Abstract
Background
Diabetes mellitus (DM) can cause severe complications, including diabetic foot ulcers (DFU). There is a significant gap in understanding the single-cell ecological atlas of DM and DFU tissues.
Methods
Single-cell RNA sequencing data were used to create a detailed single-cell ecological landscape of DM and DFU. Enrichment analysis identified pathways involved in cellular subpopulations, and pseudo-time analysis inferred cell development processes. A gene regulatory network explored the role of transcription factors in DFU progression, and a potential herbal drug-target gene interaction network was constructed.
Results
In the DFU group, immune cells were activated, with notable changes in several subpopulations. ATP5E was significantly overexpressed in Naive T cells, fibroblasts, endothelial cells, and CD8+ T cells in DM patients. Specific immune cell subsets, such as Naive T_RGCC, CTL_TYROBP_CL4, Mac_SLC40A1, and M1_CCL3L1, likely contribute to DFU formation through overactivation and proliferation, leading to tissue damage and ulcer exacerbation. Key genes TPP1, TLR4, and RIPK2 were identified, and 88 active ingredients in the herbal drug-target network showed strong correlations with these targets. Herbs like Angelica dahurica, Angelica sinensis, Boswellia carterii, liquorice, myrrh, and Semen armeniacae amarae were included.
Conclusions
This study offers insights into DM and DFU cytology. T cells in DFU are activated, attacking normal tissues and worsening tissue damage. The ATP5E gene may be related to the ecological remodeling of DM, and TPP1, TLR4, and RIPK2 are potential targets for DFU treatment.
Supplementary Information
The online version contains supplementary material available at 10.1186/s40001-024-02179-7.
Keywords: Diabetes mellitus, DFU, Single-cell RNA sequencing, Apoptosis, Immune activation, Drug targets
Introduction
Diabetes mellitus (DM) is a significant global health issue and a chronic metabolic disease characterized by hyperglycemia. It results from the destruction of β-cells, impairing insulin secretion, or the inability of insulin to bind effectively to its receptors, leading to elevated blood glucose levels. In addition to genetic predispositions, viral infections, and unhealthy lifestyles are also contributing factors to DM [1, 2]. DM encompasses several types, including type 1 diabetes (T1D), type 2 diabetes (T2D), and gestational diabetes, with T1D accounting for 5–10% of all cases. T1D is an autoimmune disease, typically manifesting in childhood, where antibodies target pancreatic islet cells, leading to β-cell destruction. T2D, on the other hand, represents the majority of DM cases and is associated with age, obesity, and other metabolic factors [3, 4]. The prevalence of DM is rising at an alarming rate, with recent studies predicting that the global prevalence in adults will reach 5.4% by 2025—an increase of 1.4% compared to 1995 statistics. The highest number of patients are projected to be in the USA, India, and China.
DM can lead to a wide range of serious and life-threatening complications, making it one of the most complication-prone diseases [5]. Diabetic foot ulcer (DFU) is a common complication of DM, affecting an estimated 30% of individuals with diabetes at some point in their lifetime. Among those with moderate to severe DFU, approximately 20% will face some degree of amputation. DFU significantly increases mortality, with affected individuals experiencing a 2.5-fold higher risk of death within 5 years compared to those with DM alone. In addition, DFU contributes to increased morbidity, a greater impact on quality of life, and imposes a severe psychosocial burden on patients [6, 7]. As a result, the burden of diabetes-related complications is a critical healthcare issue, and there is an urgent need for research into new therapies and treatments for DFU.
Single-cell RNA sequencing (scRNA-seq) enables the detailed analysis of gene structure and expression in individual cells, revealing cellular heterogeneity [8, 9]. It is now widely used to map the single-cell ecological landscape of various cancers, uncover potential molecular mechanisms underlying disease onset, and provide a theoretical framework for the development of new therapies and drugs. In this study, we apply scRNA-seq to map the single-cell ecological landscape of diabetic and DFU tissues, offering valuable insights into cellular function and disease pathophysiology.
Methods
Data sources and pre-processing
The scRNA-seq data used in this study (GSE165816) were obtained from the GEO database (https://www.ncbi.nlm.nih.gov/geo/). The data set includes 25 foot tissue samples, consisting of 5 DFU samples, 8 DM samples, and 11 healthy control samples. To maintain a balanced comparison across groups, we randomly selected five samples each from the DM and healthy control groups for subsequent analysis, ensuring equal sample sizes across the three groups. The 'sample' function in R was used to ensure randomness in the selection process, with the random seed set to 100 to allow reproducibility. To further validate the randomness and eliminate any potential bias, the selection process was repeated multiple times. These steps ensured that the five samples selected from both the DM and healthy control groups were representative and randomized, thereby supporting the fairness and scientific rigor of the analysis.
We used the IntegrateData function [10] in the Seurat package [11] to integrate scRNA-seq data and generate single-cell expression atlases (Seurat objects). Seurat tools in R were employed for data quality control and normalization, filtering out cells in the highest and lowest 1% of expression features, as well as cells with more than 10% mitochondrial gene expression.
Cell cluster analysis and subpopulation cluster analysis
The FindAllMarkers function in the Seurat package was used to identify differentially expressed genes (DEGs) in each cell cluster [10]. Subpopulation analysis was performed for each cell type using Seurat to explore the biological functions of disease-specific subpopulations. Clustering results were visualized using UMAP [12], which applies dimensionality reduction algorithms. DEGs were identified across clusters in the control, DM, and DFU groups, with a logfc.threshold of 0.25 and a p value < 0.01 considered significant.
Functional enrichment analysis
To investigate the biological pathways associated with each cell subpopulation, we performed enrichment analysis of cluster markers using the ClusterProfiler package in R [13, 14]. Pathways with a p value < 0.05 were considered significantly enriched.
Pseudo-time analysis
Throughout life, cells undergo state transitions driven by differences in gene expression. Pseudotime analysis, based on transcriptional similarity, tracks the developmental progression of cells over pseudotime. In this study, we applied the Monocle 3 algorithms [15, 16] in R to analyze the developmental trajectories of single cells.
Construction of gene regulatory network
Cellular heterogeneity within tissues arises from variations in transcriptional states, which are determined and stabilized by gene regulatory networks (GRNs) dominated by transcription factors (TFs). TFs regulate gene transcription by binding to downstream target genes, ultimately shaping the cell's phenotype [17, 18]. Single-cell regulatory network inference and clustering (SCENIC) is a method used to reconstruct GRNs from single-cell transcriptome data and identify cell states through co-expression and motif analysis. In this study, we employed the python tool pySCENIC to analyze and reconstruct TF-centered GRNs, inferring co-expression modules [19, 20]. The transcription factor binding motifs within these modules were sourced from the JASPAR database (https://jaspar.genereg.net).
Clinical sample collection
DFU and DM patients who visited the First Affiliated Hospital of Guangxi Medical University between October 2023 and May 2024 were selected for this study. We collected foot skin tissue samples from 5 DFU patients, including both ulcerated and corresponding distal normal skin tissues (Supplementary Table 1). In addition, serum samples were obtained from 10 DFU patients and 10 DM patients. All samples were collected with the informed consent of the patients and approved by the Ethics Committee of the First Affiliated Hospital of Guangxi Medical University (No. 2022-KT-NSFC-260).
Immunohistochemistry
Paraffin sections were baked overnight at 62 ℃ and then placed on a slide rack. The sections were sequentially deparaffinized using Dewaxing Solutions I, II, and III. A 20 × ethylenediaminetetraacetic acid (EDTA) antigen retrieval solution was diluted to 1 × with double-distilled water, adjusted to pH 9.0, and used for antigen retrieval in a microwave. After lightly drying the sections, a circle was drawn around the tissue using a histochemical pen, and primary and secondary antibodies were applied for incubation. The slides were washed three times with PBS on a decolorizing shaker, each wash lasting 5 min. An autofluorescence quencher was added for 5 min, followed by rinsing with running water for 20 min. Freshly prepared diaminobenzidine (DAB) chromogenic solution (100μL) was added, and once the target protein developed color without background staining, the reaction was halted by rinsing with tap water. The sections were then counterstained with hematoxylin, thoroughly washed with pure water, and dehydrated sequentially with 75%, 85%, and 95% ethanol, followed by absolute ethanol I, II, and III, each for 5 min. Finally, the sections were cleared using Clearing Solutions I, II, and III, each for 5 min. The sections were observed and imaged under an optical microscope [21].
qRT-PCR
Total RNA was extracted from tissue samples using SparKZol Reagent and reverse transcribed into cDNA. The PCR reaction was conducted for 40 cycles, with denaturation at 95 ℃ for 10 s, annealing at 60 ℃ for 20 s, and extension at 72 ℃ for 30 s. β-Actin was used as an internal reference, and each experiment was repeated at least three times. The Ct values for each gene in each sample were recorded, and the 2−ΔΔCt method was used to calculate the relative expression levels of TPP1, TLR4, and RIPK2.
Construction of a potential target gene–drug network
To construct the potential target gene–drug network, we used Cytoscape software (version 3.7.1), a platform designed for visualizing complex networks and integrating them with various types of attribute data. This network illustrates the intricate relationships between target genes and herbal compounds. The active ingredients of herbs are sourced from the Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP), and screened according to the standards of oral bioavailability not less than 30% and drug similarity not less than 0.18. Subsequently, the gene targets of these traditional Chinese medicine ingredients were downloaded and intersected with the markers of the target cell subpopulation. Finally, these ingredients and their gene targets were introduced into Cytoscape. In the network, each node represents either an herbal compound or a gene target, while the edges (lines) between the nodes represent the interactions between them, curated from literature and experimental or predicted interactions in databases. Using Cytoscape's visualization tools, we graphically represented the connections between herbal compounds and gene targets. We then analyzed network topology parameters, such as degree centrality, betweenness centrality, and closeness centrality, to identify key compounds and target genes. Nodes with high centrality scores were considered potential key players in the treatment of DFU and were validated through qRT-PCR and immunohistochemistry experiments.
Data analysis and statistics
Statistical analyses were performed using R (https://www.r-project.org/). Data were expressed as mean ± standard deviation (SD). Group comparisons were made using either a t test for two groups or one-way ANOVA followed by post hoc tests for multiple group comparisons. A p value < 0.05 was considered statistically significant. In addition, correlation analyses were performed using Pearson's or Spearman's correlation coefficients, depending on the data distribution. All statistical tests were two-tailed.
Results
Global ecological landscape of DM and DFU
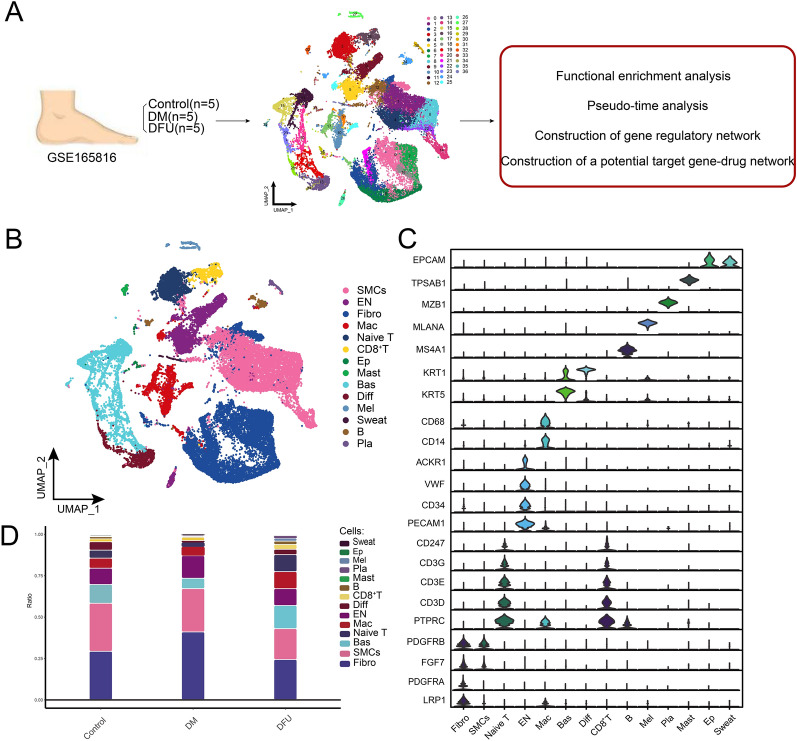
To investigate the cellular heterogeneity in DM and (DFU, we analyzed scRNA-seq data from foot skin samples of five DM patients, five DFU patients, and five healthy controls (Fig. 1A). The workflow of this study, as shown in Fig. 1A, outlines the construction of a global single-cell landscape for DM and DFU, followed by functional enrichment analysis, pseudotime analysis, gene regulatory network construction, and the development of a potential gene–drug target network.
Fig. 1.
Construction of a global single-cell ecological landscape of the diabetic and DFU. A Workflow of this study. Single-cell RNA sequencing (10X Genomics) data from the feet of 5 diabetic patients with DFU, 5 DM patients and 5 healthy controls were analyzed, and 37 clusters of a total of 34,742 cells were identified. B Single-cell atlas mapping cell types, including smooth muscle cells (SMCs), endothelial cells (EN), fibroblasts (Fibro), macrophages (Mac), Naive T cells, CD8+ T cells, epithelial cells (Ep), mast cells (Mast), basal keratin-forming cells (Bas), differentiated keratin-forming cells (Diff), melanocytes (Mel), Sweat cells (Sweat), B lymphocytes (B) and plasma cells (Pla). C Specific marker genes in different cell types. D Difference in the proportion of cellular components between the control, DM and DFU groups
After performing quality control and normalization, cluster analysis of 34,742 cells identified 37 distinct cell clusters (Fig. 1B). These clusters were further annotated into 14 different cell types based on the expression of known cell-specific marker genes (Fig. 1C and Supplementary Table 2). The identified cell types included smooth muscle cells (SMCs), endothelial cells (ENs), fibroblasts (Fibro), macrophages (Mac), Naive T cells, CD8+ T cells, epithelial cells (Ep), mast cells (Mast), basal keratinocytes (Bas), differentiated keratinocytes (Diff), melanocytes (Mel), sweat gland cells, B lymphocytes (B), and plasma cells (Pla). Each cell cluster was characterized by specific marker genes, as demonstrated in Fig. 1C, where selected marker genes such as EPCAM, CD68, and KRT5 helped define their respective cell populations.
Further analysis of cell types across different subpopulations revealed significant differences in their distribution, especially among ENs, Fibro, SMCs, Naive T cells, Mac, and CD8+ T cells (Fig. 1D). In particular, ENs were most abundant in the DM group, while Naive T cells, Mac, and CD8+ T cells were most prevalent in the DFU group. This differential distribution suggests disease-specific cellular changes, which were the focus of subsequent analyses. We performed detailed investigations on these six key cell types to understand their roles in the pathophysiology of DM and DFU (Fig. 1D and Supplementary Table 3).
In summary, we constructed a comprehensive single-cell atlas that highlights the cellular ecological differences in the foot skin of DM and DFU patients. This analysis provides a foundation for further investigations into the molecular mechanisms driving these conditions, with potential implications for targeted therapeutic interventions.
The ecological landscape of the EN cell subpopulations in DM and the DFU
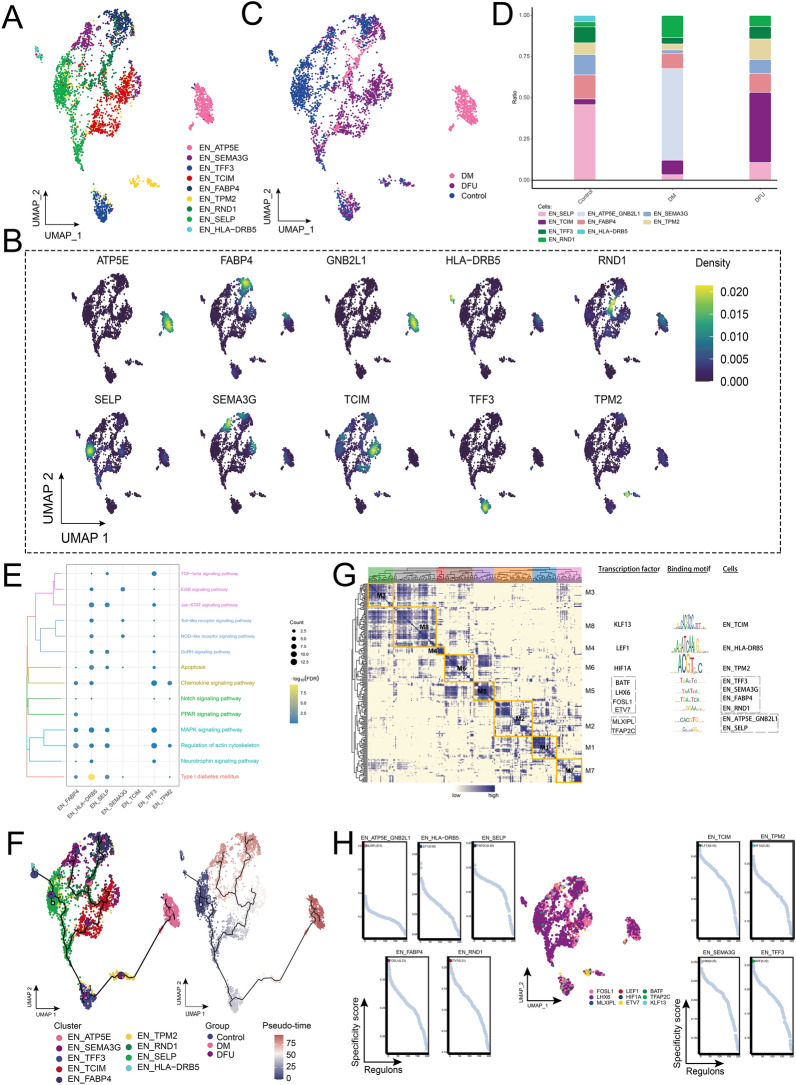
Further clustering analysis of endothelial cells identified nine distinct subpopulations (Fig. 2A). Each subpopulation was characterized by the high expression of specific marker genes, such as SELP, TCIM, ATP5E, GNB2L1, FABP4, SEMA3G, TPM2, TFF3, RND1, and HLA-DRB5 (Fig. 2B, D). Among these subpopulations, EN_ATP5E was found to be specific to DM patients, indicating a potential role in the pathophysiology of diabetes-related endothelial changes (Fig. 2B). The analysis of abundance variations among the different EN subpopulations revealed that the EN_TCIM subpopulation was significantly more prevalent in the DFU group, with the highest abundance observed in DFU patients (Fig. 2C). This finding suggests a disease-specific expansion of this subpopulation in diabetic foot ulcer pathology. To explore the biological pathways associated with these endothelial subpopulations, enrichment analysis was performed, revealing significant enrichment in pathways related to type I diabetes, apoptosis, actin regulation, and MAPK signaling (Fig. 2E). These pathways highlight the involvement of endothelial cells in key mechanisms underlying diabetes and its complications, such as DFU. Pseudotime analysis (Fig. 2F) showed that endothelial cells initially develop from the EN_SELP subpopulation and then differentiate into various other subpopulations, including EN_TCIM, EN_ATP5E, and EN_GNB2L1. This developmental trajectory further underscores the dynamic cellular changes in the endothelial landscape during disease progression.
Fig. 2.
Ecological landscape of the endothelial cell populations. A Single-cell atlas showing cell clusters of endothelial cells. B Marker genes specific to each subpopulation. C Single-cell atlas shows clusters of endothelial cells in control, DM and DFU groups. D Differences in abundance of various subpopulations of Endothelial cells in the control, DM and DFU groups. E Biological pathways involved in endothelial cells. F Trajectory and pseudo-time values of endothelial cells progression. The pie charts show the proportion of different groups in the cluster. G Gene regulatory network of the endothelial cell subpopulations. H Series of scatter plots demonstrating transcription factors regulating endothelial cell subpopulations
In addition, GRN analysis (Fig. 2G) grouped the marker genes into eight distinct modules, which were regulated by TFs such as KLF13, LEF1, HIF1A, BATF, LHX6, FOSL1, ETV7, MLXIPL, and TFAP2C. These TFs exhibited high transcriptional activity across the endothelial subpopulations (Fig. 2H), further emphasizing their potential roles in regulating key genes and pathways involved in diabetes and DFU.
In conclusion, the identification of distinct endothelial subpopulations and their associated GRNs provides new insights into the cellular and molecular mechanisms underlying DM and DFU, highlighting specific pathways and transcription factors that may serve as potential therapeutic targets for disease intervention.
The ecological landscape of fibroblast clusters
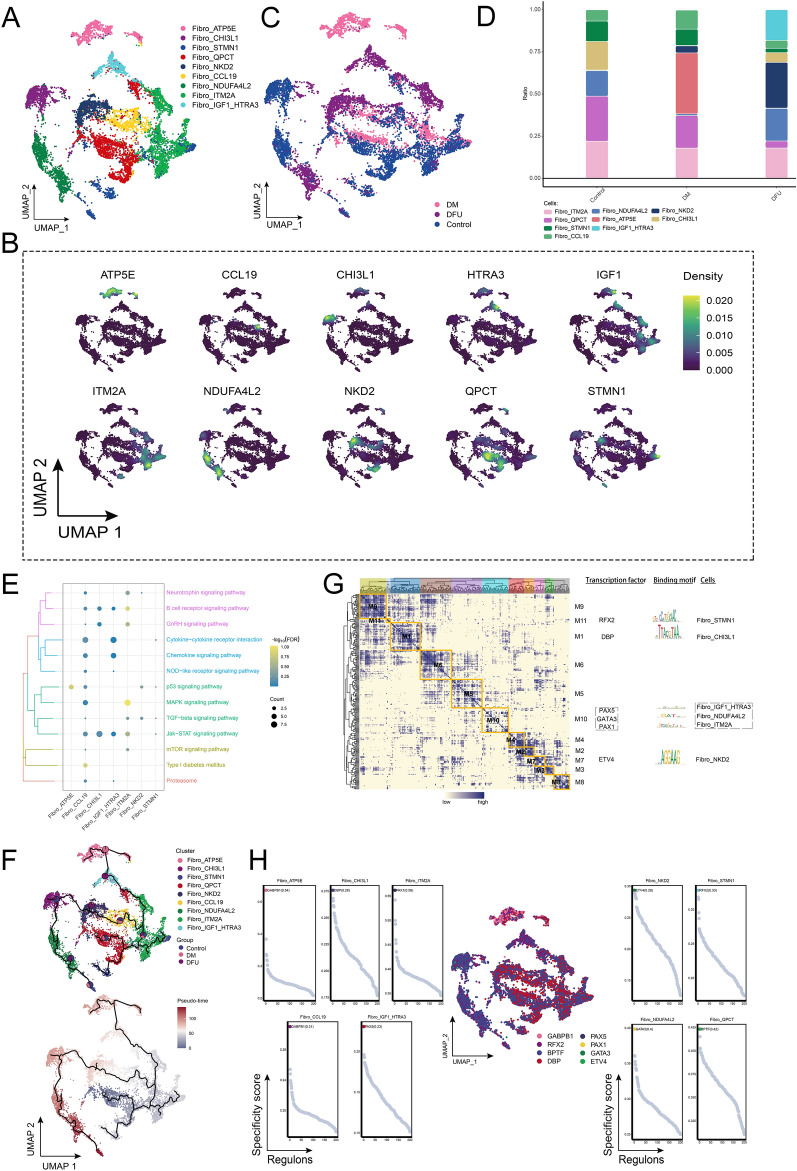
Since fibroblast abundance increased in the DM group and decreased in the DFU group compared to the control group (Fig. 1D), we conducted further clustering analysis of fibroblast (fibro) cells. This analysis identified nine distinct fibroblast subpopulations based on the expression of known marker genes within each subpopulation (Fig. 3A, B). The abundance of these subpopulations varied significantly across different patient groups (Fig. 3C). To better understand the changes in fibroblast subpopulation proportions, we visualized their relative abundances across the control, DM, and DFU groups (Fig. 3D). The results revealed that the Fibro_NKD2 subpopulation was significantly increased in the DFU group, showing the highest proportion among all fibroblast subpopulations. In contrast, the Fibro_ATP5E subpopulation was specifically enriched in the DM group, suggesting disease-specific alterations in fibroblast composition. Enrichment analysis of these fibroblast subpopulations indicated significant involvement in pathways related to type I diabetes, B-cell receptor signaling, GnRH signaling, p53 signaling, and MAPK signaling (Fig. 3E). These pathways underscore the critical roles fibroblasts play in mediating cellular responses within the DM and DFU microenvironments. To further explore the developmental trajectories of these fibroblast subpopulations, pseudotime analysis was performed. This analysis revealed that fibroblast development initiated from the Fibro_QPCT subpopulation and progressed into several distinct subpopulations, including Fibro_NKD2 and Fibro_ATP5E (Fig. 3F). In addition, GRN analysis revealed that the regulatory framework centered around TFs was organized into 11 modules (Fig. 3G). Key TFs such as RFX2, DBP, PAX5, GATA3, PAX1, and ETV4 were identified as regulators of specific gene expression patterns within these fibroblast subpopulations (Fig. 3H), suggesting their pivotal roles in shaping fibroblast heterogeneity and function in DM and DFU.
Fig. 3.
Ecological landscape of fibroblasts. A Single-cell mapping of cell clusters of fibroblasts. B Single-cell atlas showing genes specifically expressed in each subpopulation of fibroblasts. C Single-cell atlas shows clusters of fibroblasts in Control, DM and DFU groups. D Variation in the abundance of subpopulations of fibroblasts in different subpopulations. E Biological pathways involved in fibroblasts. F Developmental trajectory of fibroblasts. The pie chart shows the proportion of different groups in the cluster. G Fibro clusters of transcription factors in a co-expression pattern. Left: Heat map identifying co-expression modules; Middle: Major transcription factors and their binding sequences; Right: Cellular clusters of transcription factors. H Series of scatter plots demonstrating transcription factors regulating fibroblasts subpopulations
In summary, our analysis revealed distinct fibroblast subpopulations with disease-specific abundance patterns in DM and DFU. These fibroblast subpopulations are involved in key signaling pathways, and their developmental trajectories are governed by complex gene regulatory networks centered around specific transcription factors. These findings provide valuable insights into the role of fibroblasts in the progression of diabetes and its complications, highlighting potential therapeutic targets for further exploration.
The ecological landscape of smooth muscle cells
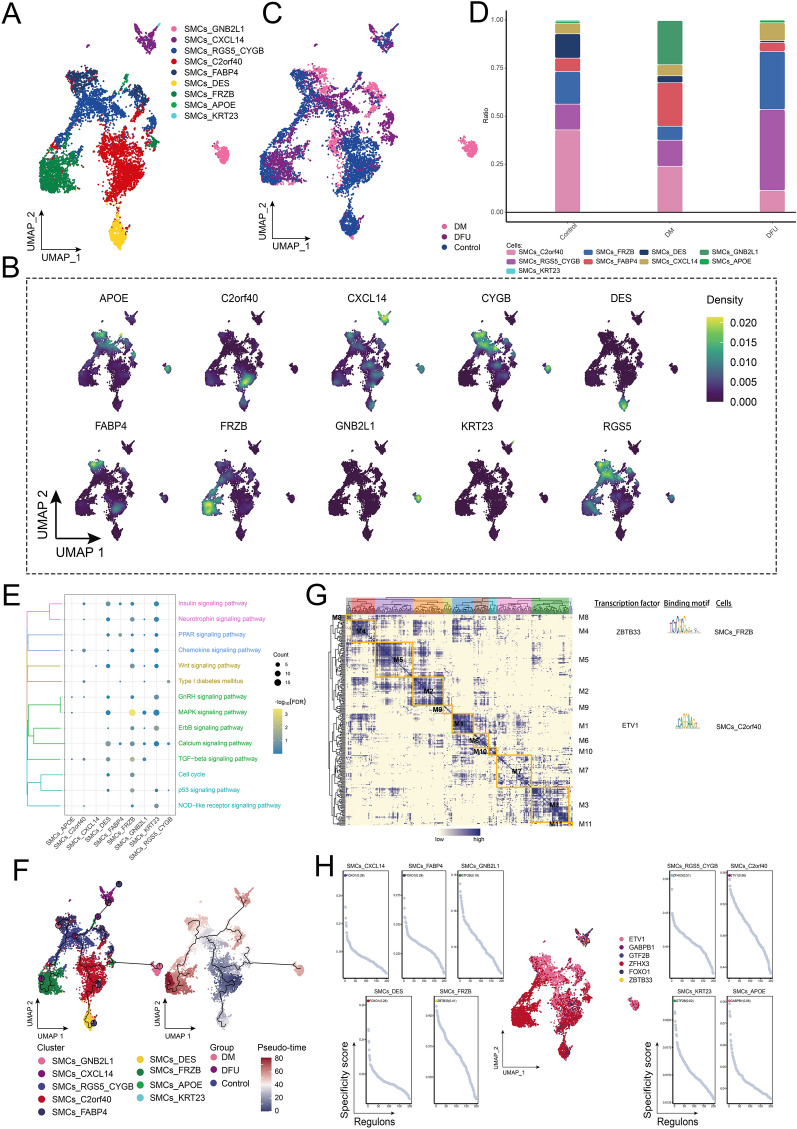
The cellular abundance of SMCs was significantly reduced in the DFU group compared to the control and DM groups (Fig. 1D). Further clustering analysis identified nine distinct SMC subpopulations (Fig. 4A), each characterized by specific marker genes mapped onto the single-cell landscape (Fig. 4B). The single-cell landscapes of the control, DM, and DFU groups revealed significant heterogeneity among these different SMC subpopulations, reflecting disease-specific alterations (Fig. 4C). Among these subpopulations, SMCs_RGS5_CYGB exhibited a significantly higher abundance in the DFU group, making it a major component of the DFU cellular microenvironment (Fig. 4D). This distinct abundance pattern likely reflects functional differences between the subpopulations. Enrichment analysis of these SMC subpopulations revealed significant involvement in several key signaling pathways, including the MAPK signaling pathway, Wnt signaling pathway, and p53 signaling pathway, suggesting their role in regulating disease progression (Fig. 4E). Pseudotime analysis (Fig. 4F) was conducted to explore the developmental trajectories of the SMC subpopulations, revealing that differentiation initiated from the SMCs_C2orf40 subpopulation and progressed into other subpopulations. In addition, GRN analysis (Fig. 4G) identified 11 regulatory modules, with TFs such as ZBTB33 and ETV1 showing significant changes in expression and activity (Fig. 4H), potentially playing critical roles in regulating SMC function and differentiation in DFU.
Fig. 4.
Ecological landscape of SMCs. A Single-cell atlas showing that SMCs comprise nine major cell subpopulations. B Single-cell atlas showing genes specifically expressed in each subpopulation of SMCs. C Single-cell atlas shows clusters of SMCs in control, DM and DFU groups. D Difference in abundance of each subpopulation of SMCs in the control, DM and DFU groups. E Signaling pathways involved in SMCs subpopulations. F Developmental trajectory of SMCs. The pie chart shows the proportion of different populations in the SMCs subpopulations. G Gene regulatory network of the SMCs subpopulations. H Transcription factors regulating SMCs subpopulations
In conclusion, the analysis revealed significant heterogeneity among SMC subpopulations in DM and DFU, with distinct disease-specific distributions. The increased abundance of the SMCs_RGS5_CYGB subpopulation in DFU highlights its potential role in the disease's cellular ecology. Enrichment of key signaling pathways, such as MAPK, Wnt, and p53, along with the identification of critical transcription factors like ZBTB33 and ETV1, provides valuable insights into the molecular mechanisms regulating SMC function and differentiation in DFU, offering potential therapeutic targets for intervention.
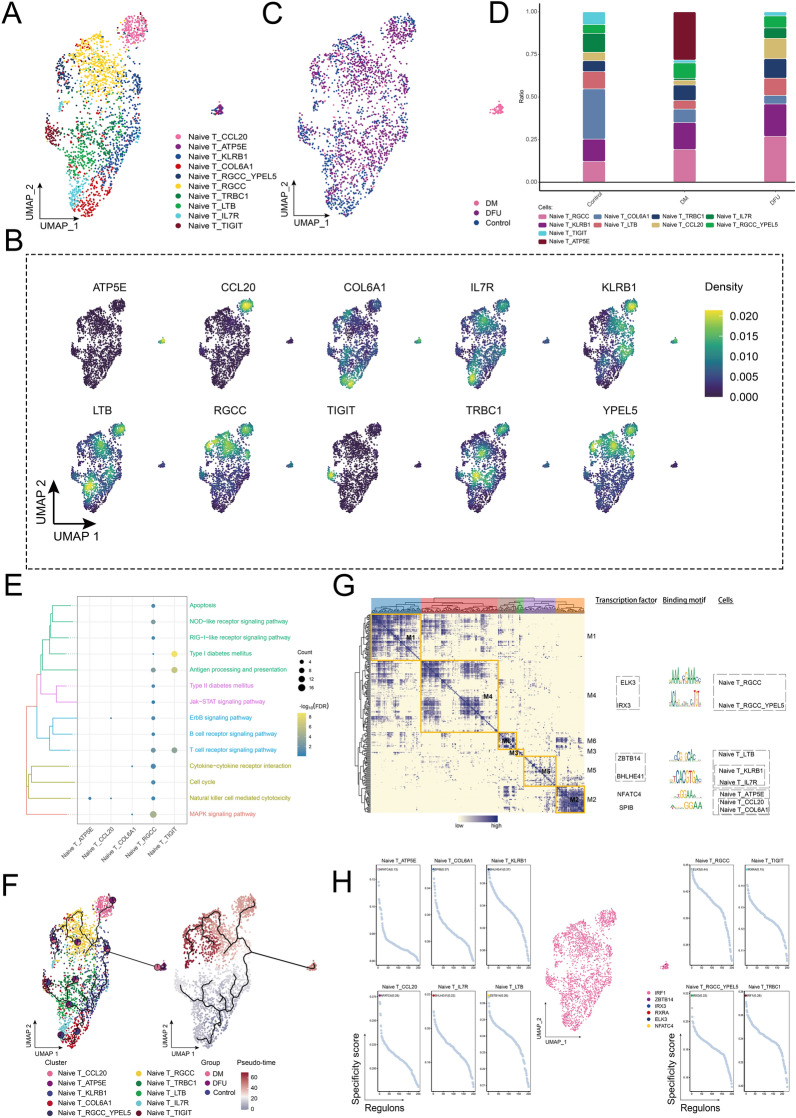
The ecological landscape of Naive T cells
The abundance of Naive T cells was significantly higher in the DFU group compared to the control and DM groups. This prompted further investigation into the dynamic changes of Naive T cells across these groups. Using the UMAP algorithm, we clustered the Naive T cells into 10 distinct subpopulations (Fig. 5A) and mapped their marker genes onto a single-cell atlas (Fig. 5B). These subpopulations displayed considerable heterogeneity among different patients (Fig. 5C). Among the subpopulations, Naive T_RGCC was significantly enriched in the DFU group, forming a major component of the DFU cellular ecology (Fig. 5D). Enrichment analysis revealed that Naive T_RGCC was primarily involved in the MAPK signaling pathway, T-cell receptor signaling pathway, antigen processing and presentation, and NOD-like receptor signaling pathway. Another subpopulation, Naive T_TIGIT, was notably associated with the T-cell receptor signaling pathway, antigen presentation, and the type 2 diabetes signaling pathway (Fig. 5E). Cell developmental trajectory analysis (Fig. 5F) showed that the differentiation of these subpopulations originated from the Naive T_COL6A1 subpopulation, which acted as the starting point for further differentiation. GRN analysis (Fig. 5G) revealed that the network, centered around TFs, was organized into six modules. Key TFs, such as ELK3, IRX3, ZBTB14, BHLHE41, NFATC4, and SPIB, were identified as regulators of Naive T cell gene expression (Fig. 5H), highlighting their roles in regulating key pathways and contributing to the functional heterogeneity of Naive T cells in DFU.
Fig. 5.
Ecological landscape of Naive T cells. A Single-cell atlas showing clusters of Naive T cells. B Single-cell atlas showing marker genes for each subpopulation of Naive T cells. C Single-cell atlas shows clusters of Naive T cells in the Control, DM and DFU groups. D Differences in the abundance of Naive T cell subpopulations in the Control, DM and DFU groups. E Signaling pathways involved in Naive T cell subpopulations. F Developmental trajectory of Naive T cells. Pie charts show the proportion of different populations in the cluster. G Gene regulatory network of the Naive T cell subpopulations. H Transcription factors regulating subpopulations of Naive T cell subpopulation
In conclusion, the analysis revealed significant enrichment of the Naive T_RGCC subpopulation in the DFU group, highlighting its involvement in key signaling pathways such as MAPK, T-cell receptor signaling, and antigen presentation. The identification of transcription factors like ELK3, IRX3, and NFATC4 as key regulators further emphasizes the functional diversity of Naive T cells in DFU, offering potential targets for therapeutic interventions aimed at modulating immune responses in diabetic complications.
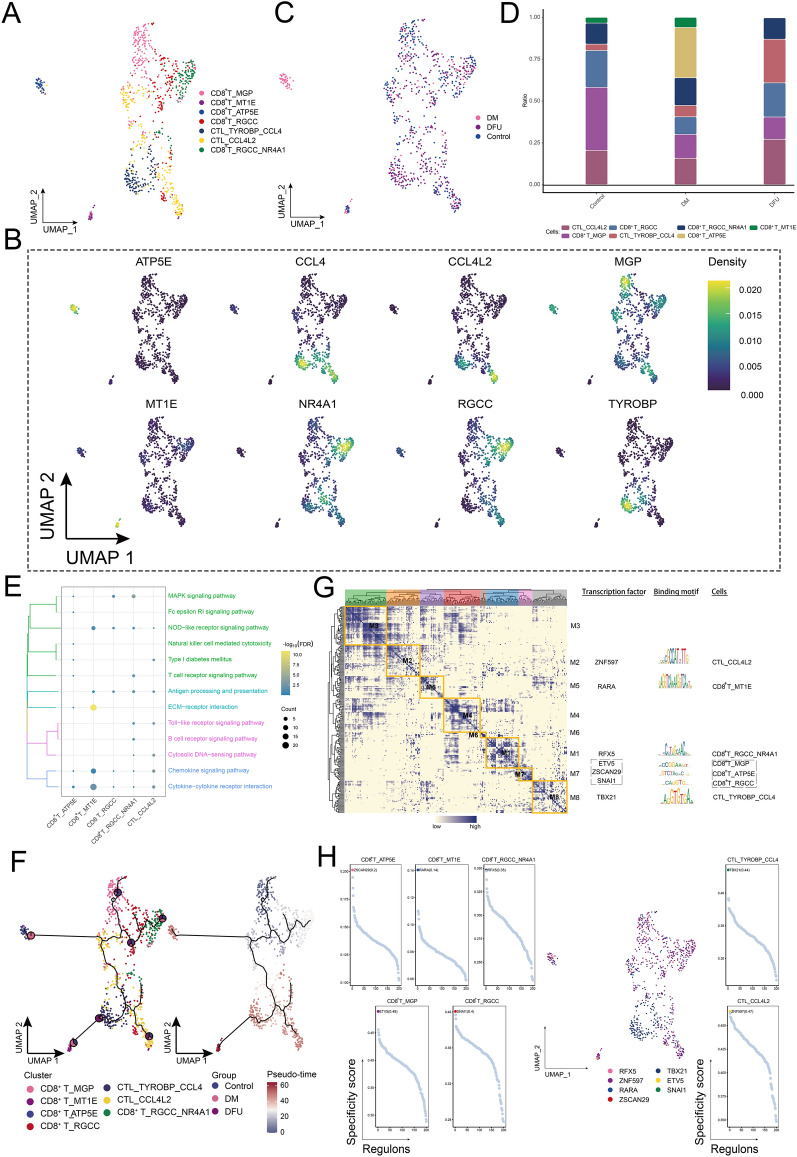
Further analysis on the ecological landscape of CD8+ T cell clusters
The abundance of CD8+ T cells was significantly higher in the DFU group compared to the control and DM groups, prompting further investigation into the heterogeneity of this cell type. We identified seven distinct CD8+ T cell subpopulations, including CTL_TYROBP_CL4, CD8+ T_RGCC_NR4A1, and CD8+ T_ATP5E (Fig. 6A). A detailed analysis revealed that subpopulations such as CTL_TYROBP_CL4, CD8+ T_RGCC_NR4A1, CD8+ T_MGP, CD8+ T_MT1E, CD8+ T_RGCC, and CTL_CCL4L2 were present across the control, DM, and DFU groups, while CD8 + T_ATP5E was specifically detected in the diabetic patient group (Fig. 6B). Further analysis of compositional changes among the subpopulations indicated that CTL_TYROBP_CL4 was significantly increased in the DFU group, exhibiting the highest enrichment among all CD8+ T cell subpopulations (Fig. 6C). Specific genes such as ATP5E, CCL4, CCL4L2, MGP, NR4A1, and RGCC were overexpressed in these subpopulations (Fig. 6D). Enrichment analysis highlighted that key pathways such as MAPK signaling, NOD-like receptor signaling, T-cell receptor signaling, antigen processing and presentation, ECM–receptor interactions, Toll-like receptor signaling, cytoplasmic DNA sensing, chemokine signaling, and cytokine–cytokine receptor interactions regulated the functional changes in CD8+ T cells (Fig. 6E). Pseudotime analysis further revealed the developmental trajectories of these CD8+ T cell subpopulations, with CD8+ T_MGP acting as the origin point for subsequent differentiation (Fig. 6F). GRN analysis identified seven distinct modules of transcriptional regulation (Fig. 6G). Transcription factors such as ZNF597, RARA, RFX5, ETV5, ZSCAN29, SNAI1, and TBX21 were recognized as key regulators of gene expression in these CD8+ T cell subpopulations (Fig. 6H), playing pivotal roles in modulating their behavior across the different disease states.
Fig. 6.
Ecological landscape of CD8+ T cells. A Single-cell mapping of CD8+ T cell subpopulations. B Single-cell atlas showing marker genes for each subpopulation of CD8+ T cells. C Single-cell atlas shows clusters of CD8+ T cells in the control, DM and DFU groups. D Differences in the abundance of CD8+ T cell subpopulations in the control, DM and DFU groups. E Signaling pathways involved in CD8+ T-cell subpopulations. F Developmental trajectory of CD8+ T-cell subpopulations. The pie chart shows the proportion of different populations in the cluster. G Gene regulatory network of the CD8+ T cell subpopulations. H Transcription factors regulating subpopulations of CD8+ T cell subpopulations
In conclusion, the analysis revealed significant heterogeneity among CD8+ T cell subpopulations, with CTL_TYROBP_CL4 being particularly enriched in the DFU group. The identification of key signaling pathways, such as MAPK, NOD-like receptor, and T-cell receptor signaling, along with the involvement of critical transcription factors like ZNF597, RARA, and RFX5, underscores the role of these subpopulations in the immune response and disease progression in DFU. These findings provide potential targets for therapeutic interventions aimed at modulating CD8+ T cell function in diabetic complications.
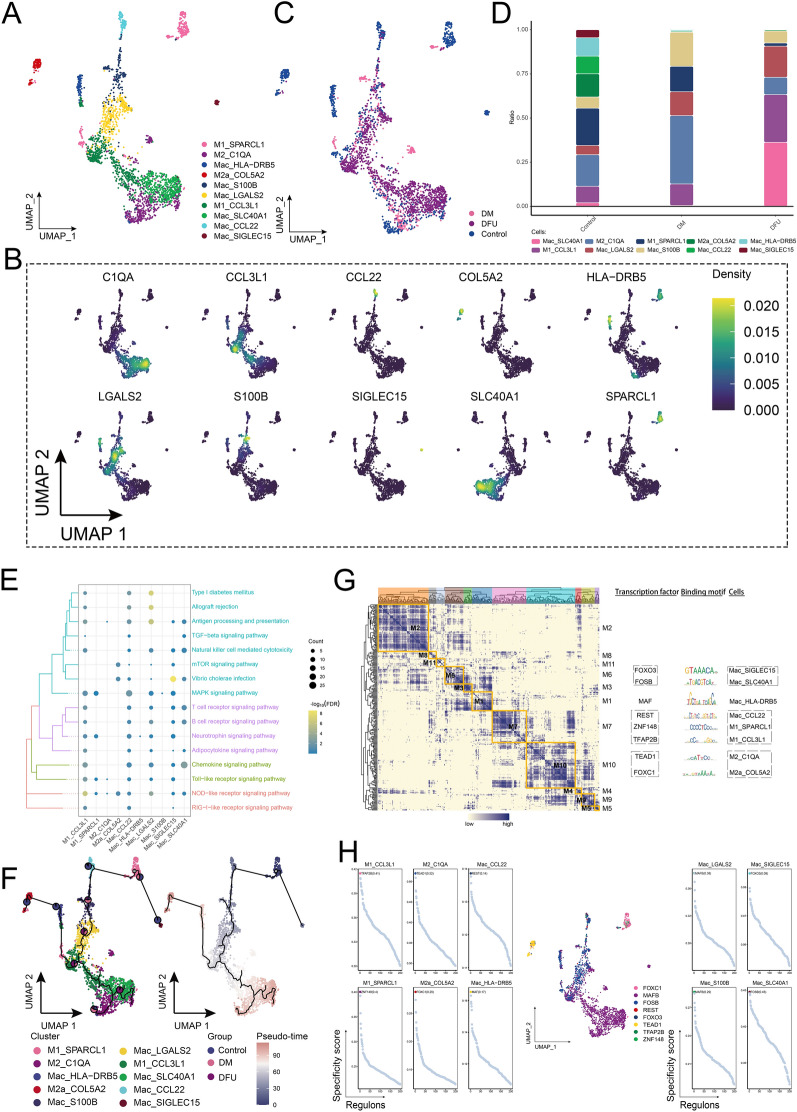
Exploration of the ecological landscape of macrophages
The abundance of Mac in the control and DM groups was comparable; however, in the DFU group, macrophage abundance was nearly double that of the DM group. This underscores the importance of studying the developmental changes of macrophages in patients at different stages of disease progression. Based on marker gene expression, we identified 10 main macrophage subgroups (Fig. 7A, B), which were further divided into detailed subclusters (Fig. 7C). Among these, Mac_SLC40A1 and M1_CCL3L1 were significantly more abundant in the DFU group compared to the control and DM groups, with Mac_SLC40A1 showing the most pronounced increase (Fig. 7D). To understand the molecular mechanisms underlying these macrophage subpopulations, KEGG pathway enrichment analysis was performed, revealing significant involvement in pathways such as type I diabetes, allograft rejection, antigen processing and presentation, TGF-β signaling, natural killer cell-mediated cytotoxicity, MAPK signaling, T-cell receptor signaling, chemokine signaling, and NOD-like receptor signaling (Fig. 7E). Cell developmental trajectory analysis (Fig. 7F) indicated that M1_SPARCL1 serves as the starting point for macrophage differentiation, giving rise to various subpopulations under different regulatory conditions. To investigate TF activation events related to cell state changes, we constructed a GRN. This analysis revealed that the GRN, centered on TFs, was organized into 11 modules (Fig. 7G). Key TFs involved in macrophage state transitions included FOXO3, FOSB, MAF, REST, ZNF148, TFAP2B, TEAD1, and FOXC1 (Fig. 7H). In addition, we observed that the ATP5E gene was exclusively expressed in the DM group (Supplementary Fig. 1A). Further analysis across various cell types demonstrated significant overexpression of ATP5E in endothelial cells, CD8+ T cells, fibroblasts, and immature T cells within the DM group (Supplementary Fig. 1B). The abundance of subpopulations expressing ATP5E increased significantly in these cell types in the DM group (Figs. 2C, 3C, 5C, 6C).
Fig. 7.
Ecological landscape of Mac. A Single-cell mapping of Mac subpopulations. B Single-cell atlas showing marker genes for each subpopulation of Mac. C Single-cell atlas shows clusters of Mac in the Control, DM and DFU groups. D Differences in the abundance of Mac subpopulations in the Control, DM and DFU groups. E Signaling pathways involved in Mac subpopulations. F Developmental trajectory of Mac subpopulations. The pie chart shows the proportion of different populations in the cluster. G Gene regulatory network of the Mac subpopulations. H Transcription factors regulating subpopulations of Mac subpopulations
In conclusion, the analysis revealed a significant increase in macrophage subpopulations, particularly Mac_SLC40A1 and M1_CCL3L1, in the DFU group, suggesting their crucial role in the disease progression. Pathway enrichment indicated involvement in key immune and inflammatory processes, such as TGF-β and MAPK signaling. The identification of transcription factors like FOXO3, FOSB, and MAF as regulators of macrophage state transitions highlights their potential as targets for therapeutic intervention in DFU. In addition, the exclusive expression of ATP5E in the DM group suggests its potential involvement in diabetic cellular mechanisms, particularly in endothelial cells, CD8+ T cells, and fibroblasts.
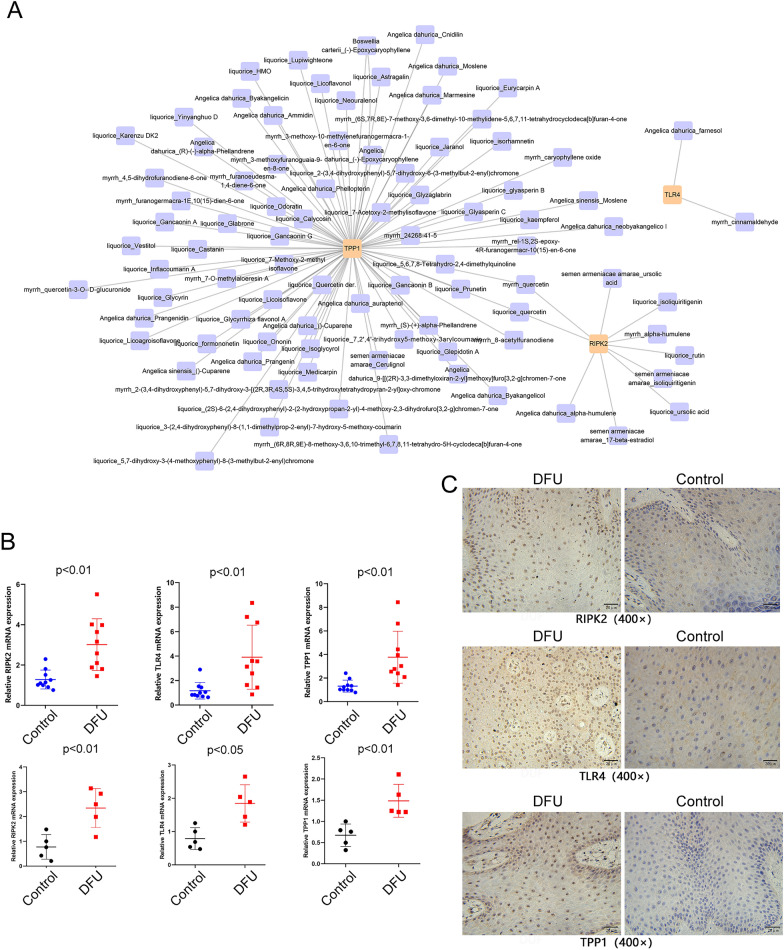
Construction and analysis of the potential herbal drug–gene network
Using cell-specific marker genes, we constructed a complex network of herbs commonly used for treating DFU. The top three marker genes identified in this network were TPP1, TLR4, and RIPK2, which showed strong correlations with several active herbal ingredients, including Angelica dahurica, Angelica sinensis, Boswellia carterii, liquorice, myrrh, and Semen armeniacae amarae (Fig. 8A). To validate the hypothesis that these genes are associated with the immune microenvironment of DFU, we examined the mRNA expression levels of TPP1, TLR4, and RIPK2 in serum and tissue samples from DM and DFU patients using qRT-PCR. Our results showed a significant increase in the expression levels of TPP1, TLR4, and RIPK2 in the serum and skin tissue of DFU patients compared to controls (Fig. 8B). Furthermore, immunohistochemistry revealed significantly higher expression of these genes in the diseased skin tissues of DFU patients compared to healthy skin (Fig. 8C).
Fig. 8.
Construction and analysis of the potential herbal drug–gene network. A Chinese medicine drug–target gene network. The size of each node in the network indicates the size of its degree value. The connection line indicates that each node is interconnected. B Expression levels of TPP1, TLR4, and RIPK2 mRNA in serum samples from 10 DFU and 10 DM patients, as well as in foot skin tissue (ulcerated and corresponding distal normal tissue) from 5 DFU patients, were detected by qRT-PCR. C Expression levels of TPP1, TLR4, and RIPK2 in foot skin tissue (ulcerated and corresponding distal normal tissue) from 5 DFU patients were assessed by immunohistochemistry
These findings suggest that TPP1, TLR4, and RIPK2 may serve as potential drug targets for DFU treatment, and targeting these genes could inhibit the progression and spread of the disease.
Discussion
In this study, we mapped a comprehensive single-cell atlas of DM and DFU, providing new insights into the cellular heterogeneity and distinct gene expression patterns associated with these conditions. Our findings significantly enhance the understanding of cellular dynamics in diabetic patients and those with DFU, establishing a robust foundation for future in-depth research. We conducted a detailed analysis of key cell types, including endothelial cells, SMCs, fibroblasts, and identified specific subsets of T lymphocytes and macrophages. In addition, we characterized the biological functions and molecular pathways involved in these subpopulations and constructed GRNs for each cell type. This integrative approach offers a novel perspective compared to previous single-cell analyses of DFU [22]. While our results align with other studies regarding the general composition of microenvironmental cell clusters, our focus on distinct patient groups and data sets highlights important differences. While the foundational data align in identifying immune cell dysregulation and impaired wound healing mechanisms, this study provides novel insights by focusing on distinct subpopulations, particularly in endothelial cells, SMCs, T cells, and macrophages, along with exploring potential therapeutic targets using a TCM pharmacology network.
Studies have shown that SMCs contribute to neo-angiogenesis and vasculogenesis, processes that are crucial for wound healing in DFU. SMCs and pericytes facilitate the formation of new blood vessels, which is essential for providing nutrients and oxygen to the healing tissues in DFU patients [23, 24]. In this study, the proportion of endothelial cells and fibroblasts in foot skin initially increased and then decreased, while the abundance of SMCs tended to decrease during the progression from a healthy state to diabetes and the formation of DFU. Conversely, the abundance of immune cells, such as Naive T cells and macrophages, decreased initially and then increased, with a progressive increase in the abundance of CD8+ T cells, indicating enhanced immune infiltration and an inflammatory environment. We hypothesize that in diabetic patients, hyperglycemia-induced inflammation leads to sustained immune system stimulation [25], resulting in hyperimmunity and the proliferation of pro-inflammatory immune cells [26, 27]. These cells become activated and attack normal tissues, causing tissue damage. In addition, excessive activation, proliferation, and repair of fibroblasts contribute to fibrosis and skin ulceration, mediating the development of DFU [28].
In this study, the analysis of SMCs, endothelial cells, T cells, and macrophages revealed distinct cellular changes during the progression from a healthy state to diabetes and the development of DFU. The abundance of SMCs, which play a role in angiogenesis and vasculogenesis, decreased significantly in DFU patients. Of particular interest was the SMCs_RGS5_CYGB subpopulation, which was highly enriched in DFU and likely contributes to cell death through the pro-apoptotic actions of RGS5 and CYGB, two genes known to induce apoptosis and lipid peroxidation [29–31].
Endothelial cells are crucial for vascular function and wound healing [23]. In DFU, the EN_TCIM subpopulation was significantly increased, suggesting a role for TCIM in promoting endothelial cell survival and proliferation through the Wnt and NF-kappaB pathways [32, 33], contributing to local tissue damage and necrosis [34].
T cells, particularly the Naive T_RGCC subpopulation, were also enriched in DFU, indicating an immune response activation. RGCC, a cell cycle regulatory gene, promotes mitosis and immune cell proliferation [35, 36], while TYROBP and CCL4 were associated with enhanced immune infiltration [37], exacerbating inflammation and contributing to tissue damage in DFU.
Macrophages, critical for all stages of wound healing, showed dysregulated polarization in DFU [23]. Two subpopulations, Mac_SLC40A1 and M1_CCL3L1, were significantly increased, suggesting their role in immune activation and excessive inflammatory responses. The overexpression of SLC40A1 promotes cell survival, while CCL3L1 drives leukocyte activation [38], both of which may contribute to the impaired healing process in DFU.
In addition, we found that ATP5E seems to play an important role in the progression of DM. ATP5E was significantly overexpressed in Naive T cells, fibroblasts, endothelial cells, and CD8+ T cells in the DM group. A related study reported that ATP5E expression is upregulated in diabetes [39], suggesting that ATP5E may be associated with ectopic remodeling in DM.
In this study, we constructed a pharmacological network of TCM to identify potential therapeutic targets for DFU. The network analysis revealed that TPP1, TLR4, and RIPK2 are highly associated with several bioactive compounds from commonly used herbs such as Angelica dahurica, Angelica sinensis, Boswellia carterii, liquorice, myrrh, and Semen armeniacae amarae. These herbs are known for their anti-inflammatory, antioxidant, and wound-healing properties, which align well with the pathophysiological processes of DFU. TPP1 is a lysosomal enzyme involved in protein degradation, and its function in DFU may relate to extracellular matrix degradation and tissue repair, which are critical for wound healing. TLR4, a pattern recognition receptor, plays a key role in recognizing pathogens and initiating immune responses. Overactivation of the TLR4 signaling pathway has been implicated in chronic inflammation in DFU, which can delay wound healing and exacerbate tissue damage [40]. RIPK2 is a kinase involved in the NOD2 signaling pathway, which regulates immune responses and inflammation. Research suggests that RIPK2 plays a role in controlling cell apoptosis and inflammatory responses, both of which are critical in ulcer healing [41].
The experimental validation of these target genes further supports their involvement in DFU pathogenesis. We observed significant upregulation of TPP1, TLR4, and RIPK2 in both serum and tissue samples from DFU patients compared to controls, as demonstrated by qRT-PCR results. Immunohistochemistry analysis confirmed that these genes are highly expressed in the diseased skin tissues of DFU patients, compared to healthy skin. These findings strongly indicate that TPP1, TLR4, and RIPK2 may serve as valuable therapeutic targets for DFU treatment. The bioactive compounds found in the identified herbs likely interact with these target genes to mediate their therapeutic effects. For instance, Angelica dahurica has been shown to promote angiogenesis and reduce inflammation through the activation of key signaling pathways such as ERK1/2, Akt, and eNOS, which are important for wound healing [23]. Similarly, Angelica sinensis enhances blood circulation and exhibits anti-inflammatory properties, both of which are beneficial in the treatment of chronic wounds like DFU [42]. Boswellia carterii and liquorice are known for their anti-inflammatory and antimicrobial activities, which can mitigate excessive immune responses and infection in DFU [24, 43]. Myrrh and Semen armeniacae amarae also possess potent anti-inflammatory and wound-healing properties, which may further support their use in DFU treatment [44]. In summary, the combined use of network pharmacology and experimental validation highlights the potential of these TCM compounds to target key pathways involved in DFU. By modulating the expression of TPP1, TLR4, and RIPK2, these herbs may help to restore proper immune function, reduce chronic inflammation, and promote tissue repair. This study provides a strong foundation for the development of novel therapeutic strategies for DFU based on the use of traditional herbs and their bioactive compounds. However, further in vivo studies and clinical trials are needed to fully validate the therapeutic efficacy of these compounds in the treatment of DFU.
This study has several limitations. First, the small sample size may not fully reflect the heterogeneity of DFU, and larger cohorts are needed for validation. Second, while bioinformatics analyses were supported by qRT-PCR and immunohistochemistry, further in vitro and in vivo experiments are required to confirm the roles of identified genes in DFU. Third, the cross-sectional design limits our ability to track dynamic changes in cell populations during DFU progression. In addition, although the study identified potential therapeutic targets through a TCM pharmacology network, experimental validation of these molecular interactions is still needed. The patient cohort lacked diversity, limiting the generalizability of the findings. Finally, other important factors in the DFU microenvironment, such as angiogenesis and oxidative stress, were not explored in detail. Future studies should address these limitations to improve clinical relevance.
Conclusion
In summary, we explored the differences in cell types and further analyzed cell subpopulations by constructing single-cell profiles of DM and DFU tissues. We found that some immune cell subsets, such as Naive T_RGCC, CTL_TYROBP_CCL4, Mac_SLC40A1, and M1_CCL3L1, may be involved in the formation of DFU. These subsets tend to overactivate, proliferate, attack normal tissues, and exacerbate skin ulcers. In addition, TPP1, TLR4, and RIPK2 have been identified as target genes for traditional Chinese medicine drugs, providing a theoretical basis for further drug discovery to treat DFU.
Supplementary Information
Supplementary material 1: Supplementary Figure 1. Ecotopic gene mapping of the DM group.. Violin plot showing that the ATP5E gene is expressed only in the DM group.. The ATP5E gene was specifically highly expressed in Naive T cells, fibroblasts, endothelial cells and CD8+ T cells in the DM group.
Supplementary material 2: Supplementary Table 1. Patient clinical information
Supplementary material 3: Supplementary Table 2. Marker genes guiding cell annotation
Supplementary material 4: Supplementary Table 3. Cell composition ratio between control, DM, and DFU groups
Acknowledgements
Not applicable.
Abbreviations
- DM
Diabetes mellitus
- T1D
Type 1 diabetes
- T2D
Type 2 diabetes
- DFU
Diabetes foot ulcer
- scRNA-seq
Single-cell RNA sequencing
- UMAP
Unified Flowform Approximation and Projection
- DEGs
Differentially expressed genes
- GRN
Gene regulatory network
- TFs
Transcription factors
- SMCs
Smooth muscle cells
- Fibro
Fibroblasts
- Mac
Macrophages
- Ep
Epithelial cells
- Bas
Basal keratin-forming cells
- Diff
Differentiated keratin-forming cells
- Mel
Melanocytes
- Pla
Plasma cells
Author contributions
Conceptualization: Lehoanganh Vu, Fei Xu, Ting Li, Steven Mo, Dengfeng Yang, Jinmin Zhao. Writing-original draft: Lehoanganh Vu, Fei Xu, Ting Li, Qikai Hua, Xiaocong Kuang, Yongqiang Jiang, Ke-Er Lu, Yan Mo. Data analysis and visualization: Yanfei Liang, Xing Niu, Yixuan Chen, Chengyu Huang, Weiliang Mo, Kejian Wang, Kaihua Tang, Jianwen Mo. Writing-review and editing: Steven Mo, Dengfeng Yang, Jinmin Zhao. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by grants from the National Natural Science Foundation of China (82260448); Natural Science Foundation of Guangxi (2023JJD140126 );Guangxi Key Research and Development Plan (2021AB11027); Clinical research climbing plan of the First Affiliated Hospital of Guangxi Medical University (YYZS2020010).
Availability of data and materials
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
All samples were obtained with the consent of patients from the First Affiliated Hospital of Guangxi Medical University and approved by the Ethics Committee (No. 2022-KT-NSFC-260).
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Lehoanganh Vu, Fei Xu and Ting Li have contributed equally to this work and share first authorship.
Contributor Information
Steven Mo, Email: Steven_Mo@ydlife.org.
Dengfeng Yang, Email: dengfengyang@163.com.
Jinmin Zhao, Email: gxzj1962@163.com.
References
- 1.Sun Y, Tao Q, Wu X, Zhang L, Liu Q, Wang L. The utility of exosomes in diagnosis and therapy of diabetes mellitus and associated complications. Front Endocrinol. 2021;12: 756581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dhankhar S, Chauhan S, Mehta DK, Nitika SK, Saini M, Das R, Gupta S, Gautam V. Novel targets for potential therapeutic use in diabetes mellitus. Diabetol Metab Syndr. 2023;15:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kanter JE, Bornfeldt KE. Impact of diabetes mellitus. Arterioscler Thromb Vasc Biol. 2016;36:1049–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashcroft FM, Rorsman P. Diabetes mellitus and the beta cell: the last ten years. Cell. 2012;148:1160–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Su M, Hu R, Tang T, Tang W, Huang C. Review of the correlation between Chinese medicine and intestinal microbiota on the efficacy of diabetes mellitus. Front Endocrinol. 2022;13:1085092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Everett E, Mathioudakis N. Update on management of diabetic foot ulcers. Ann N Y Acad Sci. 2018;1411:153–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akkus G, Sert M. Diabetic foot ulcers: a devastating complication of diabetes mellitus continues non-stop in spite of new medical treatment modalities. World J Diabetes. 2022;13:1106–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kolodziejczyk AA, Kim JK, Svensson V, Marioni JC, Teichmann SA. The technology and biology of single-cell RNA sequencing. Mol Cell. 2015;58:610–20. [DOI] [PubMed] [Google Scholar]
- 9.Wang Z, Liu Y, Niu X. Application of artificial intelligence for improving early detection and prediction of therapeutic outcomes for gastric cancer in the era of precision oncology. Semin Cancer Biol. 2023;93:83–96. [DOI] [PubMed] [Google Scholar]
- 10.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, Satija R. Comprehensive integration of single-cell data. Cell. 2019;177(1888–1902): e1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Becht E, McInnes L, Healy J, Dutertre C-A, Kwok IWH, Ng LG, Ginhoux F, Newell EW. Dimensionality reduction for visualizing single-cell data using UMAP. Nature Biotechnol. 2018. 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
- 13.Wang Z, Yao J, Dong T, Niu X. Definition of a novel cuproptosis-relevant lncRNA signature for uncovering distinct survival, genomic alterations, and treatment implications in lung adenocarcinoma. J Immunol Res. 2022;2022:2756611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ren R, Du Y, Niu X, Zang R. ZFPM2-AS1 transcriptionally mediated by STAT1 regulates thyroid cancer cell growth, migration and invasion via miR-515-5p/TUSC3. J Cancer. 2021;12:3393–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, Trapnell C. Reversed graph embedding resolves complex single-cell trajectories. Nat Methods. 2017;14:979–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32:381–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levine M. Transcriptional enhancers in animal development and evolution. Curr Biol. 2010;20:R754-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peter IS, Davidson EH. Evolution of gene regulatory networks controlling body plan development. Cell. 2011;144:970–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aibar S, Gonzalez-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, Rambow F, Marine JC, Geurts P, Aerts J, et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods. 2017;14:1083–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van de Sande B, Flerin C, Davie K, De Waegeneer M, Hulselmans G, Aibar S, Seurinck R, Saelens W, Cannoodt R, Rouchon Q, et al. A scalable SCENIC workflow for single-cell gene regulatory network analysis. Nat Protoc. 2020;15:2247–76. [DOI] [PubMed] [Google Scholar]
- 21.Guan G, Niu X, Qiao X, Wang X, Liu J, Zhong M. Upregulation of neural cell adhesion molecule 1 (NCAM1) by hsa-miR-141-3p suppresses ameloblastoma cell migration. Med Sci Monit. 2020;26: e923491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Theocharidis G, Thomas BE, Sarkar D, Mumme HL, Pilcher WJR, Dwivedi B, Sandoval-Schaefer T, Sirbulescu RF, Kafanas A, Mezghani I, et al. Single cell transcriptomic landscape of diabetic foot ulcers. Nat Commun. 2022;13:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cell Press T. Science that inspires. Cell Host Microbe. 2020;27:155–6. [DOI] [PubMed] [Google Scholar]
- 24.Miro O, Miguel AR, Herrero P, Jacob J, Martin-Sanchez FJ, Gil V, Alquezar A, Escoda R, Llorens P, Group I-SR. OBESICA study: relationship between BMI and acute heart failure outcome. Eur J Emerg Med. 2017;24:326–32. [DOI] [PubMed] [Google Scholar]
- 25.Berbudi A, Rahmadika N, Tjahjadi AI, Ruslami R. Type 2 diabetes and its impact on the immune system. Curr Diabetes Rev. 2020;16:442–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duarte LRF, Pinho V, Rezende BM, Teixeira MM. Resolution of inflammation in acute graft-versus-host-disease: advances and perspectives. Biomolecules. 2022;12:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ogbodo JO, Arazu AV, Iguh TC, Onwodi NJ, Ezike TC. Volatile organic compounds: a proinflammatory activator in autoimmune diseases. Front Immunol. 2022;13: 928379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Y, Liu Y, He W, Mu X, Wu X, Deng J, Nie X. Fibroblasts: immunomodulatory factors in refractory diabetic wound healing. Front Immunol. 2022;13: 918223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dasgupta S, Ghosh T, Dhar J, Bhuniya A, Nandi P, Das A, Saha A, Das J, Guha I, Banerjee S, et al. RGS5-TGFbeta-Smad2/3 axis switches pro- to anti-apoptotic signaling in tumor-residing pericytes, assisting tumor growth. Cell Death Differ. 2021;28:3052–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ye S, Xu M, Zhu T, Chen J, Shi S, Jiang H, Zheng Q, Liao Q, Ding X, Xi Y. Cytoglobin promotes sensitivity to ferroptosis by regulating p53-YAP1 axis in colon cancer cells. J Cell Mol Med. 2021;25:3300–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu Z, Zuo Y, Wang J, Yu Z, Peng F, Chen Y, Dong Y, Hu X, Zhou Q, Ma H, et al. Overexpression of the regulator of G-protein signaling 5 reduces the survival rate and enhances the radiation response of human lung cancer cells. Oncol Rep. 2015;33:2899–907. [DOI] [PubMed] [Google Scholar]
- 32.Su K, Huang L, Li W, Yan X, Li X, Zhang Z, Jin F, Lei J, Ba G, Liu B, et al. TC-1 (c8orf4) enhances aggressive biologic behavior in lung cancer through the Wnt/beta-catenin pathway. J Surg Res. 2013;185:255–63. [DOI] [PubMed] [Google Scholar]
- 33.Gall C, Xu H, Brickenden A, Ai X, Choy WY. The intrinsically disordered TC-1 interacts with Chibby via regions with high helical propensity. Protein Sci. 2007;16:2510–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu B, Zhou H, Zhang T, Gao X, Tao B, Xing H, Zhuang Z, Dardik A, Kyriakides TR, Goodwin JE. Loss of endothelial glucocorticoid receptor promotes angiogenesis via upregulation of Wnt/beta-catenin pathway. Angiogenesis. 2021;24:631–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vlaicu SI, Tatomir A, Rus V, Rus H. Role of C5b–9 and RGC-32 in cancer. Front Immunol. 2019;10:1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cui XB, Chen SY. Response gene to complement 32 in vascular diseases. Front Cardiovasc Med. 2018;5:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang L, Zhang M, Wang L, Li J, Yang T, Shao Q, Liang X, Ma M, Zhang N, Jing M, et al. Identification of CCL4 as an immune-related prognostic biomarker associated with tumor proliferation and the tumor microenvironment in clear cell renal cell carcinoma. Front Oncol. 2021;11: 694664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu C, Song Z, Chen Z, Lin T, Lin H, Xu Z, Ai F, Zheng S. MicroRNA-4735-3p facilitates ferroptosis in clear cell renal cell carcinoma by targeting SLC40A1. Anal Cell Pathol (Amst). 2022;2022:4213401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Urashima K, Miramontes A, Garcia LA, Coletta DK. Potential evidence for epigenetic biomarkers of metabolic syndrome in human whole blood in Latinos. PLoS ONE. 2021;16: e0259449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh K, Singh VK, Agrawal NK, Gupta SK, Singh K. Association of Toll-like receptor 4 polymorphisms with diabetic foot ulcers and application of artificial neural network in DFU risk assessment in type 2 diabetes patients. Biomed Res Int. 2013;2013. [DOI] [PMC free article] [PubMed]
- 41.Tian E, Zhou C, Quan S, Su C, Zhang G, Yu Q, Li J, Zhang J. RIPK2 inhibitors for disease therapy: Current status and perspectives. Eur J Med Chem. 2023;259:115683. [DOI] [PubMed]
- 42.Nai J, Zhang C, Shao H, Li B, Li H, Gao L, Dai M, Zhu L, Sheng H. Extraction, structure, pharmacological activities and drug carrier applications of Angelica sinensis polysaccharide. Int J Biol Macromol. 2021Jul;31(183):2337–53. [DOI] [PubMed]
- 43.Yu G, Xiang W, Zhang T, Zeng L, Yang K, Li J. Effectiveness of boswellia and boswellia extract for osteoarthritis patients: a systematic review and meta-analysis. BMC Complement Med Ther. 2020;20(1):225. [DOI] [PMC free article] [PubMed]
- 44.Weber L, Kuck K, Jürgenliemk G, Heilmann J, Lipowicz B, Vissiennon C. Anti-inflammatory and barrier-stabilising effects of myrrh, coffee charcoal and chamomile flower extract in a co-culture cell model of the intestinal mucosa. Biomolecules. 2020;10(7):1033. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1: Supplementary Figure 1. Ecotopic gene mapping of the DM group.. Violin plot showing that the ATP5E gene is expressed only in the DM group.. The ATP5E gene was specifically highly expressed in Naive T cells, fibroblasts, endothelial cells and CD8+ T cells in the DM group.
Supplementary material 2: Supplementary Table 1. Patient clinical information
Supplementary material 3: Supplementary Table 2. Marker genes guiding cell annotation
Supplementary material 4: Supplementary Table 3. Cell composition ratio between control, DM, and DFU groups
Data Availability Statement
No datasets were generated or analysed during the current study.