Abstract
Abeliophyllum distichum and Forsythia ovata are closely related species endemic to Korea and are highly valued as ornamental shrubs in the Oleaceae family. A combination of PacBio and Illumina sequencing with Hi-C scaffolding technologies was employed to develop chromosome-level genome assemblies of these species. The assembled genome sizes are 795.72 Mb for A. distichum and 1,108.53 Mb for F. ovata. The assemblies exhibit scaffold N50 lengths of 53.12 Mb and 68.97 Mb, with minimal gaps measuring 323.40 kb and 149.00 kb, and 97.71% and 98.82% BUSCO scores for Embryophyta single-copy orthologs, respectively, indicating high contiguity and completeness. The genomes contain 485.24 Mb and 691.68 Mb of repetitive sequences, 4,926 and 7,175 full-length long terminal repeat retrotransposons, and 49,414 and 57,587 protein-coding genes, respectively. The 14 pseudochromosomes encompass 93.80% of the A. distichum genome and 89.11% of the F. ovata genome, thereby demonstrating one-to-one chromosome-level collinearity. These high-quality genome assemblies serve as invaluable resources for genetic and breeding studies, facilitating a deeper understanding of the evolutionary history of these distinctive species.
Subject terms: Plant evolution, Genome duplication
Background & Summary
The Oleaceae family is comprised of 28 genera across five tribes, with approximately 700 species of temperate and tropical shrubs, trees, and occasionally lianas1. Many species from the genera Forsythia, Fraxinus, Jasminum, Ligustrum, and Syringa are widely cultivated for purposes of ornamentation, fragrance, and timber, while olive trees from the genus Olea are valued for their fruit and oil production. In the past decade, chromosome-level genome assemblies have been developed for seven Oleaceae species, including Olea europaea2–4, Fraxinus excelsior5, Osmanthus fragrans6, Fraxinus pennsylvanica7, and Syringa oblata8–10, which belong to the tribe Oleeae; Jasminum sambac11,12 in tribe Jasmineae; and Forsythia suspensa13 in tribe Forsythieae. These genome assemblies provide substantial insights into the organization and expression of genes associated with a number of important biological processes, including oil biosynthesis2, fragrance production11, disease resistance5, and the synthesis of medicinal compounds13. A comparison of these genomes suggests that a hexaploidization event likely occurred in the Oleaceae approximately 53–61 million years ago (MYA)14. Moreover, species within the tribe Oleeae underwent a shared whole-genome duplication (WGD) approximately 28 MYA2,9. It is postulated that this duplication event resulted in a doubling of chromosome pairs (n = 23) in these species, in contrast to the tribe Jasmineae (n = 13 for J. sambac) and Forsythieae (n = 14 for F. suspensa). Nevertheless, the precise nature of this WGD, whether autopolyploid or allopolyploid, and the basic chromosome number of the family remain unknown.
Endemic plants are defined as those that grow naturally within restricted habitats. A total of 373 endemic plant taxa, representing approximately 9.5% of the native flora, have been documented in the checklist of endemic plants on the Korean Peninsula. The list includes Abeliophyllum distichum, Forsythia koreana, Forsythia nakaii, and Forsythia ovata, which belong to the tribe Forsythieae15. Abeliophyllum distichum, the sole species in the monotypic genus Abeliophyllum, is distinguished by its white flowers and winged samara-type fruits (Fig. 1a). The genus Forsythia comprises 11–13 species, with a predominantly Eastern Asian distribution and one species native to southeastern Europe. Forsythia species are characterized by their yellow flowers and capsule fruits (Fig. 1a). Chromosome cytology indicates that both Abeliophyllum and Forsythia exhibit the same chromosome number (n = 14)16. The classification of Abeliophyllum as a sister group to Forsythia within the tribe Forsythieae is supported by studies of pollen morphology17 and molecular phylogeny using nuclear and chloroplast genes18,19. It has been postulated that these species may represent the basal genome structure of the common ancestor of the Oleaceae family, given their relatively small and compact genomes (ca. 1 Gb or less) with 14 chromosomes.
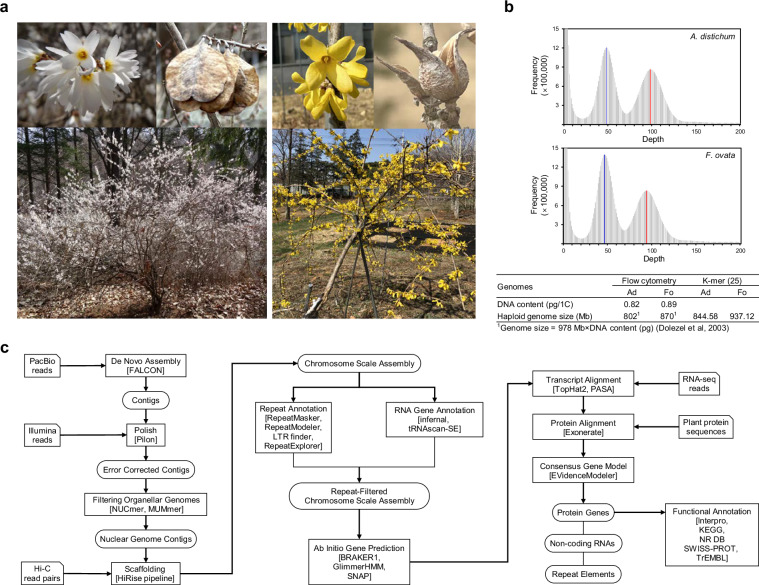
Fig. 1.
Reference accessions of Abeliophyllum distichum and Forsythia ovata used in this study. (a) Photographs of A. distichum (left) and F. ovata (right) flowers, fruits, and trees, taken between March and April 2023. (b) Estimation of genome sizes for A. distichum (Ad) and F. ovata (Fo) based on K-mer 25 distributions. The blue and red peaks correspond to heterozygous and homozygous reads, respectively. The lower panel depicts the estimated haploid genome size derived from homozygous K-mer peaks and flow cytometry analyses. (c) Workflow of genome assembly and annotation.
This study aimed to develop high-contiguity assemblies for the genomes of A. distichum and F. ovata by employing a combination of PacBio and Illumina sequencing and Hi-C scaffolding technologies. We employed RNA sequencing (RNA-seq) across various tissues and conducted whole-genome methylation profiling to annotate the assemblies and identify heterochromatic chromosome regions. Furthermore, we carried out a genome-to-genome comparison with other sequenced Oleaceae genomes. The findings from this study provide valuable resources for biology research, conservation efforts, and breeding programs for these species, as well as for evolutionary studies within the Oleaceae family.
Methods
Plant materials and nucleic acid extraction
We selected the A. distichum accession KNKB198505000391 and F. ovata accession KNKB202402200001 (Fig. 1a) at the Korea National Arboretum for genome sequencing. Leaf tissues were collected in April and May following a two-day dark treatment period. High-molecular-weight genomic DNA was extracted using established nuclear isolation methods that are suitable for long-read sequencing20–22. Total RNA was isolated from leaf, petal, carpel, and stamen tissues collected in April, using the cetyltrimethylammonium bromide method23. The RNA was subsequently used for messenger RNA (mRNA) purification and library construction. The quality and quantity of the nucleic acids were evaluated using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) and a Qubit Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA).
Library construction, genome sequencing, and genome size estimation
Library construction and sequencing were conducted by Macrogen (Seoul, Korea). For Illumina short-read sequencing, paired-end (PE) sequencing libraries with an insert size of 550 bp were generated using the Illumina TruSeq Nano DNA Kit (Illumina, San Diego, CA, USA). Single-molecule real-time DNA sequencing libraries with an insert size of 20 kb were constructed for PacBio Sequel sequencing using the SMRTbell Prep Kit 3.0 (Pacific Biosciences, Menlo Park, CA, USA). For whole-genome bisulfite sequencing (BS), fragmented DNA sized at 200 bp was treated with bisulfite using the EZ DNA Methylation-Gold Kit (Zymo Research, Irvine, CA, USA), followed by library construction using the Accel-NGS Methyl-Seq DNA Library Kit (Swift Biosciences, Inc., Ann Arbor, MI, USA). For chromosome conformation capture, leaf tissues were provided to Dovetail Genomics (Santa Cruz, CA, USA), which constructed and sequenced Hi-C libraries. RNA libraries were prepared using the Illumina TruSeq RNA v2 Kit (Illumina). As a result, we obtained a total of 203.15 Gb of Illumina raw reads, 182.52 Gb of filtered PacBio subreads, 96.79 Gb of filtered Hi-C reads, 106.72 Gb of raw BS reads, and 140.00 Gb of raw mRNA data (Table 1). Illumina raw reads with a minimum Phred quality score of 20 (Q20) were processed using Trimmomatic v0.3624 to filter out adapter contamination and low-quality regions. Polymerase chain reaction (PCR) duplicates were removed with Picard v2.2.4 (https://broadinstitute.github.io/picard) using the default parameters. The average read and N50 lengths of the filtered PacBio subreads were 17.61 and 25.47 kb, respectively.
Table 1.
Statistics of sequencing data used in the genome assemblies of Abeliophyllum distichum and Forsythia ovata.
| Type | Species | Library | Platform | Insert size (bp) | Raw data (Gb) | Clean data (Gb) | Sequence coverage (×)a |
|---|---|---|---|---|---|---|---|
| Genome | A. distichum | Illumina | HiSeqX | 550 | 98.41 | 83.45 | 98.76 |
| Hi-C | HiSeqX | 350 to 1,000 | − | 40.70 | 48.16 | ||
| PacBio | Sequel | 20,000 | − | 98.14 | 116.14 | ||
| F. ovata | Illumina | NovaSeq6000 | 550 | 104.74 | 96.32 | 102.79 | |
| Hi-C | HiSeqX | 350 to 1,000 | − | 56.09 | 59.86 | ||
| PacBio | Sequel | 20,000 | − | 84.38 | 90.05 | ||
| Methylome | A. distichum | BSb | HiSeq2000 | 200 | 53.32 | 45.79 | 54.19 |
| F. ovata | BSb | HiSeq2000 | 200 | 53.40 | 45.89 | 48.98 | |
| Transcriptome | A. distichum | mRNA-seqc | NextSeq | 350 | 74.40 | 53.85 | 63.73 |
| F. ovata | mRNA-seqc | NovaSeq6000 | 350 | 65.60 | 59.14 | 63.11 |
aSequence coverage was calculated using the genome sizes of A. distichum and F. ovata, measured at 844.58 and 937.12 Mb, respectively.
bBS, bisulfite sequencing.
cmRNA-seq, mRNA-sequencing.
The genome sizes of each species were estimated through the application of both K-mer analysis and flow cytometry. The frequency distribution of K-mer analysis using Illumina PE reads and JELLYFISH v2.1.3 software25 with a K-mer size of 25 revealed two distinct peaks at coverages of 48 × and 98 × for A. distichum and 46 × and 94 × for F. ovata, corresponding to heterozygous and homozygous reads, respectively (Fig. 1b). The maximum haploid genome sizes were estimated to be 844.58 Mb for A. distichum and 937.12 Mb for F. ovata, based on homozygous reads. Flow cytometry, conducted with a CyFlow Space system (Partec BmbH, Münster, Germany) and using diploid Raphanus sativus cv. WK10039 (1 C = 0.6 pg) as a reference, indicated that the haploid genome sizes of these species were slightly smaller.
Genome assembly and scaffolding
A schematic workflow of the genome assembly and annotation processes is presented in Fig. 1c. Initially PacBio subreads were corrected with CANU v2.1.126 and pre-assembled into contigs using the FALCON assembler v2.1.427, with parameters set as follows: length cutoff, 10 kb; max difference, 80; max coverage, 80; and minimum coverage, 2. The initial haplotype-fused contigs were refined using Pilon28 for gap filling and sequence error correction, with the parameters–fix bases,–gaps, and–diploid. Mitochondrial and plastid sequences were filtered out from the contigs using organellar genome sequences (GenBank accessions MW645067, MF407178, and MF407183) and NUCmer from the MUMmer 3 package29. Subsequently, the resulting contigs were then aligned with the filtered PE reads from the Hi-C libraries using the Dovetail HiRise scaffolding pipeline30 to produce a scaffold-level assembly. The total length of the assembled genome was 795.72 Mb for A. distichum, with a contig N50 of 373.06 kb and scaffold N50 of 53.12 Mb, and 1,108.53 Mb for F. ovata, with a contig N50 of 1.19 Mb and scaffold N50 of 68.97 Mb. The longest 14 pseudochromosomes of A. distichum and F. ovata, ranging from 40.08 to 70.22 Mb and 56.21 to 86.46 Mb, comprised 93.04% and 87.43% of the initial contig sequences, respectively (Table 2 and S1). The alignment of homologous pseudochromosomes of the A. distichum and F. ovata assemblies using MUMmer29 demonstrated global collinear synteny across the genome, with some mismatched regions due to inversion (Fig. 2a).
Table 2.
Genome assembly statistics for A. distichum and F. ovata.
| Assembly | A. distichum | F. ovata | |
|---|---|---|---|
| Contig | Number | 4,321 | 1,831 |
| Total length (bp) | 801,868,764 | 1,129,748,862 | |
| N50 number | 610 | 302 | |
| N50 length (bp) | 373,057 | 1,187,720 | |
| Scaffold | Number | 879 | 145 |
| Total length (bp) | 795,717,758 | 1,108,525,499 | |
| Gaps (bp) | 323,400 | 149,000 | |
| N50 number | 7 | 8 | |
| N50 length (bp) | 53,115,898 | 68,970,119 | |
| Repetitive sequences | 485,237,308 | 691,680,161 | |
| Protein-coding genes | 49,414 | 57,587 | |
| Average size (bp) | 3,128 | 5,027 | |
| Number of exon per gene | 4.14 | 4.65 | |
| Average exon size (bp) | 254 | 231 | |
| Average intron size (bp) | 660 | 1,084 | |
| Gene density (kb/gene) | 16.10 | 19.25 | |
| RNA genes | 4,820 | 6,071 | |
| Chromosome | Number | 14 | 14 |
| Total length (bp) | 746,393,055 | 987,838,149 | |
| Gaps (bp) | 323,400 | 149,000 | |
| GC contents (%) | 33.88 | 33.83 | |
| Repetitive sequences (bp) | 358,459,201 | 512,990,529 | |
| Protein-coding genes | 45,792 | 48,283 | |
| Gene density (kb/gene) | 16.30 | 20.50 | |
| RNA genes | 3,814 | 5,250 | |
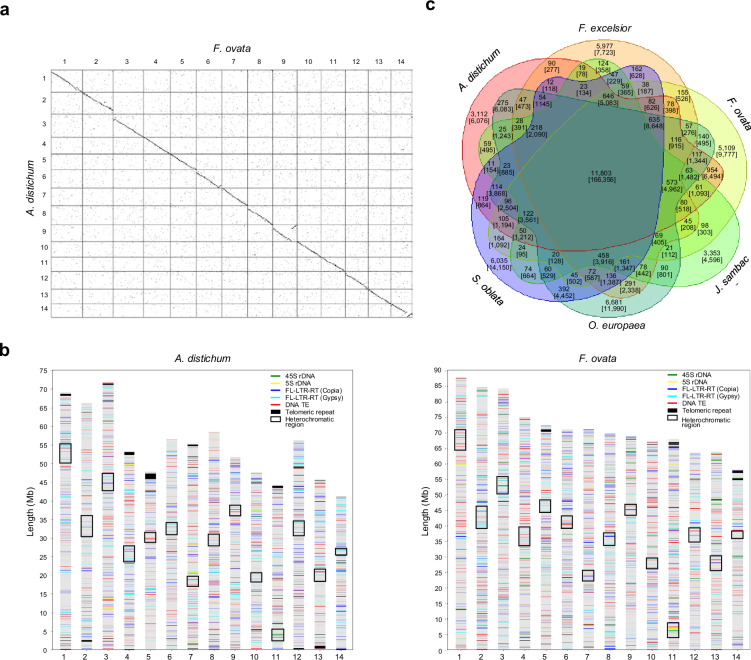
Fig. 2.
Chromosomal synteny, repetitive sequence distribution, and gene families in A. distichum and F. ovata. (a) Dot plot comparison of A. distichum and F. ovata pseudochromosome assemblies. Dots closest to the diagonal line reflect collinearity between the two assemblies. (b) Distributions of various repetitive sequences, including rDNA, FL-LTR-RTs, DNA TEs, and telomeric repeats. Black boxes indicate the location of repetitive sequence-enriched heterochromatic regions, and colored lines represent different types of repetitive sequences. (c) Venn diagram showing unique and shared gene families among six sequenced genomes of the Oleaceae family. The numbers of gene families and genes (in brackets) for each group are provided. The NCBI and China National Center for Bioinformation accession numbers are as follows: F. excelsior, GCA_900149125; J. sambac, GWHAZHY00000000; O. europaea, GCF_002742605; S. oblata, GWHBHRY00000000.
The completeness of the genome assemblies was assessed using orthologous gene analysis with BUSCO v5.4.331 in conjunction with OrthoDB v1032. The BUSCO analysis indicated that approximately 96.86% to 98.54% of Eudicots genes (comprising 2,326 single-copy orthologs) and 97.71% to 98.82% of Embryophyta genes (comprising 1,614 single-copy orthologs) were complete, while only 0.82% to 2.15% of Eudicots genes and 0.31% to1.55% of Embryophyta genes were missing (Table S2). The genome completeness estimates for the A. distichum and F. ovata assemblies were comparable to those of J. sambac and F. excelsior, and higher than those of other Oleaceae assemblies, such as those of S. oblata and O. europaea, reported thus far. By applying cutoffs of 1E−10 and >50% coverage, a BLASTN comparison of the assemblies against full-length transcript unigenes generated from mRNA-seq data using Trinity v2.2.033 with the default parameters, along with a coding region search using TransDecoder v5.5.0 (https://github.com/TransDecoder/TransDecoder), revealed that each genome assembly recovered >99% of the gene space, indicating high-quality assemblies (Table S3).
Annotation of repetitive sequences and non-coding RNAs
The genome assemblies were masked for repetitive sequences using RepeatMasker v4.0.5 (http://www.repeatmasker.org) and RepeatModeler v1.0.8 (http://www.repeatmasker.org). Retrotransposons (RTs) were identified using LTR_FINDER v1.0534. The genomes of A. distichum and F. ovata contained 485.24 Mb and 691.68 Mb of repetitive sequences, respectively, which corresponds to repetitive sequence ratios of 60.98% and 62.40% (Table 2). The most prevalent classes of repetitive sequences were RTs, accounting for 44.06% and 50.17% of the total, and DNA transposable elements (TEs), comprising 12.00% and 15.86%. The predominant RTs were Ty3/Gypsy and Ty1/Copia, while MuLE-MuDR and hAT-Ac were the most abundant DNA TEs (Table S4). Full-length long terminal repeat RTs (FL-LTR-RTs) were identified from the repetitive sequences using BLASTX searches (cutoffs of 1E−10 and >70% coverage) against mobile genetic elements in RepeatExplorer235. A total of 12,101 FL-LTR-RTs, with an average length of approximately 8 kb, were identified (Table 3). The OTA-Tat family was the most prevalent among Ty3/Gypsy FL-LTR-RTs in both genomes, whereas the Tork and SIRE families were the most abundant among Ty1/Copia FL-LTR-RTs in A. distichum and F. ovata, respectively. Tandem arrays of the telomere sequence (5′-TTTAGGG-3′) and modified units were identified at the ends of several pseudochromosomes (1, 4, 5, 7, 11, 12, 13, and 14 of A. distichum and 1, 5, 11, and 14 of F. ovata). However, the telomeres of other chromosomes remained unidentified. The investigation of repetitive sequence-enriched heterochromatic regions was conducted using whole-genome methylation data. A total of 44.3 Gb of quality-filtered Illumina BS sequences were aligned to the genome assemblies using BSMAP v2.936 with the default parameters. Only those reads that were uniquely mapped were selected, and any PCR duplicates were removed using SAMBAMBA v0.5.937. The methylation ratio at each cytosine site was extracted and partitioned based on context (CG, CHG, and CHH). The mean cytosine methylation levels for each chromosome were calculated using a 100-kb sliding window. The distribution of repetitive sequence-enriched heterochromatic regions and other repetitive sequences are illustrated in Fig. 2b.
Table 3.
Statistics of annotated FL-LTR-RTs in the A. distichum and F. ovata genomes.
| FL-LTR-RT family | A. distichum | F. ovata | ||
|---|---|---|---|---|
| Ty1/Copia | Ale | Number | 741 | 993 |
| Length (bp) | 4,019,949 | 5,598,449 | ||
| Tork | Number | 636 | 962 | |
| Length (bp) | 4,133,536 | 6,281,533 | ||
| TAR | Number | 491 | 590 | |
| Length (bp) | 3,265,422 | 4,099,906 | ||
| SIRE | Number | 332 | 674 | |
| Length (bp) | 3,334,862 | 7,047,172 | ||
| Other elements | Number | 724 | 864 | |
| Length (bp) | 5,547,945 | 6,377,486 | ||
| Total | Number | 2,924 | 4,083 | |
| Length (bp) | 20,301,714 | 29,404,546 | ||
| % Genome | 2.40 | 3.14 | ||
| Ty3/Gypsy | OTA-Tat | Number | 897 | 1,283 |
| Length (bp) | 9,789,929 | 14,573,227 | ||
| OTA-Athila | Number | 646 | 977 | |
| Length (bp) | 7,408,191 | 11,661,401 | ||
| CRM | Number | 163 | 248 | |
| Length (bp) | 1,308,303 | 1,807,119 | ||
| Tekay | Number | 134 | 287 | |
| Length (bp) | 1,321,282 | 2,727,045 | ||
| Other elements | Number | 162 | 297 | |
| Length (bp) | 983,957 | 1,764,382 | ||
| Total | Number | 2,002 | 3,092 | |
| Length (bp) | 20,811,662 | 32,533,174 | ||
| % Genome | 2.46 | 3.47 | ||
Non-coding RNAs (ncRNAs), including microRNAs (miRNAs), small nuclear RNAs (snRNAs), and other RNAs, were identified using Infernal v1.1.438. Ribosomal RNAs (rRNAs) were searched with BLASTN, while transfer RNAs (tRNAs) were predicted using tRNAscanSE39. We identified 4,820 ncRNA sequences for A. distichum and 6,071 for F. ovata. This encompasses 1,453 and 1,939 rRNAs, 727 and 953 tRNAs, 254 and 324 miRNAs, 2,285 and 2,705 snRNAs, and 101 and 150 other RNA sequences, respectively (Tables S5 and S6).
Prediction of protein-coding genes and functional annotation
We employed a combination of ab initio and evidence-based gene prediction methods for the identification of protein-coding genes. Ab initio predictions were conducted on the repetitive sequence-masked genome assemblies using BRAKER1 v1.840, GlimmerHMM v3.0.241, and SNAP42, with parameters trained on matrices from Arabidopsis thaliana and O. europaea. Genes with a coding sequence (CDS) of less than 300 bp, incomplete coding regions, or top matches to transposon-encoded proteins were excluded. For evidence-based gene prediction, we aligned approximately 112.99 Gb of quality-filtered Illumina PE mRNA-seq reads to the genome assemblies using TopHat243 with parameters set to–max-intron-length 100000 and–microexon-search. Gene predictions were further refined using the PASA v2.0.2 package44. Additionally, gene models from A. thaliana (TAIR 10, GCF_000001735), O europaea (GCF_002742605), and F. excelsior (GCA_900149125) were aligned to the genome assemblies using Exonerate v2.2.045. Finally, the ab initio gene models, transcript alignments, and protein alignments were integrated into consensus gene model sets using EVidenceModeler44. This process yielded the prediction of 49,414 protein-coding genes for A. distichum and 57,587 for F. ovata. The protein-coding genes in A. distichum averaged 3,128 bp in length, shorter than those in F. ovata, which averaged 5,027 bp. A. distichum exhibited fewer exons per gene (4.14 vs. 4.65) and shorter introns (660 bp vs. 1,084 bp) compared to F. ovata (Table 2). The average gene density was higher in A. distichum (one per 16.10 kb) compared to F. ovata (one per 19.25 kb) and other Oleaceae species (one per 18.46–20.98 kb), indicating that A. distichum had the most compact organization of gene space among sequenced Oleaceae genomes (Table S7).
All predicted protein-coding genes were functionally annotated using the SwissProt and TrEMBL databases from UniProt (www.ebi.ac.uk/uniprot) and the nucleotide databases of the National Center for Biotechnology Information (NCBI) using BLASTP with cutoffs of 1E−10 and >70% coverage. Protein motifs, domains, and Gene Ontology (GO) annotations were identified using the InterPro database (www.ebi.ac.uk/interpro). Metabolic pathways were annotated using the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database (www.genome.jp/kegg/pathway.html). Overall, 44,576 (90.21%) of the genes in A. distichum and 50,378 (87.48%) of the genes in F. ovata were successfully annotated, while the remaining 4,838 (9.79%) and 7,209 (12.52%) genes were classified as uncharacterized or hypothetical (Table 4). Transcriptional evidence was found for 98.05% of the annotated genes in A. distichum and 93.34% in F. ovata. For each gene model, the average trimmed mean of M (TMM) value was calculated from the mRNA-seq reads mapped to the CDS using STAR v2.7.9a46 with the default parameters.
Table 4.
Summary of functional annotation for the A. distichum and F. ovata genomes.
| Annotation | A. distichum | F. ovata | |||
|---|---|---|---|---|---|
| Number | Ratio (%) | Number | Ratio (%) | ||
| Database | RefSeq | 42,540 | 86.09 | 45,340 | 78.73 |
| TrEMBL | 42,420 | 85.85 | 49,695 | 86.30 | |
| InterPro | 32,793 | 66.36 | 41,751 | 72.50 | |
| GO | 32,427 | 65.62 | 43,982 | 76.37 | |
| SwissProt | 26,774 | 54.18 | 31,720 | 55.08 | |
| KEGG Pathway | 46,318 | 93.73 | 52,056 | 90.40 | |
| A. thaliana | 30,938 | 62.61 | 38,210 | 66.35 | |
| C. canephora | 32,561 | 65.89 | 40,481 | 70.30 | |
| F. excelsior | 35,230 | 71.30 | 43,647 | 75.79 | |
| O. europaea | 42,783 | 86.58 | 41,987 | 72.91 | |
| BUSCO eudicot | Complete | 2,253 | 96.86 | 2,292 | 98.54 |
| Fragmented | 23 | 0.99 | 15 | 0.64 | |
| Missing | 50 | 2.15 | 19 | 0.82 | |
| Annotated gene | 44,576 | 90.21 | 50,378 | 87.48 | |
| Uncharacterized gene | 3,873 | 7.84 | 3,375 | 5.86 | |
| Hypothetical gene | 965 | 1.95 | 3,834 | 6.66 | |
| Total | 49,414 | 100 | 57,587 | 100 | |
A comparison of the annotated genes between A. distichum and F. ovata, conducted through an all-against-all BLASTP analysis, revealed that 43,363 genes (87.75%) in A. distichum were found to correspond to 54,440 genes (94.54%) in F. ovata. There were 6,052 genes specific to A. distichum and 3,147 specific to F. ovata. Of these, 1,896 (3.84%) and 2,554 (4.44%) genes were classified as uncharacterized or hypothetical. A six-way comparison of genes from A. distichum and F. ovata with those from four other Oleaceae species (F. excelsior, J. sambac, O. europaea, and S. oblata) using OrthoFinder47 yielded 11,803 gene families (166,356 genes) that were shared across all six species. A total of 3,112 gene families (6,076 genes) were identified as unique to A. distichum, while F. ovata exhibited 5,109 unique gene families (9,777 genes) (Fig. 2c).
Data Records
All sequencing data and genome assemblies generated in this study can be retrieved from the NCBI database via Bioproject IDs PRJNA1086675 for A. distichum and PRJNA1086660 for F. ovata. The sequence read data are accessible via the NCBI Sequence Read Archive, with accession numbers SRP495333 for A. distichum48 and SRP495335 for F. ovata49. The assembled genomes have been deposited in the NCBI GenBank database under the accession numbers JBFOLK000000000.1 for A. distichum50 and JBFOLJ000000000.1 for F. ovata51. Additionally, the genome assemblies and annotation data are available on figshare52.
Technical Validation
Nucleic acid quality and quantity evaluation
For each species, nucleic acids were extracted from plant tissues of a single accession. The quality and integrity of the nucleic acids were evaluated using an Agilent 2100 Bioanalyzer (Agilent). The concentrations of nucleic acids were measured using a Qubit Fluorometer. DNA samples intended for PacBio sequencing underwent quality control using an Agilent Femto pulse system, with criteria that required >20% of the DNA to be greater than 40 kb in size. RNA samples were prepared from two biological replicates across four tissues at the same time point.
Quality control of raw sequence data
To obtain clean sequence reads for downstream analysis, we implemented a series of quality control procedures. We removed sequence reads with a quality score below Q20 or with more than 10% unidentified nucleotides (N). Adapter sequences and low-quality regions with a quality score below Q20 were trimmed from the reads, and PCR duplicates were removed. The sequence errors in the PacBio reads were corrected using the CANU v2.2.126. Additionally, PacBio subreads and Hi-C reads were quality-filtered by Macrogen and Dovetail Genomics, respectively.
Evaluation of assembled genomes
We evaluated the completeness and quality of genome assemblies by matching transcriptome data and publicly available eukaryotic orthologous genes. First, the transcript unigenes of A. distichum and F. ovata, which were assembled from the mRNA-seq data were aligned with the genome assemblies. The results indicated that >99% of the transcriptome unigenes matched the genome assemblies, suggesting sufficient coverage of the gene space. Second, a BUSCO completeness assessment was conducted using single-copy orthologous sets of plants. The BUSCO results showed that 97.71%–98.82% of Embryophyta genes and 96.86%–98.54% of Eudicots genes were complete, whereas only 1.46%–3.14% of Eudicots genes and 1.18%–2.29% of Embryophyta genes were fragmented or missing. These findings collectively demonstrate that the genome assemblies of A. distichum and F. ovata possess high quality, integrity, and annotation completeness.
Supplementary information
Acknowledgements
This work was supported by a grant from the Scientific Research Program (KNA1-1-13-14-1) of Korea National Arboretum, Korea.
Author contributions
J.H.M. planned the projects, designed the research, analyzed data, and wrote the manuscript. H.J. and A.C. performed the experiments, assemble the genomes, analyzed data, and participated in manuscript preparation. H.J.K. and J.H.K. planned the projects and participated in manuscript preparation. H.K., S.H.J., S.M.H., H.J.Y. and D.K.K. participated in plant sampling, sequencing, data analysis, and manuscript preparation.
Code availability
The software and parameters used in this study are described in the Methods section. No specific custom codes or scripts were utilized. Data processing was conducted according to the manuals and protocols provided with the respective software.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Hoyeol Jang, Ara Cho.
Contributor Information
Joo-Hwan Kim, Email: kimjh2009@gachon.ac.kr.
Jeong-Hwan Mun, Email: munjh@mju.ac.kr.
Supplementary information
The online version contains supplementary material available at 10.1038/s41597-024-04252-6.
References
- 1.Dupin, J. et al. Resolving the phylogeny of the olive family (Oleaceae): Confronting information from organellar and nuclear genomes. Genes (Basel)11, 1508 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Unver, T. et al. Genome of wild olive and the evolution of oil biosynthesis. Proc Natl Acad Sci USA114, E9413–E9422 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rao, G. et al. De novo assembly of a new Olea europaea genome accession using nanopore sequencing. Hortic Res8, 64 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang, L. et al. High-quality genome assembly of Olea europaea subsp. cuspidata provides insights into its resistance to fungal diseases in the summer rain belt in East Asia. Front Plant Sci13, 879822 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sollars, E. et al. Genome sequence and genetic diversity of European ash trees. Nature541, 212–216 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Yang, X. et al. The chromosome-level quality genome provides insights into the evolution of the biosynthesis genes for aroma compounds of Osmanthus fragrans. Hortic Res5, 72 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huff, M. et al. A high-quality reference genome for Fraxinus pennsylvanica for ash species restoration and research. Mol Ecol Res22, 1284–1302 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma, B. et al. Lilac (Syringa oblata) genome provides insights into its evolution and molecular mechanism of petal color change. Commun Biol5, 686 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang, Y. et al. A chromosome-level genome of Syringa oblata provides new insights into chromosome formation in Oleaceae and evolutionary history of lilacs. Plant J111, 836–848 (2022). [DOI] [PubMed] [Google Scholar]
- 10.Chen, L. et al. Syringa oblata genome provides new insights into molecular mechanism of flower color differences among individuals and biosynthesis of its flower volatiles. Front Plant Sci13, 1078677 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, G. et al. The Jasmine (Jasminum sambac) genome provides insight into the biosynthesis of flower fragrances and jasmonates. GPB21, 127–149 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu, S. et al. A high-quality genome assembly of Jasminum sambac provides insight into floral trait formation and Oleaceae genome evolution. Mol Ecol Resour22, 724–739 (2022). [DOI] [PubMed] [Google Scholar]
- 13.Li, Y. et al. The updated weeping forsythia genome reveals the genomic basis for the evolution and the forsythin and forsythoside A biosynthesis. Hortic Plant J9, 1149–1161 (2023). [Google Scholar]
- 14.J, W. et al. Allopolyploidization events and immense paleogenome reshuffling underlying the diversification of plants and secondary metabolites in Oleaceae. J Syst Evol10.1111/jse.13116 (2024).
- 15.Chung, G. et al. A checklist of endemic plants on the Korean Peninsula II. Korean J Pl Taxon53, 79–101 (2023). [Google Scholar]
- 16.Taylor, H. Cyto-taxonomy and phylogeny of the Oleaceae. Brittonia5, 337–367 (1945). [Google Scholar]
- 17.Lee, S. Palynological contributions to the taxonomy of family Oleaceae, with special empahsis on genus Forsythia (tribe Forsytheae). Korean J Pl Taxon41, 175–181 (2011). [Google Scholar]
- 18.Ha, Y., Kim, C., Choi, K. & Kim, J. Molecular phylogeny and dating of Forsythieae (Oleaceae) provide insight into the Miocene history of eurasian temperate shrubs. Front Plant Sci9, 299304 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim, D. & Kim, J. Molecular phylogeny of tribe Forsythieae (Oleaceae) based on nuclear ribosomal DNA internal transcribed spacers and plastid DNA trnL-F and matK gene sequences. J Plant Res124, 339–347 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Baek, S. et al. Draft genome sequence of wild Prunus yedoensis reveals massive inter-specific hybridization between sympatric flowering cherries. Genome Biol19, 127 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho, A. et al. An improved Raphanus sativus cv. WK10039 genome localizes centromeres, uncovers variation of DNA methylation and resolves arrangement of the ancestral Brassica genome blocks in radish chromosomes. Theor Appl Genet135, 1731–1750 (2022). [DOI] [PubMed] [Google Scholar]
- 22.Zhang, M. et al. Preparation of megabase-sized DNA from a variety of organisms using the nuclei method for advanced genomics research. Nat Protoc7, 467–478 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Chang, S., Puryear, J. & Cairney, J. A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep11, 113–116 (1993). [Google Scholar]
- 24.Bolger, A., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics30, 2114–2020 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marçais, G. & Kingsford, C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics27, 764–770 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koren, S. et al. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res27, 722–736 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chin, C. et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat Methods13, 1050–1054 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walker, B. et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One9, e112963 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kurtz, S. et al. Versatile and open software for comparing large genomes. Genome Biol5, R12 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Putnam, N. et al. Chromosome-scale shotgun assembly using an in vitro method for long-range linkage. Genome Res26, 342–350 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simão, F., Waterhouse, R., Ioannidis, P., Kriventseva, E. & Zdobnov, E. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics31, 3210–3212 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Kriventseva, E. et al. OrthoDB v10: sampling the diversity of animal, plant, fungal, protist, bacterial and viral genomes for evolutionary and functional annotations of orthologs. Nucleic Acids Res47, D807–D811 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grabherr, M. et al. Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat Biotechnol29, 644–652 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu, Z. & Wang, H. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res35, W265–W258 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Novák, P., Neumann, P. & Macas, J. Global analysis of repetitive DNA from unassembled sequence reads using RepeatExplorer2. Nat Protoc15, 3745–3776 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Xi, Y. & Li, W. BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinformatics10, 232 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tarasov, A., Vilella, A., Cuppen, E., Nijman, I. & Prins, P. Sambamba: fast processing of NGS alignment formats. Bioinformatics31, 2032–2034 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nawrocki, E. & Eddy, S. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics29, 2933–2935 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lowe, T. & Eddy, S. tRNAscan-SE: a program for inproved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res25, 955–964 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoff, K., Lange, S., Lomsadze, A., Borodovsky, M. & Stanke, M. BRAKER1: Unsupervised RNA-Seq-Based Genome Annotation with GeneMark-ET and AUGUSTUS. Bioinformatics32, 767–769 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Majoros, W., Pertea, M. & Salzberg, S. TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics20, 2878–2879 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Korf, I. Gene finding in novel genomes. BMC Bioinformatics5, 59 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol14, R36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haas, B. et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol9, R7 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Slater, G. & Birney, E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics6, 31 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Emms, D. & Kelly, S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol20, 238 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.NCBI Sequence Read Archivehttps://identifiers.org/ncbi/insdc.sra:SRP495333 (2024).
- 49.NCBI Sequence Read Archivehttps://identifiers.org/ncbi/insdc.sra:SRP495335 (2024).
- 50.NCBI GenBankhttps://identifiers.org/ncbi/insdc:JBFOLK000000000.1 (2024).
- 51.NCBI GenBankhttps://identifiers.org/ncbi/insdc:JBFOLJ000000000.1 (2024).
- 52.Jang, H. & Mun, J.-H. Genome assembly and annotations of A. distichum and F. ovata. figshare10.6084/m9.figshare.25539493 (2024).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- NCBI Sequence Read Archivehttps://identifiers.org/ncbi/insdc.sra:SRP495333 (2024).
- NCBI Sequence Read Archivehttps://identifiers.org/ncbi/insdc.sra:SRP495335 (2024).
- Jang, H. & Mun, J.-H. Genome assembly and annotations of A. distichum and F. ovata. figshare10.6084/m9.figshare.25539493 (2024).
Supplementary Materials
Data Availability Statement
The software and parameters used in this study are described in the Methods section. No specific custom codes or scripts were utilized. Data processing was conducted according to the manuals and protocols provided with the respective software.