Abstract
Objective
The study’s aim was to explore a novel miRNA/mRNA network mediated by circNINL in cervical cancer (CC).
Methods
Tumor tissue specimens and normal tissue specimens of 86 CC patients were collected. CircNINL, miR-2467-3p, and specificity protein 1 (SP1) expression levels in tissues were detected. In Hela cells, transfection was implemented to determine whether CircNINL, miR-2467-3p and SP1 regulated cellular progression. The interaction between circNINL, miR-2467-3p, and SP1 was explored.
Results
CircNINL and SP1 were abnormally increased whereas miR-2467-3p expression was suppressed in CC. Functionally, circNINL silencing or miR-2467-3p overexpression reduced CC cell progression, whereas circNINL overexpression did the opposite. CircNINL had a spongy effect on miR-2467-3p to regulate SP1 levels. miR-2467-3p inhibition or SP1 overexpression offset the inhibitory effect of circNINL silence on CC malignant progression.
Conclusion
CircNINL silencing can increase miR-2467-3p, thereby inducing the downregulation of SP1, thereby suppressing CC progression.
Keywords: Cervical cancer, CircNINL, miR-2467-3p, Specificity protein 1
Introduction
Cervical cancer (CC) is a prevalent tumor affecting the cervix and posing a serious threat to female reproductive health [1]. In addition to human papillomavirus (HPV) infection, environment, immune, genetic and epigenetic factors also are related to the etiology of CC [2]. HPV vaccination is an effective way to prevent infection, but the lack of timely cancer screening and vaccination has resulted in the majority of CC patients having advanced or invasive cancer at diagnosis [3]. The treatment of advanced CC is a clinical challenge that needs to be urgently solved because the prognosis of advanced CC is often poor and the possibility of metastasis is high. At present, surgery and radiotherapy are clinically applied, but lesion morphology, location and degree of infiltration are different, as well as differences in individual patient's condition, some CC patients (particular recurrent or advanced patients) are not sensitive to chemotherapy [4, 5]. Consequently, pursuing alternative therapeutic strategies requires an in-depth understanding of the molecular mechanisms related to CC progression.
Circular RNAs (circRNAs) have higher stability than linear RNAs [6] and can decoy miRNAs and therefore change mRNA expression [7, 8]. It has been demonstrated that has_circ_0000515 can promote the malignant behaviors of CC cells by binding to miR-326 [9] and circ_0031288 could exert oncogenic effects in CC [10]. circNINL is a novel oncogene. Elevated levels of circNINL in breast cancer tissues and cell lines and its high expression are associated with poor clinicopathologic features and prognosis, and significantly promote cell proliferation and migration [11]. circNINL is highly expressed in lung cancer and promotes cell proliferation, migration, invasion, and aerobic glycolysis through the miR-3918/FGFR1 axis [12]. However, the mechanism of action of circNINL in CC remains unclear.
Given the significant action of miRNAs in tumor biology [13], analysis of miRNA signature could serve as biomarkers for tumor diagnosis and progression. Some miRNAs such as miR-21 have a promoting effect on CC cells, while miR-372 has an inhibitory effect [14].
As a zinc-finger transcription factor, specificity protein 1 (Sp1) could bind to GC-rich motifs in promoters and interact with co-activating complexes of multiple signaling pathways, mediate cellular functions, and stimulate tumor malignancy [15, 16]. It has been reported that SP1 triggers ovarian cancer development [17]. Based on bioinformatics software analysis, circNINL and Sp1 had complementary binding sites to miR-2467-3p, which prompted us to speculate on the circNINL/miR-2467-3p/Sp1 network in CC. Our analysis explored the potential regulatory network of circNINL/miR-2467-3p/Sp1 in CC and provided new therapeutic targets for clinical CC patients.
Materials and methods
Clinical samples
CC tissue specimens and healthy tissue specimens were collected from 86 CC patients in Affiliated Hospital of Weifang Medical College. All were pathologically confirmed to be squamous cell carcinoma. Immediately after surgical excision, the tissues were stored in liquid nitrogen and stored at −80 °C. This study was approved by the Ethics Committee of Affiliated Hospital of Weifang Medical College. All participants signed written informed consent.
Cell culture
H8 and human CC cell lines (Caski, ME180, MS751, SiHa, HeLa) were obtained from ATCC. Caski, SiHa, HeLa, ME180 and H8 cells were cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.). MS751 was cultured in MEM (Gibco, USA). The medium was supplemented with 10% FBS (Invitrogen) and 1% penicillin/streptomycin (Gibco) [18, 19].
Cell transfection
The 3rd passage HeLa cells were digested with trypsin (Beyotime, Shanghai, China) and inoculated in 24-well plates (1 × 106 cells/well). When 85% cell confluence was reached, cell transfection was achieved by Lipofectamine 2000 (Invitrogen, USA). The synthesis of transfection vectors was commissioned to GenePharma (Shanghai, China), including sh-circNINL, circNINL overexpression vector, miR-2467-3p mimic and inhibitor, SP1 overexpression vector (pcDNA SP1), and their corresponding controls. For transient transfection, HeLa cells (1 × 106/well) were mixed with 50 nM oligonucleotides or 4.0 μg vector for 24 h at 37 °C according to the instruction of Lipofectamine 2000 (Invitrogen). Transfection efficiency was assessed by RT-qPCR.
Actinomycin D assay
Actinomycin D (Sigma, Germany) was added to HeLa cell medium at 5 µg/ml. Cells were collected at the indicated time points, and total RNA was extracted to measure circNINL expression using RT-qPCR. Untreated RNA served as a control (Mock).
RNase R treatment
RNA was digested with 3 U/μg RNase R reagent (Epicentre, USA) at 37 °C for 15 min and RNA was purified using the RNeasy MinElute Cleanup kit (Qiagen, Hilden, Germany). Subsequently, RT-qPCR was used to study circNINL expression and linear NINL.
Subcellular localization analysis
Cytoplasmic RNA and nuclear RNA of HeLa cells were isolated using the PARIS Kit (Ambion, USA), followed by RNA expression analysis.
MTT assay
HeLa cells (6 × 103/well) were collected after transfection and inoculated into 96-well plates (6 × 103 cells/well). At 24, 48, 72 or 96 h, 10 μL of prepared MTT reagent (5 mg/mL; Sigma-Aldrich) was added to each well and incubated at 37 °C for 4 h. The supernatant was aspirated. Then, 100 μl of dimethyl sulfoxide (Sigma-Aldrich) was added to each well, and the crystals were dissolved by placing the microtiter plate on a shaker for 10 min. A microplate reader (PR 4100 TSC, Bio-Rad) was used to detect the optical density value of each well at 570 nm.
Colony formation assay
HeLa cells (2 × 103/well) were inoculated into 6-well plates with DMEM containing 10% FBS and incubated in a humidified incubator (37 °C, 5% CO2) for two weeks. After colony formation was observed, the medium was removed. Cells were washed twice with PBS (Beyotime) and fixed with 1 mL of 4% paraformaldehyde (Beyotime) for 10 min. Then, 1 mL of 0.1% crystal violet staining solution was added for 10 min. Excess staining solution was rinsed with running water, cells were photographed through a microscope (Olympus Corp, Tokyo, Japan), and the number of colonies was counted.
Scratch test
Hela cells were inoculated at a density of 5 × 105 cells/well in 24-well plates and grown to 90% confluence. Confluent monolayers were scraped using a pipette tip and each well was washed with PBS (Beyotime) to remove non-adherent cells. Cells were incubated for 24 h after treatment. At 0 and 24 h, cells were observed under a light microscope (Olympus, Tokyo, Japan) and cell migration distances were measured using Image-Pro Plus analysis software (Media Cybernetics, MD, USA).
Transwell assay
Cell migration and invasion experiments were performed using Transwell chambers (Corning, NY, USA) with a pore size of 8 μm. The upper chamber of Transwell chambers (Costar, NY, USA) was coated with Matrigel. HeLa cells (8 × 104) were resuspended in the upper chamber with 200 μL of serum-free medium, and 600 μL of complete medium containing 10% FBS was added as a chemoattractant in the lower compartment. After 24 h of incubation in the incubator (37 °C, 5% CO2), cells in the upper chamber were gently wiped with a cotton swab and washed with PBS, and the cells were fixed in the lower chamber with 4% paraformaldehyde (Beyotime). The invasive cells were counted by staining with 0.5% crystal violet (Kokusai Chemical Reagent) and then observed through an inverted microscope (Olympus) with five randomly selected fields of view.
Flow cytometry
Apoptotic cells were detected by Annexin V-FITC apoptosis detection kit (Thermo Fisher Scientific). The treated HeLa cells were inoculated into 6-well plates with 5 × 104 cells per well and incubated overnight at room temperature. HeLa cells were digested with EDTA-free trypsin (0.25%, Beyotime) and centrifuged at 1200 rpm for 5 min to obtain cell precipitates. The cell precipitates were gently resuspended in 500 μl 1 × Annexin V binding buffer and stained with 10 μL Annexin V-FITC and 5 μL propidium iodide (PI) for 30 min at room temperature, protected from light. Apoptotic cells (Annexin V-positive and PI-negative) and necrotic cells (Annexin V- and PI-positive) were distinguished using a FACS Calibur flow cytometer (BD Biosciences). Apoptosis rate was the percentage of Annexin V-FITC-positive and PI-positive/negative cells.
Examination of RNA expression
Total RNA was extracted with Trizol reagent (Takarashu, Otsu, Japan), and RNA concentration was measured using a dicinchoninic acid (CC) kit (Invitrogen). For miRNA, cDNA was produced using the TaqMan™ Advanced miRNA cDNA Synthesis Kit (Takara), while PrimeScript™ RT Reagent kit (TaKaRa, Japan) was used to synthesize cDNA for mRNA and circRNA. Three replicates of qRT-PCR were completed for each sample on the First Step Plus Real-Time PCR System (Applied Biosystems) using SYBR Green PCR Kit (Takara). Gene expression was determined by 2−ΔΔCt method. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal reference gene for circRNAs and mRNAs, and U6 was used as an internal control for miRNAs to normalize the expression levels of each gene. PCR primers (Table 1) were synthesized by Takara (Dalian, China).
Table 1.
Sequences for PCR
| Sequences (5′—3′) | |
|---|---|
| CircNINL | Forward: 5'-CCTGGGGCTGCATAAGTGAT-3' |
| Reverse: 5'-TGAGTCACCAGAGCACAACG-3' | |
| miR-2467-3p | Forward: 5'-GCCGAGGGACAGGCACCTGA-3' |
| Reverse: 5'-CTCAACTGGTGTCGTGGA-3' | |
| SP1 | Forward: 5'‐GCACCTGCCCCTACTGTAAA‐3' |
| Reverse: 5'‐GTGCCTCTGTAGCTCATCCG‐3' | |
| Bcl-2 | Forward: 5'‐GGTGGGGTCATGTGTGTGG‐3' |
| Reverse: 5'‐CGGTTCAGGTACTCAGTCATCC‐3' | |
| Bax | Forward: 5'-CCCGAGAGGTCTTTTTCCGAG-3' |
| Reverse: 5'-CCAGCCCATGATGGTTCTGAT-3' | |
| U6 | Forward: 5'-GGCTGTGTTCAAGTGGTTCT-3' |
| Reverse: 5'-ACGCCTGTAATCACAGCACT-3' | |
| GAPDH | Forward: 5'- CTGCCAACGTGTCAGTGGTG-3' |
| Reverse: 5'- TCAGTGTAGCCCAGGATGCC-3' |
CircNINL Circular RNA NINL, miR-2467-3p microRNA-2467-3p, SP1 specificity protein 1, GAPDH Glyceraldehyde-3-phosphate dehydrogenase
Protein expression analysis
Cells were washed twice with pre-cooled PBS. Total proteins were then extracted with RIPA lysis buffer (Beyotime) on ice for 20 min and quantified using BCA™ protein assay kit (Pierce, Appleton, USA). Proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to polyvinylidene fluoride membranes, which was blocked with 5% skimmed milk for 2 h and interacted with primary antibodies SP1 (1:1000, ab231778), Bcl-2 (1:1000, ab32124), Bax (1:1000, ab32503), and GAPDH (1:1000, ab8245, Abcam) at 4 °C overnight. Rabbit monoclonal IgG (1:1000, ab172730) was incubated for 1 h at 37 °C as a secondary antibody for Bax, and goat Anti-Rabbit IgG HRP (1:100, 000, ab97051) for the remaining antibodies. Finally, the results were visualized with the ECL Detection Kit (Vazyme; E411-04) and examined on a FluorChem™M system.
Dual luciferase reporter assay
starBase3.0 (http://starbase.sysu.edu.cn/) predicted potential binding sites of miR-2467-3p with circNINL and Sp1. Vectors (circNINL-WT, circNINL-MUT, SP1-WT, and SP1-MUT) were generated by cloning wild-type or mutant circNINL or SP1 fragment containing the miR-2467-3p target sequence into pmirGLO luciferase reporter vectors (Promega). HeLa cells were inoculated into 96-well plates with 1 × 104 cells per well and cultured to logarithmic phase in a humidified incubator (37 °C, 5% CO2). Using Lipofectamine 2000 reagent (Invitrogen), the luciferase reporter plasmid was co-transfected with miR-2467-3p mimic to HeLa cells, and culture was continued for 48 h. Luciferase activity was detected by a dual luciferase assay system (Promega) [20].
RNA-pull down assay
Biotinylated miR-2467-3p and biotinylated miR-NC (100 pmol) were transfected into HeLa cells at 50% confluence using Lipofectamine 3000 (Invitrogen). Cells were harvested and lysed in lysis buffer at 24 h post-transfection and incubated with streptavidin magnetic beads (Life Technologies) for 3 h. The beads were washed, and RNA interacting with the miRNA was extracted using Trizol reagent. The enrichment of circNINL and SP1 in the precipitated complexes was analyzed by RT-qPCR.
Statistical analysis
All data were analyzed and plotted by GraphPad Prism8.0 software. Measurement data were reported in the form of mean ± standard deviation. After the test of normal distribution and homogeneity of variance, the data were analyzed by t-test for bilateral comparisons or one-way ANOVA or two-way ANOVA followed by Tukey's multiple comparisons for multiple comparisons. The enumeration data (rate or percentage) were assessed by chi-square test. P indicated a two-sided test, and P < 0.05 was considered statistically significant.
Results
CircNINL is overexpressed in CC
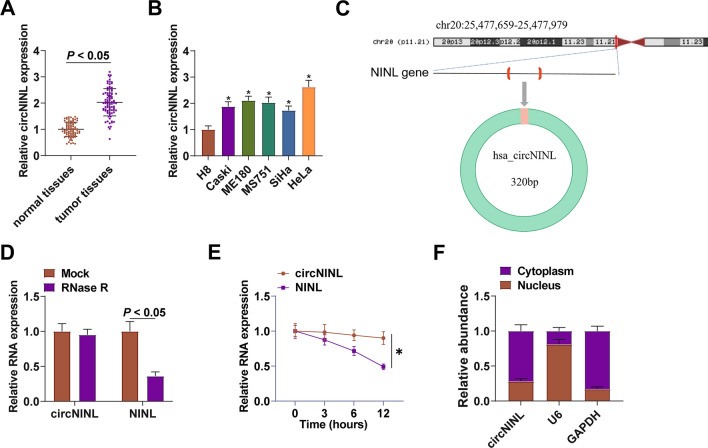
CircNINL levels in CC patients’ tissue samples (n = 86) were compared, presenting an overexpressed trend in CC tissues (Fig. 1A). The 5 CC cell lines had the same expression trend of circNINL when compared with H8 (Fig. 1B). To characterize circRNA transcripts, circbase (http://www.circbase.org/) and Primer were used to determine the stability of circNINL. circNINL was derived from “lasso splicing” of intron 16 of its NINL locus (Fig. 1C). circ-NINL was resistant to RNase R, while NINL was rapidly digested by RNase R (Fig. 1D). Furthermore, actinomycin D results showed that circ-NINL was more stable than NINL (Fig. 1E). Subcellular localization analysis validated that circNINL was mainly distributed in the cytoplasm of CC cells (Fig. 1F).
Fig. 1.
Abnormal elevation of circNINL expression in CC. circNINL in clinical samples and CC cell lines (A-B); the genomic location and structural characteristics of circNINL (C); the stability of circNINL (D); circ-NINL expression at different time points after Actinomycin D treatment (E); Subcellular localization of circNINL (F). n = 86, N = 3. Data were analyzed by t-test or by two-way ANOVA, and Tukey's multiple comparison test. * P < 0.05 vs. H8
Referring to the median of circNINL expression, there were 43 patients with circNINL high expression and 43 patients with low expression. The clinical analysis demonstrated the correlations of circNINL expression with tumor size, differentiation, interstitial infiltration, vascular invasion, FIGO stage and lymph node metastasis (Table 2). There is an upregulation of circNINL in CC, which correlates with the clinicopathological characteristics of the disease.
Table 2.
Correlation between CircNINL expression and clinicopathological features of cervical cancer
| Characteristics | Cases | CircNINL expression | P | ||
|---|---|---|---|---|---|
| High (n = 43) | Low (n = 43) | ||||
| Age | < 35 years | 19 | 11 | 8 | 0.604 |
| ≥ 35 years | 67 | 32 | 35 | ||
| Tumor size | < 3 cm | 40 | 26 | 14 | 0.017 |
| ≥ 3 cm | 46 | 17 | 29 | ||
| Differentiation | High | 58 | 34 | 24 | 0.037 |
| Low | 28 | 9 | 19 | ||
| Interstitial infiltration | < 66% | 36 | 12 | 24 | 0.016 |
| ≥ 66% | 50 | 31 | 19 | ||
| Vascular invasion | Yes | 47 | 17 | 30 | 0.009 |
| No | 39 | 26 | 13 | ||
| FIGO | I ~ II | 54 | 32 | 22 | 0.044 |
| III ~ IV | 32 | 11 | 21 | ||
| Lymph node metastasis | Yes | 61 | 36 | 25 | 0.017 |
| No | 25 | 7 | 18 | ||
The regulatory role of circNINL in CC cell biological progression
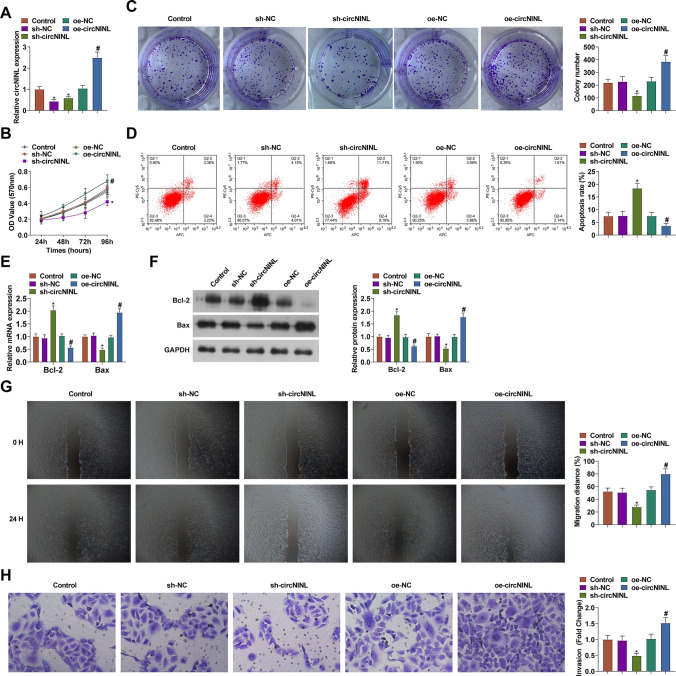
HeLa cells with the highest expression of circNINL were the study object in functional assays. sh-circNINL-1, sh-circNINL-2, and circNINL overexpression plasmid were transfected into HeLa cells, successfully altering cellular circNINL expression (Fig. 2A). Since the knockdown effect of sh-circNINL-1 was more obvious, subsequent functional experiments were performed using sh-circNINL-1, denoted as sh-circNINL. When investigating the tumor biology in CC, cellular assays were included and eventually proved that circNINL-silenced HeLa cells exhibited repressed viability and proliferation, elevated apoptosis rate, restrained Bcl-2 expression and induced Bax expression, enhanced migration and invasion abilities (Fig. 2B-H). The results were the opposite after overexpression of circNINL (Fig. 2B-H).
Fig. 2.
The regulatory role of circNINL in the biological progression of CC cells. circNINL expression (A), proliferation activity (B-C), apoptosis (D), Bcl-2 and Bax protein expression (E–F), migration and invasion (G-H) after transfection with sh-circNINL. N = 3, *P < 0.05 vs. sh-NC; #P < 0.05 vs. Vector. Data were analyzed by one-way ANOVA followed by Tukey post hoc test
A spongy effect of circNINL on miR-2467-3p
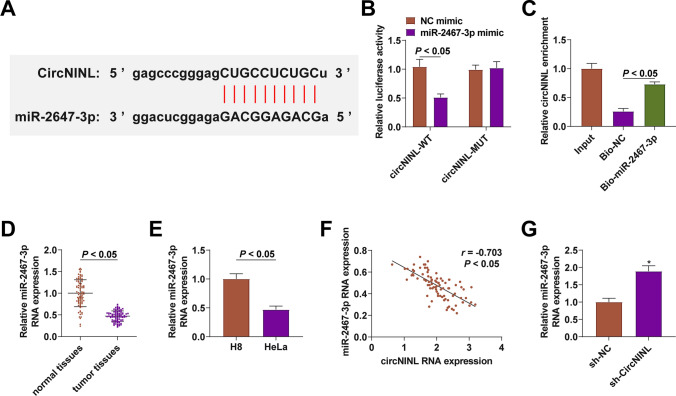
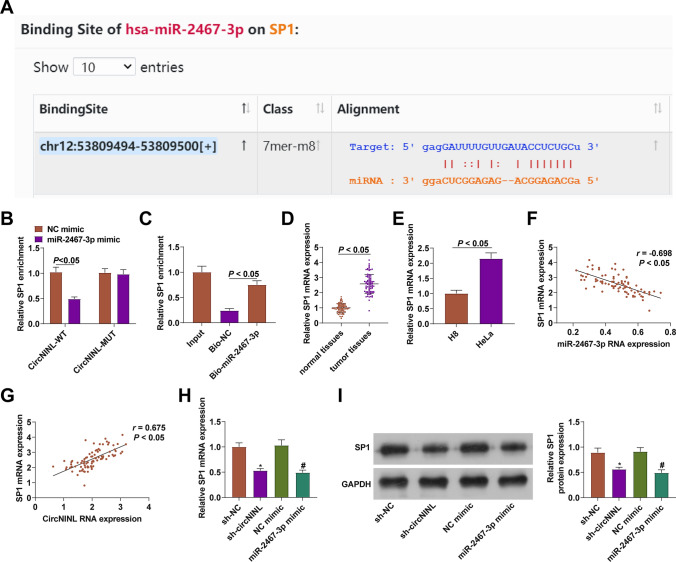
CircRNAs can regulate miRNA expression. To further study the new mechanism of circNINL regulating CC, the potential target miR-2467-3p of circNINL was analyzed through the bioinformatics software http://starbase.sysu.edu.cn/ (Fig. 3A). The dual luciferase reporter experiments showed that miR-2467-3p posed a repressive impact on the luciferase activity of circNINL-WT (Fig. 3B); RNA-pull down assay showed that biotinylated miR-2467-3p had a great promoting effect on circNINL abundance (Fig. 3C). miR-2467-3p expression was determined to be lower in CC tissues than normal tissues, and in CC cell lines than H8 cells (Fig. 3D, E). Moreover, miR-2467-3p expression had an inverse correlation with circNINL expression (Fig. 3F) and could be promoted when circNINL was silenced in HeLa cells (Fig. 3G).
Fig. 3.
A spongy effect of circNINL on miR-2467-3p. Binding site between circNINL and miR-2467-3p (A), evaluation of the binding between miR-2467-3p and circNINL (B-C), miR-2467-3p expression in CC patients (n = 86) and HeLa cells (D-E), correlation between miR-2467-3p and circNINL (n = 86) (F), miR-2467-3p expression after inhibiting circNINL (G). N = 3, *P < 0.05 vs. sh-NC group. Data were analyzed by one-way ANOVA followed by Tukey post hoc test
miR-2467-3p-induced prevention against CC malignancy
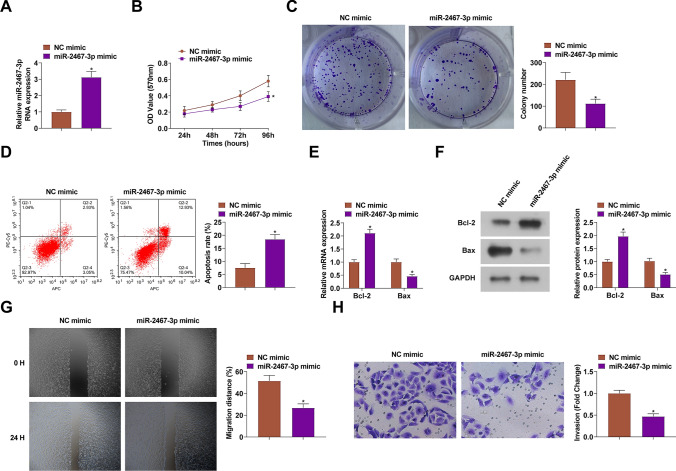
miR-2467-3p mimic transfection was conducted to probe miR-2467-3p-mediated CC progression (Fig. 4A). Then, the aforementioned experiments were repeated, figuring out that miR-2467-3p-overexpressed HeLa cells had similar malignant phenotypes of circNINL-silenced HeLa cells (Fig. 4B-H). In conclusion, miR-2467-3p inhibited CC cell progression.
Fig. 4.
The suppressive impacts of miR-2467-3p on the biological progression of CC cells. miR-2467-3p expression (A), proliferation activity (B-C), apoptosis (D), Bcl-2 and Bax protein expression (E–F), migration and invasion (G-H) after transfection with miR-2467-3p mimic. N = 3, *P < 0.05 vs. NC mimic. Data were analyzed by one-way ANOVA followed by Tukey post hoc test
miR-2467-3p-mediated targeting on SP1
The bioinformatics software http://starbase.sysu.edu.cn/ predicted miR-2467-3p targeting SP1 (Fig. 5A). The data obtained from dual-luciferase reporter assay and RNA-pull down assay also verified the binding relationship between miR-2467-3p and SP1 (Fig. 5B, C). SP1 expression was checked to be higher in CC tissues and HeLa cells (Fig. 5D, E), demonstrating a negative correlation with miR-2467-3p expression, as well as a positive correlation with circNINL expression (Fig. 5F, G). Next, it was tested that SP1 expression was reduced in HeLa cells after silencing circNINL or overexpressing miR-2467-3p (Fig. 5H, I). Briefly, miR-2467-3p could target.
Fig. 5.
A binding effect of miR-2467-3p on SP1. Binding site between miR-2467-3p and SP1 (A), evaluation of the binding between miR-2467-3p and SP1 (B-C), SP1 expression in CC patients (n = 86) and HeLa cells (D-E), correlation between miR-2467-3p and circNINL with SP1 (n = 86) (F-G), SP1 expression after inhibition of circNINL or overexpression of miR-2467-3p (H-I). N = 3, *P < 0.05 vs. sh-NC; #P < 0.05 vs. NC mimic. Data were analyzed by one-way ANOVA followed by Tukey post hoc test
circNINL/miR-2467-3p/SP1 network-mediated the biological progress of CC cells
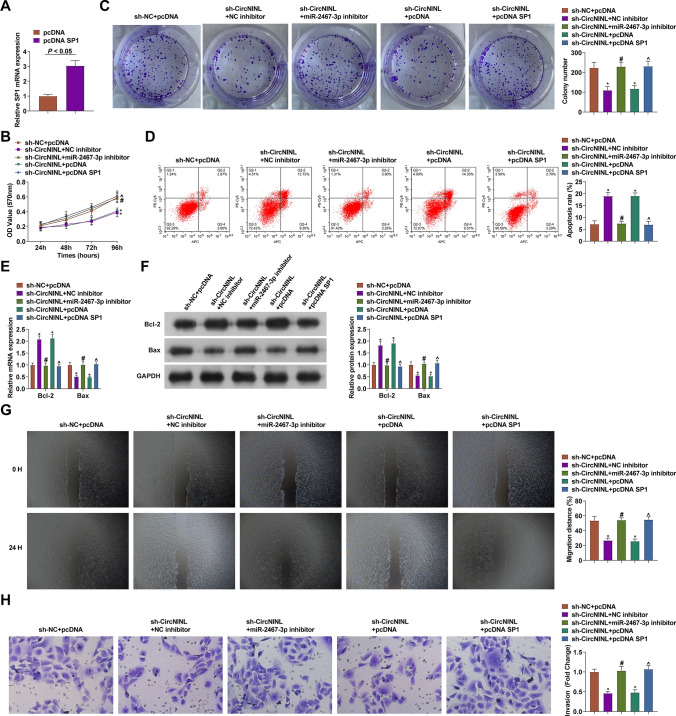
pcDNA SP1 overexpression vector or miR-2467-3p inhibitor was transfected into CC cells on the basis of sh-circNINL transfection to clarify the circNINL/miR-2467-3p/SP1 axis regulates the biological progress of CC cells. The two transfection vectors led to the restoration of SP1 mRNA expression in the pre-transfected HeLa cells (Fig. 6A). Through experimental analysis, it was turned out that after co-transfection of miR-2467-3p inhibitor or pcDNA SP1, sh-circNINL-induced alternations of HeLa cell phenotypes were mitigated (Fig. 6B-H). In conclusion, circNINL/miR-2467-3p/SP1 axis regulates CC cell biological progression.
Fig. 6.
circNINL/miR-2467-3p/SP1 network-mediated the biological progress of CC cells. SP1 expression (A), proliferation activity (B-C), apoptosis (D), Bcl-2 and Bax protein expression (E–F), migration and invasion (G-H) after co-transfection. N = 3, *P < 0.05 vs. sh-NC group; #P < 0.05 vs. sh-circNINL + NC inhibitor; ^P < 0.05 vs. sh-circNINL + pcDNA. Data were analyzed by one-way ANOVA followed by Tukey post hoc test
Discussion
Although both prevention and treatment methods have achieved rapid development at this stage, CC still faces poor prognosis such as postoperative recurrence and metastasis, and a low survival rate [21]. HPV infection and gene mutation are key pathogenic factors [22]. This work aimed to investigate the effect of circNINL on CC by sponging miR-2467-3p and upregulating SP1.
CircRNA has high stability and conservation and regulates gene expression in various organisms [23], attracting attention on genetic research. Its regulatory mechanism in cancer can be summarized as ceRNA network [24]. It has been established that CC cell invasion and proliferation could be enhanced by hsa_circRNA_101996-pivoted ceRNA network [25]. Abnormally expressed circNINL exerts functionally as to the proliferation, migration and apoptosis of breast cancer cells [6]. Concerning the novelty, the present work analyzed that circNINL was upregulated in CC and associated with tumor size, differentiation, interstitial infiltration, vascular invasion, FIGO stage and lymph node metastasis. Furthermore, silencing circNINL in HeLa cells inhibited the cellular progression while restoring circNINL caused the opposite results, indicating that circNINL may act as an oncogene in CC.
A miRNA sponge mechanism was explored, confirming the binding of circNINL to miR-2467-3p. miR-2467-3p has been considered a tumor inhibitor by directly interacting with target genes, such as non-small cell lung cancer [26] and colorectal cancer [27]. In addition, miR-2467-3p downregulation has been tested in CC previously, as well as its anti-CC effects on cellular activities [28]. Echoed with the former studies, our research tested that miR-2467-3p was downregulated in CC and that miR-2467-3p inhibited CC cell malignant activities. To investigate the mechanism thoroughly, functional rescue assay validated that knocking down miR-2467-3p released the effects of circNINL silencing on CC cells.
SP1 was shown to be a downstream target of miR-2467-3p. SP1 is closely related to tumor biology through various signaling pathways and acts aggressively in triple-negative breast cancer [29], skin squamous cell carcinoma [30], and colorectal cancer [31]. Especially, SP1 is upregulated in CC and controlling SP1 expression can inhibit the migration and invasion of human CC cells [32, 33]. Consistent with this, the present work displayed that SP1 expression was raised in CC and could block the effects of silencing circNINL on CC cells.
This study demonstrated that circNINL, a novel oncogenic RNA, regulates proliferation, apoptosis, migration and invasion of CC cell lines through miR-2467-3p/SP1. To the best of our knowledge, this is the first study to point out the role of circNINL in CC and its mechanism, providing a possible therapeutic target for CC intervention. Nevertheless, there is still a lack of sufficient clinical trials to validate the effect of the circNINL/miR-2467-3p/SP1 axis on CC treatment. Whether circNINL regulates signaling pathways other than miR-2467-3p/SP1 is also a question we need to address urgently. In addition, drug resistance in CC becomes difficult in the treatment of CC, and it is necessary to investigate the effect of circNINL/miR-3918/FGFR1 axis on drug resistance in CC cell lines in subsequent studies.
Taken together, the data suggest that circNINL is highly expressed in CC. CircNINL/miR-2467-3p/SP1 axis stimulates the malignant progression of CC cells, underlining a new regulatory pathway in CC and providing new ideas and insights for CC intervention.
Acknowledgements
Not applicable.
Author contributions
Chengcheng Cao designed the research study. Haifeng Zhang and Ting Zhang performed the research. Chengcheng Cao and Cuicui Nie provided help and advice on the experiments. Haifeng Zhang, Ting Zhang and Cuicui Nie analyzed the data. Chengcheng Cao wrote the manuscript. Cuicui Nie reviewed and edited the manuscript. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript.
Funding
Not applicable.
Data availability
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent participate
All procedures performed in this study involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. All subjects was approved by Sunshine Union Hospital (No.2017S1211).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Vu M, Yu J, Awolude OA, Chuang L. Cervicalcancer worldwide. Curr Probl Cancer. 2018;42:457–65. [DOI] [PubMed] [Google Scholar]
- 2.Yee GP, de Souza P, Khachigian LM. Current and potential treatments for cervical cancer. Curr Cancer Drug Targets. 2013;13(2):205–20. [DOI] [PubMed] [Google Scholar]
- 3.Yao Z, Shu L, Yi Y, Qiao L. Hsa_circRNA_000543 predicts poor prognosis and promotes cervical cancer cell progression through regulating miR-567/ZNF268 axis. Cancer Manag Res. 2021;30(13):5211–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang C, Liu P, Huang J, Liao Y, Pan C, Liu J, Du Q, Liu T, Shang C, Ooi S, Chen R, Xia M, Jiang H, Xu M, Zou Q, Zhou Y, Huang H, Pan Y, Yuan L, Wang W, Yao S. Circular RNA hsa_circ_0043280 inhibits cervical cancer tumor growth and metastasis via miR-203a-3p/PAQR3 axis. Cell Death Dis. 2021;12(10):888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abu-Rustum NR, Yashar CM, Bean S, Bradley K, Campos SM, Chon HS, et al. NCCN guidelines insights: cervical cancer, version 12020. J Natl Compr Canc Netw. 2020;18:660–6. [DOI] [PubMed] [Google Scholar]
- 6.Xu C, Yu H, Yin X, Zhang J, Liu C, Qi H, Liu P. Circular RNA circNINL promotes breast cancer progression through activating β-catenin signaling via miR-921/ADAM9 axis. J Biochem. 2021;169(6):693–700. [DOI] [PubMed] [Google Scholar]
- 7.Shen S, Wu Y, Chen J, Xie Z, Huang K, Wang G, Yang Y, Ni W, Chen Z, Shi P, Ma Y, Fan S. CircSERPINE2 protects against osteoarthritis by targeting miR-1271 and ETS-related gene. Ann Rheum Dis. 2019;78:826–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo WW, Feng MM, Li SF, Wei LH. Circular RNA circ_0023404 serves as a miR-636 sponge to promote malignant behaviors in cervical cancer cells through upregulation of CYP2S1. Kaohsiung J Med Sci. 2021. 10.1002/kjm2.12478. [DOI] [PubMed] [Google Scholar]
- 9.Tang Q, Chen Z, Zhao L, Xu H. Circular RNA hsa_circ_0000515 acts as a miR-326 sponge to promote cervical cancer progression through upregulation of ELK1. Aging (Albany NY). 2019;11(22):9982–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu YJ, Yu H, Liu GX. Hsa_circ_0031288/hsa-miR-139–3p/Bcl-6 regulatory feedback circuit influences the invasion and migration of cervical cancer HeLa cells. J Cell Biochem. 2020;121(10):4251–60. [DOI] [PubMed] [Google Scholar]
- 11.Xu C, Yu H, Yin X, Zhang J, Liu C, Qi H, et al. Circular RNA circNINL promotes breast cancer progression through activating β-catenin signaling via miR-921/ADAM9 axis. J Biochem. 2021;169(6):693–700. [DOI] [PubMed] [Google Scholar]
- 12.Li S, Qiu C, Sun D, Yang S, Wang L. circNINL facilitates aerobic glycolysis, proliferation, invasion, and migration in lung cancer by sponging miR-3918 to mediate FGFR1 expression. Eur J Med Res. 2024;29(1):67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol. 2009;4(1):199–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nahand J, Taghizadeh-boroujeni S, Karimzadeh M, et al. microRNAs: new prognostic, diagnostic, and therapeutic biomarkers in cervical cancer. J Cell Physiol. 2019;234(10):17064–99. [DOI] [PubMed] [Google Scholar]
- 15.Choi ES, Nam JS, Jung JY, Cho NP, Cho SD. Modulation of specificity protein 1 by mithramycin a as a novel therapeutic strategy for cervical cancer. Sci Rep. 2014;24(4):7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee HE, et al. Inhibition of specificity protein 1 by dibenzylideneacetone, a curcumin analogue, induces apoptosis in mucoepidermoid carcinomas and tumor xenografts through Bim and truncated Bid. Oral Oncol. 2014;50:189–95. [DOI] [PubMed] [Google Scholar]
- 17.Cui PH, Li ZY, Li DH, Han SY, Zhang YJ. SP1-induced lncRNA DANCR contributes to proliferation and invasion of ovarian cancer. Kaohsiung J Med Sci. 2021;37(5):371–8. [DOI] [PubMed] [Google Scholar]
- 18.Zhang C, Liu P, Huang J, Liao Y, Pan C, Liu J, et al. Circular RNA hsa_circ_0043280 inhibits cervical cancer tumor growth and metastasis via miR-203a-3p/PAQR3 axis. Cell Death Dis. 2021;12(10):888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng Q, Zhang J, Zhang T, Liu Y, Du X, Dai X, et al. Hsa_circ_0000520 overexpression increases CDK2 expression via miR-1296 to facilitate cervical cancer cell proliferation. J Transl Med. 2021;19(1):314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li X, Ma N, Zhang Y, Wei H, Zhang H, Pang X, et al. Circular RNA circNRIP1 promotes migration and invasion in cervical cancer by sponging miR-629-3p and regulating the PTP4A1/ERK1/2 pathway. Cell Death Dis. 2020;11(5):399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xia C, He Z, Cai Y. Quantitative proteomics analysis of differentially expressed proteins induced by astragaloside IV in cervical cancer cell invasion. Cell Mol Biol Lett. 2020;25:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fang J, Zhang H, Jin S. Epigenetics and cervical cancer: from pathogenesis to therapy. Tumour Biol. 2014;35:5083–93. [DOI] [PubMed] [Google Scholar]
- 23.Hsiao K-Y, Sun HS, Tsai S-J. Circular RNA–new member of noncoding RNA with novel functions. Exp Biol Med. 2017;242(11):1136–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vo JN, Cieslik M, Zhang Y, et al. The landscape of circular RNA in cancer. Cell. 2019;176(4):869–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song T, Xu A, Zhang Z, et al. CircRNA hsa_circRNA_101996 increases cervical cancer proliferation and invasion through activating TPX2 expression by restraining miR-8075. J Cell Physiol. 2019;234(8):14296–305. [DOI] [PubMed] [Google Scholar]
- 26.Duan Z, Wei S, Liu Y. Circ_0074027 contributes to non-small cell lung cancer progression through positively modulating RHOA via sequestering miR-2467-3p. J Bioenerg Biomembr. 2021;53(2):223–33. [DOI] [PubMed] [Google Scholar]
- 27.Xiao H, Liu M. Circular RNA hsa_circ_0053277 promotes the development of colorectal cancer by upregulating matrix metallopeptidase 14 via miR-2467-3p sequestration. J Cell Physiol. 2020;235(3):2881–90. [DOI] [PubMed] [Google Scholar]
- 28.Liu F, Wen C. LINC01410 knockdown suppresses cervical cancer growth and invasion via targeting miR-2467-3p/VOPP1 axis. Cancer Manag Res. 2020;5(12):855–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duan Y, Chen HL, Zhang S, Ma FX, Zhang HC, Lv XA. The curcumin analog EF24 inhibits proliferation and invasion of triple-negative breast cancer cells by targeting the lncRNA HCG11/Sp1 axis. Mol Cell Biol. 2021. 10.1128/MCB.00163-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen J, Hou SF, Tang FJ, Liu DS, Chen ZZ, Zhang HL, Wang SH. HOTAIR/Sp1/miR-199a critically regulates cancer stemness and malignant progression of cutaneous squamous cell carcinoma. Oncogene. 2022;41(1):99–111. [DOI] [PubMed] [Google Scholar]
- 31.Fang H, Ma W, Guo X, Wang J. PTPN6 promotes chemosensitivity of colorectal cancer cells via inhibiting the SP1/MAPK signalling pathway. Cell Biochem Funct. 2021;39(3):392–400. [DOI] [PubMed] [Google Scholar]
- 32.Lin CL, Ying TH, Yang SF, Wang SW, Cheng SP, Lee JJ, Hsieh YH. Transcriptional suppression of miR-7 by MTA2 induces Sp1-mediated KLK10 expression and metastasis of cervical cancer. Mol Ther Nucleic Acids. 2020;5(20):699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lv L, Wang X. MicroRNA-296 targets specificity protein 1 to suppress cell proliferation and invasion in cervical cancer. Oncol Res. 2021;28(7):835. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.