Background
Cancer patients with suspected hereditary breast and ovarian cancer (HBOC) syndrome, along with their healthy relatives, can benefit from multigene panel testing. If they are identified as carriers of pathogenic/likely pathogenic variants (PV/LPVs) in high risk cancer genes, they may be offered prevention strategies such as enhanced cancer surveillance and discuss risk reducing surgeries to lower cancer burden [1]. Multigene panel testing may reveal not only PV/LPVs in BRCA1 and BRCA2 genes, but also in other HBOC-related genes.
For over a decade, it has been recognized that germline PV/LPVs in PALB2 (partner and localizer of BRCA2) are associated with an increased risk of breast cancer (BC). Rahman et al. reported in 2007 the 2.3 fold increase in BC risk in carriers of PALB2 PV/LPV in comparison to non-carriers [2]. Germline PV/LPVs in PALB2 are reported in up to 1% of BRCA1/2 negative breast cancer patients [3, 4]. Kotnik et al. performed a large population-based study in Slovenia between years 2014 and 2022. They identified PALB2 PV/LPVs in 0.13% of 7091 individuals who were referred for exome sequencing for various rare genetic conditions other than cancer [5]. Other malignancies, such as male breast cancer [6], pancreatic cancer (PaC) [7], ovarian (OC), prostate, colorectal, and gastric cancer [8] were also reported in carriers of germline PV/LPVs in PALB2. The risk estimates are, however, based on analysis of small patients’ cohorts, the existing literature presents conflicting data, and the statistical evidence remains weak. Notably, the largest study so far by Yang et al. on 524 families from 21 countries, demonstrated a substantial association between germline PALB2 PV/LPVs and ovarian cancer (RR 2.91), pancreatic cancer (RR 2.37), and male breast cancer (RR 7.34) [9]. Nevertheless, further studies are necessary to enhance our understanding.
The PALB2 gene encodes a protein that acts as a bridge between BRCA1 and BRCA2 proteins, playing a crucial role in homology-directed recombination DNA repair [10]. There is some evidence that new targeted treatments, which are effective in BRCA1 and BRCA2 PV/LPV carriers (such as poly-ADP-ribose polymerase (PARP) inhibitors), are also effective in individuals with PALB2 PV/LPVs, which is unsurprising given the shared underlying biology [11]. Understanding an individual’s PALB2 status is therefore essential for personalised management, not only in preventive setting, but also when making treatment decisions for these cancer patients.
The prevalence and spectrum of PV/LPVs may vary across different regions due to ethnic differences. Quantifying cancer risks associated with specific PV/LPVs and understanding the biological characteristics of malignancies in carriers with these variants is important for establishment of targeted clinical guidelines.
Institute of Oncology Ljubljana (IOL), where the study was conducted, is the principal national institution that supervises programs on comprehensive cancer care in Slovenia, which is a central European country with a population of two million. IOL offers cancer genetic counselling and genetic testing of high-risk individuals at the national level and therefore serves as a referral tertiary centre for the whole country.
In the Slovene HBOC cohort the prevalence and spectrum of germline PALB2 PV/LPVs have not yet been analysed and reported. Our study aimed to describe these PV/LPVs in PALB2 and analyse the types of cancer and age of cancer diagnosis in PALB2 PV/LPV carriers.
Methods
Patients
Multigene panel testing with next generation sequencing (NGS) for HBOC-related genes was introduced at the IOL in late 2014. The latest gene panel (in use since 2022) consists of nineteen genes: ATM, BARD1, BRCA1, BRCA2, BRIP1, CDH1, CHEK2, EPCAM, MLH1, MSH2, MSH6, NF1, PALB2, PMS2, PTEN, RAD51C, RAD51D, STK11, TP53. A proband (an index case) was defined as a family member (usually an affected individual) through whom a family with a PV/LPV is ascertained. If a PV/LPV was diagnosed in more than two seemingly unrelated families, it was considered a recurrent variant for the purpose of this study. Our retrospective study cohort encompassed 5099 individuals (4564 females and 535 males) from 4610 HBOC families who underwent genetic counselling in our Cancer Genetics Clinic at the Institute of Oncology Ljubljana and were referred to germline genetic testing between January 2015 and January 2022.
Family history data was collected from all tested families and all cancer diagnoses were verified in the Slovenian Cancer Registry. The registry contains data of all cancer diagnoses since 1950, when compulsory reporting of cancer diagnosis started in Slovenia. Positive family history for HBOC syndrome was defined as a family history of at least one first- or second-degree relative with breast, ovarian, prostate, or pancreatic cancer. We disregarded cases of non-melanoma skin cancer and cervical cancer in the analysis.
Family members of carriers of PV/LPVs were offered either cascade genetic testing for known PV/LPV in the family or NGS panel testing in case they fulfilled the inclusion criteria for panel testing. If the tested individuals gave their consent to report secondary findings (reporting the finding of a PV/LPV not initially suspected), secondary findings were also reported according to the ACMG criteria [12]. Patients who harboured variants of unknown significance were not included in the analysis.
All participants provided written informed consent. The present study was approved by the National Ethics Committee and the Institutional Ethics Committee of the Institute of Oncology Ljubljana (0120–591/2020/3 on the 20th of January 2021). Research was conducted according to the 1975 Helsinki Declaration as revised in 1983 and the procedures used met the ethical standards of these bodies.
DNA extraction
DNA was isolated from blood samples according to the established laboratory protocol as previously published [13].
Next generation sequencing
Next generation sequencing (NGS) was performed on Illumina MiSeqDx Sequencing System using TruSight Cancer Panel or TruSight Hereditary Panel (Illumina, San Diego, CA, USA) to enrich and sequence all translated exons and ± 25 bp flanking intronic regions of all HBOC panel genes. Bioinformatics and copy number analysis were performed as described by our group previously [14, 15]. Germline variants were classified for their clinical importance according to ACMG/AMP guidelines [12, 16]. Variants were described according to HGVS v20.05 nomenclature [17]. All PV/LPV germline variants in HBOC genes detected by NGS were additionally confirmed by Multiplex Ligation-dependent Probe Amplification analysis or Sanger sequencing as described by our group previously [13].
Haplotype analysis
All together 21 patients were tested for the presence of a common haplotype, 11 patients from 8 families with PALB2:c.509_510delGA p.(Arg170Ilefs*14) variant and 12 patients from 7 families with PALB2:c.1451T > A p.(Leu484*) variant. Seven STR markers (21732AC23, 23037GT23, D16S412, 23622TCTA14, 23749TATG14, D16S417, D16S401) spanning a region of 3 Mb on chromosome 16 in the vicinity of PALB2 gene, were genotyped using FAM labelled forward primers. PCR products were separated by capillary electrophoresis (ABI3500, ThermoFisher Scientific, Waltham, MA, USA). Detailed description of the primers and PCR conditions were published previously by Catucci and colleagues [18]. Haplotypes were puzzled together manually based on the PCR product length.
Copy number variant detection
Detection of copy number variants (deletion of single or multiple exons) from NGS data was performed using SeqNext v4.4.0 – v.5.2.0 (JSI medical systems, Ettenheim, Germany) as previously described by Klančar et al. [15].
PMS2 variant detection
Variants detected in the PMS2 gene were confirmed using long-range PCR (LongAmp Taq 2X Master Mix, New England Biolabs, Ipswich, MA, USA) followed by Sanger sequencing. To avoid amplifying the highly homologous PMS2 pseudogenes, primers for long-range PCR were specifically designed to amplify only the PMS2 gene. The method is described in detail by Clendenning and colleagues [19].
Statistical analysis
Statistical analysis was performed using SPSS software (version 25). We used descriptive statistics to describe patients’ clinical, pathological, and genetic characteristics.
Results
Study cohort
From January 1st 2015 to January 31st 2022, 5099 individuals (535 males and 4564 females) from 4610 families were tested for germline PV/LPVs in HBOC-related genes. The median age of individuals at the time of testing was 54 years. The characteristics of the cohort are shown in Supplementary Table 1.
PV/LPV detection rate among 4610 tested probands/families
In 19.1% (883/4610) of tested families a germline PV/LPV in HBOC-related genes was detected. BRCA1 PV/LPVs were detected in 8.4% (386/4610). Additionally, PV/LPVs were detected in BRCA2 in 4.9% (224/4610) of all probands, in CHEK2 in 1.8% (83/4610), in ATM in 1.5% (69/4610) and in PALB2 in 0.9% (40/4610). The frequency of all PV/LPVs in HBOC-related genes is presented in Supplementary Fig. 1.
PALB2 PV/LPVs were detected in 1.0% of all BRCA1/2 negative families (40/4000). In 22 out of 883 (2.5%) families, probands were diagnosed with two PV/LPVs and in one family (0.1%) one proband was diagnosed with three PV/LPVs in the HBOC-related genes.
PALB2 study cohort
We identified PALB2 PV/LPVs carriers in 40 HBOC families. Within these 40 families, a total of 60 family members were identified as carriers of a PALB2 PV/LPV.
Spectrum of PALB2 PV/LPVs in the Slovenian cohort of 40 PALB2 positive families
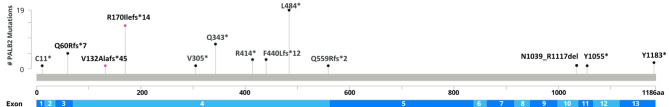
We identified 13 different PALB2 PV/LPVs, which are shown in Table 1; Fig. 1.
Table 1.
Spectrum of PALB2 PV/LPVs in the Slovenian population
| PALB2 PV/LPV type | Previously reported | ACMG/AMP and Variant class |
Variant Type | N of carriers | N of families (% of all families) |
|---|---|---|---|---|---|
|
c.1451T > A p.(Leu484*) |
yes |
PV Class 5 |
nonsense | 19 | 10 (24.5%) |
|
c.509_510del p.(Arg170Ilefs*14) |
yes |
PV Class 5 |
frameshift | 14 | 10 (24.5%) |
|
c.1027 C > T p.(Gln343*) |
yes |
PV Class 5 |
nonsense | 8 | 6 (14.7%) |
|
c.172_175del p.(Gln60Argfs*7) |
yes |
PV Class 5 |
frameshift | 5 | 4 (9.8%) |
|
c.3549 C > G p.(Tyr1183*) |
yes |
LPV Class 4 |
nonsense | 2 | 2 (4.9%) |
|
c.1317del p.(Phe440Leufs*12) |
yes |
PV Class 5 |
frameshift | 3 | 1 (2.4%) |
|
c.1240 C > T p.(Arg4*) |
yes |
PV Class 5 |
nonsense | 3 | 1 (2.4%) |
|
c.48G > A p.Cys11* |
yes |
PV Class 5 |
synonymous, splicing |
1 | 1 (2.4%) |
|
c.1676_1677delinsG p.(Gln559Argfs*2) |
yes |
PV Class 5 |
frameshift | 1 | 1 (2.4%) |
|
c.3164dup p.(Tyr1055*) |
yes |
PV Class 5 |
nonsense | 1 | 1 (2.4%) |
|
deletion of exons 11–12 c.(3113 + 1_3114-1)_(3350 + 1_3351-1)del p.? |
yes |
PV Class 5 |
multiple exon deletion | 1 | 1 (2.4%) |
|
c.395del p.(Val132Alafs*45) |
yes |
PV Class 5 |
frameshift | 1 | 1 (2.4%) |
|
c.912del p.(Val305*) |
no |
PV Class 5 |
nonsense | 1 | 1 (2.4%) |
| All PALB2 PV/LPVs | 60 | 40 (100%) |
Fig. 1.
A lollipop plot showcasing all PV/LPV in the PALB2 gene identified within our cohort. The plot was created using MutationMapper [20, 21]
PALB2 c.912 del p.(Val305*) is a novel variant and had previously not been reported in the literature. It was found in one proband (Table 1).
Four PV/LPVs were recurrent. The two most frequent were c.509_510del and c.1451T > A, detected in 10 different families each, together encompassing half (20/40, 50.0%) of all PV/LPVs detected in our population.
Haplotype analysis
Haplotype analysis was performed for both PALB2 c.509_510delGA and c.1451T > A variants. All 7 unrelated variant c.509_510delGA carriers share a common core haplotype of approximately 0.61 Mb in length. Recombination event presumably occurred between hg19 genomic coordinates chr16:23037671-chr16:23162662 (23037GT23 - D16S412) and chr16:23777202-chr16:24686016 (D16S417 - D16S401). Additionally, three of those unrelated carriers (Family 5, 7, 8) also share a larger common haplotype of at least 3 Mb (spanning over all 7 STR markers), that overlaps with a core haplotype. Neither the 3 Mb haplotype nor the 0.61 Mb core haplotype was detected in non-carriers, suggesting a common ancestor. Data is shown in Supplementary Fig. 2.
Additionally, all tested carriers of pathogenic variant c.1451T > A share a distinct haplotype spanning across 0.15 Mb. The recombination events occurred between coordinates chr16:23162662-chr16:23622400 (D16S412–23622TCTA14) and chr16:23777202-chr16:24686016 (D16S417 - D16S401), which is shown in Supplementary Fig. 3. This common haplotype suggests that the PALB2 c.1451T > A variant originated from a single ancestor.
Cancer types diagnosed in PALB2 PV/LPV carriers
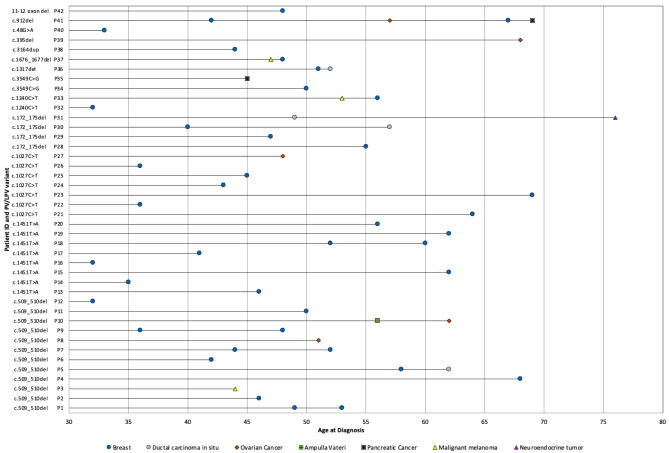
Out of the 60 PALB2-positive patients, 42 (70.5%) were diagnosed with at least one type of cancer (2 males, 40 females). The distribution of cancer types and ages at diagnosis are illustrated in Fig. 2. The median age at the diagnosis of the first malignancy was 47 years (range 32 years – 69 years).
Fig. 2.
PV/LPV variant, type of cancer and age at diagnosis in PALB2 PV/LPV carriers
Among 36 PALB2 positive BC (invasive and in situ) patients, 35 were females and one was male. The median age at BC diagnosis was 46.5 years, ranging from 32 years to 69 years. The median age at genetic testing was 51 years, ranging from 21 years – 79 years. Twelve different PV/LPVs were identified in PALB2 positive BC patients.
Clinical characteristics of double heterozygotes
Among PALB2 PV/LPV carriers, three patients were classified as double heterozygotes (DH). Their clinical characteristics are presented in Table 2.
Table 2.
Characteristics of double heterozygotes
| Patient (sex) | PALB2 PV/LPV | Other HBOC gene PV/LPV | Cancer type (age at diagnosis) |
Histopathological Characteristics |
|---|---|---|---|---|
| Patient 43 (f) |
deletion of exons 11–12 c.(3113 + 1_3114-1)_(3350 + 1_3351-1)del p.? |
ATM c.2413 C > T p.(Arg805*) |
BC (48) | poorly differentiated, bifocal, luminal B IDC |
| Patient 17 (f) |
c.1451T > A p.(Leu484*) |
CHEK2 c.1100del p.(Thr367Metfs*15) |
BC (32) | poorly differentiated, triple negative IDC |
| Patient 42 (f) |
c.912del p.(Val305*) |
BRCA1 c.181T > G p.(Cys61Gly) |
BC (42) | atypical medullary |
| OC (57) | HGSC | |||
| BC (67) | unknown histology | |||
| PaC (69) | adenocarcinoma |
Legend: PV/LPV = pathogenic variant/likely pathogenic variant, BC = breast cancer, OC = ovarian cancer, PaC = pancreatic cancer, HGSC = high-grade serous carcinoma, FIGO = the International Federation of Gynecology and Obstetrics, f = female, m = male
Multiple primary cancers
In total, 12 out of 42 (28.6%) PALB2 positive patients with cancer were diagnosed with more than one malignant tumor. Among them, 7/12 had negative family history and 3/7 had their first cancer diagnosed after the age of 50 years. Median interval between first diagnosis and a new primary cancer was 8 years (range 1–18 years). Characteristics of patients with multiple primary malignancies are presented in Table 3.
Table 3.
Characteristics of PALB2 positive patients with multiple primary malignancies
| Patient (sex) | Patient with PV/LPVs | Type of Cancer (laterality, age at diagnosis) | Family history (number of affected family members) |
|---|---|---|---|
| Patient 33 (f) | PALB2 c.1240 C > T p.(Arg414*) | melanoma (53), BC (left, 56) | positive (1) |
| Patient 10 (f) | PALB2 c.509_510del p.(Arg170Ilefs*14) | papilla Vateri (56), fallopian tube (62) | negative |
| Patient 37 (f) | PALB2 c.1676_1677delinsG p.(Gln559Argfs*2) | melanoma (47), BC (right, 48) | positive (1) |
| Patient 31 (f) | PALB2 c.172_175del p.(Gln60Argfs*7) | DCIS (right, 49), NET origo ignota (67) | negative |
| Patient 42 (f) |
PALB2 c.912del p.(Val305*) BRCA1 c.181T > G p.(Cys61Gly) |
BC (right, 42), (left, 67), OC (57), PaC (69) | positive (1) |
| Patient 1 (f) | PALB2 c.509_510del p.(Arg170Ilefs*14) | BC (left, 49, (left, 53) | positive (3) |
| Patient 7 (f) | PALB2 c.509_510del p.(Arg170Ilefs*14) | BC (left, 44), (right, 52) | negative |
| Patient 5 (f) | PALB2 c.509_510del p.(Arg170Ilefs*14) | BC (left, 58), (right, 62) | negative |
| Patient 9 (f) | PALB2 c.509_510del p.(Arg170Ilefs*14) | BC (left, 36), (right, 48) | negative |
| Patient 18 (f) | PALB2 c.1451T > A p.(Leu484*) | BC (right, 52), (left, 60) | negative |
| Patient 36 (f) | PALB2 c.1317del p.(Phe440Leufs*12) | BC (right, 51), BC (right, 52), DCIS (left, 52) | negative |
| Patient 30 (f) | PALB2 c.172_175del p.(Gln60Argfs*7) | BC (right, 40), (right, 57) | positive (2) |
Legend: PV/LPV = pathogenic variant/likely pathogenic variant, BC = breast cancer, OC = ovarian cancer, DCIS = ductal carcinoma in situ, NET = neuroendocrine tumor, PaC = pancreatic cancer; f - female
As shown in Table 3, one carrier of PALB2 PV/LPV, who also harboured a BRCA1 PV/LPV, was diagnosed with four primary cancers: twice with BC (at 42 and 67 years), a high-grade serous carcinoma of the ovary (age at diagnosis 57) and a pancreatic adenocarcinoma (age at diagnosis 69). Additionally, two PALB2 positive patients both developed BC (aged 48 and 56, respectively) and melanoma (aged 47 and 53, respectively).
PALB2 positive carriers without cancer diagnosis
Our cohort of PALB2 PV/LPV carriers without a cancer diagnosis consisted of 18 individuals from 11 families. Thirteen were female and five were male. Two of them were identified through panel testing and 16 by cascade testing. Median age at genetic testing was 44 years (range 21 – to 75 years).
Among these individuals, PALB2 c.1451T > A p.(Leu484*) PV/LPV was found in 11 individuals from six families, PALB2 c.509_510del (p.(Arg170Ilefs14)) was found in two individuals from two different families, PALB2 c.1317del p.(Phe440Leufs12) was found in two individuals from the same family, and one individual was identified with each of the following PV/LPVs: PALB2 c.1240 C > T p.(Arg414*), PALB2 c.1027 C > T p.(Gln343*), and PALB2 c.172_175del p.(Gln60Argfs*7).
Discussion
Having epidemiological data on the frequency and spectrum of germline PV/LPVs associated with different hereditary cancers is of the utmost importance for every country aiming to organize an optimal cancer prevention programme and optimize cancer patients’ management. Very little is known about the characteristics of carriers of PALB2 PV/LPVs and the clinicopathological characteristics of tumors diagnosed in these patients. This gap in knowledge underscores the importance of further research into PALB2-associated cancers, not only for specialized institutions but also for primary care physicians.
Detection rate and spectrum of PALB2 PV/LPVs
PV/LPVs in PALB2 were diagnosed in 0.9% of all individuals tested, making PALB2 the fifth most commonly mutated gene in our cohort. This was expected as it had previously been reported that 0.2–0.9% of women with BC who undergo genetic testing will carry germline PV/LPV in PALB2 [22].
In 40 families 13 different PALB2 PV/LPVs were detected. PALB2 c.912 del p.(Val305*) had not been reported previously and was found in one proband. Newly described PALB2 PV/LPVs in patients with BC are important since they can contribute to international databases and patients may benefit from prevention and treatment options.
Recurrent PV/LPVs in PALB2 in the Slovenian population
Four PV/LPVs in PALB2 were recurrent in our population, with PALB2 c.509_510del and PALB2 c.1451T > A being the two most frequent, detected in 10 different families each, and together encompassing half (20/40 or 50.0%) of all detected PV/LPVs in our population. Different recurrent PALB2 PV/LPVs have been reported in populations around the world, such as those from Argentina [23], Finland [24], Greece [25], and Poland [26]. PALB2 c.509_510del has been described by Noskowitz et al. as being present in about 1 in 400 unselected breast cancer patients from Central Europe (Germany) and Eastern Europe (Belarus, Russia) [26]. PALB2 c.509_510del has also been described as a recurrent variant in BC and OC patients from Poland [27]. We found no reports on the presence of PALB2 c.509_510del in Western European, Asian or American populations or no genotype-phenotype correlation studies for this specific PV. Of note, three patients from our cohort were diagnosed with a metachronous contralateral BC, one additional with a metachronous contralateral and ipsilateral BC. We found two cases of OC with a median age at diagnosis 56.5 years (range 51 years – 62 years), one case of carcinoma of the papilla Vateri and one malignant melanoma. Based on our data BC patients harbouring PALB2 c.509_510del are at high risk of developing a second BC and OC, making them high-risk group among PALB2 PV carriers, therefore enhanced surveillance is warranted.
PALB2 c.1451T > A has not yet been described as a recurrent variant in any population in the literature, however it has been found in nineteen individuals from ten different families in our cohort, which makes it a unique recurrent variant in the Slovenian population. Among nineteen carriers of the above-mentioned PV, eight were diagnosed with BC, with a median age at diagnosis 49 years. While it is challenging to establish genotype-phenotype correlation, this information could still be valuable in everyday clinical practice and may be included in the studies with bigger sample sizes.
Malignancies among PALB2 PV/LPV carriers
It has been known for more than a decade that PALB2 PV/LPVs increase BC risk [28]. The risk is 2–30 times higher than in the general population, depending on the type of PV/LPV, age, and family history and PALB2 is considered a high-penetrance susceptibility gene for BC [29]. As expected, the most common malignancy diagnosed in our cohort was BC in 36 patients, which confirms this association. Recent research has strengthened the correlation between PALB2 PV/LPVs and OC. Yang et al. demonstrated a substantial association between germline PALB2 PV/LPVs and OC with a risk ratio of around 3 and the lifetime risk of OC estimated to be around 3–5% [9]. OC with 5 cases was indeed the second most prevalent malignancy in our study. Two PALB2 PV/LPV carriers in our cohort (2/60, 3.3%) were diagnosed with PaC and additional carrier with carcinoma of Papilla Vateri. It is estimated that 3–4% of patients with famillial PaC are expected to harbour PALB2 PV/LPV. In the newest version of the National Comprehensive Cancer Network (NCCN) Guidelines enhanced screening not only for BC (with possible risk reducing surgeries), but also for OC (risk reducing surgery is offered as an option) and PaC is recommended [30].
Double heterozygotes
The increased use of multigene panels in the recent years has led to identification of individuals harbouring more than one PV/LPV in cancer susceptibility genes, although the data is still scarce [31]. The results of the studies on DH PV in HBOC-related genes in populations other than Ashkenazi Jews have been conflicting and inconclusive. To the best of our knowledge no research has been conducted on DH with one of the variants being PALB2 PV/LPV. There is emerging evidence of multiplicative effect of presence of PV/LPVs in more than one cancer susceptibility gene. Heidemann et al. showed that Caucasian female DH for BRCA1/2 seem to develop BC at a younger age and have more severe disease than carriers of a single BRCA1/2 PV/LPV [32]. Similarly, Sokolenko et al. pointed out that the presence of additional gene defect in female BRCA1 PV carriers may further increase their chances for cancer [33]. On the other hand, Lavie et al. suggest that DH PV in BRCA1/2 in females of Ashkenazi Jewish heritage does not seem to cause a more severe phenotype than in cases where only one of the genes is implicated [31].
Among PALB2 PV/LPV carriers, three double heterozygotes were identified, with the second PV/LPV located in one of the other hereditary BC genes. Although all three patients had early-onset BC (< 50 years), the contribution of each PV/LPV likely varied in terms of causality. PV/LPVs in BRCA1 and PALB2 are highly penetrant, associated with a strong genetic effect, and significantly increase the risk of BC and OC. In contrast, PV/LPVs in CHEK2 are considered moderately penetrant, with a much lower genetic effect compared to PALB2 and BRCA1.
In our cohort, one double heterozygote also carried the CHEK2 variant c.1100del together with PALB2 PV. For females with this variant, the odds ratio (OR) for breast cancer risk is 2.66, indicating a moderate risk. Hinić et al. suggest that individuals with biallelic CHEK2 PVs display a more severe phenotype than heterozygous carriers, with earlier onset, more cancer cases, and more instances of multiple cancers [34].
Strengths and limitations of our study
There are several limitations of our study. The expected population burden of PALB2 PV/LPVs in the Slovenian population is 0.13% [5]. The absolute number of included PV/LPV carriers was small (60). 42 (70.0%) had a cancer diagnosis. However, our cohort represents 2% of the expected population of PALB2 PV/LPV carriers in Slovenia. Also, in comparison to the literature, where mostly case series are described, this is a large cohort of PALB2 PV/LPV carriers. The subgroups of patients with different PV/LPV were very small, therefore we were not able to analyse them separately. The retrospective collection of data is always unfavourable, since data can be missing or inappropriately understood, however as it can be seen from our data, the information regarding patients’ and tumours’ characteristics was complete for patients in our study.
Its strength lies in a reliable family history, with information obtained from the Slovenian National Cancer Registry. It is one of the oldest Registries in Europe, where the diagnoses are cross-checked with histopathological reports.
In cases of rare genetic diseases large multicentric studies are required to achieve a substantial number of patients and we hope these will be able to benefit from our cohort of PALB2 PV/LPV carriers.
Conclusions
This report provides the first comprehensive insight into the genotype and phenotype of PALB2 PV/LPV carriers in Slovenia. The frequency of PALB2 PV/LPV in Slovenia is consistent with rates reported in other countries, accounting for 1.0% of all individuals tested for PVs in HBOC-related genes. Notably, we identified four recurrent PALB2 PV/LPVs within our population, collectively encompassing more than half of all affected families. Of particular interest is the PALB2 c.1451T > A variant, which has not been previously documented as a recurrent variant in any population, rendering it unique to Slovenia. The most common malignancy in our cohort was as expected BC, followed by OC and PaC, which adds to the existing evidence of PALB2 involvement in the pathogenesis of these cancer types. Despite the rarity of PALB2 carriers, who also carry PV/LPVs in other HBOC-related genes, we have identified three such individuals, and studying their disease characteristic can help elucidate the biological effect of being a DH. Overall, the results of our study provide valuable genotype and phenotype data from PALB2 positive patients which may already be utilized in a population specific assessment.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to thank all the patients and their family members who participated in this study. Also, they gratefully acknowledge the contribution made by the staff of the Department of Clinical Cancer Genetics ant the Department of Molecular Diagnostics, Institute of Oncology Ljubljana. The study was supported by the Slovenian Research Agency, program numbers: P3-0289.
Author contributions
Conceptualization: VAM, MK, Methodology: MK, KDS, SH, VŠD, VS, SN; Formal analysis and investigation: VŠD, PŠ, SN, VS, VAM, MK; Writing - original draft preparation: VAM; Writing - review and editing: MK, AB, KS, KDS, VŠD, VS, SN. All authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval
All participants provided written informed consent. The present study was approved by the National Ethics Committee and the Institutional Ethics Committee of the Institute of Oncology Ljubljana (0120–591/2020/3 on the 20th of January 2021). Research was conducted according to the 1975 Helsinki Declaration as revised in 1983 and the procedures used met the ethical standards of these bodies.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kapoor NS, Curcio LD, Blakemore CA, Bremner AK, McFarland RE, West JG, et al. Multigene Panel Testing Detects Equal Rates of Pathogenic BRCA1/2 Mutations and has a Higher Diagnostic Yield Compared to Limited BRCA1/2 Analysis Alone in Patients at Risk for Hereditary Breast Cancer. Ann Surg Oncol. 2015;22:3282–8. [DOI] [PubMed] [Google Scholar]
- 2.Rahman N, Seal S, Thompson D, Kelly P, Renwick A, Elliott A, et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007;39:165–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Woodward ER, van Veen EM, Forde C, Harkness EF, Byers HJ, Ellingford JM, et al. Clinical utility of testing for PALB2 and CHEK2 c.1100delC in breast and ovarian cancer. Genet Sci. 2021;23:1969–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu Y, Ouyang T, Li J, Wang T, Fan Z, Fan T, et al. Spectrum and clinical relevance of PALB2 germline mutations in 7657 Chinese BRCA1/2-negative breast cancer patients. Breast Cancer Res Treat. 2020;179:605–14. [DOI] [PubMed] [Google Scholar]
- 5.Kotnik U, Maver A, Peterlin B, Lovrecic L. Assessment of pathogenic variation in gynecologic cancer genes in a national cohort. Sci Rep. 2023;13:1–9. 10.1038/s41598-023-32397-8 [DOI] [PMC free article] [PubMed]
- 6.Pritzlaff M, Summerour P, McFarland R, Li S, Reineke P, Dolinsky JS, et al. Male breast cancer in a multi-gene panel testing cohort: insights and unexpected results. Breast Cancer Res Treat. 2017;161:575–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu Y, Guo M. Synthetic lethality strategies: Beyond BRCA1/2 mutations in pancreatic cancer. Cancer Sci. 2020;111:3111–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foulkes WD. Inherited Susceptibility to Common Cancers. N Engl J Med. 2008;359:2143–53. [DOI] [PubMed] [Google Scholar]
- 9.Yang X, Leslie G, Doroszuk A, Schneider S, Allen J, Decker B, et al. Cancer risks associated with germline PALB2 pathogenic variants: An international study of 524 families. J Clin Oncol. 2020;38:674–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walsh CS. Two decades beyond BRCA1/2: Homologous recombination, hereditary cancer risk and a target for ovarian cancer therapy? Gynecol Oncol. 2015;137:343–50. [DOI] [PubMed] [Google Scholar]
- 11.Castroviejo-Bermejo M, Cruz C, Llop‐Guevara A, Gutiérrez‐Enríquez S, Ducy M, Ibrahim YH, et al. A RAD 51 assay feasible in routine tumor samples calls PARP inhibitor response beyond BRCA mutation. EMBO Mol Med. 2018;10:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. May 2015;17:405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stegel V, Blatnik A, Škof E, Dragoš VŠ, Krajc M, Gregorič B, et al. Real-World Data on Detection of Germline and Somatic Pathogenic/Likely Pathogenic Variants in BRCA1/2 and Other Susceptibility Genes in Ovarian Cancer Patients Using Next Generation Sequencing. Cancers (Basel). 2022;14:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gornjec A, Novakovic S, Stegel V, Hocevar M, Pohar Marinsek Z, Gazic B, et al. Cytology material is equivalent to tumor tissue in determining mutations of BRCA 1/2 genes in patients with tubo-ovarian high grade serous carcinoma. BMC Cancer. 2019;19:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klančar G, Blatnik A, Dragoš VŠ, Vogrič V, Stegel V, Blatnik O et al. A novel germline MLH1 in-frame deletion in a Slovenian lynch syndrome family associated with uncommon isolated PMS2 loss in tumor tissue. Genes (Basel). 2020;11. [DOI] [PMC free article] [PubMed]
- 16.Plon SE, Eccles DM, Easton D, Foulkes WD, Genuardi M, Greenblatt MS, et al. Sequence variant classification and reporting: recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum Mutat. 2008;29:1282–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.den Dunnen JT, Dalgleish R, Maglott DR, Hart RK, Greenblatt MS, Mcgowan-Jordan J, et al. HGVS Recommendations for the Description of Sequence Variants: 2016 Update. Hum Mutat. 2016;37:564–9. [DOI] [PubMed] [Google Scholar]
- 18.Catucci I, Casadei S, Ding YC, Volorio S, Ficarazzi F, Falanga A, et al. Haplotype analyses of the c.1027C>T and c.2167_2168delAT recurrent truncating mutations in the breast cancer-predisposing gene PALB2. Breast Cancer Res Treat. 2016;160:121–9. [DOI] [PMC free article] [PubMed]
- 19.Clendenning M, Hampel H, LaJeunesse J, Lindblom A, Lockman J, Nilbert M, et al. Long-range PCR facilitates the identification of PMS2-specific mutations. Hum Mutat. 2006;27:490–5. [DOI] [PubMed] [Google Scholar]
- 20.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6. [DOI] [PMC free article] [PubMed]
- 21.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio Cancer Genomics Portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu C, Hart SN, Gnanaolivu R, Huang H, Lee KY, Na J, et al. A Population-Based Study of Genes Previously Implicated in Breast Cancer. N Engl J Med. 2021;384:440–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gonzalez A, Del Greco F, Vargas-Roig L, Brun B, Tabares G, Mampel A, et al. PALB2 germline mutations in a multi-gene panel testing cohort of 1905 breast-ovarian cancer patients in Argentina. Breast Cancer Res Treat. 2022;2:403–12. [DOI] [PubMed] [Google Scholar]
- 24.Erkko H, Xia B, Nikkilä J, Schleutker J, Syrjäkoski K, Mannermaa A, et al. A recurrent mutation in PALB2 in Finnish cancer families. Nature. 2007;446:316–9. [DOI] [PubMed] [Google Scholar]
- 25.Vagena A, Papamentzelopoulou M, Kalfakakou D, Kollia P, Papadimitriou C, Psyrri A, et al. PALB2 c.2257C > T truncating variant is a Greek founder and is associated with high breast cancer risk. J Hum Genet. 2019;64:767–73. [DOI] [PubMed] [Google Scholar]
- 26.Noskowicz M, Bogdanova N, Bermisheva M, Takhirova Z, Antonenkova N, Khusnutdinova E, et al. Prevalence of PALB2 mutation c.509-510delGA in unselected breast cancer patients from Central and Eastern Europe. Fam Cancer. 2014;13:137–42. [DOI] [PubMed] [Google Scholar]
- 27.Kluska A, Balabas A, Piatkowska M, Czarny K, Paczkowska K, Nowakowska D, et al. PALB2 mutations in BRCA1/2-mutation negative breast and ovarian cancer patients from Poland. BMC Med Genomics. 2017;10:2–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamdan O, Nowak KM. Gene of the month: PALB2. J Clin Pathol. 2022;76:73–5. [DOI] [PubMed] [Google Scholar]
- 29.Antoniou AC, Casadei S, Heikkinen T, Barrowdale D, Pylkäs K, Roberts J, et al. Breast-Cancer Risk in Families with Mutations in PALB2. N Engl J Med. 2014;371:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Daly MB, Pal T, Maxwell KN, Churpek J, Kohlmann W, AlHilli Z, et al. Genetic/Familial High-Risk Assessment: Breast, Ovarian, and Pancreatic, Version 2.2024 Featured Updates to the NCCN Guidelines. JNCCN J Natl Compr Cancer Netw. 2023;21:1001–10. [DOI] [PubMed] [Google Scholar]
- 31.Lavie O, Narod S, Lejbkowicz F, Dishon S, Goldberg Y, Gemer O, et al. Double heterozygosity in the BRCA1 and BRCA2 genes in the Jewish population. Ann Oncol. Apr 2011;22:964–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heidemann S, Fischer C, Engel C, Fischer B, Harder L, Schlegelberger B, et al. Double heterozygosity for mutations in BRCA1 and BRCA2 in German breast cancer patients: implications on test strategies and clinical management. Breast Cancer Res Treat. 2012;Aug:1229–39. [DOI] [PubMed] [Google Scholar]
- 33.Sokolenko AP, Bogdanova N, Kluzniak W, Preobrazhenskaya EV, Kuligina ES, Iyevleva AG, et al. Double heterozygotes among breast cancer patients analyzed for BRCA1, CHEK2, ATM, NBN/NBS1, and BLM germ-line mutations. Breast Cancer Res Treat. 2014;145:553–62. [DOI] [PubMed] [Google Scholar]
- 34.Hinić S, Van der Cybulski C, Vos JR, Schuurs-Hoeijmakers J, Brugnoletti F et al. The heterogeneous cancer phenotype of individuals with biallelic germline pathogenic variants in CHEK2. Genet Sci. 2024;26. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed during the current study.