Abstract
Oral squamous cell carcinoma (OSCC) is a prevalent oral malignancy, which poses significant health risks with a high mortality rate. Regulatory T cells (Tregs), characterized by their immunosuppressive capabilities, are intricately linked to OSCC progression and patient outcomes. The metabolic reprogramming of Tregs within the OSCC tumor microenvironment (TME) underpins their function, with key pathways such as the tryptophan-kynurenine-aryl hydrocarbon receptor, PI3K-Akt-mTOR and nucleotide metabolism significantly contributing to their suppressive activities. Targeting these metabolic pathways offers a novel therapeutic approach to reduce Treg-mediated immunosuppression and enhance anti-tumor responses. This review explores the metabolic dependencies and pathways that sustain Treg function in OSCC, highlighting key metabolic adaptations such as glycolysis, fatty acid oxidation, amino acid metabolism and PI3K-Akt-mTOR signaling pathway that enable Tregs to thrive in the challenging conditions of the TME. Additionally, the review discusses the influence of the oral microbiome on Treg metabolism and evaluates potential therapeutic strategies targeting these metabolic pathways. Despite the promising potential of these interventions, challenges such as selectivity, toxicity, tumor heterogeneity, and resistance mechanisms remain. The review concludes with perspectives on personalized medicine and integrative approaches, emphasizing the need for continued research to translate these findings into effective clinical applications for OSCC treatment.
Keywords: Oral squamous cell carcinoma, Regulatory T cells, Metabolic targeting, Immunotherapy, Tumor microenvironment
Introduction
Oral squamous cell carcinoma (OSCC) is the most prevalent malignancy of the oral cavity, accounting for over 90% of oral cancers [1]. It represents a significant global health challenge, with higher incidences reported in regions like Southeast Asia and Europe, largely attributed to risk factors such as betel quid chewing, alcohol consumption, and tobacco use [2]. According to recent epidemiological estimates, OSCC contributes to approximately 389,000 new cases and 188,000 deaths annually worldwide [3]. Despite advancements in diagnostic and therapeutic strategies, the 5-year survival rate for OSCC remains at approximately 50%, largely due to late-stage diagnosis, frequent recurrences, and resistance to conventional therapies [4, 5]. Given its aggressive progression, substantial global burden, and the limitations of current therapies, there is an urgent need to develop more effective treatment strategies.
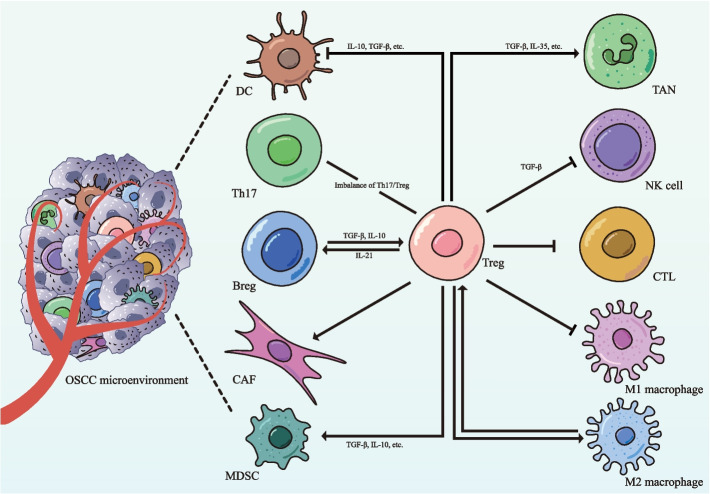
Regulatory T cells (Tregs), known for their immunosuppressive functions, play a pivotal role in the tumor microenvironment (TME) of OSCC (Fig. 1). While Tregs are essential for maintaining immune homeostasis, tumors often exploit these cells to evade immune surveillance [6]. In OSCC, Tregs suppress the the activity of various effector immune cells, including natural killer (NK) cells, dendritic cells (DCs), and B cells, simultaneously enhancing the immunosuppressive effects of cells such as tumor-associated neutrophils and myeloid-derived suppressor cells (MDSCs) [6–10]. This dual functionality makes Tregs a critical factor in the progression and immune evasion of OSCC.
Fig. 1.
Regulatory T cells as central modulators of immune dynamics in the tumor microenvironment of OSCC. The tumor microenvironment (TME) of oral squamous cell carcinoma (OSCC) represents a complex immunological landscape where regulatory T cells (Tregs) emerge as critical orchestrators of immunosuppression. Through multiple mechanisms, Tregs effectively suppress anti-tumor immune responses by inhibiting the cytotoxic functions of natural killer (NK) cells and CD8+ cytotoxic T lymphocytes (CTLs), while simultaneously suppressing the pro-inflammatory activities of M1 macrophages. By secreting immunosuppressive cytokines, primarily IL-10 and TGF-β, Tregs impair dendritic cell (DC) maturation and antigen presentation, promoting the development of tolerogenic DCs. Additionally, Tregs enhance the immunosuppressive functions of myeloid-derived suppressor cells (MDSCs) and tumor-associated neutrophils (TANs), thereby further inhibiting CTLs responses and contributing to tumor progression. This immunosuppressive network is reinforced by reciprocal interactions between Tregs, M2-polarized macrophages, and regulatory B cells (Bregs), creating self-sustaining circuits that foster tumor immune escape. Cancer-associated fibroblasts (CAFs) contribute by secreting chemokines that recruit Tregs and producing factors that stabilize their function. Additionally, the reduced Th17/Treg ratio observed in OSCC correlates with heightened immune tolerance and advanced disease progression. TME: tumor microenvironment; OSCC: oral squamous cell carcinoma; Treg: regulatory T cell; NK: natural killer; CTL: cytotoxic T lymphocyte; DC: dendritic cell; MDSC: myeloid-derived suppressor cell; TAN: tumor-associated neutrophils; Breg: regulatory B cell; CAF: cancer-associated fibroblast
Current research underscores the significant impact of Tregs on patient outcomes in OSCC, but several challenges remain. Studies have highlighted the importance of metabolic reprogramming in Tregs, which is essential for their function and survival within the TME. Key metabolic pathways, such as glycolysis, fatty acid oxidation, amino acid metabolism, and the PI3K-Akt-mTOR signaling pathway, enable Tregs to adapt to the harsh conditions of the TME, characterized by hypoxia and nutrient deprivation [11, 12]. Additionally, the oral microbiome plays a critical role in modulating Treg metabolism, adding complexity to the metabolic landscape of OSCC. Despite these insights, the precise mechanisms by which these metabolic pathways influence Treg function in OSCC remain poorly understood.
This review explores the metabolic dependencies of Tregs in OSCC and examines how targeting these pathways could pave the way for novel therapeutic strategies. By disrupting the metabolic support that sustains Treg function, it may be possible to enhance anti-tumor immunity and improve patient outcomes. However, the translation of these strategies into clinical practice is fraught with challenges. Key obstacles include achieving selectivity in targeting Tregs without impairing other immune cells, managing tumor heterogeneity, and overcoming resistance mechanisms. The future of OSCC treatment may lie in integrative and personalized approaches. By combining metabolic targeting with other therapeutic modalities, it may be possible to develop more effective and tailored interventions, offering renewed hope for improved management of this challenging malignancy.
Metabolic adaptations of regulatory T cells in the TME of OSCC
Tregs play a crucial role in shaping the immunosuppressive environment of OSCC. Their metabolic programming is essential for maintaining their function and survival within the TME. Tregs exhibit significant metabolic flexibility that allows them to adapt to the harsh conditions of the TME in OSCC. Within the TME, Tregs navigate a range of metabolic stresses such as hypoxia, nutrient competition, and the immunosuppressive influence of immune checkpoints [13, 14].
Hypoxia and glycolytic shift in tregs
Hypoxia represents a fundamental characteristic of the TME in OSCC, emerging as a critical regulator of both tumor progression and immune cell function. This oxygen-deprived environment develops due to rapid tumor proliferation and abnormal vasculature. In response, Tregs in OSCC upregulate hypoxia-inducible factors (HIFs), particularly HIF-1α, to adapt to these oxygen-deprived conditions [15]. HIF-1α plays a crucial role in reprogramming Treg metabolism, driving a shift from oxidative phosphorylation to glycolysis [16–18]. This metabolic shift not only supports Treg survival and functionality under hypoxic conditions but also enhances their immunosuppressive capabilities. By prioritizing glycolytic, Tregs can maintain their regulatory functions within the hypoxic tumor niches, contributing to immune evasion and tumor progression.
Nutrient scarcity and alternative energy source
Within the TME, Tregs face severe nutrient scarcity due to competition from tumor cells and other immune cells for glucose, amino acids, and lipids. To adapt to these challenging conditions, Tregs exhibit metabolic flexibility that allows them to utilize alternative pathways, such as fatty acid oxidation (FAO) and amino acid metabolism, to meet their energy demands [19, 20]. This metabolic plasticity enables Tregs to maintain their suppressive functions even under conditions of glucose deprivation. Additionally, Tregs can metabolize lactate, a byproduct of aerobic glycolysis prevalent in the TME, to support their energy demands and maintain stability [21, 22].
Metabolic flexibility and adaptation in tregs
The ability of Tregs to switch between different metabolic pathways is a critical aspect of their adaptability in the TME [23, 24]. Beyond glycolysis and FAO, Tregs can utilize oxidative phosphorylation when conditions permit, thus exhibiting a high degree of metabolic flexibility [25, 26]. This adaptability is regulated by key signaling pathways such as the PI3K-Akt-mTOR axis, which integrates various metabolic cues and dynamically adjusts Treg metabolic programs. By fine-tuning their metabolic states, Tregs optimize their survival and immunosuppressive functions amidst the dynamic and hostile environment of the TME.
Regulation of treg metabolism by immune checkpoints.
Immune checkpoints, such as CTLA-4 and PD-1, play pivotal roles in modulating Treg function and metabolism within the TME [27]. Tregs express checkpoint molecules, including PD-1, CTLA-4, LAG-3, ICOS, and TIM-3, making them key targets for immune checkpoint blockade therapies. These checkpoints not only reinforce Treg-mediated immunosuppression but also influence their metabolic pathways. For instance, engagement of PD-1 on Tregs enhances FAO while suppressing glycolysis, promoting more sustainable energy production under nutrient-limited conditions [26, 28]. Additionally, the activation of these checkpoints helps Tregs conserve energy and maintain their suppressive phenotype despite the metabolic stresses of the TME. Specific checkpoints further shape Treg metabolic reprogramming. LAG-3 supports Treg immunosuppressive capabilities by modulating Myc-dependent pathways, while lactate absorption can upregulate PD-1 expression on Tregs [29, 30]. Similarly, glycolytic processes in Tregs are influenced by ICOS and TIM-3, with implications for their roles in tumor immunology and potential anti-tumor therapies [31, 32]. Targeting these immune checkpoints offers dual therapeutic benefits: disrupting Treg suppressive functions and altering their metabolic programming to reduce their immunosuppressive capacity in OSCC.
Core metabolic pathways driving regulatory T cell function in OSCC
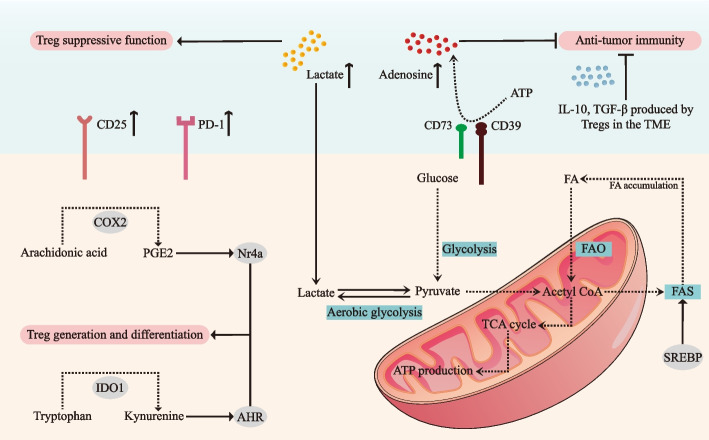
Tregs rely on several key metabolic pathways to maintain their immunosuppressive functions and adapt to the harsh conditions of the TME. In OSCC, these pathways include glycolysis, fatty acid metabolism, amino acid metabolism, PI3K-Akt-mTOR pathway and the adenosine metabolism (Fig. 2).
Fig. 2.
Metabolic and immunosuppressive roles of Treg cells in the tumor microenvironment of OSCC. The tumor microenvironment (TME) of oral squamous cell carcinoma (OSCC) is characterized by elevated levels of immunosuppressive metabolites, with lactate accumulation from enhanced aerobic glycolysis and increased adenosine generation through CD39/CD73-mediated ATP hydrolysis, suppress the anti-tumor immune functions. To maintain their suppressive capabilities, Tregs undergo significant metabolic reprogramming, characterized by enhanced fatty acid oxidation (FAO) and fatty acid synthesis (FAS) pathways, regulated by sterol regulatory element-binding proteins (SREBPs). The metabolic interplay is further complicated by competition between Tregs and tumor cells for essential nutrients, including glucose and fatty acids, creating a metabolically hostile environment for anti-tumor immune responses. Critical molecular pathways, including the indoleamine 2,3-dioxygenase 1 (IDO1)-mediated conversion of tryptophan to kynurenine, activate the aryl hydrocarbon receptor (AHR) to promote Treg differentiation, while cyclooxygenase-2 (COX-2)-derived prostaglandin E2 (PGE2) enhances Treg accumulation through nuclear receptor 4a (Nr4a) upregulation. Tregs also secrete immunosuppressive cytokines, including IL-10, TGF-β, and IL-35, which suppress effector immune responses and facilitate tumor progression. Further, Tregs employ PD-1 and CD25 to maintain their suppressive phenotype and survival under the metabolically hostile conditions of the TME. TME: tumor microenvironment; OSCC: oral squamous cell carcinoma; FAO: fatty acid oxidation; FAS: fatty acid synthesis; SREBP: sterol regulatory element-binding protein; IDO1: Indoleamine 2, 3-dioxygenase 1; AHR: aryl hydrocarbon receptor; COX-2: cyclooxygenase-2; PGE2: prostaglandin E2; Nr4a: nuclear receptor 4a
Glycolysis and its impact on treg proliferation and suppression
Glycolysis is a fundamental metabolic pathway that provides rapid energy through the breakdown of glucose into pyruvate and ATP. This process is tightly linked to the function of various immune cells, supporting tumor growth, metastasis, and chemoresistance [33]. In the hypoxic and acidic TME of OSCC, where lactate accumulates, Tregs upregulate glycolytic enzymes to meet their energy demands [22, 34]. And a spatial transcriptomics study has revealed a connection between enhanced lactate utilization, Treg infiltration, and elevated HIF1A expression in OSCC [35]. The increased glycolytic activity not only facilitates Treg proliferation but also strengthens their immunosuppressive functions, enabling effective modulation of anti-tumor immune response. Moreover, glycolysis is positively correlated with the expression of PD-L1 in OSCC, which in turn contributes to immune evasion and tumor progression [36]. These findings suggest that the glycolytic pathway plays a central role in Treg-mediated immunosuppression within the OSCC TME.
Fatty acid oxidation and the long-term survival of tregs in OSCC
FAO is another critical metabolic pathway for Tregs, providing sustained energy through the breakdown of fatty acids. FAO is particularly important in nutrient-deprived environments like the TME of OSCC. FAO provides sustained energy and essential reducing equivalents, supporting the long-term survival and stability of Tregs [25, 37–39]. Inhibition of mTOR, a key regulator of metabolism, shifts Tregs from glycolysis to FAO, further reinforcing their immunosuppressive functions [40]. High expression of the fatty acid receptor CD36 in OSCC has been linked to metastasis and mitochondrial health in Tregs, enabling adaptation to lactate-rich environments [41, 42]. CD36 might support mitochondrial health and biogenesis of Treg by providing lipid signals, and program Tregs to adapt to lactate-rich TME (Fig. 3D) [24]. Arachidonic acid (AA) metabolism, a polyunsaturated fatty acid, were significantly activated in OSCC patients. Cyclooxygenase-2 (COX-2), an enzyme that plays a crucial role in AA metabolism, has also been observed with an increase in OSCC [43]. Prostaglandin E2 (PGE2), a COX-2 product, promotes tumor progression by increasing Treg numbers through inducing nuclear receptor 4a (Nr4a) expression [44, 45]. FAO thus enables Tregs to maintain their suppressive activities even when glucose availability is limited, thus ensuring their persistence in the TME.
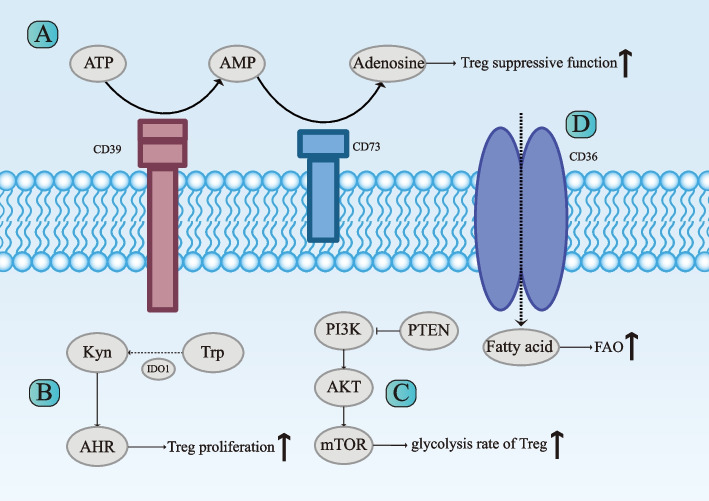
Fig. 3.
Key metabolic pathways modulating Tregs functions in the tumor microenvironment of OSCC. The suppressive function of regulatory T cells (Tregs) in the tumor microenvironment (TME) of oral squamous cell carcinoma (OSCC) is shaped by key metabolic pathways, including the CD39/CD73 adenosine axis (A), tryptophan (Trp)–kynurenine (Kyn)–aryl hydrocarbon receptor (AHR) pathway (B) and phosphatidylinositol 3‐kinase (PI3K) ‐protein kinase B (Akt)‐mammalian target of rapamycin (mTOR) Pathway (C). The CD39/CD73 cascade hydrolyzes ATP to adenosine, which enhances Treg suppressive functions and inhibits effector T cells. The Trp–Kyn–AHR pathway, mediated by indoleamine 2,3-dioxygenase 1 (IDO1), converts Trp to Kyn, activating AHR to promote Treg stability and immunosuppression. PI3K-Akt-mTOR signaling supports glucose uptake, glycolysis, and lipid metabolism to adapt to the nutrient-deprived TME. D Additionally, CD36 is upregulated in tumor-infiltrating Tregs, enhancing fatty acid uptake and fatty acid oxidation (FAO). This adaptation allows Tregs to survive and function effectively in the lactate-rich and nutrient-deprived TME. Treg: regulatory T cell; TME: tumor microenvironment; OSCC: oral squamous cell carcinoma; Trp: tryptophan; Kyn: kynurenines; AHR: aryl hydrocarbon receptor; PI3K: phosphoinositide 3-kinases; AKT: protein kinase B; mTOR: mammalian target of rapamycin; PTEN: phosphate and tensin homologue; IDO1: indoleamine 2, 3-dioxygenase 1; FAO: fatty acid oxidation
Amino acid metabolism: the kynurenine pathway and its role in treg-mediated immune evasion
Amino acid metabolism, particularly tryptophan metabolism via the kynurenine pathway, plays a significant role in Treg function in OSCC [46–48]. The enzyme indoleamine 2,3-dioxygenase 1 (IDO1) converts tryptophan to kynurenine, which activates the aryl hydrocarbon receptor (AHR), enhancing Treg immunosuppressive capabilities (Fig. 3B) [49–51]. IDO has been reported to be a significant negative prognostic factor in patients with OSCC, contributing to immune evasion by triggering MDSC activation and proliferation through Tregs [7, 52]. Other amino acids, such as arginine and glutamine, also support Treg function and proliferation [53–55]. The manipulation of amino acid metabolism offers a potential strategy to disrupt Treg-mediated immune suppression in OSCC.
PI3K-Akt-mTOR pathway: central to treg metabolic control and immune suppression
The PI3K-Akt-mTOR signaling pathway is a crucial regulator of Treg metabolism, integrating various metabolic signals to control cell growth, proliferation, and survival [29, 56]. This pathway is also involved in the cell proliferation as well as glucose, fatty acid and amino acid metabolism [57, 58]. In OSCC, this pathway modulates glucose uptake, glycolysis, and lipid metabolism, ensuring that Tregs can adapt to the dynamic conditions of the TME (Fig. 3C) [54, 59, 60]. Activation of the PI3K-Akt-mTOR pathway supports Treg survival and enhances their suppressive function by regulating key metabolic enzymes and transporters [26, 61, 62]. Moreover, this pathway influences the expression of immune checkpoints such as CTLA-4 and PD-1, further promoting the immunosuppressive environment conducive to tumor growth [63].
The pro-tumor mechanisms of PI3K-Akt-mTOR pathway include mutations of the phosphate and tensin homologue (PTEN), which is an upstream inhibitor of PI3K-Akt-mTOR signaling and plays a role in maintaining Treg cell stability [64–66]. Specifically, the targeted elimination of PTEN not only impairs Treg mitochondrial health and upregulates glycolysis but also reduces the expression of CD25 and FOXP3, collectively undermining Treg homeostasis and functional capacity [66, 67].
Nucleotide metabolism and its role in treg function
Nucleotides are the fundamental building blocks of genetic material, playing a role in metabolic biosynthesis and serving as one of forms of energy utilization within the body [68]. Tumor cells exhibit enhanced nucleotide diversity and active nucleotide biosynthesis. In OSCC, the expression of purine derivatives (e.g., hypoxanthine) and nucleosides (e.g., guanosine) are upregulated, contributing to tumor progression [69]. Targeting nucleotide metabolism, including nucleic acid biosynthesis substrates and energy sources within the TME, has the potential to enhance anti-tumor immune responses. The metabolism of folate, a key pathway for nucleotide synthesis, supports T cell activation, proliferation, and survival [70]. The expression of folate-metabolism associated methylenetetrahydrofolate dehydrogenase (MTHFD) family genes has been identified as potential biomarkers for Treg infiltration in OSCC [71].
ATP, a purine analogue, plays a pivotal role in modulating immune responses by activating immune cell receptors. In OSCC, ATP is converted to adenosine through the enzymatic actions of CD39 and CD73, especially when Tregs undergo apoptosis. The biological effects of extracellular adenosine are mediated by G-protein–coupled adenosine receptors, which are classified into A1, A2A, A2B, and A3 subtypes (Fig. 3A) [72]. Overexpression of CD73 in the OSCC microenvironment results in elevated adenosine levels, which are associated with poor patient prognosis [73]. Adenosine exerts immunosuppressive effects by reducing neutrophil adhesion and promoting macrophage production. Exogenous adenosine impairs the migratory ability of conventional T cells while enhancing Treg expansion and immunoregulatory activity via A2AR engagement [74, 75].
The pentose phosphate pathway: a critical metabolic hub in tregs
The pentose phosphate pathway (PPP) and the glycolytic pathway are two interconnected metabolic routes involved in glucose uptake and utilization. The PPP supplies essential precursors for nucleotide biosynthesis and generates NADPH, which is critical for fatty acid synthesis and redox balance. Notably, the PPP is frequently upregulated in tumor cells. Inhibitors targeting key enzymes of the PPP, such as glucose-6-phosphate dehydrogenase (G6PD), have been shown to suppress the proliferation and metastasis of OSCC in vivo [76, 77]. PPP is also regulated by the mTOR pathway, with activation of mTORC1 stimulating PPP activity [78]. Moreover, the PPP has been implicated in T cell metabolism and is thought to influence the Th17/Treg balance, potentially impacting immune regulation within the TME [79].
Influence of the oral microbiome on treg metabolism in OSCC
The oral microbiome, consisting of bacteria, fungi, and viruses, profoundly shapes the metabolic and immune landscape of Tregs within the TME of OSCC [80, 81]. Increasing attention has also been directed toward outer membrane vesicles (OMVs), spherical double-layered structures released by Gram-negative bacteria during growth or in response to environmental stress. With diameters ranging from 50 to 250 nm, OMVs carry diverse biomolecules, including proteins, lipopolysaccharides (LPS), phospholipids, and periplasmic components, playing a pivotal role in regulating the TME [82]. This section explores the interplay between the oral microbiome and Treg metabolism, emphasizing the composition of the microbiome, microbial metabolites and immune modulation.
Microbial composition and its role in OSCC development
The oral microbiome exhibits significant compositional alterations in OSCC patients compared to healthy individuals, characterized by increased microbial diversity and dysbiosis [83]. Certain microorganisms, such as Fusobacterium nucleatum, Porphyromonas gingivalis, Candida albicans, human papillomavirus (HPV) and Epstein-Barr virus (EBV) are more prevalent in OSCC and have been implicated in tumor progression [84–91]. These microorganisms contribute to OSCC progression through various mechanisms. For example, Candida induces metabolic abnormalities within tumors, causing mitochondrial damage and promoting the progression of OSCC [92, 93]. Recent studies also highlight the role of OMVs from oral bacteria in influencing OSCC behavior. OMVs derived from Fusobacterium nucleatum have been shown to activate autophagy and disrupt protein homeostasis in OSCC cells, driving metastasis [94]. Similarly, Porphyromonas gingivalis OMVs promote tumor invasion and migration [95]. Interestingly, some researches show a possible antitumorigenic effect of Aggregatibacter actinomycetemcomitans-derived OMVs in OSCC, highlighting an area for further research [96].
Microbial metabolites influencing treg metabolism
Microbial metabolites significantly impact Treg function and metabolism. Elevated level of lactate and other metabolites in the TME enhance the stability and suppressive function of Tregs [97, 98]. For instance, Candida albicans enhances metabolite production in OSCC cells, potentially influencing Treg activity indirectly [99]. EBV infection reprograms cellular metabolism, favoring the Warburg effect, which increases lactate production and supports Treg function [100]. Additionally, LPS, a major OMV component, mediates inflammation and drives Treg accumulation in OSCC, contributing to the suppression of anti-tumor immunity [101]. These findings reveal a metabolic link between microbial metabolites and Treg activity, further underscoring the complex interplay between the microbiome and TME.
Immune modulation by the oral microbiome in OSCC
The oral microbiome exerts profound effects on immune modulation in OSCC, particularly by influencing Treg activity [80, 102, 103]. Microbial dysbiosis disrupts immune homeostasis, promoting an environment that favors Treg-mediated immunosuppression [104, 105]. HPV infection is associated with metabolic reprogramming in tumor cells, including increased glycolysis, which supports Treg activity in head and neck squamous cell carcinoma (HNSCC) [106, 107]. HPV also enhances mTOR activity under hypoxic conditions and promotes HIF-1 accumulation, leading to metabolic dysregulation in OSCC. While HPV does not directly correlate with overall survival (OS) in OSCC patients, HPV-positive patients often have better prognoses, potentially due to microbiome-mediated immune modulation [108, 109].
Within the OSCC TME, bacteria such as Porphyromonas gingivalis and Fusobacterium nucleatum activate key signaling pathways, including PI3K/Akt and CDH1/β-catenin, promoting tumor proliferation and survival [110, 111]. The microbiome also contributes to tumor cell invasion, migration, and Treg differentiation and amplification through the secretion of specific metabolites. Fusobacterium nucleatum influences purine metabolism, generating metabolites that support Treg survival and function within the TME [112–114]. Additionally, it drives OSCC progression by enhancing glycolysis through GLUT1-mediated lactate production, creating an immunosuppressive environment that favors Treg activity [22, 115, 116]. Similarly, Porphyromonas gingivalis affects ATP metabolism in tumors and converts ethanol into acetaldehyde, a carcinogenic intermediate that further accelerates OSCC progression [117]. Moreover, outer membrane vesicles (OMVs), known to modulate microbial metabolism, are being investigated as a novel delivery system for cancer vaccines. Notably, OMVs have been shown to reduce Treg populations in tumor-draining lymph nodes, highlighting their potential to regulate Tregs and reshape the tumor immune microenvironment [118].
Targeting treg metabolism as a therapeutic strategy in OSCC
The metabolic targeting of Tregs in OSCC offers promising therapeutic potential by disrupting the metabolic support of Tregs, thereby reducing their suppressive function and enhancing anti-tumor immunity (Table 1). These strategies, which include the inhibition of glycolysis, modulation of fatty acid metabolism and amino acid metabolism, PI3K-Akt-mTOR pathway inhibitors, and adenosine metabolism as well as oral microbiome therapy are actively being explored in clinical trials, showing significant potential for improving patient outcomes in OSCC.
Table 1.
Clinical Trials Targeting Metabolism Pathways Related to Head and Neck Carcinoma
| Strategies | Key Molecules/Targets | Inhibitors/Drugs | NCT Number | Main Results |
|---|---|---|---|---|
| Inhibition of Glycolysis | Glycolysis/ Glucose analog | 2-DG | NCT00096707 |
11 pts (32%) had SD, 1 pts (3%) PR and 22 pts (66%) PD as best response RD of 2DG with weekly docetaxel 63 mg/kg/day [119] |
| Anaerobic glycolysis | Dichloroacetate | NCT01386632 |
No significant differences in Gr 3/4 AE rates between dichloroacetate and placebo CR rates were higher in the dichloroacetate group [120] |
|
| Modulating Fatty Acid Metabolism | CD36 | ABT-510 | NCT00113334 |
2 pts had SD, 1 pt had PD |
| 3-hydroxy-3-methyl glutaryl coenzyme A reductase | Atorvastatin | NCT04915183 | No results posted | |
| NCT02022293 | Enrollment target could not be achieved | |||
| Inhibition of Amino Acid Metabolism | IDO | Epacadostat | NCT02318277 | Phase 2 ORR 12.0%, higher in CPI-naive (16.1%) vs. previous CPI (4.1%) [123] |
| NCT02178722 | 84% pts (n = 52) experienced TRAEs, Gr 3/4 TRAEs reported in 24% pts, and 100 mg Epacadostat twice per day plus 200 mg pembrolizumab every 3 weeks recommended for phase II evaluation [124] | |||
| NCT02327078 | For pts on Epacadostat 300 mg, preliminary DCR was 70%, and 48% pts (n = 42) reported TRAEs [125, 126] | |||
| BMS-986205 | NCT03854032 | Higher pTE in radiographic responders vs. non-responders (85% vs. 5%), and higher pTE in responders in the lymph node compartment (73% vs. 23%), while CR demonstrated in 6/16 (38%) pts [127, 128] | ||
| Targeting Nucleotide Metabolism | PARP | Olaparib | NCT02882308 | Olaparib treatment modulates DNA damage response network and exerts extra antitumor effect by elevating oxidative stress in HNSCC patients [129] |
| A2aR | NIR178 | NCT03207867 | For HNSCC pts with NIR178 160 mg twice daily plus PDR001 with no immuno-oncology therapy ORR 13.3 [130] | |
| Modulating Glucose and Fatty Acid Metabolism | PI3K | Buparlisib (BKM120) | NCT02113878 | 1 of 7 pts experienced Gr 4 rash on DL1, DLTs observed (4 of 6 pts) on DL2, and 5 pts respond to buparlisib monotherapy [131] |
| NCT01816984 | Gr3 AEs reported in 10 pts, and no DLTs during dose escalation, while PR (n = 1) and SD (n = 4) reported [132] | |||
| Copanlisib | NCT02822482 | 2 of 3 pts experienced DLT on 45 mg DL, and no DLT was reported on 30 mg DL. Median PFS 2.66 months, and median OS 6.01 months [133] | ||
| Alpelisib (BYL719) | NCT02537223 | No DLT observed at 200 mg, 2 of 2 pts reported DLTs at 250 mg, and RP2D declared at 200 mg. 3-year PFS and OS rate both 77.8% [134] | ||
| NCT01602315 | 1 PR, 3 unconfirmed PRs and 5 SDs at 300 mg, and 1 PD and 1 SD at 400 mg. RP2D declared as 300 mg QD (whole tablets) [135] | |||
| Bimiralisib | NCT03740100 | ORR 17%, median PFS 5 months and median OS 7 months. 62.5% pts (n = 5) experienced 14 severe AEs [136, 137] | ||
| mTOR | Sirolimus(rapamycin) | NCT01195922 | 1 pt had a pathologic CR, while 25% pts (n = 4) reported response (1 CR, 3 PRs, 12 SDs) [138] | |
| Temsirolimus (TORISEL) | NCT01016769 | OR rate 43% (n = 13) with 1 CR, 10 confirmed PRs, and 2 unconfirmed PRs. OS 12.9 months [139] | ||
| NCT01172769 | Median PFS 56 days, median OS 152 days, and PFR 40% at 12 weeks. 87.5% pts (n = 35) experienced at least two AEs [140] | |||
| Everolimus (RAD001) | NCT01333085 | No DLT in phase I, and everolimus RD declared at 50 mg/w, while 2.6% pts experienced a CR (n = 1), 76.3% PR (n = 28), and 21% SD (n = 8). OR rate 79% [141] | ||
| NCT01051791 | CBR 28%, median PFS 1.5 months, and median OS 4.5 months. 2 SDs observed (5.5 and 4.5 months) [142] | |||
| CC-115 | NCT01353625 | 22% pts reported Gr 3 drug-related AEs. SD observed 53% [143] | ||
| EGFR | Cetuximab | NCT01252628 | Median PFS 80 days, no CR found, and 7 PR confirmed. Median OS 211 days vs. 256 days (cetuximab plus PX-866 vs. cetuximab alone) [144] | |
| NCT01256385 |
Median PFS both 3.5 months. (cetuximab plus Temsirolimus vs. Temsirolimus alone) Median OS 177 days vs. 176 days and OR rate 12.5% vs. 2.5% [145] |
|||
| Duligotuzumab | NCT01577173 | PFS 4.2 months vs. 4.0 months (duligotuzumab vs. cetuximab), OS 7.2 months vs. 8.7 months, and OR rate 12% vs. 14.5% [146] | ||
| Erlotinib | NCT00942734 |
OR rate 2.8% at 12 weeks, median PFS 11.9 weeks and median OS 10.25 months 8% pts (n = 3) achieved PR at 4 weeks [147] |
||
| Targeting Oral Microbiome | Oral microbiome | N-Acetyl Cysteine | NCT03982537 | Concept is withdrawn and a different concept will be submitted the near future |
| Oral microbiome |
Probiotic Lozenge (ProDentis Lozenge) |
NCT04925700 | No results posted | |
| Oral microbiome | MET-4 | NCT03838601 | No results posted |
Abbreviations: ORR Objective response rate, PI3K Phosphoinositide 3-kinase, CPI Immune checkpoint inhibitor, pts Patients, Gr Grade, Trp Tryptophan, Kyn Kynurenine, DCR Disease control rate, TRAE Treatment-related adverse event, pTE Pathologic treatment effect, CR Complete response, HNSCC Head and neck squamous cell carcinoma, PFR Progression-free survival rate, DL Dose level, DLT Dose limiting toxicity, AE Adverse event, PR Partial response, SD Stable disease, CI Confidence interval, PFS Progression free survival, OS Overall survival, RP2D Recommended phase 2 dose, PD Progressive disease, RD Recommended dose, OR Overall response, CBR Clinical benefit rate
Inhibition of glycolysis
Tregs in the TME of OSCC rely heavily on glycolysis for rapid energy production, particularly under hypoxic conditions. Inhibitors of glycolytic enzymes, such as 2-deoxy-D-glucose (2-DG), impair Treg function by limiting their primary energy source [148, 149]. Recent studies have demonstrated the synergistic effects of combining glycolysis inhibition with chemotherapy or radiotherapy, enhancing anti-tumor efficacy by inducing apoptosis, DNA damage, and metabolic reprogramming [150]. Importantly, glycolysis inhibitors selectively target Tregs without significantly affecting effector T cells, which can utilize alternative metabolic pathways. Ongoing clinical trials aim to evaluate the potential of these inhibitors in mitigating Treg-mediated immunosuppression to improve cancer therapy outcomes.
Modulating fatty acid metabolism
FAO is crucial for the long-term survival and suppressive function of Tregs in nutrient-deprived environments like the TME [28]. FAO inhibitors, such as etomoxir, disrupt Treg metabolism by blocking the oxidation of fatty acids, thereby reducing their suppressive capacity [40]. Targeting CD36-mediated lipid metabolism using selective inhibitors has shown potential for enhancing anti-tumor immunity in OSCC [151]. This approach takes advantage of the reliance of Tregs on FAO, making them more vulnerable to metabolic intervention. Although research on Treg-specific modulation of fatty acid metabolism remains limited, combining FAO inhibitors with immune checkpoint inhibitors may further amplify anti-tumor immune responses by weakening Treg-mediated immunosuppression.
Modulating amino acid metabolism
Amino acid metabolism, particularly the tryptophan-kynurenine- AhR pathway, is another target for disrupting Treg function [51, 152, 153]. Inhibitors of indoleamine 2,3-dioxygenase (IDO), such as epacadostat, reduce the production of kynurenine, thereby attenuating Treg proliferation and suppressive activity [154, 155]. By blocking this pathway, IDO inhibitors can decrease Treg-mediated immunosuppression and enhance the efficacy of other cancer therapies. Clinical trials are currently evaluating the effectiveness of IDO inhibitors in combination with immune checkpoint inhibitors in OSCC and other cancers, highlighting their potential to improve therapeutic outcomes.
PI3K-Akt-mTOR pathway inhibitors
The PI3K-Akt-mTOR signaling pathway is a critical regulator of Treg metabolism, controlling processes such as glucose uptake, glycolysis, and lipid metabolism. Persistent activation of the PI3K-Akt-mTOR signaling route is common in OSCC [156]. Inhibitors of this pathway, such as rapamycin, impair Treg survival and function by disrupting these metabolic processes [12]. These inhibitors reduce Treg-mediated immunosuppression and promote anti-tumor immunity. In OSCC, epidermal growth factor receptor (EGFR) overexpression, an upstream activator of the PI3K pathway, contributes to tumor progression [156]. It is noticed that EGFR-targeted antibody, such as cetuximab, increased the frequency of CD4+FOXP3+ intratumoral Treg in HNSCC, demonstrated that combining parsaclisib (a PI3K inhibitor) with itacitinib (a JAK1 inhibitor) significantly reduced Treg populations and increased the CD8/FoxP3 ratio in solid tumors [63, 157]. Additionally, trials with ipatasertib (an AKT inhibitor) in head and neck cancers aim to improve the TME by reducing hypoxia, enhancing radiotherapy efficacy [158]. These findings support the use of PI3K-Akt-mTOR inhibitors in combination therapies to improve OSCC treatment outcomes.
Targeting nucleotide metabolism
Research indicates that Foxp3 expression in Tregs is closely associated with nucleotide metabolism, glycolysis, and lipid metabolism, highlighting the metabolic flexibility of Tregs in adapting to different pathways [26]. Nucleotides, as essential components of DNA and RNA, also serve as key regulators of intracellular signaling pathways. Therapeutic strategies targeting nucleotide metabolism hold significant promise for OSCC treatment. CD39 and CD73 are essential to adenosine metabolism, which affect Treg activity. Preclinical studies and mouse tumor models have demonstrated the efficacy of agents that block CD39 and CD73 activity or employ selective A2AR inhibitors to disrupt adenosine-mediated immunosuppression [159, 160]. These inhibitors, including CD39/CD73 blockers and A2a/A2b receptor antagonist, effectively reduce the immunosuppressive capacity of Tregs mediated by adenosine [161–163]. Notably, adenosine metabolism is enhanced in OSCC, further contributing to tumor progression [164]. Although clinical trials specifically targeting nucleotide metabolism in Tregs remain limited, ongoing research underscores the potential of adenosine metabolism inhibitors to reduce tumor growth and enhance anti-tumor immune responses. Further investigations are needed to elucidate the molecular mechanisms underlying these therapies to facilitate their clinical application.
Targeting oral microbiome
The oral microbiome’s influence on Treg metabolism offers exciting opportunities for therapeutic interventions in OSCC. Targeting specific microbial populations or their metabolites can reshape the metabolic environment of the TME, reducing Treg-mediated immunosuppression and enhancing anti-tumor immunity. For instance, manipulating the abundance of Fusobacterium nucleatum or Prevotella intermedia has been shown to alter the TME, improving the efficacy of immunotherapies [165–168]. Remarkably, a combined treatment involving Fusobacterium nucleatum OMVs and oncolytic viruses significantly decreased Treg populations and upregulated PD-L1 expression in tumor tissues, thereby enhancing the effects of immune checkpoint therapies [169]. Moreover, probiotics or antibiotics could be explored as potential therapies to restore a healthy microbiome balance, thereby modulating Treg function and enhancing anti-tumor immune responses. Such approaches underscore the importance of the oral microbiome as a novel and promising target in OSCC treatment.
Challenges and future directions for targeting regulatory T cell metabolism in OSCC
Tregs in OSCC holds significant promise for improving therapeutic outcomes. However, the translation of these strategies from research to clinical application faces several challenges. Addressing these challenges while exploring future perspectives is crucial for advancing this therapeutic approach.
Selectivity and toxicity of treg metabolic inhibitors
One of the primary challenges in targeting Treg metabolism is achieving selectivity without causing toxicity to other immune cells. Many metabolic pathways that are essential for Tregs function also play crucial roles in effector T cells and other immune cells [170–172]. For instance, glycolysis and FAO are not exclusive to Tregs, and inhibiting these pathways could impair the broader immune response, potentially leading to unintended immunosuppression or other adverse effects [28, 173]. To overcome this, therapies must be designed to selectively target Tregs or be administered in a way that minimizes impact on non-target cells. Research on Treg-specific depletion has focused on targets such as CD25, CCR4, and CTLA-4, but these strategies have thus far shown limited efficacy [174]. Given that Tregs selectively rely on certain metabolic pathways, combining metabolic targets with Treg-specific approaches could help balance efficacy and toxicity in future therapies.
Tumor heterogeneity: a barrier to uniform therapeutic responses
The heterogeneity of the TME in OSCC poses a significant challenge for uniform therapeutic responses to metabolic targeting. Variations in oxygen levels, nutrient availability, and metabolic adaptations among different tumor regions can influence how Tregs and other cells respond to metabolic inhibitors [175–177]. OSCC is categorized into various subtypes, including basal, mesenchymal, classical, and atypical, each with distinct gene characteristics, protein expression, and clinical features. For example, the atypical subtype is closely associated with HPV infection, which further highlights the role of the microbiome in OSCC progression [178]. These variabilities necessitate a more personalized approach to therapy, considering the unique metabolic landscape of each tumor. Identifying biomarkers to reflect these metabolic differences could enable more tailor treatments, improving efficacy and minimizing side effects.
Resistance mechanisms to metabolic inhibitors in tregs
Tregs, like cancer cells, can develop resistance mechanisms to metabolic inhibitors [179]. In OSCC, Tregs may adapt to metabolic inhibition through mechanisms such as upregulation of alternative metabolic pathways, altered expression of metabolic enzymes, or changes in cell damage and apoptosis [8, 180–184]. For instance, Foxp3 reprograms Treg metabolism by suppressing Myc, promoting lipid metabolism over glucose metabolism [25]. Tregs also exhibit metabolic changes in mitochondrial function and lipid metabolism through fatty acid binding proteins (FABPs) mediated pathways [185]. Furthermore, Treg apoptosis can promote immunosuppressive resistance in the TME by contributing to adenosine production, which in turn shields tumor cells from anti-tumor therapies. Understanding these resistance mechanisms is critical for developing combination therapies that can prevent or overcome resistance. Continuous monitoring and adaptive treatment strategies may be required to manage resistance and sustain therapeutic efficacy. Future research should focus on elucidating these resistance mechanisms and developing combination therapies that prevent or counteract resistance. This could involve targeting multiple metabolic pathways simultaneously or using adaptive treatment strategies to stay ahead of tumor and Treg adaptations.
Managing immune-related adverse events in treg targeting therapies
Manipulating the metabolism of immune cells can lead to immune-related adverse events (irAEs), which can range from mild to severe [186, 187]. These irAEs arise because the metabolic pathways targeted in Tregs are also present in other immune cells, leading to unintended activation or suppression. Careful monitoring and management of irAEs are essential to ensure patient safety during treatment. Strategies such as dose adjustment, combining therapies with immunosuppressive drugs, and close clinical monitoring are necessary to mitigate these risks.
Personalized medicine and integrative approaches for metabolic targeting
Personalizing metabolic targeting strategies based on the unique metabolic profile of each patient’s tumor could significantly improve treatment outcomes [188]. Advances in genomics, metabolomics, and proteomics offer the potential to identify biomarkers that predict response to specific metabolic inhibitors. Tailoring treatments to individual patients’ metabolic landscapes will enhance efficacy while minimizing off-target effects, leading to more precise and effective therapies.
Combining metabolic inhibitors with existing cancer treatments, such as immune checkpoint inhibitors, chemotherapy, and radiation therapy, may further enhance therapeutic efficacy [189]. For example, CD36 antagonists have been shown to deplete Tregs by affecting lipid metabolism, which restores the therapeutic effect of PD-1 antibodies in certain tumors [190]. In addition, the interaction between ICOS and the mTOR pathway could also affect Treg metabolism, offering another potential avenue for combination strategies [31]. These integrative approaches aim to synergize the effects of different modalities, potentially overcoming the limitations of monotherapies. Preclinical studies and clinical trials should focus on identifying optimal combinations and treatment schedules to maximize anti-tumor responses while minimizing adverse effects.
Exploring novel metabolic targets in tregs and the role of the oral microbiome in shaping treg metabolism for future therapies
Exploring new metabolic targets and pathways specific to Tregs within the OSCC microenvironment is crucial for developing innovative therapies. Beyond well-known pathways like glycolysis and FAO, future research should investigate less explored metabolic pathways, such as the pentose phosphate pathway, with a focus on tumor-immune interactions. These studies could offer critical insights into Treg metabolism in tumor progression and immune evasion. High-throughput screening techniques and CRISPR-based genetic screens can aid in the discovery of novel metabolic vulnerabilities in Tregs [191].
The oral microbiome plays a significant role in modulating Treg metabolism and function. Therapeutic strategies aimed at manipulating the microbiome—using probiotics, prebiotics, or antibiotics—could influence Treg activity and enhance anti-tumor immunity [192, 193]. Further research is needed to unravel the complex interactions between the microbiome and Treg metabolism, potentially leading to microbiome-based adjunct therapies for OSCC.
Transformative potential of emerging technologies of metabolism research
Tumor metabolism and immunometabolism have become increasingly important fields for understanding tumorigenesis, progression, and therapeutic resistance. While previous research primarily focused on conventional cellular experiments and in vivo animal studies, advancements in multi-omics, single-cell sequencing, and spatial techniques have provided new opportunities for exploring metabolic dynamics in both tumor and immune microenvironments [194]. Multi-omics technologies allow the quantification of metabolites in the TME, revealing the metabolic profiles of tumor and immune cells [195]. Single-cell technologies facilitate the identification of distinct cellular populations with varying metabolic states, providing insight into Treg metabolic changes. Spatial techniques further elucidate metabolic activities across tumor tissue regions, which can help identify the metabolic preferences of tumor-infiltrating Tregs [196]. Combining these emerging technologies will provide a comprehensive understanding of the TME and offer robust support for the translational application of metabolism-targeted immunotherapy.
Conclusions
The metabolic targeting of Tregs in OSCC has emerged as a promising strategy to enhance anti-tumor immunity. This review has provided a comprehensive overview of the metabolic targeting of Tregs in OSCC. It highlights critical metabolic pathways, such as glycolysis, FAO, amino acid metabolism, nucleotide metabolism, and the PI3K-Akt-mTOR pathway, which are essential for maintaining Treg function under the nutrient- and oxygen-deprived conditions of the TME. Additionally, we have highlighted the emerging role of the oral microbiome as a crucial modulator of Treg metabolism and OSCC pathogenesis.
Therapeutic strategies targeting these metabolic dependencies show significant potential in disrupting Treg-mediated immunosuppression. These approaches include glycolysis inhibitors, FAO modulators, amino acid metabolism blockers, and innovative treatments leveraging insights from OMVs. However, the heterogeneous nature of OSCC necessitates personalized therapeutic approaches that consider tumor-specific metabolic signatures, spatial metabolic variations, and individual patient characteristics. The advancement of this field relies heavily on cutting-edge technologies, including multi-omics platforms, single-cell sequencing, spatial metabolomics, and advanced molecular imaging techniques. These tools are essential for developing effective personalized metabolic targeting strategies and understanding the complex interactions within the TME. Future research should focus on developing novel metabolic targeting approaches, understanding resistance mechanisms, optimizing combination therapies, and successfully translating preclinical findings into clinical applications.
The successful implementation of these therapeutic strategies requires a coordinated effort among researchers, clinicians, and regulatory bodies, along with rigorous clinical validation and continuous technological innovation. While challenges remain in addressing the complexity of the TME and the dynamic nature of metabolic processes, metabolic targeting of Tregs represents a potentially transformative approach in OSCC treatment. Through continued research and collaborative efforts, this emerging therapeutic paradigm offers new hope for patients battling this aggressive malignancy, potentially revolutionizing the treatment landscape of OSCC.
Acknowledgements
Not applicable.
Abbreviations
- OSCC
Oral squamous cell carcinoma
- Treg
Regulatory T cell
- TME
Tumor microenvironment
- NK
Natural killer
- DC
Dendritic cell
- MDSC
Myeloid-derived suppressor cell
- HIF
Hypoxia-inducible factor
- FAO
Fatty acid oxidation
- AA
Arachidonic acid
- COX-2
Cyclooxygenase-2
- PGE2
Prostaglandin E2
- Nr4a
Nuclear receptor 4a
- IDO
Indoleamine 2, 3-dioxygenase
- AHR
Aryl hydrocarbon receptor
- PTEN
Phosphate and tensin homologue
- MTHFD
Methylenetetrahydrofolate dehydrogenase
- PPP
Pentose phosphate pathway
- G6PD
Glucose-6-phosphate dehydrogenase
- OMV
Outer membrane vesicle
- LPS
Lipopolysaccharides
- HPV
Human papillomavirus
- EBV
Epstein-Barr virus
- EV
Extracellular vesicle
- HNSCC
Head and neck squamous cell carcinoma
- OS
Overall survival
- 2-DG
2-Deoxy-D-glucose
- EGFR
Epidermal growth factor receptors
- FABPs
Fatty acid binding proteins
- irAEs
Immune-related adverse events
Authors’ contributions
MG Writing—original draft, Visualization, Data curation, Formal analysis; NL Writing—review & editing, Data curation; WL Investigation; MC Formal analysis; ZB Data curation; DL Methodology, Writing—review & editing, Supervision; LS Methodology Writing-review &editing, Conceptualization, Funding acquisition, Supervision.All authors reviewed the manuscript.
Funding
The study was supported by the National Natural Science Foundation of China (No. 82270989 and 81902772), Natural Science Foundation of Liaoning Province (No. 2022-MS-206 and 2022-MS-200), Foundation of Liaoning Educational Committee (No. JYTMS20230115) and Shenyang Young and Middle-aged Science and Technology Innovation Talent Support Program (No.RC210038 and RC210041).
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Menglai Gan and Nanshu Liu contributed equally to this work.
Contributor Information
Dongjuan Liu, Email: dongjuanliu@cmu.edu.cn.
Sai Liu, Email: liusai@cmu.edu.cn.
References
- 1.Tan Y, Wang Z, Xu M, Li B, Huang Z, Qin S, et al. Oral squamous cell carcinomas: state of the field and emerging directions. Int J Oral Sci. 2023. 10.1038/s41368-023-00249-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Badwelan M, Muaddi H, Ahmed A, Lee KT, Tran SD. Oral squamous cell carcinoma and concomitant primary tumors, what do we know? A review of the literature. Curr Oncol. 2023. 10.3390/curroncol30040283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024. 10.3322/caac.21834. [DOI] [PubMed]
- 4.Dhanuthai K, Rojanawatsirivej S, Thosaporn W, Kintarak S, Subarnbhesaj A, Darling M, et al. Oral cancer: A multicenter study. Med Oral Patologia Oral y Cirugia Bucal. 2018;10.4317/medoral.21999. [DOI] [PMC free article] [PubMed]
- 5.Dong L, Xue L, Cheng W, Tang J, Ran J, Li Y. Comprehensive survival analysis of oral squamous cell carcinoma patients undergoing initial radical surgery. BMC Oral Health. 2024. 10.1186/s12903-024-04690-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caponio VCA, Zhurakivska K, Lo Muzio L, Troiano G, Cirillo N. The immune cells in the development of oral squamous cell carcinoma. Cancers (Basel). 2023. 10.3390/cancers15153779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kouketsu A, Haruka S, Kuroda K, Hitoshi M, Kensuke Y, Tsuyoshi S, et al. Myeloid-derived suppressor cells and plasmacytoid dendritic cells are associated with oncogenesis of oral squamous cell carcinoma. J Oral Pathol Med. 2023. 10.1111/jop.13386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu S, Liu D, Li J, Zhang D, Chen Q. Regulatory T cells in oral squamous cell carcinoma. J Oral Pathol Med. 2016. 10.1111/jop.12445. [DOI] [PubMed] [Google Scholar]
- 9.Quan H, Shan Z, Liu Z, Liu S, Yang L, Fang X, et al. The repertoire of tumor-infiltrating lymphocytes within the microenvironment of oral squamous cell carcinoma reveals immune dysfunction. Cancer Immunol Immunother. 2020. 10.1007/s00262-020-02479-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramalingam S, Shantha S, Muralitharan S, Sudhakar U, Thamizhchelvan H, Parvathi VD. Role of tissue markers associated with tumor microenvironment in the progression and immune suppression of oral squamous cell carcinoma. Med Oncol. 2023. 10.1007/s12032-023-02169-5. [DOI] [PubMed] [Google Scholar]
- 11.Solvay M, Holfelder P, Klaessens S, Pilotte L, Stroobant V, Lamy J, et al. Tryptophan depletion sensitizes the AHR pathway by increasing AHR expression and GCN2/LAT1-mediated kynurenine uptake, and potentiates induction of regulatory T lymphocytes. J Immunother Cancer. 2023. 10.1136/jitc-2023-006728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yonesi A, Tomihara K, Takatsuka D, Tachinami H, Yamazaki M, Jadidi ARY, et al. Rapamycin induces phenotypic alterations in oral cancer cells that may facilitate antitumor T cell responses. Biomedicines. 2024. 10.3390/biomedicines12051078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen H, Liu X, Yao F, Yin M, Cheng B, Yang S. Identification of metabolic signatures related to metastasis and immunotherapy resistance in oral squamous cell carcinoma. Am J Transl Res. 2023;15(1):373–91. [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang H, Zhou F, Yang Q, Huang M. Targeting the oral tumor microenvironment by nanoparticles: a review of progresses. J Drug Delivery Sci Technol. 2024. 10.1016/j.jddst.2023.105248. [Google Scholar]
- 15.Kang FW, Gao Y, Que L, Sun J, Wang ZL. Hypoxia-inducible factor-1alpha overexpression indicates poor clinical outcomes in tongue squamous cell carcinoma. Exp Ther Med. 2013. 10.3892/etm.2012.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clambey ET, McNamee EN, Westrich JA, Glover LE, Campbell EL, Jedlicka P, et al. Hypoxia-inducible factor-1 alpha-dependent induction of FoxP3 drives regulatory T-cell abundance and function during inflammatory hypoxia of the mucosa. Proc Natl Acad Sci USA. 2012. 10.1073/pnas.1202366109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miska J, Lee-Chang C, Rashidi A, Muroski ME, Chang AL, Lopez-Rosas A, et al. HIF-1alpha is a metabolic switch between glycolytic-driven migration and oxidative phosphorylation-driven immunosuppression of tregs in glioblastoma. Cell Rep. 2019. 10.1016/j.celrep.2019.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neildez-Nguyen TMA, Bigot J, Da Rocha S, Corre G, Boisgerault F, Paldi A, et al. Hypoxic culture conditions enhance the generation of regulatory T cells. Immunology. 2015. 10.1111/imm.12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lochner M, Berod L, Sparwasser T. Fatty acid metabolism in the regulation of T cell function. Trends Immunol. 2015. 10.1016/j.it.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 20.Shan Y, Xie T, Sun Y, Lu Z, Topatana W, Juengpanich S, et al. Lipid metabolism in tumor-infiltrating regulatory T cells: perspective to precision immunotherapy. Biomark Res. 2024. 10.1186/s40364-024-00588-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gu J, Zhou J, Chen Q, Xu X, Gao J, Li X, et al. Tumor metabolite lactate promotes tumorigenesis by modulating MOESIN lactylation and enhancing TGF-beta signaling in regulatory T cells. Cell Rep. 2022. 10.1016/j.celrep.2022.110986. [DOI] [PubMed] [Google Scholar]
- 22.Watson MJ, Vignali PDA, Mullett SJ, Overacre-Delgoffe AE, Peralta RM, Grebinoski S, et al. Metabolic support of tumour-infiltrating regulatory T cells by lactic acid. Nature. 2021. 10.1038/s41586-020-03045-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vilbois S, Xu Y, Ho PC. Metabolic interplay: tumor macrophages and regulatory T cells. Trends Cancer. 2024. 10.1016/j.trecan.2023.11.007. [DOI] [PubMed] [Google Scholar]
- 24.Wang H, Franco F, Tsui YC, Xie X, Trefny MP, Zappasodi R, et al. CD36-mediated metabolic adaptation supports regulatory T cell survival and function in tumors. Nat Immunol. 2020. 10.1038/s41590-019-0589-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Angelin A, Gil-de-Gomez L, Dahiya S, Jiao J, Guo L, Levine MH, et al. Foxp3 reprograms T cell metabolism to function in low-glucose high-lactate environments. Cell metabolism. 2017. 10.1016/j.cmet.2016.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerriets VA, Kishton RJ, Johnson MO, Cohen S, Siska PJ, Nichols AG, et al. Foxp3 and Toll-like receptor signaling balance T(reg) cell anabolic metabolism for suppression. Nat Immunol. 2016. 10.1038/ni.3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tay C, Tanaka A, Sakaguchi S. Tumor-infiltrating regulatory T cells as targets of cancer immunotherapy. Cancer Cell. 2023. 10.1016/j.ccell.2023.02.014. [DOI] [PubMed] [Google Scholar]
- 28.Yan Y, Huang L, Liu Y, Yi M, Chu Q, Jiao D, et al. Metabolic profiles of regulatory T cells and their adaptations to the tumor microenvironment: implications for antitumor immunity. J Hematol Oncol. 2022. 10.1186/s13045-022-01322-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, Huang T, Gu J, Lu L. Targeting the metabolism of tumor-infiltrating regulatory T cells. Trends Immunol. 2023. 10.1016/j.it.2023.06.001. [DOI] [PubMed] [Google Scholar]
- 30.Kim D, Kim G, Yu R, Lee J, Kim S, Gleason MR, et al. Inhibitory co-receptor Lag3 supports Foxp3+ regulatory T cell function by restraining Myc-dependent metabolic programming. Immunity. 2024. 10.1016/j.immuni.2024.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li DY, Xiong XZ. ICOS(+) Tregs: a functional subset of tregs in immune diseases. Front Immunol. 2020. 10.3389/fimmu.2020.02104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Banerjee H, Nieves-Rosado H, Kulkarni A, Murter B, McGrath KV, Chandran UR, et al. Expression of Tim-3 drives phenotypic and functional changes in Treg cells in secondary lymphoid organs and the tumor microenvironment. Cell Rep. 2021. 10.1016/j.celrep.2021.109699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reinfeld BI, Rathmell WK, Kim TK, Rathmell JC. The therapeutic implications of immunosuppressive tumor aerobic glycolysis. Cell Mol Immunol. 2022. 10.1038/s41423-021-00727-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takahashi H, Kawabata-Iwakawa R, Ida S, Mito I, Tada H, Chikamatsu K. Upregulated glycolysis correlates with tumor progression and immune evasion in head and neck squamous cell carcinoma. Sci Rep. 2021. 10.1038/s41598-021-97292-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Z, Zhang Z, Zhang Y, Zhou W, Zhang X, Peng C, et al. Spatial transcriptomics reveals that metabolic characteristics define the tumor immunosuppression microenvironment via iCAF transformation in oral squamous cell carcinoma. Int J Oral Sci. 2024. 10.1038/s41368-023-00267-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takahashi H, Sakakura K, Arisaka Y, Tokue A, Kaira K, Tada H, et al. Clinical and biological significance of PD-L1 expression within the tumor microenvironment of oral squamous cell carcinoma. Anticancer Res. 2019. 10.21873/anticanres.13437. [DOI] [PubMed] [Google Scholar]
- 37.Beier UH, Angelin A, Akimova T, Wang L, Liu Y, Xiao H, et al. Essential role of mitochondrial energy metabolism in Foxp3(+) T-regulatory cell function and allograft survival. FASEB J. 2015. 10.1096/fj.14-268409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Rosa V, Galgani M, Porcellini A, Colamatteo A, Santopaolo M, Zuchegna C, et al. Glycolysis controls the induction of human regulatory T cells by modulating the expression of FOXP3 exon 2 splicing variants. Nat Immunol. 2015. 10.1038/ni.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mueller DL. Mechanisms maintaining peripheral tolerance. Nat Immunol. 2010. 10.1038/ni.1817. [DOI] [PubMed] [Google Scholar]
- 40.Michalek RD, Gerriets VA, Jacobs SR, Macintyre AN, MacIver NJ, Mason EF, et al. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J Immunol (Baltimore, Md : 1950). 2011;10.4049/jimmunol.1003613. [DOI] [PMC free article] [PubMed]
- 41.Pang X, Li TJ, Shi RJ, Wan ZX, Tang YY, Tang YL, et al. IRF2BP2 drives lymphatic metastasis in OSCC cells by elevating mitochondrial fission-dependent fatty acid oxidation. Mol Carcinog. 2024. 10.1002/mc.23635. [DOI] [PubMed] [Google Scholar]
- 42.Pascual G, Avgustinova A, Mejetta S, Martin M, Castellanos A, Attolini CS, et al. Targeting metastasis-initiating cells through the fatty acid receptor CD36. Nature. 2017. 10.1038/nature20791. [DOI] [PubMed] [Google Scholar]
- 43.Thomas N, Krishnapillai R, Bindhu PR, Thomas P. Immunohistochemical expression of cyclooxygenase-2 in oral squamous cell carcinoma. Indian J Dent Res. 2019. 10.4103/ijdr.IJDR_362_17. [DOI] [PubMed] [Google Scholar]
- 44.Bell CR, Pelly VS, Moeini A, Chiang SC, Flanagan E, Bromley CP, et al. Chemotherapy-induced COX-2 upregulation by cancer cells defines their inflammatory properties and limits the efficacy of chemoimmunotherapy combinations. Nat Commun. 2022. 10.1038/s41467-022-29606-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sekiya T. Comparison between Nr4a transcription factor regulation and function in lymphoid and tumor treg cells. Front Immunol. 2022. 10.3389/fimmu.2022.866339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Platten M, Wick W, Van den Eynde BJ. Tryptophan catabolism in cancer: beyond IDO and tryptophan depletion. Can Res. 2012. 10.1158/0008-5472.CAN-12-0569. [DOI] [PubMed] [Google Scholar]
- 47.Anzai H, Yoshimoto S, Okamura K, Hiraki A, Hashimoto S. IDO1-mediated Trp-kynurenine-AhR signal activation induces stemness and tumor dormancy in oral squamous cell carcinomas. Oral Sci Int. 2021. 10.1002/osi2.1109. [Google Scholar]
- 48.Fan C, Wu J, Shen Y, Hu H, Wang Q, Mao Y, et al. Hypoxia promotes the tolerogenic phenotype of plasmacytoid dendritic cells in head and neck squamous cell carcinoma. Cancer Med. 2022. 10.1002/cam4.4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stone TW, Williams RO. Interactions of IDO and the Kynurenine pathway with cell transduction systems and metabolism at the inflammation-cancer interface. Cancers (Basel). 2023. 10.3390/cancers15112895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Han C, Ge M, Ho PC, Zhang L. Fueling T-cell antitumor immunity: amino acid metabolism revisited. Cancer Immunol Res. 2021. 10.1158/2326-6066.CIR-21-0459. [DOI] [PubMed] [Google Scholar]
- 51.Campesato LF, Budhu S, Tchaicha J, Weng CH, Gigoux M, Cohen IJ, et al. Blockade of the AHR restricts a Treg-macrophage suppressive axis induced by L-Kynurenine. Nat Commun. 2020. 10.1038/s41467-020-17750-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Laimer K, Troester B, Kloss F, Schafer G, Obrist P, Perathoner A, et al. Expression and prognostic impact of indoleamine 2,3-dioxygenase in oral squamous cell carcinomas. Oral Oncol. 2011. 10.1016/j.oraloncology.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 53.Jarrold J, Davies CC. PRMTs and arginine methylation: cancer’s best-kept secret? Trends Mol Med. 2019. 10.1016/j.molmed.2019.05.007. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Y, Guo J, Jia R. Treg: a promising immunotherapeutic target in oral diseases. Front Immunol. 2021. 10.3389/fimmu.2021.667862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ma G, Zhang Z, Li P, Zhang Z, Zeng M, Liang Z, et al. Reprogramming of glutamine metabolism and its impact on immune response in the tumor microenvironment. Cell Commun Signal. 2022. 10.1186/s12964-022-00909-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Davoodi-Moghaddam Z, Jafari-Raddani F, Delshad M, Pourbagheri-Sigaroodi A, Bashash D. Inhibitors of the PI3K/AKT/mTOR pathway in human malignancies; trend of current clinical trials. J Cancer Res Clin Oncol. 2023. 10.1007/s00432-023-05277-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kumagai S, Togashi Y, Sakai C, Kawazoe A, Kawazu M, Ueno T, et al. An oncogenic alteration creates a microenvironment that promotes tumor progression by conferring a metabolic advantage to regulatory T cells. Immunity. 2020. 10.1016/j.immuni.2020.06.016. [DOI] [PubMed] [Google Scholar]
- 58.Zhang Y, Wu Y, Su X. PLOD1 promotes cell growth and aerobic glycolysis by regulating the SOX9/PI3K/Akt/mTOR signaling pathway in gastric cancer. Front Biosci (Landmark edition). 2021. 10.52586/4946. [DOI] [PubMed] [Google Scholar]
- 59.Mayer IA, Arteaga CL. The PI3K/AKT pathway as a target for cancer treatment. Annu Rev Med. 2016. 10.1146/annurev-med-062913-051343. [DOI] [PubMed] [Google Scholar]
- 60.O’Donnell JS, Massi D, Teng MWL, Mandala M. PI3K-AKT-mTOR inhibition in cancer immunotherapy, redux. Semin Cancer Biol. 2018. 10.1016/j.semcancer.2017.04.015. [DOI] [PubMed] [Google Scholar]
- 61.Haxhinasto S, Mathis D, Benoist C. The AKT-mTOR axis regulates de novo differentiation of CD4+Foxp3+ cells. J Exp Med. 2008. 10.1084/jem.20071477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Abu-Eid R, Samara RN, Ozbun L, Abdalla MY, Berzofsky JA, Friedman KM, et al. Selective inhibition of regulatory T cells by targeting the PI3K-Akt pathway. Cancer Immunol Res. 2014. 10.1158/2326-6066.CIR-14-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jie HB, Schuler PJ, Lee SC, Srivastava RM, Argiris A, Ferrone S, et al. CTLA-4(+) Regulatory T cells increased in cetuximab-treated head and neck cancer patients suppress NK cell cytotoxicity and correlate with poor prognosis. Can Res. 2015. 10.1158/0008-5472.CAN-14-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu X, Karnell JL, Yin B, Zhang R, Zhang J, Li P, et al. Distinct roles for PTEN in prevention of T cell lymphoma and autoimmunity in mice. J Clin Investig. 2010. 10.1172/JCI42382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997. 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 66.Shrestha S, Yang K, Guy C, Vogel P, Neale G, Chi H. Treg cells require the phosphatase PTEN to restrain TH1 and TFH cell responses. Nat Immunol. 2015. 10.1038/ni.3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huynh A, DuPage M, Priyadharshini B, Sage PT, Quiros J, Borges CM, et al. Control of PI(3) kinase in Treg cells maintains homeostasis and lineage stability. Nat Immunol. 2015. 10.1038/ni.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ma J, Zhong M, Xiong Y, Gao Z, Wu Z, Liu Y, et al. Emerging roles of nucleotide metabolism in cancer development: progress and prospect. Aging. 2021. 10.18632/aging.202962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Alapati S, Fortuna G, Ramage G, Delaney C. Evaluation of metabolomics as diagnostic targets in oral squamous cell carcinoma: a systematic review. Metabolites. 2023. 10.3390/metabo13080890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Qiu Y, Xie E, Xu H, Cheng H, Li G. One-carbon metabolism shapes T cell immunity in cancer. Trends Endocrinol Metab. 2024. 10.1016/j.tem.2024.05.010. [DOI] [PubMed] [Google Scholar]
- 71.Wang W, Gu W, Tang H, Mai Z, Xiao H, Zhao J, et al. The emerging role of MTHFD family genes in regulating the tumor immunity of oral squamous cell carcinoma. J Oncol. 2022. 10.1155/2022/4867730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yegutkin GG, Boison D. ATP and adenosine metabolism in cancer: exploitation for therapeutic gain. Pharmacol Rev. 2022. 10.1124/pharmrev.121.000528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jiang L, Zhou Y, Tang S, Yang D, Zhang Y, Zhang J, et al. Nociceptive adenosine A(2A) receptor on trigeminal nerves orchestrates CGRP release to regulate the progression of oral squamous cell carcinoma. Int J Oral Sci. 2024. 10.1038/s41368-024-00308-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ohta A, Kini R, Ohta A, Subramanian M, Madasu M, Sitkovsky M. The development and immunosuppressive functions of CD4(+) CD25(+) FoxP3(+) regulatory T cells are under influence of the adenosine-A2A adenosine receptor pathway. Front Immunol. 2012. 10.3389/fimmu.2012.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sundstrom P, Stenstad H, Langenes V, Ahlmanner F, Theander L, Ndah TG, et al. Regulatory T cells from colon cancer patients inhibit effector T-cell migration through an adenosine-dependent mechanism. Cancer Immunol Res. 2016. 10.1158/2326-6066.CIR-15-0050. [DOI] [PubMed] [Google Scholar]
- 76.Qiu X, Ye H, Li X, Li D, Jiang L, Liu R, et al. IL-6/JAK2-dependent G6PD phosphorylation promotes nucleotide synthesis and supports tumor growth. Mol Metab. 2023. 10.1016/j.molmet.2023.101836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mele L, Paino F, Papaccio F, Regad T, Boocock D, Stiuso P, et al. A new inhibitor of glucose-6-phosphate dehydrogenase blocks pentose phosphate pathway and suppresses malignant proliferation and metastasis in vivo. Cell Death Dis. 2018. 10.1038/s41419-018-0635-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Düvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell. 2010. 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sun L, Fu J, Zhou Y. Metabolism controls the balance of Th17/T-regulatory cells. Front Immunol. 2017. 10.3389/fimmu.2017.01632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pandiyan P, Bhaskaran N, Zou M, Schneider E, Jayaraman S, Huehn J. Microbiome dependent regulation of T(regs) and Th17 cells in mucosa. Front Immunol. 2019. 10.3389/fimmu.2019.00426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang CY, Yeh YM, Yu HY, Chin CY, Hsu CW, Liu H, et al. Oral microbiota community dynamics associated with oral squamous cell carcinoma staging. Front Microbiol. 2018. 10.3389/fmicb.2018.00862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cecil JD, Sirisaengtaksin N, O’Brien-Simpson NM, Krachler AM. Outer Membrane vesicle-host cell interactions. Microbiol Spectr. 2019. 10.1128/microbiolspec.PSIB-0001-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.He S, Chakraborty R, Ranganathan S. Metaproteomic analysis of an oral squamous cell carcinoma dataset suggests diagnostic potential of the mycobiome. Int J Mol Sci. 2023. 10.3390/ijms24021050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Geng F, Zhang Y, Lu Z, Zhang S, Pan Y. Fusobacterium nucleatum caused DNA damage and promoted cell proliferation by the Ku70/p53 pathway in oral cancer cells. DNA Cell Biol. 2020. 10.1089/dna.2019.5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Binder Gallimidi A, Fischman S, Revach B, Bulvik R, Maliutina A, Rubinstein AM, et al. Periodontal pathogens Porphyromonas gingivalis and Fusobacterium nucleatum promote tumor progression in an oral-specific chemical carcinogenesis model. Oncotarget. 2015. 10.18632/oncotarget.4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang L, Liu Y, Zheng HJ, Zhang CP. The oral microbiota may have influence on oral cancer. Front Cell Infect Microbiol. 2019. 10.3389/fcimb.2019.00476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Inaba H, Sugita H, Kuboniwa M, Iwai S, Hamada M, Noda T, et al. Porphyromonas gingivalis promotes invasion of oral squamous cell carcinoma through induction of proMMP9 and its activation. Cell Microbiol. 2014. 10.1111/cmi.12211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhao H, Chu M, Huang Z, Yang X, Ran S, Hu B, et al. Variations in oral microbiota associated with oral cancer. Sci Rep. 2017. 10.1038/s41598-017-11779-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rahman R, Shaikh MH, Gopinath D, Idris A, Johnson NW. Human papillomavirus and Epstein-Barr virus co-infection in oral and oropharyngeal squamous cell carcinomas: a systematic review and meta-analysis. Mol Oral Microbiol. 2023. 10.1111/omi.12412. [DOI] [PubMed] [Google Scholar]
- 90.Mallika L, Augustine D, Rao RS, Patil S, Alamir AWH, Awan KH, et al. Does microbiome shift play a role in carcinogenesis? A systematic review. Transl Cancer Res. 2020. 10.21037/tcr.2020.02.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Di Spirito F, Di Palo MP, Folliero V, Cannata D, Franci G, Martina S, et al. Oral bacteria, virus and fungi in saliva and tissue samples from adult subjects with oral squamous cell carcinoma: an umbrella review. Cancers (Basel). 2023. 10.3390/cancers15235540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yang Z, Zhang S, Ji N, Li J, Chen Q. The evil companion of OSCC: Candida albicans. Oral Dis. 2024. 10.1111/odi.14700. [DOI] [PubMed] [Google Scholar]
- 93.Talapko J, Mestrovic T, Dmitrovic B, Juzbasic M, Matijevic T, Bekic S, et al. A putative role of candida albicans in promoting cancer development: a current state of evidence and proposed mechanisms. Microorganisms. 2023. 10.3390/microorganisms11061476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen G, Gao C, Jiang S, Cai Q, Li R, Sun Q, et al. Fusobacterium nucleatum outer membrane vesicles activate autophagy to promote oral cancer metastasis. J Adv Res. 2024. 10.1016/j.jare.2023.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu D, Liu S, Liu J, Miao L, Zhang S, Pan Y. sRNA23392 packaged by Porphyromonas gingivalis outer membrane vesicles promotes oral squamous cell carcinomas migration and invasion by targeting desmocollin-2. Mol Oral Microbiol. 2021. 10.1111/omi.12334. [DOI] [PubMed] [Google Scholar]
- 96.Metsäniitty M, Hasnat S, Öhman C, Salo T, Eklund KK, Oscarsson J, et al. Extracellular vesicles from Aggregatibacter actinomycetemcomitans exhibit potential antitumorigenic effects in oral cancer: a comparative in vitro study. Arch Microbiol. 2024. 10.1007/s00203-024-03976-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hoeppli RE, Wu D, Cook L, Levings MK. The environment of regulatory T cell biology: cytokines, metabolites, and the microbiome. Front Immunol. 2015. 10.3389/fimmu.2015.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013. 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang X, Zhang W, Wu W, Wu S, Young A, Yan Z. Is Candida albicans a contributor to cancer? A critical review based on the current evidence. Microbiol Res. 2023. 10.1016/j.micres.2023.127370. [DOI] [PubMed] [Google Scholar]
- 100.Heawchaiyaphum C, Yoshiyama H, Iizasa H, Burassakarn A, Tumurgan Z, Ekalaksananan T, et al. Epstein-Barr virus promotes oral squamous cell carcinoma stemness through the Warburg Effect. Int J Mol Sci. 2023. 10.3390/ijms241814072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang S, Nie F, Yin Q, Tian H, Gong P, Ju J, et al. Periodontitis promotes tumor growth and immune evasion via PD-1/PD-L1. Cancer Immunol Immunother. 2024;10.1007/s00262-024-03865-5. [DOI] [PMC free article] [PubMed]
- 102.Geng F, Liu J, Guo Y, Li C, Wang H, Wang H, et al. Persistent exposure to porphyromonas gingivalis promotes proliferative and invasion capabilities, and tumorigenic properties of human immortalized oral epithelial cells. Front Cell Infect Microbiol. 2017. 10.3389/fcimb.2017.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sami A, Elimairi I, Stanton C, Ross RP, Ryan CA. The role of the microbiome in oral squamous cell carcinoma with insight into the microbiome-treatment axis. Int J Mol Sci. 2020. 10.3390/ijms21218061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rai AK, Panda M, Das AK, Rahman T, Das R, Das K, et al. Dysbiosis of salivary microbiome and cytokines influence oral squamous cell carcinoma through inflammation. Arch Microbiol. 2021. 10.1007/s00203-020-02011-w. [DOI] [PubMed] [Google Scholar]
- 105.Frank DN, Qiu Y, Cao Y, Zhang S, Lu L, Kofonow JM, et al. A dysbiotic microbiome promotes head and neck squamous cell carcinoma. Oncogene. 2022. 10.1038/s41388-021-02137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kindt N, Descamps G, Seminerio I, Bellier J, Lechien JR, Mat Q, et al. High stromal Foxp3-positive T cell number combined to tumor stage improved prognosis in head and neck squamous cell carcinoma. Oral Oncol. 2017. 10.1016/j.oraloncology.2017.02.023. [DOI] [PubMed] [Google Scholar]
- 107.Lechien JR, Descamps G, Seminerio I, Furgiuele S, Dequanter D, Mouawad F, et al. HPV involvement in the tumor microenvironment and immune treatment in head and neck squamous cell carcinomas. Cancers (Basel). 2020. 10.3390/cancers12051060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pérez-Islas E, García-Carrancá A, Acosta-Gio E, Reynoso-Noverón N, Maldonado-Martínez HA, Guido-Jiménez M, et al. Prognostic importance of DNA from human papillomavirus in patients with oral squamous cell carcinoma. Med Oral Patologia Oral y Cirugia Bucal. 2022;10.4317/medoral.25092. [DOI] [PMC free article] [PubMed]
- 109.Chandel V, Raj S, Kumar P, Gupta S, Dhasmana A, Kesari KK, et al. Metabolic regulation in HPV associated head and neck squamous cell carcinoma. Life Sci. 2020. 10.1016/j.lfs.2020.118236. [DOI] [PubMed] [Google Scholar]
- 110.Li Z, Liu Y, Huang X, Wang Q, Fu R, Wen X, et al. F. Nucleatum enhances oral squamous cell carcinoma proliferation via E-cadherin/β-Catenin pathway. BMC Oral Health. 2024;10.1186/s12903-024-04252-3. [DOI] [PMC free article] [PubMed]
- 111.Guo Z-c, Jing S-l, Jumatai S, Gong Z-c. Porphyromonas gingivalis promotes the progression of oral squamous cell carcinoma by activating the neutrophil chemotaxis in the tumour microenvironment. Cancer Immunol Immunother. 2023;10.1007/s00262-022-03348-5. [DOI] [PMC free article] [PubMed]
- 112.Gao R, Shi GP, Wang J. Functional diversities of regulatory T cells in the context of cancer immunotherapy. Front Immunol. 2022. 10.3389/fimmu.2022.833667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li F, Huang H, Xu J, Tao L, Zhou L, Hsueh C, et al. Fusobacterium nucleatum-triggered purine metabolic reprogramming drives tumorigenesis in head and neck carcinoma. Discov Oncol. 2023. 10.1007/s12672-023-00727-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.SureshBabu SK, Godbole JH, Vaibhaw A, Chiplunkar SV. Immunosuppressive microenvironment in oral cancer: implications for cancer immunotherapy. Explor Immunol. 2021. 10.37349/ei.2021.00013. [Google Scholar]
- 115.Zhang N, Liu Y, Yang H, Liang M, Wang X, Wang M, et al. Clinical significance of fusobacterium nucleatum infection and regulatory T cell enrichment in esophageal squamous cell carcinoma. Pathol Oncol Res: POR. 2021. 10.3389/pore.2021.1609846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sun J, Tang Q, Yu S, Xie M, Zheng W, Chen G, et al. F. nucleatum facilitates oral squamous cell carcinoma progression via GLUT1-driven lactate production. EBioMedicine. 2023;10.1016/j.ebiom.2023.104444. [DOI] [PMC free article] [PubMed]
- 117.Lamont RJ, Fitzsimonds ZR, Wang H, Gao S. Role of Porphyromonas gingivalis in oral and orodigestive squamous cell carcinoma. Periodontol 2000. 2022;10.1111/prd.12425. [DOI] [PMC free article] [PubMed]
- 118.Jin M, Huo D, Sun J, Hu J, Liu S, Zhan M, et al. Enhancing immune responses of ESC-based TAA cancer vaccines with a novel OMV delivery system. J Nanobiotechnol. 2024. 10.1186/s12951-023-02273-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Raez LE, Papadopoulos K, Ricart AD, Chiorean EG, Dipaola RS, Stein MN, et al. A phase I dose-escalation trial of 2-deoxy-D-glucose alone or combined with docetaxel in patients with advanced solid tumors. Cancer Chemother Pharmacol. 2013. 10.1007/s00280-012-2045-1. [DOI] [PubMed] [Google Scholar]
- 120.Powell SF, Mazurczak M, Dib EG, Bleeker JS, Geeraerts LH, Tinguely M, et al. Phase II study of dichloroacetate, an inhibitor of pyruvate dehydrogenase, in combination with chemoradiotherapy for unresected, locally advanced head and neck squamous cell carcinoma. Invest New Drugs. 2022. 10.1007/s10637-022-01235-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cui Y, Man S, Tao J, Liu Y, Ma L, Guo L, et al. The lipid droplet in cancer: From being a tumor-supporting hallmark to clinical therapy. Acta Physiol (Oxford, England). 2024;10.1111/apha.14087. [DOI] [PubMed]
- 122.Sawyer AJ, Kyriakides TR. Matricellular proteins in drug delivery: Therapeutic targets, active agents, and therapeutic localization. Adv Drug Deliv Rev. 2016. 10.1016/j.addr.2015.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Naing A, Algazi AP, Falchook GS, Creelan BC, Powderly J, Rosen S, et al. Phase 1/2 study of epacadostat in combination with durvalumab in patients with metastatic solid tumors. Cancer. 2023. 10.1002/cncr.34512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mitchell TC, Hamid O, Smith DC, Bauer TM, Wasser JS, Olszanski AJ, et al. Epacadostat plus pembrolizumab in patients with advanced solid tumors: phase I results from a multicenter, open-label phase I/II trial (ECHO-202/KEYNOTE-037). J Clin Oncol. 2018. 10.1200/jco.2018.78.9602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Perez RP, Riese MJ, Lewis KD, Saleh MN, Daud A, Berlin J, et al. Epacadostat plus nivolumab in patients with advanced solid tumors: Preliminary phase I/II results of ECHO-204. J Clin Oncol. 2017. 10.1200/JCO.2017.35.15_suppl.3003.29236570 [Google Scholar]
- 126.Daud A, Saleh MN, Hu J, Bleeker JS, Riese MJ, Meier R, et al. Epacadostat plus nivolumab for advanced melanoma: Updated phase 2 results of the ECHO-204 study. J Clin Oncol. 2018. 10.1200/JCO.2018.36.15_suppl.9511.29283791 [Google Scholar]
- 127.Luginbuhl A, Johnson JM, Harshyne L, Tuluc M, Gargano S, Leiby B, et al. Window-of-opportunity trial of nivolumab with or without the IDO inhibitor BMS-986205 in patients with resectable squamous cell carcinoma of the head and neck (SCCHN). J Clin Oncol. 2020. 10.1200/JCO.2020.38.15_suppl.TPS6595. [Google Scholar]
- 128.Luginbuhl A, Johnson JM, Scott ER, Harshyne L, Flerova EF, Tuluc M, et al. Neoadjuvant nivolumab with or without IDO inhibitor in head and neck squamous cell carcinoma (HNSCC): Final pathologic and clinical outcomes. J Clin Oncol. 2022. 10.1200/JCO.2022.40.16_suppl.6070. [Google Scholar]
- 129.Psyrri A, Gkotzamanidou M, Papaxoinis G, Krikoni L, Economopoulou P, Kotsantis I, et al. The DNA damage response network in the treatment of head and neck squamous cell carcinoma. ESMO open. 2021. 10.1016/j.esmoop.2021.100075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Khayami R, Toroghian Y, Bahreyni A, Bahrami A, Khazaei M, Ferns GA, et al. Role of adenosine signaling in the pathogenesis of head and neck cancer. J Cell Biochem. 2018. 10.1002/jcb.27091. [DOI] [PubMed] [Google Scholar]
- 131.Lorch JH, Tishler RB, Chau NG, Hanna GJ, Rabinowits G, Schoenfeld JD, et al. A phase I study of the PI3K inhibitor buplarlisib (B) with concurrent chemoradiotherapy (CRT) in patients with high risk locally advanced squamous cell cancer of the head and neck (LASCCHN). J Clin Oncol. 2018. 10.1200/JCO.2018.36.15_suppl.6071.30285559 [Google Scholar]
- 132.Brisson RJ, Kochanny S, Arshad S, Dekker A, DeSouza JA, Saloura V, et al. A pilot study of the pan-class I PI3K inhibitor buparlisib in combination with cetuximab in patients with recurrent or metastatic head and neck cancer. Head Neck. 2019. 10.1002/hed.25910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Marret G, Isambert N, Rezai K, Gal J, Saada-Bouzid E, Rolland F, et al. Phase I trial of copanlisib, a selective PI3K inhibitor, in combination with cetuximab in patients with recurrent and/or metastatic head and neck squamous cell carcinoma. Invest New Drugs. 2021. 10.1007/s10637-021-01152-z. [DOI] [PubMed] [Google Scholar]
- 134.Day D, Prawira A, Spreafico A, Waldron J, Karithanam R, Giuliani M, et al. Phase I trial of alpelisib in combination with concurrent cisplatin-based chemoradiotherapy in patients with locoregionally advanced squamous cell carcinoma of the head and neck. Oral Oncol. 2020. 10.1016/j.oraloncology.2020.104753. [DOI] [PubMed] [Google Scholar]
- 135.Razak ARA, Ahn M-J, Yen C-J, Solomon BJ, Lee S-H, Wang H-M, et al. Phase lb/ll study of the PI3Kα inhibitor BYL719 in combination with cetuximab in recurrent/metastatic squamous cell cancer of the head and neck (SCCHN). J Clin Oncol. 2014. 10.1200/jco.2014.32.15_suppl.6044. [Google Scholar]
- 136.Johnson FM, Janku F, Gouda MA, Tran HT, Kawedia JD, Schmitz D, et al. Inhibition of the phosphatidylinositol-3 kinase pathway using bimiralisib in loss-of-function NOTCH1-Mutant Head and Neck Cancer. Oncologist. 2022. 10.1093/oncolo/oyac185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Johnson FM, Janku F, Lee JJ, Schmitz D, Streefkerk H, Frederick M. Single-arm study of bimiralisib in head and neck squamous cell carcinoma (HNSCC) patients (pts) harboring NOTCH1 loss of function (LOF) mutations. J Clin Oncol. 2020. 10.1200/JCO.2020.38.15_suppl.TPS6590.33332191 [Google Scholar]
- 138.Day TA, Shirai K, O’Brien PE, Matheus MG, Godwin K, Sood AJ, et al. Inhibition of mTOR signaling and clinical activity of rapamycin in head and neck cancer in a window of opportunity trial. Clin Cancer Res. 2019. 10.1158/1078-0432.CCR-18-2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Fury MG, Xiao H, Baxi SS, Sherman EJ, Smith-Marrone S, Ho AL, et al. A phase II trial of temsirolimus plus low-dose weekly carboplatin and paclitaxel for recurrent/metastatic HNSCC. J Clin Oncol. 2014. 10.1200/jco.2014.32.15_suppl.6019.25225434 [Google Scholar]
- 140.Grunwald V, Keilholz U, Boehm A, Guntinas-Lichius O, Hennemann B, Schmoll HJ, et al. TEMHEAD: a single-arm multicentre phase II study of temsirolimus in platin- and cetuximab refractory recurrent and/or metastatic squamous cell carcinoma of the head and neck (SCCHN) of the German SCCHN Group (AIO). Ann Oncol. 2015. 10.1093/annonc/mdu571. [DOI] [PubMed] [Google Scholar]
- 141.Raymond E, Le Tourneau C, Gatineau M, Delord J-P, Fayette J, Dreyer C, et al. CAPRA: Safety, efficacy, and translational biomarkers of weekly everolimus, carboplatin, and paclitaxel as induction therapy for locally advanced head and neck squamous cell carcinoma (HNSCC). J Clin Oncol. 2013. 10.1200/jco.2013.31.15_suppl.6036.24081937 [Google Scholar]
- 142.Geiger JL, Bauman JE, Gibson MK, Gooding WE, Varadarajan P, Kotsakis A, et al. Phase II trial of everolimus in patients with previously treated recurrent or metastatic head and neck squamous cell carcinoma. Head Neck. 2016. 10.1002/hed.24501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Munster P, Mita M, Mahipal A, Nemunaitis J, Massard C, Mikkelsen T, et al. First-in-human phase I study of a dual mTOR kinase and DNA-PK inhibitor (CC-115) in advanced malignancy. Cancer Manage Res. 2019. 10.2147/CMAR.S208720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Jimeno A, Shirai K, Choi M, Laskin J, Kochenderfer M, Spira A, et al. A randomized, phase II trial of cetuximab with or without PX-866, an irreversible oral phosphatidylinositol 3-kinase inhibitor, in patients with relapsed or metastatic head and neck squamous cell cancer. Ann Oncol. 2015. 10.1093/annonc/mdu574. [DOI] [PubMed] [Google Scholar]
- 145.Seiwert TY, Kochanny S, Wood K, Worden FP, Adkins D, Wade JL, et al. A randomized phase 2 study of temsirolimus and cetuximab versus temsirolimus alone in recurrent/metastatic, cetuximab-resistant head and neck cancer: The MAESTRO study. Cancer. 2020. 10.1002/cncr.32929. [DOI] [PubMed] [Google Scholar]
- 146.Fayette J, Wirth L, Oprean C, Udrea A, Jimeno A, Rischin D, et al. Randomized phase II study of duligotuzumab (MEHD7945A) vs. cetuximab in squamous cell carcinoma of the head and neck (MEHGAN Study). Front Oncol. 2016;10.3389/fonc.2016.00232. [DOI] [PMC free article] [PubMed]
- 147.Massarelli E, Lin H, Ginsberg LE, Tran HT, Lee JJ, Canales JR, et al. Phase II trial of everolimus and erlotinib in patients with platinum-resistant recurrent and/or metastatic head and neck squamous cell carcinoma. Ann Oncol. 2015. 10.1093/annonc/mdv194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Farooque A, Singh N, Adhikari JS, Afrin F, Dwarakanath BS. Enhanced antitumor immunity contributes to the radio-sensitization of ehrlich ascites tumor by the glycolytic inhibitor 2-deoxy-D-glucose in mice. PLoS ONE. 2014. 10.1371/journal.pone.0108131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Dwarakanath BS, Farooque A, Gupta S. Targeting regulatory T cells for improving cancer therapy: challenges and prospects. Cancer Rep (Hoboken). 2018. 10.1002/cnr2.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Pai S, Yadav VK, Kuo KT, Pikatan NW, Lin CS, Chien MH, et al. PDK1 inhibitor BX795 improves cisplatin and radio-efficacy in oral squamous cell carcinoma by downregulating the PDK1/CD47/Akt-mediated glycolysis signaling pathway. Int J Mol Sci. 2021. 10.3390/ijms222111492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Takaichi M, Tachinami H, Takatsuka D, Yonesi A, Sakurai K, Rasul MI, et al. Targeting CD36-mediated lipid metabolism by selective inhibitor-augmented antitumor immune responses in oral cancer. Int J Mol Sci. 2024. 10.3390/ijms25179438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Munn DH, Mellor AL. IDO in the tumor microenvironment: inflammation, counter-regulation, and tolerance. Trends Immunol. 2016. 10.1016/j.it.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Mezrich JD, Fechner JH, Zhang X, Johnson BP, Burlingham WJ, Bradfield CA. An interaction between kynurenine and the aryl hydrocarbon receptor can generate regulatory T cells. J Immunol (Baltimore, Md: 1950). 2010;10.4049/jimmunol.0903670. [DOI] [PMC free article] [PubMed]
- 154.Beatty GL, O’Dwyer PJ, Clark J, Shi JG, Bowman KJ, Scherle PA, et al. First-in-human phase I study of the oral inhibitor of indoleamine 2,3-dioxygenase-1 epacadostat (INCB024360) in patients with advanced solid malignancies. Clin Cancer Res. 2017. 10.1158/1078-0432.CCR-16-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Jochems C, Fantini M, Fernando RI, Kwilas AR, Donahue RN, Lepone LM, et al. The IDO1 selective inhibitor epacadostat enhances dendritic cell immunogenicity and lytic ability of tumor antigen-specific T cells. Oncotarget. 2016. 10.18632/oncotarget.9326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Molinolo AA, Hewitt SM, Amornphimoltham P, Keelawat S, Rangdaeng S, Meneses Garcia A, et al. Dissecting the Akt/mammalian target of rapamycin signaling network: emerging results from the head and neck cancer tissue array initiative. Clin Cancer Res. 2007. 10.1158/1078-0432.CCR-07-1041. [DOI] [PubMed] [Google Scholar]
- 157.Naing A, Powderly JD, Nemunaitis JJ, Luke JJ, Mansfield AS, Messersmith WA, et al. Exploring the safety, effect on the tumor microenvironment, and efficacy of itacitinib in combination with epacadostat or parsaclisib in advanced solid tumors: a phase I study. J Immunother Cancer. 2022. 10.1136/jitc-2021-004223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Mattes MD, Willers H, Lin Y, Gharzai LA, Alesi ER, Lominska CE, et al. CTEP 10492, a phase 1/1b study of the AKT inhibitor ipatasertib with chemoradiation for locally advanced head and neck squamous cell carcinoma. J Clin Oncol. 2024. 10.1200/JCO.2024.42.16_suppl.TPS6127. [Google Scholar]
- 159.Sek K, Molck C, Stewart GD, Kats L, Darcy PK, Beavis PA. Targeting adenosine receptor signaling in cancer immunotherapy. Int J Mol Sci. 2018. 10.3390/ijms19123837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Vigano S, Alatzoglou D, Irving M, Menetrier-Caux C, Caux C, Romero P, et al. Targeting adenosine in cancer immunotherapy to enhance T-cell function. Front Immunol. 2019. 10.3389/fimmu.2019.00925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Mandapathil M, Szczepanski MJ, Szajnik M, Ren J, Lenzner DE, Jackson EK, et al. Increased ectonucleotidase expression and activity in regulatory T cells of patients with head and neck cancer. Clin Cancer Res. 2009. 10.1158/1078-0432.CCR-09-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Churov A, Zhulai G. Targeting adenosine and regulatory T cells in cancer immunotherapy. Hum Immunol. 2021. 10.1016/j.humimm.2020.12.005. [DOI] [PubMed] [Google Scholar]
- 163.Hay CM, Sult E, Huang Q, Mulgrew K, Fuhrmann SR, McGlinchey KA, et al. Targeting CD73 in the tumor microenvironment with MEDI9447. Oncoimmunology. 2016. 10.1080/2162402X.2016.1208875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zhang J, Ma C, Qin H, Wang Z, Zhu C, Liu X, et al. Construction and validation of a metabolic-related genes prognostic model for oral squamous cell carcinoma based on bioinformatics. BMC Med Genomics. 2022. 10.1186/s12920-022-01417-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013. 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 166.Brennan CA, Clay SL, Lavoie SL, Bae S, Lang JK, Fonseca-Pereira D, et al. Fusobacterium nucleatum drives a pro-inflammatory intestinal microenvironment through metabolite receptor-dependent modulation of IL-17 expression. Gut microbes. 2021. 10.1080/19490976.2021.1987780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Yamamoto Y, Kamiya T, Yano M, Huyen VT, Oishi M, Nishio M, et al. Oral microbial profile analysis in patients with oral and pharyngeal cancer reveals that tumoral fusobacterium nucleatum promotes oral cancer progression by Activating YAP. Microorganisms. 2023. 10.3390/microorganisms11122957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Qin Y, Li Z, Liu T, Ma J, Liu H, Zhou Y, et al. Prevotella intermedia boosts OSCC progression through ISG15 upregulation: a new target for intervention. J Cancer Res Clin Oncol. 2024. 10.1007/s00432-024-05730-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Wang S, Song A, Xie J, Wang Y-Y, Wang W-D, Zhang M-J, et al. Fn-OMV potentiates ZBP1-mediated PANoptosis triggered by oncolytic HSV-1 to fuel antitumor immunity. Nat Commun. 2024. 10.1038/s41467-024-48032-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Lim SA, Wei J, Nguyen TM, Shi H, Su W, Palacios G, et al. Lipid signalling enforces functional specialization of T(reg) cells in tumours. Nature. 2021. 10.1038/s41586-021-03235-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Leone RD, Powell JD. Metabolism of immune cells in cancer. Nat Rev Cancer. 2020. 10.1038/s41568-020-0273-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Park J, Kim M, Kang SG, Jannasch AH, Cooper B, Patterson J, et al. Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR-S6K pathway. Mucosal Immunol. 2015. 10.1038/mi.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Cao J, Liao S, Zeng F, Liao Q, Luo G, Zhou Y. Effects of altered glycolysis levels on CD8(+) T cell activation and function. Cell Death Dis. 2023. 10.1038/s41419-023-05937-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Whiteside SK, Grant FM, Alvisi G, Clarke J, Tang L, Imianowski CJ, et al. Acquisition of suppressive function by conventional T cells limits antitumor immunity upon T(reg) depletion. Sci Immunol. 2023. 10.1126/sciimmunol.abo5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Miao Y, Wang P, Huang J, Qi X, Liang Y, Zhao W, et al. Metabolomics, transcriptome and single-cell RNA sequencing analysis of the metabolic heterogeneity between oral cancer stem cells and differentiated cancer cells. Cancers (Basel). 2024. 10.3390/cancers16020237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Mishra R. Oral tumor heterogeneity, its implications for patient monitoring and designing anti-cancer strategies. Pathol Res Pract. 2024. 10.1016/j.prp.2023.154953. [DOI] [PubMed] [Google Scholar]
- 177.Nakajima EC, Laymon C, Oborski M, Hou W, Wang L, Grandis JR, et al. Quantifying metabolic heterogeneity in head and neck tumors in real time: 2-DG uptake is highest in hypoxic tumor regions. PLoS ONE. 2014. 10.1371/journal.pone.0102452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Chai AWY, Lim KP, Cheong SC. Translational genomics and recent advances in oral squamous cell carcinoma. Semin Cancer Biol. 2020. 10.1016/j.semcancer.2019.09.011. [DOI] [PubMed] [Google Scholar]
- 179.Wang H, Franco F, Ho PC. Metabolic regulation of tregs in cancer: opportunities for immunotherapy. Trends Cancer. 2017. 10.1016/j.trecan.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 180.Ashrafizadeh M, Farhood B, Eleojo Musa A, Taeb S, Rezaeyan A, Najafi M. Abscopal effect in radioimmunotherapy. Int Immunopharmacol. 2020. 10.1016/j.intimp.2020.106663. [DOI] [PubMed] [Google Scholar]
- 181.Budi HS, Farhood B. Targeting oral tumor microenvironment for effective therapy. Cancer Cell Int. 2023. 10.1186/s12935-023-02943-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell. 2017. 10.1016/j.cell.2017.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Sarkar T, Dhar S, Sa G. Tumor-infiltrating T-regulatory cells adapt to altered metabolism to promote tumor-immune escape. Curr Res Immunol. 2021. 10.1016/j.crimmu.2021.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Saleh R, Elkord E. Treg-mediated acquired resistance to immune checkpoint inhibitors. Cancer Lett. 2019. 10.1016/j.canlet.2019.05.003. [DOI] [PubMed] [Google Scholar]
- 185.Field CS, Baixauli F, Kyle RL, Puleston DJ, Cameron AM, Sanin DE, et al. Mitochondrial integrity regulated by lipid metabolism is a cell-intrinsic checkpoint for treg suppressive function. Cell Metab. 2020. 10.1016/j.cmet.2019.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Weinmann SC, Pisetsky DS. Mechanisms of immune-related adverse events during the treatment of cancer with immune checkpoint inhibitors. Rheumatology (Oxford). 2019. 10.1093/rheumatology/kez308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Esfahani K, Elkrief A, Calabrese C, Lapointe R, Hudson M, Routy B, et al. Moving towards personalized treatments of immune-related adverse events. Nat Rev Clin Oncol. 2020. 10.1038/s41571-020-0352-8. [DOI] [PubMed] [Google Scholar]
- 188.Danzi F, Pacchiana R, Mafficini A, Scupoli MT, Scarpa A, Donadelli M, et al. To metabolomics and beyond: a technological portfolio to investigate cancer metabolism. Signal Transduct Target Ther. 2023. 10.1038/s41392-023-01380-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Sharma P, Goswami S, Raychaudhuri D, Siddiqui BA, Singh P, Nagarajan A, et al. Immune checkpoint therapy-current perspectives and future directions. Cell. 2023. 10.1016/j.cell.2023.03.006. [DOI] [PubMed] [Google Scholar]
- 190.Yang Y, Liu X, Yang D, Li L, Li S, Lu S, et al. Interplay of CD36, autophagy, and lipid metabolism: insights into cancer progression. Metabolism. 2024. 10.1016/j.metabol.2024.155905. [DOI] [PubMed] [Google Scholar]
- 191.Chen X, Zhong S, Zhan Y, Zhang X. CRISPR-Cas9 applications in T cells and adoptive T cell therapies. Cell Mol Biol Lett. 2024. 10.1186/s11658-024-00561-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Shi L, Sheng J, Chen G, Zhu P, Shi C, Li B, et al. Combining IL-2-based immunotherapy with commensal probiotics produces enhanced antitumor immune response and tumor clearance. J Immunother Cancer. 2020. 10.1136/jitc-2020-000973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Chattopadhyay I, Nandi D, Nag A. The pint- sized powerhouse: Illuminating the mighty role of the gut microbiome in improving the outcome of anti- cancer therapy. Semin Cancer Biol. 2021. 10.1016/j.semcancer.2020.07.012. [DOI] [PubMed] [Google Scholar]
- 194.Radaic A, Kamarajan P, Cho A, Wang S, Hung GC, Najarzadegan F, et al. Biological biomarkers of oral cancer. Periodontol 2000. 2024;10.1111/prd.12542. [DOI] [PMC free article] [PubMed]
- 195.Baysoy A, Bai Z, Satija R, Fan R. The technological landscape and applications of single-cell multi-omics. Nat Rev Mol Cell Biol. 2023. 10.1038/s41580-023-00615-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Vandereyken K, Sifrim A, Thienpont B, Voet T. Methods and applications for single-cell and spatial multi-omics. Nat Rev Genet. 2023. 10.1038/s41576-023-00580-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No datasets were generated or analysed during the current study.