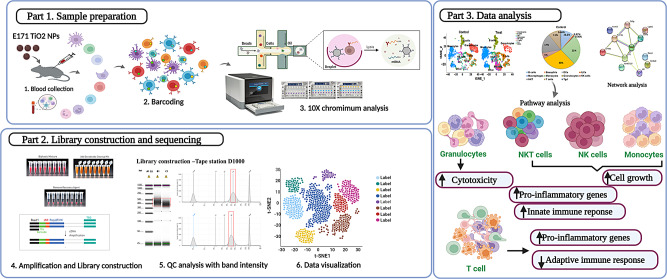
Abstract
The prospective use of food additive titanium dioxide (E171 TiO2) in a variety of fields (food, pharmaceutics, and cosmetics) prompts proper cellular cytotoxicity and transcriptomic assessment. Interestingly, smaller-sized E171 TiO2 can translocate in bloodstream and induce a diverse immunological response by activating the immune system, which can be either pro-inflammatory or immune-suppressive. Nevertheless, their cellular or immunologic responses in a heterogeneous population of the immune system following exposure of food additive E171 TiO2 is yet to be elucidated. For this purpose, we have used male Sprague-Dawley rats to deliver E171 TiO2 (5 mg/kg bw per day) via non-invasive intratracheal instillation for 13 weeks. After the 4 weeks recovery period, 3 mL of blood samples from both treated and untreated groups were collected for scRNAseq analysis. Firstly, granulocyte G1 activated innate immune response through the upregulation of genes involved in pro-inflammatory cytokine mediated cytotoxicity. Whereas NK cells resulted in heterogeneity role depending on the subsets where NK1 significantly inhibited cytotoxicity, whereas NK2 and NK3 subsets activated pro-B cell population & inhibited T cell mediated cytotoxicity respectively. While NKT_1 activated innate inflammatory responses which was confirmed by cytotoxic CD8+ T killer cell suppression. Similarly, NKT_2 cells promote inflammatory response by releasing lytic granules and MHC-I complex inhibition to arrest cytotoxic T killer cell responses. Conversely, NKT_3 suppressed inflammatory response by release of anti-inflammatory cytokines suggesting the functional heterogeneity of NKT subset. The formation of MHC-I or MHC-II complexes with T-cell subsets resulted in neither B and T cell dysfunction nor cytotoxic T killer cell inhibition suppressing adaptive immune response. Overall, our research offers an innovative high-dimensional approach to reveal immunological and transcriptomic responses of each cell types at the single cell level in a complex heterogeneous cellular environment by reassuring a precise assessment of immunological response of E171 TiO2.
Graphical Abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s12951-024-03036-9.
Keywords: E171 TiO2, scRNAseq analysis, Rat peripheral blood mononuclear cells, Immune response, Heterogeneity
Introduction
Owing to their significant photocatalytic and physiochemical properties, food additive titanium oxide (E171 TiO2) have been widely used in cosmetics, the food industry, and biomedical applications [1, 2]. However, the European Food Safety Authority (EFSA) and the Food and Drug Administration (FDA) in the United States set specific regulations to avoid the use of E171 TiO2 in excess of 1% in items for human consumption and health [2–5]. Furthermore, studies have reported that the use of food additive E171 TiO2 in toothpaste, food coloring, and skin-protective cosmetics may lead to the progression of nanomaterial toxicity [6, 7]. Furthermore, due to its multiple uses, nano- or micro-sized E171 TiO2 can be ingested, altering intestinal barrier function and translocating into the interior body [7–9]. Recent cohort study demonstrated the titanium level in the patients with active ulcerative colitis disease is higher than the healthy donors [10]. Therefore, toxicological assessments must be performed carefully to extend the industrial/ medical application of E171 TiO2 to address the increase in safety concerns.
Understanding the toxicity of E171 TiO2 on immune cells has gained particular interest as they play a vital role in the body defense mechanisms. Although toxicity assessment of E171 TiO2 was carried out in different types of cell lines and in vivo models [8, 11, 12] their results were unclear. Additionally, previous findings have demonstrated TiO2/E171 causes negative effects due to their translocation to the bloodstream, urine, or organs via various inflammatory responses [1, 13].
Due to the heterogeneity of immunological leukocytes, the immunological response in the human body can result in various biochemical reactions that can be either hyper or hypo-mediated, which can cause both organ damage and immune dysfunction [14, 15]. Additionally, smaller sized E171 TiO2 in circulating blood leukocytes can cause a variety of inflammatory allergic reactions by either activating the immune system or suppressing it due to generation of reactive oxygen species (ROS) that can be toxic to cells or the immune system [16–19]. However, there is no report on E171 TiO2 translocation in blood lymphocytes and immunological responses at the single cell level [8, 20]. Furthermore, the present accessible methods such as flow cytometry, inductively coupled plasma mass spectrometry (ICP-MS), scChIP–seq for epigenomics data, and mass cytometry or fluorescence-activated cell sorting for proteomics data may provide single cell evaluation with limitations such as panel parameter limitation, antibody fluorescence spectra overlap, and numerous batch effect experiment schedules [21–23] can only detect lymphocytes identification rather than distinct transcriptional responses at single cell level. In case of transcriptomic observation of each cell types, bulk-RNAseq approaches have been performed in various nanomaterials toward various human cell lines [24–26], but the homogeneous responses may cause misunderstanding in transcriptomic profiles since blood immune leukocytes are heterogeneous and respond diversely. As a result, an in-depth comprehension of the systemic molecular processes that regulate phenotypic changes in diverse immune cells in response after E171 TiO2 is critical and yet to be established.
Therefore, to our knowledge this is the first report to explore the immune phenotypic and subset identification in rat leukocytes after E171 TiO2 exposure. In this study we aimed to understand the heterogenous immune subsets population and their transcriptomic changes upon intratracheal instillation of E171 TiO2. Firstly, we have attempted to perform in-depth high dimensional scRNAseq analysis to uncover heterogenous immune cell subsets responsible for immune related inflammatory response. Next, we focused on exploring precise cellular phenotypic identification and their distinct cellular gene expression by 2D dimensionality reduction, including tSNE visualization. Later, the major significant immune cell types such as granulocytes, NK/NKT cells, B and T cells were subjected to PhenoGraph clustering analysis to reveal subset identification with their unique gene expression. The functional annotation and hypothesized molecular pathways of the majority of significant differentially expressed genes (DEGs) were carried out separately. Overall, the scRNAseq analysis provided a comprehensive atlas of the heterogeneous immunologic response exposed with E171 TiO2 at single-cell level that inspires translational research using animal models.
Materials and methods
Preparation of E171 TiO2 suspension
E171 TiO2 (HOMBITAN® FG; purity 99.5%) was obtained from Venator Germany GmbH (Duisburg, Germany. As previously mentioned [11], to prepare the stabilized E171 TiO2 suspension in several aqueous solutions with varying pH values, 10–640 mg of E171 TiO2 powder was added to 10 mL of pre-adjusted pH solution. To compare the stability of E171 TiO2 dispersed in 5 mM Phosphate buffer (pH 8) and deionized water (DW), the average hydrodynamic particle size was measured (Figure S1a). The Zeta potential (mV) for pre-adjusted pH solution of phosphate buffer was confirmed (Figure S1b). Furthermore, physio chemical characterization of E171 TiO2, including TEM, DLS, and zeta potential was confirmed by Han et al., [11]. Finally, the concentration of E171 TiO2 has been modified to meet the oral toxicity test condition.
Animal groups and experimental design
Further, we investigated the potential immunotoxicity profile of E171 TiO2 after 13 weeks of intratracheal instillation in Sprague-Dawley rats and assessed the reversibility of any adverse effects during a 4-week recovery period (14–17 weeks). The rats were kept in established housing at room temperature (23oC), humidity of 30–70%, and 12:12 light cycle. Each group 0, 2.5, 5, and 10 mg/mL E171 TiO2 consisted of 10 rats to replace any rat that deceased during the treatment period. A standard rat pellet diet (Lab Diet, PMI Nutritional International, USA) was provided to the animals for 7 days until the completion of the treatment and recovery periods. In this study, all animal experiments were conducted according to the National Institutes of Health Guide for the care and use of laboratory animals (NIH Publications No. 8023) and all procedures complied with the Animal Welfare and Guide for the Care and Use of Laboratory Animals and were assessed by the Institutional Animal Care and Use Committee (IACUC) (approval number N221019).
Dose administration
Animals were selected for this study based on adequate body weight and the absence of clinical signs of diseases or injuries. For each dose (2.5, 5, and 10 mg/mL bw E171 TiO2), a homogeneity test was performed prior to dosing, and the formulations were administered by non-invasive intratracheal installation once per day at the same time each day for 13 consecutive weeks. The mean concentration of each formulation was acceptable if it was within ± 15% of the nominal concentration. The animals were administered a volume of 10 mL/kg. Mortality and morbidity observations were conducted twice daily at the start and end of working hours during the treatment period.
Sample collection and preparation
The 3 mL of blood samples were randomly collected from abdominal vena cava of the 3 surviving animals of both treated (2.5, 5 and 10 mg/kg bw) and untreated group from recovery group were pooled individually to analyze the quality of PBMCs. The collected blood samples were transferred into EDTA-treated tubes (BD Vacutainer®, USA), and rPBMCs were isolated by density differential centrifugation using Ficoll-Paque PLUS (GE Health Care Bio-Sciences, Sweden) as previously reported [27, 28] and immediately stored − 80 °C. Later, the blood samples (rPBMCs) were subjected to quality check using cell counter. Finally, 5 mg/kg bw was chosen based on the various factors such as cell number, cell viability and minimal dead cells etc., Among the three different concentrations, 5 mg/kg bw showed closest value to the lethal dose was chosen for scRNAseq to assess significant immune-transcriptional changes.
scRNAseq sample preparation and analysis
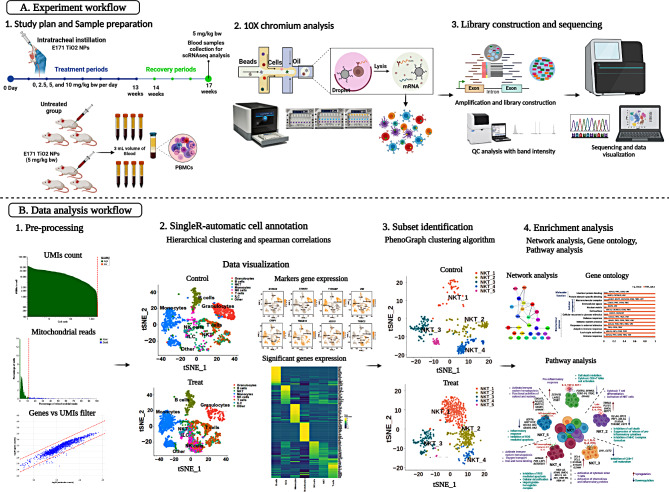
The rPBMCs were analyzed using a 10× chromium single-cell analyzer (10× Genomics Chromium, Pleasanton, CA, USA) after isolation of blood samples from the E171 TiO2 treated (5 mg/kg bw) and untreated groups. For each replicate sample, approximately 18,000 cells were processed, and the target cell recovery was selected as 10000 cells per sample. Single cell 3’ v3.1 gel beads were used for GEM generation and barcoding which are made of a primary mix with cell surface protein labels and partitioning oil added to a chromium chip (Fig. 1).
Fig. 1.
Schematic representation of experimental design. Dose dependent (2.5, 5, 10 mg/kg bw) intratracheal instillation of E171 TiO2 were administered once in a day for more than 13 weeks called as treatment periods. From 14 to 17 weeks was recovery period and blood collection were done 17th week from 5 mg/kg bw for scRNAseq analysis. 10x chromium analysis, library preparation, and sequencing were performed at the single cell level for phenotypic identification and transcriptome observation, as indicated in the figure. Additionally, data visualization and processing were performed to assess immunological response of heterogeneous immune cell types
cDNA amplification and post-GEM-RT clean-up steps were successfully performed according to the manufacturer’s instructions (GEM Single Cell Kit V3.1, Pleasanton, CA, USA). Amplified cDNA was used to create a 3’ gene expression library, which was then loaded into TruSeq read 2 using the manufacturer’s recommended methods for end repair, A-tailing, and adaptor ligation. An Illumina NextSeq 500 sequencer was used to sequence the library-constructed samples to gain insight into the heterogeneous immune leukocyte populations between the treated and untreated rats (Fig. 1). All the samples were subjected to quality control analysis using a tape station equipped with D1000 and D5000 markers (Agilent Technologies) (Figure S2).
scRNAseq data processing
A cell ranger was used to process the FASTQ files containing the datasets of treated and untreated rat groups. To exclude low-quality datasets from further processing, the total unique mapped identifier (UMI) (1000 or more than 2500 genes), abnormal mitochondrial genes (greater than 11.9% transcripts), and doublets were used to set the median line. Immediately after preprocessing, treated and untreated samples were aggregated using the IntegrateData function. The data were then combined and uploaded to the R Seurat package (4.0.6) for further analysis using R Studio and an updated version of the R programming language (R 3.3.0). The “read10x” function generated a seurat object from the barcodes, features, and matrix files. For cell annotation, an unsupervised automated clustering algorithm was used in conjunction with the SingleR package. We used the ImmGenData reference in SingleR to identify the immune cell types based on the expression of reference marker genes (Figure S3).
scRNAseq data visualization
To qualitatively observe transcriptomic changes in the treated and untreated rat groups, we used t-distributed Stochastic Neighbor Embedding (t-SNE) projections to visualize high-dimensional scRNAseq data. The position of each cell in the 2D plot is provided in the t-SNE plot. Alterations in the dot position revealed the impact of both the treated and untreated rat groups. On the integrated SEURAT object, the “RunPCA” (npcs = 30) and “RunTSNE” (dims = 1:20) functions were used. Cell types were used as labels on the t-SNE plots created using the “Dimplot” function. Initially, an overall t-SNE plot of all cells was generated with labeled major cell types, and then separate t-SNE plots were generated for each major cell type and labeled.
DEG analysis
After cell annotation using SingleR, we performed DEG analysis on each cell subtype to identify the genes that were upregulated or downregulated to examine the changes in transcriptome profiles following AgNP exposure. For the DESeq2 analysis, the “Findmarkers” function from the R Seurat package was utilized. To identify how DEGs interact and whether they have similar functions in both the treated and untreated rat groups, string network analysis (https://string-db.org/) was performed. The DAVID platform (https://david.ncifcrf.gov/home.jsp) was used to perform gene ontology (GO) analysis to reveal functional enrichment, which included biochemical processes, cellular components, and molecular functions. Gene ontologies were ranked in order of importance based on the number of input genes included in each ontology. We selected the top three to five ontologies for each category to evaluate the data in relation to each group.
Statistical analysis
Statistical significance was assessed using the Mann-Whitney U test. Statistical significance was set at p < 0.05; 0.01 < p < 0.05, 0.001 < p < 0.01 and p < 0.001 are annotated as *, **, and ***, respectively.
Results
Identification and visualization of heterogeneous immune leukocytes
Transcriptional response in heterogeneous cell types becomes more challenging, as each cell type may respond differently owing to its unique characteristics (specificity, immunological memory, and self/non-self-recognition). For the first time, we assessed the transcriptional response of E171 TiO2 in rPBMC at the single-cell level using high-dimensional scRNA-seq analysis.
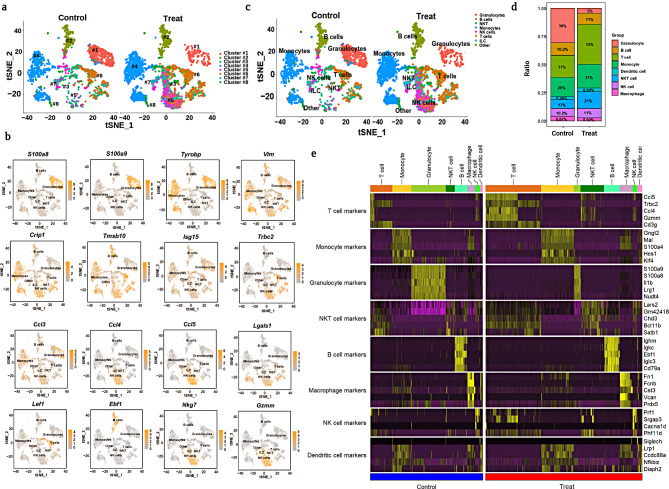
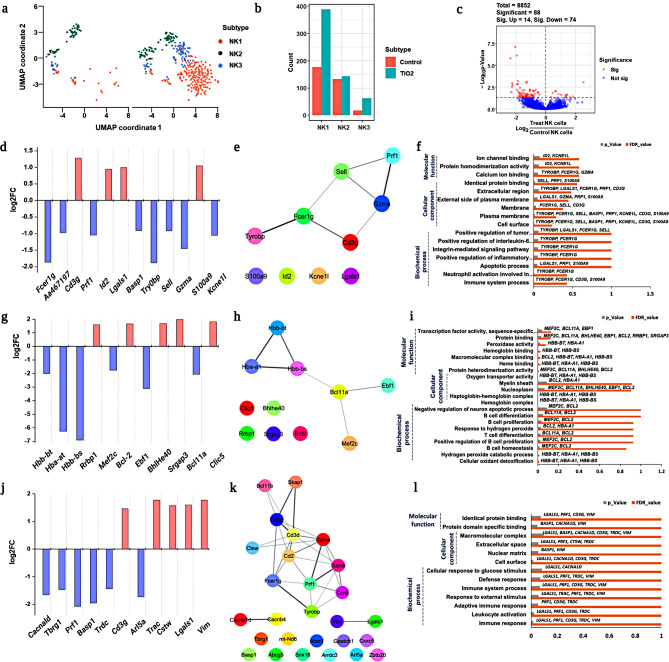
Figure 2a shows heterogeneous immune cell clusters (#1–#8) from the treated and untreated groups, which were visualized in a tSNE plot to show the differences in cell type specificity. The expression of the reference marker genes listed in Fig. 2b was used to identify various immune cell types. Granulocytes were identified using marker genes such as S100a8, S100a9, Igha, Tyrobp, Grn, Vim, Crip1, and Ccl3. Similarly, Igha, Tyrobp, Vim, and Crip1 represent monocytes, while Lgals1, Trbc2, Ccl3, Ccl4, Ccl5, Gzmm, and Nkg7 represent natural killer (NK) and natural killer T (NKT) cells; the genes Lgals1, Trbc2, Gzmm, and Lef1 represent T cells. Other minor cell types, such as dendritic cells (DCs), innate lymphoid cells (ILCs), macrophages, and gamma-delta T cells (Tgd), were also identified based on reference marker gene expression, as listed in Figure S3. Furthermore, the major cell types identified were as follows: granulocytes, B cells, (NKT), monocytes, (NK) cells, T cells, and other minor populations (Fig. 2c). Among these cell types, NK cells, NKT cells, T cells, and monocytes, showed population increment in the treated group than in the untreated group (Fig. 2d). In contrast, granulocyte population destruction was observed in the treated group compared to the untreated group (Fig. 2d). The overall gene expression in major cell types (total immune leukocytes), including B cells, granulocytes, NK cells, NKT cells, T cells, and monocytes, was visualized using a heat map (Fig. 2e). The expression levels of genes from each cell type showed substantially heterogeneous immunological responses in the treated group (Fig. 2e). Although we visualized the population differences and overall gene expression of major immune cell types in both the untreated and treated groups, further in-depth profiling is warranted to explore their functional diverse roles. Because immune cells can produce cellular heterogeneity at various molecular and phenotypic levels, depending on their reaction to foreign particles [29].
Fig. 2.
scRNAseq analysis showed identification of heterogeneous immune cells and genes expression population; a. tSNE visualization of heterogeneous clusters and differences between the treated and untreated groups; b. The expression of important immune cell type reference marker genes was visualized; c. Immune cell types were identified and visualized using tSNE in treated and untreated groups; d. Population differences between the two groups from identified immune leukocytes were observed; e. Heat map expression analysis was used to compare overall markers gene expression from all cell types between treated and untreated groups
Granulocytes and their subsets initiated innate inflammatory responses
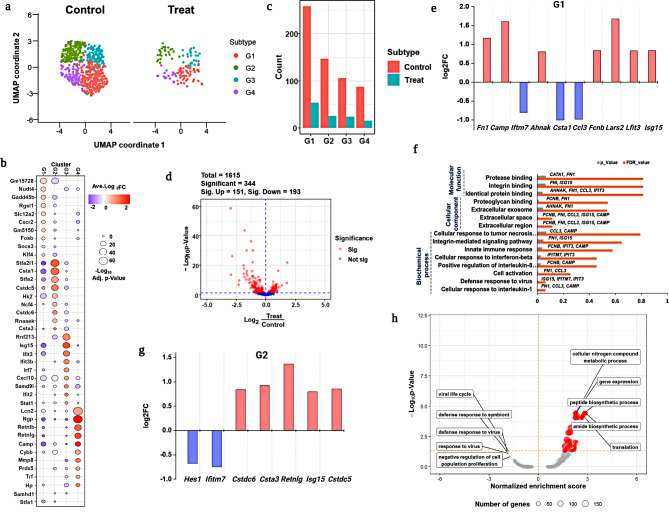
Comprehensive profiling of granulocytes revealed subset identification and population decrement in the treated and untreated groups (Fig. 3a). Furthermore, PhenoGraph clustering (PG) analysis identified four different subsets of granulocyte including G1, G2, G3 and G4 (Fig. 3a). Additionally, Fig. 3b deficit heterogeneity among the four different granulocyte subsets which was identified by significant reference marker genes. A significant decrease in the granulocyte population in the treated group (Fig. 3c) resulted might be due to the fact that the granulocytes can be destroyed when an inflammatory response (proinflammatory cytokines and chemokines) is triggered by external stimuli such as foreign substances, pathogens, or parasites [30–34].
Fig. 3.
Profiling of granulocyte subsets. (a) UMAP visualization of total granulocytes from treated and untreated groups with subset identification by PhenoGraph clustering analysis; (b) Expression of marker genes from each granulocyte subset; (c) Population differences in neutrophils and other subsets in treated and untreated groups. (d) Volcano plot of significant DEGs consists of up and downregulated genes; (e) DEG analysis from G1 with upregulation and downregulation of significant genes specified using log2fold changes; f Gene ontology enrichment analysis of neutrophils; g. DEGs of granulocyte G2 subset; h. GSEA analysis for DEGs from granulocytes to predict enrichment score based proposed pathway
The volcano plot was used to confirm the DEGs of proinflammatory responsible genes which showed a total of 1615 DEGs from granulocyte, among them 344 genes were substantially expressed with up (151) and down (193) regulation (Fig. 3d). Further, the granulocyte subset, G1 played a significant role in inducing inflammatory changes by upregulating Fn1, Camp, Ahnak, Fcnb, Lars2, Lfit3, and Isg15 genes, and by downregulating Iftm7, Csta1, and Ccl3 (Fig. 3e). Gene ontology enrichment analysis (Fig. 3f) showed that the identified DEGs specifically, Camp is primarily involved in the potent regulation of immune cell homeostasis through the activation of innate or adaptive immune responses [35], while Fn1, If1t3, and Isg15 are mostly involved in interferon-γ production [36, 37]. In our study, we demonstrated that the upregulation of those inflammatory genes Camp, Fn1, If1t3, and Isg15 involves an innate immune response by inducing pro-inflammatory cytokines such as IL-1, IL-8, and IFNγ (Fig. 3f; Table S1). Furthermore, the downregulation of Ifitm7, Ccl3, and Csta1 inhibits the expression including M2, and cystatin A1, which are responsible for the suppression of the immune system [38–40]. These findings clearly suggest that granulocyte G1 may influence the activation of early innate primary defense through the upregulation of genes involved in pro-inflammatory cytokine release; however, they also suppress the hyperimmune response by downregulating genes (IfIitm7, Ccl3, and Csta1) which may affect immunological hematopoiesis.
The other granulocyte subsets G2, G3 and G4 exhibited the least significant difference compared to G1; however, we still performed DEG analysis for G2 and discovered that the HES1 and IFITM7 genes were downregulated, whereas other genes, including Cstdc6, Csta3, Retnlg, Isg15, and Cstdc5 were significantly upregulated (Fig. 3g). Furthermore, we performed gene ontology for other granulocyte subsets and were unable to detect any clear evidence (Table S2). Furthermore, gene set enrichment analysis (GSEA) was utilized to examine statistically significant and concordant changes between the treatment and control groups to utilizing normalized enrichment score (Fig. 3h). Overall, our findings show that after being exposed to E171 TiO2, granulocytes induce immune toxicity via a hyperpro-inflammatory response, resulting in a population reduction.
NK cell subsets induced pro-inflammatory response after treatment
Significant differences in the NK cell population between the treated and untreated groups indicated that external stimuli such as E171 TiO2 influenced hyper- or hypo-inflammatory responses. Therefore, we further studied the functional role of NK cells in identifying the heterogeneous subsets responsible for immunological responses. The PG analysis visualized differences in heterogeneous populations of NK cells and identified three distinct subsets of NK cells, designated NK 1, NK 2, and NK 3 (Fig. 4a). Subsequently, the population differences in each subset of NK cells were compared between treated and untreated groups and a significant population increase in the NK1 subset was noted in the treated group when compared to that in the other subsets (Fig. 4b). This clearly demonstrates that the treated group activated NK cells response via cell surface receptors, which may induce an innate inflammatory response [41].
Fig. 4.
In depth profiling of NK cells. (a) UMAP visualization of total NK cells from the treated and untreated groups with subsets identification using PhenoGraph clustering analysis; (b) Population differences in each NK cell subset between the treated and untreated groups; (c) Volcano plot represents DEGs from NK cells with significant and non-significant entries; d, e, f. DEGs, network analysis, and gene ontology enrichment analysis for NK_1 subset were visualized; g, h, i. DEG analysis, the network and gene ontology enrichment analysis of NK_2 subset observed using log2fold changes; j, k, l. DEG analysis, network and GO profiling were observed and visualized for NK_3 subset
To identify the genes responsible for the activation of innate immune response in NK cells, complete DEGs were carried out using volcano plot. Our results identified 8852 genes in total, of which only 88 were significantly expressed, including 14 upregulated and 74 downregulated genes (Fig. 4c). Furthermore, DEG analysis for each NK cell subset was performed to identify genes with significant differences in expression between the treated and untreated groups. The significant upregulation of Cd3g, Id2, Lgals1, and S100a9 in NK1 subsets indicated the activation of the innate pro-inflammatory response via the release of inflammatory cytokines such as IL-6, IL-7, and IFNγ [42, 43], whereas downregulation of Fcer1g, Gzma, Sell (L-selectin), Prf1 inhibited cell death (Fig. 4d) [44]. This result suggests that the NK_1 subset may be involved in the induction of a pro-inflammatory response in the treated group, without exhibiting toxicological responses related to necrosis and/or cell death. Further, Granzyme A (Gzma) is a serine protease that releases cytotoxic granules and perforin (Prf1) is a glycoprotein that plays a role in pore formation during programmed cell death [45, 46]. Therefore, we searched for the Gzma and Prf1 genes and observed that the treated group showed downregulation of cell death-related genes in the NK_1 subset. These findings showed that an increase in the NK_1 subset population in the treatment group (Fig. 4b) simultaneously activated the release of cytokines by suppressing cytotoxicity response.
Furthermore, to support our findings, we performed network and GO analyses to reveal the heterogeneous functional characteristics of the DEGs that are mainly involved in biochemical, cellular, and molecular functions. In the NK_1 subset, almost all DEGs shared a common functionality, as shown by network analysis and GO performance (Fig. 4e and f). The genes Tyrobp, Fcer1g, Lgals1, Prf1, Cd3g and S100a9 involved in biochemical process demonstrated positive regulation of the inflammatory response (Fig. 4f; Table S3). Most of the genes (Tyrobp, Fcer1g, Lgals1, Prf1, Cd3g, Gzma, Sell, Basp1, Kcne1l and S100a9) involved in the cellular component were found in extracellular regions including the plasma membrane. In terms of molecular function, NK_1 subset gene, such as Id2, Kcne1l, Tyrobp, Fcer1g, Gzma, Sell, Prf1 and S100a9, are involved in calcium and protein homodimerization activity in ion channel binding (Fig. 4f; Table S3).
In the NK_2 subsets, Rrbp1, Bcl-2, Bhlhe40, Srgap3 and Clic5 were upregulated, Hbb-bt, Hba-at and Hbb-bs were mostly involved in the hepatoglobin-hemoglobin complex [45], and other genes (such as Mef2c, Ebf1 and Bcl11a) were substantially downregulated (Fig. 4g). Downregulation of the Hbb-bt and Hba-at genes may involve the evasion of ROS-mediated antioxidants in NK cells [47, 48]. To further confirm this finding, we identified that the activation of BCl2 (anti-apoptotic protein) (Fig. 4g) contributed to the inhibition of intrinsically mediated apoptosis. Hence, the treated group showed a slight increase in NK_2 subsets compared with the untreated group (Fig. 4b). Additionally, downregulation of Bcl11a and Ebf1 in NK cells may inhibit NK cell proliferation, which is indirectly responsible for the activation of pro-B cells to mediate the adaptive immune response [47, 49, 50]. In contrast, the upregulation of Rrb1, Bhlhe40 helps increase the intracellular protein, the upregulation of Srgap3 aids in NK cell activation [51, 52].
Despite the induction of an inflammatory response in NK_2 subsets similar to that in NK_1, distinct differences were observed among the genes involved in the inhibition of ROS-mediated antioxidant and intrinsic-mediated apoptotic cell death in the treated group. Correspondingly, network analysis (Fig. 4h) and GO functional enrichment analysis (Fig. 4i) revealed that DEGs from NK_2 involved two major roles, promoting B and T cell proliferation & differentiation (Bcl11a, Bcl-2, and Mef2c) (Fig. 4i) and other role was involving haptoglobin-hemoglobin complex (Hbb-bt, Hba-a1, and Hbb-bs) helpful in the removal of cellular oxidant detoxification (hydrogen peroxide catabolic process) (Fig. 4i; Table S4).
The other subset, NK_3, also showed considerable changes in gene expression similar to the other NK subsets. Genes including Cacnald, Tbrg1, Prf1, Basp1, Trdc, and Arl5a were significantly downregulated, whereas other genes including Cd3g, Trac, Ctsw, Lgals1, and Vim were upregulated, as shown in Fig. 4j. In particular, Cd3g, Lgals1, Prf1, and Basp1 showed similar trends to those of the NK_1 subset (Fig. 4d). However, the other DEGs showed unique expressions that was not found in either the NK_1 or NK_2 subsets, indicating the diverse nature of NK cells. The upregulation of Cd3g is mostly involved in the activation of T cell development and regulation ( [53] indicating that NK cells may be involved in T cell-mediated adaptive immune responses. The network (Fig. 4k) and pathway enrichment analyses (Fig. 4l) of DEGs from each NK_3 subset was primarily involved in immune system activation (adaptive immune response) (Lgals1, Prf1, Cd3g, Trdc, and Vim) through the influence of external stimuli that helped in the maintenance of defense responses (Fig. 4l; Table S5). Overall, our findings demonstrated that NK cells have a heterogeneous functional role, with distinctive subset identification upon E171 TiO2. Specifically, the NK_1 subset was mostly involved in pro-inflammatory responses by suppressing cytotoxicity, while the NK_2 subset was largely involved in inhibiting pro-B cell activation, and the NK_3 subsets were primarily engaged in activation of T cell-mediated adaptive immune response.
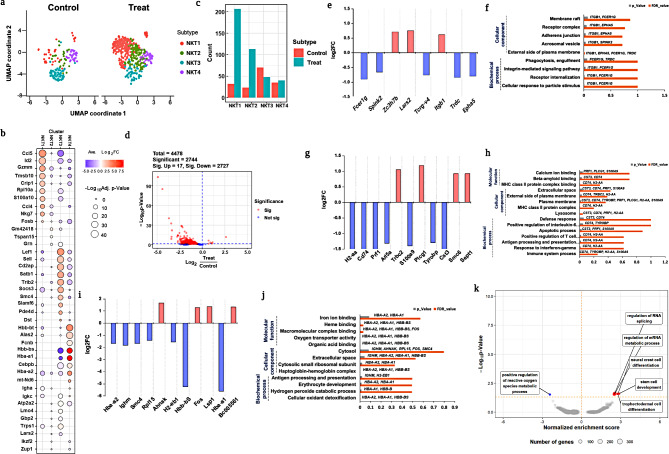
Heterogeneous NKT subsets unravels diverse immunological response
NKT cells are lymphocytes that express both NK cell surface receptors and T-cell receptors (TCR), which are part of the innate or adaptive immune response [54]. NKT cell activation may influence host defense mechanisms via innate or adaptive immune responses, which facilitate immune system protection [55, 56]. In our study, NKT cells showed a significant increase in the treated group compared to the untreated group [54]. Tupin et al. demonstrated that NKT cells have a diverse range of immune responses, including tumor monitoring, self-tolerance, and control of autoimmune diseases [52]. Therefore, subset identification of NKT cells helps in the discovery of distinct immunological responses under these circumstances. The UMAP plots of the overall NKT cell populations from both treated and untreated groups are shown in Fig. 5a. Furthermore, total NKT cells were subjected to PG clustering analysis, which revealed four different heterogeneous subsets, namely NKT_1, NKT_2, NKT_3, and NKT_4 (Fig. 5a). The heterogeneity of NKT cells was clearly indicated by the expression of markers genes that designated subset differentiation (Fig. 5b). The population comparison of treated and untreated groups among the NKT subsets is shown in Fig. 5c, and a substantial population increment in treated group was observed in NKT_1 followed by NKT_2. In contrast, a population decrement was observed in the NKT_3 subset in the treated group compared to that in the untreated group (Fig. 5c). These findings clearly indicate the diversity in NKT cell functions by expressing population differences among the subsets. DEG analysis was performed to determine gene expression at the single-cell level in each subset of NKT cells. Furthermore, DEGs of NKT cells were identified and visualized in a volcano plot as 4478 in total, with the majority of them (2727) being downregulated rather than upregulated (17) (Fig. 5d). DEGs from the NKT_1 subset showed the upregulation of the Zc3h7b, Lars2, and Itgb1, whereas other genes, including Fcerig, Spink2, Tcrg-v4, Trdc, and Epha5, were downregulated respectively (Fig. 5e). The upregulation of Lars2 (similar to G1, see Fig. 3e) may be responsible for to release of pro-inflammatory cytokines which is capable of inducing inflammatory responses [57, 58]. Similar to NK cells, downregulation of Fcer1g (see Fig. 4d) may be involved in the inhibition of cell death [44]. Additionally, downregulation of Tcrg-v4, and Trdc leads to the failure of cytotoxic CD8+ T killer cell activation [59]. Furthermore, the downregulation of Spink2 and Epha5 (which are mostly involved in disease progression) [60] leads to minimal immune activation and, prevents apoptosis-mediated avoiding apoptotic mediated cell death [59]. GO enrichment analysis revealed that DEGs from each subset had common functionality, with a possible pathway based on biochemical processes, cellular components, and molecular functions. The GO enrichment analysis revealed responsible genes, including Fcer1g, Itgb1, and Trdc which may be involved in phagocytic engulfment via receptor internalization due to the cellular response to the treatment. Along with Fcer1g and Itgb, other genes, such as Epha5, Spink2, and Trdc were mostly involved in the formation of cellular components formation, such as formation receptor complexes (Table S6). Our findings clearly showed NKT_1 subset activated innate inflammatory responses upon treatment, however, further adaptive immunological responses may have failed due to suppression of cytotoxic CD8+ T killer cell activation.
Fig. 5.
Comprehensive analysis profiling of NKT cells. a. UMAP visualization of NKT subset between the treated and untreated groups by PhenoGraph clustering analysis; b. Markers gene expression of NKT subsets; c. Population differences in each subset from NKT cells between the treated and untreated groups; d. Volcano plot depicts significant u and downregulated DEGs; e, f. DEGs from the NKT_2 subset was visualized using log2fold changes, and GO enrichment analysis were also observed; g, h. Similarly, log2fold changes, as well as GO enrichment analysis were used to visualize DEGs in the NKT-2 subset; i, j, DEG analysis for the NKT_3 subset visualized using log2fold changes, and gene ontology enrichment analysis. k. GSEA analysis to predict enrichment score of DEGs genes involved in diff pathways
Correspondingly, the NKT_2 subset revealed 11 DEGs, of which Trbc2, Plcg1, Smc6, and Sept1 were upregulated, whereas H2-aa, Cd74, Prf1, Arl5a, S100a9, Tyrobp, and Cst3 were significantly downregulated (Fig. 5g). The upregulation of Plcg1 and Trbc2 implies cytotoxic T-cell differentiation with the positive regulation of cytokine release [61, 62], whereas the highest expression of Smc6 and Sept1 indicates the activation of NKT cells, which may regulate the release of lytic granules during the phagocytic process. Downregulation of S100A9 along with other genes (Cd74 and H2-aa) may inhibit MHC I complex formation and prevent cytotoxic effects [63]. These findings are consistent with our results, in which the population of the NKT_2 subset was higher in the treated group than in the untreated group. GO enrichment analyses revealed immune response activation via positive regulation of IL-6, IFN-, and T cell differentiation, mostly through MHC complex formation and initiation of the apoptotic process (Fig. 5h). Furthermore, cellular component and molecular function analyses revealed that the genes Cd74, Tyrobp, H2-aa, Prf1, Cst3, Plcg1, and S100a9 were primarily involved in lysosome formation, MHC-II complex binding, and plasma membrane elevation (Fig. 5f; Table S7). Thus, NKT_2 cells promote inflammatory responses by releasing lytic granules, however MHC-I complex inhibition may fail to initiate cytotoxic mediate T killer cell responses.
Interestingly, the NKT_3 subset showed a population decrement compared to the other subsets in the treated group, indicating that NKT cells have diverse functional aspects (Fig. 5c). The subset NKT_3 showed downregulation of Hba-a2, Ighm, Smc4, Rpl15, H2-eb1, Hbb-bs, and Hba-a1, whereas other genes, such as Ahnak, Fos, Lsp1 and Bc005561 were upregulated (Fig. 5i). Similar to the NK_2 (see Fig. 4g), Hbb-bs, Hba-a1, and Hba-a2 were downregulated leading to ROS-mediated inhibition of apoptosis [47, 48]. Similarly, Ahnak, Fos, and Lsp1 may be involved in the activation of cytokines such as M2, and cystanin A1 that helps to suppress NK cell activation to maintain immune system hematopoiesis via initiating IL-6 secretion.
Network analysis of the NKT_3 subset revealed Hba-a1, Hba-a2, Hbb-bs, Ighm, and H2-eb1were mostly involved in the biochemical processes of cellular detoxification, hepatoglobin-hemoglobin complex formation, and antigen processing presentation (Fig. 5j). Additionally, these genes were involved in oxygen transport and iron and heme binding as cellular components and molecular functions (Fig. 5j; Table S9). GSEA analysis revealed that proposed route suggestions were less significant for all NKT subsets (Fig. 5k). The NKT_4 subset doesn’t show any significant expression impact compared to other NKT subsets (Figure S4). Overall, in our study we revealed that functional heterogeneity of NKT subset upon E171 TiO2 treatment. In particular NKT_3 mainly involving inhibition of ROS-mediated cell death.
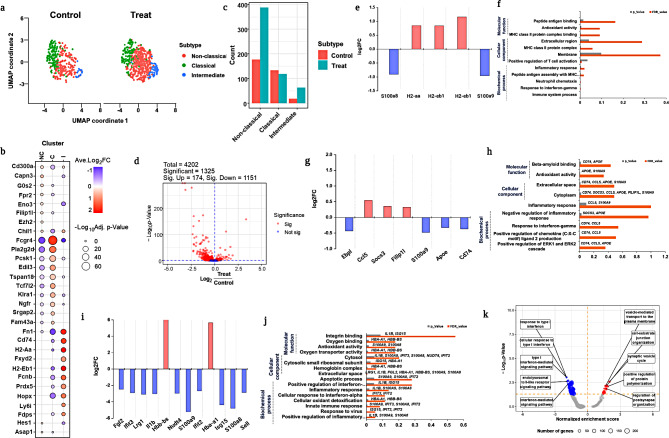
Diverse immunological response by monocytes and their subsets
Monocytes are prominent antigen-presenting cells (APCs) with a natural propensity to ingest foreign particles via phagocytosis [64, 65]. Additionally, they stimulate cellular immunity via the major histocompatibility complex, followed by exposure to nanomaterials [66, 67]. Our study revealed an increase in the monocyte population in the treated group, indicating a cellular association with reduced toxicity. We used PhenoGraph analysis to reveal the diverse role and subset identification of monocytes, and identified three distinct types of monocytes: classical, non-classical, and intermediate monocytes (Fig. 6a). The heterogeneous impact of monocytes was displayed by significant marker gene expression and (Fig. 6b) significant population differences among the monocyte subsets were found (Fig. 6c). Compared to the untreated group, the treated group showed a population decline in classical monocytes (Fig. 6c). In contrast, non-classical and intermediate monocytes demonstrated a significant population increase (Fig. 6c) in the treated group compared to that in the untreated group. Among the 1325 important DEGs, the volcano plots revealed 174 upregulated and 1151 downregulated genes (Fig. 6d). Classical monocytes showed the fewest changes in gene expression, particularly S100a8 and S100a9 were downregulated (Fig. 6e), which exhibit minimal activation with minimal inflammatory responses [68, 69]. The other genes, Ccl5, H2aa, and H2eb1, possibly involved in the formation of the MHC II complex, were upregulated to activate adaptive immune responses via T cell activation [63]. GO ontology revealed the vital involvement of both inflammatory genes (S100a8 and S100a9) and hepatoglobin-hemoglobin (H2aa and H2eb1) in the inflammatory response, positive regulation of T cells, and MHC II protein complex formation (Fig. 6f; Table S10).
Fig. 6.
T cell subset profiling. (a) UMAP visualization total T cells from the treated and untreated group were identified with various T cell subsets by PhenoGraph clustering analysis; (b) Reference markers gene expression to visualize heterogeneity of T cells; (c) Population differences of each subset from T cells between treated and untreated groups; (d) The volcano plot revealed that completely expressed genes include both upregulated and downregulated significant genes; e, f. Log2fold changes and DEGs from T1 cells were visualized in network analysis and by gene ontology enrichment analysis; g, h, Gene ontology enrichment analysis, log2fold changes, and DEGs from T2 were visualized; i, j GSEA analysis significant T cells subsets including T1 and T3 subsets
In the case of intermediate monocytes, we observed a significant population increase in cell types that might be involved in primary defense mechanisms via anti-inflammatory responses to maintain homeostasis [70], promote cell proliferation, and reduce apoptosis. However, DEG analysis showed no substantial expression of the genes (Fig. 6g). GO ontology revealed the active involvement of Cd74, Ccl5, Apoe genes that contribute to the positive regulation of chemokine ligand production and ERK1 and ERK2 cascades, respectively (Fig. 6h Table S11). In the DEG analysis, non-classical monocytes showed significant gene expression compared with both classical and non-classical monocytes. Hbb-bs and Hba-a1 were upregulated in the non-classical monocytes, whereas Fgl2, Ifit3, Lrg1, Il1b, Nudt4, S100a9, Ifit2, Isg15, S100a8, and Sell were significantly downregulated (Fig. 6i). Gene ontology results revealed that they were mostly involved in the positive regulation of the inflammatory response via the innate immune response by secreting interferon alpha and gamma, which initiate the apoptotic process (Fig. 6j). Similarly, the cellular components and molecular functions of these genes showed that they were involved in antioxidant activity, oxygen binding, and hemoglobin complex formation (Fig. 6j; Table S12). GSEA analysis revealed that inflammatory cytokines were significantly downregulated in monocyte subsets exposed to E171 TiO2 (Fig. 6k). In this section, we highlighted the diverse roles of monocyte subsets. Classical monocytes initiate an innate inflammatory response that may activate T cells. Whereas non-classical monocytes activate anti-inflammatory responses by suppressing inflammatory protein genes which may help to maintain immune homeostasis as well as reduce cytotoxicity.
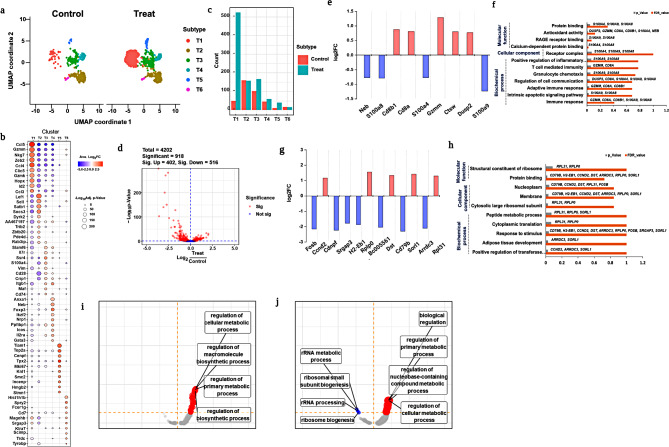
T cells mediated adaptive immune response in the treatment group
Overall, the T cell population increased significantly in the treated group compared with that in the untreated group. However, revealing T cell heterogeneity is more important for predicting population changes because of their distinct roles as T helper or killer cells. In this study, we performed PG clustering analysis of T cells, which revealed four major subsets and two minor subsets (Fig. 7a). The T cells subsets were annotated by the expression marker genes as shown in Fig. 7b and named as T1, T2, T3, T4, T5 and T5. Among the T cell subsets, T1 exhibited a significant population increase, followed by T3 (Fig. 7c). The volcano plot revealed DEGs of T cell subsets, with 402 upregulated and 516 downregulated genes out of a total of 4202 genes (Fig. 7d).
Fig. 7.
In depth profiling of monocytes. (a) UMAP visualization of total monocytes and subset identification in monocytes from treated and untreated groups; (b) Markers gene expression of monocyte subsets to reveal subsets by PhenoGraph clustering analysis; (c) Population differences of each subset of monocytes between treated and untreated groups; (d) Volcano plot diagram represents significant DEGs; e, f. DEGs with log2fold changes and gene ontology analysis of classical monocytes; g, h. DEGs with log2fold changes were observed with upregulated and downregulated genes, and gene ontology of non-classical monocytes; i. j. DEG analysis for intermediate monocytes visualized using log2fold changes, and gene ontology enrichment analysis. k. GSEA analysis to detect enrichment score for monocyte subsets
DEGs were analyzed for each T1 cell subset, and the data were visualized with upregulated and downregulated gene expression with significant fold changes. Figure 7e depicts the log2FC of significant genes from T1 cells such as Cd8a, Cd8b1, Gzmm, Ctsw, and Dusp2 were substantially upregulated, whereas Neb, S100a4, S100a8, and S100a9 were downregulated. Previous research has suggested that T cell surface proteins, such as Cd8a and Cd8b1, may be involved in the activation of the MHC I complex when exposed to foreign particles [71, 72]. Similarly, Cd84 and Cd8b1 expression in the T1 subset indicates the initiation of MHC I complex activation via E171 TiO2 specific antigen detection. However, the activation of T1 subset helps maintain hematopoiesis against inflammatory cytokine responses in both NK and NKT cells (Figs. 4 and 5). This was due to the downregulation of inflammatory proteins, such as S100a4, S100a8, and S100a9 (Fig. 7e), which may be responsible for fibrosis, inflammation, and hyper-immunological responses [68, 73]. GO functional enrichment analyses revealed that Cd8a, Cd8b1, and Gzmm shared functionality (Fig. 7f), involving T cell-mediated immunity via adaptive immune responses and intrinsic apoptotic signaling pathways (Fig. 7f). Similarly, S100a8 and S100a9 might be involved in cellular communication and the regulation of immunological responses through molecular functions, such as protein binding and antioxidant activity (Fig. 7f; Table S13).
Similarly, the gene expression population T3 substantially increased in the treated group compared to that in the untreated group. Genes from T3, such as Ccnd2, Rplp0, Dst, Sorl1, and Rpl31 were upregulated, while Fosb, Cdnpf, Srgap3, H2-eb1, Bc005561, Cd79b, and Arrdc3 were significantly downregulated (Fig. 7g). Higher expression of Ccnd2 and Rplp0 resulted in the upregulation and co-stimulation of CD8+ T cell proliferation [74, 75] through pro-inflammatory cytokine. The downregulation of H2-eb1, Bc005561, Fosb, Srgap3, and Cd79b represented MHC II complex inhibition, which resulted in the failure of specific types of T cell activation in the treated group (Fig. 7g). GO ontology analysis shown in Fig. 7h and detailed information was listed in Table S14. GSEA analysis was performed on both significant subsets, T1 and T3, and both revealed participation in the regulation of cellular metabolic processes (Fig. 7i and j). In the case of T cells subset, particularly, T1 subset mainly activates the CD8+ T cells proliferation and initiates the MHC-I complex upon initial association with E171 TiO2. But further adaptive mediate immune response was suppressed by downregulation of inflammatory protein genes. Whereas T3 subset downregulates the genes which are involved in formation of MHC-II may fail to induce further B cell mediated adaptive immune response due to B and T cell impairment.
B cell-mediated immunological response after treatment
Compared to the major immune cell types, B cells showed very little impact in the treated group; hence, no significant differences were observed between the treated and untreated groups. However, we presume that the heterogeneous functional roles of B cells might respond differently because of their unique characteristics. Therefore, we further analyzed B cell subsets to reveal their heterogeneous characteristics using PhenoGraph clustering analysis (Figure S5). Our findings demonstrated that no more significant alterations in B cell subsets denoted impairment of B and T cell activation fail further antibody mediated response to E171 TiO2.
Discussion
Despite E171 TiO2 is popularly used in various sectors including food, most of their nanotoxicological studies focused on non-oral routes of exposure especially inhalation and dermal contact. Studies have also demonstrated that E171 TiO2 accumulates in various organs such as alimentary canal, lungs, liver, heart, lung, kidneys, spleen, cardiac muscle etc., Researchers proposed that E171 TiO2 can translocate to the bloodstream, urine, or other organs with no obvious changes in immune profile particularly in GI or blood [76, 77]. That being said, repeated intratracheal administration of E171 TiO2 may affect absorption, aggregation, and tissue uptake by integrating with surrounding macromolecules [76, 78]. Another study showed intratracheal installation administration can increase number of B cells of splenocytes and blood [79]. Whereas, nano-TiO2 exposure by intratracheal installation also increased NK cells number in lung [80]. Previous contradictory findings of toxicological and overall safety of E171 TiO2 are still at the budding stage due to gut immunological homeostasis, and therefore it remains highly challenging to conclude regarding the human safety [76]. Henceforth, we would like to re-confirm the previous results of intratracheal installation of E171 TiO2 and to upgrade the understanding of adverse immunological response, we used high dimensional scRNAseq to reveal heterogeneity immune cell subsets at single cell level.
Although various in vitro and in vivo studies have demonstrated the potential E171 TiO2 immunomodulatory effects [76, 81–83] their understanding immunotoxicity at single cell level are still unknown. As a result, knowing heterogeneous immune responses at the single cell level provides in-depth knowledge that would be extremely useful in industrial applications. For the first time, we used high-dimensional scRNA sequencing to evaluate the immune-transcriptional response following intratracheal installation of 5 mg/mL E171 TiO2 after 17 weeks of exposure at the single cell level. Generally, when there is an external stimulus, granulocytes, NK/NKT cells can activate innate immune response that can further stimulate T-cell mediated adaptive immune response. Biomaterials are often considered as foreign particles that can induce pro-inflammatory response upon exposure that are truly influenced by several factors including size, coatings, surface area, charge, or geometrical arrangements etc., They can trigger various biochemical processes including activation of reactive oxygen process (ROS), and interleukin formation [84]. It is also proved that E171 TiO2 can deposit on the cellular surface or inside the cellular organelles and trigger oxidative stress signaling thus causing oxidative damage to the cell [85]. Our study identified E171 TiO2 altered granulocyte population and their respective gene expression by upregulation of pro-inflammatory responsive genes, downregulation of anti-inflammatory genes, and decreased cell number thus resulting in innate immune response. Further, exposure E171 TiO2 to the granulocytes triggered IL-1 and IL-8 mediated pro-inflammatory response. This was in line with the previous report which showed implanted biomaterials initiate the development of multiprotein to mediate pro-inflammatory cytokines such as IL-1 and IL-18 [86, 87].
Additionally, NK/NKT cell population showed increased pro-inflammatory response but also increased cell number thus exhibiting cytotoxicity inhibition. In specific, NK1/NKT1 subset is mainly involved in cytotoxic inhibition by ROS suppression. Whereas NK2 and NK3 subsets might be involved in pro-B cell activation, and inhibition of T cell development respectively. Also, NKT2 and NKT3 might be involved in inhibition of MHC-1 complex and inhibition of cytotoxic effect respectively. Furthermore, acute effect of E171 TiO2 specifically on various immune cells are limited and most of them were homogenous cell environment [2] and can often be misleading to study heterogenous cell type like PBMCs. Our study showed adaptive immune response was suppressed even after hyper innate immune response, which was confirmed by the downregulation of T cell activation. Specifically, we identified impairment of CD4 + T cells mediated MHC-II complex formation upon E171 TiO2 treatment resulted in humoral mediated immune response. Similarly, inhibition of CD8+ T cells induced MHC-1 complex formation thus resulting in cell population increase. This result indicates that E171 TiO2 didn’t exhibit cytotoxicity in T cells subsets which was shown by cell population increase.
Conclusion
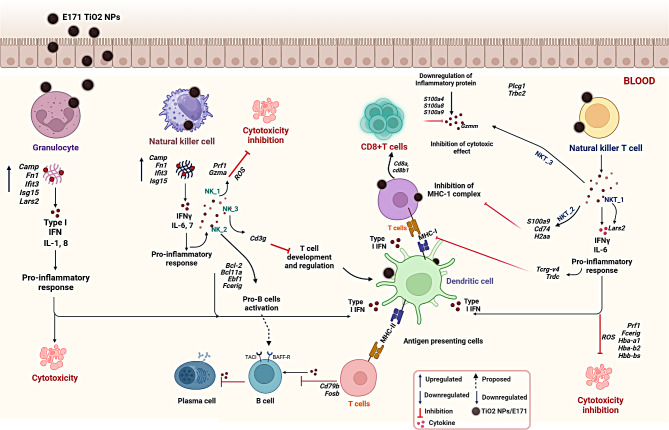
Overall, scRNAseq analysis of Sprague-Dawley Rats PBMCs exhibited heterogenous target specific immune related toxicity responses upon E171 TiO2 treatment. Generally, all immune cell subsets such as granulocytes, monocytes, NK/NKT cells activated innate immune response whereas T cells involved in suppression of adaptive immune response upon E171 TiO2 treatment (Fig. 8). Specifically, E171 TiO2 treatment prolonged hyper-inflammatory response mediated granulocyte cytotoxicity thus inhibiting cell homeostasis. Whereas, NK/NKT cells resulted in heterogeneity role depending on the subsets, nevertheless none of the subsets significantly exhibited cytotoxicity. Furthermore, pro-inflammatory response from granulocytes, NK and NKT cells triggers monocyte into antigen presenting cells that initiated MHC-I or MHC-II complexes which was confirmed by the activation of T cell subsets implying suppression of adaptive mediated immune response. Although our study using scRNA-sequencing provides comprehensive inspiring insights of immune cell types and subtypes changes upon E171 TiO2 exposure, certain limitations must be acknowledged. Firstly, proper validation assays must be done to support our findings, such as transcriptomic changes in each immune cell type and their heterogeneous subsets. Predominantly, the DEGs from significant immune subset involved in the immunological response upon E171 TiO2 exposure must be elucidated in order to confirm their inflammatory-mediated pathway. Second, our findings also didn’t reveal whether E171 TiO2 were internalized by any of the immune cell types which needs to be confirmed in future studies.
Fig. 8.
Overview of heterogeneous immunological response in treated groups at the single cell level, along with DEGs and GO results
Electronic supplementary material
Below is the link to the electronic supplementary material.
Author contributions
HP and THY conceived and designed the research study and wrote the manuscript. HP, HYK, and XX analyzed the data. HYH, JHO, SY carried out software analysis and resources: MBH and TGL contributed formal analysis and methodology. HP, THY, and HYK were involved in the related discussion. All authors reviewed and accepted the current version of the manuscript.
Funding
This work was supported by Nano Material Technology Development Program (Grant number 2021M3A7B6031397 and RS-2024-00452934 ), Basic Science Research Program (Grant number 2020R1A6A1A06046728) and International Cooperative R&D Program (grant number 2022K1A3A1A78097929) through the National Research Foundation of Korea (NRF). This work was also conducted as part of the European Commission Horizon 2020 Program (H2020) project CompSafeNano (grant agreement number 101008099) as well as the Horizon Europe program projects INSIGHT (grant agreement number 101137742 ) and CHIASMA (grant agreement number 101137613).
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethical approval and consent to participate
This study was approved by the Animal Welfare and Guide for the Care and Use of Laboratory Animals and was assessed by the Institutional Animal Care and Use Committee (IACUC) (approval number N221019).
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Baranowska-Wójcik E, Szwajgier D, Winiarska-Mieczan A. A review of research on the impact of E171/TiO2 NPs on the digestive tract. J Trace Elem Med Biol [Internet]. 2022;72:126988. https://www.sciencedirect.com/science/article/pii/S0946672X22000682 [DOI] [PubMed]
- 2.Zagal-Salinas AA, Ispanixtlahuatl-Meráz O, Olguín-Hernández JE, Rodríguez-Sosa M, García Cuéllar CM, Sánchez-Pérez Y et al. Food grade titanium dioxide (E171) interferes with monocyte-macrophage cell differentiation and their phagocytic capacity. Food Chem Toxicol [Internet]. 2024;192:114912. https://www.sciencedirect.com/science/article/pii/S0278691524004782 [DOI] [PubMed]
- 3.(FAF) EP on FA and, Younes F, Aquilina M, Castle G, Engel L, Fowler K-H et al. P,. Safety assessment of titanium dioxide (E171) as a food additive. EFSA J [Internet]. 2021;19:e06585. 10.2903/j.efsa.2021.6585 [DOI] [PMC free article] [PubMed]
- 4.Cao X, Han Y, Gu M, Du H, Song M, Zhu X et al. Foodborne Titanium Dioxide Nanoparticles Induce Stronger Adverse Effects in Obese Mice than Non-Obese Mice: Gut Microbiota Dysbiosis, Colonic Inflammation, and Proteome Alterations. Small [Internet]. 2020;16:2001858. 10.1002/smll.202001858 [DOI] [PubMed]
- 5.Heringa MB, Geraets L, van Eijkeren JCH, Vandebriel RJ, de Jong WH, Oomen AG. Risk assessment of titanium dioxide nanoparticles via oral exposure, including toxicokinetic considerations. Nanotoxicology [Internet]. 2016;10:1515–25. 10.1080/17435390.2016.1238113 [DOI] [PubMed]
- 6.Shakeel M, Jabeen F, Shabbir S, Asghar MS, Khan MS, Chaudhry AS. Toxicity of Nano-Titanium Dioxide (TiO2-NP) through various routes of exposure: a review. Biol Trace Elem Res. 2016;172:1–36. [DOI] [PubMed] [Google Scholar]
- 7.Weir A, Westerhoff P, Fabricius L, Hristovski K, von Goetz N. Titanium Dioxide Nanoparticles in Food and Personal Care Products. Environ Sci Technol [Internet]. 2012;46:2242–50. 10.1021/es204168d [DOI] [PMC free article] [PubMed]
- 8.Dorier M, Béal D, Marie-Desvergne C, Dubosson M, Barreau F, Houdeau E, et al. Continuous in vitro exposure of intestinal epithelial cells to E171 food additive causes oxidative stress, inducing oxidation of DNA bases but no endoplasmic reticulum stress. Nanotoxicology. 2017;11:751–61. [DOI] [PubMed] [Google Scholar]
- 9.Peters RJB, van Bemmel G, Herrera-Rivera Z, Helsper HPFG, Marvin HJP, Weigel S, et al. Characterization of titanium dioxide nanoparticles in food products: analytical methods to define nanoparticles. J Agric Food Chem. 2014;62:6285–93. [DOI] [PubMed] [Google Scholar]
- 10.Ruiz PA, Morón B, Becker HM, Lang S, Atrott K, Spalinger MR, et al. Titanium dioxide nanoparticles exacerbate DSS-induced colitis: role of the NLRP3 inflammasome. Gut. 2017;66:1216–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han H-Y, Yang M-J, Yoon C, Lee G-H, Kim D-W, Kim T-W, et al. Toxicity of orally administered food-grade titanium dioxide nanoparticles. J Appl Toxicol. 2021;41:1127–47. [DOI] [PubMed] [Google Scholar]
- 12.Dorier M, Tisseyre C, Dussert F, Béal D, Arnal M-E, Douki T et al. Toxicological impact of acute exposure to E171 food additive and TiO2 nanoparticles on a co-culture of Caco-2 and HT29-MTX intestinal cells. Mutat Res Toxicol Environ Mutagen [Internet]. 2019;845:402980. https://www.sciencedirect.com/science/article/pii/S1383571818302493 [DOI] [PubMed]
- 13.Ryabtseva MS, Umanskaya SF, Shevchenko MA, Krivobok VS, Kolobov AV, Nastulyavichus AA et al. Transformation of Nano-Size Titanium Dioxide Particles in the gastrointestinal tract and its role in the transfer of nanoparticles through the intestinal barrier. Int J Mol Sci. 2023;24. [DOI] [PMC free article] [PubMed]
- 14.Mikulak J, Bruni E, Oriolo F, Di Vito C, Mavilio D. Hepatic Natural Killer Cells: Organ-Specific Sentinels of Liver Immune Homeostasis and Physiopathology. Front Immunol [Internet]. 2019;10. https://www.frontiersin.org/articles/10.3389/fimmu.2019.00946 [DOI] [PMC free article] [PubMed]
- 15.Li L, Zeng Z. Live Imaging of Innate and Adaptive Immune Responses in the Liver. Front Immunol [Internet]. 2020;11. https://www.frontiersin.org/articles/10.3389/fimmu.2020.564768 [DOI] [PMC free article] [PubMed]
- 16.Shi H, Magaye R, Castranova V, Zhao J. Titanium dioxide nanoparticles: a review of current toxicological data. Part Fibre Toxicol [Internet]. 2013;10:15. 10.1186/1743-8977-10-15 [DOI] [PMC free article] [PubMed]
- 17.Gissen P, Arias IM. Structural and functional hepatocyte polarity and liver disease. J Hepatol. 2015;63:1023–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu X, Zhu Y, Bai R, Wu Z, Qian W, Yang L et al. Long-term pulmonary exposure to multi-walled carbon nanotubes promotes breast cancer metastatic cascades. Nat Nanotechnol [Internet]. 2019;14:719–27. 10.1038/s41565-019-0472-4 [DOI] [PubMed]
- 19.DeLoid G, Casella B, Pirela S, Filoramo R, Pyrgiotakis G, Demokritou P et al. Effects of engineered nanomaterial exposure on macrophage innate immune function. NanoImpact [Internet]. 2016;2:70–81. https://www.sciencedirect.com/science/article/pii/S2452074816300350 [DOI] [PMC free article] [PubMed]
- 20.Jähne EA, Eigenmann DE, Moradi-Afrapoli F, Verjee S, Butterweck V, Hebeisen S, et al. Caco-2 permeability studies and in Vitro hERG Liability Assessment of Tryptanthrin and Indolinone. Planta Med. 2016;82:1192–201. [DOI] [PubMed] [Google Scholar]
- 21.Baumgarth N, Herman OC, Jager GC, Brown LE, Herzenberg LA, Chen J. B-1 and B-2 cell-derived immunoglobulin M antibodies are nonredundant components of the protective response to influenza virus infection. J Exp Med. 2000;192:271–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adan A, Alizada G, Kiraz Y, Baran Y, Nalbant A. Flow cytometry: basic principles and applications. Crit Rev Biotechnol. 2017;37:163–76. [DOI] [PubMed] [Google Scholar]
- 23.Wu M, Cronin K, Crane JS, Biochemistry. Collagen Synthesis. Treasure Island (FL); 2021. [PubMed]
- 24.Hu H, Li L, Guo Q, Zong H, Yan Y, Yin Y, et al. RNA sequencing analysis shows that titanium dioxide nanoparticles induce endoplasmic reticulum stress, which has a central role in mediating plasma glucose in mice. Nanotoxicology. 2018;12:341–56. [DOI] [PubMed] [Google Scholar]
- 25.Wang S, Alenius H, El-Nezami H, Karisola P. A New Look at the Effects of Engineered ZnO and TiO(2) Nanoparticles: Evidence from Transcriptomics Studies. Nanomater (Basel, Switzerland). 2022;12. [DOI] [PMC free article] [PubMed]
- 26.Fuster E, Candela H, Estévez J, Vilanova E, Sogorb MA. Titanium Dioxide, but not zinc oxide, nanoparticles cause severe transcriptomic alterations in T98G human glioblastoma cells. Int J Mol Sci. 2021;22. [DOI] [PMC free article] [PubMed]
- 27.Bae J, Ha M, Perumalsamy H, Lee Y, Song J, Yoon T-H. Mass Cytometry Exploration of Immunomodulatory Responses of Human Immune Cells Exposed to Silver Nanoparticles. Pharmaceutics [Internet]. 2022;14. https://www.mdpi.com/1999-4923/14/3/630 [DOI] [PMC free article] [PubMed]
- 28.Bae J, Kim JE, Perumalsamy H, Park S, Kim Y, Jun DW et al. Mass Cytometry Study on Hepatic Fibrosis and Its Drug-Induced Recovery Using Mouse Peripheral Blood Mononuclear Cells. Front Immunol [Internet]. 2022;13. https://www.frontiersin.org/articles/10.3389/fimmu.2022.814030 [DOI] [PMC free article] [PubMed]
- 29.Satija R, Shalek AK. Heterogeneity in immune responses: from populations to single cells. Trends Immunol [Internet]. 2014;35:219–29. https://www.sciencedirect.com/science/article/pii/S1471490614000520 [DOI] [PMC free article] [PubMed]
- 30.Geering B, Stoeckle C, Conus S, Simon H-U. Living and dying for inflammation: neutrophils, eosinophils, basophils. Trends Immunol. 2013;34:398–409. [DOI] [PubMed] [Google Scholar]
- 31.Herrero-Cervera A, Soehnlein O, Kenne E. Neutrophils in chronic inflammatory diseases. Cell Mol Immunol [Internet]. 2022;19:177–91. 10.1038/s41423-021-00832-3 [DOI] [PMC free article] [PubMed]
- 32.Lee C-H, Choi EY. Macrophages and Inflammation. jrd [Internet]. 2018;25:11–8. 10.4078/jrd.2018.25.1.11
- 33.Ellis L-JA, Lynch I. Mechanistic insights into toxicity pathways induced by nanomaterials in Daphnia magna from analysis of the composition of the acquired protein corona. Environ Sci Nano [Internet]. 2020;7:3343–59. 10.1039/D0EN00625D
- 34.Lourda M, Dzidic M, Hertwig L, Bergsten H, Palma Medina LM, Sinha I et al. High-dimensional profiling reveals phenotypic heterogeneity and disease-specific alterations of granulocytes in COVID-19. Proc Natl Acad Sci [Internet]. 2021;118:e2109123118. 10.1073/pnas.2109123118 [DOI] [PMC free article] [PubMed]
- 35.Raker VK, Becker C, Steinbrink K. The cAMP Pathway as Therapeutic Target in Autoimmune and Inflammatory Diseases. Front Immunol [Internet]. 2016;7. https://www.frontiersin.org/articles/10.3389/fimmu.2016.00123 [DOI] [PMC free article] [PubMed]
- 36.Glasner A, Levi A, Enk J, Isaacson B, Viukov S, Orlanski S, et al. NKp46 receptor-mediated Interferon-γ production by Natural Killer Cells Increases Fibronectin 1 to alter Tumor Architecture and Control Metastasis. Immunity. 2018;48:107–e1194. [DOI] [PubMed] [Google Scholar]
- 37.Perng Y-C, Lenschow DJ. ISG15 in antiviral immunity and beyond. Nat Rev Microbiol [Internet]. 2018;16:423–39. 10.1038/s41579-018-0020-5 [DOI] [PMC free article] [PubMed]
- 38.Kopitar-Jerala N. The role of cystatins in cells of the immune system. FEBS Lett [Internet]. 2006;580:6295–301. 10.1016/j.febslet.2006.10.055 [DOI] [PubMed]
- 39.Schaller TH, Batich KA, Suryadevara CM, Desai R, Sampson JH. Chemokines as adjuvants for immunotherapy: implications for immune activation with CCL3. Expert Rev Clin Immunol [Internet]. 2017;13:1049–60. 10.1080/1744666X.2017.1384313 [DOI] [PMC free article] [PubMed]
- 40.Lu Y, Zuo Q, Zhang Y, Wang Y, Li T, Han J. The expression profile of IFITM family gene in rats. Intractable rare Dis Res. 2017;6:274–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mikelez-Alonso I, Magadán S, González-Fernández Á, Borrego F. Natural killer (NK) cell-based immunotherapies and the many faces of NK cell memory: A look into how nanoparticles enhance NK cell activity. Adv Drug Deliv Rev [Internet]. 2021;176:113860. https://www.sciencedirect.com/science/article/pii/S0169409X21002520 [DOI] [PubMed]
- 42.Le Garff-Tavernier M, Béziat V, Decocq J, Siguret V, Gandjbakhch F, Pautas E, et al. Human NK cells display major phenotypic and functional changes over the life span. Aging Cell. 2010;9:527–35. [DOI] [PubMed] [Google Scholar]
- 43.Freud AG, Mundy-Bosse BL, Yu J, Caligiuri MA. The Broad Spectrum of Human Natural Killer Cell Diversity. Immunity. 2017;47:820–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Crinier A, Dumas P-Y, Escalière B, Piperoglou C, Gil L, Villacreces A et al. Single-cell profiling reveals the trajectories of natural killer cell differentiation in bone marrow and a stress signature induced by acute myeloid leukemia. Cell Mol Immunol [Internet]. 2021;18:1290–304. 10.1038/s41423-020-00574-8 [DOI] [PMC free article] [PubMed]
- 45.Lieberman J. Granzyme A activates another way to die. Immunol Rev. 2010;235:93–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Osińska I, Popko K, Demkow U. Perforin: an important player in immune response. Cent J Immunol. 2014;39:109–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou G, Wang T, Zha X. RNA-Seq analysis of knocking out the neuroprotective proton-sensitive GPR68 on basal and acute ischemia-induced transcriptome changes and signaling in mouse brain. FASEB J [Internet]. 2021;35:e21461. 10.1096/fj.202002511R [DOI] [PMC free article] [PubMed]
- 48.Yang M, Fan Q, Hei TK, Chen G, Cao W, Meng G et al. Single-Cell Transcriptome Analysis of Radiation Pneumonitis Mice. Antioxidants [Internet]. 2022;11. https://www.mdpi.com/2076-3921/11/8/1457 [DOI] [PMC free article] [PubMed]
- 49.Li J, Wang X, Zhao G, Chen C, Chai Z, Alsaedi A et al. Metal–organic framework-based materials: superior adsorbents for the capture of toxic and radioactive metal ions. Chem Soc Rev [Internet]. 2018;47:2322–56. 10.1039/C7CS00543A [DOI] [PubMed]
- 50.Lukin K, Fields S, Guerrettaz L, Straign D, Rodriguez V, Zandi S, et al. A dose-dependent role for EBF1 in repressing non-b-cell-specific genes. Eur J Immunol. 2011;41:1787–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bacon C, Endris V, Rappold GA. The cellular function of srGAP3 and its role in neuronal morphogenesis. Mech Dev. 2013;130:391–5. [DOI] [PubMed] [Google Scholar]
- 52.Uyeda MJ, Freeborn RA, Cieniewicz B, Romano R, Chen PP, Liu JM-H, et al. BHLHE40 regulates IL-10 and IFN-γ production in T cells but does not interfere with Human Type 1 Regulatory T cell differentiation. Front Immunol. 2021;12:683680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mirzaei HR. Adaptive Immunity. In: Rezaei NBT-E of I and I, editor. Oxford: Elsevier; 2022. pp. 39–55. https://www.sciencedirect.com/science/article/pii/B9780128187319000288
- 54.Tupin E, Kinjo Y, Kronenberg M. The unique role of natural killer T cells in the response to microorganisms. Nat Rev Microbiol [Internet]. 2007;5:405–17. 10.1038/nrmicro1657 [DOI] [PubMed]
- 55.Kuylenstierna C, Björkström NK, Andersson SK, Sahlström P, Bosnjak L, Paquin-Proulx D, et al. NKG2D performs two functions in invariant NKT cells: direct TCR-independent activation of NK-like cytolysis and co-stimulation of activation by CD1d. Eur J Immunol. 2011;41:1913–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krijgsman D, Hokland M, Kuppen PJK. The Role of Natural Killer T Cells in Cancer—A Phenotypical and Functional Approach. Front Immunol [Internet]. 2018;9. https://www.frontiersin.org/articles/10.3389/fimmu.2018.00367 [DOI] [PMC free article] [PubMed]
- 57.Yang Z, Zhou X, Liu Y, Gong C, Wei X, Zhang T, et al. Activation of integrin β1 mediates the increased malignant potential of ovarian cancer cells exerted by inflammatory cytokines. Anticancer Agents Med Chem. 2014;14:955–62. [DOI] [PubMed] [Google Scholar]
- 58.Huang X, Shen W, Veizades S, Liang G, Sayed N, Nguyen PK. Single-cell transcriptional profiling reveals sex and age diversity of Gene expression in mouse endothelial cells. Front Genet. 2021;12:590377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu Y, Cook C, Sedgewick AJ, Zhang S, Fassett MS, Ricardo-Gonzalez RR, et al. Single-cell profiling reveals Divergent, globally patterned Immune responses in murine skin inflammation. iScience. 2020;23:101582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xue C, Zhang J, Zhang G, Xue Y, Zhang G, Wu X. Elevated SPINK2 gene expression is a predictor of poor prognosis in acute myeloid leukemia. Oncol Lett. 2019;18:2877–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li H, van der Leun AM, Yofe I, Lubling Y, Gelbard-Solodkin D, van Akkooi ACJ, et al. Dysfunctional CD8 T cells form a proliferative, dynamically regulated compartment within human melanoma. Cell. 2019;176:775–e78918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zeng L, Palaia I, Šarić A, Su X. PLCγ1 promotes phase separation of T cell signaling components. J Cell Biol. 2021;220. [DOI] [PMC free article] [PubMed]
- 63.Lee YJ, Starrett GJ, Lee ST, Yang R, Henzler CM, Jameson SC, et al. Lineage-specific Effector signatures of invariant NKT cells are Shared amongst γδ T, Innate Lymphoid, and th cells. J Immunol. 2016;197:1460–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zolnik BS, González-Fernández A, Sadrieh N, Dobrovolskaia MA. Nanoparticles and the immune system. Endocrinology [Internet]. 2009/12/16. 2010;151:458–65. https://pubmed.ncbi.nlm.nih.gov/20016026 [DOI] [PMC free article] [PubMed]
- 65.Luo Y-H, Chang LW, Lin P. Metal-Based Nanoparticles and the Immune System: Activation, Inflammation, and Potential Applications. Yu IJ, editor. Biomed Res Int [Internet]. 2015;2015:143720. 10.1155/2015/143720 [DOI] [PMC free article] [PubMed]
- 66.Curtsinger JM, Schmidt CS, Mondino A, Lins DC, Kedl RM, Jenkins MK et al. Inflammatory Cytokines Provide a Third Signal for Activation of Naive CD4 + and CD8 + T Cells. J Immunol [Internet]. 1999;162:3256–62. https://www.jimmunol.org/content/162/6/3256 [PubMed]
- 67.Gerner MY, Torabi-Parizi P, Germain RN. Strategically Localized Dendritic Cells Promote Rapid T Cell Responses to Lymph-Borne Particulate Antigens. Immunity [Internet]. 2015;42:172–85. https://www.sciencedirect.com/science/article/pii/S1074761314004919 [DOI] [PubMed]
- 68.Vogl T, Gharibyan AL, Morozova-Roche LA, Pro-Inflammatory. S100A8 and S100A9 Proteins: Self-Assembly into Multifunctional Native and Amyloid Complexes. Int J Mol Sci [Internet]. 2012;13:2893–917. https://www.mdpi.com/1422-0067/13/3/2893 [DOI] [PMC free article] [PubMed]
- 69.Madrigal A, Tan L, Zhao Y. Expression regulation and functional analysis of RGS2 and RGS4 in adipogenic and osteogenic differentiation of human mesenchymal stem cells. Biol Res [Internet]. 2017;50:43. 10.1186/s40659-017-0148-1 [DOI] [PMC free article] [PubMed]
- 70.Narasimhan PB, Marcovecchio P, Hamers AAJ, Hedrick CC. Nonclassical monocytes in Health and Disease. Annu Rev Immunol. 2019;37:439–56. [DOI] [PubMed] [Google Scholar]
- 71.Gülich AF, Preglej T, Hamminger P, Alteneder M, Tizian C, Orola MJ et al. Differential Requirement of Cd8 Enhancers E8I and E8VI in Cytotoxic Lineage T Cells and in Intestinal Intraepithelial Lymphocytes. Front Immunol [Internet]. 2019;10. https://www.frontiersin.org/articles/10.3389/fimmu.2019.00409 [DOI] [PMC free article] [PubMed]
- 72.Muntjewerff EM, Christoffersson G, Mahata SK, van den Bogaart G. Putative regulation of macrophage-mediated inflammation by catestatin. Trends Immunol. 2022;43:41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fei F, Qu J, Zhang M, Li Y, Zhang S. S100A4 in cancer progression and metastasis: a systematic review. Oncotarget. 2017;8:73219–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gotthardt D, Putz EM, Grundschober E, Prchal-Murphy M, Straka E, Kudweis P et al. STAT5 Is a Key Regulator in NK Cells and Acts as a Molecular Switch from Tumor Surveillance to Tumor Promotion. Cancer Discov [Internet]. 2016;6:414–29. 10.1158/2159-8290.CD-15-0732 [DOI] [PubMed]
- 75.Campbell AR, Regan K, Bhave N, Pattanayak A, Parihar R, Stiff AR, et al. Gene expression profiling of the human natural killer cell response to fc receptor activation: unique enhancement in the presence of interleukin-12. BMC Med Genomics. 2015;8:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Blevins LK, Crawford RB, Bach A, Rizzo MD, Zhou J, Henriquez JE et al. Evaluation of immunologic and intestinal effects in rats administered an E 171-containing diet, a food grade titanium dioxide (TiO2). Food Chem Toxicol [Internet]. 2019;133:110793. https://www.sciencedirect.com/science/article/pii/S0278691519305836 [DOI] [PMC free article] [PubMed]
- 77.MacNicoll A, Kelly M, Aksoy H, Kramer E, Bouwmeester H, Chaudhry Q. A study of the uptake and biodistribution of nano-titanium dioxide using in vitro and in vivo models of oral intake. J Nanoparticle Res [Internet]. 2015;17:66. 10.1007/s11051-015-2862-3
- 78.DeLoid GM, Wang Y, Kapronezai K, Lorente LR, Zhang R, Pyrgiotakis G et al. An integrated methodology for assessing the impact of food matrix and gastrointestinal effects on the biokinetics and cellular toxicity of ingested engineered nanomaterials. Part Fibre Toxicol [Internet]. 2017;14:40. 10.1186/s12989-017-0221-5 [DOI] [PMC free article] [PubMed]
- 79.Park E-J, Yoon J, Choi K, Yi J, Park K. Induction of chronic inflammation in mice treated with titanium dioxide nanoparticles by intratracheal instillation. Toxicology [Internet]. 2009;260:37–46. https://www.sciencedirect.com/science/article/pii/S0300483X0900136X [DOI] [PubMed]
- 80.Gustafsson Å, Lindstedt E, Elfsmark LS, Bucht A. Lung exposure of titanium dioxide nanoparticles induces innate immune activation and long-lasting lymphocyte response in the Dark Agouti rat. J Immunotoxicol. 2011;8:111–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bettini S, Boutet-Robinet E, Cartier C, Coméra C, Gaultier E, Dupuy J et al. Food-grade TiO2 impairs intestinal and systemic immune homeostasis, initiates preneoplastic lesions and promotes aberrant crypt development in the rat colon. Sci Rep [Internet]. 2017;7:40373. 10.1038/srep40373 [DOI] [PMC free article] [PubMed]
- 82.Urrutia-Ortega IM, Garduño-Balderas LG, Delgado-Buenrostro NL, Freyre-Fonseca V, Flores-Flores JO, González-Robles A, et al. Food-grade titanium dioxide exposure exacerbates tumor formation in colitis associated cancer model. Food Chem Toxicol Int J Publ Br Ind Biol Res Assoc. 2016;93:20–31. [DOI] [PubMed] [Google Scholar]
- 83.Bischoff NS, de Kok TM, Sijm DTHM, van Breda SG, Briedé JJ, Castenmiller JJM et al. Possible adverse effects of Food Additive E171 (Titanium Dioxide) related to particle specific human toxicity, including the Immune System. Int J Mol Sci. 2020;22. [DOI] [PMC free article] [PubMed]
- 84.Aljabali AA, Obeid MA, Bashatwah RM, Serrano-Aroca Á, Mishra V, Mishra Y et al. Nanomaterials and Their Impact on the Immune System. Int J Mol Sci [Internet]. 2023;24. https://www.mdpi.com/1422-0067/24/3/2008 [DOI] [PMC free article] [PubMed]
- 85.Rodríguez-Ibarra C, Medina-Reyes EI, Déciga-Alcaraz A, Delgado-Buenrostro NL, Quezada-Maldonado EM, Ispanixtlahuatl-Meráz O et al. Food grade titanium dioxide accumulation leads to cellular alterations in colon cells after removal of a 24-hour exposure. Toxicology [Internet]. 2022;478:153280. https://www.sciencedirect.com/science/article/pii/S0300483X22001925 [DOI] [PubMed]
- 86.Malik AF, Hoque R, Ouyang X, Ghani A, Hong E, Khan K et al. Inflammasome components Asc and caspase-1 mediate biomaterial-induced inflammation and foreign body response. Proc Natl Acad Sci [Internet]. 2011;108:20095–100. 10.1073/pnas.1105152108 [DOI] [PMC free article] [PubMed]
- 87.Demento SL, Eisenbarth SC, Foellmer HG, Platt C, Caplan MJ, Mark Saltzman W et al. Inflammasome-activating nanoparticles as modular systems for optimizing vaccine efficacy. Vaccine [Internet]. 2009;27:3013–21. https://www.sciencedirect.com/science/article/pii/S0264410X09004393 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed during the current study.