Abstract
Background
The visual similarities observed across various plant groups often conceal underlying genetic distinctions. This occurrence, known as cryptic diversity, underscores the key importance of identifying and understanding cryptic intraspecific evolutionary lineages in evolutionary ecology and conservation biology.
Results
In this study, we conducted transcriptome analysis of 81 individuals from 18 natural populations of a northern lineage of Picea brachytyla sensu stricto that is endemic to the Qinghai-Tibet Plateau. Our analysis revealed the presence of two distinct local lineages, emerging approximately 444.8 thousand years ago (kya), within this endangered species. The divergence event aligns well with the geographic and climatic oscillations that occurred across the distributional range during the Mid-Pleistocene epoch. Additionally, we identified numerous environmentally correlated gene variants, as well as many other genes showing signals of positive selection across the genome. These factors likely contributed to the persistence and adaptation of the two distinct local lineages.
Conclusions
Our findings shed light on the highly dynamic evolutionary processes underlying the remarkably similar phenotypes of the two lineages of this endangered species. Importantly, these results enhance our understanding of the evolutionary past for this and for other endangered species with similar histories, and also provide guidance for the development of conservation plans.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-024-05851-6.
Keywords: Cryptic diversity, Picea brachytyla, Mid-pleistocene, Positive selection, Conservation
Introduction
The worldwide decline in biodiversity poses a critical threat to the stability of ecosystems and the well-being of human populations. As such, researchers must deepen their understanding of biodiversity to include the often overlooked concept of cryptic diversity in which morphologically similar species are, in actuality, genetically distinct [1, 2]. Cryptic lineages, which often go undetected due to their subtle morphological distinctions, can offer insight into the evolutionary processes that contribute to the richness of life on Earth. This concept challenges the conventional emphasis on morphologically discernible diversity, a perspective which risks overlooking the vital genetic and phylogenetic intricacies crucial to ecosystem health and resilience [3–5]. Such oversight contributes to the “Darwinian shortfall”, a phenomenon where the depth of biodiversity is underappreciated, and may potentially lead to the compromising of conservation efforts and of ecosystem stability [6]. The reclassification of the flatwoods salamander into two distinct species serves as a prime example of the importance of recognizing and accurately categorizing cryptic lineages as a precursor to implementing appropriate conservation strategies [7]. This revelation is crucial to acknowledge in conservation biology and equips managers and policymakers with a clearer understanding of the boundaries of population units for endangered species so as to refine conservation strategies accordingly [8, 9].
The advancement of molecular techniques has been instrumental in uncovering these hidden complexities within biodiversity and has thus led to a more refined understanding of species diversity [10–13]. These technological strides have enabled the detection of cryptic speciation across a wide array of organisms including birds [14], fish [15], plants [16] and primates [17], which goes to illustrate the profound impact that geographical and climatic changes have on species evolution [12, 18, 19]. Of special urgency, alpine flora have been significantly affected by historical climate fluctuations and continue to face challenges from ongoing global warming [20–22]. These climatic changes likely promote isolation and subsequent reconnection among neighboring populations, thus leading to habitat fragmentation, range shifts, and increased speciation due to new interactions and hybridization [23, 24]. The dynamic nature of biodiversity and the variability in ecological responses to global warming further highlight the complex interplay between environmental factors and genetic diversity [25]. An enhanced understanding of biodiversity that incorporates cryptic species will be critical to adopt in the context of global climate change and habitat loss, which are both expected to negatively impact the majority of current biodiversity throughout the 21st century [26].
Against this backdrop, the surge in accessible whole-genome data and high-quality reference genomes has transformed the study of biodiversity, enabling a deeper understanding of the genetic underpinnings of species [27, 28]. These genomic tools have brought about a new level of precision in identifying distinct populations and forming conservation units, thereby significantly enhancing conservation planning efforts [9, 29]. Furthermore, the application of genotype-environment association (GEA) approaches has revealed the genetic bases that play roles in unique adaptations and responses to environmental stresses. This enhanced understanding is particularly crucial for developing effective conservation strategies. For species facing unique environmental challenges, as is the case with cryptic lineages, it is essential to design conservation strategies that specifically address these unique conditions [30–33]. Such detailed analyses are invaluable, especially for rare or endangered species where comprehensive sampling may not be feasible. Even limited whole-genome sequence data can offer essential insight into demographics, phylogenetics, and potential hybridization [29].
Picea brachytyla, an endangered species of significant ecological and economic importance, is endemic to the Qinghai-Tibet Plateau (QTP), a region known for its biodiversity hotspots, cold temperatures and strong ultraviolet (UV) radiation. Alpine environments, such as the QTP, are characterized by narrow climatic niches and the limited dispersal ability of many cold-adapted species, which makes these species particularly vulnerable to the rapid changes brought about by climate change [34, 35]. This species presents a remarkable case of polyphyly within its genetic lineage [36]. It has generally been divided into two genetically distinct lineages, with the southern lineage aligning closely with the Picea likiangensis species complex (PLSC), and the northern lineage’s unique lineage, referred to as P. brachytyla sensu stricto (s.s.), predominantly found in the mountains of Southwest China. P. brachytyla s.s. is believed to have originated from homoploid hybrid speciation events between the ancestor of the pre-diversification PLSC and P. wilsonii [37]. Research has predominantly focused on clarifying the origins of P. brachytyla s.s. via this speciation process, and its subsequent evolutionary developments have been less examined. The biodiversity in the mountains of Southwest China lends itself to cryptic speciation [38, 39], and identifying distinct lineages within P. brachytyla s.s. would be important for optimizing conservation efforts.
In this study, we conducted a transcriptomic analysis on 81 individuals derived from 18 populations of P. brachytyla s.s. spread across its known natural distribution area in Southwest China with the aim of investigating potential cryptic divergence within this endangered species. We identified two cryptic lineages geographically separated by the mountains of Southwest China, and we further investigated genes associated with local adaptation. Additionally, we assessed the role of isolation by distance (IBD) and isolation by environment (IBE) in the formation of these lineages. By revealing hidden genetic diversity within the P. brachytyla s.s. species, our research highlights the importance of integrating genomic data into biodiversity assessments, which have traditionally relied heavily on morphological traits. Such assessments will be essential for the development of effective conservation strategies that accurately reflect the true complexity of local biodiversity. This approach helps to preserve the genetic diversity that supports the stability of ecosystems and enhances the ability of natural habitats to adapt to environmental change. In summary, our study of P. brachytyla s.s. serves as a poignant reminder of the rich genetic tapestry that underlies biodiversity and of the critical role that advanced molecular techniques play in unraveling this complexity for the benefit of conservation and ecosystem management.
Materials and methods
Data preparation
In this study, most of the raw RNA-seq P. brachytyla s.s. data was obtained from NCBI (NCBI BioProject Accession ID PRJNA846694, PRJNA401149). Additionally, 24 newly generated individuals were deposited into the National Genomics Data Center (NGDC, BioProject Accession ID PRJCA028370) (Tables S1, S2), bringing the total to 81 individuals from 18 populations. In addition, two individuals of P. wilsonii were downloaded from NCBI (NCBI BioProject Accession ID PRJNA401149) for use as an outgroup. For transcriptome sequencing, total RNA was extracted using TRIzol® Reagent (Life Technologies, Thermo Fisher Scientific, Waltham, MA, USA) and was subsequently treated with DNase using the TURBO DNA-free™ Kit (Life Technologies, Thermo Fisher Scientific). The RNA was quantified on an Agilent 2100 Bioanalyzer (Agilent Technologies Inc., Santa Clara, CA, USA). Sequencing libraries for each individual were prepared using a NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (NEB, USA) and examined according to standard RNA-seq methodology [40–43]. The Illumina HiSeq 2500 platform was employed to generate 150-bp paired-end raw reads.
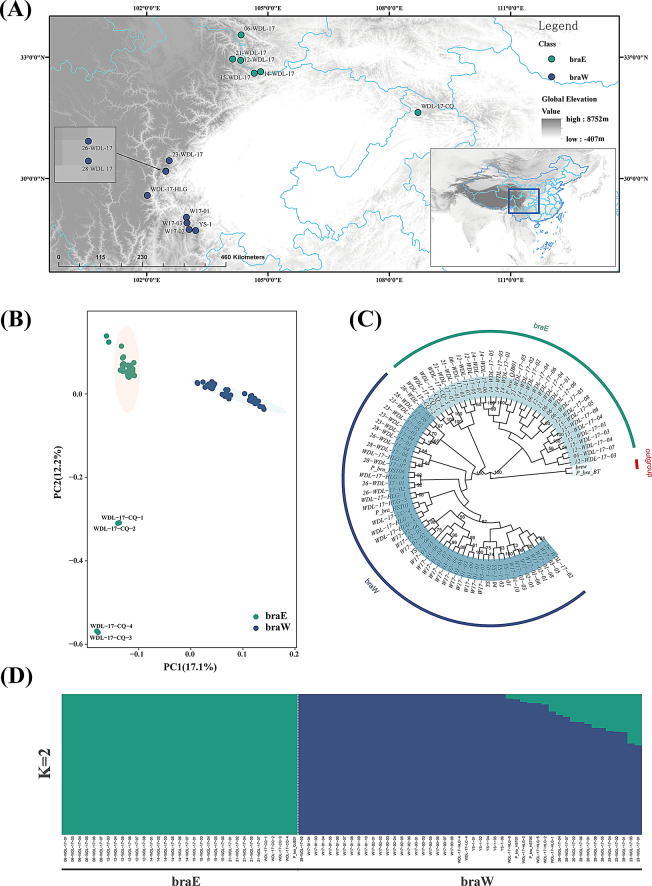
The geographical locations are shown in Fig. 1 and Table S1. For convenience, and based on geographical locations as well as the findings from Wang et al. [37], the populations were categorized into two lineages: braE and braW. The braE lineage includes the following populations: 06-WDL-17, 12-WDL-17, 14-WDL-17, 15-WDL-17, 21-WDL-17, WDL-17-CQ, and P_bra_QJB01, which is seven populations in total. The braW lineage comprises the populations 23-WDL-17, 26-WDL-17, 28-WDL-17, W17-01, W17-02, W17-03, WDL-17-HLG, P_bra_HST01, P_bra_HST06, WDL-17-LC-4, and YS-1, which is 11 populations in total.
Fig. 1.
Genetic distribution of and relationships between P. brachytyla s.s. samples. (A) Geographic distribution of P. brachytyla s.s. samples used in the analyses. (B) Principal component analysis (PCA) plots showing the first two principal components. (C) Maximum likelihood tree based on all 83 transcriptome sequences, with P. breweriana as the outgroup. (D) Bar plots indicating assignment probabilities from ADMIXTURE analysis of 81 transcriptomes, assuming two clusters (K = 2)
Read mapping and variant calling
The raw sequencing data was processed using fastp v0.20.0 [44] with the following parameters: -f 10 -F 10 -x -g -c -q 15 -u 40 -n 5. The resulting paired-end reads were then aligned to the Picea abies reference transcriptome using BWA-MEM v0.7.10 [45] with default parameters. This reference transcriptome was an updated version with fungal transcripts removed [46, 47]. The Binary Alignment/Map files were sorted using SAMtools v0.1.19 [48] and duplicate reads were identified and marked using Picard v2.18.11 (http://broadinstitute.github.io/picard/), before being excluded from downstream analyses. Local re-alignment around INDELs was performed using GATK v3.8 [49].
Single nucleotide polymorphism (SNP) calling was implemented in SAMtools v0.1.19 (mpileup and BCFtools v0.1.19) [48]. The SNP dataset was then filtered using the following criteria: (i) only genotypes with a Phred-scaled likelihood of ≥ 20 were retained (those with low-quality scores were treated as missing values) to maintain a genotyping accuracy rate of at least 99%; (ii) only biallelic loci were retained for later population-level analyses; (iii) SNPs located in INDELs (including a 5-bp buffer) were discarded to reduce the false positive rate; (iv) SNPs with a non-reference allele frequency of < 1% were removed; and (v) sites with depth (DP) < 10 per individual were considered missing, and the missing rate within each species was < 50%. After filtering, a subset of high-quality, eligible SNPs was retained for analysis. For convenience, we refer to this filtered dataset as the D-ALL dataset.
It is noteworthy that four of the 18 populations consisted of only a single individual (P_bra_QJB01, P_bra_HST01, P_bra_HST06, and WDL-17-LC-4). To improve the robustness of our analyses, these single-individual populations were excluded from the D-ALL dataset, resulting in the creation of a subdataset referred to as D-SUB.
Population structure
The population structure was assessed using ADMIXTURE analysis (K = 1–4), PCA, and a maximum-likelihood (ML) tree, based on both the D-ALL and D-SUB datasets. A model–based genetic clustering analysis was conducted using ADMIXTURE v1.30 [50] to identify genetic clusters. The cluster numbers (K) were set from 1 to 4. The optimal number of clusters was determined based on the cross-validation error (CV error) estimated for each cluster. A curve showing the change in CV error across varying K-values was created (Fig. S1), and then the K-value with the lowest CV error was selected. Meanwhile, PLINK v1.90 was employed to conduct the PCA analysis [51]. To minimize linkage effects, the PLINK command “--indep-pairwise 50 5 0.2” was applied prior to the ADMIXTURE and PCA analysis. An ML phylogeny with 1,000 bootstraps was constructed using RAxML v8.1.20 in a GTR + GAMMA model [52] with P. breweriana serving as the outgroup. Finally, we visualized the phylogenetic tree using R package ggtree v3.6.2 [53].
Demographic history
We used all individuals to reconstruct the demographic history by applying two different methods. Firstly, we employed Stairway Plot v2.1.1 [54, 55] to deduce historical fluctuations in effective population size (Ne). This methodology leverages information from the site frequency spectrum (SFS) to infer past demographic dynamics. The folded SFS was derived using the realSFS function within ANGSD v0.941-8-gcc85d8e [56], based on fourfold degenerate SNP frequency spectra for each lineage (braE and braW lineages, respectively). For the analysis, default training utilised 67% of sites, with median estimates and 95% pseudo-confidence intervals (CI) derived from 200 replicates. The following assumptions were set for both lineages: a mutation rate of 4.01 × 10− 8 mutations per site per generation and a consistent generation time of 50 years [57].
Secondly, the fastsimcoal2 v2.8.0.0 [58] approach was employed to estimate the divergence time and gene flow between the two lineages of P. brachytyla s.s. Only SNPs at fourfold degenerate sites were used to generate 2D-SFS using the realSFS function within ANGSD v0.941-8-gcc85d8e [56]. Six different demographic scenarios, varying in the presence or absence of migration, were evaluated (Fig. S2). For each scenario, 50 independent simulations were executed, with 100,000 coalescent simulations utilised for each likelihood computation. Additionally, 50 iterations of the likelihood maximisation procedure were performed to ensure accurate maximum likelihood (ML) estimates. To assess the degree of correspondence between the models and the actual data, we employed Akaike’s Information Criterion (AIC) and Akaike’s weight. The scenario that received the highest Akaike’s weight was therefore considered to be the most appropriate. The calculations incorporated the assumptions of (i) a mutation rate of 4.01 × 10− 8 per base per generation and (ii) a generational interval of 50 years [57]. To establish 95% confidence intervals, a parametric bootstrapping method was employed, and 100 separate trials were run.
Environmental variable extraction
Both to investigate the influence of the isolation by environment (IBE) and to perform the genotype-environment association analyses (GEAs), environmental variables were extracted according to the geographic coordinates. Four P. brachytyla s.s. populations downloaded from online sources were excluded from the environmental analysis due to the lack of geocoordinates and the presence of only one individual per population (Table S1). The 19 bio-climatic variables we collected were sourced from Worldclim v2.1 (https://www.worldclim.org), at a spatial resolution of 30 arc-seconds, according to the geocoordinates of the other 14 populations.
Effects of isolation by distance (IBD) and IBE on genetic structure
The influence of both IBD and IBE on the genetic structure of the remaining 14 populations of P. brachytyla s.s. (D-SUB dataset) was evaluated using two distinct analytical approaches: the partial Mantel test [59–61] and partial redundancy analysis (RDA) [62, 63].
The first approach involved the use of the partial Mantel test to identify the significance of geographic and environmental distances relative to genetic distances. This was conducted using R package Vegan v2.6-2 [64] with 9999 permutations. All distances were presented in the form of pairwise comparison matrices. The calculation of geographic distance was based on the Vincenty algorithm, which is used to compute the distance between two points on a great circle using their longitudinal and latitudinal coordinates and accounts for the ellipsoidal shape and irregularities of the Earth [65]. The environmental distance matrix was constructed based on the Euclidean distance metric. Prior to the construction of the environmental distance matrix, we calculated the Pearson correlation coefficients among 19 climate variables (Fig. S3). We removed highly collinear climate factors using an r = 0.7 threshold, and the remaining climate factors (bio1, bio2, bio12 and bio15) were retained as inputs for the matrix. Furthermore, we employed the VCFtools v0.1.14 [66] to calculate the genetic distance among populations.
In the second approach, we employed a partial RDA to ascertain the impacts of geography and the environment on genetic structure. The R package vegan provided the rda function that was employed in this study.
RDA
Given the critical importance of environmental variables in explaining genetic variation (see results), we identified only those SNPs associated with environmental factors. Using the R package Vegan v2.6-2 [64], we conducted RDA to identify candidate SNPs associated with local adaptations in response to the multivariate environment. Before conducting RDA, we transformed the longitudinal and latitudinal coordinates of individuals in the D-SUB dataset into Moran’s eigenvector maps (dbMEM1 to dbMEM6) to represent geographical factors. We adopted an RDA model recommended by Thibaut Capblancq [67], calculated eigenvalues for each constrained axis, and selected the first two axes for further investigation due to their significant contribution to genetic variation. Subsequently, we performed gene ontology (GO) enrichment analysis containing candidate SNPs. GO annotations for the corresponding protein sequences were identified using the EGGNOG-MAPPER tool (http://eggnogmapper.embl.de), followed by enrichment analysis using TBtools v2.088 [68], with an ‘evalue’ threshold of 1 × 10− 5.
Detection of the selective signals
We used the D-SUB dataset to detect selective signals. A sliding-window methodology was employed, with 10-kb windows and a 1-kb step-size, to quantify polymorphism levels (θπ, pairwise nucleotide variation) within both the braE and braW lineages. Genetic differentiation (FST) between lineages was also assessed, with both analyses conducted using VCFtools v0.1.14 [66]. For enhanced interpretability, we log2-transformed the nucleotide diversity ratios (θπ ratios, θπbraE/θπbraW). We empirically identified regions with strong selective sweep signals by selecting windows with both the highest and lowest log2(θπ ratios) (top 10%: log2(θπbraE/θπbraW) ≥ 0.695652891 or log2(θπbraE/θπbraW) ≤ -0.5275865) and high FST values (top 10%: FST ≥ 0.0400967) [69]. Subsequently, candidate genes were identified for each region, and these underwent a GO enrichment analysis to obtain functional annotations. Additionally, these candidate genes were aligned with the Arabidopsis thaliana proteome using the Basic Local Alignment Search Tool (BLASTX).
Results
SNP calling and population structure
Our P. brachytyla s.s transcriptome data includes 81 individuals, with a total size of 521.41 Gb and an average size of 6.437 Gb per individual (Table S2). We used the P. abies transcriptome [36, 46] as a reference genome for SNP identification and ultimately identified a total of 295,656 SNPs for D-ALL and 240,780 for D-SUB.
Phylogenetic analysis of the SNP loci showed that there were two lineages of P. brachytyla s.s., corresponding to the pre-categorized lineages of braE and braW (Table S1; Fig. 1). At the root of the phylogenetic tree was braE lineage, followed by braW lineage (Fig. 1C). However, two lineages of P. brachytyla s.s. (braE and braW) did not form monophyletic clades, and the bootstrap values at the internal nodes are relatively low. In addition, clustering analysis software ADMIXTURE was utilized to analyze the population structure of P. brachytyla s.s. The CV error was at its lowest when the clustering number was K = 1 (Fig. S1), which means that all the populations were confirmed to belong to one species. Nevertheless, upon contemplation of K = 2, a discernible population structure aligning with the phylogenetic tree emerged, specifically signifying the segregation of the braE and braW lineages (Fig. 1D). In the PCA results, the first principal component (PC1) explained 17.1% of the genetic variance, and the second principal component (PC2) explained 12.2%, which separated 48 individuals in braW lineage from the rest of the individuals (Fig. 2B). Accordingly, the results of the ADMIXTURE, phylogenetic analysis and PCA jointly revealed the differentiation between braE and braW lineages at the molecular level. Notably, the findings from the population structure analysis aligned with the geographic distribution of the sampling sites.
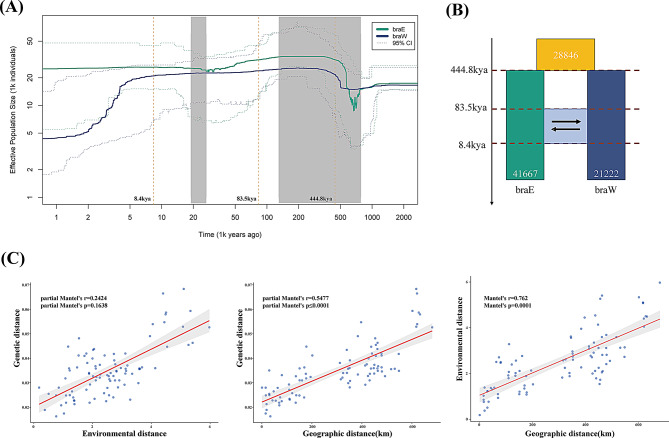
Fig. 2.
Demographic history and relationships between genetic, geographic and environmental distance of P. brachytyla s.s. (A) Changes in effective population size (Ne) over time in braE and braW lineages inferred according to the Stairway Plot method. (B) Schematic of demographic scenario modeled in fastsimcoal2. (C) Relationships from left to right: genetic distance vs. environmental distance, genetic distance vs. geographic distance, and environmental distance vs. geographic distance
Considering that four of the 18 populations consisted of only one sample (P_bra_QJB01, P_bra_HST01, P_bra_HST06, and WDL-17-LC-4), we reanalyzed the genetic structure using D-SUB dataset. This subsequent analysis confirmed that the species has diverged into two distinct lineages (Fig. S4).
Demographic history
Our Stairway Plot analysis showed that, throughout their histories, neither of the two lineages underwent significant population expansion. However, the effective population size (Ne) of braW lineage had decreased by approximately two-thirds from past levels to the present. Simulations indicated that the quantity of the two lineages split at approximately 800 thousand years ago (kya) (Fig. 2A). The braE lineage showed an initial decline in population size, followed by an increase, peaking around 400 kya before stabilizing. In contrast, while the braE lineage’s Ne remained relatively stable in recent times, the braW lineage experienced a continuous decline starting around 5 kya, and only restabilized around 2 kya.
The demographic model with the best fit, as determined by fastsimcoal2, was model 5 (Table S3; Fig. S2). This model estimated an ancestral population size of 28,846 (95% CI: ~28,277 − 29,518), with the braE and braW lineages diverging at about 444,800 years ago (95% CI: ~432,152–504,349). The current Ne was estimated to be 21,222 (95% CI: ~20,074 − 22,220) for the braW lineage and 41,667 (95% CI: ~40,528 − 43,565) for the braE lineage. Gene flow from the braE lineage to the braW lineage was higher than that in the opposite direction (5.39E-04 vs. 4.87E-04). The simulation also suggests that gene flow between the two lineages commenced at 83.5 kya (95% CI: ~66.0-118.3 kya) and concluded at 8.4 kya (95% CI: ~2.6–12.2 kya) (Table S4; Fig. 2B).
Effects of IBD and IBE on genetic structure
After removing highly collinear climate factors, four variables were retained for the Mantel test: BIO1 (Annual Mean Temperature), BIO2 (Mean Diurnal Range (Mean of monthly (max temp - min temp)), BIO12 (Annual Precipitation), and BIO15 (Precipitation Seasonality). The absolute values of the Pearson correlation coefficients between these four retained environmental factors were all below 0.7 when paired with each other (Fig. S5). The Mantel test results indicate that both geographic and environmental distances were significantly correlated with genetic distance (Table S5). However, geographic distance (Spearman’s r = 0.7847) had a stronger influence on genetic structure than environmental distance (Spearman’s r = 0.6952). Given the high degree of interaction between geography and environment (Fig. 2C), we subsequently employed partial Mantel test to mitigate any potential interaction. The partial Mantel test results indicate that genetic distance exhibited a significant relationship with geographical distance (Spearman’s r = 0.5477, p ≤ 0.0001), but not with environmental distance (Spearman’s r = 0.2424, p = 0.1638) (Fig. 2C; Table S5).
In order to circumvent the limitations of a particular model, we further investigated the independent contributions of environmental and geographic variables to genetic variation using partial RDA. As shown in Table S6, when controlling for environmental variables (partial model: gen. ~ geo. | env.), geographical variables had a significant effect on genetic variation and explained 1.32% of genetic variation on their own (adjusted R2 = 0.0132, p ≤ 0.001). Similarly, when controlling for geographical variables (partial model: gen. ~ env. | geo.), environmental variables significantly influenced genetic variation, explaining 1.27% of genetic variation alone (adjusted R2 = 0.0127, p ≤ 0.001) (Table S6). The joint geographical and environmental variables (confounded) explained 2.64% of the genetic variation. The sum of genetic variation explained by geographic and environmental variables together (model: gen. ~ geo.+env.) was 5.23% (Table S6). It can thus be deduced that, in alignment with the outcomes of the partial Mantel test, geography had a greater impact on genetics than did the environment.
As the two aforementioned models demonstrate, both geographical and environmental variables exerted a notable influence on the genetic differentiation of P. brachytyla s.s. lineages. However, geographical variables had a more pronounced impact than environmental variables.
Identification of genetic variants with local adaptation using RDA
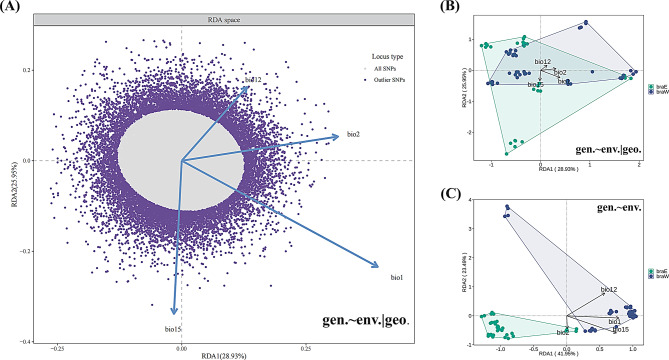
To further investigate the association between genotype and the environment, we employed RDA to examine the SNPs linked to environmental variables. The environmental data remained consistent with the previously selected four environmental factors (Fig. S5). The results of the RDA analysis indicate that, with the exception of the geographic impact on the environment, genetic variation was explained by RDA axes 1, 2, and 3 at rates of 28.93%, 25.95%, and 22.81%, respectively (Fig. S6A) (p ≤ 0.001). Bio1 and Bio2 exhibited higher loads on RDA axis 1, while Bio12 and Bio15 had significant loads on RDA axis 2 (Fig. 3A). Further, we examined the separation of these populations under the model and found that, overall, the two lineages were not significantly distinguished by the environment (Fig. 3B).
Fig. 3.
Redundancy analysis (RDA) plot for P. brachytyla s.s. (A) RDA plot for SNPs based on the first two RDA axes. (B) RDA plot for 81 P. brachytyla s.s. individuals based on the first two RDA axes, controlling for geographic variables and (C) without controlling for geographic variables
Through RDA, we identified 13,069 SNPs in total that were distributed across 4,818 genes (Table S7; Fig. 3A). These 4818 genes include one gene, comp96487_c6_seq1, that was found in a previous article and is associated with water deprivation [37]. In addition, we designated genes with an occurrence frequency exceeding twice that of the other lineage (braE or braW lineage) as prominent genes. In total, 923 genes were filtered, comprising 237 prominent genes in the braE lineage and 686 prominent genes in the braW lineage (Table S7). We performed GO enrichment analysis on this gene subset and found that some belonged to function categories associated with adaption to the environment, including “regulation of response to stimulus”, “response to cold”, “response to water”, “response to salt”, “regulation of root development”, “response to temperature stimulus” and so on (Fig. S7; Tables S8, S9).
Notably, we employed an additional RDA model to preserve the influence of geographical factors on the environment and to observe whether the divergence of these populations became distinct. In this additional model, RDA axis 1 accounted for a significantly prominent share of the explanatory power (41.95%, Fig. S6B) and roughly segregated the 14 P. brachytyla s.s. populations into either the braE or the braW lineage (Fig. 3C). In contrast to the initial RDA model, which excluded geographic impact, the clear differentiation in lineage observed in the additional RDA model provides great insight into the divergence process of this species. The additional model indicates that IBD played a primary role, while also highlighting the complex interplay between environmental and geographical factors. This finding is consistent with our deduction in the context of IBD/IBE.
Positively selected genes in P. Brachytyla s.s
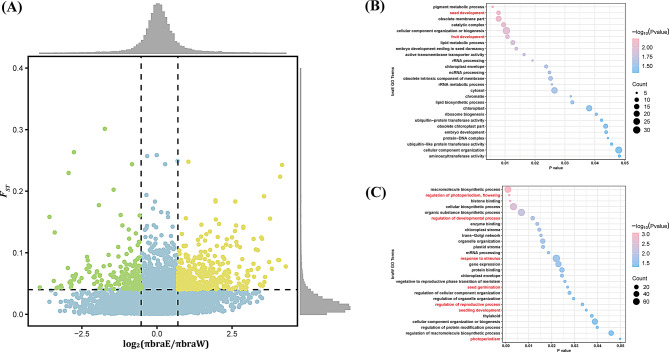
Selective sweep regions were identified from P. brachytyla s.s. SNPs (D-SUB) by combining π and FST information and selecting outliers for both metrics. A total of 547 genes were detected after annotating these regions, with 180 belonging to braE lineage and the rest belonging to braW lineage (Table S7; Fig. 4A). GO functional enrichment analysis of these 547 genes revealed 111 significantly enriched categories (Tables S10, S11), including “seed development,” “fruit development,” “regulation of photoperiodism, flowering,” “regulation of developmental process,” “response to stimulus,” “seed germination,” “regulation of reproductive process,” “seedling development” and “photoperiodism”. These nine categories related to plant growth and development directly (Fig. 4B and C). Research indicates that the GO terms related to plant growth and development often intersect with terms relevant to environmental adaptation [70, 71], highlighting the potential importance of studying how P. brachytyla s.s. adapts to climatic changes.
Fig. 4.
Genomic regions with strong selective sweep signals in the two lineages of P. brachytyla s.s. (A) Genomic regions selected in braE lineage (green dots) and braW lineage (orange dots). Black dashed lines represent the top 5% thresholds. (B) GO enrichment results for positively selected genes in braE lineage and (C) in braW lineage
To identify the proteins with the highest similarity scores, the A. thaliana proteome was searched using BLASX. We inspected the 15 genes with the highest similarity scores and found that four of them were related to plant growth and development (Table S12). Specifically, for photosynthesis, we found that comp49356_c0_seq1 and comp93775_c1_seq1 showed significant similarity to known protein sequences involved in photosystem I and photosystem II, respectively. This suggests that these two genes may encode proteins closely related to the function of photosynthesis [72, 73]. In addition, comp80667_c0_seq2 was annotated as encoding a myosin that is primarily expressed during reproductive development. It is a member of a subclass of actins composed of ACT2 and ACT8, and its mRNA is strongly expressed in leaves, roots, stems, flowers, pollen, and siliques [73]. Further, after alignment, comp41064_c0_seq1 was considered a member of the TOPLESS gene family, which plays a crucial role in the regulation of plant development and hormone signaling pathways [74]. The results of the BLASTX analysis further indicated the potential significance of environmental adaptation during the differentiation of this species.
Discussion
The present study aimed to investigate the genetic divergence within P. brachytyla s.s., an endangered plant species. This was achieved by analyzing the whole transcriptome dataset spanning the species’ distribution. The results revealed the existence of two cryptic local lineages. The divergence observed was influenced by geographic and environmental isolation during the Mid-Pleistocene epoch, which then led to the different evolutionary trajectories within the species. The braW lineage has notably declined over the last 8,000 years, while the braE lineage has remained stable. A multitude of gene variants associated with environmental adaptation and signals of positive evolution were identified, thus indicating evidence of dynamic evolution despite phenotypic similarities. These findings are pivotal for developing targeted conservation strategies to ensure the protection of this endangered species and for advancing our understanding of its adaptive mechanisms.
Cryptic divergence and demographic history after homoploid hybrid speciation
The results of our study, which employed the ADMIXTURE, PCA and phylogenetic tree analyses, indicate that P. brachytyla s.s. comprises two distinct genetic clusters (Fig. 1). However, despite this genetic differentiation, a relatively low divergence between the clusters (FST = 0.01594) was evident in the phylogenetic tree, where braE lineage and braW lineage did not form two distinct monophyletic clades and node supports were low. The phylogeny pattern mirrors that of P. crassifolia [12], and may be attributed to potential gene flow, incomplete lineage sorting, or historical introgression, phenomena commonly observed in Picea species [75, 76]. These factors are critical to consider when interpreting phylogenetic results, especially in species with complex evolutionary histories, such as P. brachytyla s.s., which originated through homoploid hybrid speciation [37]. Similarly, in the PCA, individuals such as WDL-17-CQ-1, WDL-17-CQ-2, WDL-17-CQ-3, and WDL-17-CQ-4 did not cluster tightly within the braE lineage. This is consistent with their geographical distribution, as the WDL-17-CQ population is located at a considerable distance from the rest of the braE lineage (Fig. 1A). These observations further underscore the complexity of genetic structuring within these lineages and highlight the possible role of historical demographic events or genetic exchange in shaping the current genetic landscape of P. brachytyla s.s [37]. This conclusion is further corroborated by the results of IBD/IBE and RDA (Figs. 2 and 3; Table S5). Such high genetic diversity within each lineage enhances the species’ adaptive potential for responding to environmental changes because diverse genetic resources are critical for adjusting to new environmental challenges [77, 78].
During the Pleistocene period, around 800 kya, the eastern QTP witnessed significant climatic shifts due to the largest glaciation of the era [79]. This major glaciation led to a decline in population size among the two genetic lineages. As the glaciation retreated, environmental conditions ameliorated, resulting in a subsequent increase in population size in these two lineages (Fig. 2A). The population size of the braE lineage experienced a sharp decline followed by a rapid expansion during this period. In contrast, the population size variation in the braW lineage, which is predominantly located in the Hengduan Mountains (HM), was more moderate. This is consistent with the limited glacial extensions in the HM between 460 and 710 kya [80, 81]. By approximately 440 kya (95% CI, ~ 432–504 kya; Table S4), during the Marine Isotope Stage (MIS) 12 (~ 478–424 kya) glacial [82], the two lineages began to diverge. This stage represents one of the most significant glacial intervals in the Quaternary period, characterised by the largest ice volumes observed throughout this geological epoch [83, 84]. It also saw significant vegetational shifts. For example, the cold and arid conditions of MIS 12 led to the retraction of temperate forests and the expansion of steppe vegetation [85], which likely promoted species diversification via allopatric speciation that was facilitated by geographically isolated mountains [38, 39, 86, 87]. The alteration of topography and the climatic changes associated with mountain uplifts can fragment species distributions, thus leading to reduced gene flow between isolated populations. This process initiates an allopatric divergence that can ultimately drive populations towards speciation [88, 89]. It is noteworthy that the divergence time within the P. brachytyla s.s. species predates that of P. crassifolia and P. asperata, thereby suggesting a more ancient evolutionary split [12, 90]. However, despite this ancient divergence, the relatively narrow distribution of P. brachytyla s.s. and the limited barriers to its gene flow have resulted in a lower level of genetic differentiation compared to that observed in P. crassifolia or P. asperata.
The temperatures in the QTP remained low until the late Ionian stage, approximately 300–126 kya [79]. Despite this, multiple minor glaciations occurred from 170 kya onwards, notably during the intervals of 130 − 90 kya, 75 − 60 kya, and 50 − 30 kya [91–93]. As temperatures increased, the impact of glaciations on the two genetic lineages was minimal, although the braE lineage was more adversely affected than the braW lineage. Notably, the smaller glaciers during the 75 − 60 kya and 50 − 30 kya periods appeared to have little to no impact on the lineages located along the eastern edge of the QTP and adjacent areas. It can, therefore, be reasonably concluded that the conditions on the QTP improved significantly following the glacial retreat during the 130 − 90 kya period. This environmental change facilitated a second contact between the genetic lineages and supported gene flow. Our results confirm the above assumption, that isolation between these lineages persisted until 83.5 kya, at which point gene flow commenced (Table S4; Fig. 2B). This gene flow ceased around 8.4 kya, coinciding with the arrival of hunter-gatherer groups in the high Tibetan Plateau and the onset of agriculture around 5.2–10 kya [94, 95]. This shift led to a significant decline in the braW lineage. In contrast, the braE lineage, deeply entrenched in the mountains, remained relatively unaffected by human activity. This observation is consistent with the results of the IBD and IBE analyses, which indicate that geography played a significant role in the genetic differentiation between the two lineages (Fig. 2C; Table S1).
The biodiversity of the Pleistocene epoch, which had long been relatively understudied due to cryptic diversity, is now being illuminated in recent studies. These studies reveal that significant environmental changes, such as those on “sky islands” [96, 97], and interactions with pollinators and herbivores [98, 99], have likely driven cryptic speciation processes [100, 101]. Our research underscores the significant role of allopatric speciation, as demonstrated by the two distinct metapopulation lineages in P. brachytyla s.s. (Figures 1 and 2). Additionally, our findings highlight the impact of recent Pleistocene glaciation as a substantial evolutionary force. The geographic separation and recent divergence of these lineages suggest the possibility for cryptic speciation, thus posing challenges for classification under the traditional biological species concept that emphasizes reproductive isolation and distinct morphological traits [4]. This scenario emphasizes the necessity to recognize and conserve emerging species to maintain biodiversity [102], especially in the face of ongoing global climate changes that threaten numerous species [103]. This comprehensive approach not only advances our understanding of Pleistocene biodiversity but also enhances our ability to protect genetic treasures in a rapidly changing world.
Evidence for polygenic adaptation to climate
Geographic and environmental isolation are critical in driving genetic divergence as populations adapt to specific ecological niches [104, 105]. The genetic diversity within a species, sculpted by selection pressures and its demographic history, underlies its capacity to withstand environmental change. Investigating the interplay between demography, geography, and selection illuminates the distribution of genetic variation across landscapes and the evolutionary potential of species in the context of climate change [106–109]. In the case of P. brachytyla s.s., climatic oscillations during the Quaternary likely enhanced allopatric divergence and local adaptation [39, 109–112], with populations undergoing cycles of alternating between distribution retreat and postglacial expansion [113–115]. These demographic shifts have impacted genetic variation patterns within and among populations [116].
Although the partial IBD analysis indicates that geographic factors significantly influenced the within species divergence, the initial partial IBE analysis suggests that the divergence between the two lineages was minimally impacted by environmental factors (Fig. 2C). However, further RDA analysis revealed that certain environmental variables accounted for 1.27% of the observed genetic variation, which was slightly less than the 1.32% accounted for by geographic variables (Fig. 3; Table S6). This suggests a subtle yet significant environmental influence on genetic diversity and highlights the nuanced roles that both geographic and environmental factors play in shaping the genetic landscape of this species.
In line with similar studies in other species [12, 117–120], local adaptation in P. brachytyla s.s. appears to be highly polygenic, where multiple genes contribute to a species’ adaptation to its local environment. The Mid-Pleistocene epoch was characterized by significant environmental upheavals, which, as evidenced in our study, have left a lasting imprint on the genetic makeup of species. For instance, we identified allelic variations in 4,818 genes that were correlated with environmental factors within the RDA (Table S7). The functional enrichment of these genes revealed involvement in processes related to development, catabolic, metabolic process and stress response (Tables S8, S9).
Variations in geographic and environmental selection can promote local adaptation, thus leading to specific genomic regions showing elevated FST and reduced π [121–123]. In braE lineage, our analysis highlighted positive selection in genes critical for lipid biosynthesis, pigment metabolism, and various stages of reproductive development (e.g. embryo development, fruit development and seed development) (Table S10). In contrast, braW lineage exhibited clear selection signatures in genes involved in stress response, photoperiodism, flowering, immune regulation, and other stimulus responses (Table S11). These genetic variations have driven local adaptation and divergence among lineages, thereby enhancing the species’ ability to thrive under diverse environmental conditions and leading to distinct evolutionary paths for each lineage. This study emphasized the intricate relationship between genetic diversity and environmental pressures, underscoring the necessity to conserve such diversity to maintain adaptive capacity in a changing climate.
Threats of climate change to this species and conservation implications
P. brachytyla s.s. exhibits cryptic diversity [37, 124] (Fig. 1), making it crucial to recognize and preserve these lineages as they reflect the species’ intricate genetic responses to environmental changes. However, the existence of such cryptic diversity also presents challenges for conservation. It has been demonstrated that species which have undergone unrecognized evolutionary changes are particularly vulnerable [125]. This is evident from the sharp decline of the western lineage, while the eastern lineage has remained stable (Fig. 2A). The polygenic nature of local adaptation in P. brachytyla s.s. suggests the potential for an adaptive lag due to the long generation times typical of forest trees. This lag may impede the ability of these trees to adapt to rapid climate change and suggests that current adaptive strategies may be inadequate for coping with ongoing environmental shifts [126, 127]. This scenario highlights the necessity for conservation strategies that not only preserve genetic diversity but also enhance the adaptive potential of populations in the context of accelerated climate change.
In light of these challenges, it is imperative that conservation strategies for P. brachytyla s.s. be proactive and multifaceted. First and foremost, the preservation of genetic diversity must be at the foundation of any plan. Protecting existing habitats of both lineages of P. brachytyla s.s. can achieve this goal [128, 129]. The selection of these areas should be based on ecological criteria that account for future climate scenarios, thereby ensuring that the habitats will remain suitable as the climate changes. Secondly, connectivity between fragmented populations must be enhanced. Creating new habitat corridors to facilitate gene flow can assist in maintaining genetic diversity and enhancing the species’ overall adaptability [130, 131]. This strategy is of particular importance in enabling the dissemination of adaptive traits that may emerge in response to changing environmental conditions [132]. However, it is essential to carefully monitor gene flow to ensure that local adaptations, which may be vital for each lineage’s survival in its specific environment, are not compromised [133].
In addition to in situ conservation, ex situ strategies such as the creation of germplasm banks and seed repositories are critical [134]. These banks can preserve genetic material from different populations, thus providing a resource for the restoration and reinforcement of populations suffering under climate change [135, 136]. Cultivating plants from these banks in diverse geographic settings may also help researchers understand how different populations respond to varying climatic conditions, which would provide invaluable information for future conservation efforts [137]. Furthermore, it is essential to understand the ecological roles and interactions of P. brachytyla s.s., such as pollinator relationships and competition with other species. These interactions often influence reproductive success and survival, factors that are directly impacted by climate change [138]. It is therefore imperative that conservation plans account for potential shifts in ecological dynamics and are capable of responding to real-time changes in ecosystem conditions. It is also crucial to foster public awareness and engagement in conservation efforts. It is recommended that local communities be educated about the importance of P. brachytyla s.s. and its many ecological roles so as to foster support for conservation initiatives. Community-based monitoring programs may also enhance the effectiveness of management strategies by providing early detection of ecological changes that might affect the species.
The study also underscores the need for further investigation into the functionality of the adaptive genes identified. Understanding how these genes contributed to adaptation in different environmental contexts is crucial for uncovering the mechanisms that allow species to evolve and persist in changing environments. This knowledge could inform targeted conservation strategies aimed at boosting species’ adaptive capacity [139, 140]. Additionally, exploring the genetic basis of adaptation in closely related species can reveal key evolutionary processes [141]. Comparative studies of related species facing similar environmental challenges can help identify shared and unique adaptive strategies. This comparative approach can provide a more comprehensive understanding of the genetic factors that drive species survival and evolution in dynamic ecosystems [142].
In conclusion, the conservation of P. brachytyla s.s. necessitates a comprehensive approach that integrates genetic, environmental, and ecological data. The recognition of cryptic species is of paramount importance for scientific advancement and the formulation of efficacious conservation strategies, particularly in biodiversity hotspots where the dynamics of cryptic speciation exert a significant influence [32, 97]. By implementing strategies that enhance the adaptive capacity of this species, conservationists will allow P. brachytyla s.s. to not only survive, but to thrive in the context of global climate change. This comprehensive approach to biodiversity conservation will serve as a model for the preservation of other species around the world within similarly affected mountainous regions.
It is important to note that the genetic differentiation, including all PSGs identified in this study, was based on transcriptomic data. While genetic variants in coding regions can directly influence protein function, research suggests that greater divergence often occurs in noncoding regions [143–145]. Exploring genetic divergence in these noncoding regions of P. brachytyla s.s. could provide valuable insights into adaptation. Additionally, our current analysis did not differentiate between neutral loci and potential adaptive SNPs, which limits our understanding of how natural selection versus neutral evolutionary processes contribute to speciation. Analyzing both neutral and adaptive SNPs, can reveal the genetic landscape of divergence and provide a more nuanced view of local adaptation [146, 147]. Incorporating adaptive SNPs would allow us to better assess the risk of non-adaptedness and predict the species’ capacity to respond to environmental changes [143]. Future research should involve whole-genome sequencing, a more detailed analysis of adaptive SNPs, and functional validation of candidate alleles. Additionally, common garden experiments will be crucial to thoroughly characterize the relationship between local adaptation and genetic divergence in P. brachytyla s.s., offering deeper insights into the underlying mechanisms driving these evolutionary processes and helping to inform conservation strategies.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We would like to thank Elizabeth Tokarz at Yale University for his assistance with English language and grammatical editing of the manuscript. We thank the comments on the manuscript from anonymous reviewers.
Author contributions
D.R. planned and designed the research. S.L., L.L., W.L., D.W., D.R., H.Z., Q.L., and L.M. conducted fieldwork, performed experiments and analysed data etc. D. R., S.L., and L.L. wrote the manuscript; and all authors revised and approved the final manuscript.
Funding information
National Natural Science Foundation of China (Grant Numbers: 32001085).
Data availability
Raw sequence reads have been deposited in the National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences, under BioProject accession number PRJCA028370.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Shengming Lu, Lian Liu and Weixiao Lei contributed equally to this work.
References
- 1.Pimm SL, Jenkins CN, Abell R, Brooks TM, Gittleman JL, Joppa LN, Raven PH, Roberts CM, Sexton JO. The biodiversity of species and their rates of extinction, distribution, and protection. Science. 2014;344:987. [DOI] [PubMed] [Google Scholar]
- 2.Dainese M, Martin EA, Aizen MA, Albrecht M, Bartomeus I, Bommarco R, Carvalheiro LG, Chaplin-Kramer R, Gagic V, Garibaldi LA, et al. A global synthesis reveals biodiversity-mediated benefits for crop production. Sci Adv. 2019;5:0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leys M, Keller I, Robinson CT, Räsänen K. Cryptic lineages of a common alpine mayfly show strong life-history divergence. Mol Ecol. 2017;26:1670–86. [DOI] [PubMed] [Google Scholar]
- 4.Fišer C, Robinson CT, Malard F. Cryptic species as a window into the paradigm shift of the species concept. Mol Ecol. 2018;27:613–35. [DOI] [PubMed] [Google Scholar]
- 5.Feckler A, Zubrod JP, Thielsch A, Schwenk K, Schulz R, Bundschuh M. Cryptic species diversity: an overlooked factor in environmental management? J Appl Ecol. 2014;51(4):958–67. [Google Scholar]
- 6.Diniz JAF, Loyola RD, Raia P, Mooers AO, Bini LM. Darwinian shortfalls in biodiversity conservation. Trends Ecol Evol. 2013;28:689–95. [DOI] [PubMed] [Google Scholar]
- 7.Pauly GB, Piskurek O, Shaffer HB. Phylogeographic concordance in the southeastern United States: the flatwoods salamander, Ambystoma cingulatum, as a test case. Mol Ecol. 2007;16(2):415–29. [DOI] [PubMed] [Google Scholar]
- 8.Ren YM, Zhang LS, Yang XC, Lin H, Sang YP, Feng LD, Liu JQ, Kang MH. Cryptic divergences and repeated hybridizations within the endangered living fossil dove tree (Davidia involucrata) revealed by whole genome resequencing. Plant Divers. 2024;46:169–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Funk WC, McKay JK, Hohenlohe PA, Allendorf FW. Harnessing genomics for delineating conservation units. Trends Ecol Evol. 2012;27:489–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Torres-Cambas Y, Ferreira S, Cordero-Rivera A, Lorenzo-Carballa MO. Identification of evolutionarily significant units in the Cuban endemic damselfly Hypolestes trinitatis (Odonata: Hypolestidae). Conserv Genet. 2017;18:1229–34. [Google Scholar]
- 11.Struck TH, Feder JL, Bendiksby M, Birkeland S, Cerca J, Gusarov VI, Kistenich S, Larsson KH, Liow LH, Nowak MD, et al. Finding evolutionary processes hidden in cryptic species. Trends Ecol Evol. 2018;33:153–63. [DOI] [PubMed] [Google Scholar]
- 12.Feng S, Xi E, Wan W, Ru D. Genomic signals of local adaptation in Picea Crassifolia. BMC Plant Biol. 2023; 23. [DOI] [PMC free article] [PubMed]
- 13.Feng L, Xu ZY, Wang L. Genetic diversity and demographic analysis of an endangered tree species Diplopanax Stachyanthus in subtropical China: implications for conservation and management. Conserv Genet. 2019;20:315–27. [Google Scholar]
- 14.Pulido-Santacruz P, Aleixo A, Weir JT. Morphologically cryptic Amazonian bird species pairs exhibit strong postzygotic reproductive isolation. Proceedings of the Royal Society B: Biological Sciences. 2018; 285:2017–2081. [DOI] [PMC free article] [PubMed]
- 15.Hudson AG, Lundsgaard-Hansen B, Lucek K, Vonlanthen P, Seehausen O. Managing cryptic biodiversity: fine-scale intralacustrine speciation along a benthic gradient in Alpine Whitefish (Coregonus spp). Evol Appl. 2017;10:251–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spriggs EL, Eaton DA, Sweeney PW, Schlutius C, Edwards EJ, Donoghue MJ. Restriction-site-associated DNA sequencing reveals a cryptic Viburnum species on the north American coastal plain. Syst Biol. 2019;6:187–203. [DOI] [PubMed] [Google Scholar]
- 17.Poelstra JW, Salmona J, Tiley GP, Schüßler D, Blanco MB, Andriambeloson JB, Bouchez O, Campbell CR, Etter PD, Hohenlohe PA. Cryptic patterns of speciation in cryptic primates: microendemic mouse Lemurs and the multispecies coalescent. Syst Biol. 2021;70:203–18. [DOI] [PubMed] [Google Scholar]
- 18.Shen Y, Xia H, Tu Z, Zong Y, Yang L, Li H. Genetic divergence and local adaptation of Liriodendron driven by heterogeneous environments. Mol Ecol. 2022;31:916–33. [DOI] [PubMed] [Google Scholar]
- 19.Sang Y, Long Z, Dan X, Feng J, Shi T, Jia C, Zhang X, Lai Q, Yang G, Zhang H. Genomic insights into local adaptation and future climate-induced vulnerability of a keystone forest tree in East Asia. Nat Commun. 2022;13:6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang Q, Xu X, Mao K, Wang M, Wang K, Xi Z, Liu J. Shifts in plant distributions in response to climate warming in a biodiversity hotspot, the Hengduan Mountains. J Biogeogr. 2018;45:1334–44. [Google Scholar]
- 21.Elsen PR, Tingley MW. Global mountain topography and the fate of montane species under climate change. Nat Clim Change. 2015;5:772–6. [Google Scholar]
- 22.Ding W-N, Ree RH, Spicer RA, Xing Y-W. Ancient orogenic and monsoon-driven assembly of the world’s richest temperate alpine flora. Science. 2020;369:578–81. [DOI] [PubMed] [Google Scholar]
- 23.Muellner-Riehl AN, Schnitzler J, Kissling WD, Mosbrugger V, Rijsdijk KF, Seijmonsbergen AC, Versteegh H, Favre A. Origins of global mountain plant biodiversity: testing the ‘mountain‐geobiodiversity hypothesis’. J Biogeogr. 2019;46:2826–38. [Google Scholar]
- 24.Kirschner P, Záveská E, Gamisch A, Hilpold A, Trucchi E, Paun O, Sanmartín I, Schlick-Steiner BC, Frajman B, Arthofer W. Long-term isolation of European steppe outposts boosts the biome’s conservation value. Nature Communications. 2020; 11:1968. [DOI] [PMC free article] [PubMed]
- 25.Rödder D, Schmitt T, Gros P, Ulrich W, Habel JC. Climate change drives mountain butterflies towards the summits. Sci Rep. 2021;11:14382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Habibullah MS, Din BH, Tan S-H, Zahid H. Impact of climate change on biodiversity loss: global evidence. Environ Sci Pollut Res. 2022;29:1073–86. [DOI] [PubMed] [Google Scholar]
- 27.Wang Z, Zhang X, Lei W, Zhu H, Wu S, Liu B, Ru D. Chromosome-level genome assembly and population genomics of Robinia pseudoacacia reveal the genetic basis for its wide cultivation. Commun Biology. 2023;6:797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Theissinger K, Fernandes C, Formenti G, Bista I, Berg PR, Bleidorn C, Bombarely A, Crottini A, Gallo GR, Godoy JA, et al. How genomics can help biodiversity conservation. Trends Genet. 2023;39:545–59. [DOI] [PubMed] [Google Scholar]
- 29.Hohenlohe PA, Funk WC, Rajora OP. Population genomics for wildlife conservation and management. Mol Ecol. 2021;30:62–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palsbøll PJ, Bérubé M, Allendorf FW. Identification of management units using population genetic data. Trends Ecol Evol. 2007;22:11–6. [DOI] [PubMed] [Google Scholar]
- 31.Niemiller ML, Graening GO, Fenolio DB, Godwin JC, Cooley JR, Pearson WD, Fitzpatrick BM, Near TJ. Doomed before they are described? The need for conservation assessments of cryptic species complexes using an amblyopsid cavefish (Amblyopsidae: Typhlichthys) as a case study. Biodivers Conserv. 2013;22:1799–820. [Google Scholar]
- 32.Bickford D, Lohman DJ, Sodhi NS, Ng PKL, Meier R, Winker K, Ingram KK, Das I. Cryptic species as a window on diversity and conservation. Trends Ecol Evol. 2007;22:148–55. [DOI] [PubMed] [Google Scholar]
- 33.Ahmed AS, Atickem A, Bekele A. Efficacy of cryptic rodents and challenges for conservation in Africa: a review. J Wildl Biodivers. 2024;8:343–65. [Google Scholar]
- 34.Grabherr G, Gottfried M, Pauli H. Climate change impacts in alpine environments. Geogr Compass. 2010;4:1133–53. [Google Scholar]
- 35.Ernakovich JG, Hopping KA, Berdanier AB, Simpson RT, Kachergis EJ, Steltzer H, Wallenstein MD. Predicted responses of arctic and alpine ecosystems to altered seasonality under climate change. Glob Change Biol. 2014;20:3256–69. [DOI] [PubMed] [Google Scholar]
- 36.Ru D, Mao K, Zhang L, Wang X, Lu Z, Sun Y. Genomic evidence for polyphyletic origins and interlineage gene flow within complex taxa: a case study of Picea Brachytyla in the Qinghai-Tibet Plateau. Mol Ecol. 2016;25:2373–86. [DOI] [PubMed] [Google Scholar]
- 37.Wang D, Sun Y, Lei W, Zhu H, Wang J, Bi H, Feng S, Liu J, Ru D. Backcrossing to different parents produced two distinct hybrid species. Heredity. 2023;131:145–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Y, Jin W, Wei X, Wang X. Cryptic speciation in the Chinese white pine (Pinus armandii): implications for the high species diversity of conifers in the Hengduan Mountains, a global biodiversity hotspot. Mol Phylogenet Evol. 2019;138:114–25. [DOI] [PubMed] [Google Scholar]
- 39.Liu J, Möller M, Provan J, Gao LM, Poudel RC, Li DZ. Geological and ecological factors drive cryptic speciation of yews in a biodiversity hotspot. New Phytol. 2013;199:1093–108. [DOI] [PubMed] [Google Scholar]
- 40.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang LC, Schlesinger F, Davis CA, Zhang Y, Li RH, Salit M, Gingeras TR, Oliver B. Synthetic spike-in standards for RNA-seq experiments. Genome Res. 2011;21:1543–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hansen KD, Brenner SE, Dudoit S. Biases in Illumina transcriptome sequencing caused by random hexamer priming. Nucleic Acids Res. 2010; 38. [DOI] [PMC free article] [PubMed]
- 43.Erlich Y, Mitra PP, delaBastide M, McCombie WR, Hannon GJ. Alta-Cyclic: a selfoptimizing base caller for next-generation sequencing. Nat Methods. 2008;5:679–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen SF, Zhou YQ, Chen YR, Gu J. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:884–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nystedt B, Street NR, Wetterbom A, Zuccolo A, Lin YC, Scofield DG, Vezzi F, Delhomme N, Giacomello S, Alexeyenko A, et al. The Norway spruce genome sequence and conifer genome evolution. Nature. 2013;497:579–84. [DOI] [PubMed] [Google Scholar]
- 47.Delhomme N, Sundström G, Zamani N, Lantz H, Lin YC, Hvidsten TR, Höppner MP, Jern P, Van de Peer Y, Lundeberg J et al. Serendipitous meta-transcriptomics: the fungal community of Norway spruce (Picea abies). PLoS ONE. 2015; 10. [DOI] [PMC free article] [PubMed]
- 48.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–9. 1000 Genome Project Data Processing Subgroup. [DOI] [PMC free article] [PubMed]
- 49.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alexander DH, Lange K. Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinformatics. 2011;12:246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu G, Lam TT-Y, Zhu H, Guan Y. Two methods for mapping and visualizing associated data on phylogeny using ggtree. Mol Biol Evol. 2018;35:3041–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu XM, Fu YX. Stairway plot 2: demographic history inference with folded SNP frequency spectra. Genome Biol. 2020;21:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu X, Fu Y-X. Exploring population size changes using SNP frequency spectra. Nat Genet. 2015;47:555–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Korneliussen TS, Albrechtsen A, Nielsen R. ANGSD: analysis of next generation sequencing data. BMC Bioinformatics. 2014;15:356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.De La Torre AR, Li Z, Van de Peer Y, Ingvarsson PK. Contrasting rates of molecular evolution and patterns of selection among gymnosperms and flowering plants. Mol Biol Evol. 2017;34:1363–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Excofffier L, Marchi N, Marques DA, Matthey-Doret R, Gouy A, Sousa VC. fastsimcoal2: demographic inference under complex evolutionary scenarios. Bioinformatics. 2021;37:4882–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smouse PE, Long JC, Sokal RR. Multiple-regression and correlation extensions of the mantel test of matrix correspondence. Syst Zool. 1986;35:627–32. [Google Scholar]
- 60.Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967;27:209–20. [PubMed] [Google Scholar]
- 61.Diniz-Filho JAF, Soares TN, Lima JS, Dobrovolski R, Landeiro VL, de Campos Telles MP, Rangel TF, Bini LM. Mantel test in population genetics. Genet Mol Biology. 2013;3:475–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rellstab C, Gugerli F, Eckert AJ, Hancock AM, Holderegger R. A practical guide to environmental association analysis in landscape genomics. Mol Ecol. 2015;24:4348–70. [DOI] [PubMed] [Google Scholar]
- 63.Legendre P. Numerical Ecology. In: Encyclopedia of Ecology (Second Edition). Edited by Fath B. Oxford: Elsevier; 2019: 487–493.
- 64.Oksanen J, Simpson G, Blanchet F, Kindt R, Legendre P, Minchin P, O’Hara R, Solymos P, Stevens M, Szoecs E, Wagner H et al. vegan: Community Ecology Package. R package version 2.6-2, 2022 https://github.com/vegandevs/vegan.
- 65.Vincenty T. Direct and inverse solutions of geodesics on the ellipsoid with application of nested equations. Surv Rev. 1975;23:88–93. [Google Scholar]
- 66.Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Capblancq T, Forester BR. Redundancy analysis: a Swiss army knife for landscape genomics. Methods Ecol Evol. 2021;12:2298–309. [Google Scholar]
- 68.Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020;13(8):1194–202. [DOI] [PubMed] [Google Scholar]
- 69.Li M, Tian S, Jin L, Zhou G, Li Y, Zhang Y, Wang T, Yeung CKL, Chen L, Ma J. Genomic analyses identify distinct patterns of selection in domesticated pigs and tibetan wild boars. Nat Genet. 2013;45:1431–8. [DOI] [PubMed] [Google Scholar]
- 70.Liu J-G, Han X, Yang T, Cui W-H, Wu A-M, Fu C-X, Wang B-C, Liu L-J. Genome-wide transcriptional adaptation to salt stress in Populus. BMC Plant Biol. 2019;19:367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gurung PD, Upadhyay AK, Bhardwaj PK, Sowdhamini R, Ramakrishnan U. Transcriptome analysis reveals plasticity in gene regulation due to environmental cues in Primula sikkimensis, a high altitude plant species. BMC Genomics. 2019;20:989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Friso G, Giacomelli L, Ytterberg AJ, Peltier JB, Rudella A, Sun Q, Wijk KJ. In-depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: new proteins, new functions, and a plastid proteome database. Plant Cell. 2004;16:478–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ferro M, Brugière S, Salvi D, Seigneurin-Berny D, Court M, Moyet L, Ramus C, Miras S, Mellal M, Le Gall S, et al. AT_CHLORO, a comprehensive chloroplast proteome database with subplastidial localization and curated information on envelope proteins. Mol Cell Proteom. 2010;9:1063–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Causier B, Ashworth M, Guo W, Davies B. The TOPLESS interactome: a framework for gene repression in Arabidopsis. Plant Physiol. 2012;158:423–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhu H, Lei WX, Lai Q, Sun YS, Ru DF. Comparative analysis shows high level of lineage sorting in genomic regions with low recombination in the extended Picea likiangensis species complex. Plant Divers. 2024;46(4):547–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Feng S, Ru D, Sun Y, Mao K, Milne R, Liu J. Trans-lineage polymorphism and nonbifurcating diversification of the genus Picea. New Phytol. 2019;222:576–87. [DOI] [PubMed] [Google Scholar]
- 77.Templeton AR. Biodiversity at the molecular-genetic level - experiences from disparate macroorganisms. Philosophical Trans Royal Soc B: Biol Sci. 1994;345:59–64. [DOI] [PubMed] [Google Scholar]
- 78.Schmidt SB, Brown LK, Booth A, Wishart J, Hedley PE, Martin P, Husted S, George TS, Russell J. Heritage genetics for adaptation to marginal soils in barley. Trends Plant Sci. 2023;28:544–51. [DOI] [PubMed] [Google Scholar]
- 79.Zheng D, Zhang Q, Wu S. Mountain geoecology and sustainable development of the Tibetan Plateau. Canton: Guangdong Science and Technology; 2000. [Google Scholar]
- 80.Xu L, Ou X, Lai Z, Zhou S, Wang J, Fu Y. Timing and style of late pleistocene glaciation in the Queer Shan, northern Hengduan Mountains in the eastern tibetan Plateau. J Quat Sci. 2010;25:957–66. [Google Scholar]
- 81.Wang X, Chai K, Liu SY, Wei J, Jiang Z, Liu Q. Changes of glaciers and glacial lakes implying corridor-barrier effects and climate change in the Hengduan Shan, southeastern tibetan Plateau. J Glaciol. 2017;63:535–42. [Google Scholar]
- 82.Zhou S, Wang X, Wang J, Xu L. A preliminary study on timing of the oldest pleistocene glaciation in Qinghai-Tibetan Plateau. Quatern Int. 2006;154:44–51. [Google Scholar]
- 83.Lang N, Wolff EW. Interglacial and glacial variability from the last 800 ka in marine, ice and terrestrial archives. Clim Past. 2011;7:361–80. [Google Scholar]
- 84.Koutsodendris A, Kousis I, Peyron O, Wagner B, Pross J. The Marine Isotope Stage 12 pollen record from Lake Ohrid (SE Europe): investigating short-term climate change under extreme glacial conditions. Q Sci Rev. 2019; 221.
- 85.Kafetzidou A, Fatourou E, Panagiotopoulos K, Marret F, Kouli K. Vegetation composition in a typical Mediterranean setting (Gulf of Corinth, Greece) during successive quaternary climatic cycles. Quaternary. 2023;6:30. [Google Scholar]
- 86.Smyčka J, Roquet C, Boleda M, Alberti A, Boyer F, Douzet R, Perrier C, Rome M, Valay JG, Denoeud F, et al. Tempo and drivers of plant diversification in the European mountain system. Nat Commun. 2022;13:2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu R, Wang H, Yang J, Corlett RT, Randle CP, Li D, Yu W. Cryptic species diversification of the Pedicularis siphonantha complex (Orobanchaceae) in the mountains of southwest China since the Pliocene. Front Plant Sci. 2022;13:811206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rice WR, Hostert EE. Laboratory experiments on speciation - what have we learned in 40 years. Evolution. 1993;47:1637–53. [DOI] [PubMed] [Google Scholar]
- 89.Mayr E. Animal species and evolution. Cambridge, MA and London, England: Harvard University Press; 1963. [Google Scholar]
- 90.Feng S, Wan W, Li Y, Wang D, Ren G, Ma T, Ru D. Transcriptome-based analyses of adaptive divergence between two closely related spruce species on the Qinghai-Tibet plateau and adjacent regions. Mol Ecol. 2023;32:476–91. [DOI] [PubMed] [Google Scholar]
- 91.Zhuo Z, Baoyin Y, Petit-Maire N. Paleoenvironments in China during the last glacial Maximum and the Holocene Optimum. Episodes. 1998;21:152–8. [Google Scholar]
- 92.Zheng B, Xu Q, Shen Y. The relationship between climate change and quaternary glacial cycles on the Qinghai-Tibetan Plateau: review and speculation. Quatern Int. 2002;97–8:93–101. [Google Scholar]
- 93.Shi Y. Characteristics of late quaternary monsoonal glaciation on the Tibetan Plateau and in East Asia. Quatern Int. 2002;97–8:79–91. [Google Scholar]
- 94.Zhang D, Li S-H. Comment on permanent human occupation of the central tibetan Plateau in the early Holocene. Science. 2017;357:eaam9231. [DOI] [PubMed] [Google Scholar]
- 95.Dong G, Li R, Lu M, Zhang D, James N. Evolution of human-environmental interactions in China from the late paleolithic to the bronze age. Progress Phys Geography: Earth Environ. 2020;44:233–50. [Google Scholar]
- 96.He K, Jiang XL. Sky islands of southwest China. I: an overview of phylogeographic patterns. Chin Sci Bull. 2014;59:585–97. [Google Scholar]
- 97.Chen J, Huang Y, Brachi B, Yun Q, Zhang W, Lu W, Li H, Li W, Sun X, Wang G, et al. Genome-wide analysis of Cushion willow provides insights into alpine plant divergence in a biodiversity hotspot. Nat Commun. 2019;10:5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wessinger CA. From pollen dispersal to plant diversification: genetic consequences of pollination mode. New Phytol. 2021;229:3125–32. [DOI] [PubMed] [Google Scholar]
- 99.Niu Y, Stevens M, Sun H. Commercial harvesting has driven the evolution of camouflage in an alpine plant. Curr Biol. 2021;31:446. [DOI] [PubMed] [Google Scholar]
- 100.Paudel BR, Shrestha M, Burd M, Adhikari S, Sun YS, Li QJ. Coevolutionary elaboration of pollination-related traits in an alpine ginger (Roscoea purpurea) and a tabanid fly in the Nepalese Himalayas. New Phytol. 2016;211:1402–11. [DOI] [PubMed] [Google Scholar]
- 101.Eaton DAR, Fenster CB, Hereford J, Huang S, Ree RH. Floral diversity and community structure in Pedicularis (Orobanchaceae). Ecology. 2012;93:S182–94. [Google Scholar]
- 102.De Queiroz K. Species concepts and species delimitation. Syst Biol. 2007;56:879–86. [DOI] [PubMed] [Google Scholar]
- 103.Walther GR, Post E, Convey P, Menzel A, Parmesan C, Beebee TJC, Fromentin JM, Hoegh-Guldberg O, Bairlein F. Ecological responses to recent climate change. Nature. 2002;416:389–95. [DOI] [PubMed] [Google Scholar]
- 104.Mayr E. Ecological factors in speciation. Evolution. 1947;1:263–88. [Google Scholar]
- 105.Dauphin B, Rellstab C, Wueest RO, Karger DN, Holderegger R, Gugerli F, Manel S. Re-thinking the environment in landscape genomics. Trends Ecol Evol. 2023;38:261–74. [DOI] [PubMed] [Google Scholar]
- 106.Sork VL, Nason J, Campbell DR, Fernandez JF. Landscape approaches to historical and contemporary gene flow in plants. Trends Ecol Evol. 1999;14:219–24. [DOI] [PubMed] [Google Scholar]
- 107.Orsini L, Vanoverbeke J, Swillen I, Mergeay J, De Meester L. Drivers of population genetic differentiation in the wild: isolation by dispersal limitation, isolation by adaptation and isolation by colonization. Mol Ecol. 2013;22:5983–99. [DOI] [PubMed] [Google Scholar]
- 108.Manel S, Holderegger R. Ten years of landscape genetics. Trends Ecol Evol. 2013;28:614–21. [DOI] [PubMed] [Google Scholar]
- 109.Lee C-R, Mitchell-Olds T. Quantifying effects of environmental and geographical factors on patterns of genetic differentiation. Mol Ecol. 2011;20:4631–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schorr G, Pearman PB, Guisan A, Kadereit JW. Combining palaeodistribution modelling and phylogeographical approaches for identifying glacial refugia in Alpine Primula. J Biogeogr. 2013;40:1947–60. [Google Scholar]
- 111.Li L, Abbott RJ, Liu B, Sun Y, Li L, Zou J, Wang X, Miehe G, Liu J. Pliocene intraspecific divergence and plio-pleistocene range expansions within Picea Likiangensis (Lijiang spruce), a dominant forest tree of the Qinghai-Tibet Plateau. Mol Ecol. 2013;22:5237–55. [DOI] [PubMed] [Google Scholar]
- 112.Davis MB, Shaw RG. Range shifts and adaptive responses to quaternary climate change. Science. 2001;292(5517):673–9. [DOI] [PubMed] [Google Scholar]
- 113.Searle JB. Phylogeography: the history and formation of species. Heredity. 2000;85:201–201. [Google Scholar]
- 114.Petit RJ, Aguinagalde I, de Beaulieu J-L, Bittkau C, Brewer S, Cheddadi R, Ennos R, Fineschi S, Grivet D, Lascoux M, et al. Glacial refugia: hotspots but not melting pots of genetic diversity. Science. 2003;300:1563–5. [DOI] [PubMed] [Google Scholar]
- 115.Abbott RJ, Smith LC, Milne RI, Crawford RMM, Wolff K, Balfour J. Molecular analysis of plant migration and refugia in the Arctic. Science. 2000;289:1343–6. [DOI] [PubMed] [Google Scholar]
- 116.Hewitt GM. Genetic consequences of climatic oscillations in the Quaternary. Philosophical Trans Royal Soc B: Biol Sci. 2004;359:195–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rellstab C, Zoller S, Walthert L, Lesur I, Pluess AR, Graf R, Bodénès C, Sperisen C, Kremer A, Gugerli F. Signatures of local adaptation in candidate genes of oaks (Quercus spp.) with respect to present and future climatic conditions. Mol Ecol. 2016;25:5907–24. [DOI] [PubMed] [Google Scholar]
- 118.Pina-Martins F, Baptista J, Pappas G Jr., Paulo OS. New insights into adaptation and population structure of cork oak using genotyping by sequencing. Glob Change Biol. 2019;2:337–50. [DOI] [PubMed] [Google Scholar]
- 119.Jordan R, Hoffmann AA, Dillon SK, Prober SM. Evidence of genomic adaptation to climate in Eucalyptus microcarpa: implications for adaptive potential to projected climate change. Mol Ecol. 2017;26:6002–20. [DOI] [PubMed] [Google Scholar]
- 120.Borrell JS, Zohren J, Nichols RA, Buggs RJA. Genomic assessment of local adaptation in dwarf birch to inform assisted gene flow. Evol Appl. 2020;13:161–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wang B, Mojica JP, Perera N, Lee CR, Lovell JT, Sharma A, Adam C, Lipzen A, Barry K, Rokhsar DS, et al. Ancient polymorphisms contribute to genome-wide variation by long-term balancing selection and divergent sorting in Boechera stricta. Genome Biol. 2019;20:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Guerrero RF, Hahn MW. Speciation as a sieve for ancestral polymorphism. Mol Ecol. 2017;26:5362–8. [DOI] [PubMed] [Google Scholar]
- 123.Chase MA, Ellegren H, Mugal CF. Positive selection plays a major role in shaping signatures of differentiation across the genomic landscape of two independent Ficedula flycatcher species pairs. Evolution. 2021;75:2179–96. [DOI] [PubMed] [Google Scholar]
- 124.Lyu L, Wang D, Li L, Zhu Y, Jiang D, Liu J, Xu X. Polyphyly and species delimitation of Picea brachytyla (Pinaceae) based on population genetic data. J Syst Evol. 2021;59:515–23. [Google Scholar]
- 125.Alizon S, Kucera M, Jansen VAA. Competition between cryptic species explains variations in rates of lineage evolution. Proc Natl Acad Sci USA. 2008;105:12382–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Keenan RJ. Climate change impacts and adaptation in forest management: a review. Ann for Sci. 2015;72:145–67. [Google Scholar]
- 127.Browne L, Wright JW, Fitz-Gibbon S, Gugger PF, Sork VL. Adaptational lag to temperature in valley oak (Quercus lobata) can be mitigated by genome-informed assisted gene flow. Proc Natl Acad Sci USA. 2019;116:25179–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhang X, Sun Y, Landis JB, Zhang J, Yang L, Lin N, Zhang H, Guo R, Li L, Zhang Y et al. Genomic insights into adaptation to heterogeneous environments for the ancient relictual Circaeaster agrestis (Circaeasteraceae, Ranunculales). New Phytologist. 2020; 228:285–301. [DOI] [PubMed]
- 129.Chung MY, Suh Y, López-Pujol J, Nason JD, Chung MG. Clonal and fine-scale genetic structure in populations of a restricted Korean endemic, Hosta Jonesii (Liliaceae) and the implications for conservation. Ann Botany. 2005;96:279–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gilbert-Norton L, Wilson R, Stevens JR, Beard KH. A meta-analytic review of corridor effectiveness. Conserv Biol. 2010;24:660–8. [DOI] [PubMed] [Google Scholar]
- 131.Beier P, Noss RF. Do habitat corridors provide connectivity? Conserv Biol. 1998;12:1241–52. [Google Scholar]
- 132.Ma Y, Wang J, Hu Q, Li J, Sun Y, Zhang L, Abbott RJ, Liu J, Mao K. Ancient introgression drives adaptation to cooler and drier mountain habitats in a cypress species complex. Commun Biology. 2019;2:213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sexton JP, Hangartner SB, Hoffmann AA. Genetic isolation by environment or distance: which pattern of gene flow is most common? Evolution. 2014;68:1–15. [DOI] [PubMed] [Google Scholar]
- 134.Guerrant EO, Havens K, Maunder M. Ex situ plant conservation: supporting species survival in the wild. Chicago, IL: Island; 2004. [Google Scholar]
- 135.Yang H, Li J, Milne RI, Tao W, Wang Y, Miao J, Wang W, Ju T, Tso S, Luo J, et al. Genomic insights into the genotype-environment mismatch and conservation units of a Qinghai-Tibet Plateau endemic cypress under climate change. Evol Appl. 2022;15:919–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hanson J, Ellis RH. Progress and challenges in ex situ conservation of forage germplasm: grasses, herbaceous legumes and fodder trees. Plants. 2020;9:446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hoban S, Schlarbaum S. Optimal sampling of seeds from plant populations for ex-situ conservation of genetic biodiversity, considering realistic population structure. Biol Conserv. 2014;177:90–9. [Google Scholar]
- 138.Potts SG, Biesmeijer JC, Kremen C, Neumann P, Schweiger O, Kunin WE. Global pollinator declines: Trends, impacts and drivers. Trends Ecol Evol. 2010;25:345–53. [DOI] [PubMed] [Google Scholar]
- 139.Shi C, Xie Y, Guan D, Qin G. Transcriptomic analysis reveals adaptive evolution and conservation implications for the endangered Magnolia lotungensis. Genes 2024; 15. [DOI] [PMC free article] [PubMed]
- 140.Flanagan SP, Forester BR, Latch EK, Aitken SN, Hoban S. Guidelines for planning genomic assessment and monitoring of locally adaptive variation to inform species conservation. Evol Appl. 2018;11:1035–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Brunton AJ, Farleigh K, Ogbourne SM, Rossetto M, Schoeman DS, Conroy GC. The geno-geo-climate nexus: contributions of geographic and ecological factors in shaping the genomic divergence of two closely related threatened rainforest species of Fontainea Heckel (Euphorbiaceae). Landscape Ecol. 2024; 39.
- 142.De Lafontaine G, Prunier J, Gérardi S, Bousquet J. Tracking the progression of speciation: variable patterns of introgression across the genome provide insights on the species delimitation between progenitor-derivative spruces (Picea mariana × P. Rubens). Mol Ecol. 2015;24:5229–47. [DOI] [PubMed] [Google Scholar]
- 143.Sang Y, Long Z, Dan X, Feng J, Shi T, Jia C, Zhang X, Lai Q, Yang G, Zhang H et al. Genomic insights into local adaptation and future climate-induced vulnerability of a keystone forest tree in East Asia. Nat Commun. 2022; 13. [DOI] [PMC free article] [PubMed]
- 144.Brooks AN, Turkarslan S, Beer KD, Lo FY, Baliga NS. Adaptation of cells to new environments. Wiley Interdisciplinary Reviews: Syst Biology Med. 2011;3:544–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Andolfatto P. Adaptive evolution of non-coding DNA in Drosophila. Nature. 2005;437:1149–52. [DOI] [PubMed] [Google Scholar]
- 146.Wang J, Street NR, Scofield DG, Ingvarsson PK. Variation in linked selection and recombination drive genomic divergence during allopatric speciation of European and American aspens. Mol Biol Evol. 2016;33:1754–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Savolainen O, Lascoux M, Merila J. Ecological genomics of local adaptation. Nat Rev Genet. 2013;14:807–20. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequence reads have been deposited in the National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences, under BioProject accession number PRJCA028370.