Abstract
Emerging low-emission production technologies make ethanol an interesting substrate for yeast biotechnology, but information on growth rates and biomass yields of yeasts on ethanol is scarce. Strains of 52 Saccharomycotina yeasts were screened for growth on ethanol. The 21 fastest strains, among which representatives of the Phaffomycetales order were overrepresented, showed specific growth rates in ethanol-grown shake-flask cultures between 0.12 and 0.46 h−1. Seven strains were studied in aerobic, ethanol-limited chemostats (dilution rate 0.10 h−1). Saccharomyces cerevisiae and Kluyveromyces lactis, whose genomes do not encode Complex-I-type NADH dehydrogenases, showed biomass yields of 0.59 and 0.56 gbiomass gethanol−1, respectively. Different biomass yields were observed among species whose genomes do harbour Complex-I-encoding genes: Phaffomyces thermotolerans (0.58 g g−1), Pichia ethanolica (0.59 g g−1), Saturnispora dispora (0.66 g g−1), Ogataea parapolymorpha (0.67 g g−1), and Cyberlindnera jadinii (0.73 g g−1). Cyberlindnera jadinii biomass showed the highest protein content (59 ± 2%) of these yeasts. Its biomass yield corresponded to 88% of the theoretical maximum that is reached when growth is limited by assimilation rather than by energy availability. This study suggests that energy coupling of mitochondrial respiration and its regulation will become key factors for selecting and improving yeast strains for ethanol-based processes.
Keywords: quantitative physiology, biomass composition, bioreactors, electron transport system, Hansenula polymorpha, Candida utilis
Screening >50 yeasts reveals a wide range of growth rate and biomass yields on ethanol as sole substrate, opening the path for further exploring natural diversity and regulation of bioenergetics.
Introduction
Commonly known as budding yeasts, Saccharomycotina yeasts are successfully applied as microbial cell factories in various industrial biotechnological applications (Geijer et al. 2022). Currently, these processes predominantly use (partially) refined sugars, such as cornstarch-derived glucose and sugarcane-derived sucrose, as feedstocks (van Aalst et al. 2022). While sugar-based biotechnological processes can enable lower carbon footprints than petrochemical alternatives (Farrell et al. 2006, Hermann et al. 2007), they depend on successful crop harvests and are linked with greenhouse gas (GHG) emissions from agriculture, transport, and sugar refining (Salim et al. 2019). In addition, unrestricted expansion of sugar-based industrial biotechnology may interfere with other human needs for arable land (Tilman et al. 2006). Ethanol is the largest-volume product of industrial biotechnology because of its widespread usage as a ‘drop-in’ transport fuel and, increasingly, also as renewable chemical building block (Fulton et al. 2015, Dagle et al. 2020). Current industrial ethanol production largely relies on fermentation of cornstarch hydrolysates or cane sugar by Saccharomyces cerevisiae (Jacobus et al. 2021).
Recent progress in the development of alternative, low-GHG-emission processes for ethanol production has stimulated interest in ethanol as a substrate for yeast cultivation and thereby improve the sustainability of microbial biotechnology (Geijer et al. 2022). Biotechnological conversion of agricultural and forestry residues has the potential to substantially reduce GHG emissions of ethanol production (Lynd et al. 2022). While new process modes for such ‘second generation’ ethanol production continue to be explored (Claes et al. 2020), industrial application has already moved to industrial scale (Jansen et al. 2017). Production of ethanol from syngas by acetogenic bacteria (Phillips et al. 1993, Martin et al. 2016) has also been successfully scaled up to industrial level (Heffernan et al. 2020). Although in early phases of development, direct (electro)catalytic production of CO2 to ethanol (Birdja et al. 2019) and microbial electrosynthesis (Cabau-Peinado et al. 2024) offer additional perspectives for low-GHG-emission ethanol production. Total GHG emissions of bioprocesses could be reduced if ethanol obtained from these processes were to be used as feedstock instead of sugars derived from agriculture (van Peteghem et al. 2022a).
Ethanol is readily consumed by many budding yeasts (Kurtzman et al. 2011), including species that are already applied in industrial biotechnology. Furthermore, concentrated ethanol solutions can be pumped at low temperatures and are self-sterilizing, which can contribute to water economy and reduce complexity of large-scale aerobic processes (Weusthuis et al. 2011). Recent advancements in production of low-GHG-emission ethanol, combined with the potential advantages of ethanol as feedstock for yeast-based industrial biotechnology, have led to interest in finding the ideal host for such bioprocesses.
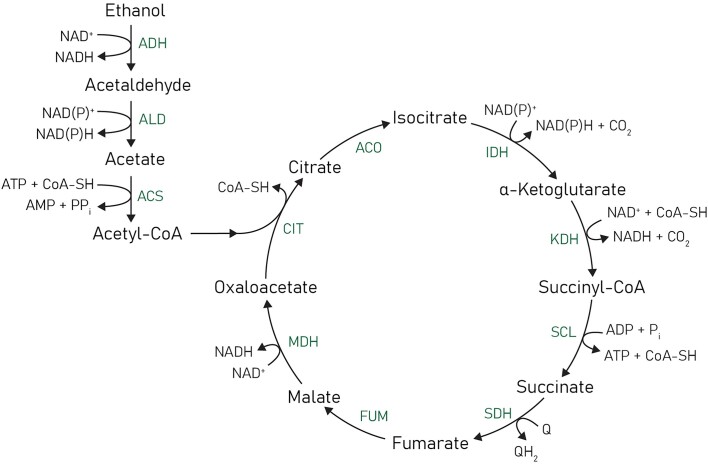
Ethanol has to be respired aerobically for the generation of adenosine triphosphate (ATP) through the mitochondrial electron transfer system (ETS), indicating that energy conservation in yeast metabolism will undoubtedly impact industrial applications. In yeasts, dissimilation of ethanol starts with its oxidation to acetaldehyde by NAD+-dependent alcohol dehydrogenases, after which acetaldehyde is oxidized to acetate by NAD(P)+-linked acetaldehyde dehydrogenases (Overkamp et al. 2002). Acetate is then activated to acetyl-coenzyme A (acetyl-CoA) by acetyl-CoA synthetases, a reaction coupled to the hydrolysis of ATP to adenosine monophosphate (AMP) and pyrophosphate. Because pyrophosphate is subsequently hydrolysed, acetate activation requires a net input of 2 ATP equivalents (Pronk et al. 1994). Complete oxidation of acetyl-CoA to CO2 in the tricarboxylic acid cycle yields 3 NADH, 1 quinol (QH2), and, through substrate-level phosphorylation, 1 ATP (Fig. 1). The overall net ATP yield from ethanol dissimilation depends on the ATP stoichiometry of mitochondrial respiration and is expressed by the P/O ratio. This ratio indicates the number of ATP molecules formed by F1Fo–ATP synthase activity upon transfer of the two electrons from NADH or QH2 to oxygen, coupled via proton translocation across the inner mitochondrial membrane by the ETS and subsequent utilization of the generated proton motive force for the synthesis of ATP molecules. For ethanol dissimilation, the net ATP yield is equal to 5 P/ONADH + 1 P/OQH2–1 mol ATP (mol ethanol)−1.
Figure 1.
Overview of the metabolic pathway for ethanol dissimilation in yeasts. Complete oxidation of 1 mol ethanol yields 2 mol CO2, 5 mol NAD(P)H, 1 mol QH2 (reduced quinone), and the net hydrolysis of 1 mol ATP. Based on the studies on S. cerevisiae (Pronk et al. 1994), hydrolysis of pyrophosphate (PPi) formed by ACS is not expected to contribute to energy conservation. In some non-Saccharomyces yeasts, NADPH generated by NADP+-dependent isoenzymes of ALD and IDH can also be oxidized by mitochondrial respiration (see e.g. Overkamp et al. 2002). Abbreviations: ADH—alcohol dehydrogenase; ALD—aldehyde dehydrogenase, ACS—acetyl-CoA synthase; CIT—citrate synthase; ACO—aconitase; IDH—isocitrate dehydrogenase; KDH—α-ketoglutarate dehydrogenase; SCL—succinyl-CoA ligase; SDH—succinate dehydrogenase; FUM—fumarase; and MDH—malate dehydrogenase.
The P/O ratio is determined by the composition of the mitochondrial ETS and, when different branches to and from the Q junction can be expressed, by the in vivo distribution of electrons over different branches. Model organism S. cerevisiae has a relatively simple ETS in which electrons from NADH and succinate are both donated to the Q pool (Bakker et al. 2001). Saccharomyces cerevisiae lacks a quinol oxidase (alternative oxidase—AOX) that, in some other yeasts, can bypass the cytochrome c-dependent pathway and the associated proton-translocating sites (Joseph-Horne et al. 2001, Guerrero-Castillo et al. 2012). In S. cerevisiae, in vivo P/ONADH and P/OQH2 ratios are both ~1 (Verduyn et al. 1991, Postmus et al. 2011), resulting in a net ATP yield of ~5 mol ATP (mol ethanol)−1. It is relevant to note that nuclear and mitochondrial genomes of many non-Saccharomyces yeasts harbour the genetic information to encode a multisubunit mitochondrial proton-pumping NADH dehydrogenase complex (‘Complex I’) (Nosek and Fukuhara 1994). The presence of a (proton-pumping) Complex I in mitochondria enables higher P/ONADH ratios and could thereby contribute to higher biomass and product yields on ethanol. In yeasts that contain a Complex I, NADH generated in the cytosol can have a different P/O ratio than NADH generated in the mitochondrial matrix (Verduyn et al. 1991). Estimation of the ATP yield of ethanol dissimilation is further complicated by the fact that, in many yeasts, Complex-I-encoding genes often occur in combination with orthologs of genes encoding the nonproton-pumping S. cerevisiae NADH dehydrogenases Ndi1 (Luttik et al. 1998, Melo et al. 2004). This implies that, depending on regulation of gene expression, electron transfer systems for NADH oxidation in these yeasts may be branched and determining a unique P/O ratio of mitochondrial NADH oxidation may not be possible.
Despite its potential as a carbon source for ethanol-based industrial biotechnology, information on specific growth rates and biomass yields of different yeast species on ethanol is limited. The goal of this study is to obtain an indication of the diversity of specific growth rates and biomass yields of Saccharomycotina yeasts when grown on ethanol as sole carbon and energy source. To this end, strains of 52 Saccharomycotina yeasts were screened and those tested positive for growth were assessed semiquantitatively in microtiter plates for their growth rates on ethanol. Specific growth rates of 21 strains with the fastest growth in microtiter plates were measured in shake-flask cultures. Finally, seven strains, the genomes of five of which harboured the genetic information to encode a Complex-I NADH dehydrogenase, were grown in ethanol-limited chemostat cultures and analysed for their biomass yield, macromolecular composition, and energetic efficiency.
Materials and methods
Strains and maintenance
Yeast strains used in this study are listed in Table S1. Saccharomyces cerevisiae CEN.PK113-7D and Yarrowia lipolytica W29 were provided by Dr P. Kötter (J.-W. Goethe Universität, Frankfurt) and Dr J.M. Nicaud (Micalis Institute, INRAE, AgroParisTech, Université Paris-Saclay), respectively. Komagataella phaffii was obtained as Pichia pastoris X-33 from Invitrogen (Thermo Fisher Scientific). All other strains were obtained from the Westerdijk Institute (Utrecht, the Netherlands). For maintenance, strains were grown in shake flasks on yeast extract peptone dextrose (YPD) medium. YPD was prepared by autoclaving a solution of 10 g l−1 yeast extract and 20 g peptone l−1 (Thermo Fisher Scientific) in demineralized water at 121°C for 20 min. A sterile 50% (w/v) glucose solution, autoclaved separately at 110°C for 20 min, was then aseptically added to achieve a glucose concentration of 20 g l−1. Sterile glycerol was added to stationary phase cultures at a final concentration of 30% (v/v) and aliquots were stored at −80°C.
Synthetic growth media
Synthetic medium (SM) for shake-flask and chemostat cultivation was prepared as described by Verduyn et al. (1992) and contained per litre: 5 g (NH4)2SO4, 3 g KH2PO4, 0.5 g MgSO4·7 H2O, 1 ml trace metals solution, and 1 ml vitamins solution. The trace elements solution (1000× final medium concentration), containing 4.5 g CaCl2·2 H2O, 4.5 g ZnSO4·7 H2O, 3 g FeSO4·7 H2O, 1 g H3BO3, 0.84 g MnCl2·2 H2O, 0.4 g NaMoO4·2 H2O, 0.3 g CoCl2·6 H2O, 0.3 g CuSO4·5 H2O, 0.1 g KI, and 15 g EDTA per litre, was added to a solution of the other mineral salts prior to autoclaving at 121°C for 20 min. For shake-flask cultivation, the pH was adjusted to 6.0 with 2 M KOH prior to autoclaving. For chemostat cultivation, 0.2 ml l−1 Pluronic 6100 PE antifoam (BASF, Ludwigshafen, Germany) was added prior to autoclaving. Ethanol and filter-sterilized vitamin solution (1000× final medium concentration, per litre: 50 mg biotin, 1 g Ca-pantothenate, 1 g nicotinic acid, 25 g myo-inositol, 1 g thiamine·HCl, 1 g pyridoxine·HCl, and 0.2 g p-aminobenzoic acid) were added aseptically after autoclaving. Unless otherwise indicated, ethanol was added to a final concentration of 7.5 g l−1 for shake-flask cultivation and 3.75 g l−1 for chemostat cultivation. Microtiter plate cultures and inocula for these cultures were grown on buffered SM described by Jensen et al. (2014) to postpone growth deceleration due to acidification. Compared with SM, this medium contained higher concentrations of (NH4)2SO4 (7.5 g l−1), KH2PO4 (14.4 g l−1) and trace elements (2 ml trace metals solution per litre). For microtiter plate cultures, 3 g l−1 ethanol was added.
Cultivation techniques
Shake-flask cultures were grown at 30°C in round-bottom flasks with a working volume of 20% of their nominal capacity and shaken at 200 rpm in an Innova incubator (Eppendorf Nederland BV, Nijmegen, the Netherlands). Late-exponential or early-stationary phase cultures were used to inoculate microtiter plates and chemostats.
Microtiter plate cultures were grown at 30°C in an image-analysis-based Growth-Profiler system (EnzyScreen BV, Heemstede, the Netherlands) equipped with 96-well plates. Culture volume was 250 µl, plates were agitated at 250 rpm and imaged at 30 min intervals. Wells were inoculated at an OD660 of ~0.1. Green values of imaged wells were corrected for well-to-well variation by calibration with a 96-well plate containing an S. cerevisiae CEN.PK113-7D cell suspension with an externally measured OD660 of 2.5. The time to reach stationary phase (TS) was determined as t2–t1, with t1 indicating the time (h) at which the corrected green value was twice as high as the value at inoculation (average of the first three measured green values after inoculation), and t2 indicating the first time point at which growth deceleration was observed (Fig. S1).
Ethanol-limited chemostat cultures were grown at 30°C and at a dilution rate of 0.10 h−1 in 1.5 l bioreactors (Getinge-Applikon, Delft, the Netherlands) with a 1-l working volume. The culture pH was maintained at 5.0 by automatic addition of 2 M KOH, controlled by an ez-Control module (Getinge-Applikon). The working volume was kept at 1.0 l by an electrical level sensor that controlled the effluent pump. The precise working volume of chemostat cultures was measured at the end of each experiment by weighing the broth. Cultures were stirred at 800 rpm and sparged with dried, compressed air (0.5 l min−1). Off-gas flow rates from bioreactors were measured with a Mass-Stream Thermal Mass Flow Meter (Bronkhorst, Vlaardingen, the Netherlands). Off-gas was cooled to 4°C in a condenser to minimize evaporation and dried with a Perma Pure dryer (Inacom Instruments, Veenendaal, the Netherlands). Concentrations of CO2 and O2 in the inlet and dried exhaust gas were measured with an analyser (Servomex MultiExact 4100, Crowborough, UK). Cultures were assumed to have reached steady state when, after a minimum of 5 volume changes, CO2 production rate, biomass concentration, and high-performance liquid chromatography (HPLC) measurements changed by less than 5% over two consecutive volume changes.
Analytical methods
Optical density of cultures was measured at 660 nm with a Jenway 7200 spectrophotometer (Jenway, Staffordshire, UK) after dilution with demineralized water to OD660 values between 0.1 and 0.3. Specific growth rates of shake-flask cultures were calculated by linear regression of the relation between ln(OD660) and time in the exponential growth phase, including at least 6 time points over at least 3 biomass doublings.
For biomass dry weight determination, duplicate culture samples of exactly 10.0 ml (or 5 ml for Ogataea parapolymorpha) were filtered over predried and preweighed membrane filters (0.45 μm pore size; Pall Corporation, Ann Arbor, MI). Filters were then washed with demineralized water, dried in a microwave oven at 320 W for 20 min, and immediately reweighed (Postma et al. 1989).
Metabolite concentrations in culture supernatants and media were measured with an Agilent 1260 Infinity HPLC system (Agilent Technologies, Santa-Clara, CA) equipped with a Bio-rad Aminex HPX-87H ion exchange column (Bio-Rad, Hercules, CA), operated at 60°C with 5 mM H2SO4 as mobile phase at a flow rate of 0.600 ml min−1. Culture supernatant was obtained by centrifuging 1 ml aliquots at 13 000 rpm for 5 min with a Biofuge Pico centrifuge (Heraeus Instruments, Hanau, Germany). Metabolite concentrations in steady-state chemostat cultures were analysed after rapid quenching of culture samples with cold steel beads (Mashego et al. 2003).
Whole-cell protein content was determined essentially as described by Verduyn et al. (1990). Exactly 20.0 ml of culture (2–4 g l−1 dry weight) was harvested from bioreactors by centrifugation (5 min, 10 000 g at 4°C) in an Avanti J-E centrifuge (Beckman Coulter Life Sciences, Indianapolis, IN), using a JA-25.50 rotor. The pellet was washed once, and the final pellet was resuspended in water to a final volume of 10.0 ml and stored at −20°C. After thawing at room temperature, samples were analysed with the alkaline copper sulphate method, using bovine serum albumin (Sigma-Aldrich, St. Louis, MO) as a standard.
Analysis of whole-cell lipid content was based on the method described by Izard and Limberger (2003). Phosphoric acid–vanillin reagent was prepared by adding 0.120 g of vanillin (Sigma-Aldrich) to 20 ml of demineralized water and adjusting the volume to 100 ml with 85% H3PO4. Biomass samples from bioreactors were obtained as described for whole-cell protein content determination. After thawing at room temperature, 100 µl of the cell suspension was added to a stoppered glass tube, together with 1 ml of concentrated H2SO4. Samples were boiled for 10 min and subsequently cooled for 5 min at room temperature in a water bath. Then, 2.5 ml of phosphoric acid–vanillin reagent were added, and samples were incubated for 15 min at 37°C. Samples were cooled for 10 min in a water bath at room temperature. Absorbance at 530 nm was measured with a Jenway 7200 spectrophotometer against a demineralized water sample. Lipid contents were calculated based on a dilution range of sunflower oil (Sigma-Aldrich) in chloroform. Prior to H2SO4 addition, glass tubes containing the standard solutions were placed in a water bath at 70°C until chloroform was fully evaporated, after which 100 µl of demineralized water was added for volume correction.
Whole-cell RNA content was quantified by determining absorbance at 260 nm of RNA degraded by alkali and extracted from the culture in HClO4 [adapted from Beck et al. (2018) and Benthin et al. (1991)]. Samples of 2 ml (4–8 mg biomass dry weight) were harvested from bioreactors and centrifuged at 4000 rpm for 10 min at 4°C in an Eppendorf 5810 R (Eppendorf Netherlands BV). Pellets were washed once with demineralized water and, after the second round of centrifugation, stored at −20°C. Pellets were thawed at room temperature and washed three times with 3 ml cold 0.7 M HClO4 and digested with 3 ml 0.3 M KOH for 60 min in a water bath set at 37°C, while vortexing briefly every 15 min. After addition 1 ml of 3 M HClO4, supernatant was collected and precipitates were washed twice with 4 ml cold 0.5 M HClO4. The collected combined supernatant was centrifuged to remove KClO4 precipitates. Absorbance at 260 nm was measured in quartz cuvettes using a Hitachi U-3010 spectrophotometer (Sysmex Europe GmbH, Norderstedt, Germany) against a standard prepared with RNA from yeast (Sigma-Aldrich).
Whole-cell total carbohydrate contents were quantified with a phenol–sulfuric acid-based method [based on Herbert et al. (1971) and Lange and Heijnen (2001)]. Briefly, 1 ml (2–4 mg biomass dry weight) samples from bioreactor cultures were harvested and frozen as described for whole-cell RNA analysis. Frozen samples were thawed on ice and resuspended in 10 ml demineralized water. 5 ml of concentrated H2SO4 was added to a mixture of 1 ml of resuspended sample and 1 ml of aqueous phenol solution (50 g l−1) in a stoppered glass tube. After boiling for 15 min and subsequent cooling to room temperature, absorbance at 488 nm was measured. Samples were measured against a 1:2 mannose:glucose standard. Results were corrected for presence of nucleic pentoses by using extinction coefficients of 0.36 and 0.26 for g l−1 RNA and DNA, respectively, and using the RNA content determined as described above and assuming a constant DNA cellular content of 0.5 wt-% (Lange and Heijnen 2001).
Phylogenetic tree construction
A phylogenetic tree of the studied yeast species was constructed by the neighbour-joining method (Saitou and Nei 1987). A bootstrap consensus tree inferred from 500 replicates was taken to represent the genealogy of the analysed taxa (Felsenstein 1985). Evolutionary distances were computed with the p-distance method (Nei and Kumar 2000) applied to LSU D1/D2 nucleotide sequences deposited in the Yeast IP database (Weiss et al. 2013). When the LSU D1/D2 sequence of a strain used in this study was not available, sequences of the type strain for the species were used instead. Ambiguous positions were removed for each sequence pair, resulting in a dataset with 635 positions in total. Evolutionary analyses were conducted in MEGA11 (Tamura et al. 2021).
Genomic analysis for genes involved in the synthesis of respiratory chain components
To infer presence and absence of genes encoding specific components of the mitochondrial respiratory chain, protein sequences of Neurospora crassa deposited at the National Center for Biotechnology Information (NCBI; https://www.ncbi.nlm.nih.gov/) or of the S. cerevisiae CEN.PK strain deposited at the Saccharomyces genome database (SGD; https://www.yeastgenome.org/) were used as queries in a tBLASTn search on the NCBI website using the default settings (Table S2). The reference sequence as denoted by NCBI was used when multiple genomic datasets were available.
Stoichiometric analysis with a core metabolic model
Growth stoichiometries of aerobic, ethanol-grown yeast cultures were analysed with a core metabolic network model based on S. cerevisiae as described by Daran-Lapujade et al. (2004). The stoichiometry matrix representing the core metabolic network (df = 2) consisted of 161 metabolites and 154 reactions divided over three compartments (Supporting Information 1). The matrix was solved assuming a steady state (S∙v = 0) with the rref function in MATLAB R2021b, expressing all metabolic fluxes as a function of growth rate and maintenance.
Data analysis
Time to stationary phase, specific growth rates, and corresponding standard deviations were calculated using Microsoft Excel. All other statistical analyses were performed with GraphPad Prism version 10.1.2.
Results
Semiquantitative comparison of growth kinetics on ethanol of a selection of budding yeast strains
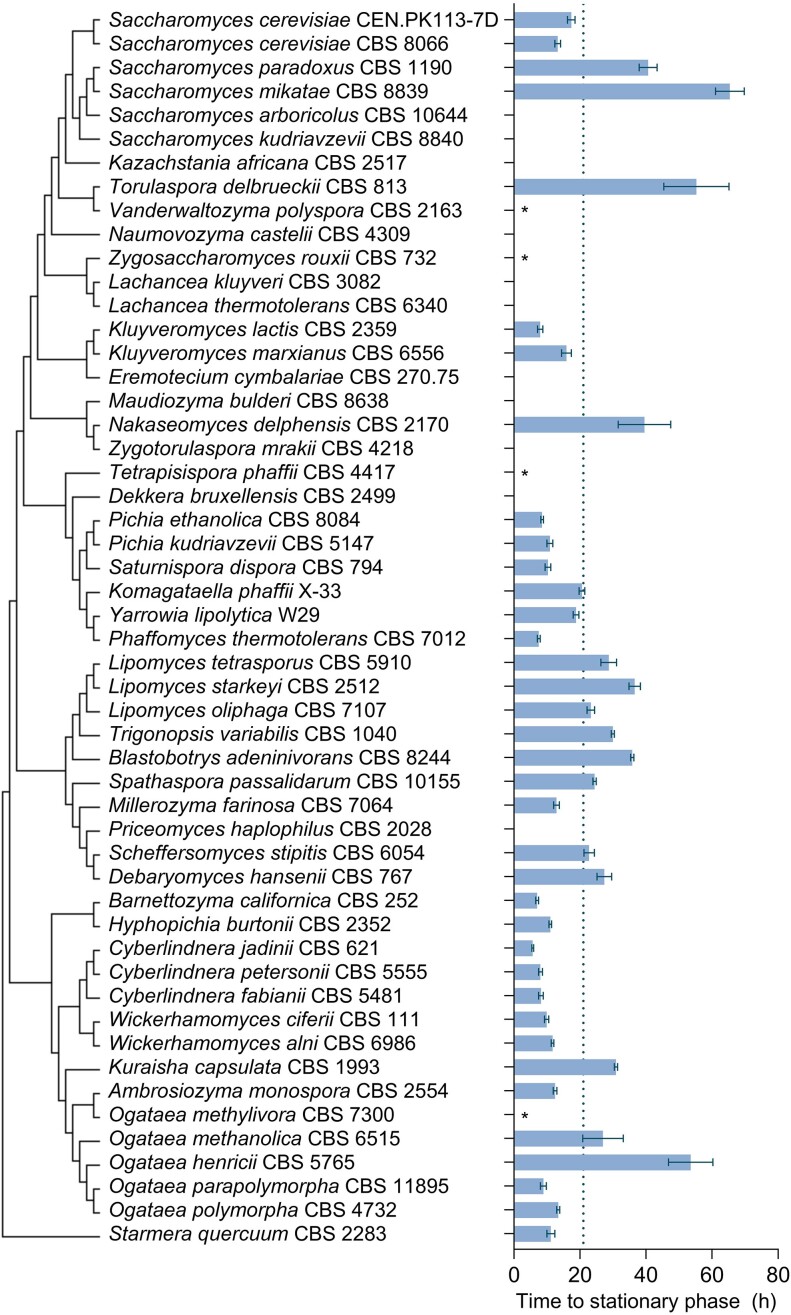
To assess diversity of ascomycete yeasts with respect to their specific growth rates on SM with ethanol, strains of 52 species covering multiple phylogenetic clades (Shen et al. 2018; Fig. 2) were selected for characterization in an automated, image-analysis-based growth profiler set-up (Duetz 2007). The selected species had previously been scored positive for growth on ethanol in taxonomic studies (Kurtzman et al. 2011). However, 8 of the 52 selected strains (Table S1) did not show significant growth on ethanol after >3 days incubation and were eliminated from further characterization (Dekkera bruxellensis CBS 2499, Kazachstania africana CBS 2517, Naumovozyma castellii CBS 4309, Priceomyces haplophilus CBS 2028, Saccharomyces arboricola CBS 10644, Maudiozyma bulderi CBS 8638, Saccharomyces kudriavzevii CBS 8840, and Zygotorulaspora mrakii CBS 4218). Three additional strains were excluded because they flocculated in ethanol-grown shake-flask precultures (Eremotecium cymbalariae CBS270.75, Lachancea kluyveri CBS 3082, and Lachancea thermotolerans CBS 6340). Growth-profiler experiments with the remaining 41 strains were performed at 30°C with SM supplemented with 3 g l−1 ethanol. Since image-analysis by the Growth Profiler set-up yields a nonlinear, strain-dependent relation between biomass concentration and read-out, TS was used as a proxy for growth rate (Fig. S1). The 41 strains showed TS values ranging from 5.7 ± 0.3 h for Cyberlindnera jadinii CBS 621 to 65 ± 4 h for Saccharomyces mikatae CBS 8839 (Fig. 2 and Fig. S2). Despite showing growth on ethanol in precultures, Zygosaccharomyces rouxii CBS 732, Vanderwaltozyma polyspora CBS 2163, Tetrapisispora phaffii CBS 4417, and Ogataea methylivora CBS 7300 showed no or extremely slow growth on ethanol in growth-profiler experiments (Fig. S2 and Table S1). Fast growth (TS values of 8 ± 2 h) was observed in all tested yeasts strains that belonged to the Phaffomycetales order (n = 8, genera Barnettozyma, Cyberlindnera, Phaffomyces, Starmera, and Wickerhamomyces).
Figure 2.
Time to stationary phase (TS) on synthetic medium with 3 g l−1 ethanol as sole carbon source of strains of different Saccharomycotina species. The data were obtained from batch cultures in microtiter plates grown at 30°C and at an initial pH of 6.0. TS was determined as the interval between the time point at which the green value measured by the Growth Profiler setup had doubled relative to its value immediately after inoculation, and the first time point at which a deceleration of the increase of the green value was observed (Figs S1 and S2). Bars represent means and standard deviation of at least four independent experiments for each yeast strain. For strains denoted with an asterisk (*) a doubling of the green value was not observed within the timeframe of the experiment (196 h). For strains without annotation, growth characteristics observed in inoculum shake flasks were the reason for exclusion from MTP screening (Table S1). Strains with a TS <21 h (cut-off value indicated with dotted line) were further characterized in shake-flask batch cultures (Fig. 3). Phylogenetic relationships are represented as a bootstrap consensus tree (500 replicates) inferred with the neighbour-joining method based on LSU D1/D2 sequences deposited in the Yeast IP database (Weiss et al. 2013).
Comparison of specific growth rates on ethanol in shake-flask cultures
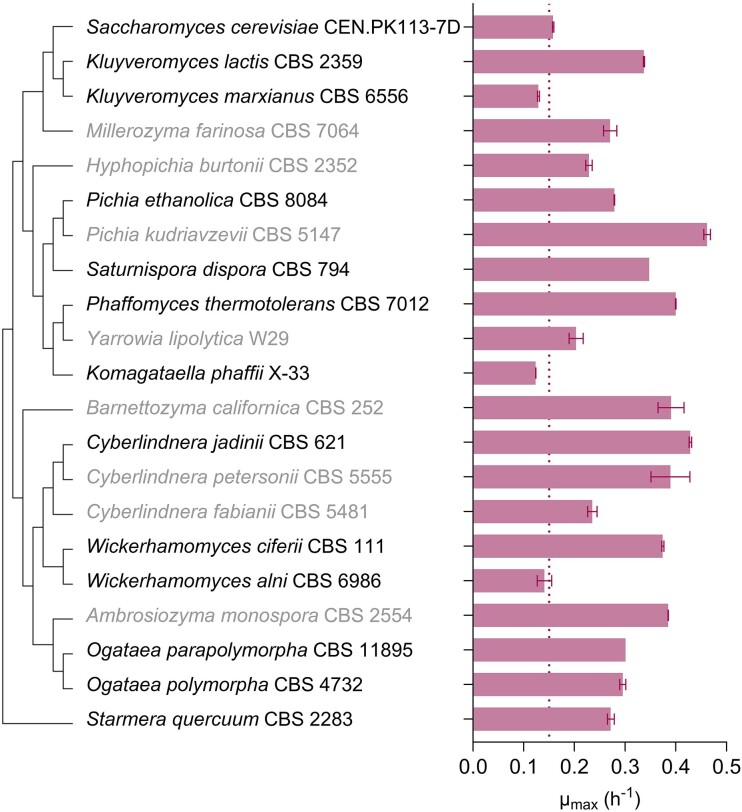
The 21 yeast strains that showed a TS below 21 h in the growth-profiler experiments were grown in shake-flask cultures for an accurate measurement of their specific growth rates on ethanol (Fig. 3). Specific growth rates of these strains on SM with 7.5 g l−1 ethanol ranged from 0.124 ± 0.001 h−1 for K. phaffii X-33 to 0.462 ± 0.007 h−1 for Pichia kudriavzevii CBS 5147 (Fig. 3 and Fig. S3, Table S1), corresponding to doubling times of 5.8 h and 1.5 h, respectively. As anticipated, specific growth rates measured in shake-flask cultures negatively correlated with growth-profiler TS values (Fig. S4). Outliers coincided with atypical growth characteristics in shake-flask cultures. For example, after inoculation, shake-flask cultures of P. kudriavzevii CBS 5147 showed an initial fast decrease in their optical density, which was followed by a short, extremely fast exponential growth phase (Fig. S3). Other ‘outlier’ strains showed a prolonged lag phase in shake-flask cultures and one exhibited pseudohyphal growth (i.e. Ambrosiozyma monospora CBS 2554), which precluded accurate growth-rate measurements (Table S1).
Figure 3.
Specific growth rates (µmax) of strains of selected Saccharomycotina species, calculated from optical density (OD660) measurements during the exponential growth phase of shake-flask batch cultures on SM with 7.5 g l−1 ethanol as sole carbon source. Cultures were grown at 30°C and at an initial pH of 6.0. Bars show means with standard deviations of specific growth rates calculated from independent duplicate experiments. Strains with µmax <0.15 h−1 (dotted line) were excluded from further characterization. Strains with names displayed in grey showed irregular growth characteristics and were also excluded from further characterization (Fig. S3 and Table S1). Phylogenetic relationships are represented as a bootstrap consensus tree (500 replicates) inferred with the neighbour-joining method based on LSU D1/D2 sequences deposited in the Yeast IP database (Weiss et al. 2013).
Based on the results from previous stage of the screening, i.e. the growth-profiler experiments (Fig. 2), all tested strains belonging to the Phaffomycetales order were selected for shake-flask characterization. Within the subgroup of faster-growing species that were selected for shake-flask characterization, the performance in terms of high growth rate of the Phaffomycetales strains was less pronounced. The average µmax of the Phaffomycetales strains was 0.33 ± 0.11 h−1, whilst the average µmax of all 21 strains tested in shake flasks was 0.29 ± 0.10 h−1. This difference is not statistically significant (unpaired two-tailed t-test, P = .42), unlike comparing the average TS of the Phaffomycetales strains (8 ± 2 h) to the average of the 37 strains, where TS could be quantified in the previous stage of the screening (21 ± 15 h, P = .02).
Selection of strains with different respiratory-chain composition based on genome data
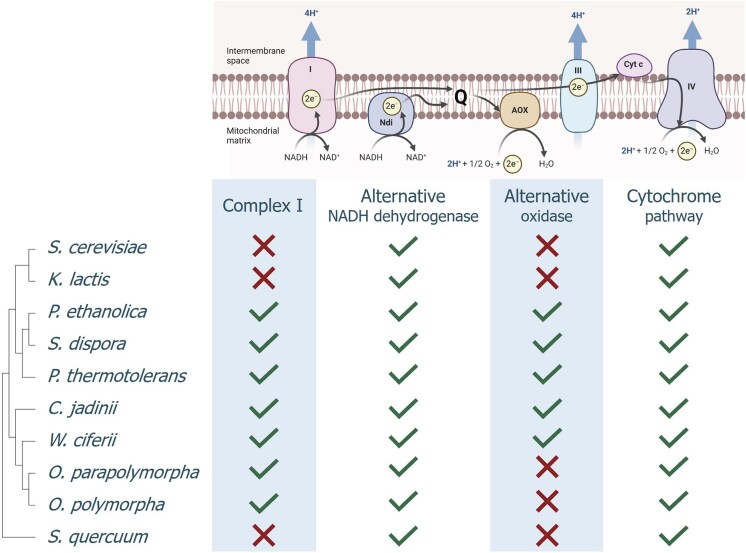
Composition of the mitochondrial ETS can significantly affect growth energetics of respiring yeast cultures. NADH oxidation by a mitochondrial proton-pumping NADH dehydrogenase complex (‘Complex I’) enables higher ATP yields from oxidative phosphorylation than NADH oxidation by non-proton-pumping, single subunit mitochondrial NADH dehydrogenases such as Ndi1 in S. cerevisiae (Juergens et al. 2020). Conversely, activity of a non-proton pumping AOX can decrease the ATP stoichiometry of mitochondrial respiration (Joseph-Horne et al. 2001, Guerrero-Castillo et al. 2012). We therefore assessed presence and absence of relevant genes in the genomes of 10 yeast species that showed specific growth rates on ethanol of at least 0.15 h−1 in shake-flask cultures (Figs 3 and 4, Table S2). Based on this analysis, three subgroups were identified: (i) absence of genes encoding Complex I and AOX (Saccharomyces cerevisiae, Kluyveromyces lactis, and Starmera quercuum), (ii) presence of genes encoding Complex I proteins and AOX (Pichia ethanolica, Saturnispora dispora, Phaffomyces thermotolerans, Cyberlindnera jadinii, and Wickerhamomyces ciferii), and (iii) presence of Complex I-encoding genes but absence of AOX genes (Ogataea parapolymorpha and Ogataea polymorpha). All three subgroups contained genetic homology with genes known to encode cytochrome c, subunits of ubiquinone:cytochrome c oxidoreductase (Complex III), and cytochrome c oxidase (Complex IV), as well as non-proton-translocating, single-subunit NADH dehydrogenases. S. cerevisiae CEN.PK113-7D and K. lactis CBS 2359 (both subgroup i), P. ethanolica CBS 8084 (formerly known as Candida ethanolica), S. dispora CBS 794, P. thermotolerans CBS 7012, and C. jadinii CBS 621 (subgroup ii, the latter two belonging to the Phaffomycetaceae family), and O. parapolymorpha CBS 11895 (subgroup iii) were selected as representatives for a quantitative analysis of biomass yields and biomass composition in chemostat cultures.
Figure 4.
Presence and absence of genes encoding key respiratory chain components in a selection of Saccharomycotina species. Phylogenetic relationships are represented as a bootstrap consensus tree (500 replicates) inferred with the neighbour-joining method based on LSU D1/D2 sequences deposited in the Yeast IP database (Weiss et al. 2013). Presence or absence of genes encoding respiratory-chain components was inferred from a tBLASTn search of genomic datasets available in the database of the NCBI (Table S2). Abbreviations: (Complex) I NADH: ubiquinone oxidoreductase type 1; Ndi NADH: ubiquinone oxidoreductase type 2; Q quinone; (Complex) III ubiquinone: cytochrome c oxidoreductase; Cyt c cytochrome c; AOX alternative oxidase; and (Complex) IV cytochrome c oxidase. Figure created with BioRender.com.
Biomass yields and biomass composition in ethanol-limited chemostat cultures
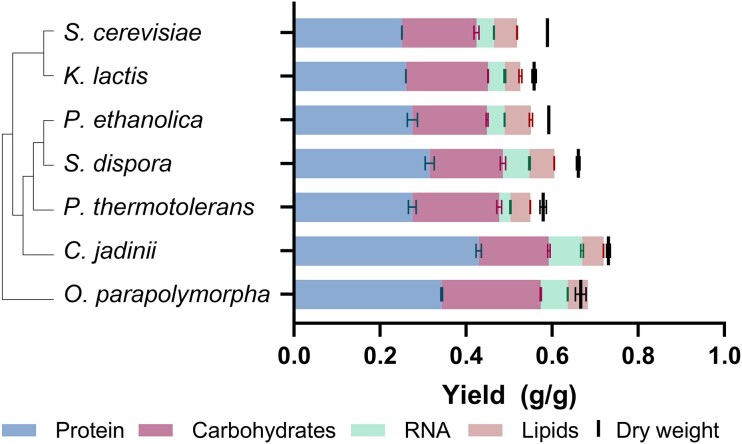
In ethanol-limited chemostat cultures, operated at a dilution rate of 0.10 h−1 (Fig. 5 and Table 1), biomass yields of the seven selected yeast strains ranged from 0.56 to 0.73 gbiomass gethanol−1. S. cerevisiae and K. lactis, both reported to lack a Complex I NADH dehydrogenase (Nosek and Fukuhara 1994; Fig. 4), showed similar biomass yields (0.589 ± 0.001 g g−1 and 0.558 ± 0.005 g g−1, respectively). Under the same conditions, higher biomass yields on ethanol were observed for the Complex-I-containing species S. dispora (0.660 ± 0.004 g g−1), O. parapolymorpha (0.67 ± 0.01 g g−1), and C. jadinii (0.730 ± 0.004 g g−1). Although genes encoding Complex-I subunits were also encountered in the genomes of P. thermotolerans and P. ethanolica, the biomass yield on ethanol of these strains at D = 0.10 h−1 (0.579 ± 0.007 g g−1 and 0.592 ± 0.001 g g−1, respectively) was more similar to that of S. cerevisiae and K. lactis than to that of S. dispora, O. parapolymorpha, and C. jadinii (Fig. S5). Biomass yields of the seven strains on oxygen ranged from 14.5 ± 0.1 to 22.1 ± 0.4 gbiomass∙[mol O2]−1 and, as anticipated, correlated positively with their biomass yields on ethanol (Table 1).
Figure 5.
Biomass yield and macromolecular biomass composition of seven different Saccharomycotina species grown in aerobic ethanol-limited chemostat cultures at 30°C, pH 5.0, and at a dilution rate of 0.10 h−1. Data are represented as mean and standard deviation of data from independent duplicate chemostat cultures for each yeast strain (Table 1). Phylogenetic relationships are represented as a bootstrap consensus tree (500 replicates) inferred with the neighbour-joining method based on LSU D1/D2 sequences deposited in the Yeast IP database (Weiss et al. 2013).
Table 1.
Physiological parameters of yeast species grown in aerobic ethanol-limited chemostat cultures at 30°C, pH 5.0, and at a dilution rate of 0.10 h−1. Residual ethanol concentrations were below the detection limit (3 mg l−1) in all cultures. The deviation in the last row indicates the difference between the overall dry weight biomass concentration and the sum of the four biomass components. Means and standard deviation were obtained from duplicate experiments.
| Species |
S. cerevisiae
CEN.PK113-7D |
K. lactis
CBS 2359 |
P. ethanolica
CBS 8084 |
S. dispora
CBS 794 |
P. thermotolerans
CBS 7012 |
C. jadinii
CBS 621 |
O. parapolymorpha
CBS 11895 |
|---|---|---|---|---|---|---|---|
| Actual dilution rate (h−1) | 0.098 ± 0.001 | 0.102 ± 0.005 | 0.100 ± 0.001 | 0.099 ± 0.004 | 0.101 ± 0.002 | 0.096 ± 0.001 | 0.103 ± 0.004 |
| YX/S (gX gS−1) | 0.589 ± 0.001 | 0.558 ± 0.005 | 0.592 ± 0.001 | 0.660 ± 0.004 | 0.579 ± 0.007 | 0.730 ± 0.004 | 0.67 ± 0.01 |
| Carbon recovery (%) | 99.2 ± 0.1 | 97.6 ± 0.7 | 100.9 ± 0.8 | 102.1 ± 0.7 | 100.1 ± 0.7 | 99.4 ± 0.2 | 99 ± 1 |
| qO2 (mmol O2 gX−1 h−1) | 6.39 ± 0.05 | 6.8 ± 0.3 | 6.3 ± 0.1 | 5.6 ± 0.1 | 6.9 ± 0.2 | 4.35 ± 0.04 | 5.33 ± 0.01 |
| qCO2 (mmol CO2 gX−1 h−1) | 3.44 ± 0.01 | 3.7 ± 0.1 | 3.60 ± 0.02 | 2.9 ± 0.1 | 3.77 ± 0.08 | 2.07 ± 0.02 | 2.67 ± 0.02 |
| Respiration quotient (mol CO2 mol O2−1) | 0.538 ± 0.005 | 0.53 ± 0.02 | 0.569 ± 0.005 | 0.521 ± 0.007 | 0.543 ± 0.001 | 0.476 ± 0.001 | 0.501 ± 0.003 |
| YX/O2 (gX mol O2−1) | 15.6 ± 0.1 | 15.1 ± 0.2 | 15.8 ± 0.4 | 17.8 ± 0.3 | 14.5 ± 0.1 | 22.1 ± 0.4 | 19.4 ± 0.7 |
| cethanol,in (g l−1) | 5.81 ± 0.01 | 3.80 ± 0.01 | 3.77 ± 0.02 | 3.76 ± 0.01 | 3.78 ± 0.01 | 3.77 ± 0.01 | 3.79 ± 0.01 |
| cbiomass (g l−1) | 3.43 ± 0.01 | 2.12 ± 0.01 | 2.23 ± 0.01 | 2.48 ± 0.02 | 2.19 ± 0.03 | 2.75 ± 0.01 | 2.53 ± 0.05 |
| Protein fraction (wt-%) | 42.6 ± 0.1 | 46.6 ± 0.5 | 46 ± 2 | 48 ± 1 | 47.5 ± 0.9 | 59 ± 2 | 51.5 ± 0.1 |
| Carbohydrate fraction (wt-%) | 29 ± 1 | 34.2 ± 0.2 | 29.3 ± 0.4 | 25.8 ± 0.8 | 35 ± 1 | 22.3 ± 0.3 | 34.5 ± 0.8 |
| RNA fraction (wt-%) | 6.86 ± 0.09 | 7.0 ± 0.2 | 6.84 ± 0.05 | 9.3 ± 0.3 | 4.5 ± 0.2 | 10.6 ± 0.4 | 9.38 ± 0.07 |
| Lipid fraction (wt-%) | 9.2 ± 0.1 | 6.5 ± 0.5 | 10.4 ± 0.6 | 8.75 ± 0.03 | 8.0 ± 0.2 | 6.86 ± 0.09 | 7.1 ± 0.1 |
| Deviation (%) | 12 ± 1 | 5.7 ± 0.1 | 7 ± 2 | 8 ± 2 | 5.2 ± 0.5 | 1.5 ± 0.6 | 3 ± 2 |
Because energy costs of protein, carbohydrates, nucleic acids, and lipid synthesis are different, comparisons of growth energetics should take into account macromolecular composition of biomass. Therefore, contents of protein, carbohydrates, RNA, and lipid were determined for biomass samples taken from chemostat cultures of the seven strains (Fig. 5). Of the four major macromolecular biomass constituents, protein is the most expensive in terms of ATP requirement (Verduyn et al. 1991). Protein content of the biomass of the seven yeasts varied from 43% to 59% (Table 1), with the highest protein content found in C. jadinii, which also showed the highest overall biomass yield. Conversely, C. jadinii biomass had the lowest carbohydrate content at 22.3 ± 0.3 wt-%, whilst the carbohydrate content for the other species ranged from 26 to 35 wt-%. The RNA content also varied among the species, with the highest content measured in C. jadinii biomass (10.6 ± 0.4 wt-%), and the lowest in P. thermotolerans (4.5 ± 0.2 wt-%). The lipid content seemed to have the smallest deviation between the species, ranging from 6.5 to 10.4 wt-%.
Theoretical analysis of growth energetics in ethanol-limited chemostat cultures
When expressed per mole of carbon, ethanol (C2H6O) has a degree of reduction (γ) of 6, while, when ammonium is used as nitrogen source (used as reference γ = 0), the generic elemental biomass composition CH1.8O0.5N0.2 corresponds to γ = 4.2 (Roels 1980). Assimilation of ethanol into biomass therefore leads to the generation of reducing equivalents (NAD(P)H and QH2), whose oxidation by mitochondrial respiration can generate ATP. A theoretical maximum biomass yield is reached when the ATP yield from respiration of the reduced cofactors generated in assimilation matches the ATP requirement in assimilation (Verduyn et al. 1991). In such a situation, assimilation generates enough redox cofactors to meet biosynthetic requirements for ATP and, consequently, complete dissimilation of additional ethanol to CO2 and water is not needed. The theoretical maximum yields of the S. cerevisiae, K. lactis, P. ethanolica, S. dispora, P. thermotolerans, C. jadinii, and O. parapolymorpha strains used in the chemostat experiments were calculated with a core stochiometric model of yeast metabolism (Table 2). Molecular and elemental biomass composition were based on the experimentally determined whole-cell contents of protein, RNA, carbohydrate, and lipid of the different yeast species when grown in ethanol-limited steady state chemostats at D = 0.10 h−1. DNA content was assumed to be 0.5 wt-% for all strains (Lange and Heijnen 2001). The oxidation of ethanol and acetaldehyde was modelled to generate cytosolic NADH. For all yeast species, a mechanistic H+/O ratio of 6 was assumed for NADH generated in the cytosol and for QH2 generated in the mitochondria. For NADH generated in the mitochondrial matrix, H+/O ratios of 6 and 10 were assumed for yeasts lacking and containing a Complex I NADH dehydrogenase, respectively. The theoretical maximum biomass scenario was simulated via minimising the H+/ATP ratio of the mitochondrial ATP synthase in the model, whilst preventing reverse electron flow predicted by the model as a lower boundary (Table S3). Growth-rate independent ATP turnover for cellular maintenance and growth-coupled ATP costs were excluded from these theoretical maximum biomass yield simulations.
Table 2.
Maximum theoretical biomass yields on ethanol as sole substrate, estimated with a stoichiometric model of the core metabolic network of yeast (Supporting Information 1). Elemental and macromolecular biomass compositions were based on measurements of protein, RNA, carbohydrates, and lipids in biomass samples from ethanol-limited chemostats (Table 1). DNA content was assumed constant at 0.5-wt% (Lange and Heijnen 2001). The third column displays for each yeast species the percentage that the experimentally determined yield makes up of the maximum theoretical yield.
| Biomass yield (g g−1) | |||
|---|---|---|---|
| Organism | Experimental | Theoretical maximum | Experimental/theoretical (%) |
| S. cerevisiae CEN.PK113-7D | 0.589 ± 0.001 | 0.826 | 71 |
| K. lactis CBS 2359 | 0.558 ± 0.005 | 0.831 | 67 |
| P. ethanolica CBS 8084 | 0.592 ± 0.001 | 0.823 | 72 |
| S. dispora CBS 794 | 0.660 ± 0.004 | 0.828 | 80 |
| P. thermotolerans CBS 7012 | 0.579 ± 0.007 | 0.824 | 70 |
| C. jadinii CBS 621 | 0.730 ± 0.004 | 0.829 | 88 |
| O. parapolymorpha CBS 11895 | 0.67 ± 0.01 | 0.835 | 80 |
Remarkably, the varying macromolecular composition of the yeast species had only little impact on the theoretical maximum yields, which range from 0.823 to 0.835 gbiomass gethanol−1(Table 2). This might indicate that the fraction of substrate dissimilated for the generation of the required amount of ATP in vivo has a much bigger impact on yields than the biomass composition itself. The Complex-I-negative yeasts S. cerevisiae and K. lactis reached 71% and 67%, respectively, of the theoretical maximum biomass yield in chemostat cultures. Similar percentages were calculated for P. ethanolica (72%) and P. thermotolerans (70%), despite the presence in its genome of genes encoding Complex-I subunits. Saturnispora dispora, O. parapolymorpha (both 80%) and, in particular, C. jadinii (88%), more closely approached the theoretical maximum biomass yield.
Discussion
The overwhelming majority of quantitative yeast physiology studies is based on the use of sugars as carbon and energy sources. By exploring a small sample of the biodiversity of ascomycete budding yeasts, this study revealed a wide range of growth rates in cultures grown on a synthetic medium with ethanol as sole carbon source. Ethanol-grown cultures of multiple tested strains matched or even exceeded the specific growth rate of ~0.4 h−1 that is observed for laboratory strains of S. cerevisiae when grown on SM with glucose (van Dijken et al. 2000). An enrichment of high growth rates was observed among all tested strains that belonged to the Phaffomycetales order, although fast growth was observed across the budding yeast subphylum. These results indicate that a broader exploration of intra- and interspecies diversity of yeasts with respect to growth rates on ethanol is highly relevant and may potentially yield even faster-growing strains. Some unexpected phenotypes observed in this study, such as the atypical growth kinetics of the fastest-growing strain P. kudriavzevii CBS 5147 (Fig. S3), merit further investigation.
K. lactis and S. cerevisiae, two yeasts whose genomes lack Complex-I genes, both showed a low biomass yield on ethanol in ethanol-limited chemostat cultures. Of five yeasts whose genomes do contain Complex-I genes, P. ethanolica and P. thermotolerans showed similarly low biomass yields, while biomass yields of C. jadinii, S. dispora, and O. parapolymorpha were significantly higher (Fig. 5 and Table 1). These results are consistent with the hypothesis that the presence of genes encoding a Complex-I proton-translocating NADH dehydrogenase is required but not sufficient for achieving high biomass yields on ethanol. Further research should reveal how growth conditions and/or medium composition affect synthesis of ETS components and biomass yields in ethanol-grown cultures and which additional levels of regulation are involved in distribution of electrons over proton-coupled and nonproton-coupled branches of yeast ETS. Such studies are not only of fundamental interest but can also identify metabolic engineering targets for improving growth energetics in industrial yeasts such as O. parapolymorpha (Juergens et al. 2020).
The biomass yield determined for C. jadinii (0.73 g g−1) in ethanol-limited cultures at a dilution rate of 0.10 h−1 is higher than previously reported (Verduyn et al. 1991). This high biomass yield, its high protein content (59 ± 2%) and its long-term application as ‘ fodder yeast’ in the previous century (Watteeuw et al. 1979, Lallemand 2024) make it a highly interesting candidate for single-cell protein from ethanol. A recent study in which biomass yields and protein contents of two bacterial and three yeast strains were compared in ethanol-grown shake-flask cultures (van Peteghem et al. 2022b) showed an even higher biomass yield (0.82 g g−1) for the related yeast species Cyberlindnera saturnus in batch cultures. The chemostat experiments in this study were performed at a relatively low growth rate (0.10 h−1). To predict performance of yeasts such as C. jadinii and O. parapolymorpha in industrial fed-batch cultures, an extended in-depth analysis of growth yield, kinetics, and energetics is required. In particular, such studies should cover a range of growth rates that are representative for the dynamic growth-rate regimes in large-scale industrial fed-batch processes.
The observed biomass yield of C. jadinii in ethanol-limited chemostat cultures approached the theoretical maximum (88%) at which all ATP requirements for biomass formation is met by respiration of the ‘excess’ reduced cofactors generated in biosynthetic reactions. This raises interesting questions on how ethanol oxidation is coupled to NADH oxidation via Complex I and on how cells allocate cellular resources to enable high fluxes through Complex I which, also in fungi, is a large, multisubunit protein complex with up to 35 subunits that has to be functionally assembled at and across the mitochondrial inner membrane (Videira 1998). Such studies are not only relevant for industrial application, but would also provide interesting models for systems biology studies on eukaryotic growth energetics and resource allocation.
This study indicates that, under a selected set of conditions, Saccharomycotina yeasts grown on ethanol exhibit considerable diversity in maximum specific growth rates, biomass yields, and biomass composition. Optimizing these parameters and, in particular, the energy coupling of ethanol dissimilation, by exploration of yeast biodiversity and/or metabolic engineering will therefore be highly relevant for optimizing the sustainability and profitability of yeast-based industrial biotechnological processes that use low-GHG-emission ethanol as sole carbon and energy source.
Supplementary Material
Acknowledgements
We kindly thank Chris Klomp for his help on the protein and RNA content determination protocols, and Niels Tiemersma for his help on the carbohydrate and lipid content determination protocols. Effie Leijten, Erik de Hulster, and Gabriela van Leersum are gratefully acknowledged for their assistance with the chemostat cultivations. Thank you to Marcel van den Broek for his advise on constructing the phylogenetic trees and to Walter van Gulik for his advise on working with the core metabolic model. Dr J.M. Nicaud is kindly acknowledged for providing the Yarrowia lipolytica W29 strain.
Contributor Information
Marieke Warmerdam, Department of Biotechnology, Delft University of Technology, Van der Maasweg 9, 2629 HZ Delft, the Netherlands.
Marcel A Vieira-Lara, Department of Biotechnology, Delft University of Technology, Van der Maasweg 9, 2629 HZ Delft, the Netherlands.
Robert Mans, Department of Biotechnology, Delft University of Technology, Van der Maasweg 9, 2629 HZ Delft, the Netherlands.
Jean Marc Daran, Department of Biotechnology, Delft University of Technology, Van der Maasweg 9, 2629 HZ Delft, the Netherlands.
Jack T Pronk, Department of Biotechnology, Delft University of Technology, Van der Maasweg 9, 2629 HZ Delft, the Netherlands.
Conflict of interest
None declared.
Funding
The PhD project of M.W. is funded from the NWO Stevin Prize awarded to J.T.P. by the Dutch Research Foundation. The work of M.A.V.L. was cofunded by DSM-Firmenich and from a supplementary grant ‘TKI-Toeslag’ for Topconsortia for Knowledge and Innovation (TKI’s) of the Netherlands Ministry of Economic Affairs and Climate Policy (CHEMIE.PGT.2021.003).
References
- Bakker BM, Overkamp KM, van Maris AJA et al. Stoichiometry and compartmentation of NADH metabolism in Saccharomyces cerevisiae. FEMS Microbiol Rev. 2001;25:15–37. 10.1111/j.1574-6976.2001.tb00570.x. [DOI] [PubMed] [Google Scholar]
- Beck AE, Hunt KA, Carlson RP. Measuring cellular biomass composition for computational biology applications. Processes. 2018;6:38. 10.3390/pr6050038. [DOI] [Google Scholar]
- Benthin S, Nielsen J, Villadsen J. A simple and reliable method for the determination of cellular RNA content. Biotechnol Tech. 1991;5:39–42. 10.1007/BF00152753. [DOI] [Google Scholar]
- Birdja YY, Pérez-Gallent E, Figueiredo MC et al. Advances and challenges in understanding the electrocatalytic conversion of carbon dioxide to fuels. In: Nature Energy. Vol. 4, London: Nature Publishing Group, 2019, 732–45. 10.1038/s41560-019-0450-y. [DOI] [Google Scholar]
- Cabau-Peinado O, Winkelhorst M, Stroek R et al. Microbial electrosynthesis from CO2 reaches productivity of syngas and chain elongation fermentations. Trends Biotechnol. 2024;42:1503–22. 10.1016/j.tibtech.2024.06.005. [DOI] [PubMed] [Google Scholar]
- Claes A, Deparis Q, Foulquié-Moreno MR et al. Simultaneous secretion of seven lignocellulolytic enzymes by an industrial second-generation yeast strain enables efficient ethanol production from multiple polymeric substrates. Metab Eng. 2020;59:131–41. 10.1016/j.ymben.2020.02.004. [DOI] [PubMed] [Google Scholar]
- Dagle RA, Winkelman AD, Ramasamy KK et al. Ethanol as a renewable building block for fuels and chemicals. Ind Eng Chem Res. 2020;59:4843–53. 10.1021/acs.iecr.9b05729. [DOI] [Google Scholar]
- Daran-Lapujade P, Jansen MLA, Daran JM et al. Role of transcriptional regulation in controlling fluxes in central carbon metabolism of Saccharomyces cerevisiae: a chemostat culture study. J Biol Chem. 2004;279:9125–38. 10.1074/jbc.M309578200. [DOI] [PubMed] [Google Scholar]
- Duetz WA. Microtiter plates as mini-bioreactors: miniaturization of fermentation methods. Trends Microbiol. 2007;15:469–75. 10.1016/j.tim.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Farrell AE, Plevin RJ, Turner BT et al. Ethanol can contribute to energy and environmental goals. Science. 2006;311:506–8. 10.1126/science.1121416. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the Bootstrap. Evolution. 1985;39:783–91. 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Fulton LM, Lynd LR, Körner A et al. The need for biofuels as part of a low carbon energy future. Biofuels Bioprod Bioref. 2015;9:476–83. 10.1002/bbb.1559. [DOI] [Google Scholar]
- Geijer C, Ledesma-Amaro R, Tomas-Pejo E. Unraveling the potential of non-conventional yeasts in biotechnology. FEMS Yeast Res. 2022;22. 10.1093/femsyr/foab071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Castillo S, Cabrera-Orefice A, Vázquez-Acevedo M et al. During the stationary growth phase, Yarrowia lipolytica prevents the overproduction of reactive oxygen species by activating an uncoupled mitochondrial respiratory pathway. Biochim Biophys Acta Bioenerget. 2012;1817:353–62. 10.1016/j.bbabio.2011.11.007. [DOI] [PubMed] [Google Scholar]
- Heffernan JK, Valgepea K, de Souza Pinto Lemgruber R et al. Enhancing CO2-valorization using Clostridium autoethanogenum for sustainable fuel and chemicals production. Front Bioeng Biotechnol. 2020;8. 10.3389/fbioe.2020.00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert D, Phipps PJ, Strange RE. Chapter III chemical analysis of microbial cells. Methods Microbiol. 1971;5:209–344. 10.1016/S0580-9517(08)70641-X. [DOI] [Google Scholar]
- Hermann BG, Blok K, Patel MK. Producing bio-based bulk chemicals using industrial biotechnology saves energy and combats climate change. Environ Sci Technol. 2007;41:7915–21. 10.1021/es062559q. [DOI] [PubMed] [Google Scholar]
- Izard J, Limberger RJ. Rapid screening method for quantitation of bacterial cell lipids from whole cells. J Microbiol Methods. 2003;55:411–8. 10.1016/S0167-7012(03)00193-3. [DOI] [PubMed] [Google Scholar]
- Jacobus AP, Gross J, Evans JH et al. Saccharomyces cerevisiae strains used industrially for bioethanol production. In: Essays in Biochemistry. Vol. 65, South Portland, ME: Portland Press Ltd, 2021, 147–61. 10.1042/EBC20200160. [DOI] [PubMed] [Google Scholar]
- Jansen MLA, Bracher JM, Papapetridis I et al. Saccharomyces cerevisiae strains for second-generation ethanol production: from academic exploration to industrial implementation. FEMS Yeast Res. 2017;17. 10.1093/femsyr/fox044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen NB, Strucko T, Kildegaard KR et al. EasyClone: method for iterative chromosomal integration of multiple genes in Saccharomyces cerevisiae. FEMS Yeast Res. 2014;14:238–48. 10.1111/1567-1364.12118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph-Horne T, Hollomon DW, Wood PM. Fungal respiration: a fusion of standard and alternative components. Biochim Biophys Acta Bioenerget. 2001;1504:179–95. 10.1016/S0005-2728(00)00251-6. [DOI] [PubMed] [Google Scholar]
- Juergens H, Hakkaart XDV, Bras JE et al. Contribution of Complex I NADH dehydrogenase to respiratory energy coupling in glucose-grown cultures of Ogataea parapolymorpha. Appl Environ Microbiol. 2020;86:86. 10.1128/AEM.00678-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtzman C, Fell JW, Boekhout T. The Yeasts: A Taxonomic Study. Amsterdam: Elsevier, 2011. [Google Scholar]
- Lallemand . Exploring Lallemand’s Torula Yeast: A Remarkable Journey. Milwaukee, WI, 2024. https://Bio-Lallemand.Com/Savory-Ingredients/Exploring-Lallemands-Torula-Yeast-Specialties/ (24 July 2024, date last accessed). [Google Scholar]
- Lange HC, Heijnen JJ. Statistical reconciliation of the elemental and molecular biomass composition of Saccharomyces cerevisiae. Biotech Bioeng. 2001;75:334–44. 10.1002/bit.10054. [DOI] [PubMed] [Google Scholar]
- Luttik MAH, Overkamp KM, Kötter P et al. The Saccharomyces cerevisiae NDE1 and NDE2 genes encode separate mitochondrial NADH dehydrogenases catalyzing the oxidation of cytosolic NADH. J Biol Chem. 1998;273:24529–34. 10.1074/jbc.273.38.24529. [DOI] [PubMed] [Google Scholar]
- Lynd LR, Beckham GT, Guss AM et al. Toward low-cost biological and hybrid biological/catalytic conversion of cellulosic biomass to fuels. In: Energy and Environmental Science. Vol. 15, London: Royal Society of Chemistry, 2022, 938–90. 10.1039/d1ee02540f. [DOI] [Google Scholar]
- Martin ME, Richter H, Saha S et al. Traits of selected Clostridium strains for syngas fermentation to ethanol. Biotechnol Bioeng. 2016;113:531–9. 10.1002/bit.25827/abstract. [DOI] [PubMed] [Google Scholar]
- Mashego MR, Van Gulik WM, Vinke JL et al. Critical evaluation of sampling techniques for residual glucose determination in carbon-limited chemostat culture of Saccharomyces cerevisiae. Biotech Bioeng. 2003;83:395–9. 10.1002/bit.10683. [DOI] [PubMed] [Google Scholar]
- Melo AMP, Bandeiras TM, Teixeira M. New insights into type II NAD(P)H:quinone oxidoreductases. Microbiol Mol Biol Rev. 2004;68:603–16. 10.1128/mmbr.68.4.603-616.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Kumar S. Molecular Evolution and Phylogenetics. Oxford: Oxford University Press, 2000. [Google Scholar]
- Nosek J, Fukuhara H. NADH dehydrogenase subunit genes in the mitochondrial DNA of yeasts. J Bacteriol. 1994;176:5622–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overkamp KM, Bakker BM, Steensma HY et al. Two mechanisms for oxidation of cytosolic NADPH by Kluyveromyces lactis mitochondria. Yeast. 2002;19:813–24. 10.1002/yea.878. [DOI] [PubMed] [Google Scholar]
- Phillips JR, Klasson KT, Clausen EC et al. Biological production of ethanol from coal synthesis gas. Appl Biochem Biotechnol. 1993;39-40:559–71. 10.1007/BF02919018. [DOI] [Google Scholar]
- Postma E, Verduyn C, Scheffers WA et al. Enzymic analysis of the crabtree effect in glucose-limited chemostat cultures of Saccharomyces cerevisiae. Appl Environ Microbiol. 1989;55:468–77. 10.1128/aem.55.2.468-477.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postmus J, Tuzun I, Bekker M et al. Dynamic regulation of mitochondrial respiratory chain efficiency in Saccharomyces cerevisiae. Microbiology. 2011;157:3500–11. 10.1099/mic.0.050039-0. [DOI] [PubMed] [Google Scholar]
- Pronk JT, Wenzel TJ, Luttik MAH et al. Energetic aspects of glucose metabolism in a pyruvate-dehydrogenase-negative mutant of Saccharomyces cerevisiae. Microbiology. 1994;140:601–10. 10.1099/00221287-140-3-601. [DOI] [PubMed] [Google Scholar]
- Roels JA. Simple model for the energetics of growth on substrates with different degrees of reduction. Biotech Bioeng. 1980;22:33–53. 10.1002/bit.260220104. [DOI] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25. 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Salim I, González-García S, Feijoo G et al. Assessing the environmental sustainability of glucose from wheat as a fermentation feedstock. J Environ Manage. 2019;247:323–32. 10.1016/j.jenvman.2019.06.016. [DOI] [PubMed] [Google Scholar]
- Shen XX, Opulente DA, Kominek J et al. Tempo and mode of genome evolution in the budding yeast subphylum. Cell. 2018;175:1533–1545.e20. 10.1016/j.cell.2018.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Kumar S. MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol Biol Evol. 2021;38:3022–7. 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilman D, Hill J, Lehman C. Carbon-negative biofuels from low-input high-diversity grassland biomass. Science. 2006;314:1598–600. 10.1126/science.1133306. [DOI] [PubMed] [Google Scholar]
- van Aalst ACA, de Valk SC, van Gulik WM et al. Pathway engineering strategies for improved product yield in yeast-based industrial ethanol production. Synth Syst Biotechnol. 2022;7:554–66. 10.1016/j.synbio.2021.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijken JP, Bauer J, Brambilla L et al. An interlaboratory comparison of physiological and genetic properties of four Saccharomyces cerevisiae strains. Enzyme Microb Technol. 2000;26:706–14. 10.1016/S0141-0229(00)00162-9. [DOI] [PubMed] [Google Scholar]
- van Peteghem L, Sakarika M, Matassa S et al. The role of microorganisms and carbon-to-nitrogen ratios for microbial protein production from bioethanol. Appl Environ Microbiol. 2022a;88:88. 10.1128/aem.01188-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Peteghem L, Sakarika M, Matassa S et al. Towards new carbon–neutral food systems: combining carbon capture and utilization with microbial protein production. Bioresour Technol. 2022b;126853:349. 10.1016/j.biortech.2022.126853. [DOI] [PubMed] [Google Scholar]
- Verduyn C, Postma E, Scheffers WA et al. Physiology of Saccharomyces cerevisiae in anaerobic glucose-limited chemostat cultures. J Gen Microbiol. 1990;136:395–403. 10.1099/00221287-136-3-395. [DOI] [PubMed] [Google Scholar]
- Verduyn C, Postma E, Scheffers WA et al. Effect of benzoic acid on metabolic fluxes in yeasts: a continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast. 1992;8:501–17. 10.1002/yea.320080703. [DOI] [PubMed] [Google Scholar]
- Verduyn C, Stouthamer AH, Scheffers WA et al. A theoretical evaluation of growth yields of yeasts. Antonie Van Leeuwenhoek. 1991;59:49–63. [DOI] [PubMed] [Google Scholar]
- Videira A. Complex I from the fungus Neurospora crassa. Biochim Biophys Acta. 1998;1364:89–100. [DOI] [PubMed] [Google Scholar]
- Watteeuw CM, Armiger WB, Ristroph DL et al. Production of single cell protein from ethanol by fed-batch process. Biotech Bioeng. 1979;21:1221–37. 10.1002/bit.260210711. [DOI] [Google Scholar]
- Weiss S, Samson F, Navarro D et al. YeastIP: a database for identification and phylogeny of Saccharomycotina yeasts. FEMS Yeast Res. 2013;13:117–25. 10.1111/1567-1364.12017. [DOI] [PubMed] [Google Scholar]
- Weusthuis RA, Aarts JMMJG, Sanders JPM. From biofuel to bioproduct: is bioethanol a suitable fermentation feedstock for synthesis of bulk chemicals?. Biofuels Bioprod Bioref. 2011;5:486–94. 10.1002/bbb.307. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.