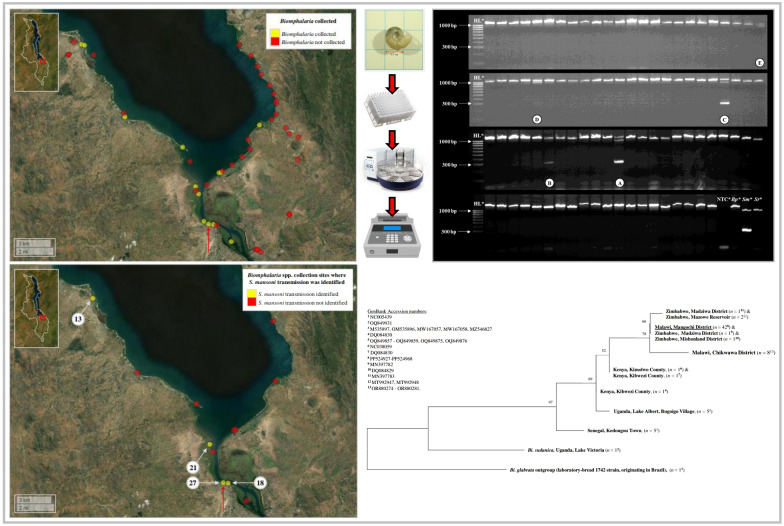
Abstract
Background
Intestinal schistosomiasis was confirmed endemic in Mangochi District, Malawi, in May of 2018 following an unexpected encounter with discreet populations of Biomphalaria spp. freshwater snails during routine malacological surveillance activities. Since then, only limited malacological surveillance of Biomphalaria has been carried out, and so the distribution of Biomphalaria populations in this area is currently unclear. Additionally, sites of active Schistosoma mansoni transmission in this area are also unknown. In the present study, through extensive malacological surveillance, we aimed to formally document the distribution of Biomphalaria in Mangochi District. We also aimed to identify active intestinal schistosomiasis transmission sites in this area through subjecting all collected Biomphalaria to a recently developed S. mansoni-specific molecular xenomonitoring PCR.
Methods
Three malacological surveys were carried out along the southern shoreline of Lake Malawi, Mangochi District, Malawi, in November 2021, July 2022 and October/November 2022. All collected Biomphalaria were subjected to cercarial shedding analysis to identify active Schistosoma infections. Shed cercariae were then genotyped to species level using a standard multi-locus PCR and Sanger sequencing protocol. Following this, a subset of Biomphalaria from each collection site were also genotyped to species level using a standard PCR and Sanger sequencing protocol. All collected Biomphalaria were then subjected to a recently developed S. mansoni-specific molecular xenomonitoring PCR to identify infected, but non-shedding, Biomphalaria.
Results
A total of 589 Biomphalaria were collected across all three surveys. One single Biomphalaria (0.17%) specimen was found to be actively shedding Schistosoma cercariae, which were molecularly confirmed as S. mansoni. All genotyped Biomphalaria (n = 42) were molecularly identified as B. pfeifferi. A further 19 Biomphalaria specimens, collected from four different surveillance sites, were found to be infected with S. mansoni through molecular xenomonitoring. Intestinal schistosomiasis transmission was therefore identified at four different foci in Mangochi District.
Conclusions
Our study highlights the importance of molecular approaches to investigate Biomphalaria populations and monitor Biomphalaria-associated intestinal schistosomiasis transmission in endemic areas. As such, the continued development and use of such approaches, in particular the development and use of molecular xenomonitoring assays that can be carried out in resource-poor schistosomiasis-endemic settings, is encouraged. The revision of ongoing schistosomiasis control programmes in Mangochi District, in line with WHO recommendations, is also encouraged.
Graphical abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s13071-024-06546-5.
Keywords: Biomphalaria pfeifferi, Schistosoma mansoni, Intestinal schistosomiasis, Malawi, Phylogenetics, Molecular xenomonitoring, Water sanitation and hygiene (WASH), Waterborne pathogens, One Health
Background
Schistosomiasis, caused by parasitic trematodes of the genus Schistosoma, is a neglected tropical disease (NTD) infecting over 230 million people worldwide [1]. Infection with these parasites can lead to debilitating morbidity and mortality. More than 90% of schistosomiasis cases occur within sub-Saharan Africa [2], and approximately one-third of these cases are deemed intestinal schistosomiasis, caused predominantly by infection with Schistosoma mansoni but also less commonly by infection with Schistosoma intercalatum and Schistosoma guineensis in some restricted areas of central Africa [3, 4]. Human infection occurs through contact with bodies of freshwater contaminated with S. mansoni cercariae that have been shed from obligate freshwater snail intermediate hosts of the genus Biomphalaria (Gastropoda: Planorbidae) [5]. Intestinal schistosomiasis is therefore often highly prevalent in rural areas lacking adequate water, sanitation and hygiene (WASH) infrastructure [6].
As each Biomphalaria species can differ in its ability to transmit S. mansoni [7, 8], malacological surveillance and accurate species identification of Biomphalaria in intestinal schistosomiasis-endemic areas is not only of taxonomic interest but can also contribute to a better understanding of disease transmission dynamics and risk of human infection. However, differentiating between Biomphalaria species based on morphological features, such as shell conchology, is difficult and requires specially trained and skilled malacologists [9]. To overcome these challenges, molecular methods such as PCR are routinely used to reliably identify and differentiate Biomphalaria species, typically though genotyping of mitochondrial cytochrome oxidase subunit 1 (cox1) DNA, mitochondrial 16S DNA and/or the nuclear internal transcribed spacer (ITS) DNA region [10].
Malacological surveillance together with the collection of Biomphalaria can also allow sites of active intestinal schistosomiasis transmission to be identified through cercarial shedding analysis [8, 11, 12]. However, this approach can be misleading owing to morphologically indistinguishable human-infecting and non-human-infecting trematode cercariae, such as S. mansoni and Schistosoma rodhaini, both of which are transmitted by Biomphalaria [13, 14]. As such, molecular methods are also needed to reliably identify the species of Schistosoma cercariae shed from Biomphalaria hosts. This is typically done through genotyping both mitochondrial (maternally inherited) and nuclear (inherited equally from both parents) DNA loci, as this multi-locus approach allows any potential Schistosoma spp. hybrids with recent multi-species ancestry to be identified [15, 16]. In addition, cercarial shedding can also be insensitive, primarily because not all freshwater snails harbouring mature, or patent, Schistosoma infections will actively shed cercariae at collection and because freshwater snails harbouring prepatent infections will also not shed cercariae [8, 11, 12]. For these reasons, the World Health Organization (WHO) recommends molecular xenomonitoring of the freshwater snail intermediate hosts of Schistosoma, particularly in low-endemicity areas, as this approach can be used to detect Biomphalaria infections with S. mansoni missed by cercarial shedding [13, 17, 18].
Lake Malawi is one of seven African Great Lakes and has an approximate water surface area of 29,600 km2. The lake’s most southern shoreline boarders Mangochi District, Malawi. Here, because WASH infrastructure is inadequate, many people rely on the lake as a source of drinking water and food (via fishing) and as a place to bathe, to tend livestock and for recreation. As such, human water-contact with the lake’s shoreline is commonplace, meaning waterborne diseases in this area, such as schistosomiasis and giardiasis, are highly prevalent [19–21]. Until recently, intestinal schistosomiasis was not considered to be locally endemic in Mangochi District as no species of Biomphalaria was known to inhabit the southernmost shoreline of Lake Malawi. However, during routine malacological surveillance in November of 2017, discreet populations of B. pfeifferi snails were unexpectedly encountered in submerged beds of Vallisneria freshwater vegetation [22]. Subsequently, in May of 2018, an outbreak of intestinal schistosomiasis was confirmed despite annual and ongoing mass drug administration (MDA) campaigns in this area used to reduce transmission of urogenital schistosomiasis [20].
One year later (in 2018), malacological surveillance and cercarial shedding of Biomphalaria was carried out in an attempt to identify intestinal schistosomiasis transmission sites in this area [22]. Despite this effort, only one of 201 collected Biomphalaria snails was found to be actively shedding S. mansoni cercariae. As such, a subset of collected Biomphalaria (n = 76) snails were screened for S. mansoni infection using a real-time molecular xenomonitoring PCR targeting the genus-specific Schistosoma ITS2 region. Whilst Schistosoma DNA was detected in a high proportion of the screened Biomphalaria (31%), it is unclear whether this was derived from invading S. mansoni or Schistosoma rodhaini trematodes, or even potentially from S. haematobium-group trematodes that had penetrated the soft tissues of Biomphalaria snails but failed to establish an infection. It is also possible that other species of non-Schistosoma trematodes may cross-react with this ITS2-targeting primer/probe combination [23]. Consequently, more extensive molecular xenomonitoring of Biomphalaria collected in this area, using a S. mansoni-specific assay, is needed to fully assess the prevalence of Biomphalaria infections with S. mansoni and identify intestinal schistosomiasis transmission sites.
In the present study, through extensive malacological surveillance, we aimed to formally document the distribution of Biomphalaria along the southern shoreline of Lake Malawi. We also aimed to identify active intestinal schistosomiasis transmission foci in this area through subjecting all collected Biomphalaria to a recently developed S. mansoni-specific molecular xenomonitoring PCR [24]. The purpose of this study was therefore to gain a more thorough understanding of Biomphalaria-associated transmission of intestinal schistosomiasis in Mangochi District 5 years post-outbreak to better inform and strengthen future disease control efforts.
Methods
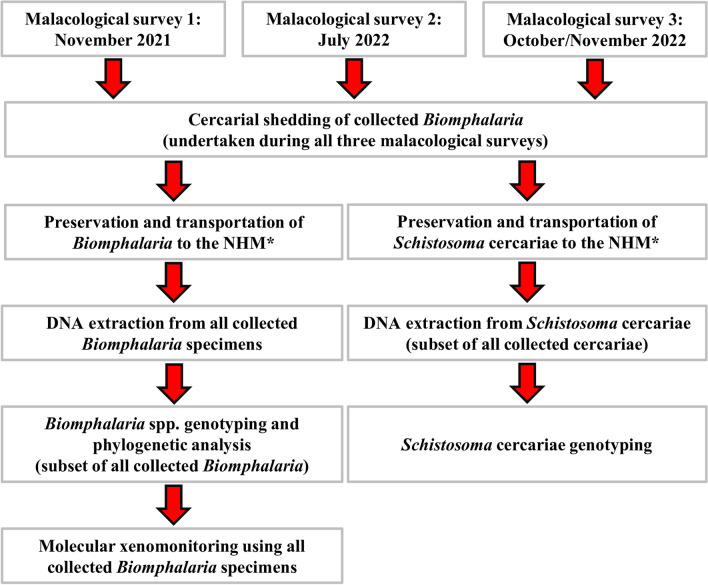
A schematic flow-diagram outlining the study design is shown in Fig. 1.
Fig. 1.
Schematic flow-diagram outlining study design. NHM, Natural History Museum, London, UK
Malacological surveys and sample sites
Three malacological surveys were carried out along, and near to, the southern shoreline of Lake Malawi, Mangochi District, Malawi on November 2021 (survey one), July 2022 (survey two) and October/November 2022 (survey three). All surveys were carried out in conjunction with human parasitological surveillance activities, which themselves were timed relative to ongoing MDA with praziquantel in the area. Both purposeful and opportunistic sampling took place; some sampling sites were selected as they are known human-water contact sites and/or Biomphalaria had been found at these sites previously, and some sampling sites were selected during surveillance activities as human-water contact was observed and/or because environmental conditions appeared favourable for the presence of Biomphalaria. In total, 41 individual sites were surveyed at least once during all three surveys (see Additional file 1: Table S1).
Collection of Biomphalaria freshwater snails
Freshwater snails were collected by scooping using handheld metal sieve scoops according to a standard protocol [18]. Collections were carried out by three trained and experienced technicians for 15 min per sampling site. Brief descriptions of all sampling sites (e.g. water depth, degree of vegetation, whether sites were temporal pools located away from the lake’s edge or were at the lake’s edge) were recorded. The same sampling protocol was applied regardless of the environmental conditions at the sample site. Biomphalaria were initially identified to genus level based on shell morphology according to previous morphological descriptions [25] and collected into small screwcap pots containing lake water and lake vegetation for short-term (< 3 h) transportation. Shortly after collection, Biomphalaria were maintained in aquaria, also containing lake water and lake vegetation, according to collection site.
Cercarial shedding of Biomphalaria and collection of Schistosoma cercariae
Cercarial shedding was carried out according to a standard protocol [26]. In brief, the day following collection, Biomphalaria were transferred from aquaria to small transparent plastic ‘shedding’ pots according to collection site. Each shedding pot contained approximately 100 ml of clean, bottled water and no more than 30 Biomphalaria specimens. Shedding pots were then placed in a shaded outdoor area (and so not in direct sunlight) between 9:00 a.m. and 4:00 p.m. to coincide with S. mansoni cercarial shedding patterns, as described previously [27]. Following light exposure, the water was decanted into a clean petri dish and examined under a dissecting microscope to check for motile, furcocercous Schistosoma cercariae according to previous morphological descriptions [28]. Any groups of Biomphalaria found to be shedding Schistosoma cercariae were recorded, and all Biomphalaria were then returned to their original aquaria.
The day following initial cercarial shedding, any groups of Biomphalaria snails found to be shedding Schistosoma cercariae were then individually separated into wells of a 12-well ‘shedding’ plate containing approximately 5 ml of clean, bottled water. As described above, shedding plates were then also placed in the same shaded outdoor area between 9:00 a.m. and 4:00 p.m. and then examined under a dissecting microscope for the presence of Schistosoma cercariae. Any individual Biomphalaria found to be shedding Schistosoma cercariae was recorded and its corresponding cercariae were individually captured using a micropipette (2.5 µl) and deposited onto Whatman FTA cards for DNA preservation (Whatman, GE Healthcare, Little Marlow, UK). A description of which Biomphalaria snail had shed which preserved cercariae was then written onto the FTA cards, and the FTA cards were then stored at ambient temperature according to manufacturer’s instructions.
A record of all shedding and non-shedding Biomphalaria was made, and any shedding Biomphalaria were individually preserved in 100% ethanol within a labelled 2 ml screwcap tube whereas any non-shedding Biomphalaria were pooled according to collection site and preserved in 100% ethanol in labelled 20 ml glass screwcap tubes. All FTA cards and ethanol-preserved Biomphalaria were transported to the Natural History Museum (NHM) under ambient conditions for molecular analyses.
Molecular characterisation of Biomphalaria
DNA extraction: ethanol-preserved Biomphalaria
DNA was individually isolated from whole snail tissues of all collected Biomphalaria using the BioSprint 96 workstation and BioSprint 96 DNA Blood Kit (QIAGEN Ltd., Manchester, UK) according to a previously outlined protocol that includes using double volume of lysis buffers and an overnight incubation lysis step [24, 29]. This magnetic bead-based DNA extraction protocol was chosen as it allows for rapid and high-throughput multiple-sample processing using a 96-well plate format.
Biomphalaria mitochondrial cox1 genotyping
Three randomly selected Biomphalaria collected from each malacological survey site across all surveys (n = 42 Biomphalaria) were genotyped, using PCR to amplify a 700-bp region of the Biomphalaria mitochondrial cox1 gene according to a standard protocol [9]. Details of all primer sequences, reaction mixes used and PCR conditions are described in Additional file 2: Tables S1A, S1B and S1C, respectively. All Biomphalaria cox1 PCRs included a positive control using B. pfeifferi DNA provided by the Schistosome and Snail Resource (SSR; an open access biomedical resource) [30] and a negative control using ddH2O in place of template DNA. Amplicons were visualised by running 4 µl of PCR products mixed with 1.5 µl of 5× loading buffer blue (Bioline Ltd., London, UK), stained with GelRed in a 1% agarose gel. The PCR products were then purified using the QIAquick PCR Purification Kit (QIAGEN Ltd.) according to the manufacturer’s instructions and Sanger sequenced in the forward direction using a dilution of the LCO_1490_FW forward primer. Sequence data were visualised, trimmed and edited as needed using Geneious Prime version 2023.01 (Biomatters Ltd., Aukland, New Zealand) before being identified using the Basic Local Alignment Search Tool (BLAST) algorithm within the National Center for Biotechnology Information (NCBI) database [31].
Phylogenetic and diversity analysis
Templeton Crandall and Sing haplotype network
To assess B. pfeifferi cox1 diversity, we performed a cox1 haplotype analysis using all generated B. pfeifferi cox1 sequence data as well as cox1 sequence data generated using eight B. pfeifferi recently collected in Chikwawa District, Malawi (approx. 250 km south of Mangochi District along the southern-flowing Shire River) (Nkolokosa et al., under review), 13 East African B. pfeifferi cox1 sequences and five West African B. pfeifferi cox1 sequences (Additional file 3: Table S1). All cox1 data were aligned using the multiple alignment with he fast Fourier transform (MAFFT) algorithm within Geneious Prime (default MAFFT parameter settings). The MAFFT alignment was then visualised, trimmed (to ensure uniform ends across all sequences), examined and edited as needed before being exported from Geneious Prime in Nexus file format and imported into PopART version 1.7 [32]. Within PopART, a Templeton Crandall and Sing (TCS) haplotype network [33] was generated to allow examination of haplotype group structuring.
Maximum likelihood phylogenetic tree
To further assess B. pfeifferi cox1 diversity and lineages, we then aligned the MAFFT alignment to a Biomphalaria glabrata cox1 outgroup sequence (laboratory-bred 1742 strain, originating in Brazil [34]), as well as to a Biophalaria sudanica cox1 sequence (originating in Uganda, Lake Albert), both downloaded from the GenBank repository (Accession numbers: NC005439 and OQ849931, respectively). The alignment was visualised, trimmed, examined and edited as described above before being exported from Geneious Prime in Nexus file format and imported into MEGA11 [35]. Within MEGA11, a bootstrapped maximum likelihood (ML) phylogenetic tree was constructed using a Hasegawa-Kishino-Yano (HKY85) evolutionary model with 10,000 iterations after having calculated the most appropriate evolutionary model for phylogenetic analysis of these data.
Molecular characterisation of Schistosoma cercariae
Individual Schistosoma cercariae were characterised using a multi-locus approach targeting both the mitochondrial (maternally inherited) cox1 and nuclear (inherited equally from both parents) ITS ribosomal RNA (rRNA) regions [36].
DNA extraction: FTA preserved cercariae
DNA was isolated from up to 20 individual Whatman FTA card-preserved Schistosoma cercariae, per shedding Biomphalaria snail, according to a standard protocol [37].
Schistosoma mitochondrial cox1 and nuclear ITS genotyping
A 956-bp region of the Schistosoma mitochondrial cox1 gene was amplified by PCR using a standard protocol [38]. Details of all primer sequences, reaction mixes used and PCR conditions are described in Additional file 2: Tables S2A, S2B and S2C, respectively. Cox1 PCRs included a positive control using S. mansoni genomic DNA (gDNA) collectively isolated from three S. mansoni adult worms provided by the SSR and a negative control using ddH2O in place of template DNA. Amplicons were visualised by running 4 µl of PCR products mixed with 1.5 µl of 5× loading buffer blue (Bioline Ltd.) stained with GelRed in a 1% agarose gel. PCR products were purified as described above and Sanger sequenced in the reverse direction using a dilution of the Schisto_3′ reverse primer. Sequence data were visualised, trimmed and edited as needed using Geneious Prime version 2023.01 (Biomatters Ltd.) before being identified using the BLAST algorithm within the NCBI database [31].
PCR then used to amplify the complete Schistosoma nuclear ITS rRNA region (approx. 1005 bp) using a standard protocol [38]. Details of all primer sequences, reaction mixes used, and PCR conditions are described in Additional file 2: Tables S3A, S3B and S3C, respectively. The ITS PCRs included one positive and one negative control as with cox1 PCRs. Amplicons were visualised as described above. PCR products were purified as described above and Sanger sequenced in the reverse direction using a dilution of the ETTS1 reverse primer. Sequence data were visualised, trimmed, edited and identified as with cox1 PCRs.
Biomphalaria molecular xenomonitoring
A recently developed high-throughput S. mansoni-specific molecular xenomonitoring PCR assay was used to detect either patent, but non-shedding, S. mansoni infections, or prepatent S. mansoni infections within Biomphalaria [24]. This assay can also be used to detect Biomphalaria infections with other trematode species, including S. rodhaini.
Molecular xenomonitoring PCRs
Molecular xenomonitoring was performed using DNA isolated from all Biomphalaria snails collected across all surveys. Details of all primer sequences, reaction mixes used and PCR conditions are described in Additional file 2: Tables S4A, S4B and S4C, respectively. Molecular xenomonitoring PCRs were carried out in batches of 92 samples in a 96-well PCR plate format. The PCR assays included one positive control using DNA) isolated from a non-infected B. pfeifferi provided by the SSR, one positive control using S. mansoni gDNA collectively isolated from three S. mansoni adult worms provided by the SSR, one positive control using S. rodhaini gDNA collectively isolated from three S. rodhaini adult worms provided by the SSR and one negative control using ddH2O in place of template DNA. PCR amplicons were visualised by running 7.5 µl PCR product mixed with 2 µl of 5× loading buffer blue (Bioline Ltd.) stained with GelRed in a 2% agarose gel.
Samples that amplified only the Biomphalaria ITS locus (considered an internal DNA extraction and PCR reaction control) were considered to be negative for Trematoda infection of any sort, including S. mansoni. Samples that successfully amplified all three target loci were considered positive for S. mansoni infection, as well as potentially for infection of other species of Trematoda. Samples that amplified only the Biomphalaria and Trematoda ITS loci were considered to be positive for non-S. mansoni Trematoda infection. Samples that failed to amplify the Biomphalaria ITS locus (regardless of Trematoda ITS and S. mansoni NADH dehydrogenase subunit 5 (ND5) amplification outcome), were considered to represent a failed PCR assay and the assay was repeated. Samples that failed to amplify this locus during the repeat screen were considered to represent a failed DNA extraction and were omitted from any further analysis.
Confirmatory S. mansoni ND5 genotyping
All samples that successfully amplified all three target loci were subjected to a secondary singleplex PCR to amplify only the S. mansoni ND5 locus for Sanger sequencing [24]. Amplicons were visualised in the same manner as for the multiplex molecular xenomonitoring PCRs. Schistosoma mansoni ND5 PCR products were purified as described above and Sanger sequenced in the forward direction using a dilution of the ND52 forward primer. Sequence data were visualised, trimmed, edited and identified as described above.
Templeton Crandall and Sing haplotype network
To assess S. mansoni ND5 diversity, we performed a haplotype analysis using all generated S. mansoni ND5 sequence data. To do this, a MAFFT alignment was performed using sequence data within Geneious Prime (default MAFFT parameter settings). The MAFFT alignment was then visualised, examined and edited as described above. The alignment was then exported from Geneious Prime in Nexus file format and imported into PopART version 1.7. Within PopART, a TCS haplotype network was generated to allow examination of haplotype group structuring.
Conformation of Biomphalaria infections with other Trematoda species
To identify Trematoda species other than S. mansoni infecting Biomphalaria, we subjected all samples that successfully amplified only Biomphalaria and Trematoda ITS loci to a secondary singleplex PCR, again to amplify only the Biomphalaria ITS and Trematoda ITS loci. To do this, the molecular xenomonitoring PCR was repeated, but with replacement of the S. mansoni ND5 forward and reverse primers with ddH2O. Amplicons were visualised in the same manner as for the multiplex molecular xenomonitoring PCRs and the approximately 1005-bp Trematoda ITS gel band was excised using a fresh scalpel. Excised gel bands were purified using the QIAquick Gel purification kit (Qiagen Ltd.) according to manufacturer’s instructions and were then purified as described above prior to Sanger sequencing in the forward direction using a dilution of the ETTS2 forward primer. Sequence data were visualised, trimmed and edited as needed using Geneious Prime version 2023.01 (Biomatters, Ltd.) before being identified using the BLAST algorithm within the NCBI database [31].
Results
Malacological surveillance and cercarial shedding of Biomphalaria
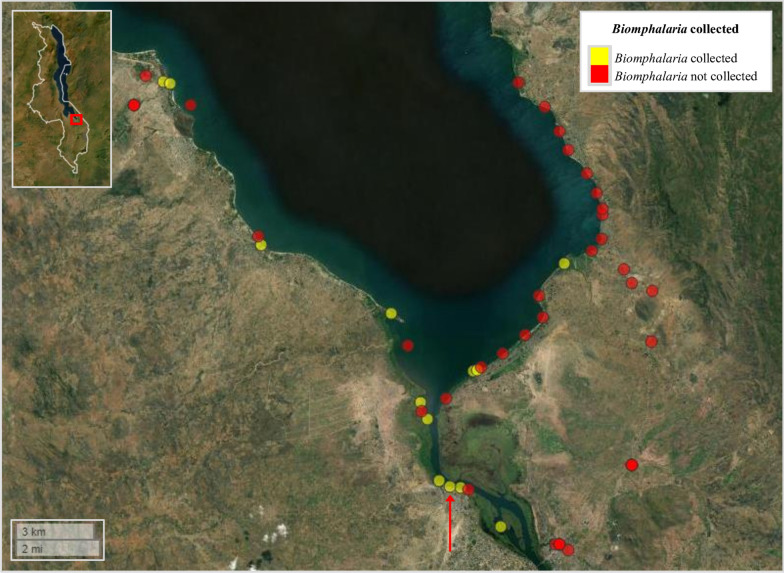
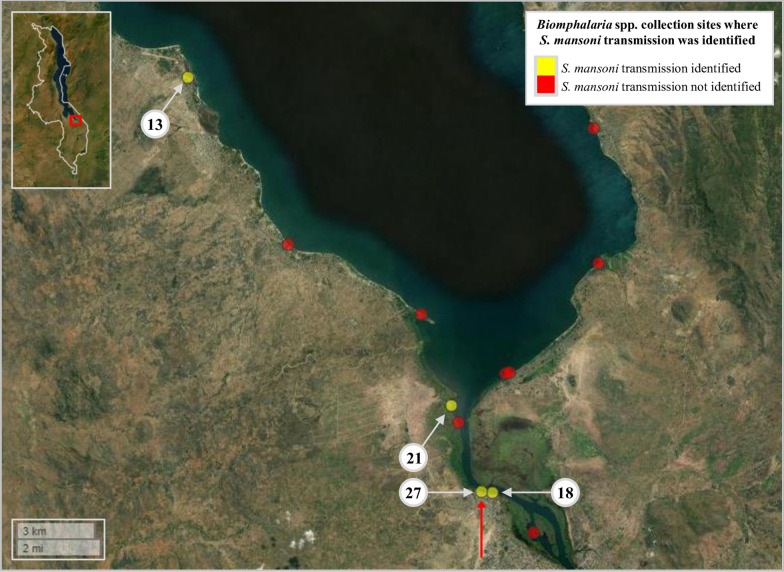
A total of 589 Biomphalaria were collected across all three surveys (survey 1: n = 285; survey 2: n = 107; survey 3: n = 197). Among all 41 sites surveyed, Biomphalaria were collected from 13 individual sites (31.7%) during at least one survey (Fig. 2; Additional file 1: Table S1), with all sites being either shallow temporal pools close to the lake’s edge or shallow waters along the shores of the lake. More sites were surveyed along the lake’s eastern shoreline owing to greater obstruction by dense vegetation on the lake’s western shoreline. The cercarial shedding analyses revealed that only one single Biomphalaria (0.17%), at survey site 27, was actively shedding Schistosoma cercariae (during survey 3) (Fig. 2; Additional file 1: Table S2). A total of 81 Schistosoma cercariae shed from this one snail were captured and preserved on a Whatman FTA card for subsequent molecular analysis.
Fig. 2.
Overview of the 41 sites surveyed during malacological surveys 1, 2 and 3, Lake Malawi, Mangochi District, Malawi. Yellow circles denote sites where Biomphalaria were found during at least one of all three surveys. Red circles denote sites where Biomphalaria snails were not found during any of the three surveys (Additional file 1: Table S1). Only one Biomphalaria was found to be actively shedding Schistosoma cercariae (later identified as S. mansoni) during cercarial shedding analyses (during survey 3) at survey site 27; denoted by red arrow. Malawi’s country border can be seen within the figure inset (upper left corner). Inset: study area is highlighted by red box. Figure was generated using the ‘mapview’ package version 2.10.0 [39] within R Studio version 2021.09.0, build 351 (Posit, Boston, MA, USA) [40]
Molecular characterisation of Biomphalaria
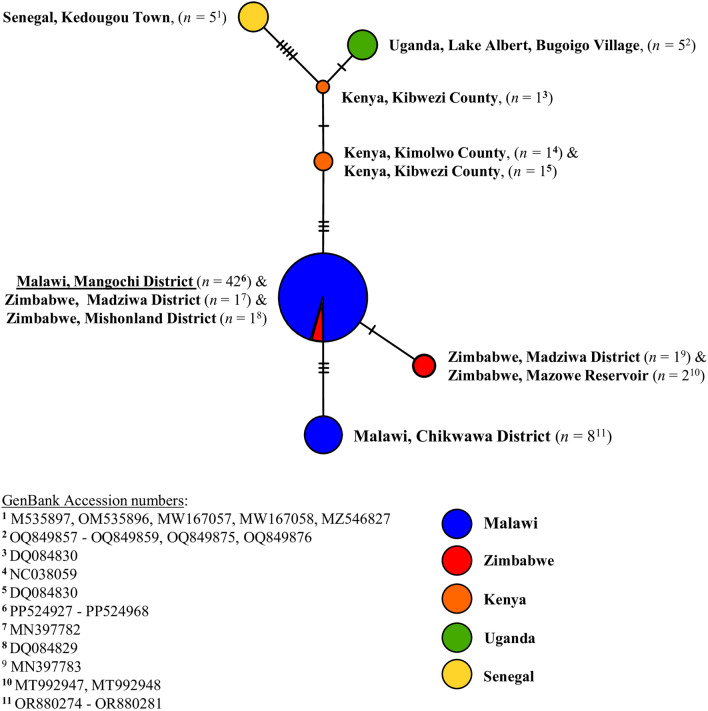
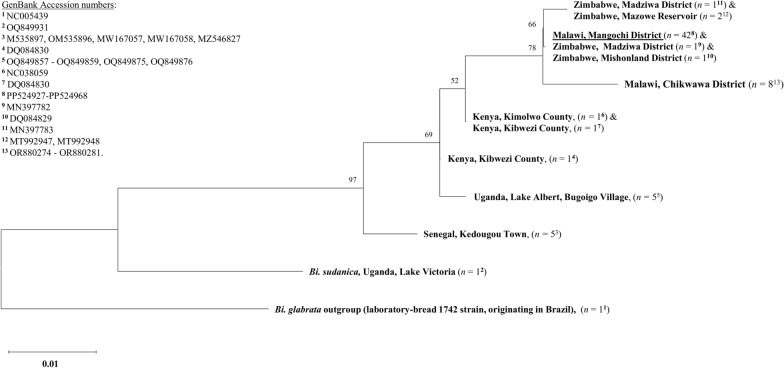
DNA was successfully extracted from all collected Biomphalaria specimens. Of these, 42 were identified as B. pfeifferi though cox1 analysis. All 42 B. pfeifferi cox1 forward sequences were uploaded to the GenBank repository, (Accession numbers: PP524927-PP524968). The B. pfeifferi cox1 TCS haplotype network is shown in Fig. 3 and the B. pfeifferi cox1 ML phylogenetic tree is shown in Fig. 4.
Fig. 3.
Biomphalaria pfeifferi cox1 TCS haplotype network. Each node (circle) represents a unique haplotype, with the size of the node proportional to the frequency of each haplotype. Each differently coloured node represents haplotype(s) from a different country, with blue denoting those haplotypes identified in Malawi; red, identified in Zimbabwe; orange, identified in Kenya; green, identified in Uganda; and yellow, identified in Senegal. The Mangochi District haplotype (current study) is underlined. Hatched lines denote the number of single nucleotide polymorphisms between nodes. cox1, Cytochrome oxidase subunit 1 gene; TCS, Templeton Crandall and Sing
Fig. 4.
Biomphalaria pfeifferi cox1 maximum likelihood phylogenetic tree. Identical sequences were collapsed. Bootstrap values can be seen at branch nodes. The scale bar for branch length indicates the mean number of nucleotide substitutions per base site. A monophyletic clade was formed that included the Mangochi District B. pfeifferi cox1 haplotype (current study), all five Zimbabwe B. pfeifferi cox1 haplotypes and the Malawi, Chikwawa District B. pfeifferi cox1 haplotype. cox1, Cytochrome oxidase subunit 1 gene
No diversity was found between any Mangochi District B. pfeifferi cox1 sequence data (current study) with all samples representing a single haplotype. This haplotype was also identified in two B. pfeifferi from Zimbabwe (Madziwa District and Mishonland District) and was most closely related to a single B. pfeifferi cox1 haplotype found in three B. pfeifferi again from Zimbabwe (Madziwa District and Mazowe Reservoir). In addition, no diversity was found between any Malawi, Chikwawa District B. pfeifferi cox1 sequence data, and this haplotype was not found in any other B. pfeifferi analysed. Three single nucleotide polymorphisms (SNPs) were found to be common between the Mangochi District B. pfeifferi cox1 haplotype (current study) and the Malawi, Chikwawa District B. pfeifferi cox1 haplotype. Overall, very little cox1 diversity was observed across all B. pfeifferi cox1 data. However, different B. pfeifferi cox1 haplotypes were found from each analysed country (Senegal, Uganda, Kenya, Zimbabwe, and Malawi). Clustering was observed between haplotypes found in Zimbabwe and haplotypes found in Malawi, whereas the greatest degree of divergence was observed between haplotypes found in Senegal (West Africa) and haplotypes found in Malawi. This structuring was also observed within the ML phylogenetic tree, which formed a monophyletic clade that included the Mangochi District B. pfeifferi cox1 haplotype, all five Zimbabwe B. pfeifferi cox1 haplotypes and the Malawi, Chikwawa District B. pfeifferi cox1 haplotype.
Molecular characterisation of Schistosoma cercariae
Five Schistosoma cercariae shed from the single shedding Biomphalaria were successfully genotyped to species level. All five were identified as S. mansoni across both cox1 and ITS loci. No genetic variation was observed between cox1 sequence data or between ITS sequence data. All five S. mansoni cox1 sequences and all five S. mansoni ITS sequences were uploaded to the GenBank repository (Accession numbers: PP529587-PP529591 and PP510205-PP510209, respectively).
Biomphalaria molecular xenomonitoring
All 589 Biomphalaria snails were screened for S. mansoni and other Trematoda species infections using molecular xenomonitoring. A total of 14 (2.4%) samples failed to amplify the Biomphalaria internal control ITS locus, and so the amplifications were repeated using the same protocol, with all 14 successfully amplifying the Biomphalaria ITS locus during the repeat molecular xenomonitoring PCR. All three target loci were amplified when using DNA extracted from the one B. pfeifferi snail shedding S. mansoni cercariae, as well when using DNA isolated from 19 additional non-shedding Biomphalaria.
The S. mansoni ND5 locus was successfully amplified during the secondary S. mansoni ND5 singleplex PCR in all 20 Biomphalaria samples that had amplified all three target loci during the initial molecular xenomonitoring PCR. All 20 ND5 amplicons were confirmed as S. mansoni through ND5 analysis, confirming infection with S. mansoni. The prevalence of S. mansoni infection in these 589 Biomphalaria was therefore increased from 0.17% based on cercarial shedding to 3.4% based on molecular xenomonitoring. Biomphalaria snails infected with S. mansoni were identified at four of the 41 (9.6%) malacological survey sites (Fig. 5). At site 13, three of 175 collected Biomphalaria (1.7%) were infected with S. mansoni; at site 18, two of 66 collected Biomphalaria (3%) were infected with S. mansoni; and at site 27, five of 151 collected Biomphalaria (4%) were infected with S. mansoni. At site 21, however, nine of 80 Biomphalaria (11.25%) were infected with S. mansoni, the highest prevalence of Biomphalaria infections across all malacological surveillance sites. The number of malacological surveillance sites where intestinal schistosomiasis transmission was identified was therefore increased from just one using cercarial shedding to four using molecular xenomonitoring.
Fig. 5.
Malacological surveillance sites where Biomphalaria were collected and where Schistosoma mansoni transmission was or was not identified using molecular xenomonitoring. Site IDs are outlined in white circles (see Additional file 1: Tables S1, S2). The cercarial shedding analyses revealed that only one Biomphalaria (during survey 3) was actively shedding Schistosoma cercariae, which was later identified as S. mansoni, at survey site 27 (denoted with the red arrow). Malawi’s country border can be seen within figure inset (upper left corner). Within the inset, the study area is highlighted by a red box. Figure was generated using the ‘mapview’ package version 2.10.0 [39] within R Studio version 2021.09.0, build 351 (Posit, Boston, MA, USA) [40]
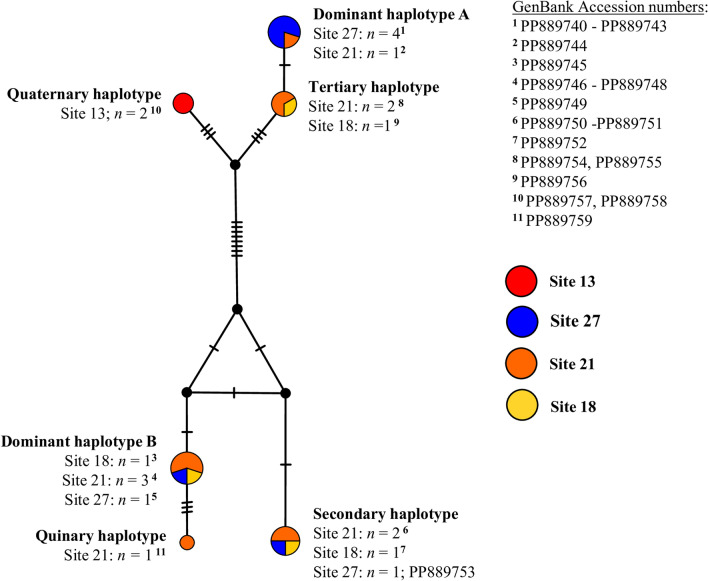
The S. mansoni ND5 TCS haplotype network is shown in Fig. 6. Two distinct clusters were formed, each comprising three unique haplotypes. One unique haplotype consisting of two S. mansoni ND5 sequences was identified at site 13; no other haplotypes were found at this site. One unique haplotype comprising one S. mansoni ND5 sequence was identified at site 21. The remaining four haplotypes were present at more than one single site, with two being present at three sites (sites 18, 21 and 27). All 20 S. mansoni ND5 sequences were uploaded to the GenBank repository (Accession numbers: PP889740-PP889759).
Fig. 6.
Schistosoma mansoni ND5 TCS haplotype network. Each node (circle) represents a unique haplotype, with the size of the node proportional to the frequency of each haplotype. Different node colours represent a specific haplotype(s) identified at that site, with red denoting the haplotype identified at site 13; blue denoting haplotypes identified at site 27; orange denoting haplotypes identified at site 21; and yellow denoting haplotypes identified at site 18. Hatched lines denote the number of SNPs between nodes. ND5, NADH dehydrogenase subunit 5; SNP, single nucleotide polymorphism; TCS, Templeton Crandall and Sing
Only the Biomphalaria ITS and Trematoda ITS loci were amplified in 12 Biomphalaria DNA samples, indicating infection with non-S. mansoni trematodes. The Trematoda ITS gel band was successfully excised and genotyped in all 12 of these samples. Of these 12 samples, six were identified as Uvulifer spp., which were all present at site 13, and the remaining six were identified as Petasiger spp., which were present at sites 18, 20, 21 and 27 (Additional file 1: Table S2), based on ITS analysis. No genetic variation was found between any ITS sequence data for Uvulifer spp. or between any ITS sequence data for Petasiger spp. All six Uvulifer spp. ITS sequences and all six Petasiger spp. ITS sequences were uploaded to the GenBank repository (Accession numbers: PP510464-PP510469 and PP510476-PP501481, respectively). An example agarose gel image of the high-throughput molecular xenomonitoring PCR assay [24] is shown in Additional file 4: Figure S1. Additional data generated during Biomphalaria molecular xenomonitoring can be found in Additional file 5: Dataset S1, Biomphalaria molecular xenomonitoring data.
Discussion
In the present study, we carried out extensive malacological surveillance along the southern shoreline of Lake Malawi, Malawi to measure the distribution and diversity of Biomphalaria freshwater snail intermediate hosts of S. mansoni. We then used a range of molecular approaches to genotype a subset of collected Biomphalaria snails to species level and to screen all collected Biomphalaria for infection with S. mansoni to identify intestinal schistosomiasis transmission sites in this area. The overall aim was to gain a more thorough understanding of Biomphalaria-associated transmission of intestinal schistosomiasis in Mangochi District five years post-outbreak to better inform and strengthen future disease control efforts.
Biomphalaria were collected at multiple surveillance sites, and all genotyped specimens were identified as B. pfefferi, which is the most widely distributed and commonly implicated intermediate host of S. mansoni across sub-Saharan Africa [25]. No cox1 variation was found between all B. pfeifferi genotyped in Mangochi District, suggesting a potential single and recent colonisation event of B. pfeifferi in this area. According to cox1 analysis, all B. pfeifferi genotyped here were most closely related to B. pfeifferi collected in Zimbabwe, with two Zimbabwean isolates from different locations sharing identical cox1 sequence data to Mangochi District B. pfeifferi; it is therefore possible that these B. pfeifferi identified in Mangochi District originated from Zimbabwe, as has been suggested previously [22]. Interestingly, sufficient genetic variation was found between the unique Malawi, Chikwawa District haplotype and the Malawi, Mangochi District haplotype to suggest that the Chikwawa District B. pfeifferi population may have in fact invaded from populations outside of Malawi, rather than migrated south along the Shire River from Mangochi District and diverged, as might have been initially assumed. Further analysis to confirm this possibility, however, is needed, particularly as the cox1 phylogenetic tree generated here suggests that all Malawian B. pfeifferi (including those from Mangochi and Chikwawa Districts) and all Zimbabwean B. pfeifferi share a common ancestor. It is worth noting that no other species of Biomphalaria, such as B. sudanica or B. choanomphala, was identified, although limited surveillance in deep water (> 1 m) took place, which is the preferred microhabitat of B. choanomphala [25].
This recent introduction of Biomphalaria into Mangochi District can likely be, in part, attributed to environmental changes caused by ongoing climate change [41, 42]. In recent years, Mangochi District—and Malawi more broadly—have been severely impacted by atypical and extreme weather conditions, such as tropical cyclones and remarkably heavy rainfall with subsequent flooding, particularly in low-altitude areas such as the upper Shire River margins [43]. Whilst flooding can transport and introduce Biomphalaria into new habitats from surrounding areas, such extreme weather events can also dramatically alter freshwater environments more generally, generating more favourable Biomphalaria habitats [41, 42]. As an example, temporal freshwater pools, which would typically evaporate during dry seasons, may instead remain as permanent waterbodies in which B. pfeifferi can propagate. Such waterbodies may therefore quickly become active sites of intestinal schistosomiasis transmission. As different Biomphalaria species, which can differ in their ability to transmit S. mansoni, have different habitat preferences, a clear understanding of how climate change can alter the environment with respect to current and potential Biomphalaria habitats is needed to better understand ongoing and future transmission of intestinal schistosomiasis. Molecular approaches to identify Biomphalaria species, such as those used here, will be crucial in these investigations.
As found previously in this area [22], just one (0.17%) of all 589 collected Biomphalaria snails was found to be actively shedding S. mansoni cercariae. Molecular xenomonitoring, however, revealed that a total of 20 (3.4%) Biomphalaria were infected with S. mansoni, 12 of which were collected from malacological surveillance sites that were not deemed intestinal schistosomiasis transmission sites based on cercarial shedding analysis. Consequently, the prevalence of Biomphalaria infection with S. mansoni increased from 0.17% to 3.4%, and the number of identified intestinal schistosomiasis transmission sites increased from one to four, based on the results of molecular xenomonitoring. Interestingly, at one such site (site 21; see Additional file 1: Table S1), not only was a relatively high number of Biomphalaria collected (n = 80), but a high proportion (11.25%) of Biomphalaria were found to be infected with S. mansoni using molecular xenomonitoring despite none of these actively shedding S. mansoni cercariae during the cercarial shedding analysis. Furthermore, of note, this site is a short distance (approx. 1.5 km) from Samama School, at which 55% of school-aged children in attendance were found to be infected with S. mansoni during recent parasitological surveillance in November 2021 [Archer et al., under review].
In addition, molecular xenomonitoring identified six unique S. mansoni mitochondrial ND5 haplotypes, which clustered into two distinct groups. A similar degree of mitochondrial DNA genetic diversity was found amongst S. mansoni miracidia cox1 haplotypes (infecting school-aged children) also recently identified in this area (Archer et al. under review). Furthermore, during these previous cox1 analyses, two genetically distinct S. mansoni cox1 linage groups (II and IV) [44, 45] were identified within the miracidia population, which may be reflected here in these ND5 haplotypes. Interestingly, it should also be noted that just one unique S. mansoni ND5 haplotype was identified at site 13, which is far removed (approx. 12–15 km) from the remaining three surveillance sites where S. mansoni transmission was found (all of which are within approx. 3 km of each other), suggesting a focal population of S. mansoni at site 13 that is genetically distinct from S. mansoni populations further south along the lake’s shoreline. At the remaining three intestinal schistosomiasis transmission sites (sites 18, 21 and 27), multiple S. mansoni haplotypes were identified and found to be present at multiple sites, suggesting mixing of S. mansoni populations in this area, likely by human hosts visiting multiple different lake water contact points but also potentially through movement of Biomphalaria between surveyed sites.
Infection with non-Schistosoma trematodes was also detected in 12 Biomphalaria collected at multiple malacological surveillance sites. Six of these were Petasiger spp., which were present at sites 18, 20, 21 and 27. Petasiger spp. trematodes have three hosts, namely, multiple genera of freshwater snails including Biomphalaria; freshwater fish and tadpoles; and piscivorous birds such as cormorants [46]. The remaining six were Uvulifer spp., which were all present at site 13. Uvulifer spp. trematodes also have three hosts: multiple genera of freshwater snails, again including Biomphalaria; freshwater sunfish; and piscivorous birds such as kingfisher [47]. Whilst there appears to be no available studies investigating whether established Uvulifer spp. or Petasiger spp. infections within Biomphalaria impact S. mansoni development and transmission, other trematode species have been found to influence S. mansoni development within Biomphalaria [48], and this may also be the case for these infecting trematodes. This possibility, however, requires further investigation to clarify.
Study limitations and future work
Given the limited diversity found across B. pfeifferi cox1 haplotypes analysed in this study, a more thorough analysis of additional B. pfeifferi DNA loci, such as the mitochondrial 16S or nuclear ITS regions, or even whole mitochondrial genome analysis, would likely provide further insights into Mangochi District B. pfeifferi phylogenies, population structuring and origins [10]. In addition, whilst molecular xenomonitoring can be used to detect Schistosoma infections in freshwater snail hosts that have been missed by cercarial shedding examination, this approach does have limitations. PCR-based molecular approaches, such as end-point PCR used here or real-time/quantitative PCR, are expensive and require specialised laboratory personnel. The continued development of more portable and easy-to-use nucleic acid amplification technologies that could be performed in schistosomiasis-endemic areas, such as loop-mediated isothermal amplification (LAMP) and recombinase polymerase/aided amplification (RPA/RAA), for molecular xenomonitoring purposes is therefore encouraged here [49, 50]. The continued development of DNA extraction technologies capable of isolating DNA from freshwater snail tissues in resource-poor settings is also encouraged [50].
Conclusions
Through extensive malacological surveillance and the use of molecular approaches, we confirm that Biomphalaria are now present at many locations along the southern shoreline of Lake Malawi, Malawi. We also confirm that many of these locations are active intestinal schistosomiasis transmission sites. When compared to the results of previous malacological surveys in this area, it would appear that B. pfeifferi populations are rapidly expanding, not only across this southern shoreline of Lake Malawi, but also further south of Malawi in Chikwawa District. As such, the revision of ongoing schistosomiasis control programmes in Mangochi District, such as the implementation of bi-annual MDA using praziquantel, significantly improving access to adequate water and sanitation hygiene (WASH) infrastructure, freshwater snail population control and continued delivery of schistosomiasis education and health programmes to promote behaviours that limit the risk of contracting and transmitting schistosomiasis, is therefore encouraged, in line with WHO recommendations. Our study also highlights the importance of molecular approaches to investigate Biomphalaria populations and monitor Biomphalaria-associated intestinal schistosomiasis transmission in endemic areas. As such, the continued development and use of such approaches, in particular the development and use of molecular xenomonitoring assays that can be carried out in resource-poor schistosomiasis-endemic settings, is also encouraged. These will be especially needed in low-endemicity areas and will aid in improving disease control efforts, significantly reducing disease-related morbidities experienced by those afflicted by intestinal schistosomiasis.
Supplementary Information
Additional file 1: Table S1. Biomphalaria collected during three malacological surveys carried out along the southern shoreline of Lake Malawi, Malawi. Table S2. Cercarial shedding and molecular xenomonitoring of collected Biomphalaria.
Additional file 2: Table S1A. Primer sequences used to detect and amplify a 700-bp fragment of the Biomphalaria spp. mitochondrial cytochrome oxidase subunit 1 (cox1) gene. Table S1B. Reaction mix used to carry out endpoint PCR to detect and amplify a 700-bp fragment of the Biomphalaria spp. mitochondrial cytochrome oxidase subunit 1 (cox1) gene. Table S1C. PCR conditions used to carry out endpoint PCR to detect and amplify a 700-bp fragment of the Biomphalaria spp. mitochondrial cytochrome oxidase subunit 1 (cox1) gene. Table S2A. Primer and probe sequences used to amplify a 956-bp region of the Schistosoma spp. mitochondrial cytochrome oxidase subunit 1 (cox1) gene. Table S2B. Reaction mix used to carry out end-point targeting a 956-bp region of the Schistosoma spp. mitochondrial cytochrome oxidase subunit 1 (cox1) gene. Table S2C. PCR conditions used to carry out end-point targeting a 956 bp region of the Schistosoma spp. mitochondrial cytochrome oxidase subunit 1 (cox1) gene. Table S3A. Primer and probe sequences used to amplify the complete Schistosoma spp. nuclear internal transcribed spacer region (inclusive of both ITS regions 1 + 2 and the nuclear 5.8S region). Table S3B. Reaction mix used to carry out end-point PCR targeting the complete Schistosoma spp. nuclear internal transcribed spacer region (inclusive of both ITS regions 1 + 2 and the nuclear 5.8S region). Table S3C. PCR conditions used to carry out end-point PCR targeting the complete Schistosoma spp. nuclear internal transcribed spacer region (inclusive of both ITS regions 1 + 2 and the nuclear 5.8S region). Table S4A. Primer sequences used to detect and amplify the complete Biomphalaria spp. and Schistosoma spp. nuclear internal transcribed spacer regions (~ 1250 bp and ~ 1005bp in length, respectively; inclusive of both ITS regions 1 + 2 and the nuclear 5.8S region). Table S4B. Molecular xenomonitoring PCR reaction mix used to detect and amplify the complete Biomphalaria spp. ITS region, the complete Trematoda ITS region and a partial region of the S. mansoni ND5 gene. Table S4C. Molecular xenomonitoring PCR cycling conditions used to detect and amplify the complete Biomphalaria spp. ITS region, the complete Trematoda. ITS region and a partial region of the S. mansoni ND5 gene.
Additional file 3: Table S1. B. pfeifferi cox1 data and GenBank accession numbers used for cox1 haplotype analysis.
Additional file 4: Figure S1. Example agarose gel image of the high-throughput molecular xenomonitoring PCR assay
Additional file 5: Dataset S1. Biomphalaria molecular xenomonitoring data.
Acknowledgements
The authors would like to thank the Schistosome Collections at the Natural History Museum (SCAN) repository as well as Dr Adam Cieplinski and Dr Fernanda Sales Coelho of the Schistosome Snail Resource (SSR) (Wellcome Trust Biomedical Resource Grant 221368/Z/20/Z (2021-2026) for provision of Schistosoma material. We are also grateful to Claire Griffin of the Natural History Museum, London’s Molecular Biology Laboratories for assistance in Sanger sequencing.
Abbreviations
- BLAST
Basic Local Alignment Search Tool
- Cox1
Cytochrome oxidase subunit 1 gene
- ITS
Internal transcribed spacer
- LAMP
Loop-mediated isothermal amplification
- MAFFT
Multiple alignment with fast Fourier transform
- ML
Maximum likelihood
- NCBI
National Center for Biotechnology Information
- ND5
NADH dehydrogenase subunit 5
- NTD
Neglected tropical disease
- RPA/RAA
Recombinase polymerase/aided amplification
- SNP
Single nucleotide polymorphism
- SSR
Schistosome and Snail Resource
- TCS network
Templeton Crandall and Sing haplotype network
- WASH
Water and sanitation hygiene
Author contributions
John Archer: conceptualisation, methodology, validation, formal analysis, investigation, resources, data curation, writing—original draft, writing—review & editing, visualisation, project administration, funding acquisition. Lucas J Cunningham: resources, writing—review & editing. Alexandra Juhász: resources, writing—review & editing. Sam Jones: resources, writing—review & editing. Amber L Reed: resources, writing—review & editing. Shi Min Yeo: methodology, validation, formal analysis, investigation, resources, data curation, writing—review & editing, visualisation. Bright Mainga: resources, writing—review & editing. Priscilla Chammudzi: resources, writing—review & editing. Donales R Kapira: resources, writing—review & editing. David Lally: resources, writing—review & editing. Gladys Namacha: resources, writing—review & editing. Peter Makaula: resources, writing—review and editing, project administration. James E LaCourse: resources, writing—review & editing, project administration. Sekeleghe A Kayuni: resources, writing—review and editing, project administration. Bonnie L Webster: conceptualisation, methodology, investigation, resources, writing—review & editing, funding acquisition. Janelisa Musaya: conceptualisation, methodology, investigation, resources, writing—review & editing, funding acquisition. J Russell Stothard: conceptualisation, methodology, investigation, resources, writing—review & editing, funding acquisition. All authors read and approved the final manuscript.
Funding
JA is supported by a Medical Research Council Doctoral Training Programme (MRC-DTP) fellowship held at the Liverpool School of Tropical Medicine. JA was also awarded a London Centre for Neglected Tropical Disease Research (LCNTDR) student travel grant. SMY was funded through a London School of Hygiene and Tropical Medicine (LSHTM) MSc Research Project fund (2023). The Hybridisations in UroGenital Schistosomiasis (HUGS) study is funded by a Wellcome Trust Joint Investigator Award 220818/Z/20/Z.
Availability of data and materials
All data generated during malacological collections, Biomphalaria cercarial shedding analyses and Biomphalaria molecular xenomonitoring can be found in Additional file 1: Table S1. All data on Biomphalaria collected during three malacological surveys carried out along the southern shoreline of Lake Malawi, Malawi can be found in Additional file 1: Table S2, cercarial shedding and molecular xenomonitoring of collected Biomphalaria. An example agarose gel image of the high-throughput molecular xenomonitoring PCR assay [24] is shown in Additional file 4: Figure S1. Additional data generated during Biomphalaria molecular xenomonitoring can be found in Additional file 5: Dataset S1, Biomphalaria molecular xenomonitoring data. All Sanger sequence data generated was uploaded to the GenBank repository as described (B. pfeifferi cox1 sequences: GenBank accession numbers PP524927-PP524968; S. mansoni cox1 sequences: GenBank accession numbers PP529587-PP529591; S. mansoni ITS sequences: GenBank accession numbers PP510205-PP510209; S. mansoni ND5 sequences: GenBank accession numbers PP889740–PP889759; Uvulifer spp. ITS sequences: GenBank accession numbers PP510464-PP510469; Petasiger spp. ITS sequences: GenBank accession numbers PP510476-PP501481).
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Janelisa Musaya and J. Russell Stothard contributed equally to this work.
References
- 1.Colley DG, Bustinduy AL, Secor WE, King CH. Human schistosomiasis. Lancet. 2014;383:2253–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McManus DP, Dunne DW, Sacko M, Utzinger J, Vennervald BJ, Zhou XN. Schistosomiasis. Nat Rev Dis Primers. 2018;4:13. [DOI] [PubMed] [Google Scholar]
- 3.Tchuem Tchuenté LA, Rollinson D, Stothard JR, Molyneux D. Moving from control to elimination of schistosomiasis in sub-Saharan Africa: time to change and adapt strategies. Infect Dis Poverty. 2017;6:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hotez PJ, Kamath A. Neglected tropical diseases in sub-Saharan Africa: review of their prevalence, distribution, and disease burden. PLoS Negl Trop Dis. 2009;3:2–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morgan JAT, Dejong RJ, Snyder SD, Mkoji GM, Loker ES. Schistosoma mansoni and Biomphalaria: Past history and future trends. Parasitology. 2001;123:211–28. [DOI] [PubMed] [Google Scholar]
- 6.Grimes JE, Croll D, Harrison WE, Utzinger J, Freeman MC, Templeton MR. The roles of water, sanitation and hygiene in reducing schistosomiasis: a review. Parasit Vectors. 2015;8:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mutuku MW, Lu L, Otiato FO, Mwangi IN, Kinuthia JM, Maina GM, et al. A Comparison of Kenyan Biomphalaria pfeifferi and B. Sudanica as vectors for Schistosoma mansoni, including a discussion of the need to better understand the effects of snail breeding systems on transmission. Parasitology. 2017;103:669–76. [DOI] [PubMed] [Google Scholar]
- 8.Tavalire HF, Blouin MS, Steinauer ML. Genotypic variation in host response to infection affects parasite reproductive rate. Int Parasitol. 2016;46:123–31. [DOI] [PubMed] [Google Scholar]
- 9.Standley CJ, Wade C, Stothard JR. A fresh insight into transmission of schistosomiasis: a misleading tale of Biomphalaria in Lake Victoria. PLoS ONE. 2011;6:e26563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jørgensen A, Kristensen TK, Stothard JR. Phylogeny and biogeography of African Biomphalaria (Gastropoda: Planorbidae), with emphasis on endemic species of the great East African lakes. Zool J Linn Soc. 2007;151:337–49. [Google Scholar]
- 11.Weerakoon KG, Gordon CA, McManus DP. DNA diagnostics for schistosomiasis control. Trop Med Infect Dis. 2018;3:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamel B, Laidemitt MR, Lu L, Babbitt C, Weinbaum OL, Mkoji GM, et al. Detecting and identifying Schistosoma infections in snails and aquatic habitats: a systematic review. PLoS Negl Trop Dis. 2021;15:e0009175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu L, Zhang SM, Mutuku MW, Mkoji GM, Loker ES. Relative compatibility of Schistosoma mansoni with Biomphalaria sudanica and B. pfeifferi from Kenya as assessed by PCR amplification of the S. mansoni ND5 gene in conjunction with traditional methods. Parasit Vectors. 2016;9:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Norton A, Rollinson D, Richards L, Webster J. Simultaneous infection of Schistosoma mansoni and S. rodhaini in Biomphalaria glabrata: impact on chronobiology and cercarial behaviour. Parasit Vectors. 2008;1:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leger E, Webster JP. Hybridizations within the Genus Schistosoma: implications for evolution, epidemiology and control. Parasitology. 2017;144:65–80. [DOI] [PubMed] [Google Scholar]
- 16.Miranda GS, Rodrigues JGM, Silva JKADO, Camelo GMA, Silva-Souza N, Neves RH, et al. New challenges for the control of human schistosomiasis: the possible impact of wild rodents in Schistosoma mansoni transmission. Acta Trop. 2022;236:106677. [DOI] [PubMed] [Google Scholar]
- 17.Andrus PS, Stothard JR, Wade CM. Seasonal patterns of Schistosoma mansoni infection within Biomphalaria snails at the Ugandan shorelines of Lake Albert and Lake Victoria. PLoS Negl Trop Dis. 2023;17:e0011506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gouvras AN, Allan F, Kinung’hi S, Rabone M, Emery A, Angelo T, et al. Longitudinal survey on the distribution of Biomphalaria sudanica and B. choanomphala in Mwanza region, on the shores of Lake Victoria, Tanzania: Implications for schistosomiasis transmission and control. Parasit Vectors. 2017;10:316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kayuni SA, O’Ferrall AM, Baxter H, Hesketh J, Mainga B, Lally D, et al. An outbreak of intestinal schistosomiasis, alongside increasing urogenital schistosomiasis prevalence, in primary school children on the shoreline of Lake Malawi, Mangochi District, Malawi. Infect Dis Poverty. 2020;9:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kayuni SA, Alharbi MH, Makaula P, Lampiao F, Juziwelo L, LaCourse EJ, et al. Male genital schistosomiasis along the shoreline of Lake Malawi: baseline prevalence and associated knowledge, attitudes and practices among local fishermen in Mangochi District, Malawi. Front Public Health. 2021;21:590695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Archer J, Cunningham LJ, Juhàsz A, Jones S, Doull F, LaCourse JE, et al. Molecular epidemiology and assemblage typing of Giardia duodenalis in school-age children situated along the Southern Shoreline of Lake Malawi, Malawi. Am J Trop Med Hyg. 2023;109:626–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alharbi MH, Condemine C, Hesketh J, Kayuni SA, Arme TM, Archer J, et al. Biomphalaria pfeifferi (Gastropoda: Planorbidae) in Lake Malawi and upper shire River, Mangochi District, Malawi: distribution, genetic diversity and pre-patent Schistosome infections. Tropical Med. 2023;8:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bakuza JS, Gillespie R, Nkwengulila G, Adam A, Kilbride E, Mable BK. Assessing S. mansoni prevalence in Biomphalaria snails in the Gombe ecosystem of western Tanzania: the importance of DNA sequence data for clarifying species identification. Parasit Vectors. 2017;10:584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Archer J, Yeo SM, Gadd G, Pennance T, Cunningham LJ, Juhàsz A, et al. Development, validation, and pilot application of a high throughput molecular xenomonitoring assay to detect Schistosoma mansoni and other trematode species within Biomphalaria freshwater snail hosts. Curr Res Parasitol Vector-Borne Dis. 2024;5:100174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown DS. Freshwater snails of Africa and their medical importance. 2nd ed. London: Taylor & Francis; 1994. [Google Scholar]
- 26.Pennance T, Ame SM, Amour AK, Suleiman KR, Muhsin MA, Kabole F, et al. Transmission and diversity of Schistosoma haematobium and S. bovis and their freshwater intermediate snail hosts Bulinus globosus and B. nasutus in the Zanzibar Archipelago, United Republic of Tanzania. PLoS Negl Trop Dis. 2022;16:e0010585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Théron A. Early and late shedding patterns of Schistosoma mansoni cercariae: ecological significance in transmission to human and murine hosts. Parasitology. 1984;70:652–5. [PubMed] [Google Scholar]
- 28.Frandsen F, Christensen NØ. An introductory guide to the identification of cercariae from African freshwater snails with special reference to cercariae of trematode species of medical and veterinary importance. Acta Trop. 1984;41:181–202. [PubMed] [Google Scholar]
- 29.Pennance T, Archer J, Lugli EB, Rostron P, Llanwarne F, Ali SM, et al. Development of a molecular snail xenomonitoring assay to detect Schistosoma haematobium and Schistosoma bovis infections in their Bulinus snail hosts. Molecules. 2020;25:4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schistosome and Snail Resource (SSR). Schistosome and Snail Resource (SSR). 2024. https://www.nhm.ac.uk/our-science/research/projects/schistosome-snail-resource.html and https://www.lshtm.ac.uk/research/centres-projects-groups/schistosome-and-snail-resource. Accessed August 2024.
- 31.Geer LY, Marchler-Bauer A, Geer RC, Han L, He J, He S, et al. The NCBI BioSystems database. Nucl Acids Res. 2009;38:492–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leigh JW, Bryant D. POPART: full-feature software for haplotype network construction. Methods Ecol Evol. 2015;6:1110–6. [Google Scholar]
- 33.Templeton AR, Crandall KA, Sing CF. A cladistic analysis of phenetypic associations with haplotypes inferred Ffom restriction endonuclease mapping and DNA sequencedData. III. Cladogram estimation. Genetics 132:619-33. [DOI] [PMC free article] [PubMed]
- 34.DeJong RJ, Emery AM, Adema CM. The mitochondrial genome of Biomphalaria glabrata (Gastrapoda: Basommatophora), the intermediate host of Schistosoma mansoni. Parasitology. 2004;90:991–7. [DOI] [PubMed] [Google Scholar]
- 35.Tamura K, Stecher G, Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 2021;38:3022–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stothard JR, Kayuni SA, Al-Harbi MH, Musaya J, Webster BL. Future schistosome hybridizations: will all Schistosoma haematobium hybrids please stand-up! PLoS Negl Trop Dis. 2020;14:e0008201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Webster BL, Rabone M, Pennance T, Emery AM, Allan F, Gouvras A, et al. Development of novel multiplex microsatellite polymerase chain reactions to enable high-throughput population genetic studies of Schistosoma haematobium. Parasit Vectors. 2015;8:432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sene-Wade M, Marchand B, Rollinson D, Webster BL. Urogenital schistosomiasis and hybridization between Schistosoma haematobium and Schistosoma bovis in adults living in Richard-Toll. Senegal Parasitol. 2018;145:1723–6. [DOI] [PubMed] [Google Scholar]
- 39.Appelhans T, Detsch F, Reudenbach C, Woellauer S. Mapview: interactive viewing of spatial data in R. R package version 2.10.0. 2021. https://CRAN.R-project.org/package=mapview. Accessed August 2024.
- 40.R Development Core Team. R: a language and environment for statistical computing. 2024. Vienna: R Foundation for Statistical Computing. https://www.gbif.org/tool/81287/r-a-language-and-environment-for-statistical-computing. Accessed August 2024.
- 41.McCreesh N, Nikulin G, Booth M. Predicting the effects of climate change on Schistosoma mansoni transmission in eastern Africa. Parasit Vectors. 2015;8:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stensgaard AS, Vounatsou P, Sengupta ME, Utzinger J. Schistosomes, snails and climate change: current trends and future expectations. Acta Trop. 2019;190:257–68. [DOI] [PubMed] [Google Scholar]
- 43.Braka F, Daniel EO, Okeibunor J, Rusibamayila NK, Conteh IN, Ramadan OPC, et al. Effects of tropical cyclone Freddy on the social determinants of health: the narrative review of the experience in Malawi. BMJ Public Health. 2024;2:e000512. [Google Scholar]
- 44.Webster BL, Webster JP, Gouvras AN, Garba A, Lamine MS, Diaw OT, et al. DNA ‘barcoding’ of Schistosoma mansoni across sub-Saharan Africa supports substantial within locality diversity and geographical separation of genotypes. Acta Trop. 2013;128:250–60. [DOI] [PubMed] [Google Scholar]
- 45.López-Hernández D, Locke SA, De Assis JCA, Drago FB, De Melo AL, Rabelo ÉML, et al. Molecular, morphological and experimental-infection studies of cercariae of five species in the superfamily Diplostomoidea (Trematoda: Digenea) infecting Biomphalaria straminea (Mollusca: Planorbidae) in Brazil. Acta Trop. 2019;199:105082. [DOI] [PubMed] [Google Scholar]
- 46.Johnston DW. Feeding ecology of pied kingfishers on Lake Malawi, Africa. Biotropica. 1989;21:275. [Google Scholar]
- 47.Laidemitt MR, Brant SV, Mutuku MW, Mkoji GM, Loker ES. The diverse echinostomes from East Africa: with a focus on species that use Biomphalaria and Bulinus as intermediate hosts. Acta Trop. 2019;193:38–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laidemitt MR, Anderson LC, Wearing HJ, Mutuku MW, Mkoji GM, Loker ES. Antagonism between parasites within snail hosts impacts the transmission of human schistosomiasis. Elife. 2019;17:e50095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gandasegui J, Fernández-Soto P, Hernández-Goenaga J, López-Abán J, Vicente B, Muro A. Biompha-LAMP: a new rapid loop-mediated isothermal amplification assay for detecting Schistosoma mansoni in Biomphalaria glabrata Snail host. PLoS Negl Trop Dis. 2016;10:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mesquita SG, Lugli EB, Matera G, Fonseca CT, Caldeira RL, Webster B. Development of real-time and lateral flow recombinase polymerase amplification assays for rapid detection of Schistosoma mansoni. Front Microbiol. 2022;18:1043596. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Biomphalaria collected during three malacological surveys carried out along the southern shoreline of Lake Malawi, Malawi. Table S2. Cercarial shedding and molecular xenomonitoring of collected Biomphalaria.
Additional file 2: Table S1A. Primer sequences used to detect and amplify a 700-bp fragment of the Biomphalaria spp. mitochondrial cytochrome oxidase subunit 1 (cox1) gene. Table S1B. Reaction mix used to carry out endpoint PCR to detect and amplify a 700-bp fragment of the Biomphalaria spp. mitochondrial cytochrome oxidase subunit 1 (cox1) gene. Table S1C. PCR conditions used to carry out endpoint PCR to detect and amplify a 700-bp fragment of the Biomphalaria spp. mitochondrial cytochrome oxidase subunit 1 (cox1) gene. Table S2A. Primer and probe sequences used to amplify a 956-bp region of the Schistosoma spp. mitochondrial cytochrome oxidase subunit 1 (cox1) gene. Table S2B. Reaction mix used to carry out end-point targeting a 956-bp region of the Schistosoma spp. mitochondrial cytochrome oxidase subunit 1 (cox1) gene. Table S2C. PCR conditions used to carry out end-point targeting a 956 bp region of the Schistosoma spp. mitochondrial cytochrome oxidase subunit 1 (cox1) gene. Table S3A. Primer and probe sequences used to amplify the complete Schistosoma spp. nuclear internal transcribed spacer region (inclusive of both ITS regions 1 + 2 and the nuclear 5.8S region). Table S3B. Reaction mix used to carry out end-point PCR targeting the complete Schistosoma spp. nuclear internal transcribed spacer region (inclusive of both ITS regions 1 + 2 and the nuclear 5.8S region). Table S3C. PCR conditions used to carry out end-point PCR targeting the complete Schistosoma spp. nuclear internal transcribed spacer region (inclusive of both ITS regions 1 + 2 and the nuclear 5.8S region). Table S4A. Primer sequences used to detect and amplify the complete Biomphalaria spp. and Schistosoma spp. nuclear internal transcribed spacer regions (~ 1250 bp and ~ 1005bp in length, respectively; inclusive of both ITS regions 1 + 2 and the nuclear 5.8S region). Table S4B. Molecular xenomonitoring PCR reaction mix used to detect and amplify the complete Biomphalaria spp. ITS region, the complete Trematoda ITS region and a partial region of the S. mansoni ND5 gene. Table S4C. Molecular xenomonitoring PCR cycling conditions used to detect and amplify the complete Biomphalaria spp. ITS region, the complete Trematoda. ITS region and a partial region of the S. mansoni ND5 gene.
Additional file 3: Table S1. B. pfeifferi cox1 data and GenBank accession numbers used for cox1 haplotype analysis.
Additional file 4: Figure S1. Example agarose gel image of the high-throughput molecular xenomonitoring PCR assay
Additional file 5: Dataset S1. Biomphalaria molecular xenomonitoring data.
Data Availability Statement
All data generated during malacological collections, Biomphalaria cercarial shedding analyses and Biomphalaria molecular xenomonitoring can be found in Additional file 1: Table S1. All data on Biomphalaria collected during three malacological surveys carried out along the southern shoreline of Lake Malawi, Malawi can be found in Additional file 1: Table S2, cercarial shedding and molecular xenomonitoring of collected Biomphalaria. An example agarose gel image of the high-throughput molecular xenomonitoring PCR assay [24] is shown in Additional file 4: Figure S1. Additional data generated during Biomphalaria molecular xenomonitoring can be found in Additional file 5: Dataset S1, Biomphalaria molecular xenomonitoring data. All Sanger sequence data generated was uploaded to the GenBank repository as described (B. pfeifferi cox1 sequences: GenBank accession numbers PP524927-PP524968; S. mansoni cox1 sequences: GenBank accession numbers PP529587-PP529591; S. mansoni ITS sequences: GenBank accession numbers PP510205-PP510209; S. mansoni ND5 sequences: GenBank accession numbers PP889740–PP889759; Uvulifer spp. ITS sequences: GenBank accession numbers PP510464-PP510469; Petasiger spp. ITS sequences: GenBank accession numbers PP510476-PP501481).