Abstract
Pangenomes are collections of annotated genome sequences of multiple individuals of a species1. The structural variants uncovered by these datasets are a major asset to genetic analysis in crop plants2. Here we report a pangenome of barley comprising long-read sequence assemblies of 76 wild and domesticated genomes and short-read sequence data of 1,315 genotypes. An expanded catalogue of sequence variation in the crop includes structurally complex loci that are rich in gene copy number variation. To demonstrate the utility of the pangenome, we focus on four loci involved in disease resistance, plant architecture, nutrient release and trichome development. Novel allelic variation at a powdery mildew resistance locus and population-specific copy number gains in a regulator of vegetative branching were found. Expansion of a family of starch-cleaving enzymes in elite malting barleys was linked to shifts in enzymatic activity in micro-malting trials. Deletion of an enhancer motif is likely to change the developmental trajectory of the hairy appendages on barley grains. Our findings indicate that allelic diversity at structurally complex loci may have helped crop plants to adapt to new selective regimes in agricultural ecosystems.
Subject terms: Genomics, Plant genetics, Agricultural genetics, Plant breeding
A pangenome analysis of 76 wild and domesticated barley accessions in combination with short-read sequence data of 1,315 barley genotypes indicates that allelic diversity at structurally complex loci may have helped crop plants to adapt to agricultural ecosystems.
Main
Reliable crop yields fuelled the rise of human civilizations. As people embraced a new way of life, cultivated plants, too, had to adapt to the needs of their domesticators. There are different adaptive requirements in a wild compared with an arable habitat. Crop plants and their wild progenitors differ in how many vegetative branches they initiate or how many seeds or fruits they produce and when. A case in point is barley (Hordeum vulgare): in six-rowed forms of the crops, thrice as many grains set as in the ancestral two-rowed forms. This change was brought about by knockout mutations3 of a recently evolved regulator4 of inflorescence development. Consequently, six-rowed barleys came to predominate in most barley-growing regions5. Taking a broader view of the environment as a set of exogeneous factors that drive natural selection, barley provides another fascinating, and economically important, example. The process of malting involves the sprouting of moist barley grains, driving the release of enzymes that break down starch into fermentable sugars. In the wild, various environmental cues can trigger germination to improve the odds of the emerging seedling encountering favourable weather conditions for subsequent growth6. In the malt house, by contrast, germination has to be fast and uniform in modern cultivars to satisfy the desired specifications of the industry. In addition to these examples, traits such as disease resistance, plant architecture and nutrient use have been a focus for plant breeders and studied intensively by barley geneticists7. Although barley genetic analysis flourished during a ‘classical’ period8 in the first half of the 20th century, it started to lag behind small-genome models because of difficulties in adapting molecular biology techniques to a large genome rich in repeats9. However, interest in barley as a diploid model for temperate cereals has surged again as DNA sequencing became more powerful. High-quality sequences of several barley genomes have been recently assembled10. New sequencing technologies have shifted the focus of barley genomics: from the modest ambition of a physical map of all genes to a ‘pangenome’, that is, near-complete sequence assemblies11 of many genomes. Jayakodi et al.10 assembled genome sequences of 20 diverse genotypes from short reads. Here we report an expanded pangenome comprising 76 chromosome-scale sequences assembled from long reads as well as short-read sequences of 1,315 barley genomes. These data in conjunction with genetic and genomic analyses provide insights into the effects of structural variation at loci related to crop evolution and adaptation.
Annotated genome sequences of 76 barleys
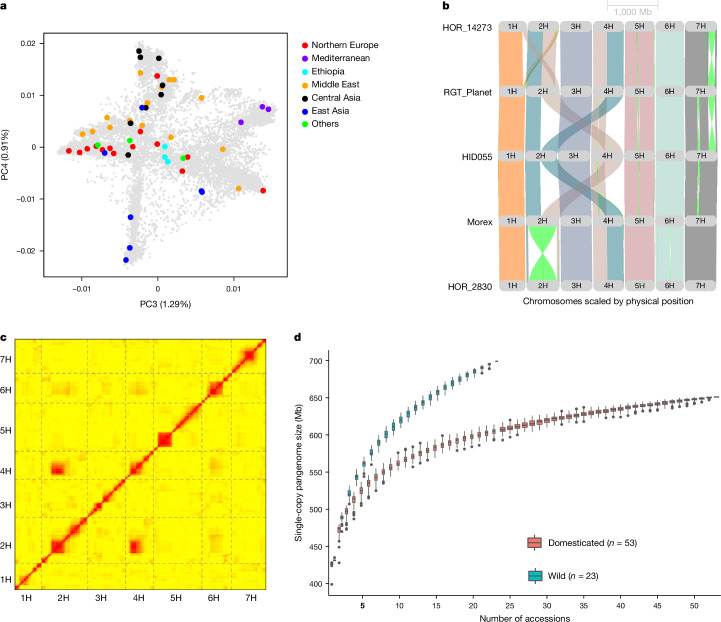
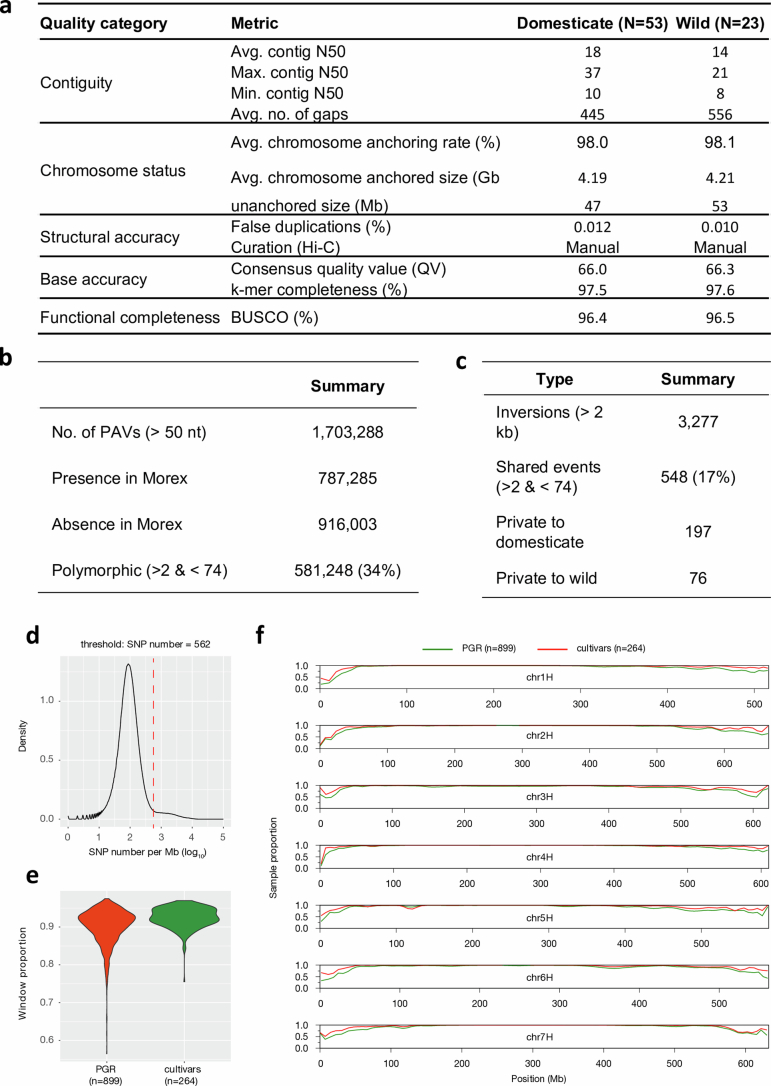
As in previous diversity studies10,12, we aimed for a judicious mix of representativeness, diversity and integration with community resources (Fig. 1a, Extended Data Fig. 1a–c and Supplementary Table 1). We selected (1) diverse domesticated germplasm with a focus on genebank accessions from barley’s centre of diversity in the Middle East; (2) 23 accessions of barley’s conspecific wild progenitor H. vulgare subsp. spontaneum from across that taxon’s geographic range (Extended Data Fig. 1d); and (3) cultivars of agronomic or scientific relevance. Examples of the last category are Bonus, Foma and Bowman, three parents of classical mutants13. Genome sequences of each accession were assembled to contig-level from PacBio HiFi accurate long reads14 and scaffolded with conformation capture sequencing (Hi-C) data15 to chromosome-scale pseudomolecules (Extended Data Fig. 2a and Supplementary Table 1). An annotation of full-length long terminal repeat retrotransposons showed that the 76 genomes had no striking difference in the composition and insertion age of their transposable elements (TEs) (Supplementary Table 2 and Supplementary Fig. 1). On average, 88% of the assembled sequence was derived from TEs. Gene models were annotated with the help of transcriptional evidence and homology. Illumina RNA sequencing (RNA-seq) and PacBio isoform sequencing of five different tissues (embryo, root, shoot, inflorescence and caryopsis; Supplementary Fig. 2 and Supplementary Table 3) were generated for 19 domesticated and one wild member of the pangenome panel. Gene models predicted in these genomes were projected onto the remaining 56 sequence assemblies (Supplementary Table 4). We ran BUSCO16 to assess the completeness of our annotations. Out of 4,896 single-copy gene models in the BUSCO ‘Poales’ set (v.5.1.2, poales_odb10), on average, fewer than 92 (1.9%) were absent from the pangenome annotations (Supplementary Table 4). Our assemblies also met the other quality metrics proposed by the EarthBiogenome project17 (Supplementary Table 1).
Fig. 1. A species-wide pangenome of H. vulgare.
a, Principal component analysis showing domesticated accessions (n = 53) in the pangenome panel in the global diversity space. Regions of origins are colour coded. The proportion of variance explained by each PC in panels is given in the axis labels. Other PCs are shown in Extended Data Fig. 1a. b, Example of large SVs including interchromosomal translocations and inversions between pangenome accessions. c, Interchromosomal LD in segregating offspring derived from a cross between HID055 and Barke. LD is indicated by the intensity of red colour. d, Size of the single-copy pangenome in wild and domesticated barleys as a function of sample size. Boxes indicate the interquartile range (IQR) with the central line indicating the median and whiskers indicating the minimum and maximum without outliers, respectively. Outliers were defined as minimum −1.5 × IQR and maximum +1.5 × IQR, respectively. LD, linkage disequilibrium; PC, principal component.
Extended Data Fig. 1. A globally representative diversity panel of domesticated and wild barley.
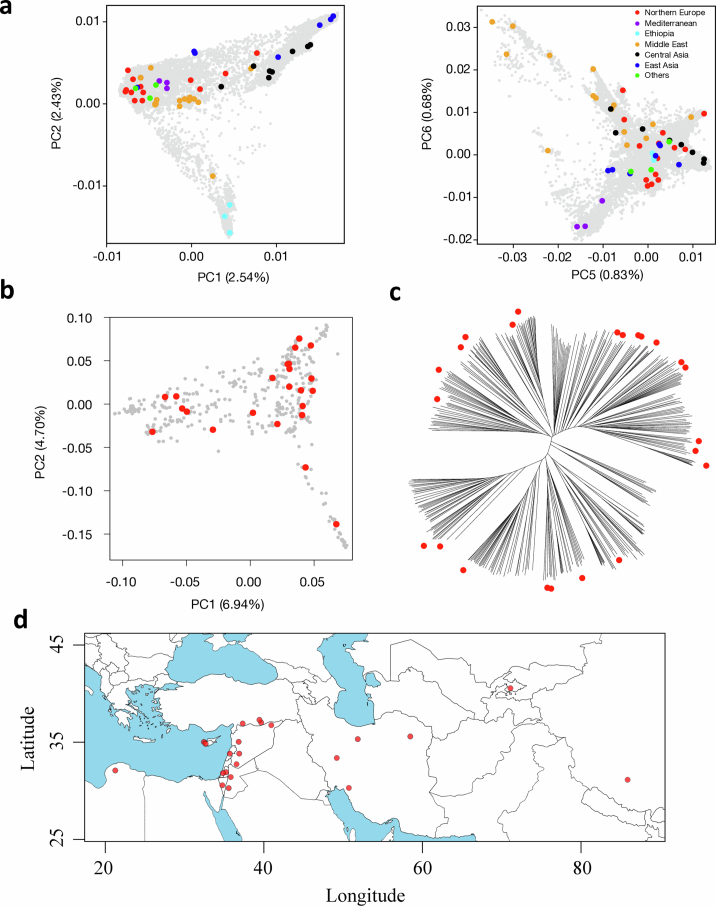
(a) Higher principal components (PC) of the barley diversity space (as defined by the genotyping-by-sequencing data of Milner et al. 5) with pangenome accessions highlighted. (b) The first two PCs of the diversity space of 412 wild barley (Hordeum vulgare subsp. spontaneum) with pangenome accessions highlighted. The underlying data were taken from Milner et al. 5 and Sallam et al. 151 (c) Neighbor-joining phylogenetic tree of those wild barleys. The branch tips corresponding to accessions selected for the pangnome are marked with red circles. The proportion of variance explained by each PC in panels (a) and (b) is given in the axis labels. (d) Map showing the collection sites of wild accessions (n = 23) included in the pangenome panel. The map was drawn in R using the package ‘mapdata’.
Extended Data Fig. 2. A pangenomic diversity map of barley.
(a) Assembly statistics of 76 chromosome-scale reference genome sequences. (b) Counts of presence/absence variants. (c) Counts of inversion polymorphisms spanning 2 kb or more. (d) Selection of threshold based on pairwise differences (number of SNPs per Mb) for the binary classification into similar/dissimilar haplotypes. (e) The proportion of samples with a close match to one of the 76 pangenome accessions is shown for plant genetic resources (PGR) and elite cultivars in sliding windows along the genome (size: 1 Mb, shift: 500 kb). (f) Distribution of the share of similar windows in individual PGR and cultivar genomes.
An atlas of structural variation
To quantify the extent of genic presence/absence variation, we constructed a gene-centric orthologous framework from the annotated pangenome. We identified a total of 95,237 hierarchical orthologous groups (HOGs), of which 16,672 were part of the ‘core genome’, that is, they contained at least one homologous gene from all 76 genotypes. Of the core HOGs, 14,736 were represented by exactly one orthologue in each of the 76 barley genotypes. A further 78,067 HOGs made up the ‘shell genome’, which consists of genes that are absent from at least one genotype but present in at least two. Finally, each single genome possessed on average 819 genes (minimum: 552, maximum: 1,790) private to it (‘cloud genes’). The proportions of genes in individual genomes that were assigned to the core, shell and cloud categories did not vary much (Supplementary Fig. 3), with on average 64.71%, 33.62% and 1.67% classified, respectively, as shell, core and cloud.
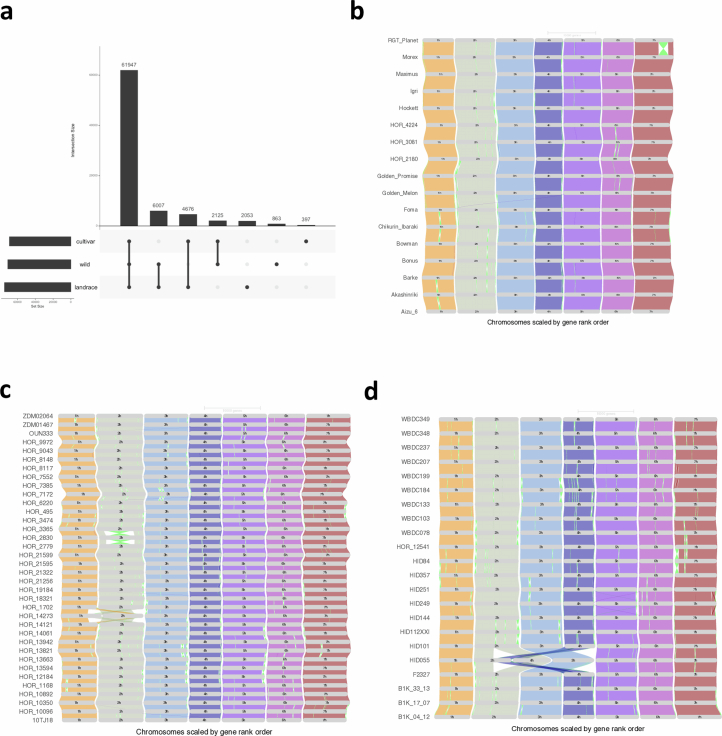
As expected for conspecific populations connected by gene flow18,19, wild and domesticated barleys were not strongly differentiated in their gene content: 61,947 HOGs were shared between both populations. A total of 863 and 397 HOGs were private to wild and domesticated barleys, respectively (Extended Data Fig. 3a). We inspected the gene ontology terms of HOGs restricted to specific gene pools (wild forms, landraces, cultivars and combinations of these groups) (Supplementary Table 5). Gene ontology terms enriched (Fisher’s exact test, Benjamini–Hochberg false discovery rate ≤ 0.05) in wild barley included ‘nutrient reservoir activity’, whereas those overrepresented in landraces included the term ‘defense response’. Note that we did not here attempt an enquiry into ‘de novo’ genes, which have recently arisen in either wild or domesticated genomes, because we had direct transcriptional evidence for only 20 of the 76 genotypes.
Extended Data Fig. 3. Gene-space collinearity.
(a) Upset plot showing the intersections between cultivars, wild forms and landraces among the shell HOGs. Individual HOGs may contain genes from e.g. all wild barleys, or any subset of wild barley genotypes down to a single wild barley genotype. (b-d) GENESPACE alignments of 76 barley genomes, grouped by cultivars (a) and landraces (c) and wild barley (d).
To expand the catalogue of presence/absence variants, insertions and deletions (indels) and polymorphic inversions, we aligned the genome sequences and detected structural variants (SVs) (Fig. 1b and Extended Data Figs. 2b–d and 3b–d). Noteworthy were two reciprocal interchromosomal translocations, the first in HOR 14273, an Iranian landrace, and the second in HID055, a wild barley from Turkey (Fig. 1b and Supplementary Fig. 4). The latter event joins the short arm of chromosome 2H with the long arm of chromosome 4H (and vice versa) and manifests itself in interchromosomal linkage in a biparental population between HID055 and Barke20 (Fig. 1c). This illustrates that inadvertent selection of germplasm with SVs can create obstacles for the use of plant genetic resources (PGRs).
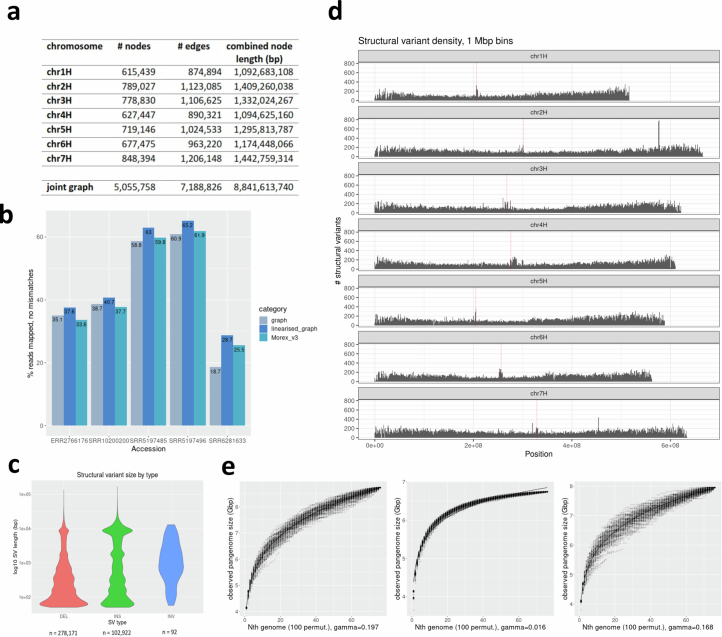
The presence of both wild and domesticated barleys in our panel made it possible to compare the levels of structural diversity in the two taxa. Graph structures tabulating the presence and absence of single-copy loci in individual genomes10 grew faster in wild than in cultivated forms (Fig. 1d): a larger amount of single-copy sequence was present in 23 wild barley genomes than in 53 genomes of the domesticate. This pattern was also seen in a whole-genome graph constructed with Minigraph21 (Extended Data Fig. 4e). The pangenome growth plots illustrate the amount of presence/absence variation present in the pangenome as a function of the number of lines added, with asymptotic curves indicating a saturation point at which addition of further lines would contribute little or no further presence/absence variation (‘closed pangenome’). To investigate whether the pangenome improved mapping rates of reads, we mapped publicly available Illumina whole-genome shotgun reads to the pangenome graph, the linear Morex V3 reference sequence, and, to exclude tool bias as a confounding factor, a linearized version of the pangenome graph. We counted perfectly matching reads only to exclude mismapped reads. Rates of mapping to the linearized graph were between 3.2% and 4% higher than for the Morex V3 reference, but rates for the graph itself were consistently lower than those for its linearized equivalent (Extended Data Fig. 4b), presumably because of algorithmic differences in the mapping tools. The bulk part of the variation in mapping rates was attributable to the identity of the sample mapped, reinforcing our finding that the current pangenome is still open, that is, not all variation within the species has been captured yet. The genome-wide distribution of SVs encapsulated in the graph matched that inferred from pairwise alignments (Extended Data Fig. 4c,d). We also computed the overlap between the two sets of SVs and found that the use of different tools and approaches was reflected by the numbers of SVs detected. Minigraph showed greater sensitivity in deletion discovery (15,584–19,878 deletions per chromosome versus 8,306–10,759 for Assemblytics), whereas Assemblytics detected greater numbers of insertions (8,409–10,467 versus 5,269–6,897 for Minigraph). The intersection of the sets of SVs in terms of size and position (on the basis of at least 70% spatial overlap) ranged from 6,253 to 8,154 per chromosome for deletions and 3,843 to 4,976 for insertions. Owing to high computational requirements22, pangenome graph construction with packages supporting small variants (less than 50 bp) is still computationally prohibitive in species with large genomes, and our own experience backs up this finding.
Extended Data Fig. 4. Graph-based pangenome analysis with Minigraph.
(a) Descriptive statistics per chromosome and for joint graph. (b) Comparative statistics of read mappings from five publicly available Illumina whole genome shotgun sequence read runs against the pan-genome graph, the MorexV3 linear reference sequence and the linearised version of the pan-genome graph. (c) Size distribution of structural variants in graph. (d) Chromosomal distribution of structural variants. Centromere positions are indicated by vertical dashed lines in red. (e) Pangenome graph growth curves generated with the odgi heaps tool. One hundred permutations were computed for each number of genomes included. Values of gamma > 0 in Heaps’ law indicate an open pangenome. Plots shown are for all accessions (left, n = 76), domesticated accessions only (cultivars + landraces, centre, n = 53) and H. spontaneum accessions (right, n = 23).
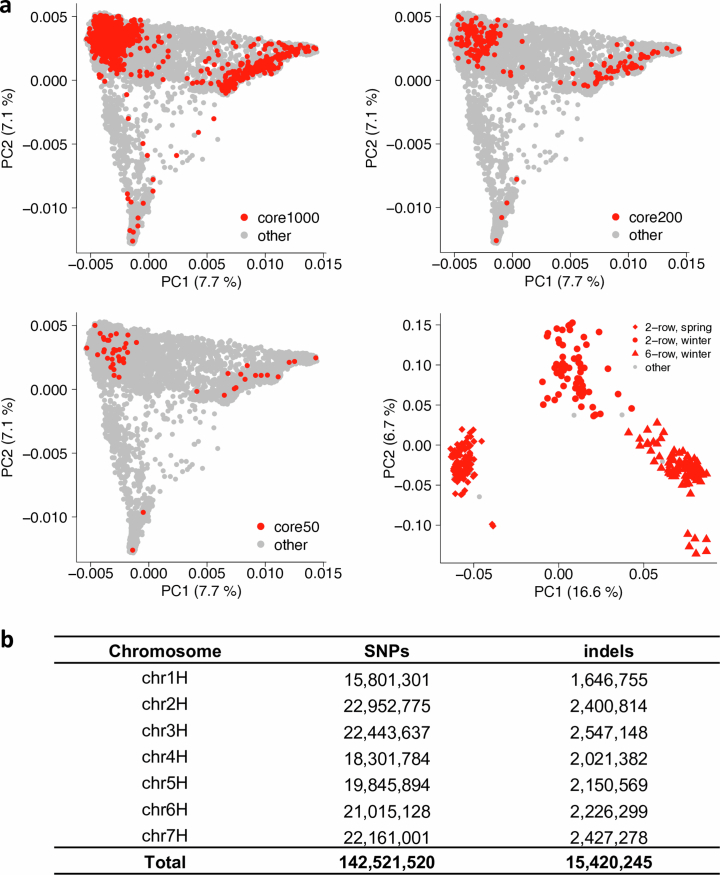
Despite domestication bottlenecks, genetic diversity is high in cultivated barley7. To quantify the completeness of the haplotype inventory of our pangenome, we compared our assemblies against short-read data of a global diversity panel (Supplementary Table 6). A core set of 1,000 genotypes selected from a collection of 22,626 barleys5 was sequenced to threefold monoploid genome coverage. Nested therein, 200 genomes10 were sequenced to tenfold depth and the gene space of 46 accessions was represented in the contigs assembled from 50-fold short-read data (Extended Data Fig. 5a and Supplementary Table 7). A total of 315 elite cultivars of European ancestry were sequenced to threefold coverage (Extended Data Fig. 5a and Supplementary Table 6). More than 157.9 million single-nucleotide polymorphisms (SNPs) and indels were detected across all panels (Extended Data Fig. 5b). Overlaying these with the pangenome showed that the 76 chromosome-scale assemblies captured almost all pericentric haplotypes of cultivated barley (Extended Data Fig. 2d–f). Coverage decreased to as low as 50% in distal regions, in which haplotypes of PGRs lacked a close relative in the pangenome more often than those of elite cultivars (Extended Data Fig. 2e,f). This suggests that, thanks to broad taxon sampling, short-read sequencing will remain indispensable for the time being, but in the future population-scale long-read sequencing23 will be as desirable in agricultural genetics as it is in medical genetics.
Extended Data Fig. 5. Short-read data complement the pangenome infrastructure.
(a) Accessions selected for short-read sequencing. Nested coresets of 1000, 200 and 50 accessions (core1000, core200, core50) are shown in the global diversity space of barley as represented by a principal component (PCA). The top-right subpanel shows a PCA of 315 elite cultivars. Accessions are according to genepool (2-rowed spring, 2-rowed winter, 6-rowed winter). The proportion of variance explained by the PCA is shown in the axis labels. (b) Counts of single-nucleotide polymorphisms (SNPs) and short insertions and deletions (indels) detected in those data.
An inventory of complex loci
Long-read sequencing has the power to resolve structurally complex genomic regions, in which repeated cycles of tandem duplication, mutation of duplicated genes and elimination by deletion or recombination have created a panoply of diverged copies of one or multiple genes in varied arrangements (Extended Data Fig. 6a). Many complex loci are intimately linked to the evolution of resistance genes24. An illustrative example is barley’s Mildew resistance locus a (Mla)25,26, which contains three families of resistance gene homologues, each with multiple members at the locus. A 40 kilobase (kb) region containing members of two families is repeated four times head-to-tail in cultivar RGT Planet, but is not present in even a single complete copy in 62 accessions of our pangenome (Supplementary Fig. 5). Mla genes sensu stricto, that is, those that have been experimentally proven to provide functional powdery mildew resistance, are among members of a subfamily that resides outside of this duplication but close to its distal border (Fig. 2a and Supplementary Fig. 5). Twenty-nine Mla alleles in the narrow sense have been defined to date27. Gene models identical to seven were identified in our pangenome (Fig. 2a). However, the sequence variation went beyond this observation: 149 unique gene models were different from, but highly similar to, known Mla alleles, with nucleotide sequences at least 98% identical. Some of these genes were present in multiple copies. HOR 8117, a landrace from Nepal, contained 11 different close homologues of Mla, two of which were present in five copies each (Supplementary Fig. 6). Genome sequences alone cannot inform us of how this sequence diversity relates to resistance to powdery mildew or other diseases28. Until the advent of long-read sequencing, it was almost impossible to resolve the structure of the Mla locus in multiple genomes at once. We expect that pangenomes will help the genomic dissection of complex resistance gene loci in barley and other crops.
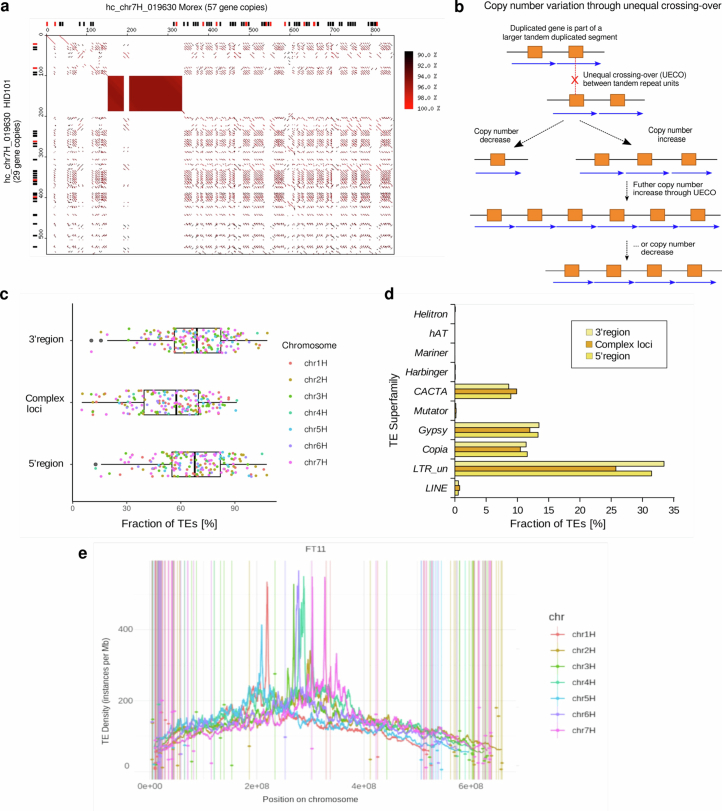
Extended Data Fig. 6. Complex loci are hot spots for copy number variation (CNV).
(a) Dot plot alignment of the example locus chr7H_019630 which contains a cluster of thionin genes. The sequences of Morex (horizontal) and wild barley HID101 (vertical) were aligned. Predicted intact genes are indicated as black boxes along the left and top axes. Predicted pseudogenes are shown in red. The axis scale is kb. The filled rectangle at positions ~150–330 kb in Morex represents an array of short tandem repeats which does not contain annotated thionin genes and does not have sequence homology to the thionin-containing tandem repeats of the locus. (b) The schematic model shows how, once an initial duplication is established, unequal homologous recombination (unequal crossing-over, UECO) between repeat units can lead to rapid expansion and contraction of the loci, thereby leading to CNV of genes. (c) TE content of complex loci. Dots represent the proportion of TEs (in %) in each to 169 complex loci. This is compared to regions of the same size (1 Mb) in the 3′ and 5′ directions. Complex loci have overall slighly lower content of annotated TEs than their flanking region, which is likely due to their higher gene content. Boxes indicate the inter-quartile range (IQR) with the central line indicating the median and whiskers indicating the minimum and maximum without outliers, respectively. Outliers were defined as minimum – 1.5 x IQR and maximum + 1.5 x IQR, respectively. (d) Contribution of TE superfamilies to complex loci and their 5′ and 3′ neigbouring regions. Complex loci contain slightly more CACTA and fewer LTR retroelements than neigbouring regions, a general characteristic of gene containing regions in barley. (e) Overall TE content along barley chromosomes (example accession B1K-04-12 [FT11]) compared to that of complex loci. TE content of complex loci is indicated by coloured dots. Due to the relatively small sizes of the loci, TE content of individual loci, in most cases, differs from that of the overall TE content in the respecive chromosomal regions.
Fig. 2. Structurally complex loci in the barley pangenome.
a, Presence/absence of known Mla alleles in the barley pangenome. Black and white squares denote presence and absence, respectively. The names of Mla alleles (y axis) and genotypes (x axis) are coloured according to, respectively, subfamily (red, 1; or black, 2; ref. 27) and domestication status (green, domesticated; orange, wild). Only the genomes containing known alleles are shown. Owing to higher SNP numbers and truncations27, members of subfamily 2 are expected to be inactive forms. b, Dot plot alignment of complex locus Chr04_015772 which contains Int-c genes. The plot shows an alignment of Morex (six-rowed barley) and Bowman (two-rowed barley). In Morex, Int-c and its surrounding sequence are present in three copies. Genes are indicated as black boxes along the axes of the plot. Individual tandem repeat units are 96–100% identical. c, CNV levels and numbers of encoded protein variants identified in 76 barley accessions. The x axis shows the level of CNV (that is, the difference between the accession with the fewest copies and that with the most copies for each locus). The y axis shows the total number of protein variants identified in all 76 barley accessions. Labels mark gene families with the highest copy numbers or the highest CNV levels. d, Complex loci are enriched in distal chromosomal regions. The seven barley chromosomes were divided into ten equally sized bins, and cumulative data for all chromosomes are shown. AQ15The bar plot indicates the number of loci, whereas the box plot shows the extent of CNV for all loci in the bin. Boxes indicate the IQR with the central line indicating the median and whiskers indicating the minimum and maximum without outliers, respectively. Outliers were defined as minimum −1.5 × IQR and maximum +1.5 × IQR, respectively.
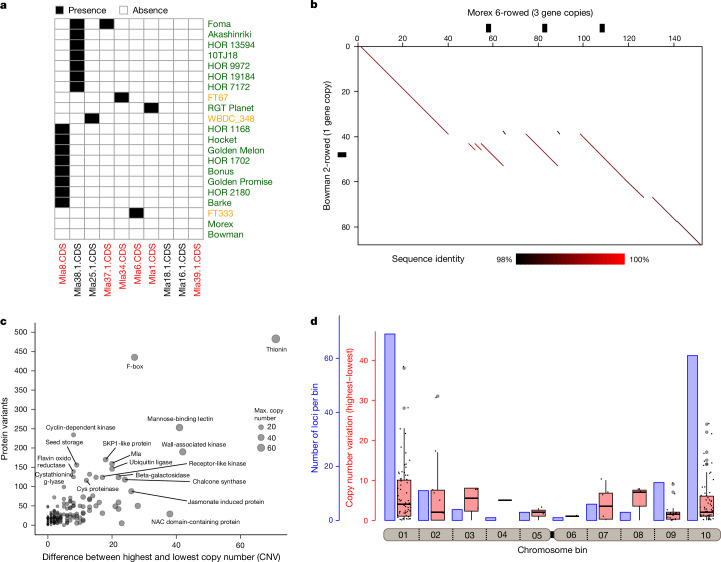
We used a gene-agnostic method29 to scan the genome sequence of Morex for structurally complex loci harbouring genes, focusing on examples that had evidently caused gene copy number variation across the pangenome via the expansion or collapse of long tandem repeats. A total of 169 loci ranging in size from 20 kb to 2.2 megabases (Mb) (median: 125 kb) matched our criteria (Fig. 2c, Supplementary Table 8 and Supplementary Figs. 7 and 8). Their copy numbers were variable in the pangenome. The most extreme case was a cluster of genes annotated by homology as thionin genes, which are possibly involved in resistance to herbivory30. The locus had as few as three thionin gene copies in the wild barley WBDC103 and up to 74 copies in WBDC199, another wild barley (Extended Data Fig. 6a). Genes associated with such complex loci possessed functional annotations suggesting involvement in various biological processes (Fig. 2c and Supplementary Table 8). Complex loci were enriched in distal chromosomal regions (Fig. 2d). In this regard, they follow the same distal-to-proximal gradient as genetic diversity and recombination frequency in barley. The latter process might have a role in their amplification and contraction owing to unequal homologous recombination between neighbouring repeat units31 (Extended Data Fig. 6b). We found no association of complex loci with specific TE types (Extended Data Fig. 6c–e). Instead, molecular dating of the tandem duplications in Morex is consistent with recent and recurring duplication/contraction cycles, leading to complex patterns of higher and lower order tandem repeats (Extended Data Fig. 7). Indeed, many gene copies seem to have been gained within the past 3 million years (Extended Data Fig. 7c), after the H. vulgare lineage split from that of its closest relative, Hordeum bulbosum32. In addition, 62 loci (36.7%) underwent at least one duplication in the past 10,000 years, that is, after domestication (Extended Data Fig. 7d). Forty-three loci expanded so recently that the genes they harboured were identical duplicates of each other. Despite high similarity of duplicated segments, TE insertions (or excisions), random deletions and mutations contribute to diversification or pseudogenization of individual gene copies over time (Fig. 3a and Supplementary Fig. 9a).
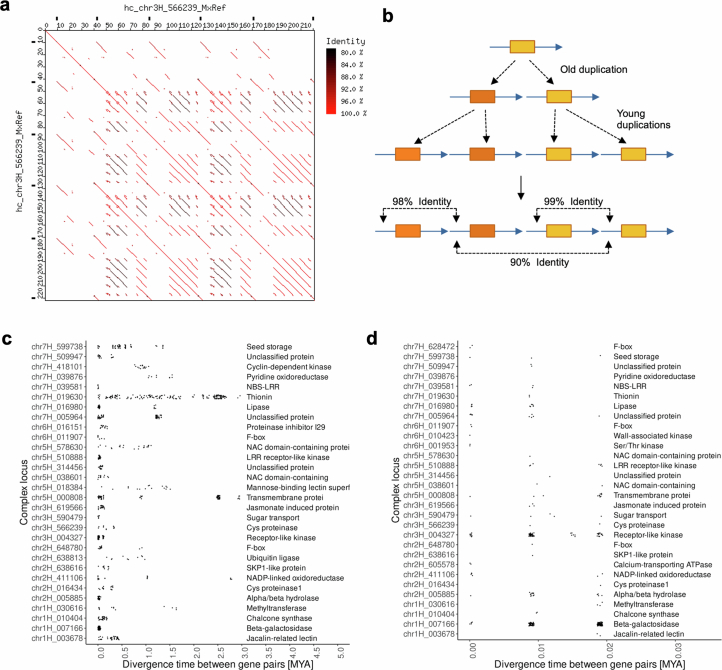
Extended Data Fig. 7. Molecular dating of divergence times between duplicated gene copies in complex loci.
(a) Dot plot example of locus hc_chr3H_566239 which underwent multiple waves of tandem duplications, which is reflected in varying levels of sequence identity between tandem repeats (color-coded). (b) Schematic mechanism for how different levels of sequence identity between tandem repeats evolve. In the example, an ancestral duplication was followed by two independent subsequent duplications, leading to varying levels of sequence identity between tandem repeat units. Genes are indicated as orange boxes while blue arrows indicate the tandem repeats they are embedded in. (c) Divergence time estimates between duplicate gene copies in complex loci. Shown are only those complex loci which have at least six tandem-duplicated genes. Each dot represents one divergence time estimate for a duplicated gene pair from the respective locus. The x-axis shows the estimated divergence time in million years. At the right-hand side, classification of proteins encoded by genes in the locus are shown. Note that several loci had multiple waves of gene duplications over the past 3 million years. (d) Subset of those loci shown in (c) that had at least one gene duplication within the past 20,000 years. The divergence time estimates appear in groups, since they represent the presence of 0, 1 and 2 nucleotide substitutions, respectively, in the approx. 4 kb of aligned sequences that were used for molecular dating.
Fig. 3. Structural diversity at the amy1_1 locus and its importance in malting.
a, Simplified structure of the amy1_1 locus in selected pangenome assemblies. A detailed depiction of the amy1_1 locus across all 76 assemblies is shown in Extended Data Fig. 9a. Identical colours indicate identical ORFs in a and d. b, Distribution of amy1_1 copy numbers (as proportion of wild or domesticated accessions) across 76 assemblies. c,d, X-ray crystal structure (PDB 1BG9, ref. 39) of α-amylase bound to acarbose as a substrate analogue (magenta and yellow spheres). In d, amy1_1 amino acid variants (found in Morex, Barke and RGT Planet; Supplementary Table 21) are added as coloured spheres. e, α-Amylase activity of micro-malted grain of RGT Planet compared to RGT Planet near-isogenic lines (NILs) containing amy1_1-Morex and Barke haplotypes. The boxes delimit the 25th and 75th percentiles, and the horizontal line inside the box represents the median. Lower and upper whiskers denote minima and maxima. Two-sided t-test was used in multiple comparison and P value was adjusted with the Holm–Bonferroni method (**P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001). n = 8 (Barke), 2 (Morex), 8 (RGT Planet) independent samples examined in 5 independent experiments or environments.
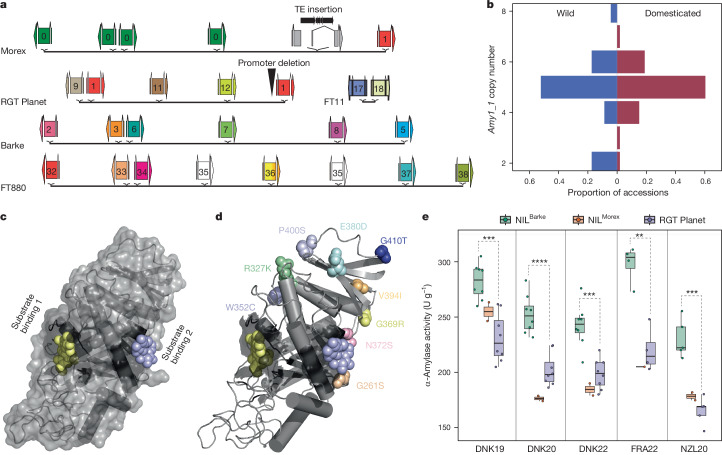
One interesting case of such recent diversification was a duplication at the HvTB1 locus (also known as INTERMEDIUM-C (INT-C) or SIX-ROWED SPIKE 5). HvTB1 is a TEOSINTE BRANCHED 1, CYCLOIDEA, PCF1 (TCP) transcription factor involved in basal branching (tillering) and other aspects of plant architecture in cereal grasses33–35. In barley, both tillering and the fertility of lateral spikelets are increased in knockout mutants35,36. Just two alleles, Int-c.a and int-c.b, dominate in six-rowed and two-rowed forms35, respectively, and HvTB1 is not genetically linked to the SIX-ROWED SPIKE 1 gene3. Both alleles of HvTB1 are thought to be functional and occur also in wild barley35,37. These patterns have defied easy explanation. Expression differences owing to regulatory variation have been postulated but not proven35. The pangenome adds another twist. HvTB1 is a single-copy gene in all 22 H. spontaneum accessions and 23 two-rowed domesticates except HOR 7385 (Supplementary Table 9). Six-rowed forms, however, have up to four copies of a 21 kb segment that contains HvTB1 and approximately 5 kb of its upstream sequence (Fig. 2b). The reference cultivar Morex has three copies, which were falsely collapsed in previous short-read assemblies of that cultivar38. On top of variable copy numbers, the pangenome revealed six hitherto unknown HvTB1 protein variants (Supplementary Fig. 9b and Supplementary Table 9). Reduced tillering in maize has been attributed to overexpression of TB1. The barley pangenome will help developmental geneticists to reveal whether copy number gains had analogous effects in six-rowed forms.
Amplification of α-amylases
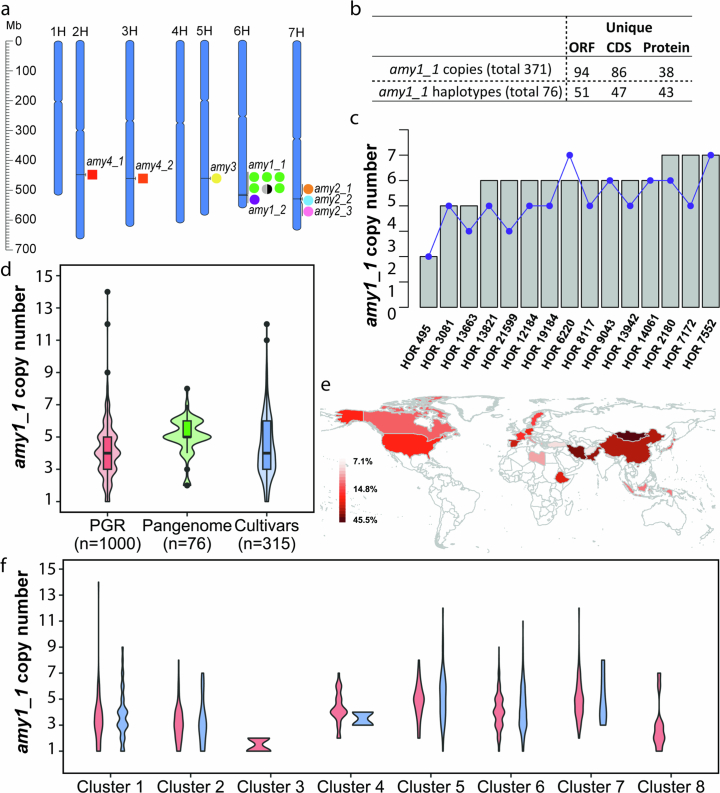
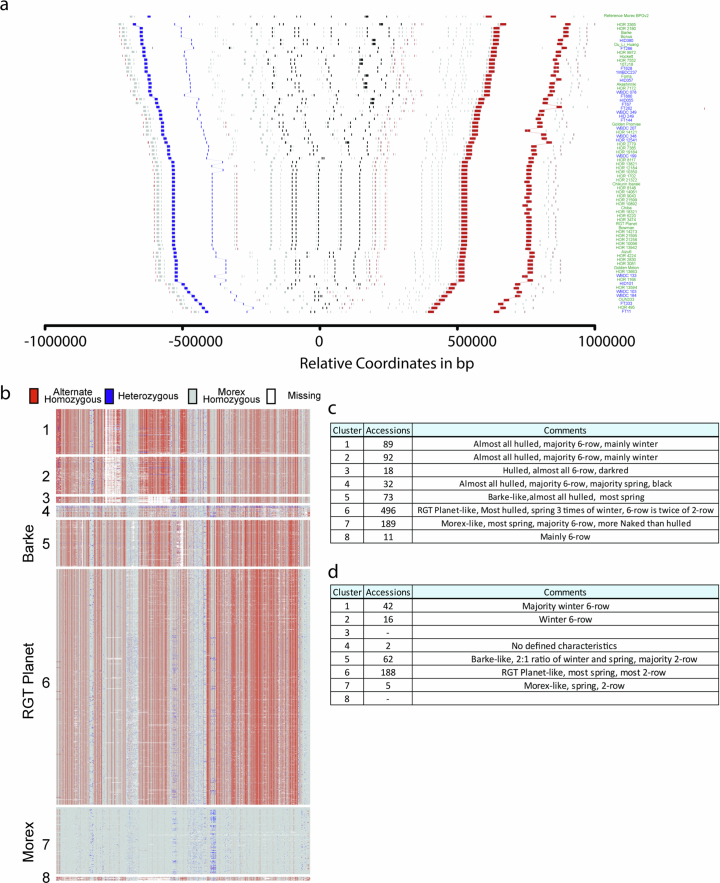
Among the complex loci we examined, the amy1_1 locus of α-amylases on chromosome 6H is arguably the one of greatest economic importance. These enzymes cleave the polysaccharide starch into short-chain forms which are then digested further into sugars39. In both wild and cultivated forms, the speed and efficiency of that process determines the energy supply to and hence the vigour and survival of the young seedling when competing for sunlight and nutrients40. In grains of domesticated barley, the enzymatic conversion of starch into fermentable sugars by α-amylases is a crucial step in malting and brewing processes. Barley α-amylases are subdivided into four families, which occupy distinct genomic loci (Extended Data Fig. 8a and Supplementary Tables 10 and 11). Owing to the large size of the amy1_1 cluster and the high similarity of its constituent gene copies (Extended Data Fig. 9a and Supplementary Table 11), earlier genomic analysis merely hinted at the presence of structural variation at the amy1_1 locus but failed to resolve its structure, including gene copy numbers and their structural arrangement41. This knowledge gap has hampered the exploitation of potentially useful variation at amy1_1 by practical breeders. It was closed only thanks to accurate long-read sequencing: all but one (HOR 8148) of our pangenome assemblies covered amy1_1 in a single contig (Extended Data Fig. 9a). We found between two and eight copies of amy1_1 in the 76 complete genomes, with substantial variation in both wild and domesticated forms (Fig. 3a,b). A local pangenome graph constructed with PGGB confirmed the complex structure locus and revealed that clustering according to structural features of the graph correlated well with amy1_1 copy numbers (Supplementary Fig. 10). Individual amy1_1 copies were addressable by 21-mers that overlap sequence variants (Extended Data Fig. 8c). We extended this analysis to 1,315 elite cultivars and PGRs by counting 21-mers in their short-read data (Extended Data Fig. 8d). SVs discovered in this wider panel hold ample potential for increasing α-amylase copy numbers in elite barleys (Extended Data Fig. 8e,f and Supplementary Tables 12 and 13). We determined SNP haplotypes around the amy1_1 locus (Extended Data Fig. 9b–d and Supplementary Tables 14–17) and defined eight clusters that were consistent with the global population structure. Among 315 European elite cultivars, clusters no. 5 and no. 6, represented by elite malting barleys RGT Planet and Barke, were most common.
Extended Data Fig. 8. amy1_1 locus structure and copy number in 76 assemblies and 1,315 whole genome sequenced accessions.
(a) Chromosomal locations of 12 α-amylase genes in the MorexV3 genome assembly. (b) Summary of amy1_1 locus sequence diversity in the 76 pangenome assemblies (Supplementary Tables 14–19). The distribution of unique amy1_1 ORFs, CDS and protein copies and haplotypes (denoting combinations of amy1_1 copies in individual accessions) across the 76 pangenome assemblies. (c) Comparison of amy1_1 copy numbers identified in the pangenome assemblies versus k-mer based estimation from raw reads (Pearson correlation coefficient r = 0.69, two-sided p-value = 0.004). Grey bars denote copy number from pangenome, blue dots denote k-mer estimated copy number. (d) amy1_1 copy number estimation in 76 pangenome assemblies (“Pangenome”), 1,000 whole-genome sequenced plant genetic resources (“PGR”), and 315 whole-genome sequenced European elite cultivars (“Cultivars”) using k-mer based methods. The boxes delimit the 25th and 75th percentile, the horizontal line inside the box represents the median. Lower and upper whiskers denote minima and maxima. (e) Distribution of accessions with amy1_1 copy numbers >5 per country (as percentage of total accessions in country for countries with ≥10 accessions). (f) amy1_1 copy number within each haplotype cluster (see Extended Data Fig. 9b). Red color refers to 1,000 plant genetic resource accessions, green refers to 76 pangenome accessions and blue refers to 315 European elite cultivars in panels d and f. Clusters #5, #6 and #7 in panel f contain Barke, RGT Planet and Morex, respectively.
Extended Data Fig. 9. Haplotype structure of the amy1_1 locus.
(a) Structural diversity in the vicinity of amy1_1 in the 76 pangenome assemblies. Each line shows the gene order in the sequence assembly of one genotype. The MorexV3 reference is shown on top. Coloured rectangles stand for gene models extracted from BLAST alignments against the corresponding gene models in MorexV3. Black rectangles represent amy1_1 homologs and grey rectangles other genes. Blue and red rectangles represent marker genes used to define the synteny, delimit the region and sort the accessions based on the distance between endpoints. Lines connect gene models between different genomes. Accession names are given on the right axis and are coloured according to type (blue – wild, green – domesticated). In HOR 8148, five copies assigned to 6H are shown. Two copies assigned to an unanchored contig are not shown. (b) SNP haplotype clusters at the amy1_1 locus among 1,315 genomes of domesticated and wild barley accessions, including genomes of 315 elite barley cultivars. The 6H:516,385,490-517,116,415 bp in the MorexV3 genome sequence is shown. Haplotype clusters #5, #6 and #7 contain the elite malting cultivars Barke, RGT Planet and Morex, respectively. (c) and (d) description of barley types in haplotype clusters #1-#8 across 315 elite cultivars (c) and 1,000 plant genetic resources (d).
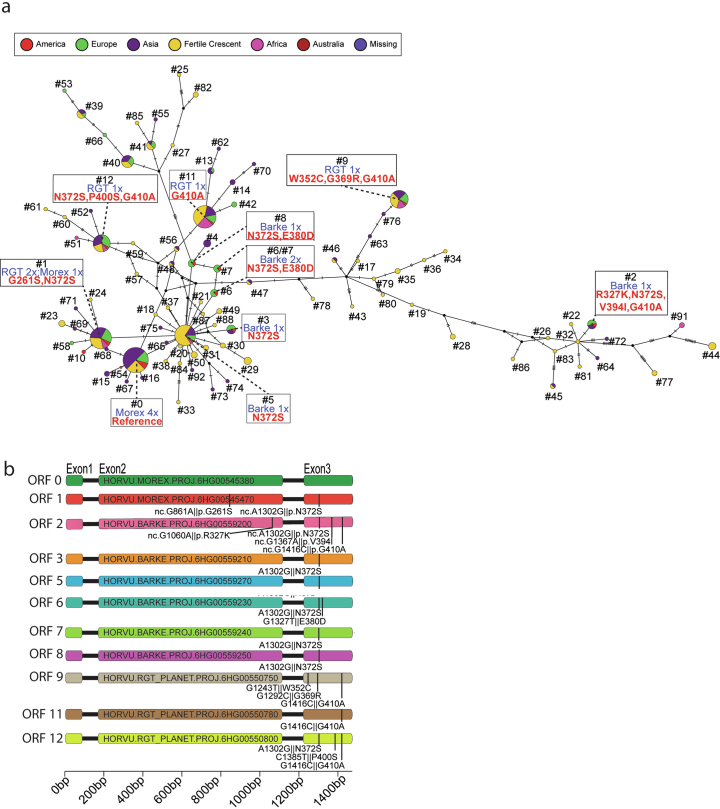
Structural diversity at amy1_1 was accompanied by differences in gene sequence owing to SNPs and indels in open reading frames (ORFs) and promoters. The 76 genome assemblies had a total of 371 amy1_1 ORFs (94 unique; Extended Data Fig. 8b). A median-joining network revealed that all major haplotypes (large nodes representing identical ORFs; see Supplementary Table 14 for gene IDs) are globally distributed and represented by the ORFs found in the elite malting barleys Barke, Morex and RGT Planet (Extended Data Fig. 10a,b). Across the AMY1_1 proteins found in Barke, RGT Planet and Morex, we identify nine individual amino acid variations (relative to reference sequence Prot0; Supplementary Table 18) that in a structural context locate both distal and proximal to the AMY1_1 substrate binding pockets (Fig. 3c,d). One amy1_1 Barke copy (ORF no. 2; Extended Data Fig. 10a,b and Supplementary Table 14) is markedly different from the remaining copies in Barke, Morex and RGT Planet. The changes in protein stability42 resulting from the amino acid variants R327K and V394I are predicted to be high-impact and could be favourable in the brewing process (Supplementary Table 20).
Extended Data Fig. 10. Sequence diversity of the amy1_1 gene.
(a) Median-joining haplotype network of amy1_1 copies in 76 pangenome assemblies. Nodes represent different ORFs and are coloured according to accession origin. The node size is proportional to the number of gene IDs a given node represents (Supplementary Table 14). Nodes containing cultivars Barke, RGT Planet and Morex amy1_1 ORFs are highlighted and the corresponding amino acid variation relative to Morex reference is shown in red. (b) Non-synonymous sequence exchanges in 12 non-redundant amy1_1 ORFs in the malting barleys Morex, Barke and RGT Planet. The positions of sequence variants and respective amino acid variations are marked by black lines. Colouring corresponds to (Fig. 3a). ORF numbers refer to Supplementary Table 14.
We investigated in more detail the elite malting barleys Morex, Barke and RGT Planet (Fig. 3, Extended Data Fig. 11 and Supplementary Tables 20 and 21). Before its use as a genome reference cultivar, Morex was a successful cultivar in North America. It had six nearly identical (greater than 99% similarity) (Supplementary Table 11) amy1_1 copies. The full-length copies were verified by PacBio amplicon sequencing. One copy was disrupted by a TE (Fig. 3a). Interestingly, 12 other amy1_1 ORFs among our assemblies had insertions of TEs and shared the same SNP haplotypes (99.9% identity) (Supplementary Table 22). These 12 accessions have the Morex amy1_1 cluster or closely related variations thereof (for example, clusters ORFHap2 and ORFHap39; Supplementary Table 15), suggesting a common ancestral insertion event. Barke, a German cultivar, also had six copies, all full-length, albeit of a different haplotype. RGT Planet, at present a successful cultivar in many barley-growing regions around the world, had five copies, one of which was likely to be inactivated by a 32 bp deletion in a pyr-box (CTTT(A/T) core) promoter binding site that is essential for α-amylase transcription43. We confirmed lower amy1_1 transcript abundance in micro-malted RGT Planet compared with a near-isogenic line (NIL) that carried the Barke amy1_1 haplotype in the genomic background of RGT Planet (Supplementary Fig. 11). The final end-use relevant α-amylase activity of a malted barley grain is the combination of its amy1_1 copy number, transcription and individual protein haplotype activity. Therefore, we tested overall α-amylase activity in micro-malting trials with RGT Planet and NILs that carried Morex and Barke amy1_1 haplotypes in the genomic background of RGT Planet. It was observed that α-amylase activity was highest in amy1_1-Barke NILs across three environments (Fig. 3e). The high-copy-number haplotype of the German cultivar Barke is common not only in cultivars favoured by European maltsters, who cater to all-malt brewing, but also among those from other regions of the world (Supplementary Table 23), where adjunct brewing is practised and barley α-amylases need to be abundant enough to cleave starch from adjuncts such as maize and rice. The patterns of sequence variation at amy1_1 uncovered by the barley pangenome pave the way for the targeted deployment, possibly even design, of amy1_1 haplotypes in breeding.
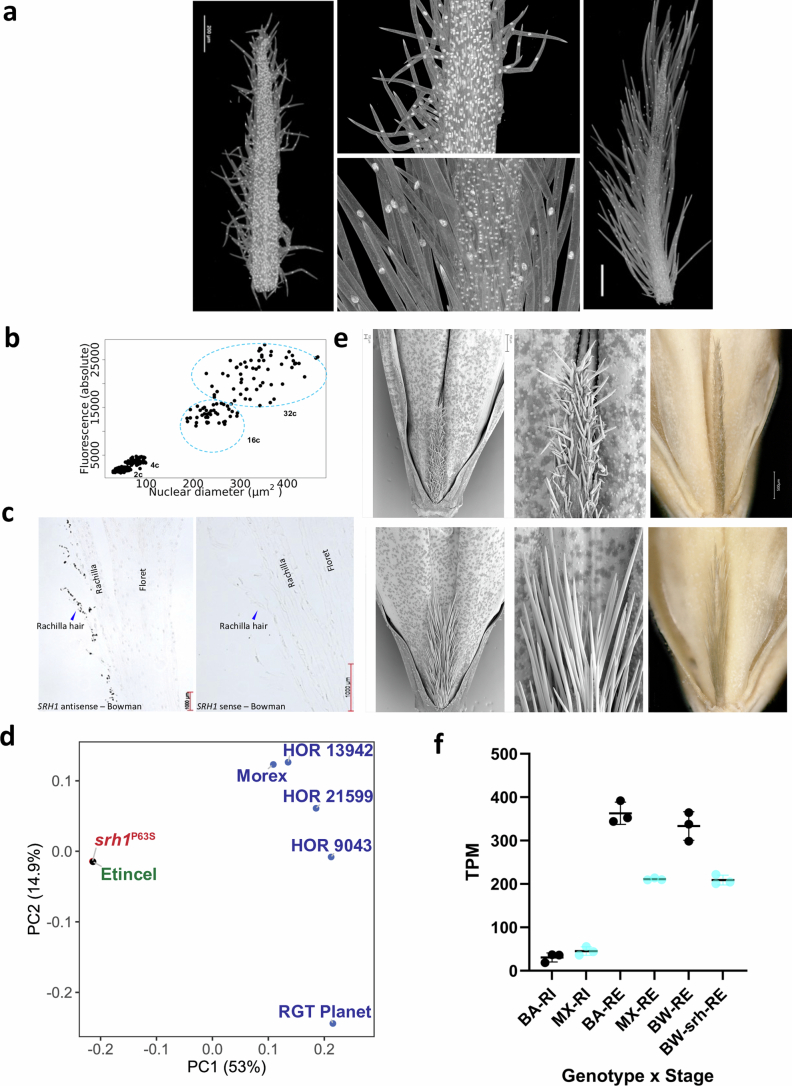
Extended Data Fig. 11. Functional dissection of the Srh1 locus.
(a) Light microscopy of short- and long-haired rachillae at Waddington developmental stage W8.5-9 using DAPI staining to visualize the nuclei. Size differences of nuclei in epidermal and trichome cells are very obvious. The shown micrographs are representative of a total of five individual spikes sampled on separate days. (b) Densitometric measurement of DNA content in epidermal and trichome cells of DAPI stained rachillae of genotypes Morex and Barke, respectively. While trichome cells in short-haired rachillae undergo only one cycle of endoreduplication, the cells in long haired trichomes show eight to sixteen-fold higher DNA contents than epidermal cells indicating three to four cycles of endoreduplication. (c) mRNA in situ hybridization of HvSRH1 in longitudinal spikelet sections of Bowman with anti-sense (left) and sense (right) probes. The blue arrow indicates the position of a rachilla hair. Representative micrographs of two independent experiments are shown. (d) Principal coordinate analysis of SNP array genotyping data of different barley genotypes. Etincel and its mutant srh1P63S cluster together, proving their isogenicity. (e) srh1 mutant discovery. FIND-IT screenings identified a mutant with short-fuzzy hairs (top) in the background of the long-haired cultivar Etincel (bottom). The mutants are a P63S non-synonymous sequence exchange. Scale bar - 1 mm. Wildtype and mutant spikes were inspected for the srh phenotype. Spikes showed either the short- or long-hair phenotype (#mutant seeds: 22, #wild type seeds: 21), respectively. Individual representative seeds wer chosen for micrographic documentation. (f) HvSRH1 transcript abundance in RNA sequencing data of rachilla tissue in Barke (BA, long-haired), Morex (MX, short-haired), Bowman (BW, long-haired) and a short-haired near-isogenic line of Bowman (BW-srh). Samples were taken at two developmental stages: rachilla hair initiation (RI) and elongation (RE). Abundance was measured as transcripts per million (TPM). Points stands for individual biological replicates (n = 3). Error bars show the mean and standard error.
An SV controls trichome development
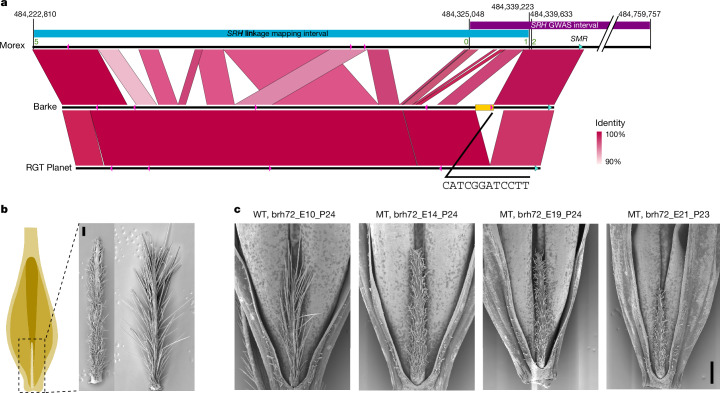
Our last example sits at the intersection of developmental genetics, breeding and domestication. Hairy appendages to grains and awns are conducive to seed dispersal in wild plants, but have lost this function in domesticates44. A pertinent example are the hairs on the rachillae of barley grains. In barley, the rachilla is the rudimentary secondary axis of the inflorescence, in which multiple grains are set in wheat45. In the single-grained spikelets of barley, the rachilla is a thin and hairy thread-like structure nested in the ventral crease of the grains. The long hairs of the rachillae of wild barleys and most cultivated forms are unicellular, whereas the short hairs of some domesticated types are multicellular and branched (Fig. 4 and Extended Data Fig. 11a). This seemingly minor difference in a vestigial organ belies its importance in variety registration trials46, for which breeders would like to predict the trait with a diagnostic marker. Short rachilla hair 1 (srh1) is also a classical locus in barley genetics47. It has been mapped genetically10,48 (Supplementary Fig. 12) and both long- and short-haired genotypes are included in our pangenome (Supplementary Table 27). Fine-mapping in a population of 2,398 recombinant inbred lines derived from a cross41,49 between cultivars Morex (short, srh1) and Barke (long, Srh1) delimited the causal variant to a 113 kb interval on the long arm of chromosome 5H (Fig. 4a and Supplementary Table 24). Outside of this interval (which is itself devoid of annotated gene models), but within 11 kb of the distal flanking marker, is a homologue of a SIAMESE-RELATED (SMR) gene of the model plant Arabidopsis thaliana50,51. Members of this family of cyclin-dependent kinase inhibitors control endoreduplication in trichomes of that species. In barley, hair cell development is likewise accompanied by endopolyploidy-dependent cell size increases (Extended Data Fig. 11a,b). The SMR homologue was expressed in the rachilla’s developing trichomes (Extended Data Fig. 10c), but there were no differences between Morex and Barke in the sequence of this otherwise plausible candidate gene (Supplementary Table 27). Despite this conflicting evidence, we proceeded with mutational analysis and obtained several mutants using FIND-IT52 (Extended Data Fig. 11d,e) and Cas9-mediated targeted mutagenesis (Fig. 4c, Supplementary Fig. 13 and Supplementary Tables 25 and 26). Mutants derived from long-haired genotypes carrying knockout variants or a non-synonymous change in a Pro phosphorylation motif (Thr62-Pro63) had short, multicellular rachillae, supporting the idea that the gene in question, HORVU.MOREX.r3.5HG0492730, is indeed HvSRH1. Sequence variation in HvSRH1 identified in the pangenome did not lend itself to easy explanation: 18 protein haplotypes caused by 23 non-synonymous variants bore no obvious relation to the phenotype (Supplementary Table 27). Thus, we then examined regulatory variation. All 14 short-haired genotypes in the pangenome lacked a 4,273 bp sequence segment (Fig. 4a), which did not contain coding sequences but was well conserved in long-haired types, with 95% overall identity to Barke. Within this sequence, we found the motif CATCGGATCCTT, matching the sequence C[ATC]T[ATC]GGATNC[CT][ATC], which is recognized by regulators of SMR expression in A. thaliana53. That sequence was repeated five times in Barke. The closest unit in long-haired types was no further than 13.6 kb from the gene, whereas the minimum distance between the gene and its putative enhancer motif in short-haired types was 22.3 kb, owing to the 4.3 kb deletion (Fig. 4a). A local pangenome graph of the Srh1 interval (Supplementary Fig. 14) showed that the paths of all of the accessions with short rachilla hairs consistently skipped these five nodes, consistent with the presence of a deletion associated with the phenotype. HvSRH1 expression during rachilla hair elongation is higher in long-haired than in short-haired genotypes (Extended Data Fig. 11f). Genome edits of the putative enhancer region will be required to obtain functional proof of its involvement in the transcriptional regulation of HvSRH1.
Fig. 4. A deletion in an enhancer motif is associated with Srh1-dependent trichome branching.
a, Top part, schematic representation of the high-resolution genetic linkage analysis at the Srh1 locus. Blue and purple horizontal bars represent the overlapping biparental and genome-wide association study (Supplementary Fig. 12) mapping intervals in reference to the 160 kb physical interval in the Morex genome (black line below the coloured bars). Note, an SMR-like gene, candidate for the srh1 mutant phenotype, sits outside the high-resolution biparental mapping interval. Bottom part, connector plot showing conserved homologous regions in the genotypes Barke (long hairs) and RGT Planet (short hairs). A region (yellow rectangle) harbouring a conserved enhancer element (pink triangle) is present in Barke, but absent in Morex and RGT Planet. b, Schematic drawing of a hulled and awned barley seed. The rachilla is the secondary axis in a cereal inflorescence, which in barley is reduced to a rudimentary structure densely covered with trichomes and attached to the base of the seed. On the right, scanning electron micrographs are shown of a short-haired and a long-haired rachilla of genotypes Morex and Barke, respectively. c, Rachilla hair phenotype of the Cas9-induced knockout mutants of the SMR-like gene. Panels from left to right show a wild-type segregant from the brhE72_E10 family (Supplementary Table 26) with long rachilla hairs; three representative mutants from three independent T1/M2 families brhE72_E14, _E19, _E24 segregating for different independent mutational events, respectively, all showing the short-hair phenotype (black bar indicates a length of 0.5 mm). MT, mutant; WT, wild type.
Discussion
The recently published human draft pangenome demonstrated how contiguous long-read sequences help to make sense of reams of sequence data54. Our study on the barley pangenome sheds light on crop evolution and breeding. The shortcomings of previous short-read assemblies made it all but impossible to see patterns that now emerge from their long-read counterparts. Here we studied the evolution of structurally complex loci of nearly identical tandem repeats. Our developmental insights are admittedly still cursory: true to the hypothesis-generating purview of genomics, and at least as many questions were raised as answered. We studied four loci—Mla, HvTB1, amy1_1 and HvSRH1—and the traits they control: disease resistance, plant architecture, starch mobilization and the hairiness of a rudimentary appendage to the grain. In two of these examples, phenotypic diversity has visibly increased in domesticated forms: there are no six-rowed or short-haired wild barleys. Malting created new selective pressures that only cultivated forms experienced. Novel allelic variation at disease resistance loci is both illustrative of the power of pangenomics and in line with our understanding of how disease resistance genes evolve. Structural variation at amy1_1 has been known for some time, but previous attempts at resolving the structure of the locus had been thwarted by incomplete genome sequences. Tandem duplications and deletions of regulatory elements, respectively, at HvTB1 and HvSRH1 were surprising because for many years barley geneticists considered the loci as monofactorial recessive. Much of the variation seems to have arisen after domestication, either because mutations that appear with clock-like regularity were absent or copy numbers were lower in the wild progenitor than in the domesticated forms. A common concern among crop conservationists is dangerously reduced genetic diversity in cultivated plants55. But crop evolution need not be a unidirectional loss of diversity. This study has shown that valuable diversity can arise after domestication. Allelic diversity at structurally complex loci may help domesticated plants to adapt to agricultural environment and fulfil the needs of farmers and breeders. More diverse crop pangenomes will help us to understand how the counteracting forces of past domestication bottlenecks and newly arisen SVs influence future crop improvement in changing climates.
Methods
Plant growth and high-molecular-weight DNA isolation
Twenty-five seeds each from the selected accessions (Supplementary Tables 1 and 7) were sown on 16-cm-diameter pots with compost soil. Plants were grown under greenhouse conditions with sodium halogen artificial 21 °C in the day for 16 h and 18 °C at night for 8 h. Leaves (8 g) were collected from 7-day-old seedlings, ground with liquid nitrogen to a fine powder and stored at −80 °C.
High-molecular-weight (HMW) DNA was purified from the powder, essentially as described56. In brief, nuclei were isolated, digested with proteinase K and lysed with SDS. Here, a standard watercolour brush with synthetic hair (size 8) was used to re-suspend the nuclei for digestion and lysis. HMW DNA was purified using phenol–chloroform extraction and precipitation with ethanol as described56. Subsequently, the HMW DNA was dissolved in 50 ml of TE (pH 8.0) and precipitated by the addition of 5 ml of 3 M sodium acetate (pH 5.2) and 100 ml of ice-cold ethanol. The suspension was mixed by slow circular movements resulting in the formation of a white precipitate (HMW DNA), which was collected using a wide-bore 5 ml pipette tip and transferred for 30 s into a tube containing 5 ml of 75% ethanol. The washing was repeated twice. The HMW DNA was transferred into a 2 ml tube using a wide-bore tip, collected with a polystyrene spatula, air-dried in a fresh 2 ml tube and dissolved in 500 µl of 10 mM Tris-Cl (pH 8.0). For quantification, the Qubit dsDNA High Sensitivity Assay Kit (Thermo Fisher Scientific) was used. The DNA size-profile was recorded using the Femto Pulse system and the Genomic DNA 165 kb kit (Agilent). In typical experiments the peak of the size-profile of the HMW DNA for library preparation was around 165 kb.
DNA library preparation and PacBio HiFi sequencing
For fragmentation of the HMW DNA into 20 kb fragments, a Megaruptor 3 device (speed: 30) was used (Diagenode). A minimum of two HiFi SMRTbell libraries were prepared for each barley genotype following essentially the manufacturer’s instructions and the SMRTbell Express Template Prep Kit (Pacific Biosciences). The final HiFi libraries were size-selected (narrow-size range: 18–21 kb) using the SageELF system with a 0.75% Agarose Gel Cassette (Sage Sciences) according to standard manufacturer protocols.
HiFi circular consensus sequencing (CCS) reads were generated by operating the PacBio Sequel IIe instrument (Pacific Biosciences) following the manufacturer’s instructions. Per genotype, about four 8M SMRT cells (average yield: 24 gigabases HiFi CCS per 8M SMART cell) were sequenced to obtain an approximate haploid genome coverage of about 20-fold. In typical experiments the concentration of the HiFi library on plate was 80–95 pM. We used 30 h movie time, 2 h pre-extension and sequencing chemistry v.2.0. The resulting raw data were processed using the CCS4 algorithm (https://github.com/PacificBiosciences/ccs).
Hi-C library preparation and Illumina sequencing
In situ Hi-C libraries were prepared from 1-week-old barley seedlings on the basis of the previously published protocol13. Dovetail Omni-C data were generated for Bowman, Aizu6, Golden Melon and 10TJ18 as per the manufacturer’s instructions (https://dovetailgenomics.com/products/omni-c-product-page/). Sequencing and Hi-C raw data processing was performed as described before57,58.
Genome sequence assembly and validation
PacBio HiFi reads were assembled using hifiasm (v.0.11-r302)59. Pseudomolecule construction was done with the TRITEX pipeline60. Chimeric contigs and orientation errors were identified through manual inspection of Hi-C contact matrices. Genome completeness and consensus accuracy were evaluated using Merqury (v.1.3)61. Levels of duplication and heterozygosity were assessed with Merqury and FindGSE (v.1.94)62. Further, we estimated heterozygosity in the HiFi reads with a k-mer approach. We selected 35,202 bi-allelic SNPs from a genebank genomic study3. For each SNP we extracted the flanking sequences (±15 bp) from the SNP positions and put either SNP in the middle to obtain 31-mers for the reference and alternative alleles. The FASTA sequences of the k-mers are available from https://bitbucket.org/ipkdg/het_estimation. We counted the occurrence of these k-mers in the HiFi FASTQ files using BBDuk (https://jgi.doe.gov/data-and-tools/software-tools/bbtools/bb-tools-user-guide/bbduk-guide/) with the parameter ‘rpkm’. Cenotype calling and the heterozygosity estimation were done in R. The full workflow is available from https://bitbucket.org/ipkdg/het_estimation.
Single-copy pangenome construction
The single-copy regions in each chromosome-level assembly were identified by filtering 31-mers occurring more than once in the genomic regions by BBDuk (BBMap_37.93, https://jgi.doe.gov/data-and-tools/software-tools/bbtools). BBMap was used to count k-mer occurrences in each genome with the parameter –mincount 2. Then, non-unique genomic regions (that is, those composed of k-mers occurring at least twice) were masked by BBDuk on the basis of k-mer counts. Single-copy regions extracted in BED format and their sequences (with the command ‘bedtools complement’) were retrieved using BEDTools (v.2.29.2)63. The single-copy sequences were clustered using MMseqs2 (Many-against-Many sequence searching)64 with the parameters ‘--cluster-mode’ and setting over 95% sequence identity. A representative from each cluster (the largest in a cluster) was selected to estimate the pangenome size.
Illumina resequencing
A total of 1,000 PGRs and 315 elite barley cultivars (Supplementary Table 6) were used for whole-genome resequencing. Illumina Nextera libraries were prepared and sequenced on an Illumina NovaSeq 6000 at IPK Gatersleben (Supplementary Table 6).
SNP and SV calling
Reciprocal genome alignment, in which each of the pangenome assemblies was aligned to the MorexV3 assembly with the latter acting either as alignment query or reference, was done with Minimap2 (v.2.20)65. From the resultant two alignment tables, indels were called by Assemblytics (v.1.2.1)66 and only deletions were selected in both alignments to convert into presence/absence variants relative to the Morex reference genome. Further, balanced rearrangements (inversions, translocations) were scanned for with SyRI67. To call SNPs, raw sequencing reads were trimmed using cutadapt (v.3.3)68 and aligned to the MorexV3 reference genome using Minimap2 (v.2.20)65. The resulting alignments were sorted with Novosort (v.3.09.01) (http://www.novocraft.com). BCFtools (v.1.9)69 was used to call SNPs and short indels. A genome-wide association study was performed in GEMMA (v.0.98.1)70 using default parameters with a mixed linear model and an estimated kinship matrix. Read depth was calculated at each complex locus in each accession. The raw HiFi reads were aligned to the respective genome using minimap2 (ref. 71) and the median depth per locus was calculated using mosdepth (v.0.2.6)72.
Linkage disequilibrium in the Barke x HID055 population
Linkage disequilibrium between each pair of SNPs (both intrachromosomal and interchromosomal) was calculated as the squared Pearson product-moment correlation between the quantitative identity-by-descent (IBD) matrix scores presented in Additional File 1 of ref. 73 (https://datadryad.org/stash/dataset/doi:10.5061/dryad.36rm1). The linkage disequilibrium plot was created with SAS PROC TEMPLATE and SGRENDER (SAS Institute) on the genetic map from ref. 18.
Preparation and Illumina sequencing of narrow-size whole-genome sequencing libraries for core50
First, 10 µg of DNA in 130 µl was sheared in tubes (Covaris microTUBE AFA Fiber Pre-Slit Snap Cap) to an average size of approximately 250 bp using a Covaris S220 focused-ultrasonicator (peak incidence power: 175 W, duty factor: 10%; cycles per burst: 200; time: 180 s) according to standard manufacturer protocols (Covaris). The sheared DNA was size-selected using a BluePippin device and a 1.5% agarose cassette with internal R2 marker (Sage Sciences). A tight size setting at 260 bp was used for the purification of fragments in the narrow range of 200–300 bp (typical yield: 1–3 µg). The size-selected DNA was used for the preparation of PCR-free whole-genome sequencing (WGS) libraries using the Roche KAPA Hyper Prep kit according to the manufacturer’s protocols (Roche Diagnostics). A total of 10–12 libraries were provided with unique barcodes, pooled at equimolar concentrations and quantified by quantitative PCR using the KAPA Library Quantification Kit for Illumina Platforms according to standard protocols (Roche Diagnostics). The pools were sequenced (2 × 151 bp, paired-end) using four S4 XP flowcells and the Illumina NovaSeq 6000 system (Illumina) at IPK Gatersleben.
Contig assembly of core50 sequencing data
Raw reads were demultiplexed on the basis of index sequences and duplicate reads were removed from the sequencing data using Fastuniq74. The read1 and read2 sequences were merged on the basis of the overlap using bbmerge.sh from bbmap (v.37.28)75. The merged reads were error-corrected using BFC (v.181)76. The error-corrected merged reads were used as an input for Minia3 (v.3.2.0)77 to assemble reads into unitigs with the following parameters, -no-bulge-removal -no-tip-removal -no-ec-removal -out-compress 9 -debloom original. The Minia3 source was assembled to enable k-mer size up to 512 as described in the Minia3 manual. Iterative Minia3 runs with increasing k-mer sizes (100, 150, 200, 250 and 300) were used for assembly generation as provided in the GATB Minia pipeline (https://github.com/GATB/gatb-minia-pipeline). In the first iteration, k-mer size of 50 was used to assemble input reads into unitigs. In the next runs, the input reads as well as the assembly of the previous iteration were used as input for the Minia3 assembler. BUSCO analysis was conducted on the contig assemblies using BUSCO (v.3.0.2) with embryophyta_odb9 dataset14. In addition, high-confidence gene models from the Morex V3 reference9 were aligned to the contig assemblies to assess completeness, with the parameters of greater than or equal to 90% query coverage and greater than or equal to 97% identity.
Pangenome accessions in diversity space
Pseudo-FASTQ paired-end reads (tenfold coverage) were generated from the 76 pangenome assemblies with fastq_generator (https://github.com/johanzi/fastq_generator) and aligned to the MorexV3 reference genome sequence assembly9 using Minimap2 (v.2.24-r1122, ref. 65). SNPs were called together with short-read data (Supplementary Table 6) using BCFtools78 v.1.9 with the command ‘mpileup -q 20 -Q20 --excl-flags 3332’. To plot the diversity space of cultivated barley, the resultant variant matrix was merged with that of 19,778 domesticated barleys from ref. 3 (genotyping-by-sequencing (GBS) data). SNPs with more than 20% missing or more than 20% heterozygous calls were discarded. Principal component analysis was done with smartpca79 v.7.2.1. To represent the diversity of wild barleys, we used published GBS and WGS data of 412 accessions of that taxon8,54. Variant calling for GBS data was done with BCFtools78 (v.1.9) using the command ‘mpileup -q 20 -Q20’. The resultant variant matrix was filtered as follows: (1) only bi-allelic SNP sites were kept; (2) homozygous genotype calls were retained if their read depth was greater than or equal to 2 and less than or equal to 50 and set to missing otherwise; (3) heterozygous genotype calls were retained if the read depth of both alleles was greater than or equal to 2 and set to missing otherwise. SNPs with more than 20% missing, more than 20% heterozygous calls or a minor allele frequency below 5% were discarded. Principal component analysis was done with smartpca79 v.7.2.1. A matrix of pairwise genetic distances on the basis of identity-by-state (IBS) was computed with Plink2 (v.2.00a3.3LM, ref. 80) and used to construct a neighbour-joining tree with Fneighbor (http://emboss.toulouse.inra.fr/cgi-bin/emboss/fneighbor) in the EMBOSS package81. The tree was visualized with Interactive Tree Of Life (iTOL)82.
Haplotype representation
Pangenome assemblies were mapped to MorexV3 as described above (‘Pangenome accessions in diversity space’). Read depth was calculated with SAMtools78 v.1.16.1. Genotype calls were set to missing if they were supported by fewer than two reads. IBS was calculated with PIink2 (v.2.000a3.3LM, ref. 80) in 1 Mb windows (shift: 0.5 Mb) using the using command ‘--sample-diff counts-only counts-cols=ibs0, ibs1’. Windows that in one of both accessions in the comparison had twofold coverage over less than 200 kb were set to missing. The number of differences (d) in a window was calculated as ibs0 + ibs1/2, where ibs0 is the number of homozygous differences and ibs1 that of heterozygous ones. This distance was normalized for coverage by the formula d/i × 1 Mb, where i is the size in bp of the region covered in both accessions in the comparison that had at least twofold coverage. In each window, we determined for each among the PGRs and cultivars panel the closest pangenome accession according to the coverage-normalized IBS distance. Only accessions with fewer than 10% missing windows due to low coverage were considered, leaving 899 PGRs and 264 cultivars.
The distance to the closest pangenome accession was plotted with the R package ggplot2 to determine the threshold for similarity (Extended Data Fig. 2d).
Transcriptome sequencing for gene annotation
Data for transcript evidence-based genome annotation were provided by the International Barley Pan-Transcriptome Consortium, and a detailed description of sample preparation and sequencing is provided elsewhere83. In brief, the 20 genotypes sequenced for the first version of the barley pangenome8 were used for transcriptome sequencing. Five separate tissues were sampled for each genotype. These were: embryo (including mesocotyl and seminal roots), seedling shoot, seedling root, inflorescence and caryopsis. Three biological replicates were sampled from each tissue type, amounting to 330 samples. Four samples failed quality control and were excluded.
Preparation of the strand-specific dUTP RNA-seq libraries and Illumina paired-end 150 bp sequencing were carried out by Novogene. In addition, PacBio Iso-Seq sequencing was carried out using a PacBio Sequel IIe sequencer at IPK Gatersleben. For this, a single sample per genotype was obtained by pooling equal amounts of RNA from a single replicate from all five tissues. Each sample was sequenced on an individual 8M SMRT cell.
De novo gene annotation
Structural gene annotation was done by combining de novo gene calling and homology-based approaches with RNA-seq, Iso-Seq and protein datasets (Extended Data Fig. 3a). Using evidence derived from expression data, RNA-seq data were first mapped using STAR84 (v.2.7.8a) and subsequently assembled into transcripts by StringTie85 (v.2.1.5, parameters -m 150-t -f 0.3). Triticeae protein sequences from available public datasets (UniProt86, https://www.uniprot.org, 10 May 2016) were aligned against the genome sequence using GenomeThreader87 (v.1.7.1; arguments -startcodon -finalstopcodon -species rice -gcmincoverage 70 -prseedlength 7 -prhdist 4). Iso-Seq datasets were aligned to the genome assembly using GMAP88 (v.2018-07-04). All assembled transcripts from RNA-seq, Iso-Seq and aligned protein sequences were combined using Cuffcompare89 (v.2.2.1) and subsequently merged with StringTie (v.2.1.5, parameters --merge -m150) into a pool of candidate transcripts. TransDecoder (v.5.5.0; http://transdecoder.github.io) was used to identify potential ORFs and to predict protein sequences within the candidate transcript set.
Ab initio annotation was initially done using Augustus90 (v.3.3.3). GeneMark91 (v.4.35) was additionally used to further improve structural gene annotation. To avoid potential over-prediction, we generated guiding hints using the above-described RNA-seq, protein and Iso-Seq datasets as described before92. A specific Augustus model for barley was built by generating a set of gene models with full support from RNA-seq and Iso-Seq. Augustus was trained and optimized following a published protocol92. All structural gene annotations were joined using EVidenceModeller93 (v.1.1.1), and weights were adjusted according to the input source: ab initio (Augustus: 5, GeneMark: 2), homology-based (10). Additionally, two rounds of PASA94 (v.2.4.1) were run to identify untranslated regions and isoforms using the above-described Iso-Seq datasets.
We used BLASTP95 (ncbi-blast-2.3.0+, parameters -max_target_seqs 1 -evalue 1e–05) to compare potential protein sequences with a trusted set of reference proteins (Uniprot Magnoliophyta, reviewed/Swissprot, downloaded on 3 August 2016; https://www.uniprot.org). This differentiated candidates into complete and valid genes, non-coding transcripts, pseudogenes and TEs. In addition, we used PTREP (release 19; http://botserv2.uzh.ch/kelldata/trep-db/index.html), a database of hypothetical proteins containing deduced amino acid sequences in which internal frameshifts have been removed in many cases. This step is particularly useful for the identification of divergent TEs with no significant similarity at the DNA level. Best hits were selected for each predicted protein from each of the three databases. Only hits with an e-value below 10 × 10−10 were considered. Furthermore, functional annotation of all predicted protein sequences was done using the AHRD pipeline (https://github.com/groupschoof/AHRD).
Proteins were further classified into two confidence classes: high and low. Hits with subject coverage (for protein references) or query coverage (transposon database) above 80% were considered significant and protein sequences were classified as high-confidence using the following criteria: protein sequence was complete and had a subject and query coverage above the threshold in the UniMag database or no BLAST hit in UniMag but in UniPoa and not PTREP; a low-confidence protein sequence was incomplete and had a hit in the UniMag or UniPoa database but not in PTREP. Alternatively, it had no hit in UniMag, UniPoa or PTREP, but the protein sequence was complete. In a second refinement step, low-confidence proteins with an AHRD score of 3* were promoted to high-confidence.
Gene projections
Gene contents of the remaining 56 barley genotypes were modelled by the projection of high-confidence genes on the basis of evidence-based gene annotations of the 20 barley genotypes described above. The approach was similar to and built upon a previously described method8. To reduce computational load, 760,078 high-confidence genes of the 20 barley annotations were clustered by cd-hit96 requiring 100% protein sequence similarity and a maximal size difference of four amino acids. The resulting 223,182 source genes were subsequently used for all downstream projections as the non-redundant transcript set representative for the evidence-based annotations. For each source, its maximal attainable score was determined by global protein self-alignment using the Needleman–Wunsch algorithm as implemented in Biopython97 v.1.8 and the blosum62 substitution matrix98 with a gap open and extension penalty of 0.5 and 10.0, respectively.
Next, we surveyed each barley genome sequence using minimap2 (ref. 65) with options ‘-ax splice:hq’ and ‘-uf’ for genomic matches of source transcripts. Each match was scored by its pairwise protein alignment with the source sequence that triggered the match. Only complete matches with start and stop codons and a score greater than or equal to 0.85 of the source self-score (see above) were retained. The source models were classified into four bins by decreasing confidence qualities: with or without pfam domains, plastid- and transposon-related genes. Projections were performed stepwise for the four qualities, starting from the highest to the lowest. In each quality group, matches were then added into the projected annotation if they did not overlap with any previously inserted model by their coding region. Insertion order progressed from the top to the lowest scoring match. In addition, we tracked the number of insertions for each source by its identifier. For the two top quality categories, we performed two rounds of projections, first inserting each source maximally only once followed by rounds allowing one source inserted multiple times into the projected annotation. To consolidate the 20 evidence-based, initial annotations for any genes potentially missed, we used an identical approach but inserted any non-overlapping matches starting from the previous RNA-seq-based annotation. A detailed description of the projection workflow, parameters and code is provided at the GitHub repository (https://github.com/GeorgHaberer/gene_projection/tree/main/panhordeum). An overview of the projection scheme can be found in the parent directory of the repository. Because complex loci contain numerous pseudogenes, the loci were searched by BLASTN99 for sequences homologous to annotated genes but not present in the set of annotated genes. Pseudogenes were accepted if they covered at least 80% of a gene homologue.
Definition of core, cloud and shell genes
Phylogenetic HOGs on the basis of the primary protein sequences from 76 annotated barley genotypes were calculated using Orthofinder100 v.2.5.5 (standard parameters). The scripts for calculation of core/shell and cloud genes have been deposited in the repository https://github.com/PGSB-HMGU/BPGv2. Core HOGs contain at least one gene model from all 76 barley genotypes included in the comparison. Shell HOGs contain gene models from at least two barley genotypes and at most 75 barley genotypes. Genes not included in any HOG (‘singletons’), or clustered with genes only from the same genotype, were defined as cloud genes. GENESPACE101 was used to determine syntenic relationships between the chromosomes of all 76 genotypes.
Annotation of TEs
The 20 barley accessions with expression data were softmasked for transposons before the de novo gene detection using the REdat_9.7_Triticeae section of the PGSB transposon library102. Vmatch (http://www.vmatch.de) was used as matching tool with the following parameters: identity > =70%, minimal hit length 75 bp, seedlength 12 bp (vmmatch -d -p -l 75 -identity 70 -seedlength 12 -exdrop 5 -qmaskmatch tolower). The percentage masked was around 84% and almost identical for all 20 accessions.
Full-length long terminal repeat retrotransposon candidate elements were detected de novo for each of the 76 barley accessions by their structural hallmarks with LTRharvest103 followed by LTRdigest104. Both programs are contained in genometools87 (http://github.com/genometools/genometools, v.1.5.10). LTRharvest identifies within the specified parameters long terminal repeats and target site duplications whereas LTRdigest was used to determine polypurine tracts and primer binding sites. The transfer RNA library needed as input for the primer binding sites was beforehand created by running tRNAscan-SE-1.3 (ref. 105) on each assembly. The parameter settings for LTRharvest were: ‘-overlaps best -seed 30 -minlenltr 100 -maxlenltr 2000 -mindistltr 3000 -maxdistltr 25000 -similar 85 -mintsd 4 -maxtsd 20 -motif tgca -motifmis 1 -vic 60 -xdrop 5 -mat 2 -mis -2 -ins -3 -del -3 -longoutput’; for LTRdigest: ‘-pptlen 8 30 -uboxlen 3 30 -pptradius 30 -pbsalilen 10 30 -pbsoffset 0 10 -pbstrnaoffset 0 30 -pbsmaxedist 1 -pbsradius 30’. The insertion age of each long terminal repeat retrotransposon instance was calculated from the divergence of its 5′ and 3′ long terminal repeat sequences using a random mutation rate of 1.3 × 10−8 (ref. 106).
Whole-genome pangenome graphs
Genome graphs were constructed using Minigraph19 v.0.20-r559. Other graph construction tools (PGGB107, Minigraph-Cactus108) turned out to be computationally prohibitive for a genome of this size and complexity, combined with the large number of accessions used in this investigation. Minigraph does not support small variants (less than 50 bp), thus graph complexity is lower than with other tools. However, even with Minigraph, graph construction at the whole-genome level was computationally prohibitive and thus graphs had to be computed separately for each chromosome, precluding detection of interchromosomal translocations.
Graph construction was initiated using the Morex V3 assembly9 as a reference. The remaining assemblies were added into the graph sequentially, in order of descending dissimilarity to Morex. SVs were called after each iteration using gfatools bubble (v.0.5-r250-dirty, https://github.com/lh3/gfatools). Following graph construction, the input sequences of all accessions were mapped back to the graph using Minigraph with the ‘--call’ option enabled, which generates a path through the graph for each accession. The resulting BED format files were merged using Minigraph’s mgutils.js utility script to convert them to P lines and then combined with the primary output of Minigraph in the proprietary RGFA format (https://github.com/lh3/gfatools/blob/master/doc/rGFA.md). Graphs were then converted from RGFA format to GFA format (https://github.com/GFA-spec/GFA-spec/blob/master/GFA1.md) using the ‘convert’ command from the vg toolkit109 v.1.46.0 ‘Altamura’. This step ensures that graphs are compatible with the wider universe of graph processing tools, most of which require GFA format as input. Chromosome-level graphs were then joined into a whole-genome graph using vg combine. The combined graph was indexed using vg index and vg gbwt, two components of the vg toolkit109.
General statistics for the whole-genome graph were computed with vg stats. Graph growth was computed using the heaps command from the ODGI toolkit110 v.0.8.2-0-g8715c55, followed by plotting with its companion script heaps_fit.R. The latter also computes values for gamma, the slope coefficient of Heap’s law which allows the classification of pangenome graphs into open or closed pangenomes, that is, a prediction of whether the addition of further accessions would increase the size of the pangenome111.
SV statistics were computed on the basis of the final BED file produced after the addition of the last line to the graph. A custom shell script was used to classify variants according to the Minigraph custom output format. This allows the extraction of simple, that is, non-nested, indels (relative to the MorexV3 graph backbone), as well simple inversions. The remaining SVs fall into the ‘complex’ category in which there can be multiple levels of nesting of different variant types and this precluded further, more fine-grained classification. To compute overlap with the SVs from Assemblytics, a custom script was used to extract the variant coordinates from both sets, and bedtools intersect63 was then used to compute their intersection on the basis of a spatial overlap of 70%.
To elucidate the effect of a graph-based reference on short-read mapping, we obtained WGS Illumina reads from five barley samples (Extended Data Fig. 4b) in the European Nucleotide Archive and mapped these onto the whole-genome graph using vg giraffe112. For comparison with the standard approach of mapping reads to a linear single genome reference, we mapped the same reads to the MorexV3 reference genome sequence assembly9 with bwa mem113 v.0.7.17-r1188. Mapping statistics were computed with vg109 stats and samtools78 stats (v.1.9), respectively.
To elucidate tool bias as a confounding factor in the comparison between the mappings, we first produced a linearized version of the pangenome graph using gfatools gfa2fa (https://github.com/lh3/gfatools) and then mapped the WGS reads from all five accessions to this new reference sequence, using BWA mem as before for the cv. Morex V3 reference sequence. This allows a more appropriate comparison between the single cultivar reference sequence and the pangenome sequence without being affected by algorithmic differences between the tools used (BWA/giraffe). Mappings were filtered to retain only reads with zero mismatches, using sambamba114. For the graph mappings, the ‘Total perfect’ statistic from the vg stats output of the GAM files was used.
To investigate the srh1 paths in the pangenome graph, we first extracted all nodes from the graph into a FASTA file and then used the enhancer region identified in cv. Barke as associated with the long-haired srh1 phenotype (chr5H:496,182,748-496,187,020) as query in a BLAST search against the nodes. This recovered five nodes with an identity percentage value of greater than 98%. We then used vg find from the vg toolkit v.1.56.0 (ref. 109) to extract a subgraph from the full graph (with a graph context of five steps either side) using the node identifiers. The subgraph was then plotted using odgi viz from the ODGI toolkit v.0.8.3-26-gbc7742ed (ref. 110).
To genotype samples from the core800 collection against the srh1 region of the graph, we first identified a small set of four samples each with either the short- or long-haired phenotype, picked at random from a group of core800 samples that all shared the same WGS read depth (5×). These samples were HOR_1102, HOR_17654, HOR_4065, HOR_1264, HOR_14704, HOR_7629, HOR_17678 and HOR_11406. We then mapped their Illumina WGS reads to the full pangenome graph using vg giraffe112 and extracted a subgraph of the mappings with vg chunk109. The subgraph was then genotyped using vg pack and vg call with cv. Barke as the reference accession, following the approach proposed in ref. 115. Variants in the resulting VCF files were identified using a simple grep command with the identifiers of the five nodes recovered with the Barke sequence as described above. Scripts used here are available at https://github.com/mb47/minigraph-barley/tree/main/scripts/srh1_analysis.
Analysis of the Mla locus
The coordinates and sequences of the 32 genes present at the Mla locus were extracted from the MorexV3 genome sequence assembly9. To find the corresponding position and copy number in each of the 76 genomes, we used BLAST95 (-perc_identity: 90, -word_size: 11, all other parameters set as default). The expected BLAST result for a perfectly conserved allele is a long fragment (exon_1) of 2,015 bp followed by a gap of approximately 1,000 bp due to the intron and another fragment (exon_2) of 820 bp. To detect the number of copies, first multiple BLAST results for a single gene were merged if two different BLAST segments were within 1.1 kb. Then only if the total length of the input was found, this was counted as a copy. To analyse the structural variation across all 76 accessions, the non-filtered BLAST results were plotted in a region of −20,000 and +500,000 base pairs around the start of the BPM gene HORVU.MOREX.r3.1HG0004540 that was used as an anchor (present in all 76 lines; Supplementary Figs. 5 and 6). To detect the different Mla alleles, three different thresholds of -Perc_identity for the BLAST were used: 100, 99 and 98.
Scan for structurally complex loci
We used a pipeline developed in ref. 27 that performs sequence-agnostic identification of long-duplication-prone regions (henceforth, complex regions) in a reference genome, followed by identification of gene families with a statistical tendency to occur within complex regions. The pipeline assumes that a candidate long, duplication-prone region will contain an elevated concentration of locally repeated sequences in the kb-scale length range. We first aligned the MorexV3 genome sequence assembly9 against itself using lastz116 (v.1.04.03; arguments: ‘--notransition --step=500 --gapped’). For practicality purposes, this was done in 2 Mb blocks with a 200 kb overlap, and any overlapping complex regions identified in multiple windows were merged. For each window, we ignored the trivial end-to-end alignment, and, of the remaining alignments, retained only those longer than 5 kb and falling fully within 200 kb of one and another. An alignment ‘density’ was calculated over the chromosome by calculating, at ‘interrogation points’ spaced equally at 1 kb intervals along the length of the chromosome, an alignment density score which is simply the sum of all the lengths of any of the filtered alignments spanning that interrogation point. A Gaussian kernel density (bandwidth 10 kb) was calculated over these interrogation points, weighted by their scores. To allow comparability between windows, the interrogation point densities were normalized by the sum of scores in the window. Runs of interrogation points at which the density surpassed a minimum density threshold were flagged as complex regions. A few minor adjustments to these regions (merging of overlapping regions, and trimming the end coordinates to ensure the stretches always begin and end in repeated sequence) yielded the final tabulated list of complex regions and their positions in the MorexV3 genome assembly (Supplementary Table 8). The method was implemented in R, making use of the package data.table. Genes in each long, duplication-prone region were clustered with UCLUST117 (v.11, default parameters) using a protein clustering distance cutoff of 0.5 and for each cluster the most frequent functional description as per the MorexV3 gene annotation9 was assigned as the functional description of the cluster. Self-alignment for characterization of evolutionary variability (Supplementary Fig. 7) was performed using lastz116 (v.1.04.03; settings ‘--self --notransition --gapped --nochain --gfextend --step=50’).
Molecular dating of divergence times of duplicated genes in complex loci
For molecular dating of gene duplications, we used segments of up to 4 kb, starting 1 kb upstream of duplicated genes in complex loci. With this, we presumed only to use intergenic sequences which are free from selection pressure and thus evolve at a neutral rate of 1.3 × 10−8 substitutions per site per year106. The upstream sequences of all duplicated genes of the respective complex locus were then aligned pairwise with the program Water from the EMBOSS package81 (obtained from Ubuntu repositories, https://ubuntu.com). This was done for all gene copies of all barley accession for which multiple gene copies were found. Molecular dating of the pairwise alignments was done as previously described118 using the substitution rate of 1.3 × 10−8 substitutions per site per year106.
Amy1_1 analysis in pangenome assemblies
The amy1_1 gene copy HORVU.MOREX.PROJ.6HG00545380 was used for BLAST against all 76 genome assemblies. Full-length sequences with identity over 95% were extracted and used for further analyses. Unique sequences were identified by clustering at 100% identity using CD-Hit96 and were aligned using MAFFT119 v.7.490. Sequence variants among amy1_1 gene copies at genomic DNA, coding sequence (CDS) and respective protein level were collected and amy1_1 haplotypes (that is, the combinations of copies) in each genotype assembly were summarized using R120 v.4.2.2. A Barke-specific SNP locus (GGCGCCAGGCATGATCGGGTGGTGGCCAGCCAAGGCGGTGACCTTCGTGGACAACCACGACACCGGCTCCACGCAGCACATGTGGCCCTTCCCTTCTGACA[A/G]GGTCATGCAGGGATATGCGTACATACTCACGCACCCAGGGACGCCATGCATCGTGAGTTCGTCGTACCAATACATCACATCTCAATTTTCTTTTCTTGTTTCGTTCATAA) for amy1_1 haplotype cluster ProtHap3 (Supplementary Table 21) was identified and used for KASP marker development (LGC Biosearch Technologies).
Comparative analysis of the amy1_1 locus structure
On the basis of the genome annotation of cv. Morex, 15 gene sequences on either side of amy1_1 gene copy HORVU.MOREX.PROJ.6HG00545440 were extracted. The 31 genes were compared against the 76 genome assemblies using NCBI-BLAST95 (BLASTN, word_size of 11 and percent identity of 90, other parameters as default). Alignment plots were generated from the BLAST result coordinates by scaling on the basis of the mid-point between HORVU.MOREX.r3.6HG0617300/HORVU.MOREX.PROJ.6HG00545250 and HORVU.MOREX.r3.6HG0617710/HORVU.MOREX.PROJ.6HG00545670. All BLAST results in the region (±1 Mb) around this mid-point were plotted using R120.
Amy1_1 PacBio amplicon sequencing
Genomic DNA from 1-week-old Morex seedling leaves was extracted with DNeasy Plant Mini Kit (QIAGEN). On the basis of the MorexV3 genome sequence assembly9, amy1_1 full-length copy-specific primers were designed using Primer3 (ref. 121) (https://primer3.ut.ee/): 6F: GTAGCAGTGCAGCGTGAAGTC; 80F: AGACATCGTTAACCACACATGC; 82F: GTTTCTCGTCCCTTTGCCTTAA; 82F: GTTTCTCGTCCCTTTGCCTTAA; 33R: GATCTGGATCGAAGGAGGGC; 79R: TCATACATGGGACCAGATCGAG; 80R: ACGTCAAGTTAGTAGGTAGCCC. All forward primers were tagged with bridge sequence (preceding T to primer name) [AmC6]gcagtcgaacatgtagctgactcaggtcac, whereas reverse primers were tagged with [AmC6]tggatcacttgtgcaagcatcacatcgtag to allow annealing to barcoding primers. These bridge sequence-tagged gene-specific primers were used in pairs with each other, targeting 1–2 copies of 3–6 kb amy1_1 genes, including upstream and downstream 500–1000 bp regions: T6F + T33R, T6F + T79R, T80F + T80R and T82F + T80R. A two-step PCR protocol was conducted. The first step PCR reaction was prepared in a 25 μl volume using 2 μl of DMSO, 0.3 μl of Q5 polymerase (New England Biolabs), 1 μl of amy1_1-specific primer pair (10 μM each), 2 μl of gDNA, 0.5 μl of dNTPs (10 mM), 5 μl of Q5 buffer and H2O. The PCR programme was as follows: initial denaturation at 98 °C/1 min followed by 25–28 cycles of 98 °C/30 s, 58 °C/30 s and 72 °C/3 min for extension, with a final extension step of 72 °C/2 min. The second PCR step (barcoding PCR) was prepared in the same way using 1 μl of the first PCR product as DNA template, barcoding primers (Pacific Biosciences) and the PCR programme reduced to 20 cycles. After quality check on 1% agarose gel, all barcoded PCR products were mixed and purified with AMPure PB (Pacific Biosciences). The SMRT bell library preparation and sequencing were carried out at BGI Tech Solutions. Sequencing data were analysed using SMRT Link v.10.2. To minimize PCR chimeric noise, CCSs were first constructed for each molecule. Second, long amplicon analysis was carried out on the basis of subreads from 50 bp windows spanning peak positions of all CCS length. Final consensus sequences for each amy1_1 were determined with the aid of size estimation from agarose gel imaging.
Amy1_1 SNP haplotype analysis and k-mer-based copy number estimation
SNP haplotypes were analysed in 1,315 PGRs and elite cultivars in the extended amy1_1 cluster region (MorexV3 chr6H: 516,385,490–517,116,415 bp). SNPs with more than 20% missing data among the analysed lines and minor allele frequency less than 0.01 were removed from downstream analyses. The data were converted to 0, 1 and 2 format using VCFtools122 and samples were clustered using the pheatmap package (https://cran.r-project.org/web/packages/pheatmap/pheatmap.pdf) from R statistical environment57. The sequential clustering approach was used to achieve the desired separation. At each step, two extreme clusters were selected and then samples from each cluster were clustered separately. The process was repeated until the desired separation was achieved on the basis of visual inspection.
K-mers (k = 21) were generated from the Morex amy1_1 gene family members’ conserved region using jellyfish123 v.2.2.10. After removing k-mers with counts from regions other than amy1_1 in the Morex V3 genome assembly, k-mers were counted in the Illumina raw reads (Supplementary Table 6) using Seal (BBtools, https://jgi.doe.gov/data-and-tools/software-tools/bbtools/). All k-mer counts were normalized to counts per MorexV3 genome and amy1_1 copy number was estimated as the median count of all k-mers from each accession in R.
Estimation ability was validated by comparing copy number from pangenome assemblies and short-read sequencing data (Extended Data Fig. 8c). For 1,000 PGRs, countries (with at least 10 accessions) were colour-shaded on the basis of their proportions of accessions with amy1_1 copy number greater than 5 on a world map using the R package maptools (https://cran.r-project.org/web/packages/maptools/index.html).
To construct a network from SNP haplotypes, all 371 amy1_1 copies (except ORF 89, 90 and 93; Supplementary Table 14) were aligned using MAFFT119 v.7.490. Median-joining haplotype networks were generated using PopART124 with an epsilon value of 0.
Local pangenome graph for amy1_1
The coordinates of amy1_1 copies in 76 genome assemblies were obtained by BLAST searches with the Morex allele of HORVU.MOREX.PROJ.6HG00545380. The genomic intervals surrounding amy1_1 from 10 kb upstream of the first copy to 10 kb downstream of the last copy were extracted from corresponding assemblies and used for further analyses. We applied PGGB (v.0.4.0, https://github.com/pangenome/pggb) for 76 amy1_1 sequences with parameters ‘-n 76 -t 20 -p 90 -s 1000 -N’. The graph was visualized using Bandage125 (v.0.8.1). ODGI (v.0.7.3, command ‘paths’)110 was used to get a sparse distance matrix for paths with the parameter ‘-d’. The resultant distance matrix was plotted with the R package pheatmap (https://cran.r-project.org/web/packages/pheatmap/pheatmap.pdf). Six representative sequences of amy1_1 were aligned against Morex by BLAST+ (v.2.13.0)99.
AMY1_1 protein structure and protein folding simulation
The published protein structure of α-amylase AMY1_1 from accession Menuet, in complex with the pseudo-tetrasaccharide acarbose (PDB: 1BG9; ref. 42), was used to simulate the structural context of the amino acid variants identified in barley accessions Morex, Barke and RGT Planet. The amino acid sequences of the crystalized AMY1_1 protein from Menuet and the Morex reference copy amy1_1 HORVU.MOREX.PROJ.6HG00545380 used in this study are identical. The protein was visualized using PyMol 2.5.5 (Schrödinger). The Dynamut2 webserver126 was used to predict changes in protein stability and dynamics by introducing amino acid variants identified in the Morex, Barke and RGT Planet genome assemblies.
Development of diverse amy1_1 haplotype barley NILs
NILs with different amy1_1 haplotypes were derived from crosses between RGT Planet as recipient and Barke or Morex amy1_1 cluster donor parents (ProtHap3, ProtHap4 and ProtHap0, respectively; Supplementary Table 21), followed by two subsequent backcrosses to RGT Planet and one selfing step (BC2S1) to retrieve homozygous plants at the amy1_1 locus. A total of four amy_1_1–Barke NILs (ProtHap3) and one amy1_1–Morex NIL (ProtHap0) were developed and tested against RGT Planet (ProtHap4) replicates. Plants were grown in a greenhouse at 18 °C under 16/8-h light/dark cycles. Foreground and background molecular markers were used in each generation to assist plant selection. Respective BC2S1 plants were genotyped with the Barley Illumina 15K array (SGS Institut Fresenius, TraitGenetics Section, Germany) and grown to maturity. Grains were collected and further propagated in field plots in consecutive years in various locations (Nørre Aaby, Denmark; Lincoln, New Zealand; Maule, France). Grains from field plots were collected and threshed using a Wintersteiger Elite plot combiner, and sorted by size (threshold, 2.5 mm) using a Pfeuffer SLN3 sample cleaner (Pfeuffer).
Micro-malting and α-amylase activity analysis
Non-dormant barley samples of RGT Planet and respective NILs with different amy1_1 haplotypes (50 g each, graded greater than 2.5 mm) were micro-malted in perforated stainless-steel boxes. The barley samples were steeped at 15 °C by submersion of the boxes in water. Steeping took place for 6 h on day one, 3 h on day two and 1 h on day three, followed by air rests, to reach 35%, 40% and 45% water content, respectively. The actual water uptake of individual samples was determined as the weight difference between initial water content, measured with a Foss 1241 NIT instrument, and the sample weight after surface water removal. During air rest, metal beakers were placed into a germination box at 15 °C. Following the last steep, the barley samples were germinated for 3 d at 15 °C. Finally, barley samples were kiln-dried in an MMK Curio kiln (Curio Group) using a two-step ramping profile. The first ramping step started at a set point of 27 °C with a linear ramping at 2 °C h−1 to the breakpoint at 55 °C using 100% fresh air. The second linear ramping was at 4 °C h−1, reaching a maximum at 85 °C. This temperature was kept constant for 90 min using 50% air recirculation. The kilned samples were then deculmed using a manual root removal system (Wissenschaftliche Station für Brauerei). α-Amylase activity was measured using the Ceralpha method (Ceralpha Method MR-CAAR4, Megazyme) modified for Gallery Plus Beermaster (Thermo Fisher Scientific).
Amy1_1 gene expression of RGT Planet and amy1_1–Barke NIL during micro-malting
Samples (50 g each, graded greater than 2.5 mm) were micro-malted as described in the previous section. During micro-malting, grains were sampled at 24 h, 48 h and 72 h. Grain samples were first freeze-dried at −80 °C and then milled at room temperature. Total RNA was isolated from 20–200 mg of flour using the Spectrum Plant Total RNA Kit (Sigma Aldrich) and cleaned using RNA Clean & Concentrator (ZYMO Research) following a published protocol127. For RNA-seq analysis, libraries were prepared and single-end sequenced with a length of 75 bp as described in ref. 127. Gene expression was quantified as transcripts per million (TPM) using kallisto128 (v.0.48.0) with 100 bootstraps.
Rachilla hair ploidy measurements
Ploidy assessment was performed on rachillae collected from barley spikes at developmental stage129 approximately Waddington 9.0. Once isolated, rachillae were fixed with 50% ethanol/10% acetic acid for 16 h after which they were stained with 1 µM DAPI in 50 mM phosphate buffer (pH 7.2) supplemented with 0.05% Triton X100. Probes were analysed with a Zeiss LSM780 confocal laser scanning microscope using a ×20 NA 0.8 objective, zoom ×4 and image size 512 × 512 pixels. DAPI was visualized with a 405 nm laser line in combination with a 405–475 nm bandpass filter. The pinhole was set to ensure the whole nucleus was measured in one scan. Size and fluorescence intensity of the nuclei were measured with ZEN black (ZEISS) software. For data normalization, small, round nuclei of the epidermal proper were used for 2C (diploid) calibration.
Scanning electron microscopy
Sample preparation and recording by scanning electron microscopy were essentially performed as described previously130. In brief, samples were fixed overnight at 4 °C in 50 mM phosphate buffer (pH 7.2) containing 2% v/v glutaraldehyde and 2% v/v formaldehyde. After washing with distilled water and dehydration in an ascending ethanol series, samples were critical-point‐dried in a Bal‐Tec critical-point dryer (Leica Microsystems, https://www.leica-microsystems.com). Dried specimens were attached to carbon‐coated aluminium sample blocks and gold‐coated in an Edwards S150B sputter coater (Edwards High Vacuum, http://www.edwardsvacuum.com). Probes were examined in a Zeiss Gemini30 scanning electron microscope (Carl Zeiss, https://www.zeiss.de) at 5 kV acceleration voltage. Images were digitally recorded.
Linkage mapping of SHORT RACHILLA HAIR 1 (HvSRH1)
Initial linkage mapping was performed using GBS data of a large ‘Morex’ x ‘Barke’ F8 recombinant inbred line (RIL) population47 (European Nucleotide Archive project PRJEB14130). The GBS data of 163 RILs, phenotyped for rachilla hair in the F11 generation, and the two parental genotypes were extracted from the variant matrix using VCFtools122 and filtered as described previously3 for a minimum depth of sequencing to accept heterozygous and homozygous calls of 4 and 6, respectively, a minimum mapping quality score of the SNPs of 30, a minimal fraction of homozygous calls of 30% and a maximum fraction of missing data of 25%. The linkage map was built with the R package ASMap131 using the MSTMap algorithm132 and the Kosambi mapping function, forcing the linkage group to split according to the physical chromosomes. The linkage mapping was done with R/qtl133 using the binary model of the scanone function with the expectation maximization method134. The significance threshold was calculated running 1,000 permutations and the interval was determined by a logarithm of the odds drop of 1. To confirm consistency between the F8 RIL genotypes and F11 RIL phenotypes, three PCR Allele Competitive Extension (PACE) markers were designed through the 3CR Bioscience free assay design service, using polymorphisms between the genome assemblies of the two parents (Supplementary Table 24), and PACE genotyping was performed as described earlier135. To reduce the Srh1 interval, 22 recombinant F8 RILs were sequenced by Illumina WGS, the sequencing reads were mapped on the MorexV3 reference genome sequence assembly9 and the SNP was called. The 100 bp region around the flanking SNPs of the Srh1 interval as well as the sequence of the candidate gene HORVU.MOREX.r3.5HG0492730 were compared with the pangenome assemblies using BLASTN99 to identify the corresponding coordinates and extract the respective intervals for comparison. Gene sequences were aligned with Muscle5 (ref. 136). Structural variation between intervals was assessed with LASTZ116 v.1.04.03. The motif search was carried out with the EMBOSS81 6.5.7 tool fuzznuc.
Cas9-mediated mutagenesis
Guide RNA (gRNA) target motifs in the ‘Golden Promise’ HvSrh1 candidate gene HORVU.GOLDEN_PROMISE.PROJ.5HG00440000.1 were selected by using the online tool WU-CRISPR137 to induce translational frameshift mutations by insertion/deletion of nucleotides leading to loss-of-function of the gene. One pair of target motifs (gRNA1a: CCTCGCTGCCCGCCGACGC; gRNA1b: GACAAGACGAAGGCCGCGG) was selected within the HvSrh1 candidate gene on the basis of their position within the first half of the coding sequence and the two-dimensional minimum free energy structures of the cognate single-gRNAs (NNNNNNNNNNNNNNNNNNNNGUUUUAGAGCUAGAAAUAGCAAGUUAAAAUAAGGCUAGUCCGUUAUCAACUUGAAAAAGUGGCACCGAGUCGGUGCUUUU) as modelled by the RNAfold WebServer138 and validated as suggested in ref. 139. gRNA-containing transformation vectors were cloned using the modular CasCADE vector system (10.15488/13200). gRNA-specific sequences were ordered as DNA oligonucleotides (Supplementary Table 25) with specific overhangs for BsaI-based cloning into the gRNA-module vectors carrying the gRNA scaffold, driven by the Triticum aestivum U6 promoter. Golden Gate assembly of gRNAs and the cas9 module, driven by the Zea mays Polyubiquitin 1 (ZmUbi1) promotor, was performed according to the CasCADE protocol to generate the intermediate vector pHP21. To generate the binary vector pHP22, the gRNA and cas9 expression units were cloned using SfiI into the generic vector140 p6i-2x35S-TE9 which harbours an hpt gene under control of a double-enhanced CaMV35S promoter in its transfer-DNA for plant selection. Agrobacterium-mediated DNA transfer to immature embryos of the spring barley Golden Promise was performed as previously described141. In brief, immature embryos were excised from caryopses 12–14 d after pollination and co-cultivated with Agrobacterium strain AGL1 carrying pHP22 for 48 h. Then, the explants were cultivated for further callus formation under selective conditions using Timentin and hygromycin, which was followed by plant regeneration. The presence of T-DNA in regenerated plantlets was confirmed by hpt- and cas9-specific PCRs (primer sequences in Supplementary Table 25). Primary mutant plants (M1 generation) were identified by PCR amplification of the target region (primer sequences in Supplementary Table 25) followed by Sanger sequencing at LGC Genomics. Double or multiple peaks in the sequence chromatogram starting around the Cas9 cleavage site upstream of the target’s protospacer-adjacent motif were considered as an indication for chimeric and/or heterozygous mutants. Mutant plants were grown in a glasshouse until the formation of mature grains. M2 plants were grown in a climate chamber under speed breeding conditions (22 h light at 22 °C and 2 h dark at 19 °C, adapted from ref. 142) and genotyped by Sanger sequencing of PCR amplicons as given above. M2 grains were subjected to phenotyping.
FIND-IT library construction
We constructed a FIND-IT library in cv. ‘Etincel’ (6-row winter malting barley; SECOBRA Recherches) as described in ref. 50. In short, we induced mutations by incubating 2.5 kg of ‘Etincel’ grain in water overnight at 8 °C following an incubation in 0.3 mM NaN3 at pH 3.0 for 2 h at 20 °C with continuous application of oxygen. After thoroughly washing with water, the grains were air-dried in a fume hood for 48 h. Mutagenized grains were sown in fields in Nørre Aaby, Denmark, and collected in bulk using a Wintersteiger Elite plot combiner. In the following generation, 2.5 kg of grain was sown in fields in Lincoln, New Zealand, and 188 pools of approximately 300 plants each were hand-harvested and threshed. A representative sample, 25% of each pool, was milled (Retsch GM200), and DNA was extracted from 25 g of the flour by LGC Genomics.
FIND-IT screening
The FIND-IT ‘Etincel’ library was screened as described in ref. 50 using a single assay for the isolation of srh1P63S variant (ID no. CB-FINDit-Hv-014). Forward primer 5′ AATCCTGCAGTCCTTGG 3′, reverse primer 5′ GAGGAGAAGAAGGAGCC 3′, mutant probe 5′6-FAM/CGTGGACGT/ZEN/CGACG/3’IABkFQ/wild-type probe/5′SUN/ACGTGGGCG/ZEN/TCGA/3′IABkFQ/ (Integrated DNA Technologies).
4K SNP chip genotyping
Genotyping, including DNA extraction from freeze-dried leaf material, was conducted by TraitGenetics. srh1P63S mutant, the corresponding wild-type ‘Etincel’ and srh1 pangenome accessions Morex, RGT Planet, HOR 13942, HOR 9043 and HOR 21599 were genotyped for background confirmation. Pairwise genetic distance of individuals was calculated as the average of their per-locus distances143 using the R package stringdist144 (v.0.9.8). Principal coordinate analysis was done with R120 (v.4.0.2) base function cmdscale on the basis of this genetic distance matrix. The first two principal components were illustrated by ggplot2 (https://ggplot2.tidyverse.org).
Sanger sequencing
gDNA of the srh1P63S variant and ‘Etincel’ was extracted from 1-week-old seedling leaves (DNeasy, Plant Mini Kit, Qiagen). Genomic DNA fragments for sequencing were amplified by PCR using gene-specific primers (forward primer 5′ TTGCACGATTCAAATGTGGT 3′, reverse primer 5′ TCACCGGGATCTCTCTGAAT 3′) and Taq DNA Polymerase (NEB) for 35 cycles (initial denaturation at 94 °C/3 min followed by 35 cycles of 94 °C/45 s, 55 °C/60 s and 72 °C/60 s for extension, with a final extension step of 72 °C/10 min). PCR products were purified using the NucleoSpin Gel and PCR Clean-Up Kit (Macherey-Nagel) according to the manufacturer’s instructions. Sanger sequencing was done at Eurofins Genomics Germany using a gene-specific sequencing primer (5′ AGAACGGAGAGGAGAGAAAGAAG 3′).
RNA preparation, sequencing and data analysis
Rachilla tissues from two contrast groups, Morex (short) and Barke (long), and Bowman (long) and BW-NIL-srh1 (short), were used for RNA-seq. The rachilla tissues were collected from the central spikelets of the respective genotypes at rachilla hair initiation (Waddington 8.0) and elongation (Waddington 9.5) stages. Total RNA was extracted using TRIzol reagent (Invitrogen) followed by 2-propanol precipitation. Genomic DNA residues were removed with DNase I (NEB, M0303L). High-throughput paired-end sequencing was conducted at Novogene (Cambridge, UK) with the Illumina NovaSeq 6000 PE150 platform. RNA-seq reads were trimmed for adaptor sequences with Trimmomatic145 (v.0.39) and the MorexV3 genome annotation was used as reference to estimate read abundance with Kallisto128. The raw read counts were normalized to TPM expression levels.
Messenger RNA in situ hybridization
In situ hybridization was conducted in longitudinal sections and cross-sections derived from whole spikelet tissues of Bowman and Morex at rachilla hair elongation developmental stage (Waddington 9.5) with HvSRH1 sense and antisense probes (124 bp). The in situ hybridization was performed as described before146 with few modifications.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Online content
Any methods, additional references, Nature Portfolio reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at 10.1038/s41586-024-08187-1.
Supplementary information
Supplementary Figs. 1–14.
Supplementary Tables 1–27.
Acknowledgements
We thank S. König, I. Walde, S. Sommerfeld, J. Fuller, N. McCallum and M. Macaulay for technical assistance and T. Münch, J. Bauernfeind and H. Miehe for IT administration. A. Börner supported the development of the core1000 diversity panel. Sequencing of Chikurin Ibaraki 1 was performed at the Institute for Clinical Molecular Biology, Competence Centre for Genomic Analysis (CCGA), Kiel University, Kiel, Germany under the supervision of J. Fuß. Sequencing of HOR 4224 was performed at Genomics & Transcriptomics Labor (BMFZ) Heinrich-Heine-Universität, Universitätsklinikum Düsseldorf, Germany under the supervision of K. Köhrer. RGT Planet and Maximus were sequenced at Genomics WA under the supervision of A. Saxena and at Novogene, respectively. N.S., M. Mascher, K.F.X.M., M. Spannagl and U.S. were supported by grants from the German Ministry of Research and Education (grant nos. BMBF, 031B0190 and 031B0884). D.P. was supported by BMBF grant no. 031B0199B and the German Federal Ministry of Food and Agriculture (grant no. 2818BIJP01). U.S.’s research is supported by the German Research Foundation (DFG, grant no. 442032008). C.D., Q.L., P.R.P. and B.S. gratefully acknowledge support from the Carlsberg Foundation to B.S. (grant nos. CF15-0236, CF15-0476, CF15-0672) and thank F. Lok, S. Knudsen, G. B. Fincher and K. G. Jørgensen for providing valuable scientific thoughts and discussions on barley α-amylases and malt quality. R.W., M.B., M. Schreiber, W.G., R.Z. and C.S. received funding from the Rural and Environment Science and Analytical Services Division (RESAS, grant no. KJHI-B1-2). W.G. and R.Z. were supported by grant no. BB/S020160/1 from the Biotechnology and Biological Sciences Research Council (BBSRC). C.J.P. acknowledges support from Genome Canada and Genome Prairie. B.K. acknowledges a grant from the Swiss National Science Foundation (grant no. 310030_204165). C.L., K.J.C. and P.L. were supported by a grant from the Grain Research and Development Corporation (grant no. UMU1806-002RTX), by the Department of Primary Industry and Regional Development Western Australia and by Pawsey Australia (for computational resources). K.P. and T.S. received a grant from DFG (HAPPAN, grant no. 433162815). G.S.B. has received support from the Saskatchewan Ministry of Agriculture (grant no. ADF20200165) and from the Saskatchewan Barley Development Commission, Western Grains Research Foundation, Alberta Barley Commission and the Manitoba Crop Alliance (grant no. ADF20210677). A.B. and W.B. are recipients of the TUGBOAT grant from the Agriculture and Agri-Food Canada–Genomics Research and Development Initiative and the grant ‘Unlocking barley endophyte microbiome to enhance plant health and grain quality’ from Agriculture and Agri-Food Canada–A-Base–Foundational Science. M.H. is supported by the Swedish Research Council (grant no. VR 2022-03858), the Swedish Research Council for Environment, Agricultural Sciences, and Spatial Planning (grant no. FORMAS 2018-01026), the Erik Philip-Sörensen Foundation and the Royal Physiographic Society in Lund. K. Sato’s research is funded by the Japan Science and Technology Agency (grant no. 18076896) and the Japan Society for the Promotion of Science (JSPS, grant no. 23H00333). K. Shirasawa is supported by the JSPS grants nos. 22H05172 and 22H05181. S.S. acknowledges the Research Support Project for the Next Generation at Tottori University. B.J.S. is supported by the Lieberman-Okinow Endowment at the University of Minnesota and S.G.K. by baseline funding at KAUST. The authors acknowledge the Research/Scientific Computing teams at The James Hutton Institute and NIAB for providing computational resources and technical support for the ‘UK’s Crop Diversity Bioinformatics HPC’ (BBSRC grant no. BB/S019669/1), use of which has contributed to the results reported in this paper.
Extended data figures and tables
Author contributions
N.S. and M. Mascher designed the study. N.S. coordinated experiments and sequencing. M. Mascher and M.J. supervised sequence assembly. M. Spannagl and K.F.X.M. supervised annotation. U.S. supervised data management and submission. A.B., W.B., G.S.B., K.J.C., Y.G., M.H., B.K., S.G.K., P.L., C.L., M. Mascher, A.M., G.J.M., D.P., K.P., C.J.P., S.S., K. Sato, T.S., B.J.S., N.S. and R.W. selected genotypes. B.B., A.H., S.I., M.K., C.L., S.P., S.S., K. Sato, T.S., M. Schreiber, K. Shirasawa, N.S., S.W. and X.Z. were responsible for genome sequencing. Sequence assembly was done by B.C., H.H., M.J., G.K.-G., M. Mascher, S.P., K. Sato, T.S. and J.F.T. Transcriptome sequencing and analysis was carried out by W.G., A.H., S.P., C.S., N.S. and R.Z. Annotation was the responsibility of H.G., G.H., N.K., T.L., K.F.X.M. and M. Spannagl. Analysis and interpretation of structural variants was done by M.B., B.C., J.-W.F., Y.G., M.J., C.L., M.P.M., A.M., S.P., H.P., K.P., T.S., M. Schreiber and P.W. Analysis of complex loci was done by B.J., B.K., M.T.R.-W., N.S. and T.W. Analysis of amy1_1 locus was done by K.B., C.D., M.E.J., B.J., M.J., S.M.K., Q.L., M.P.M., E.M., P.A.P., P.R.P., B.S., H.C.T., M.T.S.N., D.V., C.V. and M.W.R. srh1 analysis was done by C.D., M.E.J., I.H., R.E.H., M.J., R.K., J.K., Q.L., M. Melzer, H.P., L.R., P.R.P., T.R., B.S., N.S., H.C.T., C.T., C.V. and M.W.R. M.B., M.F., A.F., M.J., P.K., M. Mascher, D.S. and U.S. were responsible for data management and submission. M.B., C.D., M.E.J., M.J., Q.L., M. Mascher, N.S. and T.W. write the paper. C.D., M. Mascher and N.S. coordinated the project. All authors read and commented on the manuscript.
Peer review
Peer review information
Nature thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Data availability
All the sequence data collected in this study have been deposited at the European Nucleotide Archive147 (ENA) under BioProjects PRJEB40587, PRJEB57567 and PRJEB58554 (raw data for pangenome assemblies), PRJEB64639 (pan-transcriptome Illumina data), PRJEB64637 (transcriptome Iso-Seq data), PRJEB53924 (Illumina resequencing data), PRJEB45512 (raw data for gene space assemblies), PRJEB65284 (srh1 transcriptome data). The SNP and indel variant matrix is available at the European Variation Archive148 (EVA) under accession PRJEB70778. Accession codes for individual genotypes are listed in the Supplementary Tables: Supplementary Table 1 (pangenome assemblies and associated raw data), Supplementary Table 3 (transcriptome data), Supplementary Table 6 (Illumina resequencing), Supplementary Table 7 (gene space assemblies). Sequence assemblies and gene annotations are available from https://galaxy-web.ipk-gatersleben.de/libraries/folders/Fd071e794759ab192 and have been submitted to GrainGenes149. The orthologous framework is accessible under https://panbarlex.ipk-gatersleben.de. The variant matrix is also available for interactive browsing at https://divbrowse.ipk-gatersleben.de/barley_pangenome_v2/. Lists of structural variants and k-mers used in heterozygosity estimation have been deposited in the Plant Genomics & Phenomics Research Data Repository150 under a digital object identifier (DOI): 10.5447/ipk/2024/9.
Code availability
Scripts for pangenome graph analyses are available at https://github.com/mb47/minigraph-barley. The scripts for the definition of core/shell and cloud genes are deposited to the repository https://github.com/PGSB-HMGU/BPGv2. Scripts used for gene projection are available from https://github.com/GeorgHaberer/gene_projection/tree/main/panhordeum. The pipeline for identifying structurally complex loci is available at https://github.com/mtrw/DGS. The pipeline for the construction of the single-copy pangenome is available from https://bitbucket.org/ipk_dg_public/barley_pangenome, and that for heterozygosity estimation from https://bitbucket.org/ipkdg/het_estimation.
Competing interests
K.B., C.D., M.E.J., S.M.K., Q.L., E.M., P.R.P., B.S., H.C.T., M.T.S.N., C.V. and M.W.R. are current or previous Carlsberg A/S employees. P.A.P. and D.V. are SECOBRA Recherches employees. The other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Murukarthick Jayakodi, Qiongxian Lu, Hélène Pidon, M. Timothy Rabanus-Wallace
Contributor Information
Thomas Wicker, Email: wicker@botinst.uzh.ch.
Christoph Dockter, Email: christoph.dockter@carlsberg.com.
Martin Mascher, Email: mascher@ipk-gatersleben.de.
Nils Stein, Email: stein@ipk-gatersleben.de.
Extended data
is available for this paper at 10.1038/s41586-024-08187-1.
Supplementary information
The online version contains supplementary material available at 10.1038/s41586-024-08187-1.
References
- 1.Schreiber, M., Jayakodi, M., Stein, N. & Mascher, M. Plant pangenomes for crop improvement, biodiversity and evolution. Nat. Rev. Genet.25, 563–577 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lei, L. et al. Plant pan-genomics comes of age. Annu. Rev. Plant Biol.72, 411–435 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Komatsuda, T. et al. Six-rowed barley originated from a mutation in a homeodomain-leucine zipper I-class homeobox gene. Proc. Natl Acad. Sci. USA104, 1424–1429 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sakuma, S. et al. Divergence of expression pattern contributed to neofunctionalization of duplicated HD-Zip I transcription factor in barley. New Phytol.197, 939–948 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Milner, S. G. et al. Genebank genomics highlights the diversity of a global barley collection. Nat. Genet.51, 319–326 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Abbo, S. et al. Plant domestication versus crop evolution: a conceptual framework for cereals and grain legumes. Trends Plant Sci.19, 351–360 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Dawson, I. K. et al. Barley: a translational model for adaptation to climate change. New Phytol.206, 913–931 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Lundqvist, U. Scandinavian mutation research in barley—a historical review. Hereditas151, 123–131 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Schulte, D. et al. The international barley sequencing consortium—at the threshold of efficient access to the barley genome. Plant Physiol.149, 142–147 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jayakodi, M. et al. The barley pan-genome reveals the hidden legacy of mutation breeding. Nature588, 284–289 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mascher, M. et al. Long-read sequence assembly: a technical evaluation in barley. Plant Cell10.1093/plcell/koab077 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Russell, J. et al. Exome sequencing of geographically diverse barley landraces and wild relatives gives insights into environmental adaptation. Nat. Genet.48, 1024–1030 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Druka, A. et al. Genetic dissection of barley morphology and development. Plant Physiol.155, 617–627 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wenger, A. M. et al. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat. Biotechnol.37, 1155–1162 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Padmarasu, S., Himmelbach, A., Mascher, M. & Stein, N. In situ Hi-C for plants: an improved method to detect long-range chromatin interactions. Methods Mol. Biol.1933, 441–472 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Simao, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V. & Zdobnov, E. M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics31, 3210–3212 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Lawniczak, M. K. N. et al. Standards recommendations for the Earth BioGenome Project. Proc. Natl Acad. Sci. USA119, e2115639118 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo, Y., Himmelbach, A., Weiss, E., Stein, N. & Mascher, M. Six-rowed wild-growing barleys are hybrids of diverse origins. Plant J.111, 849–858 (2022). [DOI] [PubMed] [Google Scholar]
- 19.Kamm, A. The discovery of wild six-rowed barley and wild Hordeum intermedium in Israel. Ann. R. Agric. Coll. Sweden21, 287–320 (1954). [Google Scholar]
- 20.Maurer, A. et al. Modelling the genetic architecture of flowering time control in barley through nested association mapping. BMC Genomics16, 290 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, H., Feng, X. & Chu, C. The design and construction of reference pangenome graphs with minigraph. Genome Biol.21, 265 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andreace, F., Lechat, P., Dufresne, Y. & Chikhi, R. Comparing methods for constructing and representing human pangenome graphs. Genome Biol.24, 274 (2023). [DOI] [PMC free article] [PubMed]
- 23.De Coster, W., Weissensteiner, M. H. & Sedlazeck, F. J. Towards population-scale long-read sequencing. Nat. Rev. Genet.22, 572–587 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Michelmore, R. W. & Meyers, B. C. Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res.8, 1113–1130 (1998). [DOI] [PubMed] [Google Scholar]
- 25.Wei, F., Wing, R. A. & Wise, R. P. Genome dynamics and evolution of the Mla (powdery mildew) resistance locus in barley. Plant Cell14, 1903–1917 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bettgenhaeuser, J. et al. The barley immune receptor Mla recognizes multiple pathogens and contributes to host range dynamics. Nat. Commun.12, 6915 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seeholzer, S. et al. Diversity at the Mla powdery mildew resistance locus from cultivated barley reveals sites of positive selection. Mol. Plant Microbe Interact.23, 497–509 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Brabham, H. J. et al. Barley MLA3 recognizes the host-specificity effector Pwl2from Magnaporthe oryzae. Plant Cell36, 447–470 (2024). [DOI] [PMC free article] [PubMed]
- 29.Rabanus-Wallace, M. T., Wicker, T. & Stein, N. Replicators, genes, and the C-value enigma: high-quality genome assembly of barley provides direct evidence that self-replicating DNA forms ‘cooperative’ associations with genes in arms races. Preprint at bioRxiv10.1101/2023.10.01.560391 (2023).
- 30.Escudero-Martinez, C. M., Morris, J. A., Hedley, P. E. & Bos, J. I. B. Barley transcriptome analyses upon interaction with different aphid species identify thionins contributing to resistance. Plant Cell Environ.40, 2628–2643 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wicker, T., Yahiaoui, N. & Keller, B. Illegitimate recombination is a major evolutionary mechanism for initiating size variation in plant resistance genes. Plant J.51, 631–641 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Brassac, J. & Blattner, F. R. Species-level phylogeny and polyploid relationships in Hordeum (Poaceae) inferred by next-generation sequencing and in silico cloning of multiple nuclear loci. Syst. Biol.64, 792–808 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doebley, J., Stec, A. & Hubbard, L. The evolution of apical dominance in maize. Nature386, 485–488 (1997). [DOI] [PubMed] [Google Scholar]
- 34.Dixon, L. E. et al. TEOSINTE BRANCHED1 regulates inflorescence architecture and development in bread wheat (Triticum aestivum). Plant Cell30, 563–581 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramsay, L. et al. INTERMEDIUM-C, a modifier of lateral spikelet fertility in barley, is an ortholog of the maize domestication gene TEOSINTE BRANCHED 1. Nat. Genet.43, 169–172 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Lundqvist, U., Abebe, B. & Lundqvist, A. Gene interaction of induced intermedium mutations of two-row barley. Hereditas111, 37–47 (1989). [Google Scholar]
- 37.Youssef, H. M. et al. Natural diversity of inflorescence architecture traces cryptic domestication genes in barley (Hordeum vulgare L.). Genet. Resour. Crop Evol.64, 843–853 (2017). [Google Scholar]
- 38.Monat, C. et al. TRITEX: chromosome-scale sequence assembly of Triticeae genomes with open-source tools. Genome Biol.20, 284 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Janeček, Š., Svensson, B. & MacGregor, E. A. α-Amylase: an enzyme specificity found in various families of glycoside hydrolases. Cell. Mol. Life Sci.71, 1149–1170 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karrer, E. E., Chandler, J. M., Foolad, M. R. & Rodriguez, R. L. Correlation between α-amylase gene expression and seedling vigor in rice. Euphytica66, 163–169 (1992). [Google Scholar]
- 41.Mascher, M. et al. A chromosome conformation capture ordered sequence of the barley genome. Nature544, 427–433 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Kadziola, A., Søgaard, M., Svensson, B. & Haser, R. Molecular structure of a barley α-amylase-inhibitor complex: implications for starch binding and catalysis. J. Mol. Biol.278, 205–217 (1998). [DOI] [PubMed] [Google Scholar]
- 43.Zou, X., Neuman, D. & Shen, Q. J. Interactions of two transcriptional repressors and two transcriptional activators in modulating gibberellin signaling in aleurone cells. Plant Physiol.148, 176–186 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fuller, D. Q. Contrasting patterns in crop domestication and domestication rates: recent archaeobotanical insights from the Old World. Ann. Bot.100, 903–924 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sakuma, S. & Koppolu, R. Form follows function in Triticeae inflorescences. Breed. Sci.73, 46–56 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu, J. K. & Chung, Y. S. Plant variety protection: current practices and insights. Genes (Basel)12, 1127 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Engledow, F. Inheritance in barley: I. The lateral florets and the rachilla. J. Genet.10, 93–108 (1920). [Google Scholar]
- 48.Cockram, J. et al. Genome-wide association mapping to candidate polymorphism resolution in the unsequenced barley genome. Proc. Natl Acad. Sci. USA107, 21611–21616 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beier, S. et al. Construction of a map-based reference genome sequence for barley, Hordeum vulgare L. Sci. Data4, 170044 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kumar, N. et al. Functional conservation in the SIAMESE-RELATED family of cyclin-dependent kinase inhibitors in land plants. Plant Cell27, 3065–3080 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang, K. et al. The CDK inhibitor SIAMESE targets both CDKA;1 and CDKB1 complexes to establish endoreplication in trichomes. Plant Physiol.184, 165–175 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Knudsen, S. et al. FIND-IT: accelerated trait development for a green evolution. Sci. Adv.8, eabq2266 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nomoto, Y. et al. A hierarchical transcriptional network activates specific CDK inhibitors that regulate G2 to control cell size and number in Arabidopsis. Nat. Commun.13, 1660 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liao, W.-W. et al. A draft human pangenome reference. Nature617, 312–324 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brown, T. A. Is the domestication bottleneck a myth? Nat. Plants5, 337–338 (2019). [DOI] [PubMed] [Google Scholar]
- 56.Dvorak, J., McGuire, P. E. & Cassidy, B. Apparent sources of the A genomes of wheats inferred from polymorphism in abundance and restriction fragment length of repeated nucleotide sequences. Genome30, 680–689 (1988). [Google Scholar]
- 57.Himmelbach, A., Walde, I., Mascher, M. & Stein, N. Tethered chromosome conformation capture sequencing in Triticeae: a valuable tool for genome assembly. Bio Protoc.8, e2955 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Himmelbach, A. et al. Discovery of multi‐megabase polymorphic inversions by chromosome conformation capture sequencing in large‐genome plant species. Plant J.96, 1309–1316 (2018). [DOI] [PubMed]
- 59.Cheng, H., Concepcion, G. T., Feng, X., Zhang, H. & Li, H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods18, 170–175 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marone, M. P., Singh, H. C., Pozniak, C. J. & Mascher, M. A technical guide to TRITEX, a computational pipeline for chromosome-scale sequence assembly of plant genomes. Plant Methods18, 128 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rhie, A., Walenz, B. P., Koren, S. & Phillippy, A. M. Merqury: reference-free quality, completeness, and phasing assessment for genome assemblies. Genome Biol.21, 245 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sun, H., Ding, J., Piednoël, M. & Schneeberger, K. findGSE: estimating genome size variation within human and Arabidopsis using k-mer frequencies. Bioinformatics34, 550–557 (2017). [DOI] [PubMed] [Google Scholar]
- 63.Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Steinegger, M. & Söding, J. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat. Biotechnol.35, 1026–1028 (2017). [DOI] [PubMed] [Google Scholar]
- 65.Li, H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics1, 7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nattestad, M. & Schatz, M. C. Assemblytics: a web analytics tool for the detection of variants from an assembly. Bioinformatics32, 3021–3023 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goel, M., Sun, H., Jiao, W.-B. & Schneeberger, K. SyRI: finding genomic rearrangements and local sequence differences from whole-genome assemblies. Genome Biol.20, 277 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J10.14806/ej.17.1.200 (2011).
- 69.Li, H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics27, 2987–2993 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhou, X. & Stephens, M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet.44, 821–824 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li, H. New strategies to improve minimap2 alignment accuracy. Bioinformatics37, 4572–4574 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pedersen, B. S. & Quinlan, A. R. Mosdepth: quick coverage calculation for genomes and exomes. Bioinformatics34, 867–868 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Maurer, A., Sannemann, W., Leon, J. & Pillen, K. Estimating parent-specific QTL effects through cumulating linked identity-by-state SNP effects in multiparental populations. Heredity118, 477–485 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xu, H. et al. FastUniq: a fast de novo duplicates removal tool for paired short reads. PLoS ONE7, e52249 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bushnell, B., Rood, J. & Singer, E. BBMerge—accurate paired shotgun read merging via overlap. PLoS ONE12, e0185056 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li, H. BFC: correcting Illumina sequencing errors. Bioinformatics31, 2885–2887 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chikhi, R. & Rizk, G. Space-efficient and exact de Bruijn graph representation based on a Bloom filter. Algorithms Mol. Biol.8, 22 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Danecek, P. et al. Twelve years of SAMtools and BCFtools. Gigascience10, giab008 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Patterson, N., Price, A. L. & Reich, D. Population structure and eigenanalysis. PLoS Genet.2, e190 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chang, C. C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience4, 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rice, P., Longden, I. & Bleasby, A. EMBOSS: the European molecular biology open software suite. Trends Genet.16, 276–277 (2000). [DOI] [PubMed] [Google Scholar]
- 82.Letunic, I. & Bork, P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res.49, W293–W296 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Waugh, R. et al. A barley pan-transcriptome reveals layers of genotype-dependent transcriptional complexity. Preprint at Research Square10.21203/rs.3.rs-3787876/v1 (2024).
- 84.Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kovaka, S. et al. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol.20, 278 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Consortium, T. U. UniProt: the universal protein knowledgebase in 2023. Nucleic Acids Res.51, D523–D531 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gremme, G., Brendel, V., Sparks, M. E. & Kurtz, S. Engineering a software tool for gene structure prediction in higher organisms. Inf. Softw. Technol.47, 965–978 (2005). [Google Scholar]
- 88.Wu, T. D. & Watanabe, C. K. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics21, 1859–1875 (2005). [DOI] [PubMed] [Google Scholar]
- 89.Ghosh, S. & Chan, C. K. Analysis of RNA-seq data using TopHat and Cufflinks. Methods Mol. Biol.1374, 339–361 (2016). [DOI] [PubMed] [Google Scholar]
- 90.Stanke, M. et al. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res.34, W435–W439 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ter-Hovhannisyan, V., Lomsadze, A., Chernoff, Y. O. & Borodovsky, M. Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training. Genome Res.18, 1979–1990 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hoff, K. J. & Stanke, M. Predicting genes in single genomes with AUGUSTUS. Curr. Protoc. Bioinformatics65, e57 (2019). [DOI] [PubMed] [Google Scholar]
- 93.Haas, B. J. et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol.9, R7 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Haas, B. J. et al. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res.31, 5654–5666 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol.215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 96.Fu, L., Niu, B., Zhu, Z., Wu, S. & Li, W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics28, 3150–3152 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cock, P. J. et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics25, 1422–1423 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Henikoff, S. & Henikoff, J. G. Amino acid substitution matrices from protein blocks. Proc. Natl Acad. Sci. USA89, 10915–10919 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Camacho, C. et al. BLAST+: architecture and applications. BMC Bioinf.10, 421 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Emms, D. M. & Kelly, S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol.20, 238 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lovell, J. T. et al. GENESPACE tracks regions of interest and gene copy number variation across multiple genomes. eLife11, e78526 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Spannagl, M. et al. PGSB PlantsDB: updates to the database framework for comparative plant genome research. Nucleic Acids Res.44, D1141–D1147 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ellinghaus, D., Kurtz, S. & Willhoeft, U. LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinformatics9, 18 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Steinbiss, S., Willhoeft, U., Gremme, G. & Kurtz, S. Fine-grained annotation and classification of de novo predicted LTR retrotransposons. Nucleic Acids Res.37, 7002–7013 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lowe, T. M. & Eddy, S. R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res.25, 955–964 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ma, J. & Bennetzen, J. L. Rapid recent growth and divergence of rice nuclear genomes. Proc. Natl Acad. Sci. USA101, 12404–12410 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Garrison, E. et al. Building pangenome graphs. Nat. Methods10.1038/s41592-024-02430-3 (2024). [DOI] [PubMed]
- 108.Hickey, G. et al. Pangenome graph construction from genome alignments with Minigraph-Cactus. Nat. Biotechnol.10.1038/s41587-023-01793-w (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Garrison, E. et al. Variation graph toolkit improves read mapping by representing genetic variation in the reference. Nat. Biotechnol.36, 875–879 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Guarracino, A., Heumos, S., Nahnsen, S., Prins, P. & Garrison, E. ODGI: understanding pangenome graphs. Bioinformatics38, 3319–3326 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Park, S.-C., Lee, K., Kim, Y. O., Won, S. & Chun, J. Large-scale genomics reveals the genetic characteristics of seven species and importance of phylogenetic distance for estimating pan-genome size. Front. Microbiol.10, 834 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sirén, J. et al. Pangenomics enables genotyping of known structural variants in 5202 diverse genomes. Science374, abg8871 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at https://arxiv.org/abs/1303.3997 (2013).
- 114.Tarasov, A., Vilella, A. J., Cuppen, E., Nijman, I. J. & Prins, P. Sambamba: fast processing of NGS alignment formats. Bioinformatics31, 2032–2034 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hickey, G. et al. Genotyping structural variants in pangenome graphs using the vg toolkit. Genome Biol.21, 35 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Harris, R. S. Improved Pairwise Alignment of Genomic DNA. PhD thesis, Pennsylvania State Univ. (2007).
- 117.Edgar, R. C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics26, 2460–2461 (2010). [DOI] [PubMed] [Google Scholar]
- 118.Buchmann, J. P., Matsumoto, T., Stein, N., Keller, B. & Wicker, T. Inter-species sequence comparison of Brachypodium reveals how transposon activity corrodes genome colinearity. Plant J.71, 550–563 (2012). [DOI] [PubMed] [Google Scholar]
- 119.Kuraku, S., Zmasek, C. M., Nishimura, O. & Katoh, K. aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Res.41, W22–W28 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2022).
- 121.Untergasser, A. et al. Primer3—new capabilities and interfaces. Nucleic Acids Res.40, e115–e115 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Danecek, P. et al. The variant call format and VCFtools. Bioinformatics27, 2156–2158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Marçais, G. & Kingsford, C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics27, 764–770 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Leigh, J. W. & Bryant, D. popart: full-feature software for haplotype network construction. Methods Ecol. Evol.6, 1110–1116 (2015). [Google Scholar]
- 125.Wick, R. R., Schultz, M. B., Zobel, J. & Holt, K. E. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics31, 3350–3352 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rodrigues, C. H. M., Pires, D. E. V. & Ascher, D. B. DynaMut2: assessing changes in stability and flexibility upon single and multiple point missense mutations. Protein Sci.30, 60–69 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Betts, N. S. et al. Isolation of tissues and preservation of RNA from intact, germinated barley grain. Plant J.91, 754–765 (2017). [DOI] [PubMed] [Google Scholar]
- 128.Bray, N. L., Pimentel, H., Melsted, P. & Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol.34, 525–527 (2016). [DOI] [PubMed] [Google Scholar]
- 129.Waddington, S. R., Cartwright, P. M. & Wall, P. C. A quantitative scale of spike initial and pistil development in barley and wheat. Ann. Bot.51, 119–130 (1983). [Google Scholar]
- 130.Poursarebani, N. et al. The genetic basis of composite spike form in barley and ‘Miracle-Wheat’. Genetics201, 155–165 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Taylor, J. & Butler, D. R package ASMap: efficient genetic linkage map construction and diagnosis. J. Stat. Softw.79, 1–29 (2017).30220889 [Google Scholar]
- 132.Wu, Y., Bhat, P. R., Close, T. J. & Lonardi, S. Efficient and accurate construction of genetic linkage maps from the minimum spanning tree of a graph. PLoS Genet.4, e1000212 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Broman, K. W., Wu, H., Sen, Ś. & Churchill, G. A. R/qtl: QTL mapping in experimental crosses. Bioinformatics19, 889–890 (2003). [DOI] [PubMed] [Google Scholar]
- 134.Dempster, A. P., Laird, N. M. & Rubin, D. B. Maximum likelihood from incomplete data via the EM algorithm. J. R. Stat. Soc. Series B Stat. Methodol.39, 1–22 (1977). [Google Scholar]
- 135.Pidon, H. et al. High-resolution mapping of Rym14Hb, a wild relative resistance gene to barley yellow mosaic disease. Theor. Appl. Genet.134, 823–833 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Edgar, R. C. Muscle5: high-accuracy alignment ensembles enable unbiased assessments of sequence homology and phylogeny. Nat. Commun.13, 6968 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chen, Y. & Wang, X. Evaluation of efficiency prediction algorithms and development of ensemble model for CRISPR/Cas9 gRNA selection. Bioinformatics38, 5175–5181 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Gruber, A. R., Lorenz, R., Bernhart, S. H., Neuböck, R. & Hofacker, I. L. The Vienna RNA websuite. Nucleic Acids Res.36, W70–W74 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Koeppel, I., Hertig, C., Hoffie, R. & Kumlehn, J. Cas endonuclease technology—a quantum leap in the advancement of barley and wheat genetic engineering. Int. J. Mol. Sci.20, 2647 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Gerasimova, S. V. et al. Conversion of hulled into naked barley by Cas endonuclease-mediated knockout of the NUD gene. BMC Plant Biol.20, 255 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hensel, G., Kastner, C., Oleszczuk, S., Riechen, J. & Kumlehn, J. Agrobacterium-mediated gene transfer to cereal crop plants: current protocols for barley, wheat, triticale, and maize. Int. J. Plant Genomics2009, 835608 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Watson, A. et al. Speed breeding is a powerful tool to accelerate crop research and breeding. Nat. Plants4, 23–29 (2018). [DOI] [PubMed] [Google Scholar]
- 143.Witherspoon, D. J. et al. Genetic similarities within and between human populations. Genetics176, 351–359 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Van der Loo, M. P. The stringdist package for approximate string matching. R J.6, 111 (2014). [Google Scholar]
- 145.Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Poursarebani, N. et al. COMPOSITUM 1 contributes to the architectural simplification of barley inflorescence via meristem identity signals. Nat. Commun.11, 5138 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Burgin, J. et al. The European Nucleotide Archive in 2022. Nucleic Acids Res.51, D121–D125 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Cezard, T. et al. The European Variation Archive: a FAIR resource of genomic variation for all species. Nucleic Acids Res.50, D1216–D1220 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yao, E. et al. GrainGenes: a data-rich repository for small grains genetics and genomics. Database10.1093/database/baac034 (2022). [DOI] [PMC free article] [PubMed]
- 150.Arend, D. et al. PGP repository: a plant phenomics and genomics data publication infrastructure. Database10.1093/database/baw033 (2016). [DOI] [PMC free article] [PubMed]
- 151.Sallam, A. H. et al. Genome-wide association mapping of stem rust resistance in Hordeum vulgare subsp. spontaneum. G3 (Bethesda)7, 3491–3507 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figs. 1–14.
Supplementary Tables 1–27.
Data Availability Statement
All the sequence data collected in this study have been deposited at the European Nucleotide Archive147 (ENA) under BioProjects PRJEB40587, PRJEB57567 and PRJEB58554 (raw data for pangenome assemblies), PRJEB64639 (pan-transcriptome Illumina data), PRJEB64637 (transcriptome Iso-Seq data), PRJEB53924 (Illumina resequencing data), PRJEB45512 (raw data for gene space assemblies), PRJEB65284 (srh1 transcriptome data). The SNP and indel variant matrix is available at the European Variation Archive148 (EVA) under accession PRJEB70778. Accession codes for individual genotypes are listed in the Supplementary Tables: Supplementary Table 1 (pangenome assemblies and associated raw data), Supplementary Table 3 (transcriptome data), Supplementary Table 6 (Illumina resequencing), Supplementary Table 7 (gene space assemblies). Sequence assemblies and gene annotations are available from https://galaxy-web.ipk-gatersleben.de/libraries/folders/Fd071e794759ab192 and have been submitted to GrainGenes149. The orthologous framework is accessible under https://panbarlex.ipk-gatersleben.de. The variant matrix is also available for interactive browsing at https://divbrowse.ipk-gatersleben.de/barley_pangenome_v2/. Lists of structural variants and k-mers used in heterozygosity estimation have been deposited in the Plant Genomics & Phenomics Research Data Repository150 under a digital object identifier (DOI): 10.5447/ipk/2024/9.
Scripts for pangenome graph analyses are available at https://github.com/mb47/minigraph-barley. The scripts for the definition of core/shell and cloud genes are deposited to the repository https://github.com/PGSB-HMGU/BPGv2. Scripts used for gene projection are available from https://github.com/GeorgHaberer/gene_projection/tree/main/panhordeum. The pipeline for identifying structurally complex loci is available at https://github.com/mtrw/DGS. The pipeline for the construction of the single-copy pangenome is available from https://bitbucket.org/ipk_dg_public/barley_pangenome, and that for heterozygosity estimation from https://bitbucket.org/ipkdg/het_estimation.