Abstract
Object
The present study aimed to identify hub genes associated with the treatment and control of active and inactive Crohn’s disease (CD).
Methods
Differentially expressed genes (DEGs) were identified in normal, active CD, and inactive CD samples from GSE95095 dataset. Intersection genes screened by Venn diagram in DEGs. Subsequently, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were conducted on the intersection genes. The protein-protein interaction (PPI) network was used to screen of hub gene. The expression and mRNA levels of CXCL12 in CD and ROC curves in GSE95095 dataset. Signaling pathways of hub genes and their correlation with immune cells were analyzed by gene set enrichment analysis (GSEA), EPIC, and ESTIMATE, respectively. Finally, immunohistochemistry (IHC) and Reverse Transcription-Polymerase Chain Reaction (RT-PCR) were used to detect the expression of the hub gene in normal, inactive, and active CD tissues.
Results
In GSE95095 dataset, CXCL12 was identified as the most hub gene by limma analysis, Venn diagram and A protein-protein interaction (PPI) network. CXCL12 expression was highest in active CD (p < 0.001) followed by inactive CD (p < 0.01). Subsequently, it was validated through IHC and RT-PCR in normal intestinal mucosal, active CD, and inactive CD. CXCL12 was overexpressed in active and inactive CD (IHC: p < 0.001 and RT-PCR: p < 0.001, respectively). CXCL12 expression in active CD was determined via analysis with receiver operating characteristic (ROC) curves. The specificity and sensitivity were 0.875 and 0.625, respectively, the accuracy was 72.92%, the area under the curve (AUC) was 0.780, and the 95% confidence interval (CI) was in the range of 0.648–0.912. CXCL12 expression was closely correlated with various immune cells.
Conclusion
CXCL12 is overexpressed in active CD and is closely correlated with various immune cells. We propose that CXCL12 as a potential target genes for the treatment and management of both active and inactive CD.
Keywords: Crohn’s disease, C-X-C motif chemokine 12, gene target, immune cell
Introduction
Crohn’s disease (CD) is also known as localized or regional enteritis. The precise etiology of this intestinal inflammatory disease is unknown. This condition can occur in any part of the gastrointestinal tract.1 However, it is most often confined to the terminal ileum and right colon.2 CD and chronic nonspecific ulcerative colitis (UC) are collectively designated inflammatory bowel disease (IBD).3 In the United States, the incidence of CD is ~58/100,000 children and 119–241/100,000 adults.4,5 CD can affect people of any age and may have a negative impact on the quality of life (QoL).4,6 Clinical symptoms of CD include abdominal pain, diarrhea, intestinal obstruction, and extraintestinal manifestations such as fever and nutritional disorders.7,8 Though there is currently no specific therapy for CD, common treatment approaches include the control of disease progression and the management of symptoms.9 Nevertheless, certain patients with CD require surgical resection of the diseased intestinal tract to help prevent deterioration of overall function.10 The rate of CD recurrence is associated with the extent of the lesion, severity of disease progression, prolongation of disease course, and patient age.11 CD may be the result of exposure to a combination of environmental, immunological, and microbiological factors in genetically susceptible populations.12
Though CD is immune-related, it is not an autoimmune disease.13 At present, it is known that the inflammatory response caused by abnormal immune system reactions in the intestinal mucosa plays an important role in the pathogenesis of IBD. The immune response is related to the release of inflammatory mediators, including cytokines, interleukins, tumor necrosis factor, and the melanocortin system.14,15 Approximately half the total risk of developing CD is related to genetics, and > 70 genes have been linked to CD onset and progression.16 Lifestyle and environmental factors also have significant impacts on CD, such as smoking and regular physical activity.17,18 Smokers are twice as likely to develop CD as non-smokers, and regular physical exercise can improve the condition of mild to moderate CD.17,18 However, patients with CD often exhibit poor adherence to this type of exercise.18 CD is detected and diagnosed mainly through colonoscopy and biopsy.19 The various available treatment options aim to alleviate symptoms and reduce the risk of recurrence.1,5,8 In newly diagnosed patients, corticosteroids may rapidly alleviate inflammation-related symptoms while methotrexate or thiopurine may lower the risk of recurrence.5,8 The most important metric in CD management is the duration of therapeutic activity. Current research focuses mainly on CD therapy including the identification of CD-associated genes and drugs that target active CD.
The present study analyzed the DEGs in active CD, inactive CD, and normal tissue samples, screened the hub gene from the DEGs through a PPI network, analyzed hub gene expression, and evaluated the correlation between the hub gene and the immune cells in the GSE95095 dataset.
Methods and Materials
Data Collection and Workflow
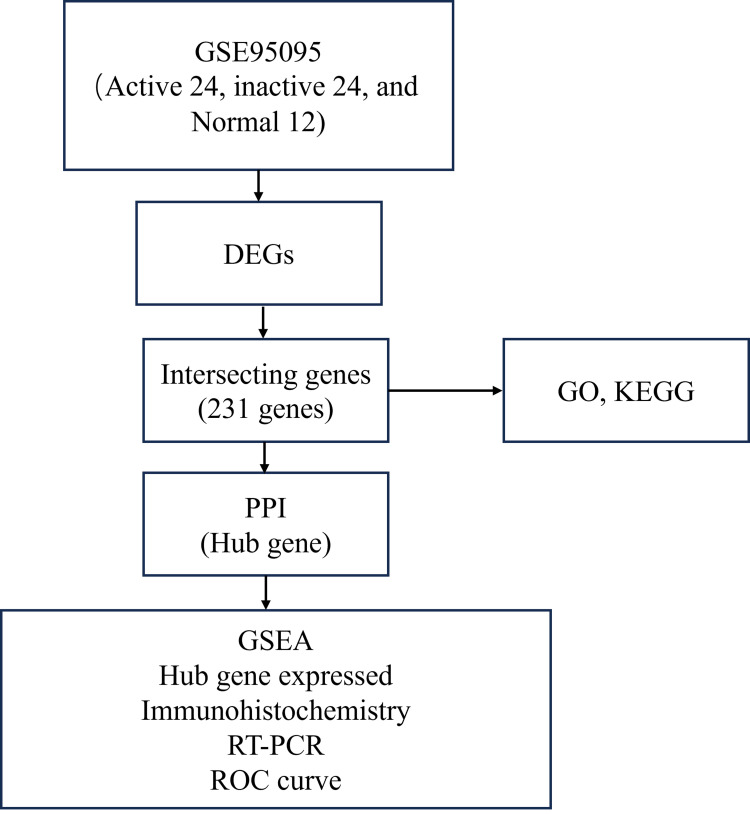
The mRNA transcriptome data for CD were obtained using the GSE95095 dataset from the GEO database (https://www.ncbi.nlm.nih.gov/geo/). The GSE95095 dataset included 6 normal, 24 inactive CD, and 24 active CD tissue samples, and the workflow is illustrated in Figure 1.
Figure 1.
The workflow is illustrated.
DEG and Intersection Gene Screening
The gene profile of the GSE95095 dataset was obtained using the cluster profile package in R v. 4.0.2 (http://www.r-project.org/). Limma v. 3.40.6 (https://bioconductor.org/packages/release/bioc/html/limma.html) was used to analyze the DEGs, and the thresholds were false discovery rate (FDR) < 0.01 and |log2 (fold change [FC])| ≥ 1.5. The intersection genes were obtained by plotting the DEGs from all three groups in a Venn diagram.
GO and KEGG Pathway Analyses
The GO functional enrichment analysis includes biological process (BP), molecular function (MF), and cellular component (CC). KEGG analyzes advanced molecular-level functions and mechanisms in biological systems. The Database for Annotation, Visualization, and Integrated Discovery (DAVID) online tool (http://david.ncifcrf.gov/) was used to perform the GO and KEGG pathway analysis of the key genes in the module. The enrichment results were then downloaded and mapped in R software, and P < 0.05 and FDR < 0.25 indicated statistical significance.
PPI Network
A PPI network of the intersection genes was constructed using the STRING protein database (http://string-db.org/). The network was analyzed and visualized with Cytoscape v. 3.6.1 (http://www.cytoscape.org/). The degree value for each gene was calculated using the network analyzer tool built into Cytoscape, and the hub gene was obtained based on the betweenness centrality (BC) value.
EPIC and ESTIMATE
The immune cell deconvolution method EPIC was performed using the IOBR v. 0.99.9 package in R (https://www.ncbi.nlm.nih.gov/pmc/articles/pmc8283787/). B cells, cancer-associated fibroblasts (CAFs), CD4_T cells, CD8_T cells, endothelial cells, macrophages, natural killer (NK) cells, and other immunocytes in the GSE95095 dataset were detected. ESTIMATE v. 1.0.13 https://bioinformatics.mdanderson.org/publicsoftware/estimate/) was then run to detect the presence of infiltrating immune cells. The stromal, immune, and ESTIMATE scores were calculated for each sample based on the hub gene expression in the GSE95095 dataset.
ROC Curve
The ROC diagnosis was conducted using pROC v. 1.18.0 (https://packages.debian.org/bookworm/r-cran-proc) and ggplot2 v. 3.3.6 (https://ggplot2.tidyverse.org/) in R v. 4.0.2. The specificity, sensitivity, AUC, and CI for the hub gene in the GSE95095 dataset were then evaluated.
Tissue Collection
Twelve active CD, 12 inactive CD, and six normal intestinal tissues were collected by colonoscopy and frozen at −80 °C until the subsequent IHC and RT-PCR. Two pathologists independently confirmed the specimens through IHC. All tissues were collected between January and June 2023 at Zhuzhou Central Hospital, Tianyuan District, Zhuzhou, Hunan, China. The tissue harvest methodology was approved by the Ethics Committee of Zhuzhou Central Hospital (No. 20231072), and each eligible participant provided written informed consent (Clinical information of 30 fresh tissue samples is provided in Supplementary Table 1).
IHC and RT-PCR
For the IHC, fresh tissue was fixed with 4% (v/v) formaldehyde, dehydrated, and subjected to high-pressure antigen repair. The samples were then blocked with goat serum and incubated overnight with primary antibody (No. ab185966; 1:400; Abcam, Cambridge, UK). The next day, the tissues were exposed to secondary antibody, and color development was performed with a diaminobenzidine (DAB) chromogen kit. CXCL12 immunohistochemical analysis: observed and photographed under a high-power microscope (20x), and the color of CD1a positive is brown. The Image J1.5 software measures the integrated option density (IOD) and area value of each image, and then calculates the mean density value (mean density=IOD/area), which reflects the unit area concentration of CXCL12 protein.
For the RT-PCR, total RNA was extracted by the TRIzol method and reverse-transcribed into cDNA with a reverse transcription kit (TaKaRa, Dalian, China) according to the manufacturer’s instructions. The cDNA was then integrated into the RT-PCR system along with the primers listed in Table 1. Fold change CXCL12 expression was compared between the experimental and control groups by the 2−ΔΔCt method.
Table 1.
RT-PCR Primer Information
| Gene name | Primer sequence (5ʹ-3ʹ) |
|---|---|
| GAPDH-F | ACAGCCTCAAGATCATCAGC |
| GAPDH-R | GGTCATGAGTCCTTCCACGAT |
| CXCL12-F | TGCCCTTCAGATTGTAGCCC |
| CXCL12-R | GCCCTTCCCTAACACTGGTT |
Statistical Analysis
Student’s t-test was conducted using the t-test function (https://www.statmethods.net/stats/ttest.html) in R to detect significant differences between the experimental and control groups, and p < 0.05 was considered statistically significant. All graphs were plotted using the ggplot2 package in R software.
Results
DEGs in Active and Inactive CD Were Screened
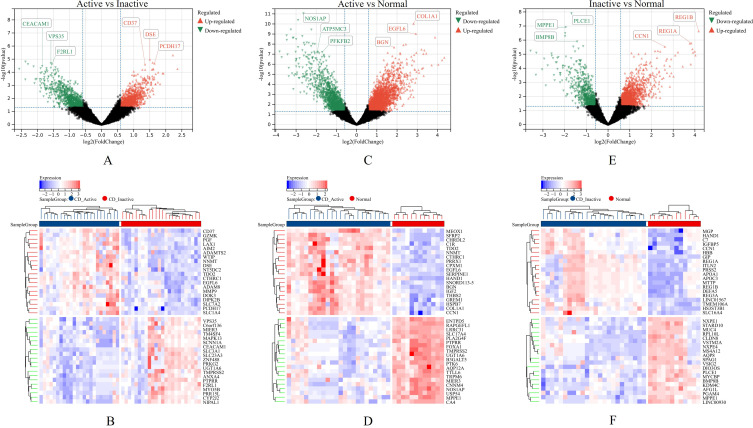
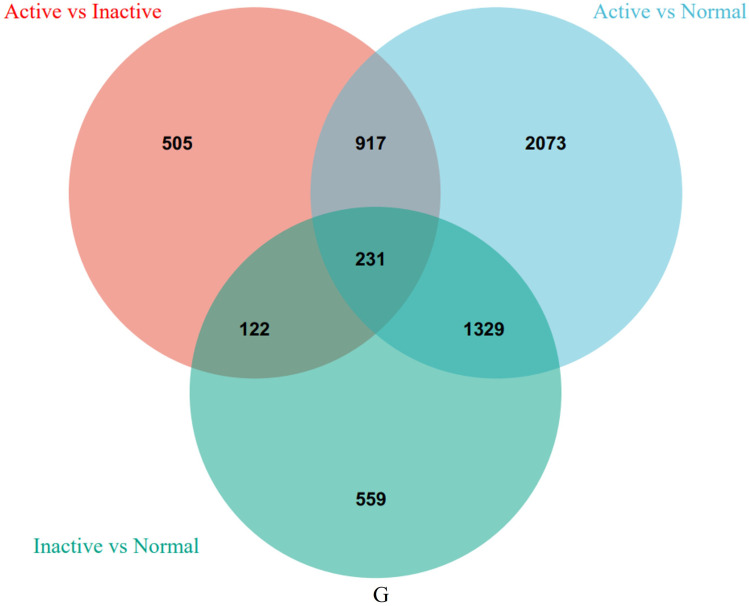
The DEGs of the active and inactive CD tissues in the GSE95095 dataset were screened. In the active CD vs inactive CD tissue sample comparison, there were 928 upregulated and 847 downregulated genes. In the inactive CD vs normal tissue sample comparison, there were 188 upregulated and 683 downregulated genes. In the active CD vs normal tissue sample comparison, there were 3,217 upregulated and 1,423 downregulated genes (Figure 2A–F). A Venn diagram analysis of the DEGs in the active CD, inactive CD, and normal tissue samples revealed 231 key intersection genes (Figure 3 and Supplementary Table 2).
Figure 2.
Screening for differential gene expression (DEGs) and Key genes in GSE95095 dataset. (A, C, and E) The volcano plot of DEGs in inflamed and uninflamed of CD from GSE179285 dataset. (B, D, and F) The heatmap of the DEGs and the top 40 genes were showed.
Figure 3.
A Venn diagram identified the intersection genes for the three DEGs groups.
GO and KEGG Pathway Analyses of Key Genes
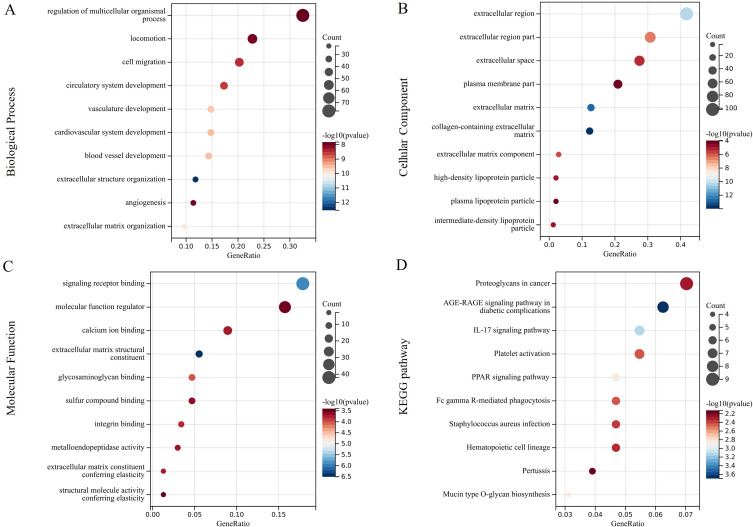
DAVID software was used to perform the GO enrichment and KEGG pathway analyses of the 231 key genes. The GO functions included multicellular organismal process, cell migration, locomotion, sulfur compound binding, molecular function regulator, and structural molecule activity conferring elasticity (p < 0.05; Figure 4A–C). The KEGG analysis included the Interleukin (IL)-17 and Peroxisome proliferator-activated receptor (PPAR) signaling pathways, Fc gamma R-mediated phagocytosis, and Proteoglycans in cancer (p < 0.05; Figure 4D).
Figure 4.
GO and KEGG pathway analysis. (A) biological process; (B) cellular component; (C) molecular function; (D) KEGG pathway.
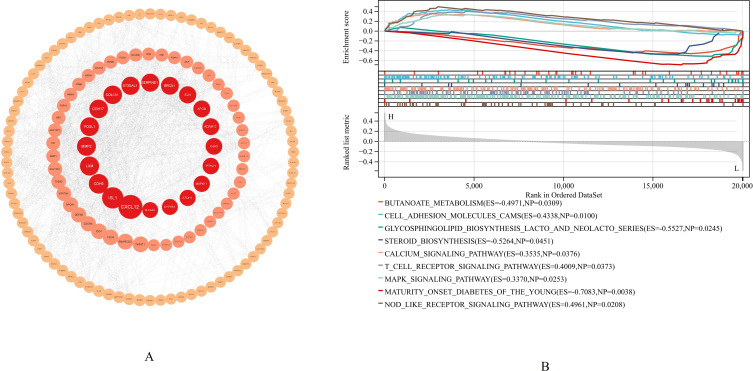
PPI Network Identification and GSEA of Hub Genes
The STRING online website analyzed the protein network nodes for 274 key genes. The PPI results were then analyzed with Cytoscape, its cytohubba plugin was used to estimate the node degree value, the latter identified CXCL12 as the hub gene in the GSE95095 dataset (Figure 5A), and CXCL12 was then subjected to GSEA. CXCL12 was significantly associated with the enrichment of BUTANOATE_METABOLISM (ES = −0.4971, NP = 0.0309), CELL_ADHESION_MOLECULES_CAMS (ES = 0.4338, NP = 0.01), GLYCOSPHINGOLIPID_BIOSYNTHESIS_LACTO_AND_NEOLACTO_SERIES = −0.5527, NP = 0.0245), STEROID_BIOSYNTHESIS (ES = −0.5264, NP = 0.0451), CALCIUM_SIGNALING_PATHWAY (ES = 0.3535, NP = 0.0376), T_CELL_RECEPTOR_SIGNALING_PATHWAY (ES = 0.4009, NP = 0.0373), MAPK_SIGNALING_PATHWAY (ES = 0.3370, NP = 0.0253), and NOD_LIKE_RECEPTOR_SIGNALING_PATHWAY (ES = 0.4961, NP = 0.0208) (Figure 5B).
Figure 5.
Protein-Protein Interaction (PPI) networks identify and gene set enrichment analysis (GSEA) of hub gene. (A) PPI networks identified CXCL12 as a hub gene. (B) CXCL12 was analyzed by GSEA in GSE95095 dataset.
CXCL12 Was Overexpressed and Closely Associated with Inflammation in CD
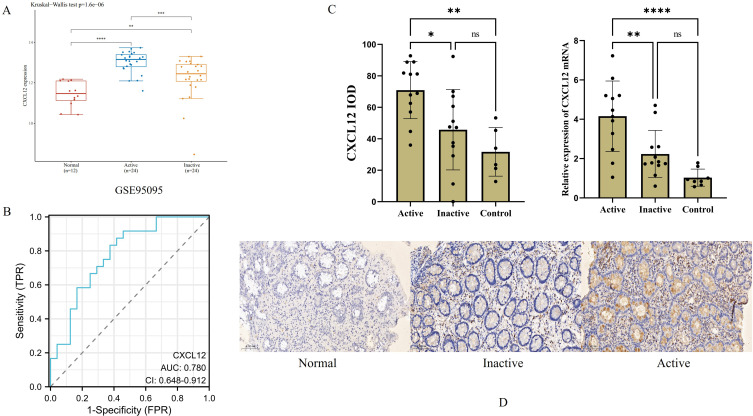
CXCL12 was overexpressed in the active CD tissue samples from the GSE95095 and GSE179285 datasets (p < 0.001, Figure 6A). The specificity and sensitivity of CXCL12 in CD were determined through ROC curves and found to be 0.875 and 0.625, respectively. The accuracy rate was 72.92%, and the AUC = 0.780 (95% CI = 0.648–0.912) (Figure 6B). Six normal intestinal mucosal and 24 CD tissues were identified by IHC and RT-PCR. CXCL12 was overexpressed in active and inactive CD (p < 0.001 and p < 0.001, respectively; Figure 6C and D).
Figure 6.
The expression and mRNA levels of CXCL12 in CD and ROC curves. (A) CXCL12 was overexpression in inflamed CD in GSE95095 dataset. (B) The specificity and sensitivity of CXCL12 was analyzed by ROC curves in inflamed CD. (C) The IOD and the mRNA levels of CXCL12 was significantly overexpression in CD tissues. (D) Immunohistochemistry of CXCL12 in CD and normal intestinal mucosal tissues (Brown color indicates expression of CXCL12). (*p<0.05; **p<0.01; ***p<0.001; ****p<0.0001).
Correlation Between CXCL12 Expression and Immune Infiltration Cell
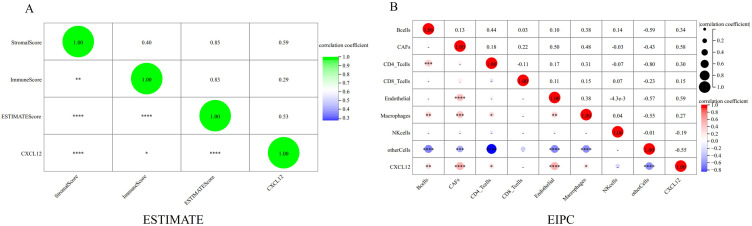
The correlation between CXCL12 and immune cell infiltration in the GSE95095 dataset was then analyzed. Significant differences were detected between CXCL12 and the stromal (p < 0.0001), immune (p < 0.05), and ESTIMATE scores (p < 0.0001) (Figure 7A). The EPIC method was used to analyze CXCL12 and the immune cells and disclosed significant correlations among them (Figure 7B). The immunocytes include B cells (p < 0.01), CAFs (p < 0.0001), CD4_T cells (p < 0.05), endothelial macrophages (p < 0.0001), and others cells (p < 0.0001) (Figure 7B).
Figure 7.
Correlation between CXCL12 and immune infiltrating cells. (A) There was a significantly negative correlation between CXCL12 and Stromal Score, Immune Score, and ESTIMATE Score. (B) The EPIC method analyzed the association between CXCL12 and immune cells, revealing significant correlations, including B cells, CAFs, CD4 T cells, endothelial macrophages, and other immune cells. (*p<0.05; **p<0.01; ***p<0.001; ****p<0.0001).
Discussion
The current therapeutic approach for CD focuses on controlling its symptoms and transforming active CD into inactive CD.20 However, it does not prevent CD-related intestinal damage.20 Up to one-half of all patients with CD in clinical remission continue to present inflammation.21 Hence, novel treatment methods for CD should primarily control inflammation rather than merely ameliorate symptoms.22
Here, we identified the DEGs of active and inactive CD in the GSE95095 dataset and screened hub genes among the DEGs. CXCL12 was deemed the hub gene common to both active and inactive CD. It was overexpressed in the active CD tissue samples from both the GSE95095 and GSE179285 datasets. IHC and RT-PCR were used to analyze 6, 12, and 12 normal intestinal mucosal, active CD, and inactive CD tissues, respectively, and validated that CXCL12 was overexpressed in CD.
CXCL12, also known as stromal-derived factor-1 (SDF-1), is a CXC subfamily chemokine member. It is designated a pre-B-cell growth-stimulating factor (PBSF) as it plays an important role in pre-B cell proliferation and differentiation.23 Unlike other inflammation-induced chemokines, CXCL12 is stably expressed in the body in the absence of any pathogen invasion.24 It is expressed in the liver, lungs, heart, kidneys, bone marrow, lymph nodes, vascular endothelial cells, interstitial fibroblasts, and osteoblasts. All the foregoing organs, tissues, and cells may secrete CXCL12 constitutively.25 CXCL12 participates in numerous pathophysiological processes.23 It regulates homeostasis, inflammation, wound healing, and tissue repair.24 Dotan et al26 reported that intestinal epithelial cells (ICES) in normal intestinal mucosae express CXCL12. CXCL12 expression is higher and more widely distributed in the ICES of patients with IBD than in those of normal controls. Zheng et al27 identified essential genes and performed drug discovery for IBD via text mining and bioinformatics analysis. They identified CXCL12 as a potential key gene implicated in CD and found that it targeted 26 existing drugs administered as CD therapy. Linares et al28 biopsied normal, slightly inflamed, and severely inflamed ileal tissues, measured their chemokine and receptor expression levels, and conducted functional enrichment analyses on them. The monoclonal antibody adalimumab and the integrin blocker vedolizumab significantly inhibited CXCL12-promoting antigen presentation by dendritic cells (DCs) as well as the initiation of leukocyte extravasation.28 The results of the foregoing studies were consistent with our observation that CXCL12 was overexpressed in active CD.
Neutrophils, monocytes, macrophages, DCs, innate lymphoid cells, and NK cells mediate intestinal innate immunity.29 Innate immune cells resist the invasion of intestinal microbial pathogens and remove them as well.30 Innate immune cells constitute part of the first line of defense and initiate adaptive immunity. Changes to the preceding equilibrium may trigger intestinal inflammation and IBD.30 Innate immune cells may produce both cytokines and chemokines such as CXCL12 which have several functions in the immune system.31
The present study disclosed a correlation between CXCL12 expression and immune cell infiltration. It also showed that CXCL12 is closely associated with various different immunocytes including B cells, CAFs, CD4 T cells, endothelial macrophages, and other immune cells. An animal experiment revealed that silencing CXCL12 fortified the immunosuppressive tumor microenvironment (TME) and increased immune cell infiltration.32 The CXCR4/CXCL12 axis biases the TME mainly towards dampening the immune responses.33 CXCL12 may inhibit T cell infiltration in tumors, and blocking the CXCL12/CXCR4 pathway improves the prophylactic and therapeutic efficacy of immune checkpoint inhibitors (ICIs).34,35 Therefore, the aforementioned combination is promising a cancer treatment strategy.
The results of the present work and those of earlier studies indicate that the chemokine CXCL12 modulates the equilibrium between microbial pathogens and the normal microflora in the gut by regulating innate immune cells. In this manner, CXCL12 may induce IBD and prolong or exacerbate the inflammation that characterizes it. We propose that CXCL12 as a potential target genes for the treatment and management of both active and inactive CD.
Funding Statement
This study was supported by the Natural Science Foundation of Hunan Province (Grant Nos. 2021JJ70076 and 2023JJ50217).
Data Sharing Statement
All data may be obtained by contacting the corresponding author (Weiming Qu) via e-mail at qwmq2q2@163.com.
Ethics Approval and Consent to Participate
The study protocol was approved by Zhuzhou Central Hospital (Tianyuan District, Zhuzhou, Hunan, China) affiliated with Xiangya Medical College, Central South University (Changsha, Hunan, China).
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors have no competing interests to declare.
References
- 1.Torres J, Bonovas S, Doherty G, et al. ECCO guidelines on therapeutics in Crohn’s disease: medical treatment. J Crohn's Colitis. 2020;14(1):4–22. [DOI] [PubMed] [Google Scholar]
- 2.Soetikno R, Sanduleanu S, Kaltenbach T. An atlas of the nonpolypoid colorectal neoplasms in inflammatory bowel disease. Gastrointest Endosc Clin N Am. 2014;24(3):483–520. doi: 10.1016/j.giec.2014.04.003 [DOI] [PubMed] [Google Scholar]
- 3.Katsandegwaza B, Horsnell W, Smith K. Inflammatory bowel disease: a review of pre-clinical murine models of human disease. Int J Mol Sci. 2022;23(16):9344. doi: 10.3390/ijms23169344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keyashian K, Dehghan M, Sceats L, Kin C, Limketkai BN, Park KT. Comparative incidence of inflammatory bowel disease in different age groups in the United States. Inflamm Bowel Dis. 2019;25(12):1983–1989. doi: 10.1093/ibd/izz092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Veauthier B, Hornecker JR. Crohn’s disease: diagnosis and management. Am Fam Physician. 2018;98(11):661–669. [PubMed] [Google Scholar]
- 6.Jones GR, Lyons M, Plevris N, et al. IBD prevalence in Lothian, Scotland, derived by capture-recapture methodology. Gut. 2019;68(11):1953–1960. doi: 10.1136/gutjnl-2019-318936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lichtenstein GR, Loftus EV, Isaacs KL, Regueiro MD, Gerson LB, Sands BE. ACG clinical guideline: management of Crohn’s disease in adults [published correction appears in Am J Gastroenterol. 2018 Jul;113(7):1101]. Am J Gastroenterol. 2018;113(4):481–517. doi: 10.1038/ajg.2018.27 [DOI] [PubMed] [Google Scholar]
- 8.Torres J, Mehandru S, Colombel JF, Peyrin-Biroulet L. Crohn’s disease. Lancet. 2017;389:1741–1755. doi: 10.1016/S0140-6736(16)31711-1 [DOI] [PubMed] [Google Scholar]
- 9.Srinivasan AR. Treat to target in Crohn’s disease: a practical guide for clinicians. World J Gastroenterol. 2024;30(1):50–69. doi: 10.3748/wjg.v30.i1.50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meima-van Praag EM, Buskens CJ, Hompes R, Bemelman WA. Surgical management of Crohn’s disease: a state of the art review. Int J Colorectal Dis. 2021;36(6):1133–1145. doi: 10.1007/s00384-021-03857-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Braun LT, Rubinstein G, Zopp S, et al. Recurrence after pituitary surgery in adult Cushing’s disease: a systematic review on diagnosis and treatment. Endocrine. 2020;70(2):218–231. doi: 10.1007/s12020-020-02432-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Majchrzak K, Fichna J. Biologic therapy in Crohn’s disease-what we have learnt so far. Curr Drug Targets. 2020;21(8):792–806. doi: 10.2174/1389450121666191218123203 [DOI] [PubMed] [Google Scholar]
- 13.Li N, Shi RH. Updated review on immune factors in pathogenesis of Crohn’s disease. World J Gastroenterol. 2018;24(1):15–22. doi: 10.3748/wjg.v24.i1.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forbes JD, Van Domselaar G, Bernstein CN. The gut microbiota in immune-mediated inflammatory diseases. Front Microbiol. 2016;7:1081. doi: 10.3389/fmicb.2016.01081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gravina AG, Pellegrino R, Durante T, et al. The melanocortin system in inflammatory bowel diseases: insights into its mechanisms and therapeutic potentials. Cells. 2023;12(14):1889. doi: 10.3390/cells12141889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Z, Liu R, Gao H, et al. Genetic architecture of the inflammatory bowel diseases across East Asian and European ancestries. Nat Genet. 2023;55(5):796–806. doi: 10.1038/s41588-023-01384-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Y, Wang Y, Shen J. Role of environmental factors in the pathogenesis of Crohn’s disease: a critical review. Int J Colorectal Dis. 2019;34(12):2023–2034. doi: 10.1007/s00384-019-03441-9 [DOI] [PubMed] [Google Scholar]
- 18.Gravina AG, Pellegrino R, Durante T, et al. Inflammatory bowel diseases patients suffer from significant low levels and barriers to physical activity: the “BE-FIT-IBD” study. World J Gastroenterol. 2023;29(41):5668–5682. doi: 10.3748/wjg.v29.i41.5668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tse CS, Nguyen HP, Singh S, et al. Correlations between gastrointestinal symptoms and endoscopic-histologic disease activity in adults with ulcerative colitis. Dig Dis Sci. 2023;68(8):3254–3258. doi: 10.1007/s10620-023-07986-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roda G, Chien Ng S, Kotze PG, et al. Crohn’s disease [published correction appears in Nat Rev Dis Primers. 2020 Apr 6;6(1):26] [published correction appears in Nat Rev Dis Primers. 2020 May 20;6(1):42] [published correction appears in Nat Rev Dis Primers. 2020 Jun 19;6(1):51]. Nat Rev Dis Primers. 2020;6(1):22. doi: 10.1038/s41572-020-0156-2 [DOI] [PubMed] [Google Scholar]
- 21.Garcia NM, Cohen NA, Rubin DT. Treat-to-target and sequencing therapies in Crohn’s disease. United European Gastroenterol J. 2022;10(10):1121–1128. doi: 10.1002/ueg2.12336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh S, Boland BS, Jess T, Moore AA. Management of inflammatory bowel diseases in older adults. Lancet Gastroenterol Hepatol. 2023;8(4):368–382. doi: 10.1016/S2468-1253(22)00358-2 [DOI] [PubMed] [Google Scholar]
- 23.Ao D, Li DJ, Li MQ. CXCL12 in normal and pathological pregnancies: a review. Am J Reprod Immunol. 2020;84(3):e13280. doi: 10.1111/aji.13280 [DOI] [PubMed] [Google Scholar]
- 24.Cambier S, Gouwy M, Proost P. The chemokines CXCL8 and CXCL12: molecular and functional properties, role in disease and efforts towards pharmacological intervention. Cell Mol Immunol. 2023;20(3):217–251. doi: 10.1038/s41423-023-00974-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janssens R, Struyf S, Proost P. The unique structural and functional features of CXCL12. Cell Mol Immunol. 2018;15(4):299–311. doi: 10.1038/cmi.2017.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dotan I, Werner L, Vigodman S, et al. CXCL12 is a constitutive and inflammatory chemokine in the intestinal immune system. Inflamm Bowel Dis. 2010;16(4):583–592. doi: 10.1002/ibd.21106 [DOI] [PubMed] [Google Scholar]
- 27.Zheng Q, Guo L, Yang R, Chen Z, Liu X. Identification of essential genes and drug discovery in bladder cancer and inflammatory bowel disease via text mining and bioinformatics analysis. Curr Comput Aided Drug Des. 2024;20(4):359–366. doi: 10.2174/1573409919666230330154008 [DOI] [PubMed] [Google Scholar]
- 28.Linares R, Gutiérrez A, Márquez-Galera Á, et al. Transcriptional regulation of chemokine network by biologic monotherapy in ileum of patients with Crohn’s disease. Biomed Pharmacother. 2022;147:112653. doi: 10.1016/j.biopha.2022.112653 [DOI] [PubMed] [Google Scholar]
- 29.Saez A, Gomez-Bris R, Herrero-Fernandez B, Mingorance C, Rius C, Gonzalez-Granado JM. Innate lymphoid cells in intestinal homeostasis and inflammatory bowel disease. Int J Mol Sci. 2021;22(14):7618. doi: 10.3390/ijms22147618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saez A, Herrero-Fernandez B, Gomez-Bris R, Sánchez-Martinez H, Gonzalez-Granado JM. Pathophysiology of inflammatory bowel disease: innate immune system. Int J Mol Sci. 2023;24(2):1526. doi: 10.3390/ijms24021526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sokol CL, Luster AD. The chemokine system in innate immunity. Cold Spring Harb Perspect Biol. 2015;7(5):a016303. doi: 10.1101/cshperspect.a016303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Portella L, Bello AM, Scala S. CXCL12 signaling in the tumor microenvironment. Adv Exp Med Biol. 2021;1302:51–70. [DOI] [PubMed] [Google Scholar]
- 33.Mezzapelle R, Leo M, Caprioglio F, et al. CXCR4/CXCL12 activities in the tumor microenvironment and implications for tumor immunotherapy. Cancers. 2022;14(9):2314. doi: 10.3390/cancers14092314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zboralski D, Hoehlig K, Eulberg D, Frömming A, Vater A. Increasing tumor-infiltrating T cells through inhibition of CXCL12 with NOX-A12 synergizes with PD-1 Blockade. Cancer Immunol Res. 2017;5(11):950–956. doi: 10.1158/2326-6066.CIR-16-0303 [DOI] [PubMed] [Google Scholar]
- 35.Guo F, Wang Y, Liu J, Mok SC, Xue F, Zhang W. CXCL12/CXCR4: a symbiotic bridge linking cancer cells and their stromal neighbors in oncogenic communication networks. Oncogene. 2016;35(7):816–826. doi: 10.1038/onc.2015.139 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data may be obtained by contacting the corresponding author (Weiming Qu) via e-mail at qwmq2q2@163.com.