ABSTRACT
Objective
In the past 5 years, a large number of serological assays for large‐scale detection of antibodies against severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) antigen emerged. Serological assays for SARS‐CoV‐2 were needed to support clinical diagnosis and epidemiological investigations. However, there were limited data on the diagnostic accuracy of these serological assays. We aimed to compare the diagnostic accuracy of 11 commercial serological assays for coronavirus disease‐2019 (COVID‐19) by taking the reverse transcriptase polymerase chain reaction (RT‐PCR) assays as the reference standard, which served as the control arm to conduct an indirect comparison of diagnostic accuracy for 11 different SARS‐CoV‐2 serological assays.
Methods
This meta‐analysis was conducted following the PRISMA 2020 reporting guideline. Electronic searches were performed using the Cochrane Library, PubMed, Embase, Web of Science, Chinese Biological Medicine Database (CBM), China National Knowledge Infrastructure (CNKI), WANFANG, and Chinese Weipu (VIP) databases. Fifty‐seven articles, including 11 serologic‐based IgG, IgM, and total antibodies assays for SARS‐CoV‐2, published before June 2024, were included in this meta‐analysis. The main outcome of this meta‐analysis used to evaluate the performance of 11 assays included pooled diagnostic odds ratio (DOR), area under the summary receiver operating characteristic (AUC), and summary receiver operating characteristic curve (SROC). The R software was used for adjusted indirect comparison to calculate the relative diagnostic odds ratio (RDOR) with corresponding 95% confidence intervals (CIs), and indirect comparison forest plots showed the results.
Results
A total of 57 articles met the eligibility criteria for inclusion in our meta‐analysis. The pooled DOR and the AUC for access SARS‐CoV‐2 IgG were 564.28 (95% CI 229.58−1386.91) and 1.00, and as for EDI novel coronavirus COVID‐19 IgG those were 85.27 (95% CI 53.99−134.68) and 0.95, for EDI novel coronavirus COVID‐19 IgM were 49.42 (95% CI 16.47−148.30) and 0.86, for iFlash‐SARS‐CoV‐2 IgG were 652.31 (95% CI 362.32−1174.41) and 0.97, for iFlash‐SARS‐CoV‐2 IgM were 36.72 (95% CI 12.42−108.54) and 0.76, for MAGLUMI 2019‐nCoV IgG were 145.44 (95% CI 59.37−356.30) and 0.90, for MAGLUMI 2019‐nCoV IgM were 21.59 (95% CI 14.27−32.67) and 0.59, for ortho‐clinical anti‐SARS‐CoV‐2 IgG were 719.46 (95% CI 262.34−1973.13) and 1.00, for ortho‐clinical anti‐SARS‐CoV‐2 total were 1104.60 (95% CI 395.64−3083.99) and 1.00, for Siemens SARS‐CoV‐2 total (COV2T) were 1143.37 (95% CI 316.49−4130.62) and 0.99, for Wantai SARS‐CoV‐2 total Ab were 1014.98 (95% CI 618.48−1665.66) and 1.00. The pooled DOR for assays‐based IgG (n = 43), assays‐based total antibody (n = 35), and assays‐based IgM (n = 20) was 242.88 (95% CI 157.66−374.16), 1215.90 (95% CI 547.14−2702.07), and 40.99 (95% CI 22.63−74.25). The diagnostic accuracy of assays‐based total antibody performed better than those of assays‐based IgG and assays‐based IgM; assays‐based IgG performed better than assays‐based IgM.
Conclusion
This study suggested that the Siemens SARS‐CoV‐2 total (COV2T), ortho‐clinical anti‐SARS‐CoV‐2 total, and Wantai SARS‐CoV‐2 total had the best overall diagnostic accuracy. The diagnostic efficacy of the assays‐based total antibody had statistically significantly higher accuracy than those of assays‐based IgG and assays‐based IgM for COVID‐19.
Keywords: adjusted indirect comparison, COVID‐19, meta‐analysis, SARS‐CoV2, serological assays
We aimed to compare the diagnostic accuracy of 11 commercial serological assays for coronavirus disease‐2019 (COVID‐19) by taking the reverse transcriptase polymerase chain reaction (RT‐PCR) assays as reference standard, which served as control arm to conduct an indirect comparison of diagnostic accuracy for 11 different SARS‐CoV‐2 serological assays.
1. Introduction
Severe acute respiratory coronavirus 2 (SARS‐CoV‐2), a novel coronavirus that caused coronavirus disease 2019 (COVID‐19), has become a pandemic threat in which serological testing from diagnosis to epidemiologic surveillance has been indispensable in the past 5 years. The molecular testing with real‐time reverse transcription polymerase chain reaction (RT‐PCR) for the detection of SARS‐CoV‐2 was the reference standard for COVID‐19 diagnosis. Besides SARS‐CoV‐2 RT‐PCR testing, serological testing comprising the detection of IgM, IgA, or IgG antibodies to SARS‐CoV‐2‐specific epitopes has the potential to play an important role in the confirmation in individual patients with suspected COVID‐19 symptoms, or for the past SARS‐CoV‐2 infections [1]. Immune response to SARS‐CoV‐2 included cell‐mediated and antibody‐mediated immunity [2]. The spike (S) glycoproteins with their receptor‐binding domain (RBD) and the nucleocapsid (N) protein were widely used as the most common antigens in commercial serological assays for the detection of specific antibodies [3, 4]. In specific contexts, serological testing might be instrumental for acute diagnostic purposes, particularly when the RT‐PCR fails to identify SARS‐CoV‐2, for example, in patients who are greater than 14 days from their onset of symptoms [5]. Freund et al. reported that serological markers as part of medical follow‐up of symptomatic COVID‐19 patients can be used for prognostication; the study found anti‐S levels were significantly associated with previous severe COVID‐19 [6]. In addition, serological testing has been reported to be significant and important for personalized vaccination plans. Vaccines are designed to induce antibodies to the S antigen or RBD [7]; vaccine‐induced antibodies may arise in response to the S antigen and are, therefore, potentially detectable by any assay using the S antigen or RBD. Freund et al. also reported that the trajectory of anti‐S IgG levels after vaccination was found to predict the response to future COVID vaccinations, and the determination of the characteristics of the humoral response to COVID‐19 vaccinations is significant in predicting the humoral response to the booster vaccines [8]. Serological testing also has potential utility for tracking the course of the SARS‐CoV‐2 pandemic in the community. Screening of individuals who may be a source for prophylactic or therapeutic neutralizing antibodies is another application of serological testing [9]. Multiple manufacturers offered various high‐throughput serological assays differing not only in their antibody isotypes (i.e., IgA, IgM, IgG, or total antibody) but also targeted SARS‐CoV‐2 antigens (i.e., the S1 subunit of the spike protein, N protein, or RBD). Due to urgency and demand in the initial days of the COVID‐19 pandemic, numerous serological assays were rapidly developed and have been validated on a limited number of samples. The diagnostic efficacy of serological assays varies greatly; few studies were conducted to compare the performance of these assays on a large scale. This study aimed to evaluate the analytic performance and diagnostic characteristics of 11 commercial serological assays for the detection of SARS‐CoV‐2 specific IgG, IgM, and total antibodies. The 11‐assays comparison included the access SARS‐CoV‐2 IgG assay from Beckman Coulter (USA), EDI novel coronavirus COVID‐19 IgG and EDI novel coronavirus COVID‐19 IgM assays from Epitope Diagnostics (San Diego, CA, USA), iFlash‐SARS‐CoV‐2 IgG and iFlash‐SARS‐CoV‐2 IgM from Shenzhen YHLO Biotech (Shenzhen, China), MAGLUMI 2019‐nCoV IgG and MAGLUMI 2019‐nCoV IgM assays from Snibe Diagnostic (Shenzhen, China), ortho‐clinical anti‐SARS‐CoV‐2 IgG and ortho‐clinical anti‐SARS‐CoV‐2 total assays from Ortho Clinical Diagnostics (France), Siemens SARS‐CoV‐2 total (COV2T) assay from Siemens (Munich, Germany), and Wantai SARS‐CoV‐2 total Ab assay from Wantai Biological Pharmacy Enterprise (Beijing, China). Meanwhile we assessed the diagnostic accuracy of antibody isotypes by meta‐analysis and indirect comparison.
2. Materials and Methods
2.1. Search Strategy
Studies were identified by searching the Cochrane Library, PubMed, Embase, Web of Science, Chinese Biological Medicine Database (CBM), China National Knowledge Infrastructure (CNKI), WANFANG, and Chinese Weipu (VIP) databases. The search terms used were (“2019‐nCoV” OR “coronavirus disease 2019 virus” OR “2019 novel coronavirus” OR “COVID‐19” OR “COVID‐19 diagnostic testing” OR “COVID‐19 serological test” OR “SARS‐CoV‐2” OR “severe acute respiratory syndrome coronavirus 2” OR “Anti‐SARS‐CoV‐2”) AND (“Access SARS‐CoV‐2 IgG” OR “EDI Novel Coronavirus COVID‐19” OR “iFlash‐SARS‐CoV‐2” OR “MAGLUMI 2019‐nCoV” OR “Ortho‐Clinical anti‐SARS‐CoV‐2” OR “Siemens SARS‐CoV‐2” OR “Siemens SARS‐CoV‐2 Total (COV2T)” OR “Wantai SARS‐CoV‐2”). The searches were limited to articles published in Chinese or English.
2.2. Inclusion and Exclusion Criteria
We included studies that evaluated the performance of the above‐mentioned 11 anti‐SARS‐CoV‐2 antibody serological assays. The 11‐assay comparison for inclusion in the data analysis met the following inclusion criteria: (1) Studies which included the COVID‐19 patients' serum samples and negative control serum samples reporting both sensitivity and specificity of serological assays for COVID‐19; (2) the diagnosis of SARS‐CoV‐2 (COVID‐19) by taking RT‐PCR as the reference standard meanwhile based on clinical symptoms and imaging diagnosis; (3) stored pre‐COVID‐19 blood samples collected from the healthy blood donors and the individuals with a history of PCR‐confirmed non‐COVID‐19 infection within the previous 6 months were used as negative control; (4) the number of true positive (TP), true negative (TN), false positive (FP), and false negative (FN) were then abstracted, or data that could transform into above information were reported.
The exclusion criteria: (1) The studies evaluated the performance of in‐house developed antibody assay for the diagnosis of COVID‐19 instead of any commercial serological assay; (2) the studies evaluated the serological assays for the detection of antibodies generated by vaccines against SARS‐CoV‐2; (3) the studies whose COVID‐19 patients were diagnosed without at least one positive RT‐PCR test carried out; (4) studies in which serological assays were evaluated without providing enough information for the immunoglobulin classes (IgG, IgA, IgM, or total antibody), the targeting antigen, manufacturer/platform or the method; (5) studies with negative control sample sizes or patients serums samples less than 30.
2.3. Data Extraction and Quality Assessment
Articles were independently assessed for inclusion by the two authors of this paper (Ying Zhao and Minjie Zhang), and data from included studies were extracted using the Quality Assessment of Diagnostic Accuracy Studies (QUADAS‐2) tool for the domains of patient selection, performance of the index test, performance of the reference test, and flow and timing (for risk of bias only). The extracted data included the name of the first author of the article, publication year, manufacturer, method, assay, immunoglobulin isotypes (IgM, IgG, or total antibody), type of antigen (S, N, or RBD), COVID‐19 patients sample size, and the negative control sample size. The TP, FP, TN, and FN results of each arm were reported separately.
2.4. Statistical Analysis
The pooled sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio (DOR), and the summary receiver operating characteristic curves (SROC) with corresponding 95% (confidence intervals, CI) were measured. The SROC curve (based on 2 × 2 contingency tables) was established to show the sensitivity and specificity for each individual arm, and the area under the curve (AUC) was used to determine diagnostic accuracy. Review Manager 5.3 (Cochrane Collaboration) analysis software was used to build the area under the SROC curve (AUC) graphics by making use of different colors for different serological assays. The relative diagnostic odds ratio (RDOR) of indirect comparison was used to compare the diagnostic accuracy of different assays and different immunoglobulin isotypes. The RDOR outcomes were summarized and exhibited in paired forest plots by R software (Parametric Technology Corporation). When the 95% CI of the RDOR contains 1, it indicates that the difference between the two comparison objects was not statistically significant; alternatively, when its 95% CI exceeds 1, suggesting that the difference between the two comparison objects was statistically significant. The Deek's test was used to evaluate whether there was publication bias.
3. Results
3.1. Study Characteristics
A PRISMA flow chart in Figure 1 was used. A total of 1781 pieces of literature were identified after the removal of duplicate articles. One hundred after full‐text review were assessed for eligibility. Fifty‐seven articles were included finally in the systematic review [10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66]. The detailed characteristics of the articles included in this study are shown in Table 1.
Figure 1.
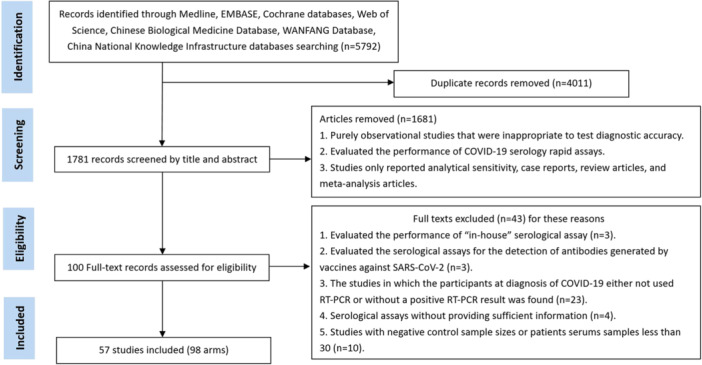
Flow diagram of selecting the literature and screening process.
Table 1.
Characteristics of the included studies.
| Author | Year | Method | Manufacturer/platform | Assay | Antigen | Antibody type | COVID‐19 patient samples (n) | Controls (n) |
|---|---|---|---|---|---|---|---|---|
| A Bown [10] | 2020 | CLIA | Siemens Atellica | Siemens SARS‐CoV‐2 total (COV2T) | S1 | Total antibody | 536 | 976 |
| Al‐Jighefee HT [11] | 2021 | ELISA | NA | EDI novel coronavirus COVID‐19 IgG | N | IgG | 291 | 119 |
| ELISA | NA | EDI novel coronavirus COVID‐19 IgM | N | IgM | 291 | 119 | ||
| Andrey DO [12] | 2021 | ELISA | DYNEX DSX | EDI novel coronavirus COVID‐19 IgG | N | IgG | 172 | 185 |
| Bundschuh C [13] | 2020 | ELISA | Serion Diagnostics | EDI novel coronavirus COVID‐19 IgG | N | IgG | 104 | 456 |
| ELISA | Serion Diagnostics | EDI novel coronavirus COVID‐19 IgM | N | IgM | 104 | 456 | ||
| Chiereghin A [14] | 2020 | CLIA | iFlash 3000 | iFlash‐SARS‐CoV‐2 IgG | N and S | IgG | 207 | 130 |
| CLIA | iFlash 3000 | iFlash‐SARS‐CoV‐2 IgM | N and S | IgM | 207 | 130 | ||
| Chua KYL [15] | 2020 | CLIA | Vitros 5600 | Ortho‐clinical anti‐SARS‐CoV‐2 IgG | S1 | IgG | 86 | 95 |
| CLIA | Beckman Unicel DxI 800 | Access SARS‐CoV‐2 IgG | S1‐RBD | IgG | 86 | 95 | ||
| Davidson N [16] | 2020 | ELISA | NA | EDI novel coronavirus COVID‐19 IgG | N | IgG | 71 | 138 |
| ELISA | NA | EDI novel coronavirus COVID‐19 IgM | N | IgM | 71 | 138 | ||
| Deng Jielun [17] | 2021 | CLIA | iFlash 3000 | iFlash‐SARS‐CoV‐2 IgG | N and S | IgG | 69 | 73 |
| CLIA | iFlash 3000 | iFlash‐SARS‐CoV‐2 IgM | N and S | IgM | 69 | 73 | ||
| Egger M [18] | 2020 | ELISA | NA | EDI novel coronavirus COVID‐19 IgG | N | IgG | 104 | 200 |
| ELISA | NA | EDI novel coronavirus COVID‐19 IgM | N | IgM | 104 | 200 | ||
| Florin L [19] | 2021 | CLIA | Siemens Atellica IM1300 | Siemens SARS‐CoV‐2 total (COV2T) | S1‐RBD | Total Ab | 175 | 90 |
| Garnett E [20] | 2020 | CLIA | Vitros 5600 | Ortho‐clinical anti‐SARS‐CoV‐2 total | S | Total antibody | 79 | 57 |
| Gdoura M [21] | 2022 | CLIA | Beckman Coulter | Access SARS‐CoV‐2 IgG | S1‐RBD | IgG | 72 | 119 |
| Han Xiaoyan [22] | 2023 | CLIA | iFlash 3000 | iFlash‐SARS‐CoV‐2 IgG | N and S | IgG | 123 | 41 |
| CLIA | iFlash 3000 | iFlash‐SARS‐CoV‐2 IgM | N and S | IgM | 123 | 41 | ||
| Harritshøj LH [23] | 2021 | CLIA | Siemens Atellica IM | Siemens SARS‐CoV‐2 total (COV2T) | S1 | Total antibody | 148 | 596 |
| CLIA | Siemens Dimension Vista 500 | Siemens SARS‐CoV‐2 total (COV2T) | S1 | Total antibody | 147 | 596 | ||
| CLIA | Maglumi 4000+ | MAGLUMI 2019‐nCoV IgG | Unspecified | IgG | 148 | 1173 | ||
| CLIA | Maglumi 800 | MAGLUMI 2019‐nCoV IgM | Unspecified | IgM | 150 | 1184 | ||
| CLIA | Vitros 3600 | Ortho‐clinical anti‐SARS‐CoV‐2 IgG | S1 | IgG | 150 | 600 | ||
| CLIA | Vitros 3600 | Ortho‐clinical anti‐SARS‐CoV‐2 total | S1 | Total antibody | 150 | 605 | ||
| ELISA | Tecan Sunrise | Wantai SARS‐CoV‐2 total Ab | S | Total antibody | 150 | 659 | ||
| CLIA | iFlash 1800 | iFlash‐SARS‐CoV‐2 IgG | N and S | IgG | 150 | 586 | ||
| CLIA | iFlash 1800 | iFlash‐SARS‐CoV‐2 IgM | N and S | IgM | 150 | 585 | ||
| Heffernan E [24] | 2021 | ELISA | Dynex DS2 | Wantai SARS‐CoV‐2 total Ab | S | Total antibody | 137 | 100 |
| Herroelen PH [25] | 2020 | ELISA | Bio‐Rad Version EIA 0_16 | Wantai SARS‐CoV‐2 total Ab | S1‐RBD | Total antibody | 169 | 57 |
| Hörber S [26]a | 2020 | CLIA | Siemens ADVIA Centaur XPT | Siemens SARS‐CoV‐2 total (COV2T) | S1 | Total antibody | 186 | 123 |
| Horn MP [27] | 2022 | ELISA | DYNEX DSX | EDI novel coronavirus COVID‐19 IgG | N | IgG | 192 | 3462 |
| Igawa G [28] | 2021 | CLIA | Siemens Dimension EXL 200 | Siemens SARS‐CoV‐2 total (COV2T) | S1‐RBD | Total Ab | 236 | 98 |
| Ikegami S [29] | 2021 | CLIA | Beckman Coulter | Access SARS‐CoV‐2 IgG | S1‐RBD | IgG | 97 | 100 |
| Irsara C [30] | 2021 | CLIA | Siemens ADVIA Centaur XPT | Siemens SARS‐CoV‐2 total (COV2T) | S1 | Total antibody | 195 | 288 |
| Kubota K [31] | 2021 | CLIA | Vitros 3600 | Ortho‐clinical anti‐SARS‐CoV‐2 IgG | S | IgG | 66 | 148 |
| CLIA | Vitros 3600 | Ortho‐clinical anti‐SARS‐CoV‐2 total | S1 | Total antibody | 66 | 148 | ||
| Kundu D [32] | 2022 | CLIA | Siemens ADVIA Centaur XPT | Siemens SARS‐CoV‐2 total (COV2T) | S1 | Total antibody | 153 | 150 |
| Lapić I [33] | 2020 | CLIA | Maglumi 800 | MAGLUMI 2019‐nCoV IgG | N and S | IgG | 42 | 48 |
| Lester SN [34] | 2024 | CLIA | Vitros ECi/ECiQ/3600 and Vitros 5600/XT 7600 | Ortho‐clinical anti‐SARS‐CoV‐2 total | S1 | Total antibody | 87 | 117 |
| Li Ping [35] | 2020 | CLIA | iFlash 3000 | iFlash‐SARS‐CoV‐2 IgG | N and S | IgG | 116 | 134 |
| CLIA | iFlash 3000 | iFlash‐SARS‐CoV‐2 IgM | N and S | IgM | 116 | 134 | ||
| Mafi S [36] | 2023 | ELISA | NA | Wantai SARS‐CoV‐2 total Ab | S1 | Total antibody | 110 | 120 |
| Mairesse A [37] | 2020 | CLIA | iFlash 1800 | iFlash‐SARS‐CoV‐2 IgG | N and S | IgG | 178 | 75 |
| CLIA | iFlash 1800 | iFlash‐SARS‐CoV‐2 IgM | N and S | IgM | 178 | 75 | ||
| Manthei DM [38] | 2021 | CLIA | Siemens Centaur XP | Siemens SARS‐CoV‐2 total (COV2T) | S1‐RBD | Total Ab | 131 | 188 |
| Marlet J [39] | 2020 | ELISA | NA | Wantai SARS‐CoV‐2 total Ab | S | Total Ab | 58 | 89 |
| Montesinos I [40] | 2020 | CLIA | Maglumi 800 | MAGLUMI 2019‐nCoV IgG | Unspecified | IgG | 126 | 72 |
| CLIA | Maglumi 800 | MAGLUMI 2019‐nCoV IgM | Unspecified | IgM | 126 | 72 | ||
| Naaber P [41] | 2020 | CLIA | Maglumi 1000 | MAGLUMI 2019‐nCoV IgG | Unspecified | IgG | 97 | 100 |
| ELISA | Dynex Agility | EDI novel coronavirus COVID‐19 IgG | N | IgG | 97 | 100 | ||
| Nedelcu I [42] | 2021 | ELISA | Dynex DSX | EDI novel coronavirus COVID‐19 IgG | N | IgG | 528 | 161 |
| ELISA | Dynex DSX | EDI novel coronavirus COVID‐19 IgM | N | IgM | 528 | 161 | ||
| Nicholson S [43] | 2021 | ELISA | NA | Wantai SARS‐CoV‐2 total Ab | S1 | Total antibody | 96 | 209 |
| Nyagwange J [44] | 2022 | ELISA | NA | Wantai SARS‐CoV‐2 total Ab | S1 | Total antibody | 149 | 327 |
| Oved K [45] | 2020 | CLIA | Beckman Coulter | Access SARS‐CoV‐2 IgG | S1‐RBD | IgG | 162 | 318 |
| 2020 | CLIA | Siemens ADVIA Centaur XPT | Siemens SARS‐CoV‐2 total (COV2T) | S1‐RBD | Total Ab | 156 | 432 | |
| Padoan A [46] | 2020 | CLIA | Vitros ECi/ECiQ/3600 and Vitros 5600/XT 7600 | Ortho‐clinical anti‐SARS‐CoV‐2 IgG | S | IgG | 130 | 54 |
| CLIA | Vitros ECi/ECiQ/3600 and Vitros 5600/XT 7600 | Ortho‐clinical anti‐SARS‐CoV‐2 total | S1 | Total antibody | 130 | 54 | ||
| Parai D [47] | 2021 | CLIA | iFlash 1800 | iFlash‐SARS‐CoV‐2 IgG | N and S | IgG | 594 | 100 |
| Pérez‐García F [48] | 2021 | CLIA | Siemens Atellica | Siemens SARS‐CoV‐2 total (COV2T) | S1 | Total antibody | 50 | 60 |
| Pflüger LS [49] | 2020 | CLIA | Siemens Atellica | Siemens SARS‐CoV‐2 total (COV2T) | S1 | Total Ab | 75 | 320 |
| ELISA | Euroimmun Analyzer I‐2 P | Wantai SARS‐CoV‐2 total Ab | S1 | Total Ab | 75 | 320 | ||
| Piec I [50] | 2021 | ELISA | Dynex Agility | EDI novel coronavirus COVID‐19 IgG | N | IgG | 43 | 152 |
| Riester E [51] | 2021 | CLIA | iFlash 1800 | iFlash‐SARS‐CoV‐2 IgG | N and S | IgG | 104 | 928 |
| CLIA | iFlash 1800 | iFlash‐SARS‐CoV‐2 IgM | N and S | IgM | 104 | 928 | ||
| Rikhtegaran Tehrani Z [52] | 2020 | ELISA | NA | EDI novel coronavirus COVID‐19 IgG | N | IgG | 97 | 288 |
| ELISA | NA | EDI novel coronavirus COVID‐19 IgM | N | IgM | 95 | 299 | ||
| Sekirov I [53] | 2021 | CLIA | Vitros XT 7600 | Ortho‐clinical anti‐SARS‐CoV‐2 IgG | S | IgG | 42 | 65 |
| CLIA | Vitros XT 7600 | Ortho‐clinical anti‐SARS‐CoV‐2 total | S1 | Total antibody | 42 | 65 | ||
| CLIA | Siemens ADVIA Centaur XPT | Siemens SARS‐CoV‐2 total (COV2T) | S1 | Total antibody | 42 | 65 | ||
| Şener B [54] | 2022 | CMIA | Beckman Coulter | Access SARS‐CoV‐2 IgG | S1‐RBD | IgG | 125 | 50 |
| CMIA | Siemens Atellica | Siemens SARS‐CoV‐2 total (COV2T) | S1‐RBD | Total Ab | 131 | 50 | ||
| Serre‐Miranda C [55] | 2021 | CLIA | NA | MAGLUMI 2019‐nCoV IgG | N and S | IgG | 117 | 35 |
| CLIA | NA | MAGLUMI 2019‐nCoV IgM | N and S | IgM | 117 | 35 | ||
| Soleimani R [56] | 2021 | CLIA | Maglumi 800 | MAGLUMI 2019‐nCoV IgG | N and S | IgG | 176 | 100 |
| 2021 | CLIA | Maglumi 800 | MAGLUMI 2019‐nCoV IgM | N and S | IgM | 176 | 100 | |
| Syre H [57] | 2022 | ELISA | DYNEX DS2 system | Wantai SARS‐CoV‐2 total Ab | S | Total antibody | 211 | 320 |
| Tan SS [58] | 2020 | CLIA | Beckman Unicel DxI 800 | Access SARS‐CoV‐2 IgG | S1‐RBD | IgG | 173 | 163 |
| CLIA | Siemens ADVIA Centaur XPT | Siemens SARS‐CoV‐2 total (COV2T) | S1 | Total antibody | 173 | 163 | ||
| CLIA | Vitros 3600 | Ortho‐clinical anti‐SARS‐CoV‐2 total | S1 | Total antibody | 173 | 163 | ||
| Theel ES [59] | 2020 | ELISA | Dynex Agility | EDI novel coronavirus COVID‐19 IgG | N | IgG | 61 | 149 |
| CLIA | Vitros 3600 | Ortho‐clinical anti‐SARS‐CoV‐2 IgG | S | IgG | 61 | 149 | ||
| Tolan NV [60] | 2023 | ELISA | NA | EDI novel coronavirus COVID‐19 IgG | N | IgG | 105 | 55 |
| ELISA | NA | EDI novel coronavirus COVID‐19 IgM | N | IgM | 103 | 57 | ||
| Van Elslande J [61] | 2020 | CLIA | Maglumi 800 | MAGLUMI 2019‐nCoV IgG | Unspecified | IgG | 223 | 113 |
| Velay A [62] | 2020 | ELISA | NA | EDI novel coronavirus COVID‐19 IgG | N | IgG | 198 | 100 |
| ELISA | NA | EDI novel coronavirus COVID‐19 IgM | N | IgM | 198 | 100 | ||
| Ward MD [63] | 2021 | CLIA | Siemens Atellica | Siemens SARS‐CoV‐2 total (COV2T) | S1 | Total antibody | 112 | 2030 |
| CLIA | Siemens EXL | Siemens SARS‐CoV‐2 total (COV2T) | S1 | Total antibody | 112 | 2030 | ||
| Wu Lianpeng [64] | 2021 | CLIA | iFlash 3000 | iFlash‐SARS‐CoV‐2 IgG | N and S | IgG | 179 | 50 |
| CLIA | iFlash 3000 | iFlash‐SARS‐CoV‐2 IgM | N and S | IgM | 179 | 50 | ||
| Yassine HM [65] | 2021 | ELISA | Epoch 2 | EDI novel coronavirus COVID‐19 IgG | N | IgG | 101 | 70 |
| Zilla M [66] | 2021 | CLIA | Beckman Coulter | Access SARS‐CoV‐2 IgG | S1‐RBD | IgG | 154 | 184 |
| CLIA | Siemens Centaur XP | Siemens SARS‐CoV‐2 total (COV2T) | S1 | Total antibody | 154 | 184 | ||
| CLIA | Siemens Vista 1500 | Siemens SARS‐CoV‐2 total (COV2T) | S1 | Total antibody | 154 | 184 |
If borderline results were considered positive.
Abbreviations: CLIA, chemiluminescent immunoassay; ELISA, enzyme immunoassay; N, nucleocapsid antigen; NA, not specified; RBD, receptor‐binding domain; S1, S1 domain of viral spike protein; S1/S2, recombinant S1 and S2 antigens.
3.2. Quality Assessment
The assessment was performed using the Review Manager Software version 5.3. Figure 2 summarized the QUADA‐2 assessment. Five articles were judged as “high” in the patient selection domain of the risk of bias (including Al‐Jighefee, Chiereghin, Ikegami, Naaber, and Nedelcu). Among the above five articles, two or three questions of a domain were answered as “no”; therefore, the level of risk of bias was judged as “high” in the patient selection domain. For the index test domain of the risk of bias, it was found that the first question of the domain was answered as “no” on account of the serological assays being evaluated not in blind; therefore, all articles were judged as “unclear”. Six articles (including Chiereghin, Harritshøj, Horn, Ikegami, Naaber, and Padoan) whose patients had been diagnosed with COVID‐19 by positive SARS‐CoV‐2 RT‐PCR regardless of clinical symptoms or contained recovered COVID‐19 patients were judged as “unclear” in the reference standard domain of the risk of bias. Thirteen articles (including Chiereghin, Chua, Davidson, Egger, Garnett, Horn, Igawa, Marlet, Riester, Sekirov, Syre, Tan, and Ward) were judged as “high” in the flow and timing domain. The applicability judgment results for the patient selection domain were consistent with the reference standard domain; six articles (including Al‐Jighefee, Chiereghin, Horn, Naaber, Padoan, and Pflüger), including asymptomatic COVID‐19 patients were judged as “high,” and nine articles (including Harritshøj, Ikegami, Irsara, Lapić, Oved, Parai, Theel, Tolan, and Yassine) including recovered COVID‐19 patients were judged as “unclear.”
Figure 2.
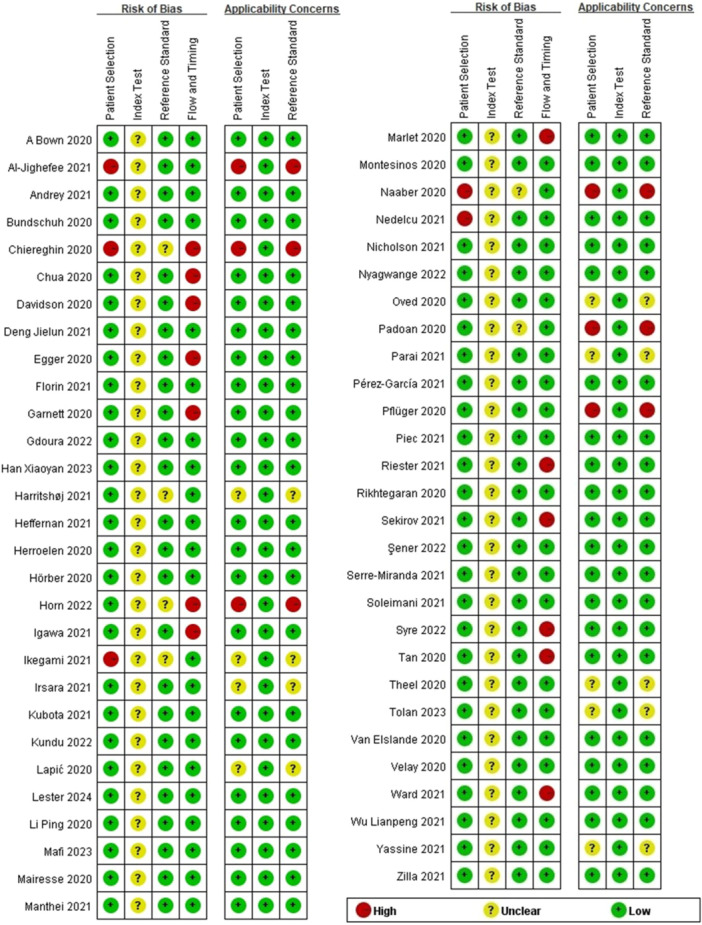
Risk of bias and applicability concerns summary.
3.3. Data Synthesis and Meta‐Analysis
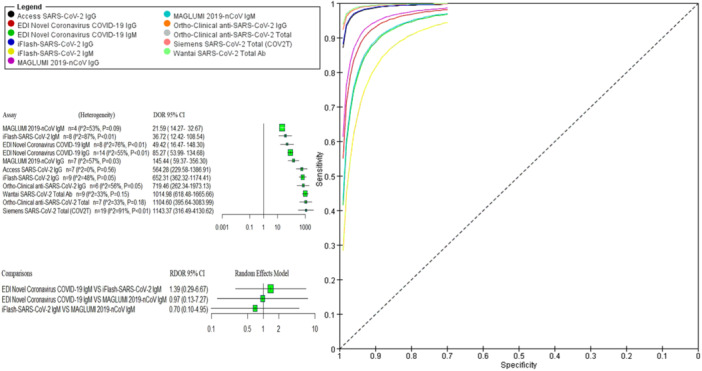
First, we evaluated the diagnostic accuracy of immunoglobulin isotypes (IgM, IgG, and total antibody). Due to significant heterogeneity in the analysis of three immunoglobulin isotypes, a random effects model was used. A forest plot of DOR with 95% CIs for immunoglobulin isotypes is shown in Figure 3A. The pooled DOR for IgG (242.88 [95% CI 157.66−374.16]), IgM (40.99 [95% CI 22.63−74.25]), and total antibody (1215.90 [95% CI 547.14−2702.07]) showed that assays‐based total antibody had the better diagnostic accuracy compared to assays‐based IgG and assays‐based IgM with significant difference. Moreover, indirect comparison results of RDOR with 95% CIs showed that assays‐based total antibody and assays‐based IgG showed significantly better diagnostic accuracy than assays‐based IgM (as shown in Figure 3B).
Figure 3.

(A) Pooled DOR with 95% CIs of immunoglobulin classes. (B) Indirect comparison forest plots of RDOR with 95% CIs for all three pairwise immunoglobulin comparisons. CIs, confidence intervals; DOR, diagnostic odds ratio; RDOR, relative diagnostic odds ratio.
As mentioned above, the diagnostic efficacy of the assays‐based total antibody had statistically significantly higher accuracy than those of assays‐based IgG and assays‐based IgM. We assessed the diagnostic accuracy of assays‐based IgM separately. Meta‐analyses evaluating the parameters of the accuracy of the reported assays were performed, and results are shown in Table 2. Forest plots of coupled sensitivity and specificity with 95% CIs for 11 serological assays are shown in Figure 4. We also constructed the SROC curves for all 11 serological assays (as shown in Figure 5A). The pooled DOR results of 11 serological assays were shown by forest plots (as shown in Figure 5B). There was no significant heterogeneity for access SARS‐CoV‐2 IgG, iFlash‐SARS‐CoV‐2 IgG, MAGLUMI 2019‐nCoV IgM, ortho‐clinical anti‐SARS‐CoV‐2 IgG, ortho‐clinical anti‐SARS‐CoV‐2 total, and Wantai SARS‐CoV‐2 total Ab, and a fixed effects model was used. The results of RDOR with 95% CIs were exhibited in the indirect comparison forest plot by R software. From the pooled DOR and the SROC curves, the overall diagnostic accuracy of the Siemens SARS‐CoV‐2 total (COV2T) (1143.37 [95% CI 316.49−4130.62]), ortho‐clinical anti‐SARS‐CoV‐2 total (1104.60 [95% CI 395.64−3083.99]), Wantai SARS‐CoV‐2 total Ab (1014.98 [95% CI 618.48−1665.66]), and ortho‐clinical anti‐SARS‐CoV‐2 IgG (719.46 [95% CI 262.34−1973.13]) performed better than the other serological assays; indirect comparison results of RDOR with 95% CIs for these four pairwise assays showed that there was no significant difference between them. Meanwhile, the RDOR results suggested that the diagnostic accuracy of these four assays was statistically significantly higher than EDI novel coronavirus COVID‐19 IgG. The RDOR value of EDI novel coronavirus COVID‐19 IgG versus iFlash‐SARS‐CoV‐2 IgG was 0.19 (95% CI 0.04−0.94), which suggested that the diagnostic accuracy of iFlash‐SARS‐CoV‐2 IgG was statistically significantly higher than EDI novel coronavirus COVID‐19 IgG (as shown in Figure 6). The diagnostic accuracy of the three IgM assays had no significant difference (as shown in Figure 5C).
Table 2.
Summary table of the diagnostic accuracy.
| Assay | Pooled analysis results (95% CI) | Heterogeneity | Deek's test | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Pooled sensitivity | Pooled specificity | Pooled PLR | Pooled NLR | AUC | DOR | I 2 (%) | p | t | p | |
| Access SARS‐CoV‐2 IgG | 0.78 (0.64−0.87) | 1.00 (0.99−1.00) | 275.20 (82.90−914.00) | 0.22 (0.13−0.38) | 1.00 | 564.28 (229.58−1386.91)a | 0 | 0.56 | −0.53 | 0.618 |
| EDI novel coronavirus COVID‐19 IgG | 0.67 (0.60−0.74) | 0.98 (0.97−0.99) | 36.10 (21.90−59.50) | 0.34 (0.27−0.41) | 0.95 | 85.27 (53.99−134.68) | 55 | < 0.01 | −0.18 | 0.859 |
|
EDI novel coronavirus COVID‐19 IgG (omitting Davidson) |
0.68 (0.60−0.74) | 0.98 (0.97−0.99) | 38.20 (24.10−60.70) | 0.33 (0.26−0.41) | 0.97 | 87.61 (67.43−113.81)a | 32 | 0.13 | 0.25 | 0.806 |
| EDI novel coronavirus COVID‐19 IgM | 0.41 (0.30−0.52) | 0.99 (0.97−1.00) | 47.20 (12.80−173.40) | 0.60 (0.49−0.73) | 0.86 | 49.42 (16.47−148.30) | 76 | < 0.01 | 0.32 | 0.758 |
|
EDI novel coronavirus COVID‐19 IgM (omitting Davidson) |
0.44 (0.33−0.55) | 0.99 (0.98−1.00) | 63.60 (18.40−219.30) | 0.57 (0.46−0.70) | 0.91 | 67.86 (25.55−180.22) | 57 | 0.03 | 1.12 | 0.315 |
| iFlash‐SARS‐CoV‐2 IgG | 0.87 (0.82−0.90) | 1.00 (0.98−1.00) | 195.10 (44.00−865.00) | 0.13 (0.10−0.18) | 0.97 | 652.31 (362.32−1174.41)a | 48 | 0.05 | −2.52 | 0.040b |
| iFlash‐SARS‐CoV‐2 IgM | 0.52 (0.49−0.55) | 0.98 (0.97−0.99) | 18.37 (5.71−59.05) | 0.52 (0.45−0.60) | 0.76 | 36.72 (12.42−108.54) | 87 | < 0.01 | −3.41 | 0.014b |
|
iFlash‐SARS‐CoV‐2 IgM (omitting Han Xiaoyan) |
0.50 (0.41−0.60) | 0.98 (0.96−0.99) | 32.30 (15.70−66.20) | 0.50 (0.42−0.61) | 0.86 | 49.19 (31.92−75.81)a | 51 | 0.06 | −2.37 | 0.064 |
| MAGLUMI 2019‐nCoV IgG | 0.69 (0.62−0.74) | 0.99 (0.97−1.00) | 71.30 (22.80−223.00) | 0.32 (0.26−0.38) | 0.90 | 145.44 (59.37−356.30) | 57 | 0.03 | −1.26 | 0.263 |
|
MAGLUMI 2019‐nCoV IgG (omitting Harritshøj) |
0.66 (0.60−0.72) | 0.99 (0.95−1.00) | 59.30 (13.40−261.50) | 0.34 (0.29−0.40) | 0.83 | 83.80 (42.86−163.85)a | 0 | 0.72 | 0.12 | 0.912 |
| MAGLUMI 2019‐nCoV IgM | 0.55 (0.51−0.59) | 0.97 (0.96−0.98) | 23.77 (4.96−113.88) | 0.45 (0.36−0.57) | 0.59 | 21.59 (14.27−32.67)a | 53 | 0.09 | 0.74 | 0.534 |
| Ortho‐clinical anti‐SARS‐CoV‐2 IgG | 0.86 (0.64−0.96) | 1.00 (0.99−1.00) | 330.20 (69.80−1562.30) | 0.14 (0.04−0.42) | 1.00 | 719.46 (262.34−1973.13)a | 56 | 0.05 | −0.68 | 0.536 |
|
Ortho‐clinical anti‐SARS‐CoV‐2 IgG (omitting Harritshøj) |
0.85 (0.54‐0.96) | 0.99 (0.98‐1.00) | 139.20 (44.90−431.50) | 0.16 (0.04−0.56) | 1.00 | 459.87 (156.30−1353.00)a | 33 | 0.20 | 1.13 | 0.340 |
| Ortho‐clinical anti‐SARS‐CoV‐2 total | 0.88 (0.70−0.96) | 1.00 (0.98−1.00) | 1036.40 (55.80−19258.80) | 0.12 (0.04−0.34) | 1.00 | 1104.60 (395.64−3083.99)a | 33 | 0.18 | −1.28 | 0.256 |
| Siemens SARS‐CoV‐2 total (COV2T) | 0.85 (0.78−0.90) | 1.00 (0.99−1.00) | 521.80 (135.30−2011.80) | 0.15 (0.10−0.22) | 0.99 | 1143.37 (316.49−4130.62) | 91 | < 0.01 | −4.11 | 0.001b |
|
Siemens SARS‐CoV‐2 total (COV2T) (omitting Kundu) |
0.85 (0.78−0.90) | 1.00 (1.00−1.00) | 301.60 (196.80−462.10) | 0.15 (0.10−0.22) | 1.00 | 1581.39 (837.31−2986.72) | 45 | 0.02 | −4.90 | 0.000b |
| Wantai SARS‐CoV‐2 total Ab | 0.93 (0.88−0.96) | 0.99 (0.98−1.00) | 107.80 (50.80−229.00) | 0.07 (0.04−0.12) | 1.00 | 1014.98 (618.48−1665.66)a | 33 | 0.15 | −0.10 | 0.919 |
The pooled DOR was calculated with a fixed effects model.
Significant publication bias was observed.
Figure 4.
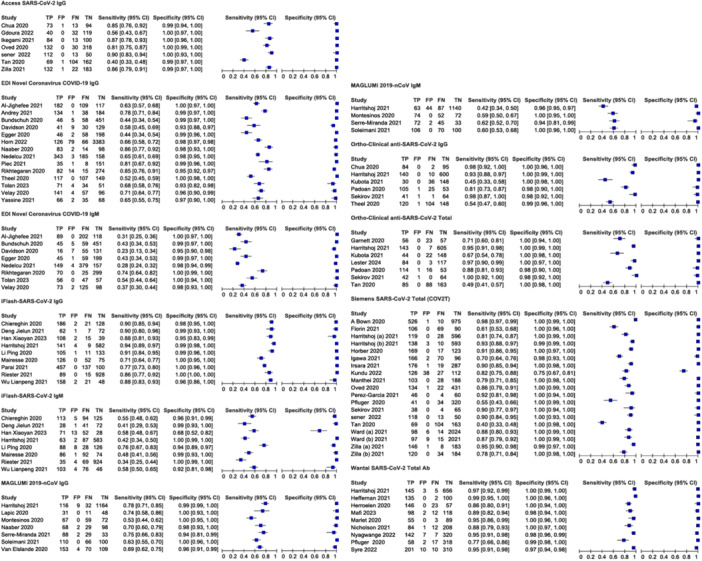
Forest plots of coupled sensitivity and specificity with 95% confidence intervals for 11 serological assays. Harritshøj (A): Siemens Vista assay was performed, Harritshøj (B): Siemens Atellica assay was performed; Ward (A): Siemens Atellica assay was used, Ward (B): Siemens EXL systems were used; Zilla (A): Siemens Centaur assay was performed; Zilla (B): Siemens Vista assay was performed.
Figure 5.
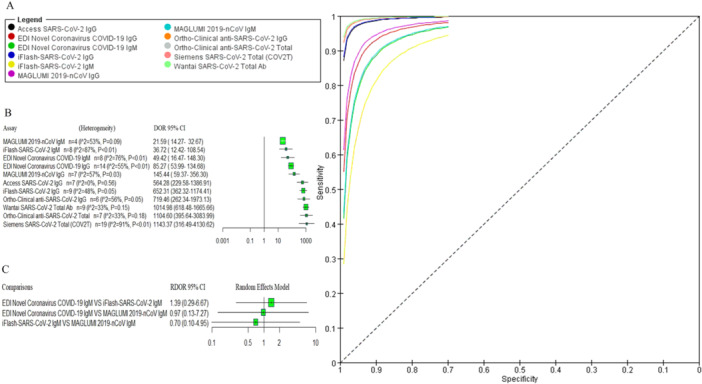
(A) SROC curve of the 11 serological assays. (B) Pooled DOR with 95% CIs of the 11 serological assays. (C) Indirect comparison forest plots of RDOR with 95% CIs for three IgM assays pairwise comparisons. CIs, confidence intervals; DOR, diagnostic odds ratio; RDOR, relative diagnostic odds ratio; SROC, summary receiver operating characteristics.
Figure 6.
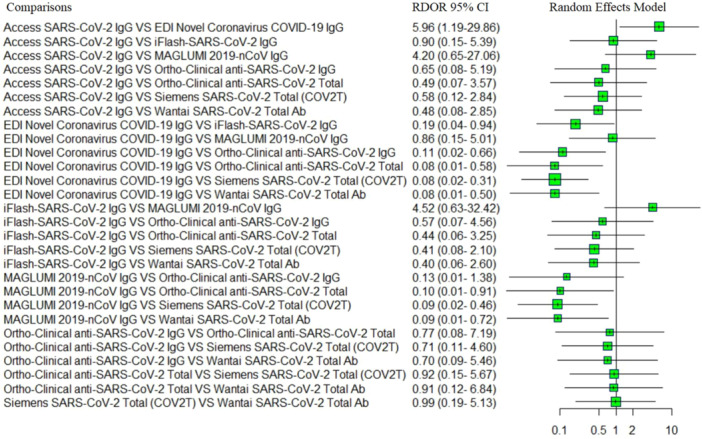
Indirect comparison forest plots of RDOR with 95% CIs for IgG and total antibody assays pairwise comparisons. CIs, confidence intervals; RDOR, relative diagnostic odds ratio.
3.4. Heterogeneity Test and Sensitivity Analysis
We investigated heterogeneity for 11 serological assays; significant high heterogeneity was observed for EDI novel coronavirus COVID‐19 IgG, EDI novel coronavirus COVID‐19 IgM, iFlash‐SARS‐CoV‐2 IgM, MAGLUMI 2019‐nCoV IgG, and Siemens SARS‐CoV‐2 total. To determine the possible source of heterogeneity, sensitivity analysis was performed. Omitting a single study did not significantly affect the pooled DOR. Nevertheless, no significant heterogeneity was observed for EDI novel coronavirus COVID‐19 IgG (p = 0.13, I 2 = 32%) when the study “Davidson 2020” was removed as well as for iFlash‐SARS‐CoV‐2 IgM (p = 0.06, I 2 = 51%) when study “Han Xiaoyan 2023” was removed and for ortho‐clinical anti‐SARS‐CoV‐2 IgG (p = 0.20, I 2 = 33%) when study “Harritshøj 20213” was removed. The heterogeneity decreased for EDI novel coronavirus COVID‐19 IgM (p = 0.03, I 2 = 57%) when study “Davidson 2020” was removed as well as for Siemens SARS‐CoV‐2 total (p = 0.02, I 2 = 45%) when study “Kundu 2022” was removed. While “Harritshøj 2021” was the primary cause of the heterogeneity for MAGLUMI 2019‐nCoV IgG (p = 0.72, I 2 = 0%, omitting “Harritshøj 2021”) (as shown in Table 2).
3.5. Risk of Bias Assessment
The Deek's test was performed to detect publication bias for iFlash‐SARS‐CoV‐2 IgG, iFlash‐SARS‐CoV‐2 IgM, and Siemens SARS‐CoV‐2 total, and the results of publication bias showed that the p value for aromatase was < 0.05, indicating that significant publication bias was observed (as shown in Table 2).
4. Discussion
Since the start of the COVID‐19 pandemic, an increasing number of serological SARS‐CoV‐2 assays have been introduced to the diagnostic market. We have demonstrated a comprehensive evaluation of 11 commercially available anti‐SARS‐CoV‐2 antibody assays. First, we evaluated the diagnostic efficiency of eight assays for detecting IgG and total antibodies against SARS‐CoV‐2. Taking into account the manufacturer's threshold, the pooled sensitivity among the evaluated eight assays ranged from 67% to 93%. Wantai SARS‐CoV‐2 total Ab and ortho‐clinical anti‐SARS‐CoV‐2 total assays had the best pooled sensitivity, followed by the iFlash‐SARS‐CoV‐2 IgG, ortho‐clinical anti‐SARS‐CoV‐2 IgG, and Siemens SARS‐CoV‐2 total (COV2T) assays. The high sensitivity of the two assays could be due to the ability of the two assays to detect all immunoglobulin classes. The pooled sensitivity of the EDI Novel Coronavirus COVID‐19 IgG and MAGLUMI 2019‐nCoV IgG assays was the lowest with a positive rate of 67% and 69% in the neglect of the three assays‐based IgM conditions, the pooled sensitivity of the three IgM assays varied from 41% to 55%. The overall sensitivity of the IgM assays was low, suggesting that there was limited utility in testing for IgM assays. Other studies have suggested development of IgM may occur earlier using a nucleocapsid antigen target compared to the spike glycoproteins [67, 68]. The pooled specificity of MAGLUMI 2019‐nCoV IgM was low compared to the other assays; all other assays demonstrated pooled specificity exceeding 98%. Besides the sensitivity and specificity, the pooled PLR (1036.40 and 521.80) and pooled NLR (0.12 and 0.15) for ortho‐clinical anti‐SARS‐CoV‐2 total and Siemens SARS‐CoV‐2 Total (COV2T) assays were also better than those for the other assays. Nevertheless, the pooled NLR (0.07) for Wantai SARS‐CoV‐2 total Ab displayed the best performance. The pooled DOR of Siemens SARS‐CoV‐2 total (COV2T), ortho‐clinical anti‐SARS‐CoV‐2 total, and Wantai SARS‐CoV‐2 total Ab assays were sharply higher compared to the other assays. We also constructed the SROC curves using RevMan 5.3 software and calculated the AUC using STATA software (version 12). The AUC was 1.00 for access SARS‐CoV‐2 IgG, ortho‐clinical anti‐SARS‐CoV‐2 IgG, ortho‐clinical anti‐SARS‐CoV‐2 total, and Wantai SARS‐CoV‐2 total Ab, 0.99 for Siemens SARS‐CoV‐2 total (COV2T). The results of the SROC curve and the AUC suggested that the diagnostic accuracy of those three assays (Siemens SARS‐CoV‐2 total, ortho‐clinical anti‐SARS‐CoV‐2 total, and Wantai SARS‐CoV‐2 total Ab) were relatively higher than the other assays.
Subsequently, we used RT‐PCR as the reference standard and conducted an indirect comparison between the 11 assays by calculating the RDOR value using R software. The adjustment indirect comparison forest plots of RDOR showed that the diagnostic accuracy of the four assays (Siemens SARS‐CoV‐2 total, ortho‐clinical anti‐SARS‐CoV‐2 total, Wantai SARS‐CoV‐2 total Ab, and ortho‐clinical anti‐SARS‐CoV‐2 IgG) had no significant difference and we also did not observe a significant difference between the other three assays (EDI novel coronavirus COVID‐19 IgM, iFlash‐SARS‐CoV‐2 IgM, and MAGLUMI 2019‐nCoV IgM) in the diagnostic accuracy of COVID‐19. We also constructed an indirect comparison to compare the diagnostic accuracy of immunoglobulin classes recognized (IgM, IgG, and total antibody). In our study, the pooled DOR of assays‐based IgM were low compared to the assays‐based IgG and the assays‐based total antibody. Independent of the serological method, the diagnostic performance of the IgM‐specific assays was lower than that of IgG and total antibody‐specific assays. Another study reported that SARS‐CoV‐2‐specific IgM is detected mostly in the early infection phase but only in rare cases [69, 70]. The antibodies assessed in these assays refer to structural antigenic proteins of SARS‐CoV‐2; the 11 serological assays differ in the type of immunoglobulin classes recognized as well as the nature of the antigen used for antibody recognition. At present, many studies have performed a structured systematic review and meta‐analysis to evaluate the diagnostic characteristics of serological testing for the detection of SARS‐CoV‐2 antibodies. Most of them provided the pooled analysis results (e.g., sensitivities and specificities) regarding the accuracy parameters of the reported serological assays. There are a limited number of comparable serological assays in the studies performed head‐to‐head comparisons. Under the condition of insufficient direct comparative study, we conducted an indirect comparison of the diagnostic efficacy of 11 assays; our data provide the overall diagnostic efficacy of 11 assays, as well as the antibody isotypes. Additionally, it is important to point out that our study took RT‐PCR as a reference standard and conducted an indirect comparison to compare the efficacy of antibody assays by calculating the RDOR value between them. To visualize results, we provide forest plots showing the RDOR with 95% CI of the 11‐assay comparison by R software.
This meta‐analysis also had some limitations. Due to the included studies differing in terms of method, manufacturer, and period of blood collection, we found high heterogeneity rates among trials. The expression change of antibodies against SARS‐CoV‐2 and the methods used for the detection of antibodies may have an effect on the overall diagnostic accuracy of serological assays. Because few articles included in this meta‐analysis have provided the data regarding the TP, FP, FN, and TN values at different sampling times, we could not directly address whether or not the sampling time affects the assay performance. The second is related to the methodological qualities of the primary studies. In the eligibility criteria, we did not set very strict definitions for the diagnosis of COVID‐19; in some studies, asymptomatic COVID‐19 and recovered COVID‐19 patient's serum samples comprise a large proportion. Specifically, the methods used for diagnosing COVID‐19 were not described in most studies. Another limitation was that this meta‐analysis had high heterogeneity, and sensitivity analysis indicated that the heterogeneity may be derived from a single study.
5. Conclusions
This study suggested that the Siemens SARS‐CoV‐2 total (COV2T), ortho‐clinical anti‐SARS‐CoV‐2 total, and Wantai SARS‐CoV‐2 total had high diagnostic efficiency. The diagnostic efficacy of the assays‐based total antibody had statistically significantly higher accuracy than those of assays‐based IgG and assays‐based IgM for COVID‐19.
Author Contributions
Ying Zhao and Minjie Zhang: conception, design, and administrative support. Weiwei Liang, Lijiang Fang, and Ying Zhao: data analysis and interpretation. Minjie Zhang, Weiwei Liang, and Lijiang Fang: manuscript writing, collection, and assembly of data. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
Data Availability Statement
Requests for any data can be made to the authors.
References
- 1. Braun J., Loyal L., Frentsch M., et al., “SARS‐CoV‐2‐Reactive T Cells in Healthy Donors and Patients With COVID‐19,” Nature 587, no. 7833 (2020): 270–274. [DOI] [PubMed] [Google Scholar]
- 2. Emmerich P., von Possel R., Hemmer C. J., et al., “Longitudinal Detection of SARS‐CoV‐2‐Specific Antibody Responses With Different Serological Methods,” Journal of Medical Virology 93, no. 10 (2021): 5816–5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Espejo A. P., Akgun Y., Al Mana A. F., et al., “Review of Current Advances in Serologic Testing for COVID‐19,” American Journal of Clinical Pathology 154, no. 3 (2020): 293–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Poh C. M., Carissimo G., Wang B., et al., “Two Linear Epitopes on the SARS‐CoV‐2 Spike Protein That Elicit Neutralising Antibodies in COVID‐19 Patients,” Nature Communications 11, no. 1 (2020): 2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hanson K. E., Caliendo A. M., Arias C. A., et al., “Infectious Diseases Society of America Guidelines on the Diagnosis of COVID‐19: Serologic Testing,” Clinical Infectious Diseases 78, no. 7 (2024): e150–e169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Freund O., Breslavsky A., Fried S., et al., “Interactions and Clinical Implications of Serological and Respiratory Variables 3 Months After Acute COVID‐19,” Clinical and Experimental Medicine 23, no. 7 (2023): 3729–3736. [DOI] [PubMed] [Google Scholar]
- 7. Valdes‐Balbin Y., Santana‐Mederos D., Paquet F., et al., “Molecular Aspects Concerning the Use of the SARS‐CoV‐2 Receptor Binding Domain as a Target for Preventive Vaccines,” ACS Central Science 7, no. 5 (2021): 757–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Freund O., Harish A., Breslavsky A., et al., “The Humoral Response to COVID‐19 Vaccinations Can Predict the Booster Effect on Health Care Workers‐Toward Personalized Vaccinations?,” Journal of Public Health 46, no. 1 (2024): e78–e83. [DOI] [PubMed] [Google Scholar]
- 9. Hayden M. K., El Mikati I. K., Hanson K. E., et al., “Infectious Diseases Society of America Guidelines on the Diagnosis of COVID‐19: Serologic Testing,” Clinical Infectious Diseases. Published ahead of print, March 15, 2024, 10.1093/cid/ciae121. [DOI] [PubMed] [Google Scholar]
- 10. Ainsworth M., Andersson M., Auckland K., et al., “Performance Characteristics of Five Immunoassays for SARS‐CoV‐2: A Head‐to‐Head Benchmark Comparison,” Lancet Infectious Diseases 20, no. 12 (2020): 1390–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Al‐Jighefee H. T., Yassine H. M., Al‐Nesf M. A., et al., “Evaluation of Antibody Response in Symptomatic and Asymptomatic COVID‐19 Patients and Diagnostic Assessment of New IgM/IgG ELISA Kits,” Pathogens 10, no. 2 (2021): 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Andrey D. O., Yerly S., Meyer B., et al., “Head‐to‐Head Evaluation of Five Automated SARS‐CoV‐2 Serology Immunoassays in Various Prevalence Settings,” Journal of Clinical Medicine 10, no. 8 (2021): 1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bundschuh C., Egger M., Wiesinger K., et al., “Evaluation of the EDI Enzyme Linked Immunosorbent Assays for the Detection of SARS‐CoV‐2 IgM and IgG Antibodies in Human Plasma,” Clinica Chimica Acta 509 (2020): 79–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chiereghin A., Zagari R. M., Galli S., et al., “Recent Advances in the Evaluation of Serological Assays for the Diagnosis of SARS‐CoV‐2 Infection and COVID‐19,” Frontiers in Public Health 8 (2020): 620222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chua K. Y. L., Vogrin S., Bittar I., et al., “Clinical Evaluation of Four Commercial Immunoassays for the Detection of Antibodies Against Established SARS‐CoV‐2 Infection,” Pathology 52, no. 7 (2020): 778–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Davidson N., Evans J., Giammichele D., et al., “Comparative Analysis of Three Laboratory Based Serological Assays for SARS‐CoV‐2 in an Australian Cohort,” Pathology 52, no. 7 (2020): 764–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Deng J., Wang Y., Keping A., et al., “Comparison of Diagnostic Efficiency of SARS‐COV‐2 Antibody Reagent Detected by Chemiluminescence Assay and Antibody Screening Strategies,” Sichuan Medical Journal 42, no. 1 (2021): 78–81. [Google Scholar]
- 18. Egger M., Bundschuh C., Wiesinger K., et al., “Comparison of the Elecsys Anti‐SARS‐CoV‐2 Immunoassay With the EDI Enzyme Linked Immunosorbent Assays for the Detection of SARS‐CoV‐2 Antibodies in Human Plasma,” Clinica Chimica Acta 509 (2020): 18–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Florin L., Maelegheer K., Vandewal W., Bernard D., and Robbrecht J., “Performance Evaluation of the Siemens SARS‐CoV‐2 Total Antibody and IgG Antibody Test,” Laboratory Medicine 52, no. 6 (2021): e147–e153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Garnett E., Jung J., Tam E., et al., “Clinical Validation and Performance Evaluation of the Automated Vitros Total Anti‐SARS‐CoV‐2 Antibodies Assay for Screening of Serostatus in COVID‐19,” American Journal of Clinical Pathology 154, no. 6 (2020): 742–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gdoura M., Halouani H., Sahli D., et al., “SARS‐CoV‐2 Serology: Utility and Limits of Different Antigen‐Based Tests Through the Evaluation and the Comparison of Four Commercial Tests,” Biomedicines 10, no. 12 (2022): 3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xiaoyan Han, “The Value of CLIA to Detect the IgM and IgG Antibody's of the New Coronavirus (SARS‐CoV‐2) in the Auxiliary Diagnosis of the New Coronavirus Pneumonia (COVID‐2019),” Medical Journal of Liaoning 37, no. 2 (2023): 55–58. [Google Scholar]
- 23. Harritshøj L. H., Gybel‐Brask M., Afzal S., et al., “Comparison of 16 Serological SARS‐CoV‐2 Immunoassays in 16 Clinical Laboratories,” Journal of Clinical Microbiology 59, no. 5 (2021): e02596–20, 10.1128/JCM.02596-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Heffernan E., Kennedy L., Hannan M. M., et al., “Performance Characteristics of Five SARS‐CoV‐2 Serological Assays: Clinical Utility in Health‐Care Workers,” Annals of Clinical Biochemistry: International Journal of Laboratory Medicine 58, no. 5 (2021): 496–504. [DOI] [PubMed] [Google Scholar]
- 25. Herroelen P. H., Martens G. A., De Smet D., Swaerts K., and Decavele A. S., “Humoral Immune Response to SARS‐CoV‐2,” American Journal of Clinical Pathology 154, no. 5 (2020): 610–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hörber S., Soldo J., Relker L., et al., “Evaluation of Three Fully‐Automated SARS‐CoV‐2 Antibody Assays,” Clinical Chemistry and Laboratory Medicine (CCLM) 58, no. 12 (2020): 2113–2120. [DOI] [PubMed] [Google Scholar]
- 27. Horn M. P., Jonsdottir H. R., Brigger D., et al., “Serological Testing for SARS‐CoV‐2 Antibodies in Clinical Practice: A Comparative Diagnostic Accuracy Study,” Allergy 77, no. 7 (2022): 2090–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Igawa G., Yamamoto T., Baba Y., et al., “Clinical Evaluation of Siemens SARS‐CoV‐2 Total Antibody Assay and IgG Assay Using the Dimension EXL 200 in the Tokyo Metropolitan Area,” Heliyon 7, no. 11 (2021): e08393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ikegami S., Benirschke R. C., Fakhrai‐Rad H., et al., “Target Specific Serologic Analysis of COVID‐19 Convalescent Plasma,” PLoS One 16, no. 4 (2021): e0249938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Irsara C., Egger A. E., Prokop W., et al., “Evaluation of Four Commercial, Fully Automated SARS‐CoV‐2 Antibody Tests Suggests a Revision of the Siemens SARS‐CoV‐2 IgG Assay,” Clinical Chemistry and Laboratory Medicine (CCLM) 59, no. 6 (2021): 1143–1154. [DOI] [PubMed] [Google Scholar]
- 31. Kubota K., Kitagawa Y., Matsuoka M., et al., “Clinical Evaluation of the Antibody Response in Patients With COVID‐19 Using Automated High‐Throughput Immunoassays,” Diagnostic Microbiology and Infectious Disease 100, no. 3 (2021): 115370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kundu D., Gautam P., Dayanand D., et al., “The Role and Diagnostic Accuracy of Serology for COVID‐19,” BMC Infectious Diseases 22, no. 1 (2022): 390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lapić I., Rogić D., Šegulja D., Kralik Oguić S., and Knežević J., “The Reliability of SARS‐CoV‐2 IgG Antibody Testing: A Pilot Study in Asymptomatic Health Care Workers in a Croatian University Hospital,” Croatian Medical Journal 61, no. 6 (2020): 485–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lester S. N., Stumpf M., Freeman B. D., et al., “Examination of SARS‐CoV‐2 Serological Test Results From Multiple Commercial and Laboratory Platforms With an In‐House Serum Panel,” Access Microbiology 6, no. 2 (2024): 000463.v4, 10.1099/acmi.0.000463.v4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li P., Li Z., Silin Z., et al., “Preliminary Study of Serum 2019‐nCoV IgM and IgG Antibodies in the Diagnosis of COVID‐19,” Chinese Journal of Laboratory Medicine 43, no. 4 (2020): 352–357. [Google Scholar]
- 36. Mafi S., Rogez S., Darreye J., Alain S., and Hantz S., “Performance of the SureScreen Diagnostics COVID‐19 Antibody Rapid Test in Comparison With Three Automated Immunoassays,” Diagnostic Microbiology and Infectious Disease 105, no. 4 (2023): 115900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mairesse A., Favresse J., Eucher C., et al., “High Clinical Performance and Quantitative Assessment of Antibody Kinetics Using a Dual Recognition Assay for the Detection of SARS‐CoV‐2 IgM and IgG Antibodies,” Clinical Biochemistry 86 (2020): 23–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Manthei D. M., Whalen J. F., Schroeder L. F., et al., “Differences in Performance Characteristics Among Four High‐Throughput Assays for the Detection of Antibodies Against SARS‐CoV‐2 Using a Common Set of Patient Samples,” American Journal of Clinical Pathology 155, no. 2 (2021): 267–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Marlet J., Petillon C., Ragot E., et al., “Clinical Performance of Four Immunoassays for Antibodies to SARS‐CoV‐2, Including a Prospective Analysis for the Diagnosis of COVID‐19 in a Real‐Life Routine Care Setting,” Journal of Clinical Virology 132 (2020): 104633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Montesinos I., Gruson D., Kabamba B., et al., “Evaluation of Two Automated and Three Rapid Lateral Flow Immunoassays for the Detection of Anti‐SARS‐CoV‐2 Antibodies,” Journal of Clinical Virology 128 (2020): 104413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Naaber P., Hunt K., Pesukova J., et al., “Evaluation of SARS‐CoV‐2 IgG Antibody Response in PCR Positive Patients: Comparison of Nine Tests in Relation to Clinical Data,” PLoS One 15, no. 10 (2020): e0237548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nedelcu I., Jipa R., Vasilescu R., et al., “Long‐Term Longitudinal Evaluation of Six Commercial Immunoassays for the Detection of IgM and IgG Antibodies Against SARS CoV‐2,” Viruses 13, no. 7 (2021): 1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nicholson S., Karapanagiotidis T., Khvorov A., et al., “Evaluation of 6 Commercial SARS‐CoV‐2 Serology Assays Detecting Different Antibodies for Clinical Testing and Serosurveillance,” Open Forum Infectious Diseases 8, no. 7 (2021): ofab239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nyagwange J., Kutima B., Mwai K., et al., “Comparative Performance of WANTAI ELISA for Total Immunoglobulin to Receptor Binding Protein and an ELISA for IgG to Spike Protein in Detecting SARS‐CoV‐2 Antibodies in Kenyan Populations,” Journal of Clinical Virology 146 (2022): 105061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Oved K., Olmer L., Shemer‐Avni Y., et al., “Multi‐Center Nationwide Comparison of Seven Serology Assays Reveals a SARS‐CoV‐2 Non‐Responding Seronegative Subpopulation,” EClinicalMedicine 29–30 (2020): 100651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Padoan A., Bonfante F., Pagliari M., et al., “Analytical and Clinical Performances of Five Immunoassays for the Detection of SARS‐CoV‐2 Antibodies in Comparison With Neutralization Activity,” EBioMedicine 62 (2020): 103101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Parai D., Dash G. C., Choudhary H. R., et al., “Diagnostic Accuracy Comparison of Three Fully Automated Chemiluminescent Immunoassay Platforms for the Detection of SARS‐CoV‐2 Antibodies,” Journal of Virological Methods 292 (2021): 114121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pérez‐García F., Pérez‐Tanoira R., Iglesias M. E., et al., “Comparative Evaluation of Six Immunoassays for the Detection of Antibodies Against SARS‐CoV‐2,” Journal of Virological Methods 289 (2021): 114047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pflüger L. S., Bannasch J. H., Brehm T. T., et al., “Clinical Evaluation of Five Different Automated SARS‐CoV‐2 Serology Assays in a Cohort of Hospitalized COVID‐19 Patients,” Journal of Clinical Virology 130 (2020): 104549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Piec I., English E., Thomas M. A., Dervisevic S., Fraser W. D., and John W. G., “Performance of SARS‐CoV‐2 Serology Tests: Are They Good Enough?,” PLoS One 16, no. 2 (2021): e0245914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Riester E., Majchrzak M., Mühlbacher A., et al., “Multicentre Performance Evaluation of the Elecsys Anti‐SARS‐CoV‐2 Immunoassay as an Aid in Determining Previous Exposure to SARS‐CoV‐2,” Infectious Diseases and Therapy 10, no. 4 (2021): 2381–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rikhtegaran Tehrani Z., Saadat S., Saleh E., et al., “Performance of Nucleocapsid and Spike‐Based SARS‐CoV‐2 Serologic Assays,” PLoS One 15, no. 11 (2020): e0237828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sekirov I., Barakauskas V. E., Simons J., et al., “SARS‐CoV‐2 Serology: Validation of High‐Throughput Chemiluminescent Immunoassay (CLIA) Platforms and a Field Study in British Columbia,” Journal of Clinical Virology 142 (2021): 104914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Şener B., Kırbaş E., Sancak B., et al., “Sensitivity Affected by Disease Severity and Serum Sampling Time: A Performance Evaluation of Six SARS‐CoV‐2 Antibody Immunoassays,” Japanese Journal of Infectious Diseases 75, no. 4 (2022): 388–394. [DOI] [PubMed] [Google Scholar]
- 55. Serre‐Miranda C., Nobrega C., Roque S., et al., “Performance Assessment of 11 Commercial Serological Tests for SARS‐CoV‐2 on Hospitalised COVID‐19 Patients,” International Journal of Infectious Diseases 104 (2021): 661–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Soleimani R., Khourssaji M., Gruson D., et al., “Clinical Usefulness of Fully Automated Chemiluminescent Immunoassay for Quantitative Antibody Measurements in COVID‐19 Patients,” Journal of Medical Virology 93, no. 3 (2021): 1465–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Syre H., Obreque M. E. B., Dalen I., et al., “The Performances of Three Commercially Available Assays for the Detection of SARS‐CoV‐2 Antibodies at Different Time Points Following SARS‐CoV‐2 Infection,” Viruses 14, no. 10 (2022): 2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tan S. S., Saw S., Chew K. L., et al., “Head‐to‐Head Evaluation on Diagnostic Accuracies of Six SARS‐CoV‐2 Serological Assays,” Pathology 52, no. 7 (2020): 770–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Theel E. S., Harring J., Hilgart H., and Granger D., “Performance Characteristics of Four High‐Throughput Immunoassays for Detection of IgG Antibodies Against SARS‐CoV‐2,” Journal of Clinical Microbiology 58, no. 8 (2020): e01243–20, 10.1128/JCM.01243-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tolan N. V., DeSimone M. S., Fernandes M. D., et al., “Lessons Learned: A Look Back at the Performance of Nine COVID‐19 Serologic Assays and Their Proposed Utility,” Clinical Biochemistry 117 (2023): 60–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Van Elslande J., Decru B., Jonckheere S., et al., “Antibody Response Against SARS‐CoV‐2 Spike Protein and Nucleoprotein Evaluated by Four Automated Immunoassays and Three ELISAs,” Clinical Microbiology and Infection 26, no. 11 (2020): 1557.e1–1557.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Velay A., Gallais F., Benotmane I., et al., “Evaluation of the Performance of SARS‐CoV‐2 Serological Tools and Their Positioning in COVID‐19 Diagnostic Strategies,” Diagnostic Microbiology and Infectious Disease 98, no. 4 (2020): 115181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ward M. D., Mullins K. E., Pickett E., et al., “Performance of 4 Automated SARS‐CoV‐2 Serology Assay Platforms in a Large Cohort Including Susceptible COVID‐19‐Negative and COVID‐19‐Positive Patients,” Journal of Applied Laboratory Medicine 6, no. 4 (2021): 942–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wu L., Zhu S., Tingting M., et al., “Application Evaluation of Chemiluminescence Immunoassay in Auxiliary Diagnosis of Novel Coronavirus Pneumonia,” Zhejiang Medical 43, no. 07 (2021): 778–780. [Google Scholar]
- 65. Yassine H. M., Al‐Jighefee H., Al‐Sadeq D. W., et al., “Performance Evaluation of Five ELISA Kits for Detecting Anti‐SARS‐CoV‐2 IgG Antibodies,” International Journal of Infectious Diseases 102 (2021): 181–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zilla M., Wheeler B. J., Keetch C., et al., “Variable Performance in 6 Commercial SARS‐CoV‐2 Antibody Assays May Affect Convalescent Plasma and Seroprevalence Screening,” American Journal of Clinical Pathology 155, no. 3 (2021): 343–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Guo L., Ren L., Yang S., et al., “Profiling Early Humoral Response to Diagnose Novel Coronavirus Disease (COVID‐19),” Clinical Infectious Diseases 71, no. 15 (2020): 778–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhao J., Yuan Q., Wang H., et al., “Antibody Responses to SARS‐CoV‐2 in Patients With Novel Coronavirus Disease 2019,” Clinical Infectious Diseases 71, no. 16 (2020): 2027–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Higgins V., Fabros A., Wang X. Y., Bhandari M., Daghfal D. J., and Kulasingam V., “Anti‐SARS‐CoV‐2 IgM Improves Clinical Sensitivity Early in Disease Course,” Clinical Biochemistry 90 (2021): 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kaduskar O., Gurav Y. K., Deshpande K., et al., “Understanding the Dynamics of IgM & IgG Antibodies in COVID‐19‐Positive Patients,” Indian Journal of Medical Research 155, no. 5&6 (2022): 565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Requests for any data can be made to the authors.


