Abstract
Background
Intercropping increases land use efficiency and farmland ecological diversity. However, little is understood about whether and how soil biota, metabolites, and nutrients change under interspecific competition among plants. Thus, this study aimed to explore the changes in the physicochemical properties, microbial communities, and metabolites of rhizosphere and bulk soils of pepper monocropping and pepper–maize intercropping systems.
Results
Intercropping significantly increased the contents of available phosphorus (AP) and available potassium (AK), and decreased the pH value, whereas it had little effect on the total nitrogen (TN) and organic matter (OM) in the rhizosphere and bulk soils, compared with those in monocropping pepper. Moreover, the OM content was higher in rhizosphere soil than in bulk soil. The microbial community structures and metabolite profiles also differed between the two systems. The diversity of bacteria and fungi increased in intercropped pepper. The relative abundances of Actinobacteria, Chloroflexi, Cyanobacteria, and Ascomycota were higher while those of Proteobacteria, Planctomycetes, Mucoromycota, and Basidiomycota were significantly lower in the rhizosphere and bulk soils from the intercropping system than in those from the monocropping system. Linear discriminant analysis revealed that the predominant bacteria and fungi in the rhizosphere soil from the intercropping system belonged to the order Sphingomonadales and genera Nitrospira, Phycicoccus and Auricularia, whereas those in the bulk soil from the intercropping system belonged to the phylum Acidobacteria and genera Calocera, Pseudogymnoascus, and Trichosporon. Intercropping promoted the secretion of flavonoids, alkaloids, and nucleotides and their derivatives in the rhizosphere soil and significantly increased the contents of organoheterocyclic compounds in the bulk soil. Furthermore, the AP and AK contents, and pH value had strong positive correlations with bacteria. In addition, co-occurrence network analysis also showed that asebogenin, trachelanthamidine, 5-methyldeoxycytidine, and soil pH were the key factors mediating root-soil-microbe interactions.
Conclusion
Intercropping can alter microbial community structures and soil metabolite composition in rhizosphere and bulk soils, enhancing soil nutrient contents, enriching soil beneficial microbes and secondary metabolites (flavonoids and alkaloids) of intercropped pepper, and provided a scientific basis for sustainable development in the pepper-maize intercropping system.
Supplementary Information
The online version contains supplementary material available at 10.1186/s40793-024-00653-7.
Keywords: Pepper, Intercropping, Rhizosphere and bulk soil, Soil nutrient, Bacterial and fungal community structures, Soil metabolite profiles
Background
Intercropping, also known as mixed cropping, involves simultaneously growing more than one species on a field. It has great potential for enhancing water and nutrient use efficiency and improving plant productivity, yield stability, and resilience to biotic and abiotic stresses, including those triggered by climate change [1]. The yield advantage of cereal and legume intercropping systems is evident in various intercropping patterns [2–5]. Intercropping pepper (Capsicum annuum L.) with other crops is an effective and economic measure to restrict the spread of Phytophthora capsici across rows in the soil [6, 7]. Understanding the critical roles of underground interspecific interactions is important to regulate the nitrogen (N) cycling of plant–soil systems and mediate the mineralisation of soil organic matter (OM) [8–10].
The rhizosphere is a key area for roots to obtain water and nutrients, and it interacts closely with soil physical, chemical, and biological components [11]. However, continuous monocropping and unreasonable fertilisation disrupt the balance of soil–microbe–plant relationships and alter soil physicochemical and biological characteristics [12, 13]. Plant roots release exudates, which are the main nutrient source driving bacterial communities and activities [14]. Thus, root–soil interactions are critical for soil health, sustainable food security, and resource use efficiency [15].
Intercropping improves soil nutrient cycling and land productivity by regulating microbial community activities [16]. In maize (Zea mays) -peanut (Arachis hypogaea) intercropping system, the diversity and richness of bacteria and fungi decrease in maize rhizosphere soil, whereas the richness of fungi increases in peanut rhizosphere soil [8]. The abundance of beneficial microbes (RB41 and Chaetomium) in the rhizosphere and the nitrogen content of maize are also increased when intercropped with peanut [8]. Apple (Malus pumila)-marigold (Tagetes erecta) intercropping significantly increases the relative abundance (RA) of plant growth-promoting bacteria, such as Rhizobiales, Pseudomonadales, and Bacillales, in rhizosphere soils. Moreover, the amount of carbohydrates is higher in intercropping systems than in monocropping systems [17]. Many studies reported that intercropping improves yield and resistance against soil-borne Phytophthora disease in pepper [6, 7]. However, few studies have clearly demonstrated the interactions between the microbial community structure and the soil metabolite profiles during intercropping. Specifically, the effects of maize–pepper intercropping on the nutrients, microbial communities, and metabolites in the soil remain unclear.
Considering that various intercropping patterns differentially affect soil physicochemical properties and microbial characteristics, we aimed to examine the changes in the bacterial and fungal communities in the pepper–maize intercropping system. We hypothesised that intercropping would greatly affect soil physicochemical properties, increase microbial diversities, and alter microbial community structures. Thus, the objectives of this study were to (1) explore the effects of pepper–maize intercropping on soil physicochemical properties and metabolite profiles; (2) compare the responses of bacterial and fungal diversities and community composition to intercropping with monoculture plantations of pepper; and (3) determine the relationships between soil microbial communities and soil physicochemical properties and metabolite profiles.
Materials and methods
Experimental site and soil sample collection
The experimental site was in Dali City, Yunnan Province, China (26°12’79” N, 99°96’91"E). This region has a subtropical climate with an annual precipitation of 719.2 mm and an average annual temperature of 14.2 °C. The soil at the test site was brown and loamy soil. Six core soil samples were collected from each cropping ridge using a soil sampler and mixed as one replication sample per group (Additional file 1: Fig. S1). Thus, 12 samples were collected and divided into four groups with three replications each: rhizosphere soil of the pepper–maize intercropping system (IPr1, IPr2, and IPr3), bulk soil of the pepper–maize intercropping system (IB1, IB2, and IB3), rhizosphere soil of the pepper monocropping system (MPr1, MPr2, and MPr3), and bulk soil of the pepper monocropping system (MB1, MB2, and MB3) (Additional file 1: Fig. S1). The effects of planting pattern on the physicochemical properties, microbiome, and metabolites of the soil samples were then evaluated. All samples were placed in an ice box and brought to the laboratory. Each soil sample was passed through a 2 mm sieve and then divided into two subsamples: one portion was used for the determination of soil physicochemical properties, whereas the remaining portion was stored at − 80 °C for subsequent microbiome and metabolome analyses.
Determination of soil properties
The physicochemical properties of the soil samples were analysed as previously described [18, 19]. Soil pH was determined using pH meter (FE28, METTLER-TOLEDO, USA) with a soil-to-water ratio of 1:5 (wt/vol). Available phosphorus (AP) was assessed using the ascorbic acid reductant method (SpectraMax 190 Microplate Reader, Molecular Devices, USA), and available potassium (AK) was determined through atomic absorption (Atomic absorption spectrometer zeenit 700Q, Jena, Gemany). Samples were combusted by high-temperature reactor (SPH120 Digestion System, Jnan Alva Instrument Co., LTD, China), after used to detect total nitrogen (TN) concentration by Kjeldahl method (KN-520 Automatic Kjeldahl Nitrogen Analyzer, Jnan Alva Instrument Co., LTD, China). The dichromate oxidation (Titrette® 50 mL Digital Bottletop Burette, Brand, Germany) and external heating method (HH-S Thermostatic Lab Digital Oil Bath, Changzhou LangYue Instrument Co., LTD, China) was applied to explore the OM content of the soil.
DNA extraction, library construction, and metagenomic sequencing
Soil DNA was extracted from 0.5 g of soil using the EZNA® Soil DNA Kit (Omega Biotek, Inc., Norcross, GA, USA) in accordance with the manufacturer’s instructions. The concentration of DNA was measured using the NanoDrop 2000-UV spectrophotometer (Thermo Scientific, Waltham, MA, USA), and the quality of DNA was monitored on 1% agarose gels. For the library construction, 1 µg of DNA per sample was used. Sequencing libraries were generated using the NEBNext® Ultra™ DNA Library Prep Kit for Illumina (NEB, USA) following the manufacturer’s recommendations. Briefly, the DNA samples were fragmented by sonication to a size of 350 bp. DNA fragments were end-polished, A-tailed, and ligated with the full-length adaptor for Illumina sequencing with further PCR amplification. Finally, the PCR products were purified (AMPure XP system, Beckman Coulter, Brea, CA, USA), and libraries were analysed for size distribution on an Agilent2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA) and quantified using real-time PCR. After cluster generation of the index-coded samples, the libraries were sequenced on an Illumina NovaSeq platform (Illumina PE150), and paired-end reads were generated.
Metagenome assembly, gene prediction, and functional annotation
To obtain high-quality clean data for subsequent analyses, we trimmed the raw data from the Illumina PE150 platform by using Readfq (Version 8.0, https://github.com/cjfields/readfq). Briefly, sequences with low-quality bases (default quality threshold value ≤ 20), N bases reaching 10 bp long, and final lengths < 50 bp were removed. The obtained clean data were assembled and analysed using MEGAHIT software (Version 1.0.4) [20]. Then, the assembled Scaftigs were interrupted from the N connection, and the Scaftigs without N were retained. Scaftigs longer than 500 bp were used to predict the open reading frame. The metagenomic DNA sequences were assigned taxonomic labels using the Kraken 2 program [21], and then the abundance of microbes in each sample at different phylogenetic levels (phylum, class, order, family, genus, and species) was estimated using Bracken (Bayesian Reestimation of Abundance after Classification with KrakEN) [22]. The clean reads were BLAST searched against the Uniref90 database using Humann2 software (based on Diamond) [23]. The annotation information and RA table from each functional database were obtained according to the corresponding relationship between Uniref90 ID and each database [24, 25].
Metabolite profiling and analysis
The soil samples were homogenised for 1.5 min at 30 Hz in a mixer mill (MM 400, Retsch, Hann, Germany) containing zirconium beads. A 100 mg sample from each replicate was weighed and extracted overnight at 4 °C with 1.2 mL of aqueous methanol (70%). After centrifugation at 12,000 rpm for 10 min, the extracts were filtered and subjected to ultra-performance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS).
The identified metabolites were annotated through the Kyoto Encyclopedia of Genes and Genomes (KEGG) database and then mapped to the KEGG pathway database. Metabolite enrichment analysis was conducted using MetaboAnalyst (Version 6.0), employing the hypergeometric test. Variable significance in projection, VIP > 1, and absolute Log2fold change > 1 were used to assess whether the metabolites were significantly regulated across the groups. VIP values were extracted from the orthogonal partial least squares discriminant analysis (OPLS-DA) results using the MetaboAnalyst (Version 6.0).
Statistical analysis
Principal coordinate analysis (PCoA) based on a Bray–Curtis distance matrix was conducted to visualize the distribution of microbial community in soils with different planting patterns. Principal Component Analysis (PCA) was performed using the data, which was scaled and logarithmically transformed by the SIMCA software package (Version 16.0.2), to investigate the changes in metabolites in soils with different planting patterns. Differential metabolites were screened using OPLS-DA. Linear discriminant analysis with effect size (LEfSe) was conducted, and heat maps were drawn using the OECloud tools (https://cloud.oebiotech.com/#/bio/tools?cat_id&tag_id&search). Spearman’s correlation analysis of metabolites and dominant microbes, and co-occurrence network analysis were performed using the OmicShare tools (http://www.omicshare.com/tools). Kruskal–Wallis tests (p < 0.05) (http://cloudtutu.com.cn/) were used to compare the abundances of dominant microbes in the samples at the phylum and genus levels. Mantel test correlation analysis of soil physicochemical properties and soil microorganisms was completed using the Wekemo Bioincloud (https://www.bioincloud.tech) [26]. Statistical analyses also were performed using SPSSAU (Version 24.0) (https://www.spssau.com.) and GraphPad Prism 8.0.
Results
Intercropping affects the physicochemical properties of rhizosphere and bulk soils
The variations in edaphic factors in the rhizosphere and bulk soils under the two cultivation systems are shown in Table 1. The nutrient contents in the rhizosphere and bulk soils were significantly higher (p < 0.01) in the intercropping system than in the monocropping system. Specifically, the contents of AP in the rhizosphere and bulk soils increased by fourfold and sixfold, respectively, and those of AK enhanced by twofold and onefold, respectively. The pH values in the rhizosphere and bulk soils were significantly lower in the intercropping system than in the monocropping system. No significant differences in OM and TN contents were found between the two cultivation systems. However, soil OM content was higher in the rhizosphere soil than in the bulk soil. These results suggested that intercropping altered the physicochemical properties and enhanced the total nutrient contents in the soils.
Table 1.
Basic physicochemical parameters in the rhizosphere and bulk soils under pepper intercropping and monocropping modes
| Samples | pH | OM (g/kg) | TN (g/kg) | AP (mg/kg) | AK (mg/kg) |
|---|---|---|---|---|---|
| IPr | 7.63 ± 0.06 B | 76.88 ± 3.91 AB | 4.68 ± 0.08 A | 229.93 ± 9.11 A | 553.51 ± 4.07 A |
| MPr | 8.10 ± 0.02 A | 79.81 ± 0.03 A | 4.60 ± 0.09 A | 46.77 ± 8.72 B | 178.93 ± 28.77 C |
| IB | 7.67 ± 0.01 B | 72.05 ± 1.77 B | 4.41 ± 0.04 A | 221.44 ± 11.01 A | 375.15 ± 15.25 B |
| MB | 8.04 ± 0.01 A | 74.19 ± 2.90 AB | 4.13 ± 0.64 A | 31.54 ± 6.97 B | 186.44 ± 0.34 C |
Data (means ± SD, n = 3) followed by different letters indicate significant differences at p < 0.01
Intercropping changes the microbial composition and structure in rhizosphere and bulk soils
The correlation coefficients of the abundance of unigenes in both rhizosphere and bulk soils between different planting patterns were close to 1, indicating that the higher the similarity of gene abundance between samples, the more reliable the experiment and the more reasonable the sample selection (Additional file 1: Fig. S2).
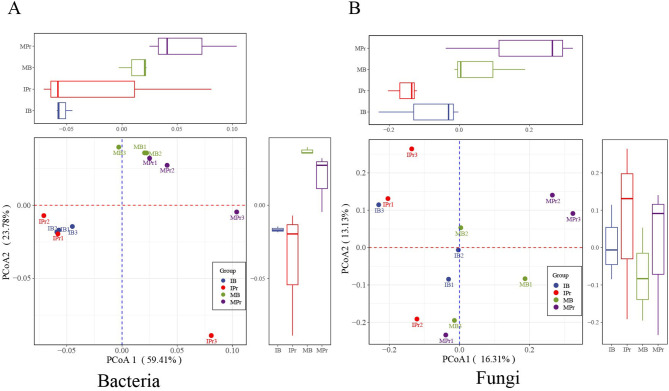
A moderate divergence in the numbers of genes was found between groups. Bacteria were the most abundant group in the rhizosphere and bulk soils from the monocropping and intercropping systems. As shown in Additional file 1: Fig. S3, the number of bacterial and fungal genes ranged from 1,092,690 (MPr) to 1,153,157 (MB) and 299 (MPr) to 315 (IPr), respectively. PCoA results showed obvious differences in the bacterial and fungal communities of both the rhizosphere soil and bulk soil between the different planting patterns (Fig. 1). The bacterial and fungal communities could be separated along the second and first coordinate axes, respectively.
Fig. 1.
Bacterial (A) and fungal (B) community composition by PCoA, based on 97% sequence similarity. IPr: intercropped pepper rhizosphere soil, IB: bulk soil of pepper-maize intercropping system, MPr: monocropped pepper rhizosphere soil, and MB: bulk soil of pepper monocropping system
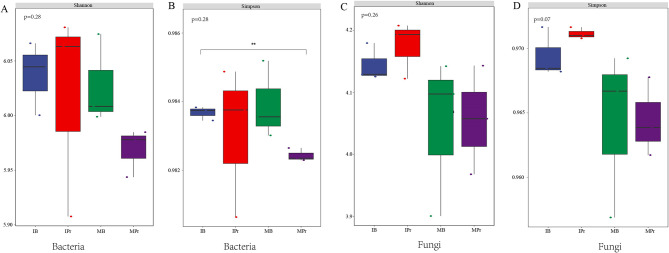
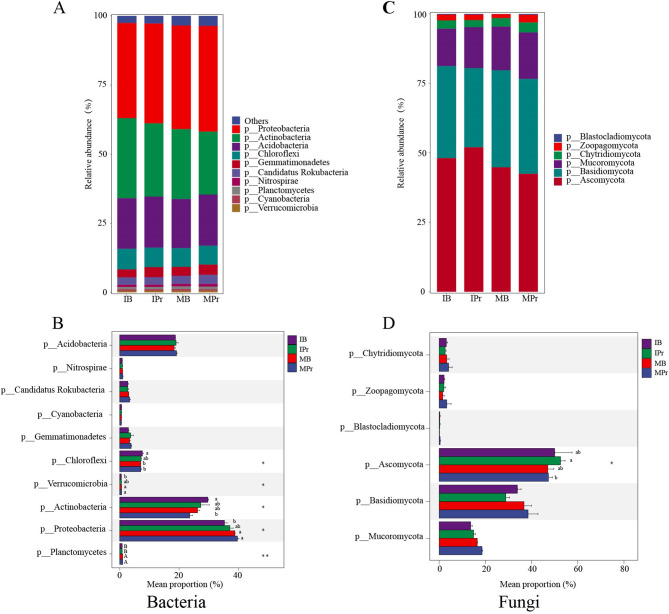
The interaction between crops increased the diversity of soil microbial communities and the abundance of some bacteria and fungi. The results of microbial diversity analyses revealed that the Shannon and Simpson index values of bacteria and fungi in the rhizosphere and bulk soils were higher under the intercropping system than under the monocropping system (Fig. 2). The RAs of the top 10 most abundant phyla and genera showed evident variations between the different planting patterns (Fig. 3A, B, Additional file 1: Fig. S4A, B). At the phylum level, Proteobacteria (34.21–38.02%), Actinobacteria (22.62–28.80%), and Acidobacteria (17.61–18.36%) were the most dominant soil bacteria identified in both cropping systems, followed by Chloroflexi (6.78–7.42%), Gemmatimonadetes (2.84–3.68%), Candidatus Rokubacteria (2.64–3.22%), Nitrospirae (0.84–0.99%), Planctomycetes (0.80–0.96%), Cyanobacteria (0.54–0.60%), and Verrucomicrobia (0.54–0.60%) (Fig. 3A). However, the RAs of Actinobacteria, Chloroflexi, and Cyanobacteria were higher in IPr and IB than in MPr and MB. Meanwhile, the RA of Planctomycetes was significantly lower (p < 0.01) in IPr and IB than in MPr and MB (Fig. 3B). Additionally, the RA of Verrucomicrobia was significantly lower (p < 0.05) in the bulk soil from the intercropping system than in that from the monocropping system. Among the top 10 bacterial genera, eight had higher RAs in IPr and IB than in MPr and MB (Additional file 1: Fig. S4A, B). Noticeably, Sphingomonas had a higher RA in IPr than in MPr. In the fungal community (Fig. 3C, D, Additional file 1: Fig. S4C, D), the RAs of the predominant phyla varied amongst the different planting patterns. The RA of Ascomycota was considerably higher in IPr and IB; those of Basidiomycota, Chytridiomycota, and Mucoromycota were higher in MPr and MB; and that of Zoopagomycota was higher in MPr and IB. The fungal genera Coniosporium and Pyrenophora had higher RAs in the intercropping system than in the monocropping system. Moreover, the RA of Penicilliopsis increased in IB (Additional file 1: Fig. S4C, D).
Fig. 2.
Diversity of the bacterial (A, B) and fungal (C, D) communities in IPr, IB, MPr, and MB.** means significantly at p < 0.01 level
Fig. 3.
RAs of the bacteria (A, B) and fungi (C, D) phyla in IPr, IB, MPr, and MB. *, and ** means significantly at p < 0.05 and p < 0.01 level, respectively. Different lower and uppercase letters means significantly at p < 0.05 and p < 0.01 level, respectively
At the species level (Additional file 1: Fig. S5), Betaproteobacteria bacterium RIFCSPLOWO2_12_FULL_65_14 and Gemmatimonadetes bacterium SCN 70 − 22 were significantly enriched in IPr. Chloroflexi bacterium RBG_16_69_14, Chloroflexi bacterium RBG_16_70_13, and Chloroflexi bacterium CSP1-4 were enriched in MPr. In the bulk soil, most Actinobacteria bacteria were the predominant species in IB, and the RAs of Actinobacteria bacterium 13_1_20CM_3_68_9 and Solirubrobacterales bacterium 70 − 9 were higher in MB than in IB. Among the fungal species (Additional file 1: Fig. S5B), several Ascomycota fungi and Linnemannia elongata were abundant in IPr, and the RAs of Mucor ambiguus, Serendipita vermifera, and Choanephora cucurbitarum were higher in MPr than in IPr. In the bulk soil, the predominant species were Spizellomyces punctatus and Fonsecaea monophora in IB, and Kockovaella imperatae and Penicillium subrubescens in MB.
These results revealed that pepper intercropping significantly changed the microbial community composition and structure in the rhizosphere and bulk soils.
Biomarker and functional analyses of soil microbial communities in rhizosphere and bulk soils under different planting patterns
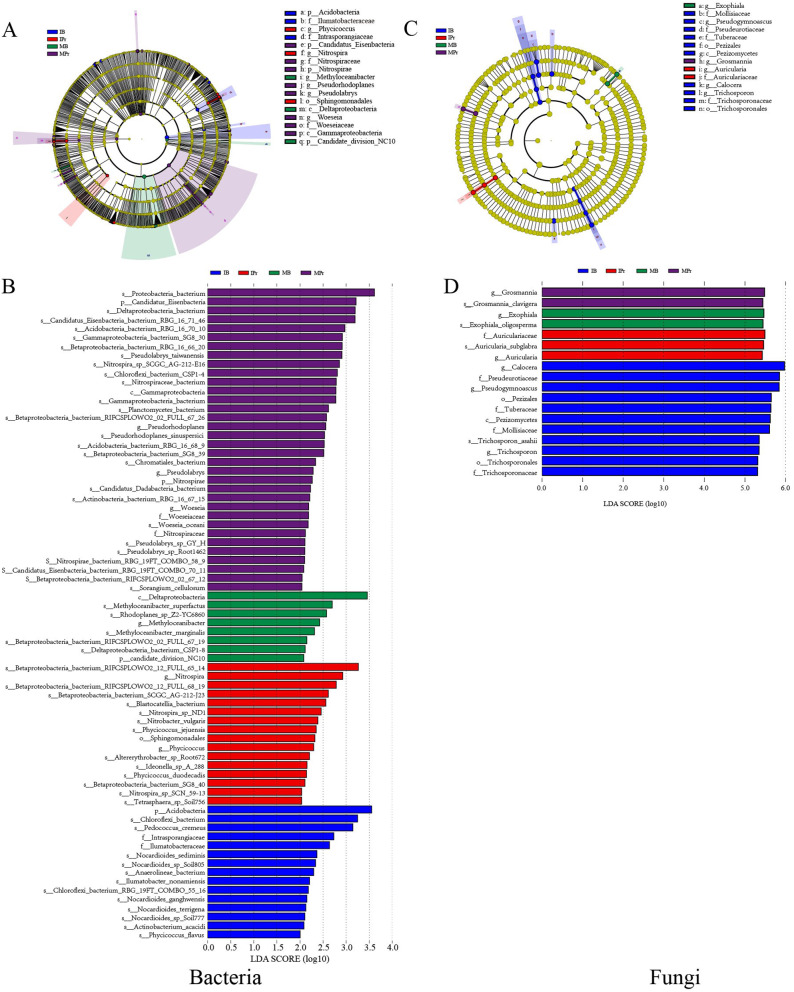
LEfSe was employed to compare the bacterial and fungal communities between the monocropping and intercropping systems (Fig. 4). At the bacterial genus level, Nitrospira and Phycicoccus were enriched in IPr (Fig. 4A, B). Meanwhile, the relative abundances of Pseudorhodoplanes, Pseudolabrys, and Woeseia increased in MPr. In addition, Methyloceanibacter was significantly enriched in MB (Fig. 4A, B). However, the phylum Candidatus Eisenbacteria and class Gammaproteobacteria were significantly enriched in MPr. Compared with MPr and MB, the relative abundances of the order Sphingomonadales and the phylum Acidobacteria were higher in IPr and IB, respectively (Fig. 4A, B). In the fungal community (Fig. 4C, D), the abundances of Auricularia, Grosmannia and Exophiala were significantly enriched in IPr, MPr, and MB, respectively, whereas Calocera, Pseudogymnoascus, and Trichosporon were more abundant in IB. Overall, LEfSe analysis identified 49 differential microbes in rhizosphere soils between the monocropping and intercropping systems, and they were used for further analyses.
Fig. 4.
LEfSe for bacterial (A, C) and fungal (B, D) taxa in IPr, IB, MPr, and MB. Significant differences are defined at p < 0.05, LDA score > 2.0 in bacterial and fungal taxa
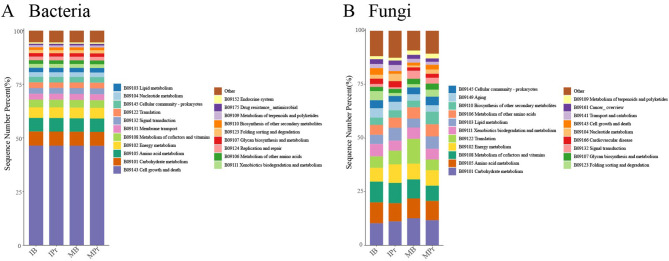
Analysis of bacterial/fungal community functional prediction
Analysis of the KEGG pathways revealed that the functions of the two levels of microbial community was mainly metabolism, and 24 major level-2 sub-systems were identified in the metagenome samples (Fig. 5). In the bacterial community, cell growth and death, carbohydrate metabolism, and amino acid metabolism were the top three pathways in level-2 sub-systems in all samples (Fig. 5A). However, in the fungal community, carbohydrate metabolism, amino acid metabolism, metabolism of cofactors and vitamins, and energy metabolism were enriched in all samples (Fig. 5B).
Fig. 5.
The Predictions of function in the bacterial (A) and fungal (B) communities
Metabolite changes in rhizosphere and bulk soils under different planting patterns
A total of 1294 peaks were detected in the chromatogram. Based on the agreement of the mass spectrum fingerprint and the retention index, 429 metabolites in the rhizosphere and bulk soils were identified and grouped into 15 classes. Of these compounds, flavonoids had the highest concentration, accounting for 16.55% of the total, followed by alkaloids (14.22%), organoheterocyclic compounds (9.32%), benzenoids (8.86%), and terpenoids (8.62%) (Additional file 1: Fig. S6A).
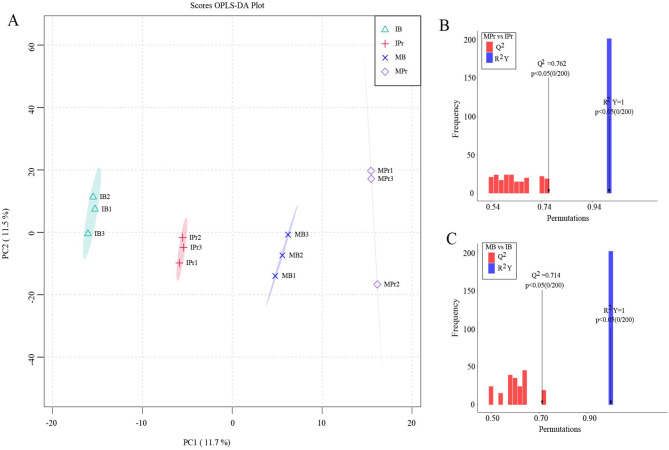
OPLS-DA results revealed a clear separation between the intercropping and monocropping systems (Fig. 6A), indicating that planting patterns significantly affected the soil metabolites. The first and second principal components accounted for 11.7% and 11.5% of the total variation, respectively. Score plots showed that the metabolites were statistically separated in IPr, IB, MPr, and MB. The OPLS-DA results also indicated that the model could better explain the differences between planting patterns (R2Y = 1, Q2 > 0.5; Fig. 6B, C), proving that the results of differential soil metabolites caused by intercropping were stable and reliable. The metabolites in MPr vs. IPr and MB vs. IB were clearly separated along the first principal component (Additional file 1: Fig. S6B, C).
Fig. 6.
OPLS-DA score plots (A) and models (B, C) of metabolites in rhizosphere and bulk soils
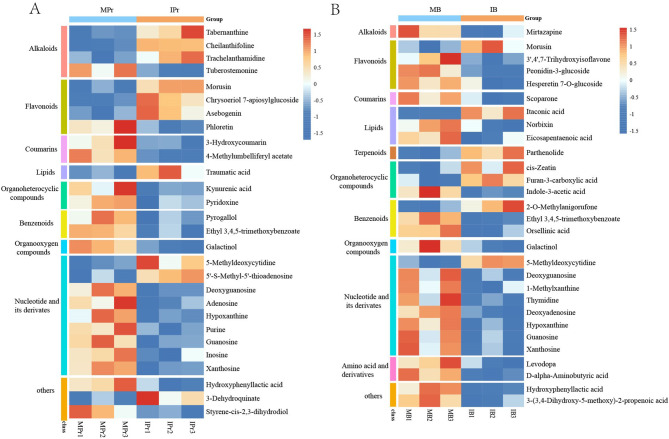
A heatmap was used to visualise the differential expressed metabolites between the intercropping and monocropping systems (Fig. 7). In total, 28 (10 upregulated and 18 downregulated) and 29 (7 upregulated and 22 downregulated) were significantly enriched in MPr vs. IPr and MB vs. IB, respectively. In rhizosphere soil, IPr mainly drives the accumulation of flavonoids (such as morusin, chrysoeriol 7-apiosylglucoside, and asebogenin), alkaloids (tabernanthine, cheilanthifoline, and trachelanthamidine), nucleotide and its derivates (5′-S-methyl-5′-thioadenosine and 5-methyldeoxycytidine), and lipids (traumatic acid) (Fig. 7A). In bulk soil, organoheterocyclic compounds (such as cis-Zeatin and furan-3-carboxylic acid) and terpenoids (parthenolide) were greatly increased in IB (Fig. 7B). Overall, the results demonstrated that intercropping can significantly alter the metabolites in the rhizosphere and bulk soils.
Fig. 7.
Heatmap analysis of the differential expressed metabolites in MPr vs. IPr (A) and MB vs. IB (B)
Pathway enrichment analysis on these differential metabolites was used to illuminate the specific changes in soil metabolic pathways. Purine metabolism was the most significantly altered process in the soils. Phenylalanine, tyrosine, and tryptophan biosyntheses were significantly impacted in the rhizosphere soil, whereas tryptophan metabolism was significantly affected in the bulk soil (Additional file 1: Fig. S7).
Correlation between the microbial community composition and soil physicochemical properties
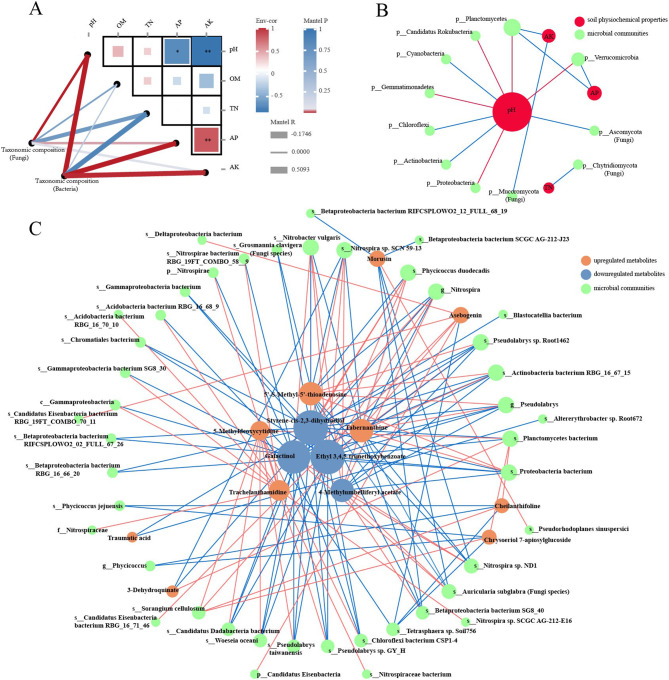
The microbial community composition was highly associated with intrinsic edaphic factors. The association between the essential edaphic factors and microbial community composition was discerned using mantel test and co-occurrence network analysis (Fig. 8A, B). The results demonstrated that the taxonomic composition of bacteria had a significantly positive correlation (p < 0.01) with soil pH, AP, and AK (Fig. 8A). Soil pH also had a positive correlation (p < 0.01) with fungal community. The co-occurrence network revealed that soil pH was a key edaphic factor regulating the bacterial community composition (Fig. 8B). Planctomycetes, Proteobacteria, Verrucomicrobia, Gemmatimonadetes, and Candidatus Rokubacteria showed a significantly positive correlation with soil pH, whereas Actinobacteria, Chloroflexi, and Cyanobacteria revealed the opposite. Soil AP and AK had a significantly positive association with Planctomycetes. The fungal phyla Ascomycota, Chytridiomycota, and Mucoromycota were significantly correlated with soil pH, TN, and AK, respectively.
Fig. 8.
The Mantel tests (A) and co-occurrence network analysis (B, C) of microbial communities with soil physiochemical properties and differential metabolites. Red and blue lines indicate positive and negative correlations, respectively
Correlations between the metabolism and microbial communities in pepper rhizosphere soil
Interactive networks were constructed to elucidate the relationship between the differential metabolites and microorganisms in rhizosphere soils from the monocropping and intercropping systems (MPr vs. IPr) (Additional file 1: Fig. S8, Fig. 8C). For upregulated metabolites, a significantly positive (p < 0.01) correlation was observed between the trachelanthamidine and bacteria related to the family Nitrospiraceae, class Gammaproteobacteria, phylum Nitrospirae, and two Acidobacteria bacterium species. Three upregulated metabolites (5-methyldeoxycytidine, cheilanthifoline, and chrysoeriol 7-apiosylglucoside) showed a significantly positive (p < 0.01) correlation with Sorangium cellulosum and Planctomycetes bacterium for MPr vs. IPr. Similarly, asebogenin also exhibited a significantly positive correlation with S. cellulosum for MPr vs. IPr. Three downregulated metabolites (ethyl 3,4,5-trimethoxybenzoate, styrene-cis-2,3-dihydrodiol, and galactinol) showed the most remarkable correlation with the rhizosphere soil bacterial and fungal taxa, being especially positively correlated with Nitrobacter vulgaris and Phycicoccus duodecadis. Sphingomonadales showed a significantly positive correlation with four downregulated metabolites, namely, inosine, adenosine, pyridoxine, and xanthosine. However, two fungal species (Auricularia subglabra and Grosmannia clavigera) displayed a significantly positive or negative correlation (p < 0.01) with 11 differential metabolites (2 upregulated and 9 downregulated).
Discussion
In this study, we characterised the effects of intercropping pepper with maize on soil microbial communities and metabolite profiles by using high-throughput sequencing and soil metabolomics. Previous studies revealed that interactions between crops affect microbial diversity [27]. Moreover, intercropping systems can promote soil microbial diversity and community composition, and biochemical property in the rhizosphere [28, 29] and non-rhizosphere soils [30]. In the present study, the diversity of the bacterial and fungal communities increased in the rhizosphere and bulk soils from the pepper intercropping system (Fig. 2). The nutrient contents (TN, AK, and AP) were also higher in the rhizosphere and bulk soils from the intercropping system than in those from the monocropping system (Table 1). However, soil OM content was higher in the rhizosphere soil than in the bulk soil.
Furthermore, intercropping systems notably affect bacterial community composition [31, 32]. In the current study, bacteria were the most abundant group in the rhizosphere soil of intercropped pepper (Additional file 1: Fig. S1). The RAs of Actinobacteria, Chloroflexi, and Cyanobacteria increased in the rhizosphere and bulk soils from the pepper intercropping system (Fig. 3B). This finding is consistent with the results of a previous study [27]. Specifically, IB had a significantly higher abundance of Actinobacteria. Actinobacteria have a ubiquitous distribution in the biosphere being a dominant taxon in soil microbial communities [33]. Actinobacteria contribute significantly to nitrogen fixation [34], decomposition of OM such as cellulose and lignin [33, 35], plant growth [36], organic acid production [37], and phosphate solubilisation [38] in soil. In addition, Sphingomonas had a higher RA in IPr than in MPr (Additional file 1: Fig. S4A, B). Sphingomonas promotes nitrogen fixation and dehydrogenation [39], which enhancing the uptake of nutrients in the rhizosphere, improving the rhizosphere soil environment of intercropped pepper, and maintaining the soil nitrogen balance.
Fungi are an important part of soil microorganisms and play a crucial role in soil ecosystems.
Compared with bacteria, fungi play critical roles as decomposers, symbionts, and pathogens in the soil [40]. Saprophytic fungi help decompose soil substrates, contributing to the soil nutrient cycle, while ectomycorrhizal fungi, such as symbiotrophs, improve the nutritional status of plants [41, 42]. In the present study, the diversity and gene number of fungal community increased in the pepper intercropping system (Fig. 2; Additional file 1: Fig. S3), consistent with the promotion of fungal community growth under intercropping [8]. The RAs of the phylum Ascomycota and genus Pyrenophora were higher in IPr than in MPr. However, the RA of the genus Rhizoctonia significantly decreased in the rhizosphere soil of intercropped pepper. The fungal community can be influenced by various biotic and abiotic factors. For instance, the diversity and amount of root exudates produced by different crops impact the abundance of the fungal community [8, 43].
Among the marker bacteria associated with pepper intercropping, Nitrospira and Phycicoccus were mostly involved in soil N and carbon (C) cycling [44–47]. In addition, diverse metabolic capabilities of Nitrospira include utilising different organic compounds, nitrite, carbon dioxide, cyanate, or urea [48]. Ammonia-oxidizing microorganisms, including ammonia-oxidizing bacteria and ammonia-oxidizing archaea, which is important regulators of the nitrification process. Specifically, they oxidize ammonia to nitrite, which is subsequently oxidized to nitrate by nitrite-oxidizing bacteria [49]. Complete ammonia oxidizers, capable of oxidizing ammonia to nitrate, were discovered in nitrite-oxidizing bacteria of the genus Nitrospira [49]. Recently, a study reported that the relative abundance of Phycicoccus increased with N addition [50]. Atmospheric N deposition leads to N enrichment in soil, inducing changes in plant growth and soil biological activity, thereby affecting global C and N cycling [51]. In this study, the abundance of Nitrospira and Phycicoccus, and the content of TN and OM increased in IPr, suggesting that intercropping may promote nitrification, ammonification and carbon dioxide assimilation pathway, which may further enhance the carbon-nitrogen cycling in pepper rhizosphere.
Soil metabolites originate from plant root exudates, microbial metabolites, and soil OM decomposition by plants, microbes, and microorganisms [52]. Differential metabolites sensitive to pepper intercropping were firstly investigated. PCA showed that the metabolites in MPr vs. IPr and MB vs. IB were significantly separated, indicating that the root interaction significantly affected the distribution of soil metabolites (Additional file 1: Fig. S6). In this research, the main differential compounds were flavonoids, alkaloids, and nucleotide and its derivates in the rhizosphere. Flavonoids, secondary metabolites secreted by plant roots, have various biological activities, such as antioxidant and antimicrobial activities [53, 54]. Flavonoids play critical roles in the nodule fixation of nitrogen and regulation of interplant and plant–microbe interactions [2]. Previous studies indicated that interspecific interaction changes the flavonoid content and proportion in the rhizosphere soil of wheat, maize, and peanut [2, 55]. In the present study, four flavonoids were identified in MPr vs. IPr, and the levels of morusin, chrysoeriol 7-apiosylglucoside, and asebogenin were upregulated in the rhizosphere of intercropped pepper. Alkaloids exert inhibitory effects on pathogenic microorganisms, specifically, the five alkaloid compounds isolated from Picrasma quassioides exhibited highly significant preventive on apple valsa canker (AVC) [56]. In vivo, two carboline alkaloids, at the concentration of 1000 µg/mL, displayed good inhibitory activity (78% and 80%) on Pytophthora blight of pepper [57]. The levels of alkaloids, including tabernanthine, cheilanthifoline, and trachelanthamidine, were upregulated in the rhizosphere of intercropped pepper. Suggesting that intercropping could enhance the resistance of pepper to P. capsici by increasing the content of alkaloids in the pepper rhizosphere. These result indicated that interspecific interactions regulated the types and contents of secondary metabolites in pepper rhizosphere.
The soil bacterial community composition is responsive to soil environmental parameters [58]. In the present study, the bacterial community composition exhibited a significantly positive correlation (p < 0.01) with soil pH, AP, and AK (Fig. 8A). Soil pH showed a significantly negative correlation with Actinobacteria, Chloroflexi, and Cyanobacteria (Fig. 8B), which enriched in the rhizosphere soil from the intercropping system. Planctomycetes, enriched in the rhizosphere soil from the monocropping system, demonstrated a significant and negtive association with soil AP and AK. This result was similar to previous reports in Sugarcane (Saccharum officinarum)-peanut and maize-peanut intercropping systems [31, 59], wherein environmental properties, such as pH, AP and AK were the principal determinant impacting bacteria dissimilarities in pepper rhizosphere soil under intercropping and monocropping systems. Thus, intercropping may regulate the composition and abundance of bacteria in the pepper rhizosphere soil by changing soil physicochemical properties. Some soil microorganisms are capable of solubilizing and mineralizing insoluble soil phosphorus (P) and potassium (K), then enhancing the content of available P and K and the growth of plants [60, 61].The contents of AP and AK were increased in the IPr and IB, indicated that intercropping increase the abundance of phosphate and potassium solubilizing microorganisms (PSMs and KSMs) in the pepper rhizosphere and buik soils, thus may promote the transformation of insoluble nutrients in soil. Three Aspergillus (PSMs and KSMs) species (A. japonicus, A. lentulus and A. sclerotioniger) were enriched in IPr. Whereas, Streptomyces (KSM) was enriched in the IB.
The significant relationship between soil metabolites and microbial communities can guide soil fertility conditions. The correlation analysis revealed that 28 different metabolites in MPr vs. IPr showed frequent significant correlations with bacterial and fungal communities, indicating that the microbiota can interact with metabolites and change to adapt to environmental stress. Flavonoids are associated with the regulation of symbiosis between plants and microbes and serve as quorum sensing inducers for communications among microbes [62]. In the present study, asebogenin was positively correlated with Candidatus Eisenbacteria, Deltaproteobacteria bacterium and Sorangium cellulosum. Morusin and chrysoeriol 7-apiosylglucoside were positively correlated with Pseudorhodoplanes sinuspersici and Planctomycetes bacterium, respectively. These results agree with those of a previous study [63]. However, in the intercropping system, three metabolites (styrene-cis-2,3-dihydrodiol, ethyl 3,4,5-trimethoxybenzoate, and galactinol) negatively regulated pepper rhizosphere microecology, and these compounds were negatively correlated with most bacteria and one fungus.
Conclusion
In this study, pepper-maize intercropping significantly increased the contents of nutrients (TN, AP, and AK) and the diversity of bacteria and fungi, as well as altered the metabolite profiles, in the rhizosphere and bulk soils. Moreover, LEfSe results showed that the RAs of beneficial microorganisms, such as Sphingomonadales, Phycicoccus and Nitrospira, were significantly enriched in the rhizosphere soil of intercropped pepper. The differential metabolites of soils were enriched in flavonoids, alkaloids, organoheterocyclic compounds, and benzenoids. The levels of flavonoids (such as morusin, chrysoeriol 7-apiosylglucoside, and asebogenin), alkaloids (tabernanthine, cheilanthifoline and trachelanthamidine), and organoheterocyclic compounds (such as cis-zeatin and furan-3-carboxylic acid) were upregulated in the intercropping system. Correlation analysis showed that asebogenin and trachelanthamidine had a significantly positive correlation with Candidatus Eisenbacteria and Nitrospirae. The results of this study demonstrate that intercropping directly or indirectly effected community structure of pepper rhizosphere microbes by enhancing soil nutrient contents and changing soil metabolites. The results provided a theoretical basis for the effects of intercropping on microbial communities and metabolites in pepper rhizosphere soil.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Abbreviations
- AP
Available P
- AK
Available K
- pH
Potential of hydrogen
- TN
Total nitrogen
- OM
Organic matter
- IPr
Intercropped pepper rhizosphere soil
- IB
Bulk soil of pepper–maize intercropping system
- MPr
Monocropped pepper rhizosphere soil
- MB
Bulk soil of pepper monocropping system
- DNA
Deoxyribonucleic acid
- PCR
Polymerase chain reaction
- NCBI
National center for biotechnology information
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- VIP
Variable importance in the projection
- OPLS-DA
Orthogonal partial least squares discriminant analysis
- PCoA
Principal coordinate analysis
- PCA
Principal component analysis
- LEfSe
Linear discriminant analysis with effect size
- RA
Relative abundance
Author contributions
CZ and MY conceived the study and wrote the initial draft of the paper. CZ, WZ and CL collected the soil samples, and analyzed the data. ZP, LZ, YX, SJ and DY performed the experiment. All authors read, revised, and approved the manuscript.
Funding
This research was funded by the Major Science and Technology Special Projects of Yunnan Province, China (202402AE090014, 202102AE090011).
Data availability
The original contributions presented in this study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Declarations
Ethical approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Rubiales D, Enjalbert J, Hohmann P, Anten NPR, Weih M. Editorial: breeding for intercropping. Front Plant Sci. 2023;14:1143653. 10.3389/fpls.2023.1143653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dong Q, Zhao X, Zhou D, Liu Z, Shi X, Yuan Y, et al. Maize and peanut intercropping improves the nitrogen accumulation and yield per plant of maize by promoting the secretion of flavonoids and abundance of Bradyrhizobium in Rhizosphere. Front Plant Sci. 2022;13:957336. 10.3389/fpls.2022.957336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Šenk M, Simić M, Milojković-Opsenica D, Brankov M, Tolimir M, Kodranov I, et al. Common millet and soybean intercropping with bio-fertilizer as sustainable practice for managing grain yield and quality. Front Nutr. 2023;10:1267928. 10.3389/fnut.2023.1267928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raseduzzaman M, Jensen ES. Does intercropping enhance yield stability in arable crop production? A meta-analysis. Eur J Agron. 2017;91:25–33. 10.1016/j.eja.2017.09.009. [Google Scholar]
- 5.Zhang WP, Gao SN, Li ZX, Xu HS, Yang H, Yang X, et al. Shifts from complementarity to selection effects maintain high productivity in maize/legume intercropping systems. J Appl Ecol. 2021;58:2603–13. 10.1111/1365-2664.13989. [Google Scholar]
- 6.Yang Y, Li Y, Mei X, Yang M, Huang H, Du F, et al. Antimicrobial terpenes suppressed the infection process of Phytophthora in fennel-pepper intercropping system. Front Plant Sci. 2022;13:890534. 10.3389/fpls.2022.890534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang M, Zhang Y, Qi L, Mei X, Liao J, Ding X, et al. Plant-plantmicrobe mechanisms involved in soil-borne disease suppression on a maize and pepper intercropping system. PLoS ONE. 2014;9:e115052. 10.1371/journal.pone. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao X, Dong Q, Han Y, Zhang K, Shi X, Yang X, et al. Maize/peanut intercropping improves nutrient uptake of side-row maize and system microbial community diversity. BMC Microbiol. 2022;22:14. 10.1186/s12866-021-02425-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J, Arafat Y, Wu L, Xiao Z, Li Q, Khan MA, et al. Shifts in soil microbial community, soil enzymes and crop yield under peanut/maize intercropping with reduced nitrogen levels. Appl Soil Ecol. 2018;124:327–34. 10.1016/j.apsoil.2017.11.010. [Google Scholar]
- 10.Guo Z, Wan S, Hua K, Yin Y, Chu H, Wang D, et al. Fertilization regime has a greater effect on soil microbial community structure than crop rotation and growth stage in an agroecosystem. Appl Soil Ecol. 2020;149:103510. 10.1016/j.apsoil.2020.103510. [Google Scholar]
- 11.Inal A, Gunes A, Zhang F, Cakmak I. Peanut/maize intercropping induced changes in rhizosphere and nutrient concentrations in shoots. Plant Physiol Biochem. 2007;45:350–6. 10.1016/j.plaphy.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 12.Lei H, Liu A, Hou Q, Zhao Q, Guo J, Wang Z. Diversity patterns of soil microbial communities in the Sophora flavescens rhizosphere in response to continuous monocropping. BMC Microbiol. 2020;20:272. 10.1186/s12866-020-01956-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng S, Bian T, Wang S, Zhang X, Li X, Zhang Y, et al. Decoupling of P from C, N, and K elements in cucumber leaves caused by nutrient imbalance under a greenhouse continuous cropping system. Horticulturae. 2021;7:528. 10.3390/horticulturae7120528. [Google Scholar]
- 14.Pii Y, Borruso L, Brusetti L, Crecchio C, Cesco S, Mimmo T. The interaction between iron nutrition, plant species and soil type shapes the rhizosphere microbiome. Plant Physiol Biochem. 2016;99:39–48. 10.1016/j.plaphy.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 15.Wang X, Whalley WR, Miller AJ, White PJ, Zhang F, Shen J. Sustainable cropping requires adaptation to a heterogeneous Rhizosphere. Trends Plant Sci. 2020;25:1194–202. 10.1016/j.tplants.2020.07.006. [DOI] [PubMed] [Google Scholar]
- 16.Sun Y, Chen L, Zhang S, Miao Y, Zhang Y, Li Z, et al. Plant interaction patterns shape the soil microbial community and nutrient cycling in different intercropping scenarios of aromatic plant species. Front Microbiol. 2022;13:888789. 10.3389/fmicb.2022.888789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xue X, Chen R, Xu C, Zhang C, Dong L, Zhao X, et al. Apple-marigold intercropping improves soil properties by changing soil metabolomics and bacterial community structures. Front Microbiol. 2023;14:1195985. 10.3389/fmicb.2023.1195985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang S, Zhang X, Li X, Shen J, Sun L, Zaman S, et al. Different changes of bacterial diversity and soil metabolites in tea plants-legume intercropping systems. Front Plant Sci. 2023;14:1110623. 10.3389/fpls.2023.1110623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tao L, Zhang C, Ying Z, Xiong Z, Vaisman HS, Wang C, et al. Long-term continuous mono-cropping of Macadamia integrifolia greatly affects soil physicochemical properties, rhizospheric bacterial diversity, and metabolite contents. Front Microbiol. 2022;13:952092. 10.3389/fmicb.2022.952092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li D, Liu CM, Luo R, Sadakane K, Lam TW. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de bruijn graph. Bioinformatics. 2015;31:1674–6. 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 21.Wood DE, Lu J, Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20:257. 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu J, Breitwieser FP, Thielen PM, Salzberg SL. Bracken: estimating species abundance in metagenomics data. PeerJ Comput Sci. 2017;3:e104. 10.7717/peerj-cs.104. [Google Scholar]
- 23.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12:59–60. 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 24.Kim J, Kim MS, Koh AY, Xie Y, Zhan X. FMAP: functional mapping and analysis pipeline for metagenomics and metatranscriptomics studies. BMC Bioinformatics. 2016;17:420. 10.1186/s12859-016-1278-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Franzosa EA, McIver LJ, Rahnavard G, Thompson LR, Schirmer M, Weingart G, et al. Species-level functional profiling of metagenomes and metatranscriptomes. Nat Methods. 2018;15:962–8. 10.1038/s41592-018-0176-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao Y, Zhang G, Jiang S, Liu Y. Wekemo Bioincloud: a user-friendly platform for meta-omics data analyses. IMeta. 2024;3:e175. 10.1002/imt2.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li S, Wu F. Diversity and co-occurrence patterns of soil bacterial and fungal communities in seven intercropping systems. Front Microbiol. 2018;9:1521. 10.3389/fmicb.2018.01521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cao X, Liu S, Wang J, Wang H, Chen L, Tian X, et al. Soil bacterial diversity changes in different broomcorn millet intercropping systems. J Basic Microbiol. 2017;57:989–97. 10.1002/jobm. [DOI] [PubMed] [Google Scholar]
- 29.Tang X, Zhong R, Jiang J, He L, Huang Z, Shi G, et al. Cassava/peanut intercropping improves soil quality via rhizospheric microbes increased available nitrogen contents. BMC Biotechnol. 2020;20:13. 10.1186/s12896-02000606-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Z, Bao X, Li X, Jin X, Zhao J, Sun J, et al. Intercropping maintains soil fertility in terms of chemical properties and enzyme activities on a timescale of one decade. Plant Soil. 2015;391:265–82. 10.1007/s11104-0152428-2422. [Google Scholar]
- 31.Pang Z, Fallah N, Weng P, Zhou Y, Tang X, Tayyab M, et al. Sugarcane-peanut intercropping system enhances bacteria abundance, diversity, and sugarcane parameters in rhizospheric and bulk soils. Front Microbiol. 2022;12:815129. 10.3389/fmicb.2021.815129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tian X, Wang C, Bao X, Wang P, Li X, Yang S, et al. Crop diversity facilitates soil aggregation in relation to soil microbial community composition driven by intercropping. Plant Soil. 2019;436:173–92. 10.1007/s11104-01803924-3928. [Google Scholar]
- 33.Bull AT. Actinobacteria of the extremobiosphere, in Extremophiles Handbook. Ed. Horikoshi K. Tokyo, Japan: Springer. 2011; 1203–1240. 10.1007/978-4-431-53898-1_58
- 34.Nouioui I, Ghodhbane-Gtari F, Del Carmen Montero-Calasanz M, Rohde M, Tisa LS, Gtari M, et al. Frankia inefficax sp. nov., an actinobacterial endophyte inducing ineffective, non nitrogen-fixing, root nodules on its actinorhizal host plants. Antonie Van Leeuwenhoek. 2017;110:313–20. 10.1007/s10482-016-0801-7. [DOI] [PubMed] [Google Scholar]
- 35.Javed Z, Tripathi GD, Mishra M, Dashora K. Actinomycetes-the microbial machinery for the organic-cycling, plant growth and sustainable soil health. Biocatal Agric Biotechnol. 2021;31:101893. 10.1016/j.bcab.2020.101893. [Google Scholar]
- 36.Mitra D, Mondal R, Khoshru B, Senapati A, Radha TK, Mahakur B, et al. Actinobacteria-enhanced plant growth, nutrient acquisition, and crop protection: advances in soil, plant, and microbial multifactorial interactions. Pedosphere. 2022;32:149–70. 10.1016/S1002-0160(21)60042-5. [Google Scholar]
- 37.Lasudee K, Rangseekaew P, Pathom-aree W. Endophytic actinobacteria associated with mycorrhizal spores and their benefits to plant growth. In: Maheshwari DK, Dheeman S, editors Endophytes: Mineral nutrient management. Sustainable Development and Biodiversity, 2021; 26: 229–246. 10.1007/978-3-030-65447-4_10
- 38.Soumare A, Boubekri K, Lyamlouli K, Hafidi M, Ouhdouch Y, Kouisni L. Efficacy of phosphate solubilizing Actinobacteria to improve rock phosphate agronomic effectiveness and plant growth promotion. Rhizosphere. 2021;17:100284. 10.1016/j.rhisph.2020.100284. [Google Scholar]
- 39.Leys NM, Ryngaert A, Bastiaens L, Verstraete W, Top EM, Springael D. Occurrence and phylogenetic diversity of Sphingomonas strains in soils contaminated with polycyclic aromatic hydrocarbons. Appl Environ Microbiol. 2004;70:1944–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qu ZL, Braima A, Liu B, Ma Y, Sun H. Soil fungal community structure and function shift during a disease-driven forest succession. Microbiol Spectr. 2022;10:e0079522. 10.1128/spectrum.00795-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bender SF, Plantenga F, Neftel A, Jocher M, Oberholzer H-R, Köhl L, et al. Symbiotic relationships between soil fungi and plants reduce N2O emissions from soil. ISME J. 2014;8:1336–45. 10.1038/ismej.2013.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Treseder KK, Lennon JT. Fungal traits that drive ecosystem dynamics on land. Microbiol Mol Biol Rev. 2015;79:243–62. 10.1128/MMBR.00001-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li WH, Liu QZ. Changes in fungal community and diversity in strawberry rhizosphere soil after 12 years in the greenhouse. J Integr Agric. 2019;18:677–87. 10.1016/S2095-3119(18)62003-9. [Google Scholar]
- 44.Han S, Huang Q, Chen W. Partitioning Nitrospira community structure and co-occurrence patterns in a long-term inorganic and organic fertilization soil. J Soil Sediment. 2021;21:1–10. 10.1007/s11368-020-02813-x. [Google Scholar]
- 45.Araya JP, González M, Cardinale M, et al. Microbiome dynamics associated with the atacama flowering desert. Front Microbiol. 2019;10:3160. 10.3389/fmicb.2019.03160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scarlett K, Denman S, Clark DR, Forster J, Vanguelova E, Brown N, et al. Relationships between nitrogen cycling microbial community abundance and composition reveal the indirect effect of soil pH on oak decline. ISME J. 2021;15:623–35. 10.1038/s41396-020-00801-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ruiz-Haddad L, Ali M, Pronk M, van Loosdrecht MCM, Saikaly PE. Demystifying polyphosphate-accumulating organisms relevant to wastewater treatment: a review of their phylogeny, metabolism, and detection. Environ Sci Ecotechnol. 2024;21:100387. 10.1016/j.ese.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vijayan A, Vattiringal Jayadradhan RK, Pillai D, Prasannan Geetha P, Joseph V, Isaac Sarojini BS. Nitrospira as versatile nitrifiers: taxonomy, ecophysiology, genome characteristics, growth, and metabolic diversity. J Basic Microbiol. 2021;61:88–109. 10.1002/jobm.202000485. [DOI] [PubMed] [Google Scholar]
- 49.Qin W, Wei SP, Zheng Y, Choi E, Li X, Johnston J, et al. Ammonia-oxidizing bacteria and archaea exhibit differential nitrogen source preferences. Nat Microbiol. 2024;9:524–36. 10.1038/s41564-023-01593-7. [DOI] [PubMed] [Google Scholar]
- 50.Ye H, Zhao Y, He S, Wu Z, Yue M, Hong M. Metagenomics reveals the response of desert steppe microbial communities and carbon-nitrogen cycling functional genes to nitrogen deposition. Front Microbiol. 2024;15:1369196. 10.3389/fmicb.2024.1369196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Püspök JF, Zhao S, Calma AD, Vourlitis GL, Allison SD, Aronson EL, et al. Effects of experimental nitrogen deposition on soil organic carbon storage in Southern California drylands. Glob Chang Biol. 2023;29:1660–79. 10.1111/gcb.16563. [DOI] [PubMed] [Google Scholar]
- 52.Ni H, Jing X, Xiao X, Zhang N, Wang X, Sui Y, et al. Microbial metabolism and necromass mediated fertilization effect on soil organic carbon after long-term community incubation in different climates. ISME J. 2021;15:2561–73. 10.1038/s41396-021-00950-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Panche AN, Diwan AD, Chandra SR. Flavonoids: an overview. J Nutr Sci. 2016;5:e47. 10.1017/jns.2016.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu Y, Ma B, Chen W, Schlaeppi K, Erb M, Stirling E, et al. Rhizobium symbiotic capacity shapes root-associated microbiomes in soybean. Front Microbiol. 2021;12:709012. 10.3389/fmicb.2021.709012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu Y, Yin X, Zheng Y. Influences of intercropping and nitrogen supply on flavonoid exudation in wheat roots. J Plant Nutr. 2020;43:2757–72. 10.1080/01904167.2020.1793189. [Google Scholar]
- 56.Wang H, Tian R, Chen Y, Li W, Wei S, Ji Z. In vivo and in vitro antifungal activities of five alkaloid compounds isolated from Picrasma quassioides (D. Don) Benn against plant pathogenic fungi. Pestic Biochem Physiol. 2022;188:105246. 10.1016/j.pestbp.2022.105246. [DOI] [PubMed] [Google Scholar]
- 57.Huang D, Zhang Z, Li Y, Liu F, Huang W, Min Y, et al. Carboline derivatives based on natural pityriacitrin as potential antifungal agents. Phytochem Lett. 2022;48:100–5. 10.1016/j.phytol.2022.02.010. [Google Scholar]
- 58.Lian T, Mu Y, Jin J, Ma Q, Cheng Y, Cai Z, et al. Impact of intercropping on the coupling between soil microbial community structure, activity, and nutrient-use efficiencies. PeerJ. 2019;7:e6412. 10.7717/peerj.6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li QS, Chen J, Wu LK, Luo XM, Li N, Arafat Y, et al. Belowground interactions impact the soil bacterial community, soil fertility, and crop yield in maize/peanut intercropping systems. Int J Mol Sci. 2018;19:622. 10.3390/ijms19020622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alori ET, Glick BR, Babalola OO. Microbial phosphorus solubilization and its potential for use in sustainable agriculture. Front Microbiol. 2017;8:971. 10.3389/fmicb.2017.00971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sharma R, Sindhu SS, Glick BR. Potassium solubilizing microorganisms as potential biofertilizer: a sustainable climate-resilient approach to improve soil fertility and crop production in agriculture. J Plant Growth Regul. 2024;43:2503–35. 10.1007/s00344-024-11297-9. [Google Scholar]
- 62.Pang Z, Chen J, Wang T, Gao C, Li Z, Guo L, et al. Linking plant secondary metabolites and plant microbiomes: a review. Front Plant Sci. 2021;12:621276. 10.3389/fpls.2021.621276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Szoboszlay M, White-Monsant A, Moe LA. The effect of root exudate 7,4’-dihydroxyflavone and naringenin on soil bacterial community structure. PLoS ONE. 2016;11:e0146555. 10.1371/journal.pone.0146555. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in this study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.