Abstract
Background
Prelingual hearing impairment (HI) is genetically highly heterogenous. Early diagnosis and intervention are essential for psychosocial development. In this study we investigated a consanguineous family from Pakistan with autosomal recessive (AR) non-syndromic sensorineural HI (NSHI).
Methods
A DNA sample from an HI member of a consanguineous Pakistani family segregating ARNSHL underwent exome sequencing. Using Sanger sequencing select variants were validated and tested for segregation using DNA samples from additional family members. We further investigated RNA expression data for the candidate gene in mouse and human inner ear and human inner ear organoids using data obtained from the gene Expression Analysis Resource.
Results
We identified thrombospondin 1 (THBS1) as a new NSHI gene. A homozygous frameshift variant [c.1470del: p.(Ile491Serfs*45)] was observed in the three hearing-impaired and in the heterozygous state in three unaffected family members. Unlike for most ARNSHI, hearing-impaired individuals had audiograms with a sloping pattern, showing more pronounced HI in the mid and high frequencies (ranging from moderate to profound) compared to the low frequencies. RNA expression data indicates THBS1 is expressed during human inner ear development. Additionally, THBS1 is expressed in the cochlear epithelium and supporting cells of the mouse inner ear during embryonic and postnatal stages. Previously, THBS1 was demonstrated to affect hearing in knockout mice by influencing the formation and function of afferent synapses in the inner ear.
Conclusions
Our findings highlight THBS1 as a potential novel candidate gene for human HI characterized by a sloping high-frequency audio profile. This discovery enhances our understanding of the genetic etiology of HI and will aid in advancing molecular diagnosis.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12920-024-02060-w.
Keywords: Autosomal recessive non-syndromic hearing impairment, Consanguinity, Exome sequencing, Inner ear, Thrombospondin, THBS1
Background
Hearing impairment (HI) is a significant global health problem that impacts speech, language, and social skills development. It is a prevalent and clinically diverse sensory disorder affecting 1–6 per 1000 newborns. It is estimated that 1% of all human genes are involved in hearing [1], highlighting the intricate genetic basis of this condition. Despite genomic advancements and with approximately 150 genes found to be implicated in human non-syndromic HI (NSHI) [2] a substantial portion of the genetic factors underlying HI remain unidentified. This is primarily due to the complexity of the auditory system that corresponds with a high level of genetic heterogeneity involved in impairment. This highlights the need to uncover the extensive array of genes contributing to hearing function. The understanding of their genetic mechanisms could lead to improved personalized interventions, genetic counseling, and improved outcomes for individuals with HI.
In this study, we describe the identification of a new NSHL candidate gene thrombospondin 1 (THBS1) (OMIM:188060). A homozygous frameshift variant in THBS1 was identified through the study of a consanguineous family from Pakistan with bilateral prelingual sensorineural autosomal recessive (AR) NSHI. THBS1 is highly intolerant to loss-of-function variants. Audiograms for the HI members are atypical for ARNSHI, in that HI was more severe in the mid and high frequencies than the low frequencies, instead of severe to profound across all frequencies.
Methods
Participant evaluation and sample collection
Family DEM4671 was ascertained from Khyber-Pakhtunkhwa (KPK) province of Pakistan. The family is consanguineous with hereditary sensorineural HI. Information obtained for the family include clinical histories, medical records, physical examinations, and pure-tone audiometry (250-8,000 Hz). Tandem gait and Romberg tests were also performed to evaluate hearing-impaired family members for gross vestibular dysfunction. Further, syndromes and HI due to infections, ototoxic medications, or trauma were assessed through detailed clinical evaluation. Peripheral blood samples were collected, and genomic DNA extracted from all participating family members [3].
Exome sequencing and variant confirmation
Sanger sequencing was performed to rule out common genetic causes of HI in the Pakistani population, i.e., GJB2 coding variants; CIB2 [p.(Phe91Ser) and p.(Cys99Trp)]; HGF (c.482 + 1986_1988delTGA and c.482 + 1991_2000delGATGATGAAA); and SLC26A4 [p.(Val239Asp) and p.(Gln446Arg)]. A DNA sample from a hearing-impaired family member (IV:5) underwent exome sequencing (Fig. 1A). Library construction and exon capture were completed using Agilent SureSelect Human All Exon V6 kit (~ 60 Mb) with short-read parallel sequencing performed to a mean on-target coverage of 58.3x. Reads were aligned to the human reference genome (GRCh38/hg38) using Burrows-Wheeler Aligner [4]. Duplicate reads were marked using Picard. Genome Analysis Toolkit (GATK) [5] was used to call single nucleotide variants and insertions/deletion (indels), following GATK’s best practices in variant calling. Annotation was performed using ANNOVAR [6]. For the analysis, exonic and splice region variants +/− 12 bp from intron-exon boundary were retained. After checking the presence of known likely pathogenic and pathogenic variants from the ClinVar database [7], rare variants with a minor allele frequency (MAF) < 0.005 in all populations of the Genome Aggregation Database (gnomAD v4.0) [8] that were consistent with an AR mode of inheritance and with a predicted effect on protein function or pre-mRNA splicing (nonsense, frameshift, missense, start-loss, splice region, etc.) were selected. Bioinformatic predictions were annotated using dbNSFP35a [9] and dbscSNV1.1 [10] which included Combined Annotation Dependent Depletion (CADD) [11] and Genomic Evolutionary Rate Profiling (GERP++) scores [12]. Variants that met the selection criteria were visualized using the Integrative Genomics Viewer (IGV2.4.3) [13] to exclude likely false-positive calls. Sanger sequencing, using DNA samples from all available family members, was performed to validate the variants and test for segregation with HI. For DEM4671 (IV:5) copy number variant (CNV) calls were generated using CONiFER (v0.2.2) [14] and annotated using the BioMart Database [15].
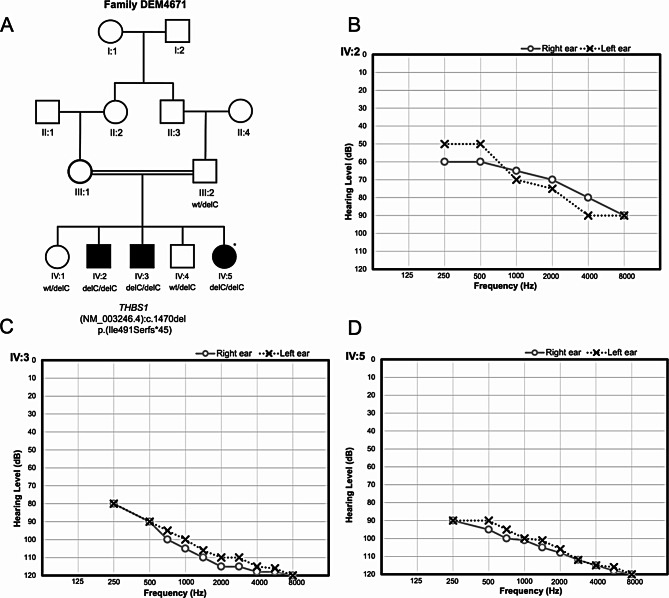
Fig. 1.
Pedigree of family DEM4671 and audiometry. (A) Pedigree of the Pakistani family DEM4671 displaying genotypes for the variant THBS1 (NM_003246.4) [c.1470del: p.(Ile491Serfs*45)] for each family member that has an available DNA sample. A star indicates the hearing-impaired family member whose DNA sample underwent exome sequencing. Females are represented by circles and males by squares. Individuals with solid symbols have NSHI while those with clear symbols are unaffected. (B–D) Air conduction thresholds for affected members IV:2, IV3 and IV:5 respectively. IV:2 has profound hearing impairment in the high frequencies and moderate to severe in the mid to low frequencies. IV:3 and IV:5 have profound bilateral HI in both ears. The audiograms display a decrease in hearing threshold at higher frequencies. Circles with smooth connecting lines represent the right ear and crosses with dotted connecting lines, the left ear
Expression analysis of THBS1
To evaluate and confirm the RNA expression of THBS1 in the inner ear, publicly available datasets in gEAR (gene Expression Analysis Resource) [16] were visualized and evaluated. RNA sequence data of hair cells and surrounding cells in the mouse cochlea at various developmental stages from E14-P7 as well as single cell expression in the cochlear floor epithelial duct cells of E14, E16, P1, and P7 wild type CD-1 mice pups (accession number GSE137299) [17] were analyzed.
We also analyzed single-nucleus RNA sequence data from two human fetal inner ears (7.5 and 9.2 weeks) and one adult inner ear (accession number GSE213796) obtained from the human inner ear atlas. Additionally, we also examined the human inner ear organoids single-cell and single-nucleus RNA sequence data which was generated by differentiating multiple human pluripotent stem cell (hPSC) lines to induce inner ear cell types (accession number GSE214099) [18].
Results
Clinical evaluation
The clinical examinations and pure-tone audiometry of the three affected members of family DEM4671 show that IV:2 (16 years-of-age) has profound hearing impairment in the high frequencies and moderate to severe in the mid to low frequencies (Fig. 1B). The individual IV:3 (12 years-of-age) and IV:5 (10 years-of-age) displays profound bilateral HI in both ears with a decrease in hearing with an increase in frequency (Fig. 1C and D). The onset of HI for all the three affected family members was prelingual and most likely congenital.
Exome and Sanger sequencing
Four homozygous rare variants were identified in the exome data obtained for individual IV:5 (DEM4671) (Supplementary Table 1). No homozygous CNVs or CNVs that were potentially compound heterozygous with either another CNV, indel, or SNV were observed. Through Sanger sequencing we confirmed that only one of these variants, a frameshift in THBS1 (NM_003246.4) [c.1470del: p.(Ile491Serfs*45)] (Supplementary Table 1) segregated with HI. The truncated mRNA resulted by this variant is predicted to likely be a target of nonsense-mediated decay (NMD). The [c.1470del: p.(Ile491Serfs*45)] variant leads to a premature termination codon (PTC) located more than 50 nucleotides upstream of the final exon-exon junction complex, marking it for NMD. This positioning aligns with NMD’s mechanism to degrade transcripts with upstream PTCs, preventing production of potentially harmful truncated proteins [19]. Therefore, this variant transcript is expected to be downregulated by NMD. This variant is absent from gnomAD v4.0 and Trans-Omics for Precision Medicine (TOPMed) Bravo [20] databases, but had a MAF of 2.0 × 10− 6 in the All of Us research program (All of Us) [21] database.
Exome data from 604 Pakistani families with individuals affected by NSHI were also screened for potential causal variants in the THBS1 gene, but no additional families identified. THBS1 was assessed for its relevance to NSHI using the framework developed by Clinical Genome Resource and was classified as “limited” evidence (score 4.5) [22].
Expression of THBS1 in human and mouse inner ear
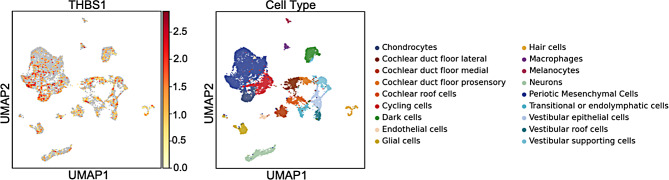
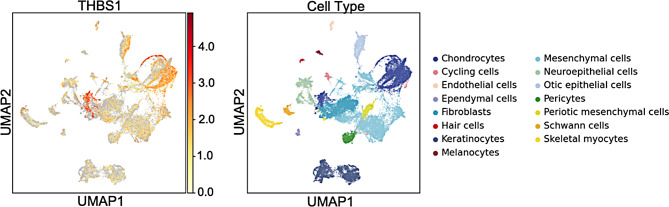
Analysis of the gEAR data shows that in the human inner ear, THBS1 is expressed ubiquitously. The human inner ear atlas (Fig. 2) and human ear organoids (Fig. 3) show moderate to high expression especially in the chondrocytes and cycling cells. THBS1 is also expressed in the otic epithelial cells, cochlear duct floor and roof cells.
Fig. 2.
Human inner ear expression of THBS1. THBS1 is expressed in the human inner ear cells with cochlear floor cells displaying the highest expression. The dataset contains three stages of human inner ear development [18]. The left panel illustrates single-nucleus RNA sequence data from two human fetal inner ears (7.5 and 9.2 weeks) and one human adult inner ear. The X axis displays the uniform manifold approximation and projection (UMAP) 1 which is used for dimension reduction and on the Y axis UMAP 2. The scale bar displays normalized gene expression levels which range from low (yellow) to high (red) that have been adjusted for sequence depth. The right panel displays the locations of specific inner ear cell types with color codes
Fig. 3.
THBS1 expression in human inner ear organoids. THBS1 is expressed in the human inner ear organoids. Chondrocytes exhibit a high expression for this gene. The dataset plotted consists of human pluripotent stem cells differentiated into complex inner ear tissue [18]. The left panel illustrates the expression of THBS1 in the single-cell and single-nucleus RNA sequence data from the differentiated inner ear organoids. The X axis displays the uniform manifold approximation and projection (UMAP) 1 which is used for dimension reduction and on the Y axis UMAP 2. The scale bar displays normalized gene expression levels which range from low (yellow) to high (red) that have been adjusted for sequence depth. The right panel displays the locations of specific inner ear cell types with color codes
In the mouse inner ear, the orthologous gene of THBS1 is expressed throughout the cochlear epithelium during E14, E16, P1, and P7 (Supplementary Figs. 1–4). There is high expression in the early Reissner’s membrane and Lateral prosensory cells at E14 and E16. There is high expression in the supporting cells (Deiters’ cells, inner phalangeal cells and inner pillar cells) at P1. Additionally, on examining the hair cell data sets, the gene is expressed in the inner hair cells (IHC) during E14-P1 (Supplementary Fig. 5) and in the outer hair cells (OHC) during E14-P7 (Supplementary Figs. 6 and 7) stages of CD-1 mice although the expression for this gene is higher in the supporting cells.
Discussion
We used exome sequencing to identify the underlying genetic defect in a consanguineous family from Pakistan with ARNSHI. A variant in THBS1 (NM_003246.4) [c.1470del: p.(Ile491Serfs*45)] was identified and segregated with HI. The audiograms from the family display a distinctive downward slope pattern, differing from what is typically observed for ARNSHI. The audiograms observed in ARNSHI are generally severe to profound observed consistently across all frequencies. In contrast, a downward sloping configuration was observed in the audiograms of the affected individuals in this study, ranging from mild to profound with a more pronounced HI in the high frequencies. Bilateral, symmetric, high-frequency HI is generally associated with progressive or age-related hearing loss. We were unable to evaluate the progression of hearing threshold with age since we could obtain only a single audiogram for each hearing-impaired family member, making it difficult to assess whether the HI due to THBS1 is progressive.
Using the criteria from Clinical Genome Resource THBS1 relevance to NSHI was classified as “limited” due to there being only a single family that segregates a frameshift variant. The “limited” evidence classification requires at least one variant that is asserted to be disease causing with or without gene-level experimental data. To increase the evidence to “moderate” three independent families or probands either with segregating or de novo likely disease-causing variants along with experimental evidence is required. For “strong” classification the finding must be replicated in independent publications with substantial genetic and experimental data. A “definitive” classification can only be met if the disease-gene relationship has been replicated and it has been approximately three or more years since the initial publication [22]. Therefore, to establish THBS1 role in the etiology of NSHI additional families\probands must be identified as well as the generation of additional experimental data.
The THBS protein family comprises of five multifunctional glycoproteins found in the extracellular matrix. These proteins have increasingly been studied for their physiological roles. Among them is THBS1, a 450-kDa homotrimeric extracellular glycoprotein made up of 1170 amino acids linked by disulfide bonds [23]. This protein is involved in several cellular processes. In the extracellular matrix, it facilitates cell adhesion, migration, and activation [24, 25]. THBS1 is highly expressed in platelets, certain types of dendritic cells, and in tissues undergoing regeneration. Importantly, THBS1 and THBS2 are present in the cochlea and contribute to the development and function of afferent synapses in the inner ear of mice [26].
Although THBS1 has not been implicated in human HI until now, there is some evidence to suggest its potential role in the auditory system. Recent studies have explored the involvement of THBS1 in the development and maintenance of the cochlea. In the mouse cochlea, expression of THBS1 is highest at birth and during early developmental stages, subsequently declining over time. Expression profiles exhibit notable variations across different developmental stages; for example, at E17 and P1, expression was seen in most areas of the cochlea including supporting cells (SCs) and hair cells whereas at P5, the expression was confined to pillar cells and to inner sulcus cells of the sensory epithelium. Expression of THBS1 was substantially higher between P0 and P29 subsequently decreasing after P60 stage, after the maturation of cochlea. Thbs1 knockout mice were shown to have fewer synapses in the IHCs and developed an elevated auditory brainstem response threshold at 12 months mainly in the high frequencies [26]. Although there is an overlap in the phenotype between mice and humans, there are some differences with the mouse developing HI in what would be equivalent to middle age in humans. It is not unusual for HI genes to display different phenotypes in mice and humans [27]. Thbs1 promotes epithelial-mesenchymal transition in mouse inner ear by activating the latent complex of TGF-beta and plays a significant role in the development of cochlear afferent synapse [26, 28]. Additionally, it promotes endothelial cell senescence in humans through the CD47 and NADPH oxidase 1 (Nox1) pathways and possesses anti-inflammatory properties [29]. Interestingly, NOX3, a homolog of NOX1, known to be specifically expressed in the inner ear at high levels is involved in sensorineural HI due to reactive-oxygen species induced cellular damage in cochlear sensory epithelium [30]. Recently, single-cell RNA sequence data has shown differential expression of thbs1b, the zebrafish analog of human THBS1, in IHC compared to neuromast cell lines between E4 and E7 [31]. Our analysis also suggests that THBS1 might be important in development and maintenance of cochlea and supporting cells. To support this further, we investigated THBS1 expression in the mouse inner ear performed using publicly available datasets [17, 18] via gEAR [16]. Our results also demonstrate the expression of THBS1 in the cochlear epithelium and supporting cells of mouse from E14 to P7 stages. In addition, this gene was found to be expressed in the human inner ear atlas and organoids as well. The expression profile provides meaningful cues to the role of THBS1 within the inner ear, and possibly elucidate the phenotype observed in our families.
For THBS1 there is a significantly low rate of missense and predicted loss-of-function (LoF) variations in the gnomAD dataset (v4.0.0). The positive Z score of 3.78 and the o/e (ratio of the observed/expected variants) value of 0.74 (0.71–0.78) indicates a high intolerance in case of missense variations. Compared to the 127.3 expected LoF mutants, only 28 were observed. The pLI (probability of being loss-of-function intolerant) for THBS1 is 1.00, and the o/e value is 0.22 (0.16–0.3) which indicates that this gene is highly LoF intolerant [32, 33].
The frameshift variant p.(Ile491Serfs*45) in family 4671 introduces a premature stop codon that is most likely to terminate the protein at the second beta-sheet after the first turn and leading to a loss of the heparin-binding region. This could result in triggering a NMD, apart from loss of one of the three major residues binding to heparin through sulfate groups at Arg29, Arg42 and Arg77 prior to the laminin G-like domain (LG) [34, 35]. Due to the premature termination of the protein and NMD, we predict a loss-of-function effect for this variant.
Although the evidence put forward in this study strongly suggests a role in the auditory system, the precise mechanisms through which THBS1 is involved in the auditory system remains unclear. Thrombospondin 1 is a multifunctional protein and is known to influence several signaling pathways, however its specific effect on the auditory system is unknown and needs further investigation.
Conclusions
In summary, we have identified THBS1 as a new candidate gene for ARNSHI. Hearing-impaired individuals which segregate a homozygous frameshift variant in this gene have high-frequency sloping audiogram. THBS1 displays expression in the inner mouse and human ear. Thbs1 knockout mice have been shown to develop an elevated auditory brainstem response threshold at 12 months. This is the first-time a gene in the thrombospondin family has been linked to human HI. This discovery contributes to a deeper understanding of the genetic basis of hearing impairment and may help improve molecular diagnosis.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank the family members for participating in this study.
Abbreviations
- THBS1
Thrombospondin 1
- HI
Hearing impairment
- ARNSHI
Autosomal recessive non-syndromic sensorineural HI
- NSHI
Non-syndromic HI
- KPK
Khyber-Pakhtunkhwa province of Pakistan
- GJB2
Gap Junction Protein, Beta 2
- CIB2
Calcium And Integrin-Binding Family Member 2
- HGF
Hepatocyte Growth Factor
- SLC26A4
Solute Carrier Family 26 Member 4
- MAF
Minor allele frequency
- gnomAD
Genome Aggregation Database
- CADD
Combined Annotation Dependent Depletion
- GERP
Genomic Evolutionary Rate Profiling
- IGV
Integrative Genomics Viewer
- CNV
Copy number variant
- gEAR
gene Expression Analysis Resource
- hPSC
Human pluripotent stem cell
- NMD
Nonsense-mediated decay
- TOPMed
Trans-Omics for Precision Medicine
- IHC
Inner hair cells
- OHC
Outer hair cells
- LoF
Loss-of-function
- NOX1
NADPH oxidase 1
- NOX3
NADPH oxidase 3
- pLI
Probability of being loss-of-function intolerant
- LG
Laminin G-like domain
Author contributions
Conceptualization, SML and WA. Material preparation, data collection and processing were performed by FK, SK, IU. Data analysis was performed by AA, TB and IS. The first draft of the manuscript was written by TB with contributions from AA. Revision and finalization, SML and IS. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Institutes of Health (NIH)-National Institute of Deafness and other Disorders grant R01 DC003594 (to S.M.L).
Data availability
The variant reported in this study was submitted to ClinVar (Accession number: SCV004697444). Expression data is available from gEAR.
Declarations
Ethics approval and consent to participate
Approval from the Institutional Review Boards (IRBs) of Quaid-i-Azam University (IRB-QAU-153), and Columbia University (IRB-AAAS2343) were obtained for the study. All adult study participant’s signed informed consent forms and parents provided consents for minors after their assent was obtained.
Consent for publication
Written informed consent were obtained from all adult participants and parents provided consents for minors after their assent was obtained for publication of this study.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Teek R, Kruustük K, Žordania R, Joost K, Kahre T, Tõnisson N, et al. Hearing impairment in Estonia: an algorithm to investigate genetic causes in pediatric patients. Adv Med Sci. 2013;58(2):419–28. [DOI] [PubMed] [Google Scholar]
- 2.Walls WD, Azaiez H, Smith RJH. Hereditary Hearing Loss Homepage. Available from: https://hereditaryhearingloss.org.
- 3.Sambrook J, Russell DW. Purification of nucleic acids by extraction with phenol:chloroform. Cold Spring Harb Protoc. 2006;2006(1):pdb.prot4455 [DOI] [PubMed]
- 4.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25(14):1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The genome analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Landrum MJ, Chitipiralla S, Brown GR, Chen C, Gu B, Hart J, et al. ClinVar: improvements to accessing data. Nucleic Acids Res. 2020;48(D1):D835–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen S, Francioli LC, Goodrich JK, Collins RL, Kanai M, Wang Q, et al. A genomic mutational constraint map using variation in 76,156 human genomes. Nature. 2024;625(7993):92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu X, Wu C, Li C, Boerwinkle E. dbNSFP v3.0: a one-stop database of functional predictions and annotations for human nonsynonymous and splice-site snvs. Hum Mutat. 2016;37(3):235–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jian X, Boerwinkle E, Liu X. In silico prediction of splice-altering single nucleotide variants in the human genome. Nucleic Acids Res. 2014;42(22):13534–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47(D1):D886–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S. Identifying a high fraction of the human genome to be under selective constraint using GERP++. Wasserman WW, editor. PLoS Comput Biol. 2010;6(12):e1001025. [DOI] [PMC free article] [PubMed]
- 13.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krumm N, Sudmant PH, Ko A, O’Roak BJ, Malig M, Coe BP, et al. Copy number variation detection and genotyping from exome sequence data. Genome Res. 2012;22(8):1525–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smedley D, Haider S, Durinck S, Pandini L, Provero P, Allen J, et al. The BioMart community portal: an innovative alternative to large, centralized data repositories. Nucleic Acids Res. 2015;43(W1):W589–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orvis J, Gottfried B, Kancherla J, Adkins RS, Song Y, Dror AA, et al. gEAR: gene expression analysis resource portal for community-driven, multi-omic data exploration. Nat Methods. 2021;18(8):843–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolla L, Kelly MC, Mann ZF, Anaya-Rocha A, Ellis K, Lemons A, et al. Characterization of the development of the mouse cochlear epithelium at the single cell level. Nat Commun. 2020;13(1):2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Der Valk WH, Van Beelen ESA, Steinhart MR, Nist-Lund C, Osorio D, De Groot JCMJ, et al. A single-cell level comparison of human inner ear organoids with the human cochlea and vestibular organs. Cell Rep. 2023;42(6):112623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewis BP, Green RE, Brenner SE. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc Natl Acad Sci. 2003;100(1):189–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kowalski MH, Qian H, Hou Z, Rosen JD, Tapia AL, Shan Y et al. Use of > 100,000 NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium whole genome sequences improves imputation quality and detection of rare variant associations in admixed African and Hispanic/Latino populations. Barsh GS, editor. PLOS Genet. 2019;15(12):e1008500. [DOI] [PMC free article] [PubMed]
- 21.Lyles CR, Lunn MR, Obedin-Maliver J, Bibbins-Domingo K. The new era of precision population health: insights for the all of Us Research Program and beyond. J Transl Med. 2018;16(1):211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strande NT, Riggs ER, Buchanan AH, Ceyhan-Birsoy O, DiStefano M, Dwight SS, et al. Evaluating the clinical validity of gene-disease associations: an evidence-based framework developed by the clinical genome resource. Am J Hum Genet. 2017;100(6):895–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Isenberg JS, Roberts DD. THBS1 (thrombospondin-1). Atlas Genet Cytogenet Oncol Haematol [Internet]. 2020 Apr [cited 2024 Feb 21](8). http://hdl.handle.net/2042/70774 [DOI] [PMC free article] [PubMed]
- 24.Hofsteenge J, Huwiler KG, Macek B, Hess D, Lawler J, Mosher DF, et al. C-mannosylation and O-fucosylation of the thrombospondin type 1 module. J Biol Chem. 2001;276(9):6485–98. [DOI] [PubMed] [Google Scholar]
- 25.Sweetwyne MT, Murphy-Ullrich JE. Thrombospondin1 in tissue repair and fibrosis: TGF-β-dependent and independent mechanisms. Matrix Biol. 2012;31(3):178–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mendus D, Sundaresan S, Grillet N, Wangsawihardja F, Leu R, Müller U, et al. Thrombospondins 1 and 2 are important for afferent synapse formation and function in the inner ear. Eur J Neurosci. 2014;39(8):1256–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carlson RJ, Avraham KB. Emerging complexities of the mouse as a model for human hearing loss. Proc Natl Acad Sci. 2022;119(35):e2211351119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He L, Wang GP, Guo JY, Chen ZR, Liu K, Gong SS. Epithelial–mesenchymal transition participates in the formation of vestibular flat epithelium. Front Mol Neurosci. 2021;14:809878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meijles DN, Sahoo S, Al Ghouleh I, Amaral JH, Bienes-Martinez R, Knupp HE, et al. The matricellular protein TSP1 promotes human and mouse endothelial cell senescence through CD47 and Nox1. Sci Signal. 2017;10(501):eaaj1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mohri H, Ninoyu Y, Sakaguchi H, Hirano S, Saito N, Ueyama T. Nox3-Derived superoxide in Cochleae induces Sensorineural hearing loss. J Neurosci. 2021;41(21):4716–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi T, Beaulieu MO, Saunders LM, Fabian P, Trapnell C, Segil N, et al. Single-cell transcriptomic profiling of the zebrafish inner ear reveals molecularly distinct hair cell and supporting cell subtypes. eLife. 2023;12:e82978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaur S, Roberts DD. Why do humans need thrombospondin-1? J Cell Commun Signal. 2023;17(3):485–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaur S, Roberts DD. Differential intolerance to loss of function and missense mutations in genes that encode human matricellular proteins. J Cell Commun Signal. 2021;15(1):93–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tan K, Duquette M, Liu J, huan, Zhang R, Joachimiak A, Wang Jhuai, et al. The structures of the thrombospondin-1 n-terminal domain and its complex with a synthetic pentameric heparin. Structure. 2006;14(1):33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kilpeläinen I, Kaksonen M, Kinnunen §∥ Tarja, Avikainen H, Fath M, Linhardt RJ, et al. Heparin-binding growth-associated molecule contains two heparin-binding β-sheet domains that are homologous to the thrombospondin type i repeat. J Biol Chem. 2000;275(18):13564–70. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The variant reported in this study was submitted to ClinVar (Accession number: SCV004697444). Expression data is available from gEAR.