Abstract
Background
The emergence and spread of Extended-Spectrum Beta-Lactamase (ESBL)-producing Escherichia coli pose significant challenges for treatment of infections globally. This challenge is exacerbated in sub-Saharan African countries, where the prevalence of ESBL-producing E. coli is high. This, combined with the lack of a strong and supportive healthcare system, leads to increased morbidity and mortality due to treatment failures. Notably, studies in Ethiopia have primarily focused on hospital settings, leaving a gap in understanding ESBL prevalence in rural communities, where human-animal proximity may facilitate microbial exchange.
Methods
We conducted a community-based study in the rural Somali region of Ethiopia, simultaneously examining the fecal carriage of ESBL-producing E. coli in children aged 2–5 years and their livestock (cattle, camel, goat). Fecal samples from 366 children and 243 animals underwent phenotypic screening for ESBL-producing E. coli. Following phenotypic confirmation, ESBL resistance genes were identified via conventional PCR. Whole-genome sequencing (WGS) was performed on a subset of isolates from human feces.
Results
We found that 43% (159/366) of children and 3.7% (9/244) of livestock harbored ESBL-producing E. coli. The ESBL gene blaCTX-M-15 was predominant in human (82.7%, 120/145) and livestock (100%) isolates. In the 48 human E. coli isolates subjected to WGS, a high diversity resulting in 40 sequence types (STs) was observed. Among these, ST-2353 was the most prevalent (5/48), followed by ST-10 and ST-48 (3/48) and ST-38, ST-450, and ST-4750 (2/48). These STs were associated with multiple resistance genes, such as blaCTX-M-15, blaTEM-1B, blaOXA-1, blaCTX-M-14 and blaTEM-35.
Conclusion
We report a high prevalence of ESBL E. coli in rural children, which outnumbers its prevalence in livestock. These isolates displayed a high diversity of sequence types (STs) with ST-2353 being the dominant ST. Our study is the first to report the association of ST-2353 with multi-drug resistance genes in Ethiopia. Further research using an integrated approach including other domains such as water and food products is needed to truly understand and combat AMR transmission and acquisition in this region.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13756-024-01502-5.
Keywords: Antimicrobial resistance, Extended spectrum beta lactamase, E. coli, One Health
Introduction
The emergence and spread of multidrug-resistant bacterial strains, particularly Extended-Spectrum Beta-Lactamase (ESBL)-producing Escherichia coli (E. coli), pose significant challenges for treatment of infections globally [1]. These are exacerbated in sub-Saharan African countries, including Nigeria, Tanzania and Ghana, where the prevalence of ESBL-producing E. coli is high, both in humans (up to 60%) and animals (up to 56%) [2–6]. This, combined with the lack of a strong and supportive healthcare system, leads to increased morbidity and mortality due to treatment failures [7]. Robust laboratory diagnostics and strong surveillance systems for antimicrobial resistance are urgently needed, yet, they are still lacking in many parts of sub-Saharan Africa [8].
ESBLs are enzymes that hydrolyze a broad range of beta-lactam antibiotics, confering resistance to multiple antibiotics [9]. CTX-M, a dominant ESBL type, has become widespread globally after the late 1990s, outpacing TEM and SHV [10]. Nowadays, its prevalence is high in infections of humans and food-producing animals, and it is considered one of the main contributing factors to multidrug resistance in E. coli in both low- and middle-income (LMICs) and Western countries [11–13].
ESBL genes, commonly found in plasmids, primarily spread through horizontal gene transfer [14, 15]. Recent findings suggest they can also exist in the chromosome, indicating potential clonal transmission [16, 17]. Transmission of ESBL-producing E. coli from animals to humans, often discussed in the context of contaminated animal-origin food, is a concern [18]. Surveillance relies on identifying similar clones, plasmids, or sequence types in human and animal populations, to infer transmission [19]. The interconnectedness of infections in both species underscores the need for a One Health approach to fully understand the dynamics and spread of resistant clones.
In Ethiopia, recent studies have revealed a high prevalence of blaCTX-M, especially the blaCTX-M-15 variant (87.7–88.4%), within hospital settings, which also conferred resistance to non-beta-lactam antibiotics, such as fluoroquinolones and aminoglycosides [20–22]. Additionally, these studies reported different E. coli phylo-groups, with several isolates linked to the ST-131 clone, which is mainly associated with blaCTX-M-15 [20–22]. Notably, studies in Ethiopia have primarily focused on hospital settings, leaving a gap in understanding ESBL prevalence in rural communities, where human-animal proximity may facilitate microbial exchange.
To fill this gap, we conducted a community-based study to investigate the fecal carriage of ESBL-producing E. coli in children aged two to five years and livestock of the same households and genetically characterized the isolated clones in the feces of children and livestock. By identifying the prevalence of ESBL-producing E. coli and their genetic characteristics, this study yields new insights to guide interventions aimed at curbing the dissemination of antimicrobial resistance and enhancing treatment strategies for infectious diseases caused by these bacteria.
Methods
Study area
The study was carried out from May 2021 until June 2021 in Adadle district in the Shebele zone, Somali Regional State (SRS), Ethiopia.
Sample size
Based on a previous study conducted in Addis Ababa, we expected the prevalence of ESBL-producing E. coli to be 42% [23] and assumed an inter-cluster correlation coefficient of 0.15. We calculated a sample size of 360 eligible children, with 180 children from pastoralist and 180 children from agro-pastoralist communities to be sufficient to estimate the prevalence with a margin of error of 10% at the 95% confidence level.
Selection criteria
Based on the nature of livelihood, eight of the thirteen Kebele in the Adadle district were randomly selected. Among the selected Kebele, four were pastoralist (Malkasalah, Todab, Harsug, Boholhagere) and four were agro-pastoralist (Bursaredo, Dabafyd, Gabal, Higlo).
A pre-enrolment screening was carried out in each Kebele to assess children between 2 and 5 years of age. All children were screened for stunting and wasting based on WHO guidelines. All stunted (height for age score, HAZ <− 2), and wasted (weight for height score, WHZ <− 2 or mid-upper arm circumference (MUAC) < 11.5 cm) were enrolled in the study. Among the screened children who were non-stunted and/or non-wasted, a random selection process was employed from an Excel file containing information on all pre-screened households/children in the community. The random selection process continued until the desired sample size was achieved. Children outside the age range (below 2 years and above 5 years) and those who had received antibiotic treatment within the last 14 days were excluded from the study. Informed consent was obtained from all participants’ legal guardians.
Anthropometric measurements
Height measurements were taken with children standing against a WHO standard wooden measuring board, ensuring the correct posture and position. Weights were recorded using a WHO standard weight scale, either the child alone or with the mother by adjusting the weight scale [24]. MUAC measurements were performed using a WHO standard tape measure. To ensure the accuracy of height, weight and MUAC measurements, each anthropometric measurement was repeated at least twice or until the measurements were within 1 mm, 100 g, or 1 mm of one another, respectively.
Data collection
A comprehensive questionnaire was administered to the mother of the child by trained field workers in the local language using Open Data Kit (ODK). The questionnaire covered different sections, including demographic characteristics, child health status, WASH behavior, breastfeeding, diet, and anthropometric measurements. As birth records were not available in the community, the child’s age was estimated based on group discussion involving the child’s mother, as well as other family and community members. These discussions considered factors such as seasonal events (floods, drought, and rainy season) and festive occasions (Ramadan, festival, and other religious events) that occurred before and after the child’s birth. Additionally, leveraging the close-knit relationships within the community, efforts were made to engage in discussions to collectively recollect and remind each other about significant events and timeframes related to the child’s birth.
Stool sample collection
Trained field workers provided mothers with instructions in the local language a day before collecting fecal samples. Mothers used sterile specimen cups, and upon receipt, samples were placed in an ice box and transported within 2–4 h to a field laboratory. There, samples were aliquoted into six cryotubes, two with glycerol (40% glycerol mixed 1:1 with the stool) and four without, stored at 4 °C. Subsequently, samples were sent to Gode within 24 h, stored at − 20 °C, and then shipped to the Armauer Hansen Research Institute in Addis Ababa, where they were preserved at − 80 °C for long-term storage.
Additionally, from the selected households that owned animals, we assessed different types of animals (camels, cattle, and goats). We randomly sampled at least one animal per species. Animal fecal samples were collected by a veterinary professional through either rectal insertion of gloved hands or aseptic collection of fresh stool (if the animals defecated in the presence of the team). Animal samples underwent a process similar to human samples—immediate shipment to the field laboratory, aliquoting, storage at − 20 °C, and final storage in Addis Ababa at − 80 °C for future analysis.
Bacterial isolation and identification
Fecal samples were aseptically inoculated onto MacConkey agar plates (Oxoid, UK) [25], and subsequently, a cefotaxime disc was placed at the center of the plate. The plates were then incubated for 24 h at 37 °C. The E. coli colony closest to the cefotaxime disc was meticulously selected based on its distinctive morphology and pigmentation characteristics. The selected colonies were re-inoculated overnight to ensure purity and, and subsequently, the colony was confirmed as E. coli through a biochemical test. Isolates were classified as E. coli if they exhibited a positive indole test, negative citrate, positive lysine decarboxylation, gas and acid production, mannitol fermentation, negative urea hydrolysis, and if they were motile.
Antibiotic susceptibility test
The disc diffusion method, was used to determine the antibiotic susceptibility using Müller-Hinton agar (Oxoid, UK) according to the recommendation of the Clinical and Laboratory Standard Institute (CLSI) guidelines [26]. Antibiotic susceptibility tests were performed for 19 antibiotics from 10 different classes, including penicillins (ampicillin (AMP) 10 µg and amoxicillin (AML) 10 μg), beta-lactamase inhibitor combinations (ampicillin-sulbactum (SAM) 30 μg and amoxicillin/clavulanic acid (AMC) 30 µg), cephalosporin (cefazolin (KZ) 30 µg, cefpodoxime (CPD) 10 µg, cefuroxime (CXM) 30 µg, ceftriaxone (CRO) 30 µg, cefotaxime (CTX) 30 µg, cefipime (FEP) 30 µg, ceftazidime (CAZ) 30 µg), carbapenem (imipenem (IPM) 10 µg and ertapenem (ETP) 10 µg), monobactams (aztreonam (ATM) 30 µg), fluoroquinolones (ciprofloxacin (CIP) 5 µg), aminoglycosides (gentamicin (CN) 30 µg), macrolides (azithromycin (AZM) 15 µg), tetracycline (TE) 30 µg), and cotrimoxazole (SXT) 25 µg (Oxoid, United Kingdom). McFarland standard 0.5 was used prior to spreading the suspension on the Müller-Hinton agar plate to ensure the bacterial suspension turbidity. Plates were incubated at 37 °C for 18 h. Based on the inhibition zone diameters, antibiotics were assigned to susceptible, intermediate, and resistant as indicated by CLSI guidelines [26]. Multidrug resistance was defined as non-susceptible to at least one antibiotic agent in three or more antimicrobial classes [27]. Extensive drug resistance (XDR) was defined as resistance to at least one agent in all but susceptible to two or fewer antimicrobial categories. Pan drug resistance (PDR) was defined as resistance to all agents in all antimicrobial categories [28].
ESBL screening and confirmatory test
According to CLSI guidelines, any strain showing resistance against cefotaxime 30 μg, ceftazidime 30 μg, or ceftriaxone 30 µg was considered as a potential ESBL producer. Confirmatory tests were performed utilizing a disc of cefotaxime 30 μg and one of ceftazidime 30 μg alone, and a disc of cefotaxime and one of ceftazidime combined with clavulanic acid 10 µg (Oxoid, United Kingdom). Isolated strains were considered ESBL-producers if an increase of inhibition zone diameter of 5 mm or greater was observed in the discs of cefotaxime or ceftazidime combined with clavulanic acid compared to the inhibition observed in the cefotaxime or ceftazidime discs without clavulanic acid. E. coli ATCC 25922 and E. coli ATCC BAA-2326 were used as negative and positive controls, respectively [29].
Molecular testing for β-lactamase genes (blaCTX-M, blaTEM, and blaSHV)
The DNA of the confirmed ESBL-producing isolates was extracted using the Wizard® HMW DNA Extraction Kit, according to the manufacturer’s instructions [30]. The presence of three ESBL genes (blaCTX-M, blaTEM, and blaSHV) was detected using a multiplex PCR approach within a single reaction tube. Details regarding the PCR reaction, cycling, and primers for amplifying the ESBL resistance genes can be found in supplementary materials 1.
Whole genome sequencing
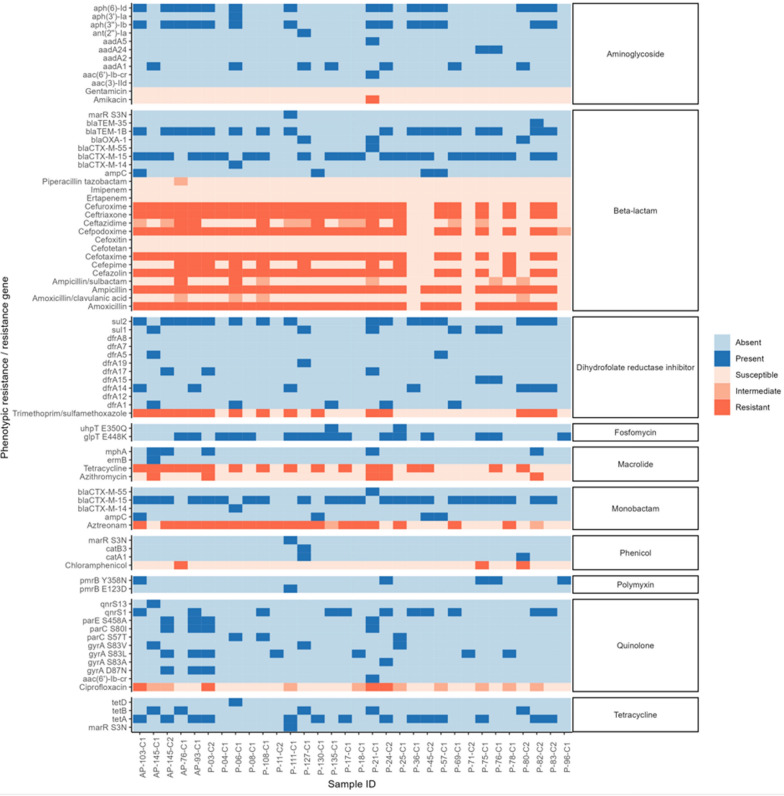
The extracted DNA from human and animal ESBL-producing isolates were shipped to the Swiss Tropical and Public Health Institute (Swiss TPH) for whole genome sequencing (WGS). However, the DNA quality of the sub-isolates, selected based on their phenotypic profiles, was insufficient for WGS. Since we did not have the original isolates in Switzerland, we decided to re-culture and perform DNA extraction on the original fecal samples that were initially shipped with the DNA. Subsequently, 48 of these samples were selected based on the phenotypic profiles obtained in Ethiopia and the phenotypic analysis was repeated for the new isolates for the data provided in Fig. 4.
Fig. 4.
Concordance and discordance between genotype and phenotype for selected ESBL-producing E. coli isolates from Adadle district, Somali region, Ethiopia
DNA concentration was quantified with the Qubit dsDNA HS Assay Kit (Invitrogen, Germany). Isolates were selected for WGS using the MinION platform (Oxford Nanopore Technologies, UK) based on DNA concentrations (> 33 ng/µL). The sequencing library was prepared according to the manufacturer’s instructions using the Native Barcoding Kit 96 (SQK-LSK114.96) and loaded onto the R10.4.1 flow cell and sequenced on the MinION Mk1C using super-accurate base calling.
Bioinformatics analysis
De novo assembly was conducted using Flye 2.9.1 at the scientific computing core facility of the University of Basel [31]. The Bacterial and Viral Bioinformatics Resource Center (BV-BRC) was used for annotation and phylogenetic analysis of the assemblies. The assemblies were annotated using the RAST 2.0 toolkit [32]. Only assemblies with less than 20 contigs were considered for further analysis. Sequence type of assembled contigs were determined using MLST [33]. Phylogenetic trees were generated by aligning protein and nucleotide sequences using MUSCLE, MAFFT and RAxML [33–35]. Resistome analysis was performed using CARD (v. 6.0.0) and ResFinder (v. 4.2.2) [34, 36]. VirulenceFinder v.2.0 and PlasmidFinder v.2.1 tools available at the Center for Genomic Epidemiology were used for the virulent detection and plasmid replicon typing [37].
Statistical analysis
R statistical software v. 4.1.3 was used to perform the statistical analysis [38]. Initial descriptive analysis of variables was performed using the gtsummary package. Multivariate analysis employed logistic regression to assess the association between ESBL-producing E. coli and independent variables. The initial model included variables (age, sex, education, sanitation practice, hygiene, source of water, owning livestock, nutritional status) based on literature knowledge, with stepwise removal of those contributing insufficient information (p > 0.2). Variables with p < 0.2 were retained. Model fitness was assessed using likelihood ratio test, AIC (Akaike Information Criterion), and adjusted R square. Significance was determined at p < 0.05. R packages ape, ggplot2, ggtree, and ggtreeExtra were used for analyzing resistance genes and visualizing phylogenetic trees.
Results
Description of study population
Out of the 366 children, only 346 children completed the questionnaire and were included in this study for further analysis. Pastoralists and agro-pastoralists were evenly represented in terms of age and sex among the enrolled children. Over half of the children showed a normal growth (61%), while a quarter (25%) of them were wasted, and 7.8% were stunted. Further, 5.2% of the children suffered concomitantly from stunting and wasting. Characteristics of the study group are summarized in Table 1.
Table 1.
Characteristic of pastoralist and agro-pastoralist in the Adadle district, Somali region, Ethiopia
| Variables | N = 346 (%) |
|---|---|
| Settlement area | |
| Pastoralist | 177 (51.2%) |
| Agro-Pastoralist | 169 (48.8%) |
| Education of the mothers | |
| Formal education | 50 (14.5%) |
| Non-formal education | 36 (10.4%) |
| Illiterate | 260 (75.1%) |
| Sex of the child | |
| Female | 176 (50.9%) |
| Male | 170 (49.1%) |
| Age group of the child | |
| 2 years | 150 (43.4%) |
| 3 years | 98 (28.3%) |
| 4–5 years | 98 (28.3%) |
| Number of children per household | |
| 1–3 children | 78 (22.5%) |
| 4–6 children | 160 (46.2%) |
| > = 7 children | 108 (31.2%) |
| Completed all required vaccinations | |
| No | 309 (89.3%) |
| Yes | 37 (10.7%) |
| Has a vaccination card | |
| No | 328 (94.8%) |
| Yes | 18 (5.2%) |
| Nutritional status | |
| Normal growth | 212 (61.3%) |
| Wasted | 88 (25.4%) |
| Stunted | 27 (7.8%) |
| Stunted and wasted | 18 (5.2%) |
| Overweight | 1 (0.3%) |
As summarized in the suppmentary Table S1, the two main sources of water for pastoralists and agro-pastoralists were rainwater/birkad (89%) and river water (95%), respectively. Most of pastoralist and agro-pastoralist (96%) used open space for defecation. The 14 households (4%) that had a toilet shared it with 28 households. Dumping waste in the street or open spaces within the compound was the predominant method of waste disposal in both pastoralist and agro-pastoralist societies (84%), while 16% opted to burn waste. Most mothers reported that they used water to wash the children’s hands (93%), while a small subfraction of mothers reported (4%) using water and soap. At the time of the sampling, more than half of the agro-pastoralists and pastoralists had soap, while 29% did not have soap very often and 13% never had soap in their house.
Nearly half of agro-pastoralist households (43%) and 12% of pastoralist households treated the water prior to consumption (mainly chlorination (98.9%)). The majority of agro-pastoralists possessed cattle (94.7%), donkeys (81.1%), goats (62.7%), sheep (53.3%), camels (10%) and chickens (10%). Among pastoralists, predominant livestock ownership included goats (87.6%), donkeys (58.8%), sheep (44.1%), camels (36.2%), and cattle (29.9%).
Phenotypic test results for ESBL E. coli carriage
A total of 609 fecal samples, comprising 366 from humans and 243 from animals (including 77 goats, 136 cows, and 30 camels) were analysed. For human isolates, 159 (43%) of the E. coli isolates were ESBL-producers (24.5% in pastoralists and 18.8% in agropastoralists). Furthermore, 7.8% of the E. coli isolates from goats (6/77) and 2.2% from cows (3/136) were ESBL-producers. Regarding animal ESBL-producing isolates, 7 (77.8%) were identified among animals of agro-pastoralists, while 2 (22.2%) were found among those of pastoralists.
Susceptibility pattern for ESBL-producing E. coli
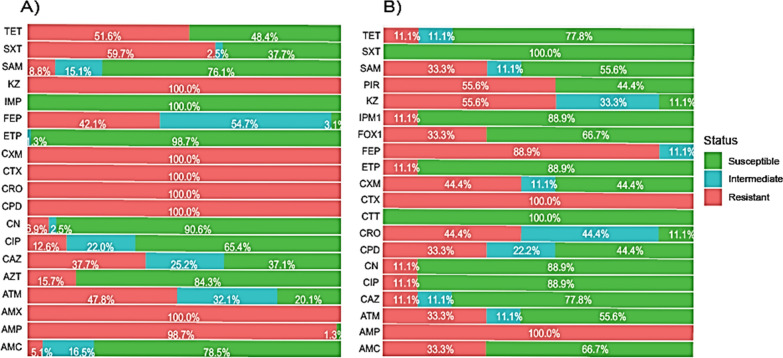
For the human isolates, ESBL-producing E. coli strains exhibited complete resistance to several commonly used antibiotics including amoxicillin, cefotaxime, cefuroxime, ceftriaxone, cefazolin, and cefpodoxime. Almost all ESBL-producing E. coli isolates (98.7%) were resistant to ampicillin and 51.6% were resistant to tetracycline.
Regarding co-trimoxazole, 57.9% of the isolates were classified as resistant, 2.5% as intermediate, and 37.7% as susceptible. Among ESBL-producing E. coli, 47.8%, 42.1%, and 27.7% were non-susceptible, and 32.1%, 54.7%, and 25.2% were intermediate to aztreonam, cefipime, and ceftazidime, respectively. In addition, 15.7% and 12.6% of the isolates were resistant to azithromycin and ciprofloxacin. Ampicillin-sulbactam (8.8%), gentamicin (6.9%), and amoxicillin-clavulanic acid (5.9%) had the lowest rates of resistance. Intermediate susceptibility was observed in 16.5% for amoxicillin-clavulanic acid, 15.1% for ampicillin-salbactum, and 2.5% for gentamicin. All carbapenems (imipenem and ertapenem), which are considered the last-resort antibiotics for infections caused by ESBL-producing E. coli, were found to be effective against the ESBL-producing E. coli isolates, except for 1.3% of the isolates that had intermediate susceptibility to ertapenem. Additionally, using whole genome sequencing (WGS), none known carbapenem or colistin resistance genes were found in the E. coli isolates.
Additonally, nine ESBL isolates obtained from animal fecal samples exhibited complete (100%) resistance to ampicillin, cefotaxime, and amoxicillin. Furthermore, an 88.9% resistance rate was observed for cefepime, followed by cefazolin (55.6%), ceftriaxone (44.4%), cefuroxime (44.4%), and aztreonam and ampicillin-sulbactam, which both exhibited an equal resistance rate of 33.3%. In ESBL-producing E. coli isolates from animals, ampicillin-clavulanic acid, ceftazidime, gentamicin, and ciprofloxacin all exhibit an equal resistance rate of 11.1%. The data is summarized in Fig. 1.
Fig. 1.
Antimicrobial resistance pattern of ESBL-producing E. coli isolated from the feces of children aged 2–5 years (A, 159/366) and from livestock (B, 9/243) in the Adadle district, Somali region, Ethiopia. Antimicrobial agents tested include: AMX (Amoxicillin), AMC (Amoxicillin-clavulanic acid), AMP (Ampicillin), ATM (Aztreonam), AZT (Azithromycin), CAZ (Ceftazidme), CIP (Ciprofloxacin), CN (Gentamicin), CPD (Cefpodoxime), CRO (Ceftriaxone), CTX (Cefotaxime), CCT (Cefotetan), ETP (Ertapenem), FEP (Cefepime), FOX (Cefoxitin), IMP (Imipenem), KZ (Cefazolin), SAM (Ampicillin-sulbactam), SXT (Co-trimoxazole), PIR (Piperacillin), TET (Tetracycline)
All ESBL-producing E. coli strains exhibited multi-drug resistance (MDR), rendering them non-susceptible to three or more antibiotic drug classes (Figure S1 in Supplementary File 1).
Thus, ESBL-carriage is very prevalent, especially in children, in a community setting and is much lower in the feces of livestock animals.
Predictors of ESBL-producing E. coli carriage
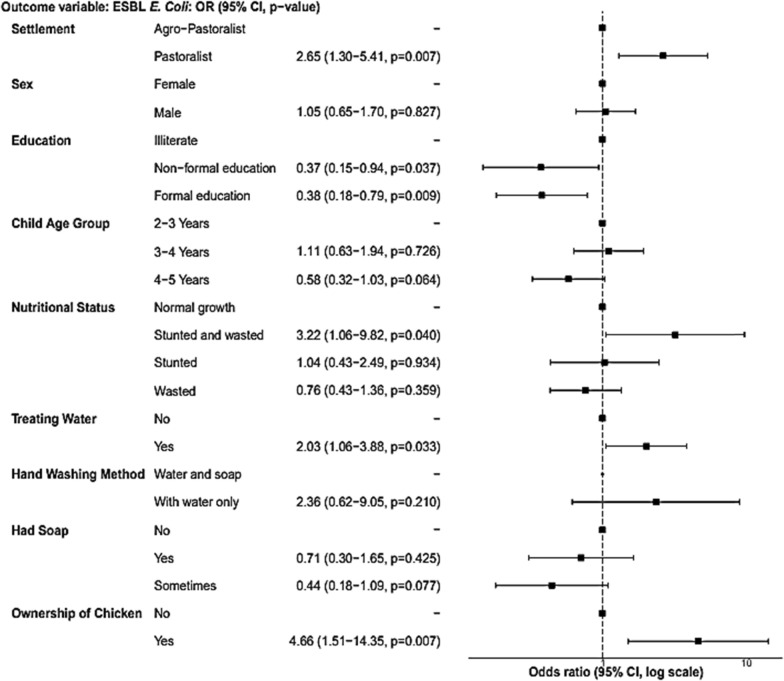
In the multivariable analysis, education was significantly associated with ESBL-producing E. coli. Children whose mothers or household heads were illiterate had twice the odds of carrying ESBL-producing E. coli compared to children whose mothers were formally educated (aOR = 2.65, 95% CI = 1.27–5.48). The odds of children who were both stunted and wasted were three time higher to harbor ESBL-producing E. coli compared to children with normal growth (aOR = 3.14, 95%CI = 1.02–9.07). Moreover, pastoralist children had 2.65 times higher odds of being colonized with ESBL producing E. coli compared to agro-pastoralist children (aOR = 2.65, 95% CI = 1.30–5.41). Counterintuitively, children who drank water treated with chlorine showed a positive association with ESBL-producing E. coli (aOR = 2.09, 95% CI = 1.10–3.98). Additionally, the possession of chicken increased the odds of infection with ESBL-producing E. coli five times (aOR = 5.13, 95% CI = 1.66–15.68). The sex and age of the child were not found to be significantly associated with infection with ESBL-producing E. coli. Similarly, although owing soap or washing hands with water and soap showed a trend to decrease the risk of contracting ESBL-producing E. coli, this association was not statistically significant (Fig. 2).
Fig. 2.
Risk factors (multivariable model) associated with fecal carriage of ESBL-producing E. coli among children living in the Adadle district, Somali region, Ethiopia
Genetic characterization of ESBL strains (conventional PCR)
During PCR screening, we found that the blaCTX-M-15 gene was the most prevalent resistance gene in both human (82.8%) and animal (100%) isolates. The prevalence of the blaCTX-M-15 resistance gene was similar in isolates from the feces of pastoralists and agro-pastoralists, both almost at 80% (Figure S2 of the Supplementary Materials 1). Comparing the nine isolates from animals alongside those from children residing in the same household, we observed that only two households within the pastoralist group demonstrated simultaneous presence of blaCTX-M-15 in both their children and livestock (Figure S3).
Multi locus sequence types (MLST), phylogenetic groups and plasmid MLST
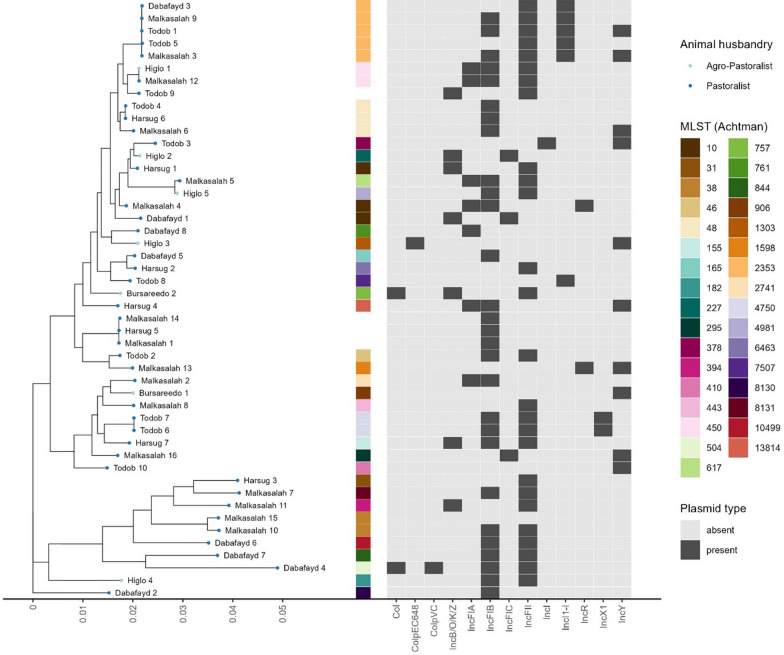
The 48 human E. coli isolate subjected to WGS analysis, were chosen based on their phenotypic profile. A sequence type (ST) by MLST using the Achtman scheme could be assigned to 44 isolates (91.7%), with five isolates having a single nucleotide polymorphism (SNP) in one gene (2 in adk, 2 in fumC, 1 in mdh). The most common ST was ST-2353 with five E. coli isolates, followed by ST-10 and ST-48 with three E. coli isolates each and ST-38, ST-450 and ST-4750 with two E. coli isolates each. All other 27 E. coli isolates had singular ST. A total of nine E. coli isolates were assigned to clonal complex ST10 (3 ST-10, 3 ST-48, 1 ST-227, 1 ST-378 and 1 ST-617) and two isolates to clonal complex ST38 (2 ST-38).
The predominant beta-lactam resistance gene among the 48 E. coli isolates was blaCTX-M-15, identified in 72.9% (35/48) of the whole-genome sequenced isolates, followed by blaTEM-1B identified in 47.9% (23/48), ampC beta-lactamase in 14.6% (7/48), blaOXA-1 in 8.3% (4/48) and blaCTX-M-55 in 4.2% of the isolates. The beta-lactam resistance genes blaCTX-M-14 and blaTEM-35 were the least prevalent, each detected in a single isolate.
Four E. coli isolates (8.3%) did not carry any known beta-lactam resistance genes, 20 E. coli isolates (41.7%) carried one, 19 E. coli isolates (39.6%) carried two and five E. coli isolates (10.4%) carried three beta-lactam resistance genes. Seventy-five percent (15/20) of E. coli isolates with one beta-lactam resistance gene carried only blaCTX-M-15, while 68.4% of E. coli isolates with two beta-lactam resistance genes carried both blaCTX-M-15 and blaTEM-1B.
Quinolone resistance conferred by mutations in the gyrA gene was detected in 33.3% (16/48) of E. coli isolates. Serine at position 83 was mutated to either Leucine (S83L, 10/16), Alanine (S83A, 3/16) or Valine (S83V, 3/16). In addition to S83L, three E. coli isolates had gyrA mutation D87N. Four E. coli isolates (8.3%) had parC mutation S57T and four E. coli isolates (8.3%) had parC mutation S80I combined with parE mutation S458A. The plasmid-encoded qnrS1 and qnrS13 genes were detected in 39.6% (19/48) and 2.1% (1/48) of the sequenced isolates, respectively.
Aminoglycoside resistance genes were present in 60.4% (29/48) of E. coli isolates. Four different aminoglycoside (3″) (9) adenylyltransferase (aadA) genes were detected in 15 E. coli isolates (31.3%), with aadA1 accounting for 66.7% (10/15), aadA2 and aadA24 for 13.3% (2/15) each and aadA5 for 6.7% (1/15) of the aadA genes found. Eighteen E. coli isolates (39.5%) had the plasmid-encoded aph(3″)-Ib and aph(6)-Id aminoglycoside resistance genes, one had only aph(3″)-Ib and one only aph(6)-Id. The aminoglycoside resistance genes aac(3)-IId, aac(6′)-Ib-cr, ant(2″)-Ia and aph(3′)-Ia were detected in one E. coli isolate each.
Sulfonamide resistance genes were detected in 62.5% of E. coli isolates (30/48), with sul1 in 30.0% (9/30), sul2 in 63.3% (19/30) and both genes in 6.7% (2/30) of the isolates. Trimethroprim resistance genes dfrA were detected in 58.3% of E. coli isolates (28/48), most commonly dfrA14 (9), followed by dfrA1 (7), dfrA5 (3), dfrA17 (3), dfrA7 (2), dfrA15 (2), dfrA8 (1) and dfrA19 (1). Tetracycline resistance genes tetA, tetB and tetD were detected in 35.4% (17/48), 16.7% (8/48) and 2.1% (1/48) of E. coli isolates, respectively. Macrolide resistance gene mphA was detected in 18.8% (9/48) of the isolates, with one isolate simultaneously carrying ermB. The results are summarized in figure S4.
Virulence genes and plasmids
A total of five different virulent genes were found in 15/48 of E. coli isolates, but none of the isolates were found to harbor Shigo-toxin genes in their genomes. The results revealed the presence of Enteroaggregative E. coli genes such as aggR (5/48), aggA (2/48) and aaiC (2/48). Additionally, both the heat-labile (elt) and heat-stable enterotoxigenic E. coli genes (est) were found in 7/48 and 8/48 E. coli isolates (Table S2).
The detection rate of IncFIB plasmid exhibited the highest frequency, found in 28/48 isolates, followed by IncFII (26/48), IncY (10/48), IncFIA (7/48) and Incl1-l (6/48). Combinations of multiple plasmids were observed, with IncFIB and IncFII being the most prevalent combination, present in 17/48 isolates. Specifically, 12 isolates carrying blaCTX-M-15 resistance genes encoded both IncFIB and IncFII plasmids. Three isolates carried a combination of three plasmids (IncFIB, IncFII, IncFIA), and single isolates carried four different plasmids (IncFIB, IncFII, IncFIA, IncY) (Fig. 3).
Fig. 3.
Plasmid replicon profiles for 48 ESBL-producing E. coli isolated from 2 to 5 year old children in the Adadle district, Somali Region, Ethiopia. The presence of plasmid is shown in dark gray and absence in light gray
Comparison of genotypes and phenotypes
Resistance to different antibiotics was inferred from WGS data. The presence of specific genes (Fig. 4, in blue) indicated resistance to specific antibiotics (Fig. 4, in red). For example, the presence of at least one of the following genes, CTX-M-type, OXA-type, TEM-type, or ampC, indicated resistance to amoxicillin, ampicillin, aztreonam, cefotaxime, ceftriaxone, cefazolin, cefuroxime, and cefepodoxime. The same procedure was used for all other antibiotics, as indicated in Fig. 4.
The resistance results inferred from WGS data were then compared with the results of the phenotypic assay. We found high concordance rate (> 90%) between genotype and phenotype for azithromycin (93%), amoxicillin (91%), and ampicillin (91%). Conversely, chloramphenicol (87%), as well as several cephalosporins including cefotaxime, ceftriaxone, cefazolin, cefuroxime, and cefepodoxime, each showed a low concordance rate of 84%. Tetracycline (81%) and trimethoprim/sulfamethoxazole (72%) showed even lower concordance rates, with ciprofloxacin (63%) registering the lowest concordance rate among them. These discordances should be analysed in more detail by assessing more antibiotics and by mining for eventual new resistance mechanisms that could be encoded in these bacterial strains. The results are summarized in Fig. 4.
Discussion
Over the past two decades, there has been a significant increase in the global prevalence of communities carrying ESBL-producing E. coli [39]. In LMICs, the colonization of ESBL-producing E. coli has risen steadily in both community and healthcare settings, with the community carriage rate approaching that of healthcare settings [40]. To our knowledge, this is the first study to employ WGS for profiling of the phylogenomic of ESBL-producing E. coli in children under the age of five in rural communities in Ethiopia.
The results show a high carriage (43%) of ESBL-producing E. coli in children under the age of five living in these communities via phenotypic analysis. This finding aligns with a study conducted among hospitalized children in Addis Ababa [23] and rural children in Ghana [41]. The elevated prevalence observed could be attributed to the widespread availability of antimicrobials and misuse of antibiotics [42]. Additionally, in rural communities, limited access to clean water and sanitation may contribute the spread of ESBL-producing E. coli resistant clones and genes [43]. On the other hand, low prevalence of ESBL-producing E. coli carriage was found in livestock, which aligns with a study performed in Kenya [44]. In rural communities, limited access to veterinary care may result in reduced antibiotic usage in livestock, thereby decreasing the selection pressure for antibiotic-resistant bacteria [45]. This situation could, in turn, contribute to the lower prevalence of ESBL-producing E. coli strains in the livestock population.
In the PCR analysis, the resistance gene blaCTX-M-15 emerged as the most frequently observed gene among E. coli isolates that exhibited an ESBL phenotype in our study, both in the children (120/159 isolates) and in livestock (9/9 isolates). Importantly, blaCTX-M-15 is known as the primary ESBL resistance gene responsible for human infections worldwide [46]. Moreover, recent research conducted in Africa corroborates this finding, highlighting blaCTX-M-15 as the dominant ESBL-producing E. coli gene in livestock populations [47]. This geographical distribution strongly suggests a significant spread of resistance genes among both humans, animals and other domains such as water and food products, highlighting the importance of understanding and addressing this issue from a One Health perspective.
Studies in Ethiopia and Ghana also demonstrated a substantial diversity of STs, accounting for over 30 of them [2, 20]. Here, ST10 and ST48 exhibited multi-drug resistance genes, primarily blaCTX-M-15 and blaTEM-1B, along with other non-beta-lactamase resistance genes. Globally, ST10 and ST48 are recognized as clonal genetic entities known to harbor multi-drug resistance genes, primarily in humans, with a notable prevalence of blaCTX-M-15 [48, 49]. Additionally, ST38 showed a lineage associated with the carriage of blaCTX-M-14 and blaCTX-M-15, which is consistent with prior research findings in Tanzania [50, 51].
Notably, ST2353, typically associated with highly pathogenic diarrheagenic E. coli strains [52, 53] and historically less reported for its resistance gene carriage, emerged as one of the predominant sequence types in our study. Remarkably, ST2353 was found to harbor multiple resistance genes, encompassing blaCTX-M-15, bla-TEM-1b, gyrA, and tetA. This observation underscores the potential for gene evolution over time, signifying the spread of resistance mechanisms across diverse clonal sequence types. This gene can impact the efficacy of antibiotics treatment on a population level. Hence, our findings underscore the imperative need for further comprehensive investigations aimed at elucidating the mechanisms underlying genetic mutations and the emergence of allelic variants associated with antibiotic resistance.
The IncFII-FIA-FIB multi-replicon plasmids have been commonly associated with blaCTX-M-15 resistance genes globally [54]. This was also found in our study along with other studies in Ethiopia, Tanzania, and Ghana [21, 55, 56]. These plasmids can maintain and transfer resistance genes among enterobacterial species independent of antibiotic exposure, which contributes to the dissemination of AMR genes with in the community [57]. Therefore, genomic surveillance is essential to tracking the evolution of resistance determinants and assessing the effectiveness of interventions aimed at controlling AMR spread in different ecological settings.
We report a high concordance rate (> 90%) between genotypic and phenotypic resistance for certain antibiotics, as similarly reported by others [58, 59]. However, lower concordance rates (< 90%) were observed for certain antibiotics classes, with fluoroquinolones showing the lowest agreement. Most cases were so-called “major errors”, i.e. genotypic resistance was observed but phenotypic susceptibility was not [60]. This phenomenon may be attributed to the suppression of gene expression through transcriptional regulation, other gene silencing mechanisms or compensatory mutations [61, 62]. Similar results were noted in other studies conducted in Singaopore and France [59, 61]. Further research is needed to fully explain this phenomenon.
In the multivariable analysis, we found that children whose mother were illiterate had higher odds of carrying ESBL-producing E. coli [63]. Furthermore, age and sex of the child were not significantly associated with ESBL-producing E. coli carriage. These results are consistent with prior studies conducted in Ethiopia, Guinea-Bissau, and Madagascar [63–65]. The observed link between maternal illiteracy and ESBL-producing E. coli colonization suggests the importance of community education in implementing effective strategies to combat this public health concern.
Children who were both stunted and wasted were significantly more likely to be colonized by ESBL-producing E. coli, with a three times higher odds compared with those without malnutrition [66–68]. This finding can be explained by the fact that malnutrition weakens the immunity system, making children more susceptible to infections, and more likely to be treated with antibiotics [69]. This connection may also involve the influence of microbiota and the creation of niches for colonization. Given the well-established link between antibiotic use and the emergence and spread of AMR, addressing malnutrition through public health interventions could contribute to the reduction of infections and colonization by AMR carrying bacteria.
Furthermore, the study revealed that there was a significant correlation between ESBL carriage and settlement type, water treatment, and chicken ownership. Pastoralists were found to have higher odds of ESBL-producing E. coli colonization compared to agro-pastoralists. The nomadic nature of pastoralists, involving frequent travels in search of water and food, may foster shared water sources and inadequate sanitation and hygiene practices. Consequently, this situation could escalate the risk of waterborne diseases like diarrhea and facilitate the transmission of ESBL-producing E. coli [70]. As infectious diseases become more prevalent in such contexts, there is an associated rise in antibiotic usage, a well-established precursor to AMR [71].
Despite chlorine being the most commonly used method for water treatment in LMICs [72], this study found a positive association between children who consumed chlorine-treated water and ESBL-producing E. coli. This finding aligns with a randomized control trial conducted in Bangladesh, which revealed that water chlorination did not significantly decrease the fecal carriage of ESBL-producing E. coli in children [73]. Other studies conducted in China and South Africa also demonstrated the tolerance of chlorine in relation to AMR [74, 75]. Furthermore, a study conducted in China highlighted that chlorination promotes horizontal gene transfer through natural transformation, thus facilitating the spread and emergence of AMR [76]. Therefore, we hypothesize that the widespread use of chlorine in LMICs for water treatment may not be as effective as previously thought in reducing the prevalence and transmission of antimicrobial-resistant E. coli strains.
In this study, possessing chickens showed a significant relationship with ESBL-producing E. coli, with the odds of colonization being five times higher. Previous studies conducted in Ethiopia, Kenya, Nigeria, Ghana, Pakistan, and the Netherlands have suggested that chickens are the primary source of ESBL-producing E. coli transmission to humans [2, 77–82]. This suggests that there is a need of effective measures to control the spread of antimicrobial resistance in animal husbandry, with a focus on poultry as the main carriers.
We attempted to collect fecal samples from children and livestock within the same households to investigate the circulation of resistance genes between the two populations. However, due to the absence of necessary kits and machines for whole genome sequencing (WGS), we were constrained to ship the DNA to Switzerland to carry out the WGS. Since the shipment was costly, we were only able to send the DNA from human and animal fecal samples, as well as the original human fecal samples.
After running a set of DNA isolates from both human and animal samples on Nanopore, we discovered that the DNA was fragmented. Fortunately, because the human fecal samples had been shipped, we were able to re-isolate and re-extract the DNA, and successfully perform WGS. However, since we did not have the animal fecal samples in Switzerland, we were unable to conduct WGS on these samples. This limitation restricted our ability to explore the genetic aspects of resistance gene transmission between human and animal populations comprehensively. Additionally, the limited sequencing data derived from human fecal isolates, along with the absence of sequenced data from animal fecal isolates and other One Health domains, has narrowed our focus and restricted our ability to conduct comprehensive analysis of mobile genetic elements, such as phages and insertion sequence, which is essential for the dissemination of an antimicrobial resistance. Instead of, we anticipate addressing this limitation in future projects by including a broader range of samples. This will enable a more comprehensive analysis and will contribute to the understanding of transmission and dissemination of AMR in different domains of One Health.
Conclusion
Our study represents the first study of molecular epidemiology of ESBL-producing E. coli isolated from rural children and livestock in Ethiopia. We found high and low prevalence of ESBL-producing E. coli in rural children and livestock, respectively, largely mediated by the gene blaCTX-M-15 encoded on a plasmid. The isolates displayed a high diversity of STs, with the predominant types being ST-2353, ST-10, ST-48, ST-38, and ST-450. Our study is the first to report that ST-2353 is associated with multi-drug resistance genes in Ethiopia. Further research including other domains such water and food products are needed to comprehensively study this diversity and the spread of antimicrobial resistance genes to better understand their acquisition. We suggest implementing an integrated One Health surveillance system, which would be able to monitor transmission events and detect resistant bacteria in a timely manner from both humans and animals.
Supplementary Information
Acknowledgements
We would like to thank pastoralist and agro-pastoralist communities, and the field team (Ahmed, Dek, Fuad, Mohammed, Mohammoud, Ramadan, and Shamil). We would also like to thank the Jigjiga One Health Initiative team at Jigjiga University and the Armauer Hansen Research Institute in Ethiopia, as well as the Human and Animal Health Unit team at the Swiss Tropical and Public Health Institute in Switzerland, and the Microbiota-targeted Interventions Group at the University of Lausanne for helpful discusions and support. We would also like to thank Prof. Andrea Endimiani for his valuable suggestions and comments.
Abbreviations
- AMR
Antimirobial resitance
- ESBL
Extended spectrum beta lactamase
- E. coli
Eschericia coli
- WGS
Whole genome sequencing
- MLST
Multilocus sequence typing
- SRS
Somali regional state
Author contributions
AM, RT, JZ, GC and PV developed the research questions. AM, AA, YO, and SH conducted laboratory work, supervised by EI and PV. SH and TS performed the bioinformatics analysis. JH contributed to data analysis. AM wrote the first draft of the manuscript and all authors (AA, SH, YO, RT, SY, TS, JH, JZ, GC, and PV) revised and approved the final text.
Funding
Open access funding provided by University of Lausanne. This work is funded by Jigjiga One Health Initiative (JOHI) (SDC) (no. 7F-09057.01.02 to JZ), Swiss Government Excellence Scholarships (ESKAS), Nutricia Research Foundation (no. 2019-20 to PV), Forschungsfonds of the University of Basel (Fellowship in 2020 to PV), Return Grant of the SNSF (no. P3P3PA_177877 to PV) and the Eccellenza Professorial Fellowship (no. PCEFP3_194545 to PV).
Availability of data and materials
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
Ethical approval was obtained from the Swiss Ethics Committee of Northwest and Central Switzerland (Ethikkommision Nordwest- und Zentralschweiz; REQ-2020-00608), the Review Committee of Armauer Hansen Research Institute in Addis Ababa, Ethiopia (AF-10-015), the Review Committee of the University of Jigjiga in Ethiopia (JJU-RERC030/2020), and National Research Ethics Review Committee (NRERC) (D2/152/533/4). A written consent was obtained from the parent or legal guardian of all participating children before study enrolment (signed or finger print). Data was recorded using the Open Data Kit and securely stored on a server at the Swiss TPH in Basel. All personally identifiable information was maintained by the local study team in Ethiopia and securely stored in a closed cupboard.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Díaz-Agero Pérez C, et al. Local prevalence of extended-spectrum beta-lactamase (ESBL) producing Enterobacteriaceae intestinal carriers at admission and co-expression of ESBL and OXA-48 carbapenemase in Klebsiella pneumoniae: a prevalence survey in a Spanish University Hospital. BMJ Open. 2019;9(3):e024879. 10.1136/bmjopen-2018-024879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Falgenhauer L, et al. Detection and characterization of ESBL-producing Escherichia coli from humans and poultry in Ghana. Front Microbiol. 2019;9:3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ojo OE, Schwarz S, Michael GB. Detection and characterization of extended-spectrum β-lactamase-producing Escherichia coli from chicken production chains in Nigeria. Vet Microbiol. 2016;194:62–8. [DOI] [PubMed] [Google Scholar]
- 4.Moremi N, et al. Faecal carriage of CTX-M extended-spectrum beta-lactamase-producing Enterobacteriaceae among street children dwelling in Mwanza city, Tanzania. PLoS ONE. 2017;12(9): e0184592. 10.1371/journal.pone.0184592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andriatahina T, et al. High prevalence of fecal carriage of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in a pediatric unit in Madagascar. BMC Infect Dis. 2010;10:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ayinla AO, Mateus AL. Extended-spectrum beta-lactamases in poultry in Africa: a systematic review. Front Antibiot. 2023;2:1140750. [Google Scholar]
- 7.Rousham EK, Unicomb L, Islam MA. Human, animal and environmental contributors to antibiotic resistance in low-resource settings: integrating behavioural, epidemiological and One Health approaches. Proc R Soc B Biol Sci. 1876;2018(285):20180332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Williams PC, Isaacs D, Berkley JA. Antimicrobial resistance among children in sub-Saharan Africa. Lancet Infect Dis. 2018;18(2):e33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bush K, Bradford PA. β-Lactams and β-lactamase inhibitors: an overview. Cold Spring Harb Perspect Med. 2016. 10.1101/cshperspect.a025247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cantón R, González-Alba JM, Galán JC. CTX-M enzymes: origin and diffusion. Front Microbiol. 2012;3:110. 10.3389/fmicb.2012.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hawkey PM. Prevalence and clonality of extended-spectrum beta-lactamases in Asia. Clin Microbiol Infect. 2008;14(Suppl 1):159–65. 10.1111/j.1469-0691.2007.01855.x. [DOI] [PubMed] [Google Scholar]
- 12.Hawser S, et al. Antimicrobial susceptibility of intra-abdominal gram-negative bacilli from Europe: SMART Europe 2008. Eur J Clin Microbiol Infect Dis. 2011;30:173–9. [DOI] [PubMed] [Google Scholar]
- 13.Wickramasinghe NH, et al. High community faecal carriage rates of CTX-M ESBL-producing Escherichia coli in a specific population group in Birmingham, UK. J Antimicrob Chemother. 2012;67(5):1108–13. 10.1093/jac/dks018. [DOI] [PubMed] [Google Scholar]
- 14.Bonnet R. Growing group of extended-spectrum β-lactamases: the CTX-M enzymes. Antimicrob Agents Chemother. 2004;48(1):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carattoli A, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58(7):3895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirai I, et al. Detection of chromosomal blaCTX-M-15 in Escherichia coli O25b-B2-ST131 isolates from the Kinki region of Japan. Int J Antimicrob Agents. 2013;42(6):500–6. [DOI] [PubMed] [Google Scholar]
- 17.Rodríguez I, et al. Chromosomal location of blaCTX-M genes in clinical isolates of Escherichia coli from Germany, The Netherlands and the UK. Int J Antimicrob Agents. 2014;43(6):553–7. [DOI] [PubMed] [Google Scholar]
- 18.Leverstein-van Hall M, et al. Dutch patients, retail chicken meat and poultry share the same ESBL genes, plasmids and strains. Clin Microbiol Infect. 2011;17(6):873–80. [DOI] [PubMed] [Google Scholar]
- 19.Liebana E, et al. Public health risks of enterobacterial isolates producing extended-spectrum β-lactamases or AmpC β-lactamases in food and food-producing animals: an EU perspective of epidemiology, analytical methods, risk factors, and control options. Clin Infect Dis. 2013;56(7):1030–7. [DOI] [PubMed] [Google Scholar]
- 20.Sewunet T, et al. Polyclonal spread of blaCTX-M-15 through high-risk clones of Escherichia coli at a tertiary hospital in Ethiopia. J Glob Antimicrob Resist. 2022;29:405–12. 10.1016/j.jgar.2021.09.017. [DOI] [PubMed] [Google Scholar]
- 21.Negeri AA, et al. Characterization of plasmids carrying bla CTX-M genes among extra-intestinal Escherichia coli clinical isolates in Ethiopia. Sci Rep. 2023;13(1):8595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Negeri AA, et al. Antimicrobial resistance profiling and molecular epidemiological analysis of extended spectrum β-lactamases produced by extraintestinal invasive Escherichia coli isolates from Ethiopia: the presence of international high-risk clones ST131 and ST410 revealed. Front Microbiol. 2021;12: 706846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Desta K, et al. High gastrointestinal colonization rate with extended-spectrum β-lactamase-producing Enterobacteriaceae in hospitalized patients: emergence of carbapenemase-producing K. pneumoniae in Ethiopia. PLoS ONE. 2016;11(8): e0161685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Onis MD. The WHO child growth standards. Pediatric nutrition in practice, 2008; pp. 254–269.
- 25.Allen ME. MacConkey agar plates protocols. American Society for Microbiology; 2005. p. 1–4. [Google Scholar]
- 26.Performance C. Standards for antimicrobial susceptibility testing, CLSI supplement M100S. Wayne: Clinical and Laboratory Standards Institute; 2016. [Google Scholar]
- 27.Magiorakos A, et al. Bacteria: an international expert proposal for interim standard definitions for acquired resistance. Microbiology. 2011;18(3):268–81. [DOI] [PubMed] [Google Scholar]
- 28.Magiorakos A-P, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–81. [DOI] [PubMed] [Google Scholar]
- 29.Humphries RM, et al. CLSI methods development and standardization working group best practices for evaluation of antimicrobial susceptibility tests. J Clin Microbiol. 2018. 10.1128/jcm.01934-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kit WHDE. Wizard® HMW DNA extraction kit. Available from: https://ch.promega.com/products/nucleic-acid-extraction/genomic-dna/high-molecular-weight-dna-extraction-kit/?gclid=CjwKCAjw9pGjBhB-EiwAa5jl3GqNYZYh3g71uZXCHQOerfMSHQDjueve5xxhApgcBryeACn9tYI4YBoCZ_8QAvD_BwE&catNum=A2920.
- 31.Kolmogorov M, et al. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol. 2019;37(5):540–6. [DOI] [PubMed] [Google Scholar]
- 32.Brettin T, et al. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep. 2015;5(1):8365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Larsen MV, et al. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol. 2012;50(4):1355–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alcock BP, et al. CARD 2023: expanded curation, support for machine learning, and resistome prediction at the comprehensive antibiotic resistance database. Nucleic Acids Res. 2022;51(D1):D690–9. 10.1093/nar/gkac920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Katoh K, et al. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30(14):3059–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bortolaia V, et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother. 2020;75(12):3491–500. 10.1093/jac/dkaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Epidemiogy, C.f.G. Center for genomics epidemiogy 2024 [cited 2024 16.06.2024]; Available from: http://www.genomicepidemiology.org/.
- 38.R Core Team, R., R: A language and environment for statistical computing. 2013.
- 39.Bezabih YM, et al. The global prevalence and trend of human intestinal carriage of ESBL-producing Escherichia coli in the community. J Antimicrob Chemother. 2021;76(1):22–9. 10.1093/jac/dkaa399. [DOI] [PubMed] [Google Scholar]
- 40.Bezabih YM, et al. Comparison of the global prevalence and trend of human intestinal carriage of ESBL-producing Escherichia coli between healthcare and community settings: a systematic review and meta-analysis. JAC-Antimicrob Resist. 2022. 10.1093/jacamr/dlac048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Akenten CW, et al. Carriage of ESBL-producing Klebsiella pneumoniae and Escherichia coli among children in rural Ghana: a cross-sectional study. Antimicrob Resist Infect Control. 2023;12(1):60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ayukekbong JA, Ntemgwa M, Atabe AN. The threat of antimicrobial resistance in developing countries: causes and control strategies. Antimicrob Resist Infect Control. 2017;6(1):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berendes D, et al. Human faeces-associated extended-spectrum β-lactamase-producing Escherichia coli discharge into sanitation systems in 2015 and 2030: a global and regional analysis. Lancet Planet Health. 2020;4(6):e246–55. 10.1016/s2542-5196(20)30099-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nüesch-Inderbinen M, et al. Antimicrobial resistant and extended-spectrum ß-lactamase (ESBL) producing Escherichia coli isolated from fecal samples of African dromedary camels. Sci Afr. 2020;7: e00274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muhummed AM, et al. Knowledge, attitudes, and practices of rural communities regarding antimicrobial resistance and climate change in Adadle District, Somali Region, Ethiopia: a mixed-methods study. Antibiotics. 2024;13(4):292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alonso C, et al. Antibiotic resistance in Escherichia coli in husbandry animals: the African perspective. Lett Appl Microbiol. 2017;64(5):318–34. [DOI] [PubMed] [Google Scholar]
- 47.Valentin L, et al. Subgrouping of ESBL-producing Escherichia coli from animal and human sources: an approach to quantify the distribution of ESBL types between different reservoirs. Int J Med Microbiol. 2014;304(7):805–16. [DOI] [PubMed] [Google Scholar]
- 48.Day MJ, et al. Extended-spectrum β-lactamase-producing Escherichia coli in human-derived and foodchain-derived samples from England, Wales, and Scotland: an epidemiological surveillance and typing study. Lancet Infect Dis. 2019;19(12):1325–35. [DOI] [PubMed] [Google Scholar]
- 49.Aibinu I, et al. Sequence type ST131 and ST10 complex (ST617) predominant among CTX-M-15-producing Escherichia coli isolates from Nigeria. Clin Microbiol Infect. 2012;18(3):E49–51. [DOI] [PubMed] [Google Scholar]
- 50.Mshana SE, et al. Predictors of blaCTX-M-15 in varieties of Escherichia coli genotypes from humans in community settings in Mwanza, Tanzania. BMC Infect Dis. 2016;16:187. 10.1186/s12879-016-1527-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moremi N, et al. Predominance of CTX-M-15 among ESBL producers from environment and fish gut from the shores of Lake Victoria in Mwanza, Tanzania. Front Microbiol. 2016;7:1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Joffré E, et al. Identification of new heat-stable (STa) enterotoxin allele variants produced by human enterotoxigenic Escherichia coli (ETEC). Int J Med Microbiol. 2016;306(7):586–94. [DOI] [PubMed] [Google Scholar]
- 53.Kwon T, et al. Comparative genomic analysis and characteristics of NCCP15740, the major type of enterotoxigenic Escherichia coli in Korea. Gut Pathog. 2017;9:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nicolas-Chanoine M-H, et al. Intercontinental emergence of Escherichia coli clone O25: H4-ST131 producing CTX-M-15. J Antimicrob Chemother. 2008;61(2):273–81. [DOI] [PubMed] [Google Scholar]
- 55.Pankok F, et al. Epidemiology of plasmids in Escherichia coli and Klebsiella pneumoniae with acquired extended spectrum beta-lactamase genes isolated from chronic wounds in Ghana. Antibiotics. 2022. 10.3390/antibiotics11050689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Minja CA, Shirima G, Mshana SE. Conjugative plasmids disseminating ctx-m-15 among human, animals and the environment in Mwanza Tanzania: a need to intensify one health approach. Antibiotics. 2021;10(7):836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bevan ER, Jones AM, Hawkey PM. Global epidemiology of CTX-M β-lactamases: temporal and geographical shifts in genotype. J Antimicrob Chemother. 2017;72(8):2145–55. [DOI] [PubMed] [Google Scholar]
- 58.Zankari E, et al. Genotyping using whole-genome sequencing is a realistic alternative to surveillance based on phenotypic antimicrobial susceptibility testing. J Antimicrob Chemother. 2013;68(4):771–7. 10.1093/jac/dks496. [DOI] [PubMed] [Google Scholar]
- 59.Moura A et al. Phenotypic and genotypic antimicrobial resistance of Listeria monocytogenes: an observational study in France. Lancet Reg Health—Europe 2024;37. [DOI] [PMC free article] [PubMed]
- 60.Yee R, Dien Bard J, Simner PJ. The genotype-to-phenotype dilemma: How should laboratories approach discordant susceptibility results? J Clin Microbiol. 2021. 10.1128/jcm.00138-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhong Y, et al. Characterization of extended-spectrum beta-lactamase-producing Escherichia coli isolates from Jurong Lake, Singapore with whole-genome-sequencing. Int J Environ Res Public Health. 2021;18(3):937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Enne VI, et al. Evidence of antibiotic resistance gene silencing in Escherichia coli. Antimicrob Agents Chemother. 2006;50(9):3003–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tola MA, et al. High prevalence of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae fecal carriage among children under five years in Addis Ababa, Ethiopia. PLoS ONE. 2021;16(10): e0258117. 10.1371/journal.pone.0258117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Herindrainy P, et al. Rectal carriage of extended-spectrum beta-lactamase-producing gram-negative bacilli in community settings in Madagascar. PLoS ONE. 2011;6(7): e22738. 10.1371/journal.pone.0022738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Isendahl J, et al. Fecal carriage of ESBL-producing E coli and K pneumoniae in children in Guinea-Bissau: a hospital-based cross-sectional study. PLoS ONE. 2012;7(12): e51981. 10.1371/journal.pone.0051981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tellevik MG, et al. High prevalence of faecal carriage of ESBL-producing enterobacteriaceae among children in Dar es Salaam, Tanzania. PLoS ONE. 2016;11(12): e0168024. 10.1371/journal.pone.0168024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Woerther P-L, et al. Massive increase, spread, and exchange of extended spectrum β-lactamase–encoding genes among intestinal Enterobacteriaceae in hospitalized children with severe acute malnutrition in Niger. Clin Infect Dis. 2011;53(7):677–85. [DOI] [PubMed] [Google Scholar]
- 68.De Lauzanne A, et al. Prevalence and factors associated with faecal carriage of extended-spectrum β-lactamase-producing Enterobacterales among peripartum women in the community in Cambodia. J Antimicrob Chemother. 2022;77(10):2658–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rytter MJ, et al. The immune system in children with malnutrition–a systematic review. PLoS ONE. 2014;9(8): e105017. 10.1371/journal.pone.0105017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cissé G. Food-borne and water-borne diseases under climate change in low- and middle-income countries: Further efforts needed for reducing environmental health exposure risks. Acta Trop. 2019;194:181–8. 10.1016/j.actatropica.2019.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Magnano San Lio R, et al. How antimicrobial resistance is linked to climate change: an overview of two intertwined global challenges. Int J Environ Res Public Health. 2023;20(3):1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nielsen A-M, et al. Chlorination for low-cost household water disinfection–a critical review and status in three Latin American countries. Int J Hyg Environ Health. 2022;244: 114004. [DOI] [PubMed] [Google Scholar]
- 73.Montealegre MC, et al. Drinking water chlorination impact on fecal carriage of extended-spectrum beta-lactamase-producing Enterobacteriaceae in Bangladeshi children in a double-blind, cluster-randomized controlled trial. Environ Health Perspect. 2022;130(11): 117702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Krige M. Molecular investigation of the chlorine and antibiotic resistance mechanisms of Escherichia coli isolated from natural water sources in the Western Cape. Stellenbosch: University of Stellenbosch; 2009. [Google Scholar]
- 75.Xiao X, et al. Chlorine tolerance and cross-resistance to antibiotics in poultry-associated salmonella isolates in China. Front Microbiol. 2021;12: 833743. 10.3389/fmicb.2021.833743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jin M, et al. Chlorine disinfection promotes the exchange of antibiotic resistance genes across bacterial genera by natural transformation. ISME J. 2020;14(7):1847–56. 10.1038/s41396-020-0656-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Aworh MK, et al. Extended-spectrum ß-lactamase-producing Escherichia coli among humans, chickens and poultry environments in Abuja, Nigeria. One Health Outlook. 2020;2(1):8. 10.1186/s42522-020-00014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Langata LM, et al. Antimicrobial resistance genes in Salmonella and Escherichia coli isolates from chicken droppings in Nairobi, Kenya. BMC Res Notes. 2019;12(1):22. 10.1186/s13104-019-4068-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liaqat Z, et al. Isolation and molecular characterization of extended spectrum beta lactamase producing Escherichia coli from chicken meat in Pakistan. PLoS ONE. 2022;17(6): e0269194. 10.1371/journal.pone.0269194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rahman MM, et al. Isolation and molecular characterization of multidrug-resistant Escherichia coli from chicken meat. Sci Rep. 2020;10(1):21999. 10.1038/s41598-020-78367-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Badr H, et al. Multidrug-resistant and genetic characterization of extended-spectrum beta-lactamase-producing E. coli recovered from chickens and humans in Egypt. Animals. 2022;12(3):346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Overdevest I, et al. Extended-spectrum β-lactamase genes of Escherichia coli in chicken meat and humans, The Netherlands. Emerg Infect Dis. 2011;17(7):1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed during the current study.