Abstract
The role of the bone marrow (BM) microenvironment in regulating the antitumor immune response in Waldenstrom macroglobulinemia (WM) remains poorly understood. Here we transcriptionally and phenotypically profiled non-malignant (CD19- CD138-) BM cells from WM patients with a focus on myeloid derived suppressive cells (MDSCs) to provide a deeper understanding of their role in WM. We found that HLA-DRlowCD11b+CD33+ MDSCs were significantly increased in WM patients as compared to normal controls, with an expansion of predominantly polymorphonuclear (PMN)-MDSCs. Single-cell immunogenomic profiling of WM MDSCs identified an immune-suppressive gene signature with upregulated inflammatory pathways associated with interferon and tumor necrosis factor (TNF) signaling. Gene signatures associated with an inflammatory and immune suppressive environment were predominately expressed in PMN-MDSCs. In vitro, WM PMN-MDSCs demonstrated robust T-cell suppression and their viability and expansion was notably enhanced by granulocyte colony stimulating factor (G-CSF) and TNFα. Furthermore, BM malignant B-cells attracted PMN-MDSCs to a greater degree than monocytic MDSCs. Collectively, these data suggest that malignant WM B cells actively recruit PMN-MDSCs which promote an immunosuppressive BM microenvironment through a direct T cell inhibition, while release of G-CSF/TNFα in the microenvironment further promotes PMN-MDSC expansion and in turn immune suppression. Targeting PMN-MDSCs may therefore represent a potential therapeutic strategy in patients with WM.
Subject terms: B-cell lymphoma, Tumour immunology
Background
Waldenstrom macroglobulinemia (WM) is a rare hematologic malignancy characterized by the accumulation of lymphoplasmacytic lymphoma (LPL) cells in the bone marrow (BM) and increased production of immunoglobulin M (IgM) [1]. Some patients may be asymptomatic or ‘smoldering’ (SMW) at the time of diagnosis, while others present with clinical symptoms due to the elevated IgM serum levels and lymphoplasmacytic cell infiltration in the BM, lymph nodes and spleen (symptomatic WM). Genetic studies have found mutations in MYD88 (86–97%) and CXCR4 (24%) frequently associated with WM patients [2, 3], however having MYD88L265P mutation is insufficient since this alteration can also be detected in other B cell malignancies such as diffuse large b cell lymphoma [4], central nervous system (CNS) [5] and testicular lymphomas [6]. Despite extensive genetic studies on WM tumor cells, the mechanistic basis of tumor development and growth remains incompletely understood.
In the last decade, there has been an increased attention to the role of the microenvironment which can profoundly affect the tumor cells, favoring their growth and survival. Using a multiomics approach, we previously identified three molecular clusters in the spectrum of WM and IgM-monoclonal gammopathy of undetermined significance (MGUS), each characterized by a distinct transcriptional, immune and metabolic profile. Specifically, we found that the group with worse prognosis displayed an immune suppressive transcriptional and phenotypic profile [7]. These findings are in line with prior studies that associated disease progression from IgM-MGUS to WM with myeloid inflammation, resulting in immune dysfunction, and immunosuppression [8].
The process of myeloid formation, or myelopoiesis, is vital for generating various immune cells like monocytes, granulocytes, and dendritic cells. However, conditions such as chronic infection or cancer can impair this process, leading to reduced peripheral myeloid cell levels. This reduction triggers an emergency response called “emergency myelopoiesis,” characterized by increased production and migration of myeloid cells, even in their immature state. This rushed replenishment can cause the accumulation of immature or dysfunctional myeloid cells with strong immunosuppressive traits, contributing to an immunosuppressive microenvironment and worsening immune dysfunction in these conditions [9, 10]. The immunosuppressive activity of monocytes was first reported 30 years ago and these cells were later named MDSCs [11]. Generally, healthy individuals have very low numbers of MDSCs which regulate immune responses and tissue repair [12]. MDSCs in humans are divided into two categories - monocytic-MDSCs (M-MDSCs) that are CD11b+ CD33+ HLA-DR lowCD14+ CD15- and polymorphonuclear MDSCs (PMN-MDSCs) characterized by a CD11b+ CD33+ HLA-DR lowCD14- CD15+/CD66b+ phenotype. The MDSCs have shown tumor promoting and immune-suppressive activity in various cancers including different hematological malignancies [9, 13–16]. In this study, we found that the immunosuppressive environment in WM is associated with the presence of increased numbers of MDSCs. To then understand the interactions better between MDSCs, T-cells, and malignant lymphoplasmacytic cells (CD19+ CD138+) in WM, we employed single-cell genomic tools, in combination with functional studies, to define the gene signature of MDSCs in WM and determine the MDSC subset likely responsible for T-cell inhibition.
Materials and methods
Patients
BM specimens from 24 patients with newly diagnosed, untreated WM were included in the study. All patients were diagnosed with WM based on the diagnostic criteria from the 11th International Workshop on Waldenstrom’s macroglobulinemia [17]. Seven patients were asymptomatic, consistent with SWM. Seventeen normal controls provided BM specimens at the time of having a hip replacement. BM specimens from 3 additional WM patients were obtained at the time of achieving a complete remission post therapy to serve as further controls. To ensure that the effect of malignant B-cells on the immune cells in the BM could be adequately evaluated, all patients had at least 10% involvement of the BM by LPL. Written informed consent was obtained from all patients as per the Mayo Clinic Institutional Review Board (IRB), IRB-118-01 requirements and the study was conducted in accordance with the Declaration of Helsinki. Clinical details of the patients are provided in Supplementary Table 1. WM patients were characterized based on their clinicopathological diagnosis that included serum IgM and ß2 microglobulin levels, treatment, and the International Prognostic Scoring System for Waldenstrom Macroglobulinemia.
BM Cell isolation and purification
BM tissue biopsy aspirates from patients with WM, SWM, and NBM were gently minced over a wire mesh screen to obtain a cell suspension. ACK lysis was performed to remove the red blood cells. CD19+ B cells and CD138+ plasma cells were isolated using positive selection with Human CD19 B-cell Enrichment Kit (StemCell Technologies, cat no. 17854) and Human CD138+ cell enrichment Kit (StemCell Technologies, cat no. 17877), respectively. For the G-CSF/TNF and T-cell suppression studies CD3+ T cells were removed from the samples, using Human CD3 positive selection kit (Stem cell technologies,17951). For the phenotypic studies, after removal of CD19+, CD138+, and CD3+ cells from the BM, CD66b + cells were then isolated by EasySep™ HLA Chimerism Whole Blood CD66b Positive Selection Kit (Stem cell technologies,17882). The migration study focused on CD11b+CD33+ cells isolated from WM bone marrow. These cells were sorted using MACS Quant Tyto for transwell assay.
Cell lines
BCWM.1 and MWCL-1 cell lines were utilized as well-established WM models [18, 19]. These cells were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum with 2 mM L-glutamine (Mediatech), 100 U/mL penicillin and 10 ug streptomycin.
Flow cytometry analysis
BM mononuclear cells from WM patients, depleted of LPL cells (CD19/CD138+ cells; n = 11), and normal controls (n = 13) were stained to identify the monocytic- or granulocytic/polymorphonuclear- MDSCs (Mono-MDSC or Gr/PMN-MDSC). The markers used included anti-human HLA-DR-v500 (Biolegend,307646), anti-human Lineage markers-APC (CD3, CD56, CD19), anti-humanCD11b-BV410 (Biolegend, 393114), anti-humanCD33-Percp 5.5 (Biolegend,303414), anti-humanCD14-PE-Cy7 (Biolegend,325618), anti-human CD66b PE (Biolegend,305106) and anti-humanCD15-FITC (Biolegend, 323004). Cells were also stained with fixable viability dye eFlourTM 780 (eBiosciences, 65-0865-14). Stained cells were analyzed on a Becton Dickinson (BD) FACS CANTO II, and data were processed by FlowJo software (V10.4).
For cytokine stimulation studies, CD19- CD138-CD3- cells from WM BM+ were treated with G-CSF (10 ng/ml) and TNF (10 ng/ml) for 24 h and 48 h and flow cytometry was performed. This study investigated three experimental groups namely: 1. CD19- CD138- CD3- cells alone, 2. CD19- CD138- CD3- cells treated with 10 ng G-CSF, and 3. CD19- CD138- CD3- cells treated with 10 ng TNF-α. The study involved assessments at three time points: 0 h, 24 h, and 48 h. Flow cytometry analysis utilized markers for viability, lineage (Lin), HLA-DR, CD11b, CD33, CD15, and CD66b. PMN-MDSCs (Lin- HLA-DR- CD11b+ CD33+ CD15+ CD66b+) were selected, and their percentages calculated. Due to a potential decrease in viability of PMN-MDSCs between 24 and 48 h (n = 4), we used the 0-h time point as the control. The objective was to investigate whether treatment with G-CSF and TNF-α could promote expansion and enhance survival of PMN-MDSCs and treated cells were compared to the 0-h baseline.
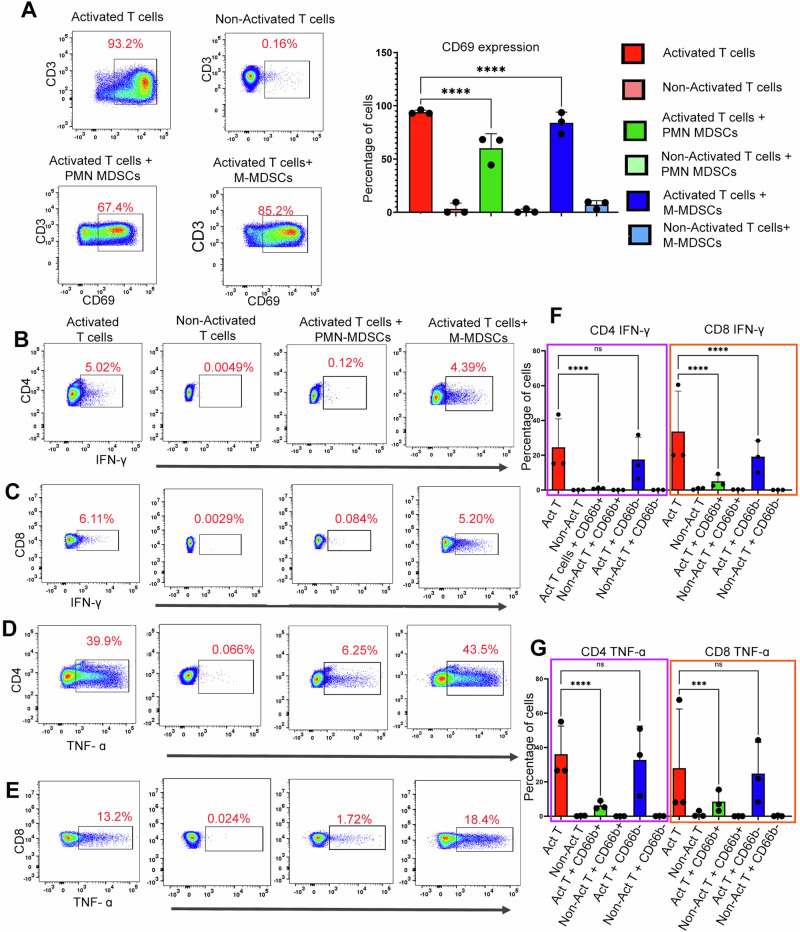
To evaluate the immunosuppressive nature of the cells, CD66b+ PMN-MDSCs were co-cultured with CD3+ T-cells (enriched from the healthy PBMCs) after CD3 and CD28 T cell activation. The ratio of CD66b+ MDSCs and CD3+ T cell was 1:1 for this study. CD69 expression was measured to monitor the T cell activation [20]. Also, IFNγ and TNFα expression were measured in CD4+ and CD8+ T cells.
Mass cytometry (CyTOF)
CyTOF was performed on 10 WM patients as previously described by Mondello et al. [21]. Patients’ samples were grouped according to their disease status (NBM, SWM, symptomatic WM and WM patients in remission post treatment). Briefly, the normalized FCS files belonging to each patient group were first concatenated using Cytobank v10.4 (cytobank.org). Concatenated files, separate for each group NBM (n = 3), SWM (n = 3), symptomatic WM (n = 7) were analyzed. Further, an additional 3 BM specimens from WM patients were analyzed at the time of achieving a complete remission post therapy. The gating strategy first removed cells with lineage markers for T-, B- and NK cells, and then focused on MDSC (CD11b+CD33+) subtypes. viSNE maps were plotted to study the different subtypes of MDSCs.
CITE-sequencing and analysis
Cellular Indexing of Transcriptomes and Epitopes by Sequencing (CITE-Seq) was performed on freshly digested NBM and WM BM cells using the Chromium Single Cell 5’ Reagent and Gel Bead Kits (10x Genomics, USA). Cells were isolated and resuspended in labeling buffer, incubated with a cocktail of 24 TotalSeq-C antibodies (BioLegend) (Supplementary Table 2), and washed before submission for single-cell partitioning. Barcoded Gel Beads were used to create Gel Beads-In-Emulsion (GEMs), which underwent various steps to generate uniquely barcoded molecules for gene expression and cell surface protein libraries. Sequencing was performed on the HiSeq 4000 platform. Gene expression and cell-surface libraries were prepared using 10× Genomics protocols and sequenced on Illumina NovaSeq 6000. Data were analyzed using CellRanger software, Seurat, and clusterProfiler package, resulting in 15 clusters spanning healthy and diseased samples. Subclustering of MDSC cells identified two major subtypes (CD66b+ and CD66b-) for further analysis.
MDSCs sorting and transwell assay
For the migration assay, MDSCs were placed in the upper chamber of the transwell. We utilized MCP-1 or 10% FBS in RPMI in the lower chamber of the transwell as positive controls. BCWM.1 cells and BCWM.1 supernatant. The supernatant from BCWM.1 cells was obtained after a 24-h culture, and this cultured medium was utilized in a 1:1 ratio with RPMI. Healthy B cells and RPMI without FBS were considered as negative controls.
MDSCs sorting and qPCR
For real-time qPCR analysis, we conducted two sets of validations: 1. Comparing WM vs NBM MDSCs (n = 3 WM and n = 3 NBM) and 2. Comparing PMN vs M-MDSCs (n = 2 WM). Sorting was performed using the MACS Quant Tyto Cell Sorter. Initially, WM bone marrow cells were sorted for CD19 and CD138 cells (Stem cell, positive isolation kit), followed by staining of the negative fraction with CD11b (BV421, Biolegend), CD33 (APC, Biolegend), CD14 (PeCy7, biolegend), and CD15 (FITC, biolegend) for 30 min on ice, with subsequent washing. The washed cells were loaded into high-speed cartridges (MACS Quant® Tyto® Cartridges HS), and CD11b+ CD33+ cells, CD11b+ CD33+ CD14+ cells, and CD11b+ CD33+ CD15+ cells were sorted. RNA (Qiagen) and cDNA were prepared (Thermo Fisher) according to the user manual instructions. Primers targeting HIF-1α, CXCR4, MPO, and AZU1 (OriGene) were used for the qPCR analysis with SYBR Green (Thermo Fisher).
Statistical analysis
Comparison of quantitative data between groups was done by Student’s t test or one-way.
ANOVA test (assuming normal distribution). Statistical analysis was done in GraphPad Prism (version 8.0.0 for Windows, GraphPad Software, San Diego, California USA). All reported P-values were two-sided, and P < 0.05 were considered statistically significant (* p < 0.05; ** p < 0.01; *** p < 0.001).
Results
PMN-MDSCs are significantly enriched in the WM bone marrow microenvironment
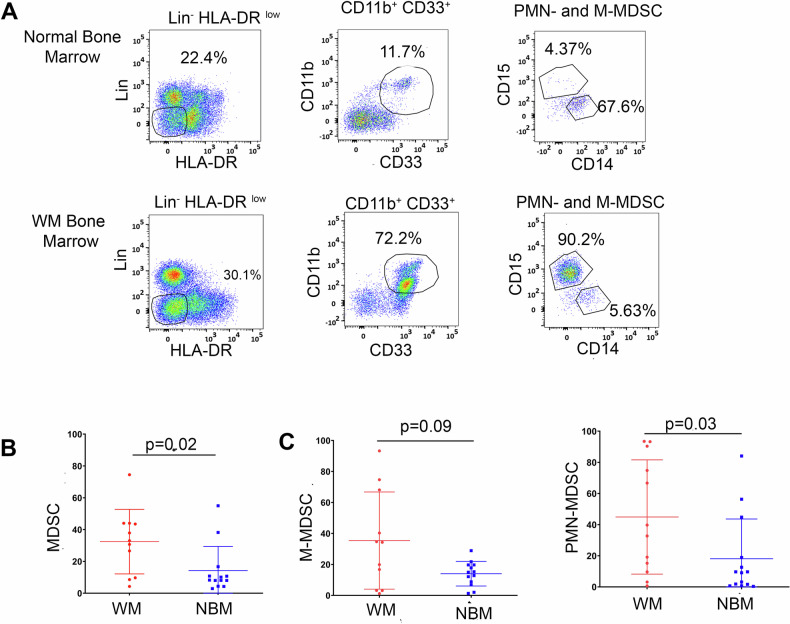
To determine the prevalence of MDSCs in the BM microenvironment of WM patients, we first performed flow cytometry (Fig. 1A) on negatively sorted CD19- CD138- BM cells from patients with WM (n = 11) and from NBM (n = 13). We analyzed the percentage of MDSCs (Lin- HLA-DRlowCD11b+CD33+) and found that MDSCs were significantly expanded in the BM of WM patients as compared to NBM (p = 0.02; Fig. 1B). CD11b+CD33+ cells were further classified by CD14 and CD15 expression to define M-MDSCs (CD14+) and PMN-MDSCs (CD15+) (Fig. 1A). We observed that both PMN-MDSCs (p = 0.03) and M-MDSCs (p = 0.09) were increased in WM-BM samples as opposed to NBM samples (Fig. 1C). The median percentage of PMN-MDSCs, relative to the total MDSCs population (i.e., CD11b+ CD33+), in WM patients was 36.2% (range:0.66–93.50%) as compared to NBM patients which was 9.3% (range:0.66–84.10%). Further, M-MDSCs, the median percentage and range in WM and NBM was 34.3% (range:1.22–93.3%) and 15% (range:1.31–28.9%) respectively (Fig. 1C).
Fig. 1. Expansion of PMN-MDSCs in WM patients.
A Gating strategy of PMN-MDSCs and M-MDSCs in NBM and WM bone marrow by flow cytometry analysis. HLA-DR-/low and Lineage- (CD3 CD19 CD138 CD56) cells were first selected, and then CD11b+ CD33+ population was further gated as CD15 (PMN-MDSCs) and CD14 (M-MDSCs). B Statistical analysis of total MDSCs (CD11b+ CD33+) in WM (n = 13) and NBM (n = 11). C Statistical analysis of PMN-MDSCs (Right side) and M-MDSCs (Left side) in NBM (n = 11) and WM (n = 13). *, P < 0.01.
Three phenotypically distinct subsets of MDSCs are identified in the WM bone marrow
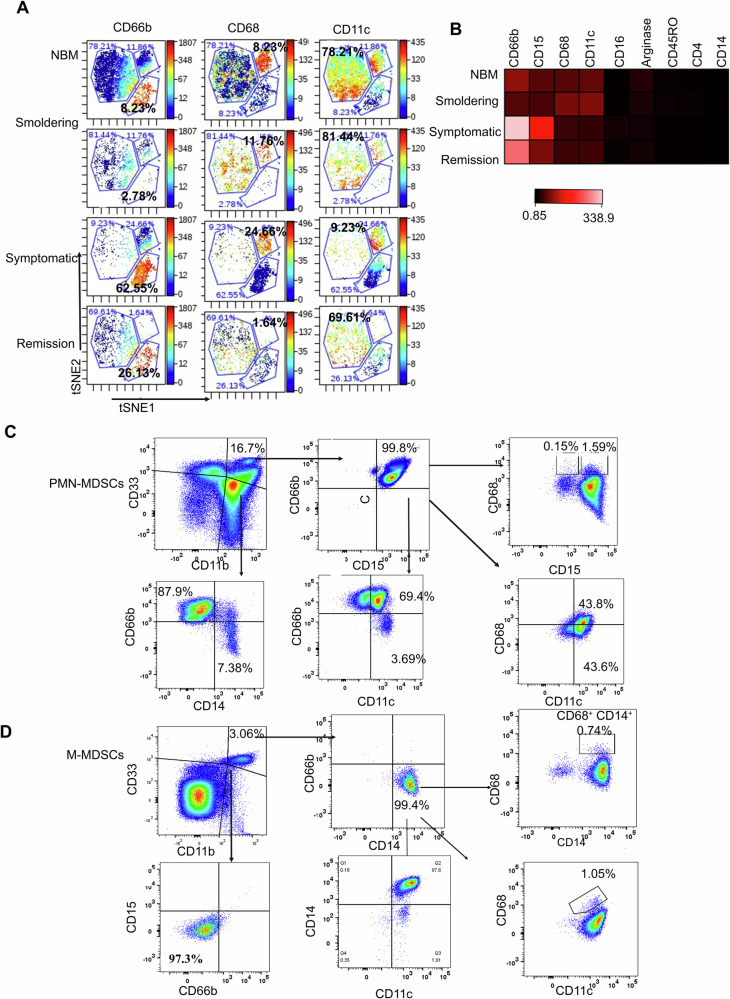
Next, we explored the phenotype of BM MDSCs using mass cytometry (CyTOF) in NBM (n = 3), SWM (n = 3), symptomatic WM (n = 7), as well as WM patients in remission post treatment (n = 3). Interestingly, 3 phenotypically distinct subsets of CD11b+CD33+ MDSCs were identified (Fig. 2A, B). One subset expressed high levels of CD66b, were CD15+ and CD16+, but lacked expression of CD68. These CD66b+ cells had a phenotype consistent with PMN-MDSCs and were more expanded in symptomatic WM patients (62.55%) compared to NBM (8.23%), SWM (2.78%) and WM patients in remission (26.13%) (Fig. 2A, B). A second MDSC subset had bright expression of CD15 and CD68, with some expression of CD45RO, but had low expression of CD66b. This subset remained quantitatively constant in NBM (8.23%), SWM (11.76%), and symptomatic WM samples (24.66%), but was less frequent in previously treated patients (1.64%). A third subset had high expression of CD11c maker (CD11c is a type I transmembrane protein that is expressed on monocytes, granulocytes, dendritic cells, and macrophages) and moderate expression of CD66b, CD15, and CD68 (CD68 is associated with inflammation with its involvement of monocytes/macrophages). This subset was highly represented in NBM (78.24%), SWM (81.44%), and WM patients in remission (69.61%), but was decreased in symptomatic WM patients (9.23%) (Fig. 2B). We then isolated CD66b+ and CD66b- cells to further define their phenotypes. The CD66b+ subset demonstrated characteristics of PMN-MDSCs with elevated CD15 expression (Fig. 2C). Conversely, the CD66b- population exhibited traits consistent with M-MDSCs exhibiting increased expression of CD14 (Fig. 2D).
Fig. 2. Identification of PMN-MDSC subtypes in the WM bone marrow.
A t-SNE plots showing the expression of three subsets of MDSCs in NBM (n = 3), Smoldering (n = 3), symptomatic (n = 5), and Remission (n = 3), namely CD66b, CD68, and CD11c. B Heatmap showing the expression of CD66b, CD15, CD68, CD11c, CD16, Arginase, CD45RO and CD14. C CD66b+ cells sorted from the WM BM showing its PMN-MDSC phenotype expressing CD15 co-expression with CD66b. D CD66b- cells sorted from the WM BM showing its M-MDSC phenotype expressing CD14.
MDSCs exhibited upregulated inflammation and metabolic signatures
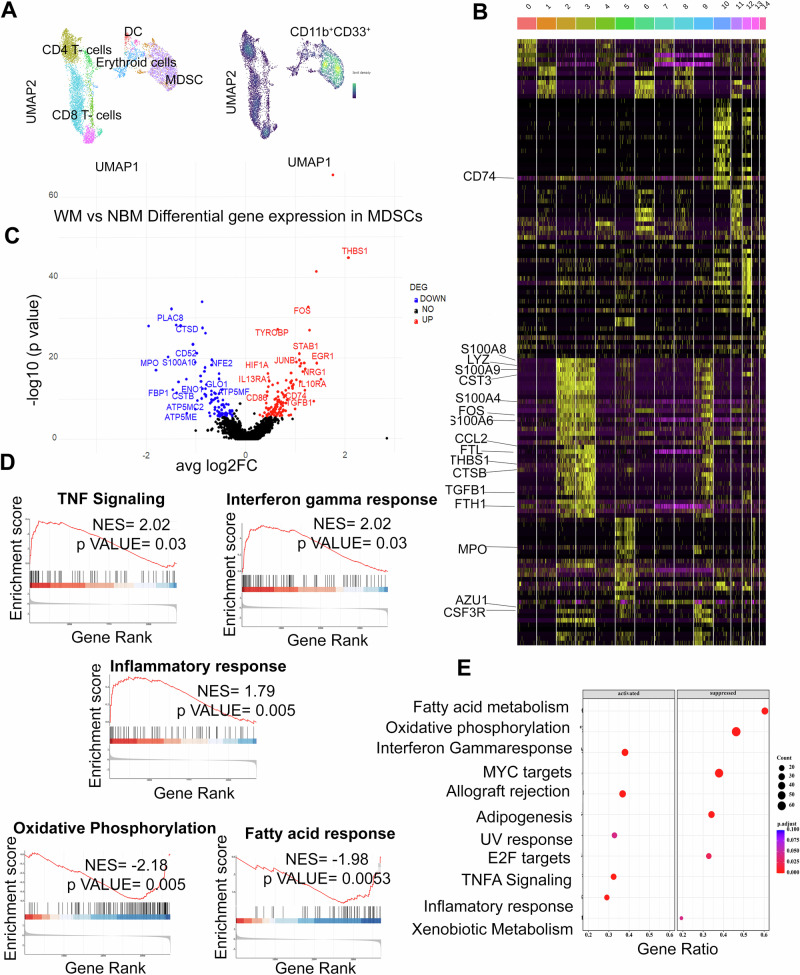
To further characterize MDSCs, we performed CITE-seq analysis on BM cells from patients with WM (n = 3) or NBM (n = 2). After B-cell and plasma cell exclusion, all mononuclear cells underwent unsupervised clustering and uniform manifold approximation and projection (UMAP) analysis [22] that identified 15 distinct clusters (Fig. 3A, B). Each cluster then underwent meticulous annotation based on their discernible phenotypes. Notably, Clusters 2, 3, and 9 displayed CD11b and CD33 expression, characteristic of the MDSCs phenotype. Further exploration highlighted a shared gene signature abundant in S100 Calcium Binding Protein A8 (S100A8), S100A9, Transforming Growth Factor Beta 1 (TGFB1), Colony Stimulating Factor 3 Receptor (CSF3R), Lysozyme (LYZ), Fos Proto-Oncogene, AP-1 Transcription Factor Subunit (FOS), and C-C Motif Chemokine Ligand 2 (CCL2) across these clusters (Fig. 3B and Supplementary Fig. 2). The detailed gene signatures within these clusters further elucidate their distinctive molecular profiles: Cluster 2 exhibited prominent expression of IL13RA1, IL17RA, IL1B, IL10RB, STAT1, STAT6, CSF1R, CSF3R. Cluster 3 displayed a distinct gene signature, including STAT2, STAT6, CSF1R, CSF3R, SOD2, and CD84. Cluster 9 expressed AZU1, MPO, CD163, FOS, CD36, LYZ, and CCL2 as part of its gene signature (Supplementary Fig. 2).
Fig. 3. Multi-omic analysis of CD11b±CD33± MDSCs in WM bone marrow.
A. UMAP plot visualization of intratumoral bone marrow (left side) and CD11b+ CD33+ MDSCs (Right side) detected in NBM (n = 2) and WM (n = 3). The clusters are colored and distinctly labeled. B Heatmap showing a relative expression of marker genes of each cluster. Cluster 2, 3, and 9 represents MDSCs (CD11b+CD33+) cells. C Volcano plot of CITE-seq transcriptome data displaying the gene expression pattern for MDSCs i.e., CD11b+ CD33+ cells in NBM (n = 2) and WM (n = 3). Significant differentially expressed genes (log2 fold change ≥ 0.25, adjusted p ≥ 10e-6) in WM vs NBM MDSCs. The highly upregulated and downregulated genes are shown in red and blue respectively. D The enrichment score curve shows alteration of pathways involving TNF signaling, Interferon-gamma response, Inflammatory response, allograft rejection, oxidative phosphorylation, and fatty acid response in MDSCs. The enrichment score normalized (NES) greater or less than zero signifies upregulation and downregulation respectively. The plots were generated using GSEA analysis. E Scatter plot of enrichment GSEA pathways. The Y-axis represents the name of the pathways and X-axis represents the gene ratio.
Differential gene expression analysis of WM MDSCs as compared to NBM MDSCs identified 146 upregulated and 94 downregulated genes (p ≤ 0.05). A detailed list of genes is provided in Supplementary Data 3. Briefly, THBS1, FOS, HIF1A, JunB, CD86, IL13RA1 and TGFBI, were significantly upregulated in WM-MDSCs. These genes have been associated with the development of MDSCs [23–27]. On the other hand, glycolysis related genes including fructose-bisphosphatase 1(FBP1) and enolase 1(ENO1), and genes regulating the citric acid cycle namely, ATP synthase membrane subunit c locus 2(ATP5MC2), ATP synthase membrane subunit e (ATP5ME) and f (ATP5MF) were downregulated, indicating inhibition of metabolic pathways [28]. Additionally, gene set enrichment analysis (GSEA) showed positive enrichment for interferon gamma signaling, TNFA signaling, and an inflammatory response in WM (Fig. 3D, E). All these pathways may promote MDSC generation and expansion [29]. On the other hand, we observed downregulation of cell metabolism signature, namely oxidative phosphorylation, and fatty acid metabolism in WM patients, which may promote tumor growth.
PMN- MDSCs exhibit distinct immune signature associated with poor outcome
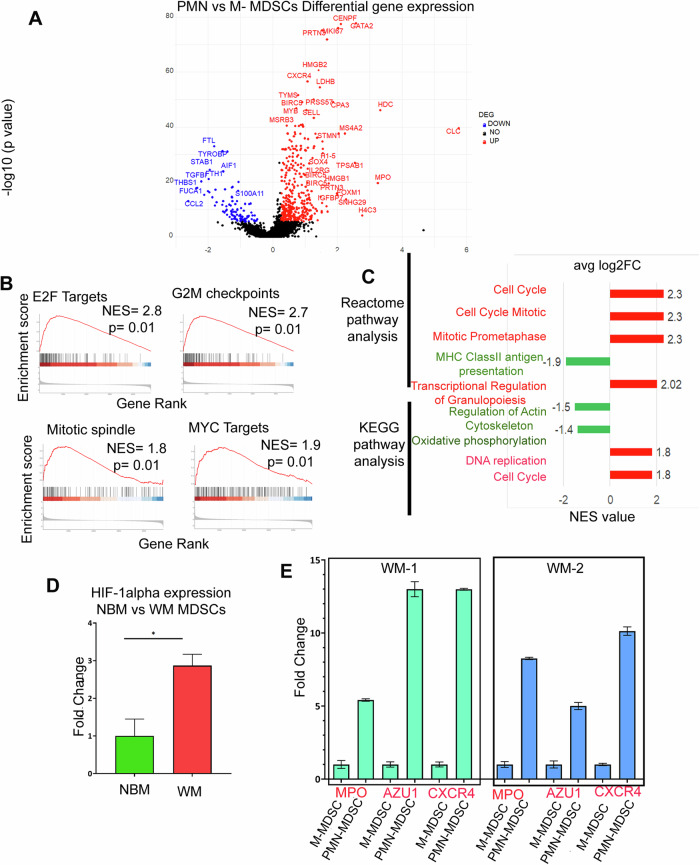
Given that PMN-MDSCs (CD66b+/CD15+) are expanded in symptomatic WM patients, we focused our analysis to better characterize the CD66b+ PMN-MDSCs cluster. Differential gene expression between CD66b+ PMN-MDSCs and CD66b- M-MDSCs identified 249 upregulated and 100 downregulated genes. CD66b+ MDSCs were characterized by genes involved in inflammation and growth, such as proteinase 3 (PRTN3), CXCR4, SOX4, IL2RG, and insulin-like growth factor-binding protein 7 (IGFBP7) (Fig. 4A). We also found upregulation of nuclear proliferative markers including MKI67, cell cycle and antiapoptotic genes including survivin, HMGB1, HMGB2, and immune suppressive markers such as myeloperoxidase (MPO), azurocidin 1 (AZU1), elastase, neutrophil elastase (ELANE) and centromere protein F (CENPF). Additionally, CD59 (an inhibitory gene for the complement system) and Lactate Dehydrogenase B (LDHB) were also significantly upregulated (Fig. 4A). Notably, these genes have been previously associated with MDSC growth and expansion [30–32]. GSEA showed a positive enrichment for gene sets related to cell cycle, mitotic spindle, and DNA replication, suggesting a growth pressure. In contrast, there was a negative correlation for antigen presentation, genes involved in cytoskeleton structure and oxidative phosphorylation, supporting putative immune escape mechanisms. Altogether, this data shows that CD66b+ PMN-MDSCs have a distinct transcriptomic profile with significant enrichment for genes associated with cell growth and proliferation, suppression of antigen presentation, and oxidative phosphorylation, all of which may collectively be part of the immune suppressive mechanisms of PMN-MDSCs.
Fig. 4. Transcriptomic profile of PMN-MDSCs and M-MDSCs.
A Volcano plot of the transcriptome profile displaying the pattern of differentially expressed genes in PMN (CD11b+CD33+ HLADR-/low CD15+/CD66b+) vs M-MDSCs (CD11b+CD33+ HLADR-/low CD14+). Significantly expressed genes (log2 fold change ≥ 0.25, adjusted p ≥ 10e-6) are shown in the plot, where red and blue represents upregulated and downregulated genes respectively. B The enrichment score curve shows alteration of pathways involving E2F Targets, G2M checkpoints, Mitotic spindle, and MYC targets. The enrichment score normalized (NES) greater or less than zero shows upregulation and downregulation respectively. The plots were generated using Hallmark GSEA analysis. C Reactome and KEGG GSEA analysis showing significant pathways up and downregulated in PMN vs M-MDSCs. The red bar signifies highly upregulated pathways with high NES values and the green bars show low NES values with downregulated pathways. D qPCR analysis of H1F-α expression in WM vs NBM. E qPCR analysis of MPO, AZU1 and CXCR4 expression in PMN-MDSCs vs M-MDSCs.
WM MDSCs express high amounts of HIF-1α, CXCR4, MPO, and AZU1
To validate our CITEseq data, we sorted MDSCs from WM patients (n = 3) and healthy BM donors (n = 2) and performed qPCR analysis. The expression of HIF1-α was calculated in WM-MDSCs vs NBM-MDSCs. The WM MDSCs exhibited a 2.87 log fold increase (p = 0.014) as compared to NBM (Fig. 4D). We also isolated PMN-MDSCs and M-MDSCs from WM patients (n = 2) and observed upregulation of CXCR4, MPO, and AZU1 compared to controls. In the first WM patient (WM1) compared to control, fold changes for MPO, CXCR4, and AZU1 were 5.41 (p = 0.0003), 12.99 (p = 0.00001), and 13.82 (p = 0.0004), respectively (Fig. 4E). Similarly, in the second WM patient (WM2), fold changes were 8.26 (p = 0.0001) for MPO, 10.13 (p = 0.087) for CXCR4, and 5.6 (p = 0.002) for AZU1 (Fig. 4E).
CD66b + PMN-MDSCs mediated T-cell suppression inhibits TNFα and IFN-γ production
To investigate the immunosuppressive role of CD66b+ PMN-MDSCs compared to CD66b- M-MDSCs in WM patients, we enriched the MDSCs extracted from 3 representative WM patients and separated them into CD66b+ PMN and CD66b- M-MDSCs. These cells were then co-cultured with sorted T-cells from healthy donors. After 24 h of co-culture with CD66b+ PMN-MDSCs, we found a substantial decrease in CD69 expression on T cells from 94.03% to 60.16% (67.74% median decrease, range 44.4–68.3% n = 3) consistent with decreased activation. While co-culture of T-cells with CD66b- MDSCs also downregulated CD69 expression, the extent of immune suppression (median: 84.85%) was less than seen with CD66b+ MDSC (p < 0.0001 - Fig. 5A).
Fig. 5. PMN-MDSCs exhibit increased T-cell suppression.
The PMN and M-MDSCs were sorted by removing CD19 + CD138 + CD3+ cells first, then the negative fraction was sorted for CD66b. Here we considered the CD66b+ fraction as PMN-MDSCs and CD66b- cells as M-MDSCs. T-cells were isolated from the healthy donors. Also, the CD3/CD28 dynabeads were added to the culture for the activation of T-cells. In the experiment, activated and non-activated T cells were cultured alone for 24 h as controls. We observed a significant decrease in the viability of PMN-MDSCs between 24 and 48 h (n = 3). Thus, we used the 0-h time point as the control for the experiment. A The representative plot shows the suppressive effect of PMN-MDSCs when cultured with T-cells from healthy donors (1:1 ratio for 24 h). CD69 expression was calculated as the mean MFI which is shown on the bar graph. B Representative plots for INF-γ suppression on CD4 T-cells by PMN-MDSCs. The activated CD4-Tcells co-cultured with PMN-MDSCs showed higher suppression. C Representative plots showing the TNF-α cytokine expression in T-cells with the PMN-MDSCs and M-MDSCs coculture; paired t-test was used here, and MFI was plotted on the bar graph. D Representative plots showing the INF-γ expression by CD8 T-cells with the co-culture with PMN and M-MDSCs. E Representative plot showing TNF-α cytokine expression by CD8 T-cells when co-cultured with PMN and M-MDSCs. F Paired t-test for INF-γ was calculated, and MFI was plotted on the bar graph was plotted. G Paired t-test for TNF-α was calculated, and MFI was plotted on the bar graph was plotted.
We analyzed cytokine production (IFN-γ and TNF-α) by CD4+ and CD8+ T cells when cocultured with either CD66b+ or CD66b- MDSCs. Our results showed that CD66b+ MDSCs significantly downregulated IFN-γ (Fig. 5B, C) and TNF-α (Fig. 5D, E) expression in both CD4+ and CD8+ T cells. Specifically, IFN-γ expression in CD4+ T cells decreased from 24.6% to 1.04% (median decrease of 1.06%, range 0.72–1.3%, n = 3, p = 0.0001) (Fig. 5F). Similarly, in CD8+ T cells, IFN-γ expression was reduced from 33.6% to 4.99% (median decrease of 3.15%, range 2.5–9.2%, n = 3, p = 0.0001) (Fig. 5F).
Furthermore, TNF-α expression in CD4+ T cells decreased from 36.12 to 6.29% (median decrease of 5.2%, range 4.65–9%, n = 3, p = 0.0001) (Fig. 5G). In CD8+ T cells, TNF-α expression was downregulated from 28 to 8.5% (median decrease of 6.93%, range 3.18–6.93%, n = 3, p = 0.001) (Fig. 5G).” A similar pattern of TNFα downregulation in CD4+ (Fig. 5D) and CD8+ (Fig. 5E) T cells was observed with the co-culture of CD66b+ MDSCs. As summarized in Fig. 5F, G, these results suggest that CD66b + PMN-MDSCs possess profound immunosuppressive properties in WM patients.
G-CSF and TNF -alpha promotes growth of CD66b+ PMN-MDSCs
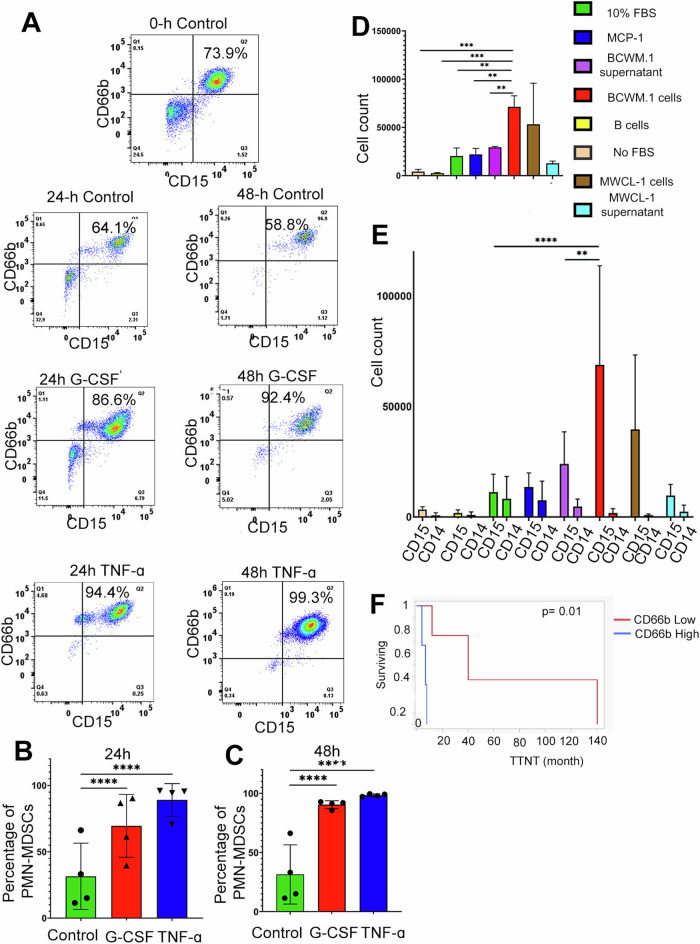
We then evaluated potential cytokines that may account for the expansion of the CD66b+ PMN-MDSC population. We had previously shown that granulocyte colony-stimulating factor (G-CSF) is increased in WM BM serum compared to that in healthy individuals [33]. Based on this finding and on our identified upregulation of TNF signaling in PMN-MDSCs, we investigated the potential role of G-CSF and TNFα in expanding CD66b+ MDSCs in the WM BM microenvironment. To do this, we enriched CD66b + cells from WM patients (n = 3), and one NBM, treated them with G-CSF and TNFα for 24 and 48 h and then measured the numbers of CD15+/CD66b+ (Lin- CD11b+ CD33+HLADR-) PMN-MDSCs by flow cytometry. After treatment with G-CSF (10 ng/ml) for 24 h, we found that the PMN-MDSCs population expanded by 38.08% (p < 0.001) (Fig. 6A, B). This population further increased by 58.95% after 48 h (p < 0.001). Similarly, TNFα treatment also increased the CD66b + PMN-MDSC population by 57.57% (p < 0.001) after 24 h, but this percentage remained about the same after 48 h of treatment (66.97%, p < 0.001; Fig. 6A, B). These findings suggests that G-CSF and TNFα promote PMN-MDSCs expansion and survival.
Fig. 6. PMN-MDSCs expand in response to cytokine stimulation and are attracted to malignant B-cells.
A PMN-MDSCs gated from the WM patient BM (CD19-CD138-CD3- CD56- HLADR-CD11b+ CD15+CD66b+). The cells isolated from the WM patient’s bone marrow at the time of processing was considered as 0 h control. (i) Representation plots showing PMN-MDSCs expansion in the presence of G-CSF (20 ng/ml) and TNF (10 ng/ml) and cultured for 24 h. (ii) Expansion was calculated (% of CD15+ CD66b+ cells) and was higher with PMN-MDSCs culture with G-CSF growth factor (B) Representation plots showing PMN-MDSCs expansion in 24 h of G-CSF and TNF culture with MDSCs from WM bone marrow. A significant expansion was observed with both the G-CSF growth factor and TNF cytokine. C Representation plots showing PMN-MDSCs expansion in 48 h of G-CSF and TNF culture with MDSCs from WM bone marrow. A significant expansion at 24 h and 48 h was observed with both the G-CSF growth factor and TNF cytokine. Expansion of MDSCs was calculated in a similar pattern with the number of CD15+ CD66b+ cells. D Transwell assay showing migration of MDSCs (CD11b+CD33+ cells) in the presence of BCWM.1 cells. (E) Transwell assay showing high migration of PMN-MDSCs in the presence of BCWM.1 cells. In the GCSF/TNF and migration analysis, paired t-test was calculated (n = 3 WM patients) and p ≤ 0.01, p ≤ 0.001, and p ≤ 0.0001 are represented by *, **, ***, and ****. F Comparison of overall time to next treatment (TTNT) in WM patients with high and low CD66b+ cells in the bone marrow [7 months (95% CI: 2.3–49 months) compared to 40 months (95% CI: 4.3–140 months) p = 0.01].
Lymphoplasmacytic cells of WM patients recruit PMN-MDSCs in the bone marrow
While we found that GCSF and TNFα promote the expansion of PMN-MDSCs, we also evaluated whether WM B-cells play a role in attracting PMN-MDSCs to the BM. Therefore, to investigate whether WM cells recruit MDSCs to the BM, we conducted a transwell migration assay of MDSCs in response to the presence of malignant BCWM.1 cells. Our results showed that BCWM.1 cells exhibited a greater capacity to attract MDSCs when compared to normal B-cells or control chemoattractants including 10% FBS and MCP-1 (p = 0.04; Fig. 6D). Notably, the physical presence of malignant B-cells had a greater capacity to recruit MDSCs than the supernatants from these cells. Additionally, we sought to determine which subtype of MDSCs was predominantly attracted by malignant B-cells. To address this, we assessed the expression of CD14 and CD15 on migrating MDSCs to delineate PMN-MDSCs and M-MDSCs. We found that BCWM.1 (p = 0.001) and MWCL-1 cells attracted PMN-MDSCs to a greater degree than M-MDSCs (Fig. 6E). These findings indicate that WM cells exhibit robust chemotactic properties for MDSCs, with a significant preference for attracting PMN-MDSCs. These observations suggest that the high prevalence of PMN-MDSCs within the BM of WM patients is in part due to recruitment of these cells to the BM by malignant B-cells.
Increased PMN-MDSCs are associated poor prognostic factors
In addition to the biological effect, the increased prevalence of CD66+ PMN-MDSCs showed clinical relevance. Clinical data from 10 WM patients was analyzed to assess the correlation of CD66b+ PMN-MDSCs with clinical features. The average percentage of CD66b+ PMN-MDSCs was employed to establish parameters for low and high levels. The median TTNT of patients with high CD66b expression was 7 months (95% CI: 2.3–49 months) compared to 40 months (95% CI: 4.3–140 months) for patients with low CD66b expression, p = 0.1. Only a 14% of patients with high CD66b expression achieved a TTNT > 12 months, compared to 71% of patients with low CD66b expression, p = 0.01.
Discussion
Our study demonstrate that the BM of symptomatic WM patients is characterized by the expansion of PMN-MDSCs and that PMN-MDSCs play a significant role in promoting an immunosuppressive microenvironment. We show that the population of PMN-MDSCs increases as the disease progresses and conclude that increased numbers of PMN-MDSCs in the BM are associated with poor prognostic clinical factors and are associated with a shorter time to the next treatment in WM patients. Overall, MDSCs are increased in WM but PMN-MDSCs are particularly expanded in BM of WM patients as confirmed by both flow cytometry and CyTOF analysis. Others have shown that the expansion of PMN-MDSCs is more pronounced in cancer as compared to infection and inflammation [34, 35] and previous studies have demonstrated the expansion of MDSCs in other hematological malignancies [9, 36–38]. However, the role and impact of MDSCs in promoting tumor development and immune suppression in WM has not been evaluated. Our findings identify the dominant presence of PMN-MDSCs expressing CD66b in the BM of WM patients and confirms their pivotal role in driving immune suppression.
We further show that the presence of malignant B-cells, as well as the cytokines secreted in the BM, both favor the expansion of PMN-MDSCs. We found that WM cells (BCWM.1 and MWCL-1) attract MDSCs to the tumor site, with PMN-MDSCs being attracted to a greater degree compared to M-MDSCs. We observed a significantly higher attraction by BCWM.1/MWCL-1 cells compared to their respective supernatants. This suggests that the physical presence of WM cells plays a pivotal role in attracting PMN-MDSCs to the tumor microenvironment. While our previous analysis of WM BM cytokines identified substantial elevations of CCL5, G-CSF, and IL-2 receptor [33], here we show the critical role of G-CSF and TNFα in promoting the expansion of CD66b+ PMN-MDSCs. Others have reported that G-CSF expands MDSCs via c-kit oncogene regulation and this results in MDSC-mediated T-cell suppression [39]. Also, tumor-derived G-CSF plays an important role in developing chemoresistance and is associated with a poor prognosis in cancer patients [40]. G-CSF, GM-CSF, and TNFα regulate the process of myelopoiesis [41] and may also facilitate the substantial accumulation of myeloid cells at tumor sites [42, 43]. The elevated levels of G-CSF in WM patient’s serum [33] and expansion of PMN-MDSCs after treatment with G-CSF underscore the crucial role of G-CSF and TNFα in promoting PMN-MDSC expansion in WM patients. Numerous chemokines secreted by cancer cells, including CXCL5, CXCL2, and CXCL1, are recognized for their ability to attract PMN-MDSCs to tumor sites [44]. Chemokine receptor-ligand interaction (such as CCL2-CCR5 interaction) are a key driving force for MDSCs migration, therefore blocking their interaction may be a rational approach to inhibit MDSC accumulation at the tumor site [45].
There is growing interest in targeting MDSCs to improve T-cell function and immunotherapy efficiency in WM. The activation of MDSCs is one of the major causes of an immune suppressive BM microenvironment due to their inhibition on T-cell function [46, 47]. However, due to the absence of specific cell markers on MDSCs, the ability to specifically target them remains a challenge. Also, MDSCs sense the changes in the environment and survive by selecting metabolic pathways to perform pro-tumorigenic and suppressive functions [48]. Our findings highlight the suppressive effects of CD66b+ PMN-MDSCs on CD4 and CD8 T-cells, resulting in decreased proliferation and suppressed secretion of TNFα and IFNγ within the T-cell population. We showed that CD66b+ PMN- MDSCs enriched from WM BM suppressed T cell activity mediated by both CD4+ and CD8+ T cells, consistent with the finding that BM PMN-MDSCs isolated from late-stage cancer patients were highly immune suppressive [49].
The distinct phenotypic and transcriptomic attributes of MDSCs are delineated within clusters 2, 3, and 9 defined by CITE-seq analysis, and express TGFB1, IL13RA1, IL17RA, IL1B, IL10RB, IL18, STAT2, STAT1, STAT6, CSF1R, CSF3R, CD84, TGFB1, and SOD2. These characteristics correlate with increased T-cell immune suppression and elevated levels of inflammatory cytokines. The changes in the MDSC subsets in WM patients are due to the overall expansion of the MDSCs population with a gene signature characterized by expression of S100A8, S100A9, S100A4, TGFB1, CSF3R, LYZ, FOS, and CCL2 [10]. These expanded MDSCs exhibited notable expression of immune regulatory genes, including CD86 [43], HIF1-α, IL13RA1 [44, 45], IL10RA [46], TGFBI [47], and CD74 [48] signifying their role in immune suppressive tumor microenvironment (TME) niche formation [12, 50, 51]. The high expression of HIF-1α in WM MDSCs compared to NBM controls was further confirmed by PCR analysis, thereby validating the CITEseq results. This increase in HIF-1α suggests that hypoxic conditions in the WM BM may induce HIF-1α expression in MDSCs, contributing in part to the suppressive phenotype [24, 52].
Further, we identified the gene signature of CD66b+ PMN-MDSCs in WM. In recent studies, the unique plasticity and heterogeneity of PMN-MDSCs in cancer and other inflammatory diseases is emerging, with a focus on diverse phenotypes and functions [53, 54]. The transcriptomic analysis by others has identified IL2RG, FOXM1, and CXCR4 as important markers for PMN- MDSCs indicating tumor promotion and suppressive activity [55]. Our findings highlight IL2RG, FOXM1, PRTN3, SOX4, IGFBP7, and CXCR4 as hallmark genes specific to CD66b+ PMN-MDSCs, signifying their distinct nature within the tumor microenvironment. This subset of MDSCs stands out due to pro-tumor growth signaling and demonstrates more pronounced suppressive features compared to other MDSC populations in the BM. This population expresses higher levels of MPO, AZU, ELANE, and LDHB, which may explain increased myelopoiesis (increasing the uncontrolled production of myeloid cells), cell proliferation [30, 56], and poorer clinical outcomes [31, 57]. Additionally, the CD66b+ PMN-MDSCs population expressed higher levels of cell cycle genes namely, CDK1, CENPF, PLK1, MKI67, and HMGN2, when compared to other MDSCs, consistent with increased proliferation. Moreover, PMN-MDSCs also showed upregulation of numerous pathways associated with DNA damage responses, apoptosis, MAPK signaling, TGFβ signaling, and several myeloid differentiation- transcripts [58]. The qPCR analysis of CXCR4, MPO, and AZU1 expression in PMN-MDSCs compared to M-MDSCs from WM patients corroborated the CITEseq findings (Fig. 4E), and further suggested that PMN-MDSCs have profoundly immunosuppressive properties.
Our study therefore provides the first evidence of a transcriptomic profile of BM-derived PMN-MDSCs from WM patients. PMN-MDSCs promote an immune suppressive BM microenvironment by inhibiting T cell activity and fostering tumor progression, particularly in the presence of G-CSF and TNF-α [39, 41, 59]. Therefore, this study provides a rationale for the development of therapeutic interventions targeting MDSCs.
Supplementary information
Acknowledgements
This work was supported by grants from the International Waldenstrom Macroglobulinemia Foundation and the Predolin Foundation.
Author contributions
VB contributed to study design, performed experiments, analyzed, and interpreted the data, reviewed the literature and wrote the manuscript. JP and JV identified the patients, data collection and data analysis, SJ performed experiments and contributed to data analysis and interpretation. RM and JW- performed CITEseq analysis, PJ, XT, JK, KW, AN- Writing–review and editing, SMA, ZZY, and PM designed the project, conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, writing–original draft, project administration., manuscript writing and editing.
Data availability
The data supporting this study is available on request.
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
All methods were carried out in strict accordance with relevant guidelines and regulations. Written informed consent was obtained from all participants in compliance with the Mayo Clinic Institutional Review Board (IRB) requirements (IRB-118-01). The study adhered to the principles outlined in the Declaration of Helsinki.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Patrizia Mondello, Email: mondello.patrizia@mayo.edu.
Stephen M. Ansell, Email: ansell.stephen@mayo.edu
Supplementary information
The online version contains supplementary material available at 10.1038/s41408-024-01173-w.
References
- 1.Gertz MA. Waldenström macroglobulinemia: 2021 update on diagnosis, risk stratification, and management. Am J Hematol. 2021;96:258–69. [DOI] [PubMed] [Google Scholar]
- 2.Treon SP, Xu L, Guerrera ML, Jimenez C, Hunter ZR, Liu X, et al. Genomic landscape of Waldenström macroglobulinemia and its impact on treatment strategies. J Clin Oncol. 2020;38:1198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Treon SP, Xu L, Yang G, Zhou Y, Liu X, Cao Y, et al. MYD88 L265P somatic mutation in Waldenström’s macroglobulinemia. N. Engl J Med. 2012;367:826–33. [DOI] [PubMed] [Google Scholar]
- 4.Chapuy B, Stewart C, Dunford AJ, Kim J, Kamburov A, Redd RA, et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med. 2018;24:679–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watanabe J, Natsumeda M, Okada M, Kobayashi D, Kanemaru Y, Tsukamoto Y, et al. High detection rate of MYD88 mutations in cerebrospinal fluid from patients with CNS lymphomas. JCO Precis Oncol. 2019;3:1–13. [DOI] [PubMed]
- 6.Oishi N, Kondo T, Nakazawa T, Mochizuki K, Tanioka F, Oyama T, et al. High prevalence of the MYD88 mutation in testicular lymphoma: Immunohistochemical and genetic analyses. Pathol Int. 2015;65:528–35. [DOI] [PubMed] [Google Scholar]
- 7.Mondello P, Paludo J, Novak JP, Wenzl K, Yang Z-Z, Jalali S, et al. Molecular clusters and tumor-immune drivers of IgM monoclonal gammopathies. Clin Cancer Res. 2022;29:957–970. [DOI] [PubMed]
- 8.Kaushal A, Nooka AK, Carr AR, Pendleton KE, Barwick BG, Manalo J, et al. Aberrant extrafollicular B cells, immune dysfunction, myeloid inflammation, and MyD88-mutant progenitors precede waldenstrom macroglobulinemia. Blood Cancer Discov. 2021;2:600–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhardwaj V, Ansell SM. Modulation of T-cell function by myeloid-derived suppressor cells in hematological malignancies. Front Cell Dev Biol. 2023;11:1129343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Veglia F, Sanseviero E, Gabrilovich DI. Myeloid-derived suppressor cells in the era of increasing myeloid cell diversity. Nat Rev Immunol. 2021;21:485–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gabrilovich DI, Bronte V, Chen SH, Colombo MP, Ochoa A, Ostrand-Rosenberg S, et al. The terminology issue for myeloid-derived suppressor cells. Cancer Res. 2007;67:425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. 2009;9:162–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Wang J, Zhu F, Wang H, Yi L, Huang K, et al. Elevated circulating myeloid-derived suppressor cells associated with poor prognosis in B-cell non-Hodgkin’s lymphoma patients. Immun Inflamm Dis. 2022;10:e616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khalifa KA, Badawy HM, Radwan WM, Shehata MA, Bassuoni MA. CD14(+) HLA-DR low/(-) monocytes as indicator of disease aggressiveness in B-cell non-Hodgkin lymphoma. Int J Lab Hematol. 2014;36:650–5. [DOI] [PubMed] [Google Scholar]
- 15.Azzaoui I, Uhel F, Rossille D, Pangault C, Dulong J, Le Priol J, et al. T-cell defect in diffuse large B-cell lymphomas involves expansion of myeloid-derived suppressor cells. Blood. 2016;128:1081–92. [DOI] [PubMed] [Google Scholar]
- 16.Tadmor T, Fell R, Polliack A, Attias D. Absolute monocytosis at diagnosis correlates with survival in diffuse large B-cell lymphoma-possible link with monocytic myeloid-derived suppressor cells. Hematol Oncol. 2013;31:65–71. [DOI] [PubMed] [Google Scholar]
- 17.Treon SP, Tedeschi A, San-Miguel J, Garcia-Sanz R, Anderson KC, Kimby E, et al. Report of consensus Panel 4 from the 11th International Workshop on Waldenstrom’s macroglobulinemia on diagnostic and response criteria. Semin Hematol. 2023;60:97–106. [DOI] [PubMed] [Google Scholar]
- 18.Ditzel Santos D, Ho AW, Tournilhac O, Hatjiharissi E, Leleu X, Xu L, et al. Establishment of BCWM.1 cell line for Waldenström’s macroglobulinemia with productive in vivo engraftment in SCID-hu mice. Exp Hematol. 2007;35:1366–75. [DOI] [PubMed] [Google Scholar]
- 19.Hodge LS, Novak AJ, Grote DM, Braggio E, Ketterling RP, Manske MK, et al. Establishment and characterization of a novel Waldenström macroglobulinemia cell line, MWCL-1. Blood. 2011;117:e190–e7. [DOI] [PubMed] [Google Scholar]
- 20.Mardiney M 3rd, Brown MR, Fleisher TA. Measurement of T-cell CD69 expression: a rapid and efficient means to assess mitogen- or antigen-induced proliferative capacity in normals. Cytometry. 1996;26:305–10. [DOI] [PubMed] [Google Scholar]
- 21.Mondello P, Paludo J, Novak JP, Wenzl K, Yang ZZ, Jalali S, et al. Molecular clusters and tumor-immune drivers of IgM monoclonal gammopathies. Clin Cancer Res. 2023;29:957–70. [DOI] [PubMed] [Google Scholar]
- 22.Becht E, McInnes L, Healy J, Dutertre C-A, Kwok IWH, Ng LG, et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol. 2019;37:38–44. [DOI] [PubMed] [Google Scholar]
- 23.Corcoran SE, O’Neill LA. HIF1α and metabolic reprogramming in inflammation. J Clin Investig. 2016;126:3699–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corzo CA, Condamine T, Lu L, Cotter MJ, Youn JI, Cheng P, et al. HIF-1α regulates function and differentiation of myeloid-derived suppressor cells in the tumor microenvironment. J Exp Med. 2010;207:2439–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jayaraman P, Parikh F, Newton JM, Hanoteau A, Rivas C, Krupar R, et al. TGF-β1 programmed myeloid-derived suppressor cells (MDSC) acquire immune-stimulating and tumor killing activity capable of rejecting established tumors in combination with radiotherapy. Oncoimmunology. 2018;7:e1490853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Drakos E, Leventaki V, Schlette EJ, Jones D, Lin P, Medeiros LJ, et al. c-Jun expression and activation are restricted to CD30+ lymphoproliferative disorders. Am J Surg Pathol. 2007;31:447–53. [DOI] [PubMed] [Google Scholar]
- 27.Mao X, Orchard G, Russell-Jones R, Whittaker S. Abnormal activator protein 1 transcription factor expression in CD30-positive cutaneous large-cell lymphomas. Br J Dermatol. 2007;157:914–21. [DOI] [PubMed] [Google Scholar]
- 28.Hammami I, Chen J, Murschel F, Bronte V, De Crescenzo G, Jolicoeur M. Immunosuppressive activity enhances central carbon metabolism and bioenergetics in myeloid-derived suppressor cells in vitro models. BMC Cell Biol. 2012;13:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Newson J, Motwani MP, Kendall AC, Nicolaou A, Muccioli GG, Alhouayek M, et al. Inflammatory resolution triggers a prolonged phase of immune suppression through COX-1/mPGES-1-derived prostaglandin E(2). Cell Rep. 2017;20:3162–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCleland ML, Adler AS, Shang Y, Hunsaker T, Truong T, Peterson D, et al. An integrated genomic screen identifies LDHB as an essential gene for triple-negative breast cancer. Cancer Res. 2012;72:5812–23. [DOI] [PubMed] [Google Scholar]
- 31.McCleland ML, Adler AS, Deming L, Cosino E, Lee L, Blackwood EM, et al. Lactate dehydrogenase B is required for the growth of KRAS-dependent lung adenocarcinomas. Clin Cancer Res. 2013;19:773–84. [DOI] [PubMed] [Google Scholar]
- 32.Jia Y, Qi Y, Wang Y, Ma X, Xu Y, Wang J, et al. Overexpression of CD59 inhibits apoptosis of T-acute lymphoblastic leukemia via AKT/Notch1 signaling pathway. Cancer Cell Int. 2019;19:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elsawa SF, Novak AJ, Ziesmer SC, Almada LL, Hodge LS, Grote DM, et al. Comprehensive analysis of tumor microenvironment cytokines in Waldenstrom macroglobulinemia identifies CCL5 as a novel modulator of IL-6 activity. Blood. 2011;118:5540–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Romano A, Parrinello NL, Vetro C, Forte S, Chiarenza A, Figuera A, et al. Circulating myeloid-derived suppressor cells correlate with clinical outcome in Hodgkin Lymphoma patients treated up-front with a risk-adapted strategy. Br J Haematol. 2015;168:689–700. [DOI] [PubMed] [Google Scholar]
- 35.Cassetta L, Bruderek K, Skrzeczynska-Moncznik J, Osiecka O, Hu X, Rundgren IM, et al. Differential expansion of circulating human MDSC subsets in patients with cancer, infection and inflammation. J Immunother Cancer. 2020;8:e001223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marini O, Spina C, Mimiola E, Cassaro A, Malerba G, Todeschini G, et al. Identification of granulocytic myeloid-derived suppressor cells (G-MDSCs) in the peripheral blood of Hodgkin and non-Hodgkin lymphoma patients. Oncotarget. 2016;7:27676–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brimnes MK, Vangsted AJ, Knudsen LM, Gimsing P, Gang AO, Johnsen HE, et al. Increased level of both CD4+FOXP3+ regulatory T cells and CD14+HLA-DR–/low myeloid-derived suppressor cells and decreased level of dendritic cells in patients with multiple myeloma. Scand J Immunol. 2010;72:540–7. [DOI] [PubMed] [Google Scholar]
- 38.Wang H, Tao Q, Wang Z, Zhang Q, Xiao H, Zhou M, et al. Circulating monocytic myeloid-derived suppressor cells are elevated and associated with poor prognosis in acute myeloid leukemia. J Immunol Res. 2020;2020:7363084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee YS, Saxena V, Bromberg JS, Scalea JR. G-CSF promotes alloregulatory function of MDSCs through a c-Kit dependent mechanism. Cell Immunol. 2021;364:104346. [DOI] [PubMed] [Google Scholar]
- 40.Kawano M, Mabuchi S, Matsumoto Y, Sasano T, Takahashi R, Kuroda H, et al. The significance of G-CSF expression and myeloid-derived suppressor cells in the chemoresistance of uterine cervical cancer. Sci Rep. 2015;5:18217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamashita M, Passegué E. TNF-α coordinates hematopoietic stem cell survival and myeloid regeneration. Cell Stem Cell. 2019;25:357–72.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karin N. The development and homing of myeloid-derived suppressor cells: from a two-stage model to a multistep narrative. Front Immunol. 2020;11:557586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strauss L, Sangaletti S, Consonni FM, Szebeni G, Morlacchi S, Totaro MG, et al. RORC1 regulates tumor-promoting “emergency” granulo-monocytopoiesis. Cancer Cell. 2015;28:253–69. [DOI] [PubMed] [Google Scholar]
- 44.Li B-H, Garstka MA, Li Z-F. Chemokines and their receptors promoting the recruitment of myeloid-derived suppressor cells into the tumor. Mol Immunol. 2020;117:201–15. [DOI] [PubMed] [Google Scholar]
- 45.Li X, Yao W, Yuan Y, Chen P, Li B, Li J, et al. Targeting of tumour-infiltrating macrophages via CCL2/CCR2 signalling as a therapeutic strategy against hepatocellular carcinoma. Gut. 2017;66:157–67. [DOI] [PubMed] [Google Scholar]
- 46.Gabrilovich DI, Velders MP, Sotomayor EM, Kast WM. Mechanism of immune dysfunction in cancer mediated by immature Gr-1+ myeloid cells. J Immunol. 2001;166:5398–406. [DOI] [PubMed] [Google Scholar]
- 47.Saio M, Radoja S, Marino M, Frey AB. Tumor-infiltrating macrophages induce apoptosis in activated CD8(+) T cells by a mechanism requiring cell contact and mediated by both the cell-associated form of TNF and nitric oxide. J Immunol. 2001;167:5583–93. [DOI] [PubMed] [Google Scholar]
- 48.Bader JE, Voss K, Rathmell JC. Targeting metabolism to improve the tumor microenvironment for cancer immunotherapy. Mol Cell. 2020;78:1019–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patel S, Fu S, Mastio J, Dominguez GA, Purohit A, Kossenkov A, et al. Unique pattern of neutrophil migration and function during tumor progression. Nat Immunol. 2018;19:1236–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bronte V, Brandau S, Chen SH, Colombo MP, Frey AB, Greten TF, et al. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat Commun. 2016;7:12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kumar V, Patel S, Tcyganov E, Gabrilovich DI. The nature of myeloid-derived suppressor cells in the tumor microenvironment. Trends Immunol. 2016;37:208–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang S, Zhao X, Wu S, Cui D, Xu Z. Myeloid-derived suppressor cells: key immunosuppressive regulators and therapeutic targets in hematological malignancies. Biomark Res. 2023;11:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Darcy CJ, Minigo G, Piera KA, Davis JS, McNeil YR, Chen Y, et al. Neutrophils with myeloid derived suppressor function deplete arginine and constrain T cell function in septic shock patients. Crit Care. 2014;18:R163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gervassi A, Lejarcegui N, Dross S, Jacobson A, Itaya G, Kidzeru E, et al. Myeloid derived suppressor cells are present at high frequency in neonates and suppress in vitro T cell responses. PLoS ONE. 2014;9:e107816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Veglia F, Hashimoto A, Dweep H, Sanseviero E, De Leo A, Tcyganov E, et al. Analysis of classical neutrophils and polymorphonuclear myeloid-derived suppressor cells in cancer patients and tumor-bearing mice. J Exp Med. 2021;218:e20201803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsuruta T, Tani K, Hoshika A, Asano S. Myeloperoxidase gene expression and regulation by myeloid cell growth factors in normal and leukemic cells. Leuk Lymphoma. 1999;32:257–67. [DOI] [PubMed] [Google Scholar]
- 57.Kumar S, Xie H, Scicluna P, Lee L, Björnhagen V, Höög A, et al. MiR-375 regulation of LDHB plays distinct roles in polyomavirus-positive and -negative Merkel cell carcinoma. Cancers. 2018;10:443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sasidharan Nair V, Saleh R, Toor SM, Alajez NM, Elkord E. Transcriptomic analyses of myeloid-derived suppressor cell subsets in the circulation of colorectal cancer patients. Front Oncol. 2020;10:1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.He K, Liu X, Hoffman RD, Shi R-Z, Lv G-Y, Gao J-L. G-CSF/GM-CSF-induced hematopoietic dysregulation in the progression of solid tumors. FEBS Open Bio. 2022;12:1268–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting this study is available on request.