Abstract
Laryngeal squamous cell carcinoma (LSCC) is a common malignant tumor of the head and neck that significantly impacts patients' quality of life, with chemotherapy resistance notably affecting prognosis. This study aims to identify prognostic biomarkers to optimize treatment strategies for LSCC. Using data from The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO), combined with mitochondrial gene database analysis, we identified mitochondrial lncRNAs associated with drug resistance genes. Key long non-coding RNAs (lncRNAs) were selected through univariate Cox regression and Lasso regression, and a multivariate Cox regression model was constructed to predict prognosis. We further analyzed the differences in immune function and biological pathway enrichment between high- and low-risk groups, developed a nomogram, and compared drug sensitivity. Results showed that the prognostic model based on seven mitochondrial lncRNAs could serve as an independent prognostic factor, with Area Under the Curve (AUC) values of 0.746, 0.827, and 0.771 at 1, 3, and 5 years, respectively, outperforming some existing models, demonstrating high predictive performance. Significant differences were observed in immune function and drug sensitivity between the high- and low-risk groups. The risk prediction model incorporating seven drug resistance-related mitochondrial lncRNAs can accurately and independently predict the prognosis of LSCC patients.
Keywords: Laryngeal squamous cell carcinoma, Mitochondria-associated lncRNAs, Chemotherapy resistance, Immune response, Prognostic model
Introduction
Laryngeal squamous cell carcinoma (LSCC) is one of the most common malignant tumors of the head and neck, accounting for approximately 20% of cases [1]. According to the 2018 Global Cancer Statistics [2], about 177,000 new cases of laryngeal cancer are diagnosed worldwide each year, leading to 95,000 deaths. The standardized incidence rate of laryngeal cancer is 2.0 per 100,000, with a mortality rate of 1.0 per 100,000 [3]. Despite a declining overall incidence of laryngeal cancer over the past 40 years, the 5-year survival rate has shown only a slight decrease from 66 to 63%, with no significant improvement [4]. Due to the lack of early symptoms, approximately 60% of patients present with advanced-stage disease at the time of diagnosis, missing the optimal treatment window [5–7]. For patients with locally advanced laryngeal cancer, surgery is typically followed by adjuvant radiotherapy or chemotherapy to reduce recurrence rates and extend survival [8]. However, the development of chemotherapy resistance significantly increases the risk of recurrence and worsens patient outcomes [9]. Postoperative laryngeal cancer often results in loss of voice, severely impacting patients' quality of life. Additionally, the side effects of chemotherapy resistance not only reduce treatment efficacy but also increase chemotherapy-related toxicities, such as nausea, vomiting, and myelosuppression, posing challenges to patients' physical, psychological, and overall quality of life [10–14]. Therefore, early assessment of patients' resistance to chemotherapeutic drugs could help avoid ineffective treatments and unnecessary side effects, ultimately improving the overall quality of life for patients.
Recent studies have highlighted the role of mitochondria as essential organelles involved in energy production, cellular metabolism, and signal transduction. Mitochondria are crucial in Adenosine triphosphate (ATP) synthesis and play a significant role in tumor progression and metastasis [15]. Mitochondrial dysfunction, including mutations in mitochondrial DNA (mtDNA) and alterations in metabolic pathways, has been closely linked to the development of drug resistance in tumors [16–18]. Moreover, the emergence, progression, and treatment response of tumors occur in close interaction with the host immune system, with many immune functions relying on intact mitochondrial metabolism [19]. Screening for the intersection of mitochondrial-related genes with drug resistance genes identifies mitochondrial-related genes associated with resistance, bridging the gap between drug resistance and mitochondrial function, and opening new avenues for understanding the interplay between mitochondrial function and drug resistance in cancer therapy [20]. Targeting mitochondrial pathways may enhance the efficacy of existing therapies and help to overcome resistance [21–23]. In summary, the intersection of mitochondrial-related genes and drug resistance genes provides a framework for understanding how cancer cells adapt to therapeutic challenges. This knowledge not only bridges the gap between mitochondrial function and drug resistance but also highlights potential strategies for improving cancer treatment outcomes. To our knowledge, the relationship between mitochondria and drug resistance in laryngeal cancer remains unexplored.
Long non-coding RNAs (lncRNAs) are RNA molecules longer than 200 nucleotides that do not encode proteins and have been found in recent years to play a crucial role in regulating gene expression [24]. lncRNAs regulate gene expression through various mechanisms, including chromatin remodeling, transcriptional regulation, RNA splicing, and stability modulation [25]. lncRNAs have been implicated in drug resistance in multiple cancers, such as hepatocellular carcinoma, melanoma, ovarian cancer, and breast cancer [26–30]. For example, lncRNA NEAT1 has been shown to increase resistance to chemotherapy by promoting cell survival and altering drug transport mechanisms [31]. lncRNA GBCDRlnc1 has been identified as a key regulator of autophagy-associated drug resistance in gallbladder cancer [32]. In recent years, the regulation of mitochondria by lncRNAs has gained attention; studies have shown that lncND5, lncND6, and lncCyt b can stabilize their corresponding mRNAs through RNA-RNA interactions, thereby regulating mitochondrial gene expression [33–36]. Investigating the regulatory role of lncRNAs on mitochondrial-related drug resistance genes can deepen our understanding of the molecular mechanisms underlying drug resistance in LSCC, provide theoretical support for elucidating the role of mitochondria in resistance, and offer potential targets for developing new therapeutic strategies.
This study aims to systematically screen and validate lncRNAs that are both mitochondrial-related and associated with drug resistance, constructing a molecular signature model capable of predicting chemotherapy response in laryngeal cancer patients. By analyzing the expression patterns of these lncRNAs in laryngeal cancer and their impact on prognosis, we aim to provide new prognostic tools for the clinical management of laryngeal cancer and support the development of personalized treatment strategies.
Materials and methods
Data collection and preparation
We obtained RNA sequencing (RNA-seq) data and associated clinical information of LSCC patients from the TCGA database (https://portal.gdc.cancer.gov/). The clinical data included patients' age, gender, survival time, survival status, histological grade, and TNM stage. After excluding samples lacking survival information, a total of 12 adjacent non-cancerous tissues and 116 LSCC tumor tissues were collected, along with the corresponding clinical data of these 116 patients. Transcriptomic data were processed and ID conversion was performed using Perl software to identify lncRNAs. Subsequently, all LSCC patients were randomly divided into a training group (n = 58) and a testing group (n = 58).
Additionally, gene expression data from 7 chemotherapy-sensitive and 4 chemotherapy-resistant patients were obtained from the GSE85607 dataset in the GEO database and analyzed using the GEO2R tool to identify drug resistance-related genes. The chi-square test was used to evaluate the differences in clinical characteristics between the two patient groups. Furthermore, a literature review identified 2,030 validated human mitochondrial-related genes [37, 38]. An intersection analysis was conducted between these genes and the identified drug resistance genes, resulting in mitochondrial-related drug resistance genes.
Construction of a risk model based on mitochondria-related drug resistance lncRNAs
The correlation between mitochondria-related drug resistance genes (MRDRGs) and lncRNA expression patterns was assessed using Pearson correlation coefficients (P < 0.001 and |R|> 0.4). Subsequently, stepwise dimensionality reduction identified 632 mitochondria-related drug resistance lncRNAs with prognostic significance. Initially, univariate Cox regression analysis was employed to select 32 lncRNAs significantly associated with the prognosis of LSCC patients. This was followed by LASSO regression for further dimensionality reduction, resulting in the identification of 15 lncRNAs associated with overall survival (OS) of the patients. Finally, multivariate Cox regression analysis determined 7 lncRNAs significantly related to prognosis, which were used to construct a prognostic model. The lncRNA risk score formula derived from the multivariate regression analysis is: Risk Score = ∑ (coefficient_i × EXP(lncRNA)_i) for i = 1 to n. Based on each patient's risk score, all LSCC patients were classified into high-risk and low-risk groups according to the median risk score.
Evaluation of the accuracy and independence of the risk model and construction of a Nomogram
To evaluate the predictive capacity of the risk model, Kaplan–Meier (K-M) survival curves were plotted. The accuracy of the model in predicting 1-year, 3-year, and 5-year survival probabilities was further assessed using Receiver Operating Characteristic (ROC) curves. Principal Component Analysis (PCA) was employed to reduce dimensionality and visualize results across different subgroups. Univariate and multivariate Cox regression analyses were conducted to verify the reliability of the risk model as an outcome predictor. Additionally, a nomogram was constructed by integrating multiple clinical features with the risk scores, and its predictive accuracy was assessed using calibration curves.
Functional enrichment analysis
Differentially expressed genes (DEGs) between the risk groups were identified based on the criteria: |log2FC|> 1.0 and p < 0.05. The identified DEGs underwent Gene Ontology (GO) analysis, encompassing three domains: Biological Processes (BPs), Cellular Components (CCs), and Molecular Functions (MFs). Additionally, the Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis was conducted to explore the signaling pathways associated with the DEGs. To further identify significant signaling pathways in the low-risk and high-risk groups, Gene Set Enrichment Analysis (GSEA) was performed.
Tumor microenvironment, somatic mutation analysis, and drug prediction
The Estimation of Stromal and Immune cells in Malignant Tumors using Expression data (ESTIMATE) algorithm was utilized to assess the differences in the tumor microenvironment (TME) between the two risk groups [39, 40]. The Cell-type Identification By Estimating Relative Subsets Of RNA Transcripts (CIBERSORT) algorithm was employed to evaluate the infiltration levels of 22 types of immune cells within the groups [41]. The single-sample Gene Set Enrichment Analysis (ssGSEA) algorithm was used to analyze immune cell infiltration and immune functions across different risk groups [42–44]. Furthermore, the Tumor Immune Dysfunction and Exclusion (TIDE) scores were calculated to assess the immune evasion capabilities of tumor cells and their response to immune checkpoint inhibitors (ICIs) in the two risk groups [45]. Somatic mutation data extracted from the TCGA database were used to analyze the tumor mutation burden (TMB) in both patient groups, which were subsequently categorized into low-TMB and high-TMB groups based on the median TMB value. To predict the response of LSCC patients in different risk groups to common anti-tumor drugs, the half-maximal inhibitory concentration (IC50) values were calculated using the "oncoPredict" R package.
Statistical analysis
All statistical analyses were performed using R software (version 4.4.1). Differences between groups were analyzed using t-tests. K-M survival analysis, combined with the log-rank test, was used to assess survival differences between the risk groups. Univariate and multivariate Cox regression analyses were conducted to identify prognostic factors associated with LSCC. Statistical significance was set at a p-value of less than 0.05.
Results
Data of LSCC patients
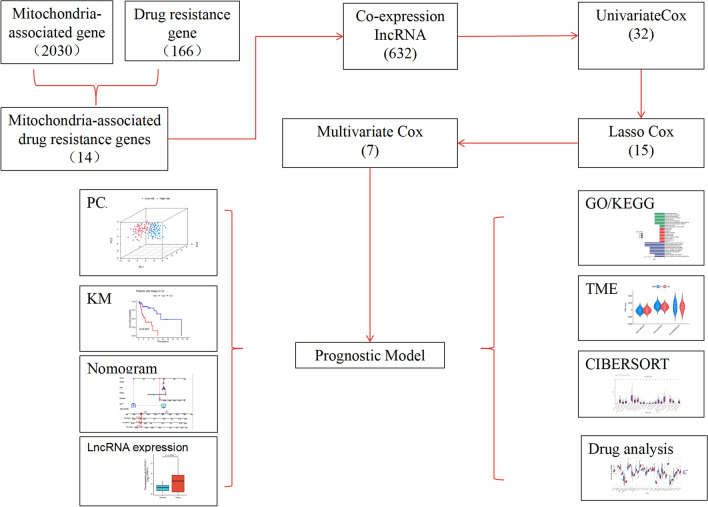
Figure 1 illustrates the study flowchart. A total of 116 LSCC patients were randomly divided into a training set (n = 58) and a test set (n = 58) in a 1:1 ratio. The training set was utilized to identify prognostic mitochondrial-related drug resistance lncRNAs and to establish the prognostic model, while the test set was used to validate the model's accuracy. There were no significant differences between the two groups in terms of clinical characteristics such as age, sex, histological grade, and TNM stage (p > 0.05), indicating good balance between the groups (Table 1).
Fig. 1.
Study Design Flowchart
Table 1.
Clinical characteristics of LSCC patients in the training, validation, and overall datasets
| Covariates | Type | Total | Test | Train | P value |
|---|---|---|---|---|---|
| Age | ≤65 | 76 (65.52%) | 38 (65.52%) | 38 (65.52%) | 1 |
| Age | > 65 | 40 (34.48%) | 20 (34.48%) | 20 (34.48%) | |
| Gender | FEMALE | 20 (17.24%) | 11 (18.97%) | 9 (15.52%) | 0.8058 |
| Gender | MALE | 96 (82.76%) | 47 (81.03%) | 49 (84.48%) | |
| Grade | G1 | 8 (6.9%) | 4 (6.9%) | 4 (6.9%) | 0.7406 |
| Grade | G2 | 71 (61.21%) | 37 (63.79%) | 34 (58.62%) | |
| Grade | G3 | 32 (27.59%) | 15 (25.86%) | 17 (29.31%) | |
| Grade | G4 | 1 (0.86%) | 0 (0%) | 1 (1.72%) | |
| Grade | unknow | 4 (3.45%) | 2 (3.45%) | 2 (3.45%) | |
| Stage | Stage I | 2 (1.72%) | 1 (1.72%) | 1 (1.72%) | 0.2865 |
| Stage | Stage II | 9 (7.76%) | 6 (10.34%) | 3 (5.17%) | |
| Stage | Stage III | 14 (12.07%) | 4 (6.9%) | 10 (17.24%) |
Risk model based on mitochondria-related drug resistance lncRNAs for predicting LSCC prognosis
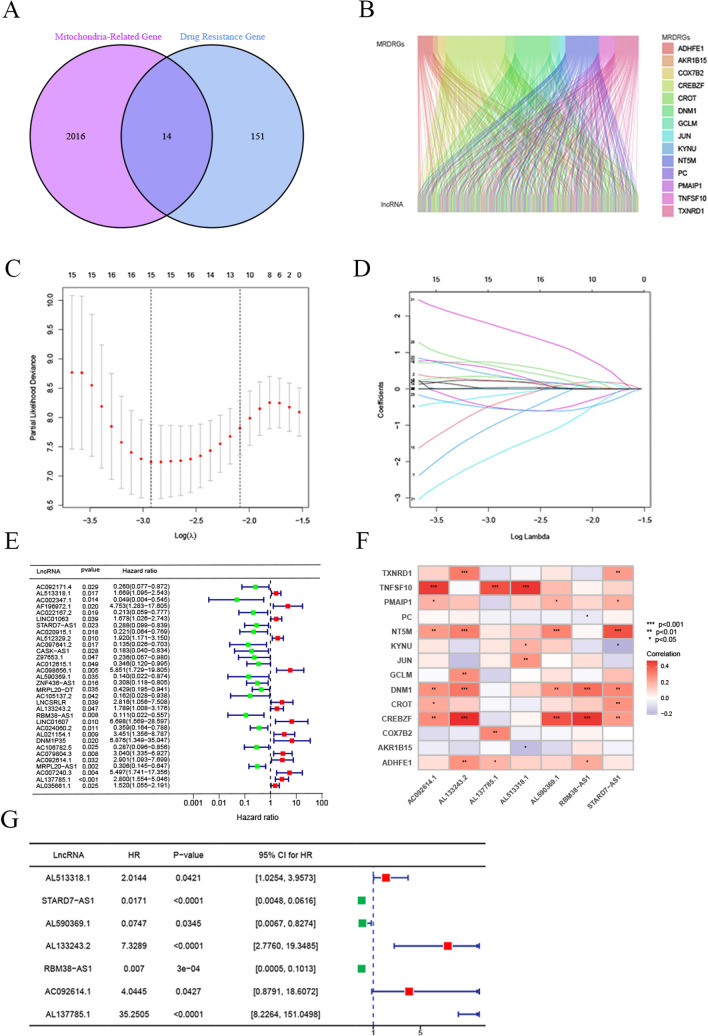
We collected 2,030 MRGs from the literature. By analyzing the GSE85607 dataset, 166 drug resistance genes were identified, and intersected with MRGs, resulting in 14 MRDRGs (Fig. 2A). Subsequently, we assessed the expression correlation between lncRNAs and the mRNAs of these MRDRGs in the TCGA database using Pearson correlation coefficients, identifying 632 mitochondria-related drug resistance lncRNAs. A Sankey diagram in Fig. 2B illustrates the co-expression network between the 14 MRDRGs and the 632 lncRNAs. In the training set, univariate Cox regression analysis of the OS data of LSCC patients identified 32 lncRNAs significantly associated with patient OS (Fig. 2E). Further screening using LASSO regression identified 15 lncRNAs highly correlated with LSCC prognosis (Fig. 2C-D). Finally, a multivariate Cox regression model was constructed based on 7 key lncRNAs: AL513318.1, STARD7-AS1, AL590369.1, AL133243.2, RBM38-AS1, AC092614.1, and AL137785.1 (Fig. 2G). The formula for calculating the risk score, based on the expression levels of these lncRNAs and their Cox regression coefficients, is as follows: Risk Score = (AL513318.1 × 0.7003) + (STARD7-AS1 × − 4.0671) + (AL590369.1 × − 2.5946) + (AL133243.2 × 1.9918) + (RBM38-AS1 × − 4.9626) + (AC092614.1 × 1.3974) + (AL137785.1 × 3.5625). This model, by integrating the expression levels of these lncRNAs, provides an effective tool for assessing the prognosis of LSCC patients.
Fig. 2.
Construction of the Risk Model Based on Mitochondria-Related Drug Resistance Gene lncRNAs. A Venn diagram showing the intersection of Mitochondria-Related Genes (MRGs) and Laryngeal Cancer Drug Resistance Genes, identifying 14 intersecting genes. B Sankey diagram illustrating the network of MRDRG lncRNAs. C, D LASSO regression analysis identified 15 MRDRG lncRNAs. E Univariate Cox regression analysis of 32 MRDRG lncRNAs. F Heatmap showing the correlation between the 7 MRDRG lncRNAs included in the multivariate Cox regression model and the MRDRGs. G The 7 MRDRG lncRNAs used to construct the multivariate Cox regression model. (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001)
Analysis of the correlation between MRDRGs and lncRNAs
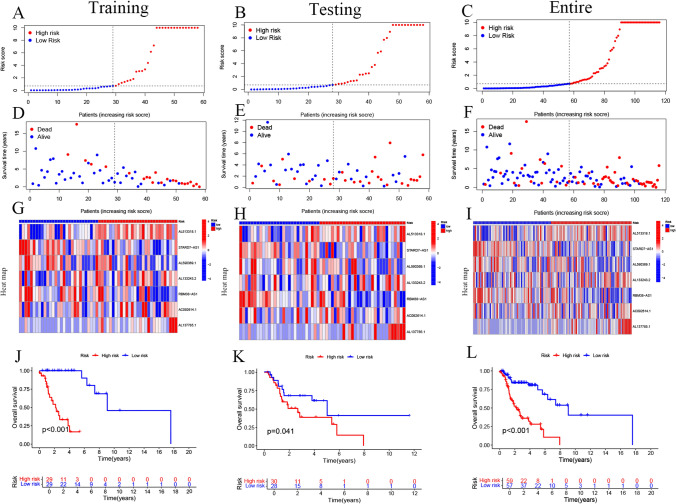
Additionally, we examined the correlation between the 14 MRDRGs and the 7 identified lncRNAs (Fig. 3A-I). The expression levels of these 7 lncRNAs in the low-risk and high-risk groups are shown in Fig. 3A-I, along with an analysis of the distribution of risk scores, survival time, and survival status in the training set. The results indicated that STARD7-AS1, AL590369.1, and RBM38-AS1 were expressed at higher levels in the low-risk group compared to the high-risk group, suggesting their potential roles as protective prognostic factors in LSCC patients. Conversely, AL513318.1, AL133243.2, and AL137785.1 were more highly expressed in the high-risk group, indicating that they may act as risk factors in LSCC. To evaluate differences in OS between different risk groups in the training set, validation set, and entire dataset, Kaplan–Meier (K-M) curves were plotted. The results demonstrated that in all datasets, the OS of the low-risk group was significantly better than that of the high-risk group (all p-values < 0.05) (Fig. 3J-L), further supporting the effectiveness of the risk model in predicting LSCC prognosis.
Fig. 3.
Validation of Mitochondria-Related Drug Resistance Gene lncRNA Features in the Training Set, Validation Set, and Entire Cohort. A–L Display the distribution of risk scores for the 7 lncRNAs, overall survival status distribution, expression heatmaps, and overall survival (OS) of patients in different risk groups across the training set, validation set, and entire cohort
Validation of the mitochondria-related drug resistance lncRNA risk model and PCA
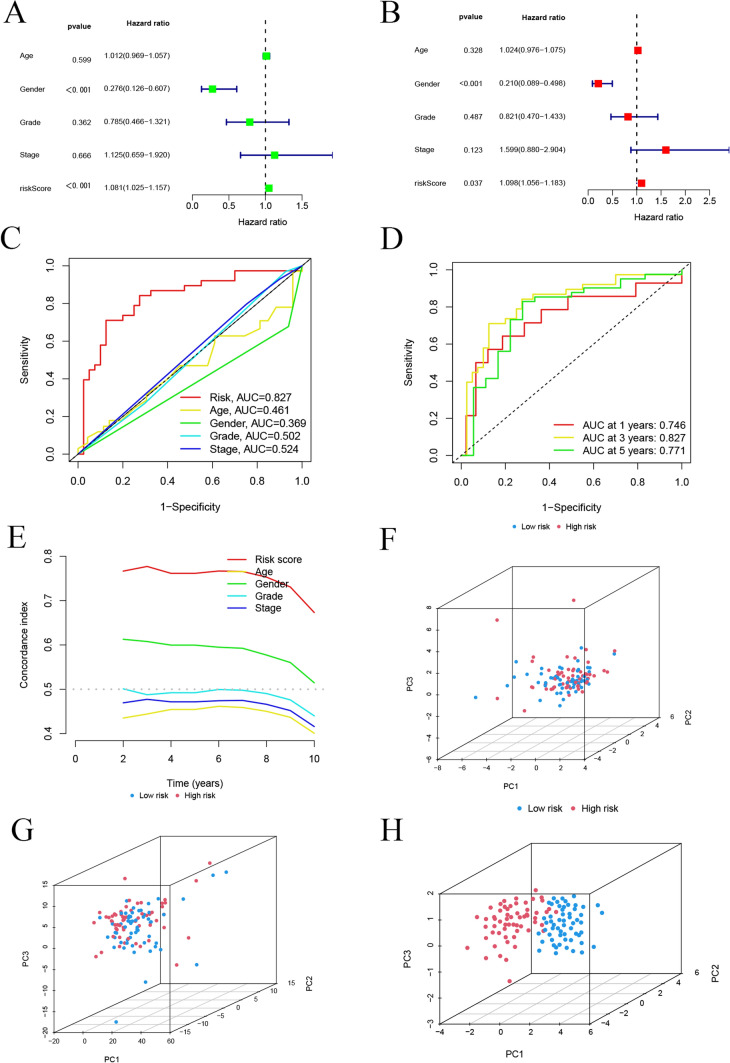
We used univariate and multivariate Cox regression analyses to assess whether the 7 lncRNAs could independently predict OS in LSCC patients. Univariate Cox regression analysis indicated that both gender and risk score were significantly associated with OS (p < 0.001) (Fig. 4A). Multivariate Cox regression analysis further confirmed that the risk score (p < 0.05) was an independent predictor of OS in LSCC patients (Fig. 4B). These results suggest that the risk model constructed from the 7 mitochondria-related drug resistance lncRNAs serves as an independent prognostic factor for LSCC. Furthermore, for the entire cohort, the AUC of the risk model (AUC = 0.827) surpassed that of other clinical risk indicators (Fig. 4C). The AUC values for predicting 1-year, 3-year, and 5-year survival probabilities were all greater than 0.7, highlighting the model’s superior accuracy and reliability in prognosis prediction (Fig. 4D). As shown in Fig. 4E-H, PCA using the 14 MRDRGs, 377 mitochondria-related drug resistance lncRNAs, and the 7 mitochondria-related drug resistance lncRNAs revealed significant differences between the two subgroups. The PCA results demonstrated that the risk model distinctly separated the low-risk and high-risk groups, with more pronounced separation in both directions. These findings underscore that the risk score effectively stratifies LSCC patients into two distinct groups: low-risk and high-risk.
Fig. 4.
Evaluation of the Prognostic Accuracy and Independence of the Risk Model. A Forest plot of univariate Cox regression analysis. B Forest plot of multivariate Cox regression analysis. C ROC curve of clinical risk indicators and risk scores for the entire cohort. D ROC curves of the risk model predicting 1-year, 3-year, and 5-year overall survival (OS) in the entire cohort. E C-index curves for risk scores and other clinicopathological variables. F PCA of the expression of 14 mitochondria-related drug resistance genes. G PCA of the expression of 632 mitochondria-related drug resistance gene lncRNAs. H PCA of the 7 prognosis-related mitochondria-related drug resistance gene lncRNA features
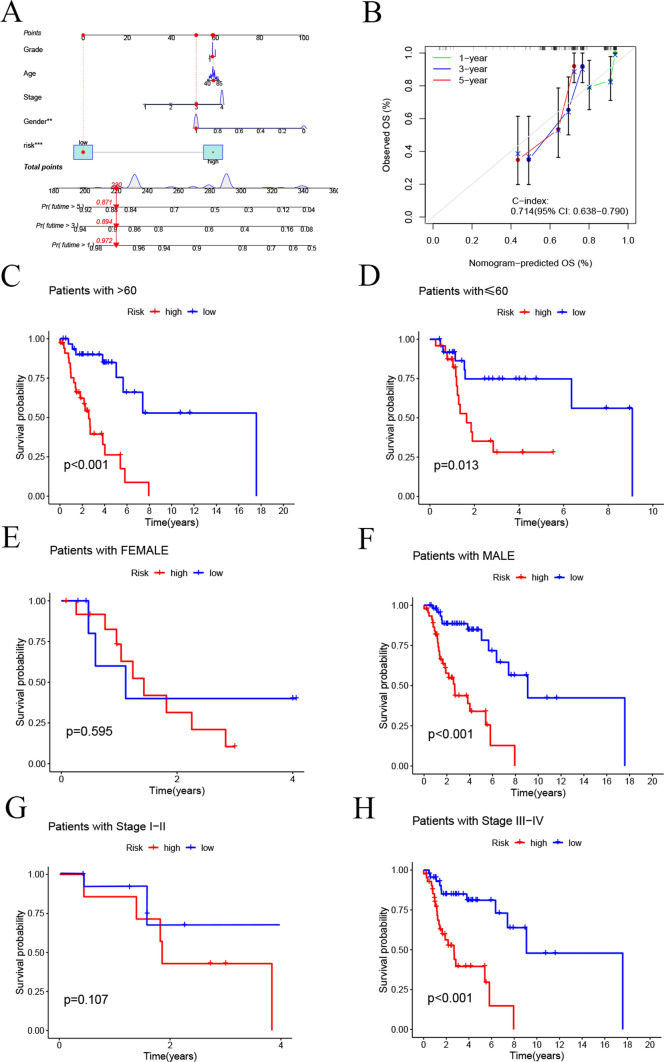
Construction of the Nomogram
By integrating multiple clinical factors and risk scores, we developed a Nomogram to predict the 1-year, 3-year, and 5-year survival rates of LSCC patients (Fig. 5A). The calibration curve of the Nomogram indicated a good concordance between the predicted outcomes and actual clinical observations (Fig. 5B). Furthermore, we plotted K-M curves to assess the performance of the risk model in predicting survival across different clinical characteristics, including age, gender, and staging. The results showed that for female patients and those with lower staging, the survival differences between the high-risk and low-risk groups were not significant. However, among patients of all age groups, male patients, and those with higher staging, the low-risk group exhibited significantly better prognosis compared to the high-risk group (Fig. 5C-H).
Fig. 5.
Construction of the Nomogram and K-M Analysis of OS Across Different Subgroups. A Nomogram. B Calibration curve of the Nomogram. K-M analysis for OS in different subgroups: C Age > 60 years. D Age ≤ 60 years. E Female. F Male. G Stage I-II. H Stage III-IV
Functional enrichment analysis
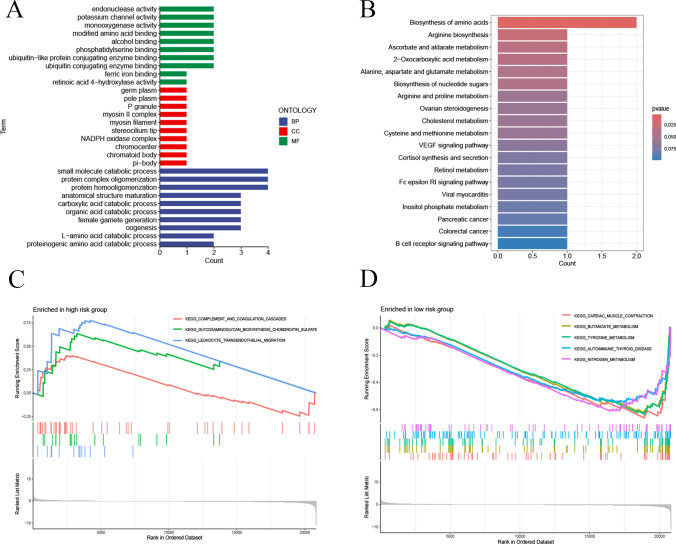
We conducted functional enrichment analysis to explore the potential biological functions of these genes in LSCC. As shown in Fig. 6A, GO analysis revealed significant enrichment in the BP related to proteinogenic amino acid catabolic processes, L-amino acid catabolic processes, and organic acid catabolic processes, suggesting these genes may be involved in metabolism-related biological processes. In terms of CC, significantly enriched items included NADPH oxidase complex, ciliary tip, myosin II complex, and P granule, indicating these genes might function within specific subcellular structures. Regarding (MF, enriched terms included retinoic acid 4-hydroxylase activity, phosphatidylserine binding, and potassium channel activity, which may relate to signal transduction, cell membrane dynamics, and ion channel regulation in LSCC cells. KEGG pathway analysis indicated significant enrichment in pathways such as B cell receptor signaling, VEGF signaling, and FcεRI signaling, which play crucial roles in tumor cell proliferation, survival, and immune evasion (Fig. 6B). GSEA further demonstrated that in the high-risk group, KEGG pathways significantly enriched included complement and coagulation cascades, glycosaminoglycan biosynthesis (e.g., chondroitin sulfate), and leukocyte transendothelial migration (Fig. 6C). In contrast, in the low-risk group, enriched KEGG pathways included cardiac muscle contraction, butanoate metabolism, tyrosine metabolism, autoimmune thyroid disease, and nitrogen metabolism (Fig. 6D). These findings provide new insights into the biological mechanisms underlying LSCC.
Fig. 6.
Functional Enrichment Analysis. A Gene Ontology (GO) analysis. B Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. C, D Gene Set Enrichment Analysis (GSEA)
Analysis of immune infiltration and immunotherapy efficacy
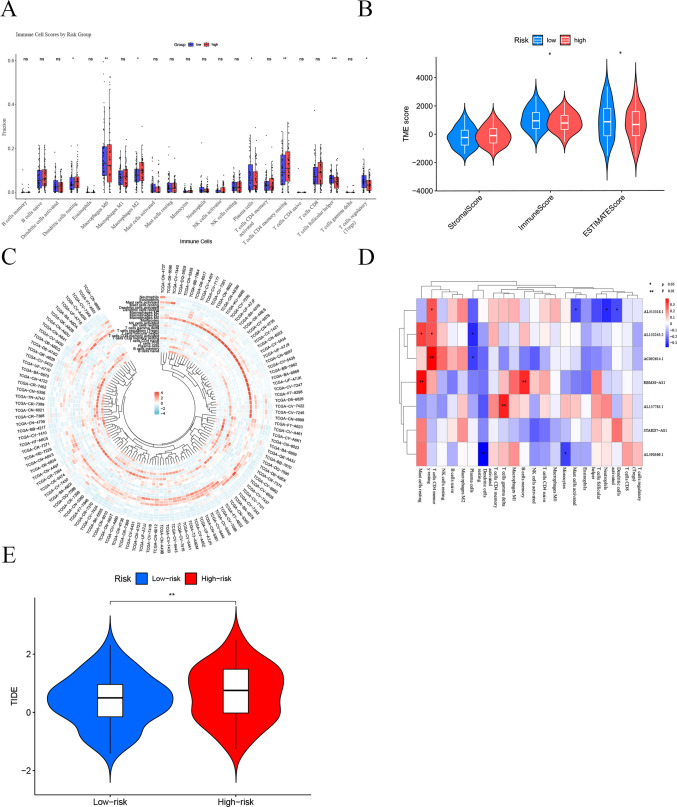
We conducted an in-depth analysis of the TME in different risk groups using multiple algorithms. The CIBERSORT algorithm was employed to evaluate the proportions of 22 types of tumor-infiltrating immune cells, revealing significant differences in immune cell distribution between the high-risk and low-risk groups (Fig. 7A). Specifically, dendritic cells in a resting state, M2 macrophages, and CD4 memory T cells in a resting state were more abundant in the high-risk group; whereas M0 macrophages, plasma cells, follicular helper T cells, and regulatory T cells were more prevalent in the low-risk group. Further analysis using the ESTIMATE algorithm showed that the immune scores and overall ESTIMATE scores in the low-risk group were significantly higher than those in the high-risk group (p < 0.05) (Fig. 7B), indicating a higher level of immune cell infiltration in the low-risk group. However, there was no significant difference in stromal scores between the two groups. Additionally, the circular cluster heatmap in Fig. 7C and D visually demonstrated the infiltration levels of various immune cell types across different samples, further confirming the differences in immune cell distribution among samples. Lastly, the TIDE algorithm was used to explore the relationship between risk scores and responses to immunotherapy. The results indicated that the low-risk group had a higher immune score (Fig. 7E). These findings suggest that patients in the low-risk group may exhibit a stronger anti-tumor immune response, potentially leading to better responses to immunotherapy; conversely, the high-risk group may face greater risks of immune evasion and reduced treatment efficacy. These results provide important reference points for optimizing risk stratification and personalizing immunotherapy strategies in clinical settings.
Fig. 7.
Analysis of Immune Infiltration and Immunotherapy Efficacy in High-Risk and Low-Risk Groups of Laryngeal Squamous Cell Carcinoma (LSCC) Based on Mitochondria-Related Drug Resistance lncRNAs. A The CIBERSORT algorithm was used to assess the differences in the abundance of 22 immune cell types between the two groups. B The ESTIMATE algorithm was employed to evaluate the differences in immune, stromal, and ESTIMATE scores between the two groups. C A circular heatmap was used to display the infiltration levels of various immune cell types across different samples. D A correlation heatmap shows the relationship between lncRNA expression and immune cell infiltration levels. E The TIDE algorithm was used to calculate TIDE scores between the two groups (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001)
Somatic mutation landscape analysis
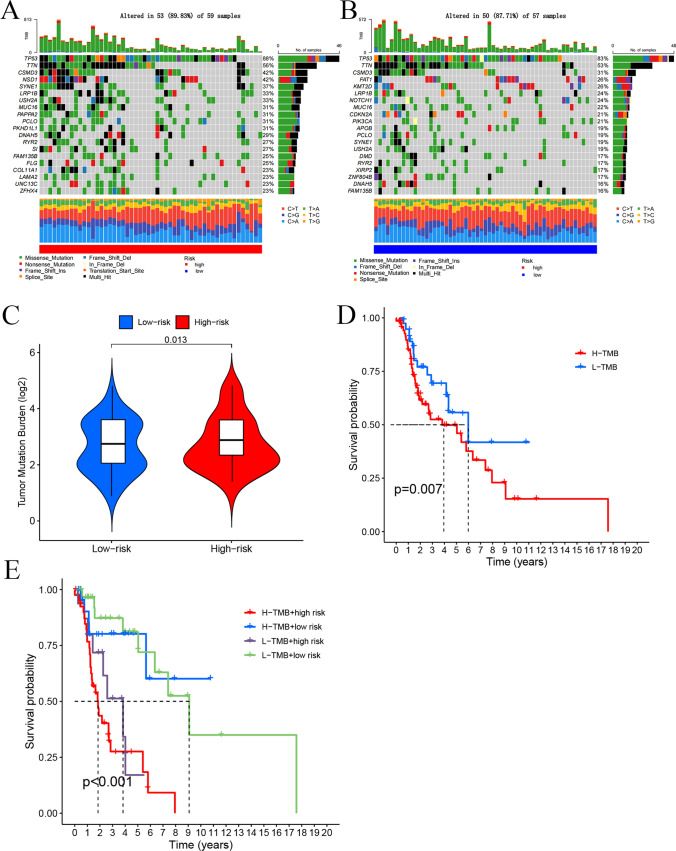
By comparing the somatic mutation rates between the two risk groups, we found that the mutation rate in the low-risk group (87.71% of samples) was lower than that in the high-risk group (89.83% of samples). The top 20 driver mutation genes are illustrated in Fig. 8A. Additionally, TMB score in the high-risk group was significantly higher than that in the low-risk group (p = 0.013) (Fig. 8B). Further analysis confirmed that the TMB score remained significantly higher in the high-risk group (p = 0.012) (Fig. 8C). Based on the median TMB, LSCC patients were divided into low-TMB and high-TMB groups. Kaplan–Meier (K-M) analysis indicated that patients in the low-TMB subgroup had a significantly better overall survival than those in the high-TMB subgroup (p = 0.007) (Fig. 8D). We combined TMB with the risk scores to compare their efficiency in predicting the prognosis of LSCC patients. K-M analysis showed that patients with low TMB and low-risk scores had the best overall survival, whereas those with high TMB and high-risk scores had the worst overall survival (p < 0.001) (Fig. 8E).
Fig. 8.
Somatic Mutation Landscape Analysis. A Mutation distribution in the high-risk group. B Mutation distribution in the low-risk group. C Differences in TMB scores between the different risk groups. D Kaplan–Meier K–M analysis of overall survival (OS) for different TMB groups. E OS K-M analysis of four groups based on TMB scores and risk scores
Drug response analysis
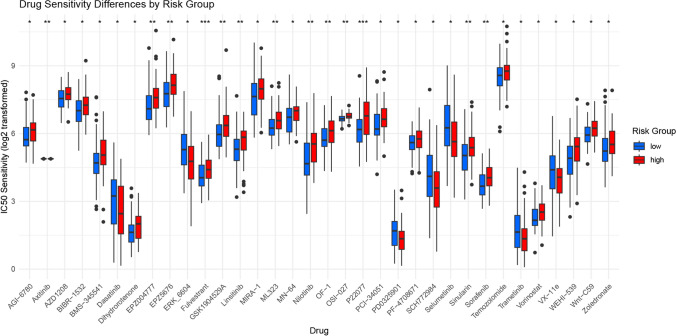
Given the significant differences in prognosis, immune microenvironment, and somatic mutation frequency between the two risk groups, we screened drugs suitable for precision therapy targeting these groups. We utilized the R package oncoPredict, which links antitumor drugs with biomarkers and predicts patient responses to various anticancer drugs [46]. Using OncoPredict, we identified antitumor drugs for LSCC patients in different risk groups (Fig. 9). Among 35 drugs, there were notable differences between the high-risk and low-risk groups. In the high-risk group, 7 drugs, including clinically used anticancer agents Dasatinib and Trametinib, were identified as having significant drug sensitivity. In contrast, drug sensitivity analysis in the low-risk group revealed 28 drugs with significant sensitivity (p < 0.05). These drugs, including commonly used agents such as Vorinostat, Nilotinib, Axitinib, and Sorafenib, had significantly lower IC50 values in the low-risk group compared to the high-risk group.
Fig. 9.
Drug Sensitivity Analysis. (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns, not significant)
Expression of lncRNAs
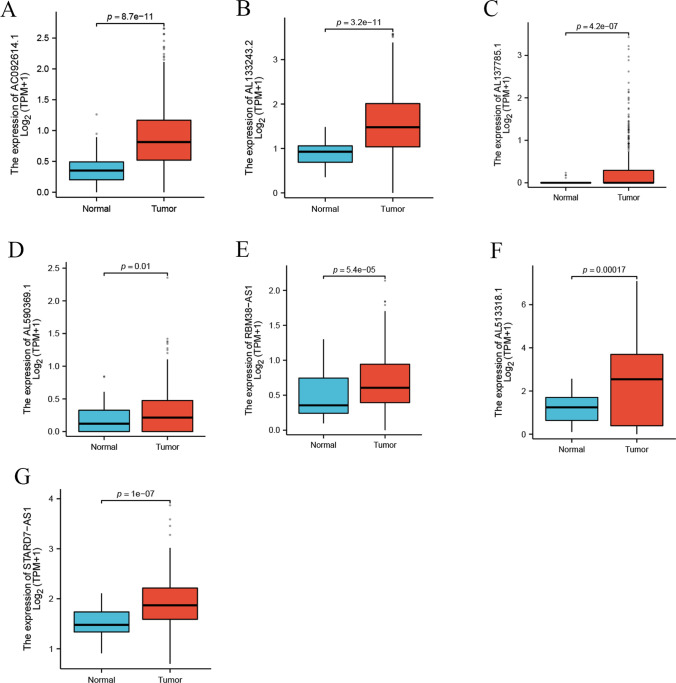
In the TCGA database, we examined the expression profiles of seven lncRNAs (AL513318.1, STARD7-AS1, AL590369.1, AL133243.2, RBM38-AS1, AC092614.1, AL137785.1) in LSCC samples compared to normal samples for the first time. As shown in Fig. 10A-G, these lncRNAs were significantly upregulated in LSCC samples relative to normal samples, suggesting that these lncRNAs may play a crucial role in the pathogenesis of LSCC.
Fig. 10.
Expression of lncRNAs in Laryngeal Squamous Cell Carcinoma (LSCC). The expression levels of seven mitochondrial-related drug resistance long non-coding RNAs (lncRNAs) (AC092614.1 (A), AL133243.2 (B), AL137785.1 (C), AL590369.1 (D), RBM38-AS1 (E), AL513318.1 (F), and STARD7-AS1 (G)) in LSCC samples compared to normal tissues
Discussion
This study developed a risk model based on mitochondrial-associated drug resistance lncRNAs to predict the prognosis and immune response of patients with LSCC. We identified seven key lncRNAs (AL513318.1, STARD7-AS1, AL590369.1, AL133243.2, RBM38-AS1, AC092614.1, and AL137785.1), which effectively distinguished high-risk from low-risk patients and demonstrated excellent predictive capabilities for 1-year, 3-year, and 5-year prognosis, with AUC values of 0.746, 0.827, and 0.771, respectively, surpassing the performance of some existing models [47–49]. PCA further validated the effectiveness of these mitochondrial-associated drug resistance lncRNA features in distinguishing between high-risk and low-risk groups. Univariate and multivariate Cox regression analyses indicated that the risk score independently predicted OS. The robustness of the model was confirmed in both the validation cohort and the overall dataset, underscoring its high reliability and effectiveness in predicting prognosis for LSCC patients. Compared to traditional clinical factors, the risk model exhibited superior prognostic predictive power. By integrating these lncRNAs with clinical information, we developed a nomogram that provides a more precise individualized survival prediction tool for LSCC patients. The results showed good concordance between predicted and actual overall survival, further demonstrating the strong predictive performance of the nomogram. Although previous studies have constructed mitochondrial-related lncRNA models and validated their prognostic significance [50–52], these studies mainly focused on other cancers, and research in laryngeal cancer remains limited. This study is the first to construct a specialized risk model for mitochondrial-associated drug resistance lncRNAs in LSCC and reveal the potential role of these lncRNAs in regulating immune response.
Our analysis revealed significant differences in the distribution of tumor-infiltrating immune cells between the high-risk and low-risk groups, which have important implications for patient prognosis and treatment response. In the high-risk group, a higher abundance of M2 macrophages was observed, which are known to be associated with immunosuppression [53]. M2 macrophages promote tumor proliferation, angiogenesis, and anti-inflammatory responses by secreting growth factors and matrix metalloproteinases within the tumor microenvironment [54]. Conversely, the low-risk group exhibited higher levels of plasma cells and follicular helper T cells, which are associated with enhanced antitumor immune responses, potentially contributing to more effective tumor control and improved prognosis [55–58]. The ESTIMATE algorithm further supported the finding that the low-risk group had higher levels of immune cell infiltration. TIDE analysis indicated that patients in the low-risk group might benefit more from immunotherapy, while the high-risk group showed a higher risk of immune escape. These findings underscore the potential clinical application of risk stratification based on immune cell characteristics and suggest that patients in the high-risk group may require comprehensive immunotherapy strategies to improve treatment outcomes.
Based on the significant immunological differences observed between the high- and low-risk groups, we hypothesize that the identified lncRNAs may regulate immune cell function by modulating mitochondrial activity, reactive oxygen species (ROS) production, autophagy, and key signaling pathways, such as PGC-1α and NRF2. ATP generated by mitochondria provides the essential energy required for T cell activation and function [59], while ROS production plays a critical role in T cell differentiation and cytotoxic responses [60]. Additionally, the polarization of macrophages (M1 or M2 phenotype) is influenced by mitochondrial metabolic pathways [61], with M1 macrophages relying on high levels of ROS and glycolysis, whereas M2 macrophages depend more on mitochondrial oxidative metabolism.
Furthermore, these lncRNAs may impact immune cell survival and function by regulating autophagy, a process crucial for clearing damaged mitochondria, maintaining cellular homeostasis, and modulating immune responses [62]. By influencing signaling pathways such as PGC-1α and NRF2, these lncRNAs can reshape immune cell metabolism and antioxidant responses [63], thereby affecting immune cell infiltration and immune evasion mechanisms within the tumor microenvironment. These mechanisms suggest that the risk stratification model based on mitochondrial-associated lncRNAs not only holds significant value in prognostic prediction but also provides theoretical support for personalized immunotherapy strategies in LSCC patients.
In the high-risk group, enrichment analysis revealed significant enrichment of pathways such as complement and coagulation cascades, glycosaminoglycan biosynthesis (e.g., chondroitin sulfate), and leukocyte transendothelial migration. The complement and coagulation cascades may promote inflammatory responses and tumor invasiveness [64], while the leukocyte transendothelial migration pathway, which is associated with immune cell infiltration [65], may lead to immune evasion, thereby enhancing tumor survival and dissemination.
Our study identified significant differences in drug sensitivity between the high-risk and low-risk groups across 35 drugs, providing new insights for personalized treatment of LSCC patients. The high-risk group showed higher sensitivity to Dasatinib and Trametinib, while the low-risk group exhibited significant sensitivity to 28 drugs, including Vorinostat, Nilotinib, Axitinib, and Sorafenib. The application of these drugs in other cancers provides a reference for exploring their potential in LSCC treatment. Dasatinib, a multi-target tyrosine kinase inhibitor, is widely used in the treatment of chronic myeloid leukemia (CML) and acute lymphoblastic leukemia (ALL) [66, 67]. It has shown potential therapeutic effects in various solid tumors such as lung cancer, colorectal cancer, and melanoma due to its inhibitory action on SRC family kinases [68]. Trametinib, a MEK inhibitor, is primarily used for BRAF-mutated melanoma and non-small cell lung cancer (NSCLC) [69, 70], which often exhibit abnormal activation of the MAPK pathway. Our findings suggest that these two drugs might offer new therapeutic strategies by targeting specific signaling pathways in high-risk LSCC patients.
In the low-risk group, Vorinostat, Nilotinib, Axitinib, and Sorafenib demonstrated significant drug sensitivity. Vorinostat, an HDAC inhibitor, has been approved for the treatment of relapsed or refractory peripheral T-cell lymphoma and exerts its anticancer effects by regulating gene expression [71]. Nilotinib, a second-generation BCR-ABL tyrosine kinase inhibitor, is mainly used for CML [72]. Axitinib and Sorafenib are VEGFR inhibitors and multi-target kinase inhibitors, respectively, widely used in angiogenesis-dependent tumors such as renal cell carcinoma and hepatocellular carcinoma [73, 74]. The sensitivity of these drugs in the low-risk group suggests their potential effectiveness against LSCC by inhibiting angiogenesis or regulating epigenetic mechanisms.
Currently, the standard treatment options for laryngeal cancer primarily include surgery, radiotherapy, and chemotherapy. Commonly used chemotherapeutic agents, such as cisplatin, fluorouracil, and paclitaxel, function by inhibiting cell division and DNA synthesis [75]. However, the efficacy of chemotherapy in laryngeal cancer is limited, and resistance often develops. Our findings suggest that exploring the application of drugs effective in other cancers could offer new treatment avenues for laryngeal cancer, especially through personalized targeted therapy tailored to high-risk and low-risk groups, potentially optimizing clinical outcomes.
Although these drugs have not yet been widely applied in laryngeal cancer, existing studies and our results indicate that drug repurposing across cancer types is a feasible strategy for achieving precision therapy [76]. These findings not only support a risk-stratified treatment approach but also provide a theoretical basis for applying treatments from other cancers to laryngeal cancer. Future clinical trials are needed to validate the efficacy of these drugs in laryngeal cancer and, combined with studies on molecular mechanisms, further refine individualized treatment protocols.
Our study is the first to construct a prognostic model based on mitochondrial-associated drug resistance lncRNAs in LSCC to evaluate patient survival, immune status, and drug sensitivity. This model demonstrates significant novelty in the context of LSCC, as prior studies on mitochondrial lncRNAs have primarily focused on other cancer types, such as liver, breast, and lung cancers [77], and its application in LSCC has not yet been explored. Moreover, compared to other existing models, the mitochondrial lncRNA-based model constructed in this study exhibits superior performance and robust predictive capabilities in prognostic evaluation.
Using data from the TCGA and GEO databases, this study developed a risk stratification model for LSCC patients based on mitochondrial-associated lncRNAs, preliminarily validating its potential in predicting drug sensitivity and immune response. However, due to the lack of direct clinical data, the practical clinical applicability of this model has yet to be fully confirmed. Additionally, the data used in the model originate from retrospective analyses, potentially introducing selection bias, which limits its broader applicability. Future research should involve multicenter prospective clinical trials to gather real-world drug response and immunotherapy outcome data in LSCC patients, thereby further evaluating and optimizing the model’s predictive accuracy. In addition, integrating multi-omics data such as genomics and proteomics may reveal the molecular mechanisms linking lncRNAs with immune regulation and drug resistance, enhancing the model’s precision and facilitating its clinical translation, ultimately providing a more effective tool for individualized treatment of LSCC.
Limitation
This study has several limitations that warrant consideration. Firstly, the analysis relies on publicly available datasets, including TCGA and GEO, with a relatively small sample size for LSCC, potentially limiting the generalizability of the findings. Additionally, while internal validation was performed using training and testing cohorts, independent external validation was not conducted, which may restrict the broader applicability of the proposed risk model across diverse populations. The retrospective nature of the study, based on pre-existing data, introduces potential selection bias, and prospective clinical studies are needed to confirm the model's predictive and prognostic value. Furthermore, although bioinformatics analyses suggest potential mechanisms for the identified mitochondrial-related lncRNAs, a lack of experimental validation limits the ability to confirm their functional roles in drug resistance and immune regulation, necessitating further in vitro and in vivo studies. The focus solely on mitochondrial-related drug resistance lncRNAs may also overlook other critical pathways or lncRNAs influencing LSCC progression and therapy response, highlighting the need for broader multi-omics analyses. Lastly, while the study explores immune infiltration and immune response, it does not include direct evidence of immunotherapy outcomes, which future studies should address through clinical data on treatment responses. Despite these limitations, this study provides novel insights and a promising foundation for understanding mitochondrial-related lncRNAs in LSCC.
Conclusion
The risk model constructed in this study, based on mitochondrial-related drug resistance lncRNAs, effectively predicts the prognosis and immune response of patients with LSCC. The seven key lncRNAs identified not only demonstrate significant prognostic predictive power but also hold potential as biomarkers for chemotherapy sensitivity. Although further validation is required, these lncRNAs present promising prospects for application in personalized therapy, with the potential to improve the survival outcomes and treatment efficacy for LSCC patients.
Acknowledgements
Not applicable.
Author contributions
All authors contributed to the study conception and design. Experimental data: ZMW, YC, DZJ and YPP; Data analysis: ZMW, TXT and YFM; Writing-original draft: ZMW, YC, TS and YFM; Experimental design: ZMW, YC, TS and YFM. All researchers attributed to this study have read and agreed to the manuscript's published version.
Funding
This work was supported by Guizhou Provincial Basic Research Program (Natural Science) (QKHJC-ZK[2022]YB434), the National Natural Science Foundation of China (82260541), the Clinical Research Project of Guizhou Medical University Affiliated Hospital (2021-GMHCT-013), the 2022 Doctoral Research Launch Fund of Guizhou Medical University Affiliated Hospital (gyfybsky-2022-17), and the Project of Guizhou Provincial Health Commission (gzwkj2021-328).
Data availability
The data used in this study are sourced from public databases. The data in this study are openly available in the GEO database (https://www.ncbi.nlm.nih.gov/geo/) and the TCGA database (https://www.cancer.gov/).
Declarations
Ethics approval and consent to participate
Not applicable as the research conducted in this study did not involve direct interactions with human subjects or the collection of primary data.
Consent for publication
All authors read the guidelines of the journal and agreed with consent for publication.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Zhimin Wu, Yi Chen and Dizhi Jiang share the first authorship.
Contributor Information
Yifei Ma, Email: 84309348@qq.com.
Tiannake Shapaer, Email: tiannakecsu@outlook.com.
References
- 1.Teng Y, Xia C, Li H, et al. Cancer statistics for young adults aged 20 to 49 years in China from 2000 to 2017: a population-based registry study. Sci China Life Sci. 2024;67(4):711–9. [DOI] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. [DOI] [PubMed] [Google Scholar]
- 3.Gormley M, Creaney G, Schache A, Ingarfield K, Conway DI. Reviewing the epidemiology of head and neck cancer: definitions, trends and risk factors. Br Dent J. 2022;233(9):780–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin X, Zhou W, Liu Z, Cao W, Lin C. Targeting cellular metabolism in head and neck cancer precision medicine era: a promising strategy to overcome therapy resistance. Oral Dis. 2023;29(8):3101–20. [DOI] [PubMed] [Google Scholar]
- 5.Cheng Z, Luo Y, Zhang Y, et al. A novel NAP1L4/NUTM1 fusion arising from translocation t(11;15)(p15;q12) in a myeloid neoplasm with eosinophilia and rearrangement of PDGFRA highlights an unusual clinical feature and therapeutic reaction. Ann Hematol. 2020;99(7):1561–4. [DOI] [PubMed] [Google Scholar]
- 6.Da Silva AC, Paterson TE, Minev IR. Electro-assisted assembly of conductive polymer and soft hydrogel into core-shell hybrids. Soft Sci. 2023;3:3. [Google Scholar]
- 7.Ruffin AT, Li H, Vujanovic L, Zandberg DP, Ferris RL, Bruno TC. Improving head and neck cancer therapies by immunomodulation of the tumour microenvironment. Nat Rev Cancer. 2023;23(3):173–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yilmaz E, Ismaila N, Bauman JE, et al. Immunotherapy and biomarker testing in recurrent and metastatic head and neck cancers: ASCO guideline. J Clin Oncol. 2023;41(5):1132–46. [DOI] [PubMed] [Google Scholar]
- 9.Shen Y, Qi Y, Wang C, Wu C, Zhan X. Predicting specific mortality from laryngeal cancer based on competing risk model: a retrospective analysis based on the SEER database. Ann Transl Med. 2023;11(4):179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sainero-Alcolado L, Liaño-Pons J, Ruiz-Pérez MV, Arsenian-Henriksson M. Targeting mitochondrial metabolism for precision medicine in cancer. Cell Death Differ. 2022;29(7):1304–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yin L, Xie S, Chen Y, et al. Novel germline mutation KMT2A G3131S confers genetic susceptibility to familial myeloproliferative neoplasms. Ann Hematol. 2021;100(9):2229–40. [DOI] [PubMed] [Google Scholar]
- 12.Guess M, Soltis I, Rigo B, Zavanelli N, Kapasi S, Kim H, Yeo WH. Wireless batteryless soft sensors for ambulatory cardiovascular health monitoring. Soft Sci. 2023;3:23. [Google Scholar]
- 13.Liu Y, Sun Y, Guo Y, et al. An overview: the diversified role of mitochondria in cancer metabolism. Int J Biol Sci. 2023;19(3):897–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin P, Jiang J, Zhou L, et al. Mitochondrial adaptation in cancer drug resistance: prevalence, mechanisms, and management. J Hematol Oncol. 2022;15(1):97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bosc C, Selak MA, Sarry JE. Resistance is futile: targeting mitochondrial energetics and metabolism to overcome drug resistance in cancer treatment. Cell Metab. 2017;26(5):705–7. [DOI] [PubMed] [Google Scholar]
- 16.Chen Y, Zhang Y, Wang Z, et al. CHST15 gene germline mutation is associated with the development of familial myeloproliferative neoplasms and higher transformation risk. Cell Death Dis. 2022;13(7):586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ke X, Mu X, Chen S, Zhang Z, Zhou J, Chen Y, Gao J, Liu J, Wang X, Ma C, Miao L. Reduced graphene oxide reinforced PDA-Gly-PVA composite hydrogel as strain sensors for monitoring human motion. Soft Sci. 2023;3:21. [Google Scholar]
- 18.Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2021;22(2):96–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sebastian-delaCruz M, Gonzalez-Moro I, Olazagoitia-Garmendia A, Castellanos-Rubio A, Santin I. The role of lncRNAs in gene expression regulation through mRNA stabilization. Noncoding RNA. 2021;7(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mattick JS, Amaral PP, Carninci P, et al. Long non-coding RNAs: definitions, functions, challenges and recommendations. Nat Rev Mol Cell Biol. 2023;24(6):430–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma L, Xu A, Kang L, et al. LSD1-demethylated LINC01134 confers oxaliplatin resistance through SP1-induced p62 transcription in HCC. Hepatology. 2021;74(6):3213–34. [DOI] [PubMed] [Google Scholar]
- 22.Zhang H, Shi Y, Ying J, et al. A bibliometric and visualized research on global trends of immune checkpoint inhibitors related complications in melanoma, 2011–2021. Front Endocrinol (Lausanne). 2023;14:1164692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim SD, Park K, Lee S, Kum J, Kim Y, An S, Kim H, Shin M, Son D. Injectable and tissue-conformable conductive hydrogel for MRI-compatible brain-interfacing electrodes. Soft Sci. 2023;3:18. [Google Scholar]
- 24.Han S, Yan Y, Ren Y, et al. LncRNA SAMMSON mediates adaptive resistance to RAF inhibition in BRAF-mutant melanoma cells. Cancer Res. 2021;81(11):2918–29. [DOI] [PubMed] [Google Scholar]
- 25.Li N, Zhan X, Zhan X. The lncRNA SNHG3 regulates energy metabolism of ovarian cancer by an analysis of mitochondrial proteomes. Gynecol Oncol. 2018;150(2):343–54. [DOI] [PubMed] [Google Scholar]
- 26.Si X, Zang R, Zhang E, et al. LncRNA H19 confers chemoresistance in ERα-positive breast cancer through epigenetic silencing of the pro-apoptotic gene BIK. Oncotarget. 2016;7(49):81452–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Long F, Li X, Pan J, et al. The role of lncRNA NEAT1 in human cancer chemoresistance. Cancer Cell Int. 2024;24(1):236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Y, Chen Y, Wang F, et al. Caveolin-1 promotes glioma progression and maintains its mitochondrial inhibition resistance. Discov Oncol. 2023;14(1):161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu C, Wang S, Feng SP, Fang NX. Portable green energy out of the blue: hydrogel-based energy conversion devices. Soft Sci. 2023;3:10. [Google Scholar]
- 30.Li Y, Li W, Hoffman AR, Cui J, Hu JF. The nucleus/mitochondria-shuttling LncRNAs function as new epigenetic regulators of mitophagy in cancer. Front Cell Dev Biol. 2021;9: 699621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang P, Liu M, Cui Y, Zheng P, Liu Y. Microsatellite instability status differentially associates with intratumoral immune microenvironment in human cancers. Brief Bioinform. 2021;22(3):bbaa180. [DOI] [PubMed] [Google Scholar]
- 32.Dong Y, Yoshitomi T, Hu JF, Cui J. Long noncoding RNAs coordinate functions between mitochondria and the nucleus. Epigenetics Chromatin. 2017;10(1):41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang J, Wu H, Wu J, et al. Constructing a novel mitochondrial-related gene signature for evaluating the tumor immune microenvironment and predicting survival in stomach adenocarcinoma. J Transl Med. 2023;21(1):191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y, Zhao S, Chen Y, et al. Vimentin promotes glioma progression and maintains glioma cell resistance to oxidative phosphorylation inhibition. Cell Oncol (Dordr). 2023;46(6):1791–806. [DOI] [PubMed] [Google Scholar]
- 35.Zhang H, Xia T, Xia Z, et al. KIF18A inactivates hepatic stellate cells and alleviates liver fibrosis through the TTC3/Akt/mTOR pathway. Cell Mol Life Sci. 2024;81(1):96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu S, Wang Y, Peng K, Lyu M, Liu F, Liu T. Establishment of a prognostic signature of stromal/immune-related genes for gastric adenocarcinoma based on ESTIMATE algorithm. Front Cell Dev Biol. 2021;9: 752023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guan M, Jiao Y, Zhou L. Immune infiltration analysis with the CIBERSORT method in lung cancer. Dis Markers. 2022;2022:3186427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang J, Xia W, Huang Y, et al. A vasculogenic mimicry prognostic signature associated with immune signature in human gastric cancer. Front Immunol. 2022;13:1016612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang P, Gu S, Pan D, et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med. 2018;24(10):1550–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang F, Wu Z, Sun S, Fu Y, Chen Y, Liu J. POEMS syndrome in the 21st century: a bibliometric analysis. Heliyon. 2023;9(10): e20612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu L, Ahn JH, Wang B. Wearable plasmonic biofluid sensors as your photonic skin. Soft Sci. 2023;3:6. [Google Scholar]
- 42.Maeser D, Gruener RF, Huang RS. oncoPredict: an R package for predicting in vivo or cancer patient drug response and biomarkers from cell line screening data. Brief Bioinform. 2021;22(6):bbab260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang M, Sun Q, Han Z, et al. Construction of a novel disulfidptosis-related lncRNAs signature for prognosis prediction and anti-tumor immunity in laryngeal squamous cell carcinoma. Heliyon. 2024;10(10): e30877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han B, Li S, Huang S, Huang J, Wu T, Chen X. Cuproptosis-related lncRNA SNHG16 as a biomarker for the diagnosis and prognosis of head and neck squamous cell carcinoma. PeerJ. 2023;11: e16197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Z, Zeinalzadeh Z, Huang T, et al. Mitochondria-related chemoradiotherapy resistance genes-based machine learning model associated with immune cell infiltration on the prognosis of esophageal cancer and its value in pan-cancer. Transl Oncol. 2024;42: 101896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu Z, Chen Y, Yu G, Ma Y. Research trends and hotspots in surgical treatment of recurrent nasopharyngeal carcinoma: a bibliometric analysis from 2000 to 2023. Asian J Surg. 2024;47(6):2939–41. [DOI] [PubMed] [Google Scholar]
- 47.Nagwade P, Parandeh S, Lee S. Prospects of soft biopotential interfaces for wearable human-machine interactive devices and applications. Soft Sci. 2023;3:24. [Google Scholar]
- 48.Zhang L, Zhang Z, Zheng X, et al. A novel microRNA panel exhibited significant potential in evaluating the progression of laryngeal squamous cell carcinoma. Noncoding RNA Res. 2023;8(4):550–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang L, Gu S, Wang L, et al. M2 macrophages promote PD-L1 expression in triple-negative breast cancer via secreting CXCL1. Pathol Res Pract. 2024;260: 155458. [DOI] [PubMed] [Google Scholar]
- 50.Sezginer O, Unver N. Dissection of pro-tumoral macrophage subtypes and immunosuppressive cells participating in M2 polarization. Inflamm Res. 2024;73(9):1411–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rubinstein A, Kudryavtsev I, Malkova A, et al. Sarcoidosis-related autoimmune inflammation in COVID-19 convalescent patients. Front Med (Lausanne). 2023;10:1271198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li J, Wu Z, Pan Y, et al. GNL3L exhibits pro-tumor activities via NF-κB pathway as a poor prognostic factor in acute myeloid leukemia. J Cancer. 2024;15(13):4072–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nam S, Park C, Sunwoo SH, Kim M, Lee H, Lee M, Kim DH. Soft conductive nanocomposites for recording biosignals on skin. Soft Sci. 2023;3:28. [Google Scholar]
- 54.Ribatti D. The discovery of plasma cells: an historical note. Immunol Lett. 2017;188:64–7. [DOI] [PubMed] [Google Scholar]
- 55.Steinert EM, Vasan K, Chandel NS. Mitochondrial metabolism regulation of T cell-mediated immunity. Annu Rev Immunol. 2021;39:395–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vasan K, Werner M, Chandel NS. Mitochondrial metabolism as a target for cancer therapy. Cell Metab. 2020;32(3):341–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang CJ, Li JM, Xu D, et al. Surface molecularly engineered mitochondria conduct immunophenotype repolarization of tumor-associated macrophages to potentiate cancer immunotherapy. Adv Sci (Weinh). 2024;11(38): e2403044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shapaer T, Chen Y, Pan Y, et al. Elevated BEAN1 expression correlates with poor prognosis, immune evasion, and chemotherapy resistance in rectal adenocarcinoma. Discov Oncol. 2024;15(1):446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang D, Sia SA, Solco SFD, Xu J, Suwardi A. Energy harvesting through thermoelectrics: topological designs and materials jetting technology. Soft Sci. 2023;3:1. [Google Scholar]
- 60.Lei Y, Zhang E, Bai L, Li Y. Autophagy in cancer immunotherapy. Cells. 2022;11(19):2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gureev AP, Shaforostova EA, Popov VN. Regulation of mitochondrial biogenesis as a way for active longevity: interaction between the Nrf2 and PGC-1α signaling pathways. Front Genet. 2019;10:435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Singh AK, Malviya R. Coagulation and inflammation in cancer: Llimitations and prospects for treatment. Biochim Biophys Acta Rev Cancer. 2022;1877(3): 188727. [DOI] [PubMed] [Google Scholar]
- 63.Chi Q, Yang Z, Liang HP. A transendothelial leukocyte transmigration model based on computational fluid dynamics and BP neural network. Front Bioeng Biotechnol. 2022;10: 881797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tokarski JS, Newitt JA, Chang CY, et al. The structure of Dasatinib (BMS-354825) bound to activated ABL kinase domain elucidates its inhibitory activity against imatinib-resistant ABL mutants. Cancer Res. 2006;66(11):5790–7. [DOI] [PubMed] [Google Scholar]
- 65.Morita K, Kantarjian HM, Sasaki K, et al. Outcome of patients with chronic myeloid leukemia in lymphoid blastic phase and Philadelphia chromosome-positive acute lymphoblastic leukemia treated with hyper-CVAD and dasatinib. Cancer. 2021;127(15):2641–7. [DOI] [PubMed] [Google Scholar]
- 66.Araujo J, Logothetis C. Dasatinib: a potent SRC inhibitor in clinical development for the treatment of solid tumors. Cancer Treat Rev. 2010;36(6):492–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Johnson DB, Flaherty KT, Weber JS, et al. Combined BRAF (Dabrafenib) and MEK inhibition (Trametinib) in patients with BRAFV600-mutant melanoma experiencing progression with single-agent BRAF inhibitor. J Clin Oncol. 2014;32(33):3697–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Paz-Cruz E, Cadena-Ullauri S, Guevara-Ramírez P, et al. Thyroid cancer in Ecuador: a genetic variants review and a cross-sectional population-based analysis before and after COVID-19 pandemic. Heliyon. 2023;10(1): e23964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Duvic M, Vu J. Vorinostat: a new oral histone deacetylase inhibitor approved for cutaneous T-cell lymphoma. Expert Opin Investig Drugs. 2007;16(7):1111–20. [DOI] [PubMed] [Google Scholar]
- 70.Ahmed K, Kaddoura R, Yassin MA. A practical guide to managing hypertension, hyperlipidemia, and hyperglycemia in patients with chronic myeloid leukemia. Front Med (Lausanne). 2022;9:1025392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rini BI, Escudier B, Tomczak P, et al. Comparative effectiveness of axitinib versus sorafenib in advanced renal cell carcinoma (AXIS): a randomised phase 3 trial. Lancet. 2011;378(9807):1931–9. [DOI] [PubMed] [Google Scholar]
- 72.Cheng AL, Kang YK, Chen Z, et al. Efficacy and safety of sorafenib in patients in the Asia-Pacific region with advanced hepatocellular carcinoma: a phase III randomised, double-blind, placebo-controlled trial. Lancet Oncol. 2009;10(1):25–34. [DOI] [PubMed] [Google Scholar]
- 73.Gao P, Gong L, Wang X. Induction chemotherapy in patients with resectable laryngeal cancer: a meta-analysis. Mol Clin Oncol. 2018;9(2):155–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Malla R, Viswanathan S, Makena S, et al. Revitalizing cancer treatment: exploring the role of drug repurposing. Cancers (Basel). 2024;16(8):1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xia W, Zeng C, Zheng Z, Huang C, Zhou Y, Bai L. Development and validation of a novel mitochondrion and ferroptosis-related long non-coding RNA prognostic signature in hepatocellular carcinoma. Front Cell Dev Biol. 2022;10: 844759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang Y, Gao S, Xu Y, Tang Z, Liu S. A mitochondrial function-related LncRNA signature predicts prognosis and immune microenvironment for breast cancer. Sci Rep. 2023;13(1):3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhou Q, Xiong J, Gao Y, et al. Mitochondria-related lncRNAs: predicting prognosis, tumor microenvironment and treatment response in lung adenocarcinoma. Funct Integr Genomics. 2023;23(4):323. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used in this study are sourced from public databases. The data in this study are openly available in the GEO database (https://www.ncbi.nlm.nih.gov/geo/) and the TCGA database (https://www.cancer.gov/).