Abstract
Background
Numerous observational studies have shown a potential association between metabolic dysfunction-associated steatotic liver disease (MASLD) and gastroesophageal reflux disease (GERD). However, causality is unclear. This study utilized genome-wide association study (GWAS) genetic data to explore the causal relationship between MASLD and GERD in European and East Asian populations.
Methods
This study utilized a bidirectional, two-sample Mendelian randomization (MR) approach. All disease data were obtained from the GWAS database, and single nucleotide polymorphisms strongly associated with exposure were selected as instrumental variables. The inverse variance weighted (IVW) method is primarily utilized to evaluate the causal relationship between exposure and outcome. Finally, sensitivity analyses were performed to ensure the robustness of the results.
Results
The IVW estimates indicated that non-alcoholic fatty liver disease (NAFLD) (odds ratio (OR) = 1.054, 95% confidence interval (CI), 0.966–1.150, p = 0.236) and percent liver fat (OR = 0.977, 95% CI, 0.937–1.018, p = 0.258) in European population were not linked to a higher risk of GERD. However, GERD in European population was associated with an increased risk of NAFLD (OR = 1.485, 95% CI, 1.274–1.729, p < 0.001) and percent liver fat (OR = 1.244, 95% CI, 1.171–1.321, p < 0.001). In addition, the IVW analysis in East Asian population showed that alanine aminotransferase (ALT) was associated with an increased risk of GERD (OR = 2.305, 95% CI, 1.241–4.281, p = 0.008), whereas aspartate aminotransferase (AST) had no causal effects on GERD risk (OR = 0.973, 95% CI, 0.541–1.749, p = 0.926). Furthermore, the associations between GERD and ALT (OR = 1.007, 95% CI, 0.998–1.015, p = 0.123) or AST (OR = 1.004, 95% CI, 0.997–1.012, p = 0.246) were not significant. After removing outliers, a significant correlation between GERD and ALT was observed (OR = 1.009, 95% CI, 1.001–1.016, p = 0.020).
Conclusion
There was reverse causality between MASLD and GERD in European population, while there was bidirectional causality between a proxie for MASLD (ALT) and GERD in East Asian population. This study can provide novel insights into cross-ethnic genetic research on MASLD and GERD.
Keywords: metabolic dysfunction-associated steatotic liver disease, gastroesophageal reflux disease, mendelian randomization, causal effect, genome-wide association studies
1 Introduction
Gastroesophageal reflux disease (GERD) is a prevalent disorder associated with gastrointestinal motility issues and can lead to complications such as reflux esophagitis (RE), esophageal stricture, Barrett’s esophagus, and esophageal adenocarcinoma (Fass, 2022; Mittal and Vaezi, 2020). Estimates suggest that GERD affects approximately 20% of adults in high-income countries (Maret-Ouda et al., 2020). Traditionally, significant risk factors for GERD include tobacco use (Kahrilas, 1992), obesity (Xie et al., 2024), and genetic predisposition (Ghoshal and Chourasia, 2011). However, preventive measures aimed at these risk factors have proven ineffective in controlling the progression of GERD (Sachar et al., 2024). Consequently, it has become essential to investigate new avenues regarding GERD risk factors. Recent epidemiological studies have indicated that metabolic diseases may also serve as risk factors for GERD (Chung et al., 2008; Wu et al., 2011).
Metabolic dysfunction-associated steatotic liver disease (MASLD), previously referred to as non-alcoholic fatty liver disease (NAFLD), is the most prevalent form of chronic liver disease globally, affecting approximately 25% of the population (Younossi et al., 2018). MASLD is a systemic metabolic disorder linked to obesity and insulin resistance (Maldonado-Rojas et al., 2024), characterized by excessive fat accumulation in liver cells (Younossi, 2019). The prognosis for MASLD is poor, as it can lead to liver fibrosis, cirrhosis, and ultimately progress to hepatocellular carcinoma (HCC) (Kanda et al., 2020; Cai et al., 2019). In recent years, numerous studies have explored the association between NAFLD and the risk of GERD. A Korean cohort study indicated a higher risk of reflux esophagitis in individuals with NAFLD compared to those without the condition (Min et al., 2018). However, this association is influenced by body mass index and other metabolic factors. Additionally, a meta-analysis that included nine observational studies and involved 185,118 participants demonstrated that NAFLD is linked to an increased risk of GERD (Xue et al., 2019). Nevertheless, the inherent limitations of traditional observational studies, such as reverse causality and confounding factors, obscure the causal relationship between MASLD and GERD. In addition, evidence underscores the genetic and environmental differences in the incidence and clinical manifestations of GERD. For example, a recent meta-analysis revealed that the prevalence of GERD is higher among Europeans (17.1%) than among Asians (10.0%) (Eusebi et al., 2018). Furthermore, Europeans with GERD may experience more severe heartburn (Spechler et al., 2002) and a higher incidence of Barrett’s esophagus and esophageal cancer compared to their Asian counterparts (Eusebi et al., 2021). Therefore, further investigation is necessary to clarify the discrepancies in the causal relationship between MASLD and GERD across different ethnic groups.
As a novel epidemiological method, Mendelian randomization (MR) leverages the random assignment and independence of genetic variants to determine causal associations between exposures and outcomes, effectively mitigating the influence of confounding factors (Davey Smith and Hemani, 2014). In addition, segregation of genetic variants occurs at conception, prior to the onset of disease, which helps to avoid reverse causation (Sanderson et al., 2022). Consequently, MR serves as a valuable alternative when conducting randomized controlled trials is constrained by ethical considerations and disease-specific characteristics. In this study, we employed a two-sample bidirectional MR analysis to evaluate the potential causal relationship between MASLD and GERD in European and East Asian populations.
2 Materials and methods
2.1 Ethical approval
All datasets used in this study were obtained from publicly available GWAS projects, each of which received the necessary ethical approval and consent. Therefore, no additional ethical approvals were required for our study.
2.2 Study design
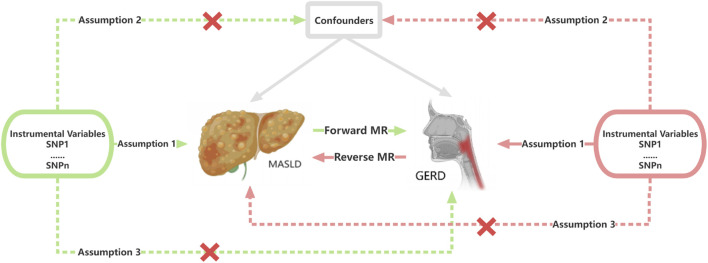
We obtained data on genetic variants related to MASLD proxies and GERD in European and East Asian populations from various GWAS datasets, which we subsequently designated as exposure factors. The specific flow of a two-sample bidirectional MR analysis is depicted in Figure 1. To ensure the reliability of our results, we strictly adhered to the following three assumptions: (1) genetic variants must be strongly associated with the selected exposure factors; (2) genetic variants should not be associated with any confounding factors affecting both the exposure and the outcome; and (3) genetic variants must influence the outcome solely through the exposure pathway (Ference, 2015). This study followed the Strengthening the Reporting of Observational Studies in Epidemiology Using Mendelian Randomization (STROBE-MR) guidelines (Skrivankova et al., 2021).
FIGURE 1.
Flowchart of the bidirectional Mendelian randomization analysis in this study. SNP, single-nucleotide polymorphism; MASLD, Metabolic dysfunction-associated steatotic liver disease; GERD, gastroesophageal reflux disease; MR, Mendelian randomization.
2.3 Data source for European population
MASLD-related indicators include NAFLD and percent liver fat. Genetic associations with NAFLD were derived from the largest GWAS meta-analysis (Ghodsian et al., 2021), which included 8,434 NAFLD cases and 770,180 controls. This analysis utilized data from the Electronic Medical Records and Genomics (eMERGE) network, the United Kingdom Biobank, the Estonian Biobank, and FinnGen. In the eMERGE cohort, NAFLD was defined based on the use of electronic health record (EHR) ICD-9 and ICD-10 codes (ICD-9: 571.5, 571.8, and 571.9; ICD-10: K75.81, K76.0, and K76.9). The United Kingdom Biobank and Estonian Biobank employed ICD-10 codes [K74.0 (fibrosis), K74.2 (fibrosis), K75.8 (non-alcoholic steatohepatitis), K76.0 (NAFLD), and K76.9 (other specified diseases of the liver)]. In the FinnGen Consortium, NAFLD was defined solely by ICD-10 code K76.0. The GWAS data for genetic associations with percent liver fat were obtained from the United Kingdom Biobank, which also included 8,434 NAFLD cases and 770,180 controls (Liu et al., 2021). At the same time, data related to MASLD can be accessed through the Medical Research Council Integrative Epidemiology Unit (IEU) Open GWAS project (https://gwas.mrcieu.ac.uk/).
In addition, the largest publicly available GWAS statistics for GERD were obtained, which included 602,604 European participants from the EBI database (Ong et al., 2022). The diagnostic criteria for GERD are based on the 10th edition of the International Classification of Diseases.
2.4 Data source for East Asian population
Due to the lack of validated MASLD GWAS datasets in East Asian population, we opted for alanine aminotransferase (ALT) and aspartate aminotransferase (AST) levels from the Biobank Japan GWAS datasets as proxies for MASLD, based on previous research (Au Yeung et al., 2023; Zhao et al., 2024). Both enzymes have been established as diagnostic markers for MASLD (Schindhelm et al., 2006; Zou et al., 2020).
The data related to GERD in East Asian population were obtained from the EBI database, which included 948 cases and 177,516 controls (Sakaue et al., 2021).
The detailed information regarding the data sources used in this study is presented in Supplementary Table 1.
2.5 Genetic instruments selection and data harmonization
The IVs were subjected to a rigorous screening process in order to meet the three hypotheses previously outlined. As the number of IVs obtained at the significance threshold (p < 5.0 × 10−8) was insufficient for NAFLD in Europeans and GERD in East Asians, we adjusted the thresholds to p < 5.0 × 10−6 and p < 5.0 × 10−5, respectively, based on previous studies (Qin et al., 2023; Xiao et al., 2022). Subsequently, single nucleotide polymorphisms (SNPs) that were strongly associated with the exposure were clustered by setting the clumping parameter in the ‘TwoSampleMR’ package to TRUE. In addition, a strict r 2 threshold (r 2 < 0.001) was implemented to minimize the issue of multicollinearity caused by linkage disequilibrium and trimmed SNPs within a window size of 10,000 kb (Pritchard and Przeworski, 2001). Previously reported confounders such as smoking (Ge et al., 2024), obesity (Ding et al., 2023), and type 2 diabetes (Younossi et al., 2024) were screened, excluded by searching the PhenoScanner database (http://www.phenoscanner.medschl.cam.ac.uk/). SNPs that were not present in the outcomes, as well as SNPs with palindromic structures and incompatible alleles, were excluded during the process of harmonizing the exposure and outcome datasets. Subsequently, we screened for weak IVs by calculating the F-value, which was defined as F = β 2/SE 2 (Burgess et al., 2011), where β represents the estimated genetic effect on exposure and SE denotes the standard error. SNPs with an F-value calculated to be less than 10 were classified as weak IVs, suggesting insufficient strength to guarantee result precision, and were therefore excluded. Finally, we do not substitute did SNPs with a proxy to maintain the quality of the integrity process.
2.6 Statistical analysis
We employed inverse variance weighting (IVW) as the primary method for our initial analyses, supplemented by MR-Egger, weighted mode, simple mode, and weighted median approaches. Currently, IVW is the most widely utilized statistic in MR analysis. It is characterized by the exclusion of the intercept term during regression calculations, with the regression fitted using the reciprocal of the outcome variance as a weight. This approach serves to minimize the influence of bias from individual SNPs on the overall results (Hemani et al., 2018). Consequently, in the absence of horizontal pleiotropy, IVW provides the most accurate estimates in MR analysis. In contrast, MR-Egger is preferred when horizontal pleiotropy is significant, as it incorporates the intercept in the regression model (Bowden et al., 2016). Subsequently, we conducted a series of sensitivity analyses. Heterogeneity among genetic variants in the IVW estimates was assessed using Cochran’s Q statistic (Bowden et al., 2019). When the Q test (p < 0.05) indicated heterogeneity of results, random effects models were applied. Additionally, the outlier test of MR-Pleiotropy Residual Sum and Outlier (MR-PRESSO) was employed to identify and exclude outliers (p < 0.05 in the outlier test; NbDistribution = 10,000), and MR analysis were was repeated with adjusted analysis of the data (Patrick et al., 2021). If significant heterogeneity in Cochran’s Q test could not be eliminated after excluding outliers, the outlier test was performed again with a more stringent p-value threshold (p < 1 in the outlier test) (Chen et al., 2020). The MR-Egger intercept (Bowden et al., 2015) and the global test of MR-PRESSO (Verbanck et al., 2018) were utilized to identify and interpret potential horizontal pleiotropy. The presence of horizontal pleiotropy was demonstrated when the p-value for each of the two tests was less than 0.05. Finally, a leave-one-out analysis was applied to evaluate the impact of each SNP on the overall MR results. All statistical analyses were conducted using the ‘TwoSampleMR’ package (version 0.5.8) and the ‘MR-PRESSO’ package (version 1.0) in R software (version 4.2.3). The effect estimates of MR analysis were expressed as odds ratios (ORs) with 95% confidence intervals (CIs), and significant heterogeneity was defined as p < 0.05.
3 Results
3.1 Causal associations between MASLD and GERD in European population
Screening for MASLD and GERD-related indicators in European population, including NAFLD, percent liver fat, and GERD, was conducted following the screening process for genetic markers as outlined above. The F-statistics of all SNPs screened were greater than 10, indicating the absence of weak instrumental bias. A comprehensive list of all SNPs is provided in Supplementary Tables 2-5.
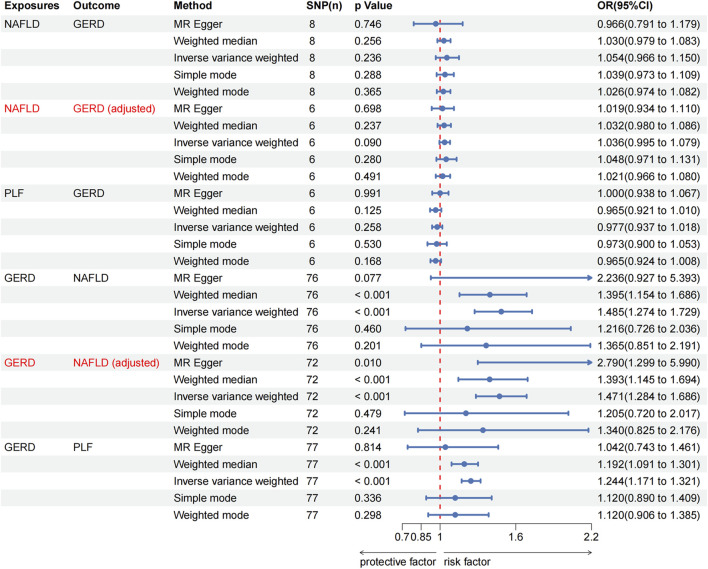
No significant causal association was observed between NAFLD and GERD according to IVW estimates (OR = 1.054, 95% CI, 0.966–1.150, p = 0.236). Moreover, no causal effect of percent liver fat on GERD was found (IVW: OR = 0.977, 95% CI, 0.937–1.018, p = 0.258). These findings were consistently supported by other supplementary methods. However, genetically predicted GERD was associated with an increased risk of NAFLD (IVW: OR = 1.485, 95% CI, 1.274–1.729, p < 0.001) and percent liver fat (IVW: OR = 1.244, 95% CI, 1.171–1.321, p < 0.001), and consistent results were obtained with weighted median (GERD on NAFLD: OR = 1.395, 95% CI, 1.154–1.686, p < 0.001; GERD on percent liver fat: OR = 1.192, 95% CI, 1.091–1.301, p < 0.001). All MR analysis results in European population were presented in Figures 2, 3A–F.
FIGURE 2.
The Mendelian randomization analysis results of MASLD with GERD in European population. MASLD, Metabolic dysfunction-associated steatotic liver disease; GERD, gastroesophageal reflux disease; NAFLD, non-alcoholic fatty liver disease; PLF, percent liver fat; OR, odds ratio.
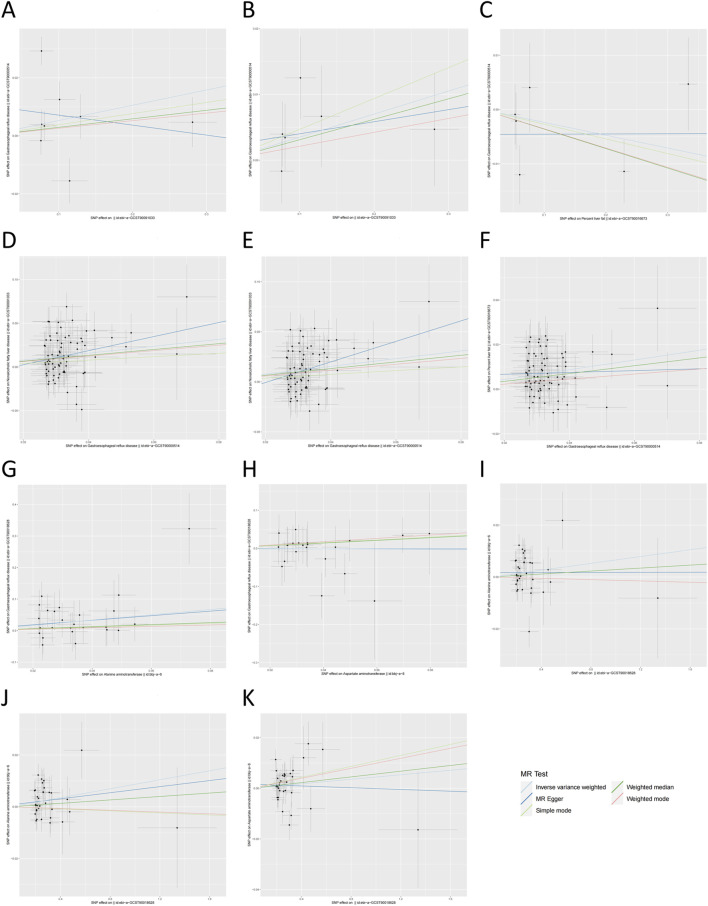
FIGURE 3.
The scatter plot of Mendelian randomization analysis. (A) NAFLD on GERD; (B) NAFLD on GERD (adjusted); (C) percent liver fat on GERD; (D) GERD on NAFLD; (E) GERD on NAFLD (adjusted); (F) GERD on percent liver fat; (G) ALT on GERD; (H) AST on GERD; (I) GERD on ALT; (J) GERD on ALT (adjusted); (K) GERD on AST. NAFLD, non-alcoholic fatty liver disease; GERD, gastroesophageal reflux disease; ALT, alanine aminotransferase; AST, aspartate aminotransferase.
The results of the sensitivity analysis in European population are summarized in Table 1. The Cochran’s Q statistic using the IVW method revealed no observed heterogeneity, except for the MR analysis between NAFLD and GERD (NAFLD on GERD, p < 0.001; GERD on NAFLD, p = 0.002). Subsequently, we conducted the MR-PRESSO outlier test to address bias resulting from significant heterogeneity and then repeated the MR analyses (p < 0.05 in the outlier test). Specifically, heterogeneity disappeared after removing the outliers (rs7144175 and rs9922619) in the forward analysis (p IVW = 0.645). In addition, since no outliers were detected in the reverse analysis, we no longer observed heterogeneity (p IVW = 0.156) after excluding outliers with p-values less than 1 (rs2023878, rs569356, rs7685686, and rs9940128). Importantly, the exclusion of outliers did not change the results of the preliminary MR analyses between NAFLD and GERD [NAFLD on GERD, p = 0.090; GERD on NAFLD, p < 0.001; Figures 2, 3B,E] and was corrected for horizontal pleiotropy in the global test (NAFLD on GERD, p = 0.690; GERD on NAFLD, p = 0.170). Significant pleiotropy was not detected in European population, as indicated by the Egger intercept (all p > 0.05). Furthermore, when the leave-one-out method was applied to remove any single SNP, none of the IVW effect values were affected (Supplementary Figure 1A-F). The forest plot and funnel plot are shown in Supplementary Figures 2A-F, 3A-F.
TABLE 1.
Results of sensitivity analysis in Mendelian randomization analysis.
| Exposure | Outcome | Ancestor | Method | nSNP | Cochran’s Q test | Intercept term | Global test | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Q | Q_df | p | Intercept | SE | p | RSSobs | p | |||||
| NAFLD | GERD | European | MR Egger | 8 | 33.706 | 6 | 7.67E-06 | 0.011 | 0.011 | 0.377 | ||
| IVW | 8 | 38.817 | 7 | 2.12E-06 | ||||||||
| MR-PRESSO | 8 | 48.486 | <0.001 | |||||||||
| NAFLD | GERD (adjusted) | European | MR Egger | 6 | 3.173 | 4 | 0.529 | 0.002 | 0.005 | 0.689 | ||
| IVW | 6 | 3.358 | 5 | 0.645 | ||||||||
| MR-PRESSO | 6 | 4.992 | 0.690 | |||||||||
| Percent liver fat | GERD | European | MR Egger | 6 | 6.174 | 4 | 0.186 | −0.005 | 0.005 | 0.399 | ||
| IVW | 6 | 7.547 | 5 | 0.183 | ||||||||
| MR-PRESSO | 6 | 14.770 | 0.337 | |||||||||
| GERD | NAFLD | European | MR Egger | 76 | 114.444 | 74 | 0.002 | −0.014 | 0.015 | 0.357 | ||
| IVW | 76 | 115.772 | 75 | 0.002 | ||||||||
| MR-PRESSO | 76 | 118.842 | 0.001 | |||||||||
| GERD | NAFLD (adjusted) | European | MR Egger | 72 | 79.812 | 70 | 0.198 | −0.021 | 0.013 | 0.100 | ||
| IVW | 72 | 82.980 | 71 | 0.156 | ||||||||
| MR-PRESSO | 72 | 85.289 | 0.170 | |||||||||
| GERD | Percent liver fat | European | MR Egger | 38 | 81.940 | 75 | 0.273 | 0.006 | 0.006 | 0.299 | ||
| IVW | 38 | 83.132 | 76 | 0.269 | ||||||||
| MR-PRESSO | 38 | 85.376 | 0.315 | |||||||||
| ALT | GERD | East Asian | MR Egger | 25 | 21.554 | 23 | 0.547 | 0.004 | 0.034 | 0.909 | ||
| IVW | 25 | 21.568 | 24 | 0.605 | ||||||||
| MR-PRESSO | 25 | 23.326 | 0.614 | |||||||||
| AST | GERD | East Asian | MR Egger | 22 | 11.327 | 20 | 0.937 | −0.001 | 0.033 | 0.977 | ||
| IVW | 22 | 11.328 | 21 | 0.956 | ||||||||
| MR-PRESSO | 22 | 12.578 | 0.953 | |||||||||
| GERD | ALT | East Asian | MR Egger | 34 | 54.175 | 29 | 0.008 | 0.002 | 0.004 | 0.674 | ||
| IVW | 34 | 54.480 | 30 | 0.011 | ||||||||
| MR-PRESSO | 34 | 57.828 | 0.013 | |||||||||
| GERD | ALT (adjusted) | East Asian | MR Egger | 33 | 39.567 | 31 | 0.139 | 0.001 | 0.003 | 0.834 | ||
| IVW | 33 | 39.624 | 32 | 0.166 | ||||||||
| MR-PRESSO | 33 | 42.202 | 0.187 | |||||||||
| GERD | AST | East Asian | MR Egger | 34 | 45.051 | 32 | 0.063 | 0.002 | 0.004 | 0.656 | ||
| IVW | 34 | 45.335 | 33 | 0.075 | ||||||||
| MR-PRESSO | 34 | 48.131 | 0.056 | |||||||||
SNPs, single nucleotide polymorphisms; SE, standard error; p, p-value; IVW, inverse variance weighted; PRESSO, Pleiotropy RESidual Sum and Outlier; NAFLD, non-alcoholic fatty liver disease; GERD, gastroesophageal reflux disease; ALT, alanine aminotransferase; AST, aspartate aminotransferase.
3.2 Causal associations between MASLD and GERD in East Asian population
We effectively screened for genetic variants associated with MASLD and GERD, including ALT, AST, and GERD in East Asian population through the process described above. All F-statistics exceeded the threshold of 10, indicating the absence of weak instrumental bias. The detailed information for all SNPs is available in Supplementary Tables 6-9.
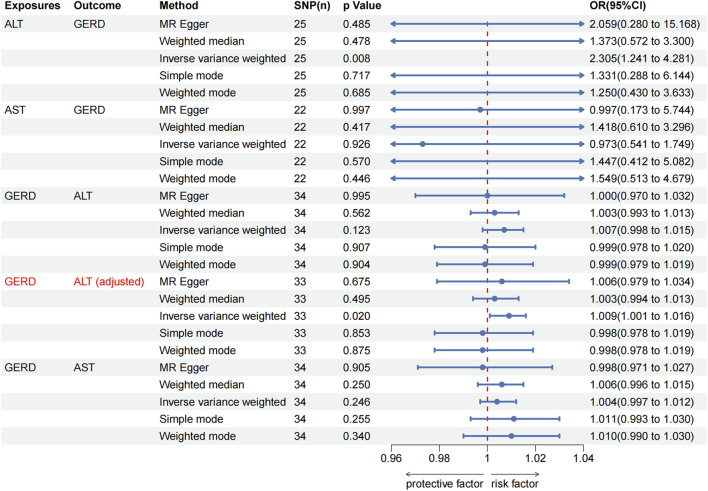
As shown in Figure 4, the IVW analysis indicated that genetically predicted ALT was positively associated with GERD (OR = 2.305, 95% CI, 1.241–4.281, p = 0.008) in East Asian population. Surprisingly, the IVW analysis indicated no evidence of a causal link between AST and the risk of GERD (OR = 0.973, 95% CI, 0.541–1.749, p = 0.926) in East Asian population. Furthermore, no evidence of a causal link between GERD and the risk of ALT (IVW: OR = 1.007, 95% CI, 0.998–1.015, p = 0.123) and AST (IVW: OR = 1.004, 95% CI, 0.997–1.012, p = 0.246) in East Asian population was observed during this study. These findings were consistently supported by other supplementary methods. Detailed information on all MR analysis in East Asian population is presented in Figures 3G–K.
FIGURE 4.
The Mendelian randomization analysis results of MASLD with GERD in East Asian population. MASLD, Metabolic dysfunction-associated steatotic liver disease; GERD, gastroesophageal reflux disease; ALT, alanine aminotransferase; AST, aspartate aminotransferase; OR, odds ratio.
We conducted a sensitivity analysis to assess the robustness of the MR results in East Asian population (Table 1). The Cochran’s Q test revealed the presence of heterogeneity in the causal relationship between GERD and ALT in East Asian population (p IVW = 0.011). Subsequently, we identified outliers through the MR-PRESSO outlier test (rs6022197). Heterogeneity was no longer detected when outliers were eliminated (p IVW = 0.166), and horizontal pleiotropy in the global test was also eliminated (p = 0.187). Interestingly, following the adjustment, GERD was associated with high levels of ALT (IVW: OR = 1.009, 95% CI, 1.001–1.016, p = 0.020; Figures 4, 3J). Conversely, Cochran’s Q statistic showed no significant heterogeneity in the remaining MR analyses in East Asian population (all p > 0.05). In addition, the MR-Egger intercept did not provide evidence of horizontal pleiotropy in any of the MR analyses (all p > 0.05). Finally, the robustness of the MR results is further emphasized by the leave-one-out sensitivity analysis (Supplementary Figure 1G-K). The forest plot and funnel plot are shown in Supplementary Figures 2G-K, 3G-K.
4 Discussion
This study is the first to establish a causal relationship between MASLD and GERD through large-scale MR analysis. Specifically, for European population, our results indicated that factors related to MASLD had no effect on the risk of developing GERD. However, GERD was associated with an increased risk of MASLD. Furthermore, for East Asian population, ALT, a surrogate marker for MASLD, was positively associated with GERD; however, there was no evidence of a causal relationship between GERD and altered ALT and AST levels. In addition, sensitivity analyses demonstrate the robustness of our findings.
MASLD has been characterized as a systemic metabolic disease closely associated with metabolic disorders and insulin resistance (Al Hashmi et al., 2024). In 2020, a panel of international experts from 22 countries renamed NAFLD to MASLD through a Delphi process, as NAFLD does not accurately reflect the etiology of the disease. The new definition incorporates cardiometabolic criteria and underscores the importance of obesity, diabetes, and Metabolic Syndrome (MetS) in the development of MASLD (Eslam et al., 2020). Numerous observational studies have confirmed that MASLD is a risk factor for GERD. A meta-analysis conducted by Xue et al., which included 185,118 subjects, demonstrated a significant association between NAFLD and an increased risk of GERD, although the findings may have been influenced by severe publication bias (Xue et al., 2019). Furthermore, a large cohort study of Korean adults revealed a higher incidence of reflux esophagitis among participants with NAFLD, even after adjusting for age and sex. Notably, the correlation between the two conditions diminished when the researchers further adjusted for confounding factors, including smoking status, alcohol consumption, regular exercise, education level, and body mass index (BMI) (Yang et al., 2017). Additionally, Qiu et al. found that NAFLD significantly elevated the risk of RE by strictly controlling for the subjects’ BMI. Interestingly, the study also indicated a higher prevalence of NAFLD in patients with RE compared to those without RE, and this difference was statistically significant (Qiu et al., 2023).
It is noteworthy that a significant number of studies have focused on East Asian population. Our research indicates a positive correlation between the proxy for MASLD (ALT) and GERD in this demographic. Unfortunately, due to the unavailability of a MASLD dataset specifically for East Asian populations, we were constrained to seek proxy data that exhibited a high degree of correlation with MASLD for the purpose of our analysis. It is important to acknowledge that this practice is still limited by several potential risks. First, transaminases do not fully capture the complex pathomechanisms of MASLD, which include alterations such as dysregulation of fatty acid metabolism and impaired bile acid cycling (Feng et al., 2024). Additionally, the proxy status of transaminases may be more relevant to individuals with metabolic abnormalities, potentially leading to bias in our conclusions (Clark et al., 2003). Furthermore, it cannot be ruled out that patients with MASLD may present with normal aminotransferase levels, raising concerns about the potential overestimation of aminotransferases as proxy indicators. Nevertheless, there is substantial evidence supporting the feasibility of using transaminases as proxies for MASLD. A significant body of research demonstrated a strong correlation between transaminase levels and MASLD risk factors, including obesity, dyslipidemia, and elevated insulin levels (Lin et al., 2022; Ma et al., 2019). Additionally, a large-scale epidemiological study indicated that ALT and AST could serve as accurate predictors of MASLD prevalence when common chronic liver disease conditions are excluded (Clark et al., 2003). However, the development of a comprehensive MASLD dataset specifically for East Asian populations will be essential for enhancing our understanding of the current findings and for improving future research.
In clinical practice, the risk factors for GERD are often numerous and can be modified (Sadafi et al., 2024). However, the systemic metabolic characteristics of MASLD have obscured the potential mechanisms by which it affects a single system. Currently, advances in the study of new biological mechanisms may provide insights into the link between MASLD and GERD in East Asian populations. First, MASLD leads to the activation of pro-inflammatory mediators, including IL-1β, IL-6, and hydrogen peroxide, which in turn triggers neurogenic esophageal muscle contractions, resulting in frequent dilation of the esophagus to the point of relaxation (Cao et al., 2004). Secondly, leptin may serve as another link connecting MASLD to GERD. MASLD is associated with elevated leptin levels (Jiménez-Cortegana et al., 2021), and increased leptin can disrupt the acidic environment of the esophageal lumen, leading to the initiation and progression of esophageal mucosal damage (Pardak et al., 2021). Finally, MASLD activates systemic oxidative stress levels (Bae et al., 2023), which diminishes the esophagus’s ability to repair damage to its mucosa (Han and Zhang, 2022).
It is important to note that studies elucidating the causal relationship between MASLD and GERD are not confined to East Asia. Observational studies conducted in Europe have also documented this association. A cross-sectional study by an Italian internal medicine team thoroughly analyzed the prevalence and clinical characteristics of GERD symptoms in individuals with NAFLD. The findings indicated that the prevalence of gastroesophageal reflux symptoms was significantly higher in the NAFLD group compared to the non-NAFLD group (Miele et al., 2012). Furthermore, another cross-sectional study by Catanzaro et al. reported that the prevalence of GERD symptoms was significantly greater in NAFLD patients than in the control group (Catanzaro et al., 2014). However, these findings contradict those observed in European population in this study, where no significant association was found between MASLD and GERD risk. Interestingly, reverse MR analysis suggested a causal relationship between GERD and MASLD. Potential factors contributing to this discrepancy may include several scenarios. Firstly, observational studies, particularly cross-sectional studies, can only demonstrate associations. Their ability to investigate causality is limited, and they cannot completely eliminate subtle biases introduced by confounding factors, even if some covariates are adjusted for in the study. Second, the association between the two diseases in European population observed in the study may be due to reverse causation, as suggested by the results of our MR. The potential relationship between GERD and MASLD in European population should be further validated in future research, including an examination of clinical features and biological mechanisms. Third, the association between MASLD and GERD in European populations may be mediated by specific metabolic pathways. Evidence suggests that MASLD is strongly linked to insulin resistance (IR) within these populations. In particular, hepatic steatosis can impair insulin action in the liver (Barber et al., 2023). Additionally, a pilot study involving European and African American obese women indicated that insulin resistance contributes to both the prevalence and severity of GERD in this demographic (Pointer et al., 2016). However, the precise mechanisms through which IR serves as an intermediary in this relationship remain unclear.
It is noteworthy that our study found different results between European and East Asian populations. In particular, we observed a unidirectional association between MASLD and GERD in European population. In contrast, the East Asian population showed bidirectional causality, and this discrepancy requires further discussion. A potential factor contributing to these inconsistent results could be the use of MASLD proxies among East Asians. Although we demonstrated the reliability of ALT and AST as proxies, the results should still be interpreted with caution. On the other hand, genetic differences between races can also play an important role. Several studies have shown that Caucasians with a genetic predisposition to MASLD predominantly express the PNPLA3, GCKR, and PPP1R3B genes (Oddy et al., 2013), while East Asians are characterized primarily by polymorphisms in the APOC3 gene (Hsu and Kao, 2012). Regarding metabolic predisposition, the metabolic mechanisms of MASLD in Caucasians are primarily influenced by IR, whereas in East Asians, they are more closely associated with BMI levels (Al Rifai et al., 2015). In addition, variations in dietary habits, including the amount of alcohol consumed, high-fat diets, and coffee intake, may also play a significant role (Fan and Cao, 2013; Festi et al., 2009). It is therefore imperative that further investigation be conducted into the underlying causes and mechanisms of these racial disparities.
Our study has several strengths that merit discussion. First, we employed a rigorous design concept of MR analysis, which enhances the authority and credibility of our exploration into the causal association between MASLD and GERD. Second, our study utilized bidirectional MR analysis, allowing for a comprehensive examination of the directionality of the causal relationship between these diseases and assessing the potential for reverse causation. Third, the data sources for all diseases included multiple traits rather than a single trait, enriching the breadth of our findings. Finally, this study investigated differences in causal associations across racial groups, aiming to address the oversight of racial disparities in disease prevalence and outcomes.
However, it is important to acknowledge some inherent limitations in our study. First, due to the lack of MASLD datasets in East Asian population, we substituted MASLD with liver enzyme levels. Although ALT and AST are well-established markers of MASLD, this substitution was based on similar previous studies. Furthermore, liver enzymes still lack absolute specificity for MASLD; therefore, caution must be exercised when interpreting the results. Second, publicly available GWAS meta-analyses lacked specific information on age and gender, which hindered our ability to stratify the results using appropriate subgroup analyses. Finally, MR analyses can only provide linear assessments of associations between diseases and do not elucidate the specific molecular mechanisms underlying the association between MASLD and GERD.
In summary, our study provided novel clinical insights into the cross-racial association between GERD and MASLD. Based on these findings, clinicians should pay closer attention to the management of GERD when evaluating European patients to prevent and mitigate the progression of MASLD. Furthermore, for the clinical management of East Asian patients, we advocate for a more integrated treatment approach that considers both GERD and MASLD. Meanwhile, the specific mechanisms underlying this bidirectional relationship warrant further investigation to optimize treatment strategies for East Asian patients.
5 Conclusion
In conclusion, our evidence supports a reverse causality relationship between MASLD and GERD in European population. However, our study provides novel evidence suggesting a bidirectional causality between a proxie for MASLD (ALT) and GERD in East Asian population. Future research should include larger prospective studies and basic research to further elucidate the underlying mechanisms of the association between MASLD and GERD.
Acknowledgments
We thank the above databases for making their data publicly available.
Funding Statement
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was supported by the Hebei Provincial Key Research and Development Program Project (21377767D).
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics statement
Ethical approval was not required for the study involving humans in accordance with the local legislation and institutional requirements. Written informed consent to participate in this study was not required from the participants or the participants’; legal guardians/next of kin in accordance with the national legislation and the institutional requirements.
Author contributions
CS: Conceptualization, Data curation, Methodology, Writing–original draft. ZL: Investigation, Software, Supervision, Writing–review and editing. HL: Formal Analysis, Methodology, Project administration, Writing–review and editing. YP: Data curation, Formal Analysis, Methodology, Resources, Writing–review and editing. ZW: Resources, Validation, Writing–review and editing. JaL: Resources, Visualization, Writing–review and editing. JnL: Funding acquisition, Writing–review and editing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2024.1428334/full#supplementary-material.
Leave-one-out sensitivity analysis of Mendelian randomization analysis. (A) NAFLD on GERD; (B) NAFLD on GERD (adjusted); (C) percent liver fat on GERD; (D) GERD on NAFLD; (E) GERD on NAFLD (adjusted); (F) GERD on percent liver fat; (G) ALT on GERD; (H) AST on GERD; (I) GERD on ALT; (J) GERD on ALT (adjusted); (K) GERD on AST. NAFLD, non-alcoholic fatty liver disease; GERD, gastroesophageal reflux disease; ALT, alanine aminotransferase; AST, aspartate aminotransferase.
The forest plot of Mendelian randomization analysis. (A) NAFLD on GERD; (B) NAFLD on GERD (adjusted); (C) percent liver fat on GERD; (D) GERD on NAFLD; (E) GERD on NAFLD (adjusted); (F) GERD on percent liver fat; (G) ALT on GERD; (H) AST on GERD; (I) GERD on ALT; (J) GERD on ALT (adjusted); (K) GERD on AST. NAFLD, non-alcoholic fatty liver disease; GERD, gastroesophageal reflux disease; ALT, alanine aminotransferase; AST, aspartate aminotransferase.
The funnel plot of Mendelian randomization analysis. (A) NAFLD on GERD; (B) NAFLD on GERD (adjusted); (C) percent liver fat on GERD; (D) GERD on NAFLD; (E) GERD on NAFLD (adjusted); (F) GERD on percent liver fat; (G) ALT on GERD; (H) AST on GERD; (I) GERD on ALT; (J) GERD on ALT (adjusted); (K) GERD on AST. NAFLD, non-alcoholic fatty liver disease; GERD, gastroesophageal reflux disease; ALT, alanine aminotransferase; AST, aspartate aminotransferase.
References
- Al Hashmi K., Giglio R. V., Pantea Stoian A., Patti A. M., Al Waili K., Al Rasadi K., et al. (2024). Metabolic dysfunction-associated fatty liver disease: current therapeutic strategies. Front. Nutr. 11, 1355732. 10.3389/fnut.2024.1355732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Rifai M., Silverman M. G., Nasir K., Budoff M. J., Blankstein R., Szklo M., et al. (2015). The association of nonalcoholic fatty liver disease, obesity, and metabolic syndrome, with systemic inflammation and subclinical atherosclerosis: the Multi-Ethnic Study of Atherosclerosis (MESA). Atherosclerosis 239 (2), 629–633. 10.1016/j.atherosclerosis.2015.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au Yeung S. L., Borges M. C., Wong T. H. T., Lawlor D. A., Schooling C. M. (2023). Evaluating the role of non-alcoholic fatty liver disease in cardiovascular diseases and type 2 diabetes: a Mendelian randomization study in Europeans and East Asians. Int. J. Epidemiol. 52 (3), 921–931. 10.1093/ije/dyac212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae S. J., Lee W. Y., Bak S. B., Kim Y. E., Kim M. J., Kim Y. W. (2023). Unraveling the antioxidant capacity of spatholobi caulis in nonalcoholic fatty liver disease: a multiscale network approach integrated with experimental validation. Antioxidants (Basel) 12 (5), 1097. 10.3390/antiox12051097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber T. M., Kabisch S., Pfeiffer A. F. H., Weickert M. O. (2023). Metabolic-associated fatty liver disease and insulin resistance: a review of complex interlinks. Metabolites 13 (6), 757. 10.3390/metabo13060757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden J., Davey Smith G., Burgess S. (2015). Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 44 (2), 512–525. 10.1093/ije/dyv080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden J., Del Greco M. F., Minelli C., Davey Smith G., Sheehan N. A., Thompson J. R. (2016). Assessing the suitability of summary data for two-sample Mendelian randomization analyses using MR-Egger regression: the role of the I2 statistic. Int. J. Epidemiol. 45 (6), 1961–1974. 10.1093/ije/dyw220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden J., Del Greco M. F., Minelli C., Zhao Q., Lawlor D. A., Sheehan N. A., et al. (2019). Improving the accuracy of two-sample summary-data Mendelian randomization: moving beyond the NOME assumption. Int. J. Epidemiol. 48 (3), 728–742. 10.1093/ije/dyy258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S., Thompson S. G. CRP CHD Genetics Collaboration (2011). Avoiding bias from weak instruments in Mendelian randomization studies. Int. J. Epidemiol. 40 (3), 755–764. 10.1093/ije/dyr036 [DOI] [PubMed] [Google Scholar]
- Cai J., Zhang X. J., Li H. (2019). Progress and challenges in the prevention and control of nonalcoholic fatty liver disease. Med. Res. Rev. 39 (1), 328–348. 10.1002/med.21515 [DOI] [PubMed] [Google Scholar]
- Cao W., Cheng L., Behar J., Fiocchi C., Biancani P., Harnett K. M. (2004). Proinflammatory cytokines alter/reduce esophageal circular muscle contraction in experimental cat esophagitis. Am. J. Physiol. Gastrointest. Liver Physiol. 287 (6), G1131–G1139. 10.1152/ajpgi.00216.2004 [DOI] [PubMed] [Google Scholar]
- Catanzaro R., Calabrese F., Occhipinti S., Anzalone M. G., Italia A., Milazzo M., et al. (2014). Nonalcoholic fatty liver disease increases risk for gastroesophageal reflux symptoms. Dig. Dis. Sci. 59 (8), 1939–1945. 10.1007/s10620-014-3113-7 [DOI] [PubMed] [Google Scholar]
- Chen X., Kong J., Diao X., Cai J., Zheng J., Xie W., et al. (2020). Depression and prostate cancer risk: a Mendelian randomization study. Cancer Med. 9 (23), 9160–9167. 10.1002/cam4.3493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S. J., Kim D., Park M. J., Kim Y. S., Kim J. S., Jung H. C., et al. (2008). Metabolic syndrome and visceral obesity as risk factors for reflux oesophagitis: a cross-sectional case-control study of 7078 Koreans undergoing health check-ups. Gut 57 (10), 1360–1365. 10.1136/gut.2007.147090 [DOI] [PubMed] [Google Scholar]
- Clark J. M., Brancati F. L., Diehl A. M. (2003). The prevalence and etiology of elevated aminotransferase levels in the United States. Am. J. Gastroenterol. 98 (5), 960–967. 10.1111/j.1572-0241.2003.07486.x [DOI] [PubMed] [Google Scholar]
- Davey Smith G., Hemani G. (2014). Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum. Mol. Genet. 23 (R1), R89–R98. 10.1093/hmg/ddu328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Deng Q., Yang M., Niu H., Wang Z., Xia S. (2023). Clinical classification of obesity and implications for metabolic dysfunction-associated fatty liver disease and treatment. Diabetes Metab. Syndr. Obes. 16, 3303–3329. 10.2147/dmso.S431251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eslam M., Newsome P. N., Sarin S. K., Anstee Q. M., Targher G., Romero-Gomez M., et al. (2020). A new definition for metabolic dysfunction-associated fatty liver disease: an international expert consensus statement. J. Hepatol. 73 (1), 202–209. 10.1016/j.jhep.2020.03.039 [DOI] [PubMed] [Google Scholar]
- Eusebi L. H., Cirota G. G., Zagari R. M., Ford A. C. (2021). Global prevalence of Barrett's oesophagus and oesophageal cancer in individuals with gastro-oesophageal reflux: a systematic review and meta-analysis. Gut 70 (3), 456–463. 10.1136/gutjnl-2020-321365 [DOI] [PubMed] [Google Scholar]
- Eusebi L. H., Ratnakumaran R., Yuan Y., Solaymani-Dodaran M., Bazzoli F., Ford A. C. (2018). Global prevalence of, and risk factors for, gastro-oesophageal reflux symptoms: a meta-analysis. Gut 67 (3), 430–440. 10.1136/gutjnl-2016-313589 [DOI] [PubMed] [Google Scholar]
- Fan J. G., Cao H. X. (2013). Role of diet and nutritional management in non-alcoholic fatty liver disease. J. Gastroenterol. Hepatol. 28 (Suppl. 4), 81–87. 10.1111/jgh.12244 [DOI] [PubMed] [Google Scholar]
- Fass R. (2022). Gastroesophageal reflux disease. N. Engl. J. Med. 387 (13), 1207–1216. 10.1056/NEJMcp2114026 [DOI] [PubMed] [Google Scholar]
- Feng X., Zhang R., Yang Z., Zhang K., Xing J. (2024). Mechanism of metabolic dysfunction-associated steatotic liver disease: important role of lipid metabolism. J. Clin. Transl. Hepatol. 12 (9), 815–826. 10.14218/jcth.2024.00019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ference B. A. (2015). Mendelian randomization studies: using naturally randomized genetic data to fill evidence gaps. Curr. Opin. Lipidol. 26 (6), 566–571. 10.1097/mol.0000000000000247 [DOI] [PubMed] [Google Scholar]
- Festi D., Scaioli E., Baldi F., Vestito A., Pasqui F., Di Biase A. R., et al. (2009). Body weight, lifestyle, dietary habits and gastroesophageal reflux disease. World J. Gastroenterol. 15 (14), 1690–1701. 10.3748/wjg.15.1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X., Lu J., Yu C., Guo W., Tian T., Xu X., et al. (2024). Associations between active, passive smoking and the risk of nonalcoholic fatty liver disease. J. Clin. Transl. Hepatol. 12 (1), 113–118. 10.14218/jcth.2023.00165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghodsian N., Abner E., Emdin C. A., Gobeil É., Taba N., Haas M. E., et al. (2021). Electronic health record-based genome-wide meta-analysis provides insights on the genetic architecture of non-alcoholic fatty liver disease. Cell Rep. Med. 2 (11), 100437. 10.1016/j.xcrm.2021.100437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoshal U. C., Chourasia D. (2011). Genetic factors in the pathogenesis of gastroesophageal reflux disease. Indian J. Gastroenterol. 30 (2), 55–62. 10.1007/s12664-011-0095-7 [DOI] [PubMed] [Google Scholar]
- Han D., Zhang C. (2022). The oxidative damage and inflammation mechanisms in GERD-induced Barrett's esophagus. Front. Cell Dev. Biol. 10, 885537. 10.3389/fcell.2022.885537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemani G., Bowden J., Davey Smith G. (2018). Evaluating the potential role of pleiotropy in Mendelian randomization studies. Hum. Mol. Genet. 27 (R2), R195–r208. 10.1093/hmg/ddy163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu C. S., Kao J. H. (2012). Non-alcoholic fatty liver disease: an emerging liver disease in Taiwan. J. Formos. Med. Assoc. 111 (10), 527–535. 10.1016/j.jfma.2012.07.002 [DOI] [PubMed] [Google Scholar]
- Jiménez-Cortegana C., García-Galey A., Tami M., Del Pino P., Carmona I., López S., et al. (2021). Role of leptin in non-alcoholic fatty liver disease. Biomedicines 9 (7), 762. 10.3390/biomedicines9070762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahrilas P. J. (1992). Cigarette smoking and gastroesophageal reflux disease. Dig. Dis. 10 (2), 61–71. 10.1159/000171345 [DOI] [PubMed] [Google Scholar]
- Kanda T., Goto T., Hirotsu Y., Masuzaki R., Moriyama M., Omata M. (2020). Molecular mechanisms: connections between nonalcoholic fatty liver disease, steatohepatitis and hepatocellular carcinoma. Int. J. Mol. Sci. 21 (4), 1525. 10.3390/ijms21041525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M. S., Lin H. S., Chang M. L., Tsai M. H., Hsieh Y. Y., Lin Y. S., et al. (2022). Alanine aminotransferase to aspartate aminotransferase ratio and hepatitis B virus on metabolic syndrome: a community-based study. Front. Endocrinol. (Lausanne) 13, 922312. 10.3389/fendo.2022.922312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Basty N., Whitcher B., Bell J. D., Sorokin E. P., van Bruggen N., et al. (2021). Genetic architecture of 11 organ traits derived from abdominal MRI using deep learning. Elife 10, e65554. 10.7554/eLife.65554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L. N., Du R., Cheng D., Lin L., Wu X. Y., Hu C. Y., et al. (2019). Serum alanine aminotransferase is associated with metabolic syndrome and 10-year risk of cardiovascular disease. Biomed. Environ. Sci. 32 (2), 121–125. 10.3967/bes2019.016 [DOI] [PubMed] [Google Scholar]
- Maldonado-Rojas A. D. C., Zuarth-Vázquez J. M., Uribe M., Barbero-Becerra V. J. (2024). Insulin resistance and Metabolic dysfunction-associated steatotic liver disease (MASLD): pathways of action of hypoglycemic agents. Ann. Hepatol. 29 (2), 101182. 10.1016/j.aohep.2023.101182 [DOI] [PubMed] [Google Scholar]
- Maret-Ouda J., Markar S. R., Lagergren J. (2020). Gastroesophageal reflux disease: a review. Jama 324 (24), 2536–2547. 10.1001/jama.2020.21360 [DOI] [PubMed] [Google Scholar]
- Miele L., Cammarota G., Vero V., Racco S., Cefalo C., Marrone G., et al. (2012). Non-alcoholic fatty liver disease is associated with high prevalence of gastro-oesophageal reflux symptoms. Dig. Liver Dis. 44 (12), 1032–1036. 10.1016/j.dld.2012.08.005 [DOI] [PubMed] [Google Scholar]
- Min Y. W., Kim Y., Gwak G. Y., Gu S., Kang D., Cho S. J., et al. (2018). Non-alcoholic fatty liver disease and the development of reflux esophagitis: a cohort study. J. Gastroenterol. Hepatol. 33 (5), 1053–1058. 10.1111/jgh.14042 [DOI] [PubMed] [Google Scholar]
- Mittal R., Vaezi M. F. (2020). Esophageal motility disorders and gastroesophageal reflux disease. N. Engl. J. Med. 383 (20), 1961–1972. 10.1056/NEJMra2000328 [DOI] [PubMed] [Google Scholar]
- Oddy W. H., Herbison C. E., Jacoby P., Ambrosini G. L., O'Sullivan T. A., Ayonrinde O. T., et al. (2013). The Western dietary pattern is prospectively associated with nonalcoholic fatty liver disease in adolescence. Am. J. Gastroenterol. 108 (5), 778–785. 10.1038/ajg.2013.95 [DOI] [PubMed] [Google Scholar]
- Ong J. S., An J., Han X., Law M. H., Nandakumar P., Schumacher J., et al. (2022). Multitrait genetic association analysis identifies 50 new risk loci for gastro-oesophageal reflux, seven new loci for Barrett's oesophagus and provides insights into clinical heterogeneity in reflux diagnosis. Gut 71 (6), 1053–1061. 10.1136/gutjnl-2020-323906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardak P., Filip R., Woliński J., Krzaczek M. (2021). Associations of obstructive sleep apnea, obestatin, leptin, and ghrelin with gastroesophageal reflux. J. Clin. Med. 10 (21), 5195. 10.3390/jcm10215195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick M. T., Stuart P. E., Zhang H., Zhao Q., Yin X., He K., et al. (2021). Causal relationship and shared genetic loci between psoriasis and type 2 diabetes through trans-disease meta-analysis. J. Invest. Dermatol. 141 (6), 1493–1502. 10.1016/j.jid.2020.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pointer S. D., Rickstrew J., Slaughter J. C., Vaezi M. F., Silver H. J. (2016). Dietary carbohydrate intake, insulin resistance and gastro-oesophageal reflux disease: a pilot study in European- and African-American obese women. Aliment. Pharmacol. Ther. 44 (9), 976–988. 10.1111/apt.13784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J. K., Przeworski M. (2001). Linkage disequilibrium in humans: models and data. Am. J. Hum. Genet. 69 (1), 1–14. 10.1086/321275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin S., Wang C., Wang X., Wu W., Liu C. (2023). Causal association of gastroesophageal reflux disease with obstructive sleep apnea and sleep-related phenotypes: a bidirectional two-sample Mendelian randomization study. Front. Neurol. 14, 1283286. 10.3389/fneur.2023.1283286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu P., Du J., Zhang C., Li M., Li H., Chen C. (2023). Increased risk of reflux esophagitis in non-obese individuals with nonalcoholic fatty liver disease: a cross-sectional study. Ann. Med. 55 (2), 2294933. 10.1080/07853890.2023.2294933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachar M., Mautner Wizentier M., Risner E., Asmail H., Omara M., Chablaney S., et al. (2024). Risk factors associated with functional esophageal disorders (FED) versus gastroesophageal reflux disease (GERD). Surg. Endosc. 38, 2842–2849. 10.1007/s00464-024-10714-0 [DOI] [PubMed] [Google Scholar]
- Sadafi S., Azizi A., Pasdar Y., Shakiba E., Darbandi M. (2024). Risk factors for gastroesophageal reflux disease: a population-based study. BMC Gastroenterol. 24 (1), 64. 10.1186/s12876-024-03143-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaue S., Kanai M., Tanigawa Y., Karjalainen J., Kurki M., Koshiba S., et al. (2021). A cross-population atlas of genetic associations for 220 human phenotypes. Nat. Genet. 53 (10), 1415–1424. 10.1038/s41588-021-00931-x [DOI] [PubMed] [Google Scholar]
- Sanderson E., Glymour M. M., Holmes M. V., Kang H., Morrison J., Munafò M. R., et al. (2022). Mendelian randomization. Nat. Rev. Methods Prim. 2, 6. 10.1038/s43586-021-00092-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindhelm R. K., Diamant M., Dekker J. M., Tushuizen M. E., Teerlink T., Heine R. J. (2006). Alanine aminotransferase as a marker of non-alcoholic fatty liver disease in relation to type 2 diabetes mellitus and cardiovascular disease. Diabetes Metab. Res. Rev. 22 (6), 437–443. 10.1002/dmrr.666 [DOI] [PubMed] [Google Scholar]
- Skrivankova V. W., Richmond R. C., Woolf B. A. R., Yarmolinsky J., Davies N. M., Swanson S. A., et al. (2021). Strengthening the reporting of observational studies in Epidemiology using mendelian randomization: the STROBE-MR statement. Jama 326 (16), 1614–1621. 10.1001/jama.2021.18236 [DOI] [PubMed] [Google Scholar]
- Spechler S. J., Jain S. K., Tendler D. A., Parker R. A. (2002). Racial differences in the frequency of symptoms and complications of gastro-oesophageal reflux disease. Aliment. Pharmacol. Ther. 16 (10), 1795–1800. 10.1046/j.1365-2036.2002.01351.x [DOI] [PubMed] [Google Scholar]
- Verbanck M., Chen C. Y., Neale B., Do R. (2018). Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 50 (5), 693–698. 10.1038/s41588-018-0099-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P., Ma L., Dai G. X., Chen Y., Tong Y. L., Wang C., et al. (2011). The association of metabolic syndrome with reflux esophagitis: a case-control study. Neurogastroenterol. Motil. 23 (11), 989–994. 10.1111/j.1365-2982.2011.01786.x [DOI] [PubMed] [Google Scholar]
- Xiao G., He Q., Liu L., Zhang T., Zhou M., Li X., et al. (2022). Causality of genetically determined metabolites on anxiety disorders: a two-sample Mendelian randomization study. J. Transl. Med. 20 (1), 475. 10.1186/s12967-022-03691-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M., Deng L., Fass R., Song G. (2024). Obesity is associated with higher prevalence of gastroesophageal reflux disease and reflux related complications: a global healthcare database study. Neurogastroenterol. Motil. 36 (4), e14750. 10.1111/nmo.14750 [DOI] [PubMed] [Google Scholar]
- Xue J., Xin H., Ren N., Zhou C., Yang J., Song L., et al. (2019). Nonalcoholic fatty liver disease increases the risk of gastroesophageal reflux disease: a systematic review and meta-analysis. Eur. J. Clin. Invest. 49 (9), e13158. 10.1111/eci.13158 [DOI] [PubMed] [Google Scholar]
- Yang H. J., Chang Y., Park S. K., Jung Y. S., Park J. H., Park D. I., et al. (2017). Nonalcoholic fatty liver disease is associated with increased risk of reflux esophagitis. Dig. Dis. Sci. 62 (12), 3605–3613. 10.1007/s10620-017-4805-6 [DOI] [PubMed] [Google Scholar]
- Younossi Z., Anstee Q. M., Marietti M., Hardy T., Henry L., Eslam M., et al. (2018). Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat. Rev. Gastroenterol. Hepatol. 15 (1), 11–20. 10.1038/nrgastro.2017.109 [DOI] [PubMed] [Google Scholar]
- Younossi Z. M. (2019). Non-alcoholic fatty liver disease - a global public health perspective. J. Hepatol. 70 (3), 531–544. 10.1016/j.jhep.2018.10.033 [DOI] [PubMed] [Google Scholar]
- Younossi Z. M., Golabi P., Price J. K., Owrangi S., Gundu-Rao N., Satchi R., et al. (2024). The global Epidemiology of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis among patients with type 2 diabetes. Clin. Gastroenterol. Hepatol. 22, 1999–2010.e8. 10.1016/j.cgh.2024.03.006 [DOI] [PubMed] [Google Scholar]
- Zhao Y., Li D., Shi H., Liu W., Qiao J., Wang S., et al. (2024). Associations between type 2 diabetes mellitus and chronic liver diseases: evidence from a Mendelian randomization study in Europeans and East Asians. Front. Endocrinol. (Lausanne) 15, 1338465. 10.3389/fendo.2024.1338465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y., Zhong L., Hu C., Sheng G. (2020). Association between the alanine aminotransferase/aspartate aminotransferase ratio and new-onset non-alcoholic fatty liver disease in a nonobese Chinese population: a population-based longitudinal study. Lipids Health Dis. 19 (1), 245. 10.1186/s12944-020-01419-z [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Leave-one-out sensitivity analysis of Mendelian randomization analysis. (A) NAFLD on GERD; (B) NAFLD on GERD (adjusted); (C) percent liver fat on GERD; (D) GERD on NAFLD; (E) GERD on NAFLD (adjusted); (F) GERD on percent liver fat; (G) ALT on GERD; (H) AST on GERD; (I) GERD on ALT; (J) GERD on ALT (adjusted); (K) GERD on AST. NAFLD, non-alcoholic fatty liver disease; GERD, gastroesophageal reflux disease; ALT, alanine aminotransferase; AST, aspartate aminotransferase.
The forest plot of Mendelian randomization analysis. (A) NAFLD on GERD; (B) NAFLD on GERD (adjusted); (C) percent liver fat on GERD; (D) GERD on NAFLD; (E) GERD on NAFLD (adjusted); (F) GERD on percent liver fat; (G) ALT on GERD; (H) AST on GERD; (I) GERD on ALT; (J) GERD on ALT (adjusted); (K) GERD on AST. NAFLD, non-alcoholic fatty liver disease; GERD, gastroesophageal reflux disease; ALT, alanine aminotransferase; AST, aspartate aminotransferase.
The funnel plot of Mendelian randomization analysis. (A) NAFLD on GERD; (B) NAFLD on GERD (adjusted); (C) percent liver fat on GERD; (D) GERD on NAFLD; (E) GERD on NAFLD (adjusted); (F) GERD on percent liver fat; (G) ALT on GERD; (H) AST on GERD; (I) GERD on ALT; (J) GERD on ALT (adjusted); (K) GERD on AST. NAFLD, non-alcoholic fatty liver disease; GERD, gastroesophageal reflux disease; ALT, alanine aminotransferase; AST, aspartate aminotransferase.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.