Abstract
Illumina-based BeadChip arrays have revolutionized genome-wide DNA methylation profiling, pushing it into diagnostics. However, comprehensive quality assessment remains challenging within a wide range of available tissue materials and sample preparation methods. This study tackles two critical issues: differentiating between biological effects and technical artefacts in suboptimal quality samples and the impact of the first sample on the Illumina-like normalization algorithm. We introduce three quality control scores based on global DNA methylation distribution (DB-Score), bin distance from copy number variation analysis (BIN-Score) and consistently methylated CpGs (CM-Score) that rely on biological features rather than internal array controls. These scores, designed to be adjustable for different analysis tools and sample cohort characteristics, were explored and benchmarked across independent cohorts. Additionally, we reveal deviations in beta values caused by different sample rankings with the Illumina-like normalization algorithm, verified these with whole-genome methylation sequencing data and showed effects on differential DNA methylation analysis. Our findings underscore the necessity of consistently utilizing a pre-defined normalization sample within the ranking process to boost reproducibility of the Illumina-like normalization algorithm. Overall, our study delivers valuable insights, practical recommendations and R functions designed to enhance reproducibility and quality assurance of DNA methylation analysis, particularly for challenging sample types.
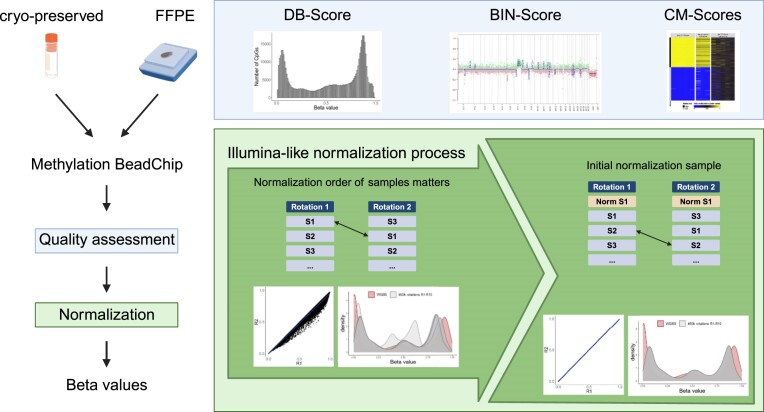
Graphical Abstract
Graphical Abstract.
Introduction
DNA methylation represents one of the main epigenetic modifications that plays a crucial role not only in developmental processes but also in cancer (1). Due to its regulatory significance, DNA methylation has gained increasing attention in research, particularly in exploring DNA methylomes across various malignancies. Furthermore, DNA methylation has emerged as a potential biomarker in the field of diagnostics, being utilized for assessing epigenetic age in lifestyle medicine (2) and is now employed in tumor classification e.g. for brain tumors according to the World Health Organization (3–5). Within this DNA methylation-based diagnostic approaches not only global changes in the DNA methylome are taken into account, but in part also modest effects at particular loci. These minor deviations are often due to biological effects, but in some cases are merely artefacts of bioinformatic analyses and algorithms, so that a comprehensive and reproducible procedure for quality assessment and normalization is required.
Several high-throughput arrays for genome-wide DNA methylation analysis have been developed over the past few years. One of the most used array technologies for DNA methylation analyses was established and launched by Illumina (https://emea.illumina.com/techniques/microarrays/methylation-arrays.html). While the Infinium HumanMethylation27k BeadChip measured DNA methylation at only 27 578 methylation sites, in the meantime the Infinium HumanMethylation450 (450k) and Infinium MethylationEPIC (EPIC) BeadChip Kit were launched interrogating >450 000 and 850 000 methylation sites, respectively (6). Currently, >935 000 methylation sites are included in the EPIC v2.0 (7).
The pre-processing and normalization procedures are crucial and sometimes underestimated steps when analysing high-throughput array data. Pre-processing steps include the correction of dye bias, probe type and background. In the last decade, not only has the number of measurable CpGs raised, but also the number of algorithms for normalization. Widely used methods for normalizing Illumina-based arrays comprise e.g. the subset-quantile within array normalization (8), quantile normalization (9,10), noob (11), functional normalization (12) and beta-mixture quantile method (13). However, no gold-standard pipeline has been established yet.
Besides pre-processing and normalization, several quality controls and filtering steps can be applied to array based DNA methylation data. Most quality controls rely on the control probes integrated on the array, such as those for bisulfite conversion, G/C mismatch, extension, hybridization, non-polymorphic sequences, specificity, target removal and hybridization (14). Plots based on these integrated control probes can be assessed using Illumina’s GenomeStudio software or alternative software packages, e.g. the minfi package’s qcReport function (10). Within these plots, the intensity values or their log2 of these integrated control probes are shown for each sample; however, appropriate data interpretation is still challenging. Another widely used quality control is the filtering for reliability of signals. Therefore, detection P-values can be calculated using e.g. GenomeStudio or minfi, whereby the total signal for each probe is compared to the background signal, which is estimated from the negative control probes (14). While low P-values indicate a reliable signal, high P-values point out poor quality signals. In general, thresholds for P-values for loci to be included in subsequent analyses are often set to <0.01 or 0.05. Overall, all these quality parameters are solely dependent on technical aspects referring to the array design, lacking controls that take biological variables into account.
In various studies, we conducted exploration and validation of DNA methylation data derived from numerous Illumina-based BeadChip arrays (15–19). Thereby, we experienced challenges when dealing with low-quality samples, particularly those derived from formalin-fixed paraffin-embedded (FFPE) tissue. Distinction between a true biological effect and technical failure due to low DNA content or poor DNA quality proved to be a challenging task. In addition, we investigated the central role of the initial sample in the normalization process when applying the algorithm used by numerous software packages, including Illumina’s GenomeStudio software (normalization to internal controls), which is subsequently referred to as the Illumina-like normalization algorithm (14).
Thus, within this study, we aimed to improve the quality control strategies by implementing adjustable quality control scores (QC-Scores) based on biological features. To this end, we established three novel QC-Scores: a distribution score (DB-Score), a bin distance score (BIN-Score) and three measures related to highly consistent DNA methylation levels at specific CpGs (CM-Scores). Furthermore, we encourage for the consistent utilization of a pre-defined normalization sample within the sample ranking to enhance the reproducibility of the Illumina-like normalization algorithm.
Materials and methods
Samples
Six paired Burkitt lymphoma samples, each as FFPE tumor tissue (named as: FFPE_S1-FFPE_S6) and as cryo-preserved tumor tissue (named as: Cryo_S1-Cryo_S6), were used for the DNA methylation analysis by the Infinium HumanMethylation450 BeadChip, and whole genome bisulfite sequencing (WGBS) was performed on the cryo-preserved samples (20). In addition, five non-neoplastic samples (germinal center B populations) were used for the identification of differentially methylated CpG loci. These samples were part of the ICGC MMML-Seq project, which has been approved by Ethics Committee of the Medical Faculty of the University of Kiel (A150/10) and Ulm University (349/11) and of the recruiting centres. The normalization sample was collected from the blood of a healthy donor. Its use in this study has been approved by Ethics Committee of the Medical Faculty of Ulm University (102/15). The samples are summarized in the Supplemental Table S1.
We further collected publicly available DNA methylation array data derived from primary material including various malignancies, non-malignant samples and tissues as well as cell lines (n = 465) to investigate consistency of our identified stable high and low methylated CpGs (21–23). For benchmarking of our QC-Scores, we used DNA methylation array hybridizations from the ICGC MMML-Seq project (450k, EPIC) and hybridizations derived from FFPE tissue from Salmeron-Villalobos et al. (EPIC) (24).
DNA methylation analysis
Infinium-based methylation BeadChips
For DNA methylation analysis 0.5 to 1 μg genomic DNA was bisulfite converted using the EZ DNA Methylation kit (ZymoResearch, Irvine, CA, USA) according to the protocol supplied by the manufacturer and subsequently hybridized onto the Infinium Methylation EPIC v1.0 and HumanMethylation450 BeadChip (Illumina Inc., San Diego, CA, USA). The arrays were scanned using an iScan or NextSeq 550 device (Illumina Inc.).
Raw idat files were read into the statistical program R (version 4.3.0) using the minfi package (version 1.46.0) (10). Normalization was performed with the preprocessIllumina function against the intrinsic controls and without background correction. For downstream analysis, beta values were calculated, the 65 rs loci (annotated with ‘rs’ in the Illumina manifest column IlmnID), loci on gonosomes as well as loci with a detection P-value > 0.01 were excluded. Finally, 473 864 CpGs entered the analysis.
Whole genome bisulfite sequencing
WGBS data were published and processed as described in Kretzmer et al. (20).
ACE-Seq
Allele specific copy number estimation (ACE-Seq, version 1.2.6–2, https://github.com/DKFZ-ODCF/ACEseqWorkflow) was performed as previously described (25).
Statistical analyses
All statistical analyses were performed in R (www.R-project.org, version 4.3.0). Pearson correlation was used for the correlation of beta values. The copy number variation (CNV) analysis was performed with the R package conumee (version 1.34.0) (26). The ‘normalization samples’ were used as reference samples for the CNV analysis. For data visualization for intersecting CpGs sets, the R package UpSetR (version 1.4.0) was used (27).
Significant differentially methylated CpGs were identified with the limma package (version 3.58.1) (28). Differentially methylated CpGs with a false discovery rate (FDR) < 0.01 and a |Δβ| > 0.3 were considered as significant. For comparison of DNA methylation array and WGBS data, the WGBS data were reduced to the CpGs located on the array.
Results
Implementation of QC-scores
We want to highlight that the QC-Scores are specifically designed for use with the current DNA methylation BeadChip arrays from Illumina (450k and EPIC). It is important to note that samples with functional knockouts particularly targeting epigenetic writers or erasers or those subjected to specific treatments may exhibit deviations in the QC-Scores. These exceptions should be considered when interpreting the results, as the scores may not fully capture the unique DNA methylation patterns induced by such experimental conditions.
Workflow of the proposed DNA methylation analysis pipeline
Our proposed quality assessment strategy aims to provide a comprehensive distinction between samples with DNA methylation profiles that are due to a biological effect (e.g. disease-related changes in the DNA methylation profile or global DNA hypomethylation) and those samples where altered DNA methylation values are due to suboptimal DNA quality (‘technical failure’).
To this end, three QC-Scores were implemented: First, the Distribution Score (DB-Score), which indicates a shift from a bimodal distribution of the beta values towards a normal-like distribution. Second, the Bin Distance Score (BIN-Score), which measures the heterogeneity of the intensity values within defined genomic segments. Third, three measures based on deviations from consistently high or low methylated CpGs (CM-Scores). Cut-offs for the QC-Scores for data filtering represent suggestions based on our experience (Supplemental Table S2and Figure 1) and might be optimized depending on the type of samples used.
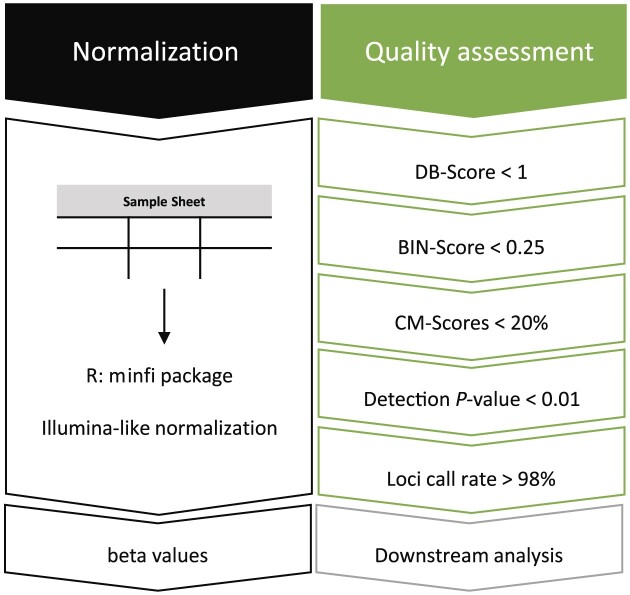
Figure 1.
Workflow DNA methylation analysis pipeline – normalization and quality assessment. Samples are normalized using the Illumina-like process with the minfi package in R. Afterwards, beta values are calculated. The quality of each sample is assessed by the newly implemented DB-Score, BIN-Score and the three measures based on deviations from consistently high or low CM-Scores. Loci with a detection P-value > 0.01 were filtered out and samples with a loci call rate (LCR) < 98% are excluded.
Our DNA methylation pipeline is implemented within the statistic program R and is built upon the Illumina-like normalization process using the R package minfi (10). In addition to the conventional detection P-value filtering (<0.01), we make use of a further parameter, termed the LCR, computed as the percentage of CpGs with a P-value < 0.01 for an individual sample (29). In the following, the design of the scores is described.
DB-scores assess the ‘bimodality’ of global DNA methylation
After normalization, average beta values are calculated from signal intensities ranging from 0 to 1, whereby 0 indicates 0% and 1 represents 100% DNA methylation at a given CpG. When visualizing all measured beta values in a histogram or density plot, a bimodal distribution is expected in normal somatic samples, exhibiting two distinct peaks within the ranges 0–0.3 (representing unmethylated CpGs) and 0.7–1 (representing methylated CpGs).
In contrast, a prevalent phenomenon in tumor samples is the genome-wide loss of DNA methylation often co-occurring with specific hypermethylation of CGI promoters, resulting in a deviation from the bimodal distribution shifting the peaks towards 0.5 (1). However, deviations from the bimodal distribution, potentially even resulting in a Gaussian-like curve, might also be derived from technical issues. Consequently, with the implementation of the DB-Score, we first aim to identify cases exhibiting deviations from the expected bimodal distribution, thereby signifying potentially insufficient DNA quality.
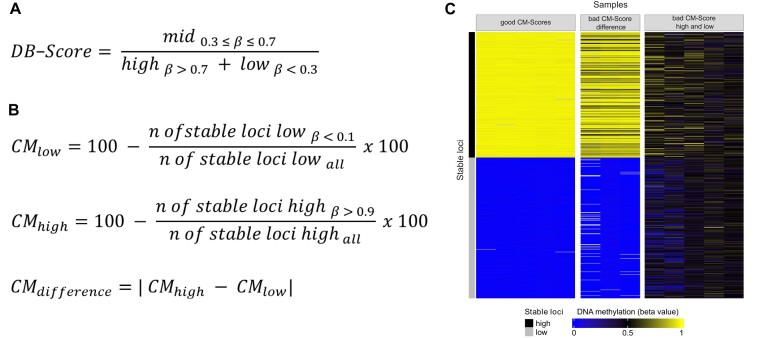
The DB-Score is defined as the ratio between the count of CpGs with beta values falling within the range of 0.3–0.7 (mid) and the count of CpGs with beta values less than or equal to 0.3 (low) or greater than or equal to 0.7 (high), as depicted in Figure 2A. According to the DB-Score, samples can be divided into three categories: Samples having a DB-Score < 1 show a bimodal distribution, and are considered of good quality. Samples in which the DB-Score is >1 and <8 (deviating from a bimodal distribution) are interpreted as being of problematic quality and should be further investigated (Supplemental Figure S1). And finally, we detected a series of samples with a DB-Score > ∼8, which usually show in our data a normal-like distribution, frequently in combination with incorrect even CNVs. These are considered as technical failures, likely due to globally equal levels of fluorescence signals of the methylated and unmethylated beads.
Figure 2.
Implementation of the DB-Score and CM-Scores. (A) Formula for calculating the DB-Score. Mid: Number of CpGs with a beta value between 0.3 and 0.7; high: Number of CpGs with a beta value higher 0.7; low: Number of CpGs with a beta value smaller 0.3. (B) Formulas of the three measures for the CM-Scores. (C) Heatmap of stable high and low methylated CpGs showing outstanding examples for samples with a good CM-Score high and low (left panel), samples with a bad CM-Score difference (middle panel) and samples with a bad CM-Score high and low (right panel). CpGs are listed per row, samples are listed per column.
BIN-Score reveals technical failure
As a second measure, we established a QC-Score, termed BIN-Score. The computation of the BIN-Score relies on the utilization of the R package ‘conumee,’ which facilitates CNV calling through the analysis of DNA methylation data (26). This score uses the distribution of the intensity values across defined genomic fragments as a measure for good or questionable quality.
The genome is segmented into fragments, termed bins, each delineated by a specified minimum size and a requisite minimum number of CpGs. These bins, represented as points in the plot, serve to visually capture gains and deletions across the entire genome, shifting the segment line to the positive (gain) or negative (loss) along the y-axis. Overall, a sample’s BIN-Score is the median of the absolute deviations between bins and their respective segment lines. It is pertinent to note that the BIN-Score does not solely account for samples from malignant tissue, where CNVs are predominantly expected. Instead, the calculation relies on the distribution span of the bins along the segment line and the y-axis, rather than the CNVs per se (see Supplemental Figure S2 for a schematic representation of the calculation). Thus, the same threshold (<0.25) applies to malignant and non-malignant samples. Consequently, substantial deviations of the points from their respective segments are indicative of technical variation, potentially induced by suboptimal DNA quality.
CM-Scores safeguard the detection of biological effects
Third, we implemented three measures summarized under the term CM-Scores. This involved the identification of CpGs (termed stable CpG loci) characterized by a consistent DNA methylation pattern across diverse tissues, various malignancies, non-malignant samples and distinct sample preparation methods.
These stable CpG loci were further categorized into highly methylated CpGs (beta value >0.9; 450k: 279, EPIC: 249), and lowly methylated CpGs (beta value <0.1; 450k: 313, EPIC: 299) (Supplemental Table S3). In Supplemental Figure S3, beta values from 465 samples are displayed as an example of the stable CpG loci. Although it is well known that cell lines may manifest differences in their DNA methylation profiles when compared to primary samples (30), they also demonstrate notably uniform methylation patterns across these stable CpG loci.
On the basis of the stable CpG loci, we calculated three related measures: ‘CM-Score low’ based on the lowly methylated stable CpGs; ‘CM-Score high’ based on the highly methylated stable CpGs; and ‘CM-Score difference’ representing the absolute difference between CM-Score high and low (Figure 2B). Overall, the CM-Score high and low give the percentage of CpGs exhibiting a deviation from their defined methylated thresholds (beta value >0.9 or <0.1). The threshold for all CM-Scores is set at 20%. If both CM-Scores exhibit >20%, thus showing a deviation of the DNA methylation levels in >20% of the stable CpG loci high and 20% of the stable CpG loci low, this sample is categorized as a technical failure.
Using selected samples, the three possible scenarios are shown as a heatmap in Figure 2C: good CM-Score high and low, bad CM-Score high and low, bad CM-Score difference. As an example of good CM-Score high and low samples, we used blood-derived samples from healthy donors (e.g. the normalization sample), which clearly show high DNA methylation for the stable high methylated CpGs (in yellow) and a low DNA methylation for the stable low methylated CpGs (in blue). In contrast, samples featuring a bad CM-Score high and low have an average DNA methylation of about 0.5 (depicted in black) across all usually stable methylated CpGs, indicating a technical failure. Lastly, samples exclusively manifesting deviations for stable highly methylated CpGs, possibly due to global DNA methylation loss, are illustrated, resulting in an elevated CM-Score difference and thus may indicate a biological effect.
While we have provided a set of consistently methylated CpGs based on our diverse dataset, we acknowledge that users may have specific research contexts or sample types that differ from those in our study. Thus, researchers may adapt the CM-Scores to their specific needs following the described concept to identify consistently methylated CpGs within their own datasets.
Importance of the sample ranking using Illumina-like normalization process
According to Illumina’s GenomeStudio Methylation Module User Guide (chapter: Applying Methylation Algorithms, Normalization to Internal Controls) the first sample within the sample sheet serves to calculate a normalization factor which is applied to all other listed samples (14). However, the extent to which the first sample impacts the calculated beta values remains unclear, and many users may not be aware of this factor. To investigate this further, we conducted the following ‘ranking experiment’.
Twelve samples from six patients, one from cryo-preserved and one from FFPE material, were run on the 450k array (Burkitt lymphoma from the ICGC MMML-Seq). When choosing the samples, we ensured both high-quality (as determined by our QC-Scores) and low-quality samples (e.g. FFPE samples and the sample S4) were part of our cohort and used as the first sample (Supplemental Figure S4). This allowed us to investigate the impact of sample quality and sample ranking during normalization.
For the normalization procedure, the order within the sample sheet was randomly rotated 10 times (rankings: R1–R10). It was ensured that in each ranking the first sample differs. Subsequently, beta values were computed for each ranking and compared across other rankings (afterwards referred to as ‘without normalization sample’, Supplemental Table S4). In order to compare the rankings with each other, beta values of each sample from one ranking were correlated to the beta values of the same sample to the nine other rankings. Given the influence of the initial sample in the sheet, we explored whether the inclusion of a ‘normalization sample’ with good quality (LCR > 98%, DB-Score < 1, BIN-Score < 0.25, CM-Scores < 20%) listed at the top lead to overall reproducible beta values for all samples across all rankings (afterwards referred to as ‘with normalization sample’). The same sample sheet rankings were normalized both with and without the inclusion of the normalization sample. The experimental design is depicted in Supplemental Figure S5.
Impact of the first sample
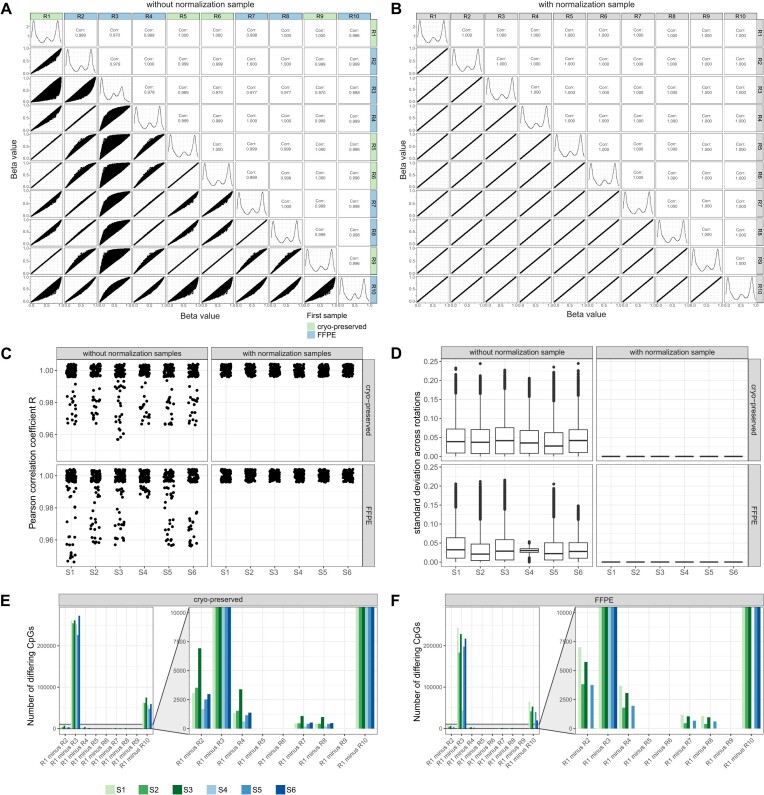
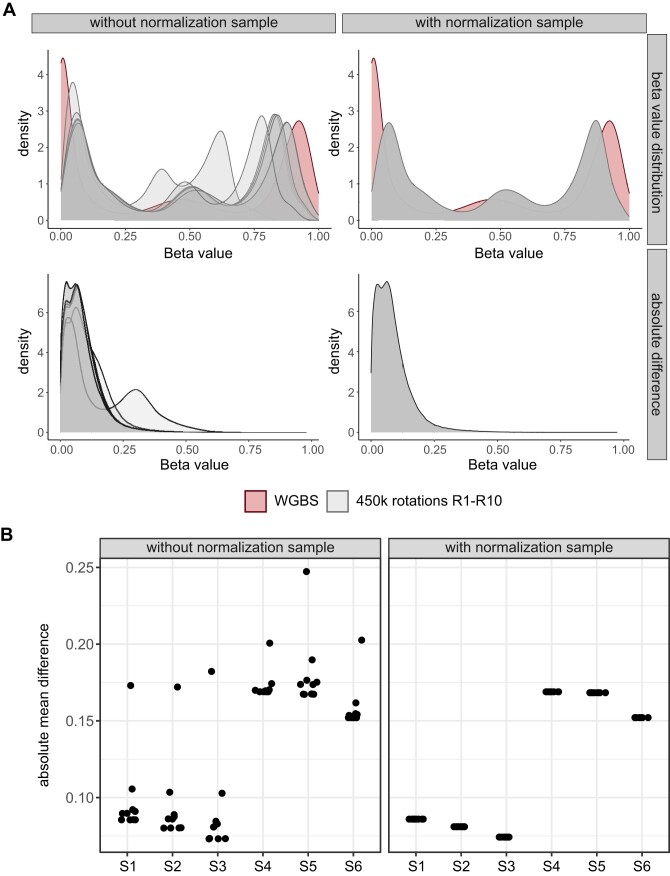
To investigate the effect of the first sample on the results of the overall analysis, we ran the calculations several times, varying the order of the samples. Preceding this analysis, CpGs with a P-value > 0.01 (Supplemental Table S4), loci on gonosomes, and rs loci were excluded. By correlating beta values of each sample across all rankings, variations in various CpGs became apparent, exemplified by the cryo-preserved sample S1 in Figure 3A. The correlation plots show higher discrepancies for R3 and R10, although to a lesser extent. Interestingly, both rankings are determined by a low-quality sample (FFPE S4 and S6) listed as the first samples within the sample sheet. Interestingly, almost exact correlations are observed when the normalization sample for both rankings is either cryo-preserved or FFPE material. However, the incorporation of the normalization sample effectively minimized discrepancies between all rankings, as illustrated in Figure 3B. Considering all samples across all rankings we detected Pearson correlation coefficients ranging from 0.95 to 1 (Figure 3C) and discrepancies in beta values with standard deviations reaching 0.25 (Figure 3D).
Figure 3.
Influence of the first listed sample using Illumina-like normalization algorithm. A total of 12 samples from 6 patients, of which one sample was cryo-preserved and one FFPE, were selected. Samples were 10 times randomly rotated to generate different sample sheet orders. Special care was taken to ensure that the initial sample within each sample sheet was different to investigate its influence on the normalization of the following samples listed within the sample sheet. For every ranking (R1–R10) beta values were calculated using the Illumina-like normalization process (against intrinsic controls). Afterwards, beta values of each sample from one ranking (e.g. R1) were correlated to the beta values of the other nine rankings (e.g. R2–R10). (A) Correlation matrix (Pearson correlation) of the beta values of sample S1 (cryo-preserved) from R1–R10 without a normalization sample revealed differences for some rankings. The rankings labels are colored according the material (cryo-preserved, FFPE) of the first listed sample in the sample sheet. (B) Correlation matrix (Pearson correlation) of the beta values of the sample S1 (cryo-preserved) from R1–R10 with a normalization sample placed at the beginning of the sample sheet yield in reproducible beta values across all rankings. (C) Correlation coefficients were calculated for all samples across all normalization rankings with and without the use of a normalization sample. (D) The standard deviation for each CpG was calculated of all samples across all normalization rankings with and without the use of a normalization sample. (E) Number of CpGs with an absolute difference of >0.1 between R1 and R2–R10 for the cryo-preserved samples. (F) Number of CpGs with an absolute difference of >0.1 between R1 and R2–R10 for the FFPE samples.
Next, we conducted a detailed investigation of the CpGs exhibiting variations among rankings. Initially, we first computed the number of CpGs exhibiting a difference (>0.1) for each sample between the rankings. To showcase, the differences between R1 and R2–R10 (e.g. R1 minus R2, R1 minus R3 etc.) are illustrated in Figure 3E and F. Differences in beta values higher than 0.1 were detected in R1 against six other rankings, with R3 revealing disparities in over 200 000 CpGs in cryo-preserved samples (Figure 3E) and 150 000 CpGs in FFPE samples (Figure 3F). The remaining rankings exhibited differences in beta values within the range of 3–75 018 CpGs. In general, the absolute average mean for each ranking remained low, ranging from 0.01 to 0.15 (Supplemental Figure S6A). Nevertheless, these differences could be mitigated by incorporation of the normalization sample (Supplemental Figure S6B). Furthermore, a comparison of the different CpGs from R1 to R2–R10 elucidated that mostly the same CpGs were consistently affected across all rankings (Supplemental Figure S6C and D). As these findings were solely collected on the older 450k array, we conducted a shortened version of the ranking experiment (R1–R5) using a publicly available EPIC dataset (GSE169643). This analysis also revealed differences using varying normalization samples, as illustrated in Supplemental Figure S7.
To validate these observations, beta values derived from the 450k array, normalized without and with the inclusion of the normalization sample, were compared with matched WGBS data. WGBS is widely acknowledged as the gold-standard method for DNA methylation analyses (31), and in this context, WGBS data were exclusively available for the cryo-preserved samples within each pair. Density plots were generated to compare beta values from all rankings (450k array) with WGBS data from the corresponding sample (Figure 4). While some rankings exhibited better concordance between 450k and WGBS data, others displayed more pronounced discrepancies like ranking R3, characterized with a bad quality sample at the beginning, with an absolute mean difference above 0.15 for all samples [Figure 4A (left panel) and Figure 4B (left panel)]. Notably, the inclusion of the normalization sample within the sample sheets resulted in uniform beta value distributions and absolute mean differences across all array rankings [Figure 4A (right panel) and Figure 4B (right panel)], taking into account the general shift of the array data compared to the WGBS data due to the array technology.
Figure 4.
Validation of the use of a normalization sample against WGBS data. Beta values from all sample sheet rankings (R1–R10 including 12 samples) normalized with Illumina-like normalization process and generated with the 450k array were compared with beta values received from WGBS of the cryo-preserved sample. (A) Density plots showing beta values normalized without (upper left panel) and with (upper right panel) the use of a normalization sample compared to WGBS data from the same sample and CpGs. Results are depicted for the sample S1. Absolute difference between matched CpGs on the 450k array and WGBS data was calculated for each sample sheet ranking normalized without (lower left panel) and with (lower right panel) the use of a normalization sample. (B) Mean was calculated summarizing the absolute difference for each sample within each sample sheet ranking from 450k array compared to the WGBS data (only cryo-preserved samples).
In summary, our detailed examination of beta value disparities across different sample sheet rankings, particularly prominent in R3, highlights the significance of normalization sample in attenuating these variations.
Influence of the use of normalization sample on downstream analysis
Due to this deviation in beta values based on the sample order, our subsequent investigation focused on elucidating its impact on further downstream analysis. Thus, we performed a differential DNA methylation analysis on the 12 lymphoma samples from the ranking experiment, comparing them against five non-neoplastic samples derived from germinal center B cells.
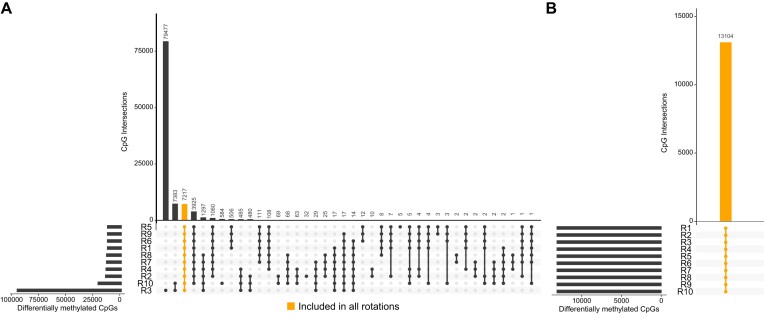
Without normalization sample, varying numbers of significant differentially methylated CpGs (FDR < 0.01, mean |Δβ| > 0.3) were identified comparing the 10 sample sheet rankings ranging between 12 997 CpGs and 96 416 CpGs with an overlap of only 7 212 CpGs (Figure 5A). Remarkably, using the normalization sample, the same set of 13 104 significant CpGs was consistently detected across all 10 rankings (Figure 5B). We compared our differentially methylated CpGs to unreliable probes identified in the literature and found only minimal overlap, indicating that these CpGs are unlikely to be affected by quality issues (Supplemental Figure S8).
Figure 5.
Identification of differentially methylated CpGs while normalizing without and with a normalization sample. Six paired samples (n = 12), once cryo-preserved and once FFPE, were selected and randomly mixed to set up 10 different sample sheet rankings (R1–R10). Each ranking was once normalized without and with a normalization sample. Student t-test was applied to the 12 samples (cryo-preserved and FFPE) against 5 non-neoplastic samples for the 10 sample sheet rankings. (A) Intersections of differentially methylated CpGs (FDR < 0.01, mean |Δβ| > 0.3) normalized without a normalization sample. In addition to varying numbers of significant CpGs, only 7217 significant CpGs were detected in all sample sheet rankings. (B) Intersections of differentially methylated CpGs normalized with a normalization sample. The same 13 104 significant CpGs were detected for all sample sheet rankings.
The findings suggest that differential DNA methylation analysis on the 450k data, particularly when influenced by different sample sheet rankings, resulted in varying numbers of significant differentially methylated CpGs. This underscores the importance of carefully considering sample sheet order and the potential benefits of including a normalization sample in ensuring robust and consistent DNA methylation analysis.
Influence of the use of normalization sample on QC-Scores
Given the observed variations in beta values associated with different samples listed at the top, we sought to investigate the potential influence on the introduced QC-Scores. Thus, we compared the QC-Score for each sample and each ranking to one another (Supplemental Table S5).
Since the CNV analysis, and consequently, the calculation of the BIN-Score, relies on the methylated and unmethylated intensity values and the set of reference samples used for CNV analysis, no or only minor differences were expected and observed based on the first sample or the utilization of a normalization sample. Consistently, nearly identical BIN-Scores were computed for each sample across all rankings (Supplemental Figure S9A). Notably, BIN-Scores were consistently higher for almost all FFPE samples when compared to their corresponding cryo-preserved counterparts, with the highest BIN-Score for FFPE sample S6 at 0.28.
DB-Scores exhibit slight fluctuations among rankings for all samples between 0.23 and 0.55 (not considering FFPE sample S4), with ranking R3 consistently displaying a notably higher value between 0.51 and 1.27 (Supplemental Figure S9B). R3 is a good example of the impact of a low-quality sample, namely the FFPE sample S4 used as a normalization sample. Its use increases the DB-Score for all subsequent normalized samples. However, the utilization of the normalization sample ensures consistent DB-Scores between 0.23 and 0.46 for all samples despite FFPE sample S4 with a DB-Score > 20.
The same holds for the CM-Score high; however, although the CM-Score high of all samples for the R3 is >50%, the CM-Score high of all other rankings is <20%, underpinning the influence of the first sample. Once again, applying the normalization sample facilitates the calculation of consistent CM-Scores for all samples across all rankings (Supplemental Figure S9D).
Overall, for the FFPE sample S4 (previously known for its poor quality), consistently poor DB-Scores (ranging from 6.7 to 25.0) and deficient CM-Score high and low, both at 100%, resulting in a CM-Score difference of zero, were computed across all rankings (Supplemental Figure S9B–E).
It is important to acknowledge that numerous other quality metrices exist for assessing DNA methylation array data quality, either to detect unreliable probes or bad quality samples. We evaluated some of those in the light of the 12 samples used in the ranking experiment. Results are provided in the Supplemental Figure S10.
Significance of DB-score and BIN-Score: application to different sample types
To validate our findings and approaches, we applied our DNA methylation analysis pipeline to four distinct cohorts, one cryo-preserved-based, one FFPE-based, on 5-azacytidine-treated samples and on a large cohort of cell lines, specifically highlighting the utility of the DB-Score and BIN-Score. All datasets were normalized with the inclusion of a normalization sample.
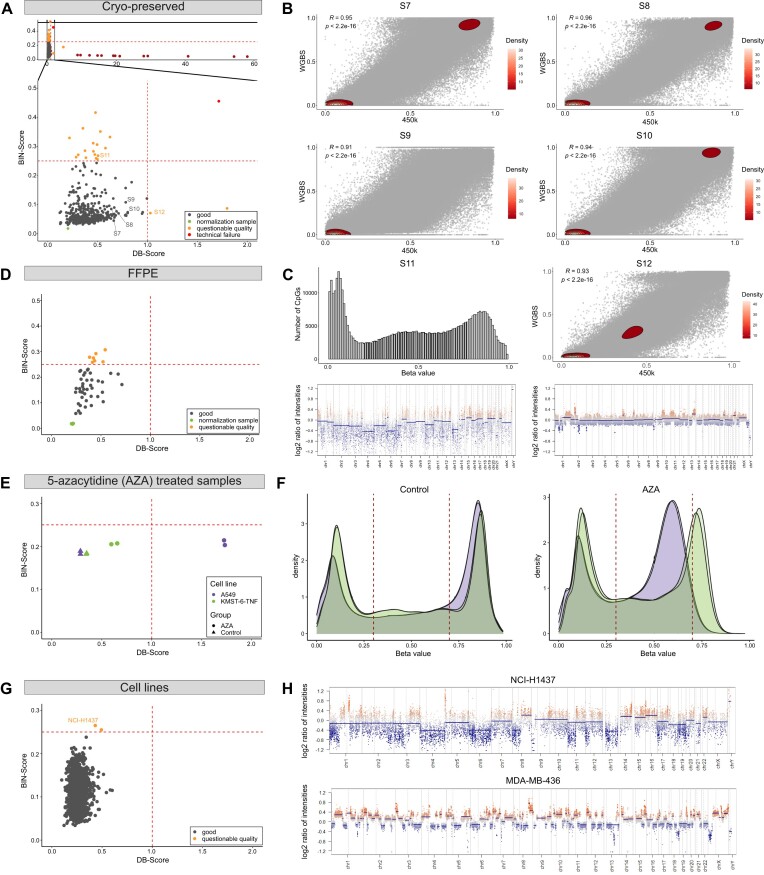
In the first cohort, we assessed the quality of 413 samples (including replicates) from the 450k and EPIC array of cryo-preserved samples (ICGC MMML-Seq cohort). Overall, the majority of the samples exhibit a good DB-Score (<1) and BIN-Score (<0.25) (Figure 6A), as further evidenced by array and WGBS data alignment for four representative samples (Pearson correlation R = 0.91–0.96, Figure 6B). Among the hybridizations, we identified 13 lacking CNVs exhibiting a nearly normal-like distribution and, thus, a high DB-Score (>8), 21 with questionable quality, and one as a technical failure (Supplemental Table S6). We further investigated two cases with questionable quality in more detail (Figure 6C, labelled in orange). Case S11 displayed a good DB-Score but an unfavorable BIN-Score. In general, a higher BIN-Score indicates problems with the quality. Intriguingly, analysis of whole genome sequencing data with the bioinformatic tool ACE-Seq (Figure 6C, Supplemental Figure S11andSupplemental Table S7) revealed a large number of small duplications, which could explain the heterogeneity of the intensity values and hence the heightend BIN-Score. Conversely, case S12 exhibited a poor DB-Score but a good BIN-Score, placing it in the category of cases with questionable quality. However, WGBS data demonstrated a robust concordance with the BeadChip array data (Pearson correlation R= 0.93) for this case as both methods show a global loss of DNA methylation (Figure 6C, right).
Figure 6.
Application of the DNA methylation analysis pipeline. We applied our DNA methylation pipeline on four distinct cohorts, one cryo-preserved-based, one FFPE-based, on 5-azacytidine-treated samples and a large cohort of predominately cancer cell lines (n = 1080), specifically highlighting the utility of the DB-Score and BIN-Score. (A) Correlation of DB-Score and BIN-Score from cryo-preserved samples of the ICGC MMML-Seq cohort (including 450k and EPIC array data). As the ICGC MMML-Seq cohort comprises 450k and EPIC array data, two normalization samples are displayed. Good quality: DB-Score < 1 and BIN-Score < 0.25; questionable quality: DB-Score > 1 or BIN-Score > 0.25; technical failure: DB-Score > 1 and BIN-Score > 0.25. (B) Comparison of 450k array and WGBS data from samples with a good DB- and BIN-Score. Pearson correlation was applied. See samples S7-S10 in panel (A). (C) Left panel (case S11: histogram and CNV-plot): Example with a questionable quality due to a good DB-Score but bad BIN-Score likely because of a large number of small duplications. Sample S11 in panel (A). Right panel (case 12: density plot and CNV-plot): Example of a sample with a questionable quality due to a bad DB-Score but good BIN-Score. Correlation with WGBS data demonstrated a robust concordance with array data demonstrated by Pearson correlation, suggesting an authentic biological effect. See sample S12 in panel (A). (D) Correlation of DB-Score and BIN-Score from FFPE tissues (EPIC array data). (E) Correlation of DB-Score and BIN-Score from 5-azacytidine (AZA)-treated samples. Cells from two cell lines (A549, KMST-6-TNF) were treated with AZA (duplicates). (F) Density plots of the AZA-treated samples and corresponding controls. (G) Correlation of DB-Score and BIN-Score from predominately cancer cell lines (n = 1 080, GSE68379). (H) CNV-Plots of two distinct cell lines, one with a high BIN-Score (upper plot, BIN-Score: 0.265) and a cell line showing extensive genomic aberrations (lower plot).
For the second cohort, data was collected from a publicly available study, comprising 48 FFPE samples of pediatric nodal marginal zone lymphoma and pediatric-type follicular lymphoma (24). Within this cohort we observed that although all samples exhibit a good DB-Score they show higher BIN-Scores with the majority above 0.1 (Figure 6D and Supplemental Table S8). This trend is likely attributed to the fixation method, as FFPE material is known to exhibit more fragmented DNA.
For the third cohort, we investigated the effects of 5-azacytidine (AZA) treatment on DNA methylation patterns in two cell lines, A549 and KMST-6TNF. Cells from each cell line were treated with AZA in duplicate. Our analysis revealed that AZA-treated samples exhibited a global decrease in DNA methylation levels, which was reflected in higher DB-Scores (Figure 6E and F). This effect was more pronounced in the A549 cell line compared to KMST-6TNF. Interestingly, we also observed that AZA-treated samples showed slightly elevated BIN-Scores.
In our fourth and final cohort, we conducted a large-scale analysis of 1 080 (predominately) cancer cell lines, combining in-house samples with publicly available data from GSE68379. We selected a cohort of cancer cell lines to increase for the frequency of samples with high genomic instability, which could potentially influence the calculation of the BIN-Score. Despite this inherent high frequency of samples with chromosomal instability, our analysis revealed that only two out of the 1 080 cell lines exhibited a BIN-Score slightly exceeding (BIN-Score: 0.255, 0.265) our established threshold of 0.25 (Figure 6G and H, upper plot). This finding underscores the robustness of our BIN-Score metric, even in the context of potentially unstable genomic landscapes. To showcase an extreme example of genomic instability within this large cohort, we have included a CNV-plot of a cell line (MDA-MB-436: DB-Score = 0.197; BIN-Score = 0.176) with particularly high genomic instability in Figure 6H (lower plot).
Discussion
Here, we present three novel QC-Scores that provide a more comprehensive quality assessment, particularly when dealing with suboptimal quality samples like FFPE tissue. The main advantage of these QC-Scores is that they are independent of the internal array control probes, which serve as the basis for the conventional quality monitoring of several experimental procedures (14), accessible through various R packages like minfi or Enmix (10,32). While existing tools provide various quality assessment measures, our QC-Scores are specifically tailored to address the unique challenges posed by suboptimal quality samples. Importantly, our principles underlying our scores allow adjustment towards study specific features, enabling researchers to adapt them to different analysis tools and the specific characteristics of their sample cohorts. By offering a complementary approach to current quality control methods, these scores contribute to enhancing the overall reliability of DNA methylation data analysis, particularly in scenarios where sample quality may be compromised.
The DB-Score, which reflects the distribution of beta values in a histogram, is intended to indicate samples of doubtful quality. Interestingly, most packages (10,32–34) allow to create histograms and density plots, but the concept of a threshold and why the sample should be excluded are not explicitly specified or described. Thus, the statistical summary of the beta value distribution in the DB-Score in combination with the BIN-Score gives the user the opportunity to assess the quality of the samples more precisely and to distinguish technical issues from possible biological effects. However, it is important to note that certain biological conditions, such as rare cancers characterized by extensive DNA hypomethylation cells at certain times during development (e.g. primordial germ cells) or samples with extensive alterations due to distinct defects in epigenetic writers or erasers, as well as those treated with demethylation agents (e.g. 5-azacytidine), may present exceptions where a high DB-Score could reflect underlying biological phenomenon rather than technical artifacts. As long as only one score alone has a high value, this indicates questionable quality, but if both scores are high (DB-Score > 1 and BIN-Score > 0.25), this is likely due to a technical complication.
For the BIN-Score calculation we chose the conumee package due to its widespread use and established clinical relevance in CNV analysis (35). We acknowledge that alternatives like ChAMP (36), cnAnalysis450K (37) and Epicopy (38) exist. Taking into account the underlying principle of the BIN-Score approach these alternatives could also be adapted by researchers favoring these packages. Regarding the BIN-Score, it is crucial to highlight that a significant proportion of tumors have a high number of chromosomal imbalances or mutation hotspots (e.g. chromothripsis or kataegis), which could have an impact on the computation of the BIN-Score (39). Within this study we explored one case showing multiple small duplications leading to a high BIN-Score (>0.25). Furthermore, it should be noted that FFPE tissue samples, in particular, have a higher BIN-Score, as the fixation results in more fragmented DNA, which can lead to problems during hybridisation of the DNA to the array probes (40). Thus, we want to emphasize again, whereas the scores can be applied widely, that the precise thresholds of the here presented QC-Scores are determined based on our experience and might require adjustment to suit the specific characteristics of individual cohorts and laboratories.
Utilizing the Illumina-like normalization algorithm (against intrinsic controls), the initial sample listed within the sample sheet serves to calculate a normalization factor, which is applied to all subsequent samples. Although Illumina points out to this fact in the GenomeStudio Methylation Module User Guide (14), it is not widely recognized. Thus, we here demonstrated by comparing beta values observed from different sample sheet rankings, especially differing in the first sample, significant changes. We made two observations through a comparative analysis of beta values derived from the same samples subjected to different sample sheet rankings. First, we detected a considerable amount of varying CpGs for one sample across different sample sheet rankings. Subsequently, these variations in beta values exerted an impact on further downstream analyses, leading to the identification of distinct differentially methylated CpGs. The observed fluctuation in beta values underscores the importance of implementing stringent criteria when identifying significant CpGs. In particular, filtering based on absolute delta beta values is crucial. While P-values can indicate statistical significance, they do not guarantee biological relevance. A CpG site may show a low P-value, which may also fall out of the detection limits of the array, but have a negligible difference in beta values, indicating no meaningful biological impact. By applying an absolute delta beta threshold, we aim at ensuring that the differences reported are beyond technical noise or the result of variations introduced during pre-processing and normalization steps, preventing false positives in downstream analyses (e.g. gene ontology enrichment analysis) and enhancing the robustness of the findings. This approach allows to focus on CpG sites that demonstrate both statistical and biological significance, improving the reliability of the results.
Illumina’s approach of using negative control probes for P-value detection filtering has been a standard practice in DNA methylation data analysis. However, recent studies have demonstrated that this method is insufficient for accurately capturing signal background (41,42). The limitations of negative control probes in estimating background noise levels can lead to suboptimal filtering of low-quality probes. An emerging alternative is the use of Infinium-type-I probe out-of-band signal for background correction and normalization (41).
While the ‘first sample’ method for dye bias correction is used by Illumina GenomeStudio, more advanced and effective methods have been developed in recent years. These include, e.g. RELIC (REgression on Logarithm of Internal Control probes) (43), or approaches used in the Funnorm method (12) and the dasen option in the wateRmelon R package (44). Researchers might therefore consider alternative methods when addressing dye bias in DNA methylation array data analysis.
With this study, we aim to underline the significance of a high-quality assessment in the analysis of array-based DNA methylation data since it affects the functional downstream analysis. To assure correctness and better reproducibility, we strongly recommend using a normalization sample in combination with the Illumina-like normalizing algorithm.
Supplementary Material
Acknowledgements
We would like to thank the members of the cancer epigenetics and genetics laboratory of the Institute of Human Genetics in Ulm for expert support. The authors acknowledge the members of the ICGC MMML-Seq project which was essential for generating major parts of the experimental data analysed here.
Contributor Information
Selina Glaser, Institute of Human Genetics, Ulm University and Ulm University Medical Center, Albert-Einstein-Allee 11, Ulm 89081, Germany.
Helene Kretzmer, Department of Genome Regulation, Max Planck Institute for Molecular Genetics, Ihnestraße 63-73, Berlin 14195, Germany; Digital Health Cluster, Hasso Plattner Institute for Digital Engineering, Digitial Engineering Faculty, University of Potdsdam, Prof.-Dr.-Helmert-Str. 2-3, Potsdam 14482, Germany.
Iris Tatjana Kolassa, Clinical and Biological Psychology, Institute of Psychology and Education, Ulm University, Albert-Einstein-Allee 47, Ulm 89081, Germany.
Matthias Schlesner, Biomedical Informatics, Data Mining and Data Analytics, Faculty of Applied Computer Science and Medical Faculty, University of Augsburg, Alter Postweg 101, Augsburg 86159, Germany.
Anja Fischer, Institute of Human Genetics, Ulm University and Ulm University Medical Center, Albert-Einstein-Allee 11, Ulm 89081, Germany.
Isabell Fenske, Institute of Human Genetics, Ulm University and Ulm University Medical Center, Albert-Einstein-Allee 11, Ulm 89081, Germany.
Reiner Siebert, Institute of Human Genetics, Ulm University and Ulm University Medical Center, Albert-Einstein-Allee 11, Ulm 89081, Germany.
Ole Ammerpohl, Institute of Human Genetics, Ulm University and Ulm University Medical Center, Albert-Einstein-Allee 11, Ulm 89081, Germany.
Data availability
Required scripts for the calculation of the established QC-Scores as well as for generating Figures 2–6 are available on the following GitHub page: https://github.com/GlaserSe/DNAm-qc-scores. The information is stored and released on Zenodo (DOI: 10.5281/zenodo.14189846).
WGBS data are available at the European Genome-phenome Archive (EGA, http://www.ebi.ac.uk/ega/) under accession number EGAS00001001067. HumanMethylation450 BeadChip has been deposited in the Gene Expression Omnibus (GEO) under accession number GSE269421.
Supplementary data
Supplementary Data are available at NARGAB Online.
Funding
German Research Foundation (DFG) [1074 (B9N/B9)]; German Ministry of Science and Education (BMBF) [01KU1002A-J, 01KU1505G and 01KU1505E]; Deutsches Zentrum für Lungenforschung [82DZL001C5].
Conflict of interest statement. None declared.
References
- 1. Lakshminarasimhan R., Liang G.. Jeltsch A., Jurkowska R.Z.. The role of DNA methylation in cancer. DNA Methyltransferases - Role and Function, Advances in Experimental Medicine and Biology. 2016; 945:Cham: Springer International Publishing; 151–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Franzago M., Pilenzi L., Di Rado S., Vitacolonna E., Stuppia L.. The epigenetic aging, obesity, and lifestyle. Front. Cell Dev. Biol. 2022; 10:985274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Capper D., Jones D.T.W., Sill M., Hovestadt V., Schrimpf D., Sturm D., Koelsche C., Sahm F., Chavez L., Reuss D.E.et al.. DNA methylation-based classification of central nervous system tumours. Nature. 2018; 555:469–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Galbraith K., Vasudevaraja V., Serrano J., Shen G., Tran I., Abdallat N., Wen M., Patel S., Movahed-Ezazi M., Faustin A.et al.. Clinical utility of whole-genome DNA methylation profiling as a primary molecular diagnostic assay for central nervous system tumors—A prospective study and guidelines for clinical testing. Neurooncol. Adv. 2023; 5:vdad076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rodriguez F.J. The WHO classification of tumors of the central nervous system-finally here, and welcome!. Brain Pathol. 2022; 32:e13077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bibikova M., Barnes B., Tsan C., Ho V., Klotzle B., Le J.M., Delano D., Zhang L., Schroth G.P., Gunderson K.L.et al.. High density DNA methylation array with single CpG site resolution. Genomics. 2011; 98:288–295. [DOI] [PubMed] [Google Scholar]
- 7. Noguera-Castells A., García-Prieto C.A., Álvarez-Errico D., Esteller M.. Validation of the new EPIC DNA methylation microarray (900K EPIC v2) for high-throughput profiling of the human DNA methylome. Epigenetics. 2023; 18:2185742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Maksimovic J., Gordon L., Oshlack A.. SWAN: subset-quantile within array normalization for Illumina Infinium HumanMethylation450 BeadChips. Genome Biol. 2012; 13:R44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Touleimat N., Tost J.. Complete pipeline for Infinium ® Human Methylation 450K BeadChip data processing using subset quantile normalization for accurate DNA methylation estimation. Epigenomics. 2012; 4:325–341. [DOI] [PubMed] [Google Scholar]
- 10. Aryee M.J., Jaffe A.E., Corrada-Bravo H., Ladd-Acosta C., Feinberg A.P., Hansen K.D., Irizarry R.A.. Minfi: a flexible and comprehensive bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014; 30:1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Triche T.J., Weisenberger D.J., Van Den Berg D., Laird P.W., Siegmund K.D.. Low-level processing of Illumina Infinium DNA methylation BeadArrays. Nucleic Acids Res. 2013; 41:e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fortin J.-P., Labbe A., Lemire M., Zanke B.W., Hudson T.J., Fertig E.J., Greenwood C.M., Hansen K.D.. Functional normalization of 450k methylation array data improves replication in large cancer studies. Genome Biol. 2014; 15:503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Teschendorff A.E., Marabita F., Lechner M., Bartlett T., Tegner J., Gomez-Cabrero D., Beck S.. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics. 2013; 29:189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Illumina 2011; GenomeStudio® Methylation Module v1.8 User Guide.
- 15. Vogt J., Wagener R., Montesinos-Rongen M., Ammerpohl O., Paulus W., Deckert M., Siebert R.. Array-based profiling of the lymphoma cell DNA methylome does not unequivocally distinguish primary lymphomas of the central nervous system from non-CNS diffuse large B-cell lymphomas. Genes Chromosomes Cancer. 2019; 58:66–69. [DOI] [PubMed] [Google Scholar]
- 16. Bens S., Kolarova J., Beygo J., Buiting K., Caliebe A., Eggermann T., Gillessen-Kaesbach G., Prawitt D., Thiele-Schmitz S., Begemann M.et al.. Phenotypic spectrum and extent of DNA methylation defects associated with multilocus imprinting disturbances. Epigenomics. 2016; 8:801–816. [DOI] [PubMed] [Google Scholar]
- 17. Goldmann T., Schmitt B., Müller J., Kröger M., Scheufele S., Marwitz S., Nitschkowski D., Schneider M.A., Meister M., Muley T.et al.. DNA methylation profiles of bronchoscopic biopsies for the diagnosis of lung cancer. Clin Epigenet. 2021; 13:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kouroukli A.G., Fischer A., Kretzmer H., Chteinberg E., Rajaram N., Glaser S., Kolarova J., Bashtrykov P., Mathas S., Drexler H.G.et al.. The DNA methylation status of the TERT promoter differs between subtypes of mature B-cell lymphomas. Blood Cancer J. 2023; 13:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lopez C., Schleussner N., Bernhart S.H., Kleinheinz K., Sungalee S., Sczakiel H.L., Kretzmer H., Toprak U.H., Glaser S., Wagener R.et al.. Focal structural variants revealed by whole genome sequencing disrupt the histone demethylase KDM4C in B-cell lymphomas. Haematologica. 2022; 108:543–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. ICGC MMML-Seq project; BLUEPRINT project Kretzmer H., Bernhart S.H., Wang W., Haake A., Weniger M.A., Bergmann A.K., Betts M.J., Carrillo-de-Santa-Pau E.et al.. DNA methylome analysis in Burkitt and follicular lymphomas identifies differentially methylated regions linked to somatic mutation and transcriptional control. Nat. Genet. 2015; 47:1316–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kulis M., Merkel A., Heath S., Queirós A.C., Schuyler R.P., Castellano G., Beekman R., Raineri E., Esteve A., Clot G.et al.. Whole-genome fingerprint of the DNA methylome during human B cell differentiation. Nat. Genet. 2015; 47:746–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee S.-T., Xiao Y., Muench M.O., Xiao J., Fomin M.E., Wiencke J.K., Zheng S., Dou X., De Smith A., Chokkalingam A.et al.. A global DNA methylation and gene expression analysis of early human B-cell development reveals a demethylation signature and transcription factor network. Nucleic Acids Res. 2012; 40:11339–11351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Oakes C.C., Seifert M., Assenov Y., Gu L., Przekopowitz M., Ruppert A.S., Wang Q., Imbusch C.D., Serva A., Koser S.D.et al.. DNA methylation dynamics during B cell maturation underlie a continuum of disease phenotypes in chronic lymphocytic leukemia. Nat. Genet. 2016; 48:253–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Salmeron-Villalobos J., Egan C., Borgmann V., Müller I., Gonzalez-Farre B., Ramis-Zaldivar J.E., Nann D., Balagué O., López-Guerra M., Colomer D.et al.. A unifying hypothesis for PNMZL and PTFL: morphological variants with a common molecular profile. Blood Adv. 2022; 6:4661–4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kleinheinz K., Bludau I., Hübschmann D., Heinold M., Kensche P., Gu Z., López C., Hummel M., Klapper W., Möller P.et al.. ACEseq – allele specific copy number estimation from whole genome sequencing. 2017; bioRxiv doi:29 October 2017, preprint: not peer reviewed 10.1101/210807. [DOI]
- 26. Hovestadt V., Conumee Z.M.. Enhanced copy-number variation analysis using Illumina DNA methylation arrays. R package version 1.9.0(13 October 2024, date last accessed)http://bioconductor.org/packages/conumee/.
- 27. Conway J.R., Lex A., Gehlenborg N.. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics. 2017; 33:2938–2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K.. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015; 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wagener R., López C., Kleinheinz K., Bausinger J., Aukema S.M., Nagel I., Toprak U.H., Seufert J., Altmüller J., Thiele H.et al.. IG-MYC+ neoplasms with precursor B-cell phenotype are molecularly distinct from Burkitt lymphomas. Blood. 2018; 132:2280–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nestor C.E., Ottaviano R., Reinhardt D., Cruickshanks H.A., Mjoseng H.K., McPherson R.C., Lentini A., Thomson J.P., Dunican D.S., Pennings S.et al.. Rapid reprogramming of epigenetic and transcriptional profiles in mammalian culture systems. Genome Biol. 2015; 16:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chapin N., Fernandez J., Poole J., Delatte B.. Anchor-based bisulfite sequencing determines genome-wide DNA methylation. Commun. Biol. 2022; 5:596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xu Z., Niu L., Li L., Taylor J.A.. ENmix: a novel background correction method for Illumina HumanMethylation450 BeadChip. Nucleic Acids Res. 2016; 44:e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhou W., Triche T.J., Laird P.W., Shen H.. SeSAMe: reducing artifactual detection of DNA methylation by Infinium BeadChips in genomic deletions. Nucleic Acids Res. 2018; 46:e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Morris T.J., Butcher L.M., Feber A., Teschendorff A.E., Chakravarthy A.R., Wojdacz T.K., Beck S.. ChAMP: 450k chip analysis methylation pipeline. Bioinformatics. 2014; 30:428–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Daenekas B., Pérez E., Boniolo F., Stefan S., Benfatto S., Sill M., Sturm D., Jones D.T.W., Capper D., Zapatka M.et al.. Conumee 2.0: enhanced copy-number variation analysis from DNA methylation arrays for humans and mice. Bioinformatics. 2024; 46:e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Feber A., Guilhamon P., Lechner M., Fenton T., Wilson G.A., Thirlwell C., Morris T.J., Flanagan A.M., Teschendorff A.E., Kelly J.D.et al.. Using high-density DNA methylation arrays to profile copy number alterations. Genome Biol. 2014; 15:R30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Knoll M., Debus J., Abdollahi A.. cnAnalysis450k: an R package for comparative analysis of 450k/EPIC Illumina methylation array derived copy number data. Bioinformatics. 2017; 33:2266–2272. [DOI] [PubMed] [Google Scholar]
- 38. Cho S., Kim H.-S., Zeiger M.A., Umbricht C.B., Cope L.M.. Measuring DNA copy number variation using high-density methylation microarrays. J. Comput. Biol. 2019; 26:295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Koltsova A.S., Pendina A.A., Efimova O.A., Chiryaeva O.G., Kuznetzova T.V., Baranov V.S.. On the complexity of mechanisms and consequences of chromothripsis: an update. Front. Genet. 2019; 10:393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jasmine F., Rahaman R., Roy S., Raza M., Paul R., Rakibuz-Zaman M., Paul-Brutus R., Dodsworth C., Kamal M., Ahsan H.et al.. Interpretation of genome-wide infinium methylation data from ligated DNA in formalin-fixed, paraffin-embedded paired tumor and normal tissue. BMC Res. Notes. 2012; 5:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhou W., Triche T.J., Laird P.W., Shen H.. SeSAMe: reducing artifactual detection of DNA methylation by Infinium BeadChips in genomic deletions. Nucleic Acids Res. 2018; 46:e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lehne B., Drong A.W., Loh M., Zhang W., Scott W.R., Tan S.-T., Afzal U., Scott J., Jarvelin M.-R., Elliott P.et al.. A coherent approach for analysis of the Illumina HumanMethylation450 BeadChip improves data quality and performance in epigenome-wide association studies. Genome Biol. 2015; 16:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xu Z., Langie S.A.S., De Boever P., Taylor J.A., Niu L.. RELIC: a novel dye-bias correction method for Illumina Methylation BeadChip. BMC Genomics. 2017; 18:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pidsley R., Wong Y., Volta C.C., Lunnon M., Mill K.J., Schalkwyk L.C.. A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genomics. 2013; 14:293. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Required scripts for the calculation of the established QC-Scores as well as for generating Figures 2–6 are available on the following GitHub page: https://github.com/GlaserSe/DNAm-qc-scores. The information is stored and released on Zenodo (DOI: 10.5281/zenodo.14189846).
WGBS data are available at the European Genome-phenome Archive (EGA, http://www.ebi.ac.uk/ega/) under accession number EGAS00001001067. HumanMethylation450 BeadChip has been deposited in the Gene Expression Omnibus (GEO) under accession number GSE269421.