Graphical abstract
Keywords: Bone metastasis, Bone marrow microenvironment, Breast cancer, Tomography, X-ray computed, Radiomics
Highlights
-
•
Biological alterations in bone marrow occur before bone metastasis.
-
•
CT-based radiomics quantifies pre-metastatic bone marrow changes.
-
•
Radiomics model of bone marrow changes predicts bone metastasis risk.
Background
Bone metastasis from breast cancer significantly elevates patient morbidity and mortality, making early detection crucial for improving outcomes. This study utilizes radiomics to analyze changes in the thoracic vertebral bone marrow microenvironment from chest computerized tomography (CT) images prior to bone metastasis in breast cancer, and constructs a model to predict metastasis. Methods: This study retrospectively gathered data from breast cancer patients who were diagnosed and continuously monitored for five years from January 2013 to September 2023. Radiomic features were extracted from the bone marrow of thoracic vertebrae on non-contrast chest CT scans. Multiple machine learning algorithms were utilized to construct various radiomics models for predicting the risk of bone metastasis, and the model with optimal performance was integrated with clinical features to develop a nomogram. The effectiveness of this combined model was assessed through receiver operating characteristic (ROC) analysis as well as decision curve analysis (DCA). Results: The study included a total of 106 patients diagnosed with breast cancer, among whom 37 developed bone metastases within five years. The radiomics model’s area under the curve (AUC) for the test set, calculated using logistic regression, is 0.929, demonstrating superior predictive performance compared to alternative machine learning models. Furthermore, DCA demonstrated the potential of radiomics models in clinical application, with a greater clinical benefit in predicting bone metastasis than clinical model and nomogram. Conclusion: CT-based radiomics can capture subtle changes in the thoracic vertebral bone marrow before breast cancer bone metastasis, offering a predictive tool for early detection of bone metastasis in breast cancer.
1. Introduction
Bone metastasis is a significant focus in clinical research due to its profound impact on increasing the disease burden and the risk of diminished quality of life for patients [1]. Additionally, it is closely associated with poor prognosis and higher mortality rates [2], [3]. Investigating the processes involved in bone metastasis related to breast cancer, along with improving early detection methods, is crucial for developing effective treatment strategies and improving patient outcomes [4], [5], [6]. Investigating the changes in the microenvironment that occur before bone metastasis can lead to the development of new insights and theoretical frameworks aimed at preventing, diagnosing early, and specifically treating breast cancer bone metastasis, a factor that is crucial for enhancing patient survival rates [7], [8].
Recent progress in studying the metastasis of breast cancer and the microenvironment of bone marrow has revealed the complex mechanisms underlying the interactions between tumor cells and the bone marrow milieu [9], [10]. Recent studies are aimed at investigating the ways in which the microenvironment of bone marrow facilitates the colonization and spread of breast cancer cells, involving the interactions among cytokines, immune cells, and stromal cells within the bone marrow [11], [12]. Simultaneously, growing evidence indicates that modifications in the bone marrow microenvironment could significantly influence the initial phases of breast cancer bone metastasis, thereby providing new pathways for creating preventive and therapeutic strategies [13], [14]. Through an in-depth exploration of the evolving dynamics within the bone marrow microenvironment, we can enhance our comprehension of the biological foundations underlying breast cancer metastasis, ultimately leading to improved early diagnosis and personalized treatment outcomes. Subtle changes in the bone marrow microenvironment are difficult to visually identify on computerized tomography (CT) images. Although CT imaging may not yet show visually observable bone abnormalities, metastasis may have already occurred. The early detection of metabolic abnormalities is achievable through positron emission tomography-computed tomography (PET-CT), whereas bone changes detectable on CT are typically observed at a later stage [15]. With the rapid advancement of radiomics, researchers can extract more detailed and quantitative information from imaging data [16], [17]. By applying radiomics to CT image analysis, it may be possible to capture early changes in the bone marrow microenvironment before the onset of bone metastasis, thereby offering new possibilities for early diagnosis and intervention.
Therefore, this study combines routine chest CT scans performed on breast cancer patients with radiomics techniques to compare and identify the bone marrow of thoracic vertebrae in patients prior to breast cancer bone metastasis with those in patients without bone metastasis. The aim is to explore imaging techniques that can forecast the risk of developing potential bone metastases in breast cancer patients.
2. Methods
2.1. Study participants
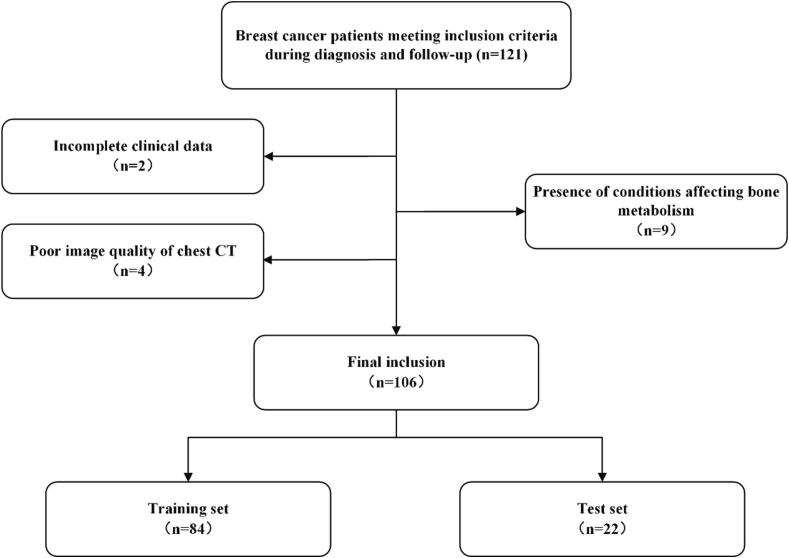
Approval for this study was obtained from the institutional review board, and the necessity for informed consent was waived because of the retrospective nature of the research. This study retrospectively gathered data from breast cancer patients diagnosed and monitored over a period of five years, spanning from January 2013 to September 2023. Data were gathered from The First Affiliated Hospital of Zhejiang Chinese Medical University and The Second Affiliated Hospital of Fujian Medical University. The inclusion criteria required: (1) a PET-CT scan must have been completed at the time of diagnosis for breast cancer, bone metastasis, or during the fifth year of follow-up; and (2) a non-contrast chest CT must have been conducted within one week prior to or following the PET-CT scan at the breast cancer diagnosis. Exclusion criteria encompassed: (1) incomplete clinical data; (2) poor image quality, such as artifacts affecting the analysis; and (3) conditions impacting bone metabolism, including severe osteoporosis, hyperparathyroidism, secondary bone disease from chronic kidney disease, or multiple myeloma. The data inclusion and exclusion details are presented in Fig. 1. During the follow-up period, patients were categorized into two groups: those with bone metastasis and those without, according to the PET-CT findings. The study also collected clinical data, including age, human epidermal growth factor receptor 2 (Her-2), estrogen receptor (ER), progesterone receptor (PR), Ki-67, and molecular subtypes of breast cancer.
Fig. 1.
Summary of patient recruitment and exclusions.
2.2. Image acquisition
Non-contrast chest CT imaging was acquired using three different multi-slice CT scanners: Brilliance 64 CT (Philips Healthcare, Eindhoven, the Netherlands), Brilliance iCT (Philips Healthcare, Eindhoven, the Netherlands), and the Somatom Definition AS (Siemens Healthineers, Erlangen, Germany). Across all three scanners, the imaging parameters were standardized, including a tube voltage of 120 kVp, automatic tube current modulation, a matrix size of 512 × 512, and a slice thickness of 5 mm. Image reconstruction was performed using the filtered back-projection technique for the Philips scanners, while the Siemens scanner utilized the “B70f” reconstruction kernel.
2.3. Image segmentation
In this study, CT image segmentation was performed using ITK-SNAP (version 4.0.0). The region of interest (ROI) for manual segmentation was chosen as the bone marrow of the sixth thoracic vertebral body. If this vertebral body showed fractures or other pathological conditions, the subsequent thoracic vertebra was selected as the ROI. The initial segmentation of all ROIs was conducted by a musculoskeletal imaging radiologist with three years of expertise. A second radiologist, with over ten years of experience in the same field, then reviewed and refined the ROIs.
2.4. Radiomic features extraction
A total of 1197 features were extracted from each ROI. These features were organized into 12 categories. The extracted feature set includes 234 first-order statistical features, 286 features derived from the Gray-Level Co-occurrence Matrix (GLCM), 182 features from the Gray Level Dependence Matrix (GLDM), 208 features from the Gray Level Run Length Matrix (GLRLM), 208 features from the Gray Level Size Zone Matrix (GLSZM), 65 features from the Neighboring Gray Tone Difference Matrix (NGTDM), and 14 shape-related features.
2.5. Machine learning models construction
Data preprocessing involved standardizing units, imputing missing values, and addressing outliers. The features with a Pearson correlation coefficient exceeding 0.90 were eliminated to mitigate redundancy. The minimum redundancy maximum relevance (mRMR) method was subsequently employed to select the top 20 features most closely associated with categorical variables, facilitating the identification of key imaging features. The mRMR method functions as a tool for selecting features, aimed at determining the most significant subset of features associated with the target variable. A variety of algorithms were utilized to construct predictive models, such as logistic regression, K-Nearest Neighbors (KNN), Support Vector Machines (SVM), Random Forest, Extremely Randomized Trees (ExtraTrees), Light Gradient Boosted Machine (LightGBM), and Multilayer Perceptron (MLP). The model demonstrating the highest area under the curve (AUC) on the test set was selected to compute the bone metastasis risk score, referred to as the radiomics score (Rad-score), which was subsequently used for nomogram development.
2.6. Clinical model and nomogram construction
The variable with the highest AUC from the radiomics model, referred to as the Rad-score, will be used alongside clinical features to build the nomogram. In this study, both the clinical model and the nomogram were created using multivariable logistic regression. A stepwise regression approach, guided by the Akaike information criterion (AIC), was used, combining forward selection and backward elimination techniques to optimize the model. The workflow of this study is presented in Fig. 2.
Fig. 2.
Overview of the processing workflow for this study.
2.7. Statistical analyses
Analyses of statistical data were conducted using R software (version 3.6.0; R Foundation). To compare categorical variables among groups with bone metastasis and those without, either Fisher's exact test or Pearson's χ2 test was employed. Meanwhile, the Kruskal-Wallis test was utilized for the evaluation of continuous variables. The models' predictive ability was evaluated by calculating the AUC and examining metrics including sensitivity, specificity, accuracy, negative predictive value (NPV), and positive predictive value (PPV). To assess the goodness of fit and identify potential overfitting, the Hosmer-Lemeshow test was utilized. Decision curve analysis (DCA) served to evaluate the practical application of the clinical model, radiomics model, and nomogram within a clinical environment. A P-value of less than 0.05 was established as the criterion for statistical significance across all analyses.
3. Results
3.1. Clinical features
In this study, 106 patients diagnosed with breast cancer were analyzed, among which 37 experienced the development of bone metastases within a five-year timeframe. The training set consisted of 29 individuals with bone metastases, whereas the testing set included 8 such patients. Analysis revealed no significant age differences, nor any disparities in ER, PR, Her-2, Ki-67, or molecular subtypes between the groups with and without bone metastases (all P > 0.05). The distribution of clinical features in both the training and testing sets is illustrated in Table 1, which also includes the P-values that compare the groups with and without bone metastases.
Table 1.
Clinical features in training and external test sets.
| Clinical Feature | Training Set (n = 84) | Test Set (n = 22) | P-value |
|---|---|---|---|
| Age, yr | 50.3 ± 9.5 | 51.4 ± 11.2 | 0.720 |
| ER | 0.098 | ||
| Positive | 52 | 12 | |
| Negative | 32 | 10 | |
| PR | 0.156 | ||
| < 20 % | 46 | 13 | |
| > 20 % | 38 | 9 | |
| Her-2 | 0.106 | ||
| Positive | 45 | 13 | |
| Negative | 39 | 9 | |
| Ki-67 | 0.797 | ||
| < 14 % | 67 | 20 | |
| > 14 % | 17 | 2 | |
| Molecular subtypes | 0.348 | ||
| HER-2 Positive (HR Positive) | 18 | 3 | |
| HER-2 Positive (HR Negative) | 19 | 10 | |
| Luminal A | 13 | 0 | |
| Luminal B | 21 | 7 | |
| Triple Negative | 11 | 1 | |
| Special | 2 | 1 |
ER estrogen receptor, Her-2 human epidermal growth factor receptor 2, PR progesterone receptor.
3.2. Evaluation of machine learning models
With the exception of the LightGBM model, all other machine learning models accurately predicted breast cancer bone metastasis within five years using thoracic vertebral bone marrow data (Table 2). The Hosmer-Lemeshow test results indicate that none of the machine learning models exhibit signs of overfitting (all P > 0.05). Among the various models examined, the model utilizing logistic regression demonstrated the highest AUC values within the test set. This model was constructed using twelve radiomic features, as illustrated in Fig. 3. The AUC values recorded for the logistic regression model were 0.959 (95 % CI: 0.923–––1.00) in the training set and 0.929 (95 % CI: 0.814–––1.00) in the test set. In both training and testing sets, a statistically significant difference in Rad-score was observed between the bone metastasis and non-bone metastasis groups (P both < 0.05; Fig. 4). Consequently, the logistic regression model was chosen for the subsequent development of the nomogram.
Table 2.
Predictive Performance of Each Model.
| Model | Set | ACC | AUC | 95 % CI | SEN | SPE | PPV | NPV |
|---|---|---|---|---|---|---|---|---|
| Logistic Regression | Train | 0.893 | 0.959 | 0.923–––0.996 | 0.828 | 0.927 | 0.857 | 0.911 |
| Logistic Regression | Test | 0.864 | 0.929 | 0.814–––1.000 | 0.750 | 0.929 | 0.857 | 0.867 |
| SVM | Train | 0.940 | 0.991 | 0.978–––1.000 | 0.966 | 0.927 | 0.875 | 0.981 |
| SVM | Test | 0.818 | 0.875 | 0.719–––1.000 | 0.625 | 0.929 | 0.833 | 0.812 |
| KNN | Train | 0.905 | 0.977 | 0.955–––1.000 | 0.828 | 0.945 | 0.889 | 0.912 |
| KNN | Test | 0.773 | 0.853 | 0.671–––1.000 | 0.500 | 0.929 | 0.800 | 0.765 |
| RandomForest | Train | 0.952 | 0.989 | 0.975–––1.000 | 0.897 | 0.982 | 0.963 | 0.947 |
| RandomForest | Test | 0.864 | 0.875 | 0.702–––1.000 | 0.750 | 0.929 | 0.857 | 0.867 |
| ExtraTrees | Train | 0.857 | 0.955 | 0.918–––0.993 | 0.931 | 0.818 | 0.730 | 0.957 |
| ExtraTrees | Test | 0.773 | 0.830 | 0.655–––1.000 | 0.625 | 0.857 | 0.714 | 0.800 |
| LightGBM | Train | 0.869 | 0.950 | 0.909–––0.991 | 0.931 | 0.836 | 0.750 | 0.958 |
| LightGBM | Test | 0.727 | 0.781 | 0.578–––0.984 | 0.750 | 0.714 | 0.600 | 0.833 |
| MLP | Train | 0.905 | 0.959 | 0.916–––1.000 | 0.828 | 0.945 | 0.889 | 0.912 |
| MLP | Test | 0.818 | 0.893 | 0.758–––1.000 | 0.750 | 0.857 | 0.750 | 0.857 |
| Clinical Model | Train | 0.631 | 0.654 | 0.528–––0.780 | 0.655 | 0.618 | 0.475 | 0.773 |
| Clinical Model | Test | 0.591 | 0.500 | 0.202–––0.780 | 0.375 | 0.714 | 0.429 | 0.667 |
| Nomogram | Train | 0.905 | 0.962 | 0.928–––0.997 | 0.862 | 0.927 | 0.862 | 0.927 |
| Nomogram | Test | 0.818 | 0.830 | 0.597–––1.000 | 0.750 | 0.857 | 0.750 | 0.857 |
AUC area under the curve, SEN Sensitivity, SPE Specificity, NPV negative predictive value, PPV positive predictive value.
Fig. 3.
Features and corresponding weights of the radiomics model constructed using the logistic regression algorithm.
Fig. 4.
Waterfall plots of the Rad-score distribution in the training set (A) and test set (B).
3.3. Comparison of predictive models
Through multivariate logistic regression analysis, variables including age, Ki-67, ER, HER-2, PR, and breast cancer molecular subtypes were incorporated into both the clinical model and the nomogram (Fig. 5). In the test set, the radiomics model developed with the logistic regression approach demonstrated the most elevated AUC value (Table 2, Fig. 6). DCA indicated that the radiomics model designed with the logistic regression method provides considerably greater clinical benefits compared to both the clinical model and the nomogram (Fig. 7).
Fig. 5.
Nomogram for predicting the 5-year risk of bone metastasis in breast cancer patients. ER estrogen receptor, Her-2 human epidermal growth factor receptor 2, PR progesterone receptor, Rad-score radiomics score.
Fig. 6.
ROC curves of the clinical model, radiomics model, and nomogram in the training and testing sets. ROC receiver operating characteristic.
Fig. 7.
Decision curve analysis curves for the clinical model, radiomics model, and nomogram.
4. Discussion
Micro-level changes may occur in the bone marrow environment of the thoracic spine in breast cancer patients prior to bone metastasis [13], [14]. These changes can be detected on CT images using radiomics techniques. This study found that the radiomics model constructed using logistic regression could accurately predict the occurrence of bone metastasis within the next five years in breast cancer patients, even without incorporating clinical data.
Bone metastasis commonly occurs and poses a severe complication for patients with breast cancer, greatly elevating the burden of the disease and the risk of mortality [18]. Increasing evidence suggests that significant biological changes take place in the bone marrow microenvironment prior to the emergence of bone metastasis [6], [19]. These alterations involve immune cell remodeling, cytokine secretion, and the interactions that take place between bone marrow stromal cells and tumor cells, collectively creating an environment conducive to tumor cell growth and metastasis [20], [21], [22]. The bone marrow microenvironment is often referred to as “fertile ground” for the migration and colonization of tumor cells [23], [24]. During the initial stages of bone metastasis in breast cancer, tumor cells infiltrate the bone marrow microenvironment via the bloodstream and are influenced by factors within this environment that enable their survival and proliferation [25]. Various cell types in the bone marrow, including stromal cells, osteoblasts, and osteoclasts, along with secreted growth factors and cytokines, interact with tumor cells through complex signaling pathways, facilitating their colonization and expansion [11], [12]. This study selected a single thoracic vertebral bone marrow as the subject of analysis because the microenvironmental changes in bone marrow that occur prior to bone metastasis are systemic.
Currently, radiomics research in breast cancer mainly emphasizes analyzing the tumor itself and its surrounding tissue features, including size, shape, density, and texture, to predict treatment response and patient prognosis [26], [27]. However, studies focusing on the thoracic spinal bone marrow in breast cancer patients are relatively limited. The thoracic spinal bone marrow, identified as a potential site for bone metastasis, may play a critical role in the development of breast cancer bone metastasis due to alterations in its microenvironment. Radiomics can differentiate between bone islands and metastases in the vertebrae [28]. It can also identify changes in vertebral bone density based on CT [29]. Radiomics has demonstrated the capability to detect microscopic bone changes in non-contrast CT scans. This study shows that radiomics based on chest CT can detect subtle vertebral alterations prior to the onset of bone metastasis in breast cancer patients, as well as forecast the risk of bone metastasis occurring within a five-year period through capturing quantitative features.
Radiomics provides an innovative method for comprehending the pathophysiological alterations occurring in the bone marrow microenvironment preceding bone metastasis in breast cancer. By extracting high-dimensional features from imaging data of thoracic vertebral bone marrow, radiomics can identify these microstructural changes, particularly subtle alterations in bone density and texture, which often represent early manifestations of bone marrow remodeling [29]. Radiomics can help capture details related to trabecular changes, reflecting stromal activation, immune cell dynamics, and cytokine network reorganization within the bone marrow [25], [28]. These quantitative features provide new insights into how tumor cells survive and proliferate within the “fertile ground” of the bone marrow and offer a non-invasive method for early risk assessment of bone metastasis. This capability has promising potential for patient management by identifying high-risk individuals before metastasis develops, thereby enabling timely interventions to delay or prevent bone metastasis.
The study has several limitations that should be acknowledged. First, the relatively small sample size may have hindered the identification of some potentially important radiomic features, which could restrict the applicability of the results to a broader demographic. Second, as a retrospective study, we were only able to determine whether bone metastasis occurred within the five-year follow-up period, but not the exact timing, due to the lack of regular PET-CT scans for all patients. The key to addressing this limitation is to conduct prospective studies with regular imaging follow-ups to accurately capture the timing of metastasis.
This study utilized radiomics to identify changes in features of the thoracic vertebral bone marrow environment on CT images before bone metastasis in breast cancer patients, consistent with the concept that the bone marrow microenvironment undergoes alterations before the occurrence of metastasis. Additionally, the logistic regression model based on radiomic features from routine chest CT scans accurately predicted the occurrence of bone metastasis within five years, demonstrating its potential as a tool for assessing bone metastasis risk.
CRediT authorship contribution statement
Hao-Nan Zhu: Writing – review & editing, Writing – original draft. Yi-Fan Guo: Writing – review & editing, Writing – original draft, Visualization. Ying-Min Lin: Validation, Data curation.Zhi-Chao Sun: Validation, Supervision, Methodology. Xi Zhu: Writing – review & editing, Writing – original draft, Methodology, Formal analysis. YuanZhe Li: Writing – review & editing, Writing – original draft, Validation, Supervision, Software, Resources, Project administration, Methodology, Investigation, Formal analysis, Data curation, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Contributor Information
Zhi-Chao Sun, Email: 20103004@zcmu.edu.cn.
Xi Zhu, Email: yzdxlcyxyzhuxi@qq.com.
YuanZhe Li, Email: ctmr@fjmu.edu.cn.
References
- 1.Mundy G.R. Metastasis to bone: causes, consequences and therapeutic opportunities. Nat. Rev. Cancer. 2002;2:584–593. doi: 10.1038/nrc867. [DOI] [PubMed] [Google Scholar]
- 2.Weilbaecher K.N., Guise T.A., McCauley L.K. Cancer to bone: a fatal attraction. Nat. Rev. Cancer. 2011;11:411–425. doi: 10.1038/nrc3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xiong Z., Deng G., Huang X., et al. Bone metastasis pattern in initial metastatic breast cancer: a population-based study. Cancer Manag. Res. 2018;10:287–295. doi: 10.2147/cmar.S155524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ibragimova M.K., Tsyganov M.M., Kravtsova E.A., Tsydenova I.A., Litviakov N.V. Organ-specificity of breast cancer metastasis. Int. J. Mol. Sci. 2023;24 doi: 10.3390/ijms242115625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liang Y., Zhang H., Song X., Yang Q. Metastatic heterogeneity of breast cancer: Molecular mechanism and potential therapeutic targets. Semin. Cancer Biol. 2020;60:14–27. doi: 10.1016/j.semcancer.2019.08.012. [DOI] [PubMed] [Google Scholar]
- 6.Clézardin P., Coleman R., Puppo M., et al. Bone metastasis: mechanisms, therapies, and biomarkers. Physiol. Rev. 2021;101:797–855. doi: 10.1152/physrev.00012.2019. [DOI] [PubMed] [Google Scholar]
- 7.Coleman R.E., Croucher P.I., Padhani A.R., et al. Bone metastases. Nat. Rev. Dis. Primers. 2020;6:83. doi: 10.1038/s41572-020-00216-3. [DOI] [PubMed] [Google Scholar]
- 8.Futakuchi M., Fukamachi K., Suzui M. Heterogeneity of tumor cells in the bone microenvironment: mechanisms and therapeutic targets for bone metastasis of prostate or breast cancer. Adv. Drug Deliv. Rev. 2016;99:206–211. doi: 10.1016/j.addr.2015.11.017. [DOI] [PubMed] [Google Scholar]
- 9.Hofbauer L.C., Bozec A., Rauner M., et al. Novel approaches to target the microenvironment of bone metastasis. Nat. Rev. Clin. Oncol. 2021;18:488–505. doi: 10.1038/s41571-021-00499-9. [DOI] [PubMed] [Google Scholar]
- 10.Gdowski A.S., Ranjan A., Vishwanatha J.K. Current concepts in bone metastasis, contemporary therapeutic strategies and ongoing clinical trials. J. Exp. Clin. Cancer Res. 2017;36:108. doi: 10.1186/s13046-017-0578-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang P., Gao W., Ma T., et al. CD137 promotes bone metastasis of breast cancer by enhancing the migration and osteoclast differentiation of monocytes/macrophages. Theranostics. 2019;9:2950–2966. doi: 10.7150/thno.29617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li X., Liang Y., Lian C., et al. CST6 protein and peptides inhibit breast cancer bone metastasis by suppressing CTSB activity and osteoclastogenesis. Theranostics. 2021;11:9821–9832. doi: 10.7150/thno.62187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yip R.K.H., Rimes J.S., Capaldo B.D., et al. Mammary tumour cells remodel the bone marrow vascular microenvironment to support metastasis. Nat. Commun. 2021;12:6920. doi: 10.1038/s41467-021-26556-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bado I.L., Zhang W., Hu J., et al. The bone microenvironment increases phenotypic plasticity of ER(+) breast cancer cells. Dev. Cell. 2021;56:1100–1117.e1109. doi: 10.1016/j.devcel.2021.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Catalano O.A., Nicolai E., Rosen B.R., et al. Comparison of CE-FDG-PET/CT with CE-FDG-PET/MR in the evaluation of osseous metastases in breast cancer patients. Br. J. Cancer. 2015;112:1452–1460. doi: 10.1038/bjc.2015.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lambin P., Rios-Velazquez E., Leijenaar R., et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur. J. Cancer. 2012;48:441–446. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gillies R.J., Kinahan P.E., Hricak H. Radiomics: images are more than pictures, they are Data. Radiology. 2016;278:563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hofbauer L.C., Rachner T.D., Coleman R.E., Jakob F. Endocrine aspects of bone metastases. Lancet Diabetes Endocrinol. 2014;2:500–512. doi: 10.1016/s2213-8587(13)70203-1. [DOI] [PubMed] [Google Scholar]
- 19.Ma X., Yu J. Role of the bone microenvironment in bone metastasis of malignant tumors - therapeutic implications. Cell. Oncol. (Dordr.) 2020;43:751–761. doi: 10.1007/s13402-020-00512-w. [DOI] [PubMed] [Google Scholar]
- 20.Haider M.T., Ridlmaier N., Smit D.J., Taipaleenmäki H. Interleukins as mediators of the tumor cell-bone cell crosstalk during the initiation of breast cancer bone metastasis. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms22062898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Puppo M., Valluru M.K., Clézardin P. MicroRNAs and their roles in breast cancer bone metastasis. Curr. Osteoporos. Rep. 2021;19:256–263. doi: 10.1007/s11914-021-00677-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shin E., Koo J.S. The role of adipokines and bone marrow adipocytes in breast cancer bone metastasis. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21144967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peinado H., Zhang H., Matei I.R., et al. Pre-metastatic niches: organ-specific homes for metastases. Nat. Rev. Cancer. 2017;17:302–317. doi: 10.1038/nrc.2017.6. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y., Hu Y., Wang M., Wang M., Xu Y. The role of breast cancer cells in bone metastasis: suitable seeds for nourishing soil. Curr. Osteoporos. Rep. 2024;22:28–43. doi: 10.1007/s11914-023-00849-9. [DOI] [PubMed] [Google Scholar]
- 25.Hanahan D., Coussens L.M. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell. 2012;21:309–322. doi: 10.1016/j.ccr.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 26.Fowler A.M., Strigel R.M. Clinical advances in PET-MRI for breast cancer. Lancet Oncol. 2022;23:e32–e43. doi: 10.1016/s1470-2045(21)00577-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi Z., Huang X., Cheng Z., et al. MRI-based quantification of intratumoral heterogeneity for predicting treatment response to neoadjuvant chemotherapy in breast cancer. Radiology. 2023;308:e222830. doi: 10.1148/radiol.222830. [DOI] [PubMed] [Google Scholar]
- 28.Hong J.H., Jung J.Y., Jo A., et al. Development and validation of a radiomics model for differentiating bone islands and osteoblastic bone metastases at abdominal CT. Radiology. 2021;299:626–632. doi: 10.1148/radiol.2021203783. [DOI] [PubMed] [Google Scholar]
- 29.Jiang Y.W., Xu X.J., Wang R., Chen C.M. Radiomics analysis based on lumbar spine CT to detect osteoporosis. Eur. Radiol. 2022;32:8019–8026. doi: 10.1007/s00330-022-08805-4. [DOI] [PMC free article] [PubMed] [Google Scholar]