Abstract
Asian water buffalo (Bubalus bubalis) is the fundamental livestock resource for local rural populations and holds a promising prospect of their milk and meat. Xuyi mountain (XYM) and Haizi (HZ) buffaloes from Yangtze valley comprises of species diversity of Asian water buffaloes. Current multi-omics enables identification of causal genes and elucidation of genetic regulatory mechanisms underlying complex traits in buffaloes. Here, we conducted the integrated analysis of metabolome and metagenome of rumen fluid, transcriptome and metabolome of blood, and whole genome sequence data from XYM (n = 7) and HZ (n = 10) male buffaloes. Our results revealed the apparent diversity of multi-layer omics profiles between two buffalo species. The built-up multi-omics database supports the discoveries of diversity in Asian water buffalo and potentially serves valuable resources for studying causal regulatory variants and their mechanisms.
Subject terms: Next-generation sequencing, Animal breeding
Background & Summary
Asian water buffaloes (Bubalus bubalis), acclaimed by the Food and Agriculture Organization (FAO) as the livestock species that sustain the most human population (http://www.fao.org/faostat/), are primarily distributed in Asia1,2. In China, buffaloes play the crucial roles in farming activities and transportation as the power providers and in meat and milk as the food supplies in the rice-growing regions of the Yangtze valley3–5. As the typical extant swamp buffaloes of Asian water buffaloes, Xuyi mountain (XYM) buffaloes are bred in the areas of Jiangsu province along Yangtze valley characterized by numerous hills and fewer plains, and developed through long-term selection by local mountain farmers. Haizi (HZ) buffaloes are raised in similar regions to XYM buffaloes but in coastal areas5. Higher phylogeographic genetic diversity but lower phenotype differentiations are observed in such buffalo populations1. Therefore, accurate elucidation of the domestic buffalo phenotypes holds the significant importance in understanding the genetic regulatory mechanisms underlying their important economic traits for enhancing their further productivity6–8.
Currently, multi-omics technologies have been employed to study buffaloes for muscle development, embryonic development, and spermatogenesis9–12. Wu et al.13 utilized a combination strategy of transcriptomics and metabolomics to explore the potential molecular mechanisms regulating buffalo meat quality traits. Wang et al.14 investigated the nutritional metabolism mechanisms of rumen bacteria under heat stress in buffaloes using metagenomics and metabolomics technologies. Huang et al.15 generated a high-resolution single-cell transcriptomic atlas of sperms and somatic cells and revealed the normal spermatogenesis maintenance within buffalo testicular microenvironment. However, the handicap of understanding the regulatory mechanisms of domestic buffaloes still remains because of the limited multi-omics at the moment.
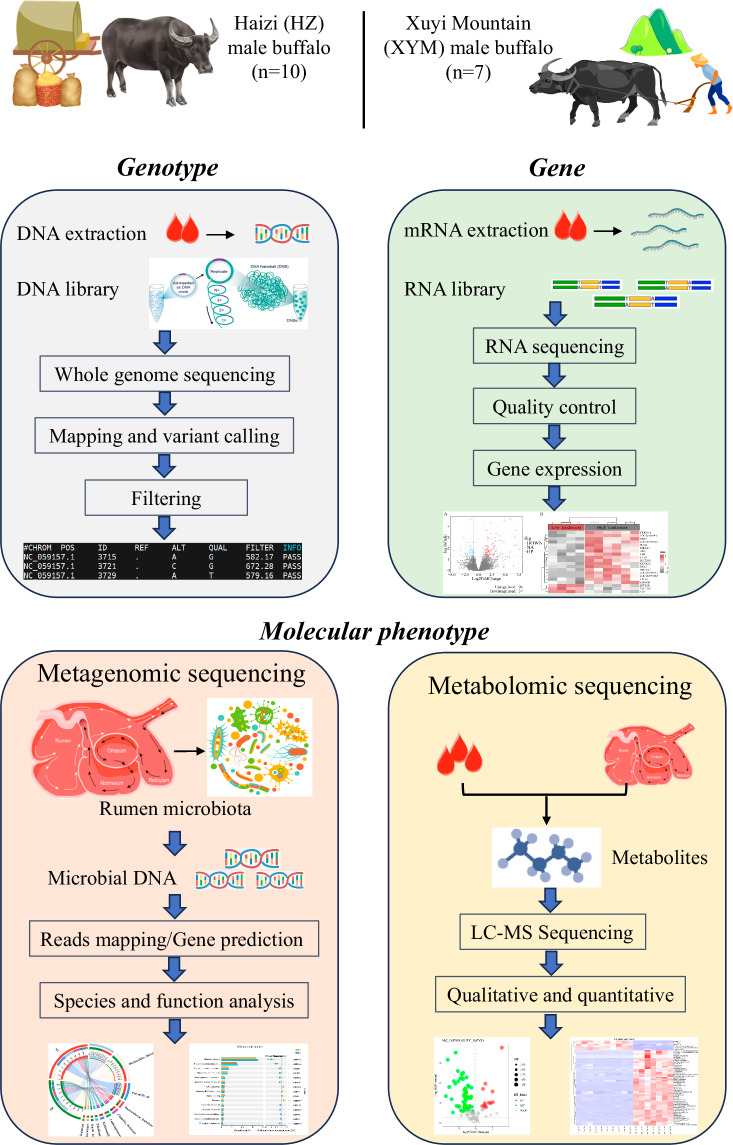
To explore genetic resources and decipher regulatory mechanisms underlying complex traits in domestic buffaloes, we selected XYM (n = 7 individuals) and HZ (n = 10 individuals) male buffaloes from two similar management farms (Table 1). Integrated analysis of metabolomics and metagenomics of rumen fluid, transcriptomics and metabolomics of blood samples, and whole genome sequence (WGS) data were comprehensively performed (Fig. 1). Consequently, our study generated a high-quality database including five datasets of the genome, transcriptome, metabolome, and metagenome above, the preliminary results of measured growth phenotype and identified genes, metabolites, and microorganisms, and the analysis pipelines with all programming scripts. The population genetic structure was also investigated using WGS data of HZ and XYM buffaloes with the other three local buffaloes that are Dehong (DH), Wenzhou (WZ), and Yibing (YB) buffaloes. Based on the 119.52 gigabyte (GB) of blood transcriptome of HZ and XYM buffaloes, we identified 23,703 and 23,216 genes, respectively, with 1,739 differentially expressed genes (DEGs) between them. Subsequently, 508 rumen fluid and 851 plasma metabolites in HZ buffalo and 846 rumen fluid and 897 plasma metabolites in XYM buffalo were identified using untargeted metabolomics sequencing. We revealed the species-level composition of rumen microbiota and differences in KEGG enrichments based on 69.15 GB and 45.76 GB of rumen metagenome from HZ and XYM buffaloes, respectively. Our database reveals the diversities of multi-layer omics profiles between two Asian water buffaloes and offers the valuable multi-omics resources for further investigation of molecular mechanisms.
Table 1.
Description of breed, gender, age and weight of all 17 buffaloes.
| Sample ID | Breed | Gender | Age (Month) | Weight (Kg) |
|---|---|---|---|---|
| XYM1 | Xuyi mountain buffalo | Male | 26 | 440 |
| XYM2 | Xuyi mountain buffalo | Male | 40 | 640 |
| XYM3 | Xuyi mountain buffalo | Male | 42 | 700 |
| XYM4 | Xuyi mountain buffalo | Male | 42 | 679 |
| XYM5 | Xuyi mountain buffalo | Male | 40 | 632 |
| XYM6 | Xuyi mountain buffalo | Male | 30 | 470 |
| XYM7 | Xuyi mountain buffalo | Male | 28 | 462 |
| HZ1 | Haizi buffalo | Male | 30 | 550 |
| HZ2 | Haizi buffalo | Male | 28 | 430 |
| HZ3 | Haizi buffalo | Male | 26 | 540 |
| HZ4 | Haizi buffalo | Male | 27 | 490 |
| HZ5 | Haizi buffalo | Male | 26 | 510 |
| HZ6 | Haizi buffalo | Male | 30 | 520 |
| HZ7 | Haizi buffalo | Male | 28 | 440 |
| HZ8 | Haizi buffalo | Male | 28 | 430 |
| HZ9 | Haizi buffalo | Male | 28 | 440 |
| HZ10 | Haizi buffalo | Male | 26 | 480 |
Fig. 1.
Integrated analysis of genome-transcriptome-metabolome-metagenome multi-omics database of Xuyi mountain and Haizi male buffaloes.
Methods
Animals and sample collection
Seventeen healthy buffaloes were selected from two local buffalo breeding farms using similar managing systems in Jiangsu Province, China, including 7 XYM and 10 HZ male individuals (Table 1). Their ages range from 26 to 42 months and weights range from 430 to 700 kilograms (Kg). The feeding and management of both HZ and XYM buffaloes followed the local guidelines and it included the free access to an exercise yard, feeding twice daily, regular vaccination. The weight description of all 17 buffaloes is shown in Table 1.
The whole blood sample of each individual was collected from the tail vein using sodium heparin anticoagulant tubes. Those samples were subsequently centrifuged at 3000 rpm for 10 minutes at 4 °C and the supernatant plasma was transferred to 1.5 mL centrifuge tubes. The plasma collections were flash-frozen in liquid nitrogen for 15 minutes. In the meantime, the surplus of the whole blood samples was used for DNA and RNA extractions. Rumen fluid was collected using a negative-pressure oral stomach tube. To avoid saliva contamination, the initial 50 mL of the collected fluid was discarded and the subsequent 20 mL of them was obtained and filtered through gauze to remove feed residues. The filtered rumen fluid was aliquoted into 5 mL sterile tubes and stored in liquid nitrogen. All sample collections were simultaneously performed 2 hours after the first morning feeding.
Whole genome sequencing and data processing
Genomic DNA was extracted from the whole blood samples using the standard phenol-chloroform protocol. For each individual, at least 5 μg of DNA was used to construct paired-end libraries. The whole genome sequencing (WGS) was performed at ~30 × coverage of 2 × 150 bp paired-end reads on the DNBSEQ platform by MGI Tech Co., Ltd. After preprocess by Fastp (v0.19.5)16, all clean data were mapped to the buffalo reference genome (NDDB_SH_1, Gene bank: GCA_019923935.1) using BWA-mem2 (v2.2.1)17 with default parameters. The delivered mapping reads in SAM format were then transferred to BAM format and sorted using Samtools (v1.10)18. GATK (v4.1.6)19 Haplotypecaller and GenotypeGVCFs were used to generate an intermediate GVCF of each sample for the joint genotyping of multiple samples. GATK VariantFiltration was used for the efficient high-quality SNP identification after removing the potential false-positive variant calls following the parameters of QD < 2.0, MQ < 40.0, FS > 60.0, SOR > 3.0, MQRankSum < −12.5, and ReadPosRankSum < −8.0. Genotype density distribution was plotted using R package CMplot (https://github.com/YinLiLin/CMplot). Population genetic structure was investiaged by principal components analysis (PCA) and ADMIXTURE (v1.3.0)20 using the WGS data of HZ and XYM buffaloes and the other three local buffaloes that are Dehong (DH, n = 5), Wenzhou (WZ, n = 4) and Yibing (YB, n = 5) buffaloes21 obtained from Genome Sequence Archive (GSA) of China National Center for Bioinformation with the accession number CRA001463 (https://ngdc.cncb.ac.cn/gsa/browse/CRA001463)21.
RNA sequencing and data processing
Total RNA was extracted from the whole blood samples using TRIzol reagent, followed by an assessment of RNA concentration, purity, and integrity using Nanodrop 2000 and Agilent 2100 bioanalyzer systems. Subsequently, mRNA was enriched from total RNA using magnetic beads with Oligo(dT) to fragment and reverse-transcribe them into cDNA using random hexamers. Double-stranded cDNA was synthesized using DNA polymerase I and RNase H purified with AMPure XP beads and subjected to end repair, A-tailing, adapter ligation, and size selection with AMPure XP beads. The constructed libraries were sequenced using the BGISEQ DNBSEQ-T7 platform by Beijing Novogene Technology Co., Ltd.
Quality control was performed using Fastp (v0.19.5) software to trim adapters, polyG tails, and polyX tails and filter out reads with uncertain N content > 10% and low-quality (Qphred < 20). High-quality reads were aligned to the buffalo reference genome (NDDB_SH_1) using HISAT2 (v2.0.5)22 and all transcripts were quantified by fragments per kilobase of transcript per million mapped reads (FPKM) values to measure each gene expression level. R package DESeq2 (v1.44.0)23 was used for differential expression analysis with the screening criteria of |log2(FoldChange)| ≥ 2 and Padj < 0.05. R package clusterProfiler (v4.12.0)24 was applied to identify significant GO terms and KEGG pathways associated with differentially expressed genes (DEGs) with a significant threshold of Padj value < 0.05.
Liquid chromatography–mass spectrometry and data processing
According to the previous metabolic profiling method25, 100 μL of each plasma and rumen fluid sample and 400 μL of 80% methanol aqueous solution were mixed for 1 minute and incubated for 30 minutes at −20 °C. After incubation, the mixture was centrifuged at 12,000 rpm for 20 minutes at 4 °C and 150 μL of supernatant was resuspended with 150 μL of 80% methanol solution containing 2-chlorobenzylalanine and filtered through a 0.22 μm sterile membrane to obtain the test sample. A QC sample was created by mixing 50 μL of each test sample to ensure the consistency of processing and detection. Liquid chromatography–mass spectrometry (LC-MS) analysis was performed and every five samples accompanying with QC sample was designed to evaluate the detection stability during the analysis.
The processed samples undergo chromatographic separation using the Vanquish Ultra-High Performance Liquid Chromatography (UHPLC) system (Thermo Fisher, Germany) with a Hypesil Gold C18 column (100 mm × 2.1 mm, 1.9 μm, Thermo Fisher, USA). Chromatographic conditions include a column temperature of 40 °C, a flow rate of 0.2 mL/min, and a mobile phase consisting of 0.1% formic acid (A) and methanol (B) for positive ion modes and 5 mmol/L ammonium acetate (A) and methanol (B) for negative ion modes. The chromatographic peaks were analyzed using the Q Exactive™HF-X mass spectrometer (Thermo Fisher, Germany) with a scan range of m/z 100–1500. The ESI source settings include a spray voltage of 3.5 kV, sheath gas flow rate of 35 psi, auxiliary gas flow rate of 10 L/min, capillary temperature of 320 °C, S-lens RF level of 60, and auxiliary gas heater temperature of 350 °C. MS/MS secondary scan is performed using data-dependent scans.
The data files were processed using the Compound Discoverer26 software (v3.1) to conduct retention time and mass-to-charge ratio screening for each metabolite. Peak alignment was performed with a retention time deviation of 0.2 minutes and a mass deviation of 5 ppm across different samples to enhance identification accuracy. Peak extraction was conducted with a mass deviation of 5 ppm, a signal intensity deviation of 30%, and a signal-to-noise ratio of 3. Based on molecular ion peaks and fragment ions, target ions were integrated and molecular formulas were predicted to compare with the mzCloud, mzVault, and Masslist databases. The background ions were removed using blank samples and the original quantitative results were standardized using the formula of sample original quantitative value/(total quantitative value of sample metabolites/total quantitative value of QC sample metabolites) to obtain the relative peak areas. After removing compounds with a coefficient of variation (CV) of relative peak areas >30% in all QC samples, metabolite identification and relative quantification results were finally obtained.
PCA and orthogonal partial least squares discriminant analysis (OPLS-DA) were conducted using R package ropes (v1.6.2) with 7-fold cross-validation to assess model stability. Significant differential metabolites are selected based on the student’s t-test and differential multiple analysis using OPLS-DA model with variable importance in the projection (VIP) values > 1 and P values < 0.05. Metabolic pathway annotation is performed using the KEGG database (https://www.kegg.jp/kegg/pathway.html) based on differential metabolites.
Microbial community and function profiling
The extraction procedures of total metagenome DNA followed the FastDNA® Spin Kit for Soil protocol (MP Biomedicals). Quality assessment evaluations included DNA purity, concentration, and integrity using NanoDrop 2000, TBS-380, and 1% agarose gel electrophoresis. Ultrasonication with a Covaris M220 instrument fragmented samples into approximately 400 bp fragments. Library construction was executed using the NEXTFLEX Rapid DNA-Seq Kit. Next, adapter trimming of 3′ and 5′ ends and removal of reads shorter than 50 bp, with an average base quality below 20, and containing N bases were performed for high-quality clean reads using the Fastp (v0.19.5) tool16. The BWA software was used to align clean reads to host DNA sequences and to filter out contaminating reads with high alignment similarity. Subsequent open reading frames (ORFs) prediction on the selected contigs was performed using the Prodigal tool (v2.6.3)27.
All predicted gene sequences were clustered using CD-HIT (https://sites.google.com/view/cd-hit)28 (90% identity and 90% coverage), where the longest gene from each cluster formed a non-redundant gene set. The SOAPaligner29 was used to align the high-quality reads from each sample to the non-redundant gene set (95% similarity threshold) to quantify gene abundance in each sample. Diamond software (v0.8.35)30 was conducted for BLASTP comparisons of amino acid sequences of the non-redundant gene sets against KEGG databases (e-value threshold of 1 × 10−5) to obtain species information and KEGG functional annotations. Species α-diversity differences were assessed using one-way analysis of variance (ANOVA). Principal coordinate analysis (PCoA) was generated with inter-group and analysis of similarity (ANOSIM) using R package QIIME31. Linear discriminant analysis (LDA) effect size (LEfSe) was estimated with an LDA threshold of 2 for differential species and functional analysis. Spearman correlation analysis was used to assess the correlation among differentially identified species, functional genes and rumen carbohydrate-degrading enzyme activity.
Data Records
Three datasets were deposited into NCBI Sequence Read Archive with BioProject ID of PRJNA1173791 with the SRP accession number SRP53962932 for genome, blood transcriptome, and rumen metagenome of 7 Xuyi mountain (XYM) and 10 Haizi (HZ) buffaloes. Two datasets of raw rumen fluid and plasma metabolome of those buffaloes were deposited into Figshare with the accession number 2616163333. In addition, the whole five datasets above together with the procedures of quality control and statistical analysis, processed data, and preliminary results were uploaded to an online database (https://shenyy01.github.io/shenyy.github.io/) that can be easily and freely accessed.
Technical Validation
Whole genome alignments and population structure
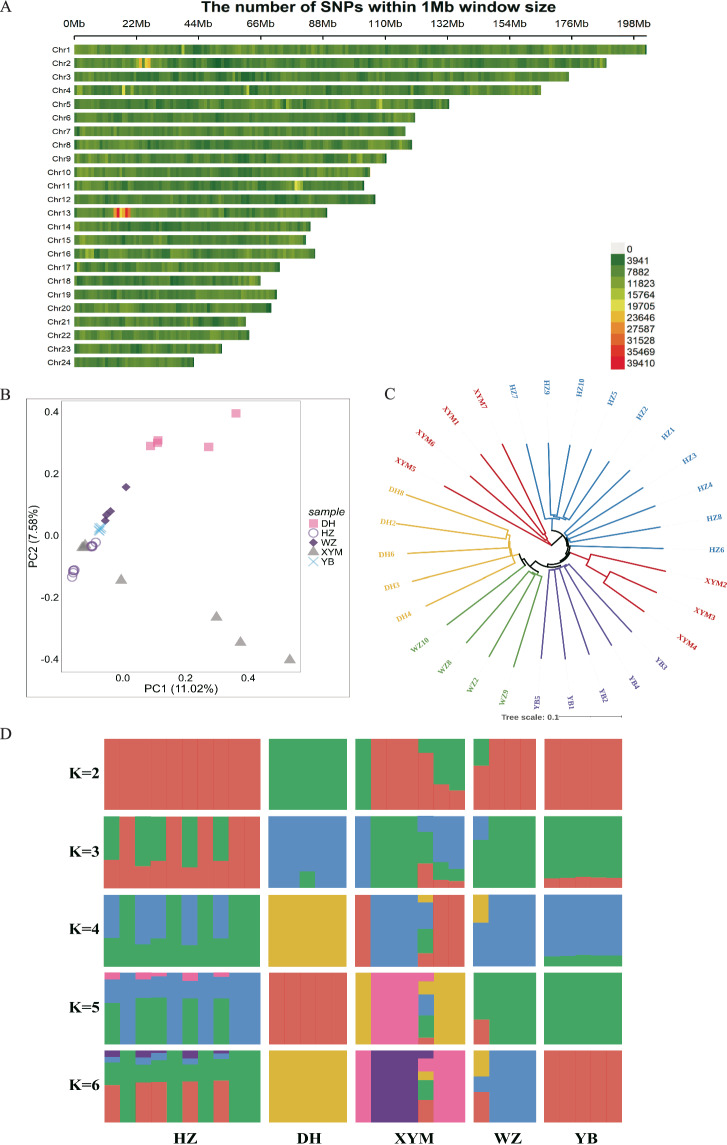
The HZ and XYM buffaloes yielded 108.90 GB and 95.53 GB of raw reads and 108.78 GB and 95.42 GB of clean reads, respectively. An overall moderate marker density was distributed across the whole genome using a 1 Mb sliding window (Fig. 2A). The PCA results demonstrated a good clustering among five local buffaloes, where first and second principal components explained 11.02% and 7.5% of variances, respectively (Fig. 2B). The Neighbor-Joining (NJ) tree indicated a close genetic distance between HZ and XYM buffaloes, but XYM buffaloes was divided into two clades. YB buffalo was found to have an earlier evolutionary timeline compared to the other four local buffaloes (Fig. 2C). The ancestral components for five local buffalo populations are shown in Fig. 2D.
Fig. 2.
Population structure analysis based on whole genome sequencing data of five local buffaloes including Xuyi mountain (XYM, n = 7), Haizi (HZ, n = 10), Dehong (DH, n = 5), Wenzhou (WZ, n = 4) and Yibing (YB, n = 5) buffaloes. (A) Distribution map of whole-genome SNP marker density across chromosomes calculated using a 1 Mb sliding window. The color gradient from green to red indicates increasing SNP density in the respective regions. (B) Principal component analysis (PCA) of five local buffaloes. (C) Evolutionary tree of five local buffaloes based on genetic distance. (D) Ancestry compositions with the assumed number of ancestries at K value from 2 to 6. K is an adjustable parameter representing the number of possible ancestral varieties, where K = 2 is the best value after calculation of the cross-validation error.
Transcriptome and differentially expressed genes
RNA sequencing (RNA-Seq) generated an average of 119.52 GB of raw data and 113.98 GB of clean data (Supplementary table S1). A total of 23,703 and 23,216 genes were identified in HZ and XYM buffaloes, respectively and 1,739 DEGs were identified between them, where EGR1, SKI, and PHF13 were listed as the top three DEGs.
Metabolites profiling and pathway enrichment
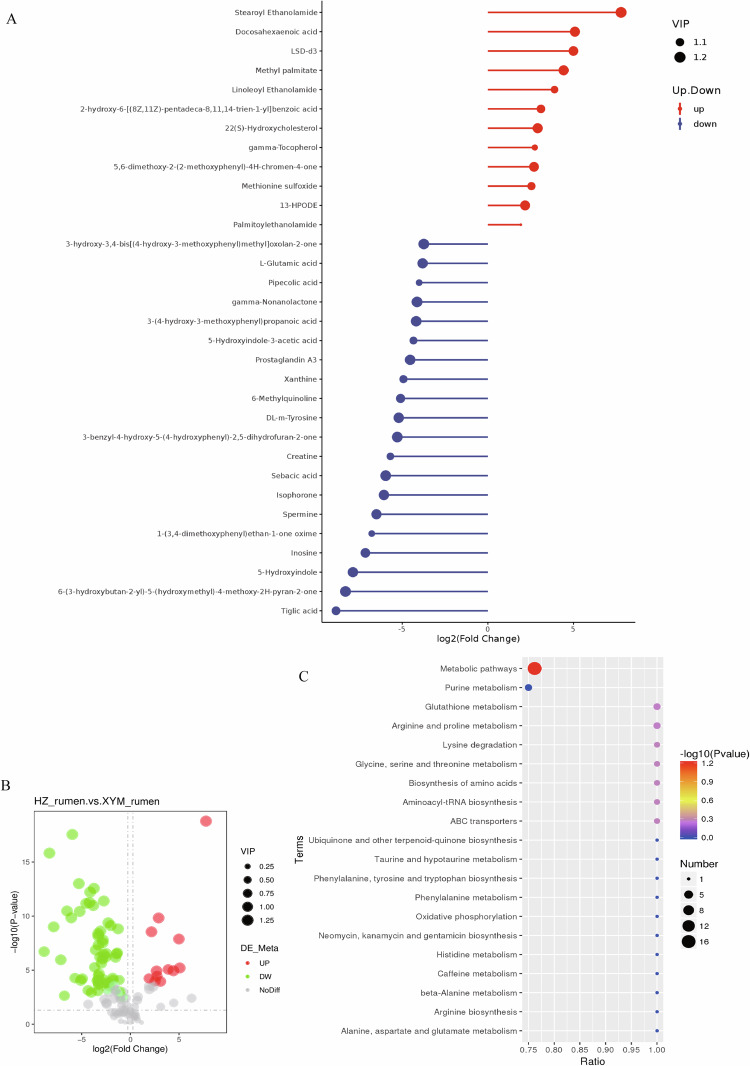
For rumen liquid and plasma, around 203~569 metabolites were identified (Supplementary figure S1). We identified 12 and 6 upregulated metabolites and 58 and 48 downregulated metabolites in the positive and negative ion modes, respectively, for rumen liquid comparing HZ buffalo to XYM buffalo (Fig. 3A,B, Supplementary figure S2A, and Supplementary figure S2B). The top significant KEGG pathways included the metabolic pathways, the glutathione metabolism, and the arginine and proline metabolism (Fig. 3C and Supplementary figure S2C).
Fig. 3.
Differential metabolite identifications of rumen liquids between Haizi and Xuyi mountain buffaloes in the positive ion mode. (A) VIP values and Fold change of metabolites. (B) Volcano plots of differential metabolites based on log2Fold change and -log10P-value. (C) KEGG pathway enrichment of differential metabolites. The size of the black points indicates the number of differential metabolites enriched.
Metagenomic profiling and taxonomic classification
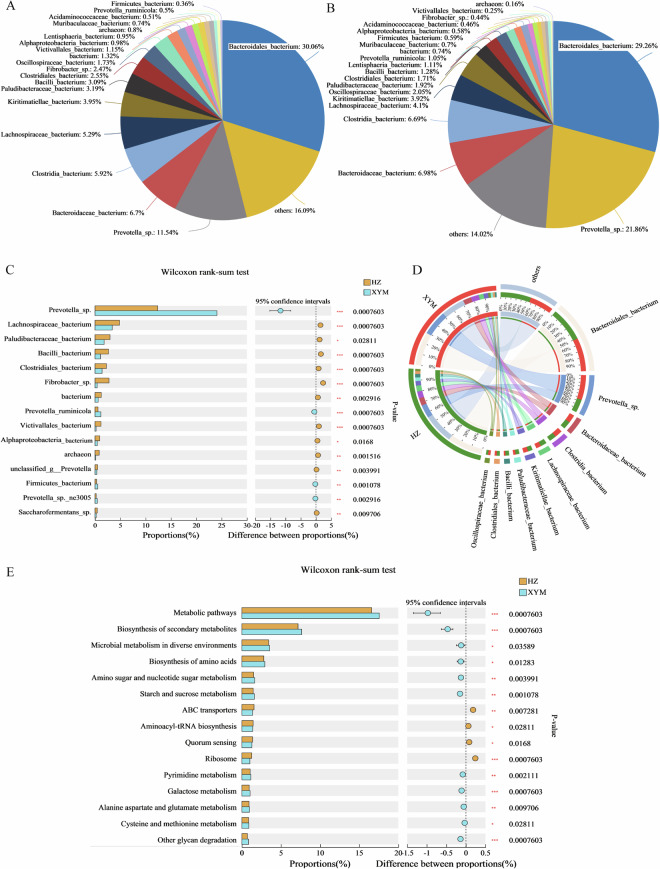
The rumen metagenome generated 69.15 GB and 45.76 GB of raw data and 68.96 GB and 45.65 GB of clean data for HZ and XYM buffaloes, respectively (Supplementary table S2). Compared to XYM buffaloes, HZ buffaloes exhibited a significant increasing index but a significant decreasing in Simpson index (Supplementary figure S3). We identified 13 kingdoms, 221~222 phyla, 437~447 classes, 897~941 orders, 1725~1890 families, 4165 ~4630 genera, and 13827~15448 species (Supplementary figure S3). The top three taxa included Bacteroidaceae_bacterium (6.70% and 6.98%), Clostridia_bacterium (5.92% and 6.69%), and Lachnospiraceae_bacterium (5.29% and 4.10%) (Fig. 4A,B). The significant differences of taxa between two buffalo species were found in Prevotella_sp (P-value = 7.60 × 10−4) (Fig. 4C). The relationship of species with two buffalo species was visualized in the Circos plots (Fig. 4D). Based on the KEGG database of level 3, the metabolic pathways were enriched as the most significant pathway (Fig. 4E).
Fig. 4.
Comparative analysis of microbial composition and function at species level of Haizi (HZ) and Xuyi mountain (XYM) buffaloes. (A) The abundance percentages of HZ buffalo. (B) The abundance percentages of XYM buffalo. (C) The average relative abundance of the same species between HZ and XYM buffaloes. The vertical coordinate indicates the species under different classification levels. The horizontal coordinate indicates the abundance proportions of a certain species. Red asterisk indicates * of 0.01 < P-value ≤ 0.05, ** of 0.001 < P-value ≤ 0.01, *** of P-value ≤ 0.001. (D) Circos plots of species in relationship with HZ and XYM buffaloes. The abundance distribution of different species is connected through the inner color bands. The left semicircle (smaller circle) indicates the composition of species, while the right semicircle (larger circle) indicates the distribution of species. (E) Comparison of microbial functions based on the KEGG database at KEGG level 3. The vertical coordinate indicates the KEGG function names under different classification levels. The horizontal coordinate indicates the abundance proportions of a certain KEGG function.
Ethics declarations
All experimental procedures and protocols were approved by the Research Committee of Jiangsu Academy of Agricultural Sciences following the regulations for the Administration of Affairs Concerning Experimental Animals (Decree No. 63 of Jiangsu Academy of Agricultural Science on 8 July 2014). All experiments were performed in accordance with the ARRIVE (Animal Research: Reporting In Vivo Experiments) guidelines.
Supplementary information
Acknowledgements
This work was supported by grants from the National Key R&D Program of China (2021YFD1200404, 2021YFF1000701-06, and 2023ZD0404901-05), Taishan Scholar Foundation of Shandong Province (tsqnz20231240), Shandong Provincial Natural Science Foundation (ZR2021MC070 and ZR2023QC252), the Earmarked Fund for CARS-36, and Technology innovation group project of Foshan 2019 (FS0AA-KJ919-4402-0027). Thanks to Cassie Zhao (GemPharmatech Co., Ltd.) for improving the English of this manuscript. We acknowledge China National Center for Bioinformation for data sharing.
Author contributions
Y.S. and X.W. conceived the study and constructed the database. Y.S., Z.A. and X.S. performed the data analysis. X.S., Q.D., K.C., Y.M., J.L. and J.Z. collected the data. Y.S., Z.A. and X.S. prepared the figures, Y.S. wrote the original manuscript. X.W., H.W., Y.M., T.W. and K.C. revised the original manuscript. K.C., J.Z. and H.W. managed laboratory work and supervised the project.
Code availability
All software and tools used in this study are accessible from their published journals and have been carefully described in the Methods section. The scripts for whole analysis of genome, transcriptome, metagenome, and metabolome were deposited in GitHub (https://github.com/Shenyy01/omics). The datasets and preliminary results were deposited into an online database (https://shenyy01.github.io/shenyy.github.io/) for the easier access.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xiao Wang, Email: xiaowangzntc@163.com.
Huili Wang, Email: wanghuili318@163.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41597-024-04246-4.
References
- 1.Zhang, Y., Colli, L. & Barker, J. S. F. Asian water buffalo: domestication, history and genetics. Anim Genet51, 177–191, 10.1111/age.12911 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Rehman, S. U., Hassan, F. U., Luo, X., Li, Z. & Liu, Q. Whole-Genome Sequencing and Characterization of Buffalo Genetic Resources: Recent Advances and Future Challenges. Animals (Basel)1110.3390/ani11030904 (2021). [DOI] [PMC free article] [PubMed]
- 3.Sun, T. et al. Multiple domestication of swamp buffalo in China and South East Asia. J Anim Breed Genet137, 331–340, 10.1111/jbg.12445 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Sun, T. et al. Selection signatures of Fuzhong Buffalo based on whole-genome sequences. BMC Genomics21, 674, 10.1186/s12864-020-07095-8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang, Y., Sun, D., Yu, Y. & Zhang, Y. Genetic diversity and differentiation of Chinese domestic buffalo based on 30 microsatellite markers. Anim Genet38, 569–575, 10.1111/j.1365-2052.2007.01648.x (2007). [DOI] [PubMed] [Google Scholar]
- 6.Du, C. et al. Systematic analyses for candidate genes of milk production traits in water buffalo (Bubalus Bubalis). Anim Genet50, 207–216, 10.1111/age.12739 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Lazaro, S. F. et al. Genomic studies of milk-related traits in water buffalo (Bubalus bubalis) based on single-step genomic best linear unbiased prediction and random regression models. J Dairy Sci104, 5768–5793, 10.3168/jds.2020-19534 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Singh, B., Mal, G., Kues, W. A. & Yadav, P. S. The domesticated buffalo - An emerging model for experimental and therapeutic use of extraembryonic tissues. Theriogenology151, 95–102, 10.1016/j.theriogenology.2020.04.003 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Li, H. et al. Comparison of Long Non-Coding RNA Expression Profiles of Cattle and Buffalo Differing in Muscle Characteristics. Front Genet11, 98, 10.3389/fgene.2020.00098 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwoji, I. D., Aiyegoro, O. A., Okpeku, M. & Adeleke, M. A. ‘Multi-omics’ data integration: applications in probiotics studies. NPJ Sci Food7, 25, 10.1038/s41538-023-00199-x (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khongdee, T., Sripoon, S. & Vajrabukka, C. The effects of high temperature and roof modification on physiological responses of swamp buffalo (Bubalus bubalis) in the tropics. Int J Biometeorol57, 349–354, 10.1007/s00484-012-0557-3 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Huang, L. et al. Comparative transcriptome analysis reveals potential testosterone function-related regulatory genes/pathways of Leydig cells in immature and mature buffalo (Bubalus bubalis) testes. Gene802, 145870, 10.1016/j.gene.2021.145870 (2021). [DOI] [PubMed] [Google Scholar]
- 13.Wu, G. et al. Integrated Analysis of Transcriptome and Metabolome Profiles in the Longissimus Dorsi Muscle of Buffalo and Cattle. Curr Issues Mol Biol45, 9723–9736, 10.3390/cimb45120607 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang, Z. et al. Heat Stress Induces Shifts in the Rumen Bacteria and Metabolome of Buffalo. Animals (Basel)1210.3390/ani12101300 (2022). [DOI] [PMC free article] [PubMed]
- 15.Huang, L. et al. Single-cell RNA sequencing uncovers dynamic roadmap and cell-cell communication during buffalo spermatogenesis. iScience26, 105733, 10.1016/j.isci.2022.105733 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics34, i884–i890, 10.1093/bioinformatics/bty560 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aluru, M. V. S. M. H. L. S. Efficient Architecture-Aware Acceleration of BWA-MEM for Multicore Systems. IEEE International Parallel and Distributed Processing Symposium (IPDPS), 314–324 (2019).
- 18.Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics25, 2078–2079, 10.1093/bioinformatics/btp352 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res20, 1297–1303, 10.1101/gr.107524.110 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alexander, D. H., Novembre, J. & Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res19, 1655–1664, 10.1101/gr.094052.109 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo, X. et al. Understanding divergent domestication traits from the whole-genome sequencing of swamp- and river-buffalo populations. Natl Sci Rev7, 686–701, 10.1093/nsr/nwaa024 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim, D., Paggi, J. M., Park, C., Bennett, C. & Salzberg, S. L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol37, 907–915, 10.1038/s41587-019-0201-4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol15, 550, 10.1186/s13059-014-0550-8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu, T. et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (Camb)2, 100141, 10.1016/j.xinn.2021.100141 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dunn, W. B. et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat Protoc6, 1060–1083, 10.1038/nprot.2011.335 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Cooper, B. & Yang, R. An assessment of AcquireX and Compound Discoverer software 3.3 for non-targeted metabolomics. Sci Rep14, 4841, 10.1038/s41598-024-55356-3 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hyatt, D. et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics11, 119, 10.1186/1471-2105-11-119 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fu, L., Niu, B., Zhu, Z., Wu, S. & Li, W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics28, 3150–3152, 10.1093/bioinformatics/bts565 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo, R. et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience1, 18, 10.1186/2047-217X-1-18 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buchfink, B., Xie, C. & Huson, D. H. Fast and sensitive protein alignment using DIAMOND. Nat Methods12, 59–60, 10.1038/nmeth.3176 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Bolyen, E. et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol37, 852–857, 10.1038/s41587-019-0209-9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.NCBI Sequence Read Archivehttps://identifiers.org/ncbi/insdc.sra:SRP539629 (2024).
- 33.Figshare.10.6084/m9.figshare.26161633.v1 (2024).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- NCBI Sequence Read Archivehttps://identifiers.org/ncbi/insdc.sra:SRP539629 (2024).
- Figshare.10.6084/m9.figshare.26161633.v1 (2024).
Supplementary Materials
Data Availability Statement
All software and tools used in this study are accessible from their published journals and have been carefully described in the Methods section. The scripts for whole analysis of genome, transcriptome, metagenome, and metabolome were deposited in GitHub (https://github.com/Shenyy01/omics). The datasets and preliminary results were deposited into an online database (https://shenyy01.github.io/shenyy.github.io/) for the easier access.