Abstract
Background
Aberrant embryo implantation and suboptimal placentation can lead to (severe) complications such as preeclampsia and fetal growth restriction later in pregnancy. Current identification of high-risk pregnancies relies on a combination of risk factors, biomarkers, and ultrasound examinations, a relatively inaccurate approach.
Previously, aberrant DNA methylation due to placental hypoxia has been identified as a potential marker of placental insufficiency and, hence, potential (future) pregnancy complications. The goal of the Early Prediction of prEgnancy Complications Testing, or the ExPECT study, is to validate a genome-wide, cell-free DNA (cfDNA) methylation strategy to diagnose preeclampsia accurately. More importantly, the predictive potential of this strategy is also explored to reliably identify high-risk pregnancies early in gestation. Furthermore, a longitudinal study was conducted, including sequential blood samples from pregnant individuals experiencing both uneventful and complicated gestations, to assess the methylation dynamics of cfDNA throughout these pregnancies.
A significant strength of this study is its enzymatic digest, which enriches CpG-rich regions across the genome without the need for proprietary reagents or prior selection of regions of interest. This makes it useful for the cost-effective discovery of novel markers.
Results
Investigation of methylation patterns throughout pregnancy showed different methylation trends between unaffected and affected pregnancies. We detected differentially methylated regions (DMRs) in pregnancies complicated with preeclampsia as early as 12 weeks of gestation, with distinct differences in the methylation profile between early and late pregnancy. Two classification models were developed to diagnose and predict preeclampsia, demonstrating promising results on a small set of validation samples.
Conclusions
This study offers valuable insights into methylation changes at specific genomic regions throughout pregnancy, revealing critical differences between normal and complicated pregnancies. The power of noninvasive cfDNA methylation profiling was successfully proven, suggesting the potential to integrate this noninvasive approach into routine prenatal care.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13148-024-01798-5.
Keywords: Preeclampsia, Placental insufficiency, Cell-free DNA, CfDNA methylation, Epigenetics, Differentially methylated regions, DMRs, Prediction, Prenatal
Background
The placenta is a dynamic and temporary organ that plays a vital role in fetal development, involving finely tuned mechanisms such as appropriate cytotrophoblast invasion with spiral artery remodeling, extensive placental angiogenesis, and crucial fetal–maternal interface development. It also acts as a protective barrier against infections and teratogens and produces hormones essential for maintaining and regulating various stages of pregnancy. Placentation begins with the implantation of the blastocyst into the endometrium, inducing its transformation into the decidua. Extravillous trophoblasts from the blastocyst migrate into this decidua, modifying the spiral arteries of the uterus, a crucial step in establishing uteroplacental circulation. At the end of the first trimester, maternal blood flows through the placental villi, introducing a drastic increase in oxygen concentration. Adverse pregnancy outcomes such as preeclampsia and fetal growth restriction are assumed to result from dysfunction in one of these processes, leading to suboptimal uteroplacental perfusion and oxidative stress [1–3]. Several factors are known to increase the risk of developing preeclampsia, such as chronic hypertension, assisted reproduction, increased maternal BMI, and advanced age [2–6]. Additionally, it is hypothesized that couple-specific immune maladaptation, an important risk factor for primiparous individuals or a subsequent pregnancy with a new partner, could also play a significant role in causing insufficient trophoblast invasion, leading to the development of preeclampsia [7].
Preeclampsia is a pregnancy-related hypertensive disorder with a highly variable clinical presentation induced by maternal endothelial dysfunction, including proteinuria and/or maternal end-organ dysfunctions such as neurological complications, pulmonary edema, hematological issues, acute kidney injury, liver involvement, and uteroplacental dysfunction [2, 8, 9]. These complications pose a substantial risk for preterm birth, fetal growth restriction, and neurodevelopmental delay. After childbirth, a considerably increased risk for a maternal cardiovascular disorder remains, necessitating lifelong follow-up. This is particularly crucial for those who experienced early onset and/or severe preeclampsia [10]. Although this disease affects 2–5% of pregnancies worldwide [8, 11, 12] and the extensive research already conducted on preeclampsia, the exact pathophysiology remains unclear. Moreover, most preeclampsia patients did not exhibit a prior increased risk. Preeclampsia can be categorized into an early and late-onset disease [13], even though this division is still a topic of debate given the many diverse definitions of this disease and the incomplete knowledge of the underlying pathology. Early stage preeclampsia can be linked to poor placentation and fetal growth restriction, characterized by placental dysfunction involving incomplete spiral artery remodeling and undetectable abnormal placentation. In the later stage, this is followed by the onset of maternal symptoms. Mainly due to endoplasmic reticulum stress and oxidative stress in the syncytiotrophoblast, the ischemic placenta releases vasoactive molecules, proinflammatory cytokines, toxins, and syncytial fragments into the maternal bloodstream, leading to systemic endothelial dysfunction, intravascular inflammation, and activation of the hemostatic system [1, 2, 14–16]. Early onset preeclampsia is acknowledged as the severe type, often leading to fetal morbidity, mortality, and iatrogenic preterm delivery. Late-onset preeclampsia is associated with factors related to the growth and aging of the placenta, such as maternal obesity, environmental factors, multiple pregnancies, and large-for-gestational-age fetuses, leading to constrained intervillous perfusion [12].
The primary preventive treatment is a daily low dose of acetylsalicylic acid (aspirin), which lowers the incidence of early onset preeclampsia by half when administered before the 16th week of pregnancy [17–21]. Additionally, hypertension medication can be prescribed to manage maternal blood pressure. Fetal delivery is currently the only known ‘cure’ for severe preeclampsia, often resulting in a preterm birth.
A wide variety of clinical prediction tools and diagnostic approaches have been introduced worldwide, each incorporating variations of ultrasound observations, maternal characteristics, and medical history [22, 23]. Additionally, circulating angiogenic factors in plasma can be used as noninvasive biomarkers of placental health. An imbalance between the anti-angiogenic factor soluble fms-like tyrosine kinase 1 (sFlt-1) and the pro-angiogenic factor placental growth factor (PlGF) has been demonstrated, with their ratio tending to be elevated just before the onset of preeclampsia (1–4 weeks) [24–28]. Unfortunately, this latter approach is a short-term prediction tool, mainly applicable late in pregnancy, providing little opportunity for early preventive or therapeutic treatment. Instead, it serves as a tool to determine whether to proceed with urgent interventions, like hospital admission or delivery of the baby.
Postnatal investigations on placental tissue have been conducted to elucidate the underlying pathology of placental insufficiency. Microscopic examinations have identified localized areas of ischemic villous necrosis, diagnosing placental hypoxia [29]. The processes of placentation have been shown to correlate with altered epigenetic mechanisms such as histone modification, microRNAs, and DNA methylation, hereby inducing changes in placental gene expression. Indeed, altered gene expression profiles and dysregulation of biological pathways associated with preeclampsia have been identified in placental cells [30–34]. Genome-wide DNA methylation studies have revealed genome-wide hypo- and hypermethylation as well as specific imprinting patterns necessary to adapt to different intrinsic and environmental factors [35–39]. Furthermore, studies focusing on placental dysfunction have shown that placental disease can induce aberrant methylation patterns and, consequently, altered gene expression, primarily due to oxidative stress [40–46].
Although obtaining placental tissue during pregnancy would be invasive and therefore not feasible for routine screening, these investigations have provided valuable insights into placental pathology, indicating the potential for exploring the placental epigenome as a prenatal diagnostic or, preferably, a prenatal screening tool for pregnancy complications. A solution to tackle this issue is to analyze cell-free RNA (cfRNA) and cell-free DNA (cfDNA) present in maternal blood and originating from placental syncytiotrophoblast cells. Recently, liquid biopsies have been applied to explore adverse pregnancy outcomes resulting from placental dysfunction [47–55]. This approach allows for the noninvasive examination of placental-derived transcription profiles and (aberrant) methylation patterns.
The Early Prediction of prEgnancy Complications Testing or ExPECT study aims to develop a noninvasive screening test early in pregnancy to predict future placenta-related complications based on cfDNA methylation profiling. The two main objectives of the ExPECT study are (1) to confirm the feasibility of using cfDNA methylation data for the accurate diagnosis of pregnancies complicated by preeclampsia, and (2), with a greater clinical significance, to explore the predictive capacity of cfDNA methylation profiling during the early stages of pregnancy, to reliably identify high-risk pregnancies and thereby enable an improved, personalized prenatal care. To achieve the objectives of the ExPECT study, cfDNA from individuals with uneventful and complicated pregnancies was analyzed to develop the optimal prediction model based on unsupervised clustering of significant differentially methylated regions (DMRs). Furthermore, a longitudinal study was conducted, including sequential blood draws from individuals experiencing both uncomplicated and complicated gestations, to assess the methylation dynamics of cfDNA throughout their pregnancies.
Methods
Participants and sample collection
Since April 1st, 2019, all pregnant people followed at Ghent University Hospital have been asked to consent to four-weekly blood sampling throughout gestation and one sample after delivery. Additionally, all patients admitted to the maternal intensive care unit were asked for a blood sample at admission, and after delivery. Once delivered, the pregnancies of the participants were categorized as normal, complicated by preeclampsia with or without fetal growth restriction, complicated by fetal growth restriction without preeclampsia, or complicated by other pathologies (e.g., preterm premature rupture of the membranes or spontaneous preterm labor). Plasma samples were stored in a dedicated biobank, and relevant patient characteristics were recorded, including preconception BMI, pre-existing or chronic hypertension, prior history of preeclampsia, and maternal cardiovascular disease. Additionally, pregnancy details, such as fertility treatment, ultrasound findings, laboratory tests, noninvasive prenatal testing (NIPT), and aspirin treatment, were documented. Standardized definitions were applied to enhance the validity of its findings, addressing the issue of variability in preeclampsia definitions observed in other studies [8]. Data registration was done in REDCap (Research Electronic Data Capture) [56, 57], a web-based platform that facilitates research data capture with an intuitive interface, audit trails, automated export, and interoperability with external sources.
Patient and sample selection during symptomatic and presymptomatic stages
For the symptomatic cohort (diagnosing disease approach), 23 samples were chosen from 23 severe preeclampsia cases (e.g., gestational hypertension accompanied with maternal end-organ dysfunction). All samples were taken between 24 and 37 weeks of gestation (with an average gestational age (GA) of 31.6 weeks). Furthermore, 49 samples were selected from 49 controls (uneventful pregnancies without an increased risk), with an average GA of 31.0 weeks (Fig. 1B). The cases and controls were matched by selecting gestational age over 20 weeks, with a similar average GA at sampling per category.
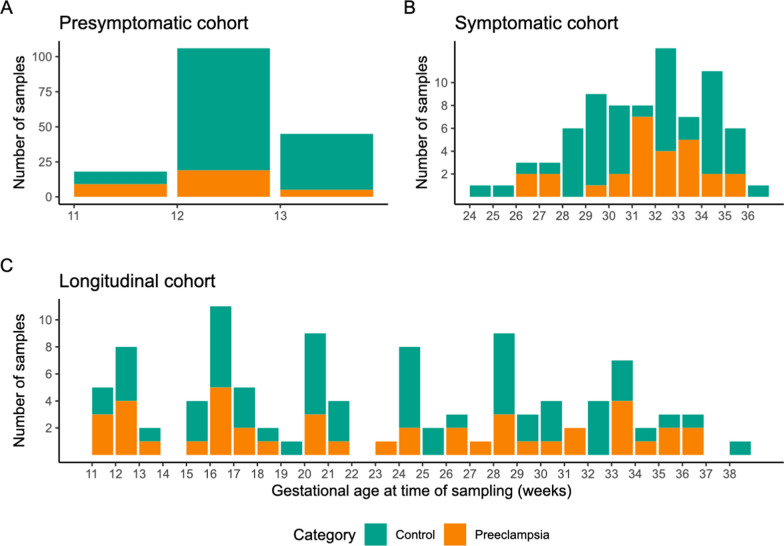
Fig. 1.
Overview of the sample collection per analysis cohort. A stacked bar chart showing the number of samples collected at given time points during pregnancy. Each bar represents a group of samples collected during the specified week of gestation. Colors show the sample type, controls in green, and preeclampsia (PE) cases in orange. Separate charts show the numbers of samples used in the different analyses, presymptomatic (A), symptomatic (B) and longitudinal (C) study
For the presymptomatic cohort (predicting disease approach), 33 samples from 33 cases were selected (with an average GA at blood sampling of 11.9 weeks), of which 25 later developed severe preeclampsia and eight a less severe form. 136 samples from 136 controls with term-birth were selected, again GA-matched at sampling (with an average GA of 12.2 weeks) (Fig. 1A).
For the longitudinal analysis, sequential blood samples of 25 individuals were investigated (Fig. 1C). A total of 104 samples, taken between 11 and 38 weeks of gestation, were analyzed, comprising 43 samples from 10 preeclamptic patients and 61 samples from 15 individuals with uncomplicated pregnancies.
All included participants had singleton pregnancies and were of predominantly Caucasian ethnicity (Supplementary Table 1). The samples in the different cohorts were not fully independent; most patients in the longitudinal cohort were also included in the presymptomatic cohort (n = 15). Overlap between the symptomatic and presymptomatic cohort is limited, with only three patients having samples included in both cohorts. (Supplementary Fig. 1).
Genome-wide methylation profiling through cfDNA reduced representation bisulfite sequencing (cf-RRBS)
Blood was collected in EDTA tubes and processed within 2–4 h post-collection. When not feasible, such as during weekends, blood was collected in PAXgene tubes to stabilize the maternal blood cells. Plasma separation was achieved through centrifugation at 1900 g for 15 min. Subsequently, the plasma samples were stored in a biobank at − 80 °C, aliquoted to prevent freeze–thaw cycles. Cell-free DNA (cfDNA) was extracted using the Maxwell RSC LV ccfDNA kit (Promega) according to the manufacturer’s instructions.
The cf-RRBS protocol is a cost-effective and automatable approach for methylation profiling of highly fragmented cfDNA, initially described by De Koker et al. [58] and adapted by Van Paemel et al. [59], using the EZ DNA Lightning kit (ZymoResearch). CpG-rich regions are targeted through restriction enzyme activity, and subsequent hairpin-shaped adaptors generate circular cfDNA with dual cut-sites. This process effectively removes non-circular undesired fragments, amplifies CpG-rich genomic regions, and preserves crucial methylation information, guaranteeing sufficient genome coverage and accurate quantification of the methylation status. The library concentrations were assessed by qPCR using the KAPA Library Quantification Kit for Illumina platforms (Kapa Biosystems), and samples were pooled equimolarly. Single read sequencing (100 cycles) was done using the NovaSeq 6000 instrument (Illumina), with dual indexing and a 2% PhiX spike-in. On average, 26.76 million reads [19.83–33.69] were obtained per sample.
Raw sequencing data were first demultiplexed using BCL Convert (v4.0.3; Illumina) and then processed with the nf-core/methylseq (v2.4.0) [60] pipeline. The Bismark [61] workflow of the pipeline was used for mapping and methylation extraction, with the additional RRBS flag to ensure correct trimming of the reads and the omission of the deduplication step. Reads were mapped with bowtie2 [62] to the GRCh38 reference genome. Bismark coverage files from the methylation extraction output were used to further analyze the CpG site methylation. From these coverage files, CpG clusters were defined based on location with a custom script. This script gathers coordinates for all CpG sites that were covered in all samples and orders these into groups. CpG clusters are defined based on specific criteria. First, each cluster must contain at least two CpG sites. Additionally, every CpG site in the cluster should be covered at least once, and the total coverage of the site in the cluster should be at least five. The maximum distance between CpG sites in a single cluster is 50 base pairs. Finally, the created clusters must be present in at least 50% of the samples included in the study. The methylation for these clusters was computed as a mean of the CpG methylation values per cluster after local likelihood smoothing with BSmooth [63]. The smoothed methylation values for these clusters were then used as an input for differentially methylated region (DMR) calculation with the R packages limma (v3.45.2) [64] and bsseq (v1.38.0) [63].
Cohort-level DMR investigation
Genome-wide methylation differences were calculated by generating a sample-wise mean methylation value for all included clusters, as described above. Wilcoxon rank-sum tests were performed to compute differences between sample groups. Samples from patients included in both cohorts were removed from the symptomatic dataset to assure independence (four control samples and one case sample). DMRs were computed with limma, and a linear model, including the category of the sample (case versus control), fetal sex, sequencing batch, and gestational age (in weeks) as main effects, was created based on the methylation clusters described above. Clusters that were significantly correlated with the category factor of the limma model were then extracted and further investigated. Significant DMRs were filtered based on their absolute log fold change (> 0.1) and Benjamini and Hochberg’s adjusted p value (< 0.01). This approach obtained the best results for the symptomatic sample cohort and was used on this cohort. These DMRs were visualized with the R package pheatmap (v1.0.12).
DMRs were annotated with overlapping genes, and a gene ontology enrichment analysis was performed using the goana function from the limma package with a false discovery rate cutoff of 0.05. Mean methylation values for genes of interest, such as soluble fms-like tyrosine kinase-1 (sFLT-1) and placental growth factor (PlGF), were computed by taking the mean across overlapping CpG clusters. For the longitudinal cohort, mean methylation levels were computed across the CpG clusters on a whole genome basis as well as on clusters overlapping the genes of interest. The DMRs computed on the symptomatic and presymptomatic cohorts, as described above, were also investigated in the longitudinal cohort. For this, DMRs were clustered using hierarchical clustering with complete linkage. Clustering trees were cut into four groups for visualization purposes. Mean methylation values were then computed on a sample-wise base for each of these four groups.
Prediction modeling
Exploratory machine learning prediction models were created with the R package tidymodels (v1.1.0). For this, the symptomatic and presymptomatic cohort were split into a training and testing set. DMRs were then recalculated with DMRfinder based on the samples in the training datasets. Only CpG sites with a minimum coverage of two in at least two samples of the control and preeclampsia set were included. DMRs were filtered based on size (> = 3 CpG sites), absolute log fold change (> 0.1), and q-cutoff (> 0.99 or < 0.01). The top 25 most significant DMRs from these training datasets were further used for model creation. Three classification models were evaluated: random forest, support vector machine, and logistic regression (with the ranger, kernlab and glmnet packages, respectively, as backends). Methylation values of training DMRs were used as predictors for disease status. Hyperparameter tuning was performed with a fivefold cross-validation using random oversampling examples (ROSE) to adjust for class imbalance in the analysis set of each validation fold. The final model performance was calculated using the original test set with sensitivity, specificity, and accuracy as quality metrics.
Code availability
All codes used for data processing and analysis are available on GitHub. A rendered report of the code can also be found in the same repo. https://github.com/CenterForMedicalGeneticsGhent/ExPECT-analysis
Results
Patient characteristics
Significant differences in primiparity of the pregnancy (p = 0.022) were found when comparing all pregnancies affected by preeclampsia (n = 56) versus unaffected controls (n = 183) (Supplementary Table 1). The mean GA at birth was 34.2 ± 3.45 standard deviation (SD) for complicated pregnancies and 39 ± 1.39 SD weeks for normal pregnancies (p < 0.001, Wilcoxon rank-sum test), indicating the associated higher incidence of preterm birth in the severe preeclampsia cohort. Significant differences between the two groups could be observed in fertility treatments (e.g., insemination, IVF/ICSI) and BMI (p = 0.066 and p = 0.056).
Methylation profiling
To explore the effects of established preeclampsia on DNA methylation, cfDNA blood samples from cases (n = 27) were collected after the onset of maternal symptoms. Control samples (n = 50) were collected at a similar GA. A total of 4,517,769 unique methylation loci were detected, with an average of 4,010,419 unique CpG sites per sample. To increase the overlap in covered methylation loci between samples, methylation values were smoothed with BSmooth. To further improve the power of the analysis, the methylation loci were clustered based on their position, as the methylation of most CpG loci is heavily correlated with that of nearby located sites [65]. 367,429 unique CpG clusters could be identified using a custom coordinate-based clustering method.
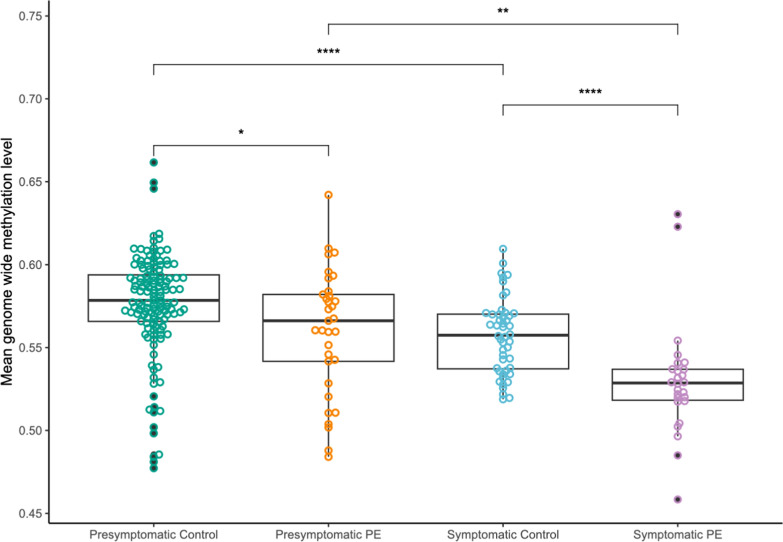
After batch correction, an overall hypomethylation was observed in symptomatic preeclampsia samples compared to controls, with an estimated difference of 0.032 (95% confidence interval (CI) 0.020–0.043) (Fig. 2).
Fig. 2.
General decrease in methylation during pregnancy, with a significant genome-wide hypomethylation in preeclampsia patients near late pregnancy. Box plots showing the mean genomic methylation level of controls and preeclampsia (PE) cases. Means were compared with Wilcoxon rank-sum tests using Bonferroni multiple testing correction. Methylation levels from presymptomatic samples, sampled between 11 and 14 weeks of gestation, show no significant difference. Methylation levels between cases and controls sampled after 20 weeks of gestation show a significantly lower methylation level of cases compared to controls. Significant methylation differences between presymptomatic and symptomatic groups can be observed for both preeclampsia cases and controls
Within the early pregnancy cohort, a significant difference was also observed, with preeclampsia cases having an estimated hypomethylation of 0.014 (95% CI 0.027–0.0033).
Identification of differentially methylated regions (DMRs)
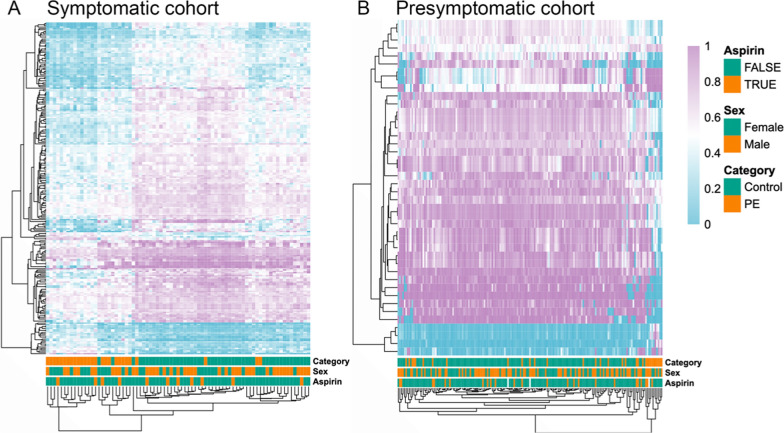
We observed a clear impact of the gestational age on the methylation patterns and, therefore, computed DMRs separately in the different groups, using the diagnostic approach using symptomatic samples and the predicting approach using presymptomatic samples. After correcting for batch, fetal sex, and gestational age, 53,994 DMRs between symptomatic preeclampsia patients and controls were discovered (p < 0.05). Of these, 21,499 had an absolute methylation difference larger than 10% between controls and preeclamptic patients. Gene set enrichment analysis of these regions identified multiple groups of genes related to developmental pathways. However, no gene ontology (GO) terms related to placental development were found to be significantly overrepresented. To test the power of the identified DMRs in differentiating between the sample categories, a hierarchical clustering of the samples is performed using the top 250 most significant DMRs (Fig. 3A). The sex of the fetus and aspirin treatment were not utilized in the clustering method. They were, however, included as annotations to facilitate the interpretation of whether these parameters influenced the clustering patterns and, consequently, had an impact on the methylation. We found no differences in clustering patterns based on fetal sex or aspirin treatment.
Fig. 3.
Significantly differently methylated regions (DMRs) enable distinct clustering of preeclampsia cases and controls during third trimester pregnancies. Heatmap figures show a hierarchical clustering of samples (columns) from the symptomatic analysis set (A) and the presymptomatic analysis set (B). The clustering is based on the methylation level of the top 250 significant DMRs for the symptomatic set and all 42 DMRs for the presymptomatic set. The sample category, fetal sex, and aspirin treatment are included as additional annotations per sample
In the presymptomatic case–control group, a similar approach yielded 67 DMRs (p < 0.05), of which 42 had an absolute methylation difference above 10% when compared to matched controls (p < 0.01). To assess the differentiation power, a clustering with these DMRs was performed (Fig. 3B), again with annotation of fetal sex and aspirin treatment, with, once more, no differences in clustering patterns based on each of these parameters.
When we compare the 21,499 DMRs from the symptomatic group with the 42 DMRs identified in the presymptomatic group, only two are present in both, representing, respectively, 0.0001% and 4.8% of the DMRs, with an overlap of 0.0001%.
Classification performance for preeclampsia diagnosis
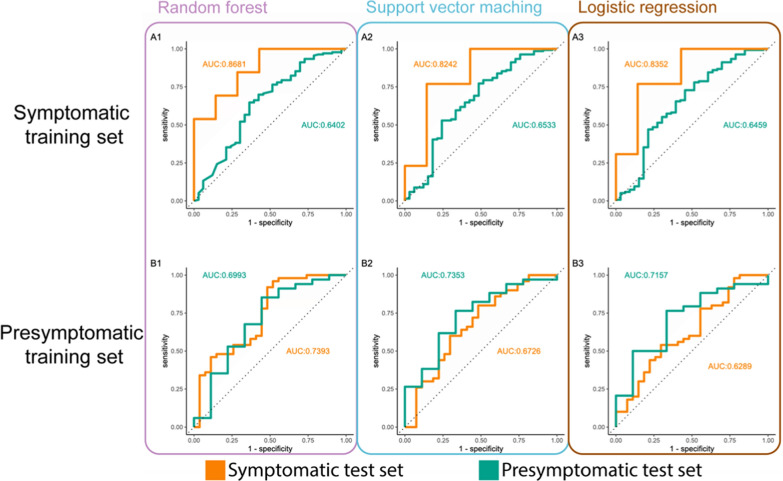
For the diagnostic approach, three preliminary classification models, based on the methylation of the top 25 most significant DMRs, were created to test the potential diagnostic power: random forest, support vector machine, and binomial logistic regression (Fig. 4A). For these models, the methylation values of the selected DMRs were the only used predictors for disease status. The training subset contained 57 samples (split in 5 validation folds of 45 and 12 samples for hyperparameter tuning), and models were tested using a validation set of 20 samples. Areas under the curve (AUC) of 0.868, 0.824, and 0.835 were obtained for the random forest, support vector machine, and regression model after receiver operator characteristic (ROC) analysis. Optimal models were selected based on the highest accuracy. The random forest and support vector machine models obtain a specificity of 100% and sensitivity of 57.1%, while the logistic regression model obtains a specificity of 61.5% and sensitivity of 85.7%.
Fig. 4.
Multiple machine learning classifiers allow successful classification and prediction of preeclampsia patients. Three different types of classification models were tested: random forest (1), support vector machine (2), and a weighted logistic regression model with elastic net regularization (3). Models in the top row (A) are trained on a symptomatic sample set (> 20 weeks gestational age, n = 57). Models in the bottom row (B) are trained on a presymptomatic sample set (between 11 and 14 weeks of gestation, n = 126). Curves are based on a symptomatic test set (n = 20) in orange and a presymptomatic test set in green (n = 43)
Prediction performance for preeclampsia risk stratification during early pregnancy
Identical model training was performed for the predictive approach using a training set of 126 first and early second-trimester samples (Fig. 4B). A test set of 43 samples was used for validation, and AUCs of 0.699, 0.735, and 0.716 were obtained for the random forest, support vector machine, and regression model, respectively. Optimal models were selected based on the highest accuracy. The random forest model obtained the highest specificity of 97% and a sensitivity of only 22%. The support vector machine achieved a specificity of 91.1% and a sensitivity of 33.3%, while the logistic regression model obtained a similar specificity of 88.2% and an equal sensitivity of 33.3%.
Additionally, models trained on early samples did not perform well in classifying late-pregnancy samples, and vice versa. Models trained on the presymptomatic data performed slightly better on the symptomatic cohort than the models trained on symptomatic data performed on the presymptomatic cohort. The first set of three models obtained AUCs of 0.739, 0.673, and 0.629, respectively, while the second set obtained AUCs of 0.640, 0.653, and 0.646, respectively (Fig. 4).
Longitudinal methylation profiling
When investigating the mean genomic methylation of sequential samples from pregnancies affected by preeclampsia compared to healthy pregnancies, no significant differences could be observed in the methylation trends across the pregnancy (Fig. 5A). Global genomic methylation appears to follow a similar pattern in both cases and controls, namely a decrease during the pregnancy.
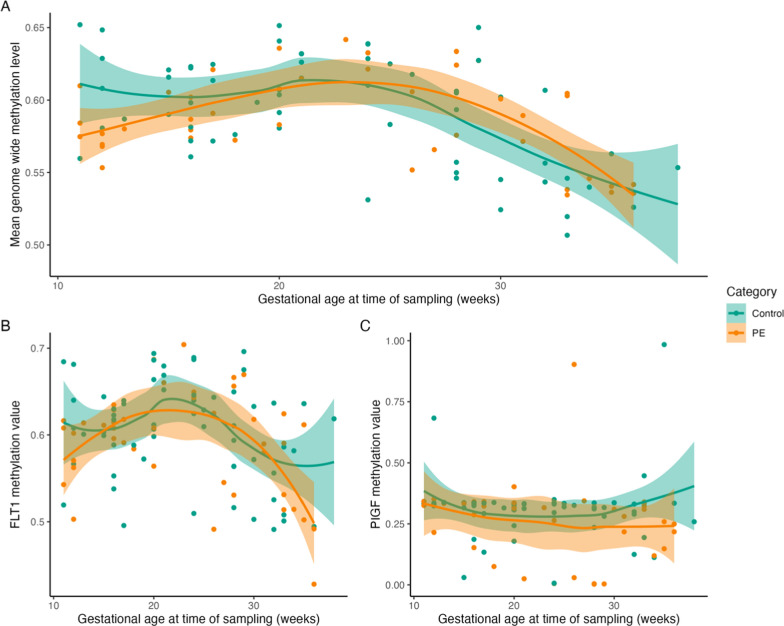
Fig. 5.
General genome-wide methylation trends across pregnancy differ between preeclampsia cases and controls. Mean whole genome methylation follows a similar trend for preeclampsia patients and controls across pregnancy duration. At 11–13 weeks of gestation, a lower global methylation is noted for preeclampsia cases (A). The methylation pattern of two preeclampsia-related genes, sFLT-1 (B) and PlGF (C), is slightly different for controls and preeclampsia patients. Smoothing was performed with local polynomial regression fitting, and standard error (SE) is shown as a band
The methylation trends of sFLT-1 and PIGF were also investigated (Fig. 5B, C). We observed a consistent methylation of PIGF throughout pregnancy for both pregnancies unaffected and affected by preeclampsia (Fig. 5B, C). Pregnancies affected by preeclampsia appear to maintain stable methylation. For sFLT-1, no significant changes in methylation could be observed in either cases or controls of the longitudinal cohort. However, when looking at the presymptomatic and symptomatic cohorts, a significant decrease in methylation could be observed for both cases and controls in the later stages of the pregnancy (Supplementary Fig. 4). No methylation differences could be observed for PIGF in these cohorts. (Supplementary Fig. 5).
However, when comparing the methylation trends of the DMRs identified during both the predictive and diagnostic stages of this study, there are apparent differences in the evolution of the methylation. To investigate if multiple genomic regions followed similar methylation trends across the duration of pregnancy, hierarchical clustering of DMRs discovered in symptomatic and presymptomatic samples was performed. The methylation values of regions contained in these clusters were plotted across the pregnancy duration (Supplementary Figs. 2 and 3). Trends in the methylation patterns of symptomatic DMRs seemed to diverge at the end of pregnancy duration, while the methylation patterns of presymptomatic DMRs seemed to converge. This further illustrates the difference between DMRs present early in pregnancy versus later in pregnancy. However, due to limited data points, the significance of these trends remains to be seen.
Lastly, the DMR calculation was reevaluated, employing the methodology as previously described, but now all longitudinal samples were included, meaning multiple samples per participant. Novel DMRs were computed with GA as a factor in the DMR modeling procedure with limma; subsequent samples from the same patient were not considered. This analysis identified a limited number (73) of DMRs between preeclampsia cases and healthy pregnancies that were statistically independent of GA. These DMRs prove insufficient for the creation of a classification model that would perform well throughout the duration of the pregnancy.
Discussion
This study revealed important insights into the epigenetic landscape of placental development, with a primary focus on diagnosing and predicting preeclampsia. Our findings offer a significant understanding of DMRs linked to pregnancy complications, providing crucial knowledge about methylation changes at specific genomic regions. By revealing critical differences between normal and complicated pregnancy, this study underscores the potential for further refinement of these regions through an expanded sample set, which appears to be a worthwhile prospective goal for additional analysis.
Genome-wide DNA methylation studies on placental tissue have shown that placental disease can induce aberrant methylation patterns and, hence, inferred gene expression, primarily due to oxidative stress [40, 46]. In general, genome-wide hypomethylation is observed, with hypo- and hypermethylation of specific promotor regions. This suggests that these regions are associated with disease progression and play a role in regulating early onset preeclampsia-specific gene expression [36, 40–42, 44, 46, 66–68].
Recent developments in liquid biopsies have revealed important biomarker discoveries for noninvasive disease exploration. These advancements have enabled early cancer detection, tumor identification, and monitoring of treatment responses [69]. The potential of noninvasive cfDNA profiling to predict preeclampsia has been investigated in several studies [47, 49, 51–54, 66]. Typically, there is a gradual decrease in cfDNA methylation levels throughout both normal and complicated pregnancies, particularly noticeable in the latter. In this prEgnancy Complications Testing or ExPECT study, we conducted analyses in both early, mid, and late stages of pregnancy for 25 pregnancies, allowing for a longitudinal assessment of the genome-wide methylation status. Indeed, we observed a general decrease in genome-wide methylation throughout pregnancy (Fig. 5A). Furthermore, we observe a significant reduction within both control and complicated pregnancies when distinguishing between general genome-wide methylation patterns during early and late pregnancy (Fig. 2). Importantly, we noted a substantial hypomethylation in late pregnancy among pregnancies complicated by preeclampsia compared to their early pregnancy stage and compared to the entire pregnancy duration of the controls. Interestingly, within the early pregnancy cohort, we detected a significant difference in methylation patterns between pregnancies that later developed preeclampsia and those that remained uncomplicated. This observation indicates that methylation changes associated with preeclampsia may be detectable early in pregnancy, potentially before clinical symptoms manifest.
The diagnostic precision of our developed test is promising. It could serve as a screening test for high-risk pregnancies during their third trimester, guiding decision-making concerning timely interventions. It could enhance disease identification and the distinction of preeclampsia from other less severe pregnancy-related disorders, such as pregnancy-induced hypertension (PIH), that might not progress to preeclampsia. A clinical application could be retesting at several time points starting from the onset of maternal symptoms, allowing for more precise monitoring of (suspected) preeclampsia.
More importantly, a key finding was the identification of 42 distinct DMRs during early pregnancy in cfDNA from patients who subsequently developed severe preeclampsia. We employed unsupervised clustering to analyze a vast dataset using a limited set of predefined data groups without relying on prior knowledge or labeled information. This approach aims to maximize the similarity within each cluster while simultaneously maximizing dissimilarity between different clusters. This process reveals valuable insights and relationships that may not be readily apparent; using various machine learning techniques, preliminary diagnostic models were created with a sensitivity of 71.4% and a specificity of 100%.
In this study’s diagnostic and predictive approaches, we identified different, significant DMRs during late and early pregnancy, with a limited overlap of 0.0001%. This disparity is also evident when applying the symptomatic prediction models to the presymptomatic sample cohort and vice versa, leading to low AUC values. This finding is significantly lower than previously reported data [51], where a 7.4% overlap of significant DMRs during and proceeding preeclampsia was observed. This indicates that the DMRs associated with preeclampsia early in pregnancy do not correspond to those during maternal symptoms. The higher percentage of overlapping DMRs in previous studies is most likely due to their targeted focus on placenta-driven regions. The difference in detecting significant DMRs in cfDNA during early and late pregnancy can be attributed to several factors. Different gene expression levels in the placenta throughout pregnancy depend on several fetal development stages, maternal health, and environmental influences. Therefore, methylation changes in the placenta over time are a natural phenomenon. Additionally, maternal blood flow through the placental villi typically starts at the end of the first trimester, significantly increasing oxygen levels and leading to placental stress, potentially initiating aberrant placentation. As a result, subtle methylation changes in the placenta may appear during this period, making it difficult to detect noninvasively due to the limited amount of placental cfDNA present in maternal circulation at that time point. In contrast, as pregnancy progresses and maternal disease develops, there is a more apparent association between cfDNA shed from multiple tissues, including the placenta, liver, and kidneys, and altered methylation patterns. This increased relevant cfDNA provides a more robust signal for detecting methylation changes, making it easier to observe significant preeclampsia-related changes compared to the earlier stages of pregnancy.
A significant strength of this ExPECT study lies in the non-selection approach, which overcomes the limitations of studies with predefined target regions, covering ~ 3% of the genome and ~ 80% of relevant CpG islands. This approach, enriching regions across the genome, might also explain the significantly reduced overlap between DMRs observed early in pregnancy and those observed during maternal symptoms, as we do not focus only on specific placental-driven regions. The success of our developed models can be attributed to the evaluation of DMRs from methylation data across the genome, without preselection of regions and the diverse set of patients in the study. However, due to a limited testing set of samples, rigorous validation of the obtained model is mandatory. A more extensive validation set of samples from presymptomatic preeclampsia patients should be used to confirm our findings and determine the screening test’s positive and negative predictive values.
Several other studies focused on the analysis of cfRNA, identifying aberrant gene expression profiles associated with GA, infection, preterm birth, and preeclampsia, and independent of maternal characteristics, like BMI and race [47, 48, 70–72]. As epigenetic changes are directly linked to gene expression, it is unsurprising that significant differences can also be measured here.
However, it is important to mention that our analysis process using cf-RRBS is considerably more straightforward than complex cfRNA analyses.
A detailed evaluation of genes and associated pathways was not the focus of our study. However, given the significance of the biomarkers PlGF and sFLT-1 in clinical prediction models, we did evaluate the methylation status of these two genes. sFLT-1 and PlGF biomarkers are able to predict preeclampsia ahead of symptom onset and prognostically characterize symptom severity after diagnosis [73, 74]. We found a slightly increased methylation of the anti-angiogenic factor PlGF in normal pregnancies after 30 weeks of gestation. A relatively stable methylation was observed in preeclampsia cases, indicating that more PlGF will be expressed late in these pregnancies compared to normal controls. However, the methylation values of PIGF are less reliable in our data due to the limited number of CpG sites we pick up related to the gene (three clusters). For the pro-angiogenic factor sFLT-1, a drop in methylation was observed in preeclampsia cases after 30 weeks of gestation; in control pregnancies, a smaller decrease was observed, which could result in an increased sFLT-1 gene expression late in preeclamptic pregnancies. The methylation values of sFLT-1 are more reliable due to a higher number of data points (23 clusters). Hence, expression values of sFLT-1 could lie significantly higher in preeclampsia cases than in control pregnancies. At the same time, we cannot draw significant conclusions about PIGF based on our data.
Gestational age is an important parameter influencing cfDNA methylation levels. Obesity, associated with a lower fetal fraction, increases methylation during the first trimester [66]. However, other variables like maternal age, fetal gender, smoking, alcohol and drug use, chemical exposure, and nutrition are also associated with changed methylation patterns [50, 68]. We matched cases and controls for GA to address the potential bias from naturally induced changes in placental and cfDNA methylation. Other patient characteristics such as BMI, aspirin treatment, chronic hypertension, previous history of preeclampsia, and maternal cardiovascular disease were not matched. De Borre et al. [51] conducted an elaborate matching strategy and found no significant effect of BMI, parity, a positive history of preeclampsia, or disease severity. Therefore, we opted against matching for these patient characteristics, creating a model that requires minimal patient information. This approach reduces the risk of errors or delays due to incomplete patient data and is objective by excluding operator bias, such as those from ultrasound examinations or inaccurate blood pressure measurements. The clinical parameters BMI, primiparity of the pregnancy, and fertility treatment were included as annotations in the unsupervised clustering but showed no clear relevance (data not shown). This lack of significance supports our decision to exclude them from further analysis. An important positive influencing factor on placental development is aspirin treatment [75]. However, for the predictive power of our models, it is important to note that blood sampling occurs before 14 weeks when no or little effect of this aspirin treatment has occurred yet. For this reason, we did not include aspirin treatment in our models.
The National Institute for Health and Care Excellence (NICE, UK) and the American College of Obstetricians and Gynecologists (ACOG) have made recommendations based on maternal risk factors such as hypertensive disorder during a previous pregnancy and high BMI [76, 77]. However, these risk factors have a poor predictive performance, with a detection rate of only 41% when following the NICE scoring system [78]. Numerous studies aimed to enhance risk assessment, leading to the development of various models [22, 23, 79–83]. The Fetal Medicine Foundation (FMF) developed an algorithm that improved first trimester risk assessment by combining maternal factors, biomarkers (PlGF and Pregnancy-associated plasma protein A or PAPP-A), ultrasound-based measurements (mean uterine artery resistance) and mean arterial blood pressure further. This approach resulted in an improved detection rate of 82% [84–88]. De Borre et al. [51] included the extended prior risk model (ePRM) [89] into their prediction model, improving the sensitivity of their model. We did not integrate an existing clinical model into our prediction model because, in practice, obstetricians use various parameters and guidelines. These clinical screening models rely on different combinations of maternal characteristics and medical history, which can lack sufficient power or reliability for effective risk prediction. Additionally, the interpretation of specialized ultrasound examinations, blood biomarkers, and blood pressure measurements can be subjective and may not accurately reflect the actual condition, often influenced by operator bias. To facilitate the implementation of our prediction model into obstetric health care, we aimed to develop a genetic model that can easily be introduced as an independent risk factor determination, guiding each obstetrician in the prenatal follow-up of their patients.
cf-RRBS is a cost-efficient, open-source technology that is easy to implement and can be automated. It requires minimal high-cost investments, primarily an Illumina sequencing platform. The library preparation is a single-tube procedure and does not rely on expensive target probes or commercial kits. Additionally, our data analysis workflow is freely available and straightforward to implement, making it accessible for various applications in research and clinical settings. Another advantage of the cf-RRBS method is the ability to perform copy number analysis [59]. This would allow for a more comprehensive prenatal screening to simultaneously assess the risk of preeclampsia and to screen for fetal aneuploidies. This would broaden the clinical application and is worthwhile to explore further. However, large-scale validation studies must be conducted before this method can be implemented in the context of noninvasive prenatal testing (NIPT).
While cf-RRBS effectively assesses cfDNA methylation, novel long-read sequencing technologies now offer a more holistic view. This expands our understanding beyond methylation by incorporating end motif analysis, and fragment size evaluation, and tissue of origin determination, ultimately enabling more comprehensive characterization of complex pathologies [90, 91]. For example, long-read sequencing by Oxford Nanopore Technologies (ONT) has been introduced in liquid biopsy studies to identify cell type, cancer-specific methylation changes, and cancer-associated fragmentation signatures [92, 93]. This comprehensive analysis enables a deeper insight into the biological processes underlying various diseases. Despite its potential, the application of fragmentomics in routine diagnostics requires further investments to manage the complex data analysis.
Conclusions
Improving the accuracy of identifying pregnant individuals at risk of developing placental dysfunction would enable improved and more precise prenatal care, leading to a reduction or even prevention of adverse pregnancy outcomes, and could avoid overtreatment. Previous studies have established the identification of disease-associated methylation changes in the placenta. We have translated this identification process into a noninvasive strategy via cfDNA methylation profiling. Our approach can be used to identify and monitor preeclampsia during third-trimester pregnancy to enable more targeted prenatal interventions that would lead to improved pregnancy outcomes. The focus of the ExPECT study was to improve the reliability of identifying high-risk pregnancies early in gestation, a significant step forward in prenatal care. Our findings reveal promising results. However, we acknowledge the need to expand the test set and include additional hospitals to further validate and improve the model’s performance. Nonetheless, our noninvasive cfDNA analysis has demonstrated its potential to be introduced into daily diagnostic practice as a diagnostic and predictive tool for placental dysfunction, aiming to minimize adverse pregnancy outcomes and reduce unnecessary treatments.
Supplementary Information
Acknowledgements
We express our gratitude for the exceptional support provided in patient counseling and inclusion, as well as in the administration, processing, and laboratory work: Isabel Brisard, Dimitri Broucke, Geert De Vos, Eveline Debals, Peter Degrave, Liesbet Demaegd, Charlotte Opsomer, midwives and study team Women’s Clinic.
Abbreviations
- ACOG
American College of Obstetricians and Gynecologists
- AUC
Area under the curve
- BMI
Body mass index
- cfDNA
Cell-free DNA
- cfRNA
Cell-free RNA
- cf-RRBS
Cell-free DNA Reduced Representation Bisulfite Sequencing
- CI
Confidence interval
- DMR
Differentially methylated region
- ePRM
Extended prior risk model
- ExPECT
Early Prediction of Pregnancy Complications Testing
- FMF
Fetal Medicine Foundation
- GA
Gestational age
- GO
Gene ontology
- IVF/ICSI
In vitro fertilization/intracytoplasmic sperm injection
- NICE
National Institute for Health and Care Excellence
- NIPT
Noninvasive prenatal testing
- PAPP-A
Pregnancy-associated plasma protein A
- PE
Preeclampsia
- PlGF
Placental growth factor
- REDCap
Research Electronic Data Capture
- ROC
Receiver operator characteristic
- ROSE
Random oversampling examples
- SD
Standard deviation
- SE
Standard error
- sFlt-1
Soluble fms-like tyrosine kinase 1
Author contributions
MB and BVG prepared the manuscript. MB organized the sample collection, designed and coordinated the patient database, supervised the experimental processes in the lab, and assisted with the interpretation of the data. BVG performed the analysis and interpretation of the data. ID and SD assisted with the collection and interpretation of the clinical information. ID and BM are the project’s principal investigators. All authors reviewed and approved the final manuscript.
Funding
Industrieel Onderzoeksfonds (IOF): F2020IOF STarTT035.
Availability of data and materials
All codes used for data processing and analysis are available on GitHub. A rendered report of the code can also be found in the same repo. https://github.com/CenterForMedicalGeneticsGhent/ExPECT-analysis.
Declarations
Ethics approval and consent to participate
The Ethical Committee of the University Hospital Ghent approved all research included in this study (2019/0056). Written informed consent was obtained from all participants under the approval of the Ethical Committee.
Consent for publication
Applicable.
Competing interests
All authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Shared first authors: Baetens Machteld and Van Gaever Bram.
Shared last authors: Dehaene Isabelle and Menten Björn.
References
- 1.Redman CWG, Staff AC. Preeclampsia, biomarkers, syncytiotrophoblast stress, and placental capacity. Am J Obstet Gynecol. 2015;213:S9.e1-S9.e4. [DOI] [PubMed] [Google Scholar]
- 2.Dimitriadis E, et al. Pre-eclampsia. Nat Rev Dis Prim. 2023;9:1–22. [DOI] [PubMed] [Google Scholar]
- 3.Burton GJ, Redman CW, Roberts JM, Moffett A. Pre-eclampsia: pathophysiology and clinical implications. BMJ. 2019. 10.1136/bmj.l2381. [DOI] [PubMed]
- 4.Sites CK, et al. Embryo cryopreservation and preeclampsia risk. Fertil Steril. 2017;108:784–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Santos S, et al. Impact of maternal body mass index and gestational weight gain on pregnancy complications: an individual participant data meta-analysis of European, North American and Australian cohorts. BJOG An Int J Obstet Gynaecol. 2019;126:984–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mayrink J, et al. Incidence and risk factors for Preeclampsia in a cohort of healthy nulliparous pregnant women: a nested case-control study. Sci Rep. 2019;9:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robillard PY, Dekker G, Scioscia M, Saito S. Progress in the understanding of the pathophysiology of immunologic maladaptation related to early-onset preeclampsia and metabolic syndrome related to late-onset preeclampsia. Am J Obstet Gynecol. 2022;226:S867–75. [DOI] [PubMed] [Google Scholar]
- 8.Magee LA, et al. The 2021 International Society for the Study of Hypertension in Pregnancy classification, diagnosis & management recommendations for international practice. Pregnancy Hypertens. 2022;27:148–69. [DOI] [PubMed] [Google Scholar]
- 9.Roberts JM. Preeclampsia epidemiology(ies) and pathophysiology(ies) James. Best Pract Res Clin Obstet Gynaecol. 2024;94:102480. [DOI] [PubMed] [Google Scholar]
- 10.Pittara T, Vyrides A, Lamnisos D, Giannakou K. Pre-eclampsia and long-term health outcomes for mother and infant: an umbrella review. BJOG An Int J Obstet Gynaecol. 2021;128:1421–30. [DOI] [PubMed] [Google Scholar]
- 11.Poon LC, et al. The International Federation of Gynecology and Obstetrics (FIGO) initiative on pre-eclampsia: a pragmatic guide for first-trimester screening and prevention. Int J Gynecol Obstet. 2019;145:1–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lisonkova S, Joseph KS. Incidence of preeclampsia: Risk factors and outcomes associated with early-versus late-onset disease. Am J Obstet Gynecol. 2013;209(544):e1-544.e12. [DOI] [PubMed] [Google Scholar]
- 13.Saito S. Preeclampsia—basic, genomic, and clinical. Berlin: Springer; 2018. [Google Scholar]
- 14.Burton GJ, Yung HW. Endoplasmic reticulum stress in the pathogenesis of early-onset pre-eclampsia. Pregnancy Hypertens. 2011;1:72–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu XQ, Zhang L. Mitochondrial dysfunction in the pathogenesis of endothelial dysfunction. Curr Hypertens Rep. 2022;24:157–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Phipps EA, Thadhani R, Benzing T, Karumanchi SA. Pre-eclampsia: pathogenesis, novel diagnostics and therapies. Nat Rev Nephrol. 2019;15:275–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rolnik DL, et al. Aspirin versus placebo in pregnancies at high risk for preterm preeclampsia. N Engl J Med. 2017;377:613–22. [DOI] [PubMed] [Google Scholar]
- 18.Robillard P-Y, et al. Preeclampsia in 2023: time for preventing early onset- and term preeclampsia: The paramount role of gestational weight gain. J Reprod Immunol. 2023;158:103968. [DOI] [PubMed] [Google Scholar]
- 19.Rolnik DL, et al. Early screening and prevention of preterm pre-eclampsia with aspirin: time for clinical implementation. Ultrasound Obstet Gynecol. 2017;50:551–6. [DOI] [PubMed] [Google Scholar]
- 20.Rolnik DL, Nicolaides KH, Poon LC. Prevention of preeclampsia with aspirin. Am J Obstet Gynecol. 2022;226:S1108–19. [DOI] [PubMed] [Google Scholar]
- 21.Rolnik DL, et al. Aspirin for evidence-based preeclampsia prevention trial: effects of aspirin on maternal serum pregnancy-associated plasma protein A and placental growth factor trajectories in pregnancy. Am J Obstet Gynecol. 2023. 10.1016/j.ajog.2023.12.031. [DOI] [PubMed]
- 22.Al-Rubaie ZTA, Askie LM, Ray JG, Hudson HM, Lord SJ. The performance of risk prediction models for pre-eclampsia using routinely collected maternal characteristics and comparison with models that include specialised tests and with clinical guideline decision rules: a systematic review. BJOG Int J Obstet Gynaecol. 2016;123:1441–52. [DOI] [PubMed] [Google Scholar]
- 23.Elawad T, et al. Risk factors for pre-eclampsia in clinical practice guidelines: comparison with the evidence. BJOG Int J Obstet Gynaecol. 2024;131:46–62. [DOI] [PubMed] [Google Scholar]
- 24.Zeisler H, et al. Predictive value of the sFlt-1: PlGF ratio in women with suspected preeclampsia. N Engl J Med. 2016;374:13–22. [DOI] [PubMed] [Google Scholar]
- 25.Verlohren S, et al. Clinical interpretation and implementation of the sFlt-1/PlGF ratio in the prediction, diagnosis and management of preeclampsia. Pregnancy Hypertens. 2022;27:42–50. [DOI] [PubMed] [Google Scholar]
- 26.Verlohren S, Dröge LA. The diagnostic value of angiogenic and antiangiogenic factors in differential diagnosis of preeclampsia. Am J Obstet Gynecol. 2022;226:S1048–58. [DOI] [PubMed] [Google Scholar]
- 27.Melo DCS, et al. The role of the soluble fms-like tyrosine kinase-1/placental growth factor (sFlt-1/PIGF) - Ratio in clinical practice in obstetrics: diagnostic and prognostic value. J Perinat Med. 2023;51:896–903. [DOI] [PubMed] [Google Scholar]
- 28.Klein E, et al. Influence of the sFlt-1/PlGF ratio on clinical decision-making in women with suspected preeclampsia. PLoS One. 2016;11:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roberts JM. Pathophysiology of ischemic placental disease. Semin Perinatol. 2014;38:139–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winn VD, Gormley M, Fisher SJ. The impact of preeclampsia on gene expression at the maternal-fetal interface. Pregnancy Hypertens. 2011;1:100–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kang JH, et al. Preeclampsia leads to dysregulation of various signaling pathways in placenta. J Hypertens. 2011;29:928–36. [DOI] [PubMed] [Google Scholar]
- 32.Várkonyi T, et al. Microarray profiling reveals that placental transcriptomes of early-onset HELLP syndrome and preeclampsia are similar. Placenta. 2011;32:S21–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benton SJ, Leavey K, Grynspan D, Cox BJ, Bainbridge SA. The clinical heterogeneity of preeclampsia is related to both placental gene expression and placental histopathology. Am J Obstet Gynecol. 2018;219(604):e1-604.e25. [DOI] [PubMed] [Google Scholar]
- 34.Leavey K, et al. Unsupervised placental gene expression profiling identifies clinically relevant subclasses of human preeclampsia. Hypertension. 2016;68:137–47. [DOI] [PubMed] [Google Scholar]
- 35.Schroeder DI, et al. The human placenta methylome. Proc Natl Acad Sci U S A. 2013;110:6037–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Novakovic B, et al. Evidence for widespread changes in promoter methylation profile in human placenta in response to increasing gestational age and environmental/stochastic factors. BMC Genomics. 2011. 10.1186/1471-2164-12-529. [DOI] [PMC free article] [PubMed]
- 37.Nelissen ECM, van Montfoort APA, Dumoulin JCM, Evers JLH. Epigenetics and the placenta. Hum Reprod Update. 2011;17:397–417. [DOI] [PubMed] [Google Scholar]
- 38.Norton C, et al. Altered epigenetic profiles in the placenta of preeclamptic and intrauterine growth restriction patients. Cells. 2023;12:1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ashraf UM, Hall DL, Rawls AZ, Alexander BT. Epigenetic processes during preeclampsia and effects on fetal development and chronic health. Clin Sci. 2021;135:2307–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lim JH, et al. Epigenome-wide DNA methylation profiling of preeclamptic placenta according to severe features. Clin Epigenetics. 2020;12:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anton L, Brown AG, Bartolomei MS, Elovitz MA. Differential methylation of genes associated with cell adhesion in preeclamptic placentas. PLoS One. 2014;9:100148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blair JD, et al. Widespread DNA hypomethylation at gene enhancer regions in placentas associated with early-onset pre-eclampsia. Mol Hum Reprod. 2013;19:697–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van den Berg CB, et al. Early- and late-onset preeclampsia and the DNA methylation of circadian clock and clock-controlled genes in placental and newborn tissues. Chronobiol Int. 2017;34:921. [DOI] [PubMed] [Google Scholar]
- 44.Yuen RKC, Pẽaherrera MS, Von Dadelszen P, McFadden DE, Robinson WP. DNA methylation profiling of human placentas reveals promoter hypomethylation of multiple genes in early-onset preeclampsia. Eur J Hum Genet. 2010;18:1006–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Almomani SN, et al. Identification and validation of DNA methylation changes in pre-eclampsia. Placenta. 2021;110:16–23. [DOI] [PubMed] [Google Scholar]
- 46.Chu T, et al. Comprehensive analysis of preeclampsia-associated DNA methylation in the placenta. PLoS One. 2014;9:107318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moufarrej MN, et al. Early prediction of preeclampsia in pregnancy with cell-free RNA. Nature. 2022;602:689–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moufarrej MN, Bianchi DW, Shaw GM, Stevenson DK, Quake SR. Noninvasive prenatal testing using circulating DNA and RNA: advances, challenges, and possibilities. Annu Rev. 2023;6:397–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moufarrej MN, Winn VD, Quake SR. Cell-free nucleic acids for early prediction of preeclampsia. Curr Hypertens Rep. 2023;26:175. [DOI] [PubMed] [Google Scholar]
- 50.Del Vecchio G, et al. Cell-free DNA methylation and transcriptomic signature prediction of pregnancies with adverse outcomes. Epigenetics. 2021;16:642–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De Borre M, et al. Cell-free DNA methylome analysis for early preeclampsia prediction. Nat Med. 2023;29:2206–15. [DOI] [PubMed] [Google Scholar]
- 52.He W, et al. Epigenetic phenotype of plasma cell-free DNA in the prediction of early-onset preeclampsia. J Obstet Gynaecol. 2023. 10.1080/01443615.2023.2282100. [DOI] [PubMed]
- 53.Jensen TJ, et al. Whole genome bisulfite sequencing of cell-free DNA and its cellular contributors uncovers placenta hypomethylated domains. Genome Biol. 2015;16:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spinelli M, et al. Hypertensive disorders of pregnancy share common cfDNA methylation profiles. Sci Rep. 2022;12:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chu T, Shaw P, McClain L, Simhan H, Peters D. High-resolution epigenomic liquid biopsy for noninvasive phenotyping in pregnancy. Prenat Diagn. 2021;41:61–9. [DOI] [PubMed] [Google Scholar]
- 56.Harris PA, et al. Research electronic data capture (REDCap)—a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inf. 2009;11:687–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Harris P, et al. The REDCap consortium: Building an international community of software platform partners. J Biomed Inform. 2019;95:103208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.De Koker A, van Paemel R, De Wilde B, De Preter K and Callewaert N. A versatile method for circulating cell-free DNA methylome profiling by reduced representation bisulfite sequencing. bioRxiv. 2019;663195.
- 59.Van Paemel R, et al. Minimally invasive classification of pediatric solid tumors using reduced representation bisulfite sequencing of cell-free DNA: a proof-of-principle study. Epigenetics. 2021;16:196–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ewels P et al. nf-core/methylseq: Huggy mollusc (10.5281/zenodo.1343417). (2024).
- 61.Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Langmead B, Salzber S. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hansen KD, Langmead B, Irizarry RA. BSmooth: from whole genome bisulfite sequencing reads to differentially methylated regions. Genome Biol. 2012. 10.1186/gb-2012-13-10-r83. [DOI] [PMC free article] [PubMed]
- 64.Ritchie ME, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Angeloni A, Bogdanovic O. Sequence determinants, function, and evolution of CpG islands. Biochem Soc Trans. 2021;49:1109–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cirkovic A, et al. Systematic review supports the role of DNA methylation in the pathophysiology of preeclampsia: a call for analytical and methodological standardization. Biol Sex Differ. 2020;11:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Herzog EM, et al. Early- and late-onset preeclampsia and the tissue-specific epigenome of the placenta and newborn. Placenta. 2017;58:122–32. [DOI] [PubMed] [Google Scholar]
- 68.Wilson SL, Robinson WP. Utility of DNA methylation to assess placental health. Placenta. 2018;64:S23–8. [DOI] [PubMed] [Google Scholar]
- 69.Lone SN, et al. Liquid biopsy: a step closer to transform diagnosis, prognosis and future of cancer treatments. Mol Cancer. 2022;21:1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rasmussen M, et al. RNA profiles reveal signatures of future health and disease in pregnancy. Nature. 2022;601:422–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Moufarrej MN, Wong RJ, Shaw GM, Stevenson DK, Quake SR. Investigating pregnancy and its complications using circulating cell-free RNA in women’s blood during gestation. Front Pediatr. 2020;8:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tsui NBY, et al. Maternal plasma RNA sequencing for genome-wide transcriptomic profiling and identification of pregnancy-associated transcripts. Clin Chem. 2014;60:954. [DOI] [PubMed] [Google Scholar]
- 73.Levine RJ, et al. Circulating angiogenic factors and the risk of preeclampsia. N Engl J Med. 2004;49:672–83. [DOI] [PubMed] [Google Scholar]
- 74.Thadhani R, et al. Circulating angiogenic factor levels in hypertensive disorders of pregnancy. NEJM Evid. 2022;1:1–13. [DOI] [PubMed] [Google Scholar]
- 75.Aerden M, De Borre M, Thienpont B. Cell-free DNA methylation-based preeclampsia prediction: a journey to improve maternal health. Prenat Diagn. 2023;44:418. [DOI] [PubMed] [Google Scholar]
- 76.NICE. National collaboration Centre of Women’s and Children’s Health (UK). Hypertens. Pregnancy Manag. Hypertens. Disorders During Pregnancy London; RCOG. (2010).
- 77.O’Gorman N, et al. Multicenter screening for pre-eclampsia by maternal factors and biomarkers at 11–13 weeks’ gestation: comparison with NICE guidelines and ACOG recommendations. Ultrasound Obstet Gynecol. 2017;49:756–60. [DOI] [PubMed] [Google Scholar]
- 78.Tan MY, et al. Comparison of diagnostic accuracy of early screening for pre-eclampsia by NICE guidelines and a method combining maternal factors and biomarkers: results of SPREE. Ultrasound Obstet Gynecol. 2018;51:743–50. [DOI] [PubMed] [Google Scholar]
- 79.O’Gorman N, et al. Competing risks model in screening for preeclampsia by maternal factors and biomarkers at 11–13 weeks gestation. Am J Obstet Gynecol. 2016;214(103):e1-103.e12. [DOI] [PubMed] [Google Scholar]
- 80.Wright D, Syngelaki A, Akolekar R, Poon LC, Nicolaides KH. Competing risks model in screening for preeclampsia by maternal characteristics and medical history. Am J Obstet Gynecol. 2015;213(62):e1-62.e10. [DOI] [PubMed] [Google Scholar]
- 81.Poon LCY, Kametas NA, Chelemen T, Leal A, Nicolaides KH. Maternal risk factors for hypertensive disorders in pregnancy: a multivariate approach. J Hum Hypertens. 2010;24:104–10. [DOI] [PubMed] [Google Scholar]
- 82.Poon LCY, Kametas NA, Maiz N, Akolekar R, Nicolaides KH. First-trimester prediction of hypertensive disorders in pregnancy. Hypertension. 2009;53:812–8. [DOI] [PubMed] [Google Scholar]
- 83.Chaemsaithong P, Sahota DS, Poon LC. First trimester preeclampsia screening and prediction. Am J Obstet Gynecol. 2022;226:S1071–97. [DOI] [PubMed] [Google Scholar]
- 84.De Castro Rezende KB, et al. Performance of fetal medicine foundation software for pre-eclampsia prediction upon marker customization: cross-sectional study. J Med Internet Res. 2019;21:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Riishede I, et al. Pre-eclampsia screening in Denmark (PRESIDE): national validation study. Ultrasound Obstet Gynecol. 2023;61:682–90. [DOI] [PubMed] [Google Scholar]
- 86.Zwertbroek EF, et al. Performance of the FMF first-trimester preeclampsia-screening algorithm in a high-risk population in the Netherlands. Fetal Diagn Ther. 2021;48:103–11. [DOI] [PubMed] [Google Scholar]
- 87.Guizani M, et al. First-trimester combined multimarker prospective study for the detection of pregnancies at a high risk of developing preeclampsia using the fetal medicine foundation-algorithm. Fetal Diagn Ther. 2018;43:266–73. [DOI] [PubMed] [Google Scholar]
- 88.Lee NMW, Chaemsaithong P, Poon LC. Prediction of preeclampsia in asymptomatic women. Best Pract Res Clin Obstet Gynaecol. 2023;92:102436. [DOI] [PubMed] [Google Scholar]
- 89.Wright D, Wright A, Nicolaides KH. The competing risk approach for prediction of preeclampsia. Am J Obstet Gynecol. 2020;223:12–23. [DOI] [PubMed] [Google Scholar]
- 90.Qi T, et al. Cell-free DNA fragmentomics: the novel promising biomarker. Int J Mol Sci. 2023;24:1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ding SC, Lo YMD. Cell-Free DNA fragmentomics in liquid biopsy. Diagnostics. 2022;12:978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Katsman E, et al. Detecting cell-of-origin and cancer-specific methylation features of cell-free DNA from nanopore sequencing. Genome Biol. 2022;23:1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.van der Pol Y, et al. Real-time analysis of the cancer genome and fragmentome from plasma and urine cell-free DNA using nanopore sequencing. EMBO Mol Med. 2023;15:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All codes used for data processing and analysis are available on GitHub. A rendered report of the code can also be found in the same repo. https://github.com/CenterForMedicalGeneticsGhent/ExPECT-analysis.