Abstract
Bacterial infections and antibiotic resistance represent significant global public health challenges, necessitating the development of innovative antibacterial agents with targeted delivery capabilities. Our study utilized macrophages’ natural ability to recognize bacteria and the increased reactive oxygen species (ROS) at infection sites to develop a novel nanoparticle for targeted delivery and controlled release. We prepared bacteria-activated macrophage membranes triggered by Staphylococcus aureus (Sa-MMs), which showed significantly higher expression of Toll-like receptors (TLRs), compared to normal macrophage membranes (MMs). These Sa-MMs were then used to coat vancomycin-loaded amphiphilic nanoparticles with ROS responsiveness (Van-NPs), resulting in the novel targeted delivery system Sa-MM@Van-NPs. Studies both In vitro and in vivo demonstrated that biocompatible Sa-MM@Van-NPs efficiently targeted infected sites and released vancomycin to eliminate bacteria, facilitating faster wound healing. By combining targeted delivery to infected sites and ROS-responsive antibiotic release, this approach might represent a robust strategy for precise infection eradication and enhanced wound healing.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12951-024-03056-5.
Keywords: Bacteria-activation, Macrophage membrane, ROS-responsive nanoparticle, Targeted delivery
Introduction
Since the beginning of the 21st century, bacterial infections have become a significant threat to global public health. The widespread use of antibiotics has worsened bacterial resistance, leading to substantial social and economic burdens. In 2019, deaths directly attributed to antibiotic resistance were reported to be equal to the combined deaths from acquired immunodeficiency syndrome (AIDS) and malaria. Antibiotic resistance is now the third leading cause of death globally, after ischemic heart disease and stroke [1]. By 2025, it is estimated that there will be 10 million deaths every year worldwide due to antibiotic-resistant infections [2]. Currently, antibiotics remain the primary treatment for bacterial infections. However, the development of novel antibiotics presents significant challenges. Therefore, the most effective strategy is to efficiently deliver existing antibiotics to the infection site, ensuring precise release to enhance local therapeutic effectiveness, reduce toxicity, and mitigate the evolution of antibiotic resistance [3].
Biomimetic drug delivery systems, especially those utilizing cell membrane-coated nanoparticles, have garnered considerable interest for their improved circulation, targeting capabilities, and biocompatibility [4]. Cell membrane coating technology, employing cell membranes as carriers, facilitates the targeted delivery of core nanoparticles, irrespective of the nature of the core nanomaterials [5]. This approach has employed a diverse range of cell membranes [6], such as those from erythrocytes [7, 8], leukocytes [9], platelets [10], tumor cells [11], stem cells [12, 13], and hybrid membranes [14, 15]. For example, Wang et al. developed neutrophil membrane-encapsulated nanoparticles capable of delivering antibiotics precisely to inflammatory sites and extending their half-life [16]. Similarly, Hu et al. used M1 macrophage membrane (M1-MM)-coated nanoparticles to delay phagocytosis-mediated clearance, increasing circulation time, and effectively concentrating drugs at tumor sites, thereby inhibiting primary tumor growth, recurrence, and metastasis without significant side effects [17]. Additionally, hybrid biomimetic nanoparticles were developed for ovarian cancer therapy by fusing membranes from murine-derived ID8 ovarian cancer cells with red blood cell membranes. These hybrid nanoparticles exhibited targeted self-recognition in both in vitro and in vivo studies, as well as prolonged circulation time [15]. Although cell membrane coating has improved the targeting of nanoparticles, achieving on-demand drug release at the disease site has emerged as a major challenge.
Wound healing is a multifaceted biological process comprising three stages: inflammation, proliferation, and tissue remodeling [18–20]. Bacterial infections can trigger intense inflammatory responses, which make it difficult to heal the defective tissue, and even cause systemic complications such as sepsis, threatening the life and safety of patients. In response to bacterial infection and immune reactions, wounds often develop unique microenvironments distinct from healthy tissue. For instance, the inflammatory response elevates reactive oxygen species (ROS) levels at the site of infection. Therefore, employing ROS-responsive nanosystem to deliver antibiotics to the infected site may be an effective strategy [21, 22]. Borates and derivatives, especially phenylborate, serve as a key chemical functional group in cells with ROS reactivity [23, 24]. Yang et al. covalently modified maltodextrin (MD) with 4-(hydroxymethyl) phenylboronic acid pinacol ester to prepare ROS-responsive MD-prodrugs, which were internalized and degraded back to MD, reactivating dormant bacteria and increasing their sensitivity to rifampicin [25]. Zhao et al. developed and synthesized a polymeric prodrug, polyglutamic acid modified with 4-hydroxymethyl phenylboronic acid caffeic acid ester (PBC). The PBC can break the boronic acid ester bond in response to high ROS levels, releasing two antioxidant molecules, caffeic acid and 4-hydroxybenzyl alcohol, which can scavenge the localized excess of ROS on-demand [26]. Although nanoscale ROS-responsive prodrugs facilitate controlled drug release, nanoparticles must navigate through a complex physiological environment before reaching the target tissue, during which they are prone to being cleared by the immune system [27–29].
Macrophages are essential to innate immunity, functioning as the main defense against bacterial infections. Through pattern recognition receptors (PRRs), mainly toll-like receptors (TLRs), they detect bacterial pathogen-associated molecular patterns (PAMPs) to trigger an inflammatory response [30]. TLRs are located on the cell surface and within cells to detect various PAMPs. Surface TLRs, including TLR1, TLR2, and TLR6, identify bacterial lipoproteins, whereas TLR4 detects lipopolysaccharides (LPS) and specific viral membrane proteins. TLR5 recognizes flagellin, a protein found in bacterial flagella. Intracellular TLRs, such as TLR3, TLR7, TLR8, and TLR9, detect nucleic acids from viruses and bacteria [31].
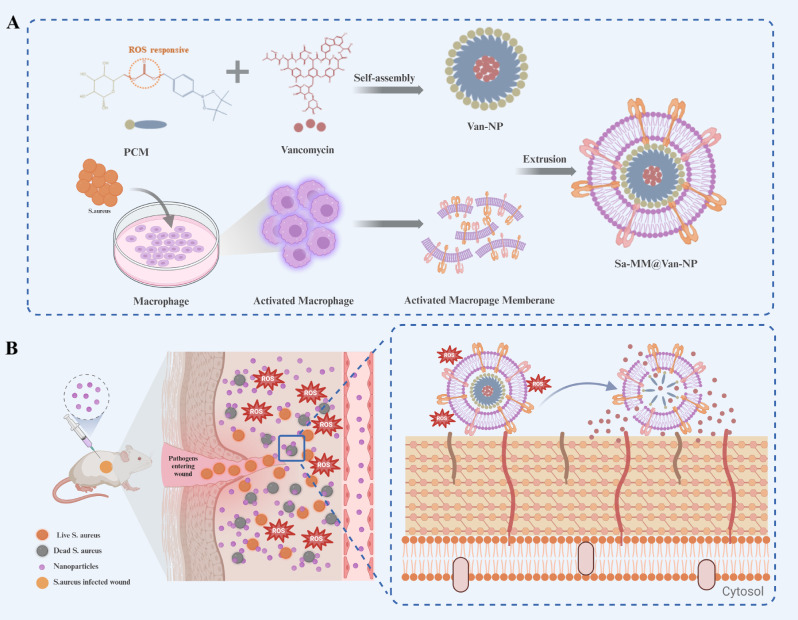
Leveraging the altered microenvironment of bacterially infected tissues and the bacterial recognition capabilities of macrophages, we engineered bacterial pre-activated macrophage membranes to encapsulate antibiotic-loaded ROS-responsive nanoparticles. These innovative nanoparticles are designed to target infection sites and release antibiotics in response to ROS, facilitating the healing of infected wounds (Sch. 1). Staphylococcus aureus (S. aureus), a leading cause of wound infections [32–34], was selected as the model bacterium, while vancomycin, the last line of defense against Gram-positive bacterial infections [35, 36], served as the model drug. The design of our nanoparticles involves two key stages: (1) S. aureus triggered macrophages to acquire membranes with enhanced bacterial recognition capabilities, termed Sa-MMs; (2) Sa-MMs were integrated onto the surface of vancomycin-loaded ROS-responsive nanoparticles (Van-NPs) using extrusion, resulting in Sa-MM@Van-NPs. The integration of MMs endowed the nanoparticles with biomimetic properties, allowing them to evade immune clearance. The presence of bacterial-specific receptors on Sa-MMs facilitated preferential accumulation at bacterial infection sites compared to normal tissues, thereby minimizing drug toxicity. With elevated ROS levels at the infected sites, Sa-MM@Van-NPs degraded, releasing vancomycin and effectively eradicating bacteria while promoting wound healing. We anticipated that this nanoparticle, Sa-MM@Van-NPs, may represent a promising strategy for highly targeted and efficient antibacterial therapy.
Sch. 1.
Schematic illustration of bacteria-activated macrophage membranes coated ROS-responsive nanoparticle for infected wound healing: A. Schematic of Sa-MM@Van-NPs synthesis. B. Schematic of the antibacterial mechanism of Sa-MM@Van-NPs
Results and discussions
Bacteria-activated macrophage characterization
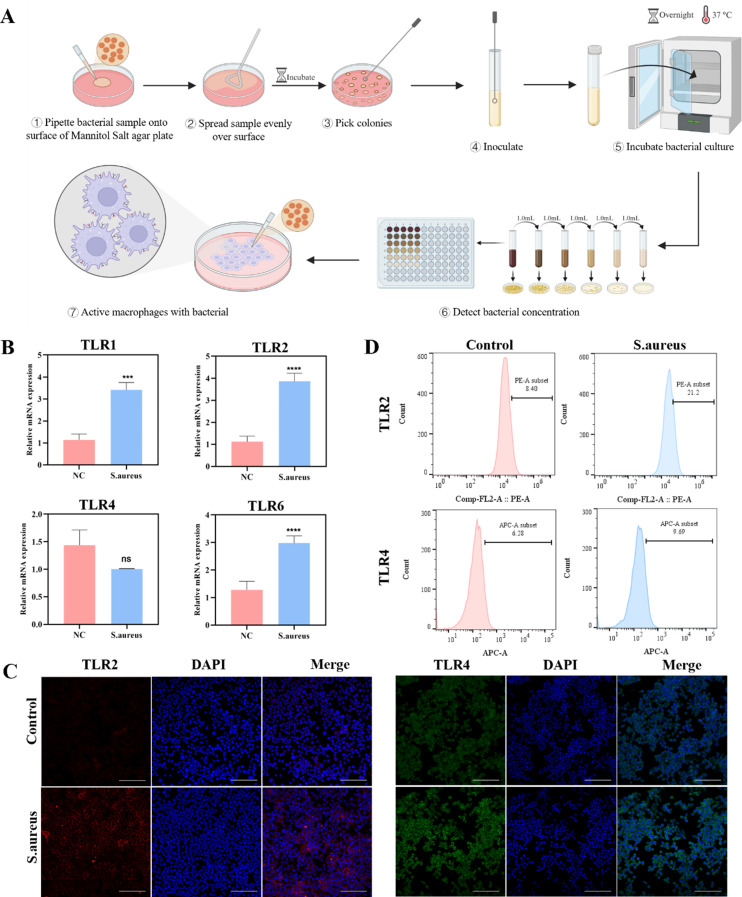
To obtain the Sa-MM, RAW264.7 cells were co-cultured with S. aureus to activate macrophages (Fig. 1A). TLR2 and TLR4 on macrophage membrane are crucial for identifying extracellular pathogens [37, 38]. Initially, we assessed the expression of these receptors on macrophages. Quantitative real-time polymerase chain reaction (qPCR) revealed significant upregulation of TLR1, TLR2, and TLR6 mRNA expressions in macrophages after bacterial stimulation (Fig. 1B). The TLR1 or TLR6 can form heterodimers with TLR2 to recognize lipoproteins [39]. The TLR2 /TLR1 heterodimer can recognize triacylated lipoproteins, whereas the TLR2 /TLR6 heterodimer is associated with the recognition of diacylated lipoproteins and lipoteichoic acid [40, 41]. This upregulation aligned with their established functions in detecting Gram-positive bacteria. In contrast, TLR4 mRNA levels did not show a significant change, aligning with its primary role in detecting LPS from Gram-negative bacteria [42–44]. Immunofluorescence staining further confirmed the upregulation of TLR2 and TLR4 protein expressions (Fig. 1C). Statistical analysis indicated a 2.2-fold increase in TLR2 fluorescence intensity and only a 1.5-fold increase in TLR4 fluorescence intensity (Figure S1). These findings were corroborated by flow cytometry analysis (Fig. 1D). The differences in TLR4 mRNA expression and protein expression may be due to post-translational modifications [45, 46]. Consequently, incorporating MMs with these specific receptor expressions onto nanoparticles may enhance their ability to recognize and bind to specific pathogenic bacteria.
Fig. 1.
Characterization of bacteria-activated macrophages triggered by S. aureus: A. Schematic representation of bacteria-activated macrophages triggered by S. aureus. B. qPCR analysis of RAW264.7 cells revealed mRNA expression levels for TLR1, TLR2, TLR4, and TLR6 (n = 3; mean ± SD; ***p < 0.001, ****p < 0.0001). C. Immunofluorescence images of TLR2 (red) and TLR4 (green) expressions in RAW264.7 cells, with nuclei stained in blue (scale bar = 100 μm). D. Flow cytometry analysis of TLR2 and TLR4 expressions in RAW264.7 cells
Synthesis and characterization of Sa-MM@Van-NPs
Amphiphilic nanoparticles responsive to ROS were synthesized through the self-assembly of PBAP-CDI-Mannose (PCM), where 4-(hydroxymethyl) phenylboronic acid pinacol ester (PBAP) served as hydrophobic element and D-mannose as hydrophilic element (Fig. 2A). And the mechanism of ROS responsiveness [24, 47] was shown in Figure S2. The chemical structure of the PCM was confirmed using 1H NMR [48, 49] (Fig. 2B). Fourier transform infrared (FT-IR) spectral was conducted to further determine the successful synthesis of PCM (Fig. 2C). The successful reaction of PBAP with CDI (PBAP-CDI) is evidenced by the loss of the O-H stretching vibrational absorption peak at 3325 cm− 1 and the emergence of the C = O stretching vibrational absorption peak of the ester group at 1760 cm− 1 [50], alongside the C = C and C = N double bonding stretching vibrational absorption peaks on CDI (1,1’-carbonyldiimidazole) at 1515 cm− 1. The infrared spectrogram of PCM showed a strengthened O-H stretching vibrational absorption peak at 3414 cm− 1, compared to the water peak on PBAP-CDI, indicating a successful reaction with Mannose [51, 52]. Subsequently, vancomycin was encapsulated in the hydrophobic core of the nanoparticles via nano-precipitation, resulting in a drug encapsulation efficiency (DEE) of about 80% and a drug loading efficiency (DLE) of approximately 7%. Sa-MM@Van-NPs were synthesized by coating Sa-MM onto Van-NPs using an extrusion technique. Transmission electron microscopy (TEM) showed that Van-NPs had a roughly spherical shape, whereas Sa-MM@Van-NPs displayed a corona layer about 20 nm in diameter, due to the functionalized Sa-MM coating [53, 54] (Fig. 2D). Dynamic light scattering (DLS) revealed that the average hydrodynamic diameter of Sa-MM@Van-NPs increased from 127.6 ± 4.98 nm to 178.0 ± 3.51 nm compared to uncoated Van-NPs (Fig. 2E). And the polymer dispersity index (PDI) was 0.2866, indicating a relatively uniform particle size distribution [55, 56]. The zeta potential decreased to a level comparable to MM, indicating the successful synthesis of Sa-MM@Van-NPs (Fig. 2F). The enhanced electrostatic repulsion between nanoparticles helps to prevent aggregation, thus improving stability [57]. Furthermore, Western blot analysis verified that key membrane antigens including CD36, CCR2, and TNFR2, were present on both MMs and Sa-MM@Van-NPs surface, indicating that the macrophage membranes on the Sa-MM@Van-NPs resembled those on macrophages (Fig. 2I). Dual fluorescent labeling experiment was performed to further confirm the successful encapsulation of Sa-MM by labeling the core nanoparticles (NPs) with red fluorescent dye DiI (DiI-NPs) and Sa-MM with green fluorescent dye DiO (DiO-Sa-MM). Then, DiO-Sa-MM@DiI-NPs were incubated with macrophages. As depicted in Fig. 2J, the red fluorescence surrounding the nucleus extensively overlapped with the green fluorescence, confirming the successful encapsulation of Sa-MM components onto the NPs. Considering that the effect of proteins in vivo is an important factor, the particle size of Sa-MM@Van-NPs was measured in simulating body fluids with 10% fetal bovine serum (FBS) solution (Figure S3), indicating relatively stable over 5 days.
Fig. 2.
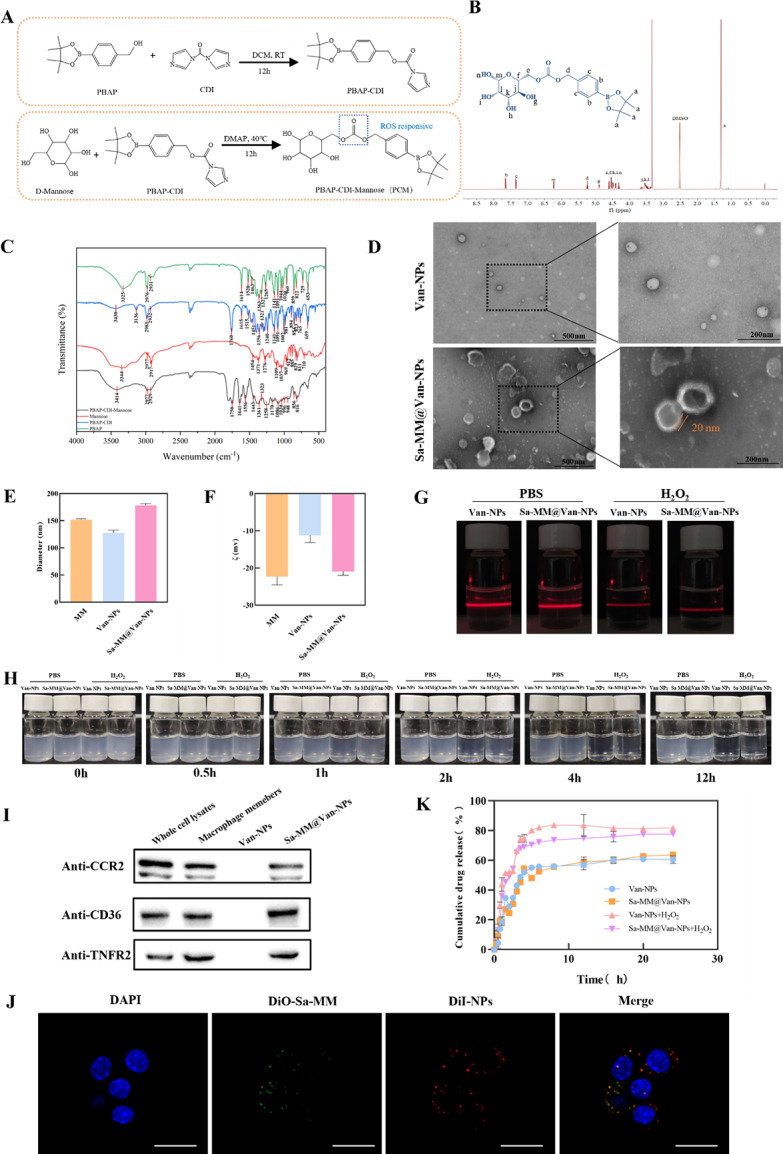
Preparation and characterization of Sa-MM@Van-NPs: A. Synthetic process of PCM. B. 1H NMR spectra of PCM. C. FT-IR spectra of PCM. D. TEM images of Van-NPs (up) and Sa-MM@Van-NPs (down). E. Particle sizes of Van-NPs and Sa-MM@Van-NPs were measured using DLS, with results presented as mean ± SD (n = 3). F. The zeta potentials of Van-NPs and Sa-MM@Van-NPs were evaluated using DLS, with results presented as mean ± SD (n = 3). G. Tyndall effect observed in Van-NPs and Sa-MM@Van-NPs after 12 h of hydrolysis without or with 100 µM H2O2. H. Hydrolysis kinetics of Van-NPs and Sa-MM@Van-NPs in PBS buffer without or with 100 µM H2O2 at different time points. I. Specific protein analysis of macrophage members, Van-NPs, and Sa-MM@Van-NPs by Western blotting. J. Co-localization images of dual fluorescent labeled DiO-Sa-MM@DiI-NPs in RAW264.7 cells. Cell nuclei were stained with DAPI (blue), NPs with DiI (red), and Sa-MMs with DiO (green), with a scale bar of 20 μm. K. In vitro drug release profiles of Van-NPs and Sa-MM@Van-NPs were evaluated with and without 100 µM H2O2 (n = 3, mean ± SD)
H2O2 was selected to evaluate the responsiveness of the nanoparticles to ROS due to its relative stability, accessibility and abundance in vivo [58, 59]. They were exposed to 100 µM H2O2 solution (the pathological oxidative microenvironment) [58, 60] for 12 h followed by laser irradiation to observe the Tyndall effect. As shown in Fig. 2G, both Van-NPs and Sa-MM@Van-NPs suspensions exhibited a weakened Tyndall effect light path, indicating structural instability of the nanoparticles after exposure to the ROS-mimicking H2O2 solution. There was visible movement of solid particles in the light path, suggesting disruption or degradation of the nanoparticles under ROS conditions. In the H2O2-treated group the blue color of the suspensions significantly faded within 1 h and almost disappeared after 12 h, while the control group showed no noticeable color change (Fig. 2H). The ROS-responsive drug release profile was evaluated by monitoring vancomycin release from Sa-MM@Van-NPs in Phosphate Buffered Saline (PBS) with and without 100 µM H2O2 using a microplate reader. As shown in Fig. 2K, the cumulative percentage of drug release from Sa-MM@Van-NPs increased from approximately 63% to around 78% in the presence of H2O2, a response similar to that observed for Van-NPs. The above results indicated the successful preparation of Sa-MM coated ROS-responsive amphiphilic nanoparticle.
Immune escape and active targeting
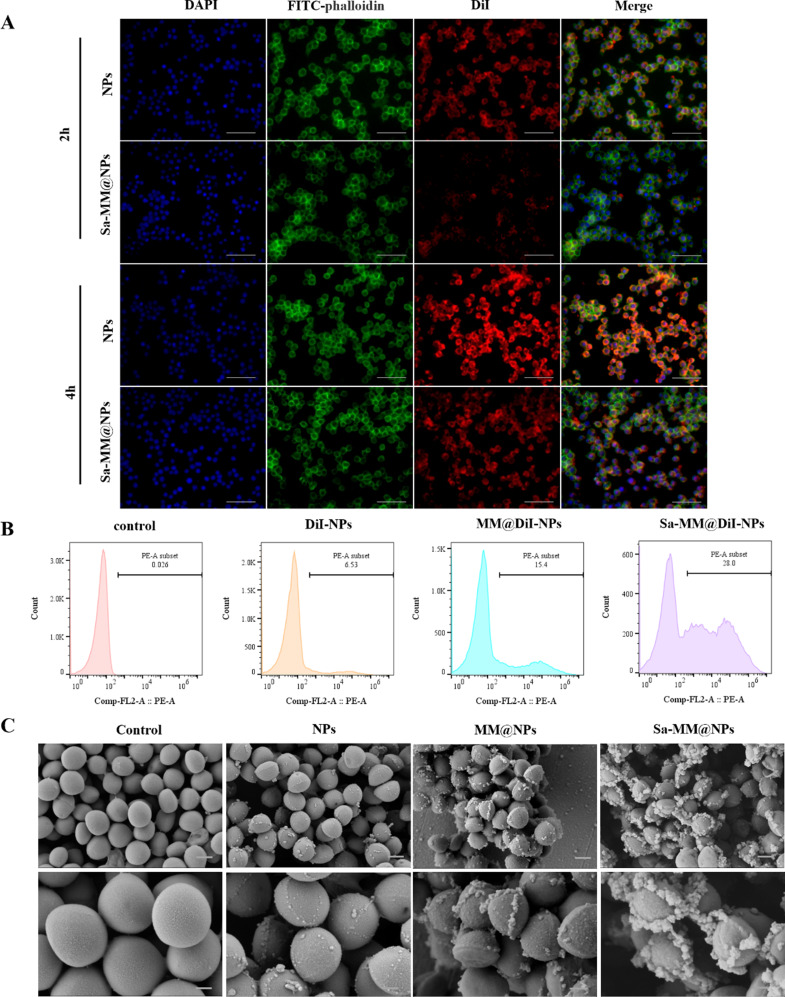
One significant challenge with current delivery systems is the frequent clearance of nanoparticles by the mononuclear phagocyte system before reaching their intended target sites [29, 61, 62]. Encapsulating MMs on the surface of nanoparticles serves as an autologous camouflage, thereby reducing immune clearance. Therefore, we investigated the phagocytosis of Sa-MM@NPs by macrophages. DiI was encapsulated in the cores of nanoparticles (DiI-NPs) and Sa-MM@NPs (Sa-MM@DiI-NPs) to monitor the cellular uptake process. As shown in Fig. 3A, macrophages uptake increased over time in both the DiI-NPs and Sa-MM@DiI-NPs groups. However, the red fluorescence intensity of the Sa-MM@DiI-NPs group was lower than that of the DiI-NPs group at the same time points, suggesting that the drug delivery system encapsulating MMs may represent an effective strategy to evade macrophage clearance.
Fig. 3.
Reduction of macrophage uptake and enhancement of S. aureus adhesion by Sa-MM@NPs: A. Fluorescence images of DiI-NPs and Sa-MM@DiI-NPs uptaken by RAW 264.7 cells after 2 and 4 h (scale bar = 100 μm). B. Flow cytometry analysis of the adherence of different DiI-labeled nanoparticles to S. aureus. C. SEM images depicting different nanoparticles adhering to S. aureus (top: scale bar = 500 nm, bottom: scale bar = 200 nm)
The targeting of Sa-MM@NPs to bacteria was assessed through flow cytometry and scanning electron microscopy (SEM). As shown in Fig. 3B, bare NPs exhibited a binding rate of only 6.53% with S. aureus, whereas NPs coated with MMs (MM@NPs) showed a higher binding rate (15.4%). Surprisingly, NPs incorporating bacteria-stimulated MMs (Sa-MM@NPs) demonstrated an even stronger binding rate (28%). SEM images supported these results. Figure 3C showed that the NPs, MM@NPs, and Sa-MM@NPs groups adhered numerous particles to the S. aureus surface compared to the control group. Moreover, the Sa-MM@NPs group exhibited the highest particle adhesion, followed by the MM@NPs group. These results together underscored that encapsulating MMs enhanced the NPs’ ability to adhere to bacteria and that bacteria-activated MMs further improved their bacterial recognition capability.
In summary, these findings demonstrated the encapsulation of MMs effectively mitigated the immune clearance, while bacteria-stimulated MMs additionally enhanced their adhesion and targeting efficacy towards bacteria.
In vitro antibacterial activity
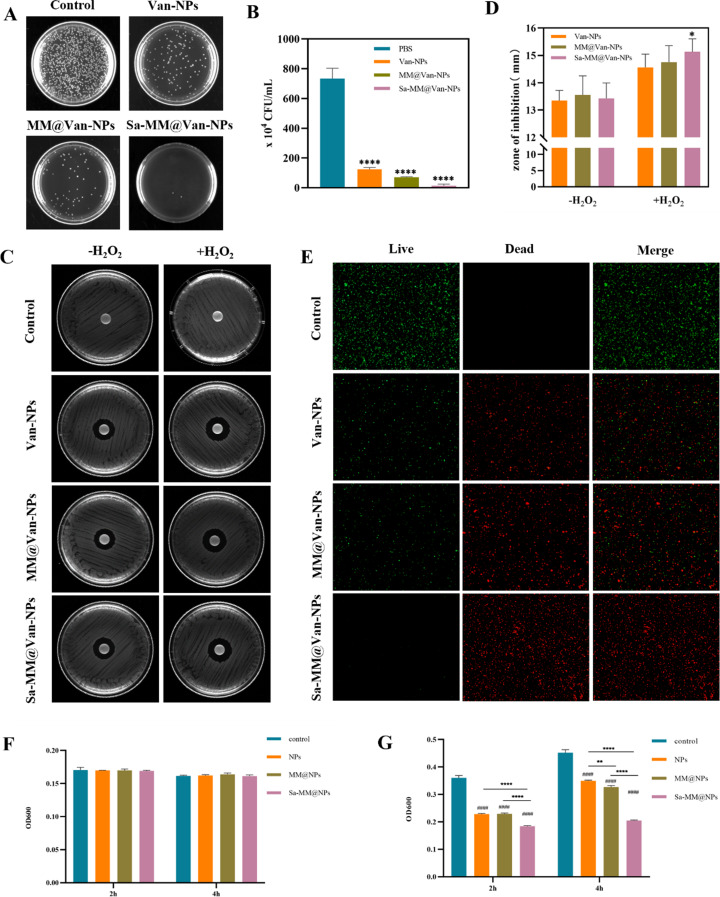
The antibacterial properties of Sa-MM@Van-NPs were assessed in vitro using the spread plate method, inhibition zone experiment and bacterial live/dead staining assay. The antibacterial effects of various concentrations of Van-NPs were firstly compared in Figure S4. At concentrations above 200 µg/mL, the antibacterial rate exceeded 98%. Therefore, we selected 200 µg/mL for the in vitro antibacterial experiments to evaluate the effects of different treatments. The Van-NPs, MM@Van-NPs, and Sa-MM@Van-NPs groups demonstrated significantly fewer colonies and higher antibacterial rates than the control group, as evidenced by colony images (Fig. 4A), quantitative colony analysis (Fig. 4B), and antibacterial rate data (Figure S5). Specifically, the antibacterial rate of Van-NPs was 83.1%, while those of MM@Van-NPs and Sa-MM@Van-NPs were 90.4% and 98.1%, respectively, suggesting that the MM coating improved its antibacterial efficacy. Remarkably, there were only few bacterial colonies on Sa-MM@Van-NPs agar plates, likely due to the high expression of TLRs on Sa-MM. The live/dead bacterial staining assay yielded comparable results. Red fluorescence signified dead bacteria, while green fluorescence signified live bacteria. Figure 4E demonstrated that Sa-MM@Van-NPs treatment significantly increased red fluorescence with minimal green fluorescence, while Van-NPs and MM@Van-NPs showed a slight presence of green fluorescence. This further confirmed that the Sa-MM coating significantly enhanced the antibacterial effect of the nanoparticles.
Fig. 4.
In vitro antibacterial activity of Sa-MM@Van-NPs: A. Representative colony images of S. aureus treated with different formulations. B. Quantification of the number of colonies with different treatments (n = 3; mean ± SD). C. Antibacterial efficacy of various nanoparticles with or without 100 µM H2O2 assessed by disk diffusion assay. D. Quantification of inhibition zone diameters (n = 3; mean ± SD). E. Fluorescent images from live/dead staining of S. aureus exposed to different nanoparticles. F. OD600 values of S. aureus exposed to different vancomycin unloaded nanoparticles for 2 h and 4 h in PBS solution and LB medium (G) (n = 3; mean ± SD; * p < 0.05, **p < 0.01, ****p < 0.0001, #### p < 0.0001 vs. control)
To investigate the role of ROS responsiveness in antibacterial activity, Van-NPs, MM@Van-NPs, and Sa-MM@Van-NPs were incubated with 100 µM H2O2 for 4 h, and their antibacterial efficacy was evaluated in vitro by measuring the diameter of the inhibition zone using the agar disk diffusion method. As shown in Fig. 4C and D, both with and without H2O2, Van-NPs, MM@Van-NPs, and Sa-MM@Van-NPs groups displayed distinct inhibition zones. In contrast, NPs lacking vancomycin did not exhibit inhibition zones (Figure S6), likely due to their limited diffusion ability on the agar plate. Following the addition of H2O2, the diameter of the inhibition zone in the Sa-MM@Van-NPs group increased from 13.4 mm to 15.13 mm, nearly equivalent to the antibacterial effect of free vancomycin at the same concentration (inhibition zone diameter: 15.54 mm). This finding suggested that vancomycin encapsulated within the hydrophobic core was released to exert its primary antibacterial effect.
Next, we measured the OD600 values of bacteria co-cultured with nanoparticles unloaded with vancomycin (NPs, MM@NPs, Sa-MM@NPs) in lysogeny broth (LB) medium and PBS solution to evaluate their antibacterial function. S. aureus can proliferate rapidly in LB medium whereas its growth is severely inhibited in PBS. This was confirmed by the fact that the OD values of the control group in LB medium increased significantly at 4 h compared to 2 h, whereas no difference was shown in PBS. Figure 4F showed that the OD600 values for the NPs, MM@NPs, and Sa-MM@NPs groups remained consistent with control group in PBS solution. In contrast, the OD600 values for NPs, MM@NPs, and Sa-MM@NPs were significantly reduced in LB medium in Fig. 4G. These results may reveal that the nanoparticles unloaded with vancomycin may exert their antibacterial effect by inhibiting bacterial proliferation (bacteriostatic). Moreover, Sa-MM@NPs exhibited the most potent bacteriostatic effect, followed by MM@NPs, suggesting that the coating of MMs may enhance the recognition and adhesion of NPs to bacteria and the overexpression of TLRs on the Sa-MM cell membranes further augmented targeting efficacy.
In vitro biocompatibility of Sa-MM@Van-NPs
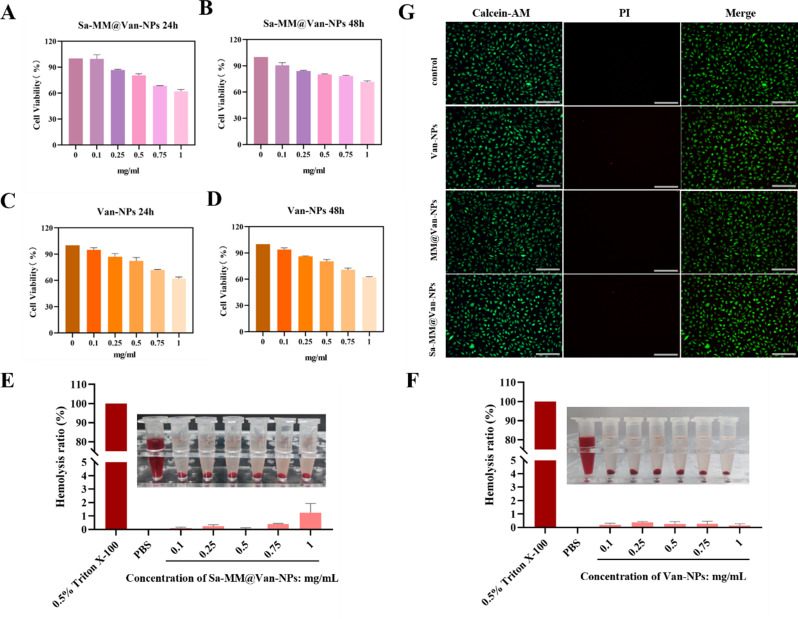
Toxicity assessment is crucial for the application of nanomaterials. The cytotoxicity of Sa-MM@Van-NPs was assessed in vitro using a cell counting kit 8 (CCK8) assay. Results showed that at increasing doses of Sa-MM@Van-NPs (up to 0.5 mg/mL), cell viability remained above 80% (Fig. 5A and B), with no significant cytotoxic effects observed on L929 fibroblasts after 24 and 48 h of incubation. Similar findings were observed in groups treated with Van-NPs (Fig. 5C and D). The hemolysis assay is a well-established method for evaluating the in vitro hemocompatibility of nanomaterials [62]. As per the ASTM F756-2008, biomaterials are considered acceptable if the hemolysis rate is below 5% [63]. Figure 5E demonstrated that erythrocytes exposed to Sa-MM@Van-NPs at concentrations of 0.1, 0.25, 0.5, 0.75 and 1 mg/mL showed no significant hemolysis, with 0.5% Triton X-100 and PBS serving as positive and negative controls, respectively. Quantitative analysis confirmed that the hemolysis rate was consistently below 5% across all groups, demonstrating the excellent hemocompatibility of Sa-MM@Van-NPs. Similarly, hemocompatibility tests conducted for Van-NPs also showed a hemolysis rate below 5%, indicating their favorable hemocompatibility as well (Fig. 5F).
Fig. 5.
In vitro biocompatibility assessment of Sa-MM@Van-NPs: A. The CCK8 assay was used to assess the cell viability of Sa-MM@Van-NPs co-cultured with L929 cells for 24 and 48 h (B) (n = 3; mean ± SD). C. Assessment of cell viability for various concentrations of Van-NPs co-cultured with L929 cells at 24 and 48 h (D) using the CCK8 assay (n = 3; mean ± SD). E. Representative images and quantitative analysis of hemolysis ratio for various concentrations of Sa-MM@Van-NPs and Van-NPs (F) (n = 3; mean ± SD). G. Fluorescent images from live/dead staining of L929 cells exposed to various nanoparticles for 24 h (scale bar = 200 μm)
Additionally, a live-dead cell staining assay was performed to evaluate the cell viability of L929 fibroblasts after 24 h treatment with Van-NPs, MM@Van-NPs, and Sa-MM@Van-NPs. The concentration of 500 µg/mL (cell viability > 80%) was chosen based on CCK8 assay findings (Fig. 5A-D). Green fluorescence marked live cells, whereas red fluorescence marked dead cells. As shown in Fig. 5G, most cells in all treatment groups exhibited green staining, with only a very small number of cells showing red staining, indicating good biocompatibility of Van-NPs, MM@Van-NPs, and Sa-MM@Van-NPs with L929 fibroblast cells.
Accumulation in infection sites and anti-subcutaneous infection
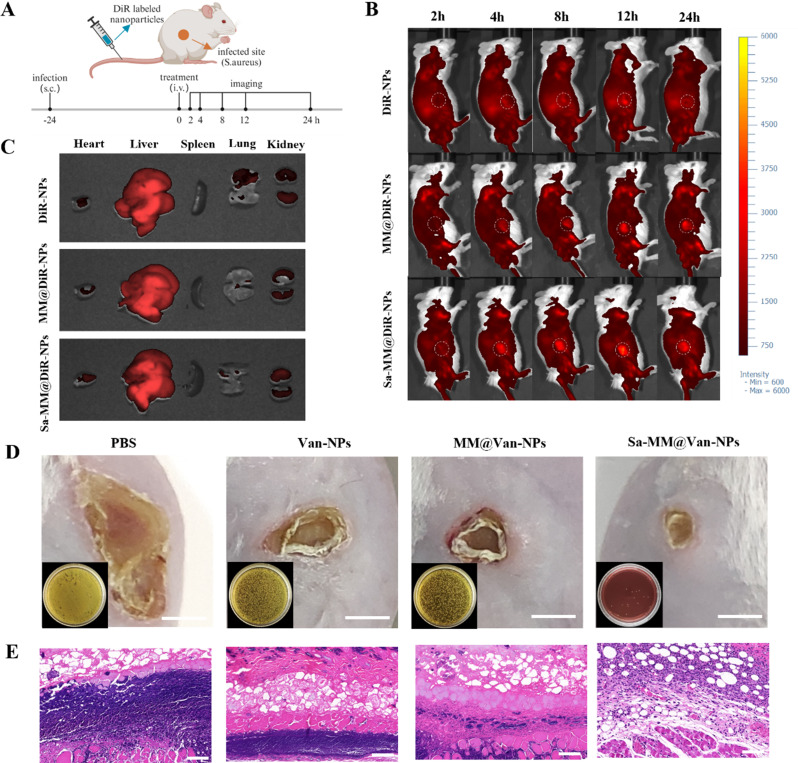
A subcutaneous infection model was used to evaluate the in vivo targeting characteristics of Sa-MM@NPs by tracking their real-time distribution. DiR, encapsulated in the hydrophobic cores of the NPs, was used to prepare DiR-NPs, MM@DiR-NPs, and Sa-MM@DiR-NPs for in vivo imaging observation. As depicted in Fig. 6A, S. aureus was injected into the subcutaneous area of the upper right leg of mice for 24 h. Then, the mice were imaged using an in vivo imaging system (IVIS) at 2, 4, 8, 12, and 24 h following intravenous injection of DiR-NPs, MM@DiR-NPs, and Sa-MM@DiR-NPs. Figure 6B illustrated that in the DiR-NPs group, the red fluorescent signal at the infection site increased over 12 h and diminished by 24 h. In contrast, the MM@DiR-NPs and Sa-MM@DiR-NPs groups sustained elevated fluorescent signals at 24 h. At each time point, the red fluorescent signal intensity was highest for Sa-MM@DiR-NPs, followed by MM@DiR-NPs, and lowest for DiR-NPs. This result demonstrated that the MM coating improved accumulation at the injection site, and the overexpression of TLRs on Sa-MMs further enhanced targeting efficacy. Ex vivo imaging at 8 h post-injection revealed that the liver was the predominant off-target site for the nanoparticle (Fig. 6C).
Fig. 6.
Evaluation of infection site accumulation and anti-subcutaneous infection: A. Schematic representation of experiments assessing the targeting capability of Sa-MM@Van-NPs in a mouse model of subcutaneous abscess infected by S. aureus. B. In vivo optical imaging of mice with subcutaneous abscesses following various treatments at 2, 4, 8, 12, and 24 h post-injection. C. Ex vivo imaging of DiR-labeled nanoparticles in major organs at 8 h post-injection. D. Representative images of skin lesions and bacterial colonies (lower left corner) in infected tissues under different treatments on day 4 (scale bar = 5 mm). E. H&E staining of skin tissue sections on day 4 (scale bar = 100 μm)
Furthermore, we evaluated the therapeutic effect by observing the size of skin lesions. On the 4th day of treatment, it was observed that the areas of skin lesions in the Van-NPs, MM@Van-NPs, and Sa-MM@Van-NPs treatment groups were obviously smaller than those in the PBS group. Sa-MM@Van-NPs group exhibited the smallest skin lesion size, indicating the most effective therapeutic outcome (Fig. 6D). The spread plate method confirmed the antibacterial efficacy, showing minimal bacterial colonies in the Sa-MM@Van-NPs group, aligning with the in vitro results. Histological analysis of day 4 skin samples using hematoxylin and eosin (H&E) staining revealed a significant reduction in inflammatory infiltration following Sa-MM@Van-NPs treatment (Fig. 6E). These results collectively underscored the favorable therapeutic efficacy of Sa-MM@Van-NPs in treating subcutaneous infections.
In vivo infected wound healing
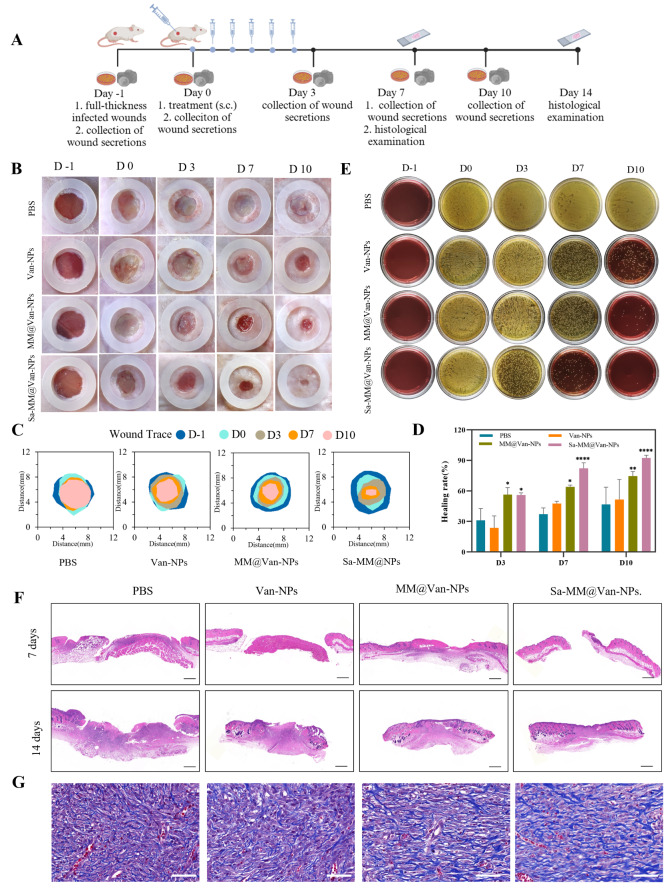
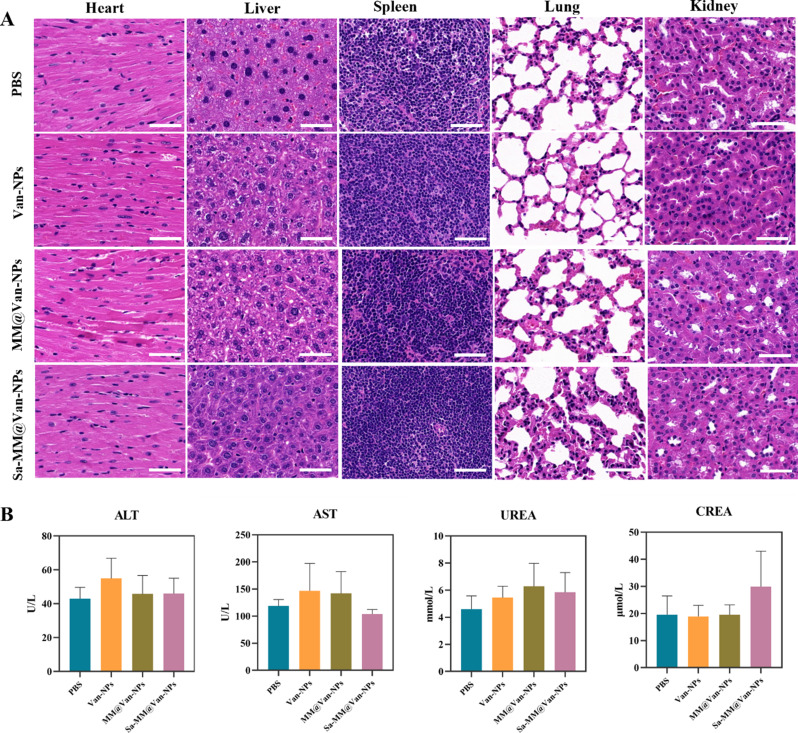
We developed a full-thickness S. aureus-infected wound model (6 mm in diameter) on the dorsal side of mice to assess the in vivo antibacterial and wound healing efficacy of Sa-MM@Van-NPs (Fig. 7A). The infected wounds were randomly assigned to one of four groups: PBS, Van-NPs, MM@Van-NPs, and Sa-MM@Van-NPs. Photographs were captured on days − 1, 0, 3, 7, and 10 to monitor wound healing, and wound exudates were collected for spread plate assays to assess antibacterial effects. Histopathological examination of wound tissues from each group was performed on days 7 and 14. As shown in Fig. 7B-D, the wound healing effects in the MM@Van-NPs and Sa-MM@Van-NPs groups were significantly superior to that in the Van-NPs group. Specifically, Sa-MM@Van-NPs consistently maintained the highest wound healing rates, reaching 82.15% and 92.46% on days 7 and 10, respectively, whereas the MM@Van-NPs group showed rates of 63.93% and 74.52% on the same days. The spread plate assay results aligned with the wound healing trends (Fig. 7E), showing a progressive decrease in colony formation in the Van-NPs, MM@Van-NPs, and Sa-MM@Van-NPs groups compared to the PBS group. By day 10, colony formation in the MM@Van-NPs and Sa-MM@Van-NPs groups was significantly lower than that in the Van-NPs group, indicating that the MM coating enhanced the antibacterial efficacy. Furthermore, almost no colony formation was observed in the Sa-MM@Van-NPs group by day 10, suggesting that the high expression of TLRs on Sa-MM promoted the targeted recognition of bacteria and further enhanced sterilization. Subsequently, the wound healing status was further examined by histologic changes of the skin on days 7 and 14. H&E staining showed that the Sa-MM@Van-NPs group had the smallest scars on day 7 and had been fully epithelialized on day 14 (Fig. 7F). Masson staining revealed increased collagen deposition in the treatment groups (Van-NPs, MM@Van-NPs and Sa-MM@Van-NPs) compared to the control, and the Sa-MM@Van-NPs group had the most collagen content with a relatively uniform arrangement (Fig. 7G). After 14 days of treatment, H&E staining (Fig. 8A) showed no evident pathological damage to the heart, liver, spleen, lungs, or kidneys. Liver function biomarkers, including alanine aminotransferase (ALT) and aspartate aminotransferase (AST), as well as kidney function biomarkers, such as urea nitrogen (UREA) and creatinine (CREA), exhibited no abnormalities (Fig. 8B). These findings collectively indicated that Sa-MM@Van-NPs had a favorable in vivo biosafety.
Fig. 7.
In vivo wound healing in an infected wound model: A. Diagrammatic representation of a full-thickness wound model infected with S. aureus and its treatment procedure. B. Representative images of wounds treated with different formulations on days − 1, 0, 3, 7, and 10. C. Traces of wound-bed closure by day 10 for all treatment groups. D. Quantification of wound healing rates among different treatments (n = 3; mean ± SD). E. Images of S. aureus colonies on Mannitol Salt Agar plates derived from infected wound exudates. F. H&E staining of skin tissue sections on days 7 and 14 (scale bar = 500 μm). G. Masson staining of skin tissue sections on day 14 (scale bar = 50 μm)
Fig. 8.
Assessment of the in vivo adverse effects from various treatments: A. Histological analysis of major organs (heart, liver, spleen, lung, and kidney) after 14 days of treatment (scale bar = 50 μm). B. Levels of kidney and liver function markers (ALT, AST, UREA, CREA) in mice after 14 days of treatment (n = 3; mean ± SD)
Conclusion
In summary, this study successfully developed a novel nanoparticle based on ROS-responsive amphiphilic nanoparticles coated with bacteria-activated MMs. This nanoparticle demonstrated targeted recognition and effective sterilization capabilities. The bacteria-activated MM coating facilitated specific targeting of infected sites through receptor-bacteria interactions, followed by the release of antibiotics in response to high ROS levels to eradicate bacteria. Moreover, the nanoparticle exhibited good biocompatibility. In vitro experiments confirmed that Sa-MM@Van-NPs had good bacterial adhesion and exerted potent antibacterial effects. In vivo experiments further revealed that Sa-MM@Van-NPs accumulated at infection sites, effectively eliminating bacteria and promoting wound healing. Therefore, leveraging bacteria-activated MMs to enhance bacterial recognition in conventional amphiphilic nanoparticles represents a practical and effective strategy for combating bacterial infections, reducing inflammation, and facilitating wound healing. This innovative approach holds promise for achieving significant advancements in targeted bacterial clearance.
Materials and methods
Cell culture
RAW264.7 and L929 cell lines were sourced from the American Type Culture Collection (ATCC), both authenticated through short tandem repeat (STR) profiling and confirmed to be free of mycoplasma contamination. The cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% FBS and 1% Penicillin-Streptomycin, and incubated at 37 °C in a humidified atmosphere containing 5% CO2. LB broth was used to cultivate S. aureus at 37 °C.
mRNA expression of TLRs in RAW264.7 cells
To detect mRNA expression levels of TLRs, qPCR was performed. A suitable density of RAW264.7 cells was inoculated in 24-well plates overnight. Afterward, S. aureus was added to co-incubate with cells for 3 h. Total RNA extraction was carried out using the FastPure Cell/Tissue Total RNA Isolation Kit V2 (RC112, Vazyme) according to the manufacturer’s instructions. cDNA synthesis was performed with HiScript II Q RT SuperMix for qPCR (+ gDNA wiper) (R223, Vazyme), and qPCR was conducted with Cham Q SYBR qPCR Master Mix (Without ROX) (Q321, Vazyme) on a LightCycler 480II machine (Roch, US).
All primer sequences were listed as follows: TLR1-forword: GTCAAAGCTTGGAAAGAATCTGAAG, TLR1-reverse: TLR2-forward: CGTTGTTCCCTGTGTTGCT, TLR2-reverse: AAAGTGGTTGTCGCCTGCT; TLR4-forward: GAATGAGGACTGGGTGAGAAATGAG, TLR4-reverse: TGGATGATGTTGGCAGCAATGG; TLR6-forward: CCTGTGATACTGTTCTGCTGACTG, TLR6-reverse: CCTGTGCCTGGTCTGTGTCC: GAPDH-forward: CAATCTGTCCGTCGTGGATC, GAPDH-reverse: CCTGCTTCACCACCTTCTTG.
Flow cytometry of TLRs
At a density of 2 × 106 cells per well, RAW264.7 cells were inoculated into 6-well plates. S. aureus was introduced to the cells at a concentration of 2 × 107 CFU/well for 3 h, following a 24-hour period. The cells were washed thrice with PBS and resuspended in 100 µL of PBS. The cells were subsequently stained with 1 µg of APC anti-mouse CD284 (TLR4) antibody (Cat#145406, Biolegend) or 0.5 µg of PE anti-mouse CD282 (TLR2) antibody (Cat#148604, Biolegend) and incubated for 30 min on ice, protected from light. After three PBS washes, the cells were resuspended in 500 µL of PBS for flow cytometry analysis using the Accuri C6 Plus (BD Biosciences).
Immunofluorescence staining of TLRs
RAW264.7 cells were inoculated at an appropriate density in 24-well plates with coverslips placed. After 24 h, S. aureus was added to treat for 3 h. Cells were washed thrice with PBS, fixed and permeabilized using prechilled 100% methanol at -20 °C for 5 min, and subsequently blocked with 5% goat serum for 1 h at room temperature. Samples were incubated overnight at 4 °C with either anti-TLR2 antibody (ab209216, Abcam) or anti-TLR4 antibody (sc-293072, Santa Cruz). Following three PBS washes, TLR2 was incubated with Cy3 goat anti-rabbit IgG (H + L) (AS007, ABclonal), and TLR4 was incubated with FITC goat anti-mouse IgG (H + L) (AS001, ABclonal). The coverslips were sealed with an anti-fluorescence quenching solution containing DAPI (P0131, Beyotime) and examined using laser scanning confocal microscopy (LSM880, Carl Zeiss).Fluorescence intensity was quantified using ImageJ software.
Synthesis of Van-NPs
1 g of PBAP and 1.4 g of CDI were completely dissolved in 6 mL DCM, followed by stirring and reaction overnight at room temperature. The mixture was treated with an additional 6 mL of DCM, washed thrice with ultrapure water, and dried over anhydrous magnesium sulfate (MgSO4) for 4 h before filtration. Then, the collected products were subjected to rotary evaporator to fully remove the DCM until white solid, i.e. PBAP-CDI, was obtained. Subsequently, 0.5 g of PBAP-CDI, 90 mg of D-mannose, and 183 mg of DMAP were dissolved in 4 mL DMF and stirred overnight at 40 ℃. The product was enclosed in a dialysis bag (MWCO 500 Da) and subjected to dialysis in ultrapure water for 24 h. The dialyzed products were lyophilized by vacuum freeze-drying (CTFD-12 S-U, CREATRUST) to obtain PBAP- CDI- D-mannose (PCM) power. The chemical structure of PBAP-CDI and PCM were characterized by 1H NMR spectroscopy (600 MHz, DMSO-d6).
Van-NPs were prepared using the dialysis method. A solution of 10 mg PCM and 1 mg vancomycin in 200 µL DMF was gradually dropped into 5 mL of rapidly stirred ultrapure water. Van-NPs were acquired by dialyzing with a 1000 Da MWCO dialysis bag for 12 h. The dialyzed products were lyophilized by vacuum freeze-drying (CTFD-12 S-U, CREATRUST).
Synthesis of Sa-MM@Van-NPs
RAW264.7 cells were inoculated in 10 cm dishes. S. aureus was introduced and co-incubated with cells for 3 h after a 24-hour period. Cells were collected in a centrifuge tube, and membranes were isolated using the Membrane and Cytosol Protein Extraction Kit (P0033, Beyotime). Briefly, cells were incubated on ice for 15 min with Reagent A containing protease inhibitor. Cells were lysed through three freeze-thaw cycles using liquid nitrogen, followed by centrifugation at 700 g for 10 min at 4ºC. The supernatant was then transferred to a new tube. The supernatant underwent centrifugation at 14,000 g for 30 min at 4ºC, resulting in the collection of cell membrane fragments as the precipitate.
Extrusion method was used to synthesize Sa-MM@Van-NPs.1 mL Van-NPs was mixed with Sa-MM and transferred to a syringe. The suspension underwent 16 extrusions through a 200 nm polycarbonate membrane using an Avestin LiposoFast extruder. The suspension underwent centrifugation at 12,000 rpm for 10 min to eliminate surplus macrophage membranes.
Characterization of Sa-MM@Van-NPs
Nanoparticle morphology and structure were examined using a Hitachi HT7800 TEM. The hydrodynamic diameter and zeta potential were analyzed using Malvern Zetasizer Nano ZS. The proteins, CCR2 (MA5-41175, Invitrogen), CD36 (ab252922, Abcam) and TNFR2 (MA5-32618, Invitrogen) on Sa-MM@Van-NPs, were detected using Western blot.
DLE and DEE
Vancomycin standard solutions at different concentrations were prepared in DMF, and their OD280 values were measured using a Biotek Synergy H4 microplate reader. A standard curve of concentration versus absorbance was then plotted. The Van-NPs powder was dissolved in DMF, and its OD280 value was measured. The vancomycin concentration in Van-NPs was determined using the standard curve. In conclusion, DLE and DEE were determined using the specified equations.
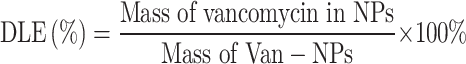 |
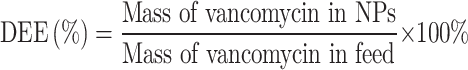 |
Release profiles of vancomycin from nanoparticles
The release profiles of vancomycin from Van-NPs and Sa-MM@Van-NPs were investigated with or without 100 µM H2O2 using a microplate reader at 280 nm. Solutions of Van-NPs and Sa-MM@Van-NPs (10 mg/mL, 5 mL) were placed in a dialysis bag (MWCO 3500 Da) and submerged in 100 mL of buffer solution. The dialysis bags were shaken with 30 g at 37 °C to mimic the in vivo environment. At various intervals, 5 mL of buffer was extracted and replaced with an equal volume of fresh solution. After the measurement of OD280 values, the drug release profiles were plotted by calculating the percentage of drug release.
Co-localization study
DiI was utilized to label the NPs, while the Sa-MM was labeled with DiO to obtain dual fluorescent labeled DiO-Sa-MM@DiI-NPs. RAW64.7 cells were treated with DiO-Sa-MM@DiI-NPs for 2–4 h, followed by three PBS washes. Nuclei were stained with DAPI following fixation with 4% paraformaldehyde (PFA). The fluorescence was observed using CLSM.
Cellular uptake by RAW 267 cells
To track the phagocytosis process, the red fluorescent dye DiI was encapsulated into hydrophobic core to obtain the DiI-NPs, MM@DiI-NPs and Sa-MM@DiI-NPs. RAW264.7 cells were cultured in 24-well plates for 24 h and then incubated with 100 µg of DiI-labeled nanoparticles for either 2 or 4 h. The cells were fixed with 4% PFA, stained using FITC Phalloidin and DAPI, and examined via CLSM.
Sa-MM@NPs adhered to S. aureus study
For flow cytometry, DiI-labeled nanoparticles were used for tracking. S. aureus was fixed with 4% PFA for 15 min at room temperature, followed by three PBS washes. It was then incubated for 1 h with either DiI-NPs, MM@DiI-NPs, or Sa-MM@DiI-NPs at a concentration of 20 µg, using PBS as a control. S. aureus was ultimately examined with a flow cytometer (Accuri C6 Plus, BD Biosciences).
For scanning electron microscopy (SEM) experiment, S. aureus was inoculated on silicon wafers and subjected to treatments with PBS, NPs, MM@NPs, and Sa-MM@NPs. Subsequently, samples were fixed in 2.5% glutaraldehyde and progressively dehydrated using a series of ethanol concentrations (30%, 50%, 70%, 90%, 95%, 100%), concluding with tert-butanol. Finally, they were vacuum-dried and observed on SEM (Sigma 300, ZEISS).
Antibacterial study in vitro
1mL suspensions of S. aureus were incubated with 200 µg of different nanoparticles (PBS as control) for 4 h. For the spread plate assay, the suspensions were spread evenly on agar plates, followed by incubation for 18 h at 37℃. Then, the images were acquired and the number of colonies was recorded. For the live/dead staining assay (L7007, Thermo), SYTO 9 was used to stain live bacteria, whereas Propidium Iodide (PI) was used to stain dead bacteria.
For the bacteriostatic and bactericidal activity of vancomycin-unloaded nanoparticles, the cultured bacteria were centrifuged and resuspended in LB medium and sterile PBS, respectively. NPs, MM@NPs, and Sa-MM@NPs at a concentration of 200 µg were separately added and incubated with S. aureus on a shaker at 37 ℃ for 2 or 4 h. Finally, their OD600 values were measured using a microplate reader (Synergy H4, Biotek).
For the inhibition zone experiment, the different nanoparticles (same concentration of vancomycin as positive control, PBS as negative control) were incubated in the solutions with or without H2O2 for 4 h. A total of 20 µL of the solutions were dropped onto the blank drug-sensitive papers, air-dried in a fume hood, and then placed on agar plates containing S. aureus. Images were captured, and the inhibition zone diameter was measured after 18 h.
Biocompatibility and hemocompatibility assay
CCK8 and cell live-dead staining assays were performed to assess the biocompatibility. Briefly, L929 fibroblasts were inoculated in 96-well plates for 24 h at a density of 8,000 per well, followed by co-incubation with various concentrations of Van-NPs or Sa-MM@Van-NPs for 24 or 48 h. Then, OD450 was determined using a microplate reader (Synergy H4, Biotek) after incubation for 2 h with 10% cell counting kit-8 (CCK8) reagent (C6005, NCM). For Calcein-AM/PI cell viability/cytotoxicity assay (C2015L, Beyotime), the cells were treated with different nanoparticles at a concentration of 0.5 mg/mL for 24 h, followed by incubation with the staining solution at 37℃ in a 5% CO2 incubator for 30 min. Finally, fluorescence images were obtained by an inverted fluorescence microscope (Axio Vert A1, ZEISS).
For the hemocompatibility Assay, 5% erythrocyte solutions were incubated with 0.5% (v/v) Triton X-100 (positive control), PBS (negative control), and various concentrations of Van-NPs or Sa-MM@Van-NPs (0.1, 0.25, 0.5, 0.75, 1 mg/mL), respectively, at 37 °C for 2 h. After centrifugation at 3500 rpm for 5 min, the supernatants were collected to measure released hemoglobin at OD540 using a microplate reader (Synergy H4, Biotek).
In vivo infection site accumulation, antibacterial and wound healing of Sa-MM@Van-NPs
Animal experiments received approval from the Institutional Animal Care and Use Committee at the Zhejiang Center of Laboratory Animals (ZJCLA-IACUC-20010842). The study utilized male BALB/c mice aged 4 to 6 weeks. For in vivo targeting of Sa-MM@NPs, a subcutaneous abscess model was created, and DiR-labeled nanoparticles were used for in vivo tracing. Briefly, bacterial suspension was injected subcutaneously in the back of mice. Then the tail vein was injected with PBS, DiR-NPs, MM@DiR-NPs, and Sa-MM@DiR-NPs, respectively. A small animal live imager (VISQUE® InVivo ART 100, Vieworks) was used for observation at 2, 4, 8, 12, and 24 h after injection. And major organs were taken for imaging observation at 8 h.
Subsequently, we evaluated the efficacy of Sa-MM@Van-NPs in full-thickness S. aureus-infected wounds. The mice were anesthetized and their backs shaved to create wounds. Then, a full-thickness round wound was produced using 6-mm diameter skin biopsy punches. The wound was subsequently inoculated with 10 µL of a 107 CFU S. aureus suspension for 24 h. The wound was treated with different interventions: control (PBS), Van-NPs, MM@Van-NPs, or Sa-MM@Van-NPs. On days - 1, 0, 3, 7, and 10, wound secretions were collected using sterile swabs. Following incubation at 37℃ with shaking, the samples were evenly distributed on agar plates for recording and analysis of bacterial colonies. Images were taken on days - 1, 0, 3, 7, and 10 to track wound healing, with the wound area quantified using Image J.
Statistical analysis
All data are presented as mean ± standard deviation (SD). Statistical differences among groups were assessed by one-way analysis of variance (ANOVA). *p < 0.05 was considered statistically significant, while **p < 0.01, ***p < 0.001 and ****p < 0.0001 indicated increasing levels of statistical significance.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
N/A.
Author contributions
All authors prepared the figures. All wrote and reviewed the manuscript. G. L. and M. X. supervised the project.
Funding
There was no funding.
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
All animal experiments were approved by the Institutional Animal Care and Use Committee of the Zhejiang Center of Laboratory Animals (ZJCLA-IACUC-20010842).
Consent for publication
All authors agree with the publication.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ying Luo and Xiaoli Jia contributed equally to this work.
Contributor Information
Malcolm Xing, Email: malcolm.xing@umanitoba.ca.
Guozhong Lyu, Email: luguozhong@jiangnan.edu.cn.
References
- 1.Global burden of bacterial antimicrobial resistance. In 2019: a systematic analysis. Lancet. 2022;399(10325):629–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’NEILL J. Tackling drug-resistant infections globally: final report and recommendations. London: Review on Antimicrobial Resistance; 2016. [Google Scholar]
- 3.Hussain S, Joo J, Kang J, Kim B, Braun GB, She ZG, Kim D, Mann AP, Mölder T, Teesalu T, Carnazza S, Guglielmino S, Sailor MJ, Ruoslahti E. Antibiotic-loaded nanoparticles targeted to the site of infection enhance antibacterial efficacy. Nat Biomed Eng. 2018;2(2):95–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Q, Zou J, Chen Z, He W, Wu W. Current research trends of nanomedicines. Acta Pharm Sin B. 2023;13(11):4391–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fang RH, Kroll AV, Gao W, Zhang L. Cell Membrane Coat Nanatechnol Adv Mater. 2018;30(23):e1706759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Y, Luo J, Chen X, Liu W, Chen T. Cell membrane Coating Technology: a promising strategy for Biomedical Applications. Nanomicro Lett. 2019;11(1):100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu CM, Fang RH, Luk BT, Zhang L. Nanoparticle-detained toxins for safe and effective vaccination. Nat Nanotechnol. 2013;8(12):933–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang F, Fang RH, Luk BT, Hu CJ, Thamphiwatana S, Dehaini D, Angsantikul P, Kroll AV, Pang Z, Gao W, Lu W, Zhang L. Nanoparticle-based Antivirulence Vaccine for the management of Methicillin-resistant Staphylococcus aureus skin infection. Adv Funct Mater. 2016;26(10):1628–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parodi A, Quattrocchi N, van de Ven AL, Chiappini C, Evangelopoulos M, Martinez JO, Brown BS, Khaled SZ, Yazdi IK, Enzo MV, Isenhart L, Ferrari M, Tasciotti E. Synthetic nanoparticles functionalized with biomimetic leukocyte membranes possess cell-like functions. Nat Nanotechnol. 2013;8(1):61–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhuang J, Gong H, Zhou J, Zhang Q, Gao W, Fang RH, Zhang L. Targeted gene silencing in vivo by platelet membrane-coated metal-organic framework nanoparticles. Sci Adv. 2020;6(13):eaaz6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu JY, Zheng DW, Zhang MK, Yu WY, Qiu WX, Hu JJ, Feng J, Zhang XZ. Preferential Cancer Cell Self-Recognition and Tumor Self-Targeting by Coating nanoparticles with Homotypic Cancer cell membranes. Nano Lett. 2016;16(9):5895–901. [DOI] [PubMed] [Google Scholar]
- 12.Yang N, Ding Y, Zhang Y, Wang B, Zhao X, Cheng K, Huang Y, Taleb M, Zhao J, Dong WF, Zhang L, Nie G. Surface functionalization of polymeric nanoparticles with umbilical cord-derived mesenchymal stem cell membrane for Tumor-targeted therapy. ACS Appl Mater Interfaces. 2018;10(27):22963–73. [DOI] [PubMed] [Google Scholar]
- 13.Ren N, Liang N, Dong M, Feng Z, Meng L, Sun C, Wang A, Yu X, Wang W, Xie J, Liu C, Liu H. Stem cell membrane-encapsulated Zeolitic Imidazolate Framework-8: a targeted Nano-platform for osteogenic differentiation. Small. 2022;18(26):e2202485. [DOI] [PubMed] [Google Scholar]
- 14.Dehaini D, Wei X, Fang RH, Masson S, Angsantikul P, Luk BT, Zhang Y, Ying M, Jiang Y, Kroll AV, Gao W, Zhang L. Erythrocyte-Platelet Hybrid Membrane Coating for Enhanced Nanoparticle Functionalization, Adv Mater 29(16) (2017). [DOI] [PMC free article] [PubMed]
- 15.Xiong J, Wu M, Chen J, Liu Y, Chen Y, Fan G, Liu Y, Cheng J, Wang Z, Wang S, Liu Y, Zhang W. Cancer-Erythrocyte Hybrid membrane-camouflaged magnetic nanoparticles with enhanced photothermal-immunotherapy for ovarian Cancer. ACS Nano. 2021;15(12):19756–70. [DOI] [PubMed] [Google Scholar]
- 16.Wang K, Lei Y, Xia D, Xu P, Zhu T, Jiang Z, Ma Y. Neutrophil membranes coated, antibiotic agent loaded nanoparticles targeting to the lung inflammation. Colloids Surf B Biointerfaces. 2020;188:110755. [DOI] [PubMed] [Google Scholar]
- 17.Hu C, Lei T, Wang Y, Cao J, Yang X, Qin L, Liu R, Zhou Y, Tong F, Umeshappa CS, Gao H. Phagocyte-membrane-coated and laser-responsive nanoparticles control primary and metastatic cancer by inducing anti-tumor immunity. Biomaterials. 2020;255:120159. [DOI] [PubMed] [Google Scholar]
- 18.Sun L, Huang Y, Bian Z, Petrosino J, Fan Z, Wang Y, Park KH, Yue T, Schmidt M, Galster S, Ma J, Zhu H, Zhang M. Sundew-inspired adhesive hydrogels combined with adipose-derived stem cells for Wound Healing. ACS Appl Mater Interfaces. 2016;8(3):2423–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gurtner GC, Werner S, Barrandon Y, Longaker MT. Wound repair and regeneration. Nature. 2008;453(7193):314–21. [DOI] [PubMed] [Google Scholar]
- 20.Hua C, Yang F, Jia X, Lu Y, Li X, Zhao P, Xing M, Lyu G. Multi-comparted microgels delivering human derived probiotics and deferoxamine for multidrug-resistant infection and healing. Chem Eng J. 2024;483:148432. [Google Scholar]
- 21.Quek JY, Uroro E, Goswami N, Vasilev K. Design principles for bacteria-responsive antimicrobial nanomaterials. Mater Today Chem. 2022;23:100606. [Google Scholar]
- 22.Wang X, Shan M, Zhang S, Chen X, Liu W, Chen J, Liu X. Stimuli-Responsive Antibacterial materials: Molecular structures, Design principles, and Biomedical Applications. Adv Sci (Weinh). 2022;9(13):e2104843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang P, Gong Q, Hu J, Li X, Zhang X. Reactive oxygen species (ROS)-Responsive Prodrugs, Probes, and Theranostic Prodrugs: applications in the ROS-Related diseases. J Med Chem. 2021;64(1):298–325. [DOI] [PubMed] [Google Scholar]
- 24.Pu M, Cao H, Zhang H, Wang T, Li Y, Xiao S, Gu Z. ROS-responsive hydrogels: from design and additive manufacturing to biomedical applications. Mater Horiz. 2024;11(16):3721–46. [DOI] [PubMed] [Google Scholar]
- 25.Yang X, Tang X, Yi S, Guo T, Liao Y, Wang Y, Zhang X. Maltodextrin-derived nanoparticles resensitize intracellular dormant Staphylococcus aureus to rifampicin. Carbohydr Polym. 2025;348:122843. [DOI] [PubMed] [Google Scholar]
- 26.Zhao J, Fu J-y, Jia F, Li J, Yu B, Huang Y, Ren K-f, Ji J. -s. Fu, Precise Regulation of inflammation and oxidative stress by ROS-Responsive Prodrug Coated Balloon for preventing vascular restenosis. Adv Funct Mater. 2023;33(30):2213993. [Google Scholar]
- 27.Wen R, Umeano AC, Kou Y, Xu J, Farooqi AA. Nanoparticle systems for cancer vaccine. Nanomed (Lond). 2019;14(5):627–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitragotri S, Burke PA, Langer R. Overcoming the challenges in administering biopharmaceuticals: formulation and delivery strategies. Nat Rev Drug Discov. 2014;13(9):655–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blanco E, Shen H, Ferrari M. Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nat Biotechnol. 2015;33(9):941–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma L, Li W, Zhang Y, Qi L, Zhao Q, Li N, Lu Y, Zhang L, Zhou F, Wu Y, He Y, Yu H, He Y, Wei B, Wang H. FLT4/VEGFR3 activates AMPK to coordinate glycometabolic reprogramming with autophagy and inflammasome activation for bacterial elimination. Autophagy. 2022;18(6):1385–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fitzgerald KA, Kagan JC. Toll-like receptors and the control of immunity. Cell. 2020;180(6):1044–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spaan AN, Vrieling M, Wallet P, Badiou C, Reyes-Robles T, Ohneck EA, Benito Y, de Haas CJ, Day CJ, Jennings MP, Lina G, Vandenesch F, van Kessel KP, Torres VJ, van Strijp JA, Henry T. The staphylococcal toxins γ-haemolysin AB and CB differentially target phagocytes by employing specific chemokine receptors. Nat Commun. 2014;5:5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, Zhang J, Chen W, Angsantikul P, Spiekermann KA, Fang RH, Gao W, Zhang L. Erythrocyte membrane-coated nanogel for combinatorial antivirulence and responsive antimicrobial delivery against Staphylococcus aureus infection. J Control Release. 2017;263:185–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hogan PG, Mork RL, Thompson RM, Muenks CE, Boyle MG, Sullivan ML, Morelli JJ, Williams CV, Sanchez N, Hunstad DA, Wardenburg JB, Gehlert SJ, Burnham CD, Rzhetsky A, Fritz SA. Environmental methicillin-resistant Staphylococcus aureus Contamination, Persistent colonization, and subsequent skin and soft tissue infection. JAMA Pediatr. 2020;174(6):552–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Czuban M, Srinivasan S, Yee NA, Agustin E, Koliszak A, Miller E, Khan I, Quinones I, Noory H, Motola C, Volkmer R, Di Luca M, Trampuz A, Royzen M. Mejia Oneto, Bio-orthogonal Chemistry and Reloadable Biomaterial Enable Local Activation of Antibiotic Prodrugs and enhance treatments against Staphylococcus aureus infections. ACS Cent Sci. 2018;4(12):1624–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rong M, Zheng X, Ye M, Bai J, Xie X, Jin Y, He X. Phenotypic plasticity of Staphylococcus aureus in Liquid Medium containing vancomycin. Front Microbiol. 2019;10:809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brodsky I, Medzhitov R. Two modes of ligand recognition by TLRs. Cell. 2007;130(6):979–81. [DOI] [PubMed] [Google Scholar]
- 38.Jin MS, Kim SE, Heo JY, Lee ME, Kim HM, Paik SG, Lee H, Lee JO. Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell. 2007;130(6):1071–82. [DOI] [PubMed] [Google Scholar]
- 39.Noreen M, Arshad M. Association of TLR1, TLR2, TLR4, TLR6, and TIRAP polymorphisms with disease susceptibility. Immunol Res. 2015;62(2):234–52. [DOI] [PubMed] [Google Scholar]
- 40.Takeuchi O, Sato S, Horiuchi T, Hoshino K, Takeda K, Dong Z, Modlin RL, Akira S. Cutting Edge: Role of Toll-Like receptor 1 in Mediating Immune response to Microbial Lipoproteins1. J Immunol. 2002;169(1):10–4. [DOI] [PubMed] [Google Scholar]
- 41.Takeuchi O, Kawai T, Mühlradt PF, Morr M, Radolf JD, Zychlinsky A, Takeda K, Akira S. Discrimination of bacterial lipoproteins by toll-like receptor 6. Int Immunol. 2001;13(7):933–40. [DOI] [PubMed] [Google Scholar]
- 42.Kang JY, Nan X, Jin MS, Youn SJ, Ryu YH, Mah S, Han SH, Lee H, Paik SG, Lee JO. Recognition of lipopeptide patterns by toll-like receptor 2-Toll-like receptor 6 heterodimer. Immunity. 2009;31(6):873–84. [DOI] [PubMed] [Google Scholar]
- 43.Schenk M, Belisle JT, Modlin RL. TLR2 looks at lipoproteins. Immunity. 2009;31(6):847–9. [DOI] [PubMed] [Google Scholar]
- 44.Fournier B, Philpott DJ. Recognition of Staphylococcus aureus by the innate immune system. Clin Microbiol Rev. 2005;18(3):521–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luo S, Liao C, Zhang L, Ling C, Zhang X, Xie P, Su G, Chen Z, Zhang L, Lai T, Tang J. METTL3-mediated m6A mRNA methylation regulates neutrophil activation through targeting TLR4 signaling. Cell Rep. 2023;42(3):112259. [DOI] [PubMed] [Google Scholar]
- 46.Gao S, Chen T, Li L, Liu X, Liu Y, Zhao J, Lu Q, Zeng Z, Xu Q, Huang D, Tu K. Hypoxia-inducible Ubiquitin Specific Peptidase 13 contributes to Tumor Growth and Metastasis via enhancing the toll-like receptor 4/Myeloid differentiation primary response gene 88/Nuclear Factor-κB pathway in Hepatocellular Carcinoma. Front Cell Dev Biology 8 (2020). [DOI] [PMC free article] [PubMed]
- 47.Liu Y, Liu Y, Zang J, Abdullah AAI, Li Y, Dong H. Design strategies and applications of ROS-Responsive Phenylborate Ester-based nanomedicine. ACS Biomaterials Sci Eng. 2020;6(12):6510–27. [DOI] [PubMed] [Google Scholar]
- 48.Gao C, Huang Q, Liu C, Kwong CHT, Yue L, Wan JB, Lee SMY, Wang R. Treatment of atherosclerosis by macrophage-biomimetic nanoparticles via targeted pharmacotherapy and sequestration of proinflammatory cytokines. Nat Commun. 2020;11(1):2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu B, Yan W, Luo L, Wu S, Wang Y, Zhong Y, Tang D, Maruf A, Yan M, Zhang K, Qin X, Qu K, Wu W, Wang G. Macrophage membrane camouflaged reactive oxygen species responsive nanomedicine for efficiently inhibiting the vascular intimal hyperplasia. J Nanobiotechnol. 2021;19(1):374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang C, Aslan H, Zhang P, Zhu S, Xiao Y, Chen L, Khan N, Boesen T, Wang Y, Liu Y, Wang L, Sun Y, Feng Y, Besenbacher F, Zhao F, Yu M. Carbon dots-fed Shewanella oneidensis MR-1 for bioelectricity enhancement. Nat Commun. 2020;11(1):1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y, Schwendeman SP. Minimizing acylation of peptides in PLGA microspheres. J Control Release. 2012;162(1):119–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun XY, Wang JM, Ouyang JM, Kuang L. Antioxidant Activities and Repair Effects on Oxidatively Damaged HK-2 Cells of Tea Polysaccharides with Different Molecular Weights, Oxid Med Cell Longev 2018 (2018) 5297539. [DOI] [PMC free article] [PubMed]
- 53.Zhang C, Peng SY, Hong S, Chen QW, Zeng X, Rong L, Zhong ZL, Zhang XZ. Biomimetic carbon monoxide nanogenerator ameliorates streptozotocin induced type 1 diabetes in mice. Biomaterials. 2020;245:119986. [DOI] [PubMed] [Google Scholar]
- 54.Li Y, Liu Y, Ren Y, Su L, Li A, An Y, Rotello V, Zhang Z, Wang Y, Liu Y, Liu S, Liu J, Laman JD, Shi L, van der Mei HC, Busscher HJ. Coating of a Novel Antimicrobial nanoparticle with a macrophage membrane for the selective entry into infected macrophages and Killing of Intracellular Staphylococci. Adv Funct Mater 30(48) (2020). [DOI] [PMC free article] [PubMed]
- 55.Guo P, Liu D, Subramanyam K, Wang B, Yang J, Huang J, Auguste DT, Moses MA. Nanoparticle elasticity directs tumor uptake. Nat Commun. 2018;9(1):130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kang S, Duan W, Zhang S, Chen D, Feng J, Qi N. Muscone/RI7217 co-modified upward messenger DTX liposomes enhanced permeability of blood-brain barrier and targeting glioma. Theranostics. 2020;10(10):4308–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ji Y, Wang Y, Wang X, Lv C, Zhou Q, Jiang G, Yan B, Chen L. Beyond the promise: exploring the complex interactions of nanoparticles within biological systems. J Hazard Mater. 2024;468:133800. [DOI] [PubMed] [Google Scholar]
- 58.Sies H, Jones DP. Reactive oxygen species (ROS) as pleiotropic physiological signalling agents. Nat Rev Mol Cell Biol. 2020;21(7):363–83. [DOI] [PubMed] [Google Scholar]
- 59.Agapouda A, Butterweck V, Hamburger M, de Beer D, Joubert E, Eckert A. Honeybush extracts (Cyclopia spp.) rescue mitochondrial functions and bioenergetics against oxidative Injury. Oxid Med Cell Longev. 2020;2020:1948602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang S, Zheng H, Zhou L, Cheng F, Liu Z, Zhang H, Wang L, Zhang Q. Nanoenzyme-Reinforced Injectable Hydrogel for Healing Diabetic wounds infected with Multidrug resistant Bacteria. Nano Lett. 2020;20(7):5149–58. [DOI] [PubMed] [Google Scholar]
- 61.Barua S, Mitragotri S. Challenges associated with penetration of nanoparticles across cell and tissue barriers: a review of current status and future prospects. Nano Today. 2014;9(2):223–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jia X, Hua C, Yang F, Li X, Zhao P, Zhou F, Lu Y, Liang H, Xing M, Lyu G. Hydrophobic aerogel-modified hemostatic gauze with thermal management performance. Bioact Mater. 2023;26:142–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen J, Liu Y, Cheng G, Guo J, Du S, Qiu J, Wang C, Li C, Yang X, Chen T, Chen Z. Tailored hydrogel delivering Niobium Carbide boosts ROS-Scavenging and antimicrobial activities for Diabetic Wound Healing. Small. 2022;18(27):e2201300. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed during the current study.