Abstract
Angioimmunoblastic T-cell lymphoma (AITL) is a kind of aggressive T-cell lymphoma with significant enrichment of non-malignant tumor microenvironment (TME) cells. However, the complexity of TME in AITL progression is poorly understood. We performed single-cell RNA-Seq (scRNA-seq) and imaging mass cytometry (IMC) analysis to compare the cellular composition and spatial architecture between relapsed/refractory AITL (RR-AITL) and newly diagnosed AITL (ND-AITL). Our results showed that the malignant T follicular helper (Tfh) cells showed significantly increased proliferation driven by transcriptional activation of YY1 in RR-AITL, which is markedly associated with the poor prognosis of AITL patients. The CD8+ T cell proportion and cytotoxicity decreased in RR-AITL TME, resulting from elevated expression of the inhibitory checkpoints such as PD-1, TIGIT, and CTLA4. Notably, the transcriptional pattern of B cells in RR-AITL showed an intermediate state of malignant transformation to B-cell-lymphoma, and contributed to immune evasion by highly expressing CD47 and PD-L1. Besides, compared to ND-AITL samples, myeloid-cells-centered spatial communities were more prevalent but showed reduced phagocytic activity and impaired antigen processing and presentation in RR-AITL TME. Furthermore, specific inhibitory ligand-receptor interactions, such as CLEC2D-KLRB1, CTLA4-CD86, and MIF-CD74, were exclusively identified in the RR-AITL TME. Our study provides a high-resolution characterization of the immunosuppression ecosystem and reveals the potential therapeutic targets for RR-AITL patients.
Subject terms: Cell biology, T-cell lymphoma, Cancer microenvironment
Introduction
Angioimmunoblastic T-cell lymphoma (AITL) is a subtype of mature T-cell lymphoma, representing ~1–2% of the non-Hodgkin’s lymphomas and about 20% of the peripheral T-cell lymphoma (PTCL) [1]. The majority of newly diagnosed AITL (ND-AITL) patients are at an advanced stage and characterized by generalized lymphadenopathy, hepatosplenomegaly, bone marrow involvement, and symptoms related to immune activation such as high fever and skin rash [2–5]. AITL is aggressive and has a poor prognosis, with a median survival time of less than three years [3, 5]. Currently, there are no standard treatment strategies for AITL [6]. Chemotherapy is the most commonly used regimen for AITL patients, despite its limitations in terms of primary progression or short duration of remission [7, 8]. The median overall survival (OS) of relapsed/refractory AITL (RR-AITL) patients is about six months [9]. Thus, it is urgent to elucidate the underlying mechanisms and identify potential therapeutic targets for AITL, particularly in RR-AITL patients.
According to gene expression profiles of AITL tissue and isolated tumor cells, previous studies have confirmed that the cellular origin of AITL is follicular helper T (Tfh) cells [10]. It should be noticed that the malignant cells only occupy a minority of the AITL ecosystem but with massive enrichment of microenvironment cells with multiple lineages, including CD4+ or CD8+ T cells, B cells, plasma cells, macrophages, follicular dendritic cells (fDCs) and high endothelial venules (HEVs) [6, 11]. Due to the low tumor cell burden, AITL has been designated as a kind of immuno-dysplastic disease rather than neoplasm for a long time [4, 6]. It has been reported that some pathways or signatures of the immune microenvironment were related to AITL pathogenesis and prognosis [3, 12]. And the immunomodulatory strategies may have promising potential for the treatment of AITL [4]. However, the conventional tissue-based technology such as bulk RNA-seq or microarray limited the power of dissecting complicated tumor microenvironment (TME) for AITL. Further analyses based on high-resolution technologies are anticipated to identify novel immunomodulatory targets for AITL.
The emergence of single-cell technology provides an unprecedented opportunity to investigate the microenvironment components of AITL. Pritchett et al. utilized mass cytometry and single-cell transcriptome analysis to compare the TME between AITL patients and normal control (NC) samples, revealing the remodeling of CD8+ T cell and B cell populations in AITL TME [13]. Study from Fujisawa et al. observed the marked expansion of abnormal germinal center B (GCB) cells could promote AITL development through the CD40-CD40LG axis in a mouse model, highlighting the important role of B cells in AITL occurrence [14]. A recent study investigated the tumor heterogeneity, clonal evolution, and immunoevasion phenotypes in T follicular cell lymphoma at single-cell resolution, further suggesting the importance of TME in the progression of T follicular cell lymphoma [15]. However, previous studies primarily focused on the inter-group differences in proportions and/or phenotypes of cell types, while the spatial organization and molecular interactions of cell-cell communications, and their roles in therapy resistance and tumor progression remain poorly understood.
Here, we applied single-cell RNA sequencing (scRNA-seq) and imaging mass cytometry (IMC) techniques for ND-AITL and RR-AITL samples to investigate the specific cellular and molecular alterations in RR-AITL. Our findings demonstrate that RR-AITL samples exhibit a substantial increase in proliferative malignant cells and an immunosuppressive TME, primarily driven by the depletion of CD8+ T cells and dysfunction of myeloid and B cells. This comprehensive study reveals the distinctive cellular states and spatial architectures presented in RR-AITL patients, highlighting potentially effective therapeutic targets for RR-AITL patients.
Results
Single-cell transcriptomic and spatial analyses deconstruct the cellular ecosystem of ND- and RR-AITL patients
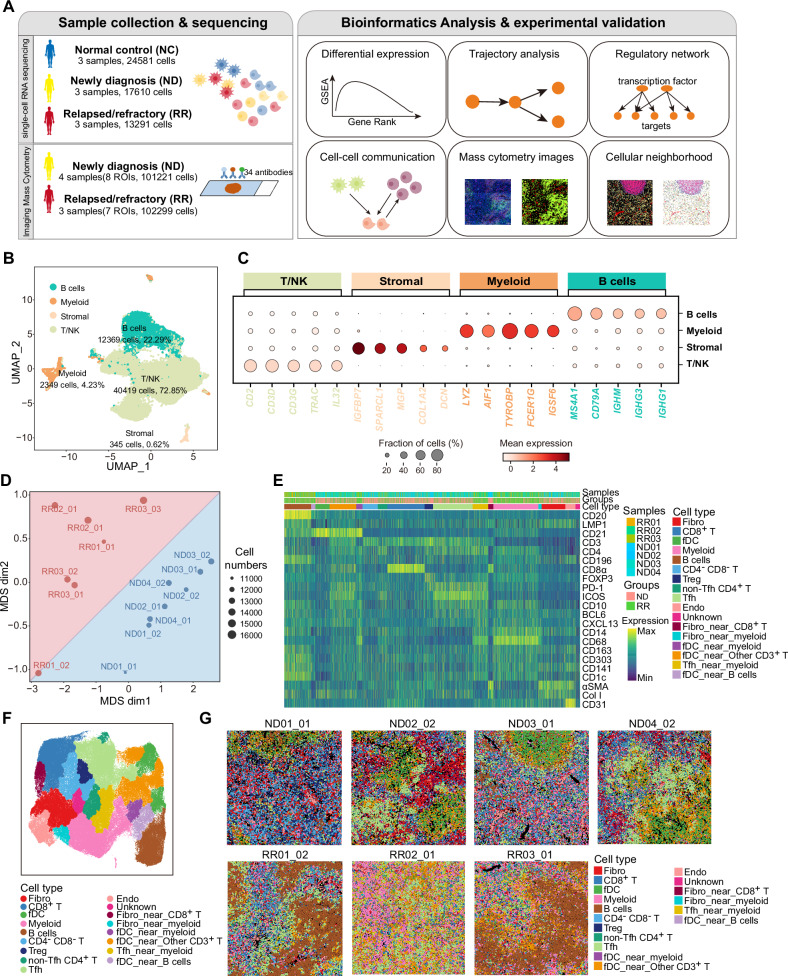
Based on the scRNA-seq and IMC data, we comprehensively dissected and compared the TME of lymphoid tissues from NC, ND-AITL, and RR-AITL patients (Fig. 1A). The detailed clinical characteristics of these individuals were summarized in Supplementary Table 1. After quality control (see Methods), we got transcriptomic data for a total of 55,482 single cells, including 24,581 cells from NC, 17,610 cells from ND-AITL, and 13,291 cells from RR-AITL. Additionally, a total of 101,221 cells from ND-AITL and 102,299 cells from RR-AITL were collected within the IMC dataset, respectively (Fig. 1A).
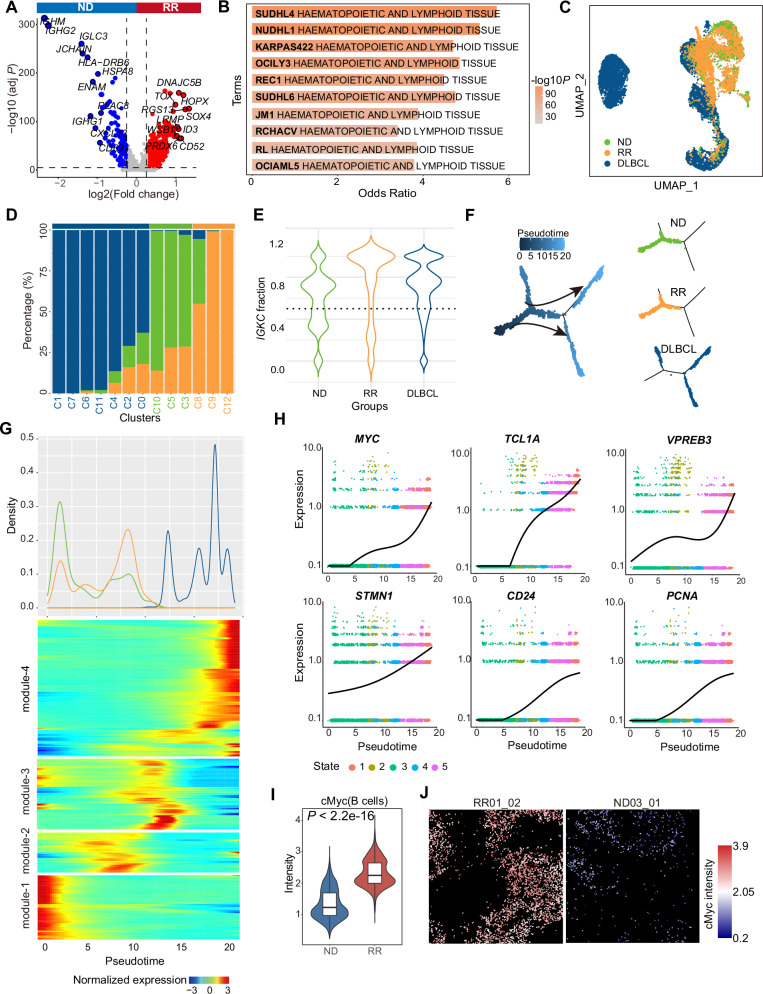
Fig. 1. Comparison of the TME composition between ND-AITL and RR-AITL at the single-cell level.
A Overview of sample collection, computational analysis and experimental validation workflow. B UMAP plot of 55482 cells obtained from scRNA-seq profiles from 9 samples, colored by four main cell types. The cell counts and cell proportions for each cell type are labeled on the graph. C Dot plot depicting the expression of representative markers for four distinct cell types. Dot size is indicative of the proportion of cells within each cell type expressing these markers, and dot color represents the mean expression levels of these markers within each cell type. D MDS analysis demonstrates the inter- and intragroup differences among different ROIs. The dots were color-coded to indicate groups, with red representing the RR-AITL group and blue representing the ND-AITL group. The size of the dots was proportional to the number of cells within the ROIs. E The heatmap illustrates the protein levels of classical marker genes within individual cell. For the sake of clarity, a random sample of 5000 cells was selected for visualization. F The UMAP plot illustrates the identified cell types within the IMC dataset. G The cell types were visually represented in the images, with different colors denoting distinct cell types. One representative ROI was shown for each sample. The upper panel comprises four images from ND-AITL, while the lower panel displays three images from RR-AITL.
We integrated the transcriptomic data of single cells and performed unsupervised clustering, resulting in 26 clusters (Supplementary Fig. S1A–C). Based on the expression of well-established gene markers, four cell types were identified, including T/NK, B cells, myeloid, and stromal (Fig. 1B, C, Supplementary Fig. S1D). To further validate the accuracy of cell type annotations, we applied the SingleR method to perform unbiased cell type recognition. Results showed a high consistency between the cell type definitions based on these two methods (Supplementary Fig. S1E). Besides, the correlations of top five highly expressed genes in each cluster also validated the robustness of cell categories (Supplementary Fig. S1F, Supplementary Table 2). Furthermore, we found that the cellular components among three normal samples were identically consistent, whereas the samples from AITL patients showed a relatively higher inter-tumoral heterogeneity presented in the UMAP visualization (Supplementary Fig. S1G). Nonetheless, the percentage of these four main cell types identified in the scRNA-seq data had no obvious differences between ND-AITL and RR-AITL patients (Supplementary Fig. S1H), provoking us to further dissect and identify the detailed cellular and molecular plasticity in RR patients.
Besides, we employed IMC to comprehensively evaluate the cellular heterogeneity and spatial organization of the TME. Based on prior knowledge [6], we developed an IMC panel of three distinct categories of biomarkers, including cell types, immune status, and cellular state. This panel comprised 35 proteins specific to AITL histology (Supplementary Fig. S2A). These proteins enabled the identification of major cell types present in the AITL TME, including T cells, Tfh cells, macrophage, dendritic cells (DCs), fDCs, endothelial cells (Endo), B cells, and fibroblast (Fibro). Then we applied the panel to four ND-AITL and three RR-AITL samples, including eight and seven regions of interest (ROIs), respectively. The detailed clinical characteristics of these individuals were summarized in Supplementary Table 1.
We conducted single-cell segmentation and quality control to obtain high-quality spatial data at the single-cell level (see Methods, Supplementary Fig. S2B). Furthermore, we performed multidimensional scaling (MDS) analysis on the average expression of different markers in each ROI. The results showed a clear separation between ROIs from ND-AITL and RR-AITL (Fig. 1D). After batch correction and unsupervised clustering, we obtained 54 clusters of 16 distinct cell types (Supplementary Fig. S2C–F, Supplementary Table 3). The majority of these cell types were clearly separated based on the expression levels of their unique markers (Fig. 1E, F, Supplementary Fig. S2G). Similar to the previous IMC study [16], we also observed that some clusters co-expressed markers of different cell types simultaneously, such as fDC_near_myeloid. This phenomenon might be explained by the spatial co-localization of different cell types. The cellular composition varied between the RR group and the ND group across different ROIs and samples (Fig. 1G, Supplementary Fig. S2H). Taken together, these findings suggested the disparities in cellular proportions and states between RR-AITL and ND-AITL.
Malignant Tfh cells showed increased proliferation capacity in RR-AITL patients
Our initial emphasis was placed on delineating the distinct cellular states of malignant cells within the RR-AITL samples compared to ND-AITL samples. Hence, we re-clustered the T/NK populations to further investigate the cellular components of NC, ND-AITL, and RR-AITL. Ten clusters were identified, including T_C0~T_C9 (Fig. 2A). Based on the expression level of canonical markers, six subtypes were annotated (Fig. 2A, Supplementary Fig. S3A, B, Supplementary Table 4), including naïve T cells (Tnaive, including T_C0, T_C5, T_C7, and T_C8), CD4+ effector memory T cells (CD4+ Tem, T_C1), regulatory T cells (Treg, T_C6), follicular helper T (Tfh, including T_C2 and T_C4), CD8+ T (T_C3), and NK cells (T_C9). Tfh cells were further confirmed by analyzing the expression level of 7 antigens proposed by 2016 WHO classification for Tfh lymphomas diagnosis, including CXCL13, MME (CD10), ICOS, PDCD1 (CD279), BCL6, SH2D1A (SAP), and CXCR5 [17] (Supplementary Fig. S3A).
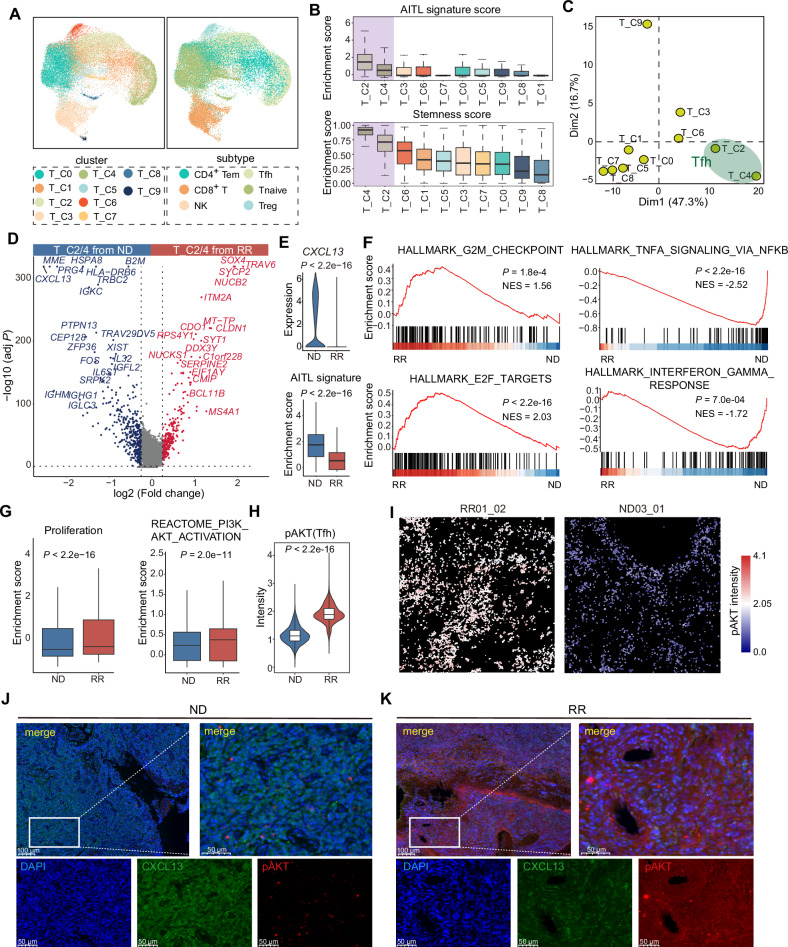
Fig. 2. Differences in the cellular states of malignant Tfh cells between ND-AITL and RR-AITL.
A UMAP visualization of various T/NK cell clusters (left) and subtypes (right). B Boxplot showing enrichment scores of AITL signature and stemness inferred by CytoTRACE across different T cells clusters. The boxplot represents the mean value and the error bar represents the standard error value. C Principal component analysis (PCA) of the clusters according to the mean transcription factor activity of each cluster. D Volcano plot of gene expression changes of Tfh cells between ND-AITL and RR-AITL. Upregulated (red) and downregulated (blue) genes in RR-AITL are highlighted. E Violin plot showing expression levels of CXCL13 in Tfh cells from ND-AITL and RR-AITL samples (upper panel). Violin plot showing enrichment score of AITL signatures in Tfh cells from ND-AITL and RR-AITL samples (bottom panel). P values were determined by two-sided unpaired Wilcoxon test. The boxplot represents the mean value and the error bar represents the standard error value. F GSEA plot showing the hallmarks that are significantly enriched in RR-AITL and ND-AITL, respectively. G Boxplot showing enrichment score of proliferation signature and PI3K/AKT pathway. P values were determined by two-sided unpaired Wilcoxon test. H Boxplot showing intensities of pAKT in IMC data. P values were determined by two-sided unpaired Wilcoxon test. I The IMC images showing the protein activity of pAKT within malignant Tfh cells. The colors represent protein activity levels, ranging from lower (blue) to higher (red). J, K Immunofluorescence analysis for ND-AITL and RR-AITL samples stained by an-ti-DAPI (blue), anti-CXCL13 (green), anti-pAKT (red) antibodies. Scale bar, 100 μm.
We found that Tfh cell clusters (T_C2 and T_C4) exhibit the lowest proportion of cells derived from NC samples (8.4% and 8.6%, respectively), and they showed the highest scores of AITL signature, stemness, and proliferation among all the T/NK clusters (Fig. 2B, Supplementary Fig. S3C, D), indicating the malignant phenotype of Tfh cells in AITL. Our findings revealed that a large number of transcription factors were specifically activated in T_C2 and T_C4 compared to other T cell clusters (Supplementary Fig. S3E). We performed principal component analysis (PCA) based on the activity of transcription factors (TFs) in single cells (see Methods), and observed that T_C2 and T_C4 clusters were separated from other T/NK clusters on the first two principal components (PCs), implying a distinct pattern of transcription regulation in malignant Tfh cells (Fig. 2C). In addition, we evaluated the activity of metabolism pathways among various T/NK clusters and found that Tfh clusters T_C2 and T_C4 had higher activity in a majority of metabolism pathways compared to other non-malignant clusters (Supplementary Fig. S3F). Overall, the analysis of transcriptional regulation and biological characteristics at the single-cell level provides evidence suggesting the involvement of Tfh in the pathogenesis of AITL, which is in agreement with prior knowledge of AITL.
Furthermore, we identified the significantly differently expressed genes between Tfh cells from ND- and RR- AITL (Fig. 2D, Supplementary Table 5). Notably, the classical Tfh markers CXCL13 and MME (CD10) were markedly downregulated in RR-AITL in comparison to ND-AITL (Fig. 2D, E). Immunohistochemistry (IHC) staining revealed that the proportion of CD10 and CXCL13 positive patients in the RR-AITL group was lower compared to the ND-AITL group, with a particularly notable decrease in CXCL13 positivity (Supplementary Fig. S3G). By exploring the expression of seven canonical AITL markers, we observed that the percentage of cells expressing these markers was significantly lower in the RR-AITL group compared to the ND-AITL group (Supplementary Fig. S3H). The enrichment score of AITL signature was also significantly lower in Tfh cells from RR-AITL (Wilcoxon rank sum test P < 2.2e-16; Fig. 2E). These results implied that the Tfh phenotype may lost in RR-AITL patients, which is similar to the erosion of chronic lymphocytic leukemia (CLL) cell identity observed by Rendeiro et al. [18].
As an essential transcriptional factor promoting tumor cell proliferation and stemness [19], SOX4 was markedly upregulated in RR-AITL (Fig. 2D). Moreover, the genes highly expressed in malignant Tfh cells from RR-AITL showed enrichment of proliferation-related hallmarks such as E2F targets (Normalized Enrichment Score (NES) = 2.03, P < 2.2e-16) and G2M checkpoint (NES = 1.56, P = 0.0018) (Fig. 2F, Supplementary Fig. S3I, Supplementary Table 6). Additionally, the proliferation signature score in malignant Tfh cells from RR-AITL was found to be higher compared to that in ND-AITL. This observation may be attributed to the significantly activated PI3K/AKT pathway in malignant Tfh cells from RR-AITL (Fig. 2G, Supplementary Fig. S3I). Protein levels of pAKT, as measured by IMC, were also found to be elevated in Tfh cells from RR-AITL patients in comparison to those from ND-AITL patients (Fig. 2H, I, Supplementary Fig. S3J). The immunofluorescence analysis further validated the loss of AITL phenotype and elevated pAKT level in RR-AITL (Fig. 2J, K).
Notably, pathways related to immune response such as IFN-γ and TNF-α signaling were significantly downregulated within malignant Tfh cells from RR-AITL compared to those from ND-AITL (Fig. 2F, Supplementary Fig. S3I, Supplementary Table 6), indicating the escape of immune surveillance.
Transcriptional factor YY1 is involved in the proliferation and immune evasion in RR-AITL
To elucidate the core TFs that govern the proliferation and modulating the immune response of RR-AITL, we compared the regulatory activity and expression level of TFs between RR-AITL and ND-AITL (see Methods). Our findings revealed significant upregulation in the activity and expression of four TFs, including YY1, REST, RUNX3, and ZNF91, in RR-AITL compared to ND-AITL (Fig. 3A, B, Supplementary Fig. S4A, Supplementary Table 7). Furthermore, YY1 and REST protein were found to be more abundant in malignant Tfh cells of RR-AITL compared to those in ND-AITL (Fig. 3C, Supplementary Fig. S4B). To comprehensively investigate the expression level of YY1, we compared the YY1 expression in Tfh cells and other T/NK subtypes derived from three different sample sources (Supplementary Fig. S4C). Results showed that malignant Tfh cells had significantly higher YY1 expression levels compared to other T cell types (including Tfh cells in NC samples). The proportion of YY1-expressing cells was also substantially higher in RR-AITL (Supplementary Fig. S4D). Additionally, malignant Tfh cells had significantly higher YY1 activity compared to other T/NK cell types including Tfh cells in NC samples (Fig. 3B, Supplementary Fig. S4E). The upregulation of YY1 in RR-AITL was validated by IMC dataset and immunofluorescence (Fig. 3D, F, Supplementary Fig. S4F). Meanwhile, the immunoreactive score (IRS) derived from IHC was significantly higher in RR-AITL, further supporting the activation of YY1 in RR-AITL (Supplementary Fig. S4G, H). These results collectively demonstrate the specific activation of YY1 in malignant Tfh cells from RR-AITL.
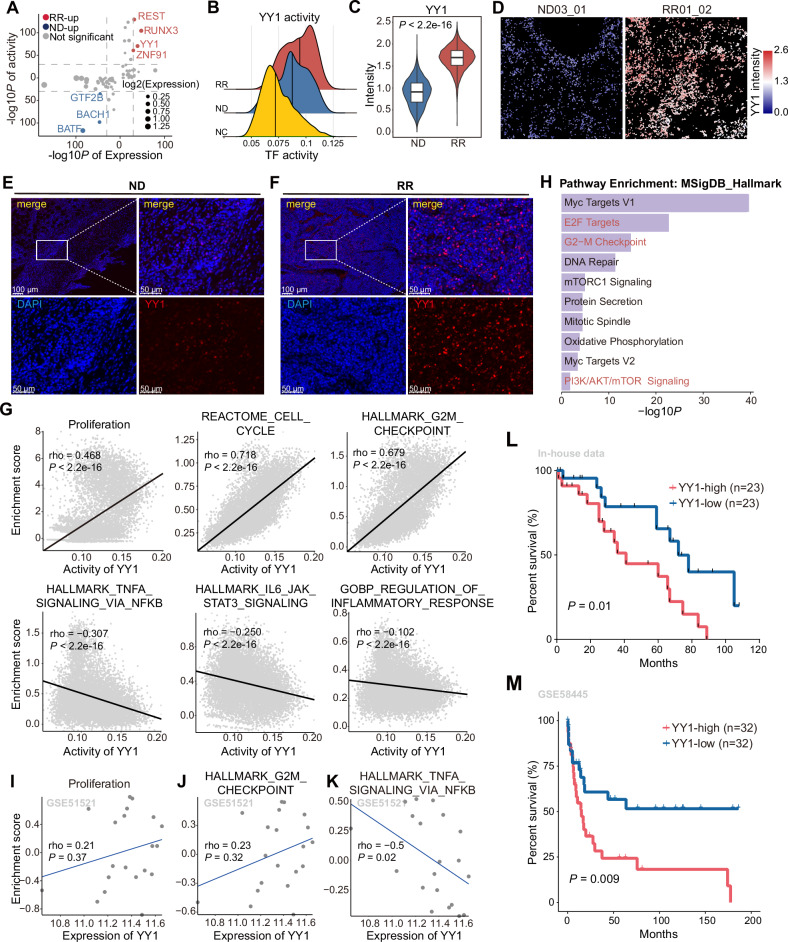
Fig. 3. The distinct transcriptional regulatory mechanisms of malignant Tfh cells in RR-AITL.
A Dot plot shows TFs that significantly activated in Tfh cells from RR-AITL compared to ND-AITL. Y axis represents -log10 transformation of adjusted P values of the activity of TFs inferred by pySCENIC, x axis represents -log10 transformation of adjusted P values of the expression of TFs. B Density plot shows the activity of YY1 across Tfh from NC (yellow), ND-AITL (blue) and RR-AITL (red). The vertical line represents the median of YY1 activity. C The intensities of YY1 between Tfh cells from ND-AITL and RR-AITL. The y axis represents the protein levels of YY1 in IMC dataset. D The protein activity of YY1 within malignant Tfh cells shown on IMC images. The colors represent protein activity levels, ranging from lower (blue) to higher (red). E, F Immunofluorescence analysis for ND-AITL (E) and RR-AITL (F) samples stained by anti-DAPI (blue) and anti-YY1(red) antibodies. Scale bar, 100 μm. G The spearman correlation between activity of YY1 and the enrichment score of pathways in malignant Tfh cells. Each data point represents an individual cell. H The functional enrichment analysis of YY1 target genes inferred by pySCENIC. The spearman correlation between YY1 expression and enrichment scores of proliferation (I), G2M checkpoint (J), TNFα signaling via NFκB (K) in GSE51521 dataset. Each data point represents individual sample. L Overall survival of AITL patients in our in-house cohort through Kaplan–Meier analysis. All patients were categorized into two groups based on the median YY1 expression level estimated by IHC. The significance was evaluated by the log-rank test. M Kaplan–Meier analysis was performed to assess overall survival in PTCL patients within the GSE58445 dataset. Only samples with available follow-up data were included in the analysis. Patients were stratified into four quartiles based on YY1 expression levels. For further comparison, the top 25% and bottom 25% of patients were specifically analyzed. The statistical significance of survival differences between these two groups was evaluated using the log-rank test.
In addition, we found that YY1 activity was positively correlated with proliferation-related pathways (e.g., cell cycle and G2M checkpoint) and negatively correlated with immune responses pathways (e.g., TNFα signaling, IL6/JAK/STAT3 signaling and regulation of inflammatory response) (Fig. 3G). In line with the aforementioned findings, YY1 target genes predicted by pySCENIC showed significant enrichment in proliferation-related hallmarks, including Myc targets, E2F targets, G2M checkpoint, and the PI3K/AKT/mTOR signaling pathway (Fig. 3H). By analyzing the overlap between PI3K/AKT/mTOR signaling pathway genes and YY1 target genes, we found that ~34% of the genes in the PI3K/AKT/mTOR signaling pathway were directly regulated by YY1 (Supplementary Fig. S4I). Furthermore, the expression levels of these YY1 target genes showed significant positive correlations with YY1 activity (Supplementary Fig. S4J). Moreover, we identified an enrichment of proliferation-related pathways in malignant Tfh cells exhibiting higher YY1 activity, while immune response pathways were enriched in those with lower YY1 activity (Supplementary Fig. S4K). By using a publicly available bulk RNA-seq dataset, we further validated the significantly positive correlation between YY1 expression and proliferation-related signatures (Fig. 3I, J). Meanwhile, we also observed a significant negative correlation with TNFα signaling in this dataset (Fig. 3K).
To elucidate the downstream mechanisms by which YY1 facilitates malignant Tfh proliferation, we investigated to recognize potential target genes. Based on intersection of known and predicted targets genes of YY1 (see Methods), four genes were identified, including CBX5, PCBP1, SMC3, and RAC1 (Supplementary Fig. S4L, M, Supplementary Table 8). Additionally, the expression level of YY1 was positively correlated with the expression of these genes in bulk RNA-seq data (Supplementary Fig. S4N). A large number of studies have shown that these genes are involved in cellular proliferation processes [20–23]. Notably, RAC1 is a member of the Rho family of GTPases and regulates multiple signaling pathways, such as NFκB and PI3K/AKT [24], implying that YY1 may activate PI3K/AKT pathway through an RAC1-dependent mechanism.
In addition, our in-house cohort containing 46 tumor tissue samples from AITL patients were collected and analyzed using immunohistochemistry to determine YY1 protein levels. The evaluation revealed a significant association between elevated YY1 expression and a poorer prognosis in AITL patients (P = 0.010; Fig. 3L). We further investigated the association between YY1 and prognosis in a public PTCL dataset. We found that PTCL patients with higher YY1 expression also had significantly poor prognosis (P = 0.009; Fig. 3M), indicating that YY1 may play an important role in tumor progression of PTCL patients.
The depletion of cytotoxic CD8+ T cells in RR-AITL compared to ND-AITL
Through a sophisticated interplay of signaling pathways, tumor cells actively remodel the TME to evade immune surveillance and support tumor progression. Considering the pivotal role of T cells in targeting and eliminating tumors within the TME, our study delves deeper into the cellular states of T cells in RR-AITL. We observed marked heterogeneity in the T/NK populations among NC, ND-, and RR-AITL (Fig. 4A). We analyzed the differential enrichment of T/NK subpopulations in these three groups (Fig. 4B, Supplementary Table 9). Tfh cells were significantly overrepresented in both ND-AITL and RR-AITL compared to NC. Besides, we observed a relative depletion of CD4+ Tem and CD8+ T cells in both ND-AITL and RR-AITL compared to NC. Moreover, the comparative results between ND-AITL and RR-AITL showed that Tregs were enriched in the RR group compared to the ND group with an odds ratio of 1.795 (P = 1.487e-13); while the CD8+ T cells had a significantly lower enrichment in RR group with an odds ratio of 0.831 (P = 7.967e-06) (Fig. 4B). The alterations in intergroup proportions of Tregs and CD8+ T cells were further validated by immunofluorescence staining (Fig. 4C). We also observed the diminishment of CD8+ T cells within the IMC dataset for RR-AITL (Fig. 4D, E, Supplementary Fig. S5A). Given the immune inhibitory role of Tregs and anti-tumor capabilities of CD8+ T cells and CD4+ Tem [25–27], these results indicated the immunosuppressive microenvironment in RR-AITL.
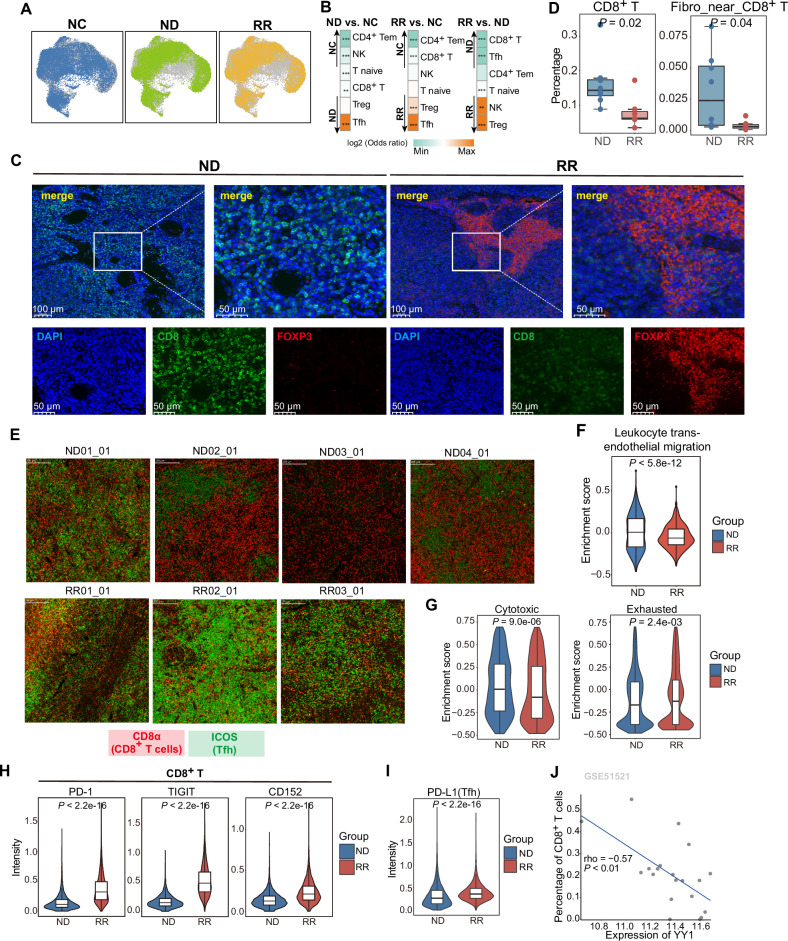
Fig. 4. The exclusion and dysfunction of CD8+ T cells in RR-AITL.
A T/NK cells from NC, ND-AITL and RR-AITL illustrated in UMAP plots, respectively. B Group preference of T cells subtypes, with color encoded by odds ratio estimated by one-sided Fisher’s exact test. *P value < 0.05, **P value < 0.01,***P value < 0.001. The orange color represents enrichment of subpopulation in the sample, while green color represents depletion of subpopulation in the sample. C Immunofluorescence analysis for ND-AITL (left) and RR-AITL (right) samples stained by anti-DAPI (blue), anti-CD8 (green), anti-FOXP3(red) antibodies. Scale bar, 100 μm. D The percentage of selected cell types within IMC dataset between ND-AITL and RR-AITL. E The representative markers for Tfh cells (ICOS; green) and CD8+ T cells (CD8α; red) stained in each ROI. F Enrichment scores of pathways related to T cell migration capacity in CD8+ T cells from ND-AITL and RR-AITL samples. P values were determined by two-sided unpaired Wilcoxon test. G Enrichment scores of cytotoxic and exhausted signatures of CD8+ T cells from ND-AITL and RR-AITL samples. ***P value < 0.001. P values were determined by two-sided unpaired Wilcoxon test. H Violin plots depict the intensities of immune checkpoint molecules in CD8+ T cells between seven and eight ROIs from RR-AITL and ND-AITL, respectively. P values were determined by two-sided unpaired Wilcoxon test. I Violin plots showing the intensities of PD-L1 in malignant Tfh cells across seven and eight ROIs in RR-AITL and ND-AITL, respectively. P values were determined by two-sided unpaired Wilcoxon test. J The spearman correlation between YY1 expression and the percentage of CD8+ T cells estimated by CIBERSORTx in GSE51521 dataset. Each data point represents individual sample.
Furthermore, we found that T cell migration-related pathways were more enriched in CD8+ T of ND-AITL compared to RR-AITL (Fig. 4F, Supplementary Fig. S5B), which may explain the relatively higher CD8+ T abundance in ND-AITL. Besides, we investigated the cellular states of CD8+ T cells and observed a significant reduction in their cytotoxic capacity in RR-AITL as compared to ND-AITL (Wilcoxon rank sum test, P = 8.984e-6). Conversely, CD8+ T cells exhibited a greater degree of exhaustion in RR-AITL compared with ND-AITL (Wilcoxon rank sum test, P = 2.376e-3; Fig. 4G) where the exhaustive markers, including PD-1, TIGIT and CD152 (encoded by CTLA4), were elevated in CD8+ T cells from RR-AITL (Fig. 4H), as observed in the IMC dataset. Meanwhile, malignant Tfh cells in RR-AITL had higher intensities of PD-L1 compared to ND-AITL (Fig. 4I). Besides, YY1 expression negatively correlates with the percentage of CD8+ T cells (Fig. 4J), suggesting a potential role for YY1 in excluding CD8+ T cells.
B cells in RR-AITL are in an intermediate state of malignant transformation to B-cell-lymphoma
Under physiological conditions, the interaction between Tfh and B cells is a central event in the formation of the germinal center (GC). Accompanying the malignant transformation of Tfh cells in AITL, B cells in AITL may exhibit Epstein-Barr virus (EBV) infection, undergo clonal expansion, and display specific genetic mutations, such as TET2 [6]. The crosstalk between Tfh and B cells promotes AITL progression [14]. Hence, we next attempted to elucidate the role of B cells in AITL progression by comparing the expression profiles of B cells from RR-AITL and ND-AITL patients. The 469 upregulated genes and 126 downregulated genes were identified in RR-AITL (Fig. 5A, Supplementary Table 10). Notably, RGS13, a canonical marker of GCB cells, is highly expressed in RR-AITL. Previous studies showed that RGS13 contributes to reduced responsiveness to chemokines in GCB cells and B lymphoma [28, 29]. In contrast, the markers for plasma cells, such as MZB1, IGHG2 and JCHAIN, were highly expressed in ND-AITL. Functional enrichment analysis showed that genes upregulated in RR-AITL were enriched in VEGFA-VEGFR2 signaling pathway and Fc-gamma receptor signaling pathway. B cell-derived vascular endothelial growth factor A can partly contribute to the proliferative HEVs, a key histologic feature of AITL [11, 30] (Supplementary Fig. S6A). Fc-gamma receptors on B lymphocytes negatively regulate B cell receptor (BCR)-dependent activation [31]. Notably, genes highly expressed in B cells from RR-AITL were enriched in B cell lymphoma, especially in diffuse large B-cell lymphoma (DLBCL) cell lines, including SUDHL4, NUDHL1, KARPAS422, SUDHL6 and RL, suggesting the propensity for malignant transforming of B cells in RR-AITL (Fig. 5B).
Fig. 5. Integrative analysis for B cells from ND-AITL, RR-AITL and DLBCL.
A Volcano plot of expression changes between B cells from ND-AITL and RR-AITL samples. Upregulated genes in RR-AITL (red) and ND-AITL (blue) are highlighted. B The enrichment analysis of upregulated genes in B cells from RR-AITL compared to ND-AITL. The Cancer Cell Line Encyclopedia gene set is derived from Enrichr website. C The UMAP plot illustrates B cells colored according to sample classification, including B cells from ND-AITL (green), RR-AITL (orange), and DLBCL (blue). D Barplot showing the fraction of B cells from ND-AITL (green), RR-AITL (orange) and DLBCL (blue) samples in each cluster. E The violin plot showing IGKC fraction in ND-AITL (green), RR-AITL (orange) and DLBCL (blue) samples. The IGKC fraction, IGKC/(IGKC + IGLC2), was calculated for each B cell. F The trajectory plots showing the pseudotime (left) and group (right) of each cell. G Density plot (upper panel) of cell number along the pseudotime from ND-AITL (green), RR-AITL (orange) and DLBCL patients (blue). Heatmap (bottom panel) showing gene expression alterations along the pseudotime. H The plot showing expression of MYC, TCL1A, STMN1, VPREB3, CD24 and PCNA along the pseudotime. Line indicates smoothed mean expression of genes in the state across pseudotime. I The protein levels of cMyc in B cells. P values were determined by two-sided unpaired Wilcoxon test. J The IMC images showing the protein activity of cMyc within B cells. The colors represent protein activity levels, ranging from lower (blue) to higher (red).
We further integrated the scRNA-seq data of B cells from ND-AITL, RR-AITL, and DLBCL to explore the alterations of cellular state of B cells (see Methods, Fig. 5C). After unsupervised clustering, 13 clusters were identified, including B_C0~B_C12 (Supplementary Fig. S6B). According to the relative fraction of B cells from ND-AITL, RR-AITL and DLBCL in each cluster (Fig. 5D), we classified these clusters into three groups: ND (B_C3, B_C5 and B_C10), RR (B_C8, B_C9 and B_12), and DLBCL (B_C1, B_C7, B_C6, B_C11, B_C4, B_C2 and B_C0). Based on the average expression of top 30 highly expressed genes in each cluster, we calculated the Pearson correlation between these B cell clusters. Hierarchical clustering showed that the RR clusters had higher similarity with the DLBCL clusters (Supplementary Fig. S6C). We also evaluated the activity of metabolic pathways of each B cell cluster (Supplementary Fig. S6D). Results showed that B-cell clusters of ND-AITL had relatively lower metabolic activity, and DLBCL B-cell clusters had the highest metabolic activity across multiple pathways. Notably, B-cell clusters of RR-AITL showed significantly high activity of pathways which were also activated in DLBCL, such as glycerophospholipid metabolism and inositol phosphate metabolism [32, 33]. According to a previously established method that identifies malignant B cells based on the expression of immunoglobulin κ constant (IGKC) and immunoglobulin λ constant 2 (IGLC2) [34], we calculated the IGKC fraction for each B cell (see Methods, Fig. 5E). Results showed that ND-AITL patients contained both κ- and λ-expressing B cells, indicating the normal status of B cells. In contrast, the majority of B cells from RR-AITL and DLBCL homogeneously expressed the κ light chain, suggesting the propensity for malignant transformation of B cells in RR-AITL.
Furthermore, pseudotime analysis was used to uncover the potential developmental trajectory of B cells (Fig. 5F). We found that B cell clusters from various groups were distributed differentially along the trajectory, with ND-AITL clusters located at the early branch, RR-AITL clusters located at early/middle branches, and DLBCL clusters located at the end of the trajectory (Fig. 5F, Supplementary Fig. S6E). This observation implied the potential evolution trajectory of B cells from ND-AITL, RR-AITL to malignant B-cell lymphoma. The density plot of cells along the pseudotime also showed that most of the B-cells from ND-AITL located at the beginning of the pseudotime, and a majority of B cells from RR-AITL located between the ND-AITL and DLBCL, implying that B cells in RR-AITL might represent an intermediate state during the process of malignant transformation (Fig. 5G, upper panel). Next, we identified gene modules that had differential expression along with the single-cell trajectory, including module-1 specifically expressed in ND-AITL, module-2 and module-3 highly activated in RR-AITL, and module-4 expressed in DLBCL (Fig. 5G, bottom panel). Significantly, we observed a progressive increase in the expression levels of several genes associated with B-cell lymphoma tumorigenesis along with the trajectory from ND-AITL to RR-AITL, and eventually to DLBCL. These genes include MYC, STMN1, CD24, PCNA, TCL1A, and VPREB3 (Fig. 5H, Supplementary Fig. S6F). As an important oncogene of multiple cancer types, MYC was confirmed to regulate the initiation and aggressive progression of DLBCL [35]. Consistent with the observation in scRNA-seq data, we found that B cells in RR-AITL had higher levels of cMyc, further proving the propensity of malignant transformation in RR-AITL patients (Fig. 5I, J, Supplementary Fig. S6G). Besides, PCNA was also reported to involve in DNA replication and cell proliferation of cancer, and is a prognosis factor for non-Hodgkin’s lymphoma [36]. Remarkably, akin to malignant Tfh cells, B cells in RR-AITL exhibited a heightened protein expression of PD-L1, suggestive of the evasion of CD8+ T cell cytotoxicity (Supplementary Fig. S6H).
Further, we aimed to identify the potential drugs to suppress the malignant transformation of B cells (see Methods). We screened drugs that could potentially inhibit the expression of genes in module-2, -3 and -4 (see Methods). Results showed that resveratrol, etoposide, and methotrexate might be used to suppress the process of malignant transformation of B cells in AITL patients (Supplementary Fig. S6I).
Higher prevalence yet dysfunctional myeloid-centered interaction network in RR-AITL
Tumor cells and TME typically establish a distinctive spatial architecture that promotes tumor recurrence and drug resistance [37]. We further compared the spatial architecture between RR-AITL and ND-AITL by performing cellular neighborhood (CN) analysis. Ten CNs (CN1-10) were identified and recapitulated the spatial structures of the TME architecture in AITL (see Methods, Fig. 6A, Supplementary Fig. S7A). Our observations indicated notable disparities in CN distribution between the RR-AITL and ND-AITL cohorts (Fig. 6A, B). Notably, ROIs from RR-AITL had relatively lower CN2 and CN7, and relatively higher CN4 and CN6 (Fig. 6C, Supplementary Fig. S7B), suggesting the existence of T cell exclusion and the prevalence of myeloid-centered structure. It is noteworthy that there is a significant variation in the distribution of cell types across different CNs (Fig. 6D, Supplementary Fig. S7C). Results showed that CN5 and CN8 were enriched for fDC, and CN1 was enriched for endothelial cells. These CNs represent unique histologic features in AITL, including proliferation of fDCs and a prominent arborization of HEVs [38]. T cells and fibroblast was enriched to CN10, CN2, and CN7, suggesting fibroblast may hinder T cells infiltration. CN4 and CN6 were myeloid-centered structures. Then, CN3 and CN9 might suggest B cells region and malignant Tfh region, respectively. Notably, B cell region and malignant Tfh region were distinct from other CNs and represent spatial exclusion of other cells (Fig. 6D, Supplementary Fig. S7C). This might be the potential mechanism of malignant Tfh cells evading immune surveillance and killing.
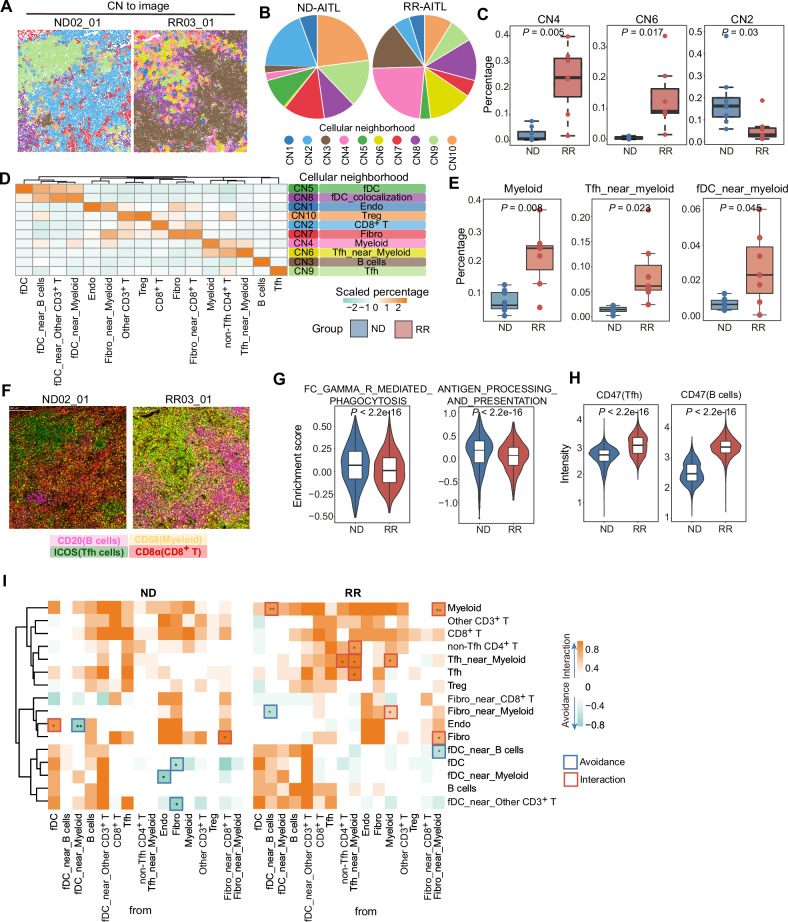
Fig. 6. The dysfunctional myeloid cells in RR-AITL.
A The CNs were depicted in the representative images from RR-AITL and ND-AITL, with colors indicating different CNs. B Pie chart showing the CNs distribution within ND-AITL and RR-AITL. C Boxplots depict the percentage of selected CNs within all cells between ROIs from ND-AITL and RR-AITL. D The heatmap illustrates the scaled percentage of cell types in each cellular neighborhood. The predominant cell type in each CN is presented in the right. E The percentage of selected cell types between ROIs from ND-AITL and RR-AITL. P values were determined by two-sided unpaired Wilcoxon test. F four markers representing B cells, Tfh cells, myeloid cells, and CD8+ T cells were visualized in the representative images. G The enrichment score of phagocytosis and antigen presentation in myeloid cells based on scRNA-seq data. H Violin plot showing the protein levels of CD47 in malignant Tfh cells and B cells. P values were determined by two-sided unpaired Wilcoxon test. I The heatmap illustrates significant pairwise cell-cell interactions (in orange) or avoidance (in green) between different cell types. Boxes highlight significant interactions (in red) or avoidance (in blue) between RR-AITL and ND-AITL. P-values were determined using a two-sided unpaired Wilcoxon test. *P values < 0.05, **P values < 0.01.
Myeloid cells play a critical role in the progression of cancer. We observed that compared to ND-AITL, ROIs from RR-AITL had significantly higher fraction of myeloid, Tfh_near_myeloid and fDC_near_myeloid in RR-AITL (Fig. 6E, F, Supplementary Fig. S8). Despite our observation of an augmentation in myeloid cells and myeloid-dominated CNs in RR-AITL, their capacity to phagocytose tumor cells has yet to be evaluated. Our observations revealed a notable reduction in the phagocytic and antigen presentation capacities of myeloid cells in RR-AITL (Fig. 6G). Simultaneously, we observed that B cells and malignant Tfh cells all had higher expression of CD47 (Fig. 6H), a transmembrane protein known as “don’t eat me” signal, suggesting that B cells and malignant Tfh cells in the RR group may evade phagocytosis by myeloid cells [39]. Previous studies demonstrated that CD47 expression was increased in malignant cells from B cell lymphoma, which correlates with a worse clinical prognosis [40, 41]. Through a comparative analysis of myeloid cell expression between RR-AITL and ND-AITL, we observed an enrichment of myeloid cells in RR-AITL within pathways including T cell receptor (TCR), CTLA4, and IL4 (Supplementary Fig. S7D, E). IL4 is a pivotal molecule that induces M2 macrophage polarization and facilitates tumor progression [42]. Malignant cells can educate myeloid cells, thereby facilitating tumor progression. We investigated the relationship between malignant cells and the functional dysregulation of myeloid cell. Results indicated that YY1 expression negatively correlates with phagocytic and antigen presentation functions (Supplementary Fig. S7F), suggesting a crucial role for YY1 in the suppression of myeloid cell function. To conduct a detailed analysis of the functional changes across different myeloid cell subtypes, We re-clustered myeloid cells and identified 10 distinct subtypes (Supplementary Fig. S9A, B). Hallmark enrichment scores revealed significant functional differences between subtypes and functional variations within the same subtype between ND-AITL and RR-AITL samples (Supplementary Fig. S9C). For instance, APOE+ macrophages in the RR-AITL exhibited diminished fatty acid metabolism compared to the ND-AITL. Furthermore, we calculated enrichment scores for phagocytosis and antigen presentation at the single-cell level. A comparison of enrichment scores between the ND-AITL and RR-AITL samples for each subtype revealed a marked reduction in the phagocytic and antigen-presenting capacities of APOE+ macrophages in the RR-AITL, while MAST cells exhibited the opposite trend (Supplementary Fig. S9D).
To comprehensively analyze the interactions between different cell types, we investigated the spatial organization of cells using permutation tests to quantify the co-localization between pairs of cell types and detect their potential interaction or avoidance behavior (Fig. 6I, Supplementary Table 11). We observed that fDC_near_B cells and fibro_near_myeloid exhibit significant co-localization with myeloid cells in RR-AITL. Moreover, in RR-AITL, malignant Tfh cells and myeloid cells exhibit more closely intertwined spatial interactions compared to ND-AITL. For example, the spatial distances between Tfh_near_myeloid and Tfh cells, as well as myeloid cells and Tfh_near_myeloid, are significantly reduced in RR-AITL.
In brief, these results indicate that despite the greater abundance of myeloid cells in RR-AITL and their closer spatial proximity to tumor cells, their ability to effectively eliminate tumor cells has seemingly diminished.
The immunosuppressive ligand-receptors interactions activated in RR-AITL
To elucidate the influence of malignant Tfh cells on shaping TME conducive to RR-AITL, we conducted a high-resolution dissection of cell-to-cell ligand-receptor interactions between malignant Tfh and immune cells using scRNA-seq data. Based on alterations of cell proportions and cellular states, we specifically compared the molecular interactions among Tfh, Treg, B cells, CD8+ T cells, and myeloid cells in ND-AITL and RR-AITL. We noted a significant reduction in the count of ligand-receptor interactions originating from RR-AITL across almost all cell types, in comparison to those from ND-AITL. However, the number of interactions between B cell and Tfh/Treg cells increased in RR-AITL (Fig. 7A, B). The most substantial reduction in the count of interactions was observed between myeloid cells and other cell types in RR-AITL (Fig. 7B), providing further evidence of myeloid cell dysfunction (Fig. 6E).
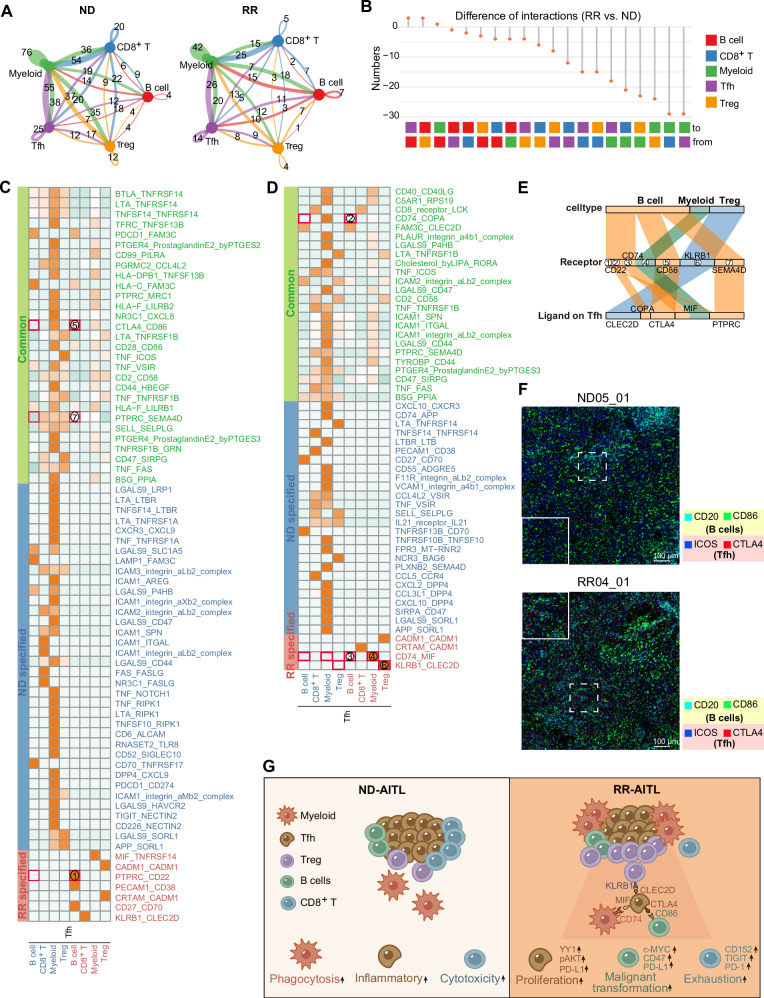
Fig. 7. The alterations of cell-cell communications in RR-AITL compared to ND-AITL.
A Circos plots showing the interaction numbers among various cell types in ND-AITL(left) and RR-AITL(right) samples. These interactions were predicted based on scRNA-seq data. B The number of significant ligand-receptor pairs between the RR-AITL and ND-AITL, as depicted in a lollipop plot. C, D Heatmap depicting the intricate ligand-receptor interactions between Tfh cells and other cell types. The ‘Common’ category (green) signifies the presence of ligand-receptor pairs in both ND-AITL and RR-AITL. The ‘ND-specified’ category (blue) and ‘RR-specified’ category (red) indicate the occurrence of ligand-receptor pairs exclusively in ND-AITL or RR-AITL, respectively. Numbers enclosed in red boxes indicate the interacting pairs displayed in Figure E. E Sankey plot illustrating the essential ligand-receptor pairs exclusive to RR-AITL, with ligands expressed on Tfh cells. The color scheme indicates different cell types expressing the receptors. F The IMC images showing the protein activity of CD20, CD86 within B cells and ICOS, CTLA4 within Tfh cells in ND-AITL and RR-AITL samples. The colors represent corresponding proteins. The solid line frame represents a magnified version of the dashed line frame. Scale bar, 100 μm. G Schematic representation of TME alterations between RR-AITL and ND-AITL.
Ligand-receptor interactions that commonly or specifically presented in RR-AITL and ND-AITL samples were identified (Fig. 7C, D, Supplementary Table 12). Specifically, our results revealed that certain inhibitory interactions, such as CLEC2D-KLRB1, MIF-CD74, and CTLA4-CD86 interactions, were exclusively detected between Tfh and Treg/Myeloid/B cells in RR-AITL (Fig. 7E). Previous study showed that KLRB1 could inhibit activation of T cell populations when bound to CLEC2D [43]. Besides, MIF-CD74 signaling was confirmed to impede microglial M1 polarization and result in a M2 shift [44–46]. Notably, a recent study showed that Tfh could suppress B cells responses through CTLA4-CD86 interaction [47]. The IMC analysis further validated that compared to ND-AITL, Tfh cells with elevated CTLA4 level are found in close proximity to CD86+ B cells in RR-AITL (Fig. 7F), indicating B cells may be inhibited in their normal functions through CTLA4-CD86 interactions mediated by Tfh cells. Therefore, these results suggest that malignant Tfh cells in RR-AITL may reconfigure the TME into an immunosuppressive milieu through intercellular interactions (Fig. 7G).
Discussion
AITL is an aggressive PTCL subtype with poor prognosis, largely because of primary progression or short duration of remission [11]. The intricate complexity of the TME leads to the difficulty of establishing effective treatment strategies for AITL patients [6]. The advancement of scRNA-seq enables researchers to decipher the TME comprehensively. Previous studies mainly focused on the distinct alterations of TME between AITL samples and non-tumor samples [13, 14]. However, the TME in RR-AITL remains to be thoroughly elucidated. By performing scRNA-seq and IMC analysis, this study aims to elucidate the distinctive cellular and molecular characteristics of RR-AITL and to identify potential therapeutic strategies. Firstly, our study demonstrated that malignant Tfh cells in RR-AITL exhibit an elevated proliferation capacity in a YY1-dependent manner. The increased YY1 activity is also associated to the suppression of immune-related pathways, such as the TNFα signaling pathway, and the decreased proportion of CD8+ T cells. These factors are further linked to the poor prognosis of AITL patients. Secondly, our results showed that compared to ND-AITL, RR-AITL had a more immunosuppressive TME characterized by the decreased proportion of CD8+ T cells along with a decline in their cytotoxic activity. The activation of YY1 may play an important role in restraining CD8+ T cells infiltration. Thirdly, our single-cell trajectory analysis revealed the propensity of B cells in RR-AITL to undergo malignant transformation towards DLBCL, characterized by the upregulation of factors (such as MYC) that play important roles in B-cell malignancy. Finally, we further found the higher prevalence yet dysfunctional myeloid-centered interaction network in RR-AITL compared to ND-AITL. Collectively, our study suggests the critical role of TME reprogramming and cellular plasticity in RR-AITL (Fig. 7F).
CD8+ T cells were relatively depleted in RR-AITL compared to ND-AITL. Previous studies have highlighted three cell types known to impede T cell infiltration within tumors, namely cancer-associated fibroblasts, myeloid-derived suppressor cells, and the M2 subtype of tumor-associated macrophages (TAMs) [48]. Hence, the increased proportion of myeloid cells may promote the CD8+ T cells exclusion in RR-AITL. Besides, there was a substantial decline in the cytotoxic capacity of CD8+ T cells against tumor cells in RR-AITL. Exhausted markers including PD-1, TIGIT and CD152, were highly expressed in CD8+ T cells from RR-AITL, suggesting the decreased cytotoxicity. PD-L1 can dampen the cytotoxic activity of T cells via its interaction with the PD-1 protein [49]. The elevated protein levels of PD-L1 in malignant Tfh cells and B cells from RR-AITL may possess notable implications for the impairment of CD8+ T cells.
Additionally, both IMC and scRNA-seq data underscore the importance of myeloid cells in the TME. The scRNA-seq data reveals a reduction in the intrinsic capacity of myeloid cells to eliminate malignant cells, including diminished phagocytosis and antigen presentation, in RR-AITL. Although an increased proportion of myeloid cells and the prevalence of myeloid-centered interaction networks in RR-AITL are evident from the IMC data, their ability to eradicate malignant cells is suppressed. Besides, B cells and malignant Tfh cells all had relatively higher protein levels of CD47 in RR-AITL, further limiting the phagocytosis capacity of myeloid cells [50]. Additionally, the shrinkage of cell-cell communications in RR-AITL, especially myeloid cell interacting with other cell types, may be a potential mechanism for malignant cells to evade immune killing.
YY1 was negatively correlated with the proportion of CD8+ T cells and the function of myeloid cells. Previous studies have reported that YY1 may influence the immune microenvironment through its regulation of the PI3K/AKT signaling pathway, which plays a critical role in immunosuppression [51]. Activation of PI3K/AKT in tumor cells promotes the secretion of immunosuppressive molecules and upregulates immune checkpoint proteins like PD-L1, thereby impairing CD8+ T cell activity and myeloid cell function [52–55]. Our analysis revealed a negative correlation between YY1 expression and CD8+ T cell proportions, as well as altered myeloid cell function, supported by pathway enrichment findings linking YY1 to PI3K/AKT activation. These results suggest that YY1 contributes to an immunosuppressive TME by enhancing PI3K/AKT-driven modulation of immune cell behavior, highlighting its potential role in tumor-mediated immune evasion.
Malignant Tfh cells had significantly higher YY1-dependent proliferation capacity in RR-AITL. YY1 is a critical TF associated with unchecked cellular proliferation, resistance to apoptotic stimuli, tumorigenesis and metastatic potential across many cancer types, such as DLBCL and hepatocellular carcinoma [56, 57]. YY1 could regulate the PI3K/AKT pathway, which is also activated in RR-AITL [51]. Besides, RAC1, a member of the Rho family of GTPases involved in cell proliferation, was regulated by YY1. Hence, our results indicate that in RR-AITL, YY1 can promote the proliferation of malignant Tfh cells by regulating the PI3K/AKT signaling and RAC1. Additionally, YY1 can activate both endogenous and exogenous cMyc promoters [58]. Casey et al. suggested that cMyc can regulate the expression of PD-L1 and CD47 [59]. Hence, the high protein levels of PD-L1 and CD47 in malignant Tfh cells may attribute to YY1-dependent cMyc activation. Also, we observed that YY1 was closely linked to the reduced activity of immune-related pathways, such as TNFα signaling pathway, in the TME of AITL. However, the specific mechanisms are still to be explored. In addition, we observed the erosion of Tfh cell identity in RR-AITL. The erosion of cell identity was also observed in the RR patients with CLL [18], suggesting this might be a common pathological characteristic in malignant lymphocytic disease. However, further experiments are required to elucidate the detailed mechanisms.
The B cells in RR-AITL exhibited elevated expression of GCB cell markers, and reduced expression of plasma cell markers compared to ND-AITL. Consistently, Fujisawa et al. also found significant expansion of aberrant B cells with GCB cells phenotype in AITL, which supporting the development of Tfh-like lymphomas [14]. These observations underscore the significant role of abnormal B cells with a GCB phenotype in AITL onset and progression. Notably, we found that B cells in RR-AITL tend to obtain a B-cell-lymphoma-like transcription patterns, such as the activation of MYC and STMN1. This may explain the clinical observations that AITL patients had higher risk of developing DLBCL [60, 61]. Furthermore, the elevated protein levels of cMyc may potentially regulate the expression of CD47 and PD-L1, akin to malignant Tfh cells. This, in turn, could further exacerbate the dysfunction of CD8+ T cells and enable evasion from macrophage phagocytosis. Therefore, the targeted therapies specifically addressing the potential malignant transformation of B cells may be profoundly significant in AITL patients.
We also observed the reduction of cell-cell interactions in RR-AITL compared to ND-AITL. Cell-cell interactions are important for immune cells to recognize and eliminate cancer cells, particularly by recognizing tumor-specific antigens presented on the surface of cancer cells [62]. The decrease in intercellular interactions may indicate a decline in the anti-tumor capabilities of immune cells.
Due to the rarity of AITL cases and the stringent rigorous sample prerequisites of single-cell experiments, our research incorporates a restricted number of AITL samples. It is important to acknowledge that this limitation may potentially impact the conclusion of this study. However, our findings have effectively characterized the distinctive cellular state and spatial architecture within the TME of RR-AITL. These observations need to be validated with a larger sample size of clinical specimens in the future. Besides, further experiments are required to elucidate the detailed mechanisms. However, it is unfortunate that, to date, no cell repositories (such as National Science and Technology Infrastructure, the National Biomedical Cell-Line Resource (NSTI-BMCR), European Collection of Authenticated Cell Cultures (ECACC), American Type Culture Collection (ATCC)) have been able to provide stable and well-established cell lines of AITL for the purpose of validating intracellular biological mechanisms through experimentation.
In summary, we characterized the distinct alterations of the TME ecosystem in RR-AITL compared to ND-AITL through scRNA-seq and IMC analysis. Our results reveal the immunosuppressive TME in RR-AITL mediated by the depletion of cytotoxic T cells, the dysfunction of myeloid cells, and the potential malignant transition of B cells. Furthermore, the programs regulated by YY1 are found to play important roles in the aggressive proliferation and immune escape of AITL malignant cells, significantly associated with the poor prognosis of AITL patients. Our single-cell transcriptomic and spatial analysis will provide important information for understanding the complex TME and the development of novel therapeutic strategies for RR-AITL patients.
Methods
Clinical samples
Fresh lymph node samples (n = 9), encompassing three specimens from ND-AITL, three from RR-AITL, and three from non-tumor sources, were employed for single-cell RNA sequencing. Formalin-fixed, paraffin-embedded (FFPE) sections (n = 7) of lymph node tissue, including four specimens from ND-AITL and three from RR-AITL, were subjected to IMC experiments. Besides, several FFPE sections of lymph node tissue from AITL patients were applied to multiplex immunofluorescence and immunohistochemistry. The ages of these patients diagnosed with AITL ranged from 43 to 75 years. The specimens were collected at Jiangsu Province Hospital, located in Nanjing, Jiangsu, China, during the period spanning June 2019 to December 2020. Diagnosis of AITL was determined mainly according to the revised 4th edition of the WHO Classification of Hematolymphoid Tumors and the diagnosis was reviewed and approved by two specialists in the Department of Pathology in the Jiangsu Province Hospital (Nanjing, Jiangsu, China). OS was set in the period from the date of diagnosis to death or to the most recent clinic visit. Relapsed AITL was defined as patients that received complete response after first-line treatment but relapsed finally. Refractory AITL was defined as patients that failed to receive partial response after 4 cycles of standard first-line treatment.
Libraries preparation and sequencing for scRNA-seq
The lymph node samples were conserved in the GEXSCOPE Tissue Preservation Solution (Singleron) and shipped to the Singleron lab with ice pack. The specimens were washed 3 times with Hanks Balanced Salt Solution (HBSS, Cat. No.14025-076, Gibco) and shredded into 1–2 mm pieces. Then the tissue debris was submitted to the digestion with 2 ml GEXSCOPE Tissue Dissociation Solution at 37 °C for 15 min in 15 ml centrifuge tube (Falcon, Cat. No.352095) with sustained agitation. Cells were filtered through 40-micron sterile strainers (Falcon, Cat. No.352340) and centrifuged (eppendorf, 5810R) at 300 × g for 5 min. Then the supernatant was removed, and the pellets were resuspended in 1 ml PBS (Hyclone, Cat. No.SA30256.01). To remove the red blood cells, which were frequently a significant portion of the cells produced, a 2 ml RBC lysis buffer (Roche, Cat. No. 11814389001) was added to the cell suspension according to the manufacturer’s protocol. Centrifuge the cells at 500 × g for 5 min in a microfuge at 15–25 °C and resuspend in PBS (Hyclone, Cat. No.SA30256.01). Samples were counted with an automated cell counter (JSY-FL-049), and once the cell viability exceeded 80%, subsequent sample processing could be performed.
Single-cell suspension was of 1 × 105 cells/ml in PBS and loaded onto a microfluidic chip according to the Singleron GEXSCOPE Single Cell RNA-seq Library Kit (Singleron). Single-cell RNA libraries were constructed following the GEXSCOPE protocol. After construction, the libraries were initially quantified using a Qubit 2.0 Fluorometer and diluted to 1.5 ng/µl. All libraries were more than 2 nM, examined by qRT-PCR, and pooled for sequencing. The pools were sequenced on the Illumina HiSeq X Ten and Illumina Novaseq 6000 platform with 150 bp paired-end reads.
Raw reads were processed to generate gene expression profiles using the standard internal pipeline based on the CeleScope. After read 1 without poly T tails were removed, cell barcodes and unique molecular identifiers (UMIs) were extracted. Adapters and poly-A tails were trimmed using cutadapt. Then STAR was used to align the read 2 to GRCh38 Ensembl build 92 genomes. Reads with the same cell barcode, UMIs, and genes were grouped to calculate the number of UMIs per gene per cell. The UMI count tables of each cellular barcode were used for further analysis.
Data preprocessing, cell type annotation for scRNA-seq
Seurat package was used to perform the scRNA-seq analysis. Firstly, expression matrix of each sample was converted to seurat object by the CreateSeuratObject function. Genes expressed in less than 3 cells were excluded from the following analysis. Then, we only retained cells that expressed at least 200 genes, not more than 6000 genes, and less than 20% of mitochondrial transcripts. These cutoffs were determined through quality control inspection of each library. Then, seurat objects of nine samples were integrated using canonical correlation analysis algorithm to remove potential technical noise and batch effect. The FindAllMarkers function was performed to identify the specific genes of each cluster. Genes with expression percentage greater than 10% and log fold change greater than 0.25 were defined as differential expressed genes.
To identify the cell identity, mean expression of representative markers for main cell types and differentially expressed genes for clusters was calculated. The following markers were used: (1) T/NK cells: CD2, CD3D, CD3G, TRAC, (2) Stromal cells: IGFBP7, SPARCL1, MGP, COL1A2, (3) myeloid cells: LYZ, CST3, TYROBP, FCER1G, (4) B cells: MS4A1, CD79A, IGHM, IGHG3, IGLC2, IGHG1, respectively. Additionally, we also annotated cells using the SingleR package with the MonacoImmuneData database. Fisher’s exact test was employed to identify overrepresented cell types in a particular group. We applied the CytoTRACE with default parameters to evaluate the differentiation status for each T cell in the scRNA-seq dataset.
Functional enrichment analysis
The clusterProfiler package was used to perform gene set enrichment analysis (GSEA). The Enrichr website (https://maayanlab.cloud/Enrichr/) was used to perform functional enrichment analysis based on the differentially expressed genes between cells in RR-AITL and ND-AITL. VISION was applied to quantify the activity of important pathways in individual cells in scRNA-seq data. For bulk RNA-seq or microarray data, the ssGSEA method included in the GSVA package was used to score the gene set in each sample.
Transcriptional regulatory network inference
The pySCENIC module was used to infer the TF activity in individual cells. Firstly, seurat objects were converted to loom files as the input matrix of downstream analysis using the loomR package. Next, regulons were inferred using the GRNBoost2 algorithm. Finally, regulon activity was quantified by AUCell. The differentially activated and differentially expressed TFs were identified by the Wilcoxon rank-sum test.
Pseudotime analysis
We used the Monocle2 algorithm to conduct pseudotime analysis. The differentially expressed genes between clusters were calculated by the differentialGeneTest function. Dimension reduction was performed using the DDRTree algorithm and the top 500 differentially expressed genes were used to sort cells in a temporal differentiation order.
Metabolism activity inference based on scRNA-seq data
To characterize the metabolic heterogeneity of different clusters at single-cell resolution, we followed the workflow previously described by Xiao et al. [63]. Data imputation and normalization was applied to gene expression profiles. Then the global structure of single-cell metabolic programs was characterized by clustering. Cell type-specific metabolic programs were identified and metabolic heterogeneity was quantified.
Cell type abundance estimation
We used the CIBERSORTx deconvolution algorithm to characterize the immune infiltration within the bulk tissue of AITL. Briefly, we used the LM22 signature as signature matrix file. Then, B-mode batch correction and quantile normalization were applied to expression profiles. Meanwhile, 500 permutations were set for statistical analysis. Finally, we obtained the relative fractions of 22 immune cell types within bulk tissues.
Drug prediction
To predict potential drugs, we followed the established framework provided by Du et al. [64]. The genes within module two, three, and four that were recognized from pseudotime analysis were considered as potential therapeutic targets. Then, we screened drugs that could downregulate these genes within the cMAP database and calculated P values through hypergeometric distribution test.
Intercellular ligand-receptor analysis
To elucidate the communication networks between distinct cell populations, we employed CellPhoneDB, a computational tool that utilizes the expression of ligands and receptors across cell types to infer potential cell-cell interactions. We focused on characterizing the cell-cell interactions between malignant Tfh cells and various immune cells, including B cells, CD8+ T cells, myeloid cells, and Tregs. Ligand-receptor pairs with P values less than 0.05 were considered significant interactions.
Tissue preparation, antigen retrieval and imaging mass cytometry
Bake the slides for 2 h at 60 °C in the slide oven/dryer. Alternatively, if no oven is available, a heat block set at 60 °C can be used. In either case, ensure that all visible wax is removed. Turn the heating block on to 96 °C during the baking step. Prior to dewaxing, prepare 50 ml conical tubes containing 40 ml of antigen retrieval solution (10× diluted to 1×) and put the tubes in the heating block (96 °C) with loose lids. Dewax the slides in fresh xylene in the fume hood for 20 min with loose lids. Hydrate the slides in descending grades of ethanol (100%, 95%, 80%, 70%), 5 min each. Wash the slides in Maxpar Water for 5 min in a Coplin jar placed on an orbital shaker plate with gentle agitation. Insert the slides with tissues into preheated antigen retrieval solution and incubate them for 30 min, leaving the lids loose. Following incubation, remove the slides from the heating block, place the tubes containing the antigen retrieval solution and slides on a lab bench, and cool them to 70 °C by monitoring the temperature of the antigen retrieval solution for about 10 min. Wash the slides with Maxpar Water for 10 min in a Coplin jar with gentle agitation (orbital shaker). Wash the slides with Maxpar PBS for 10 min with gentle agitation. Use a PAP pen to encircle the sample. Block with 3% BSA in Maxpar PBS for 45 min at room temperature in a hydration chamber. To prepare the antibody cocktail, calculate the total volume of antibodies at concentrations specific for the assay and bring the volume up to a final volume of 0.5% BSA in Maxpar PBS. Place the slides in a hydration chamber and pipette the antibody master mix onto the section. Incubate overnight with the antibody cocktail at 4 °C in a hydration chamber. Wash the slides in 0.2% Triton X-100 in Maxpar PBS for 8 min with slow agitation in Coplin jars. Repeat. Wash the slides in Maxpar PBS for 8 min with gentle agitation. Repeat the washing. Stain the tissue with Intercalator-Ir in Maxpar PBS (300–500 μl/section for a 20 mm2 section of 1:400 solution) for 30 min at room temperature in a hydration chamber. Wash the slides in Maxpar Water for 5 min with gentle agitation. Air-dry the slides for at least 20 min at room temperature. Observe and photograph with Hyperion Imaging System.
Cell segmentation, cell type identification and spatial analysis
We utilized the workflow previously established by Windhager et al. [65] to analyze the IMC data. Firstly, we employed steinbock to extract multiple images from ROIs stained with different antibodies. Then, deepCell, a deep learning library for single-cell analysis of biological images, was used to obtain cells by segmenting images. Finally, we measured the intensities of different antibodies for each cell and the spatial distance among cells. We evaluated the signal quality of each channel and marker by calculating the signal-to-noise ratio (SNR). The SNR is defined as the ratio of foreground signal intensity to background noise intensity, given by the formula:
where Is represents the mean intensity of foreground pixels (true signal), and In represents the mean intensity of background pixels (noise). To identify foreground (signal) and background (noise) pixels, we applied the Otsu thresholding method. Using this approach, the SNR was calculated as the mean intensity of the foreground pixels divided by the mean intensity of the background pixels. Afterwards, we only retained the antibodies listed in Supplementary Fig. S2B for downstream analysis. Thus, Vimentin, DNA1 and DNA2 were excluded. Moreover, we eliminated cells with a size below 5 pixels from our analysis to ensure the integrity and reliability of our results.
To remove technical noise and batch effect, we employed the FastMNN algorithm included in the batchelor package. Comprehensive clustering analysis was conducted on all cells using the PhenoGraph algorithm. Cluster annotations were assigned to specific cell types based on the expression profiles of canonical markers. The clusters co-expressed the markers of two cell types were annotated as A_near_B, such as fDC_near_myeloid. To identify spatially resolved CN, we constructed spatial graphs by detecting the k-nearest neighbors (k = 10) based on the cell distances between cells measured by Steinbock. Then we performed cellular neighborhood analysis through k-means clustering with k = 10. To identify significant interaction and avoidance between different cell types, we first computed the averaged cell type interaction count and then performed permutation tests of single-cell interactions as previously described [66]. Interactions or avoidances with P values less than 0.01 were considered statistically significant.
Multiplex immunofluorescence assay and image analysis
The FFPE sections were baked for 1 h at 63 °C followed by dewaxing in a fully automatic dyeing machine. The dewaxing time is as follows: twice in Xylene, 15 min each; twice in 100% alcohol, 7 min each; once in 90% alcohol, 5 min; once in 80% alcohol, 5 min; once in 70% alcohol, 5 min. Repair solution was heated to boil before slides were put in for 15–20 min for antigen retrieval, followed by natural cooling in room temperature. Commercial H2O2 was used to remove the endogenous peroxidase and blocking buffer was added dropwise for blocking. Then the samples were stained with primary antibodies at 4 °C overnight followed by incubation with fluorescently labeled secondary antibodies for 1 h at room temperature protected from light. Antifade solution containing DAPI was dropped and coverslips were mounted. Finally, cells were observed and photographed with a Stellaris STED laser confocal microscope (Stellaris STED, Leica) or Thunder Imager Fast High Resolution Inverted Fluorescence Imaging System (Thunder DMi8, Leica).
IHC staining
IHC analysis of lymph node tissues was conducted in AITL patients according to standard procedure. Briefly, paraffin-embedded tissue was sectioned into 4-µm slides and incubated with anti-YY1 (#ab109237, Abcam) at 1:250 dilution overnight at 4 °C. Subsequently, slides were incubated with horseradish peroxidase (HRP)-coupled secondary antibody (#ab6721, Abcam, 1:1000) for 1 h at 20–22 °C, followed by incubation with DAB (#SK-4100, Vector Laboratories, Burlingame, CA). Expression of YY1 was evaluated based on IRS: the staining degree (0-3 points) and positive rate (0–4 points) in the immunohistochemical section were scored and multiplied to obtain the comprehensive score (0–12 points) as follows. The staining degree was scored according to the staining characteristics of the target cells: 0 point for no staining, 1 point for light yellow, 2 points for brown, and 3 points for brown. The positive rate is also scored: 0 to 5% is 0 point, 6 to 25% is 1 point, 26 to 50% is 2 points, 51 to 75% is 3 points, and >75% is 4 points.
Supplementary information
Author contributions
MYZ, NL, RW: data collection, data analysis, results interpretation. LF, WS: pathological diagnosis and clinical sample collection. LC, YR, CL: collection of clinical samples and statistical analysis of clinical data. MYZ, NL, RW and KNL: manuscript writing and revision. XYL, JYL, RZC, CW, LY: experiment, sequencing and patient follow-up. LSZ, YC, BH, RH, YS, ZL, SZ, XH, WZ, DY: data collection, data analysis. LXW, WW, QZL, PX: data collection, software and code. RHZ, SLL: manuscript revision, results interpretation. JYL, QHW, KNL, HJ: conceptualization, supervision, funding acquisition, manuscript revision. All authors have read and agreed to the published version of the manuscript.
Funding information
This study was supported by the National Natural Science Foundation of China Grant Nos. 32200590 (K.L.), 82170185 (H.J.), 82370193 (J.L), 81972358 (Q.W.), 91959113 (Q.W.), 82372897 (Q.W.), Natural Science Foundation of Jiangsu Province Grant No. BK20210530 (K.L.), BK20211376 (H.J.), The Social Development Project of Jiangsu Province Science and Technology Plan Grant No. BE2023775 (J.L), and Jiangsu Province Capability Improvement Project through Science, Technology and Education Grant No. ZDXK202209 (J.L).
Data availability
The processed count matrices have been deposited in the OMIX database (id: OMIX004773). Original code has been deposited at Github (https://github.com/zhumengyan/AITL2023) and is publicly available as of the date of publication. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The study was reviewed and approved by the Ethical Committee of the Nanjing Medical University (Nanjing, Jiangsu, China). The study was performed after receiving written informed consent from patients, by the principles outlined in the Declaration of Helsinki.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Mengyan Zhu, Ning Li, Lei Fan, Rongrong Wu, Lei Cao.
Contributor Information
Jianyong Li, Email: lijianyong@jsph.org.cn.
Qianghu Wang, Email: wangqh@njmu.edu.cn.
Kening Li, Email: likening@njmu.edu.cn.
Hui Jin, Email: jinhui@jsph.org.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41408-024-01199-0.
References
- 1.Wang Y, Zhang M, Song W, Cai Q, Zhang L, Sun X, et al. Chidamide plus prednisone, etoposide, and thalidomide for untreated angioimmunoblastic T-cell lymphoma in a Chinese population: a multicenter phase II trial. Am J Hematol. 2022;97:623–9. 10.1002/ajh.26499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nguyen TB, Sakata-Yanagimoto M, Fujisawa M, Nuhat ST, Miyoshi H, Nannya Y, et al. Dasatinib is an effective treatment for angioimmunoblastic T-cell lymphoma. Cancer Res. 2020;80:1875–84. 10.1158/0008-5472.CAN-19-2787. [DOI] [PubMed] [Google Scholar]
- 3.Federico M, Rudiger T, Bellei M, Nathwani BN, Luminari S, Coiffier B, et al. Clinicopathologic characteristics of angioimmunoblastic T-cell lymphoma: analysis of the international peripheral T-cell lymphoma project. J Clin Oncol. 2013;31:240–6. 10.1200/JCO.2011.37.3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dunleavy K, Wilson WH, Jaffe ES. Angioimmunoblastic T cell lymphoma: pathobiological insights and clinical implications. Curr Opin Hematol. 2007;14:348–53. 10.1097/MOH.0b013e328186ffbf. [DOI] [PubMed] [Google Scholar]
- 5.Odejide O, Weigert O, Lane AA, Toscano D, Lunning MA, Kopp N, et al. A targeted mutational landscape of angioimmunoblastic T-cell lymphoma. Blood. 2014;123:1293–6. 10.1182/blood-2013-10-531509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiba S, Sakata-Yanagimoto M. Advances in understanding of angioimmunoblastic T-cell lymphoma. Leukemia. 2020;34:2592–606. 10.1038/s41375-020-0990-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mohammed Saleh MF, Kotb A, Abdallah GEM, Muhsen IN, El Fakih R, Aljurf M. Recent advances in diagnosis and therapy of angioimmunoblastic T cell lymphoma. Curr Oncol. 2021;28:5480–98. 10.3390/curroncol28060456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moskowitz AJ. Practical treatment approach for angioimmunoblastic T-cell lymphoma. J Oncol Pr. 2019;15:137–43. 10.1200/JOP.18.00511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mak V, Hamm J, Chhanabhai M, Shenkier T, Klasa R, Sehn LH, et al. Survival of patients with peripheral T-cell lymphoma after first relapse or progression: spectrum of disease and rare long-term survivors. J Clin Oncol. 2013;31:1970–6. 10.1200/JCO.2012.44.7524. [DOI] [PubMed] [Google Scholar]
- 10.Ahearne MJ, Allchin RL, Fox CP, Wagner SD. Follicular helper T-cells: expanding roles in T-cell lymphoma and targets for treatment. Br J Haematol. 2014;166:326–35. 10.1111/bjh.12941. [DOI] [PubMed] [Google Scholar]
- 11.Lunning MA, Vose JM. Angioimmunoblastic T-cell lymphoma: the many-faced lymphoma. Blood. 2017;129:1095–102. 10.1182/blood-2016-09-692541. [DOI] [PubMed] [Google Scholar]
- 12.Iqbal J, Wright G, Wang C, Rosenwald A, Gascoyne RD, Weisenburger DD, et al. Gene expression signatures delineate biological and prognostic subgroups in peripheral T-cell lymphoma. Blood. 2014;123:2915–23. 10.1182/blood-2013-11-536359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pritchett JC, Yang ZZ, Kim HJ, Villasboas JC, Tang X, Jalali S, et al. High-dimensional and single-cell transcriptome analysis of the tumor microenvironment in angioimmunoblastic T cell lymphoma (AITL). Leukemia. 2022;36:165–76. 10.1038/s41375-021-01321-2. [DOI] [PubMed] [Google Scholar]
- 14.Fujisawa M, Nguyen TB, Abe Y, Suehara Y, Fukumoto K, Suma S, et al. Clonal germinal center B cells function as a niche for T-cell lymphoma. Blood. 2022;140:1937–50. 10.1182/blood.2022015451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suma S, Suehara Y, Fujisawa M, Abe Y, Hattori K, Makishima K, et al. Tumor heterogeneity and immune-evasive T follicular cell lymphoma phenotypes at single-cell resolution. Leukemia. 2024;38:340–50. 10.1038/s41375-023-02093-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoch T, Schulz D, Eling N, Gomez JM, Levesque MP, Bodenmiller B. Multiplexed imaging mass cytometry of the chemokine milieus in melanoma characterizes features of the response to immunotherapy. Sci Immunol. 2022;7:eabk1692 10.1126/sciimmunol.abk1692. [DOI] [PubMed] [Google Scholar]
- 17.Swerdlow SH, Campo E, Pileri SA, Harris NL, Stein H, Siebert R, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127:2375–90. 10.1182/blood-2016-01-643569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rendeiro AF, Krausgruber T, Fortelny N, Zhao F, Penz T, Farlik M, et al. Chromatin mapping and single-cell immune profiling define the temporal dynamics of ibrutinib response in CLL. Nat Commun. 2020;11:577 10.1038/s41467-019-14081-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramezani-Rad P, Geng H, Hurtz C, Chan LN, Chen Z, Jumaa H, et al. SOX4 enables oncogenic survival signals in acute lymphoblastic leukemia. Blood. 2013;121:148–55. 10.1182/blood-2012-05-428938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prieto C, Nguyen DTT, Liu Z, Wheat J, Perez A, Gourkanti S, et al. Transcriptional control of CBX5 by the RNA binding proteins RBMX and RBMXL1 maintains chromatin state in myeloid leukemia. Nat Cancer. 2021;2:741–57. 10.1038/s43018-021-00220-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng Y, Zhou Z, Wei R, Xiao C, Zhang H, Fan T, et al. The RNA-binding protein PCBP1 represses lung adenocarcinoma progression by stabilizing DKK1 mRNA and subsequently downregulating beta-catenin. J Transl Med. 2022;20:343 10.1186/s12967-022-03552-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gastonguay A, Berg T, Hauser AD, Schuld N, Lorimer E, Williams CL. The role of Rac1 in the regulation of NF-kappaB activity, cell proliferation, and cell migration in non-small cell lung carcinoma. Cancer Biol Ther. 2012;13:647–56. 10.4161/cbt.20082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Archibald A, Mihai C, Macara IG, McCaffrey L. Oncogenic suppression of apoptosis uncovers a Rac1/JNK proliferation pathway activated by loss of Par3. Oncogene. 2015;34:3199–206. 10.1038/onc.2014.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bosco EE, Mulloy JC, Zheng Y. Rac1 GTPase: a “Rac” of all trades. Cell Mol Life Sci. 2009;66:370–4. 10.1007/s00018-008-8552-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Philip M, Schietinger A. CD8(+) T cell differentiation and dysfunction in cancer. Nat Rev Immunol. 2022;22:209–23. 10.1038/s41577-021-00574-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nunez NG, Tosello Boari J, Ramos RN, Richer W, Cagnard N, Anderfuhren CD, et al. Tumor invasion in draining lymph nodes is associated with Treg accumulation in breast cancer patients. Nat Commun. 2020;11:3272 10.1038/s41467-020-17046-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Church SE, Jensen SM, Antony PA, Restifo NP, Fox BA. Tumor-specific CD4+ T cells maintain effector and memory tumor-specific CD8+ T cells. Eur J Immunol. 2014;44:69–79. 10.1002/eji.201343718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bourne CM, Mun SS, Dao T, Aretz ZEH, Molvi Z, Gejman RS, et al. Unmasking the suppressed immunopeptidome of EZH2-mutated diffuse large B-cell lymphomas through combination drug treatment. Blood Adv. 2022;6:4107–21. 10.1182/bloodadvances.2021006069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ly A, Liao Y, Pietrzak H, Ioannidis LJ, Sidwell T, Gloury R, et al. Transcription factor T-bet in B cells modulates germinal center polarization and antibody affinity maturation in response to malaria. Cell Rep. 2019;29:2257–69 e6. 10.1016/j.celrep.2019.10.087. [DOI] [PubMed] [Google Scholar]
- 30.Gaulard P, de Leval L. The microenvironment in T-cell lymphomas: emerging themes. Semin Cancer Biol. 2014;24:49–60. 10.1016/j.semcancer.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 31.Tzeng SJ, Bolland S, Inabe K, Kurosaki T, Pierce SK. The B cell inhibitory Fc receptor triggers apoptosis by a novel c-Abl family kinase-dependent pathway. J Biol Chem. 2005;280:35247–54. 10.1074/jbc.M505308200. [DOI] [PubMed] [Google Scholar]
- 32.Xiong J, Wang L, Fei XC, Jiang XF, Zheng Z, Zhao Y, et al. MYC is a positive regulator of choline metabolism and impedes mitophagy-dependent necroptosis in diffuse large B-cell lymphoma. Blood Cancer J. 2017;7:e0 10.1038/bcj.2017.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leivonen SK, Icay K, Jantti K, Siren I, Liu C, Alkodsi A, et al. MicroRNAs regulate key cell survival pathways and mediate chemosensitivity during progression of diffuse large B-cell lymphoma. Blood Cancer J. 2017;7:654. 10.1038/s41408-017-0033-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roider T, Seufert J, Uvarovskii A, Frauhammer F, Bordas M, Abedpour N, et al. Dissecting intratumour heterogeneity of nodal B-cell lymphomas at the transcriptional, genetic and drug-response levels. Nat Cell Biol. 2020;22:896–906. 10.1038/s41556-020-0532-x. [DOI] [PubMed] [Google Scholar]
- 35.Dang CV. Sex, life, and death in MYC-driven lymphomagenesis. Mol Cell. 2021;81:3886–7. 10.1016/j.molcel.2021.09.009. [DOI] [PubMed] [Google Scholar]
- 36.Sun C, Cheng X, Wang C, Wang X, Xia B, Zhang Y. Gene expression profiles analysis identifies a novel two-gene signature to predict overall survival in diffuse large B-cell lymphoma. Biosci Rep. 2019;39. 10.1042/BSR20181293. [DOI] [PMC free article] [PubMed]
- 37.Fu T, Dai LJ, Wu SY, Xiao Y, Ma D, Jiang YZ, et al. Spatial architecture of the immune microenvironment orchestrates tumor immunity and therapeutic response. J Hematol Oncol. 2021;14:98. 10.1186/s13045-021-01103-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mourad N, Mounier N, Briere J, Raffoux E, Delmer A, Feller A, et al. Clinical, biologic, and pathologic features in 157 patients with angioimmunoblastic T-cell lymphoma treated within the Groupe d’Etude des Lymphomes de l’Adulte (GELA) trials. Blood. 2008;111:4463–70. 10.1182/blood-2007-08-105759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takimoto CH, Chao MP, Gibbs C, McCamish MA, Liu J, Chen JY, et al. The Macrophage ‘Do not eat me’ signal, CD47, is a clinically validated cancer immunotherapy target. Ann Oncol. 2019;30:486–9. 10.1093/annonc/mdz006. [DOI] [PubMed] [Google Scholar]
- 40.Chao MP, Alizadeh AA, Tang C, Myklebust JH, Varghese B, Gill S, et al. Anti-CD47 antibody synergizes with rituximab to promote phagocytosis and eradicate non-Hodgkin lymphoma. Cell. 2010;142:699–713. 10.1016/j.cell.2010.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Izykowska K, Rassek K, Korsak D, Przybylski GK. Novel targeted therapies of T cell lymphomas. J Hematol Oncol. 2020;13:176. 10.1186/s13045-020-01006-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Czimmerer Z, Halasz L, Daniel B, Varga Z, Bene K, Domokos A, et al. The epigenetic state of IL-4-polarized macrophages enables inflammatory cistromic expansion and extended synergistic response to TLR ligands. Immunity. 2022;55:2006–26.e6. 10.1016/j.immuni.2022.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mathewson ND, Ashenberg O, Tirosh I, Gritsch S, Perez EM, Marx S, et al. Inhibitory CD161 receptor identified in glioma-infiltrating T cells by single-cell analysis. Cell. 2021;184:1281–98.e26. 10.1016/j.cell.2021.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ghoochani A, Schwarz MA, Yakubov E, Engelhorn T, Doerfler A, Buchfelder M, et al. MIF-CD74 signaling impedes microglial M1 polarization and facilitates brain tumorigenesis. Oncogene. 2016;35:6246–61. 10.1038/onc.2016.160. [DOI] [PubMed] [Google Scholar]
- 45.Figueiredo CR, Azevedo RA, Mousdell S, Resende-Lara PT, Ireland L, Santos A, et al. Blockade of MIF-CD74 signalling on macrophages and dendritic cells restores the antitumour immune response against metastatic melanoma. Front Immunol. 2018;9:1132. 10.3389/fimmu.2018.01132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Du Y, Cai Y, Lv Y, Zhang L, Yang H, Liu Q, et al. Single-cell RNA sequencing unveils the communications between malignant T and myeloid cells contributing to tumor growth and immunosuppression in cutaneous T-cell lymphoma. Cancer Lett. 2022;551:215972. 10.1016/j.canlet.2022.215972. [DOI] [PubMed] [Google Scholar]
- 47.Sage PT, Paterson AM, Lovitch SB, Sharpe AH. The coinhibitory receptor CTLA-4 controls B cell responses by modulating T follicular helper, T follicular regulatory, and T regulatory cells. Immunity. 2014;41:1026–39. 10.1016/j.immuni.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Joyce JA, Fearon DT. T cell exclusion, immune privilege, and the tumor microenvironment. Science. 2015;348:74–80. 10.1126/science.aaa6204. [DOI] [PubMed] [Google Scholar]
- 49.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–64. 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu X, Pu Y, Cron K, Deng L, Kline J, Frazier WA, et al. CD47 blockade triggers T cell-mediated destruction of immunogenic tumors. Nat Med. 2015;21:1209–15. 10.1038/nm.3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guo Q, Wang T, Yang Y, Gao L, Zhao Q, Zhang W, et al. Transcriptional factor Yin Yang 1 promotes the stemness of breast cancer cells by suppressing miR-873-5p transcriptional activity. Mol Ther Nucleic Acids. 2020;21:527–41. 10.1016/j.omtn.2020.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Deng S, Cheng D, Wang J, Gu J, Xue Y, Jiang Z, et al. MYL9 expressed in cancer-associated fibroblasts regulate the immune microenvironment of colorectal cancer and promotes tumor progression in an autocrine manner. J Exp Clin Cancer Res. 2023;42:294. 10.1186/s13046-023-02863-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen J, Jiang CC, Jin L, Zhang XD. Regulation of PD-L1: a novel role of pro-survival signalling in cancer. Ann Oncol. 2016;27:409–16. 10.1093/annonc/mdv615. [DOI] [PubMed] [Google Scholar]
- 54.Fernandez JG, Rodriguez DA, Valenzuela M, Calderon C, Urzua U, Munroe D, et al. Survivin expression promotes VEGF-induced tumor angiogenesis via PI3K/Akt enhanced beta-catenin/Tcf-Lef dependent transcription. Mol Cancer. 2014;13:209 10.1186/1476-4598-13-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Long X, Wong CC, Tong L, Chu ESH, Ho Szeto C, Go MYY, et al. Peptostreptococcus anaerobius promotes colorectal carcinogenesis and modulates tumour immunity. Nat Microbiol. 2019;4:2319–30. 10.1038/s41564-019-0541-3. [DOI] [PubMed] [Google Scholar]
- 56.Ramkumar C, Cui H, Kong Y, Jones SN, Gerstein RM, Zhang H. Smurf2 suppresses B-cell proliferation and lymphomagenesis by mediating ubiquitination and degradation of YY1. Nat Commun. 2013;4:2598. 10.1038/ncomms3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Han J, Meng J, Chen S, Wang X, Yin S, Zhang Q, et al. YY1 complex promotes quaking expression via super-enhancer binding during EMT of hepatocellular carcinoma. Cancer Res. 2019;79:1451–64. 10.1158/0008-5472.CAN-18-2238. [DOI] [PubMed] [Google Scholar]
- 58.Vella P, Barozzi I, Cuomo A, Bonaldi T, Pasini D. Yin Yang 1 extends the Myc-related transcription factors network in embryonic stem cells. Nucleic Acids Res. 2012;40:3403–18. 10.1093/nar/gkr1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Casey SC, Tong L, Li Y, Do R, Walz S, Fitzgerald KN, et al. MYC regulates the antitumor immune response through CD47 and PD-L1. Science. 2016;352:227–31. 10.1126/science.aac9935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chihara D, Dores GM, Flowers CR, Morton LM. The bidirectional increased risk of B-cell lymphoma and T-cell lymphoma. Blood. 2021;138:785–9. 10.1182/blood.2020010497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang QX, Pei XJ, Tian XY, Li Y, Li Z. Secondary cutaneous Epstein-Barr virus-associated diffuse large B-cell lymphoma in a patient with angioimmunoblastic T-cell lymphoma: a case report and review of literature. Diagn Pathol. 2012;7:7. 10.1186/1746-1596-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hwang I. Cell-cell communication via extracellular membrane vesicles and its role in the immune response. Mol Cells. 2013;36:105–11. 10.1007/s10059-013-0154-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xiao Z, Dai Z, Locasale JW. Metabolic landscape of the tumor microenvironment at single cell resolution. Nat Commun. 2019;10:3763. 10.1038/s41467-019-11738-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Du Y, Li K, Wang X, Kaushik AC, Junaid M, Wei D. Identification of chlorprothixene as a potential drug that induces apoptosis and autophagic cell death in acute myeloid leukemia cells. FEBS J. 2020;287:1645–65. 10.1111/febs.15102. [DOI] [PubMed] [Google Scholar]
- 65.Windhager J, Zanotelli VRT, Schulz D, Meyer L, Daniel M, Bodenmiller B, et al. An end-to-end workflow for multiplexed image processing and analysis. Nat Protoc. 2023;18:3565–613. 10.1038/s41596-023-00881-0. [DOI] [PubMed]
- 66.Schapiro D, Jackson HW, Raghuraman S, Fischer JR, Zanotelli VRT, Schulz D, et al. histoCAT: analysis of cell phenotypes and interactions in multiplex image cytometry data. Nat Methods. 2017;14:873–6. 10.1038/nmeth.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The processed count matrices have been deposited in the OMIX database (id: OMIX004773). Original code has been deposited at Github (https://github.com/zhumengyan/AITL2023) and is publicly available as of the date of publication. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.