Abstract
Skeletal muscles are predominantly composed of long, multinucleated muscle fibers, classified according to their metabolic and contractile phenotype. The determination of fiber types is influenced by various factors (e.g., innervation, hormones, physical demand). Our laboratory and others showed that resolvins, lipid mediators derived from omega‐3 fatty acids, promote muscle regeneration and function after an injury or in models of muscular dystrophies; however, the effect of resolvins on the determination of muscle phenotype remains unknown. Here, we investigated the impact of lipid mediators on muscle phenotype during myogenesis. Transcriptomics analysis of single‐nuclei RNAseq data sets revealed that the enzymes responsible for bioactive lipids biosynthesis are differentially expressed in slow fibers versus fast fibers. Lipidomics analysis of slow‐twitch muscle (soleus) versus fast‐twitch muscle (tibialis anterior) showed that the levels of lipids derived from arachidonic acid are similar between muscle groups, but lipids derived from alpha‐linolenic acid, linoleic acid, eicosapentaenoic acid, and docosahexaenoic acid are enriched in slow‐twitch muscle. Screening for different lipids in vitro showed that resolvin‐D2 enhances the formation of myotubes expressing the slow myosin heavy chain isoform. In vivo, the administration of resolvin‐D2 enhances muscle strength, increases myofiber size, and affects fiber typing in injured muscles but not in uninjured muscles. Resolvin‐D2 promoted the transition toward the dominant fiber types in regenerating muscle (i.e., type I in the slow‐twitch soleus and type IIB in the fast‐twitch tibialis anterior muscle), suggesting its participation in fiber typing in conjunction with other factors. Overall, these findings identified new roles of bioactive lipids in the regulation of fiber typing, which could have therapeutic applicability in muscle injuries or dystrophies.
Keywords: bioactive lipids, fiber types, myogenesis, resolvin, satellite cells, skeletal muscle
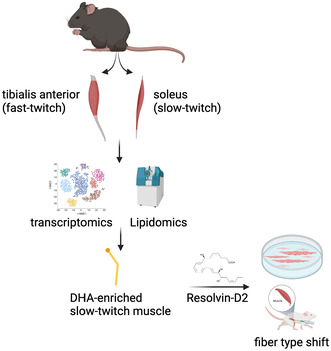
Transcriptomics (bulk and single‐nuclei RNAseq) and lipidomics (mass spectrometry) analysis showed that slow‐twitch muscle (soleus) is enriched in bioactive lipids derived from docosahexaenoic acid (DHA) and their biosynthesizing enzymes compared to fast‐twitch muscle (tibialis anterior). In vitro screening revealed that the DHA‐derived mediator resolvin‐D2 promotes myogenesis and the switch to the slow phenotype. In vivo, resolvin‐D2 administration increased muscle strength, fiber size, and phenotype switch in regenerative muscle, but the transition to the slow phenotype requires a permissive environment.
Abbreviation
- 4E‐BP
4E‐ binding protein
- AA
Arachidonic acid
- ALA
Alpha‐ linolenic acid
- ALOX12
12‐ lipoxygenase
- ALOX15
15‐ lipoxygenase
- ALOX5
5‐ lipoxygenase
- AMPK
AMP‐ activated protein kinase
- bFGF
Basic Fibroblast Growth Factor
- CHU
Centre hospitalier universitaire
- CIBPAR
Comité institutionnel des bonnes pratiques animales en recherche
- COX2
Cyclooxygenase 2
- CTX
Cardiotoxin
- CYP
Cytochromes P450
- DHA
Docosahexaenoic acid
- DiHETE
Dihydroxy‐eicosatetraenoic acid
- DiHOME
Dihydroxy‐octadecenoic acid
- EDL
Extensor digitorum longus
- EPA
Eicosapentaenoic acid
- EpDPA
Epoxydocosapentaenoic Acid
- Ephx2
Epoxide hydrolase 2
- EpOME
Epoxyoctadecenoic Acid
- GLA
Gamma‐linoleic acid
- Gpr18
G Protein‐Coupled Receptor 18
- Gpx4
Glutathione peroxidase 4
- HDHA
Hydroxydocosahexaenoic Acid
- HEPE
Hydroxyeicosapentaenoic Acid
- HETE
Hydroxyeicosatetraenoic Acid
- HETrE
Hydroxyeicosatrienoic Acid
- HODE
Hydroxyoctadecadienoic Acid
- i.m.
Intramuscular
- i.p.
Intraperitoneal
- iPF2a‐VI
Isoprostane F2‐alpha VI
- LA
Linoleic acid
- Lgr6
Leucine‐Rich Repeat‐Containing G‐Protein Coupled Receptor 6
- LOD
Limit of detection
- LOQ
Limit of quantification
- LPS
Lipopolysaccharide
- Lta4h
Leukotriene A4 hydrolase
- LTB4
Leukotriene B4
- LxA4
Lipoxin A4
- MaR1
Maresin‐ 1
- MFR
Minimal feret diameter
- MGST2
Microsomal glutathione S‐ transferase 2
- mTOR
Mammalian target of rapamycin
- MyHC
Myosin heavy chain
- MyHCemb
Myosin heavy chain embryonic
- NADPH
Nicotinamide Adenine Dinucleotide Phosphate
- NOX2
NADPH Oxidase 2
- NOX4
NADPH Oxidase 4
- OxoETE
Oxo‐Eicosatetraenoic Acid
- OxoODE
Oxo‐Octadecadienoic Acid
- PGC‐1a
PPARγ‐coactivator 1‐alpha
- PGD2
Prostaglandin D2
- PGF2a
Prostaglandin F2‐alpha
- PGF3a
Prostaglandin F3‐alpha
- Pla2g5
Phospholipase A2 group‐ V
- Pla2g7
Phospholipase A2 group‐ VII
- PPAR
Peroxisome proliferator‐ activated receptor
- Ptges2
Prostaglandin E synthase‐ 2
- Ptges3
Prostaglandin E synthase‐ 3
- PUFAs
Polyunsaturated fatty acids
- ROS
Reactive oxygen species
- RvD1
Resolvin‐ D1
- RvD2
Resolvin‐D2
- RvE1
Resolvin‐ E1
- S6K
Ribosomal protein S6 kinase
- TA
Tibialis anterior
- Tbx1
T‐box transcriptional factor 1
- TXB2
Thromboxane B2
- Veh
Vehicle
- WT
Wild‐type
1. INTRODUCTION
Skeletal muscles make up approximately 30%–40% of the total body mass and are predominantly composed of long multinucleated muscle fibers. 1 , 2 These fibers are classically classified according to their metabolic and contractile phenotype (myosin heavy chain type; MyHC) as type I (oxidative slow fibers), IIA (oxidative fast fibers) or IIX and IIB (glycolytic fast fibers). 3 Noteworthy, while MyHC is a good marker for fiber typing it is not a direct predictor of the metabolic profile. 4 Nevertheless, type I fibers are described as resistant fibers that are enriched in muscles involved in activities requiring low‐level sustained efforts (e.g., postural muscles), whereas type II fibers are powerful but fatigable fibers that are prevalent in muscles utilized for movements that demand high levels of explosiveness (e.g., jumping, sprinting). 5
The determination of fiber type depends on several signaling pathways and extrinsic factors such as innervation, hormones, and physical demand. 3 Therefore, changes in the microenvironment can affect the fiber type composition. For instance, endurance exercise leads to a shift in the composition of muscle fibers toward those that have a slower phenotype accompanied by other phenotypic changes such as mitochondrial biogenesis and angiogenesis. 6 Moreover, different pathological conditions also affect fiber typing. Conditions such as disuse or detraining are characterized by a shift from type I to type II fibers, 7 while cancer cachexia and aging are associated with type II to I transition. 8 Therefore, a better understanding of the factors regulating fiber typing could have therapeutic applications in different muscular disorders.
Muscle stem cells, called satellite cells, have also been suggested to participate in the process of fiber type determination. These quiescent cells are activated after an injury to become myoblasts that will fuse to form multinucleated myotubes, which will transiently express embryonic/neonatal MyHC isoforms that will be replaced by fast or slow MyHC isoforms as the myofiber matures. 2 Some studies suggest that satellite cells form all fiber types irrespective of their muscle of origin, 9 whereas other studies indicate that satellite cells are predisposed to generate muscle fibers with a phenotype reflecting their original fiber type. 10 , 11 , 12 This discrepancy could be explained by different experimental setups. When cultured on collagen‐coated plates, satellite cells from the tibialis anterior (fast) and soleus (slow) muscles generate myotubes that express only fast MyHC; however, when cultured on matrigel‐coated plates, satellite cells from the soleus led to the formation of myotubes expressing both fast and slow MyHC. 13 In vivo experiments involving slow (soleus) or fast (extensor digitorum longus, EDL) regenerating muscles, which were denervated and stimulated with the same impulse pattern, revealed a significant difference in MyHC expression between these muscles, thereby suggesting intrinsically different satellite cells. 12 Fast‐muscle‐derived myoblasts were shown to express higher levels of the T‐box transcriptional factor Tbx1, which modulates myotube fiber type and oxidative metabolism. 14 These findings suggested that satellite cells could be pre‐programmed to preferentially form one type of fiber reflecting the original fiber type; however, this intrinsic program can be overcome by extrinsic factors.
Several studies have shown that bioactive lipids derived from omega‐3 or omega‐6 polyunsaturated fatty acids (PUFAs) play a key role in regulating myogenesis. 15 , 16 , 17 , 18 During the initial inflammatory phase postinjury, arachidonic acid (AA), an omega‐6‐derived PUFAs, is converted into prostaglandins and leukotrienes by the respective action of enzymes such as cyclooxygenases 2 (COX2) and 5‐lipoxygenase (ALOX5). 19 During the resolution phase, omega‐3‐derived PUFAs, such as alpha‐linolenic acid (ALA), docosahexaenoic acid (DHA), eicosapentaenoic acid (EPA), are converted by enzymes such as 12‐ and 15‐lipoxygenases (ALOX12 and ALOX15) and ALOX5 into pro‐resolving lipids like resolvin‐Ds (DHA‐derived), resolvin‐Es (EPA‐derived), protectins, and maresins. 20 In addition to reducing muscle inflammation, 21 these pro‐resolution lipid mediators promote muscle regeneration and improve muscle function following injury. 15 , 16 , 17 , 22 , 23 For instance, Maresin‐1 was shown to increase myoblast proliferation through the Lgr6 receptor. 22 Administration of resolvin‐D1 (RvD1) increases myoblasts differentiation and the cross‐sectional area of regenerating muscle fibers postinjury in vivo. 16 Resolvin‐E1 (RvE1) treatment increased the diameter of the myotubes in vitro in a model of LPS‐induced atrophy. 24 Moreover, our laboratory recently showed in a model of Duchenne muscular dystrophy that resolvin‐D2 (RvD2) increases the differentiation and fusion of myoblasts in vitro and in vivo. 15 These studies show that pro‐resolving lipids impact myogenesis in different conditions; however, their effect on the determination of muscle phenotype is unknown.
Here, we investigate the influence of bioactive lipids on muscle phenotype during myogenesis. Our transcriptomics and lipidomics analysis revealed that slow muscles are enriched in enzymes and omega‐3 PUFAs derived from DHA, EPA, and ALA. We demonstrate that RvD2 enhances the formation of slow myotubes in vitro and increases muscle strength, myofiber size, and regulates fiber typing in vivo. These findings provide novel insight into the effect of RvD2 and support its therapeutic potential for conditions characterized by impaired myogenesis, muscle weakness/wasting, and alterations in muscle fiber typing.
2. METHODS
2.1. Animals and experimental design
Male wild‐type (WT; C57BL/6) mice of 6–8 weeks old were obtained from the Jackson Laboratory (Bar Harbor, ME) and were sacrificed at 10 weeks of age. Only male mice were used due to limited resources and to minimize confounding factors. Mice were housed in pathogen‐free cages within the Animal Holding Facility at the CHU Sainte‐Justine (Montreal, CA) and maintained under controlled environmental conditions, including a 12:12 h light–dark cycle, a temperature of 21°C, a humidity level of 40%, and free access to food (Harlan Teklad #2918) and tap water. To induce standardized muscle injury, an intramuscular (i.m.) injection of cardiotoxin (50 μL at 10 μM, CTX, Latoxan) was administered into the tibialis anterior (TA) muscle of one leg (the contralateral leg was used as uninjured control). 25 , 26 These mice were randomly assigned to different experimental groups that received either a 10 μL i.m. injection containing a dose of 10 ng of RvD2 (the stock solution of 100 ng/μl in ethanol was diluted 1:100 in saline to get the working solution, Cayman Chemicals) or vehicle (10 μL of a solution containing 1:100 ethanol diluted in saline) into both TA muscles. The injections were repeated twice a week for a period of 1 week or 3 weeks. Mice were anesthetized by inhalation with isoflurane (Fresenius Kabi) during the i.m. injections. On the day of sacrifice, the mice were anesthetized using intraperitoneal (i.p.) injection of buprenorphine (0.1 mg/kg) and pentobarbital sodium (50 mg/kg). The TA (fast‐twitch muscle) and soleus (slow‐twitch muscle) muscles from both legs were collected. Mice were euthanized by cervical dislocation under anesthesia.
In another set of experiments in which the systemic effect of RvD2 on muscle regeneration was investigated, the mice were injured by i.m. injection of CTX into the TA muscle of one leg. These mice received daily i.p. injection containing a 100 ng dose of RvD2 diluted in 10 μL (5 μg/kg, RvD2 stock solution in ethanol diluted 1:10 in saline) or vehicle (10 μL injection of 1:10 ethanol diluted in saline) for 21 days. 15 At the end of the 21‐day treatment period, TA and soleus muscles from both legs were collected and the mice were euthanized by cervical dislocation under anesthesia. Muscles collected from the different experimental designs were weighed, embedded in a freezing medium (OCT), rapidly frozen in liquid nitrogen‐cooled isopentane, and stored at −80°C until sectioning and staining were performed. All animal experiments performed in this study were approved by the Comité Institutionnel des Bonnes Pratiques Animales en Recherche (CIBPAR; approval number 2022‐3608) in accordance with the guidelines of the Canadian Council on Animal Care.
2.2. Ex vivo muscle contractility
Isometric contractile properties were assessed as previously described, 15 according to the standard operating procedures described by TREAT‐NMD (protocol DMD_M.1.2.002). Prior to the measurements of muscle contractility, mice received a dose of buprenorphine (0.1 mg/kg, i.p.) and pentobarbital sodium (50 mg/kg, i.p.). Then, the distal tendon of the TA muscle was surgically severed, isolating it from the adjacent muscles (the proximal part of the muscle remained attached to the tibial bone). 27 A suture silk was attached to the distal tendon. The mouse was placed on a heating plate, maintaining a temperature of 27°C, and the knee was stabilized using a clamp. The suture silk attached to the lower tendon of the TA muscle was attached at the other end to a force lever arm (300C‐LR dual‐mode lever; Aurora Scientific, Canada). Two electrodes were inserted in the TA muscle near the knee for muscle stimulation. Optimal muscle length (L0) was determined and gradually adjusted until the maximum isometric twitch tension was achieved. Following a 2‐min rest period, the contractile properties of the muscle were assessed. A force–frequency curve was established by stimulating the muscle at different frequencies (25, 50, 80, and 100 Hz), with a 2‐min rest between each stimulation. Thereafter, muscle length and weight were measured to assess specific muscle force (N/cm2) by using a mathematical approximation of the cross‐sectional area based on the following formula: Specific force (N/cm2) = (absolute force (N) × fiber length (0.6 × L0 for the TA muscle) × muscle density (1.06 g/cm3))/muscle mass (g).
2.3. Satellite cell isolation and culture
Hind limb muscles from WT mice were collected and dissociated in collagenase B/dispase II (Sigma) using the gentle MACS dissociator (Miltenyi Biotech). The resulting cell suspension was filtered through a 30 μm cell strainer, and the cells were stained with antibodies for 30 min on ice in the dark. Satellite cells were isolated by FACS (fluorescence‐activated cell sorting) using a gating strategy based on forward scatter and side scatter profiles, cell viability (7‐AAD; 1:40; Biolegend), negative selection with FITC‐conjugated antibodies anti‐Sca‐1 (Clone D7; 1:30; Miltenyi Biotech), anti‐CD45 (clone 30F11; 1:30; Miltenyi Biotech), anti‐CD31 (Clone 390; 1:30; Miltenyi Biotech), anti‐CD11b (M1/70.15.11.5; 1:30; Miltenyi Biotech), and positive selection for APC‐conjugated anti‐Itgb1 (clone HMβ1‐1; 1:15; Miltenyi Biotech), and PE‐conjugated anti‐Itga7 (clone 3C12, 1:100; Miltenyi Biotech). 15 Isolated satellite cells were cultured in collagen‐coated Petri dishes with Ham's F10 media (GIBCO), supplemented with 20% fetal bovine serum (Wisent), 1% penicillin–streptomycin (GIBCO), and 2.5 ng/ mL bFGF (basic‐Fibroblast Growth Factor, Wisent). Cells were incubated at 37°C until reaching a confluence of 80%. Then, a low‐serum differentiation medium (50% Ham's F10, 50% DMEM low glucose, 1% penicillin–streptomycin, 5% horse serum) was added to the cells to induce differentiation and myotube formation.
2.4. Immunofluorescence
Immunofluorescence was performed on cultured myoblasts or skeletal muscle sections. 28 For myoblast immunostaining, the cells were fixed with 2% paraformaldehyde solution for 10 min and permeabilized with 0.2% Triton X‐100 and 0.1% glycine solution in PBS for 10 min. Samples were blocked with blocking buffer (5% goat serum, 2% bovine serum albumin, and 3% donkey serum in PBS) for 60 min at room temperature and incubated overnight at 4°C with the following primary antibodies: mouse anti‐MYH7 (clone BA‐F8; 1:10; Developmental study hybridoma bank, DSHB, created by the NICHD of the NIH and maintained at The University of Iowa, Department of Biology, Iowa City, IA 52242), mouse monoclonal antibody myosin heavy chain fast (clone WB‐MHCf; 1:20; Leica) or mouse anti‐MYH3 (clone F1.652; 0.3 μg/mL; DSHB). Samples were washed with PBS and incubated with appropriate cross‐adsorbed secondary antibodies for 1 h at room temperature protected from light. The secondary antibodies used included goat anti‐mouse IgG1 (Alexa Fluor 488, 1:1000) or goat anti‐mouse IgG2B (Alexa Fluor 647, 1:1000). Samples were washed with PBS and counterstained with DAPI. Images were viewed and captured using an epifluorescence EVOS M5000 (Thermo Fisher Scientific).
For immunostaining on skeletal muscle, the samples were sectioned at 10 μm thick using a cryostat (ThermoFisher) from both the proximal and distal portions of the TA and soleus muscles. These sections were mounted on Superfrost Plus slides (Thermo Fisher Scientific, Canada) and kept at −20°C. The samples were blocked for 90 min at room temperature with the blocking buffer described previously to which mouse on mouse‐blocking reagent (1:8; Vector labs) was added. Samples were incubated for 45 min at 37°C with the following primary antibodies: mouse anti‐MYH4 (clone BF‐F3; 1:40; DSHB), mouse anti‐MYH2 (clone SC‐71; 1:10; DSHB), mouse anti‐MYH7 (clone BA‐F8; 1:10; DSHB), mouse anti‐MYH3 (clone F1.652; 0.3 μg/mL; DSHB), and rabbit anti‐Laminin (cat. no. ab11575; 1:1000; Abcam). Considering that there were not enough fluorescence channels available to perform all the MyHC staining on the same slide, the staining was performed on two consecutive slides. For the soleus muscle, the first slide was stained with SC‐71, BA‐F8, and laminin antibodies. The second slide was stained with BF‐F3 and laminin antibodies. For the TA muscle, the first slide was stained with SC‐71, BF‐F3, and laminin antibodies. The second slide was stained with BA‐F8 and laminin antibodies. Samples were washed with PBS and incubated with appropriate cross‐adsorbed secondary antibodies (Invitrogen, Thermo Fisher) for 45 min at 37°C protected from light: goat anti‐mouse IgM (Alexa Fluor 488, 1:1000), goat anti‐mouse IgG H + L (Alexa Fluor 647, 1:1000), goat anti‐rabbit IgG H + L (Alexa Fluor 555, 1:1000), goat anti‐mouse IgG2B (Alexa Fluor 647, 1:1000) or goat anti‐mouse IgG1 (Alexa Fluor 488, 1:1000). Samples were washed with PBS, counterstained with DAPI and mounted with PermaFluor (Thermo Fisher Scientific). Images were viewed and captured on the entire sections using an epifluorescence Leica DM5000 B (Leica Microsystems, Canada). The images of each muscle section were subjected to unbiased analysis and quantification with Fiji using the Myosight plugin. 29 The plugin allows for unbiased semi‐automated analysis of different parameters such as the minimal Feret diameter (MFD) and fiber type count. A minimum of 2500 fibers for TA sections and 700 fibers for soleus sections was analyzed.
2.5. Western blot
Myotubes were washed with sterile PBS and lysed with ice‐cold RIPA buffer containing 1% of protease inhibitor cocktail (Roche). The lysates were centrifuged for 10 min at 10 000× g and the resulting supernatant was collected, aliquoted, and the protein content was quantified using the BCA Assay Kit (Thermo Scientific, Canada). A volume corresponding to 40 μg of protein was diluted with sample buffer (containing 125 mM Tris buffer at pH 6.8, 4% sodium dodecyl sulfate, 20% glycerol, 0.05% bromophenol blue, and 200 mM dithiothreitol), heated 5 min at 100°C, and electroseparated on 8% sodium dodecyl sulfate‐polyacrylamide gel. Subsequently, the proteins were transferred to a polyvinylidene difluoride membrane, which was blocked with 5% nonfat milk for 60 min at room temperature. The membranes were incubated at 4°C overnight with primary antibodies diluted in 5% nonfat milk including mouse anti‐MYH4 (clone BF‐F3; 1:100; DSHB), mouse anti‐MYH2 (clone SC‐71; 1:100; DSHB), mouse anti‐MYH7 (clone BA‐D5; 1:80; DSHB), mouse anti‐MYH1 (clone 6H1; 1:50; DSHB) mouse anti‐MYH3 (clone F1.652; 1:50; DSHB), and β‐actin (4967; 1000; New England Biolabs). After washing, the membranes were incubated with goat anti‐mouse (H + L) or goat anti‐rabbit horseradish peroxidase‐conjugated secondary antibodies (1:3000; Abcam) for 1 h at room temperature. The protein bands were revealed using an ECL‐plus Western blotting reagent (PerkinElmer Life and Analytical Sciences, USA) and visualized with the GeneSys image software (Syngene).
2.6. Transcriptomics analyses
Different transcriptomic data sets already published were explored. Bulk RNAseq of soleus and TA muscles from mouse samples can be found in the Gene Expression Omnibus database GSE226117. 30 Single‐nuclei RNAseq data sets from mouse TA and soleus muscles or human gastrocnemius muscles can be found in the Gene Expression Omnibus database GSE147127 and GSE233882, respectively. 31 , 32 Rstudio software (R‐4.3.1) as well as DESeq2, Seurat v5, and Harmony packages were used for the reanalysis of the data sets.
2.7. Mass spectrometry
Bioactive lipid extraction and analysis were carried out as described elsewhere. 33 Briefly, 10 μL of a deuterated standard was combined with 15 mg of soleus or 30 mg of tibialis anterior (see Table S1 for details regarding which deuterated standards were used for each analyte). Muscle tissues were crushed using a potter with a volume of 289 μL of an ethanolic solution (41%). The latter were incubated with acetonitrile (2:1) at room temperature and then at −20°C in order to precipitate the proteins. After centrifugation, supernatants were removed and combined with 600 μL of an ammonium hydroxide solution (0.01 M) before being loaded on solid phase extraction columns. A portion of the unwanted compounds were washed from the cartridges with the aforementioned solution of ammonium hydroxide and then a mixture of acetonitrile and methanol (8:2). Compounds were eluted using an acidified version of the latter organic solution, nitrogen dried, and reconstituted in 60 μL (30% acetonitrile with 0.01% acetic acid). In order to enhance the precision of the quantification, a spiked calibration curve was generated. This was achieved by using a pool of muscular tissues that had been added with pure standards and extracted through the same steps as those used for the sample. Analysis was performed with a high‐performance liquid chromatography–tandem mass spectrometry system operated in negative mode and using specific transitions in scheduled multiple reaction monitoring previously described (Table S1). 34 A Shimadzu Prominence system (Columbia, MD, USA) linked to a 3200 QTRAP LC/MS/MS system from AB Sciex (Concord, ON, Canada) was operated in the negative mode. A Kinetex C18 100 Å column from Phenomenex (Torrance, CA, USA) with a ternary mobile phase gradient described elsewhere was used. 34 The limit of detection (LOD) is defined as a signal‐to‐noise ratio above 3:1 within a retention time of ±0.03 min from the commercial standard using algorithm of MultiQuant 3.0.2 software (AB Sciex). The limit of quantification (LOQ) of all analytes presented according to the muscle tested is defined in Table S2. LOQ signal‐to‐noise ratio of 1:10 with a 20% coefficient of variation using a standard curve in the investigated matrix. Please note that values between LOD and LOQ were used as a conservative method to avoid using “0.”
2.8. Statistical analysis
Data were analyzed using the GraphPad Prism 9 Software. Different statistical tests, including unpaired Student's t‐test and two‐way ANOVA uncorrected Fisher's LSD test, were utilized and are specified in the figure legends. Data were analyzed by an experimenter blinded to the identification of the samples. Data are presented as mean ± SEM (standard error of the mean). A 95% confidence interval was employed for all data analysis. Statistical significance is indicated as *p < .05, **p < .01, ***p < .001.
3. RESULTS
3.1. Transcriptomics analysis reveals different profiles in the expression of bioactive lipids enzymes in slow versus fast fibers
To determine how bioactive lipids are regulated across the different fiber types, we first investigated if the enzymes regulating their expression are differentially expressed in fast versus slow muscle. To do so, a publicly available bulk RNAseq data set comparing soleus versus TA muscles was analyzed for the expression of enzymes responsible for the biosynthesis of omega‐3 and ‐6 PUFAs. 30 Analysis revealed that the fast‐twitch TA muscle expresses higher levels of genes involved in AA conversion or membrane release such as glutathione peroxidase 4 (Gpx4), leukotriene A4 hydrolase (Lta4h), phospholipase A2 group‐V and ‐VII (Pla2g5 and Pla2g7), prostaglandin E synthase‐2 and ‐3 (Ptges2 and Ptges3) (Figure 1A). On the other hand, type I fibers express higher levels of Soluble epoxide hydrolase (Ephx2) that contributes to the conversion of linoleic acid (LA), as well as Alox5 and Alox12 that are involved in the conversion of various PUFAs such as DHA, EPA, and AA to form pro‐resolving lipids (resolvins, maresins, and lipoxins). Next, we investigated another superfamily of enzymes that play key roles in bioactive lipids metabolism, the cytochromes P450 (CYP) that act mainly as monooxygenases. Results showed that some Cyp genes are preferentially expressed in slow fibers (e.g., Cyp2j6, Cyp2e1, Cyp2f2), whereas others are more expressed in fast fibers (e.g., Cyp4f39, Cyp2u1, Cyp4f13, Cyp27a1, Cyp39a1) (Figure 1B).
FIGURE 1.
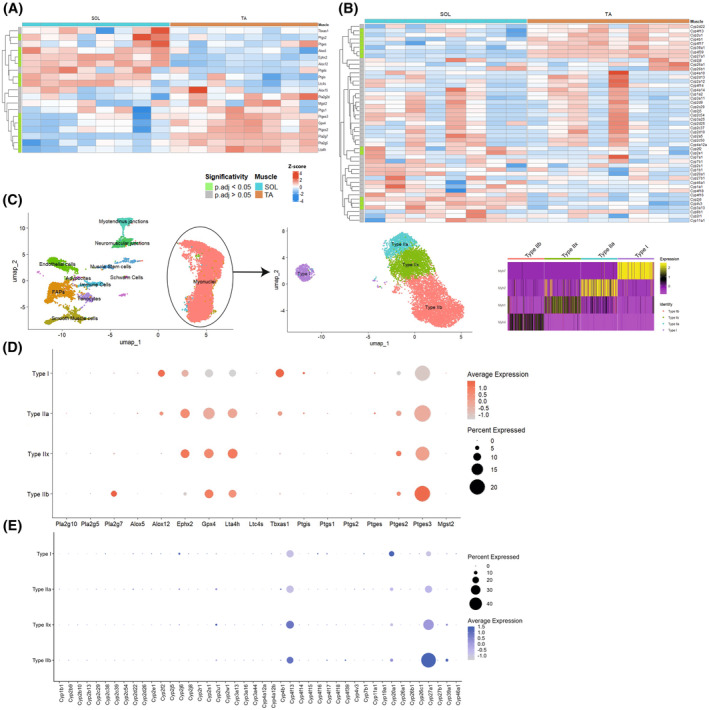
Differential expression patterns of lipid mediator enzymes in slow and fast myonuclei. (A, B) Heatmap from bulk RNAseq analysis of soleus and TA muscles showing the expression of different bioactive lipid biosynthesizing enzymes involved in the conversion of AA, LA, DHA, and EPA (A) as well as Cyp enzymes (B). (C) Single‐nuclei RNAseq data set of TA and soleus muscle was used to study gene expression in type I, IIA, IIX, and IIB fibers. (D, E) Dot plots showing the expression of different bioactive lipid biosynthesizing enzymes involved in the conversion of AA, LA, DHA, and EPA (D) as well as Cyp enzymes (E) across the different fiber types.
This bulk RNAseq analysis revealed that slow and fast muscles differentially express genes involved in bioactive lipids biosynthesis; however, considering that it contains a mix of different fiber types as well as other cell types, it does not allow us to determine if specific fiber types express different bioactive lipid signatures. Therefore, we explored single‐nuclei RNAseq (snRNAseq) data sets from the mouse TA and soleus muscle containing myonuclei from type I, IIA, IIX, and IIB fibers (Figure 1C). 31 The analysis confirmed that type II fibers express higher levels of genes encoding for AA membrane release like Pla2g7 or converting enzymes such as Gpx4, Lta4h, Ptges2, and Ptges3 (Figure 1D). Moreover, Alox12 is also expressed predominantly in type I fibers, whereas Ephx2 is expressed in type I, IIA, and IIX fibers at higher levels than type IIB. Analysis of the Cyp genes also showed a similar pattern to what was observed in the bulk RNAseq with genes expressed at higher levels in type I fibers (e.g., Cyp2j6, Cyp4f16, Cyp20a1), and others overexpressed in type II (e.g., Cyp2u1, Cyp4f13, Cyp27a1, Cyp39a1) (Figure 1E). Globally, the snRNAseq data set showed very similar findings to the bulk RNAseq data set indicating that the bioactive lipid enzyme expression is an intrinsic signature of each fiber type.
Other cell types in the skeletal muscles, such as immune cells, satellite cells, fibroadipogenic progenitors, and vascular cells, also express some of these bioactive lipid enzymes (Figure S1). However, myonuclei represent the highest proportion of nuclei in the skeletal muscle (Figure 1C) and should represent a large proportion of the bioactive lipid enzyme expression. Altogether, these findings suggest that myofibers express enzymes involved in the metabolism of bioactive lipids at different levels between slow versus fast fibers.
Further, we investigated if similar differences were observed in humans by exploring a snRNAseq data set from gastrocnemius muscle biopsies (Figure S2A). 32 Analysis of the human dataset revealed a similar pattern of expression compared to what was observed in mice, with enzymes predominantly expressed in type I fiber (e.g., EPHX2, ALOX12) and others in type II (e.g., PTGES2, PTGES3, GPX4) (Figure S2B). One notable exception is that, contrarily to mouse muscle, LTA4H is expressed at higher levels in slow fibers of human muscle. This finding in human samples is also accompanied by an increase, especially in type I fibers, in microsomal glutathione S‐transferase 2 (MGST2), that converts leukotriene A4 into leukotriene C4, suggesting that the leukotriene pathway might be differentially regulated in human versus mouse myofibers. Analysis of the CYP genes also revealed some similarities between the mouse and human data sets (e.g., CYP27A1 overexpressed in type II fibers); however, considering that the number of CYP genes varies considerably between mice and humans, 35 it was expected to observe differences in CYP expression patterns in the human compared to mouse samples.
3.2. Targeted lipidomics analysis reveals a different profile in the expression of bioactive lipids in slow versus fast fibers
To determine if the transcriptomic changes in biosynthesis enzymes are associated with changes in the bioactive lipids, we performed targeted lipidomics analysis by mass spectrometry analysis of soleus versus TA muscles. Out of the 104 bioactive lipids tested (Table S1), 44 were expressed over the limit of detection (Figure S2). Regarding the omega‐6 PUFAs, the levels of most arachidonic acid‐derived lipids (8‐iso‐PGE2, PGD2, PGE2, PGF2α, TXB2, 5–8,12‐iso‐iPF2a‐VI, 5(S)‐HETE, 5‐OxoETE, 8(S)‐HETE, 9(R)‐HETE, 12(S)‐HETE, 12‐OxoETE) were not significantly different between soleus and TA muscles (Figures 2A and S3A). Only 15(S)‐HETE and 15‐OxoETE were enriched in the soleus muscle. Regarding other types of omega‐6 PUFAs, enrichment was observed in almost all bioactive lipids derived from LA or gamma‐LA (±12(13)‐EpOME, ±12(13)‐DiHOME, ±9(10)‐EpOME, ±9(10)‐DiHOME, 9(S)‐HODE, 9‐OxoODE, 13(S)‐HODE, 13‐OxoODE) in the soleus muscle (Figures 2B and S3B). Only 12(S)‐HETrE and 13(S)‐HOTrE were not significantly different between soleus and TA muscles.
FIGURE 2.
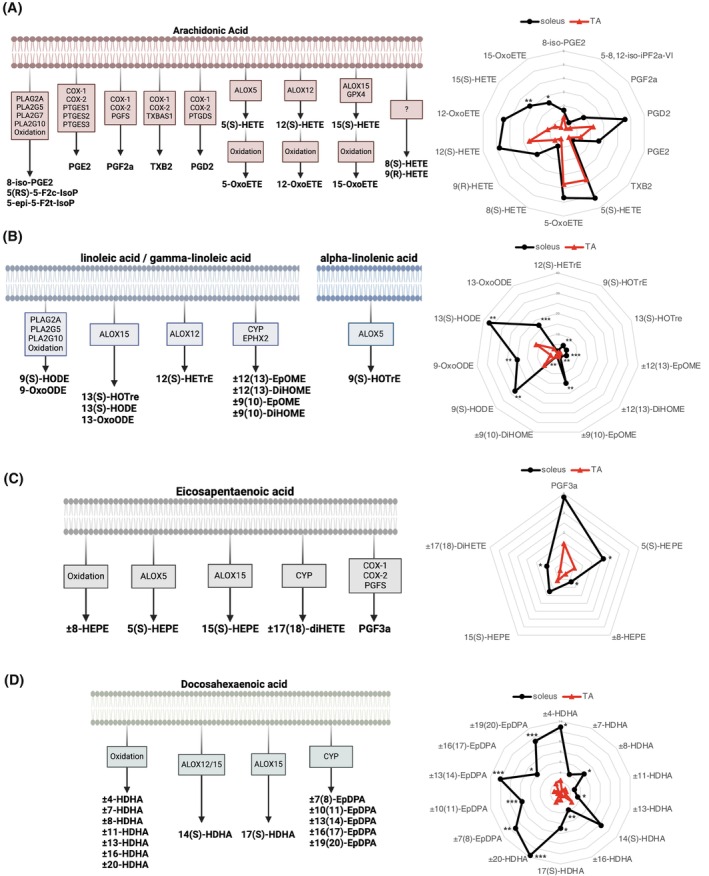
Differential pattern of expression of bioactive lipid mediators in slow‐ and fast‐twitch muscle. Mass spectrometry analysis was performed on slow‐twitch muscle (soleus) versus fast‐twitch muscle (tibialis anterior). Schematics showing the biosynthesis pathway and spider charts showing the expression of bioactive lipids derived from (A) Arachidonic acid (AA), (B) linoleic acid (LA)/gamma‐linoleic acid (GLA)/alpha‐linolenic acid (ALA), (C) eicosapentaenoic acid (EPA), and (D) docosahexaenoic acid (DHA). Data are expressed as pg/mg of muscle. Statistical analysis was performed using unpaired Student's t‐test. *p < .05, **p < .01, ***p < .001.
Analysis of the omega‐3 PUFAs also revealed many differences between slow‐ and fast‐twitch muscles. The ALA‐derived lipid 9(S)‐HOTrE was enriched in the soleus muscle (Figures 2B and S3B). Some bioactive lipids derived from EPA were also enriched in the soleus muscle, especially 5(S)‐HEPE, ±8‐HEPE, and ±17(18)‐DiHETE, whereas others were not significantly different such as PGF3α and 15(S)‐HEPE (Figures 2C and S3C). However, the strongest differences between soleus and TA muscles were observed in DHA‐derived bioactive lipids such as ±4‐HDHA, ±8‐HDHA, ±13‐HDHA, ±16‐HDHA, 17(S)‐HDHA (resolvin‐Ds precursor), ±20‐HDHA, ±7(8)‐EpDPA, ±10(11)‐EpDPA, ±13(14)‐EpDPA, ±16(17)‐EpDPA, and ±19(20)‐EpDPA (Figures 2D and S3D).
Overall, fast and slow muscles express similar levels of arachidonic acid‐derived lipids, whereas slow muscles are especially enriched in DHA‐derived lipids compared to fast muscles. These findings are consistent with the transcriptomics analyses and indicate that the bioactive lipid profile varies depending on the phenotype of the muscle.
3.3. RvD2 induces fiber type remodeling during myogenesis in vitro
Transcriptomics and lipidomics analyses revealed significant differences in bioactive lipid profiles between slow and fast muscles. Next, to investigate if these bioactive lipids can affect fiber typing, a screening experiment was conducted in vitro. Primary myoblasts were isolated from the skeletal muscle of wild‐type mice and cultured in vitro until ~80% confluence before inducing differentiation into myotubes for 4 days. Bioactive lipids derived from AA: PGE2, lipoxin A4 (LXA4), and leukotriene B4 (LTB4) as well as bioactive lipids derived from DHA: RvD2, 17‐HDHA (resolvin‐D precursor), and maresin‐1 (MaR1) were added to the culture medium at a concentration of 200 nM (vehicle was used as control condition) (Figure 3A). These bioactive lipids were chosen in part based on our lipidomic results showing that DHA was particularly enriched in the slow‐twitch soleus muscle compared to AA, as well as on our transcriptomics analysis showing that biosynthesizing enzymes such as Ptges2 and Lta4h (enriched in fast‐twitch muscle) or Alox5 and Alox12 (higher in slow‐twitch muscle) are differentially expressed depending on the fiber type. While some of these specialized resolving mediators were not detected by mass spectrometry in the uninjured muscle, they were shown to be expressed during the regeneration process postinjury. 16 , 17
FIGURE 3.
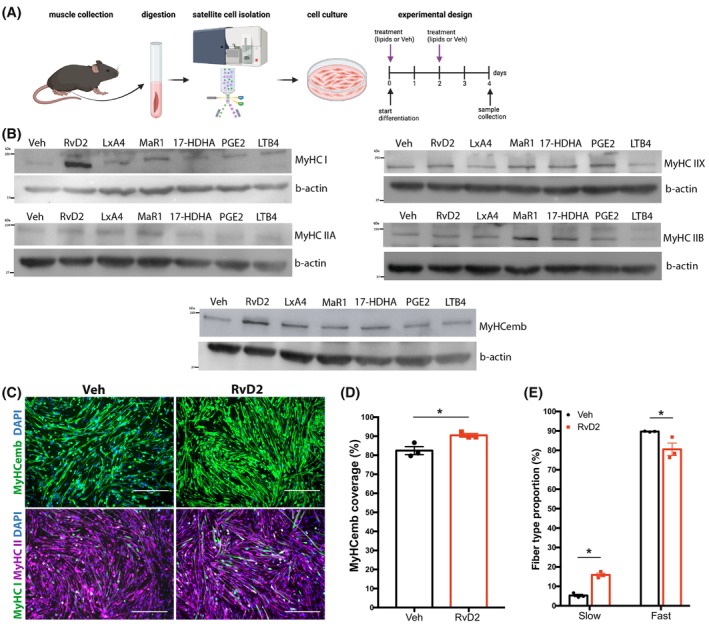
Resolvin‐D2 induces fiber type shifting in vitro. (A) Graphical overview of the experimental design (image created with BioRender.com) showing satellite cell isolation, culture, differentiation, and treatment. (B) Myoblasts were differentiated and treated with different bioactive lipids: Resolvin‐D2 (RvD2), Lipoxin‐A4 (LxA4), Maresin‐1 (MaR1), 17‐HDHA, prostaglandin‐E2 (PGE2), leukotriene‐B4 (LTB4), or vehicle. Representative Western blots showing the expression of different MyHC isoforms (I, IIA, IIX, IIB, embryonic) following the different treatments. β‐actin was used as a loading control. (C) Representative images of immunostaining for MyHCemb (green, upper panels) as well as MyHC I (green, lower panels) and MyHC II (purple, lower panels) on RvD2‐treated myotubes and controls. Scale bars = 300 μm. (D) Quantification of area coverage of MyHCemb myotubes. (E) Proportion of fibers expressing slow or fast MyHC isoform versus total. Data are presented as mean ± SEM, n = 3 (except n = 2 for B) independent biological samples. Experiments were conducted with technical duplicates. Statistical analysis was performed using unpaired Student's t‐test. *p < .05.
Western blot was performed to determine the impact of the treatment on the different fiber types (Figures 3B and S4). Results showed that RvD2 treatment has the most pronounced effects on the different isoforms of MyHC (Figure 3B), especially MyHC I and MyHCemb (MYH3) (Figure 3B). Next, the impact of RvD2 on the myotube phenotype was further investigated in vitro by immunofluorescence for the markers MyHCemb, MyHC slow, and MyHC fast (Figure 3C). Treatment with RvD2 increased the number of myotubes expressing MyHCemb (Figure 3D). Moreover, RvD2 increased the proportion of myotubes expressing MyHC I and reduced MyHC II‐expressing myotubes (Figure 3E).
Next, we investigated the expression of the RvD2 receptor, Gpr18. Previous findings from our lab indicate that Gpr18 is expressed particularly when myoblasts enter differentiation. 15 , 36 Further analysis of the transcriptomics data set of soleus and TA muscle shows that Gpr18 is expressed in slow and fast muscles in mice (Figure S5).
Altogether, these results indicate that RvD2 promotes myogenesis and the transition toward slow fiber type in vitro.
3.4. RvD2 improves strength in early stages of muscle regeneration in vivo
To determine the effects of RvD2 on muscle phenotype at the early stages of muscle regeneration in vivo, muscle injury was induced in WT mice by cardiotoxin (CTX) injection into the TA muscle of one limb (contralateral limb is used as uninjured control). The injured and noninjured hind limbs of these WT mice were treated with intramuscular (i.m.) injections of RvD2 or vehicle into the TA muscle twice a week (on days 2 and 5 postinjury) for a duration of 1 week. On the day of sacrifice, the TA muscles from both the injured and noninjured limbs were isolated to assess the contractile property ex vivo and their muscle phenotype (Figure 4A). In the uninjured TA, the treatment with RvD2 did not significantly affect muscle force (Figures 4B and S6A). However, in the injured TA, an increase in the absolute (mN) and specific strength maximum (N/cm2) was observed in the RvD2‐treated group compared to vehicle‐treated mice (Figures 4C and S6B). Immunofluorescence analysis of skeletal muscle sections from the TA revealed no significant changes in the size of the myofibers between the RvD2‐treated and control mice for both noninjured and injured limbs (Figure 4D–G). The proportion of slow and fast (IIA, IIX, IIB) fibers was also unaffected in the TA from uninjured and injured limbs (Figures 4H,I and S7). Altogether, these findings show that local injections of RvD2 increased muscle strength postinjury but did not affect muscle phenotype in the short term.
FIGURE 4.
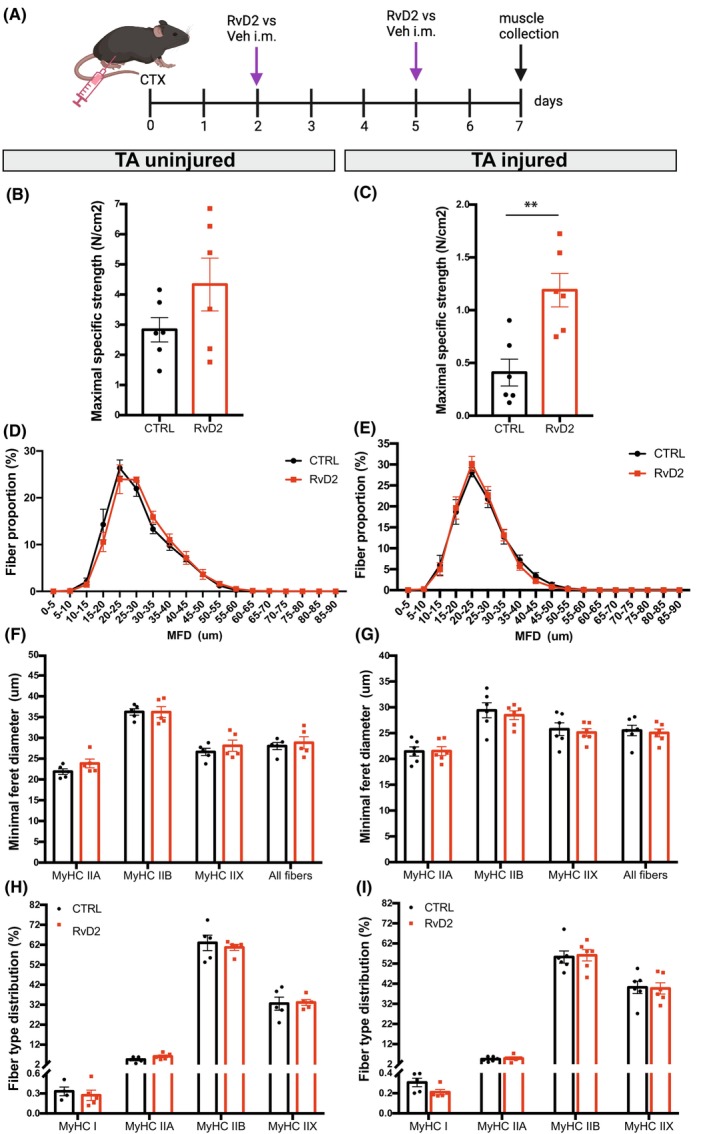
RvD2 enhances muscle strength at an early stage of regeneration in vivo. (A) Graphical overview of the experimental design (image created with BioRender.com) showing cardiotoxin injury (CTX), Resolvin‐D2 (RvD2) or vehicle (Veh) i.m. injections, and samples collection at 7 days postinjury. (B, C) Maximal specific strength (N/cm2) of the uninjured (B) and injured (C) TA muscles. (D, E) Distribution curve of the minimal Feret diameter (MFD) of the uninjured (D) and injured (E) TA muscles. (F, G) MFD of the uninjured (F) and injured (G) TA muscles expressed per fiber type. (H, I) Proportion of fibers expressing type I, IIA, IIX, IIB isoforms of MyHC. Data are presented as mean ± SEM, n = 5–6 mice per group. Immunostaining experiments were conducted with technical duplicates. Statistical analysis was performed using unpaired Student's t‐test (panels B, C) and two‐way ANOVA uncorrected Fisher's LSD test (for panels D–I). **p < .01.
3.5. RvD2 improves muscle strength and myofiber size at late stages of muscle regeneration in vivo
To determine the effects of RvD2 on the later stages of muscle regeneration in vivo, mice were treated twice weekly with i.m. injection of RvD2 versus vehicle for 21 days post‐CTX injury (Figure 5A). On the day of sacrifice, the contractile properties of the TA muscles from both legs were isolated to assess the contractile property ex vivo. In the uninjured TA, the treatment with RvD2 did not affect the absolute or specific maximal muscle strength (Figures 5B and S6C); however, a significant increase was observed in the RvD2‐treated injured TA compared to control (Figures 5C and S6D). Immunofluorescence analysis on TA muscle sections revealed that RvD2 induces a mild shift in the myofiber size in the uninjured muscle, which was not attributable to a specific fiber type (Figure 5D,F). This increase in myofiber size induced by RvD2 was stronger in the injured muscle and was caused predominantly by a specific increase in the size of type IIB fibers (Figure 5E,G). No significant changes in the proportion of fiber types were observed between the groups for both uninjured and injured TA (Figure 5H,I).
FIGURE 5.
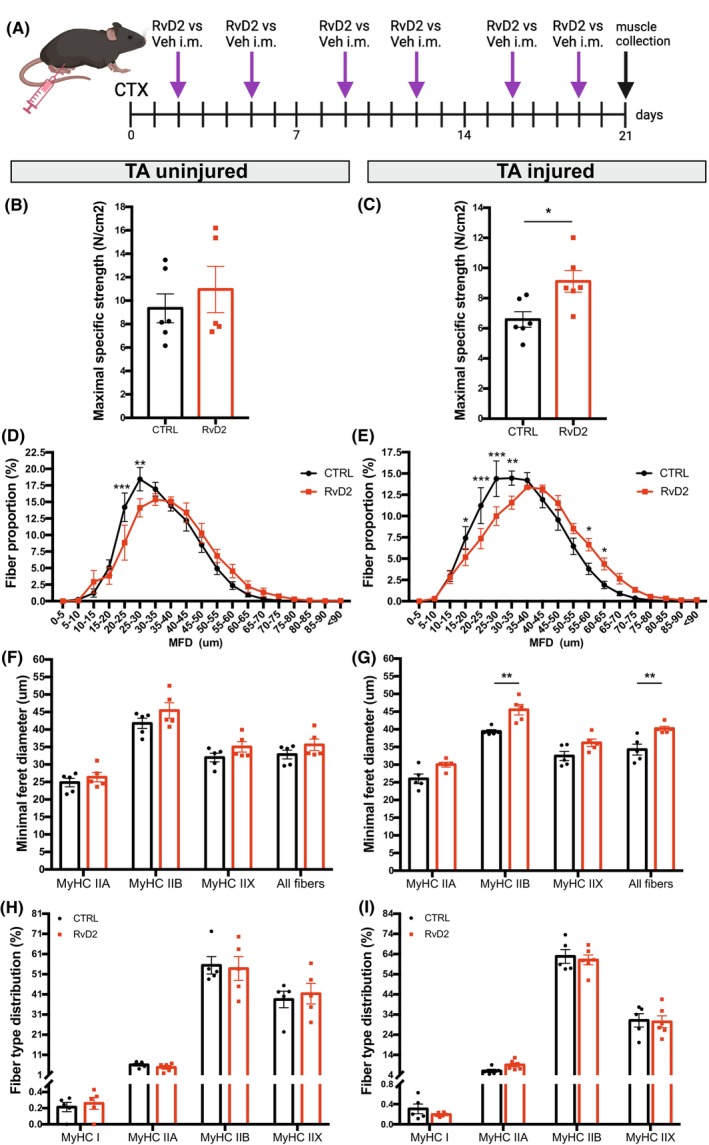
RvD2 enhances muscle strength and fiber size at a later stage of regeneration in vivo. (A) Graphical overview of the experimental design (image created with BioRender.com) showing cardiotoxin injury (CTX), Resolvin‐D2 (RvD2) or vehicle (Veh) i.m. injections, and samples collection at 21 days postinjury. (B, C) Maximal specific strength (N/cm2) of the uninjured (B) and injured (C) TA muscles. (D, E) Distribution curve of the minimal Feret diameter (MFD) of the uninjured (D) and injured (E) TA muscles. (F, G) MFD of the uninjured (F) and injured (G) TA muscles expressed per fiber type. (H, I) Proportion of fibers expressing type I, IIA, IIX, IIB isoforms of MyHC. Data are presented as mean ± SEM, n = 5–6 mice per group. Immunostaining experiments were conducted with technical duplicates. Statistical analysis was performed using unpaired Student's t‐test (panels B, C) and two‐way ANOVA uncorrected Fisher's LSD test (for panels D–I). *p < .05, **p < .01, ***p < .001.
3.6. Systemic RvD2 administration induces fiber type remodeling at late stages of muscle regeneration in vivo
To determine if a higher frequency of RvD2 administration could have a stronger impact on muscle phenotype, RvD2 was administered daily using systemic intraperitoneal (i.p.) injections (as described before 15 ) (Figure 6A). Intraperitoneal injections were chosen instead of daily i.m. injections to avoid repeated microinjuries to the muscle and to obtain systemic effects on multiple muscle types. At 21 days postinjury, TA and soleus muscles were collected to assess their muscle phenotype. In the uninjured limb, treatment with RvD2 did not affect the fiber type distribution of the TA or soleus muscles (Figure 6B,C,F,G). However, in the injured limb, administration of RvD2 led to significant remodeling of fiber types in both the TA and soleus muscles (Figure 6D,E,H,I). In the TA muscle, RvD2 increased the proportion of fiber type IIB while decreasing the proportion of fiber type IIX (Figure 6D,E). This change in muscle phenotype was associated with a faster twitch contraction velocity (Figure S8). In the soleus muscle, RvD2 increased the proportion of fiber type I while decreasing the proportion of fiber type IIA (Figure 6H,I). These findings indicate that when injured, the daily systemic injections of RvD2 promote the formation of the predominant fiber types in both slow‐ and fast‐twitch‐regenerating muscles.
FIGURE 6.
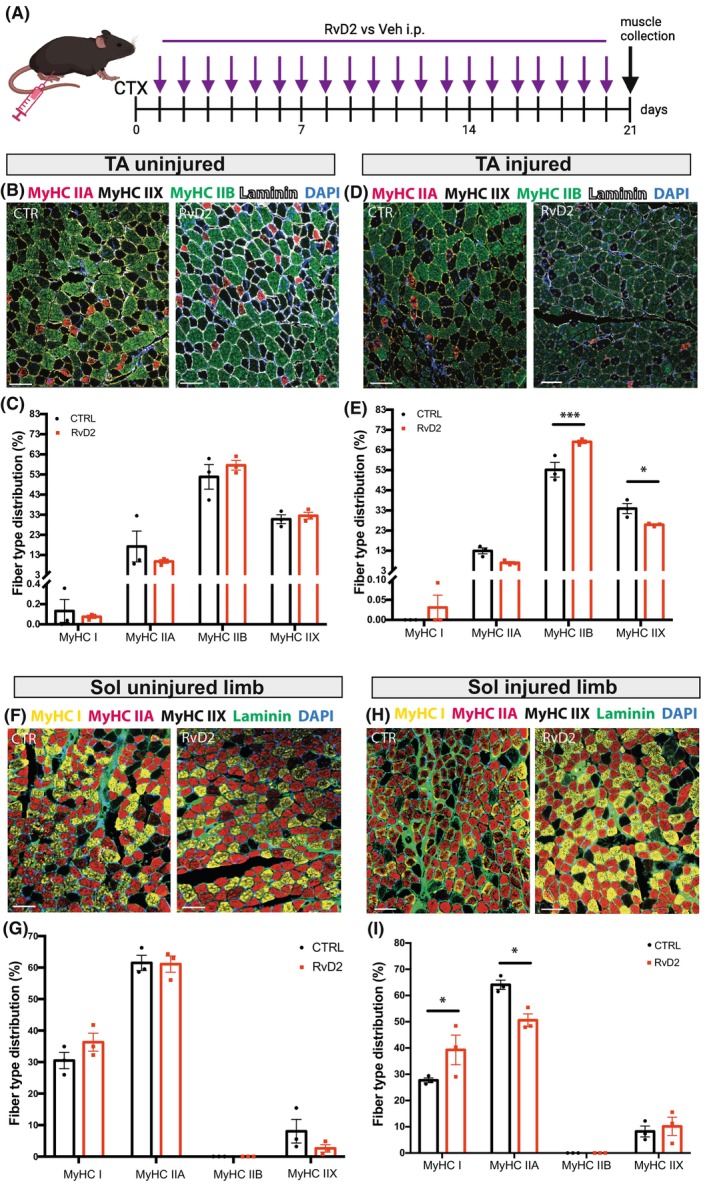
Resolvin‐D2 induces fiber type shifting at a later stage of regeneration in vivo. (A) Graphical overview of the experimental design (image created with BioRender.com) showing cardiotoxin injury (CTX), Resolvin‐D2 (RvD2) or vehicle (Veh) i.p. injections, and samples collection at 21 days postinjury. (B–E) Representative micrographs of MyHC IIA (red), MyHC IIX (black), MyHC IIB (green), laminin (white), and DAPI (blue) immunofluorescence; and quantification of the proportion of each fiber type in the uninjured (B, C) and injured (D, E) TA muscles. (F–I) Representative micrographs of MyHC I (yellow), MyHC IIA (red), MyHC IIX (black), laminin (green), and DAPI (blue) immunofluorescence; and quantification of the proportion of each fiber type in the soleus muscle from the uninjured (F, G) and injured limb (H, I). Scale bars = 100 μm. Data are presented as mean ± SEM, n = 3 mice per group. Experiments were conducted with technical duplicates. Data were analyzed using two‐way ANOVA uncorrected Fisher's LSD test. *p < .05, ***p < .001.
4. DISCUSSION
Slow and fast muscle fibers are well known to have different mitochondrial, lipid, and metabolic profiles. Slow fibers contain higher levels of neutral lipids such as triglycerides and cholesterol than fast fibers 37 ; however, the impact of bioactive lipids on muscle phenotype was still elusive. Our transcriptomic analysis shows that type I, IIA, IIX, and IIB fibers display an inherently different profile in their expression of bioactive lipids biosynthesis enzymes. Our lipidomics data indicate that slow‐twitch muscle expresses higher levels of DHA, EPA, and LA‐derived PUFAs, but similar levels of AA‐derived PUFAs, than fast‐twitch muscle. This is consistent with the higher expression in the TA muscle of Pla2g7 that preferentially releases AA. These findings are similar to another study showing in a rat model of critical illness that the slow‐twitch soleus muscle contains higher levels of the free fatty acids DHA and EPA as well as many omega‐3 PUFA‐derived fatty acids (e.g., 5‐HEPE, 12‐HEPE, 14‐HDHA, 17‐HDHA), than the fast‐twitch extensor digitorum longus muscle (EDL) at rest or following LPS injection. 38 However, contrary to our findings, this previous study also observed an increase in AA levels and their downstream bioactive lipids (e.g., PGE2, LTB4) in the slow‐twitch soleus muscle, which could be related to the model (mouse vs. rats) or muscles used (EDL vs. TA). Another study investigating the impact of muscle phenotype, diet, and sex on oxylipin profiles showed that the soleus muscle had higher expression of oxylipins than white gastrocnemius, which is similar to our findings. 39 Most of these bioactive lipids were formed by the lipoxygenase pathway. The authors also show that dietary DHA, EPA, or ALA increases the expression of their respective downstream oxylipins in all muscle types while reducing the expression of AA‐derived oxylipins.
Slow fibers also express higher activity and expression levels of NADPH oxidases NOX2 and NOX4 that are the main sources of reactive oxygen species (ROS). 40 Increased ROS levels could contribute to the oxidation of bioactive lipids observed at higher levels in slow‐twitch muscles compared to fast‐twitch muscles. The higher levels of markers of nonenzymatic oxidation such as 8‐HEPE or 4‐HDHA in the soleus muscle support this hypothesis. Noteworthy, some enzymes, such as Pla2g7, preferentially metabolize the oxidized form of AA. 41 This could also explain in part why the AA‐derived PUFA levels are similar between slow‐ and fast‐twitch muscles despite that many biosynthesizing enzymes of this pathway are expressed at higher levels in type II fibers. Moreover, the availability of the substrate in the lipid membrane also plays an important role in the production of the downstream bioactive lipids.
Our findings indicate that bioactive lipids content is not only a consequence of the fiber type but also a regulatory factor controlling fiber type determination. In vitro analysis showed that the DHA‐derived bioactive lipid, RvD2, promotes slow fiber type switching in myogenic cells. RvD2 is known to target the Gpr18 receptor, which was shown to be expressed at a higher level in slow‐twitch muscles compared to fast‐twitch muscles in a rat model of diet‐induced obesity, 42 although our findings in mice showed similar expression between slow and fast muscles at the RNA level. Gpr18 agonist was shown to activate the AMP‐activated protein kinase (AMPK), mammalian target of rapamycin (mTOR), and peroxisome proliferator‐activated receptor (PPAR) pathways. 36 The mTOR pathway stimulates the activation of 4E‐BP (4E‐binding protein) and S6K (ribosomal protein S6 kinase), thereby promoting the synthesis of contractile proteins, as well as mitochondrial and cytosolic proteins in both human and mouse muscles, 43 which is coherent with the increased myofiber size and strength that we observed in late regeneration. The AMPK is known to activate PPARγ‐coactivator 1‐alpha (PGC‐1α) which acts as a co‐activator for PPAR resulting in increased mitochondrial biogenesis and higher oxidative capacity. 44 Moreover, other pro‐resolving lipid mediators, such as RvD1, RvE1, and MaR1, have been shown to enhance mitochondrial metabolism in macrophages via the activation of AMPK. 45 Another bioactive lipid derived from omega‐9 fatty acids, oleic acid (released by Pla2g5, among others), was shown to activate PPARδ and increase the expression of slow MyHC and mitochondrial respiration in C2C12 myotubes in vitro. 46 Similarly, EPA supplementation to L6 myotubes activated the PPARδ and AMPK pathways, increased the expression of fatty acid metabolism genes and reduced the expression of fast MyHC isoforms. 47 Altogether, our findings showing that specialized pro‐resolving mediators derived from DHA promote slow fiber type contribute to the emerging evidence showing the regulatory role of bioactive lipids on fiber type determination.
In vivo results showed that RvD2 affects muscle strength, fiber size, and phenotype during muscle regeneration, but not in uninjured muscle. This is coherent with other groups showing that pro‐resolving mediators, such as RvD2, are mainly expressed during muscle regeneration. 16 , 17 Our lipidomics analysis confirmed that the precursors are expressed in the uninjured muscles, but most of the pro‐resolving mediators themselves were below the limit of detection (which does not mean that they are completely unexpressed). Moreover, our previous findings indicate that the expression of the RvD2 receptor, Gpr18, is increased during myogenesis when myoblasts enter differentiation. 15 , 36 These findings suggest that RvD2 mediates its effect during muscle regeneration.
Assessment of muscle function revealed that RvD2 increases maximal force at early stages of regeneration before the increase in muscle size. This could be due to faster maturation of the newly formed muscle fibers or other neuromuscular components such as the excitation–contraction coupling. The increase in muscle strength was also maintained at a later time point postinjury together with an increase in fiber size. These results are in accordance with our previous study in a model of Duchenne Muscular Dystrophy. 15 This increase in strength was also observed in a mouse model of delayed muscle regeneration, where i.m. injection of RvD2 3 days after injury resulted in enhanced strength on days 8 and 14 postinjury. 17 Other bioactive lipids were also shown to impact muscle strength and fiber size. For instance, RvD1 and MaR1 have also demonstrated their potential to improve muscle strength and fiber size in different models of injury. 16 , 22 Overall, our findings are coherent with the literature in the field and now demonstrate that bioactive lipids also affect fiber typing.
In vivo findings show that RvD2 changes fiber typing during the later stages of muscle regeneration and especially if it is administered systemically and daily. In the soleus muscle, RvD2 increased the proportion of type I fibers, which is consistent with what was observed in vitro. These findings are in accordance with another study showing that 4‐week EPA supplementation promotes transition to type I fiber in the rat plantaris muscle. 47 However, our results from the TA muscle showed that RvD2 stimulates the switch in fiber type toward type IIB fibers. These findings suggest that RvD2 promotes fiber type plasticity but other factors such as innervation and physical demand also participate in fiber type determination. 48 Altogether the in vitro and in vivo findings suggest that RvD2 promotes the slow fiber type, which can only be expressed in a permissive environment. Similar findings were observed in a model of dietary intake of oleic acid which increased type I and IIX fibers in the slow‐twitch soleus muscle, but only the type IIX in the fast‐twitch EDL muscle. 46
The ability to promote fiber type switch can have therapeutic advantages. Many disorders affecting skeletal muscle such as obesity/type 2 diabetes, 49 , 50 heart failure and chronic obstructive pulmonary disease, 51 and disuse 7 are known to selectively affect the type I fibers. Moreover, in dystrophic mdx mice, slow muscles are more resistant to degeneration than fast muscles 52 ; and a specific inhibitor of fast skeletal myosin was shown to protect against muscle degeneration. 53
This study also has limitations to consider. First, the systemic administration of RvD2 does not allow to determine if the effect on fiber typing is mediated directly on skeletal muscle or through other mechanisms; however, the i.m. injection of RvD2 indicates that it can mediate its effect directly on skeletal muscle. Moreover, the experiments were carried out only in male mice. Sex differences have been reported in DHA contents and synthesis in different tissues. 39 , 54 Particularly, in skeletal muscle, higher levels of oxylipins were observed in females, especially in the soleus or red gastrocnemius muscle. 39 Therefore, follow‐up experiments on female mice should be performed in the future to validate that our findings in male mice could be extrapolated to females.
Overall, our transcriptomics and lipidomics analysis revealed that slow and fast fibers have different signatures for biosynthesizing enzymes and bioactive lipids derived from AA, LA, DHA, and EPA. Moreover, in vitro and in vivo findings demonstrate that RvD2 stimulates myogenesis and influences fiber typing. RvD2 stimulates the slow phenotype by default, although this signature can be overcome by other environmental factors. In conjunction with previous study from our lab and others regarding the effect of bioactive lipids on muscle regeneration and strength, these findings provide a novel insight into the role of RvD2 and highlight its therapeutic potential for the treatment of muscle disorders.
AUTHOR CONTRIBUTIONS
Nicolas A. Dumont conceptualized the study. Lupann Rieger and Nicolas A. Dumont designed the experiments. Lupann Rieger, Thomas Molina, Paul Fabre, Karine Greffard, Ornella Pellerito, and Junio Dort collected and analyzed data. Lupann Rieger and Nicolas A. Dumont wrote the manuscript. Nicolas A. Dumont and Jean‐François Bilodeau obtained funding, coordinated the project, supervised trainees, and data collection. All authors critically reviewed and revised the manuscript.
FUNDING INFORMATION
LR was supported by fellowships of the FRQNT (Fonds de recherche du Québec—Nature et Technologies). TM and PF were supported by fellowships of the FRQS (Fonds de recherche du Québec—Santé). NAD was supported by a FRQS Junior‐2 award. This research was funded by a research grant to NAD by the Natural Sciences and Engineering Research Council (NSERC; RGPIN‐2018‐05979).
DISCLOSURES
The authors declare no conflicts of interest.
Supporting information
Data S1.
ACKNOWLEDGMENTS
We would like to thank Sonja Lesperance for animal husbandry. We acknowledge the support of the Quebec Cell, Tissue and Gene Therapy Network—ThéCell (a thematic network supported by the FRQS).
Rieger L, Molina T, Fabre P, et al. Transcriptomic and lipidomic profiling reveals distinct bioactive lipid signatures in slow and fast muscles and highlights the role of resolvin‐D2 in fiber type determination during myogenesis. The FASEB Journal. 2024;38:e70250. doi: 10.1096/fj.202401747R
DATA AVAILABILITY STATEMENT
Data are available from the Gene Expression Omnibus database GSE226117, GSE147127, and GSE233882.
REFERENCES
- 1. Janssen I, Heymsfield SB, Wang ZM, Ross R. Skeletal muscle mass and distribution in 468 men and women aged 18‐88 yr. J Appl Physiol (1985). 2000;89:81‐88. [DOI] [PubMed] [Google Scholar]
- 2. Dumont NA, Bentzinger CF, Sincennes M‐C, Rudnicki MA. Satellite cells and skeletal muscle regeneration. Compr Physiol. 2015;5:1027‐1059. [DOI] [PubMed] [Google Scholar]
- 3. Schiaffino S, Reggiani C. Fiber types in mammalian skeletal muscles. Physiol Rev. 2011;91:1447‐1531. [DOI] [PubMed] [Google Scholar]
- 4. Blemker SS, Brooks SV, Esser KA, Saul KR. Fiber‐type traps: revisiting common misconceptions about skeletal muscle fiber types with application to motor control, biomechanics, physiology, and biology. J Appl Physiol (1985). 2024;136:109‐121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mukund K, Subramaniam S. Skeletal muscle: a review of molecular structure and function, in health and disease. Wiley Interdiscip Rev Syst Biol Med. 2020;12:e1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yan Z, Okutsu M, Akhtar YN, Lira VA. Regulation of exercise‐induced fiber type transformation, mitochondrial biogenesis, and angiogenesis in skeletal muscle. J Appl Physiol. 2011;110:264‐274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vikne H, Strøm V, Pripp AH, Gjøvaag T. Human skeletal muscle fiber type percentage and area after reduced muscle use: a systematic review and meta‐analysis. Scand J Med Sci Sports. 2020;30:1298‐1317. [DOI] [PubMed] [Google Scholar]
- 8. Ciciliot S, Rossi AC, Dyar KA, Blaauw B, Schiaffino S. Muscle type and fiber type specificity in muscle wasting. Int J Biochem Cell Biol. 2013;45:2191‐2199. [DOI] [PubMed] [Google Scholar]
- 9. Edom F, Mouly V, Barbet JP, Fiszman MY, Butler‐Browne GS. Clones of human satellite cells can express in vitro both fast and slow myosin heavy chains. Dev Biol. 1994;164:219‐229. [DOI] [PubMed] [Google Scholar]
- 10. Barjot C, Cotten ML, Goblet C, Whalen RG, Bacou F. Expression of myosin heavy chain and of myogenic regulatory factor genes in fast or slow rabbit muscle satellite cell cultures. J Muscle Res Cell Motil. 1995;16:619‐628. [DOI] [PubMed] [Google Scholar]
- 11. Rosenblatt JD, Parry DJ, Partridge TA. Phenotype of adult mouse muscle myoblasts reflects their fiber type of origin. Differentiation. 1996;60:39‐45. [DOI] [PubMed] [Google Scholar]
- 12. Kalhovde JM, Jerkovic R, Sefland I, et al. “Fast” and “slow” muscle fibres in hindlimb muscles of adult rats regenerate from intrinsically different satellite cells. J Physiol. 2005;562:847‐857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Düsterhöft S, Pette D. Satellite cells from slow rat muscle express slow myosin under appropriate culture conditions. Differentiation. 1993;53:25‐33. [DOI] [PubMed] [Google Scholar]
- 14. Motohashi N, Uezumi A, Asakura A, et al. Tbx1 regulates inherited metabolic and myogenic abilities of progenitor cells derived from slow‐ and fast‐type muscle. Cell Death Differ. 2019;26:1024‐1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dort J, Orfi Z, Fabre P, et al. Resolvin‐D2 targets myogenic cells and improves muscle regeneration in Duchenne muscular dystrophy. Nat Commun. 2021;12:6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Markworth JF, Brown LA, Lim E, et al. Resolvin D1 supports skeletal myofiber regeneration via actions on myeloid and muscle stem cells. JCI Insight. 2020;5:e137713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Giannakis N, Sansbury BE, Patsalos A, et al. Dynamic changes to lipid mediators support transitions among macrophage subtypes during muscle regeneration. Nat Immunol. 2019;20:626‐636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ho ATV, Palla AR, Blake MR, et al. Prostaglandin E2 is essential for efficacious skeletal muscle stem‐cell function, augmenting regeneration and strength. Proc Natl Acad Sci U S A. 2017;114:6675‐6684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang B, Wu L, Chen J, et al. Metabolism pathways of arachidonic acids: mechanisms and potential therapeutic targets. Sig Transduct Target Ther. 2021;6:1‐30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Serhan CN. Pro‐resolving lipid mediators are leads for resolution physiology. Nature. 2014;510:92‐101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Serhan CN. Novel lipid mediators and resolution mechanisms in acute inflammation. Am J Pathol. 2010;177:1576‐1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Castor‐Macias JA, Larouche JA, Wallace EC, et al. Maresin 1 repletion improves muscle regeneration after volumetric muscle loss. elife. 2023;12:e86437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Markworth JF, Brown LA, Lim E, et al. Metabolipidomic profiling reveals an age‐related deficiency of skeletal muscle pro‐resolving mediators that contributes to maladaptive tissue remodeling. Aging Cell. 2021;20:e13393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Baker LA, Martin NRW, Kimber MC, Pritchard GJ, Lindley MR, Lewis MP. Resolvin E1 (Rv E1) attenuates LPS induced inflammation and subsequent atrophy in C2C12 myotubes. J Cell Biochem. 2018;119:6094‐6103. [DOI] [PubMed] [Google Scholar]
- 25. Hardy D, Besnard A, Latil M, et al. Comparative study of injury models for studying muscle regeneration in mice. PLoS One. 2016;11:e0147198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dumont NA, Wang YX, von Maltzahn J, et al. Dystrophin expression in muscle stem cells regulates their polarity and asymmetric division. Nat Med. 2015;21:1455‐1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fabre P, Molina T, Orfi Z, Dumont NA. Assessment of muscle function following hiPSC‐derived myoblast transplantation in dystrophic mice. Curr Protoc. 2022;2(1):e356. doi: 10.1002/cpz1.356 [DOI] [PubMed] [Google Scholar]
- 28. Dumont NA, Rudnicki MA. Characterizing satellite cells and myogenic progenitors during skeletal muscle regeneration. In: Pellicciari C, Biggiogera M, eds. Histochemistry of Single Molecules. Springer, New York, NY, USA; 2017:179‐188. [DOI] [PubMed] [Google Scholar]
- 29. Babcock LW, Hanna AD, Agha NH, Hamilton SL. MyoSight‐semi‐automated image analysis of skeletal muscle cross sections. Skelet Muscle. 2020;10:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shavlakadze T, Xiong K, Mishra S, et al. Age‐related gene expression signatures from limb skeletal muscles and the diaphragm in mice and rats reveal common and species‐specific changes. Skelet Muscle. 2023;13:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Petrany MJ, Swoboda CO, Sun C, et al. Single‐nucleus RNA‐seq identifies transcriptional heterogeneity in multinucleated skeletal myofibers. Nat Commun. 2020;11:6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pass CG, Palzkill V, Tan J, et al. Single‐nuclei RNA‐sequencing of the gastrocnemius muscle in peripheral artery disease. Circ Res. 2023;133:791‐809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Robb JL, Boisjoly F, Machuca‐Parra AI, et al. Representative of consortium. Blockage of ATGL‐mediated breakdown of lipid droplets in microglia alleviates neuroinflammatory and behavioural responses to lipopolysaccharides. Brain Behav Immun. 2025;123:315‐333. doi: 10.1016/j.bbi.2024.09.027 [DOI] [PubMed] [Google Scholar]
- 34. Bilodeau J‐F, Gevariya N, Larose J, et al. Long chain omega‐3 fatty acids and their oxidized metabolites are associated with reduced prostate tumor growth. Prostaglandins Leukot Essent Fatty Acids. 2021;164:102215. [DOI] [PubMed] [Google Scholar]
- 35. Nelson DR, Zeldin DC, Hoffman SMG, Maltais LJ, Wain HM, Nebert DW. Comparison of cytochrome P450 (CYP) genes from the mouse and human genomes, including nomenclature recommendations for genes, pseudogenes and alternative‐splice variants. Pharmacogenetics. 2004;14:1‐18. [DOI] [PubMed] [Google Scholar]
- 36. Dort J, Orfi Z, Fiscaletti M, Campeau PM, Dumont NA. Gpr18 agonist dampens inflammation, enhances myogenesis, and restores muscle function in models of Duchenne muscular dystrophy. Front Cell Dev Biol. 2023;11:1187253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fiehn W, Peter JB. Lipid composition of muscles of nearly homogeneous fiber type. Exp Neurol. 1973;39:372‐380. [DOI] [PubMed] [Google Scholar]
- 38. Miyoshi M, Usami M, Nishiyama Y, et al. Soleus muscle contains a higher concentration of lipid metabolites than extensor digitorum longus in rats with lipopolysaccharide‐induced acute muscle atrophy. Clin Nutr ESPEN. 2023;57:48‐57. [DOI] [PubMed] [Google Scholar]
- 39. Penner AL, Waytt V, Winter T, Leng S, Duhamel TA, Aukema HM. Oxylipin profiles and levels vary by skeletal muscle type, dietary fat and sex in young rats. Appl Physiol Nutr Metab. 2021;46:1378‐1388. [DOI] [PubMed] [Google Scholar]
- 40. Loureiro ACC, Do Rêgo‐Monteiro IC, Louzada RA, et al. Differential expression of NADPH oxidases depends on skeletal muscle fiber type in rats. Oxidative Med Cell Longev. 2016;2016:6738701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stremler KE, Stafforini DM, Prescott SM, Zimmerman GA, McIntyre TM. An oxidized derivative of phosphatidylcholine is a substrate for the platelet‐activating factor acetylhydrolase from human plasma. J Biol Chem. 1989;264:5331‐5334. [PubMed] [Google Scholar]
- 42. Simcocks AC, O'Keefe L, Jenkin KA, et al. The role of atypical cannabinoid ligands O‐1602 and O‐1918 on skeletal muscle homeostasis with a focus on obesity. Int J Mol Sci. 2020;21:5922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Philp A, Hamilton DL, Baar K. Signals mediating skeletal muscle remodeling by resistance exercise: PI3‐kinase independent activation of mTORC1. J Appl Physiol. 2011;110:561‐568. [DOI] [PubMed] [Google Scholar]
- 44. Hood DA. Invited review: contractile activity‐induced mitochondrial biogenesis in skeletal muscle. J Appl Physiol. 2001;90:1137‐1157. [DOI] [PubMed] [Google Scholar]
- 45. Calderin EP, Zheng J‐J, Boyd NL, et al. Exercise‐induced specialized proresolving mediators stimulate AMPK phosphorylation to promote mitochondrial respiration in macrophages. Mol Metab. 2022;66:101637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Watanabe N, Komiya Y, Sato Y, Watanabe Y, Suzuki T, Arihara K. Oleic acid up‐regulates myosin heavy chain (MyHC) 1 expression and increases mitochondrial mass and maximum respiration in C2C12 myoblasts. Biochem Biophys Res Commun. 2020;525:406‐411. [DOI] [PubMed] [Google Scholar]
- 47. Komiya Y, Sakazaki Y, Goto T, et al. Eicosapentaenoic acid increases proportion of type 1 muscle fibers through PPARδ and AMPK pathways in rats. iScience. 2024;27:109816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ausoni S, Gorza L, Schiaffino S, Gundersen K, Lømo T. Expression of myosin heavy chain isoforms in stimulated fast and slow rat muscles. J Neurosci. 1990;10:153‐160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Oberbach A, Bossenz Y, Lehmann S, et al. Altered fiber distribution and fiber‐specific glycolytic and oxidative enzyme activity in skeletal muscle of patients with type 2 diabetes. Diabetes Care. 2006;29:895‐900. [DOI] [PubMed] [Google Scholar]
- 50. Tanner CJ, Barakat HA, Dohm GL, et al. Muscle fiber type is associated with obesity and weight loss. Am J Physiol Endocrinol Metab. 2002;282:E1191‐E1196. [DOI] [PubMed] [Google Scholar]
- 51. Gosker HR, Wouters EF, van der Vusse GJ, Schols AM. Skeletal muscle dysfunction in chronic obstructive pulmonary disease and chronic heart failure: underlying mechanisms and therapy perspectives. Am J Clin Nutr. 2000;71:1033‐1047. [DOI] [PubMed] [Google Scholar]
- 52. Webster C, Silberstein L, Hays AP, Blau HM. Fast muscle fibers are preferentially affected in Duchenne muscular dystrophy. Cell. 1988;52:503‐513. [DOI] [PubMed] [Google Scholar]
- 53. Russell AJ, DuVall M, Barthel B, et al. Modulating fast skeletal muscle contraction protects skeletal muscle in animal models of Duchenne muscular dystrophy. J Clin Invest. 2023;133:e153837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kitson AP, Smith TL, Marks KA, Stark KD. Tissue‐specific sex differences in docosahexaenoic acid and Δ6‐desaturase in rats fed a standard chow diet. Appl Physiol Nutr Metab. 2012;37:1200‐1211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1.
Data Availability Statement
Data are available from the Gene Expression Omnibus database GSE226117, GSE147127, and GSE233882.


