Abstract
Type IV pili of the opportunistic pathogen Pseudomonas aeruginosa mediate twitching motility and act as receptors for bacteriophage infection. They are also important bacterial adhesins, and nonpiliated mutants of P. aeruginosa have been shown to cause less epithelial cell damage in vitro and have decreased virulence in animal models. This finding raises the question as to whether the reduction in cytotoxicity and virulence of nonpiliated P. aeruginosa mutants are primarily due to defects in cell adhesion or loss of twitching motility, or both. This work describes the role of PilT and PilU, putative nucleotide-binding proteins involved in pili function, in mediating epithelial cell injury in vitro and virulence in vivo. Mutants of pilT and pilU retain surface pili but have lost twitching motility. In three different epithelial cell lines, pilT or pilU mutants of the strain PAK caused less cytotoxicity than the wild-type strain but more than isogenic, nonpiliated pilA or rpoN mutants. The pilT and pilU mutants also showed reduced association with these same epithelial cell lines compared both to the wild type, and surprisingly, to a pilA mutant. In a mouse model of acute pneumonia, the pilT and pilU mutants showed decreased colonization of the liver but not of the lung relative to the parental strain, though they exhibited no change in the ability to cause mortality. These results demonstrate that pilus function mediated by PilT and PilU is required for in vitro adherence and cytotoxicity toward epithelial cells and is important in virulence in vivo.
Type IV pili of Pseudomonas aeruginosa are significant adhesins for interaction with mammalian cells (reviewed in references 15 and 25), contributing to this organism’s ability to cause opportunistic infections in humans. They also are required for motility of the bacteria across a solid surface (twitching motility) (8) and for the binding and entry of bacteriophages (6, 7). P. aeruginosa pili are polymers of a single gene product, called PilA or pilin (32), but their assembly and function requires the products of at least 30 additional genes (reviewed in reference 1). Three of these genes, pilB, pilT, and pilU, encode proteins that contain motifs common among nucleotide-binding proteins (Walker box A domains [37]) and thus are postulated to contribute energy to pili formation or function (1, 18, 40). Inactivation of pilB results in a nonpiliated phenotype (24), while strains with defects in pilT or pilU overexpress surface pili but are no longer motile on a solid surface (10, 40). These observations, in addition to electron microscopy studies comparing phage binding of pilT mutant and wild-type strains (9), suggested that PilB enables pili extension whereas PilT and PilU catalyze pili retraction (15, 23). However, the precise functions of PilT and PilU in this process remain obscure, and pilU mutants, unlike pilT mutants, remain sensitive to bacteriophage infection (40). The proposed dynamic nature of pili could promote the entry of bacteriophage into the bacterium and provide the impetus for twitching motility, but its importance in P. aeruginosa adherence to mammalian cells and virulence has not been examined. Importantly, since pilT and pilU mutants retain (hyper)expression of surface pili, such mutants provide an opportunity to dissect the role of pili as adhesins and as the mediators of twitching motility in the process of P. aeruginosa infection. This may be also relevant to infection caused by a wide range of other type IV piliated bacteria, including Neisseria spp., Moraxella spp., Aeromonas spp., and Legionella spp. (4, 22, 30, 35, 41).
The pilT and pilU genes are contiguous on the P. aeruginosa chromosome but reside in a locus separated from other genes involved in pilus biogenesis and function. Northern blot and complementation analyses suggest that the two genes are transcribed independently and thus are not part of the same operon (40). The encoded proteins are closely related to each other (39% amino acid identity and 61% similarity) but are more distantly related to PilB and to other putative bacterial nucleotide-binding proteins involved in fimbrial biogenesis, protein secretion, or DNA transfer (39, 40). Direct homologs of PilT that are involved in type IV pili function have been identified in enteropathogenic Escherichia coli (EPEC) (31), Myxococcus xanthus (43), and Neisseria gonorrheae (11). Examination of the unfinished genome sequences of N. gonorrhoeae and N. meningitidis indicates that these species at least also contain homologs of PilU (40a). An EPEC mutant defective in the pilT-like bfpF gene, despite having morphologically wild-type bundle-forming pili, displayed increased adherence to epithelial cells in culture, formed irregular bacterial aggregates, and had decreased virulence in humans (2, 5). An in-frame deletion of the M. xanthus pilT gene did not affect the expression of pili or cell to cell agglutination but did cause a loss of pilus-dependent social gliding motility (43). Furthermore, the natural competence and twitching motility of a N. gonorrhea pilT mutant were abolished, but this strain retained wild-type pilus expression and adherence to epithelial cells (41–43). Inactivation of pilT was also shown to rescue the pilus production defect of pilC mutants, further demonstrating a role for N. gonorrheae PilT in pilus function (42).
A number of studies have implicated P. aeruginosa pili as factors important for adherence to epithelial cells in vitro (reviewed in references 15 and 25) and for virulence at several sites of infection (13, 14, 27). These studies have all used nonpiliated mutants of P. aeruginosa except for one investigation which demonstrated that a pilT mutant of P. aeruginosa PAO (DB2) and two other hyperpiliated isolates were noninfectious in a mouse corneal infection model (17). This study suggested that the presence of pili (as adhesins) per se is insufficient to support virulence in vivo. Here we sought to examine further the importance of PilT and PilU in causing epithelial cell injury in vitro and in vivo by comparing the adherence, cytotoxicity, and virulence of the parental strain PAK to those of isogenic pilT and pilU mutants and of isogenic nonpiliated strains.
PilT and PilU contribute to PAK-mediated cytotoxicity toward multiple epithelial cell lines.
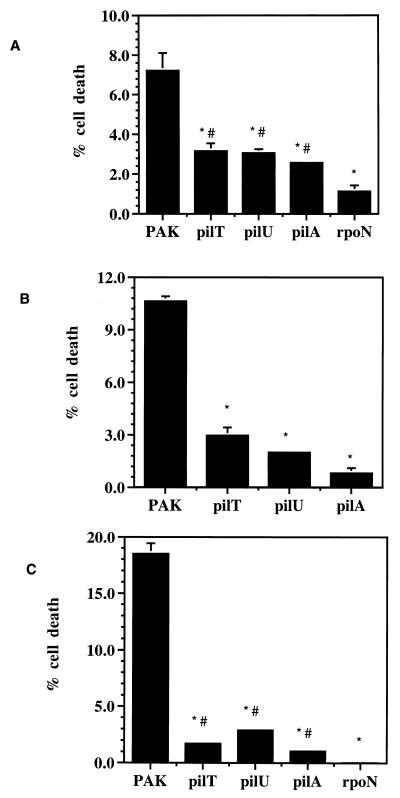
Previous studies have demonstrated that the addition of P. aeruginosa PAK to the apical surface of a highly polarized epithelial cell monolayer consisting of Madin-Darby canine kidney (MDCK) cells resulted in significant cytotoxicity in vitro (3). Comparison of isogenic mutants and different strains demonstrated that the amount of cell damage observed in vitro correlated well to relative virulence in an animal model of acute pneumonia (16, 28). We used this MDCK system to determine if PilT or PilU were required for cytotoxicity in vitro. MDCK type II cells (5 × 106) were grown as a confluent monolayer on Transwell filters (Corning) for 3 days as previously described (20), washed, and placed in minimal essential medium Eagle (MEM) supplemented with 20 mM HEPES buffer pH 7.4 (MEM-lite). Approximately 107 CFU (as determined by dilution plating) of a stationary-phase culture of PAK wild-type or isogenic mutant strain (Table 1) grown for 16 h in Luria broth without shaking at 37°C was added to the apical surface of the MDCK cell monolayer (multiplicity of infection of 2). After incubation for 9 h at 37°C in room air, aliquots of the apical and basal medium were removed and assayed for lactate dehydrogenase (LDH) activity as instructed by the manufacturer (Sigma Chemical Co.). The percentage of cell death was calculated by comparison of the total LDH released to that released from uninfected cells lysed with 0.25% Triton X-100 in MEM-lite. The strains lacking PilT or PilU showed reduced cytotoxicity relative to PAK, causing 44 and 42%, respectively, as much cytotoxicity as the wild type, while a pilA mutant damaged 36% as many cells as the wild type did (Fig. 1A). A strain with a defect in rpoN had even less cytotoxic capability, 16% of the wild-type level, presumably because of pleiotropic effects on a range of genes (36). These findings are not the result of differences in bacterial growth rates since the mutant and wild-type strains had similar rates of growth in minimal or complex medium (data not shown). Furthermore, the mutant strains did not achieve wild-type cytotoxicity even with longer incubation times (up to 11 h; data not shown).
TABLE 1.
Strains used
| Strain | Relevant characteristics | Source or reference |
|---|---|---|
| PAK | Wild type; pili+ and flagella+ | J. Mattick |
| PAK-N1 | Tn5G insertion in rpoN; pili− and flagella− | 19 |
| PAKpilT::Tn5 | Tn5B21 insertion in pilT (mutant R364); hyperfimbriated, phage resistant | 40 |
| PAKpilU::Tn5 | Tn5B21 insertion in pilU (mutant S34); hyperfimbriated, phage sensitive | 40 |
| PAKpilA::TcR | Tetracycline resistance cassette inserted into pilA; nonpiliated | 38 |
FIG. 1.
P. aeruginosa pilT and pilU mutants have decreased ability to damage epithelial cells. Cytotoxicity of wild-type PAK, hyperpiliated pilT or pilU mutants, or nonpiliated pilA or rpoN mutants was assayed by incubation of the bacteria with the cell type indicated. Cell death was quantitated by LDH release and is expressed as a percentage of the total LDH released by lysis of cells not exposed to bacteria. Each assay was performed in triplicate, and error bars represent SEM. (A) Cytotoxicity of MDCK cells after incubation with bacteria for 9 h. ∗, P < 0.003 compared to PAK; #, P < 0.003 compared to rpoN. (B) Cytotoxicity observed on A549 cells after incubation with bacteria for 8 h. ∗, P < 0.001 compared to PAK. (C) Cytotoxicity observed on HeLa cells after incubation with bacteria for 5 h. ∗, P < 0.003 compared to PAK; #, P < 0.05 compared to rpoN. Statistical analysis was performed by using Student’s two-tailed t test with unequal variance.
These strains were tested for cytotoxicity toward two additional cell lines, A549 lung pneumocytes and HeLa cells, with similar results (Fig. 1B and C). In preparation for this assay, 2 × 106 A549 cells were grown in Waymouth’s 752/1 medium without insulin (Gibco BRL) containing 10% fetal calf serum for 3 days on Transwell filters at 37°C with 5% CO2. HeLa cells (2 × 106) were maintained in Dulbecco’s modified essential medium containing 5% glucose (UCSF cell culture facility) and 5% fetal calf serum for 2 days in a tissue culture-treated 24-well plate at 37°C with 5% CO2. As with MDCK cells, cytotoxicity assays were performed by the addition of approximately 107 CFU of the appropriate strain (prepared as described above), but incubation times with these cell types were shortened to 8 h for A549 cells and to 5 h for HeLa cells due to the greater susceptibility of these cells to P. aeruginosa-mediated cytotoxicity. Cell death was calculated as described above. When incubated with A549 or HeLa cells, the PAK strains with either the pilT or pilU gene inactivated were less cytotoxic than the wild type (Fig. 1B and C). Cell death caused by the pilT and pilU mutants was reduced to 28 and 19%, respectively, of the level for the parental strain with respect to A549 cells and to 9 and 16% with respect to HeLa cells. The pilA mutant exhibited 8 and 6% of the wild-type cytotoxicity toward A549 and HeLa cells, respectively. As with MDCK cells, the rpoN mutant had the most severe phenotype, causing only 0.2% of the wild-type cytotoxicity toward HeLa cells. Even with extended incubation (up to 11 h [data not shown]), the pilT and pilU mutants did not achieve wild-type cytotoxicity levels.
These results indicate that the putative nucleotide-binding proteins PilT and PilU are required for full P. aeruginosa-mediated epithelial cell damage in vitro. In this assay, the result of inactivation of pilT or pilU on bacterially mediated cytotoxicity is not reproducibly distinguishable. It should be noted that the transposon insertion into pilT is not polar to pilU (40), and pilus-related defects in each mutant could be rescued by complementation with the appropriate gene (40). Importantly, these results show that the presence of pili on the bacterial surface of the hyperpiliated pilT and pilU mutants is not sufficient to enable normal Pseudomonas-epithelial cell interaction and subsequent wild-type levels of cytotoxicity. As more cytotoxicity is caused by either the pilT and pilU mutant than by the nonpiliated pilA mutant, the overexpression or presence of abnormally functioning pili on the surface of these strains appears to make a minor contribution to epithelial cell injury. RpoN-dependent gene products, such as the previously identified nonpilus adhesins (26), also appear to play a role in cytotoxicity since the rpoN mutant caused significantly less cell damage than the pilA mutant in MDCK cells and HeLa cells (Fig. 1A and C).
PilT and PilU contribute to the association of PAK with epithelial cells.
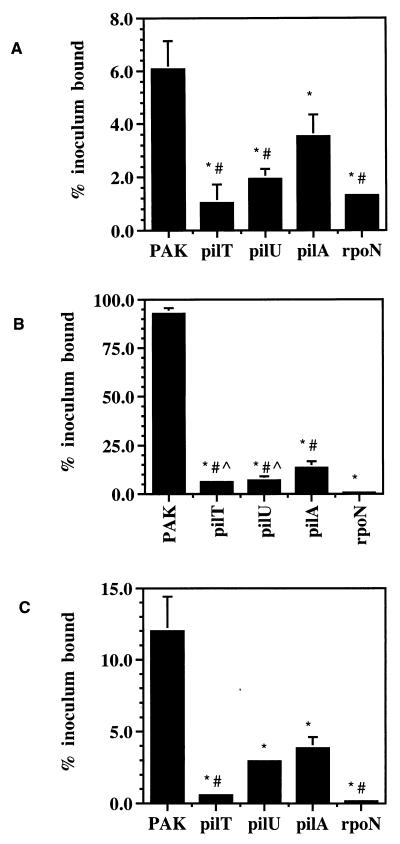
To determine if the loss of the cytotoxic capability of the pilT or pilU mutant corresponded to a defect in adherence to epithelial cells, the association of the mutant strains to the three cell types used was measured. Approximately 107 CFU of each of the various strains (grown and prepared as for the cytotoxicity assays) was incubated with MDCK, A549, or HeLa cells cultured and plated on Transwell filters exactly as described above. Association was assayed by excising the filters (to eliminate bacteria that adhered to plastic) followed by washing the filter-bound cells three times in MEM-lite. The cells were then lysed by treatment with 0.25% Triton X-100 in MEM-lite for 30 min followed by vortexing with glass beads. The bacteria contained within the lysate were quantified by dilution plating on LB agar. Association assays were performed at time points prior to detectable cytotoxicity (3 h for MDCK and A549 cells and 1 h for HeLa cells) to avoid potential bacterial adherence to cell debris. Adherent bacteria were not distinguished from those internalized, but the fraction of internalized bacteria was less than 1% of the fraction of adherent bacteria with all cell types (data not shown). Using this measure, we calculated that 6% of the added wild-type bacteria associated with MDCK monolayers (Fig. 2A), 92% associated with A549 monolayers (Fig. 2B), and 12% associated with HeLa cells (Fig. 2C). The relative binding of PAK to these cell lines is consistent with that previously reported for these cell types (12).
FIG. 2.
Association of P. aeruginosa with epithelial cells is reduced by inactivation of pilT or pilU. Wild-type PAK, hyperpiliated pilT or pilU mutants, or nonpiliated pilA or rpoN mutants were incubated with the indicated cell type, and cell-associated bacteria were quantitated after the amount of time indicated. Each assay was performed in triplicate and normalized to the number of CFU initially added to the assay. SEM is represented by the error bars. (A) Fraction of input bacteria associated with MDCK cells after 3 h. *, P < 0.04 compared to PAK; #, P < 0.04 compared to pilA. (B) Fraction of input bacteria associated with A549 cells after 3 h. *, P < 0.0001 compared to PAK; #, P < 0.003 compared to rpoN; ^, P < 0.05 compared to pilA. (C) Fraction of input bacteria associated with HeLa cells for 1 h. *, P < 0.006 compared to PAK; #, P < 0.003 compared to pilA. Statistical analysis was performed by using the Student’s two-tailed t test with unequal variance.
The two nonpiliated and two hyperpiliated mutants exhibited reduced adherence to each cell type compared to the parental strain (Fig. 2A to C). Of the nonpiliated mutants, the rpoN-defective strain adhered less than the pilA mutant to each cell type, again reflecting a likely role of additional gene products in this interaction. Unexpectedly, the hyperpiliated pilT or pilU mutant also showed reduced binding relative to the pilA mutant. The effect was most marked with MDCK monolayers, where the pilA mutant bound approximately threefold more effectively than the pilT mutant and approximately twofold better than the pilU mutant (Fig. 2A). The adherence of each of the pilT and pilU mutants to A549 cells was roughly twofold less than that of the pilA mutant (Fig. 2B), while the pilT strain appeared to bind to HeLa cells significantly less than the pilA mutant (Fig. 2C). This trend in the adherence assays is in obvious contrast that observed with in vitro cytotoxicity, where the pilT and pilU mutant strains displayed a greater ability to kill epithelial cells than the pilA mutant. Importantly, the greater cytotoxicity of the hyperpiliated mutants cannot be accounted for simply by increased adherence due to the overexpression of abnormally functioning pili on the surface of the these strains. Although the explanation for the greater adherence but reduced cytotoxicity of the pilA mutant is unclear, it is possible that the lack of pili on the pilA mutant uncovers alternate adhesins. Alternatively, the nonfunctional pili on the surface of the pilT or pilU strains may act in a dominant negative manner and interfere with these other adhesins, thus inhibiting intimate contact between the bacteria and the host cells.
Since both hyperpiliated and nonpiliated mutants of P. aeruginosa show markedly reduced adherence to epithelial cells relative to the wild type (12, 41), the expression of surface pili cannot solely account for the observed cell adherence, suggesting that the extension and retraction of pili or surface translocation by twitching motility may be a relevant factor in adherence to and colonization of host cells. Additional, uncharacterized functions of pili may also contribute to these processes.
In summary, the inactivation of pilT or pilU decreased both bacterial adherence and cytotoxicity, indicating their importance to these processes. Compared to the pilA mutant, however, adhesion of the pilT and pilU mutant strains was decreased to a greater extent than cytotoxicity. The effect of inactivation of pilT in P. aeruginosa differs from that in N. gonorrhoeae or EPEC with regard to their adherence to epithelial cells. Although not without a phenotype, an N. gonorrhoeae pilT mutant bound as well as the wild type to epithelial cells though in a more localized fashion (41), while an EPEC bfpF null mutant adhered better to mammalian cells than the parental strain (2). This finding suggests that although all of these bacteria possess homologous proteins related to pili function, their absence has slightly different outcomes in each system.
PilT and PilU are required for full virulence in a mouse model of acute pneumonia.
To assess the contribution of PilT or PilU to P. aeruginosa-induced acute pneumonia, we assayed the virulence of the wild-type and mutant strains in a mouse model of acute pneumonia. Virulence was assessed by two methods, the first using mortality caused by the different isogenic strains as an endpoint and the second measuring the relative ability of the strains to colonize the lung and liver.
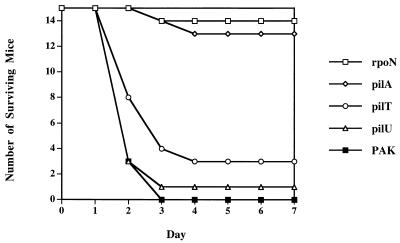
For P. aeruginosa-induced mouse mortality, approximately 5 × 107 CFU of bacteria (grown for 17 h in MINS medium [16] at 37°C with shaking and then washed and resuspended in 50 ml of phosphate-buffered saline) were instilled into the nares of methoxyfluorane-anesthetized 6- to 12-week-old BALB/c mice (B&K Laboratories), using a pipette tip. Viable counts were determined by dilution plating on LB plates. For 50% lethal dose calculations, five mice each were infected with twofold dilutions of bacteria. Mice were monitored over the subsequent 7 days in compliance with guidelines of the Animal Care Committee of the University of California, San Francisco. Statistical significance was assigned by the chi-square test. As shown in Fig. 3, none of the 15 mice infected with the PAK wild type survived to the 7-day time point. In contrast, the pilA mutant strain showed little virulence, and 13 of 15 animals survived for the length of the experiment (P < 0.0001). Results with the rpoN mutant were similar in that 14 of 15 mice survived for 7 days (P < 0.0001). These data are in agreement with previously published results for a neonatal mouse model of acute pneumonia which demonstrated a loss of virulence associated with a lack of surface pili (33). Surprisingly, in this assay the virulence of the pilT and pilU mutant strains was statistically indistinguishable from that of the wild type; only 3 of 15 mice receiving pilT and 1 of 15 mice inoculated with pilU survived for the duration of the experiment. The 50% lethal dose of the pilU mutant, the pilT mutant, and PAK differed twofold at most (data not shown).
FIG. 3.
Virulence of wild-type PAK, hyperpiliated pilT or pilU mutants, or nonpiliated pilA or rpoN mutants in a mouse model of acute pneumonia. Approximately 5 × 107 CFU of each strain was used to intranasally infect 15 mice, each of which was monitored for viability over 7 days.
Using colonization as a more sensitive measure of virulence, we performed a competition assay (34) comparing the efficiency of colonization of the lung or liver of the hyperpiliated strains to that of the wild type. Individual strains were grown and prepared as described above for the mortality experiments. Equal numbers (as determined measuring A600 and by plating for viable counts) of PAK and pilU mutant (either 2.5 × 107 or 5.0 × 107 CFU of each strain), PAK and pilT mutant (either 5.0 × 107 or 7.5 × 107 CFU of each strain), or PAK and pilA mutant (7.5 × 107 CFU of each strain) were resuspended in a total of 50 μl of phosphate-buffered saline and instilled into the nares of anesthetized 6-to-12-week old BALB/c mice. Twenty-four hours later, the animals were sacrificed. The right lobes of the lung and the liver were removed under sterile conditions and placed in 1 ml of LB, and serial dilutions were plated onto LB plates containing no antibiotic or 50 μg of tetracycline per ml (PAK is tetracycline sensitive, whereas each of the mutants carried a tetracycline resistance cassette). A competitive index was calculated by generating the ratio of mutant strain to wild-type bacteria recovered from the lung or liver and comparing it to the same ratio in the infecting inoculum (roughly 1.0 but calculated precisely for each experiment). A competitive index greater than 1.0 indicates that the mutant strain colonized better than wild type, and a competitive index less than 1.0 indicates that the mutant strain was less efficient than the wild type in colonization. The results of two to three experiments using a total of 5 to 10 mice were combined for each sample.
The competitive index of the pilU mutant in the lung was 2.3 (standard error of the mean [SEM] 0.73, n = 5), suggesting that this mutant strain may colonize the lung better than the wild type. In contrast, the liver competitive index was 0.10 (SEM 0.04, n = 10), indicating that the pilU mutant exhibited a 25-fold decrease in its ability to spread to and colonize the liver 24 h postinoculation. Likewise, the pilT mutant was approximately 20-fold less efficient in colonizing the liver compared to the lung; the lung index was 1.08 (SEM 0.79, n = 6), while in the liver it was 0.07 (SEM 0.03, n = 8). For either the PAK-pilU or PAK-pilT mixture, the total CFU recovered from the livers was at least 2 orders of magnitude less than the quantity of bacteria in the lungs (data not shown). The recovered pilU and pilT strains were indistinguishable from the input bacteria and failed to exhibit twitching motility on Vogel-Bonner medium plates. When mice were infected with a combination of PAK and the pilA mutant, no bacteria were recovered from the liver, even when a larger inoculum of 7.5 × 107 was used. This precluded calculating a competitive index for the liver, though in the same experiment, the competitive index in the lungs was 2.14 (SEM 0.70, n = 5). Similarly, recovery of bacteria (wild type or mutant) from the lungs and livers of PAK-pilT-infected mice required use of a larger inoculum of each strain compared to mice inoculated with PAK-pilU. The negative effect of the mutant strains on the wild-type infection implies that the bacteria present in a mixture act synergistically to colonize the lung or the liver, possibly at the stage of initial interaction with the nasal epithelium and upper airways. It may be that the pilU mutant strain is more efficient at this process than either the pilT or pilA mutant. Alternatively, the mutant strains may differ in the ability to provide protection from simultaneous infection with the parental strain.
Thus, in the mouse model of acute pneumonia, the virulence of the pilT, pilU, and pilA mutants as measured by the competition assay mirrored their in vitro cytotoxicity. Although the pilT and pilU strains colonize the lungs as well as or better than the parental strain, the hyperpiliated mutants appear defective in the ability to spread to or colonize the liver. This observation suggests that death caused by the pilU or pilT strain in this animal model results from localized lung damage, although our data do not rule out bacteremia and colonization of distant organs at later times during infection.
In summary, inactivation of P. aeruginosa pilT and pilU genes caused a reduction in adherence to and cytotoxicity toward three epithelial cell types and in ability to colonize the liver following intranasal inoculation in an animal model of acute pneumonia. PilT and PilU thus contribute to P. aeruginosa-induced cell damage in vitro and in vivo. As one probable function of PilT and PilU is to catalyze the retraction of pili, the results indicate that this process is likely necessary for full cytotoxic capability. Since pili are adhesins, they may be necessary to cause initial interactions between the bacteria and epithelial cells, and the proposed retraction of pili could then allow a formation of a more intimate association via other nonpilus adhesins. Evidence suggests that EPEC use such a mechanism of retraction of their bundle-forming pili, catalyzed by the PilT homolog BfpF, to interact with host cells. The bundle-forming pili of EPEC allow an initial, weak interaction of the bacteria with epithelial cells (29) that may be necessary for intimate association involving the adhesin intimin and the bacterium-produced receptor Tir (21). In EPEC, and possibly in P. aeruginosa, this intimate association may be necessary for subsequent translocation of effectors by type III secretion. It should be noted that the hyperpiliated P. aeruginosa pilT and pilU mutants still retain more cytotoxicity and virulence than nonpiliated strains. This finding implies that the overexpression of abnormally functioning pili can partially correct for their lack of retraction. However, our results also indicate that this compensation is due to an uncharacterized mechanism other than increased adherence to host cells.
Acknowledgments
We thank members of the Engel laboratory for reading the manuscript and for scientific advice.
This work was supported by grants from the University Wide AIDS Research Program (J.N.E.), the NIH (J.N.E. [R01 AI42806] and A.R.H. [K08 AI001524]), the American Lung Association (J.N.E.), the Cancer Research Fund of the Damon Runyon-Walter Winchell Foundation Fellowship (A.R.H.), the Bank of America-Gianinni Foundation (J.C.C.), the Australian National Health and Medical Research Council (C.B.W. and J.S.M.), and the Australian Research Council (J.S.M.). J.N.E. is a Career Investigator of the American Lung Association.
REFERENCES
- 1.Alm R, Mattick J S. Genes involved in the biogenesis and function of type-4 fimbriae in Pseudomonas aeruginosa. Gene. 1997;192:89–98. doi: 10.1016/s0378-1119(96)00805-0. [DOI] [PubMed] [Google Scholar]
- 2.Anantha R P, Stone K D, Donnenberg M S. Role of BfpF, a member of the PilT family of putative nucleotide-binding proteins, in type IV pilus biogenesis and in interactions between enteropathogenic Escherichia coli and host cells. Infect Immun. 1998;66:122–131. doi: 10.1128/iai.66.1.122-131.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Apodaca G, Bomsel M, Lindstedt R, Engel J, Frank D, Mostov K, Wiener-Kronish J. Characterization of Pseudomonas aeruginosa-induced MDCK cell injury: glycosylation defective host cells are resistant to bacterial killing. Infect Immun. 1995;63:1541–1551. doi: 10.1128/iai.63.4.1541-1551.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnett T C, Kirov S M, Strom M S, Sanderson K. Aeromonas spp. possess at least two distinct type IV pilus families. Microb Pathog. 1997;23:241–247. doi: 10.1006/mpat.1997.0152. [DOI] [PubMed] [Google Scholar]
- 5.Bieber D, Ramer S W, Wu C Y, Murray W J, Tobe T, Fernandez R, Schoolnik G K. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science. 1998;280:2114–2118. doi: 10.1126/science.280.5372.2114. [DOI] [PubMed] [Google Scholar]
- 6.Bradley D E. The adsorption of Pseudomonas aeruginosa pilus-dependent bacteriophages to a host mutant with nonretractile pili. Virology. 1974;51:489–492. doi: 10.1016/0042-6822(74)90150-0. [DOI] [PubMed] [Google Scholar]
- 7.Bradley D E. Evidence for the retraction of Pseudomonas aeruginosa RNA phage pili. Biochem Biophys Res Commun. 1972;47:142–149. doi: 10.1016/s0006-291x(72)80021-4. [DOI] [PubMed] [Google Scholar]
- 8.Bradley D E. A function of Pseudomonas aeruginosa PAO pili: twitching motility. Can J Microbiol. 1980;26:146–154. doi: 10.1139/m80-022. [DOI] [PubMed] [Google Scholar]
- 9.Bradley D E. Shortening of Pseudomonas aeruginosa pili after RNA-phage adsorption. J Gen Microbiol. 1972;72:303–319. doi: 10.1099/00221287-72-2-303. [DOI] [PubMed] [Google Scholar]
- 10.Bradley D E, Pitt T L. Pilus-dependence of four Pseudomonas aeruginosa bacteriophages with non-contractile tails. J Gen Virol. 1974;23:1–15. doi: 10.1099/0022-1317-24-1-1. [DOI] [PubMed] [Google Scholar]
- 11.Brossay L, Paradis G, Fox R, Koomey M, Hébert J. Identification, localization, and distribution of the PilT protein in Neisseria gonorrhoeae. Infect Immun. 1994;62:2302–2308. doi: 10.1128/iai.62.6.2302-2308.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chi E, Mehl T, Nunn D, Lory S. Interaction of Pseudomonas aeruginosa with A549 pneumocyte cells. Infect Immun. 1991;59:822–828. doi: 10.1128/iai.59.3.822-828.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farinha M A, Conway B D, Glasier L M G, Ellert N W, Irvin R T, Sherburne R, Paranchych W. Alteration of the pilin adhesin of Pseudomonas aeruginosa PAO results in normal pilus biogenesis but a loss of adherence to human pneumocyte cells and decreased virulence in mice. Infect Immun. 1994;62:4118–4123. doi: 10.1128/iai.62.10.4118-4123.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feldman M, Bryan R, Rajan S, Scheffler L, Brunnert S, Tang H, Prince A. Role of flagella in pathogenesis of Pseudomonas aeruginosa pulmonary infection. Infect Immun. 1998;66:43–51. doi: 10.1128/iai.66.1.43-51.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hahn H P. The type-4 pilus is the major virulence-associated adhesin of Pseudomonas aeruginosa—a review. Gene. 1997;192:99–108. doi: 10.1016/s0378-1119(97)00116-9. [DOI] [PubMed] [Google Scholar]
- 16.Hauser A R, Kang P J, Engel J. PepA, a novel secreted protein of Pseudomonas aeruginosa, is necessary for cytotoxicity and virulence. Mol Microbiol. 1998;27:807–818. doi: 10.1046/j.1365-2958.1998.00727.x. [DOI] [PubMed] [Google Scholar]
- 17.Hazlett L D, Moon M M, Singh A, Berk R S, Rudner X L. Analysis of adhesion, piliation, protease production and ocular infectivity of several P. aeruginosa strains. Curr Eye Res. 1991;10:351–362. doi: 10.3109/02713689108996341. [DOI] [PubMed] [Google Scholar]
- 18.Hobbs M, Mattick J S. Common components in the assembly of type 4 fimbriae, DNA transfer systems, filamentous phage and protein-secretion apparatus: a general system for the formation of surface-associated protein complexes. Mol Microbiol. 1993;10:233–243. doi: 10.1111/j.1365-2958.1993.tb01949.x. [DOI] [PubMed] [Google Scholar]
- 19.Ishimoto K S, Lory S. Formation of pilin in Pseudomonas aeruginosa requires the alternative ς factor (RpoN) of RNA polymerase. Proc Natl Acad Sci USA. 1989;86:1954–1957. doi: 10.1073/pnas.86.6.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang P J, Hauser A R, Apodaca G, Fleiszig S, Wiener-Kronish J, Mostov K, Engel J N. Identification of Pseudomonas aeruginosa genes required for epithelial cell injury. Mol Microbiol. 1997;24:1249–1262. doi: 10.1046/j.1365-2958.1997.4311793.x. [DOI] [PubMed] [Google Scholar]
- 21.Kenny B, DeVinney R, Stein M, Reinsheid J, Frey E A, Finaly B B. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell. 1997;91:511–20. doi: 10.1016/s0092-8674(00)80437-7. [DOI] [PubMed] [Google Scholar]
- 22.Liles M R, Viswanathan V K, Cianciotto N P. Identification and temperature regulation of Legionella pneumophila genes involved in type IV pilus biogenesis and type II protein secretion. Infect Immun. 1998;66:1776–1782. doi: 10.1128/iai.66.4.1776-1782.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mattick J S, Alm R A. Common architecture of type 4 fimbriae and complexes involved in macromolecular traffic. Trends Microbiol. 1995;3:411–413. [Google Scholar]
- 24.Nunn D, Bergman S, Lory S. Products of three accessory genes, pilB, pilC, and pilD, are required for biogenesis of Pseudomonas aeruginosa pili. J Bacteriol. 1990;172:2911–2919. doi: 10.1128/jb.172.6.2911-2919.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prince A. Adhesins and receptors of Pseudomonas aeruginosa associated with infection of the respiratory tract. Microb Pathog. 1992;13:251–260. doi: 10.1016/0882-4010(92)90035-m. [DOI] [PubMed] [Google Scholar]
- 26.Ramphal R, Koo L, Ishimoto K, Totten P A, Lara J C, Lory S. Adhesion of Pseudomonas aeruginosa pilin-deficient mutants to mucin. Infect Immun. 1991;59:1307–1311. doi: 10.1128/iai.59.4.1307-1311.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sata H, Okinda K, Saiton H. Role of pilin in the pathogenesis of Pseudomonas aeruginosa burn infection. Microbiol Immunol. 1988;32:131–139. doi: 10.1111/j.1348-0421.1988.tb01372.x. [DOI] [PubMed] [Google Scholar]
- 28.Sawa T, Ohara M, Kurahashi K, Twining S, Frank D, Doroques D, Long T, Gropper M, Wiener-Kronish J. In vitro cellular cytotoxicity predicts Pseudomonas aeruginosa virulence in lung infections. Infect Immun. 1998;66:3242–3249. doi: 10.1128/iai.66.7.3242-3249.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sohel K, Puente J, Murray W, Vuopio-Varkila J, Schoolnk G. Cloning and characterization of the bundle-forming pilin gene of enteropathogenic Escherichia coli and its distribution in Salmonella serotypes. Mol Microbiol. 1993;7:563–575. doi: 10.1111/j.1365-2958.1993.tb01147.x. [DOI] [PubMed] [Google Scholar]
- 30.Stone B J, Abu Kwaik Y. Expression of multiple pili by Legionella pneumophila: identification and characterization of a type IV pilin gene and its role in adherence to mammalian and protozoan cells. Infect Immun. 1998;66:1768–1775. doi: 10.1128/iai.66.4.1768-1775.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stone K D, Zhang H Z, Carlson L K, Donnenberg M S. A cluster of fourteen genes from enteropathogenic Escherichia coli is sufficient for the biogenesis of a type IV pilus. Mol Microbiol. 1996;20:325–337. doi: 10.1111/j.1365-2958.1996.tb02620.x. [DOI] [PubMed] [Google Scholar]
- 32.Strom M S, Lory S. Structure-function and biogenesis of the type IV pili. Annu Rev Microbiol. 1993;47:565–596. doi: 10.1146/annurev.mi.47.100193.003025. [DOI] [PubMed] [Google Scholar]
- 33.Tang H, Kays M, Prince A. Role of Pseudomonas aeruginosa pili in acute pulmonary infection. Infect Immun. 1995;63:1278–1285. doi: 10.1128/iai.63.4.1278-1285.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taylor R K, Miller V L, Furlong D B, Mekalanos J J. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc Natl Acad Sci USA. 1987;84:2822–2837. doi: 10.1073/pnas.84.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tennent J M, Mattick J S. Type 4 fimbriae. In: Klemm P, editor. Fimbriae: aspects of adhesion, genetics, biogenesis and vaccines. Boca Raton, Fla: CRC Press; 1994. pp. 127–146. [Google Scholar]
- 36.Totten P A, Lara J C, Lory S. The rpoN gene product of Pseudomonas aeruginosa is required for expression of diverse genes, including the flagellin gene. J Bacteriol. 1990;172:389–396. doi: 10.1128/jb.172.1.389-396.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequences in the a- and b-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watson A A, Mattick J S, Alm R A. Functional expression of heterologous type 4 fimbriae in Pseudomonas aeruginosa. Gene. 1996;175:143–150. doi: 10.1016/0378-1119(96)00140-0. [DOI] [PubMed] [Google Scholar]
- 39.Whitchurch C B, Hobbs M, Livingston S P, Krishnapillai V, Mattick J S. Characterization of a Pseudomonas aeruginosa twitching motility gene and evidence for a specialised protein export system widespread in eubacteria. Gene. 1991;101:33–44. doi: 10.1016/0378-1119(91)90221-v. [DOI] [PubMed] [Google Scholar]
- 40.Whitchurch C B, Mattick J S. Characterization of a gene, pilU, required for twitching motility but not phage sensitivity in Pseudomonas aeruginosa. Mol Microbiol. 1994;13:1079–1081. doi: 10.1111/j.1365-2958.1994.tb00499.x. [DOI] [PubMed] [Google Scholar]
- 40a.Whitchurch, C. B., and J. S. Mattick. Unpublished data.
- 41.Wolfgang M, Lauer P, Park H-S, Brossay L, Hebert J, Koomey M. PilT mutations lead to simultaneous defects in competence for natural transformation and twitching motility in piliated Neisseria gonorrhoeae. Mol Microbiol. 1998;29:321–330. doi: 10.1046/j.1365-2958.1998.00935.x. [DOI] [PubMed] [Google Scholar]
- 42.Wolfgang M, Park H S, Hayes S F, van Putten J P, Koomey M. Suppression of an absolute defect in type IV pilus biogenesis by loss-of-function mutations in pilT, a twitching motility gene in Neisseria gonorrhoeae. Proc Natl Acad Sci USA. 1998;95:14973–14978. doi: 10.1073/pnas.95.25.14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu S S, Wu J, Kaiser D. The Myxococcus xanthus pilT locus is required for social gliding motility although pili are still produced. Mol Microbiol. 1997;23:109–121. doi: 10.1046/j.1365-2958.1997.1791550.x. [DOI] [PubMed] [Google Scholar]