Abstract
Background
Systemic lupus erythematosus (SLE) has numerous symptoms across organs and an unpredictable flare-remittance pattern. This has made it challenging to understand drivers of long-term SLE outcomes. Our objective was to identify whether changes in DNA methylation over time, in an actively flaring SLE cohort, were associated with remission and whether these changes meaningfully subtype SLE patients.
Methods
Fifty-nine multi-ethnic SLE patients had clinical visits and DNA methylation profiles at a flare and approximately 3 months later. Methylation was measured using the Illumina EPIC array. We identified sites where methylation change between visits was associated with remission at the follow-up visit using limma package and a time x remission interaction term. Models adjusted for batch, age at diagnosis, time between visits, age at flare, sex, medications, and cell-type proportions. Separately, a paired T-test identified Bonferroni significant methylation sites with ≥ 3% change between visits (n = 546). Methylation changes at these sites were used for unsupervised consensus hierarchical clustering. Associations between clusters and patient features were assessed.
Results
Nineteen patients fully remitted at the follow-up visit. For 1,953 CpG sites, methylation changed differently for remitters vs. non-remitters (Bonferroni p < 0.05). Nearly half were within genes regulated by interferon. The largest effect was at cg22873177; on average, remitters had 23% decreased methylation between visits while non-remitters had no change. Three SLE patient clusters were identified using methylation differences agnostic of clinical outcomes. All Cluster 1 subjects (n = 12) experienced complete flare remission, despite similar baseline disease activity scores, medications, and demographics as other clusters. Methylation changes at six CpG sites, including within immune-related CD45 and IFI genes, were particularly distinct for each cluster, suggesting these may be good candidates for stratifying patients in the future.
Conclusions
Changes in DNA methylation during active SLE were associated with remission status and identified subgroups of SLE patients with several distinct clinical and biological characteristics. DNA methylation patterns might help inform SLE subtypes, leading to targeted therapies based on relevant underlying biological pathways.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13148-024-01792-x.
Keywords: DNA methylation, Disease subtyping, Systemic lupus erythematosus, Unsupervised machine learning, Hierarchical clustering, Longitudinal, Lupus flare, Lupus remission
Introduction
Systemic lupus erythematosus (SLE) is a complex autoimmune disease that follows a relapsing (flare)-remitting pattern with systemic symptoms that vary greatly from person to person. The disease activity of SLE involves a dynamic interplay between molecular processes, particularly interferon pathways. How exactly this relates to flares, remission, and organ damage are largely unknown. Importantly, clinical characteristics are not strongly correlated with response to treatment, disease activity, or future prognosis. Given the diversity of symptoms and flare-remittance patterns of illness, it has been challenging to predict long-term SLE outcomes.
Patterns of DNA methylation (DNAm) have emerged as potential mechanisms for and biomarkers of SLE risk, disease activity, and heterogeneity [1–7]. Identifying differences in DNAm by SLE phenotypes or flare remission status might also illuminate mechanistic pathways responsible for SLE heterogeneity. Recently, much effort has been devoted to identifying SLE patient subtypes to better characterize disease heterogeneity. These efforts have primarily been based on bulk and single cell RNA sequencing, clinical symptoms, autoantibodies, and gene sequencing [8–17]. The strong involvement of type I interferon signaling has consistently emerged as one SLE patient subtype, while others have varied. Despite this consistency and the great need to stratify SLE patients, these studies have predominantly been cross sectional with participants not necessarily experiencing active disease flares. To the best of our knowledge, no studies have subtyped SLE patients based on changes in DNAm at different points of disease activity. Additionally, there has been limited inclusion of non-white participants in efforts to subtype SLE, a significant limitation given the worse disease outcomes and higher incidence among African American, Hispanic, and Asian individuals [18, 19].
In this study, we utilized 59 multi-ethnic SLE patients recruited during an active flare who have DNAm profiles at baseline flare and approximately three months later. Our objectives were to: (1) identify whether changes in DNAm between visits were associated with complete remission status at follow-up, and (2) perform unsupervised clustering of changes in DNAm to identify clinically or biologically relevant SLE subtypes.
Methods
Study subjects and design
Individuals experiencing an active SLE flare were recruited to the California Lupus Epidemiology Study (CLUES) Flare Cohort during routine clinical care at the University of California, San Francisco (UCSF). SLE diagnoses were confirmed by study physicians based upon one of the following definitions: (a) meeting ≥ 4 of the 11 American College of Rheumatology (ACR) revised criteria for the classification of SLE as defined in 1982 and updated in 1997, (b) meeting 3 of the 11 ACR criteria plus a documented rheumatologist’s diagnosis of SLE, or (c) a confirmed diagnosis of lupus nephritis, defined as fulfilling the ACR renal classification criterion (> 0.5 g of proteinuria per day or 3 + protein on urine dipstick analysis) or having evidence of lupus nephritis on kidney biopsy. A flare was established by the treating physician and characterized by the SELENA Systemic Lupus Erythematosus Disease Activity Index (SLEDAI) [20]. A SLEDAI ≥ 3 was required for inclusion and physician determination that a flare was significant enough to warrant a treatment change. Approximately three months later, participants returned to the clinic as part of routine clinical care. Individuals with blood samples and complete clinical data at each visit were included in our sample. This resulted in a total of 122 paired samples (61 individuals).
This study was approved by the Institutional Review Board of UCSF. All participants signed a written informed consent to participate.
DNA methylation data
Genome-wide DNAm was measured using the Illumina Infinium MethylationEPIC BeadChip v1.0 from DNA extracted from whole blood at each visit. Both samples from an individual were run on the same plate/array. DNAm data was processed using the minfi package in R version 4.3.3 [21, 22]. Signal intensities were background subtracted using the noob function and quantile normalized. We excluded both paired samples from individuals if at least one of their samples had ≥ 5% detection p < 0.01, self-reported sex did not match sex predicted from DNAm (n = 1), or if paired samples had mismatched genotypes detectable from the methylation array (n = 1). CpG sites were removed if ≥ 5% detection p < 0.01, overlapped with annotated single nucleotide polymorphisms (SNPs) from any ancestral population in the single base extension or CpG, overlapped with previously identified cross-reactive probes, or were on sex chromosomes [23, 24]. This resulted in 606,337 CpG sites across 118 samples for analyses.
Global cell-type proportions for 12 leukocyte subtypes (neutrophils, eosinophils, basophils, monocytes, naive and memory B cells, naive and memory CD4 + and CD8 + T-cells, natural killer, and T regulatory cells) were estimated at each visit using estimateCellCounts2() with EPIC IDOL-Ext library CpGs in the FlowSorted.Blood.EPIC package in R [25]. Input DNAm data were normalized using preprocessNoob().
Clinical data
Clinic visits included the collection and review of medical records, a SLE history and physical examination conducted by a physician specializing in SLE, and completion of a structured interview by an experienced research assistant. Data collected included sex, self-reported race, Hispanic ethnicity, age, and SELENA SLEDAI components and score at each visit. Medication assessment included whether individuals were on the following at the time of each blood draw: prednisone, cyclophosphamide, mycophenolate mofetil or mycophenolic acid, rituximab, belimumab, azathioprine, cyclosporine, tacrolimus, hydroxychloroquine, methotrexate, and solumedrol. One person had missing medication for the follow-up visit so their medications were imputed with the baseline visit medications. Multiple correspondence analysis (MCA), similar to principle component analysis but for categorical variables, was conducted separately at each time point on binary medication variables to reduce the dimensionality of the medication data using the FactoMineR R package [26]. The EpiSmokEr R package was used to predict smoking status (current, past, never) using DNA methylation data [27].
Statistical analyses
Differential methylation position analyses for remission
Our primary objective was to identify CpG sites where changes in DNAm between visits were associated with complete remission status at follow-up. Because we were limited to two timepoints and without additional longitudinal clinical data, it was not possible to determine if patients who only had serologic activity (presence of double-stranded DNA antibodies and/or low complement) were clinically quiescent. Therefore, we conservatively defined “remitter” as a patient having a SELENA SLEDAI score = 0 at the follow-up visit. For each CpG site that reached quality control thresholds, we used a linear model with a time x remission interaction term in limma R package [28]. Time represented baseline flare (time = 0) or follow-up (time = 1) visits. The paired design was accounted for using the duplicatecorrelation() function. DNAm m-values were used to obtain respective p-values while DNAm beta-values were reported for coefficient interpretation [29]. Interaction coefficients were interpreted as the average difference in percent change in DNAm between baseline and follow-up comparing the remitters to non-remitters at each CpG site. The primary model (Model 1) included plate batch, sex, diagnosis age, baseline age, and days between visits. CpG sites with p < 0.05/606,337 were considered Bonferroni significant. For sites that reached this threshold and had an interaction coefficient absolute value ≥ 0.1, we additionally tested for the effect of medications (Model 2) and cell -type proportions (Model 3) on observed associations. Model 2 included Model 1 covariates plus the first medication MCA component. Additional components were not included because they were not associated (p < 0.05) with remission status and/or DNAm principal components in bivariate logistic or linear regression models. Model 3 included all covariates from Models 1 and 2 plus seven estimated cell-type proportions for SLE-relevant cell types, e.g., neutrophils, naïve and memory CD4 + T-cells, naïve and memory CD8 + T-cells, and naïve and memory B cells. Only one patient was predicted to be a current smoker at baseline and none were predicted to be current smokers at follow-up. Because we were interested in whether changes in DNA methylation were associated with remission status, we wouldn’t expect unchanged smoking behaviors to affect change in methylation. For this reason, smoking status was not included as a covariate.
Unsupervised clustering of DNA methylation difference data
We sought to identify whether changes in DNAm between visits might be able to subtype patients into clinically or biologically relevant subgroups. We used a paired t-test in limma to identify sites where DNAm changed between visits, agnostic of clinical outcomes. DNAm m-values were used to obtain respective p-values while DNAm beta-values were used to obtain interpretable coefficients. CpGs with false discover rate (FDR) q < 0.05 and ≥ 0.03 DNAm change between visits were used for clustering (“cluster input CpGs”). The standardized difference in DNAm beta-values between baseline flare and follow-up visits were input into the consensus hierarchical clustering algorithm using ConsensusClusterPlus R package [30]. Pearson distance with average linkage was used. Agglomerative hierarchical clustering was repeated 1,000 times, with 80% of CpG sites and 80% of participants resampled per iteration. The optimal cluster number was determined based on the following criteria: a relatively low-variation coefficient within the cluster, relatively high consistency, and no obviously increased area under the cumulative distribution function curve.
Associations with SLE methylation subtypes
To determine whether SLE patient clusters identified from DNAm data had distinct demographic or clinical characteristics, we used chi-square and ANOVA tests. Demographics included sex, self-reported race, and Hispanic ethnicity. Clinical features, at both visits when relevant, included age at visit, diagnosis age, SELENA SLEDAI total score and components, remission status, and medications. Estimated proportions of 12 cell types derived from DNAm data at each visit were also assessed.
To determine which cluster input CpG sites were most strongly contributing to cluster assignments, we first used an ANOVA to identify DNAm changes that were different between any clusters. For CpGs with ANOVA p < 0.05/# input CpGs, we used a bivariate linear regression model to estimate associations between DNAm differences at each cluster input CpG (outcome) and cluster (predictor). Cluster was represented as a categorical variable with one cluster used as a common reference category. A CpG site was considered significant for a particular cluster if the respective cluster category regression term had p < 0.05/# cluster input CpGs significant from ANOVA.
Pathway analyses
Pathway analyses were conducted to test for enrichment of Reactome terms among several sets of CpG sites from analyses. This was conducted using the methylgometh function in the methylGSA R package [31]. Pathways with FDR q < 0.05 were considered significantly enriched. Because of the strong involvement of interferon signaling in SLE, we annotated CpG sites according to whether they were within interferon regulated genes (IRGs) (identified in experiments when cells or organisms were treated with an interferon) using the Interferome Database v2.01 [32].
Results
Characteristics of participants
Nineteen participants fully remitted by the follow-up visit (Table 1). On average, remitters were five years older than non-remitters and diagnosed in their mid-30s, compared to mid-20s. Within this multi-ethnic cohort, remitters and non-remitters did not have significantly different sex, race, or ethnicity. SLEDAI at the baseline flare were similar among remitters and non-remitters (mean = 11.4 (standard deviation (sd) = 5.7) and 11.5 (sd = 5.9), respectively). At the follow-up visit, non-remitters had a mean SLEDAI = 5.7 (sd = 4.1). At baseline, both groups had similar presence of SLEDAI proteinuria (42% each), an indicator of lupus nephritis, a severe manifestation of SLE. All SLEDAI components and estimated cell-type proportions were similar among remitters and non-remitters, except for CD8 + naïve T-cells at baseline and follow-up and natural killer cells at baseline (Supplementary Table 1). Patients were not treatment-naïve at baseline and medication use at the baseline and follow-up visits were similar between remitters and non-remitters (Supplementary Table 1).
Table 1.
Clinical and demographic features of 59 systemic lupus erythematosus participants by remission status at follow-up visit
| Remission status | |||
|---|---|---|---|
| Yes | No | P-value | |
| N | 19 | 40 | |
| Female, n (%) | 18 (94.74) | 34 (85.00) | 0.52 |
| Self-reported race, n (%) | 0.79 | ||
| White | 1 (5.26) | 6 (15.00) | |
| Black | 3 (15.79) | 6 (15.00) | |
| Asian | 7 (36.84) | 11 (27.50) | |
| Other | 0 (0.00) | 2 (5.00) | |
| Multiple racesa | 1 (5.26) | 2 (5.00) | |
| Not reportedb | 7 (36.84) | 13 (32.50) | |
| Hispanic, n (%) | 7 (36.84) | 15 (37.50) | 1.00 |
| Age at diagnosis (years), mean (sd) | 35.68 (16.34) | 27.05 (10.57) | 0.02 |
| Age at baseline flare visit (years), mean (sd) | 40.68 (13.64) | 35.88 (12.20) | 0.18 |
| Days between visits, median [IQR] | 96.00 [57.00, 267.50] | 78.50 [48.50, 163.25] | 0.48 |
| SELENA SLEDAI, mean (sd) | |||
| Baseline | 11.42 (5.65) | 11.53 (5.85) | 0.95 |
| Follow-up | 0.00 (0.00) | 5.72 (4.08) | < 0.001 |
| SLEDAI Proteinuria, n (%) | |||
| Baseline | 8 (42.11) | 17 (42.50) | 1.00 |
| Follow-up | 0 (0.00) | 6 (15.00) | 0.19 |
| Medications at follow-up (yes/no), n (%) | |||
| Prednisone | 15 (78.95) | 31 (77.50) | 1.00 |
| Hydroxychloroquine | 17 (89.47) | 32 (80.00) | 0.59 |
| Mycophenolate | 7 (36.84) | 15 (37.50) | 1.00 |
| Azathioprine | 0 (0.00) | 7 (17.50) | 0.13 |
| Belimumab | 1 (5.26) | 5 (12.50) | 0.69 |
| Cyclophosphamide | 1 (5.26) | 3 (7.50) | 1.00 |
| Solumedrol | 1 (5.26) | 0 (0.00) | 0.70 |
| Rituximab | 1 (5.26) | 3 (7.50) | 1.00 |
| Tacrolimus | 0 (0.00) | 2 (5.00) | 0.82 |
| Methotrexate | 3 (15.79) | 5 (12.50) | 1.00 |
Remission defined as SLEDAI = 0 at follow-up visit
IQR, interquartile range; sd, standard deviation; SLE, Systemic Lupus Erythematosus; SELENA SLEDAI, SLE Disease Activity Index—the Safety of Estrogens in Lupus Erythematosus: National Assessment trial
aAdditional race categories not solely indicated by participants included American Indian and Pacific Islander
bMajority of participants not reporting race identified as Hispanic
Changes in DNA methylation among active SLE patients were associated with remission status
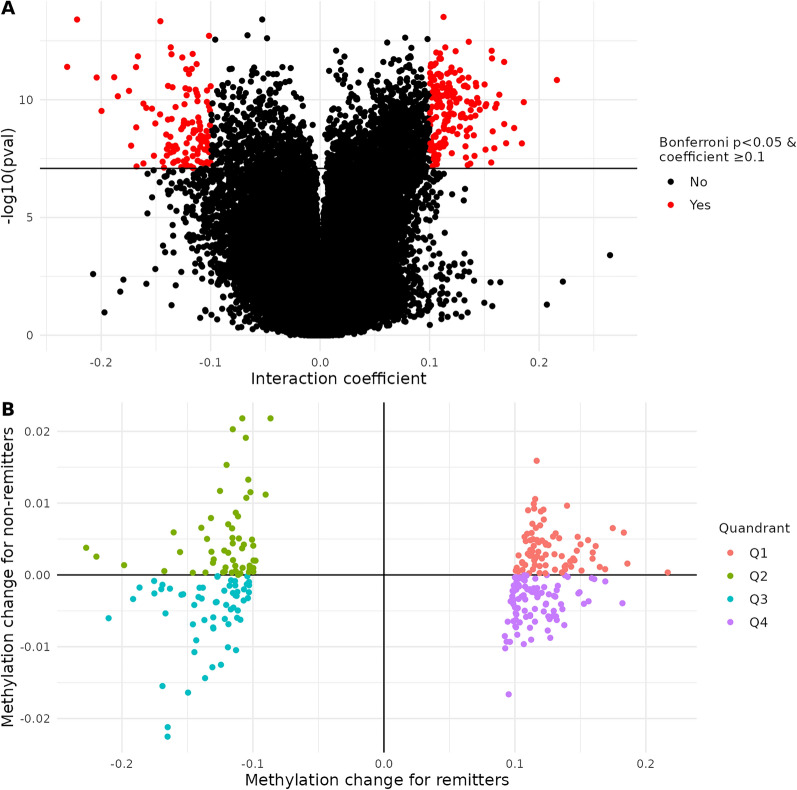
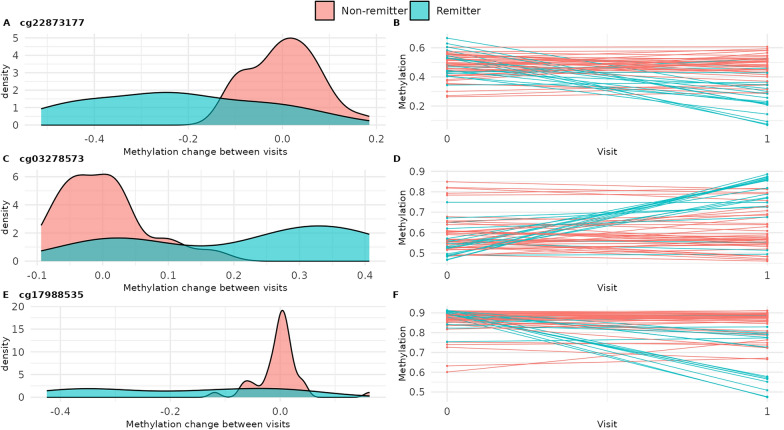
Our primary objective was to identify CpG sites where DNAm changed between baseline flare and follow-up visits differently depending on remission status. After adjusting for batch, sex, diagnosis age, baseline age, and days between visits (Model 1), we identified 1,953 significant CpG sites (Fig. 1A). Of these, 291 had absolute value ≥ 0.10 (provided in Supplementary Table 2 and subsequently described). DNAm at these sites changed little between visits for non-remitters (mean absolute change of 0.00) but more for remitters (mean absolute change of 0.12) (Fig. 1B). For remitters, approximately 60% (n = 176) of these sites had higher DNAm at the follow-up visit compared to the baseline flare (Fig. 1B); 131 were annotated to IRGs (Supplementary Table 2) [33]. These CpGs were not significantly enriched in pathways at FDR q < 0.05 (Supplementary Table 3). The 25 CpGs with the largest absolute interaction coefficient effect size are shown in Table 2. After adjusting for medications (Model 2) and cell-type proportions (Model 3), these 25 results remained Bonferroni significant and interaction effect sizes did not appreciably change (Supplementary 2). For three of these sites within immune-related genes, distributions of DNAm change and patients’ DNAm trajectories between visits are shown in Fig. 2. Site cg22873177, upstream of URB2, had the largest effect (interaction coefficient − 0.23, 95% confidence interval (CI): − 0.29, − 0.17). At this site, DNAm decreased 23%, on average, from baseline flare to follow-up for remitters and did not change for non-remitters (Fig. 2A and B, Supplementary Table 2). For the second, cg03278573, within the body of death associated protein (DAP), DNAm increased 22%, on average, from baseline flare to follow-up for remitters and was unchanged for non-remitters (Fig. 2C and D). For the third, cg17988535, within the body of RASA3 (involved in T-cell homeostasis), DNAm decreased 19%, on average, from baseline flare to follow-up for remitters and was unchanged for non-remitters (Fig. 2E and F).
Fig. 1.
Changes in DNA methylation among active SLE patients were associated with remission status. A Differential methylation change between baseline flare and follow-up visits by remission status, with Model 1 covariates (in red, Bonferroni p < 0.05 and coefficient absolute value ≥ 0.1). Black line indicates Bonferroni significance. B Among CpGs with Bonferroni p < 0.05 and coefficient absolute value ≥ 0.1, average methylation change from baseline flare to follow-up visit for remitters (x-axis) and non-remitters (y-axis). Coefficients shown were from Model 1. Quadrants one (red) and three (blue) represent methylation changes in the same direction of effect (hypo- or hyper-methylation) for remitters and non-remitters. Quadrants 2 (green) and four (purple) represent opposite direction of methylation change between groups
Table 2.
Difference in DNA methylation change from baseline flare to follow-up visit comparing remitters to non-remitters (visit x remission interaction) for 25 CpGs genome-wide significant from Model 1 with largest interaction coefficient
| Model 1 | ||||||||
|---|---|---|---|---|---|---|---|---|
| Names | chr | pos | Gene | Group | Gene’s relevance to immune function, SLE, or autoimmunity | Coef | 95% CI | P-value |
| cg22873177 | chr1 | 229,761,584 | URB2; TAF5L | TSS1500; 5’UTR | Expression associated with cell cycle and TGF beta, ERBB, RIG I-like receptor, and P53 signaling pathways. PMID: 37033651 | − 0.23 | − 0.29, − 0.17 | 4.04E-12 |
| cg14543959 | chr3 | 113,557,658 | GRAMD1C | TSS200 | Expression associated with IL-6 levels. PMID: 25311648 | − 0.22 | − 0.28, − 0.17 | 3.94E-14 |
| cg03278573 | chr5 | 10,741,861 | DAP | Body | Multiple eQTLs downregulate its transcription in immune cells; expression associated with higher autoantibody titers. PMID: 33213505 | 0.22 | 0.16, 0.28 | 1.46E-11 |
| cg04038932 | chr9 | 135,286,214 | C9orf171 | Body | − 0.20 | − 0.26, − 0.15 | 1.14E-11 | |
| cg00102561 | chr9 | 138,799,260 | CAMSAP1 | TSS1500 | Expression associated with cell cycle and TGF beta, ERBB, and T-cell receptor signaling pathways. PMID: 36212130 | − 0.20 | − 0.26, − 0.14 | 3.00E-10 |
| cg23945273 | chr4 | 120,987,944 | MAD2L1 | 5’UTR; 1stExon | − 0.19 | − 0.24, − 0.13 | 1.11E-11 | |
| cg17988535 | chr13 | 114,808,856 | RASA3 | Body | T-cell homeostasis; promotes pathogenic T helper 17 cell generation. PMID: 37545505, 30446383 | − 0.19 | − 0.24, − 0.13 | 7.14E-11 |
| cg08323960 | chr10 | 45,684,508 | 0.19 | 0.13, 0.24 | 1.27E-10 | |||
| cg24566341 | chr7 | 98,909,383 | − 0.18 | − 0.22, − 0.13 | 4.22E-11 | |||
| cg22615071 | chr3 | 156,432,778 | 0.18 | 0.12, 0.23 | 1.59E-09 | |||
| cg10369197 | chr17 | 70,815,226 | SLC39A1 | Body | Expression associated with cell cycle and TGF beta, NOD-like receptor, and MAPK signaling pathways. PMID: 35211427 | 0.18 | 0.12, 0.24 | 7.18E-09 |
| cg09241617 | chr16 | 7,473,688 | RBFOX1 | Body | Implicated in SLE GWAS. PMID: 28246883 | − 0.17 | − 0.21, − 0.13 | 1.44E-12 |
| cg26118675 | chr2 | 96,256,841 | TRIM43 | TSS1500 | Antiviral defense mechanisms. PMID: 30420784 | 0.17 | 0.13, 0.21 | 2.49E-12 |
| cg08399134 | chr6 | 126,344,877 | TRMT11 | Body | T-cell proliferation; associated with lupus nephritis. PMID: 36168063, 34923866 | − 0.17 | − 0.21, − 0.12 | 4.12E-12 |
| cg17264064 | chr18 | 47,322,166 | ACAA2 | Body | Downregulated in T-cells of lupus-prone mice. PMID: 37216123 | 0.17 | 0.12, 0.22 | 1.08E-09 |
| cg11986223 | chr7 | 26,824,073 | SKAP2 | Body | myeloid cell activation and migration. PMID: 34172489 | − 0.17 | − 0.22, − 0.12 | 1.49E-09 |
| cg12615557 | chr1 | 109,046,245 | 0.17 | 0.12, 0.22 | 7.06E-09 | |||
| cg24600355 | chr20 | 34,329,943 | RBM39 | 5’UTR; 1stExon; Body | Splicing factor highly expressed in immune cells such as CD4+ and CD8+ T cells. PMID: 34389703 | − 0.17 | − 0.23, − 0.12 | 8.96E-09 |
| cg06089892 | chr2 | 128,101,198 | MAP3K2 | TSS1500 | Kinase dysregulated in SLE. PMID: 36309313 | − 0.17 | − 0.22, − 0.11 | 6.86E-08 |
| cg00470768 | chr15 | 41,285,147 | INO80 | Body | Chromatin remodeling; cell fate. PMID: 34139016 | 0.16 | 0.12, 0.21 | 6.13E-11 |
| cg00350932 | chr2 | 86,335,912 | PTCD3 | Body | − 0.16 | − 0.21, − 0.12 | 1.46E-10 | |
| cg06176124 | chr3 | 105,588,009 | CBLB | TSS200 | May contribute to the deregulated activation of T lymphocytes observed in SLE. PMID: 21558139 | 0.16 | 0.12, 0.20 | 1.51E-10 |
| cg05062679 | chr10 | 94,000,186 | CPEB3 | 5'UTR | Associated with immune biomarkers and infiltrating immune cell types. PMID: 38476610 | 0.16 | 0.12, 0.20 | 2.20E-10 |
| cg12981595 | chr17 | 39,254,427 | KRTAP4-8 | TSS200 | 0.16 | 0.11, 0.21 | 1.15E-08 | |
| cg13529755 | chr1 | 149,138,398 | − 0.16 | − 0.22, − 0.10 | 5.09E-08 | |||
chr, chromosome; CI, confidence interval; coef, interaction coefficient; pos, hg37 position; UTR, untranslated region; SLE, systemic lupus erythematosus; TSS, transcription start site
Bolded genes are interferon regulated genes
Fig. 2.
Distributions of three genome-wide significant methylation sites. Density plot of change in methylation between visits and methylation trajectories for remitters and non-remitters for cg22873177 (a and b), cg03278573 (c and d), and cg17988535 (e and f)
Within-subject changes in DNA methylation stratify SLE patients into three clinically relevant subgroups
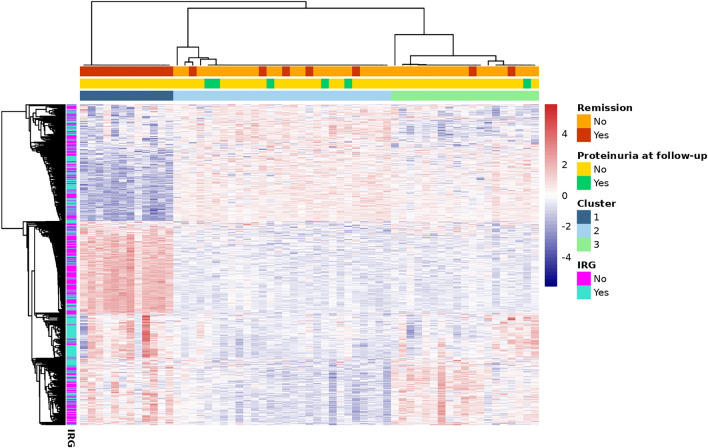
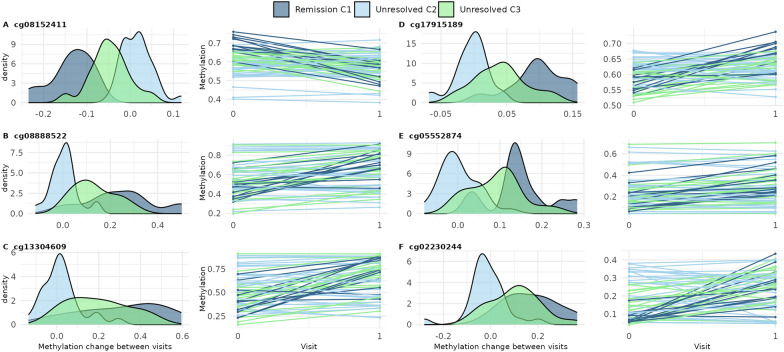
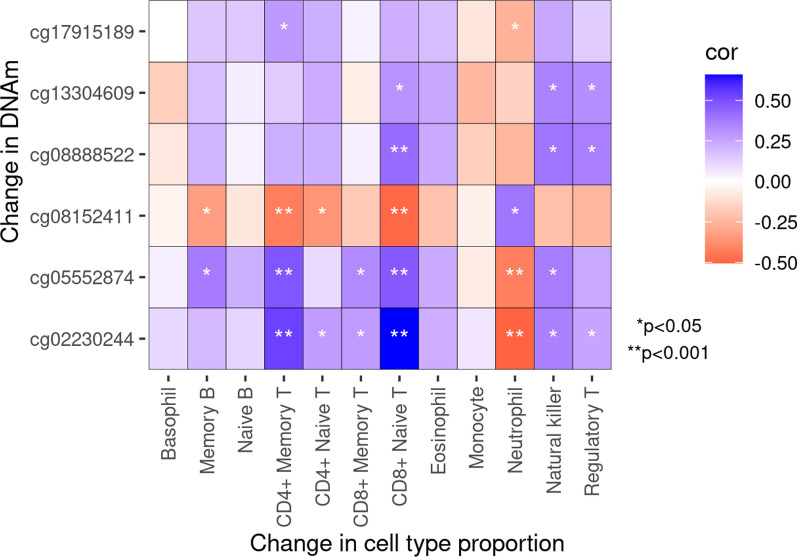
Our second objective was to identify whether changes in DNAm might subtype patients into clinically or biologically relevant subgroups. We identified 546 CpG sites that changed between visits, agnostic of clinical outcomes (FDR q < 0.05 and absolute change ≥ 0.03) (Supplementary Table 4). These were significantly enriched in three Reactome pathways (FDR q < 0.05) involved in interferon signaling (Supplementary Fig. 1); 263 were annotated to IRGs. Using DNAm change at these sites, we identified three clusters of SLE patients (Fig. 3). All Cluster 1 patients (n = 12) remitted at follow-up. We called this the “remission” cluster. SLEDAI at baseline flare was similar across clusters (mean 12.3 (sd = 5.8), 10.3 (sd = 5.7), and 12.7 (sd = 5.7) for remission, cluster 2, and cluster 3, respectively) (Table 3). For clusters 2 and 3, disease was still active at follow-up (mean SLEDAI 5.3 (sd = 5.1) and 4.3 (sd = 2.8), respectively). We called these the “unresolved” C2 and C3 clusters. No clusters differed significantly by sex, race, ethnicity, or age at visits. Average age at SLE diagnosis was youngest for the unresolved C2 cluster (mean 26.9 years) and oldest for the remission cluster (mean 36.3 years). Median time between visits was largest for the remission cluster (mean 127 days) and less for the others (mean 80 days, each). Several cell-type proportions estimated from DNAm were different across clusters (Supplementary Table 5). At baseline flare, CD4 + memory T-cell proportion was highest for unresolved C2 (mean 0.08 (sd = 0.05)) and lowest for the remission cluster (mean 0.03 (sd = 0.03)) (Table 3). A similar trend was observed for CD8 + naïve T-cells. At baseline flare, average neutrophil proportion was highest for the remission cluster (mean 0.76 (sd = 0.10)) and lowest for unresolved C2 (mean 0.57 (sd = 0.14)). Medication use was similar across clusters (Supplementary Table 5). Figure 3 shows a distinct patterning of DNAm changes across clusters, particularly for the remission cluster. The remission cluster had pronounced strong changes in DNAm in the opposite direction of the weak changes observed in Clusters 2 and 3. To better characterize these patterns, we used ANOVA (global test) and linear regression (cluster-specific tests) to identify which cluster input CpGs were most strongly contributing to cluster assignments. ANOVA identified 411 input CpGs that were Bonferroni significant (Supplementary Table 6). Of these, linear regression identified 371 with significantly different DNAm change over time comparing remission to unresolved C2 clusters and 116 comparing the unresolved C3–C2 clusters (Supplementary Table 6). Each of these cluster-specific CpG sets were not significantly enriched in Reactome pathways (FDR q < 0.05). Both cluster-specific CpG sets included many IRGs, 43 and 41% for remission and unresolved C3 cluster comparisons, respectively. There were six CpGs that were significant in both remission and unresolved C3 comparisons (vs. C2) and had coefficients at least 50% different from each other (Fig. 4). For example, at cg08152411, within the body of PTPRC (CD45 antigen), there was a 15% smaller (95% CI: − 0.18, − 0.12) DNAm change between visits among the remission cluster compared to the unresolved C2 cluster but only a 6% smaller DNAm change among the unresolved C3 comparing to the C2 cluster (Fig. 4A and Supplementary Table 6). The other five cluster differentiating CpGs were within IFIH1, IFI44L, IFIT1, and ENO2. All six CpGs showed similar patterns where the remission cluster had the most DNAm change, unresolved C3 had intermediate change, and unresolved C2 individuals had the least change. Change in DNAm at these six sites were significantly correlated with change in several cell-type proportions (Fig. 5). For example, an increase in DNAm between visits at cg08152411 was correlated with a decreased proportion of memory B cells, CD4 + memory T-cells, CD4 + naïve T-cells, and CD8 + naïve T-cells and an increased proportion of neutrophils. Oppositely, increases in DNAm at cg05552874 and cg02230244 were correlated with an increased proportion of several T-cell types and decreased proportion of neutrophils. This suggests DNAm at these six sites and the identities of the methylation clusters might be driven my cell-type proportions.
Fig. 3.
Three SLE clusters were identified from methylation changes over time from 546 cluster input CpGs. Rows represent CpG sites and were annotated as being within an interferon regulated gene (IRG). Red represents increased methylation from baseline to follow-up while blue denotes decreased. Columns represent participants and were annotated with cluster, remission status, and SLEDAI proteinuria at follow-up visit
Table 3.
Characteristics of participants in SLE patient clusters identified from consensus hierarchical clustering of DNA methylation changes between baseline flare and follow-up visits
| Remission–C1 | Unresolved–C2 | Unresolved–C3 | p-value | |
|---|---|---|---|---|
| n | 12 | 28 | 19 | |
| Female, n (%) | 12 (100.0) | 24 (85.71) | 16 (84.21) | 0.36 |
| Self-reported race, n (%) | 0.27 | |||
| Asian | 5 (41.67) | 5 (17.86) | 8 (42.11) | |
| Black | 3 (25.00) | 5 (17.86) | 1 (5.26) | |
| White | 1 (8.33) | 2 (7.14) | 4 (21.05) | |
| Multiple races | 1 (8.33) | 1 (3.57) | 1 (5.26) | |
| Other | 0 (0.00) | 2 (7.14) | 0 (0.00) | |
| Missing | 2 (16.67) | 13 (46.43) | 5 (26.32) | |
| Hispanic, n (%) | 2 (16.67) | 14 (50.00) | 6 (31.58) | 0.11 |
| Age (years) at SLE diagnosis, mean (sd) | 36.33 (18.68) | 26.86 (10.98) | 30.11 (11.24) | 0.11 |
| Age (years) at baseline flare visit, mean (sd) | 41.83 (14.72) | 36.04 (12.02) | 36.68 (12.62) | 0.41 |
| Time between visits (days), median [IQR] | 127.00 [83.00, 290.25] | 80.00 [47.25, 276.50] | 80.00 [45.50, 139.50] | 0.22 |
| Remission, n (%) | 12 (100.0) | 5 (17.86) | 2 (10.53) | 1.13E-7 |
| SLEDAI-SELENA, mean (sd) | ||||
| Baseline flare visit | 12.33 (5.82) | 10.29 (5.70) | 12.74 (5.68) | 0.31 |
| Follow-up visit | 0.00 (0.00) | 5.29 (5.08) | 4.26 (2.75) | 8.51E-4 |
| Proteinuria, n (%) | ||||
| Baseline | 7 (58.33) | 10 (35.71) | 8 (42.11) | 0.42 |
| Follow-up | 0 (0.00) | 5 (17.86) | 1 (5.26) | 0.16 |
| Cell-type proportions, mean (sd) | ||||
| CD4 + Memory T-cells (baseline) | 0.03 (0.03) | 0.08 (0.05) | 0.04 (0.02) | 1.13E-3 |
| CD4 + Memory T-cells (follow-up) | 0.05 (0.04) | 0.07 (0.05) | 0.07 (0.04) | 0.43 |
| CD8 + Naive T-cells (baseline) | 0.02 (0.01) | 0.07 (0.04) | 0.04 (0.03) | 1.69E-4 |
| CD8 + Naive T-cells (follow-up) | 0.03 (0.04) | 0.05 (0.04) | 0.06 (0.04) | 0.06 |
| Neutrophils (baseline) | 0.76 (0.10) | 0.57 (0.14) | 0.72 (0.10) | 1.61E-5 |
| Neutrophils (follow-up) | 0.70 (0.12) | 0.63 (0.15) | 0.63 (0.13) | 0.27 |
Fig. 4.
Six CpGs had different methylation change over time depending on methylation cluster. Density plot of change in methylation between visits and methylation trajectories for remission, unresolved C2, and unresolved C3 clusters for a cg08152411, b cg08888522, c cg13304609, d cg17915189, e cg05552874, and f cg02230244. These were defined as CpGs meeting all the following criteria: significant from ANOVA (p < 0.05/546), p < 0.05/411 for remission vs. unresolved C2 cluster comparison, p < 0.05/411 for unresolved C3 vs. C2 comparison, and cluster comparison coefficients were at least 50% different from each other
Fig. 5.
Correlation between change in cell-type proportions and change in DNAm between visits at six CpG sites that strongly differentiated patient clusters. Color represents strength and direction of Spearman correlation coefficient. P-values denoted by “*” p < 0.05, “**” p < 0.001, or empty p ≥ 0.05
Discussion
Using DNAm data from a multi-ethnic cohort of SLE patients with active disease, we identified 1,953 methylation sites whose change over time significantly differed depending on whether a patient fully remitted at follow-up or not. Of these, the CpG sites most differentially changed over time were located within genes relevant to immune function and autoimmunity. In addition, we identified three SLE patient clusters based on DNAm changes between visits. There were few distinct clinical features of the clusters, apart from one cluster where all members remitted. Significant DNAm changes between visits at six CpG sites, including CD45 and IFI genes, were particularly different among the clusters, suggesting these may be good candidates for stratifying patients in the future. The identification of these clinically relevant clusters using DNAm data alone may help inform future disease subtyping and treatment decisions. Overall, these findings support the ability of serial DNAm profiling to uncover biologically meaningful differences beyond what conventional clinical tools can reveal.
Our study identified many CpG sites that had significantly different changes in DNAm between baseline flare and follow-up visits depending on remission status. For most of these sites, DNAm did not change for non-remitters but did change for remitters. Changes over time were upwards of 15% among remitters at 38 CpG sites. This suggests a robust effect of treatment at these sites for these 19 individuals. The most significant effect was observed for cg22873177, in the 1500 transcription start site (TSS) of URB2. DNAm at this site decreased 23% between visits, suggesting a shift in the chromatin state between baseline flare and follow-up from less to more permissive for translation. URB2 is involved in ribosomal biogenesis and interacts with several proteins including interferon-inducible protein IFI16, which is a critical antiviral factor and sensor of viral DNA [34]. Autoantibodies against IFI16 have been identified in people with SLE, and a recent study showed that expression of IFI16 was associated with SLEDAI and prognosis in lupus nephritis [35, 36].
When examining the other significantly differentially methylated CpG sites between remitters and non-remitters, most were within or near genes relevant to the immune system. For example, the second most significant effect was observed for cg14543959, in the 200TSS of GRAMD1C, which is associated with IL-6 levels, an important inflammatory biomarker for the SLE severity and risk of progression [37]. The third most significant CpG was within the body of DAP, death associated protein. A previous study found that SLE patients with the DAP1 risk allele exhibited significantly higher autoantibody titers and altered expression of immune system, autophagy, and apoptosis pathway transcripts [38]. Interestingly, a recent study found both GRAMD1C and DAP1 to have significantly higher expression in SLE patients in remission compared to SLE patients with active disease [39]. Our findings do not directly compare to others because none have investigated change in DNAm over time between flare and post-flare timepoints. Another immune-related gene with a significant CpG (cg17900535) was RASA3, part of the RAS P21 protein activators. Ras is important for T-cell function and dysfunction has been shown to be associated with SLE [40]. Additionally, cg08399134 is within TRMT11, transfer RNA methyltransferase, which has been shown to selectively enhance protein translation to drive T-cell proliferation in vivo when methylated and is associated with lupus nephritis [41, 42].
Results from consensus hierarchical clustering of DNAm changes identified three SLE patient clusters. Interestingly, a remission cluster emerged, despite solely relying on DNAm data for clustering. It’s important to note that our sample had 19 people fully remit over the follow-up period, but only 12 were classified into the “remission” cluster. This highlights that, while the clusters were clinically relevant, they did not completely correspond to clinical features. The two “unresolved” disease activity clusters only differed in proportion of neutrophils at the baseline flare visit (57% for C2 and 72% for C3 clusters). Neutrophil proportions were similar for these clusters at follow-up. Neutrophils have been implicated in SLE pathogenesis and organ damage [43].
An important finding from our cluster analysis was that, at six CpG sites, the average DNAm change between visits was distinct for each cluster. For example, at cg08888522, within the body of IFIH1, DNAm increased, on average, from flare to follow-up visit 25% for the remission cluster, 13% for the C3 unresolved cluster, and only 1% for the unresolved C2 cluster. IFIH1 senses double-stranded RNA and activates type I interferon signaling [44]. Several studies have found an association between IFIH1 and SLE, including a de novo rare gain-of-function genetic variant identified in a severe SLE patient and several common variants associated with SLE susceptibility, IL-18 and granzyme B serum levels, and autoantibodies in SLE patients [45–48]. Another “cluster differentiating CpG” included cg13304609 in the TSS1500 of IFI44L. Methylation in the promoter of IFI44L has been proposed as a diagnostic biomarker for SLE [49, 50]. Additionally, cg08152411, within the body of PTPRC (CD45 antigen), was a “cluster differentiating CpG”. Autoantibodies to CD45 have been found in SLE [51, 52]. Our results also showed these methylation changes were correlated with changes in SLE-relevant cell-type proportions. This suggests that a shift in immune cell composition is reflected in change in DNAm in immune relevant genes. Single cell genomics approaches should be used to determine the relevance of this correlation to better understand disease heterogeneity. Altogether, this cluster analysis showcases the heterogeneity of the immune response of SLE and indicates that DNAm data may be useful for subtyping SLE patients who cannot be distinguished from each other using clinical data alone.
This study has several strengths and limitations. The majority of SLE studies are cross-sectional and have patients with relatively low disease activity. Our study has two timepoints and only includes individuals actively experiencing a physician-diagnosed flare and warranting a treatment change at study enrollment. This sheds light on mechanisms underpinning active disease, which might be missed in other studies. Our sample is also ethnically and racially diverse. SLE patients from non-European populations, such as Hispanics, African Americans, and Asians, develop SLE at a younger age and experience worse disease manifestations than patients of European descent. Yet, these groups are underrepresented in translational studies of SLE. Despite having a highly informative set of individuals, our sample size was modest (118 samples). This is partly due to the challenge of recruiting and studying active SLE flaring patients in clinics. Still, we were able to find many CpG sites where DNAm changed differentially depending on remission status and identify meaningful patient clusters. Additionally, our study involved a real-world uncontrolled medication setting, making it difficult to study treatment response. While we adjusted for medications using MCA, we may not have captured differences in medication use such as adherence, dosage, or combinations. Patients also had different follow-up times depending on when they were seen at outpatient visits. This is relevant for medications that take longer to garner an immunological response. Ideally, in the future, DNAm would be profiled longitudinally in clinical trials for prediction of treatment response and identification of mechanistic pathways underlying response. This would also improve our ability to determine whether serologically active, clinically quiescent patients have different patterns of DNAm change compared to those whose remission included serological inactivity. Finally, we were not able to replicate our findings in an independent cohort because, to the best of our knowledge, such longitudinal, repeated sampling of DNAm among active SLE cohorts does not exist. However, we did use consensus hierarchical clustering, which conducts clustering 1000 times with slightly different resampling of patients and data, to build confidence in the replicability of findings.
Conclusions
Changes in DNAm during active SLE were associated with remission status and identified subgroups of SLE patients with several distinct clinical and biological characteristics. Knowledge of DNAm patterns might help better inform SLE subtypes, leading to targeted therapies based on relevant underlying biological pathways. Further studies are needed to elucidate the potential importance of the associated CpG sites and genes in immune function and to validate the prognostic or diagnostic value of the individual CpGs or panels. Additionally, another longitudinal timepoint would be useful for identifying whether changes in DNAm might predict future disease status and damage.
Supplementary Information
Acknowledgements
We are grateful to the study participants and all study/clinic staff for providing their valuable time and data for this study.
Author contributions
M.K.H. conducted all data analysis and drafted the manuscript. L.F.B. generated the DNA methylation data. C.L., C.H., J.Y., M.D., C.J.Y., and P.K. recruited the cohort and collected participant data. P.K., J.Y., M.D., L.A.C, C.J.Y, and C.L. oversaw the funding for the project. L.A.C. and C.L. conceived the study, supervised the data collection and analysis, and edited the manuscript. J.N., K.E.T., and M.K.H. performed sample quality control and data analysis. All authors approved the final version of the manuscript.
Funding
Open access funding provided by the National Institutes of Health. This work was funded by U01DP006701 CDC and NIH/NIAMS P30 AR070155. This work was supported in part by the Intramural Research Program of the National Human Genome Research Institute, NIH.
Availability of data and materials
Data is available upon request.
Declarations
Ethics approval and consent to participate
This study was approved by the Institutional Review Board of the University of California, San Francisco (IRB #14–14429). All participants signed a written informed consent to participate.
Consent for publication
NA.
Competing interests
The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: M.K.H., J.N., K.E.T., P.K., M.D., C.H., L.F.B., L.A.C., and C.L. declare no conflict of interest. C.J.Y. has received consulting fees from Maze Therapeutics, HiBio, Santa Ana, TRex Bio, ImYoo, and DeciBio; received payment for honoraria at Renji Hospital School of Medicine, American College of Medic al Genetics and Genomics, St Jude’s Children’s Research Hospital, Yonsei University College of Medicine, and the Lupus Research Alliance; received support for attending meetings from Icahn School of Medicine at Mount Sinai, Parker Institute for Cancer Immunotherapy, Renji Hospital School of Medicine, Northwestern University Feinberg School of Medicine, and St Jude’s Children’s Research Hospital; participated on advisory/data safety monitoring board for DropPrint Genomics and Related Sciences; leadership role in other boards/societies/groups including Survey Genomics, Arc Institute, and DropPrint Genomics; has stock in Survey Genomics, Related Sciences, Maze Therapeutics, and DropPrint Genomics; has received equipment or gifts/service from Illumina; and has other financial or non-financial interests at Chan Zuckerberg Initiative, Genentech, ScaleBio, Parker Institute for Cancer Immunotherapy, Chan Zuckerberg Biohub, BioLegend, and Illumina. J.Y. has grant/contracts from Gilead, Astra Zeneca, and BMS Foundation; has received consulting fees from Astra Zeneca, Aurinia, Pfizer, Immpact Bio, and Adelphi Values; and is the research lead for the ACR RISE registry.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Coit P, Ortiz-Fernandez L, Lewis EE, McCune WJ, Maksimowicz-McKinnon K, Sawalha AH. A longitudinal and transancestral analysis of DNA methylation patterns and disease activity in lupus patients. JCI Insight. 2020;5(22):e143654. 10.1172/jci.insight.143654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang B, Liu L, Zhou T, et al. A simple and highly efficient method of IFI44L methylation detection for the diagnosis of systemic lupus erythematosus. Clin Immunol. 2020;221:108612. 10.1016/j.clim.2020.108612. [DOI] [PubMed] [Google Scholar]
- 3.Xie S, Zeng Q, Ouyang S, Liang Y, Xiao C. Bioinformatics analysis of epigenetic and SNP-related molecular markers in systemic lupus erythematosus. Am J Transl Res. 2021;13(6):6312–29. [PMC free article] [PubMed] [Google Scholar]
- 4.He Z, Zhou S, Yang M, et al. Comprehensive analysis of epigenetic modifications and immune-cell infiltration in tissues from patients with systemic lupus erythematosus. Epigenomics. 2022;14(2):81–100. 10.2217/epi-2021-0318. [DOI] [PubMed] [Google Scholar]
- 5.Lanata CM, Nititham J, Taylor KE, et al. Dynamics of methylation of CpG Sites associated with systemic lupus erythematosus subtypes in a longitudinal cohort. Arthritis Rheumatol Hoboken Nj. 2022;74(10):1676–86. 10.1002/art.42237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang W, Liang G, Zhou H, et al. Identification of potential biomarkers for systemic lupus erythematosus by integrated analysis of gene expression and methylation data. Clin Rheumatol. 2023;42(5):1423–33. 10.1007/s10067-022-06495-3. [DOI] [PubMed] [Google Scholar]
- 7.Sung WY, Lin YZ, Hwang DY, et al. Methylation of TET2 promoter is associated with global hypomethylation and hypohydroxymethylation in peripheral blood mononuclear cells of systemic lupus erythematosus patients. Diagnostics. 2022;12(12):3006. 10.3390/diagnostics12123006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu L, Su X, Liu Z, Zhou A. Predicted immune-related genes and subtypes in systemic lupus erythematosus based on immune infiltration analysis. Dis Markers. 2022;2022:8911321. 10.1155/2022/8911321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cui M, Li T, Yan X, et al. Blood genomics identifies three subtypes of systemic lupus erythematosus: “IFN-High”, “NE-High”, and “Mixed.” Mediators Inflamm. 2021;2021:e6660164. 10.1155/2021/6660164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toro-Domínguez D, Martorell-Marugán J, Martinez-Bueno M, et al. Scoring personalized molecular portraits identify systemic lupus erythematosus subtypes and predict individualized drug responses, symptomatology and disease progression. Brief Bioinform. 2022;23(5):bbac332. 10.1093/bib/bbac332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guthridge JM, Lu R, Tran LTH, et al. Adults with systemic lupus exhibit distinct molecular phenotypes in a cross-sectional study. eclinicalMedicine. 2020;20:100291. 10.1016/j.eclinm.2020.100291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perez RK, Gordon MG, Subramaniam M, et al. Single-cell RNA-seq reveals cell type–specific molecular and genetic associations to lupus. Science. 2022;376(6589):eabf1970. 10.1126/science.abf1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lanata CM, Paranjpe I, Nititham J, et al. A phenotypic and genomics approach in a multi-ethnic cohort to subtype systemic lupus erythematosus. Nat Commun. 2019;10(1):3902. 10.1038/s41467-019-11845-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rogers JL, Eudy AM, Pisetsky D, et al. Using clinical characteristics and patient-reported outcome measures to categorize systemic lupus erythematosus subtypes. Arthritis Care Res. 2021;73(3):386–93. 10.1002/acr.24135. [DOI] [PubMed] [Google Scholar]
- 15.Diaz-Gallo LM, Oke V, Lundström E, et al. Four systemic lupus erythematosus subgroups, defined by autoantibodies status, differ regarding HLA-DRB1 genotype associations and immunological and clinical manifestations. ACR Open Rheumatol. 2022;4(1):27–39. 10.1002/acr2.11343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Artim-Esen B, Çene E, Şahinkaya Y, et al. Cluster analysis of autoantibodies in 852 patients with systemic lupus erythematosus from a single center. J Rheumatol. 2014;41(7):1304–10. 10.3899/jrheum.130984. [DOI] [PubMed] [Google Scholar]
- 17.Sandling JK, Pucholt P, Hultin Rosenberg L, et al. Molecular pathways in patients with systemic lupus erythematosus revealed by gene-centred DNA sequencing. Ann Rheum Dis. 2021;80(1):109–17. 10.1136/annrheumdis-2020-218636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dall’Era M, Cisternas MG, Snipes K, Herrinton LJ, Gordon C, Helmick CG. The incidence and prevalence of systemic lupus erythematosus in San Francisco County, California: the California lupus surveillance project. Arthritis Rheumatol. 2017;69(10):1996–2005. 10.1002/art.40191. [DOI] [PubMed] [Google Scholar]
- 19.Lewis MJ, Jawad AS. The effect of ethnicity and genetic ancestry on the epidemiology, clinical features and outcome of systemic lupus erythematosus. Rheumatology. 2017;56(suppl_1):i67–77. 10.1093/rheumatology/kew399. [DOI] [PubMed] [Google Scholar]
- 20.Petri M, Kim MY, Kalunian KC, et al. Combined oral contraceptives in women with systemic lupus erythematosus. N Engl J Med. 2005;353(24):2550–8. 10.1056/NEJMoa051135. [DOI] [PubMed] [Google Scholar]
- 21.Aryee MJ, Jaffe AE, Corrada-Bravo H, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30(10):1363–9. 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing. Published online 2020. https://www.R-project.org/
- 23.Pidsley R, Zotenko E, Peters TJ, et al. Critical evaluation of the Illumina MethylationEPIC Beadchip microarray for whole-genome DNA methylation profiling. Genome Biol. 2016;17(1):208. 10.1186/s13059-016-1066-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCartney DL, Walker RM, Morris SW, McIntosh AM, Porteous DJ, Evans KL. Identification of polymorphic and off-target probe binding sites on the Illumina Infinium MethylationEPIC BeadChip. Genomics Data. 2016;9:22–4. 10.1016/j.gdata.2016.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Enhanced cell deconvolution of peripheral blood using DNA methylation for high-resolution immune profiling | Nature Communications. Accessed March 31, 2023. https://www.nature.com/articles/s41467-021-27864-7 [DOI] [PMC free article] [PubMed]
- 26.Lê S, Josse J, Husson F. FactoMineR: an R package for multivariate analysis. J Stat Softw. 2008;25:1–18. 10.18637/jss.v025.i01. [Google Scholar]
- 27.Bollepalli S, Korhonen T, Kaprio J, Anders S, Ollikainen M. EpiSmokEr: a robust classifier to determine smoking status from DNA methylation data. Epigenomics. 2019;11(13):1469–86. 10.2217/epi-2019-0206. [DOI] [PubMed] [Google Scholar]
- 28.Ritchie ME, Phipson B, Wu D, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47–e47. 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis | BMC Bioinformatics | Full Text. Accessed 3 April 2024. https://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-11-587 [DOI] [PMC free article] [PubMed]
- 30.Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26(12):1572–3. 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.methylGSA: a Bioconductor package and Shiny app for DNA methylation data length bias adjustment in gene set testing. Bioinformatics. Oxford Academic. Accessed 3 April 2024. https://academic.oup.com/bioinformatics/article/35/11/1958/5140219 [DOI] [PubMed]
- 32.INTERFEROME v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Research. Oxford Academic. Accessed 27 February 2024. https://academic.oup.com/nar/article/41/D1/D1040/1068970 [DOI] [PMC free article] [PubMed]
- 33.Villicaña S, Castillo-Fernandez J, Hannon E, et al. Genetic impacts on DNA methylation help elucidate regulatory genomic processes. Genome Biol. 2023;24(1):176. 10.1186/s13059-023-03011-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Diner BA, Lum KK, Javitt A, Cristea IM. Interactions of the antiviral factor interferon gamma-inducible protein 16 (IFI16) mediate immune signaling and herpes simplex virus-1 immunosuppression. Mol Cell Proteomics MCP. 2015;14(9):2341–56. 10.1074/mcp.M114.047068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Antiochos B, Trejo-Zambrano D, Fenaroli P, et al. The DNA sensors AIM2 and IFI16 are SLE autoantigens that bind neutrophil extracellular traps. Elife. 2022;11:e72103. 10.7554/eLife.72103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X, Fu S, Yu J, et al. Renal interferon-inducible protein 16 expression is associated with disease activity and prognosis in lupus nephritis. Arthritis Res Ther. 2023;25:112. 10.1186/s13075-023-03094-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin H, Joehanes R, Pilling LC, et al. Whole blood gene expression and interleukin-6 levels. Genomics. 2014;104(6):490–5. 10.1016/j.ygeno.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raj P, Song R, Zhu H, et al. Deep sequencing reveals a DAP1 regulatory haplotype that potentiates autoimmunity in systemic lupus erythematosus. Genome Biol. 2020;21(1):281. 10.1186/s13059-020-02184-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang FQ, Shao L, Dang X, et al. Unraveling transcriptomic signatures and dysregulated pathways in systemic lupus erythematosus across disease states. Arthritis Res Ther. 2024;26(1):99. 10.1186/s13075-024-03327-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mor A, Philips MR, Pillinger MH. The role of Ras signaling in lupus T lymphocytes: biology and pathogenesis. Clin Immunol. 2007;125(3):215–23. 10.1016/j.clim.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 41.Marchingo JM. tRNA methylation - a new level of control for T cell immunity. Nat Immunol. 2022;23(10):1401–2. 10.1038/s41590-022-01317-9. [DOI] [PubMed] [Google Scholar]
- 42.Liang Y, Zhang J, Qiu W, et al. Dysregulation of tRNA-derived small RNAs and their potential roles in lupus nephritis. Lupus. 2021;30(14):2248–55. 10.1177/09612033211061482. [DOI] [PubMed] [Google Scholar]
- 43.Kaplan MJ. Neutrophils in the pathogenesis and manifestations of SLE. Nat Rev Rheumatol. 2011;7(12):691–9. 10.1038/nrrheum.2011.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goubau D, Deddouche S. Reis e Sousa C Cytosolic sensing of viruses. Immunity. 2013;38(5):855–69. 10.1016/j.immuni.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van Eyck L, De Somer L, Pombal D, Bornschein S, Frans G, Humblet-Baron S, Moens L, de Zegher F, Bossuyt X, Wouters C, Liston A. Brief report: IFIH1 mutation causes systemic lupus erythematosus with Selective IgA deficiency. Arthritis Rheumatol. 2015;67(6):1592–7. 10.1002/art.39110. [DOI] [PubMed] [Google Scholar]
- 46.Zhang J, Liu X, Meng Y, et al. Autoimmune disease associated IFIH1 single nucleotide polymorphism related with IL-18 serum levels in Chinese systemic lupus erythematosus patients. Sci Rep. 2018;8(1):9442. 10.1038/s41598-018-27782-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Molineros JE, Maiti AK, Sun C, et al. Admixture mapping in lupus identifies multiple functional variants within IFIH1 associated with apoptosis, inflammation, and autoantibody production. PLOS Genet. 2013;9(2):e1003222. 10.1371/journal.pgen.1003222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang C, Ahlford A, Laxman N, et al. Contribution of IKBKE and IFIH1 gene variants to SLE susceptibility. Genes Immun. 2013;14(4):217–22. 10.1038/gene.2013.9. [DOI] [PubMed] [Google Scholar]
- 49.Zhao M, Zhou Y, Zhu B, et al. IFI44L promoter methylation as a blood biomarker for systemic lupus erythematosus. Ann Rheum Dis. 2016;75(11):1998–2006. 10.1136/annrheumdis-2015-208410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang J, Dang X, Wu X, et al. DNA methylation of IFI44L as a potential blood biomarker for childhood-onset systemic lupus erythematosus. Pediatr Res. 2024. 10.1038/s41390-024-03135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mimura T, Fernsten P, Jarjour W, Winfield JB. Autoantibodies specific for different isoforms of CD45 in systemic lupus erythematosus. J Exp Med. 1990;172(2):653–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.CD45 autoantibodies mediate neutralization of activated T cells from lupus patients through anergy or apoptosis - A Mamoune, S Kerdreux, V Durand, A Saraux, P Le Goff, P Youinou, R Le Corre, 2000. Accessed 3 April 2024. https://journals.sagepub.com/doi/abs/10.1191/096120300678828776 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is available upon request.