbial dark matter. Nature 499, 431–437 (2013). [DOI] [PubMed] [Google Scholar]
- 110.Yarza P., Yilmaz P., Pruesse E., Glöckner F. O., Ludwig W., Schleifer K.-H., Whitman W. B., Euzéby J., Amann R., Rosselló-Móra R., Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 12, 635–645 (2014). [DOI] [PubMed] [Google Scholar]
- 111.Baker B. J., Appler K. E., Gong X., New microbial biodiversity in marine sediments. Ann. Rev. Mar. Sci. 13, 161–175 (2021). [DOI] [PubMed] [Google Scholar]
- 112.Tully B. J., Wheat C. G., Glazer B. T., Huber J. A., A dynamic microbial community with high functional redundancy inhabits the cold, oxic subseafloor aquifer. ISME J. 12, 1–16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Edwards K. J., Wheat C. G., Sylvan J. B., Under the sea: Microbial life in volcanic oceanic crust. Nat. Rev. Microbiol. 9, 703–712 (2011). [DOI] [PubMed] [Google Scholar]
- 114.S. E. Ruff, in Marine Hydrocarbon Seeps - Microbiology and Biogeochemistry of a Global Marine Habitat, A. P. Teske, V. Carvalho, Eds. (Springer, 2020), pp. 1–19; 10.1007/978-3-030-34827-4_1). [DOI]
- 115.Crespo-Medina M., Bowles M. W., Samarkin V. A., Hunter K. S., Joye S. B., Microbial diversity and activity in seafloor brine lake sediments (Alaminos Canyon block 601, Gulf of Mexico). Geobiology 14, 483–498 (2016). [DOI] [PubMed] [Google Scholar]
- 116.Le Bris N., Yücel M., Das A., Sievert S. M., LokaBharathi P., Girguis P. R., Hydrothermal energy transfer and organic carbon production at the deep seafloor. Front. Mar. Sci. 5, 531 (2019). [Google Scholar]
- 117.Mouser P. J., Borton M., Darrah T. H., Hartsock A., Wrighton K. C., Hydraulic fracturing offers view of microbial life in the deep terrestrial subsurface. FEMS Microbiol. Ecol. 92, fiw166 (2016). [DOI] [PubMed] [Google Scholar]
- 118.Plümper O., King H. E., Geisler T., Liu Y., Pabst S., Savov I. P., Rost D., Zack T., Subduction zone forearc serpentinites as incubators for deep microbial life. Proc. Natl. Acad. Sci. U.S.A. 114, 4324–4329 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Brazelton W. J., Nelson B., Schrenk M. O., Metagenomic evidence for H2 oxidation and H2 production by serpentinite-hosted subsurface microbial communities. Front. Microbiol. 2, 268 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Motamedi S., Orcutt B. N., Früh-Green G. L., Twing K. I., Pendleton H. L., Brazelton W. J., Microbial residents of the Atlantis Massif’s shallow serpentinite subsurface. Appl. Environ. Microbiol. 86, e00356-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Macalady J. L., Lyon E. H., Koffman B., Albertson L. K., Meyer K., Galdenzi S., Mariani S., Dominant microbial populations in limestone-corroding stream biofilms, Frasassi cave system, Italy. Appl. Environ. Microbiol. 72, 5596–5609 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Inagaki F., Hinrichs K. U., Kubo Y., Bowles M. W., Heuer V. B., Hong W. L., Hoshino T., Ijiri A., Imachi H., Ito M., Kaneko M., Lever M. A., Lin Y. S., Methé B. A., Morita S., Morono Y., Tanikawa W., Bihan M., Bowden S. A., Elvert M., Glombitza C., Gross D., Harrington G. J., Hori T., Li K., Limmer D., Liu C. H., Murayama M., Ohkouchi N., Ono S., Park Y. S., Phillips S. C., Prieto-Mollar X., Purkey M., Riedinger N., Sanada Y., Sauvage J., Snyder G., Susilawati R., Takano Y., Tasumi E., Terada T., Tomaru H., Trembath-Reichert E., Wang D. T., Yamada Y., Exploring deep microbial life in coal-bearing sediment down to ∼2.5 km below the ocean floor. Science. 349, 420–424 (2015). [DOI] [PubMed] [Google Scholar]
- 123.Lau M. C. Y., Kieft T. L., Kuloyo O., Linage-Alvarez B., van Heerden E., Lindsay M. R., Magnabosco C., Wang W., Wiggins J. B., Guo L., Perlman D. H., Kyin S., Shwe H. H., Harris R. L., Oh Y., Yi M. J., Purtschert R., Slater G. F., Ono S., Wei S., Li L., Sherwood Lollar B., Onstott T. C., An oligotrophic deep-subsurface community dependent on syntrophy is dominated by sulfur-driven autotrophic denitrifiers. Proc. Natl. Acad. Sci. U.S.A. 113, E7927–E7936 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lin L.-H., Slater G. F., Sherwood Lollar B., Lacrampe-Couloume G., Onstott T. C., The yield and isotopic composition of radiolytic H2, a potential energy source for the deep subsurface biosphere. Geochim. Cosmochim. Acta 69, 893–903 (2005). [Google Scholar]
- 125.Pedersen K., Exploration of deep intraterrestrial microbial life: Current perspectives. FEMS Microbiol. Lett. 185, 9–16 (2000). [DOI] [PubMed] [Google Scholar]
- 126.F. S. Colwell, R. P. Smith, in Geophysical Monograph Series (Wiley, 2004), vol. 144, pp. 355–367; 10.1029/144GM22. [DOI]
- 127.Rebata-Landa V., Santamarina J. C., Mechanical limits to microbial activity in deep sediments. Geochem. Geophys. Geosyst. 7, Q11006 (2006). [Google Scholar]
- 128.D’Hondt S., Inagaki F., Zarikian C. A., Abrams L. J., Dubois N., Engelhardt T., Evans H., Ferdelman T., Gribsholt B., Harris R. N., Hoppie B. W., Hyun J.-H., Kallmeyer J., Kim J., Lynch J. E., McKinley C. C., Mitsunobu S., Morono Y., Murray R. W., Pockalny R., Sauvage J., Shimono T., Shiraishi F., Smith D. C., Smith-Duque C. E., Spivack A. J., Steinsbu B. O., Suzuki Y., Szpak M., Toffin L., Uramoto G., Yamaguchi Y. T., Zhang G., Zhang X.-H., Ziebis W., Presence of oxygen and aerobic communities from sea floor to basement in deep-sea sediments. Nat. Geosci. 8, 299–304 (2015). [Google Scholar]
- 129.Ruff S. E., Schwab L., Vidal E., Hemingway J., Kraft B., Murali R., Widespread occurrence of dissolved oxygen anomalies, aerobic microbes, and oxygen-producing metabolic pathways in apparently anoxic environments. FEMS Microbiol. Ecol. 100, fiae132 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Brantley S. L., Goldhaber M. B., Ragnarsdottir K. V., Crossing disciplines and scales to understand the critical zone. Elements 3, 307–314 (2007). [Google Scholar]
- 131.Küsel K., Totsche K. U., Trumbore S. E., Lehmann R., Steinhäuser C., Herrmann M., How deep can surface signals be traced in the critical zone? Merging biodiversity with biogeochemistry research in a central German Muschelkalk landscape. Front. Earth Sci. 4, 32 (2016). [Google Scholar]
- 132.Steele J. H., Brink K. H., Scott B. E., Comparison of marine and terrestrial ecosystems: Suggestions of an evolutionary perspective influenced by environmental variation. ICES J. Mar. Sci. 76, 50–59 (2019). [Google Scholar]
- 133.Carr M. H., Neigel J. E., Estes J. A., Andelman S., Warner R. R., Largier J. L., Comparing marine and terrestrial ecosystems: Implications for the design of coastal marine reserves. Ecol. Appl. 13, 90–107 (2003). [Google Scholar]
- 134.Zhang Y., Horne R. N., Hawkins A. J., Primo J. C., Gorbatenko O., Dekas A. E., Geological activity shapes the microbiome in deep-subsurface aquifers by advection. Proc. Natl. Acad. Sci. U.S.A. 119, e2113985119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bornemann T. L. V., Adam P. S., Turzynski V., Schreiber U., Figueroa-Gonzalez P. A., Rahlff J., Köster D., Schmidt T. C., Schunk R., Krauthausen B., Probst A. J., Genetic diversity in terrestrial subsurface ecosystems impacted by geological degassing. Nat. Commun. 13, 284 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gleeson T., Moosdorf N., Hartmann J., van Beek L. P. H., A glimpse beneath earth’s surface: GLobal HYdrogeology MaPS (GLHYMPS) of permeability and porosity. Geophys. Res. Lett. 41, 3891–3898 (2014). [Google Scholar]
- 137.Edwards K. J., Fisher A. T., Wheat C. G., The deep subsurface biosphere in igneous ocean crust: Frontier habitats for microbiological exploration. Front. Microbiol. 3, 8 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Schrenk M. O., Huber J. A., Edwards K. J., Microbial provinces in the subseafloor. Ann. Rev. Mar. Sci. 2, 279–304 (2010). [DOI] [PubMed] [Google Scholar]
- 139.Longhurst A., Sathyendranath S., Platt T., Caverhill C., An estimate of global primary production in the ocean from satellite radiometer data. J. Plankton Res. 17, 1245–1271 (1995). [Google Scholar]
- 140.Galla G., Praeg N., Rzehak T., Sprecher E., Colla F., Seeber J., Illmer P., Hauffe H. C., Comparison of DNA extraction methods on different sample matrices within the same terrestrial ecosystem. Sci. Rep. 14, 8715 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Nelson M. C., Morrison H. G., Benjamino J., Grim S. L., Graf J., Analysis, optimization and verification of illumina-generated 16S rRNA gene amplicon surveys. PLOS ONE 9, e94249 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Topçuoğlu B. D., Stewart L. C., Morrison H. G., Butterfield D. A., Huber J. A., Holden J. F., Hydrogen limitation and syntrophic growth among natural assemblages of thermophilic methanogens at deep-sea hydrothermal vents. Front. Microbiol. 7, 1240 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Callahan B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., Holmes S. P., DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods. 13, 581–583 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., Glöckner F. O., The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Bray J. R., Curtis J. T., An ordination of the upland forest communities of Southern Wisconsin. Ecol. Monogr. 27, 325–349 (1957). [Google Scholar]
- 146.Kruskal J. B., Nonmetric multidimensional scaling: A numerical method. Psychometrika 29, 115–129 (1964). [Google Scholar]
- 147.J. Oksanen, F. G. Blanchet, R. Kindt, P. Legendre, P. R. Minchin, R. B. O’Hara, G. L. Simpson, P. Solymos, M. H. H. Stevens, H. Wagner, vegan: Community Ecology Package (2012); http://cran.r-project.org/package=vegan).
- 148.Nurk S., Meleshko D., Korobeynikov A., Pevzner P. A., MetaSPAdes: A new versatile metagenomic assembler. Genome Res. 27, 824–834 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hyatt D., Chen G.-L., LoCascio P. F., Land M. L., Larimer F. W., Hauser L. J., Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11, 119 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Aramaki T., Blanc-Mathieu R., Endo H., Ohkubo K., Kanehisa M., Goto S., Ogata H., KofamKOALA: KEGG Ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics 36, 2251–2252 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Probst A. J., Elling F. J., Castelle C. J., Zhu Q., Elvert M., Birarda G., Holman H.-Y. N., Lane K. R., Ladd B., Ryan M. C., Woyke T., Hinrichs K.-U., Banfield J. F., Lipid analysis of CO2-rich subsurface aquifers suggests an autotrophy-based deep biosphere with lysolipids enriched in CPR bacteria. ISME J. 14, 1547–1560 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Eddy S. R., Accelerated profile HMM searches. PLOS Comput. Biol. 7, e1002195 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Menzel P., Ng K. L., Krogh A., Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat. Commun. 7, 11257 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wagner G. P., Kin K., Lynch V. J., Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 131, 281–285 (2012). [DOI] [PubMed] [Google Scholar]
- 155.Gruber-Vodicka H. R., Seah B. K. B., Pruesse E., PhyloFlash: Rapid small-subunit rRNA profiling and targeted assembly from metagenomes. mSystems 5, e00920-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.H. Wickham, ggplot2: Elegant Graphics for Data Analysis. (Springer-Verlag, 2009); https://ggplot2.tidyverse.org.
- 157.Clarke K. R., Non-parametric multivariate analyses of changes in community structure. Austral. J. Ecol. 18, 117–143 (1993). [Google Scholar]
- 158.Marlow J. J., Colocci I., Jungbluth S. P., Weber N. M., Gartman A., Kallmeyer J., Mapping metabolic activity at single cell resolution in intact volcanic fumarole sediment. FEMS Microbiol. Lett. 367, fnaa031 (2020). [DOI] [PubMed] [Google Scholar]
- 159.Crognale S., Venturi S., Tassi F., Rossetti S., Rashed H., Cabassi J., Capecchiacci F., Nisi B., Vaselli O., Morrison H. G., Sogin M. L., Fazi S., Microbiome profiling in extremely acidic soils affected by hydrothermal fluids: The case of the Solfatara Crater (Campi Flegrei, southern Italy). FEMS Microbiol. Ecol. 94, fiy190 (2018). [DOI] [PubMed] [Google Scholar]
- 160.Dutta A., Sar P., Sarkar J., Dutta Gupta S., Gupta A., Bose H., Mukherjee A., Roy S., Archaeal communities in deep terrestrial subsurface underneath the Deccan Traps, India. Front. Microbiol. 10, 1362 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Bekaert D. V., Gazel E., Turner S., Behn M. D., de Moor J. M., Zahirovic S., Manea V. C., Hoernle K., Fischer T. P., Hammerstrom A., Seltzer A. M., Kulongoski J. T., Patel B. S., Schrenk M. O., Halldórsson S. A., Nakagawa M., Ramírez C. J., Krantz J. A., Yücel M., Ballentine C. J., Giovannelli D., Lloyd K. G., Barry P. H., High 3He/4He in central Panama reveals a distal connection to the Galápagos plume. Proc. Natl. Acad. Sci. U.S.A. 118, e2110997118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Magnabosco C., Timmers P. H. A., Lau M. C. Y., Borgonie G., Linage-Alvarez B., Kuloyo O., Alleva R., Kieft T. L., Slater G. F., van Heerden E., Sherwood Lollar B., Onstott T. C., Fluctuations in populations of subsurface methane oxidizers in coordination with changes in electron acceptor availability. FEMS Microbiol. Ecol. 94, fiy089 (2018). [DOI] [PubMed] [Google Scholar]
- 163.Etiope G., Vadillo I., Whiticar M. J., Marques J. M., Carreira P. M., Tiago I., Benavente J., Jiménez P., Urresti B., Abiotic methane seepage in the Ronda peridotite massif, southern Spain. Appl. Geochem. 66, 101–113 (2016). [Google Scholar]
- 164.Meyer-Dombard D. R., Osburn M. R., Cardace D., Arcilla C. A., The effect of a tropical climate on available nutrient resources to springs in ophiolite-hosted, deep biosphere ecosystems in the Philippines. Front. Microbiol. 10, 761 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Cardace D., Meyer-Dombard D. R., Woycheese K. M., Arcilla C. A., Feasible metabolisms in high pH springs of the Philippines. Front. Microbiol. 6, 10 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Vigni L. L., Daskalopoulou K., Calabrese S., Kyriakopoulos K., Parello F., Brugnone F., D’Alessandro W., Geochemical characterisation of the thermo-mineral waters of Greece. Environ. Geochem. Health 44, 2111–2133 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Daskalopoulou K., Calabrese S., Grassa F., Kyriakopoulos K., Parello F., Tassi F., D’Alessandro W., Origin of methane and light hydrocarbons in natural fluid emissions: A key study from Greece. Chem. Geol. 479, 286–301 (2018). [Google Scholar]
- 168.Bomberg M., Lamminmäki T., Itävaara M., Microbial communities and their predicted metabolic characteristics in deep fracture groundwaters of the crystalline bedrock at Olkiluoto, Finland. Biogeosciences 13, 6031–6047 (2016). [Google Scholar]
- 169.Edwards R. A., Rodriguez-Brito B., Wegley L., Haynes M., Breitbart M., Peterson D. M., Saar M. O., Alexander S., Alexander E. C., Rohwer F., Using pyrosequencing to shed light on deep mine microbial ecology. BMC Genomics 7, 57 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Magnabosco C., Tekere M., Lau M. C. Y., Linage B., Kuloyo O., Erasmus M., Cason E., van Heerden E., Borgonie G., Kieft T. L., Olivier J., Onstott T. C., Comparisons of the composition and biogeographic distribution of the bacterial communities occupying South African thermal springs with those inhabiting deep subsurface fracture water. Front. Microbiol. 5, 679 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Lavalleur H. J., Colwell F. S., Microbial characterization of basalt formation waters targeted for geological carbon sequestration. FEMS Microbiol. Ecol. 85, 62–73 (2013). [DOI] [PubMed] [Google Scholar]
- 172.Koppers A. A. P., Yamazaki T., Geldmacher J., IODP Expedition 330: Drilling the louisville Seamount trail in the SW pacific. Sci. Drill. 15, 11–22 (2013). [Google Scholar]
- 173.Sylvan J. B., Wankel S. D., LaRowe D. E., Charoenpong C. N., Huber J. A., Moyer C. L., Edwards K. J., Evidence for microbial mediation of subseafloor nitrogen redox processes at Loihi Seamount, Hawaii. Geochim. Cosmochim. Acta 198, 131–150 (2017). [Google Scholar]
- 174.Zinke L. A., Glombitza C., Bird J. T., Røy H., Jørgensen B. B., Lloyd K. G., Amend J. P., Reese B. K., Microbial organic matter degradation potential in Baltic Sea sediments is influenced by depositional conditions and in situ geochemistry. Appl. Environ. Microbiol. 85, e02164-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Zinke L. A., Reese B. K., McManus J., Wheat C. G., Orcutt B. N., Amend J. P., Sediment microbial communities influenced by cool hydrothermal Fluid migration. Front. Microbiol. 9, 1249 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Goordial J., D’Angelo T., Labonté J. M., Poulton N. J., Brown J. M., Stepanauskas R., Früh-Green G. L., Orcutt B. N., Microbial diversity and function in shallow subsurface sediment and oceanic lithosphere of the Atlantis Massif. mBio 12, e0049021 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Teske A., McKay L. J., Ravelo A. C., Aiello I., Mortera C., Núñez-Useche F., Canet C., Chanton J. P., Brunner B., Hensen C., Ramírez G. A., Sibert R. J., Turner T., White D., Chambers C. R., Buckley A., Joye S. B., Soule S. A., Lizarralde D., Characteristics and evolution of sill-driven off-axis hydrothermalism in Guaymas Basin—The Ringvent site. Sci. Rep. 9, 13847 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Brandt L. D., House C. H., Marine subsurface microbial community shifts across a hydrothermal gradient in Okinawa trough sediments. Archaea 2016, 2690329 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Dowell F., Cardman Z., Dasarathy S., Kellermann M., Lipp J. S., Ruff S. E., Biddle J. F., McKay L., MacGregor B. J., Lloyd K. G., Albert D. B., Mendlovitz H., Hinrichs K.-U., Teske A., Microbial communities in methane- and short chain alkane-rich hydrothermal sediments of Guaymas Basin. Front. Microbiol. 7, 17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Lecoeuvre A., Ménez B., Cannat M., Chavagnac V., Gérard E., Microbial ecology of the newly discovered serpentinite-hosted Old City hydrothermal field (southwest Indian ridge). ISME J. 15, 818–832 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kleindienst S., Grim S., Sogin M., Bracco A., Crespo-Medina M., Joye S. B., Diverse, rare microbial taxa responded to the Deepwater Horizon deep-sea hydrocarbon plume. ISME J. 10, 400–415 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Klasek S. A., Torres M. E., Loher M., Bohrmann G., Pape T., Colwell F. S., Deep-sourced fluids from a convergent margin host distinct Subseafloor microbial communities that change upon mud flow expulsion. Front. Microbiol. 10, 1436 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Angermeyer A., Crosby S. C., Huber J. A., Decoupled distance-decay patterns between dsrA and 16S rRNA genes among salt marsh sulfate-reducing bacteria. Environ. Microbiol. 18, 75–86 (2016). [DOI] [PubMed] [Google Scholar]
- 184.Trembath-Reichert E., Butterfield D. A., Huber J. A., Active subseafloor microbial communities from Mariana back-arc venting fluids share metabolic strategies across different thermal niches and taxa. ISME J. 13, 2264–2279 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Angermeyer A., Crosby S. C., Huber J. A., Salt marsh sediment bacterial communities maintain original population structure after transplantation across a latitudinal gradient. PeerJ. 6, e4735 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Roguet A., Eren A. M., Newton R. J., McLellan S. L., Fecal source identification using random forest. Microbiome. 6, 185 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Newton R. J., Huse S. M., Morrison H. G., Peake C. S., Sogin M. L., McLellan S. L., Shifts in the microbial community composition of Gulf coast beaches following beach oiling. PLOS ONE 8, e74265 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Feng S., Bootsma M., McLellan S. L., Human-associated lachnospiraceae genetic markers improve Detection of fecal pollution sources in urban waters. Appl. Environ. Microbiol. 84, e00309-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Bradley J. A., Arndt S., Amend J. P., Burwicz-Galerne E., LaRowe D. E., Sources and fluxes of organic carbon and energy to microorganisms in global marine sediments. Front. Microbiol. 13, 910694 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Supplementary Methods
Figs. S1 to S24
Legends for datasets S1 to S12
Legends for movies S1 and S2
References
Datasets S1 to S12
Movies S1 and S2