Abstract
Stigma exsertion rate (SER) is a crucial trait that influences the seed production of hybrid rice by determining the outcrossing ability of male sterile lines (MSLs). However, the molecular genetic mechanisms underlying SER are still poorly understood. In this study, we identified 14 quantitative trait loci (QTLs) using a recombinant inbred line (RIL) population derived from B805D-MR-16-8-3 (B805D) and Hua6S. Two major QTLs, qSE1 and qSE9, were validated for their effects in the residual heterozygous line (RHL) background. The RHL carrying homozygous qSE1 region from Hua6S increased dual stigma exsertion rate (DSE) by 14.67% and 15.04%, and increased total stigma exsertion rate (TSE) by 11.73% and 13.04%, in F10 and F11 progeny, respectively. Conversely, the RHL carrying homozygous qSE9 region from B805D showed a substantial increase of 22.72% and 14.45% in single stigma exsertion rate (SSE), an increase of 13.46% and 8.30% in TSE, and an increase in percentage of spikelets with exserted stigma (PSE) by 24.82% and 15.57%, respectively, in F10 and F11 progeny. Furthermore, examination of floral organ traits revealed that both the Hua6S allele of qSE1 and the B805D allele of qSE9 increased pistil size to improve SER, but they had contrasting effects on spikelet shape. Subsequently, qSE1 and qSE9 were fine-mapped to intervals of 246.5 kb and 341.4 kb, respectively. A combination of sequencing, expression and haplotype analysis revealed that a single nucleotide variation (T to C) in the 5’UTR region of LOC_Os01g72020 (OsBOP1) was likely to be the functional variation for qSE1. Collectively, our work has laid a foundation for cloning the genes responsible for SER, and demonstrated that the Hua6S allele of qSE1 and the B805D allele of qSE9 can effectively increase SER, which could make important contributions to the genetic improvement of MSLs aimed at improving hybrid seed production.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12284-024-00752-6.
Keywords: Stigma exsertion rate, QTL, Recombinant inbred line, Residual heterozygous line, Fine mapping, Rice
Introduction
Rice (Oryza sativa L.) is one of the most important cereal crops in the world. A landmark achievement in the history of rice breeding occurred with the adoption of hybrid rice, which greatly enhanced rice yield and made significant contributions to global food security (Qian et al. 2016; Yuan. 2014). Nevertheless, a key issue in the practice of hybrid rice is the poor production and high cost of hybrid seeds, which depend on the outcrossing rate of MSLs (Virmani et al. 1982; Tian et al. 2004; Xie and Peng. 2016; Malik and Baba. 2018). SER is one of the key factors that influence the outcrossing ability of MSLs (Marathi and Jena. 2015; El-Namaky. 2018), and enhancing SER has been proved to be a practical approach for improving hybrid seed production by increasing the outcrossing rate of MSLs (Hasan et al. 2014; Dang et al. 2020, 2022; Prahalada et al. 2021; Tan et al. 2022a, b; Zhu et al. 2023). Therefore, SER plays a crucial role in hybrid rice seed production by influencing the outcrossing ability of MSLs.
SER is a complex quantitative trait that is controlled by polygenes and influenced by environmental conditions. Extensive identification of QTLs responsible for SER has been achieved through the utilization of diverse segregating populations and continuous advancements in molecular marker technologies over recent decades. To investigate the genetic basis of SER, wild rice and maintainer lines with high SER were frequently utilized for detecting QTLs associated with SER. Some SER QTLs were detected from wild rice germplasm resources (Xiong et al. 1999; Li et al. 2001; Uga et al. 2003; Miyata et al. 2007; Huang et al. 2012; Khumto et al. 2017; Bakti and Tanaka. 2019; Tan et al. 2020; Zou et al. 2020; Tan et al. 2022a, b). From the common wild rice (O. rufipogon, P16), es1 on chromosome 6 for exserted stigma was identified (Xiong et al. 1999). Three major loci and two minor loci responsible for exserted stigma were identified from O. rufipogon, W1943, and overlapped with the loci related to rice domestication (Huang et al. 2012). Using single-segment substitution lines (SSSLs) derived from the wild rice species O. glumaepatula, seven QTLs were detected and distributed on chromosomes 1, 3, 5, 9, and 10 (Tan et al. 2020). The qSER-5, qSER-1b and qSER-8b loci were fine-mapped to regions of 92.5 kb, 333.0 kb and 107.5 kb, respectively, using SSSLs derived from the wild rice species O. glaberrima (Tan et al. 2022a, b). Likewise, multiple QTLs for SER were identified from rice maintainer lines (Li et al. 2014a, b; Rahman et al. 2017a, 2017b; Zhang et al. 2018; Liu et al. 2019; Tan et al. 2021). From the maintainer line XieqingzaoB, qSE7 was detected and further fine-mapped to a 322.9 kb region on chromosome 7 (Zhang et al. 2018), and qSE11 was fine-mapped to a 350.7 kb region on chromosome 11 (Rahman et al. 2017b). Additionally, from the maintainer line II-32B, qSER-7 was identified and narrowed down to a 28.4 kb region on chromosome 7, and LOC_Os07g15370 (OsNRAMP5) was confirmed as a candidate gene (Liu et al. 2019). Furthermore, utilizing the MSL DaS, a total of 21 QTLs were detected for SER (Wang et al. 2017). Among those, qSE4 was subsequently fine-mapped to a 410.4 kb region where LOC_Os04g43910 (OsARF10) was identified as a potential candidate gene (Guo et al. 2022). In summary, although numerous QTLs for SER have been reported in the rice genome, only a limited number of them have been fine-mapped within 350 kb, and even fewer have been successfully cloned.
SER is closely associated with both spikelet and pistil traits in rice (Miyata et al. 2007; Yan et al. 2009; Uga et al. 2010; Liu et al. 2015; Marathi and Jena. 2015; Zhou et al. 2017; Khumto et al. 2018; Jiang et al. 2021; Prahalada et al. 2021; Wei et al. 2021; Tan et al. 2023). Spikelet (grain) shape is one of the important factors that determine the yield and quality of rice (Ren et al. 2023). A series of genes related to grain shape has been cloned and functionally characterized (Fan and Li. 2019). Three major grain shape genes, GS3, GW5 and GW2 had pleiotropic effects on SER (Zhou et al. 2017). GS3 regulated stigma length by controlling the number of cells, which in turn affected the SER (Takano-Kai et al. 2011). By knocking out the grain shape genes GS3, GW8, and GS9, it was possible to significantly improve SER and increase the outcrossing rate, thereby facilitating the hybrid seed production of MSLs (Zhu et al. 2023). Similarly, it was found that alleles gs3, gw5, GW7 and gw8 in constructed SSSLs had pleiotropic effects on grain shape and SER (Tan et al. 2023). In addition, the SER exhibited a close correlation with pistil morphology (Li et al. 2001; Yan et al. 2009; Marathi and Jena. 2015; Dang et al. 2022), particularly demonstrating a high degree of correlation with stigma length (Takano-Kai et al. 2011; Liu et al. 2015; Zhou et al. 2017; Khumto et al. 2018; Zhu et al. 2023). Dozens of QTLs associated with pistil morphology have been identified (Yan et al. 2009; Uga et al. 2010; Liu et al. 2015; Marathi and Jena. 2015; Dang et al. 2016, 2020; Khumto et al. 2018; Tan et al. 2022a, b). Among those, qSTL3/SYL3 (LOC_Os03g14850), encoding a MADS box family gene, was proved to be responsible for the length of stigma and style (Liu et al. 2015; Dang et al. 2022). Furthermore, a genome-wide association study on pistil characteristics identified a new gene locus, OsSYL2, which controlled style length (Dang et al. 2020). In conclusion, spikelet and pistil are flower organs in rice that share typical developmental mechanisms, which collectively determine SER.
To address the challenge posed by the limited understanding of molecular basis of SER, we investigated the genetic basis of SER using the RIL population developed from B805D-MR-16-8-3 (B805D) and Hua6S, and constructed RHL populations to validate genetic effects of two QTLs, qSE1 and qSE9. Furthermore, qSE1 was narrowed down to an interval of 246.5 kb. A combination of expression analysis, sequencing and haplotype analysis revealed that a single nucleotide variation (T to C) in the 5’UTR of the candidate gene LOC_Os01g72020 (OsBOP1) was likely to be the functional variation of qSE1. Additionally, qSE9 was fine-mapped to a 341.4 kb region. These findings present new QTLs and potential genes that could be useful in developing MSLs with high out-crossing rate, contributing to increasing seed production and decreasing seed cost for hybrid rice.
Materials and Methods
Population and Cultivation
The detailed process of population development was illustrated in Fig. S1a. A population consisting of 135 RILs was derived from a cross between B805D-MR-16-8-3 (B805D) and Hua6S (Zhou et al. 2021). B805D-MR-16-8-3 is an indica variety sourced from the rice core germplasm collection (Zhao et al. 2014), and exhibits high SER values (Fig. S2a–d; Table S2). Hua6S is a photoperiod-thermo-sensitive genic male sterile line developed by Huazhong Agricultural University. The RHLs carrying heterozygous qSE1 and qSE9 regions were developed through a series of self-crosses via maker-assisted selection (MAS) respectively (Fig. S1a).
The RIL populations were planted in year 2017, while RHL populations of qSE1 and qSE9, and recombinants were planted from year 2019 to 2021, during the normal rice growing seasons at the Experimental Farm of Huazhong Agricultural University in Wuhan, China (N 30.49°, E 114.36°).
Trait Evaluation
To determine the degree of SER, ten main stem panicles were collected from each line after the end of flowering for RILs, while three panicles were collected from each plant for RHLs and recombinants. By counting the number of spikelets with single exserted stigma (SES), dual exserted stigma (DES), and no exserted stigma (NES), the SER was calculated (Fig. S1c). The SER was further divided into four traits: single stigma exsertion rate (SSE), dual stigma exsertion rate (DSE), total stigma exsertion rate (TSE) and percentage of spikelets with exserted stigma (PSE). The four SER traits were calculated using the following formulas:
For the observation of pistil morphology, ten panicles were collected from each line during the flourishing florescence for RHLs. Flowering spikelets were randomly selected to carefully separate pistils from glumes, and then photos were taken under a microscopes system (Eclipse Ni-E, Nikon, Tokyo, Japan). Pistil morphology was illustrated in Fig. S1b, and it was divided into stigma length, style length, stigma width, stigma angle, and style angle. All traits were measured using the ImageJ software (Schneider et al. 2012).
QTL Analysis and Marker Development
The genetic linkage map used for QTL mapping was constructed in a previous study (Zhou et al. 2021). QTL mapping was performed by the composite interval mapping (CIM) method using WinQTLCart2.5 software (Wang et al. 2012) with logarithm of odds (LOD) values over 2.5 as the threshold. The graphic of QTL distribution on the rice genome was drawn by R package RIdeogram (Hao et al. 2020). To validate the QTLs detected from RILs, SNP makers flanking the QTLs were replaced with closely linked InDel markers designed using the RiceVarMap (Zhao et al. 2014) and Primer Premier 6 software (Premier Biosoft Interpairs, Palo Alto, CA). Molecular markers for fine mapping of qSE1 and qSE9 were also designed using the same approach. Relevant primer sequences were shown in Table S1.
Total RNA Extraction and qRT-PCR
Total RNA was extracted from pistil separated from young panicles at the pre-heading stage of rice using Trizol reagent (Invitrogen, Carlsbad, CA, USA). Total RNA was reverse transcribed to generate cDNA using the HiScript II 1st Strand cDNA Synthesis Kit (Vazyme biotech co, Itd, Nanjing, China). Gene expression was measured by qRT-PCR using the OsActin (LOC_Os03g50885) as an internal control. PCR was performed via the Applied Biosystems Quant Studio 6 Real-Time PCR System (Thermo Fisher Scientific, USA). All primers used for qRT-PCR were shown in Table S1.
Haplotype and Statistical Analysis
SNP and InDel variation information for LOC_Os01g72020 (OsBOP1) were obtained from RiceVarMap (Zhao et al. 2014). The LD heatmap and gene haplotypes were constructed using SNPs and InDels (frequencies > 0.03) from the 2 kb promoter and coding regions using Haploview software (Barrett et al. 2005). Phenotypic data used for haplotype analysis was derived from a previous study (Zhou et al. 2017). The data analysis and figure making were finished with a combination of software Microsoft Excel (Microsoft Corporation, Redmond, Washington, United States), GraphPad Prism 8.0 (GraphPad Software Inc., La Jolla, CA, USA), SPSS 22.0 (IBM Corp. Released 2013. IBM SPSS Statistics for Windows, Version 22.0. Armonk, NY: IBM Corp) and R (Ihaka and Gentleman. 1996).
Results
Phenotypic Description of SER in the Parents and the RIL Population
The SER performance of parents and the RIL population was assessed, which was subdivided into four traits, namely DSE, SSE, TSE and PSE. B805D displayed significantly higher values than Hua6S for all four traits. The DSE, SSE, TSE and PSE values of B805D were 6.89%, 40.36%, 27.07% and 47.25% respectively, while that of Hua6S were 3.92%, 25.84%, 16.83% and 29.75% respectively (Table S2). The phenotypic in the RIL population ranged from 50.02 to 0.44% for DSE (Fig. S2a), from 52.81 to 9.55% for SSE (Fig. S2b), from 79.46 to 5.42% for TSE (Fig. S2c) and from 94.58 to 10.40% for PSE (Fig. S2d). The SSE, TSE and PSE followed a normal distribution, while the DSE displayed deviation from the normal distribution, based on Q–Q plots (quantile–quantile plot) and Kolmogorov-Smirnov (K-S) tests (Fig. S3; Table S2) (Ghasemi and Zahediasl. 2012).
The correlation coefficients among SSE, DSE, TSE and PSE were shown in Fig. S2e. The four traits were significantly positively correlated with each other. The highest correlation coefficient was 0.98 between TSE and PSE, while the lowest was 0.39 between DSE and SSE.
QTL Mapping of SER in the RIL Population
A total of 14 QTLs associated with SER were identified, distributed on chromosomes 1, 2, 4, 5, 6, 8 and 9 respectively (Table 1; Fig. S4). The phenotypic variance explained by each QTL ranged from 20.15 to 6.63%, with LOD scores ranging from 7.47 to 2.52 (Table 1).
Table 1.
Putative QTLs for SER detected in the RIL population
| Trait | QTL | Chr | Physical interval (bp) | LOD | Add | PVE (%) |
|---|---|---|---|---|---|---|
| DSE | qDSE1/qSE1 | 1 | 41,923,017–42,574,450 | 3.6 | −4.63 | 9.64 |
| qDSE4/qSE4 | 4 | 25,401,580–29,142,744 | 3.3 | −4.04 | 8.35 | |
| qDSE8 | 8 | 3,316,744–3,602,540 | 2.52 | 3.59 | 6.63 | |
| qDSE9/qSE9 | 9 | 19,239,288–21,383,102 | 5.65 | 5.5 | 15.47 | |
| SSE | qSSE2/qSE2 | 2 | 10,659,278–18,494,768 | 4.07 | 3.22 | 10.3 |
| qSSE5 | 5 | 2,297,474–5,361,872 | 3.86 | −3.16 | 9.89 | |
| qSSE9/qSE9 | 9 | 19,248,045–21,615,415 | 7.47 | 4.37 | 20.15 | |
| TSE | qTSE1/qSE1 | 1 | 42,328,822–42,574,450 | 3.22 | −4.73 | 8.05 |
| qTSE4/qSE4 | 4 | 28,358,979–30,075,347 | 4 | −5.13 | 9.97 | |
| qTSE9/qSE9 | 9 | 19,2392,88–21,615,415 | 6.02 | 6.41 | 15.69 | |
| PSE | qPSE2/qSE2 | 2 | 10,618,982–17,246,109 | 4.15 | 7.09 | 10.02 |
| qPSE4/qSE4 | 4 | 25,401,580–30,011,860 | 3.86 | − 5.92 | 9.17 | |
| qPSE6 | 6 | 12,795,997–19,609,502 | 3.83 | 5.97 | 9.2 | |
| qPSE9/qSE9 | 9 | 19,239,288–21,383,102 | 5.6 | 7.43 | 13.87 |
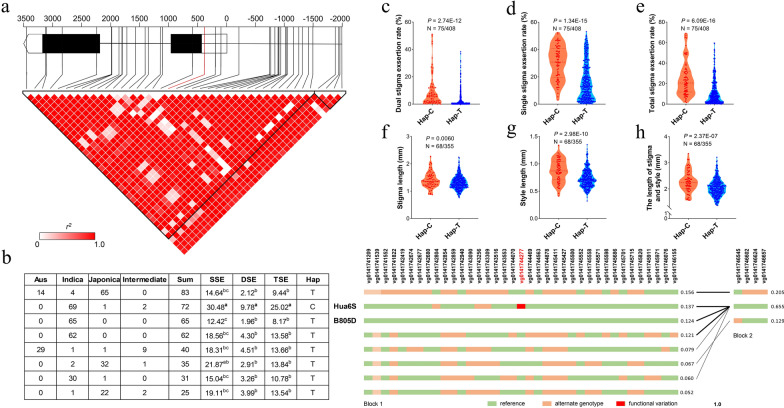
In the column of QTL, the co-located QTLs for SER were renamed as qSE, and different renamed QTLs were shown in different color. Chr, chromosome; LOD, logarithm of odds; Add, additive effect; PVE, the phenotypic variance explained by each QTL
Four QTLs were detected for DSE on chromosomes 1, 4, 8 and 9 (Table 1; Fig. S4). Among them, the QTL qDSE9 explained the highest phenotypic variance of 15.47% with a LOD value of 5.65. qDSE8 explained 6.63% of the phenotypic variance with a LOD value of 2.52. Alleles from B805D increased DSE for qDSE9 and qDSE8, with additive effects of 5.50% and 3.59%, respectively. In contrast, alleles from Hua6S increased DSE for qDSE1 and qDSE4, exhibiting additive effects of 4.63% and 4.04%, and explaining 9.64% and 8.35% of the phenotypic variation, respectively.
Three QTLs were identified for SSE on chromosomes 2, 5, and 9 (Table 1; Fig. S4). The alleles from B805D increased SSE for qSSE2 and qSSE9. The QTL qSSE9 explained 20.15% of the phenotypic variation with a LOD value of 7.47. The QTL qSSE2 exhibited a phenotypic variance of 10.30% with a LOD value of 4.07. Additionally, qSSE5 explained 9.89% of the phenotypic variation with a LOD value of 3.86, with the allele from Hua6S increasing SSE.
Three QTLs for TSE and four QTLs for PSE were identified on chromosomes 1, 2, 4, 6 and 9, respectively (Table 1; Fig. S4). qTSE9 and qPSE9 explained 15.69% and 13.87% of the TSE and PSE variation respectively, and coincided with the genomic region of qDSE9 and qSSE9 (Table 1). This locus had the highest impact on SER and was renamed as qSE9. The qPSE2 region accounted for 10.02% of the variation in PSE, and overlapped with the qSSE2 region, which was renamed as qSE2 (Table 1). qTSE4 and qPSE4 accounted for 9.17% and 9.97% of the variation in TSE and PSE, respectively, and were co-located with the qDSE4 region, which was renamed as qSE4 (Table 1). The qTSE1 locus accounted for 8.05% of the variation in TSE and co-localized with qDSE1 within a QTL cluster, which was renamed as qSE1 (Table 1).
Validation of qSE1 and qSE9 Using RHL Populations
To confirm the genetic effects of identified QTLs, we developed residual heterozygous line (RHL) populations for each QTL through a series of self-crosses and maker-assisted selection (MAS), and then reconducted QTL analysis (Fig. S1a). Two QTLs, qSE1 and qSE9, significantly influenced SER in RHL populations (Fig. 1; Table S3, S4).
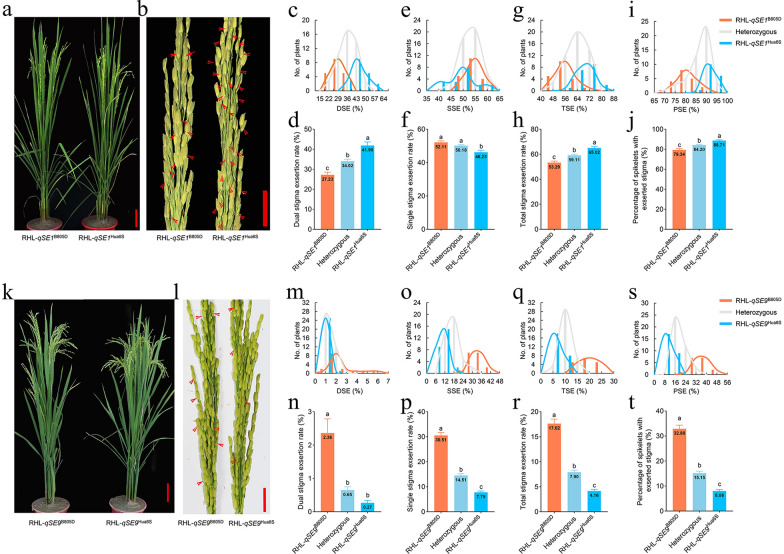
Fig. 1.
The phenotype of RHLs for qSE1 and qSE9. Plant type of RHL-qSE1B805D and RHL-qSE1Hua6S (a), RHL-qSE9B805D and RHL-qSE9Hua6S (k) at heading stage. Scale bar, 10 cm. Panicle of RHL-qSE1B805D and RHL-qSE1Hua6S (b), RHL-qSE9B805D and RHL-qSE9Hua6S (l). The red arrows pointed to exserted stigmas. Scale bar, 2 cm Frequency distribution of dual stigma exsertion rate (DSE) (c, m), single stigma exsertion rate (SSE) (e, o), total stigma exsertion rate (TSE) (g, q), percentage of spikelets with exserted stigma (PSE) (i, s) in the RHL population. Comparison of DSE (d, n), SSE (f, p), TSE (h, r), and PSE (j, t) in the RHL population. Different letters indicated significant differences at p < 0.05 (one-way analysis of variance followed by Tukey’s multiple-comparison test)
qSE1 explained 31.77% of the variation for DSE, 12.49% for SSE, 33.45% for TSE, and 29.94% for PSE in the RHL population (Table S3). Significant difference was observed among alleles for all traits, except for the SSE between lines carrying the B805D genotype and the heterozygous genotype (Fig. 1c–j; Table S4). RHL-qSE1B805D (homozygous for the B805D qSE1 allele in the RHL background) and RHL-qSE1Hua6S (homozygous for the Hua6S qSE1 allele in the RHL background) exhibited differences of 14.67% for DSE, 11.74% for TSE, 8.81% for PSE. However, the SSE of RHL-qSE1B805D was only 5.86% higher than that of RHL-qSE1Hua6S, which could be attributed to our calculation method based on counting spikelets with exerted stigma on rice panicle. RHL-qSE1Hua6S showed a relatively high number of spikelets with dual-exerted stigma, resulting in a decreased number of spikelets with single-exerted stigma per panicle. These findings suggested that qSE1 is a genetic locus primarily impacting DSE, thereby influencing TSE and PSE.
qSE9 explains 65.39% of the variation for DSE, 10.66% for SSE, 11.07% for TSE, and 11.37% for PSE in the RHL population (Table S3). Significant difference was observed among alleles for all traits (Fig. 1m–t; Table S4). RHL-qSE9Hua6S (homozygous for the Hua6S qSE9 allele in the RHL background) and RHL-qSE9B805D (homozygous for the B805D qSE9 allele in the RHL background) displayed differences of 2.09% for DSE, 22.73% for SSE, 13.45% for TSE, and 24.82% for PSE.
Floral Characteristics in the RHL Populations
To characterize the role of qSE1 and qSE9 in influencing SER, we examined the spikelet and pistil traits in RHLs of qSE1 and qSE9.
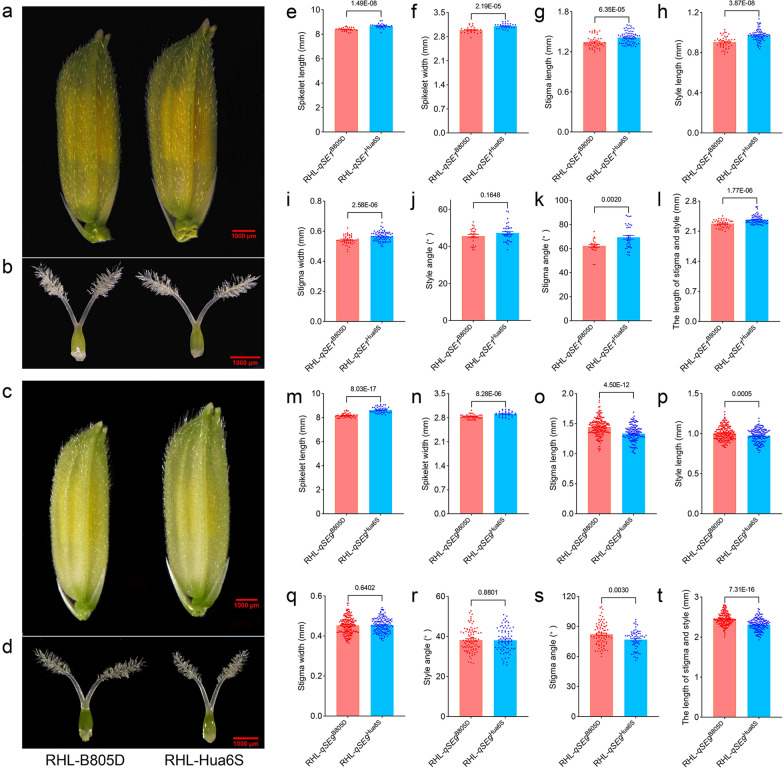
For qSE1, the spikelet length and width of RHL-qSE1Hua6S were higher than those of RHL-qSE1B805D (Fig. 2a, e and f; Table S5). Correspondingly, RHL-qSE1Hua6S, which had higher SER values except for SSE (Fig. 1c–j; Table S4), displayed increased style length and stigma length as well as width, along with an increase in stigma angle and the length of stigma and style (Fig. 2b and g–l; Table S5). Correlation analysis showed significant positive correlation between SER traits (except for SSE) and floral organ traits (Fig. S5a). These findings clearly demonstrate that qSE1 synchronously regulates spikelet size, pistil growth and the SER in rice.
Fig. 2.
Floral morphology of RHLs for qSE1 and qSE9. Spikelet morphology of RHLs for qSE1 (a) and qSE9 (c). Pistil morphology of RHLs for qSE1 (b) and qSE9 (d). Scale bar, 1000 um. Spikelet length (e, m), spikelet width (f, n), stigma length (g, o), style length (h, p), stigma width (i, q), style angle (j, r), stigma angle (k, s), and the length of stigma and style (i, t) of RHLs for qSE1 and qSE9, respectively. P values were based on two-tailed Student’s t test
For qSE9, the spikelet length and width of RHL-qSE9B805D were lower than those of RHL-qSE9Hua6S (Fig. 2c, m and n; Table S5). The pistil traits of the RHL- qSE9B805D, including stigma length, style length, stigma angle and the length of stigma and style, were significantly higher than those of RHL-qSE9Hua6S (Fig. 2d and o–t; Table S5). There was a significant positive correlation between SER traits and pistil length, but negative correlation with spikelet length and width (Fig. S5b). These results indicate that the qSE9 allele from B805D has positive effects on pistil traits to increase SER, but negatively affects spikelet size simultaneously.
Fine Mapping of qSE1
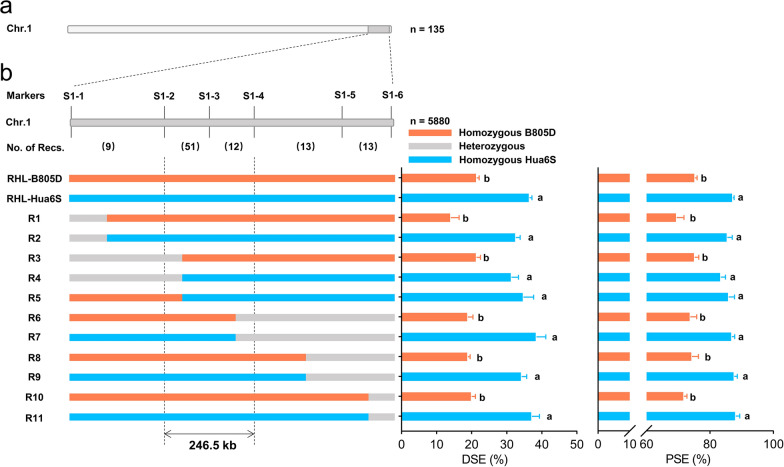
To fine map qSE1, a segregating population consisting of 5880 individuals was developed from several F10 plants carrying heterozygous qSE1 region, and 217 recombinant individuals were identified by DNA markers S1-1 and S1-6 (Table S1). The effect of qSE1 was further confirmed using 360 RHLs at F11 generation. Significant difference was observed among alleles for DSE, TSE and PSE in RHLs (Table S4; Fig. S6). For individuals with homozygous B805D genotype, DSE ranged from 29.80 to 12.37%, with a mean value of 21.41%; TSE ranged from 55.64 to 37.63%, with a mean value of 48.35%; PSE ranged from 83.42 to 62.90%, with a mean value of 75.28%. For individuals with homozygous Hua6S genotype, DSE ranged from 49.78 to 27.04%, with a mean value of 36.45%; TSE ranged from 71.65 to 53.62%, with a mean value of 61.84%; PSE ranged from 94.35 to 80.19% with a mean value of 87.24% (Table S4; Fig. S6).
In order to accurately deduce the genotype of qSE1, recombinants with DSE values less than 20.00% and TSE values less than 48.00% were tentatively defined to carry homozygous qSE1 regions from B805D, while recombinants with DSE values greater than 35.00% and TSE values greater than 60.00% were tentatively defined to carry homozygous qSE1 regions from Hua6S. Thus, 98 out of 217 recombinants were selected for further study (Table S6). Subsequently, four novel InDel markers were developed in the qSE1 region (Table S1), and used to conduct genotyping of 98 recombinants. 11 recombinant types were classified based on the genotype of six markers (Fig. 3b; Table S6). The mean phenotypic values of all recombinants belonging to the same recombinant type were calculated and defined as the values of each recombinant type, which was exploited to accurately deduce the genotype of qSE1. Based on the analysis of marker genotype and qSE1 genotype of 11 recombinant types, qSE1 was finally delimited to a region of 246.5 kb between markers S1-2 and S1-4 (Fig. 3b).
Fig. 3.
Fine mapping of qSE1. a The qSE1 locus was initially mapped on the long arm of chromosome 1 using 135 RILs, with the dark grey region indicating its approximate interval. bqSE1 was finely mapped to a region of 246.5 kb. Six markers were used to perform genotyping of recombinants screened from 5880 F11 plants. The numbers in brackets indicated the number of recombinant crossover events occurred between flanking markers. Graphical genotypes and phenotype statistics were shown for different recombinant types. Different letters indicated significant differences at p < 0.05 (one-way analysis of variance followed by Tukey’s multiple-comparison test)
Fine Mapping of qSE9
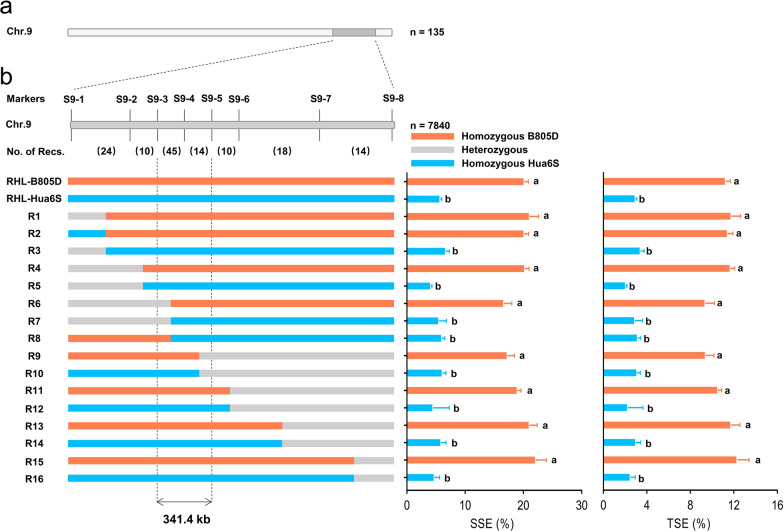
To narrow down the qSE9 interval, a segregating population consisting of 7840 individuals was developed from several F10 plants carrying heterozygous qSE9 region, and 238 recombinant individuals were identified by DNA markers S9-1 and S9-8 (Table S1). The effect of qSE9 was further confirmed using 354 RHLs at F11 generation (Table S4; Fig. S7). For individuals with homozygous B805D genotype, SSE ranged from 31.28 to 15.37%, with a mean value of 20.15%; TSE ranged from 17.57 to 8.08%, with a mean value of 11.24%; PSE ranged from 33.21 to 15.65%, with a mean value of 21.38%. For individuals with homozygous Hua6S genotype, SSE ranged from 9.24 to 2.02%, with a mean value of 5.70%; TSE ranged from 4.96 to 1.01%, with a mean value of 2.94%; PSE ranged from 9.58 to 2.02%, with a mean value of 5.81% (Table S4; Fig. S7).
Taking that no overlap was observed for each SER trait between individuals with B805D genotype and Hua6S genotype into consideration, 135 out of 238 recombinants with tentatively defined homozygous qSE9 regions from B805D or Hua6S were selected for further study (Table S7). Subsequently, six novel InDel markers were developed in the qSE9 region (Table S1), and used to conduct genotyping of 135 recombinants. 16 recombinant types were classified based on the genotype of eight markers (Fig. 4b; Table S7). Based on the analysis of marker genotype and qSE1 genotype of 16 recombinant types, qSE9 was finally delimited to an interval between S9-3 and S9-5 with an estimated length of 341.4 kb (Fig. 4b).
Fig. 4.
Fine mapping of qSE9. a The qSE9 locus was initially mapped on chromosome 9 using 135 RILs, with the dark grey region indicating its approximate interval. b Fine mapping qSE9 to a 341.4 Kb region. Eight markers were used to perform genotyping of recombinants screened from 7840 F11 plants. The numbers in brackets indicated the number of recombinant crossover events occurred between flanking markers. Graphical genotypes and phenotype statistics were shown for different recombinant types. Different letters indicated significant differences at p < 0.05 (one-way analysis of variance followed by Tukey’s multiple-comparison test)
Identification of Candidate Genes for qSE1 and qSE9
In the candidate region of qSE1, thirty-one predicted ORFs were annotated in the reference genome of Nipponbare (http://rice.uga.edu/, MSU-version_7.0), excluding four encoding retrotransposon or transposon protein (Table S8). The levels of mRNA expression of thirty-one genes in pistil of RHL-qSE1Hua6S and RHL-qSE1B805D were examined by qRT-PCR (Fig. S8). The results showed that twelve genes displayed differential expression between the two RHLs, while ten genes did not express in pistil (Fig. S8; Table S8). Notably, among the thirty-one genes, LOC_Os01g72020 (OsBOP1) was found to be involved in the development of leaves and spikelet organ in rice (Toriba et al. 2019). Comparison of genomic sequences revealed four variants between the parental alleles of OsBOP1, including three in the intron and a SNP (T for B805D and C for Hua6S) in the 5’UTR (Fig. 5b). Moreover, a significant differential expression of OsBOP1 was observed between the two RHLs (Fig. S8). Consequently, LOC_Os01g72020 (OsBOP1) emerged as a promising candidate gene underlying qSE1.
Fig. 5.
Haplotype analysis of the candidate gene LOC_Os01g72020 (OsBOP1). a Gene structure and linkage disequilibrium (LD) display of OsBOP1. The predicted functional variation (marked in red) was observed within the 5’UTR of OsBOP1. The color scheme represented the LD relationships between DNA polymorphisms, based on r2 values. Two triangles indicated the two haplotype blocks divided by software Haploview. b Haplotypes of OsBOP1 in the rice core germplasm collection were depicted on the left panel. Different letters indicated significant differences among phenotypic values at p < 0.05 (one-way analysis of variance followed by Tukey’s multiple-comparison test). Variation information was displayed on the right panel. Variation IDs from website RiceVarMap2 were shown on the top (Zhao et al. 2014), with the predicted functional variation highlighted in red. Block1 and 2 correspond to the two triangles in (a). c–h Phenotypic comparison between two haplotypes for dual stigma exsertion rate (DSE) (c), single stigma exsertion rate (SSE) (d), total stigma exsertion rate (TSE) (e), stigma length (f), style length (g), the length of stigma and style (h). p value was calculated based on two-tailed Student’s t tests
The genomic sequence of OsBOP1 was further analyzed using the rice germplasm collection consisting of 533 accessions (Table S9) (Zhao et al. 2014). A total of forty-one polymorphisms were identified in the 2 kb promoter and coding regions of OsBOP1, with high linkage disequilibrium (LD) observed among these variants (Fig. 5a). Four variations were located in the coding regions, while none were identified in the promoter region between B805D and Hua6S (Fig. 5b). Eight major alleles were identified, and the Hua6S allele displayed significantly higher SER values than the other alleles (Fig. 5b), suggesting that variations vg0141742804 and vg0141744277 may serve as functional variants. Notably, variation vg0141744277 was the SNP observed in the 5’UTR between the parent alleles. Based on these findings, we classified the eight alleles into two haplotypes: Hap-T and Hap-C. These two haplotypes exhibited significant differences both in terms of SER values and pistil traits (Fig. 5c–h). Therefore, the SNP in the 5’UTR of LOC_Os01g72020 is likely to be the functional variant underlying qSE1.
Within the qSE9 region, a total of forty-three predicted ORFs were annotated in the reference genome of Nipponbare (http://rice.uga.edu/, MSU-version_7.0), excluding five encoding retrotransposon, transposon and hypothetical protein respectively (Table S10). Among those, nineteen genes displayed differential expression levels in the pistil of RHL-qSE9Hua6S and RHL-qSE9B805D (Fig. S9). Notably, the gene LOC_Os09g32944 (OsSPL18) was previously reported to positively regulate grain width and thickness by influencing cell proliferation to modulate glume development (Yuan et al. 2019). OsSPL18 exhibited significant differential expression in the pistil of the two RHLs. The gene LOC_Os09g32540 (LGD1), which encodes a protein containing a von Willebrand factor type A (vWA) domain, was reported to be involved in pleiotropic regulating of rice vegetative growth and development (Thangasamy et al. 2012). LGD1 exhibited high expression levels in the pistil and showed higher expression in RHL-qSE9B805D than in RHL-qSE9Hua6S (Fig. S9). Additionally, LOC_Os09g32948 encoding a MADS-box protein was reported to play a role in regulating floral organ development (Shen et al. 2023). It showed high expression level in the pistil but no difference was observed between two RHLs (Fig. S9). LOC_Os09g32680 encoding a cyclin protein showed high expression level in the pistil as well (Fig. S9), suggesting its potential role in modulating cell development and division for affecting pistil growth. More work was required to further narrow down the interval of qSE9 and to ascertain the functional gene.
Discussion
Understanding the genetic architecture of SER in rice could facilitate the genetic improvement of SER in MSLs, further enhancing the outcrossing ability of MSLs and hybrid rice seed production. In the past two decades, numerous QTLs for SER and related traits have been identified and are distributed across all 12 chromosomes. In the present study, a total of 14 QTLs were detected from the RIL population derived from a cross betweeen parent Hua6S and B805D, including 4 QTLs for DSE, 3 QTLs for SSE, 3 QTLs for TSE and 4 QTLs for PSE (Table 1). Among those, the QTL qSE9 was close to qTSSL9, a locus influencing the length of stigma and style (Dang et al. 2020). The qSE4 region encompassed the intervals of qSER-4.1 (Xu et al. 2019) and qPSES4 (Wang et al. 2017), and it was suggested that LOC_Os04g43910 (ARF10) may serve as a potential target gene within this genomic region (Guo et al. 2022). The qSE2 region overlapped with the region of qSTL2, a locus conferring stigma length (Dang et al. 2020). qSSE5 coincided with the regions of qTSE-5a (Li et al. 2014a), qSER-5.1 (Xu et al. 2019), and qSER-5 (Tan et al. 2022a, b). qDSE8 was co-located with qTSE8 (Liu et al. 2022). The qPSE6 region overlapped with that of qSER-6.1 (Xu et al. 2019), qPES-6 (Lou et al. 2015), and qTSSL6, a locus influencing the length of stigma and style (Dang et al. 2020). In addition, qSE1 was novel.
Despite numerous QTLs associated with SER and related traits in rice have been identified, cloning of genes conferring high SER remains a great challenge. The utilization of secondary mapping populations, such as near-isogenic line (NIL) population and RHL population, represents an effective approach for fine mapping and cloning of genes for SER(Guo et al. 2022; Liu et al. 2019; Rahman et al. 2017b; Tan et al. 2021, 2022a, b). In the present study, RHL populations of F10 and F11 generations were exploited to validate genetic effects of qSE1 and qSE9, and conduct fine mapping of the two QTLs (Fig. S1). It should be noted that RHL populations were developed from RHLs selected from RILs of F7 generation, due to the great difficulty in identifying RHLs from RILs of higher generations. Thus, RILs of low generations from F5 to F7 are valuable genetic materials for two reasons. On one hand, they carry homozygous regions of more than 93% genome that could facilitate accurate evaluation of target traits for QTL mining. On the other hand, they carry heterozygous regions of less than 7% genome that could facilitate the screening of RHLs of target QTLs for effect validation and fine mapping. In short, a combination of RILs of low generations and derived RHLs provided solid material foundation for fine mapping of qSE1 and qSE9 in this study, which could be of great use in fine mapping of other QTLs of interest.
Previous studies have revealed that SER is influenced by multiple factors, with spikelet shape and pistil size being two key factors. SER related traits are positively correlated with spikelet length, stigma length, style length and stigma angle, but negatively correlated with spikelet width, stigma width and style angle (Dang et al. 2016, 2020, 2022; Virmani et al. 1973; Virmani et al. 1974, 1987; Uga et al. 2003; Yan et al. 2009; Zhou et al. 2017; Zhu et al. 2023). In the present study, both the qSE1 allele from Hua6S and the qSE9 allele from B805D significantly increased stigma length, style length, the length of stigma and style, and stigma angle, and the qSE1 allele from Hua6S increased stigma width additionally (Fig. 2). Thus, both qSE1 and qSE9 increase pistil size to improve SER. In contrast, the qSE1 allele from Hua6S significantly increased spikelet length and width, while the qSE9 allele from B805D decreased spikelet length and width. In addition, several grain shape genes, namely gs3, gw8, gs9, gw5 and gw7, could effectively enhance SER through decreased spikelet width and increased spikelet length (Tan et al. 2023; Zhu et al. 2023). These results suggest that different genes conferring high SER have different effects on spikelet shape, all of which may be likely to increase pistil size. Therefore, we can boldly hypothesize that pistil size rather than spikelet shape is the key factor in determining SER. Cloning the functional genes underlying qSE1 and qSE9 and elucidating their molecular mechanisms will contribute to a better understanding of the relationship among SER, pistil size and spikelet shape.
In the present study, LOC_Os01g72020 (OsBOP1) was identified as the candidate gene for qSE1 with the combination of sequencing, expression and haplotype analysis (Fig. 5). OsBOP1 encodes a Broad Complex BTB domain with Ankyrin repeat region protein, with a high expression level in pistil (http://rice.uga.edu/, MSU-version_7.0). In a previous study, BOP1 and its homologous genes was reported to influence flower development in Arabidopsis (Hepworth et al. 2005) and rice (Toriba et al. 2019). Significant differences in spikelet shape and pistil size were observed between the B805D allele and Hua6S allele of qSE1 in the RHL population (Fig. 2; Table S5). The Hua6S allele exhibited higher expression level in the pistil than the B805D allele (Fig. S8), possibly due to the SNP in the 5’UTR of OsBOP1. As the 5'UTR is of great importance to the regulation of transcription and translation of downstream coding genes (Ryczek et al. 2023), the SNP is likely to be the functional variation underlying qSE1. It is exciting to note that the allele frequency of Hap-C (Hua6S allele) with higher SER is far lower than that of Hap-T (B805D allele) in diverse cultivated rice (Fig. 5b), suggesting that the Hua6S allele of qSE1 could be a promising target in the genetic improvement of male sterile lines.
Conclusion
In this study, we detected 14 QTLs for SER using the RILs derived from B805D and Hua6S, and confirmed the effects of two major QTLs, qSE1 and qSE9, using RHLs. Both qSE1 and qSE9 improved SER by increasing pistil size and were fine mapped to intervals of 246.5 kb and 341.4 kb, respectively. A combination of sequencing, expression and haplotype analysis revealed that LOC_Os01g72020 (OsBOP1) was likely to be the candidate gene for qSE1. The Hua6S allele of qSE1 and B805D allele of qSE9 will be favorable for molecular breeding of MSLs with high outcrossing ability, contributing to the development of hybrid rice and ensuring global food security.
Supplementary Information
Acknowledgements
Not applicable.
Abbreviations
- MSL
Male sterile line
- SER
Stigma exsertion rate
- SSSLs
Single-segment substitution lines
- RIL
Recombinant inbred line
- QTL
Quantitative trait loci
- RHL
Residual heterozygous line
- SES
Spikelets with single exserted stigma
- DES
Spikelets with dual exserted stigma
- NES
Spikelets with no exserted stigma
- DSE
Dual stigma exsertion rate
- SSE
Single stigma exsertion rate
- TSE
Total stigma exsertion rate
- PSE
Percentage of spikelets with exserted stigma
- MSL
Male sterile line
- MAS
Maker-assisted selection
- SNP
Single nucleotide polymorphism
Author Contributions
Professor Yuqing He designed and supervised the study; Mr Hanyuan Yang performed the experiments; Miss Yin Zhou, Miss Enyu Liu, Miss Ping Sun,Dr Rongjia Liu, Mr Haozhou Gao, Mr Zerui Xu, Miss Ping Yang, Miss Xingyue Wang participated in the experiments. Miss Yiting Ao provided the plant materials. Mr Guanjun Gao, Mr Qinglu Zhang and Dr Lizhong Xiong help to field management. Mr Hanyuan Yang and Miss Yin Zhou analyzed the date; Mr Hanyuan Yang wrote the manuscript. Mr Yin Zhou, Dr Pingbo Li and Professor Yuqing He revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by grants from STI 2030-Major Project(2023ZD04069), National Natural Science Foundation of China (U21A20211, 31821005), the Ministry of Science and Technology (2022YFD1200100, 2021YFF1000200), AgroST Project (NK20220501) and the Earmarked fund of China Agriculture Research System (CARS-01-01).
Data Availability
No datasets were generated or analysed during the current study.
Declarations
Ethics Approval and Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Competing Interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Bakti C, Tanaka J (2019) Detection of dominant QTLs for stigma exsertion ratio in rice derived from Oryza rufipogon accession “W0120.” Breed Sci 69(1):143–150. 10.1270/jsbbs.18139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21(2):263–265. 10.1093/bioinformatics/bth457 [DOI] [PubMed] [Google Scholar]
- Dang X, Liu E, Liang Y, Liu Q, Breria CM, Hong D (2016) QTL detection and elite alleles mining for stigma traits in Oryza sativa by association mapping. Front Plant Sci 7:1188. 10.3389/fpls.2016.01188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang X, Yang Y, Zhang Y, Chen X, Fan Z, Liu Q-m, Ji J, Li D, Li Y, Fang B, Wu ZF, Liu E, Hu X, Zhu S, She D, Wang H, Li Y, Chen S, Wu Y, Hong D (2020) OsSYL2AA, an allele identified by gene-based association, increases style length in rice (Oryza sativa L.). Plant J 104:1491–1503. 10.1111/tpj.15013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang X, Zhang Y, Li Y, Chen S, Liu E, Fang B, Liu Q, She D, Dong Z, Fan Z, Li D, Wang H, Zhu S, Hu X, Li Y, Jiang J, Hong D (2022) SYL3-k increases style length and yield of F1 seeds via enhancement of endogenous GA4 content in Oryza sativa L. pistils. Theor Appl Genet 135(1):321–336. 10.1007/s00122-021-03968-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Namaky R (2018) The genetic variability of floral and agronomic characteristics of newly-bred cytoplasmic male sterile rice. Agriculture 8:1–11. 10.3390/agriculture8050068 [Google Scholar]
- Fan Y, Li Y (2019) Molecular, cellular and Yin-Yang regulation of grain size and number in rice. Mol Breed 39(12):163. 10.1007/s11032-019-1078-0 [Google Scholar]
- Ghasemi A, Zahediasl S (2012) Normality tests for statistical analysis: a guide for non-statisticians. Int J Endocrinol Metab 10(2):486–489. 10.5812/ijem.3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo N, Wang Y, Chen W, Tang S, An R, Wei X, Hu S, Tang S, Shao G, Jiao G, Xie L, Wang L, Sheng Z, Hu P (2022) Fine mapping and target gene identification of qSE4, a QTL for stigma exsertion rate in rice (Oryza sativa L.). Front Plant Sci. 10.3389/fpls.2022.959859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Z, Lv D, Ge Y, Shi J, Weijers D, Yu G, Chen J (2020) RIdeogram: drawing SVG graphics to visualize and map genome-wide data on the idiograms. PeerJ Comput Sci 6:e251. 10.7717/peerj-cs.251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan MJ, Kulsum U, Rahman MH, Akter A, Shamsuddin AKM (2014) Comparative study of floral characteristics in the component lines of hybrid rice (Oryza sativa L.). Bangladesh J Bot 43(1):1–8. 10.3329/bjb.v43i1.19739 [Google Scholar]
- Hepworth SR, Zhang Y, McKim S, Li X, Haughn GW (2005) BLADE-ON-PETIOLE–dependent signaling controls leaf and floral patterning in arabidopsis. Plant Cell 17(5):1434–1448. 10.1105/tpc.104.030536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Kurata N, Wei X, Wang Z-X, Wang A, Zhao Q, Zhao Y, Liu K, Lu H, Li W, Guo Y, Lu Y, Zhou C, Fan D, Weng Q, Zhu C, Huang T, Zhang L, Wang Y, Feng L, Furuumi H, Kubo T, Miyabayashi T, Yuan X, Xu Q, Dong G, Zhan Q, Li C, Fujiyama A, Toyoda A, Lu T, Feng Q, Qian Q, Li J, Han B (2012) A map of rice genome variation reveals the origin of cultivated rice. Nature 490(7421):497–501. 10.1038/nature11532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihaka R, Gentleman R (1996) R: a language for data analysis and graphics. J Comput Graph Stat 5(3):299–314. 10.1080/10618600.1996.10474713 [Google Scholar]
- Jiang J, Xu L, Xiao M, Hu C, Zhang Y, Wang D, Dang X (2021) Genetic analysis and QTLs identification of stigma traits in japonica rice (Oryza sativa L.). Euphytica 217(5):82. 10.1007/s10681-021-02813-z [Google Scholar]
- Khumto S, Pusadee T, Olsen KM, Jamjod S (2017) Genetic relationships between anther and stigma traits revealed by QTL analysis in two rice advanced-generation backcross populations. Euphytica 214(1):5. 10.1007/s10681-017-2091-1 [Google Scholar]
- Khumto S, Sreethong T, Pusadee T, Rerkasem B, Jamjod S (2018) Variation of floral traits in Thai rice germplasm (Oryza sativa). Genet Resour Crop Evol 65(4):1123–1132. 10.1007/s10722-017-0600-7 [Google Scholar]
- Li C, Sun C, Mu P, Chen L, Wang X (2001) QTL analysis of anther length and ratio of stigma exsertion, two key traits of classification for cultivated rice (Oryza sativa L) and common wild rice (O. rufipogon Griff.). Acta Genet Sin 28:746–751 [PubMed] [Google Scholar]
- Li P, Feng F, Zhang Q, Chao Y, Gao G, He Y (2014a) Genetic mapping and validation of quantitative trait loci for stigma exsertion rate in rice. Mol Breed 34(4):2131–2138. 10.1007/s11032-014-0168-2 [Google Scholar]
- Li P, Su G, Feng F, Wang P, Yu S, He Y (2014b) Mapping of minor quantitative trait loci (QTLs) conferring fertility restoration of wild abortive cytoplasmic male sterility and QTLs conferring stigma exsertion in rice. Plant Breed 133(6):722–727. 10.1111/pbr.12220 [Google Scholar]
- Liu Q, Qin J, Li T, Liu E, Fan D, Edzesi WM, Liu J, Jiang J, Liu X, Xiao L, Liu L, Hong D (2015) Fine mapping and candidate gene analysis of qSTL3, a stigma length-conditioning locus in rice (Oryza sativa L.). PLoS ONE 10(6):e0127938. 10.1371/journal.pone.0127938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhang A, Wang F, Kong D, Li M, Bi J, Zhang F, Wang J, Luo X, Pan Z, Yu X, Liu G, Luo L (2019) Fine mapping a quantitative trait locus, qSER-7, that controls stigma exsertion rate in rice (Oryza sativa L.). Rice 12(1):46. 10.1186/s12284-019-0304-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Fu D, Kong D, Ma X, Zhang A, Wang F, Wang L, Xia H, Liu G, Yu X, Luo L (2022) Linkage mapping and association analysis to identify a reliable QTL for stigma exsertion rate in rice. Front Plant Sci. 10.3389/fpls.2022.982240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou J, Yue GH, Yang WQ, Mei H, Luo L, Lu HJ (2014) Mapping QTLs influencing stigma exertion in rice. Bulg J Agric Sci 20:1450–1456 [Google Scholar]
- Malik T-U-H, Baba A (2018) Techniques hybrid seed production in rice. J Pharmacogn Phytochem 7:962–967 [Google Scholar]
- Marathi B, Jena KK (2015) Floral traits to enhance outcrossing for higher hybrid seed production in rice: present status and future prospects. Euphytica 201(1):1–14. 10.1007/s10681-014-1251-9 [Google Scholar]
- Miyashita Y, Takasugi T, Ito Y (2010) Identification and expression analysis of PIN genes in rice. Plant Sci 178(5):424–428. 10.1016/j.plantsci.2010.02.018 [Google Scholar]
- Miyata M, Yamamoto T, Komori T, Nitta N (2007) Marker-assisted selection and evaluation of the QTL for stigma exsertion under japonica rice genetic background. Theor Appl Genet 114(3):539–548. 10.1007/s00122-006-0454-4 [DOI] [PubMed] [Google Scholar]
- Prahalada GD, Marathi B, Vinarao R, Kim SR, Diocton R, Ramos J, Jena KK (2021) QTL mapping of a novel genomic region associated with high out-crossing rate derived from Oryza longistaminata and development of new CMS lines in Rice, O Sativa l. Rice 14(1):80. 10.1186/s12284-021-00521-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Q, Guo L, Smith SM, Li J (2016) Breeding high-yield superior quality hybrid super rice by rational design. Natl Sci Rev 3(3):283–294. 10.1093/nsr/nww006 [Google Scholar]
- Rahman MH, Zhang YX, Sun LP, Zhang KQ, Rahman MS, Wu WX, Zhan XD, Cao LY, Cheng SH (2017a) Genetic mapping of quantitative trait loci for the stigma exsertion rate in rice (Oryza sativa L.). J Integrat Agric 16(7):1423–1431. 10.1016/S2095-3119(16)61540-X [Google Scholar]
- Rahman MH, Zhang Y, Zhang K, Rahman MS, Barman HN, Riaz A, Chen Y, Wu W, Zhan X, Cao L, Cheng S (2017b) Genetic dissection of the major quantitative trait locus (qSE11), and its validation as the major influence on the rate of stigma exsertion in rice (Oryza sativa L.). Front Plant Sci 8:1818. 10.3389/fpls.2017.01818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D, Ding C, Qian Q (2023) Molecular bases of rice grain size and quality for optimized productivity. Sci Bull 68(3):314–350. 10.1016/j.scib.2023.01.026 [DOI] [PubMed] [Google Scholar]
- Ryczek N, Łyś A, Makałowska I (2023) The functional meaning of 5’UTR in protein-coding genes. Int J Mol Sci. 10.3390/ijms24032976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito H, Okumoto Y, Yoshitake Y, Inoue H, Yuan Q, Teraishi M, Tsukiyama T, Nishida H, Tanisaka T (2011) Complete loss of photoperiodic response in the rice mutant line X61 is caused by deficiency of phytochrome chromophore biosynthesis gene. Theor Appl Genet 122(1):109–118. 10.1007/s00122-010-1426-2 [DOI] [PubMed] [Google Scholar]
- Sasaki A, Yamaji N, Yokosho K, Ma JF (2012) Nramp5 is a major transporter responsible for manganese and cadmium uptake in rice. Plant Cell 24(5):2155–2167. 10.1105/tpc.112.096925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9(7):671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C, Zhang Y, Li G, Shi J, Wang D, Zhu W, Yang X, Dreni L, Tucker MR, Zhang D (2023) MADS8 is indispensable for female reproductive development at high ambient temperatures in cereal crops. Plant Cell 36(1):65–84. 10.1093/plcell/koad246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sripinyowanich S, Klomsakul P, Boonburapong B, Bangyeekhun T, Asami T, Gu H, Buaboocha T, Chadchawan S (2013) Exogenous ABA induces salt tolerance in indica rice (Oryza sativa L.): the role of OsP5CS1 and OsP5CR gene expression during salt stress. Environ Exp Bot 86:94–105. 10.1016/j.envexpbot.2010.01.009 [Google Scholar]
- Sun J, Song W, Chang Y, Wang Y, Lu T, Zhang Z (2022) OsLMP1, encoding a deubiquitinase, regulates the immune response in rice. Front Plant Sci. 10.3389/fpls.2021.814465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano-Kai N, Doi K, Yoshimura A (2011) GS3 participates in stigma exsertion as well as seed length in rice. Breed Sci 61(3):244–250. 10.1270/jsbbs.61.244 [Google Scholar]
- Tan Q, Zou T, Zheng M, Ni Y, Luan X, Li X, Yang W, Yang Z, Zhu H, Zeng R, Liu G, Wang S, Fu X, Zhang G (2020) Substitution mapping of the major quantitative trait loci controlling stigma exsertion rate from Oryza glumaepatula. Rice 13(1):37. 10.1186/s12284-020-00397-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Q, Wang C, Luan X, Zheng L, Ni Y, Yang W, Yang Z, Zhu H, Zeng R, Liu G, Wang S, Zhang G (2021) Dissection of closely linked QTLs controlling stigma exsertion rate in rice by substitution mapping. Theor Appl Genet 134(4):1253–1262. 10.1007/s00122-021-03771-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Q, Bu S, Chen G, Yan Z, Chang Z, Zhu H, Yang W, Zhan P, Lin S, Xiong L, Chen S, Liu G, Liu Z, Wang S, Zhang G (2022a) Reconstruction of the high stigma exsertion rate trait in rice by pyramiding multiple QTLs. Front Plant Sci. 10.3389/fpls.2022.921700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Q, Haitao Z, Hui L, Yuerong N, Shengze W, Xin L, Junwei L, Yang W, Zifeng Y, Ruizhen Z, Guifu L, Wang S, Zhang G (2022b) Fine mapping of QTLs for stigma exsertion rate from Oryza glaberrima by chromosome segment substitution. Rice Sci 29:55–66. 10.1016/j.rsci.2021.12.005 [Google Scholar]
- Tan Q, Chen S, Gan Z, Lu Q, Yan Z, Chen G, Lin S, Yang W, Zhao J, Ba Y, Zhu H, Bu S, Liu G, Liu Z, Wang S, Zhang G (2023) Grain shape is a factor affecting the stigma exsertion rate in rice. Front Plant Sci. 10.3389/fpls.2023.1087285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thangasamy S, Chen PW, Lai MH, Chen J, Jauh GY (2012) Rice LGD1 containing RNA binding activity affects growth and development through alternative promoters. Plant J 71(2):288–302. 10.1111/j.1365-313X.2012.04989.x [DOI] [PubMed] [Google Scholar]
- Tian D, Huang S, Duan Y, Wang Y (2004) The relationship between flowering and pollination time and outcrossing rate of male sterile lines in hybrid rice seed production. Hybrid Rice 19(3):50–54 [Google Scholar]
- Toriba T, Tokunaga H, Shiga T, Nie F, Naramoto S, Honda E, Tanaka K, Taji T, Itoh J-I, Kyozuka J (2019) BLADE-ON-PETIOLE genes temporally and developmentally regulate the sheath to blade ratio of rice leaves. Nat Commun 10(1):619. 10.1038/s41467-019-08479-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uga Y, Fukuta Y, Cai HW, Iwata H, Ohsawa R, Morishima H, Fujimura T (2003) Mapping QTLs influencing rice floral morphology using recombinant inbred lines derived from a cross between Oryza sativa L. and Oryza rufipogon Griff. Theor Appl Genet 107(2):218–226. 10.1007/s00122-003-1227-y [DOI] [PubMed] [Google Scholar]
- Uga Y, Siangliw M, Nagamine T, Ohsawa R, Fujimura T, Fukuta Y (2010) Comparative mapping of QTLs determining glume, pistil and stamen sizes in cultivated rice (Oryza sativa L.). Plant Breed 129(6):657–669. 10.1111/j.1439-0523.2009.01765.x [Google Scholar]
- Virmani SS, Athwal DS (1973) Genetic variability in floral characteristics influencing outcrossing in Oryza sativa L. Crop Sci 13:66–67. 10.2135/cropsci1973.0011183X001300010019x [Google Scholar]
- Virmani SS, Athwal DS (1974) Inheritance of floral characteristics influencing outcrossing in rice. Crop Sci 14:350–353. 10.2135/cropsci1974.0011183X001400030002x [Google Scholar]
- Virmani SS, Aquino RC, Khush GS (1982) Heterosis breeding in rice (Oryza sativa L.). Theor Appl Genet 63(4):373–380. 10.1007/BF00303911 [DOI] [PubMed] [Google Scholar]
- Wang JR, Hu H, Wang GH, Li J, Chen JY, Wu P (2009) Expression of PIN genes in rice (Oryza sativa L.): tissue specificity and regulation by hormones. Mol Plant 2(4):823–831. 10.1093/mp/ssp023 [DOI] [PubMed] [Google Scholar]
- Wang J, Shi L, Hu P, Tang S, Li W, Wei X, Wu Y, Sheng Z, Zhu Z (2017) QTL mapping of japonica rice stigma exsertion rate. Chin J Rice Sci 31(1):23–30. 10.16819/j.1001-7216.2017.6043 [Google Scholar]
- Wang S, Basten CJ, Zeng ZB (2012) Windows QTL cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC. http://statgen.ncsu.edu/qtlcart/WQTL Cart.htm
- Xie F, Peng S (2016) History and prospects of hybrid rice development outside of China. Chin Sci Bull 61(35):3858–3868. 10.1360/N972016-01018 [Google Scholar]
- Xiong LZ, Liu KD, Dai XK, Xu CG, Zhang Q (1999) Identification of genetic factors controlling domestication-related traits of rice using an F2 population of a cross between Oryza sativa and O. rufipogon. Theor Appl Genet 98(2):243–251. 10.1007/s001220051064 [Google Scholar]
- Xu S, Zheng Y, Liu Y, Guo X, Tan Y, Qian Q, Shu Q, Huang J (2019) Identification of a major quantitative trait locus and its candidate underlying genetic variation for rice stigma exsertion rate. Crop J 7(3):350–359. 10.1016/j.cj.2018.11.006 [Google Scholar]
- Yan WG, Li Y, Agrama HA, Luo D, Gao F, Lu X, Ren G (2009) Association mapping of stigma and spikelet characteristics in rice (Oryza sativa L.). Mol Breed 24(3):277–292. 10.1007/s11032-009-9290-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Zhang Y, Zhang L, Hu J, Zhang X, Lu K, Dong H, Wang D, Zhao F-J, Huang C-F, Lian X (2014) OsNRAMP5 contributes to manganese translocation and distribution in rice shoots. J Exp Bot 65(17):4849–4861. 10.1093/jxb/eru259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Li J, Li H, Ding Y, Wu W, Qin R, Ni J, Xu R, Wei P, Yang J (2023) OsGSTU5 and OsGSTU37 encoding glutathione reductases are required for cadmium tolerance in rice. Int J Environ Sci Technol 20:10253–10260. 10.1007/s13762-022-04550-9 [Google Scholar]
- Yuan L (2014) Development of hybrid rice to ensure food security. Rice Sci 21(1):1–2. 10.1016/S1672-6308(13)60167-5 [Google Scholar]
- Yuan H, Qin P, Li Hu, Zhan S, Wang S, Gao P, Li J, Jin M, Zhengyan Xu, Gao Q, Anping Du, Bin Tu, Chen W, Ma B, Wang Y, Li S (2019) OsSPL18 controls grain weight and grain number in rice. J Genet Genom 46(1):41–51. 10.1016/j.jgg.2019.01.003 [DOI] [PubMed] [Google Scholar]
- Zhang K, Zhang Y, Wu W, Zhan X, Anis GB, Rahman MH, Hong Y, Riaz A, Zhu A, Cao Y, Sun L, Yang Z, Yang Q, Cao L, Cheng S (2018) qSE7 is a major quantitative trait locus (QTL) influencing stigma exsertion rate in rice (Oryza sativa L.). Sci Rep 8(1):14523. 10.1038/s41598-018-32629-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Yao W, Ouyang Y, Yang W, Wang G, Lian X, Xing Y, Chen L, Xie W (2014) RiceVarMap: a comprehensive database of rice genomic variations. Nucleic Acids Res 43(D1):D1018–D1022. 10.1093/nar/gku894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Li P, Xie W, Hussain S, Li Y, Xia D, Zhao H, Sun S, Chen J, Ye H, Hou J, Zhao D, Gao G, Zhang Q, Wang G, Lian X, Xiao J, Yu S, Li X, He Y (2017) Genome-wide association analyses reveal the genetic basis of stigma exsertion in rice. Mol Plant 10(4):634–644. 10.1016/j.molp.2017.01.001 [DOI] [PubMed] [Google Scholar]
- Zhou H, Xia D, Li P, Ao Y, Xu X, Wan S, Li Y, Wu B, Shi H, Wang K, Gao G, Zhang Q, Wang G, Xiao J, Li X, Yu S, Lian X, He Y (2021) Genetic architecture and key genes controlling the diversity of oil composition in rice grains. Mol Plant 14(3):456–469. 10.1016/j.molp.2020.12.001 [DOI] [PubMed] [Google Scholar]
- Zhu X, Gou Y, Heng Y, Ding W, Li Y, Zhou D, Li X, Liang C, Wu C, Wang H, Shen R (2023) Targeted manipulation of grain shape genes effectively improves outcrossing rate and hybrid seed production in rice. Plant Biotechnol J 21(2):381–390. 10.1111/pbi.13959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou T, Zhao H, Li X, Zheng M, Zhang S, Sun L, He N, Pan X, Liu Z, Fu X (2020) QTLs detection and pyramiding for stigma exsertion rate in wild rice species by using the single-segment substitution lines. Mol Breed 40(8):74. 10.1007/s11032-020-01157-1 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed during the current study.