Abstract
Invasive meningococcal disease, caused by Neisseria meningitidis (N. meningitidis), is a critical global health issue, necessitating swift and precise diagnostics for effective management and control. Here, we introduce a novel diagnostic assay, NM-RT-MCDA, that combines multiple cross displacement amplification (MCDA) with real-time fluorescence detection, targeting a specific ctrA gene region in the N. meningitidis genome. The assay utilizes a primer set designed for high specificity and incorporates a fluorophore-quencher pair with a restriction endonuclease site for real-time monitoring. Optimized at 65 °C for 40 min, NM-RT-MCDA demonstrates exceptional specificity, with no cross-reactivity observed with nontarget species. It achieves a remarkable sensitivity, detecting as low as 100 fg of genomic DNA per reaction, and has been successfully applied to clinical sputum samples, matching the sensitivity of nanoparticle-based lateral flow biosensors. The assay’s rapid turnaround time, completed within an hour including DNA extraction and amplification, positions NM-RT-MCDA as a promising diagnostic tool for various clinical scenarios, potentially facilitating timely diagnosis and intervention in invasive meningococcal disease management.
1. Introduction
Neisseria meningitidis (N. meningitidis), a Gram-negative diplococcal bacterium exclusively found in humans, is a major etiological agent of meningitis.1 This opportunistic pathogen colonizes the nasopharynx asymptomatically in approximately 10% of the population during nonepidemic periods.2 During acute infections, N. meningitidis can invade the bloodstream and cerebrospinal fluid, leading to meningitis. Infants under one year of age and adolescents exhibit the highest incidence of bacterial meningitis.3 Globally, bacterial meningitis is responsible for an estimated 200,000 deaths annually, with case fatality rates reaching 60% in certain regions of sub-Saharan Africa and developing countries.4,5 In Europe, the notification rate for meningococcal disease was 1 per 100,000 in 2007, with Ireland and the UK reporting the highest rates at 3.8 and 2.5 per 100,000, respectively.6
Based on the chemical composition of its capsular polysaccharides, N. meningitidis is classified into 13 serogroups: A, B, C, D, E, H, I, K, L, W, X, Y, and Z.7 Serogroups A, B, C, Y, and W-135 are primarily responsible for invasive human infections, with serogroups A and C being the most frequent causes of meningitis.7−10 Serogroups B, Y, and W-135 have been implicated in severe epidemics in certain regions.11,12 The widespread implementation of meningococcal vaccines has led to a global decline in meningitis cases in recent decades.13 Polysaccharide vaccines targeting serogroups A and C, often combined with components targeting serogroups Y and W-135, have been extensively used. However, these vaccines exhibit limited immunogenicity in infants and young children owing to their inability to elicit robust anamnestic immune responses against the meningococcal capsule.6 Despite treatment, the mortality rate of meningococcal disease remains around 10%, and survivors may experience severe sequelae, including limb amputation, neurological deficits, and other debilitating conditions. This underscores the persistent public health significance of meningococcal disease.14
While traditional culture methods are considered the gold standard for N. meningitidis diagnosis, they are time-consuming and lack sensitivity, particularly in patients who have received antibiotic treatment.6 Microscopy, although useful, can also suffer from suboptimal sensitivity, and fluorescence microscopy requires specialized equipment that may be cost-prohibitive.4 Consequently, molecular methods have emerged as promising alternatives for rapid diagnoses. Polymerase chain reaction (PCR) is widely employed because of its high sensitivity, specificity, and relative ease of use.15−19 However, despite advancements compared to culture methods, PCR-based assays typically require several hours for completion, which can hinder timely reporting of clinical results.
Despite advances in diagnostic techniques, there remains a need for assays that offer rapid, sensitive, and specific detection of N. meningitidis, particularly in resource-limited settings. Although PCR-based methods are highly sensitive and specific, they require specialized equipment and infrastructure, limiting their accessibility. Isothermal amplification techniques, such as multiple cross-displacement amplification (MCDA), provide a compelling alternative.20 MCDA reactions proceed at a constant temperature, eliminating the need for thermal cyclers and are therefore suitable for a wider range of settings. The results can be visualized through colorimetric changes or using nanoparticle-based lateral flow biosensors (LFBs), offering a user-friendly approach for diagnosis.21,22 However, colorimetric and LFB-based detection methods have certain limitations.23 They lack the ability to provide semiquantitative results without specialized reading equipment.24 Additionally, matrix components in samples can interfere with these assays, leading to nonspecific adsorption on the membrane and potentially affecting the interpretation of results. Furthermore, nanoparticle stability can be influenced by environmental conditions, necessitating improvements in long-term storage and stability during use.25
To address the limitations of existing diagnostic methods, we developed a novel assay, NM-RT-MCDA, for the rapid, sensitive, and specific detection of N. meningitidis. This assay combines the efficiency of MCDA with the precision of the real-time fluorescence detection. The NM-RT-MCDA assay uses a set of primers targeting conserved regions within the N. meningitidisctrA gene, which encodes an outer membrane protein involved in capsule transport.26,27ctrA has been demonstrated to be a specific marker for N. meningitidis identification, enabling highly sensitive detection without cross-reactivity with other bacterial species. This assay holds promise as a valuable diagnostic tool for accurate identification of N. meningitidis in various clinical settings.
2. Materials and Methods
2.1. Reagents and Instruments
The DNA extraction kits were obtained from Beijing Transgen Biotech Co., Ltd. (Beijing, China). The disposable lateral flow biosensor (LFB), Isothermal Amplification Kit, biotin-14-dCTP, and color indicator were obtained from Huidexin Biotech Co., Ltd. (Tianjin, China). Restriction endonuclease (Nb.BsrDI) was purchased from New England Biolabs (America). All primers were synthesized by Beijing DIA-UP Biotech Co. Ltd. (Beijing, China). The real-time turbidimeter LA-320C was supplied by Eiken Chemical Co., Ltd. (Japan). Applied Biosystems Inc. (USA) was used as the source for the Applied Biosystems 7500 Fast Real-Time PCR System.
2.2. Making a Gold Nanoparticle-Based Dipstick Biosensor
An LFB (4 mm × 60 mm) was made, slightly altering it from earlier reports.28,29 In summary, the absorbent pad, conjugate pad, sample pad, and NC membrane were laminated onto a plastic backing card using glue. The anti-FITC Ab (0.2 mg/mL) and biotin-BSA (2.5 mg/mL) were sprayed onto the NC membrane to create the test line (TL) and control line (CL). Five millimeters separated the TL and CL. SA-Gs were put on the conjugate pad of the strip after being diluted in 0.01 M PBS (pH 7.4). The constructed cards were cut into 4 mm-wide strips (Deli No. 8012), sealed with desiccant gel, and stored at room temperature in a plastic box.
2.3. Primer Design
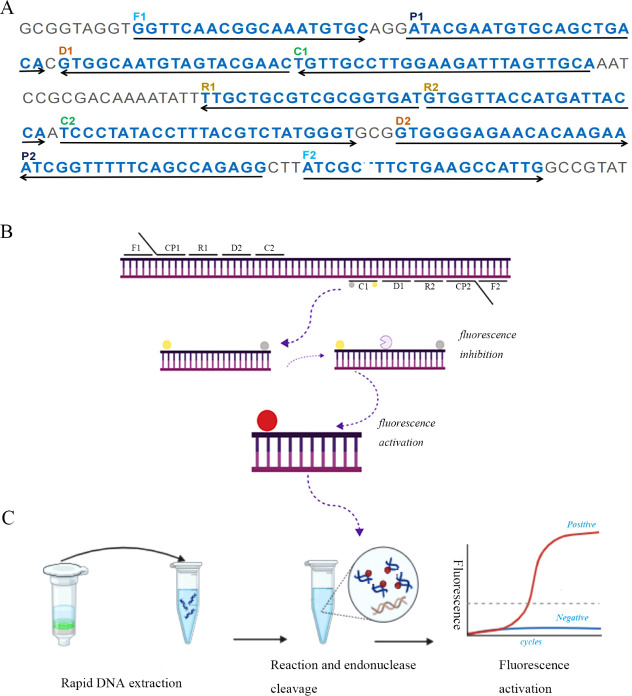
Using Primer Premier 5.0 software, ten primers were built based on the sequence of the ctrA gene (GenBank NZ_CP021520) of Neisseria meningitidis in accordance with the MCDA method principle. Six amplification primers (C1, C2, D1, D2, R1, and R2), two displacement primers (F1 and F2), and two cross-primers (CP1 and CP2) were included in the primer set. Black Hole Quencher 1 (BHQ1) was inserted in the center of primer C1 (referred to as C1*) along with a short sequence (TGCAATG) and a fluorophore FAM at the 5′end for real-time fluorescence detection. Furthermore, primer D1 was labeled with FAM at the 5′ end (referred to as D1*) for LFB detection. The sequences, locations, and alterations in each primer are listed in Table 1 and Figure 1, respectively.
Table 1. Primers Used in the Studya.
| Primer name | Sequence and modifications | Length |
|---|---|---|
| NM-F1 | GGTTCAACGGCAAATGTGC | 19 |
| NM-F2 | CAATGGCTTCAGAAAGCGAT | 20 |
| NM-CP1 | TGCAACTAAATCTTCCAAGGCAACA-ATACGAATGTGCAGCTGACA | 46 |
| NM-CP2 | TCCCTATACCTTTACGTCTATGGGT-CCTCTGGCTGAAAAACCGAT | 46 |
| NM-C1 | TGCAACTAAATCTTCCAAGGCAACA | 25 |
| NM-C2 | TCCCTATACCTTTACGTCTATGGGT | 25 |
| D1 | GTTCGTACTACATTGCCAC | 19 |
| D2 | GTGGGGAGAACACAAGAA | 18 |
| R1 | ATCACCGCGACGCAGCAA | 18 |
| R2 | GTGGTTACCATGATTACCA | 19 |
| NM-C1* | FAM-TGCAATG-T(BHQ1)GCAACTAAATCTTCCAAGGCAACA | 43 |
| NM-D1* | FAM-GTTCGTACTACATTGCCAC | 23 |
NM-C1*, the modified primer for theNM-RT-MCDAassay; NM-D1*, the modified primer for the NM-MCDA-LFB assay.
Figure 1.
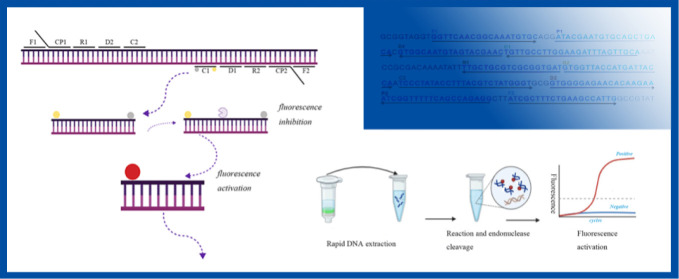
Diagrammatic representation of the NM-RT-MCDA assay’s foundation. (A) The study’s primers’ locations and sequences that target the N. meningitis ctrA gene. Sense and complementary sequences are indicated by the right and left arrows, respectively. The primer positions are indicated by the yellow colored text. There are six amplification primers (C1, C2, D1, D2, R1 and R2), two cross primers (CP1 and CP2), and two displacement primers (F1 and F2). (B) MCDA reactions schematic diagram using the modified primers. A quencher (BHQ1, gray ball) was inserted in the middle of primer C1, which was extended with an endonuclease recognition site at the 5′ end and a fluorophore (FAM, yellow ball) at the 5′ end. Double-stranded target amplification containing the restriction endonuclease recognition site were produced following amplification. The fluorescent signal was produced upon recognition and cleavage by restriction endonuclease. (C) The NM-RT-MCDA detection system in its entire form. The entire procedure, which includes real-time fluorescence detection, MCDA reaction, endonuclease cleavage, and real-time DNA extraction.
2.4. Standard NM-MCDA Assay and Optimal Reaction Temperature of the NM-MCDA Assay
According to earlier studies, MCDA reactions were conducted in a 25 μL reaction apparatus, in summary, each reaction contained 12.5 μL of 2× reaction mix (Isothermal Amplification kit), 1 μL of Bst DNA polymerase (8 U), 0.4 μM of displacement primers F1 and F2, 0.8 μM of amplification primers C1, C2, R1, R2, D1, and D2, and 1.6 μM of cross primers CP1 and CP2,1 μL of template (or 5 μL of clinical templates).30,31 And appropriate distilled the optimal amplification temperature was initially determined by performing NM-MCDA reactions at temperatures ranging from 62 to 67 °C at 1 °C intervals in order to maximize the performance of the NM-RT-MCDA assay. Using double-distilled water (DW) as the blank control and extracted DNA of N. meningitidis as the positive control, reactions were carried out and continuously observed using a PCR turbidimeter. The procedures were run for 40 min at the appropriate temperature, and then the reaction was stopped for 5 min at 90 °C. Optimal temperature was defined as the temperature at which the best results were obtained. The NM-MCDA-LFB and NM-RT-MCDA experiments that followed made used the optimal reaction temperatures that had been reached.
2.5. Standard NM-RT-MCDA Assay
The NM-RT-MCDA assay reaction mixture (25 μL) was identical to the previously described NM-MCDA reaction with a few modifications: C1 was replaced with C1* and an additional 1.0 μL of restriction endonuclease Nb. Then, BsrDI was added to the reaction mixture. Using double-distilled water (DW) as the blank control and extracted DNA from N. meningitidis as the positive control, the NM-RT-MCDA experiment was conducted using a real-time PCR machine for 40 min at 65 °C. Fluorescence generation served as a basis for assessing the performance of the NM-RT-MCDD reaction.
2.6. Standard NM-MCDA-LFB Assay
The NM-MCDA-LFB assay’s reaction mixture, which was 25 μL, was identical to the previously described NM-MCDA reaction with a few modifications: D1 was replaced by D1*, 0.5 μL of biotin-14-dCTP, 1.2 μL of color indicator.20−22,25,32 Either A real-time turbidimeter or a conventional PCR device was used to carry out the amplification process, which was run for 40 min at 65 °C. The experimental results can be observed by the color change of the visual indicator interpreted using the LFB. For LFB, 100 μL of running buffer (containing 10 mM PBS, PH 7.4, and 1% Tween 20) and 5 μL of MCDA amplicons were then placed onto the sample pad. The positive result then showed two red bands at the testing line (TL) and the control line (CL) after 2 min, whereas the negative result showed one red band at CL.
2.7. Sensitivity Evaluation of the NM-RT-MCDA Assay
To determine the detection limit of the NM-RT-MCDA assay, DNA templates extracted from pure cultures of N. meningitidis (strainATCC 13102) were continuously diluted to 1 ng, 100 pg, 10 pg, 1 pg, 100 fg, and 50 fg per microliter. Accordingly, three replicates were conducted for each serial dilution, as well as for a blank control of distilled water.
2.8. Specificity Evaluation of the NM-RT-MCDA Assay
For specificity analysis, the assay was performed with DNA templates of seven N. mengingitidis strains and 19 non- N. mengingitidis strains (Table 2). The results were displayed using a real-time fluorescence detection platform as well as the LFB platform for comparison.
Table 2. Bacterial Strains Used in This Studya.
| No. | Bacteria | Serogroup/species | Strain name (source of strain) | No. of strains |
|---|---|---|---|---|
| 1 | N. meningitidis | A | ATCC13077 | 1 |
| 2 | N. meningitidis | B | ATCC 13090 | 1 |
| 3 | N. meningitidis | C | ATCC 13102 | 1 |
| 4 | N. meningitidis | D | ATCC 13113 | 1 |
| 5 | N. meningitidis | W-135 | ATCC 35559 | 1 |
| 6 | N. meningitidis | X | ATCC 35560 | 1 |
| 7 | N. meningitidis | Y | ATCC 35561 | 1 |
| 8 | Bordetella pertussis | u | Isolated strains (CDC) | 1 |
| 9 | Haemophilus influenzae | u | Isolated strains (CDC) | 1 |
| 10 | Staphylococcus amber | u | Isolated strains (CDC) | 1 |
| 11 | Shigella sonnei | u | Isolated strains (CDC) | 1 |
| 12 | Staphylococcus epidermidis | u | Isolated strains (CDC) | 1 |
| 13 | Salmonella spp. | u | Isolated strains (CDC) | 1 |
| 14 | Citrobater spp. | u | Isolated strains (CDC) | 1 |
| 15 | EnteroinvasiveEscherichia coli | u | Isolated strains (CDC) | 1 |
| 16 | EnterotoxicEscherichia coli | u | Isolated strains (CDC) | 1 |
| 17 | Staphylococcus hemolyticus | u | Isolated strains (CDC) | 1 |
| 18 | Stenotrophomonas maltophilia | u | Isolated strains (CDC) | 1 |
| 19 | Streptococcus suis | u | Isolated strains (CDC) | 1 |
| 20 | Moraxella catarrhalis | u | Isolated strains (CDC) | 1 |
| 21 | Corynebacterium striatum | u | Isolated strains (CDC) | 1 |
| 22 | Streptococcus salivarius | u | Isolated strains (CDC) | 1 |
| 23 | Candida albicans | u | Isolated strains (CDC) | 1 |
| 24 | Pseudomonas aeruginosa | u | Isolated strains (CDC) | 1 |
| 25 | Klebsiella pneumoniae | u | Isolated strains (CDC) | 1 |
| 26 | Bordetella parapertussis | u | Isolated strains (CDC) | 1 |
Abbreviations: U, unidentified serotype; ATCC, American Type Culture Collection; CDC, Chinese Center for Disease Control and Prevention.
2.9. Clinical Feasibility Evaluation of the NM-RT-MCDA Assay
The clinical viability of the NM-RT-MCDA assay was assessed by retrospectively analyzing nucleic acid extractions from 34 cerebrospinal fluid specimens and nasal swab samples. The clinical samples, which were 5 N. mengingitidis samples taken from individuals suspected of having an N. meningitidis infection based on their clinical symptoms, were identified using the traditional PCR method. The DNA extracted from these cerebrospinal fluid specimens was kept at −20 °C. The NM-RT-MCDA and NM-MCDA-LFB assay protocols were carried out using aliquots of 5 μL of the DNA template.
3. Results
3.1. Formulation of the Effective NM-RT-MCDA-RT Assay
A novel NM-RT-MCDA assay was developed by integrating multiple cross displacement amplification (MCDA), restriction endonuclease (Nb.BsrDI) cleavage, and real-time fluorescence detection. The assay targeted the ctrA gene of N. meningitidis using a specifically designed primer set (Figure 1A). The MCDA reaction generates a large number of amplicons containing the recognition sequence for the restriction endonuclease Nb.BsrDI incorporated within primer C1*. Upon cleavage by Nb.BsrDI (restriction endonuclease recognition site TGCAATG), the fluorophore (FAM) was released from the quenching effect of BHQ1, generating a detectable fluorescence signal (Figure 1B). This enabled the detection of N. meningitidis in a single tube using real-time fluorescence monitoring. The entire process can be completed within 1 h, encompassing rapid DNA extraction (15 min), MCDA amplification and endonuclease cleavage (40 min), and real-time fluorescence detection (2 min) (Figure 1C).
3.2. The Assay’s Optimum Reaction Temperature for NM-RT-MCDA
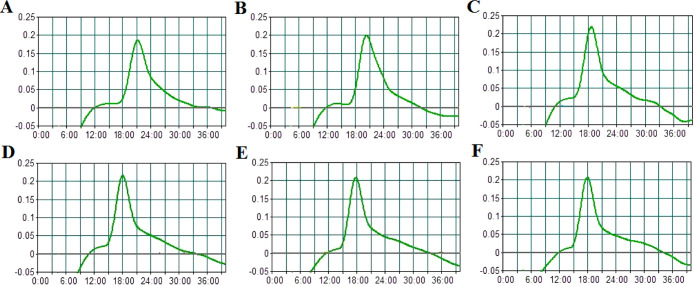
To determine the optimal reaction temperature for the NM-RT-MCDA assay, we performed MCDA reactions on a real-time turbidimeter across a temperature gradient ranging from 63 to 68 °C in 1 °C increments. The real-time amplification kinetics were monitored and recorded. An optimal temperature of 65 °C was selected based on analysis of the peak fluorescence value and the inflection point of the amplification curves (Figure 2A–F).Subsequent experiments were conducted at 65 °C.
Figure 2.
Temperature optimization for NM-RT-MCDA experiment. The temperature range for the NM-MCDA reactions was 63 to 68°C, and the real-time tubidimeter was utilized to obtain the kinetic curves at various temperatures (A-F). The experiment was repeated three times.
3.3. Verification of the NM-RT-MCDA Test’s Performance
To evaluate primer efficiency and validate the NM-RT-MCDA assay, the N. meningitidis MCDA assay was performed using the selected primer set under optimized conditions. Amplification was performed at 65 °C for 40 min, followed by NM-RT-MCDA analysis. As shown in Figure 3A, reactions containing N. meningitidis DNA or positive control DNA exhibited robust fluorescence signals, whereas negative and blank controls showed no amplification. Consistent with these results, reactions with N. meningitidis DNA or the positive control DNA displayed a clear increase in turbidity, as measured by a real-time turbidimeter (Figure 3B). Additionally, a green color change was observed in these positive reactions using a visual indicator (Figure 3C). Lateral flow biosensor (LFB) analysis of positive reactions revealed the presence of both control (CL) and test (TL) lines (Figure 3D). In contrast, the negative and blank controls for the LFB assay showed no turbidity increase (Figure 3B), no color change (Figure 3C), and only a single red band corresponding to the control line (Figure 3D). The results of the NM-RT-MCDA assay were concordant with those of the NM-MCDA-LFB assay, confirming the ability of the selected primer set to detect N. meningitidis.
Figure 3.
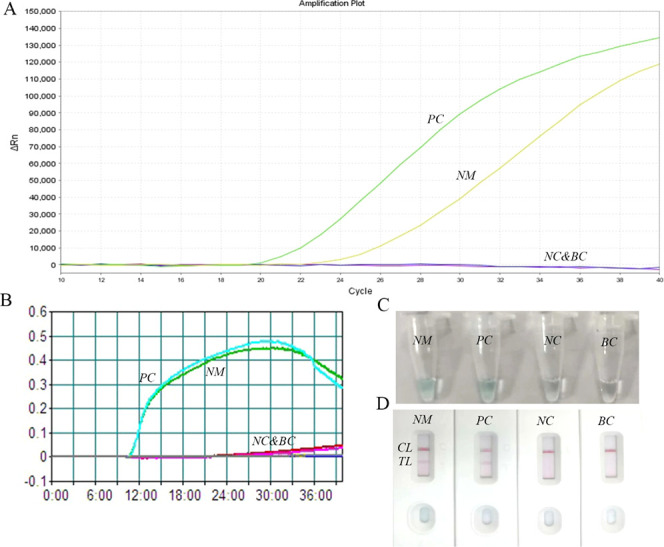
Verification of the NM-RT-MCDA assay. (A) The NM-RT-MCDAassay was performed at 65 °C for 40 min using real-time fluorescence detector monitoring the results. (B) For primer verification, NM-MCDA assay was carried out at 65 °C for 40 min and the results were reported using PCR turbidimeter. (C) Amplification products of the NM-MCDA assay were visually analyzed by observation of the color change. (D) A lateral flow biosensor was applied for visual detection of NM-MCDA products. NM: N. meningitis, PC:positive control, NC: negative control, BC: blank control,TL: testing line, CL: control line.
3.4. Assay Sensitivity for NM-RT-MCDA
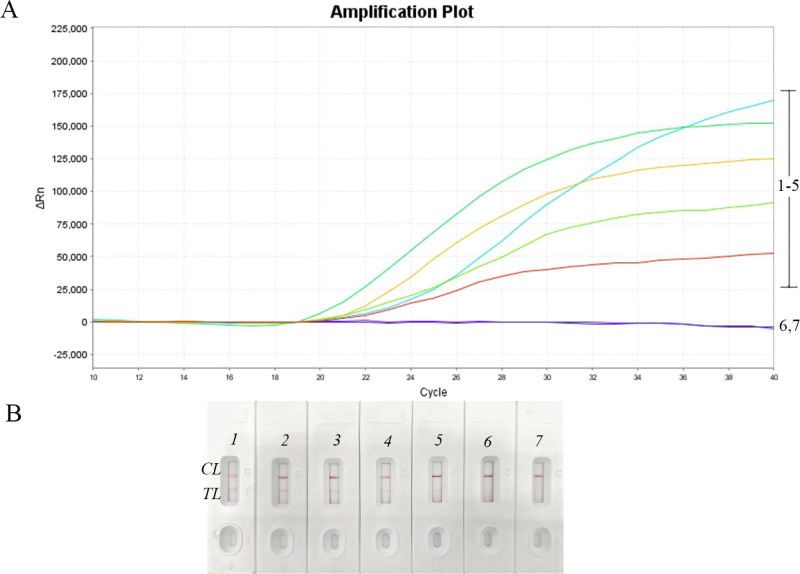
To determine the limit of detection (LOD) of the NM-RT-MCDA assay, we tested serial dilutions of N. meningitidis genomic DNA in triplicates. As shown in Figure 4A, the NM-RT-MCDA assay consistently generated detectable fluorescence signals, with a minimum detectable concentration of 100 fg of N. meningitidis DNA per reaction. These results are concordant with those obtained using the NM-MCDA-LFB assay (Figure 4B). Therefore, the LOD of the NM-RT-MCDA assay was established to be 100 fg of N. meningitidis DNA per reaction.
Figure 4.
Verification of the NM-RT-MCDA assay’s sensitivity. (A) DNA template that was isolated from pure N. meningitidis strain was diluted in order to examine the sensitivity of the NM-RT-MCDA test. Three separate tests were conducted on the NM-MCDA reactions. Signals 1 through 7 are as follows: 1 ng/mL, 100 pg/mL, 10 pg/mL, 1 pg/mL, 100 fg/mL, 50 fg/mL, and blank control. (B) For comparison and confirmation, the NM-MCDA-LFB assay was also run in parallel. Strips1 through 7 are as follows: 1 ng/mL, 100 pg/mL, 10 pg/mL, 1 pg/mL, 100 fg/mL, 50 fg/mL, and blank control. TL: Testing line, CL: control line.
3.5. Assay Specificity for NM-RT-MCDA
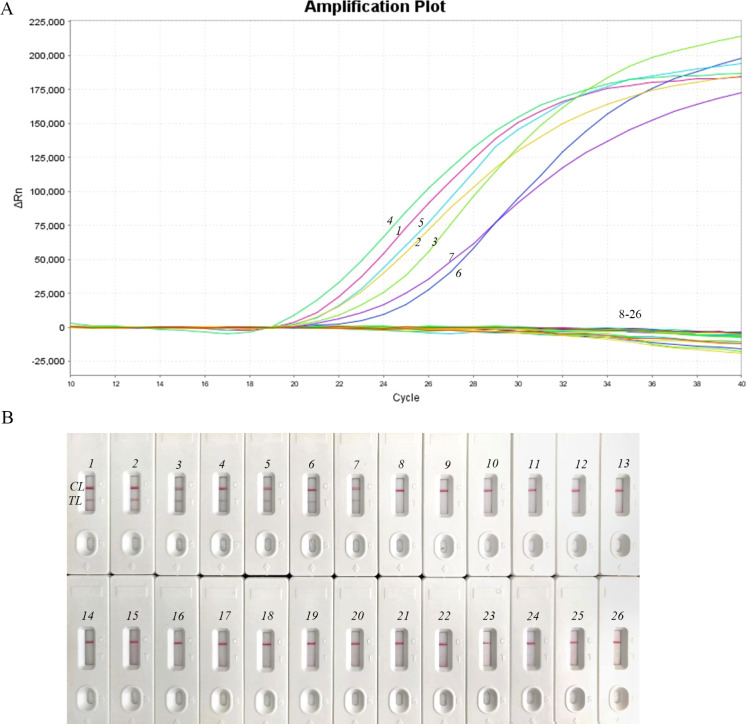
The specificity of the NM-RT-MCDA assay was evaluated using a panel of 14 bacterial strains, including 7 N. meningitidis strains and 19 non- N. meningitidis strains (Table 2). As expected, only the N. meningitidis strains yielded positive results, whereas all non-N. meningitidis strains yielded negative results (Figure 5A). These findings were consistent with those obtained using the NM-MCDA-LFB assay (Figure 5B), demonstrating the 100% specificity of the NM-RT-MCDA assay for N. meningitidis detection.
Figure 5.
Verification of the NM-RT-MCD Aassay’s specificity (A) To verify the NM-RT-MCDAassay’s specificity, DNA templates from 7 N. meningitidis strains and 19 non-N. meningitidis strains were examined. Signals 1 to 7 represented N. meningitidis strains, and 8–26 represented non-N. meningitidis strains. (B) Furthermore, the NM-MCDA-LFB assay was run concurrently for validation and comparison. Strips 1 to 7 represented N. meningitidis strains, and 8–26 represented non-N. meningitidis strains. TL: testing line, CL: control line.
3.6. Feasibility of NM-RT-MCDA in Clinical Specimens
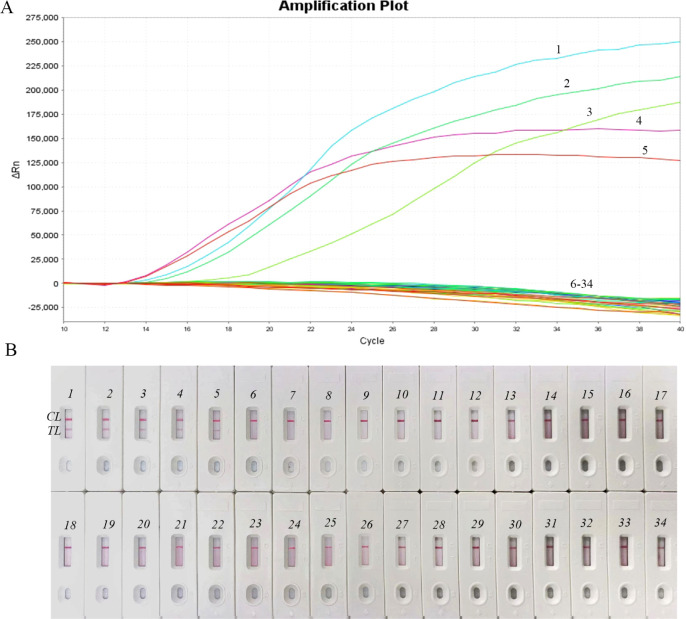
To assess the clinical utility of the NM-RT-MCDA assay, we analyzed 34 sputum samples. Five samples tested positive for N. meningitidis, whereas 29 tested negative (Figure 6A). The NM-RT-MCDA assay identified every sample that tested positive for N. meningitidis compared to the previous clinical diagnosis made using the traditional PCR test. These results are concordant with those obtained using the NM-MCDA-LFB assay (Figure 6B), demonstrating the clinical applicability of the NM-RT-MCDA assay. Taken together, these findings suggest that the NM-RT-MCDA assay has potential as a valuable diagnostic tool for N. meningitidis detection in clinical settings.
Figure 6.
NM-RT-MCDAassay application in clinical specimens. (A) 34 sputum samples were used to identify DNA templates using the NM-RT-MCDAtest. Samples 1 through 5 showed positive test findings; the remaining samples showed negative results. (B) 34 sputum samples were used to identify DNA templates using the NM-MCDA-LFB test. Samples 1 through 5 showed positive test findings; the remaining samples showed negative results. TL: testing line, CL: control line.
4. Discussions
This study presents a novel nucleic acid detection assay, termed NM-RT-MCDA, which combines fluorescence detection with MCDA amplification for the highly sensitive and specific identification of N. meningitidis. The assay achieved a remarkable detection limit of 100 fg of target nucleic acids and exhibited no cross-reactivity with other pathogens. Optimized for rapid execution, the NM-RT-MCDA assay requires only 40 min of isothermal amplification at 65 °C, followed by real-time PCR analysis, enabling completion of the entire procedure within an hour. The assay’s efficiency, sensitivity, and specificity are valuable diagnostic tools for detecting N. meningitidis in various clinical scenarios.
Traditional diagnostic methods for N. meningitidis include culture, agglutination assays, and lateral flow assays (LFA) (Table 3). While cerebrospinal fluid (CSF) culture has long been considered the gold standard,33 it has several limitations. Culture results can take up to 48 h, and prior antibiotic use can render bacteria nonviable, leading to false-negative results.34 Agglutination assays are simple to perform but are susceptible to reduced sensitivity due to prior antibiotic treatment.6 Additionally, cross-reactivity from polyagglutination can complicate serogroup identification.35 LFAs, such as the meningoSpeed test, offer rapid, point-of-care results by utilizing immunochromatographic strips to detect meningococcal antigens in the CSF.36 However, their sensitivity and specificity may be lower than those of other techniques, and they can be affected by prior antibiotic use owing to reduced antigen levels.37,38 LFAs also provide only qualitative results, lacking the ability to quantify the antigen concentration. To address these limitations, molecular diagnostic techniques have been developed, including traditional PCR, real-time PCR (RT-PCR), and loop-mediated isothermal amplification (LAMP) assays. These methods target bacterial DNA, circumventing the need for cultivable organisms and overcoming the many drawbacks of culture-based techniques (Table 3). The World Health Organization (WHO) recommends RT-PCR for detecting meningococcus in suspected meningitis cases.34
Table 3. Overview of the Most Used Methods for the Detection of N. meningitidis.
| Assay | Turnaround time | Antimicrobial treatment/antibiotic on outcomes | Quantitative measure |
|---|---|---|---|
| Culture | 24–48h or longer | Yes | No |
| Agglutination | 15 min-1h | Yes | No |
| LFA | 15–30 min | Yes | No |
| Traditional PCR | 3h | No | No |
| Real-time PCR | 3h | No | Yes |
| HM-MCDA-LFB | 1h | No | No |
| HM-RT-MCDA | 1h | No | Yes |
The NM-RT-MCDA assay developed in this study offers a significant advantage in turnaround time compared to traditional methods such as culture, lateral flow assays, and PCR-based techniques, which require hours or days for completion.20−22,32 This rapid assay enables the timely reporting of results to clinicians, facilitating prompt diagnosis and treatment decisions. The entire procedure, including rapid DNA extraction (15 min), isothermal amplification, endonuclease cleavage (40 min), and real-time fluorescence detection (2 min), can be completed within 1 h (Figure 1C).
Unlike the culture, agglutination, and LFA methods, which are susceptible to reduced sensitivity and specificity in patients who have received prior antibiotic treatment, the NM-RT-MCDA assay targets bacterial DNA, circumventing the limitations associated with detecting viable organisms. This study focused on the N. meningitidisctrA gene, a conserved gene present in all subtypes, as a target for the MCDA reaction. This target has demonstrated excellent sensitivity and specificity in previous studies.26 The assay utilizes ten primers, each targeting a distinct region within the ctrA gene, to enhance amplification specificity. Primer design was optimized using specialized software and algorithms to maximize the accuracy and efficiency (Figure 1). The NM-RT-MCDA assay effectively detected genomic DNA from N. meningitidis strains with high accuracy and efficiency under optimized conditions. Importantly, no cross-reactivity was observed with the non-N. meningitidis pathogens or blank controls (Figures 3–6), demonstrating the exceptional specificity of this assay. The assay achieved a detection limit of 100 fg/μL, which is comparable to that of the previously developed NM-MCDA-LFB assay (Figure 3). Clinical validation using 34 samples yielded five positive and 29 negative results, consistent with both the NM-MCDA-LFB assay results (Figure 6) and prior clinical diagnoses established via traditional PCR. These findings highlight the potential clinical utility of NM-RT-MCDA assay as a diagnostic tool for N. meningitidis.
Although molecular diagnostic techniques provide significant advantages, they also face challenges. Implementing these assays in low- and middle-income countries (LMICs) remains difficult because of factors such as extended turnaround times, limited access to skilled personnel, and variations in laboratory infrastructure. The NM-MCDA assay addresses some of these challenges by employing a simplified isothermal amplification approach that can be performed using a basic water bath or incubator, thereby eliminating the need for a thermal cycler. This method rapidly amplified the target ctrA gene, generating 109 to 1010 copies at an optimal temperature of 65 °C (Figure 2). Amplification results can be visualized using a lateral flow biosensor (LFB) or by observing the color change in a preadded chromogenic indicator (Figure 3).20,32 However, conventional NM-MCDA assays provide only qualitative or semiquantitative results, which are limited by the subjective interpretation of color changes and the potential for contamination inherent in biosensor-based detection (Table 3). To address this limitation, the present study introduces the NM-RT-MCDA assay, which combines MCDA with real-time fluorescence detection, enabling the quantitative analysis of target DNA. This quantitative capability is particularly valuable for monitoring the disease progression in severe cases.
In conclusion, we successfully developed a novel HM-RT-MCDA assay for rapid and accurate detection of N. meningitidis. This assay, targeting the conserved ctrA gene, displays a high specificity and sensitivity of 100 fg of target DNA per reaction. The entire procedure, encompassing rapid DNA extraction, isothermal amplification at 65 °C for 40 min, and real-time fluorescence detection, can be completed within 1 h. The speed, specificity, and sensitivity of the NM-RT-MCDA assay make it a promising diagnostic tool for invasive meningococcal disease, facilitating timely diagnosis and improving clinical management across diverse healthcare settings.
Data Availability Statement
The original contributions presented in the study are includedin the article/Supporting Information. Further inquiries can be directed to the corresponding authors.
Author Contributions
Y.W. and R.H. contributed to the design and implementation of the research, R.H. performed the experiments, analyzed the results and drafted the manuscript. C.S., J.Z., N.J., F.X. and X.H. helped the experiments and analysis of the data. S.L. and D.H. provided clinical sputum samples and materials, and supervised clinical guidance. Y.W. and S.L. conceived the original and supervised the project.
This study was funded by Beijing Nova Program (Grant Nos. Z211100002121042[Yi Wang]) and National Natural Science Foundation of China (Grant Nos. 82200115[Yi Wang]).
Ethical review and approval was not required for the study onhuman participants in accordance with the local legislation and institutional requirements. Written informed consent from the participants was not required to participate in this study in accordance with the national legislation and the institutional requirements.
The authors declare no competing financial interest.
References
- Takahashi H.; Haga M.; Sunagawa T.; Saitoh T.; Kitahara T.; Matsumoto S. Meningococcal carriage rates in healthy individuals in Japan determined using Loop-Mediated Isothermal Amplification and oral throat wash specimens. J. Infect. Chemother. 2016, 22 (7), 501–504. 10.1016/j.jiac.2015.12.016. [DOI] [PubMed] [Google Scholar]
- Caugant D. A.; Tzanakaki G.; Kriz P. Lessons from meningococcal carriage studies. FEMS Microbiol. Rev. 2007, 31 (1), 52–63. 10.1111/j.1574-6976.2006.00052.x. [DOI] [PubMed] [Google Scholar]
- Borrow R.; Alarcón P.; Carlos J.; Caugant D. A.; Christensen H.; Debbag R. The Global Meningococcal Initiative: Global epidemiology, the impact of vaccines on meningococcal disease and the importance of herd protection. Expert Rev. Vaccines 2017, 16 (4), 313–328. 10.1080/14760584.2017.1258308. [DOI] [PubMed] [Google Scholar]
- Dou M.; Sanjay S. T.; Dominguez D. C.; Liu P.; Xu F.; Li X. Multiplexed instrument-free meningitis diagnosis on a polymer/paper hybrid microfluidic biochip. Biosens. Bioelectron. 2017, 87, 865–873. 10.1016/j.bios.2016.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihret W.; Sletbakk Brusletto B.; Øvstebø R.; Siebke Troseid A. M.; Norheim G.; Merid Y. Molecular studies of meningococcal and pneumococcal meningitis patients in Ethiopia. Innate Immun. 2019, 25 (3), 158–167. 10.1177/1753425918806363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison O. B.; Brueggemann A. B.; Caugant D. A.; van der Ende A.; Frosch M.; Gray S.; Heuberger S.; Krizova P.; Olcen P.; Slack M.; et al. Molecular typing methods for outbreak detection and surveillance of invasive disease caused by Neisseria meningitidis, Haemophilus influenzae and Streptococcus pneumoniae, a review. Microbiology 2011, 157 (8), 2181–2195. 10.1099/mic.0.050518-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H.; Morita M.; Yasuda M.; Ohama Y.; Kobori Y.; Kojima M.; Shimuta K.; Akeda Y.; Ohnishi M. Detection of Novel US Neisseria meningitidis Urethritis Clade Subtypes in Japan. Emerg. Infect. Dis. 2023, 29 (11), 2210–2217. 10.3201/eid2911.231082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens D. S. Biology and pathogenesis of the evolutionarily successful, obligate human bacterium Neisseria meningitidis. Vaccine 2009, 27 (Suppl 2), B71–B77. 10.1016/j.vaccine.2009.04.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison O. B.; Claus H.; Jiang Y.; Bennett J. S.; Bratcher H. B.; Jolley K. A. Description and nomenclature of Neisseria meningitidis capsule locus. Emerging Infect. Dis. 2013, 19 (4), 566–573. 10.3201/eid1904.111799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisier P.; Nicolas P.; Djibo S.; Taha M. K.; Jeanne I.; Maïnassara H. B. Meningococcal meningitis: Unprecedented incidence of serogroup X-related cases in 2006 in Niger. Clin. Infect. Dis. 2007, 44 (5), 657–663. 10.1086/511646. [DOI] [PubMed] [Google Scholar]
- Schwartz B.; Moore P. S.; Broome C. V. Global epidemiology of meningococcal disease. Clin. Microbiol. Rev. 1989, 2 (Suppl), S118–S124. 10.1128/CMR.2.Suppl.S118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. A.; Kim D. W.; Dong B. Q.; Kim J. S.; Anh D. D.; Kilgore P. E. An expanded age range for meningococcal meningitis: Molecular diagnostic evidence from population-based surveillance in Asia. BMC Infect. Dis. 2012, 12, 310. 10.1186/1471-2334-12-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memish Z. A.; Goubeaud A.; Bröker M.; Malerczyk C.; Shibl A. M. Invasive meningococcal disease and travel. J. Infect. Public Health 2010, 3 (4), 143–151. 10.1016/j.jiph.2010.09.008. [DOI] [PubMed] [Google Scholar]
- Booy R.; Gentile A.; Nissen M.; Whelan J.; Abitbol V. Recent changes in the epidemiology of Neisseria meningitidis serogroup W across the world, current vaccination policy choices and possible future strategies. Hum. Vaccines Immunother. 2019, 15 (2), 470–480. 10.1080/21645515.2018.1532248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taha M. K.; Alonso J. M.; Cafferkey M.; Caugant D. A.; Clarke S. C.; Diggle M. A. Interlaboratory comparison of PCR-based identification and genogrouping of Neisseria meningitidis. J. Clin. Microbiol. 2005, 43 (1), 144–149. 10.1128/JCM.43.1.144-149.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrow R.; Findlow J.; Gray S.; Taylor S.; Kaczmarski E. Safe laboratory handling of Neisseria meningitidis. J. Infect. 2014, 68 (4), 305–312. 10.1016/j.jinf.2014.01.003. [DOI] [PubMed] [Google Scholar]
- Corless C. E.; Guiver M.; Borrow R.; Edwards-Jones V.; Fox A. J.; Kaczmarski E. B. Simultaneous detection of Neisseria meningitidis, Haemophilus influenzae, and Streptococcus pneumoniae in suspected cases of meningitis and septicemia using real-time PCR. J. Clin. Microbiol. 2001, 39 (4), 1553–1558. 10.1128/JCM.39.4.1553-1558.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiducci S.; Moriondo M.; Nieddu F.; Ricci S.; De Vitis E.; Casini A. Culture and Real-time Polymerase Chain reaction sensitivity in the diagnosis of invasive meningococcal disease: Does culture miss less severe cases?. PLoS One 2019, 14 (3), e0212922 10.1371/journal.pone.0212922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraisier C.; Stor R.; Tenebray B.; Sanson Y.; Nicolas P. Use of a new single multiplex PCR-based assay for direct simultaneous characterization of six Neisseria meningitidis serogroups. J. Clin. Microbiol. 2009, 47 (8), 2662–2666. 10.1128/JCM.02415-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.; Liu C.; Liu Y.; Ma Q.; Wang Y.; Wang Y. Development of a multiple cross displacement amplification combined with nanoparticles-based biosensor assay to detect Neisseria meningitidis. Infect. Drug. Resist. 2019, 12, 2077–2087. 10.2147/IDR.S210735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L.; Gu R.; Li X.; Song M.; Huang X.; Mu D. Multiple Cross Displacement Amplification Coupled with Lateral Flow Biosensor (MCDA-LFB) for rapid detection of Legionella pneumophila. BMC Microbiol. 2022, 22 (1), 20. 10.1186/s12866-021-02363-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.; Zhou Q.; Dong S.; Wang S.; Liu R.; Wu X. Multiple Cross Displacement Amplification Linked with Nanoparticles-Based Lateral Flow Biosensor in Screening of Hepatitis B Virus in Clinical Application. Infect. Drug. Resist. 2021, 14, 1219–1229. 10.2147/IDR.S297645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada-González D.; Sena-Torralba A.; Wicaksono W. P.; de la Escosura-Muñiz A.; Ivandini T. A.; Merkoçi A. Iridium oxide (IV) nanoparticle-based lateral flow immunoassay. Biosens. Bioelectron. 2019, 132, 132–135. 10.1016/j.bios.2019.02.049. [DOI] [PubMed] [Google Scholar]
- Xiao F.; Zhou J.; Sun C.; Huang X.; Zheng B.; Fu J. Loop-Mediated Isothermal Amplification Coupled With Nanoparticle-Based Biosensor: A Rapid and Sensitive Method to Detect Mycoplasma pneumoniae. Front. Cell. Infect. Microbiol. 2022, 12, 882855. 10.3389/fcimb.2022.882855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada-González D.; Merkoçi A. Nanoparticle-based lateral flow biosensors. Biosens. Bioelectron. 2015, 73, 47–63. 10.1016/j.bios.2015.05.050. [DOI] [PubMed] [Google Scholar]
- Whiley D. M.; Crisante M. E.; Syrmis M. W.; Mackay I. M.; Sloots T. P. Detection of Neisseria Meningitidis in clinical samples by a duplex real-time PCR targeting the porA and ctrA genes. Mol. Diagn. 2003, 7 (3), 141–145. 10.1007/BF03260030. [DOI] [PubMed] [Google Scholar]
- Diene S. M.; Bertelli C.; Pillonel T.; Jacquier N.; Croxatto A.; Jaton K.; Greub G. Comparative genomics of Neisseria meningitidis strains: New targets for molecular diagnostics. Clin. Microbiol. Infect. 2016, 22 (6), 568.e1–e7. 10.1016/j.cmi.2016.03.022. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Sun L.; Li J. Q.; Wang Z. M.; Jiao W. W.; Xiao J.; Shen C.; Xu F.; Qi H.; Wang Y.-H.; et al. Label-Free Cross-Priming Amplification Coupled With Endonuclease Restriction and Nanoparticles-Based Biosensor for Simultaneous Detection of Nucleic Acids and Prevention of Carryover Contamination. Front. Chem. 2019, 7, 322. 10.3389/fchem.2019.00322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Wang Y.; Wang H.; Xu J.; Ye C. A label-free technique for accurate detection of nucleic acid-based self-avoiding molecular recognition systems supplemented multiple cross-displacement amplification and nanoparticles based biosensor. Artif. Cells, Nanomed., Biotechnol. 2018, 46 (8), 1671–1684. 10.1080/21691401.2017.1389748. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Wang Y.; Ma A. J.; Li D. X.; Luo L. J.; Liu D. X.; Jin D.; Liu K.; Ye C.-Y. Rapid and Sensitive Isothermal Detection of Nucleic-acid Sequence by Multiple Cross Displacement Amplification. Sci. Rep. 2015, 5, 11902. 10.1038/srep11902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Wang Y.; Zhang L.; Liu D.; Luo L.; Li H. Multiplex, Rapid, and Sensitive Isothermal Detection of Nucleic-Acid Sequence by Endonuclease Restriction-Mediated Real-Time Multiple Cross Displacement Amplification. Front. Microbiol. 2016, 7, 753. 10.3389/fmicb.2016.00753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L.; Tang C.; Cai Y.; Nong J.; Zhang K.; Zhu L. Ultra-efficient multiple cross displacement amplification-lateral flow biosensor (MCDA-LFB) for serogroup identification of prevalent Neisseria meningitidis. Anal. Biochem. 2022, 654, 114740. 10.1016/j.ab.2022.114740. [DOI] [PubMed] [Google Scholar]
- van de Beek D.; Cabellos C.; Dzupova O.; Esposito S.; Klein M.; Kloek A. T. ESCMID guideline: Diagnosis and treatment of acute bacterial meningitis. Clin. Microbiol. Infect. 2016, 22, S37–62. 10.1016/j.cmi.2016.01.007. [DOI] [PubMed] [Google Scholar]
- World Health Organization & Centers for Disease Control and Prevention (U.S.). Laboratory methods for the diagnosis of meningitis caused by neisseria meningitidis, Streptococcus pneumoniae and Haemophilus influenzae, 2nd ed.; World Health Organization, 2011. [Google Scholar]
- Rishishwar L.; Katz L. S.; Sharma N. V.; Rowe L.; Frace M.; Dolan Thomas J. Genomic basis of a polyagglutinating isolate of Neisseria meningitidis. J. Bacteriol. 2012, 194 (20), 5649–5656. 10.1128/JB.06604-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddar C. H.; Terrade A.; Verhoeven P.; Njanpop-Lafourcade B. M.; Dosso M.; Sidikou F.; Mahamane A. E.; Lombart J.-P.; Razki A.; Hong E.; et al. Validation of a New Rapid Detection Test for Detection of Neisseria meningitidis A/C/W/X/Y Antigens in Cerebrospinal Fluid. J. Clin. Microbiol. 2020, 58 (3), 10–128. 10.1128/JCM.01699-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung S. F.; Cheng S. K.; Kamei D. T. Paper-Based Systems for Point-of-Care Biosensing. J. Lab. Autom. 2015, 20 (4), 316–333. 10.1177/2211068215577197. [DOI] [PubMed] [Google Scholar]
- Posthuma-Trumpie G. A.; Korf J.; van Amerongen A. Lateral flow (immuno)assay: Its strengths, weaknesses, opportunities and threats. A literature survey. Anal. Bioanal. Chem. 2009, 393 (2), 569–582. 10.1007/s00216-008-2287-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original contributions presented in the study are includedin the article/Supporting Information. Further inquiries can be directed to the corresponding authors.