Abstract
Background
Pancreatic neuroendocrine tumors (pNETs) are genomically diverse tumors. The management of newly diagnosed well‐differentiated pNETs is limited by a lack of sensitivity of existing biomarkers for prognostication. Our goal was to investigate the potential utility of genetic markers as a predictor of progression‐free survival (PFS) and recurrence‐free survival (RFS).
Methods
Whole‐exome sequencing of resected well‐differentiated, low and intermediate‐grade (G1 and G2) pNETs and normal adjacent tissue from patients who underwent resection from 2005 to 2015 was performed. Genetic alterations were classified using pan‐genomic and oncogenic pathway classifications. Additional samples with genetic and clinicopathologic data available were obtained from the publicly available International Cancer Genome Consortium (ICGC) database and included in the analysis. The prognostic relevance of these genomic signatures on PFS and RFS was analyzed.
Results
Thirty‐one patients who underwent resection for pNET were identified. Genomic analysis of mutational, copy number, cytogenetic, and complex phenomena revealed similar patterns to prior studies of pNETs with relatively few somatic gene mutations but numerous instances of copy number changes. Analysis of genomic and clinicopathologic outcomes using the combined data from our study as well as the ICGC pNET cohort (n = 124 patients) revealed that the recurrent pattern of whole chromosome loss (RPCL) and metastatic disease were independently associated with disease progression. When evaluating patients with local disease at the time of resection, RPCL and alterations in the TGFβ oncogenic pathway were independently associated with the risk of recurrence.
Conclusions
Well‐differentiated pNETs are genomically diverse tumors. Pathway signatures may be prognostic for predicting disease progression and recurrence.
Keywords: genomic biomarker, pancreatic neuroendocrine tumor, prognosis, recurrence, whole‐exome sequencing
1. INTRODUCTION
The incidence and detection of pancreatic neuroendocrine tumors (pNETs) is increasing. 1 , 2 The majority of WD‐pNETs are low‐grade and indolent. However, a small proportion of patients with G1 and G2 WD‐pNETs have an aggressive, treatment‐resistant form of the disease. 3 Current clinicopathologic predictors of disease recurrence and progression lack sensitivity, resulting in conjecture for which patients should undergo resection or active surveillance. 4 , 5 , 6 , 7 Genomic signatures have demonstrated promise in risk stratifying disease progression and recurrence in other solid tumors. 8 , 9
The genetics of pNETs is marked by a wide assortment of genomic dysregulation without well‐established prototypical or canonical alterations. 10 , 11 , 12 Studies to date have found pNETs to have a relatively low rate of recurrent somatic mutations compared to other cancer types. 11 In fact, the genomic landscape is instead dominated by copy number changes and aneuploidy. 12 Studies of RNA expression and epigenetics have also not revealed canonical alterations. 12 , 13 Furthermore, studies have found large differences in the genetics of pNETs from different populations worldwide. 14 Despite the diversity of individual genomic alterations, patterns have emerged regarding the key pathways and mechanisms driving pNET tumorigenesis. 15
The combination of indolent clinical course, small sample sizes, and relative lack of recurrent alterations has limited the ability to define genomic alterations that might prove useful in guiding the surgical management of WD‐pNET patients. Thus, while IHC studies looking at Ki67, DAXX, ATRX, and alternative lengthening and shortening of telomeres (ALT) have been studied for the risk of liver metastases 16 and survival, 17 whole exome or genome approaches have not been tailored to evaluate for surgically relevant alterations and have resulted in conflicting findings. 10 In this study, we aimed to identify genomic alterations and oncogenic pathway signatures in our institutional cohort of pNET patients who underwent resection and combine these findings with recent pNET sequencing studies and assess for associations with progression and recurrence free survival.
2. METHODS
2.1. Patient selection and sample collection
Patients who underwent resection for pNET at our institution from 2005 to 2015 and for which fresh frozen specimens were available in the UCLA Pancreas Tissue Bank were identified and obtained under an Institutional Review Board (IRB)‐approved protocol at the University of California, Los Angeles (IRB#13‐001646). Pathologic diagnosis, staging and quality control on biobanked tissues was performed by a gastrointestinal pathologist with extensive experience with pancreas pathology (DWD). Inclusion criteria ≥18 years of age, G1 or G2 pNETs, and further availability of patient‐matched normal adjacent tissue for sequencing. G3 pNETs or neuroendocrine tumors of non‐pancreatic origin on pathologic examination were excluded. Patients with available clinicopathologic and genomic data from the International Cancer Genome Consortium (ICGC) pNET study meeting the same inclusion/exclusion criteria were included to increase the power of the analyses.
2.2. DNA processing and sequencing
DNA sequencing was performed by NantOmics, LLC as a service. At least 100 μg of genomic DNA was extracted from fresh‐frozen tissue using the QIAsymphony DSP DNA Minikit (Qiagen) per the manufacturer's protocol. Tissue from the surrounding normal pancreas was used for control samples and to evaluate for germline mutations. Genomic quality was assessed using Fragment Analyzer and analyzed with PROSize 3.0 software (Agilent). Exomic DNA libraries were prepared using the xGen Exome Research Panel v1.0 (IDT), and libraries were prepared using the KAPA Hyper prep kit (Kapa Biosystems) per the manufacturer's protocol. DNA libraries were sequenced to a target depth of 200 reads for tumor sample and 100 reads for normal samples on the Illumina HiSeq platform (Illumina). The methods for exome variant calling, copy number analysis, and genetic variable analysis are described in Supporting Information S2: supplemental methods.
2.3. Statistical analysis
Categorical variables were summarized as frequencies and percentages while continuous variables were summarized as medians and interquartile ranges (IQRs). Chi‐square test was used for categorical variables when appropriate. The primary statistical endpoint was investigator‐assessed progression‐free survival (PFS). PFS was defined as the time from surgery until first evidence of objective tumor progression or recurrence. Secondary analysis for recurrence‐free survival (RFS) was performed on patients without metastatic disease at the time of resection. Kaplan–Meier methods were used to estimate differences in survival. p Values less than 0.05 were considered statistically significant. The multivariable model included variables with p values less than 0.2 on univariable analysis. Hazard ratios (HRs) and 95% confidence intervals (CIs) were estimated using Cox proportional hazard regression. The 2010 World Health Organization (WHO) neuroendocrine neoplasm grading was used. Statistical analyses were performed in SPSS (v28). Oncoplots and graphics were created using SPSS, Prism (v9.1.0), and Microsoft Excel.
3. RESULTS
3.1. Institutional cohort
A total of 40 patients who underwent resection for pNETs from 2005 to 2015 with frozen tissue available for analysis were identified. Three NETs initially classified as of pancreatic origin were subsequently determined to be of duodenum or ampulla origin and were excluded. An additional six patients were excluded (four patients with G3 tumors and two patients with unassigned grades). Of the 31 patients analyzed, the median age was 65, and nearly 75% were male. Twenty patients had G2 tumors. The median follow‐up time for the cohort was 93 months. The clinicopathologic characteristics are show in Table 1.
Table 1.
Clinicopathologic features of institutional cohort.
| N = 31 (%) | |
|---|---|
| Age, median (IQR) | 65 (51–78) |
| Sex | |
| Female | 8 (25.8) |
| Male | 23 (74.2) |
| Familial syndrome | 2 (6.5) |
| Functional tumor | 3 (9.7) |
| Pathology | |
| Median tumor size, cm (IQR) | 3 (2.3–6.1) |
| Node Positive | 11 (35.5) |
| Metastatic | 5 (16.1) |
| WHO grade | |
| G1 | 11 (35.5) |
| G2 | 20 (64.5) |
| Perineural invasion | 9 (29) |
| Lymphovascular invasion | 24 (64.9) |
| Median follow‐up, months (IQR) | 93 (44–110) |
| Mortality | 4 (12.9) |
Abbreviations: IQR, interquartile range; WHO, World Health Organization.
Whole‐exome sequencing was performed on all 31 patients' primary tumors as well as matched normal germline tissue. We obtained a median of 201 million reads (IQR: 150–239 million reads) from the tumor tissue and a median of 95 million reads (IQR: 72–127 million reads) from the matched normal tissue. There was a low frequency of somatic single nucleotide variants (SNVs) and indels detected. The median exonic mutation rate was 0.62 mutations per megabase (range: 0–12.41). The most frequently mutated gene was MEN1 (n = 13; 42%). Few other recurrent mutations were identified as demonstrated in Figure 1. The individual gene mutations from our institutional cohort largely correlated with the findings from the ICGC cohort. However, ATRX, which has been implicated in low‐grade pNETs, was only mutated in one patient in the institutional cohort (3.2%) versus 39.8% in the ICGC cohort. Additionally, the institutional cohort demonstrated mutations in MLL3 (19.4%) DIS3L2 (12.9%), and DEPDC5 (12.9%) while no patients had mutations of these genes in the ICGC cohort (data not shown).
Figure 1.
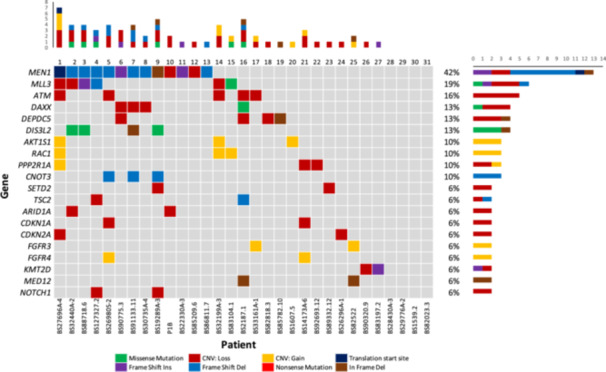
Oncoplot of the most frequently mutated genes in the institutional cohort.
In contrast to the low somatic mutation rate, CNVs and chromosomal abnormalities were almost universally found. Loss of heterozygosity (LOH) occurred in 18 patients (58.1%), with a median of at least one chromosome demonstrating an arm‐length or longer LOH event (range: 0–12 events) across all pNETs. Additionally, tumor cytogenetic analysis revealed four distinct groups of copy number changes previously identified in the ICGC pNET study: a group with numerous high level copy number gains and polyploidy (n = 6), a group with significant aneuploidy (n = 6), a group with fewer total copy number events (n = 14), and a group with a recurrent pattern of whole chromosome loss (RPCL, n = 5). Supporting Information S1: Figure 1 demonstrates a cytogenetic plot of inferred integral copy number for a patient exhibiting RPCL.
To better understand the genetic alterations driving pNET progression and recurrence we evaluated SNVs, indels, and somatic CNVs involving recurrently altered oncogenic pathways developed from the pan‐cancer TCGA data 18 as well as known chromatin remodeling genes. A total of 73 alterations were found, involving all 11 identified pathways. The chromatin pathway was the most frequently affected (61.3% of samples), followed by the RAS (32.3%) and WNT (22.6%) pathways (Figure 2A). The frequencies of oncogenic pathway alterations in the ICGC data set were similar to the institutional cohort with the exception of the chromatin pathway, which had a significantly higher alteration rate than our institutional cohort (80.6% vs. 61.3%; p = 0.029, data not shown).
Figure 2.
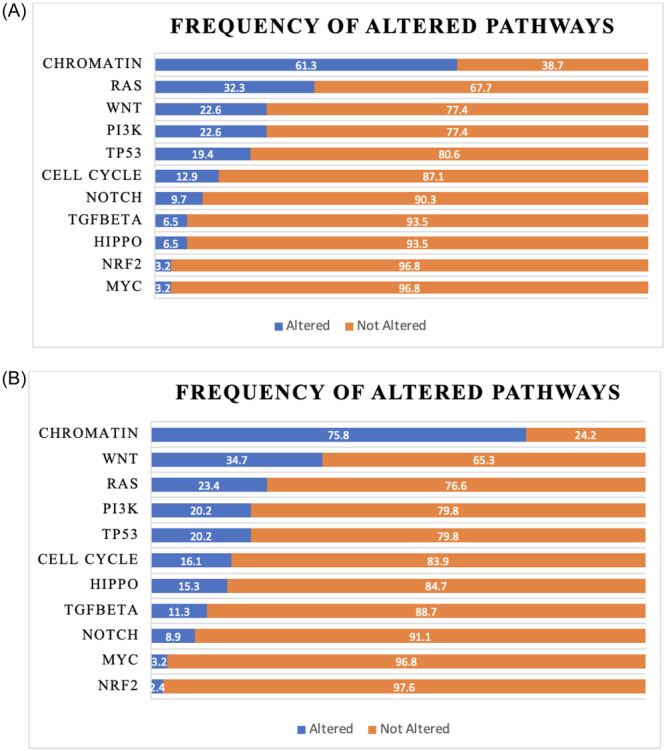
(A, B) Frequency of altered pathways.
3.2. Combined cohort
To better understand the drivers and clinical importance of genetic alterations in pNETs we combined genetic and clinicopathologic data from the ICGC pNET study (n = 93) with our institutional cohort (n = 124 patients) (Table 2). 11 Using this combined data set, we developed a classification model that incorporated clinicopathologic variables, the most frequent recurrent somatic alterations (n = 55 genes), copy number subtypes, and pathway alterations for all patients.
Table 2.
Clinicopathologic features of combined cohort.
| N = 124 (%) | |
|---|---|
| Age, median (IQR) | 65 (51–78) |
| Sex | |
| Female | 42 (33.9) |
| Male | 82 (66.1) |
| Familial syndrome | 5 (4) |
| Functional tumor | 14 (11.3) |
| Pathology | |
| Median tumor size, cm (IQR) | 3 (2.3–6.1) |
| Node positive | 53 (42.7) |
| Metastatic | 20 (16.1) |
| WHO grade | |
| G1 | 47 (37.9) |
| G2 | 77 (62.1) |
| Perineural invasion | 40 (32.3) |
| Lymphovascular invasion | 70 (56.5) |
| Median follow‐up, months (IQR) | 38 (22–84) |
| Mortality | 14 (11.3) |
Abbreviations: IQR, interquartile range; WHO, World Health Organization.
3.3. Progression‐free survival
In the combined cohort, 32.3% of patients (n = 40) had progression and/or recurrence of their disease. Of patients with progressive disease, 40% were noted to have already had metastatic disease at the time of surgery. Additionally, 80% of patients with metastases had disease progression compared with 23% in those without metastatic disease (p < 0.001). On univariate analysis comparing clinicopathologic characteristics and oncogenic pathways in those who progressed with those who did not, metastatic disease, positive lymph nodes, RPCL, PI3K, and the cell cycle pathway were more frequent in patients who had disease progression (Table 3). On multivariable Cox Regression, cell cycle pathway (OR: 4.64, 95% CI: 1.78–12.11), metastatic disease (OR: 4.20, 95% CI: 1.54–11.43), and RPCL (OR: 2.61, 95% CI: 1.11–6.12) were independent predictors of disease progression (Figure 3).
Table 3.
Progression‐free survival univariable Cox regression.
| Hazard ratio | 95.0% CI for Exp(B) | p Value | ||
|---|---|---|---|---|
| Tumor size | 1.12 | 0.96 | 1.28 | 0.11 |
| Positive lymph nodes | 2.87 | 1.34 | 6.15 | 0.007 |
| Grade | 0.76 | 0.51 | 1.15 | 0.19 |
| Metastatic | 4.65 | 2.23 | 9.68 | <0.001 |
| RPCL | 3.67 | 1.79 | 7.52 | <0.001 |
| Cell cycle pathway | 3.64 | 1.67 | 7.87 | 0.001 |
| Chromatin pathway | 2.57 | 0.89 | 7.41 | 0.08 |
| PI3K pathway | 2.51 | 1.17 | 5.41 | 0.018 |
| TGF beta pathway | 2.42 | 0.99 | 5.95 | 0.053 |
Abbreviations: CI, confidence interval; PI3K, phosphoinositide 3‐kinase; RPCL, recurrent pattern of whole chromosome loss; TGF, transforming growth factor.
Figure 3.
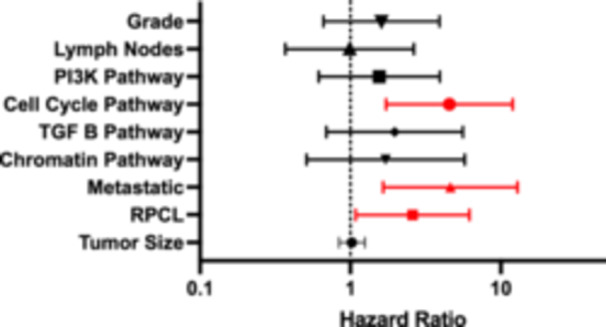
Progression‐free survival multivariable Cox regression. PI3K, phosphoinositide 3‐kinase; RPCL, recurrent pattern of whole chromosome loss; TGF, transforming growth factor.
3.4. Recurrence‐free survival
Given the high proportion of metastatic patients who developed disease progression, the analysis was repeated excluding patients with metastatic disease at presentation. Of 104 patients without metastatic disease at the time of surgery, 24 patients (23.1%) had disease recurrence. On univariable analysis, RPCL, PI3K, cell cycle pathway, and the TGFβ pathway were associated with disease recurrence (Table 4). For the multivariable analysis, tumor size, tumor grade, chromatin pathway, and the NOTCH pathway were also included. The TGFβ pathway (OR: 5.5, 95% CI: 1.36–22.23) and RPCL (OR: 8.73, 95% CI: 2.63–28.99) were independent predictors associated with disease recurrence (Figure 4, Supporting Information: Figure 2).
Table 4.
Recurrence‐free survival univariable Cox regression.
| Hazard ratio | 95.0% CI for Exp(B) | p Value | ||
|---|---|---|---|---|
| Tumor size | 1.12 | 0.973 | 1.37 | 0.1 |
| Grade | 3.05 | 0.88 | 10.58 | 0.08 |
| RPCL | 6.3 | 2.38 | 16.73 | <0.001 |
| Cell cycle pathway | 4.99 | 1.94 | 12.83 | <0.001 |
| Chromatin pathway | 2.35 | 0.67 | 8.25 | 0.18 |
| NOTCH pathway | 2.15 | 0.7 | 6.58 | 0.18 |
| PI3K pathway | 3.15 | 1.22 | 8.16 | 0.018 |
| TGF beta pathway | 5.29 | 1.97 | 14.2 | <0.001 |
Abbreviations: CI, confidence interval; PI3K, phosphoinositide 3‐kinase; RPCL, recurrent pattern of whole chromosome loss; TGF, transforming growth factor.
Figure 4.
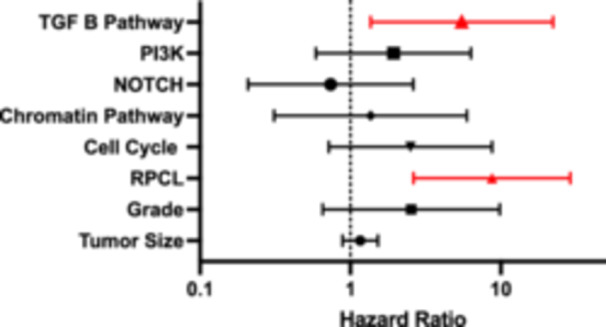
Recurrence‐free survival multivariable Cox regression. PI3K, phosphoinositide 3‐kinase; RPCL, recurrent pattern of whole chromosome loss; TGF, transforming growth factor.
4. DISCUSSION
In this study, we provide a detailed analysis of the genomic profile of G1 and G2 pNETs and their association with disease progression and recurrence. Herein, we demonstrated that WD‐pNETs in our cohort are genomically heterogenous with similar mutation profiles seen in prior studies. 11 , 16 We then demonstrated that certain genomic aberrations were associated with decreased PFS and RFS while demonstrating certain clinicopathologic characteristics, such as lymph node metastases and tumor size, did not affect these outcomes. The refinement of oncogenic pathway membership has allowed researchers to expand beyond single genes to classifying tumors based on the pathways involved. 18 , 19 By focusing on the larger pool of potential alterations, we hoped to use these new genomic features to look for prognostic predictive and potentially actionable genomic alterations and targetable pathways.
Not surprisingly, of the clinicopathologic characteristics, metastatic disease at the time of surgical resection was a strong predictor of decreased PFS. The significance of lymph node metastases in relation to disease progression and recurrence is controversial with several conflicting studies. 20 , 21 , 22 In our study, positive lymph nodes were not associated with RFS or PFS in our multivariable analyses. However, the effect of lymph node metastases on these outcomes may be limited by an insufficient follow‐up time.
In addition to synchronous metastases, cell cycle pathway alterations and RPCL were associated with decreased PFS. Prior studies have associated cell cycle pathway dysregulation with a higher proliferative index in NETs. 23 A single‐arm phase II study evaluating the effect of Palbociclib (cyclin‐dependent kinase 4/6 inhibitor) monotherapy on metastatic G1 and G2 pNETs (PALBONET trial) did not demonstrate a measurable decrease in disease burden. However, all study participants were molecularly unselected and had already failed other systemic therapy regimens. 24 About half of the cohort had no disease progression during the study period, although this could not be attributed to the Palbociclib treatment. The researchers concluded that predictive biomarkers are important and may improve patient selection for CDK4/6 therapy. The association of cell cycle pathway alteration with decreased PFS gives credence to whole exome or targeted sequencing to identify patients who may benefit from targeted cell‐cycle modulators.
Additionally, RPCL has been identified in pNETs previously. 11 , 25 The chromosomes most frequently affected are 1, 2, 3, 6, 8, 10, 11, 15, 16, and 22 (Supporting Information S1: Figure 1). Thirty‐three patients in our combined cohort had this aneuploid abnormality (26.6%), including 5 from our institutional cohort (16.1%). Associations between RPCL and the mutational state of MEN1‐DAXX‐ATRX (MAD+) have been established and associated with advanced disease. 25 In one study, MAD+ mutational status was associated with centromere cohesion failure, leading to selective chromosomal loss and the a loss of heterozygosity pattern seen in metastatic pNETs. 25
Alterations in the Pi3K/Akt/mTOR signaling pathway has been implicated in proliferation of pNETs. 26 Everolimus, an mTOR inhibitor, is approved by the FDA for pNETs and often used as a second‐line agent based on a PFS benefit for advanced low‐ and intermediate‐grade pNETS observed in the RADIANT‐3 trial. 27 , 28 In our study, the Pi3K was associated with disease progression on univariable analysis. However, in our multivariable model, this association was not significant. This discrepancy may be explained in part by the inclusion of primarily patients that did not meet the criteria that RADIANT‐3 trial considered “advanced” disease (i.e., unresectable or diffuse metastatic disease).
Recurrence free survival was negatively associated with TGFβ pathway alteration in addition to RPCL. TGFβ has demonstrated tumor suppressing and tumor promoting effects in several cancers. In NETs, neutralization of TGFβ at the protein level has been shown to induce cell proliferation. 29 Other in vitro studies demonstrated that the growth suppressor effect of TGF‐B is mediated by the somatostatin signaling pathway. However, when the TGFβ‐somatostatin connection is disrupted, TGFβ signaling causes cell proliferation. 30 , 31 In our study, tumors in 12 patients without metastatic disease harbored TGFβ mutations of which 6 (50%) had disease recurrence. Despite the strong association of TGFβ alteration with recurrence, only 2 of the 20 patients (10%) with metastatic disease at the time of surgical resection (synchronous metastases) had alterations in the TGFβ pathway. This suggests differences in pathway signaling alterations in synchronous and metachronous metastases exist and warrants further investigation.
The prognostic findings of our study can be used to inform clinicians on decision‐making in the preoperative setting as well as postoperatively for surveillance purposes. Clinicopathologic characteristics of pNETs have long been used to predict recurrence and progression. 32 For example, the National Comprehensive Cancer Network guidelines provide recommendations for surgical resection based on size and other radiographic characteristics. The guidelines recommend the generally accepted, yet controversial, cutoff of 2 cm when deciding to pursue observation versus resection of pNETs. However, one meta‐analysis including 1491 resected pNETs demonstrated a survival benefit for smaller tumors. 33 Preoperative sequencing of pNETs can bridge this gap for prognostication and better inform physicians when counseling patients. Several studies have demonstrated that endoscopic ultrasound with fine needle or core needle aspiration of pancreatic lesions provides a sufficient DNA yield for next generation sequencing. 34 Additionally, understanding the molecular mechanisms underlying the genomic patterns observed in pNETs is important for the development of strategic therapeutic options for clinically actionable mutations and altered pathways.
In summary, we performed whole‐exome sequencing of tumors from 31 patients with WD‐pNETs to identify the landscape and diversity of genomic alterations. This data was then combined with genomic data from additional patients in the ICGC pNET cohort to increase our sample size and evaluate for clinically relevant genomic alterations. By focusing on pathways and pan‐genomic variables as opposed to individual genes, we attempted to overcome the genomic diversity of pNETs. We identified several prognostic genomic variables that outperformed the clinicopathologic variables of grade and tumor rise in predicting PFS and DFS. Our findings are limited by the high cost of exome sequencing and data analysis required at this time. However, these costs will likely continue to decrease in the coming years. Additionally, our study is limited by a relatively small sample size and diversity of sample types included in the analysis. Given these limitations and large number of genomic variables studied, our results are largely exploratory. Further studies are needed to confirm these findings and to better characterize their clinical implications.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
ETHICS STATEMENT
This study was conducted under our Institutional Review Board (IRB) approved protocol at the University of California, Los Angeles (IRB#11‐002112 and IRB#13‐001646). All of the authors have seen and approved the manuscript being submitted.
SYNOPSIS
Pancreatic neuroendocrine tumors (pNETs) are genomically diverse tumors, and current biomarkers lack sensitivity for prognostication. This study aimed to evaluate the potential of genetic markers in predicting progression‐free and recurrence‐free survival by performing whole‐exome sequencing on resected pNETs as well as analyzing previously published pNET genomic data from the International Cancer Genome Consortium. The results indicated that recurrent patterns of whole chromosome loss (RPCL) and TGFβ pathway alterations were associated with disease progression and recurrence, suggesting that pathway signatures may serve as valuable prognostic tools.
Supporting information
Supporting information.
Supporting information.
ACKNOWLEDGMENTS
Thomas G. Graeber is supported by the NCI/NIH (P01CA168585, R01CA222877, and R01CA227089), UCLA SPORE in Prostate Cancer (NIH P50CA092131), and the W.M. Keck Foundation. David W. Dawson and the UCLA Pancreas Tissue Bank are supported by the Hirshberg Foundation for Pancreatic Research.
Mederos MA, Court CM, Dipardo BJ, et al. Oncogenic pathway signatures predict the risk of progression and recurrence in well‐differentiated pancreatic neuroendocrine tumors. J Surg Oncol. 2024;130:1070‐1077. 10.1002/jso.27830
Michael A. Mederos and Colin M. Court contributed equally to this study.
Contributor Information
Michael A. Mederos, Email: mmederos.md@gmail.com.
James S. Tomlinson, Email: jtomlinson@mednet.ucla.edu.
DATA AVAILABILITY STATEMENT
The data sets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Sonbol MB, Mazza GL, Mi L, et al. Survival and incidence patterns of pancreatic neuroendocrine tumors over the last 2 decades: a SEER database analysis. Oncologist. 2022;27(7):573‐578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang J, Liu J, He C, et al. Trends in incidence and survival of patients with pancreatic neuroendocrine neoplasm, 1987‐2016. J Oncol. 2021;2021:4302675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Halfdanarson TR, Rabe KG, Rubin J, Petersen GM. Pancreatic neuroendocrine tumors (PNETs): incidence, prognosis and recent trend toward improved survival. Ann Oncol. 2008;19(10):1727‐1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bilimoria KY, Bentrem DJ, Merkow RP, et al. Application of the pancreatic adenocarcinoma staging system to pancreatic neuroendocrine tumors. J Am Coll Surg. 2007;205(4):558‐563. [DOI] [PubMed] [Google Scholar]
- 5. Bilimoria KY, Talamonti MS, Tomlinson JS, et al. Prognostic score predicting survival after resection of pancreatic neuroendocrine tumors: analysis of 3851 patients. Ann Surg. 2008;247(3):490‐500. [DOI] [PubMed] [Google Scholar]
- 6. Sho S, Court CM, Winograd P, et al. A prognostic scoring system for the prediction of metastatic recurrence following curative resection of pancreatic neuroendocrine tumors. J Gastrointest Surg. 2019;23(7):1392‐1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sadot E, Reidy‐Lagunes DL, Tang LH, et al. Observation versus resection for small asymptomatic pancreatic neuroendocrine tumors: a matched case‐control study. Ann Surg Oncol. 2016;23(4):1361‐1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jones GD, Brandt WS, Shen R, et al. A genomic‐pathologic annotated risk model to predict recurrence in early‐stage lung adenocarcinoma. JAMA Surg. 2021;156(2):e205601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ribeiro IP, Caramelo F, Esteves L, et al. Genomic predictive model for recurrence and metastasis development in head and neck squamous cell carcinoma patients. Sci Rep. 2017;7(1):13897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jiao Y, Shi C, Edil BH, et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011;331(6021):1199‐1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Scarpa A, Chang DK, Nones K, et al. Whole‐genome landscape of pancreatic neuroendocrine tumours. Nature. 2017;543(7643):65‐71. [DOI] [PubMed] [Google Scholar]
- 12. Lawrence B, Blenkiron C, Parker K, et al. Recurrent loss of heterozygosity correlates with clinical outcome in pancreatic neuroendocrine cancer. NPJ Genom Med. 2018;3:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stefanoli M, La Rosa S, Sahnane N, et al. Prognostic relevance of aberrant DNA methylation in g1 and g2 pancreatic neuroendocrine tumors. Neuroendocrinology. 2014;100(1):26‐34. [DOI] [PubMed] [Google Scholar]
- 14. Yuan F, Shi M, Ji J, et al. KRAS and DAXX/ATRX gene mutations are correlated with the clinicopathological features, advanced diseases, and poor prognosis in Chinese patients with pancreatic neuroendocrine tumors. Int J Biol Sci. 2014;10(9):957‐965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mafficini A, Scarpa A. Genomic landscape of pancreatic neuroendocrine tumours: the International Cancer Genome Consortium. J Endocrinol. 2018;236(3):R161‐R167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pea A, Yu J, Marchionni L, et al. Genetic analysis of small well‐differentiated pancreatic neuroendocrine tumors identifies subgroups with differing risks of liver metastases. Ann Surg. 2020;271(3):566‐573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Singhi AD, Liu TC, Roncaioli JL, et al. Alternative lengthening of telomeres and loss of DAXX/ATRX expression predicts metastatic disease and poor survival in patients with pancreatic neuroendocrine tumors. Clin Cancer Res. 2017;23(2):600‐609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sanchez‐Vega F, Mina M, Armenia J, et al. Oncogenic signaling pathways in The Cancer Genome Atlas. Cell. 2018;173(2):321‐337.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Doherty GJ, Petruzzelli M, Beddowes E, Ahmad SS, Caldas C, Gilbertson RJ. Cancer treatment in the genomic era. Annu Rev Biochem. 2019;88:247‐280. [DOI] [PubMed] [Google Scholar]
- 20. Casadei R, Ricci C, Pezzilli R, et al. Are there prognostic factors related to recurrence in pancreatic endocrine tumors? Pancreatology. 2010;10(1):33‐38. [DOI] [PubMed] [Google Scholar]
- 21. Krampitz GW, Norton JA, Poultsides GA, Visser BC, Sun L, Jensen RT. Lymph nodes and survival in pancreatic neuroendocrine tumors. AArch Surg. 2012;147(9):820‐827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tan Q, Wang X, Yang L, et al. Analysis of recurrence after resection of well‐differentiated non‐functioning pancreatic neuroendocrine tumors. Medicine. 2020;99(24):e20324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shi Y, Qian ZR, Zhang S, et al. Cell cycle protein expression in neuroendocrine tumors: association of CDK4/CDK6, CCND1, and phosphorylated retinoblastoma protein with proliferative index. Pancreas. 2017;46(10):1347‐1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Grande E, Teulé A, Alonso‐Gordoa T, et al. The PALBONET trial: a phase II study of palbociclib in metastatic grade 1 and 2 pancreatic neuroendocrine tumors (GETNE‐1407). Oncologist. 2020;25(9):745‐e1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Quevedo R, Spreafico A, Bruce J, et al. Centromeric cohesion failure invokes a conserved choreography of chromosomal mis‐segregations in pancreatic neuroendocrine tumor. Genome Med. 2020;12(1):38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Missiaglia E, Dalai I, Barbi S, et al. Pancreatic endocrine tumors: expression profiling evidences a role for AKT‐mTOR pathway. J Clin Oncol. 2010;28(2):245‐255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yao JC, Shah MH, Ito T, et al. Everolimus for advanced pancreatic neuroendocrine tumors. N Engl J Med. 2011;364(6):514‐523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yao JC, Pavel M, Lombard‐Bohas C, et al. Everolimus for the treatment of advanced pancreatic neuroendocrine tumors: overall survival and circulating biomarkers from the randomized, phase III RADIANT‐3 study. J Clin Oncol. 2016;34(32):3906‐3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wimmel A. Autocrine growth inhibition by transforming growth factor‐1 (TGF‐1) in human neuroendocrine tumour cells. Gut. 2003;52(9):1308‐1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Leu FP, Nandi M, Niu C. The effect of transforming growth factor β on human neuroendocrine tumor BON cell proliferation and differentiation is mediated through somatostatin signaling. Mol Cancer Res. 2008;6(6):1029‐1042. [DOI] [PubMed] [Google Scholar]
- 31. Ungefroren H, Kunstner A, Busch H, et al. Differential effects of somatostatin, octreotide, and lanreotide on neuroendocrine differentiation and proliferation in established and primary NET cell lines: possible crosstalk with TGF‐beta signaling. Int J Mol Sci. 2022;23(24):15868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Krogh S, Grønbæk H, Knudsen AR, Kissmeyer‐Nielsen P, Hummelshøj NE, Dam G. Predicting progression, recurrence, and survival in pancreatic neuroendocrine tumors: a single center analysis of 174 patients. Front Endocrinol. 2022;13:925632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Finkelstein P, Sharma R, Picado O, et al. Pancreatic Neuroendocrine Tumors (panNETs): analysis of overall survival of nonsurgical management versus surgical resection. J Gastrointest Surg. 2017;21(5):855‐866. [DOI] [PubMed] [Google Scholar]
- 34. Kandel P, Nassar A, Gomez V, et al. Comparison of endoscopic ultrasound‐guided fine‐needle biopsy versus fine‐needle aspiration for genomic profiling and DNA yield in pancreatic cancer: a randomized crossover trial. Endoscopy. 2021;53(4):376‐382. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.
Supporting information.
Data Availability Statement
The data sets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.


