Abstract
Marbling is a key indicator of the meat quality of ruminants. Gastrointestinal microbiota may regulate the formation of marbling by influencing the nutritional metabolism of animals. This study analyzed the composition and functional differences of microbiota in the rumen and cecum, the differences in volatile fatty acids (VFAs) content in the longissimus thoracis muscle, and the differences in protein abundance in the longissimus thoracis muscle of ruminants with different marbling grades through microbiome-proteome analysis. The results showed that the diversity of gastrointestinal microbiota in high-marbling ruminants was significantly higher than that in low-marbling ruminants. The relative abundance of Firmicutes and Akkermansia in the gastrointestinal of high-marbling ruminants was higher than that in low-marbling ruminants, while the relative abundance of Bacteroidetes and Prevotella was lower. In addition, PICRUST2 functional prediction results of the microbiota revealed that the gastrointestinal microbiota of high-marbling ruminants was mainly involved in the biosynthesis pathways of fat and lipids. The metabolomics results showed that the content of VFAs (acetic acid, propionic acid, butyric acid, isovaleric acid, valeric acid, and hexanoic acid) in the rumen of high-marbling ruminants was significantly higher than that in low-marbling ruminants. The proteome analysis results indicated that the differential proteins in the longissimus thoracis muscle of high-marbling ruminants were mainly involved in lipid transport and metabolism compared to low-marbling ruminants. In summary, the differences in the composition and function of the gastrointestinal microbiota led to higher levels of VFAs in the gastrointestinal tract of high-marbling ruminants, which provides the basis for lipid/fat synthesis. The proteome results of the longissimus thoracis muscle support the view that high-marbling ruminants have richer lipid transport and metabolic functions in their muscle.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12917-024-04417-w.
Keywords: Cattle, Marbling, Microbiome, Intestinal microbiota, Proteomics
Introduction
Marbled meat is a crucial indicator of animal meat quality. With the continuous improvement of meat production efficiency in recent decades, the market niche for marbled meat has been steadily expanding [1, 2]. The formation of marbling in meat is a complex process that involves multiple chemical reactions, as well as the metabolic changes in muscle fibers that facilitate intramuscular fat deposition, and is regulated by various hormones, enzymes, and transcription factors [3]. A marble-like pattern is an intuitive manifestation of intramuscular fat deposition, directly proportional to its fat content. Fat content remains the primary indicator for assessing meat quality in most countries and regions [4, 5]. Previous research on animal fat and muscle fiber usually focused on factors such as breed, age, nutrition, environment, and feeding management. However, recent studies have discovered a certain correlation between gastrointestinal microbiota and animal meat quality [6, 7].
The study found that the improvement of marbling score and the increase of lipid content in longissimus thoracis muscle were highly correlated with the increase of microbial richness, suggesting that the higher fat content of this muscle may be related to the more abundant bacterial species in the rumen [8]. High-fat animals have a higher abundance of methanogenic bacteria, leading to more efficient fat deposition [9]. In addition, studies have shown that aflatoxin can affect the gut microbiota and degrade the meat quality of ruminants [10]. Feeding rapeseed increased the content of linolenic acid and various amino acids in muscle, while also increasing the abundance of cellulose-decomposing bacteria in the rumen. This revealed a close relationship between the final body weight, loin eye muscle area, and microbial composition of ruminants, providing a clue for strategies to improve growth performance and carcass traits through microbial intervention [11]. A study using multi-omics analysis demonstrated that the structure of the microbiota was significantly correlated with the metabolome and gene expression in ruminant muscle [12]. Clearly, the gut microbiota plays a crucial role in nutrient utilization by the host, regulating fat deposition, and enhancing meat quality.
Although the genetic background, age, feed nutrition and living environment of cattle are the same, the marbled meat of the longissimus thoracis muscle of cattle often shows great differences. Some studies have shown that the marbled meat of longissimus thoracis muscle can be improved by changing the composition of cattle diet [11]. Therefore, it is speculated that the difference of gastrointestinal microbiota may be one of the important factors affecting marbled meat, because gastrointestinal microbiota plays an important role in nutrient digestion and health, and the disturbance of gastrointestinal microbiota usually has some adverse effects. However, to date, it is unclear which microorganisms are involved in the formation of high-grade marbled meat. The rapid development of high-throughput sequencing technology has greatly promoted the study of human and animal gut microbiota. Several studies have been conducted to explore the impact of gut microbiota composition on the host by using 16S rDNA gene sequencing and proteome sequencing [13, 14]. In this study, high-throughput sequencing technology (16S rRNA gene sequencing and protein sequencing technology) was used to analyze the differential microbiota (microbiota) in the rumen and cecum of the longissimus thoracis muscle with high- and low-grade marbling, as well as the differential proteins in the longissimus thoracis muscle. In depth study on the mechanism of ruminant gastrointestinal microbiota involved in meat quality formation has positive significance for understanding the core microbiota of ruminant marbled meat quality formation and exploring new measures to improve meat quality.
Materials and methods
Experimental design and animal management
The experiment was conducted in June 2022 at Wang Zhongwang Agriculture Co., Ltd. in Luoyang City, Henan Province for feeding and slaughtering. The experimental process follows the operating procedures for experimental animals at Luoyang Normal University. Forty-eight Wagyu (26 months old) were randomly selected for this experiment and were randomly assigned to 12 fences (4 cattle per fence). The feeding process designed according to the NRC (2016) standard to meet the nutritional requirements of beef cattle at each stage [15]. The composition of the diet was shown in Supplementary Table S1. During the experiment, all cattle were free to access food and water.
Sample collection and grouping
Grouping: Marble Pattern Score: After slaughtering the experimental cattle (Electrodes were placed on the head and back waist of the cow to form circuits on its body. When power was turned on, the current entered the body of the cow from one electrode and flowed out from the other electrode. High voltage electricity passed through the cow’s body, causing it to become paralyzed and dizzy, followed by bloodletting through the arteries, ultimately resulting in death), the entire surface of the rib interface of the 12th to 13th thoracic vertebrae on the left side of the carcass was immediately cut, and the quality level was determined based on the quality level of the United States Department of Agriculture (a = moderately rich, b = mildly rich, c = moderate, d = moderate, e = small, f = mild). The marbling grades can be specified as follows: Prime (a and b): This grade represents the highest level of marbling. The meat has abundant intramuscular fat with a fine and evenly distributed pattern. Choice (c and d): A good level of marbling. The intramuscular fat is less abundant than prime but still provides good flavor and tenderness. Select (e and f): Lower marbling compared to prime and choice. The cattle were divided into a high-grade group (marble pattern score is a-c grade) and a low-grade group (marble pattern score is e-f grade) (Fig. S1A). After determining the quality levels, a, b, c, d, e, and f were converted into values of 6, 5, 4, 3, 2, and 1, respectively, for statistical analysis (Fig. S1B). Initial body weight of the cattle was shown in Fig. S1C.
Sample collection: After the cattle was euthanized, the abdominal cavity was dissected and tissues such as the rumen and cecum were removed. The rumen fluid was carefully collected, filtered through four layers of gauze, and divided into 15 mL sterile cryogenic vials. Subsequently, the cecum was transected using aseptic methods and the cecal contents were divided into 5 mL sterile cryovials. The longest thoracis muscle tissue was collected and placed in enzyme-free, sterile cryovials. All collected samples were temporarily stored in liquid nitrogen and then transferred to -80 °C for long-term storage.
Rumen and cecal microbiota sequencing
DNA extraction and quality control
Take twelve samples (six for high-grade group and six for low-grade group) along with cattle rumen content and caecum content from − 80 °C. Extract DNA from both rumen content and caecum content according to the instructions of the Tiangen fecal DNA extraction kit. Use 0.8% agarose gel electrophoresis (Beijing Liuyi Biotechnology Co., Ltd.) to judge the molecular size, and employ ultraviolet spectrophotometer (Thermo Fisher Scientific Co., Ltd., NanoDrop2000) for quantitative analysis.
High throughput sequencing
PCR was performed using universal primers (F: 5’- CCTACGGGNGGCWGCAG-3’ and R: 5’- GGACTACHVGGTATCTAAT-3’) to amplify the 16S rRNA V3-V4 region. The PCR amplification system: 10×KOD Buffer 5 μL. dNTPs (concentration of 2.5 mmol/L) 5 μL. Upstream and downstream primers (concentration of 5 μmol/L) 1.5 μL each. KOD polymerase 1 μL. DNA template 100 ng, complete with ddH2O to 50 μL. The PCR amplification program: 95 ℃ pre denaturation for 2 min; 98 ℃ denaturation for 10 s, 62 ℃ annealing for 30 s, 68 ℃ extension for 30 s, 27 cycles; Extend at 68 ℃ for 10 min. The PCR products were then cut and recovered, quantified, and sent to Guangzhou Jidio Biotechnology Co., Ltd. for library construction and sequencing on the HiSeq2500 PE250 platform.
Sequencing data processing and bioinformatics analysis
Quantitative insights into microbial ecology 2 (QIIME 2) was used to perform alpha diversity analysis of rumen and cecal microbiota, which calculates the simpson and shannon indices for sample microorganisms. R language was used to conduct beta diversity analysis of rumen and cecal microbiota. QIIME2 was employed to visualize the composition and distribution of microorganisms at six classification levels: phylum, class, order, family, genus, and species. Microbial community function prediction analysis was performed through PICRUSt2.
Protein sequencing of the longissimus thoracis muscle
Protein extraction and peptide enzymatic hydrolysis
Take six samples (three each for high-grade group and low-grade group) of the longissimus thoracis muscle tissue from back of the cattle, grind them in a mortar, and then add 30 mg of the sample to 900 μL SDT lysis solution (4% [w/v] SDS, 100 mM Tris HCL [pH 7.6], 0.1 M DTT). Subject the lysis solution to ultrasound treatment (80 W for 10 s, with a 15-second interval, for a total of 10 cycles). Boil the solution for 15 min and centrifuge at 12 000 rpm for 40 min to obtain uterine tissue protein samples. The protein concentration was detect using the BCA protein assay kit (Bio-Rad, USA), and the protein was detected using 10% SDS-PAGE and trypsin digestion was performed using the Filter-Aided Sample Preparation method.
LC-MS/MS analysis
Each graded sample was separated using an HPLC liquid chromatography system Easy nLC. The chromatographic column was equilibrated with 95% A solution (0.1% formic acid aqueous solution). Subsequently, the sample was transferred from the automatic sampler to the loading column (Thermo Scientific Acclaim PepMap 100 μm * 2 cm, nanoViper C18), and then separated through the analytical column (Thermo Scientific EASY column, 10 cm, ID 75 μm, 3 μm, C18-A2) at a flow rate of 300 nL/min. After the peptide fragments underwent nano-electrospray ionization, mass spectrometry analysis was performed using the Q-Exactive (ThermoFisher Scientific, USA) mass spectrometer to complete the mass-to-charge ratio (m/z) collection of the peptides and peptide fragments.
Data analysis
Annotate and analyze the functions of differential proteins using the cluster of orthologous groups of proteins (COG) database, which is a database developed by NCBI for homologous protein annotation. By comparing proteins with databases, the function of proteins can be accurately predicted. The process involves several steps: (1) Functional annotation of unknown sequences using known proteins; (2) By checking the number, presence, and absence of proteins corresponding to the specified COG number, it is possible to infer whether a specific metabolic pathway exists; (3) Each COG number represents a type of protein, and by performing multiple sequence alignment between the query sequence and the proteins aligned with the COG number, conserved sites can be identified and their evolutionary relationships can be analyzed.
Detection of SCFAs in the rumen
Take sample and place them into the 2 mL EP tubes. Extracted each tube with 1 mL H2O, then vortex mixing for 10 s. Homogenized the samples in ball mill for 4 min at 40 Hz. Subsequently, treated them with ultrasound for 5 min (incubated in ice water), and repeat 3 times. Centrifuge the tubes for 20 min at 5000 rpm and 4 °C. Transfer 0.8 mL of the supernatant into a fresh 2 mL EP tube. Add 0.1 mL of 50% H2SO4 and 0.8 mL of an extracting solution (25 mg/L stock in methyl tert-butyl ether), which serves as an internal standard. Vortex mix for 10 s, then oscillate for 10 min. Treat the mixture with ultrasound for 10 min while incubating in ice water. Centrifuge for 15 min at 10 000 rpm and 4 °C. Keep the samples at -20 °C for 30 min. Finally, transfer the supernatant into a fresh 2 mL glass vial for GC-MS analysis.
Statistical analysis
The experimental data were statistically analyzed using the student’s t test of SPSS 24.0 software between the two groups, *P < 0.05 indicating significant differences and ** P < 0.01 indicating extremely significant differences.
Results
Rumen and cecal microbiota alpha and beta diversity
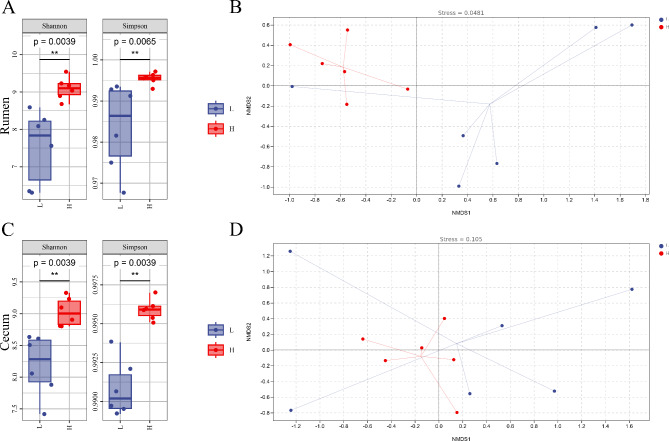
The alpha diversity results showed that the Simpson index and Shannon index of rumen and cecal microbiota in high-grade cattle were significantly higher than those in low-grade cattle (P < 0.01) (Fig. 1A&C). The beta diversity results showed that the composition and structure of the rumen and cecal microbiota in high-grade cattle were more similar, while the composition and structure of the rumen and cecal microbiota in low-grade cattle were farther apart (Fig. 1B&D).
Fig. 1.
Alpha and beta diversity. A Alpha diversity of rumen microbiota. B Beta diversity of rumen microbiota. C Alpha diversity of cecal microbiota. D Beta diversity of cecal microbiota
Composition of rumen and cecal microbial communities
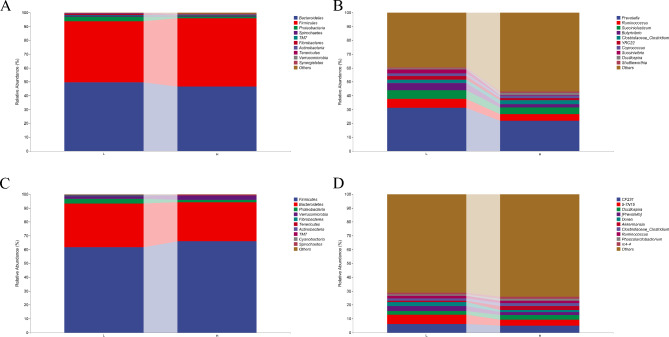
At the phylum level, the relative abundance of Firmicutes in the rumen and cecum of high-grade cattle was higher than that of low-grade cattle, while the relative abundance of Bacteroidetes and Proteobacteria was lower than that of low-grade cattle (Fig. 2A&C). At the genus level, the relative abundance of Prevotella in the rumen and cecum of high-grade cattle was lower than that of low-grade cattle, while the relative abundance of Akkermansia in the cecum of high-grade cattle was higher than that of low-grade cattle (Fig. 2B&D).
Fig. 2.
Species composition. A The composition and distribution of rumen microbiota at the phylum level. B The composition and distribution of rumen microbiota at the genus level. C The composition and distribution of cecal microbiota at the phylum level. D The composition and distribution of cecal microbiota at the genus level
Functional prediction of rumen and cecal microbial communities
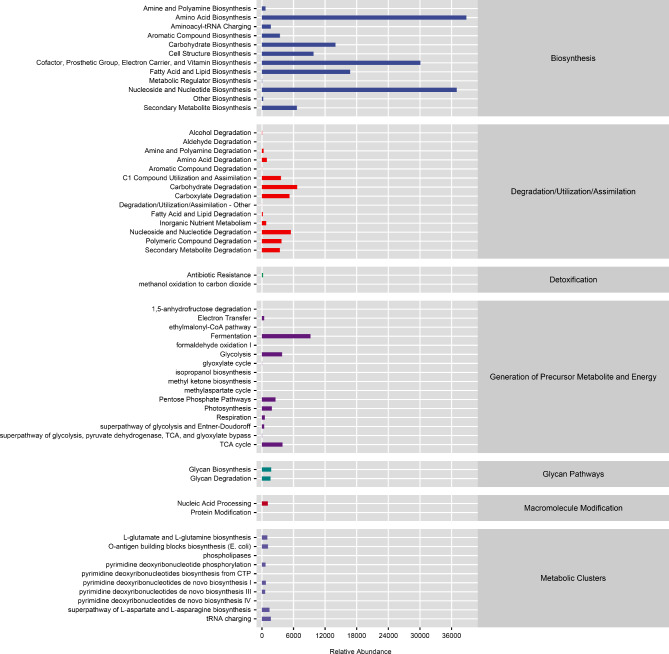
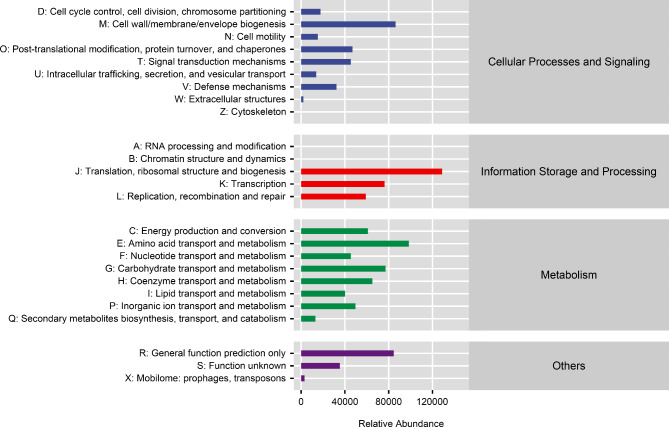
The MetaCyc analysis results showed that microorganisms mainly participate in amino acid biosynthesis, nucleoside and nucleotide biosynthesis, cofactor, prosthetic group, electron carrier, and vitamin biosynthesis, as well as fatty acid and lipid biosynthesis pathways (Fig. 3). The COG analysis results showed that microorganisms mainly participate in pathways such as translation, ribosomal structure and biogenesis, amino acid transport and metabolism, translation, carbohydrate transport and metabolism, lipid transport and metabolism (Fig. 4).
Fig. 3.
MetaCyc functional prediction of microbial communities
Fig. 4.
Cluster of orthologous groups of proteins functional prediction of microbial communities
Differential protein abundance analysis
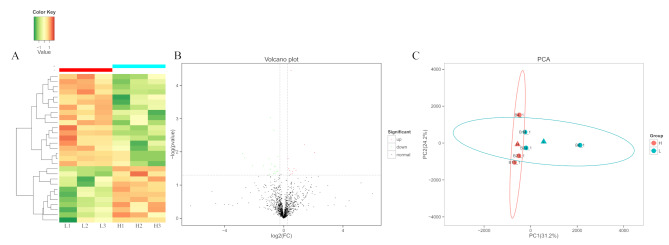
The hierarchical clustering method was used to perform clustering analysis on the differentially expressed proteins in the comparison group, and the data were presented in the form of a heatmap. The similarity of the data within each group was high, and the differences between groups were significant, allowing for effective distinction between them. The results of cluster analysis indicate that the differentially expressed proteins identified in this experiment are reasonable and accurate (Fig. 5A). We screened for differentially expressed proteins with a fold change greater than 1.2 (up regulation greater than 1.2 times or down regulation less than 0.83) and P-value less than 0.05. Compared with lower-grade cattle, there were 29 differentially expressed proteins in high-grade cattle, of which 11 were significantly upregulated and 18 were significantly downregulated (Fig. 5B). We used a sample difference distance matrix combined with principal coordinate analysis (PCA) to analyze sample functional differences in low dimensions. The closer the projection distance between two points on the coordinate axis, the more similar the functions of these two samples in the corresponding dimensions. The results showed that compared with low-grade cattle, the protein functions of the longissimus thoracis muscle in the samples of high-grade cattle were more similar (Fig. 5C).
Fig. 5.
Analysis of differentially abundance proteins. A Cluster diagram of differentially expressed proteins in the longissimus thoracis muscle. B Volcano plot of differential abundance analysis of proteins identified in the longissimus thoracis muscle. C Principal component analysis analysis of differential proteins in the longissimus thoracis muscle
Functional analysis of differentially expressed proteins
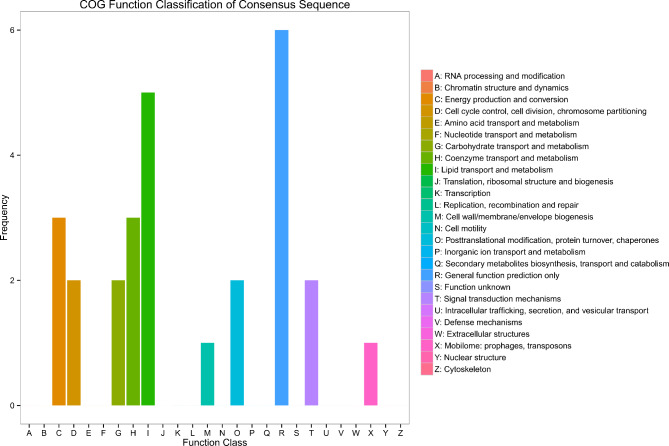
The functional analysis results of differential proteins revealed that proteins from the longissimus thoracis of high-grade cattle were more frequently involved in the biosynthesis, transport, and metabolism of secondary metabolites, lipid transport and metabolism, as well as energy production and conservation, compared to those from low-grade cattle (Fig. 6).
Fig. 6.
Cluster of orthologous groups of proteins functional analysis of differentially abundance proteins
Rumen short chain fatty acids
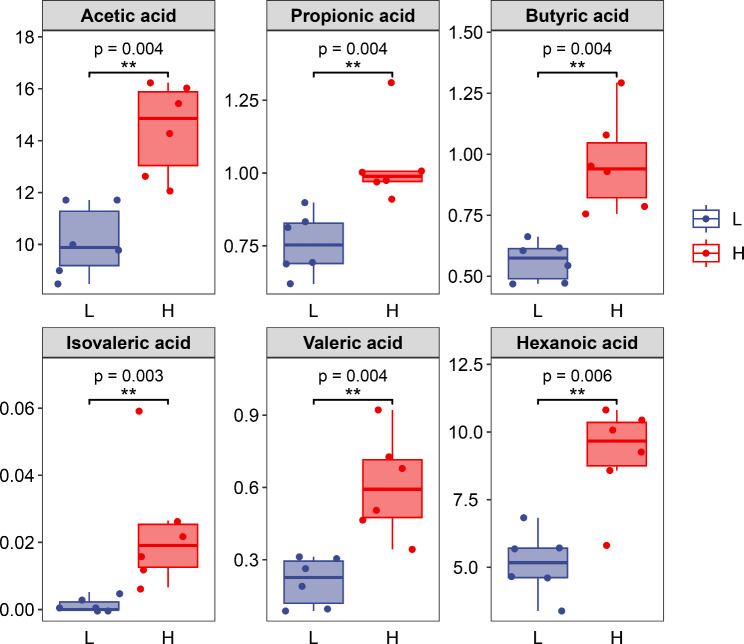
The results of short chain fatty acids in the rumen showed that the content of acetic acid, propionic acid, butyric acid, isovaleric acid, valeric acid, and caproic acid in the muscle of high-grade cattle were significantly higher than that of low-grade cattle (Fig. 7).
Fig. 7.
Volatile fatty acids content in the rumen
Discussion
With the improvement of people’s living quality and consumption level, the dietary structure has changed from subsistence to quality-oriented. Consumers have higher and higher requirements for meat quality. Intramuscular fat (IMF) content is one of the key factors affecting meat quality and determines the sensory qualities such as tenderness, juiciness and flavor of meat [16]. Intramuscular fat deposition is mainly affected by factors such as the age, gender, breed, nutritional level and genetics of animals. However, more and more research reports have confirmed that intestinal flora can also regulate the deposition of intramuscular fat in animals [17]. Therefore, elucidating the relationship between the gastrointestinal microbiota of ruminants and meat quality can provide a theoretical basis for fully exploring and improving the superior traits of ruminants, and provide technical means for improving the meat quality traits of ruminants. The majority of microorganisms in the gastrointestinal tract of ruminant animals are bacteria, with Firmicutes, Bacteroidetes, Proteobacteria, and Fibrobacteres accounting for about 60% of all bacteria in the gastrointestinal tract of ruminants. Other bacterial phyla include a small number of Tenerices and Actinobacteria [18].
There are significant differences in gastrointestinal microbiota among ruminants with different fat related traits [8, 19, 20]. Krause et al. compared and analyzed the differences in rumen microbiota and meat quality among 24 Angus cattle fed the same diet [8]. The results showed that the Chao1 index, which reflects rumen microbial richness, and the number of operational classification units, as well as the Pielou’s evenness index, which reflects rumen microbial diversity, were positively correlated with marbling score and longissimus thoracis muscle fat content; Zhang et al. compared and analyzed the differences and correlations between rumen microbial communities and fat deposition in Hu sheep fed the same diet [20]. The results showed that there were significant differences in the Shannon and Simpson indices of rumen microorganisms between the lower fat deposition group and the higher fat deposition group of Hu sheep, both representing microbial diversity. The abundance of Prevotellaceae and Lachnospiraceae in the rumen of lower fat deposition group Hu sheep is higher than that of high fat deposition group, while the abundance of Ruminococcus in the rumen of high fat deposition group Hu sheep is higher. In this study, the alpha diversity and the relative abundance of Prevotella in the rumen and cecum of them was significantly lower than that in the low-grade group. Prevotella is ubiquitous in the intestine and can improve glucose and insulin tolerance by fermenting dietary fiber to produce succinic acid [21, 22]. Some studies have compared the differences in gut microbiota between the obese group and the control group and found that in addition to abnormal carbohydrate metabolism in the obese group, there is a significant increase in the abundance of Prevotella copri in the gut microbiota. In the study by Chen et al., a systematic correlation study was conducted on the gut microbiota and lean meat percentage of pigs, and it was found that Prevotella copri was significantly correlated with fat accumulation in pigs [23]. The main products of Prevotella are butyryl phosphate transferase and butyrate kinase, which are key enzymes for the formation of butyrate. Butyrate can increase feed intake and body weight of cattle, promote the proliferation of gastrointestinal epithelial cells, and enhance rumen fermentation. Prevotella uses the cellulose degradation products of other cellulolytic bacteria as an energy source. The main fermentation products are acetic acid and succinic acid and including a small amount of isobutyric acid, isovaleric acid and lactic acid. Acetic acid is absorbed through the rumen epithelium and is a raw material for de novo synthesis of fatty acids, which may relate to fat deposition in the body [24].
The proteomic results of this study showed that differentially expressed proteins are mainly involved in lipid transport and metabolism, energy production and conversion, as well as carbohydrate transport and metabolism. The gastrointestinal microbiota interacts with the host. The microbiota not only receives energy from the host to maintain normal growth, but also provides energy to the host by releasing enzymes and metabolites such as short chain fatty acids, amino acids, bile acids, caseinolytic protein B, and lipopolysaccharides [25]. Gastrointestinal microbiota can help hosts utilize fibrous substances and ferment them to produce volatile fatty acids (VFAs) and hydrogenated fatty acids [26]. Volatile fatty acids are the main source of energy for hosts and have a direct impact on their metabolism, while the gastrointestinal is the main site for microbial fermentation in the digestive tract of ruminants to produce VFAs [5]. Ruminant animals have fewer gut microbiota than rumen microbiota, and they also have functions such as producing VFAs and hydrogenated fatty acids, but they are more involved in polysaccharide and amino acid metabolism [27, 28]. In addition, gastrointestinal microbiota can help the host establish the intestinal immune system and maintain gastrointestinal health [29]. Fat is a triglyceride formed from glycerol and long-chain fatty acids, which is synthesized in the liver and adipose tissue of ruminants and stored in adipose tissue, thereby affecting the marble pattern, tenderness, and shear strength of meat products [30, 31]. Both the fatty acids synthesized by animals themselves and those ingested through feed can be used for the synthesis of triglyceride (TG), and VFAs, the main metabolic products of ruminant gastrointestinal microbiota, especially rumen and cecal microbiota, are one of the important raw materials for the synthesis of TG [32]. In summary, the gastrointestinal microbiota of ruminant animals, especially the VFAs produced by gastrointestinal microbiota fermentation, can be used as long-chain fatty acid synthesis materials to participate in fat regulation, which may contribute to fat deposition.
Conclusion
The rumen and cecal microbiota of high-grade cattle are significantly different from those of low-grade cattle, mainly reflected in the higher abundant Firmicutes, Akkermansia, and lower abundant Bacteroidetes and Prevotella in the rumen and cecum of the high-grade cattle. The functional prediction results of the microbiota, along with proteomic results, indicated that the gastrointestinal microbiota of high-grade cattle was more involved in the biosynthesis and transport of fats and lipids. The differences in microbiota and their functions may further contribute to variations in the protein function related to lipid transport and metabolism in the longissimus thoracis muscle.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Not applicable.
Abbreviations
- PCA
Principal component analysis
- PICRUSt
Phylogenetic investigation of communities by reconstruction of unobserved states
- SCFAs
Short-chain fatty acid
- TG
Triglyceride
- VFAs
Volatile fatty acids.
Author contributions
MS designed the experiment and draft the manuscript; ZL and JZ analyzed the data and revised the manuscript; SH and LH managed the cattle; PZ and SM collected the slaughter data; JZ collected the experimental samples. ZM revised the manuscript. All authors reviewed the manuscript.
Funding
This study was supported by the Provincial Key Technology Research and Development Program of Henan (232102110079, 232102111055, 222102110021, 202102110241); Core Technology Research Projects in Luoyang City (2202036 A); Innovative Research Team (in Science and Technology) at the University of Henan Province (22IRTSTHN026); the Key Scientific Research Projects of Colleges and Universities in Henan Province (Henan Province, China; 23B230003); West Henan Yellow River Wetland Ecosystem Observation and Research Station, Engineering Research Center for Wetland Ecological Restoration in the Middle-Lower Reaches of Yellow River.
Data availability
The datasets used and analysed during the current study can be accessed at https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1190918/.
Declarations
Ethics approval and consent to participate
The cattle selected in this study obtained informed consent from the owners, who agreed that we can use these animals in this study. All animal experiments were performed according to protocols and guidelines approved by the Institutional Animal Care and Use Committee of Luoyang Normal University, China.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Zhiguo Miao, Email: zhiguomiao2023@163.com.
Jinzhou Zhang, Email: zhangjz69@126.com.
References
- 1.Salter AM. The effects of meat consumption on global health. Rev Sci Tech. 2018;37(1):47–55. [DOI] [PubMed] [Google Scholar]
- 2.Lee S, Jo K, Jeong SK, Jeon H, Choi YS, Jung S. Recent strategies for improving the quality of meat products. J Anim Sci Technol. 2023;65(5):895–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li X, Zhang D, Ren C, Bai Y, Ijaz M, Hou C, Chen L. Effects of protein posttranslational modifications on meat quality: a review. Compr Rev Food Sci Food Saf. 2021;20(1):289–331. [DOI] [PubMed] [Google Scholar]
- 4.Picard B, Gagaoua M. Muscle fiber properties in cattle and their relationships with meat qualities: an overview. J Agric Food Chem. 2020;68(22):6021–39. [DOI] [PubMed] [Google Scholar]
- 5.Campos CF, Duarte MS, Guimarães SE, Verardo LL, Wei S, Du M, Jiang Z, Bergen WG, Hausman GJ, Fernyhough-Culver M, et al. Review: animal model and the current understanding of molecule dynamics of adipogenesis. Animal. 2016;10(6):927–32. [DOI] [PubMed] [Google Scholar]
- 6.Yan H, Diao H, Xiao Y, Li W, Yu B, He J, Yu J, Zheng P, Mao X, Luo Y, et al. Gut microbiota can transfer fiber characteristics and lipid metabolic profiles of skeletal muscle from pigs to germ-free mice. Sci Rep. 2016;6:31786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiao Y, Kong F, Xiang Y, Zhou W, Wang J, Yang H, Zhang G, Zhao J. Comparative biogeography of the gut microbiome between Jinhua and Landrace pigs. Sci Rep. 2018;8(1):5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krause TR, Lourenco JM, Welch CB, Rothrock MJ, Callaway TR, Pringle TD. The relationship between the rumen microbiome and carcass merit in angus steers. J Anim Sci. 2020;98(9). [DOI] [PMC free article] [PubMed]
- 9.Zhao G, Xiang Y, Wang X, Dai B, Zhang X, Ma L, Yang H, Lyu W. Exploring the possible link between the gut microbiome and fat deposition in pigs. Oxid Med Cell Longev. 2022;2022:1098892. [DOI] [PMC free article] [PubMed]
- 10.Cao QQ, Lin LX, Xu TT, Lu Y, Zhang CD, Yue K, Huang SC, Dong HJ, Jian FC. Aflatoxin B1 alters meat quality associated with oxidative stress, inflammation, and gut-microbiota in sheep. Ecotoxicol Environ Saf. 2021;225:112754. [DOI] [PubMed] [Google Scholar]
- 11.Du E, Guo W, Zhao N, Chen F, Fan Q, Zhang W, Huang S, Zhou G, Fu T, Wei J. Effects of diets with various levels of forage rape (Brassica napus) on growth performance, carcass traits, meat quality and rumen microbiota of Hu lambs. J Sci Food Agric. 2022;102(3):1281–91. [DOI] [PubMed] [Google Scholar]
- 12.Wu J, Yang D, Gong H, Qi Y, Qiu X. Multiple omics analysis reveals that high fiber diets promote gluconeogenesis and inhibit glycolysis in muscle. BMC Genomics. 2020;21(1):660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Xu L, Sun X, Wan X, Sun G, Jiang R, Li W, Tian Y, Liu X, Kang X. Characteristics of the fecal microbiota of high- and low-yield hens and effects of fecal microbiota transplantation on egg production performance. Res Vet Sci. 2020;129:164–73. [DOI] [PubMed] [Google Scholar]
- 14.Ge S, Lian W, Bai Y, Wang L, Zhao F, Li H, Wang D, Pang Q. TMT-based quantitative proteomics reveals the targets of andrographolide on LPS-induced liver injury. BMC Vet Res. 2023;19(1):199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Agriculture BO. Nutrient requirements of beef cattle: eighth revised edition (2016). 2016.
- 16.Heyer A, Lebret B. Compensatory growth response in pigs: effects on growth performance, composition of weight gain at carcass and muscle levels, and meat quality. J Anim Sci. 2007;85(3):769–78. [DOI] [PubMed] [Google Scholar]
- 17.Fang S, Xiong X, Su Y, Huang L, Chen C. 16S rRNA gene-based association study identified microbial taxa associated with pork intramuscular fat content in feces and cecum lumen. BMC Microbiol. 2017;17(1):162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cholewińska P, Czyż K, Nowakowski P, Wyrostek A. The microbiome of the digestive system of ruminants - a review. Anim Health Res Rev. 2020;21(1):3–14. [DOI] [PubMed] [Google Scholar]
- 19.Kim M, Park T, Jeong JY, Baek Y, Lee HJ. Association between rumen microbiota and marbling score in korean native beef cattle. Anim (Basel). 2020;10(4). [DOI] [PMC free article] [PubMed]
- 20.Zhang Y, Zhang X, Li F, Li C, Zhang D, Li X, Zhao Y, Wang W. Exploring the ruminal microbial community associated with fat deposition in lambs. Anim (Basel). 2021;11(12). [DOI] [PMC free article] [PubMed]
- 21.Vandeputte D, Kathagen G, D’Hoe K, Vieira-Silva S, Valles-Colomer M, Sabino J, Wang J, Tito RY, De Commer L, Darzi Y, et al. Quantitative microbiome profiling links gut community variation to microbial load. Nature. 2017;551(7681):507–11. [DOI] [PubMed] [Google Scholar]
- 22.Vieira-Silva S, Sabino J, Valles-Colomer M, Falony G, Kathagen G, Caenepeel C, Cleynen I, van der Merwe S, Vermeire S, Raes J. Quantitative microbiome profiling disentangles inflammation- and bile duct obstruction-associated microbiota alterations across PSC/IBD diagnoses. Nat Microbiol. 2019;4(11):1826–31. [DOI] [PubMed] [Google Scholar]
- 23.Chen C, Fang S, Wei H, He M, Fu H, Xiong X, Zhou Y, Wu J, Gao J, Yang H, et al. Prevotella copri increases fat accumulation in pigs fed with formula diets. Microbiome. 2021;9(1):175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carberry CA, Kenny DA, Han S, McCabe MS, Waters SM. Effect of phenotypic residual feed intake and dietary forage content on the rumen microbial community of beef cattle. Appl Environ Microbiol. 2012;78(14):4949–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fetissov SO. Role of the gut microbiota in host appetite control: bacterial growth to animal feeding behaviour. Nat Rev Endocrinol. 2017;13(1):11–25. [DOI] [PubMed] [Google Scholar]
- 26.Kim M, Park T, Yu Z. Metagenomic investigation of gastrointestinal microbiome in cattle. Asian-Australas J Anim Sci. 2017;30(11):1515–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson KA, Johnson DE. Methane emissions from cattle. J Anim Sci. 1995;73(8):2483–92. [DOI] [PubMed] [Google Scholar]
- 28.O’Hara E, Neves ALA, Song Y, Guan LL. The role of the gut microbiome in cattle production and health: driver or passenger? Annu Rev Anim Biosci. 2020;8:199–220. [DOI] [PubMed] [Google Scholar]
- 29.Malmuthuge N, Guan LL. Understanding host-microbial interactions in rumen: searching the best opportunity for microbiota manipulation. J Anim Sci Biotechnol. 2017;8:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wretlind A. Future perspectives in parenteral nutrition. Infusionstherapie. 1989;16(5):192–7. [PubMed] [Google Scholar]
- 31.Hudson NJ, Reverter A, Greenwood PL, Guo B, Cafe LM, Dalrymple BP. Longitudinal muscle gene expression patterns associated with differential intramuscular fat in cattle. Animal. 2015;9(4):650–9. [DOI] [PubMed] [Google Scholar]
- 32.Gao J, Cheng BB, Liu YF, Li MM, Zhao GY. Effects of red cabbage extract rich in anthocyanins on rumen fermentation, rumen bacterial community, nutrient digestion, and plasma indices in beef bulls. Animal. 2022;16(5):100510. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and analysed during the current study can be accessed at https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1190918/.