Abstract
Ionic liquids (ILs) are structurally tunable salts with applications ranging from chemical synthesis to batteries, novel materials and medicine. Despite their potential, the toxicity of ILs poses significant environmental and biological challenges. This study introduces a comprehensive dataset of cytotoxicity of 1227 ILs, compiled from 151 research papers and encompassing 3837 data entries. For each entry, the following information is provided: substance name, empirical formula, CAS, SMILES, molecular weight, cytotoxicity value, details on the experimental setup (incubation time, cell line, assay used, etc.), and reference to the original publication. This dataset can be used for deriving structure‒activity relationships and establishing the major structural elements that govern the cytotoxic effects of ILs on eukaryotic cells. The dataset is available freely to all researchers.
Subject terms: Cheminformatics, Environmental monitoring, Toxicology
Background & Summary
Ionic liquids (ILs) are famous organic salts with low melting points and unique physicochemical and biochemical properties1–3. Members of this class of substances have applications in diverse scientific and industrial fields, such as organic synthesis4, catalysis5, biomass processing6, and biological7–9 and medical research10,11. The wide range of applications of ILs is related to their extraordinary structural changeability. The number of hypothetical combinations of cations and anions in a given IL is virtually unlimited, suggesting that any desired properties can be combined in one molecule12.
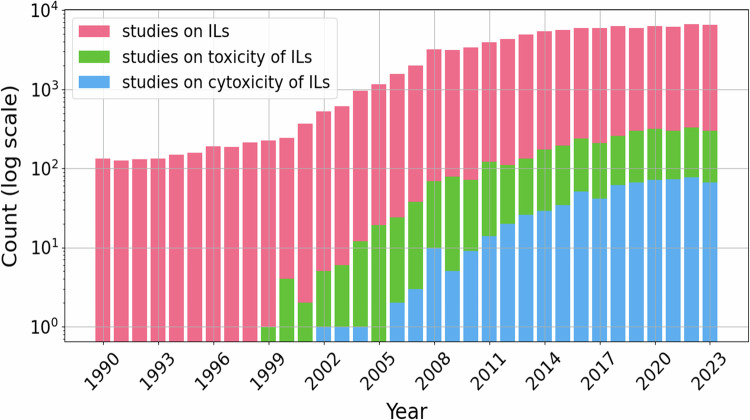
Since the beginning of the 21st century, the investigation and application of ILs have progressed steadily, representing notable advancements in the fields of chemistry and materials science13,14. According to the Scopus database15, since the early 1990s, there has been pronounced growth in the number of scientific studies on the toxicity of ILs (see Fig. 1).
Fig. 1.
The evolution of research on ionic liquids over the last three decades (according to the Scopus database as of March 2024). The y-axis corresponds to the number of individual articles presented on a log scale. The following search queries were used: “ionic liquids” OR “molten salts” (studies on ILs, pink), “ionic liquids” AND *toxic* (studies on the toxicity of ILs, green), and “ionic liquids” AND cytotoxic* (studies on the cytotoxicity of ILs, blue).
Because of their low flammability, very low vapor pressure, and high thermal stability, some ILs have been considered ‘green’ solvents by the scientific community. Nevertheless, many ILs can have a significant impact on living organisms because of their considerable water solubility and ability to spread into the environment16,17. In addition, ILs demonstrate the ability to penetrate lipid bilayers, which constitute the cellular membranes of all living organisms18–21. This amphiphilic nature can be the primary factor determining the rather high biological activity of many ILs. Several other structural factors that can govern the biological activity of ILs have been established thus far22. In particular, the length of the alkyl side chain in the cation is thought to be the major attribute to which the toxicity can be related.
Understanding the mode of the biological effects of ILs is a key requirement for their approval for widespread practical use. It is also important to develop efficient models for the prediction of IL toxicity toward various organisms. According to the Scopus database, in the last two decades, several thousand studies concerning some type of biological activity of ILs have been published (see Fig. 1). Because of the very large number of ILs available, the development of reliable methods for predicting their toxicity is essential. Considering the time required to carry out experimental studies, computational modeling (in silico) methods can become a convenient and less expensive alternative for the assessment of IL-related environmental risks. Computer-aided calculations are indispensable tools in modern research, offering invaluable assistance in the exploration of the vast array of chemical structures found in ILs23. By leveraging computational methods, scientists can efficiently analyze and understand the intricate properties and behaviors of ILs, facilitating the development of predictive models24–26. These models, in turn, can serve as powerful instruments for predicting the IL behavior under different conditions and in diverse applications, thus driving innovation and advancement in the fields ranging from chemistry to materials science and beyond. For this purpose, databases on IL-related biological activity are in high demand. Systematization of the already obtained data on IL toxicity will improve the knowledge of the potential hazards of ILs and help define the directions of future research on this topic.
There are some databases on the physicochemical properties of ILs, among which ILThermo27, the IPE Ionic Liquid Database28, and DDBST GmbH (Dortmund data bank)29 should be noted. However, only a few dedicated databases on the toxicity of ILs have been developed thus far, and currently, only the ILTOX30 database is available to researchers. ILTOX contains information on the cytotoxicity of ILs measured in five cell lines – that is, the CaCo-2, HeLa, HepG2, IPC-81, and HT-29 cell lines - but it does not provide full details on the methodology, which is essential for comparing the cytotoxicity values presented in different works. The whole dataset for this database cannot be downloaded, limiting its usage to the functionality of its web interface. According to the website, the last update of ILTOX was in February 2022; thus, it does not provide data published in the last two years.
Studies on the cytotoxicity of chemical compounds are usually considerably faster and more affordable than experiments on multicellular organisms. Thus, there are significantly more scientific publications on the effects of ILs on eukaryotic cells than on other biological objects. Moreover, cytotoxicity data are used at the initial steps of the development of chemical agents with both high and low biological activities. Therefore, accumulating and organizing the available data on the cytotoxicity of ILs will be necessary for designing ILs with high target activity and low toxicity.
In this study, we present a comprehensive dataset of experimental data related to the cytotoxicity of ILs. A total of 3837 entries for 1227 individual ILs were obtained by annotating 151 research papers. For each entry, the following information is provided: substance name, empirical formula, Chemical Abstract Service (CAS) number, simplified molecular input line entry system (SMILES) presentation, molecular weight, cytotoxicity value and its range (standard error of the mean, confidence interval, etc.), incubation time, cell line, cytotoxicity measurement method, and reference to the original publication. This dataset can be used for deriving structure‒activity relationships and establishing the major structural elements that govern the cytotoxic impact of ILs on eukaryotic cells.
Methods
All annotated cytotoxicity data were manually extracted from scientific papers, which were selected according to a literature search carried out in the Scopus database by using the following search query: “ionic liquids” AND cytotoxic* (Search within Article title, Abstract, Keywords). A total of 750 papers published before March 2024 were selected and filtered according to the following conditions: (1) full-text accessibility; (2) unambiguous information on the structure of a particular ionic liquid and the experimental conditions provided; and (3) the exact cytotoxicity value present. Note that papers on API-ILs (active pharmaceutical compounds – ionic liquids) were not considered because the active pharmaceutical ingredient present in the IL structure would, in most cases, determine the cytotoxic effect observed and would eclipse any effects of other structural elements in a given IL.
Finally, 151 articles were selected for annotation. For each IL, we extracted the following information: compound name, empirical formula, CAS, SMILES, molecular weight, cytotoxicity value and range (standard error of the mean, confidence interval, etc.), incubation time, cell line, cytotoxicity measurement method, and bibliographic information. Additional tools and databases, such as ChemBioDraw Ultra 13.031, SciFinder32, ACD/Labs 10.0 (ChemSketch)33, and PubChem34, were used to determine the molecular weight, SMILES and CAS.
Annotation rules
The annotation process was performed as follows
The abstracts and metadata from the Scopus database were retrieved via the following search query: “ionic liquids” AND cytotoxic* (Search within Article title, Abstract, Keywords);
Upon screening the abstracts, the candidate papers for annotation were selected (750 in total);
The full texts of the selected papers were obtained and subjected to secondary screening for the ILs fitting the scope of the dataset (see criteria below; after this stage, approximately 20% of the initially identified publications remained for further processing).
The relevant information was extracted from the publications;
The annotated data were cross-validated manually by human curators (100% of the entries reviewed).
Data Records
A complete version of the described dataset has been placed at and is publicly available for download from figshare35. The general dataset forms are described in Table 1. Each data point contains a number of fields related to the chemical and biological properties of ILs.
Table 1.
Structure of entries in the dataset.
| Section | Column | Description | Data type |
|---|---|---|---|
| Chemical | Compound name | Generated in ChemBioDraw | string |
| Empirical formula | Generated in ChemBioDraw | string | |
| CAS | Chemical Abstracts Service number (where available) | string | |
| SMILES | Simplified Molecular Input Line Entry System, a chemical format that allows encoding a 3-D structure of a chemical in a string of symbols. Regular SMILES can have multiple valid representations for the same molecule. | string | |
| Canonical SMILES | A unique or standardized SMILES representation ensuring that each molecule is assigned a distinct SMILES string. The specific output, however, depends on the canonicalization algorithm employed, which may influence the final representation. | string | |
| Mw | Molecular weight | numeric | |
| Biological | IC50, EC50, CC50* | Half-maximal inhibitory, effective, or cytotoxicity concentration | numeric |
| Range of IC50, EC50, or CC50 | Confidence intervals, standard deviations, standard errors of the mean, etc., for the cytotoxicity values provided in the previous field | numeric | |
| Incubation time | Time period, for which cells were exposed to a given IL | numeric | |
| Cell line | Name of the cell culture used in a particular study | string | |
| Method | Assay used for measuring IC50, EC50, or CC50 | string | |
| Notes | Additional information on biological activity of a particular IL, if provided | string | |
| Bibliographic | Reference | Author, year, and journal | text |
| DOI | Digital Object Identifier | text |
* In the papers annotated for the dataset, the designation of the value – IC50, EC50, or CC50 – depended mainly on the authors of a particular study. Here, the assay determines the type of value, and we can compare the values from different studies obtained by the same assay. Thus, in most cases, we made no marks in the dataset on the particular designations used by the authors.
Structure of the dataset
In the current project, the source data on ILs include three main types of information: chemical, biological, and bibliographic. Thus, each entry includes 14 sections, each of which contains verified information on a particular IL.
The chemical section includes: name; empirical formula; CAS; SMILES; canonical SMILES; and molecular weight. The names, empirical formulas and molecular weights of the ILs were obtained using ChemBioDraw software (CambridgeSoft Corporation)31. CAS numbers were obtained from the SciFinder database (American Chemical Society)32. SMILES were generated in ChemBioDraw31 or obtained from SciFinder32. Canonical SMILES were generated using RDKit36, an open-source toolkit designed for cheminformatics and machine learning tasks.
The biological section includes: IC50, EC50, or CC50 values (half-maximal inhibitory, effective, or cytotoxic concentrations, respectively) and their ranges (standard errors of the mean, confidence intervals, etc.); the time of exposure of a given cell line to a given IL; the cell line in which the effect was studied; the method of study; and notes. This information was obtained from original research papers.
The bibliographic section includes: information on the corresponding publication, including the first author’s name, publication year, journal title, and DOI (digital object identifier), where applicable. This section serves as an unambiguous reference to the original research.
The datasets are provided in XLSX and CSV formats. In the XLSX file, the chemical information is color-coded for enhanced clarity. For the compounds highlighted in green, the relevant chemical data were extracted from ChemBioDraw31. The SMILES notations that correspond to the associated CAS numbers are highlighted in pink, whereas the CAS numbers that do not correspond to the SMILES are highlighted in blue. In addition, detailed data on the experimental methodologies and cell lines are provided in the supplementary tabs.
Technical Validation
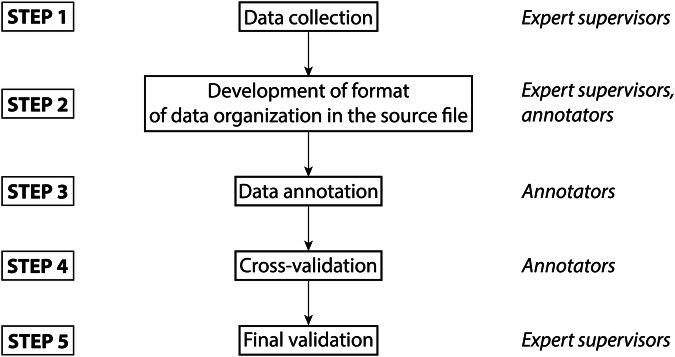
The key objective of this study was to provide the scientific community with a reliable, high-quality dataset on the cytotoxicity of ionic liquids. Thus, the obtained source data file was subjected to two-step validation by expert curators. The scheme of the verification procedure is shown in Fig. 2.
Fig. 2.
Stepwise validation of the dataset.
Here, cross-validation is the error checking upon which a particular annotator checks the dataset entries added by other annotators and compares them with the original publications, and the final validation is the error checking performed by expert chemists and biologists (not annotators) using the original publications and additional services (such as SciFinder). Both of these procedures were carried out via point-by-point comparisons of the fields in a given dataset entry with the corresponding original paper. All the entries in the dataset were subjected to this two-step verification.
Revealing the main trends
In scientific work, statistical representation is crucial for understanding the data distribution and patterns. It involves organizing, analyzing, and presenting the data by means of charts (e.g., histograms, scatter plots, etc.) and measures such as averages and standard deviations. Charts serve as indispensable tools for elucidating the relationships within datasets, facilitating clearer interpretation and establishing general trends.
In our study, we employed PostgreSQL37, an open-source relational database management system, for storing and managing the structured data. PostgreSQL offers robust features for data organization, querying, and transaction management. Additionally, we utilized Matplotlib38,39, a comprehensive data visualization library in Python, to generate plots and charts for data analysis and presentation.
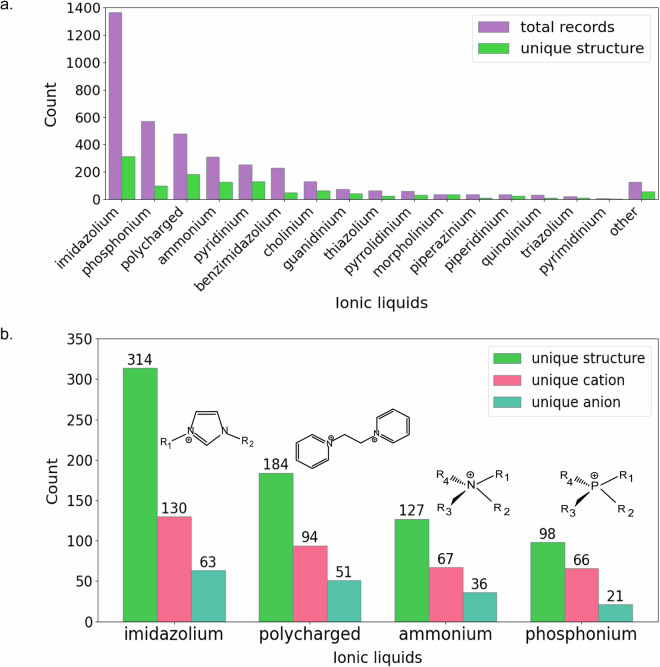
First, we determined the total number of entries and unique structures in the dataset (Fig. 3a). Figure 3b shows the distributions for the four most populated IL groups, which include imidazolium, phosphonium, and ammonium ILs, as well as polycharged ILs.
Fig. 3.
Total records and unique structures for each group of ILs in the dataset (a) and unique structure, cation and anion counts for imidazolium, polycharged, ammonium, and phosphonium ILs in the dataset (b). Count = number of records in the dataset.
Second, we estimated the counts of unique IL structures, specific anions and cations, cell lines and experimental methods across all the groups of ILs and in the four most populated groups (see Table S1 in the Supporting Information). Information about the remaining IL groups is provided in Table S2. Among these groups, imidazolium ILs constitute 36% of the entries in the dataset, representing 314 unique molecular structures, which consist of 130 unique cations and 63 unique anions. The cytotoxicity of these imidazolium ILs was studied in 104 cell lines by using 13 different methods. Phosphonium ILs constitute 15% of the entries; there are 98 unique structures consisting of 66 cations and 21 anions that were tested in 104 cell lines by 11 different methods. Polycharged compounds constitute 12% of the entries; they are represented by 184 unique structures including 94 cations and 51 anions. Polycharged materials were tested in 27 cell lines by seven methods. Finally, ammonium ILs constituted 8% of the entries (127 unique structures). These ILs consist of 67 unique cations and 36 unique anions. The cytotoxicity of the ammonium ILs was studied in 93 cell lines by 11 methods.
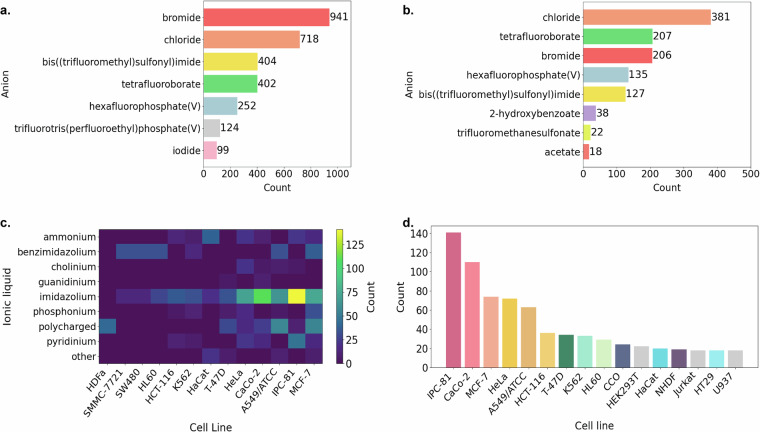
At the next stage, we assessed the distribution of the unique anions in the ILs present in the dataset. For this purpose, a dedicated dataset containing information on anions and their respective counts was created. In particular, we detected anions with counts exceeding 99, thereby identifying the most prevalent compounds within the dataset. The obtained distribution was visualized by means of Matplotlib (see Fig. 4a). The most prevalent anions were those of the halide family, namely, bromide and chloride. Other popular anions included bis((trifluoromethyl)sulfonyl)imide, tetrafluoroborate, hexafluorophosphate, trifluorotris(perfluoroethyl)phosphate, and iodide.
Fig. 4.
Most common anions found in (a) all groups of ILs present in the dataset and (b) imidazolium ILs. (c) Cell lines most commonly used in studies on the cytotoxicity of ILs. (d) Cell lines most commonly used in studies on the cytotoxicity of imidazolium ILs. Count = number of records in the dataset.
The imidazolium ILs, the most populated subclass in the dataset, predominantly contained chloride or bromide anions (see Fig. 4b). In addition, a significant number of imidazolium ILs contained tetrafluoroborate, hexafluorophosphate, and bis((trifluoromethyl)sulfonyl)imide anions.
We also analyzed the distributions of the cell lines and methods utilized for studies on the cytotoxicity of ILs. The choice of a particular cell line and experimental method is important for evaluating the cytotoxicity of chemical substances. Using appropriate cell lines is crucial for ensuring the relevance and applicability of the findings to more complex biological systems40. Furthermore, the method employed for the cytotoxicity assessment, whether it involves cell viability assays, cell proliferation assays, or other techniques, significantly influences the accuracy and reliability of the results. Therefore, careful consideration of both cell lines and methodologies is essential for robust and unambiguous conclusions regarding the cytotoxic effects of ILs.
The complete dataset comprises 17 groups of ILs and 155 cell lines. Figure 4c shows the distribution of ILs by the cell lines, in which the cytotoxicity tests were conducted more than five times. The colors in the figure represent the number of measurements. Consequently, our analysis revealed nine groups of ILs and identified 13 types of cell lines that were most commonly used for these experiments. All these cell lines were found in the entries of imidazolium ILs, with IPC-81 being the most frequently utilized cell line (see Fig. 4d; only those cell lines, in which the tests were carried out more than 16 times, are shown).
The appropriate selection of the method is fundamental in the cytotoxicity assessment because it can have a considerable impact on the comprehensiveness and accuracy of the results40. Various assays, such as cell viability assays (e.g., 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay and lactate dehydrogenase (LDH) assay), cell proliferation assays (e.g., bromodeoxyuridine (BrdU) assay and carboxyfluorescein succinimidyl ester (CFSE) assay), and apoptosis assays (e.g., propidium iodide (PI)-Annexin V assay and terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay), offer distinct insights into different aspects of the cellular response to chemical substances41. The choice of the most suitable assay depends on the research objectives, the mechanisms of action of the tested compounds, and the desired endpoints. Thus, by carefully selecting both the appropriate cell line and methodology, researchers can gain deeper insights into the cytotoxic effects of chemical substances and better understand their potential impact on biological systems.
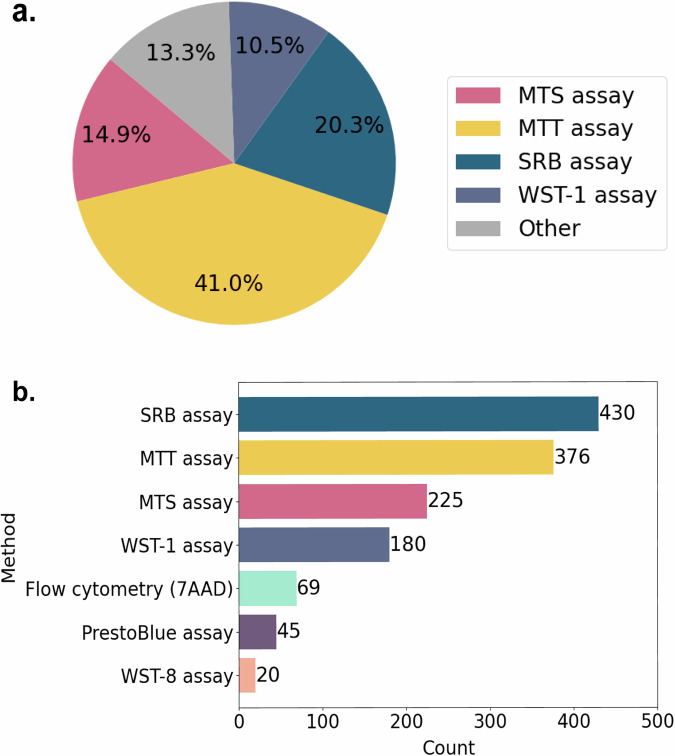
Figure 5a shows the experimental approaches most commonly used in studies on the cytotoxicity of ILs. These assays include the MTT, 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS), Sulforhodamine B (SRB) and water-soluble tetrazolium salt (WST-1) assays. Figure 5b shows the methods most commonly used in studies on the cytotoxicity of imidazolium ILs. Among the two most popular assays, the SRB assay relies on the binding of a dye to cellular proteins under acidic conditions, whereas the MTT assay measures the reduction of a yellow reagent to a purple formazan product by mitochondrial enzymes41. The SRB assay is generally more sensitive and faster than the MTT assay, making it suitable for high-throughput screening and drug discovery studies. Additionally, the SRB assay provides straightforward quantification using spectrophotometry42. The MTT assay, while widely used and reproducible, may be less sensitive and requires additional steps for solubilization of the formazan product.
Fig. 5.
Methods most commonly used for studying the cytotoxicity of (a) ILs in whole and (b) imidazolium ILs. Count = number of records in the dataset.
Usage Notes
Before 2019, most attempts24,43–51 to predict the biological activity of ILs using QSAR models relied on the UFT Merck Ionic Liquids Biological Effects Database, which has been discontinued. In some works, authors also used the OCHEM (Online Chemical Modeling Environment)52. OCHEM is a collection of chemical structures and measured biological activities and properties of ILs. In this repository, the structure of ILs can be entered as a mixture of separate ions encoded by SMILES. The database contains some ILs with antimicrobial data points (minimum inhibitory concentration (MIC) values). It is used mainly for QSAR predictions53,54 but not as a multipurposed repository of information. The results of predictions using QSAR modeling showed that three types of molecular fingerprints were able to represent the structural features of ILs properly55. Furthermore, deep learning models did not demonstrate better predictive ability than traditional machine learning models, which was attributed mainly to the relatively small datasets used30.
Thus, the main aim of this work is to provide researchers in the field with a new multipurposed data repository on standardized experimental values of the cytotoxicity of common ILs used in chemistry and biotechnology. This repository will be useful both as a source of information on studies on IL cytotoxicity carried out thus far and as a basis for building prediction models. The secondary aim concerns the need to understand the biological activity of ILs in more detail. Unraveling the mechanisms of action of ILs in living systems is a very complex undertaking. At the moment, this area is in stagnation. Further analysis of the available data will require the use of artificial intelligence algorithms, which will possibly lead to a prominent and perceptible revolution. However, no publicly available database exists to be processed by artificial intelligence. For the first time, we present such a repository with full access to the annotated data.
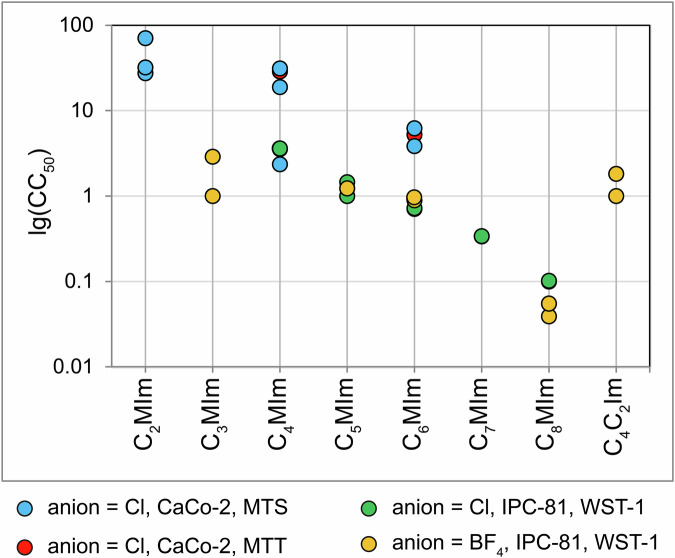
An overview of the “cell line-method” combinations derived from the dataset is provided in Table S3 in the Supporting Information. Note that, according to Fig. 6, there are pronounced differences between the measurements carried out for the same ILs by different research groups. Since the experimental setup – even such as cultural media and plastics used – can have a significant impact on the results, such discrepancies are inevitable. This is why we think it is necessary to supply the measured IC50/EC50/CC50 values with appropriate statistics, preferably confidence intervals. In each doubtful case, we advise the users to refer to original publications to gain their own impression of the quality of the data.
Fig. 6.
Comparison of 24-h CC50 values of 3-alkyl-1-methylimidazolium (CnMIm) ILs with chloride or tetrafluoroborate anions measured by various scientific groups using various assays. The y-axis corresponds to the Log10 of the CC50/IC50/EC50 values in mM. CaCo-2, CaCo-2 cell line; IPC-81, IPC-81 cell line; MTS, MTS assay; MTT, MTT assay; WST-1, WST-1 assay (see the legend below the plot).
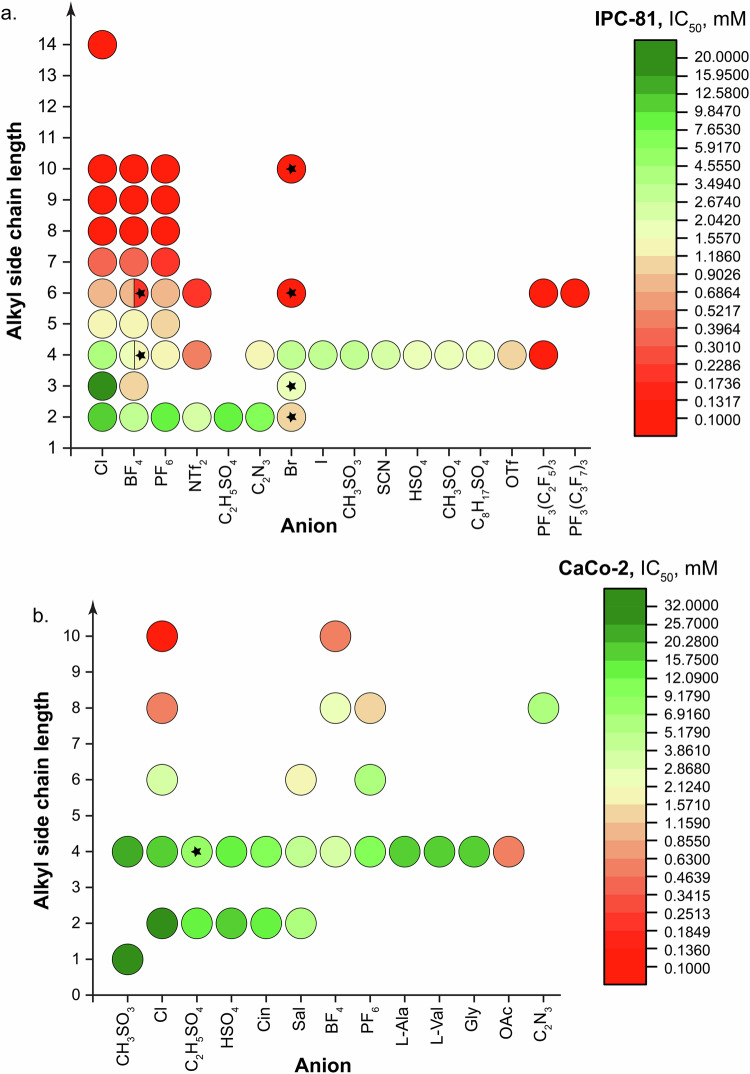
The effects of the alkyl side chain length and anion are demonstrated by the example of 3-alkyl-1-methylimidazolium ILs studied in the IPC-81 and CaCo-2 cell lines (see Fig. 7a and b, respectively). According to the data obtained in IPC-81 cells, long alkyl side chains in the cation (six carbon atoms or more) were the major structural factor governing the cytotoxicity, whereas the impact of the anion was lower in comparison (see Fig. 7a). However, there were two anions – that is, trifluorotris(perfluoroethyl)phosphate(V) (PF3(C2F5)3) and trifluorotris(perfluoropropyl)phosphate(V) (PF3(C3F7)3) – which rendered the ILs highly cytotoxic. On the contrary, the chloride anion seemed the least harmful among all the anions tested.
Fig. 7.
Impact of the alkyl side chain length and anion on the IL cytotoxicity by the example of 3-alkyl-1-methylimidazolium ILs tested in (a) IPC-81 using the WST-1 assay and (b) CaCo-2 cells using the MTT or MTS assays. The color of the circles corresponds to the CC50/IC50/EC50 value for a given IL (see the legends on the right). ILs with 3-alkyl-1-ethylimidazolium cations are designated by asterisks.
Similar observations could be made on the basis of the data obtained in CaCo-2 cells, in which case the acetate anion had the greatest impact on the cytotoxicity of the 3-butyl-1-methylimidazolium ILs studied, whereas the methyl sulfate anion contributed to the least toxic IL (see Fig. 7b).
Supplementary information
Acknowledgements
This work was funded by the Russian Science Foundation grant #24-63-00002.
Author contributions
L.A.A. developed the dataset format, prepared the dataset, annotated the papers, uploaded the dataset to a public repository, and prepared the draft of the manuscript; D.M.A. and M.M.S. annotated the papers and validated the records; A.V.V., L.T.S. and S.K.K. annotated the papers; A.V.P. validated and corrected the cell line information; K.S.E. designed the concept of the study, coordinated the works on the project, developed the dataset format, validated the records, and prepared the draft of the manuscript; V.P.A. developed the idea of this project, supervised the project, designed the concept of the study, coordinated the works on the project, and was involved in the preparation of the draft of the manuscript. All the authors were involved in the preparation of the final version of the manuscript.
Code availability
No custom code was used for the curation and validation of the dataset. Entries in the dataset do not require additional code for accessing the data.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ksenia S. Egorova, Email: egorova-ks@ioc.ac.ru
Valentine P. Ananikov, Email: val@ioc.ac.ru
Supplementary information
The online version contains supplementary material available at 10.1038/s41597-024-04190-3.
References
- 1.Egorova, K. S., Gordeev, E. G. & Ananikov, V. P. Biological Activity of Ionic Liquids and Their Application in Pharmaceutics and Medicine. Chem. Rev.117, 7132–7189, 10.1021/acs.chemrev.6b00562 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Welton, T. Ionic liquids: a brief history. Biophys. Rev.10, 691–706, 10.1007/s12551-018-0419-2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaur, G., Kumar, H. & Singla, M. Diverse applications of ionic liquids: A comprehensive review. J. Mol. Liq.351, 118556, 10.1016/j.molliq.2022.118556 (2022). [Google Scholar]
- 4.Shi, J. & Bent, S. F. Bridging the Synthesis Gap: Ionic Liquids Enable Solvent-Mediated Reaction in Vapor-Phase Deposition. ACS Nano15, 3004–3014, 10.1021/acsnano.0c09329 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Liu, Y. et al. Degradation of poly(ethylene terephthalate) catalyzed by metal-free choline-based ionic liquids. Green Chem.22, 3122–3131, 10.1039/d0gc00327a (2020). [Google Scholar]
- 6.Liu, L. Production of chemicals from marine biomass catalysed by acidic ionic liquids. Green Chem.23, 9800–9814, 10.1039/d1gc03249f (2021). [Google Scholar]
- 7.Egorova, K. S., Posvyatenko, A. V., Larin, S. S. & Ananikov, V. P. Ionic liquids: prospects for nucleic acid handling and delivery. Nucleic Acids Res.49, 1201–1234, 10.1093/nar/gkaa1280 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fatima, U., Yadav, N. & Venkatesu, P. Sustainable combination of ionic liquid and deep eutectic solvent for protecting and preserving of the protein structure: The synergistic interaction of enzymes and eco-friendly hybrid ionic fluids. Int. J. Biol. Macromol.268, 131997, 10.1016/j.ijbiomac.2024.131997 (2024). [DOI] [PubMed] [Google Scholar]
- 9.Sindhu, A., Kumar, S. & Venkatesu, P. Contemporary Advancement of Cholinium-Based Ionic Liquids for Protein Stability and Long-Term Storage: Past, Present, and Future Outlook. ACS Sustainable Chem. Eng.10, 4323–4344, 10.1021/acssuschemeng.1c08595 (2022). [Google Scholar]
- 10.Curreri, A. M., Mitragotri, S. & Tanner, E. E. L. Recent Advances in Ionic Liquids in Biomedicine. Adv. Sci. 8, 10.1002/advs.202004819 (2021). [DOI] [PMC free article] [PubMed]
- 11.Shamshina, J. L. & Rogers, R. D. Ionic Liquids: New Forms of Active Pharmaceutical Ingredients with Unique, Tunable Properties. Chem. Rev.123, 11894–11953, 10.1021/acs.chemrev.3c00384 (2023). [DOI] [PubMed] [Google Scholar]
- 12.Koutsoukos, S., Philippi, F., Malaret, F. & Welton, T. A review on machine learning algorithms for the ionic liquid chemical space. Chem. Sci.12, 6820–6843, 10.1039/d1sc01000j (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Armand, M., Endres, F., MacFarlane, D. R., Ohno, H. & Scrosati, B. Ionic-liquid materials for the electrochemical challenges of the future. Nat. Mater.8, 621–629, 10.1038/nmat2448 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Matuszek, K. et al. Unexpected energy applications of ionic liquids. Adv. Mater., 10.1002/adma.202313023 (2024). [DOI] [PubMed]
- 15.Guz, A. N. & Rushchitsky, J. J. Scopus: A system for the evaluation of scientific journals. Int. J. Appl. Mech.45, 351–362, 10.1007/s10778-009-0189-4 (2009). [Google Scholar]
- 16.Egorova, K. S. & Ananikov, V. P. Toxicity of Ionic Liquids: Eco(cyto)activity as Complicated, but Unavoidable Parameter for Task‐Specific Optimization. ChemSusChem7, 336–360, 10.1002/cssc.201300459 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Ma, J., Li, X., Cui, M., Li, W. & Li, X. Negative impact of the imidazolium-based ionic liquid [C8mim]Br on silver carp (Hypophthalmichthys molitrix): Long-term and low-level exposure. Chemosphere213, 358–367, 10.1016/j.chemosphere.2018.09.075 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Egorova, K. S., Kibardin, A. V., Posvyatenko, A. V. & Ananikov, V. P. Mechanisms of Biological Effects of Ionic Liquids: From Single Cells to Multicellular Organisms. Chem. Rev.124, 4679–4733, 10.1021/acs.chemrev.3c00420 (2024). [DOI] [PubMed] [Google Scholar]
- 19.Kumar, S. et al. Effect of the Alkyl Chain Length of Amphiphilic Ionic Liquids on the Structure and Dynamics of Model Lipid Membranes. Langmuir35, 12215–12223, 10.1021/acs.langmuir.9b02128 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Kumari, P., Pillai, V. V. S., Rodriguez, B. J., Prencipe, M. & Benedetto, A. Sub-Toxic Concentrations of Ionic Liquids Enhance Cell Migration by Reducing the Elasticity of the Cellular Lipid Membrane. J. Phys. Chem. Lett.11, 7327–7333, 10.1021/acs.jpclett.0c02149 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Benedetto, A. Ionic liquids meet lipid bilayers: a state-of-the-art review. Biophys. Rev.15, 1909–1939, 10.1007/s12551-023-01173-3 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumari, P., Pillai, V. V. S. & Benedetto, A. Mechanisms of action of ionic liquids on living cells: the state of the art. Biophys. Rev.12, 1187–1215, 10.1007/s12551-020-00754-w (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Varnek, A. & Baskin, I. Machine Learning Methods for Property Prediction in Chemoinformatics:Quo Vadis? J. Chem. Inf. Model.52, 1413–1437, 10.1021/ci200409x (2012). [DOI] [PubMed] [Google Scholar]
- 24.Cao, L., Zhu, P., Zhao, Y. & Zhao, J. Using machine learning and quantum chemistry descriptors to predict the toxicity of ionic liquids. J. Hazard. Mater.352, 17–26, 10.1016/j.jhazmat.2018.03.025 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Ghanem, O. B., Mutalib, M. I. A., Lévêque, J.-M. & El-Harbawi, M. Development of QSAR model to predict the ecotoxicity of Vibrio fischeri using COSMO-RS descriptors. Chemosphere170, 242–250, 10.1016/j.chemosphere.2016.12.003 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Ranjan, P. et al. Appraisal of 1‐Butylimidazole‐Derived Ionic Liquids as Anthelmintic Agents: An Experimental and In Silico Approach. ChemistrySelect3, 7518–7526, 10.1002/slct.201800402 (2018). [Google Scholar]
- 27.Dong, Q. et al. ILThermo: A Free-Access Web Database for Thermodynamic Properties of Ionic Liquids. J. Chem. Eng. Data52, 1151–1159, 10.1021/je700171f (2007). [Google Scholar]
- 28.Zhang, S., Sun, N., He, X., Lu, X. & Zhang, X. Physical Properties of Ionic Liquids: Database and Evaluation. J. Phys. Chem. Ref. Data35, 1475–1517, 10.1063/1.2204959 (2006). [Google Scholar]
- 29.Onken, U., Rarey-Nies, J. & Gmehling, J. The Dortmund Data Bank: A computerized system for retrieval, correlation, and prediction of thermodynamic properties of mixtures. Int. J. Thermophys.10, 739–747, 10.1007/bf00507993 (1989). [Google Scholar]
- 30.Yan, J. et al. ILTox: A Curated Toxicity Database for Machine Learning and Design of Environmentally Friendly Ionic Liquids. Environ. Sci. Technol. Lett.10, 983–988, 10.1021/acs.estlett.3c00106 (2023). [Google Scholar]
- 31.Cousins, K. R. ChemDraw Ultra 9.0. CambridgeSoft, 100 CambridgePark Drive, Cambridge, MA 02140. J. Am. Chem. Soc.127, 4115–4116, 10.1021/ja0410237 (2005). [Google Scholar]
- 32.Gabrielson, S. W. SciFinder. Bull. Med. Libr. Assoc. 106, 10.5195/jmla.2018.515 (2018).
- 33.Eller, G. A. Improving the Quality of Published Chemical Names with Nomenclature Software. Molecules11, 915–928, 10.3390/11110915 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim, S. et al. PubChem 2023 update. Nucleic Acids Res.51, D1373–D1380, 10.1093/nar/gkac956 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arakelyan, L. A., et alA comprehensive dataset on cytotoxicity of ionic liquids, 10.6084/m9.figshare.27979307 (2024). [DOI] [PMC free article] [PubMed]
- 36.Bento, A. P. et al. An open source chemical structure curation pipeline using RDKit. J. Cheminf. 12, 10.1186/s13321-020-00456-1 (2020). [DOI] [PMC free article] [PubMed]
- 37.Viloria, A. et al. Integration of Data Mining Techniques to PostgreSQL Database Manager System. Procedia Comput. Sci.155, 575–580, 10.1016/j.procs.2019.08.080 (2019). [Google Scholar]
- 38.Chauhan, R. & Oberoi, A. Visualizing data using Matplotlib and Seaborn libraries in Python for data science. IJSRP9, 10.29322/IJSRP.9.03.2019.p8733 (2019).
- 39.Stancin, I. & Jovic, A. in 2019 42nd International Convention on Information and Communication Technology, Electronics and Microelectronics (MIPRO) 977–982 (2019).
- 40.Riss, T. L., Moravec, R. A. & Niles, A. L. Cytotoxicity testing: Measuring viable cells, dead cells, and detecting mechanism of cell death. Mammalian Cell Viability: Methods and Protocols740, 103–114, 10.1007/978-1-61779-108-6_12 (2011). [DOI] [PubMed] [Google Scholar]
- 41.Kamiloglu, S., Sari, G., Ozdal, T. & Capanoglu, E. Guidelines for cell viability assays. Food Front.1, 332–349, 10.1002/fft2.44 (2020). [Google Scholar]
- 42.Keepers, Y. P. et al. Comparison of the sulforhodamine B protein and tetrazolium (MTT) assays for in vitro chemosensitivity testing. Eur. J. Cancer Clin. Oncol.27, 897–900, 10.1016/0277-5379(91)90142-z (1991). [DOI] [PubMed] [Google Scholar]
- 43.Cho, C. W. et al. In silico modelling for predicting the cationic hydrophobicity and cytotoxicity of ionic liquids towards the Leukemia rat cell line,Vibrio fischeri and Scenedesmus vacuolatus based on molecular interaction potentials of ions. SAR QSAR Environ. Res.24, 863–882, 10.1080/1062936x.2013.821092 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Fatemi, M. H. & Izadiyan, P. Cytotoxicity estimation of ionic liquids based on their effective structural features. Chemosphere84, 553–563, 10.1016/j.chemosphere.2011.04.021 (2011). [DOI] [PubMed] [Google Scholar]
- 45.Ma, S. et al. Predicting the ecotoxicity of ionic liquids towards Vibrio fischeri using genetic function approximation and least squares support vector machine. J. Hazard. Mater.283, 591–598, 10.1016/j.jhazmat.2014.10.011 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Cruz-Monteagudo, M. & Cordeiro, M. N. D. S. Chemoinformatics Profiling of Ionic Liquids—Uncovering Structure-Cytotoxicity Relationships With Network-like Similarity Graphs. Toxicol. Sci.138, 191–204, 10.1093/toxsci/kft210 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Melo, E. B. d. A structure–activity relationship study of the toxicity of ionic liquids using an adapted Ferreira–Kiralj hydrophobicity parameter. Phys. Chem. Chem. Phys.17, 4516–4523, 10.1039/c4cp04142a (2015). [DOI] [PubMed] [Google Scholar]
- 48.Zhao, Y. et al. Toxicity of ionic liquids: Database and prediction via quantitative structure–activity relationship method. J. Hazard. Mater.278, 320–329, 10.1016/j.jhazmat.2014.06.018 (2014). [DOI] [PubMed] [Google Scholar]
- 49.Das, R. N. & Roy, K. Predictive modeling studies for the ecotoxicity of ionic liquids towards the green algae Scenedesmus vacuolatus. Chemosphere104, 170–176, 10.1016/j.chemosphere.2013.11.002 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Farahani, S. R., Sohrabi, M. R. & Ghasemi, J. B. A detailed structural study of cytotoxicity effect of ionic liquids on the leukemia rat cell line IPC-81 by three dimensional quantitative structure toxicity relationship. Ecotoxicol. Environ. Saf.158, 256–265, 10.1016/j.ecoenv.2018.04.040 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Izadiyan, P., Fatemi, M. H. & Izadiyan, M. Elicitation of the most important structural properties of ionic liquids affecting ecotoxicity in limnic green algae; a QSAR approach. Ecotoxicol. Environ. Saf.87, 42–48, 10.1016/j.ecoenv.2012.10.005 (2013). [DOI] [PubMed] [Google Scholar]
- 52.Sushko, I. et al. Online chemical modeling environment (OCHEM): web platform for data storage, model development and publishing of chemical information. J. Comput.-Aided Mol. Des.25, 533–554, 10.1007/s10822-011-9440-2 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hodyna, D. et al. Imidazolium ionic liquids as effective antiseptics and disinfectants against drug resistant S. aureus: In silico and in vitro studies. Comput. Biol. Chem.73, 127–138, 10.1016/j.compbiolchem.2018.01.012 (2018). [DOI] [PubMed] [Google Scholar]
- 54.Trush, M. M. et al. New 1,3-oxazolylphosphonium Salts as Potential Biocides: QSAR Study, Synthesis, Antibacterial Activity and Toxicity Evaluation. Lett. Drug Des. Discovery15, 1259–1267, 10.2174/1570180815666180219164334 (2018). [Google Scholar]
- 55.Abramenko, N. et al. A review of recent advances towards the development of QSAR models for toxicity assessment of ionic liquids. J. Hazard. Mater. 384, 10.1016/j.jhazmat.2019.121429 (2020). [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Arakelyan, L. A., et alA comprehensive dataset on cytotoxicity of ionic liquids, 10.6084/m9.figshare.27979307 (2024). [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
No custom code was used for the curation and validation of the dataset. Entries in the dataset do not require additional code for accessing the data.