Abstract
Multidrug-resistant (MDR) Acinetobacter baumannii (A. baumannii) is currently recognized not only as a significant nosocomial pathogen but also is an emerging bacterial infection in food-producing animals, posing a critical threat to global health. However, this is a hindrance to detailed bioinformatic studies of MDR A. baumannii of chicken origin due to the lack of its complete genome sequence. Here, we report whole-genome sequencing analysis of MDR A. baumannii Y03 isolated from chickens. The Y03 genome consists of 1 circular chromosome and 4 circular plasmids, The Y03 chromosome harbors 41 antimicrobial resistance genes conferring resistance to major classes of antibiotics, including β-lactams, phenicols, macrolides, lincosamides, aminoglycosides, and nitrofurans, as well as 135 virulence factors involved in effector delivery system, immune modulation, adherence, stress survival, biofilm, exotoxin, and nutritional/metabolic factor. The in vivo infection experiments certificated that Y03 was virulent to chickens. Meanwhile, we used PCR amplification method to detect 10 antimicrobial resistance genes including abeM, adeB, adeH, adeK, blaapmC, blaOXA−90, catB9, macB, folP, and parE, as well as 14 virulence genes including lpxC, pilO, fimT, ompA, basA, bauA, gspL, csu, pgaC, plc2, tssA, tviB, bap, and vgrG. Whole-genome sequencing analysis revealed that Y03 contained 46 horizontal gene transfer elements, including 11 genomic islands, 30 transposons, and 5 prophages, as well as 518 mutations associated with reduced virulence and 44 mutations resulting in loss of pathogenicity. Furthermore, there were 22 antibiotic targets and 28 lethal mutations on the Y03 chromosome that could be used as potential targets to prevent, control, and treat infections caused by MDR A. baumannii Y03. Therefore, this study contributes to the development of strategies for the prevention, control, and treatment of A. baumannii infections and their spread in chickens.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12866-024-03694-7.
Keywords: Multidrug-resistant Acinetobacter baumannii, Whole-genome sequencing, Antimicrobial resistance, Virulence factors
Introduction
Acinetobacter baumannii (A. baumannii) is a strictly aerobic, non-fermenting, Gram-negative coccobacillus with pili and capsule, but no flagella [1–3]. It has emerged as an important opportunistic pathogen of nosocomial infections responsible for high mortality and morbidity rates during the last decades due to its ability for survival in the hospital environment on a wide range of dry and moist surfaces [1, 4–6]. A. baumannii could cause various hospital-acquired infections, including pneumonia, endocarditis, bacteremia, urinary tract infections, meningitis, peritonitis, skin and wound infections, particularly among severely ill patients in the intensive care unit (ICU) and immunocompromised individuals [3, 4, 6–8]. These infections are closely connected with the implantation of the ventilators, vascular catheters, and cerebrospinal fluid shunts, on which A. baumannii colonizes and adheres [5, 9]. Therefore, A. baumannii has drawn the attention of medical professionals worldwide as a public health threat.
The ability of A. baumannii to survive in the hospital environment and inside the host for extended periods is due to its possession of a series of virulence factors, including capsular polysaccharides, lipopolysaccharides, type I fimbriae, P fimbriae, curli fiber, chaperone-usher type I pili, type IV pili, invasins, biofilm-associated protein, outer membrane proteins, outer membrane vesicles, serum resistance, phospholipases, acinetobactin, fimsbactin, baumannoferrin, etc [5, 10–16]. These virulence factors are categorized into four main groups: adhesion and invasion, toxins, protein secretion systems, and iron acquisition, which facilitate A. baumannii to adhere and colonize host tissues and organs, disseminate, invade host cells, and evade the host immune response [16–18].
Antibiotics nowadays are used to prevent, control and treat A. baumannii infections and outbreaks [1, 19]. However, the continued overuse and misuse of antibiotics have enabled A. baumannii to develop different types of resistance mechanisms, e.g., the production of degradative enzymes, a change in metabolic status, a decrease in bacterial membrane permeability, the alteration of antibiotic targets, the overexpression of efflux pumps, and the formation of biofilms [5, 10, 12, 20, 21]. Documented data revealed that the A. baumannii isolates harbored the high prevalence of antibiotic resistance against a wide range of clinically effective antibiotics [3, 5]. Accordingly, they are classified into three categories: multidrug-resistant (MDR) strains, extensively drug-resistant (XDR) strains, and even pan-drug-resistant (PDR) strains, representing a significant challenge for therapy in clinics [3, 11, 16, 20].
Whole-genome sequencing provides an unprecedented level of information on species identification, antimicrobial resistance, and the molecular epidemiological typing of microorganisms to researchers, and it also helps in the identification of new drug targets and the development of new drugs [16, 22–25]. More studies about the pathogenicity, the prevalence and antibiotic resistance of A. baumannii isolated from human clinical isolates have been reported [2, 3, 7]. However, studies on their pathogenicity, antibiotic resistance and genomes in food-producing animals are somewhat limited. The presence of A. baumannii in food-producing animals is currently considered a major public health problem since it could be disseminated to community and hospital settings through the food chain [2]. Chickens are the most widely kept food-producing animals in the world due to their abilities to providing high quality protein and income for the rural households [26]. Upon infection of chickens with A. baumannii, the bacteria could be transmitted directly from chickens and their products (i.e., meat and eggs) to farm workers and consumers, and indirectly to a larger population through contaminated food, water, and soil [26].
The aim of this study was to investigate the whole genome, pathogenicity and antibiotic resistance of A. baumannii, and to understand the pathogenicity and antibiotic resistance mechanisms of A. baumannii in chickens and the differences between it and A. baumannii isolates from other sources. Therefore, this study contributes to the development of strategies for the prevention, control and treatment of A. baumannii infections and their spread in chickens.
Materials and methods
Isolation and identification of A. Baumannii
A bacterial disease broke out in a chicken farm in Linyi, Shandong province, China. The diseased chickens presented obvious clinical symptoms of listlessness, tachypnea, anorexia, runny nose, hypothermia, and lethargy with closed eyes. For the isolation of bacteria, the chickens were euthanized by injecting intravenous sodium pentobarbital (100 mg/kg) into the wing vein and dissected. The pathological changes of the chicken autopsy showed slight bleeding in the lungs, mottled bleeding in the livers and heart, tumefaction in the spleen and kidneys. And then approximately 5 g of heart, livers, spleen, lungs, and kidneys were collected and homogenized in a stomacher (Scientz, Ningbo, Zhejiang, China) for 2 min, respectively. Each sample was added in 10 mL of the sterile phosphate-buffered saline (PBS) buffer (pH 7.4) and carried out 10-fold serial dilutions through 4 microfuge tubes containing 900 µL of PBS. 100 µL of the above each dilutions were then dropped and spread onto Acinetobacter chromogenic media (CHROMagar, Paris, France), incubated at 37 °C for 16–24 h, and examined for the growth of typical red colonies of the genus Acinetobacter. Subsequently, one typical colony morphological representative of the genus Acinetobacter was inoculated on Luria-Bertani (LB) agar plates and further confirmed by microbiological and biochemical tests such as gram staining, oxidase, catalase, urea urease, simon citrate, MR-VP, motility and indole production, and PCR amplification using the specific primer parC-F/R of A. baumannii. The confirmed A. baumannii was tentatively named Y03 and stored in LB broth supplemented with 25% sterile glycerol at -80 °C. Before performing each assay, pure culture of A. baumannii Y03 was accomplished by inoculation on a LB agar plate and incubated overnight at 37 °C.
Whole-genome sequencing and library construction
Genomic DNA of Y03 was extracted using the TIANamp Bacteria DNA kit (Tiangen, Beijing, China) according to the manufacturer’s instructions. The quality, purity, integrality, and yield of the extracted genomic DNA were detected using 0.35% agarose gel electrophoresis and a NanoDrop2000 spectrophotometer (Thermo Fisher, Pittsburg, PA, USA), and quantified using a Qubit 4.0 fluorometer (Invitrogen, Carlsbad, CA, USA). Whole-genome sequencing was performed on the Illumina NovaSeq platform and the Oxford Nanopore ONT platform. Libraries were constructed for sequencing using TruSeqTM DNA Sample Prep Kit (Illumina, San Diego, CA, USA) for Illumina TruSeq Nano DNA LT, and quantified using a Qubit 4.0 fluorometer (Invitrogen). All of the above processes were performed at Shanghai Personal Biotechnology Co. Ltd (Shanghai, China). After the completion of the genome assembly, the whole-genome sequence of Y03 was compared with that of A. baumannii published using the NCBI Genbank nucleotide sequence database (https://www.ncbi.nlm.nih.gov/).
Genome functional component analysis
Genome functional component analysis of Y03 was performed to predict the protein coding genes, non-coding RNA (ncRNA), clustered regularly interspaced short palindromic repeats (CRISPRs), and the repetitive sequences. The protein coding genes were predicted using GeneMarkS software (http://topaz.gatech.edu/GeneMark/). ncRNA includes small RNA (sRNA), ribosome RNA (rRNA), transfer RNA (tRNA), small nuclear RNA (snRNA), small nucleolar RNA (snoRNA), and microRNA (miRNA). tRNA was predicted using tRNAscan-SE software (http://lowelab.ucsc.edu/tRNAscan-SE/), and rRNA was predicted using Barrnap software (http://www.vicbioinformatics.com/software.barrnap.shtml). The perdiction of the other ncRNA was obtained by comparison with the Rfam database (http://rfam.xfam.org/). CRISPRs was predicted using CRISPR finder (http://crispr.i2bc.paris-saclay.fr/Server/). The interspersed repeats were predicted by comparison with the Repbase database (https://www.girinst.org/repbase/).
Genome subsystem analysis
Genome subsystem analysis of Y03 was performed to predict prophage, genomic islands (GIs), virulence factors of pathogenic bacteria (VFDB, http://www.mgc.ac.cn/VFs/main.htm), the comprehensive antibiotic resistance database (CARD, https://card.mcmaster.ca/), and the carbohydrate-active enzymes database (CAZy, http://www.cazy.org/). The prophage on the genome was predicted using the PHASTER tool (http://phaster.ca/), GIs using the IslandViewer 4 database (http://www.pathogenomics.sfu.ca/islandviewer/), VFDB and CARD using the BLAST tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi), and CAZy using the hmmscan software (http://hmmer.org/).
Genome functional annotation analysis
The various databases were utilized to analyze the protein coding gene functions of Y03, including the non-redundant protein database databases (NR, https://ftp.ncbi.nih.gov/blast/db/), the clusters of orthologous groups (COG, http://eggnogdb.embl.de/#/app/home/), the kyoto encyclopedia of genes and genomes (KEGG, http://www.genome.jp/kegg/), the gene ontology (GO, http://www.geneontology.org/), the transporter classification database (TCDB, http://www.tcdb.org/), and the pathogen host interactions (PHI, http://www.phi-base.org/). Besides, the secretory proteins were also predicted using the SignalP tool (http://www.cbs.dtu.dk/services/SignalP/) and the TMHMM tool (http://www.cbs.dtu.dk/services/TMHMM/).
Antimicrobial susceptibility testing
Antimicrobial susceptibility profile of Y03 was assessed by the Kirby-Bauer (KB) disk diffusion method according to the Clinical and Laboratory Standards Institute guidelines (CLSI, 2021) using the Mueller-Hinton (MH) agar (Haibo, Qingdao, Shandong, China). The antimicrobial disks included ampicillin (10 µg), aztreonam (30 µg), penicillin G (10 µg), piperacillin (100 µg), oxacillin (1 µg), cefalotin (30 µg), cefazolin (30 µg), cefotaxime (30 µg), cefoxitin (30 µg), cefuroxime (30 µg), ceftazidime (30 µg), cefoperazone (75 µg), cefepime (30 µg), cotrimoxazole (23.75/1.25 µg), chloramphenicol (30 µg), florfenicol (30 µg), clarithromycin (15 µg), erythromycin (15 µg), midecamycin (30 µg), clindamycin (2 µg), ciprofloxacin (5 µg), levofloxacin (5 µg), ofloxacin (5 µg), norfloxacin (10 µg), tetracycline (30 µg), minocycline (30 µg), amikacin (30 ± 7.5 µg), tobramycin (10 µg), streptomycin (10 µg), gentamicin (10 ± 2.5 µg), kanamycin (30 µg), spectinomycin (100 µg), nitrofurantoin (300 µg), and polymyxin B (300 µg) in antimicrobial susceptibility determination test.
Animals
One-day-old chicks were purchased from Rizhao Langya Chicken Co. Ltd. These chicks were adequately fed food and water (a complete diet without antibiotics) and a 12 h illumination period per day. Healthy 7-day-old chicks were selected for the animal infection experiments. Euthanasia of all chicks was performed by intravenous injection of pentobarbital sodium in wing vein at a dose three times higher than the anesthetic dose. The loss of consciousness was rapid, followed by cessation of respiration and heartbeat, and exsanguination, confirming euthanasia. All procedures were carried out in accordance with the guidelines of the NIH Guide for the Care and Use of Laboratory Animals, as well as the regulations of the American Veterinary Medical Association (AVMA) regarding euthanasia, and they were approved by the Committee of Linyi University (Approval No. LYU20240206).
Animal infection experiments
After 7 days of feeding, healthy chicks were selected for the animal infection experiments to evaluate the virulence of Y03. They were divided randomly into 6 groups (group A, B, C, D, E, and F), with 8 chicks in each group. Firstly, the overnight cultures of Y03 were scraped down from LB agar plates, washed three times and resuspended in PBS, and adjusted to 4.0 × 1010, 2.0 × 1010, 2.0 × 109, 2.0 × 108, and 2.0 × 107 CFU/mL, respectively. Next, chicks in group A, B, C, D, and E, were intramuscularly injected with 0.5 mL of 4.0 × 1010, 2.0 × 1010, 2.0 × 109, 2.0 × 108, and 2.0 × 107 CFU/mL of Y03, respectively. The group F was intramuscularly injected with 0.5 mL of PBS, which was the negative control. The clinical signs of infected chicks, such as lethargy, anorexia and hypothermia, were observed, and the survival and death of the chicks were recorded until 7 days post-infection. The survival curve was drawn to evaluate the virulence of Y03. After 7 days, all chicks were euthanized by injecting intravenous sodium pentobarbital (100 mg/kg) into the wing vein. Chicks in group E and F were dissected and collected their heart, liver, spleen, lung, and kidney, and these organs were fixed in 4% paraformaldehyde for histological observation.
Detection of virulence genes and antimicrobial resistance genes
We used genomic DNA of Y03 as template to amplify virulence genes and antibiotic resistance genes by PCR using the corresponding primers. The sequences of primers used in this study were listed in Table 1. PCR products were electrophoresed using 1.0% agarose gel (Sangon, Shanghai, China) containing DNA Green Stain (Vazyme, Nanjing, Jiangsu, China) and were observed by gel documentation system (Clinx, Shanghai, China).
Table 1.
Oligonucleotide primers used in this study
| Primer name | Oligonucleotide sequence (5′-3′) | Product size/bp |
|---|---|---|
| parC-F | TCGCGGAAAGCTCATCTTGT | 410 |
| parC-R | TGATGCGGAAGCAGTGATGA | |
| abeM-F | GAATGTCACGTCGTTTCGGT | 1011 |
| abeM-R | CGGCCTAGAGCAATAAAAGA | |
| adeB-F | GCACGTTTTCCAAGTGTGGC | 1604 |
| adeB-R | CCCTGATCTTCCTCTGGCAT | |
| adeH-f | CATCAAAACAAAACTGGTTG | 1092 |
| adeH-R | CATCAAATAAAGGTAACGAC | |
| adeK-F | TGCAAAAAGTATGGTCTATTTCAGG | 1110 |
| adeK-R | CAGACAATGCAATTTTCTGATCAGT | |
| blaapmC-F | TTTTAGTACCTCAATTTATGCGGAC | 864 |
| blaapmC-R | AACGTTGCCGGATAAGAAAA | |
| blaOXA−90-F | ACAAGCGCTATTTTTATTTCAGCCT | 634 |
| blaOXA−90-R | CCCAACCACTTTTTGCGTATATTTT | |
| catB9-F | CCTCTAATTAACACACCAGT | 489 |
| catB9-R | CTGAGGAAAACGGTATTTAA | |
| folP-F | TACCAAAGCAGATTTTACAG | 643 |
| folP-R | GGATAACCAAGTTCATTAAG | |
| macB-F | TAACTCGCTTGCGACAAGAG | 1048 |
| macB-R | ACATCAAGGTCAATTGGAGC | |
| parE-F | GTGACACAATATACGGCTCA | 1413 |
| parE-R | GTCTACACCAATTGCAATCG | |
| vgrG-F | TCTTACAACGTATAGAAGGC | 2077 |
| vgrG-R | ATGTCTTGAATAAAGCCACC | |
| lpxC-F | ATGGTGAAACAGCGTACTCT | 681 |
| lpxC-R | ACCTTCTTCGTTCACCACAC | |
| pilO-F | ATGAACAACTACGGTAGTTG | 560 |
| pilO-R | CCTACGTATCGGTATGTTTT | |
| fimT-F | TGAGCTCACTATAACACTCG | 400 |
| fimT-R | GATAAAACTGCCATTCGACT | |
| ompA-F | CACTTGCTACTATGCTTGTT | 803 |
| ompA-R | ACCAGTGTTATCTGTGTGAC | |
| basA-F | TTTGAGTCAAAATTTAGGGC | 1405 |
| basA-R | GCTATTTAGGCGAGTAGTTT | |
| bauA-F | AAGGTCTTTCAATGAACAAC | 1728 |
| bauA-R | AATCAACCTTCAAACCAAGT | |
| gspL-F | TTTGGCATTGGTCTAACGGA | 868 |
| gspL-R | TGCTGGCTCATTCTTAAATG | |
| csu-F | CATTCAGAAATCACTTCTTG | 402 |
| csu-R | TGTGGTTGGTTTACAACATA | |
| pgaC-F | TTGCCTTTTTATATCCGTTG | 936 |
| pgaC-R | AATCCACAAACAATCCCACA | |
| plc2-F | GCTGCAGCGTTAGCGGCTTT | 1685 |
| plc2-R | GCTGCTTGCGTACCCGTATT | |
| tssA-F | TTTCTGAACTACTTAAACCC | 821 |
| tssA-R | GATTTGTACAGGCACTTGAT | |
| tviB-F | AATAGCAATTATTGGGTTGG | 956 |
| tviB-R | TTTCTTTAAAGCTAAGGCCC | |
| bap-F | TCAGTGTGGCTGATAATACT | 2228 |
| bap-R | CCAATTCATATCCACAACTT |
Statistical analysis
The IBM SPSS statistics 25.0 (IBM, Armonk, New York, USA) and GraphPad Prism 8.0.1 (GraphPad, San Diego, California, USA) were used for statistical analysis. The test results were shown as means ± SDs.
Results
Identification of A. Baumannii
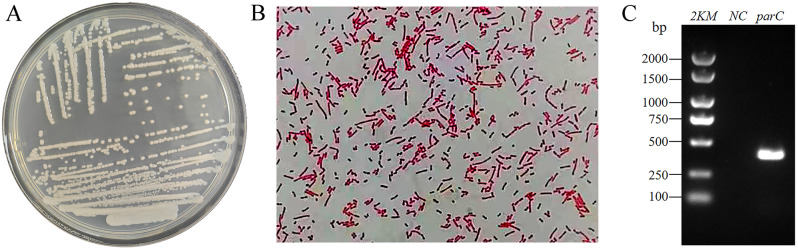
The colony morphology of this one isolate was white, circular, moist, smooth, opaque colonies on LB agar plates (Fig. 1A). Gram staining showed that this one isolate was a red coccobacillus under a 100-fold oil lens of microscope, determining that it was a Gram-negative coccobacillus (Fig. 1B). The biochemical tests showed that it was oxidase-negative, indole-negative, citratee-negative, MR, and VP-negative, catalase-positive, urease-positive, and absence of motility (data not shown). As shown in Fig. 1C and Figure S1, the gel electrophoresis band was bright, with the PCR product size around 410 bp by PCR amplification using the specific primer parC-F/R of A. baumannii. These results confirmed that this one isolate was an A. baumannii, which was known as Y03.
Fig. 1.
Isolation and identification of A. baumannii Y03. A A. baumannii was cultured on LB agar plates; B Observe the results under a 100-fold oil lens of microscope after gram staining; and C Electropherograms of parC in A. baumannii Y03. 2KM, 2 000 bp Marker; NC, blank control
Quality assessment of whole-genome sequence assembly
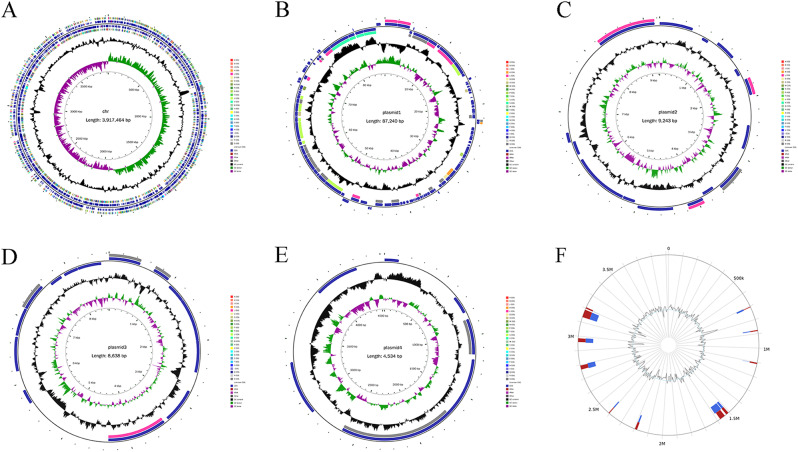
We found that Y03 owned 1 chromosome and 4 plasmids by assessing the quality of whole-genome sequence assembly. As shown in Fig. 2A–E; Table 2, the Y03 chromosome consisted of 3 917 464 bp in length with a GC content of 38.92%; the 4 plasmids included 87 240 bp of plasmid 1 in length with a GC content of 34.01%, 9 243 bp of plasmid 2 in length with a GC content of 34.70%, 8 638 bp plasmid 3 in length with a GC content of 31.86%, and 534 bp of plasmid 4 in length with a GC content of 37.21%. Additionally, the genomic comparative analysis showed that the identity of the Y03 genome sequence was greater than 98% when compared to the genome sequence of A. baumannii published in the NCBI Genbank nucleotide sequence database (Table S1). Hence, it was again determined that the Y03 isolate was an A. baumannii.
Fig. 2.
Overview of the complete A. baumannii Y03 genome. A Chromosome; B Plasmid 1; C Plasmid 2; D Plasmid 3; and E Plasmid 4. From inner circle to outer circle, the first circle is the scale, the second circle is GC skew, the third circle is the GC content, the fourth and seventh circles are each ORF belonging to the COG classifications, and the fifth and sixth circles are the positions of ORF, tRNA, and rRNA on the genome. F Genomic islands (GIs) on the A. baumannii Y03 chromosome. Red is the predicted result by at least one method, blue is the predicted result by IslandPath-DIMOB, and yellow is the predicted result of SIGI-HMM
Table 2.
General features obtained from A. Baumannii Y03 using whole-genome sequencing
| Genomic parametera | Value for parameter |
|---|---|
| Chromosome | |
| Accession number | CP163382 |
| Sequence length/bp | 3 917 464 |
| GC content/% | 38.92 |
| ORF number | 3 665 |
| tRNA copy number | 73 |
| rRNA copy number | 6 |
| ncRNA copy number | 34 |
| Prophage number | 4 |
| Short interspersed repeats number | 9 |
| Long interspersed repeats number | 31 |
| Long terminal repeats number | 72 |
| Transposons number | 30 |
| Unclassified interspersed repeats number | 9 |
| Satellites RNA number | 4 |
| Simple-repeats number | 0 |
| Plasmid 1 | |
| Accession number | CP163383 |
| Sequence length/bp | 87 240 |
| GC content/% | 34.01 |
| ORF number | 107 |
| tRNA copy number | 0 |
| rRNA copy number | 0 |
| ncRNA copy number | 0 |
| Prophage number | 1 |
| Short interspersed repeats number | 0 |
| Long interspersed repeats number | 0 |
| Long terminal repeats number | 0 |
| Transposons number | 0 |
| Unclassified interspersed repeats number | 0 |
| Satellites RNA number | 0 |
| Simple-repeats number | 0 |
| Plasmid 2 | |
| Accession number | CP163384 |
| Sequence length/bp | 9 243 |
| GC content/% | 34.70 |
| ORF number | 15 |
| tRNA copy number | 0 |
| rRNA copy number | 0 |
| ncRNA copy number | 0 |
| Prophage number | 0 |
| Short interspersed repeats number | 0 |
| Long interspersed repeats number | 0 |
| Long terminal repeats number | 0 |
| Transposons number | 0 |
| Unclassified interspersed repeats number | 0 |
| Satellites RNA number | 0 |
| Simple-repeats number | 0 |
| Plasmid 3 | |
| Accession number | CP163385 |
| Sequence length/bp | 8 638 |
| GC content/% | 31.86 |
| ORF number | 14 |
| tRNA copy number | 0 |
| rRNA copy number | 0 |
| ncRNA copy number | 0 |
| Prophage number | 0 |
| Short interspersed repeats number | 0 |
| Long interspersed repeats number | 0 |
| Long terminal repeats number | 0 |
| Transposons number | 0 |
| Unclassified interspersed repeats number | 0 |
| Satellites RNA number | 0 |
| Simple-repeats number | 0 |
| Plasmid 4 | |
| Accession number | CP163386 |
| Sequence length/bp | 534 |
| GC content/% | 37.21 |
| ORF number | 7 |
| tRNA copy number | 0 |
| rRNA copy number | 0 |
| ncRNA copy number | 0 |
| Prophage number | 0 |
| Short interspersed repeats number | 0 |
| Long interspersed repeats number | 0 |
| Long terminal repeats number | 0 |
| Transposons number | 0 |
| Unclassified interspersed repeats number | 0 |
| Satellites RNA number | 0 |
| Simple-repeats number | 0 |
ORF open reading frame, ncRNA non-coding RNA
aBased on NCBI Prokaryotic genomic annotation pipeline
Genome functional component analysis
The protein coding genes of Y03 were predicted using the GeneMarkS software. The results showed that the protein coding genes of Y03 were 3 665 on the chromosome, 107 on the plasmid 1, 15 on the plasmid 2, 14 on the plasmid 3, and 7 on the plasmid 4, respectively (Table 2). The number of tRNA on the chromosome was 73, rRNA was 6, and ncRNA was 34. The number of CRISPRs on the chromosome was 3. Additionally, the interspersed repeats of Y03 on the chromosome included 9 SINEs (short interspersed repeats), 31 LINEs (long interspersed repeats), 72 L (long terminal repeats), 30 transposons, 9 unclassified interspersed repeats, and 4 satellites RNA (Table 2).
Genome subsystem analysis
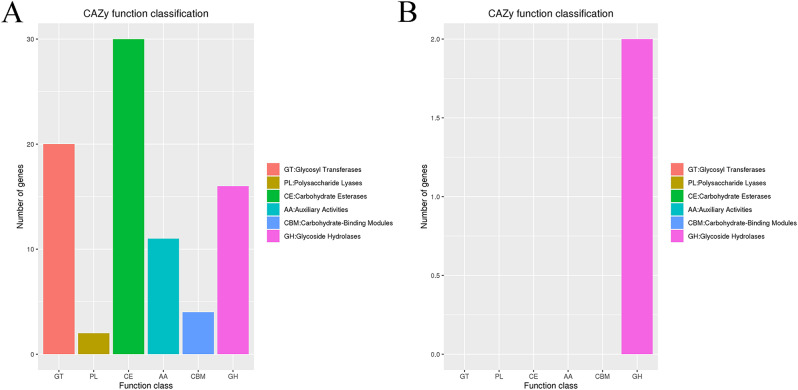
Prophages of Y03 were predicted using the PHASTER tool. The results showed that there were 4 prophages on the chromosome, 1 prophage on the plasmid 1, and the plasmid 2, 3, and 4 had no prophage (Table 2). The number of GIs were 11 on the chromosome by comparison with the IslandViewer 4 database, and no GIs were predicted on the plasmid (Fig. 2F). The number of the virulence factors on the chromosome were 135 by BLAST analysis using the VFDB database, which were related with effector delivery system, immune modulation, adherence, stress survival, biofilm, exotoxin, and nutritional/metabolic factor (Table S2). There were 41 antibiotic resistance genes, 22 antibiotic target genes, and 3 antibiotic biosynthesis genes on the chromosome by BLAST analysis using the CARD database (Table S3). No antibiotic resistance genes were predicted on the plasmid. Additionally, a total of 85 carbohydrate-active enzymes on the Y03 genome were annotated using the CAZy database (Fig. 3 and Tables S4, 5), including 16 glycoside hydrolases (GHs), 20 glycosyl transferases (GTs), 2 polysaccharide lyases (PLs), 30 carbohydrate esterases (CEs), 11 auxiliary activities (AAs), and 4 carbohydrate-binding modules (CBMs) on the chromosome, and 2 GHs on the plasmid 1.
Fig. 3.
The functional classifications annotated using CAZy on the A. baumannii Y03 genome. A Chromosome; and B Plasmid 1
Genome functional annotation analysis
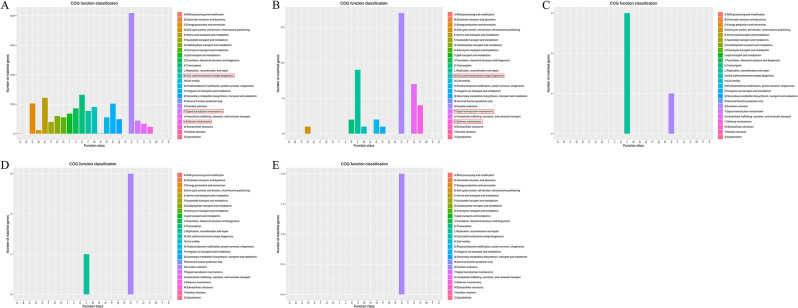
The protein coding genes were annotated using various databases. As shown in Table 3, the number of the protein coding genes annotated using the NR database was 3 657 on the chromosome, 106 on the plasmid 1, 13 on the plasmid 2, 14 on the plasmid 3, and 6 on the plasmid 4. By comparative analysis in the COG database, a total of 3 131 genes were annotated on the chromosome, 44 genes on the plasmid 1, 4 genes on the plasmid 2, 4 genes on the plasmid 3, and 2 genes on the plasmid 4 (Fig. 4; Table 3). As shown in Fig. 4A, 181 genes were related with cell wall/membrane/envelope biogenesis, 89 genes were related with signal transduction mechanisms, and 46 genes were related with defense mechanisms on the chromosome. 1 gene was related with cell wall/membrane/envelope biogenesis, 0 gene was related with signal transduction mechanisms, and 4 genes were related with defense mechanisms on the plasmid 1 (Fig. 4B).
Table 3.
Overview on the functional annotation of the protein coding genes in A. Baumannii Y03
| Sequence ID | Annotation in database | Genes number |
|---|---|---|
| Chromosome | NR | 3 657 |
| COG | 3 131 | |
| Plasmid 1 | NR | 106 |
| COG | 44 | |
| Plasmid 2 | NR | 13 |
| COG | 4 | |
| Plasmid 3 | NR | 14 |
| COG | 4 | |
| Plasmid 4 | NR | 6 |
| COG | 2 |
Fig. 4.
The functional classifications annotated using COG on the A. baumannii Y03 genome. A Chromosome; B Plasmid 1; C Plasmid 2; D Plasmid 3; and E Plasmid 4
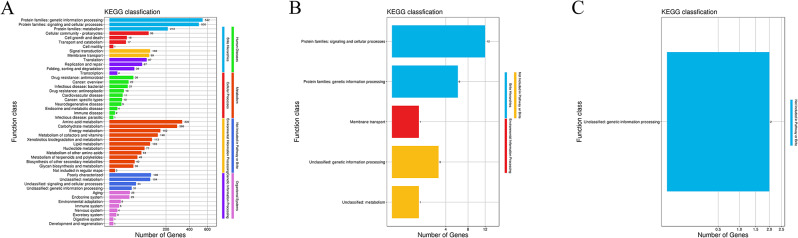
The protein encoding genes on the Y03 genome were annotated and mapped into 8 KEGG categories by KO (KEGG ortholog) annotation and the KEGG pathway annotation. As shown in Fig. 5A, a total of 47 pathways on the chromosome were covered 8 KEGG categories, including brite hierarchies (3), cellular processes (4), environmental information processing (2), human diseases (10), genetic information processing (4), metabolism (12), not included in pathway or brite (4), and organismal systems (8). Similarly, 5 pathways on the plasmid 1 were covered 3 KEGG categories, including brite hierarchies (2), environmental information processing (2), and not included in pathway or brite (2); and 1 pathways on the plasmid 2 was not included in pathway or brite (Fig. 5B, C). The 4 most represented pathways on the chromosome related with survival were antimicrobial drug resistance (36), bacterial infectious diseases (21), environmental adaptation (8), and the host immune system (6) (Fig. 5A).
Fig. 5.
The functional classifications annotated using KEGG on the A. baumannii Y03 genome. A Chromosome; B Plasmid 1; and C Plasmid 2
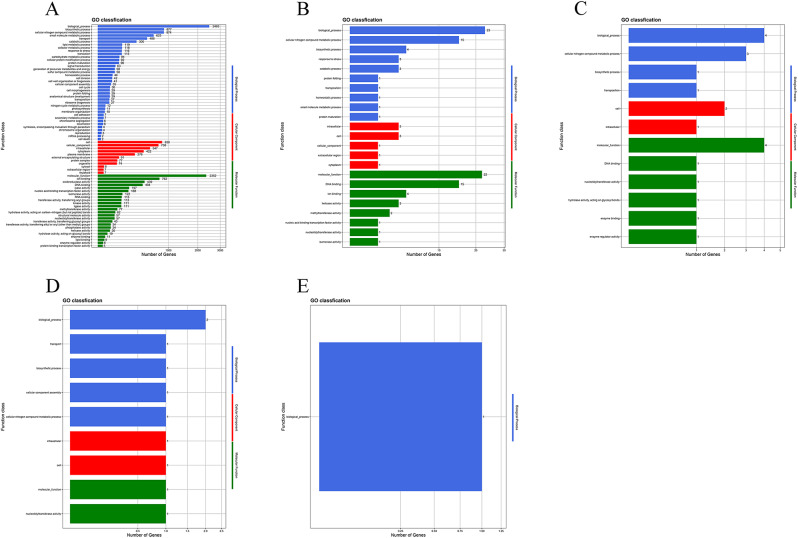
GO is a functional classification system fully describing the properties of genes and gene products [27]. In this study, GO classification statistics revealed that a total of 73 GO terms on the chromosome were functionally characterized into 3 ontologies, including biological processes (38), cellular components (11), and molecular functions (24); 12 GO terms on the plasmid 1 included biological processes (4), cellular components (2), and molecular functions (5); 12 GO terms on the plasmid 2 included biological processes (4), cellular components (2), and molecular functions (6); 9 GO terms on the plasmid 3 included biological processes (5), cellular components (2), and molecular functions (2); and 1 GO terms on the plasmid 3 included biological processes (1) (Fig. 6). As shown in Fig. 6A, GO term related with stress response had 116 protein encoding genes, GO term related with cell wall organization or biogenesis had 41 protein encoding genes, and GO term related with cell adhesion had 7 protein encoding genes, etc.
Fig. 6.
The functional classifications annotated using GO on the A. baumannii Y03 genome. A Chromosome; B Plasmid 1; C Plasmid 2; D Plasmid 3; and E Plasmid 4
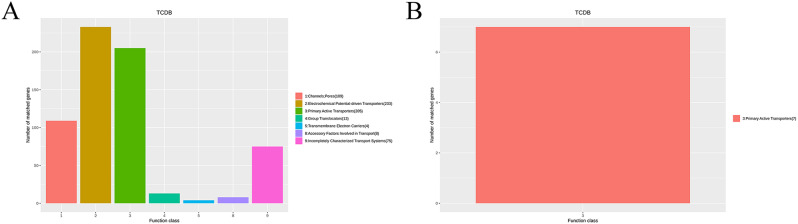
A total of 647 transport proteins on the chromosome were obtained by comparative analysis of the TCDB database, including 109 channels or pores, 205 primary active transporters, 233 electrochemical potential-driven transporters, and 13 group translocators, 4 transmembrane electron carriers, 4 accessory factors involved in the transport, and 75 incompletely characterized transport systems (Fig. 7A). Moreover, there were 7 primary active transporters on the plasmid 1 (Fig. 7B).
Fig. 7.
The functional classifications annotated using TCDB on the A. baumannii Y03 genome. A Chromosome; and B Plasmid 1
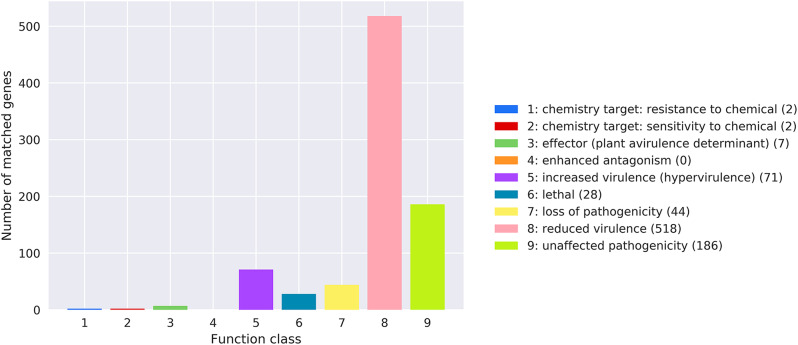
The bacterial virulence and pathogenicity were annotated using the PHI database. The results revealed that 4 mutations were related with chemistry targets, 71 mutations were associated with enhanced virulence, 518 mutations leaded to reduced virulence, 44 mutations resulted in loss of pathogenicity, and 28 lethal mutations contributed to the death of Y03 on the chromosome (Fig. 8 and Table S6). The prediction results for the secreted proteins in Y03 indicated that a total of 264 secreted proteins were located on the chromosome, 12 secreted proteins on the plasmid 1, 1 secreted proteins on the plasmid 2, and no secreted proteins on the plasmid 3 and 4 (Table 4).
Fig. 8.
The functional classifications annotated using PHI on the A. baumannii Y03 chromosome
Table 4.
The prediction results of the secreted proteins in A. Baumannii Y03
| Sequence ID | Secreted proteins number | Percentagea/% |
|---|---|---|
| Chromosome | 264 | 7.20 |
| Plasmid 1 | 12 | 11.21 |
| Plasmid 2 | 1 | 6.67 |
| Plasmid 3 | 0 | 0 |
| Plasmid 4 | 0 | 0 |
aThe secreted protein coding genes are as a percentage of the total protein coding genes
Antimicrobial susceptibility analysis
The antimicrobial susceptibility pattern of Y03 to the tested 34 antimicrobial agents was presented in Table 5 and Table S7. Y03 was resistant to penicillin G, ampicillin, oxacillin, cefalotin, cefazolin, cefoxitin, chloramphenicol, florfenicol, midecamycin, clindamycin, spectinomycin, and nitrofurantoin; intermediate susceptible to aztreonam, cefotaxime, cefuroxime, clarithromycin, erythromycin, and tetracycline; and susceptible to piperacillin, cefoperazone, ceftazidime, cefepime, cotrimoxazole, ciprofloxacin, levofloxacin, ofloxacin, norfloxacin, minocycline, amikacin, tobramycin, streptomycin, gentamicin, kanamycin, and polymyxin B. These results demonstrated that Y03 was resistant to at least one agent from three or more antimicrobial classes and was considered to be a MDR A. baumannii.
Table 5.
Susceptibility of A. Baumannii Y03 to antimicrobial drugs
| Antibiotics classes | Antimicrobial drugs | Inhibition zone diameter/mm |
Susceptibility | |
|---|---|---|---|---|
| β-lactams | Penicillins | Penicillin G | 10.00 ± 1.00 | R |
| Ampicillin | 11.67 ± 0.58 | R | ||
| Oxacillin | 6.00 ± 0.00 | R | ||
| Piperacillin | 21.67 ± 1.53 | S | ||
| Monobactams | Aztreonam | 16.33 ± 1.15 | I | |
| First-generation cephalosporins | Cefalotin | 6.00 ± 0.00 | R | |
| Cefazolin | 6.00 ± 0.00 | R | ||
| Second-generation cephalosporins | Cefoxitin | 12.33 ± 0.58 | R | |
| Cefuroxime | 16.00 ± 1.00 | I | ||
| Third-generation cephalosporins | Cefotaxime | 17.00 ± 1.00 | I | |
| Cefoperazone | 18.00 ± 2.00 | S | ||
| Ceftazidime | 18.33 ± 0.58 | S | ||
| Fourth-generation cephalosporins | Cefepime | 21.00 ± 2.00 | S | |
| Sulfonamides | Cotrimoxazole | 23.33 ± 2.89 | S | |
| Phenicols | Chloramphenicol | 8.00 ± 1.73 | R | |
| Florfenicol | 15.00 ± 0.00 | R | ||
| Macrolides | Midecamycin | 6.00 ± 0.00 | R | |
| Clarithromycin | 17.00 ± 2.00 | I | ||
| Erythromycin | 18.67 ± 0.58 | I | ||
| Lincosamides | Clindamycin | 6.00 ± 0.00 | R | |
| Fluoroquinolones | Ciprofloxacin | 28.00 ± 1.00 | S | |
| Levofloxacin | 26.33 ± 2.08 | S | ||
| Ofloxacin | 27.00 ± 2.00 | S | ||
| Norfloxacin | 21.00 ± 1.00 | S | ||
| Tetracyclines | Tetracycline | 14.00 ± 2.00 | I | |
| Minocycline | 22.33 ± 2.51 | S | ||
| Aminoglycosides | Amikacin | 21.33 ± 2.89 | S | |
| Tobramycin | 23.67 ± 0.58 | S | ||
| Streptomycin | 20.00 ± 0.00 | S | ||
| Gentamicin | 20.33 ± 0.58 | S | ||
| Kanamycin | 21.00 ± 1.73 | S | ||
| Spectinomycin | 12.67 ± 1.15 | R | ||
| Nitrofurans | Nitrofurantoin | 6.00 ± 0.00 | R | |
| Polymyxins | Polymyxin B | 12.67 ± 1.53 | S | |
S susceptible, I intermediate susceptible, R resistant
Animal infection analysis
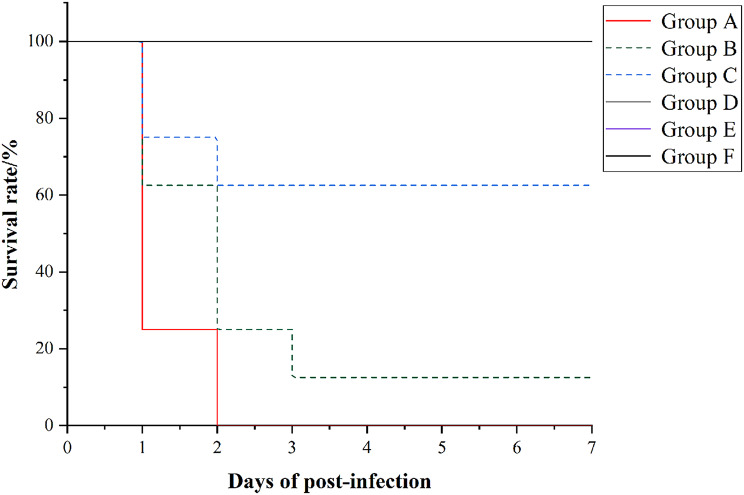
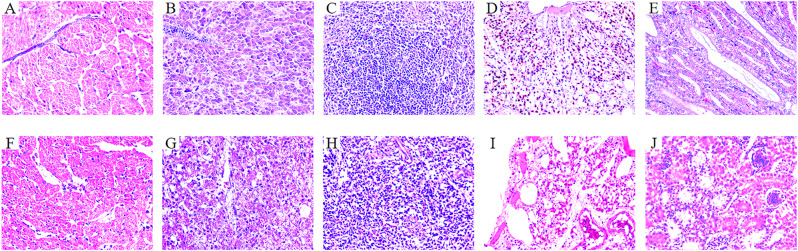
The virulence of Y03 was evaluated in the chick models. Chicks were intramuscularly infected with 2.0 × 1010, 1.0 × 1010, 1.0 × 109, 1.0 × 108, and 1.0 × 107 CFU of Y03, and the chick mortality was observed for 7 days post-infection. On the first day, they began to show lethargy, anorexia and hypothermia in the test groups (group A, B, C, D and E) with Y03. Meanwhile, the mortality of chicks was observed in group A, B, and C, but not in group D, E, and the negative controls. As shown in Fig. 9, the mortality of group A was 100% (8/8), group B was 87.5% (7/8), and group C was 37.5% (3/8). Moreover, the histopathological sections of the Y03-infected group of chicks revealed that the myocardial fibers exhibited vacuolar degeneration, the interstitium was congested and edematous, and there was significant infiltration of heterophilic granulocytes in the heart (Fig. 10F). The hepatocytes were degenerated and necrotic, and numerous blue-staining granules in the hepatic sinusoids were suspected to be bacterial colonies in the liver (Fig. 10G). The necrosis and loss of lymphocytes in the white pulp were observed, along with infiltration of heterophilic granulocytes in the spleen (Fig. 10H). Congestion and inflammatory exudate in the bronchioles were observed, and the interstitium was widened and edematous in the lung (Fig. 10I). The tubular epithelial cells were degenerated and necrotic, and the interstitium was congested and hemorrhaged in the kidney (Fig. 10J).
Fig. 9.
The survival rates of chicks infected by different concentrrations of A. baumannii Y03 were detected using animal infection experiments. Seven-day-old chicks in the test groups were intramuscularly injected with 2.0 × 1010, 1.0 × 1010, 1.0 × 109, 1.0 × 108, and 1.0 × 107 CFU of MDR A. baumannii Y03. Seven-day-old chicks in the control groups were intramuscularly injected with PBS. Survival was monitored until 7 days post-infection
Fig. 10.
Histological observation of chick lesions (400×). A–E Heart, liver, spleen, lung, and kidney of group F, which was the control group of chicks intramuscularly injected with PBS; and F–J Heart, liver, spleen, lung, and kidney of group E, which was the Y03-infected group of chicks intramuscularly injected with 1.0 × 107 CFU of MDR A. baumannii Y03
Antimicrobial resistance genes and virulence genes
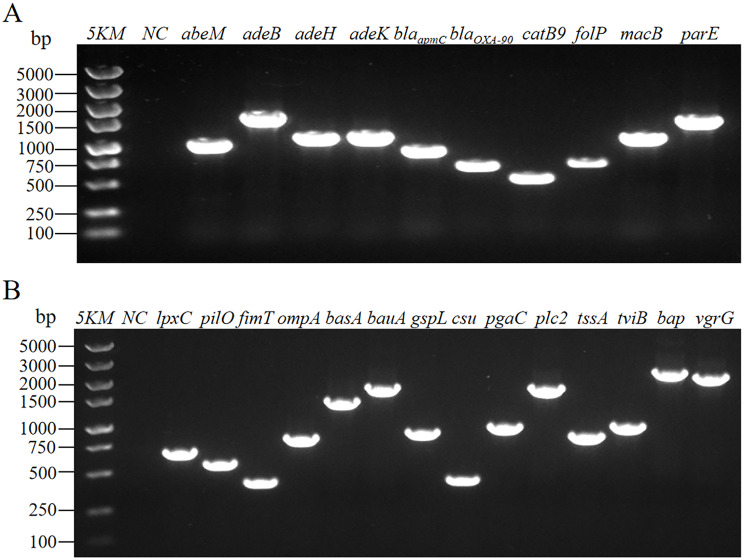
The antimicrobial resistance genes abeM, adeB, adeH, adeK, blaapmC, blaOXA−90, catB9, macB, folP, and parE were detected using PCR amplification method according to the results of antimicrobial susceptibility testing. As shown in Fig. 11A and Figure S1, Y03 contained abeM, adeB, adeH, adeK, blaapmC, blaOXA−90, catB9, folP, macB, and parE. The virulence genes included the lpxC, pilO, fimT, ompA, basA, bauA, gspL, csu, pgaC, plc2, tssA, tviB, bap, and vgrG on the Y03 chromosome (Fig. 11B and Figure S2).
Fig. 11.
Electropherograms of antimicrobial resistance genes and virulence genes in MDR A. baumannii Y03. A Electropherograms of antimicrobial resistance genes in A. baumannii Y03. 5KM, 5 000 bp Marker; NC, blank control. B Electropherograms of virulence genes in A. baumannii Y03. 5KM, 5 000 bp Marker; NC, blank control
Discussion
Multiple investigations have shown that the antimicrobial resistance mechanism and the pathogenic mechanism of A. baumannii as a nosocomial pathogen [4, 5, 28]. A. baumannii has been isolated from a variety of wild and domestic animals, in some cases, carbapenemase-producing isolates have been described [10, 29, 30]. However, a study on the antimicrobial susceptibility profile and the pathogenicity of A. baumannii of chicken origin is rare, especially studies focusing on its genome. Therefore, understanding the genomic characteristics and the protein coding gene functions of A. baumannii could provide important insights into the adaptation and evolution of this bacterium of chicken origin.
Horizontal gene transfer refers to the transfer of the genetic material from one organism to another, significantly contributing to genome rearrangements and evolution [27, 31, 32]. The common horizontal gene transfer elements include transposons, prophages, plasmids, and GIs, which assist in the spread of antimicrobial resistance genes and virulence factors between bacteria, further increasing their antimicrobial resistance and pathogenicity [33–35]. There were 30 transposons, 5 prophages, 4 plasmids, and 11 GIs annotated on the genome of MDR A. baumannii Y03 using whole-genome sequencing, indicating that Y03 possessed the potential of these mobile genetic elements transferred to other bacteria and might be a significant threat to public health.
Whole-genome sequencing revealed that 3 CRISPRs were identified on the chromosome of MDR A. baumannii Y03, indicating that it could acquire some kind of adaptive immunity by integrating exogenous DNA into the chromosome via the spacer regions of CRISPRs [36, 37]. In addition, there are 3 131 protein encoding genes on the chromosome and 52 protein encoding genes on the plasmid, both of which have been annotated and functionally classified using the COG database. Among them, 842 protein encoding genes encode hypothetical proteins with unknown functions, which is a major hindrance in inferring their involvement in metabolism, proliferation, development, virulence, and antimicrobial resistance. Hence, further studies are needed to elucidate the precise function of these proteins.
Antimicrobial resistance is one of the most important characters of the A. baumannii isolates [6]. We found that MDR A. baumannii Y03 was resistant to several β-lactams, phenicols, macrolides, lincosamides, aminoglycosides, and nitrofurans. There were 41 antimicrobial resistance genes identified on the Y03 chromosome using whole-genome sequencing, including (i) β-lactamses: blaAmpC and blaOXA−90; (ii) the ATP-binding cassette (ABC) transporters: macB; (iii) the resistance nodulation cell division (RND) family: adeAB, adeFGH and adeIJK; (iv) the small multidrug resistance (SMR) family efflux pump: abeS; (v) the multidrug and toxic compound extrusion (MATE) family: abeM; (vi) chloramphenicol acetyltransferase: catB9; (vii) the resistance genes to fluoroquinolones: gyrA, parE, and parC; and others. Various studies have demonstrated that many multidrug efflux pump systems are correlated with resistance to a range of antibiotics in other A. baumannii strains, including aminoglycosides, β-lactams, tetracyclines, and fluoroquinolones [38–40]. However, Y03 was susceptible to most of aminoglycosides, tetracyclines, fluoroquinolones, sulfonamides, polymyxins, third-generation and fourth-generation cephalosporins. This suggests that the resistance mechanisms of Y03 are different from those of other A. baumannii strains, and some antimicrobial resistance genes of Y03 might be less active or expressed at lower levels compared to their homologs in other A. baumannii strains, and they only constitute the intrinsic resistome of Y03. Furthermore, there were 22 antibiotic target genes on the Y03 chromosome encoding the B subunit of DNA topoisomerase IV, the β subunit of RNA polymerase, the A subunit of DNA gyrase, and others. This implies that certain antibiotics could be used to kill MDR A. baumannii Y03, including fluoroquinolone, cycloserine, and rifamycin.
A total of 135 virulence genes belonging to 17 classes of virulence factor associating with the pathogenic potential of Y03 were identified using whole-genome sequencing, including adhesins, invasion, biofilms, acinetobactin, secretion proteins, toxins, cell surface appendages, immune evasion, serum resistance, stress adaptation, and others. And we certificated that Y03 was virulent to chickens in the in vivo infection experiments; however, it did not exhibit comparable hypervirulence, which is defined as ≤ 1.0 × 108 CFU of bacteria causing the death of experimental animals. Although A. baumannii ATCC 19606 only contained 69 virulence genes associated with adherence, biofilm formation, phospholipase, iron uptake, and immune evasion, 1.9 × 106 CFU of ATCC 19606 resulted in a mortality of 100%, illustrating that it exhibited hypervirulence in a mouse infection model [38, 41, 42]. Therefore, this suggests that the pathogenicity and the pathogenic mechanisms of MDR A. baumannii Y03 in a chickens infection model is complex and differs from that of A. baumannii ATCC 19606 in a mouse infection model. Besides, there were 518 mutations leading to reduced virulence, 44 mutations resulting in loss of pathogenicity, and 28 lethal mutations contributing to the death of Y03 using the PHI database annotation. This implies that certain mutants could reduce virulence or lose pathogenicity, and even cause the death of MDR A. baumannii Y03.
Our study has some limitations. Our results from the study demonstrated that MDR A. baumannii Y03 contained 41 antimicrobial resistance genes on its chromosome, but Y03 was only resistant to 6 classes of antimicrobial agents, including several β-lactams, phenicols, macrolides, lincosamides, aminoglycosides, and nitrofurans. Consequently, the discrepancies between the phenotype and genotype of antimicrobial resistance remains to be fully explored. Despite the high carriage rate of virulence factors, Y03 did not exhibit comparable hypervirulence in the in vivo chickens infection experiments. Therefore, it is necessary to confirm whether inherent or chromosomal mechanisms underlie the observed virulence heterogeneity in the MDR A. baumannii Y03 in future experiments.
In reference to future perspectives, extensive epidemiological researches are essential to comprehensively investigate the distribution of A. baumannii isolates in food-producing animals, their antimicrobial resistome, their virulence factor and the pathogenicity, and their association with A. baumannii of human origin.
Conclusion
This study reported that MDR A. baumannii Y03 isolated from chickens contained 1 circular chromosome harboring 41 antimicrobial resistance genes and 135 virulence genes, as well as 4 circular plasmids. Y03 poses a critical threat to global health due to its virulence and resistance to 6 classes of antibiotics, including several β-lactams, phenicols, macrolides, lincosamides, aminoglycosides, and nitrofurans. Interestingly, whole-genome sequencing analysis revealed that 22 antibiotic targets and 28 lethal mutations on the Y03 chromosome could serve as potential targets to prevent, control, and treat infections caused by MDR A. baumannii Y03.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Professor Ting Xue (Anhui Agricultural University) for kindly providing much helps in the experiment and writing.
Abbreviations
- A. baumannii
Acinetobacter baumannii
- MDR
Multidrug-resistant
- XDR
Extensively drug-resistant
- PDR
Pan-drug-resistant
- ncRNA
Non-coding RNA
- rRNA
Ribosome RNA
- sRNA
Small RNA
- rRNA
Ribosome RNA
- tRNA
Transfer RNA
- snRNA
Small nuclear RNA
- snoRNA
Small nucleolar RNA
- miRNA
microRNA
- NR
Non-Redundant Protein Database
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- COG
Clusters of Orthologous Groups
- TCDB
Transporter Classification Database
- CAZy
Carbohydrate-Active Enzymes Database
- PHI
Pathogen Host Interactions Database
Author contributions
LY: investigation, methodology, experiments, writing original draft, writing review and editing, data curation and funding acquisition. SZ, WN and YZ: methodology and experiments. LZ, CX and PK: formal analysis, methodology. XZ: conceptualization, supervision and funding acquisition. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant Number 32202810) and the Natural Science Foundation of Shandong Province, China (Grant Number ZR2022QC115).
Data availability
The complete genome sequences of MDR A. baumannii Y03 including 1 chromosome sequence and 4 plasmids sequences using whole-genome sequencing have been deposited into the NCBI Genbank nucleotide sequence database under accession numbers CP163382, CP163383, CP163384, CP163385, and CP163386. The raw sequencing data of MDR A. baumannii Y03 are available in the NCBI Sequence Read Archive (SRA) database with the SRA accession number SRR30247697.
Declarations
Ethics approval and consent to participate
This study was carried out in compliance with the ARRIVE guidelines. Animal experiments were conducted under animal protocols approved by the Institutional Animal Care and Use Committee (IACUC) of Linyi University (Approval No. LYU20240206). All animal work was carried out following accordance within the guidelines of the Laboratory Animal Research Center of Linyi University.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Lumin Yu, Email: yulumin@lyu.edu.cn.
Xinglin Zhang, Email: zhangxinglin@lyu.edu.cn.
References
- 1.Li P, Zhang S, Wang J, Al-Shamiri MM, Han B, Chen Y, et al. Uncovering the secretion systems of Acinetobacter baumannii: structures and functions in pathogenicity and antibiotic resistance. Antibiot (Basel). 2023;12(2). 10.3390/antibiotics12020195. [DOI] [PMC free article] [PubMed]
- 2.Carvalheira A, Casquete R, Silva J, Teixeira P. Prevalence and antimicrobial susceptibility of Acinetobacter spp. isolated from meat. Int J Food Microbiol. 2017;243:58–63. 10.1016/j.ijfoodmicro.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 3.Lucidi M, Visaggio D, Migliaccio A, Capecchi G, Visca P, Imperi F, et al. Pathogenicity and virulence of Acinetobacter baumannii: factors contributing to the fitness in healthcare settings and the infected host. Virulence. 2024;15(1):2289769. 10.1080/21505594.2023.2289769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh S, Singh S, Trivedi M, Dwivedi M. An insight into MDR Acinetobacter baumannii infection and its pathogenesis: potential therapeutic targets and challenges. Microb Pathog. 2024;192:106674. 10.1016/j.micpath.2024.106674. [DOI] [PubMed] [Google Scholar]
- 5.Tavakol M, Momtaz H, Mohajeri P, Shokoohizadeh L, Tajbakhsh E. Genotyping and distribution of putative virulence factors and antibiotic resistance genes of Acinetobacter baumannii strains isolated from raw meat. Antimicrob Resist Infect Control. 2018;7:120. 10.1186/s13756-018-0405-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Askari N, Momtaz H, Tajbakhsh E. Prevalence and phenotypic pattern of antibiotic resistance of Acinetobacter baumannii isolated from different types of raw meat samples in Isfahan, Iran. Vet Med Sci. 2020;6(1):147–53. 10.1002/vms3.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jeong GJ, Khan F, Tabassum N, Kim YM. Motility of Acinetobacter baumannii: regulatory systems and controlling strategies. Appl Microbiol Biotechnol. 2024;108(1):3. 10.1007/s00253-023-12975-6. [DOI] [PubMed] [Google Scholar]
- 8.Gurung M, Nam HM, Tamang MD, Chae MH, Jang GC, Jung SC, et al. Prevalence and antimicrobial susceptibility of Acinetobacter from raw bulk tank milk in Korea. J Dairy Sci. 2013;96(4):1997–2002. 10.3168/jds.2012-5965. [DOI] [PubMed] [Google Scholar]
- 9.Gedefie A, Demsis W, Ashagrie M, Kassa Y, Tesfaye M, Tilahun M, et al. Acinetobacter baumannii Biofilm formation and its role in Disease Pathogenesis: a review. Infect Drug Resist. 2021;14:3711–9. 10.2147/idr.S332051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghaffoori Kanaan MH, Al-Shadeedi SMJ, Al-Massody AJ, Ghasemian A. Drug resistance and virulence traits of Acinetobacter baumannii from Turkey and chicken raw meat. Comp Immunol Microbiol Infect Dis. 2020;70:101451. 10.1016/j.cimid.2020.101451. [DOI] [PubMed] [Google Scholar]
- 11.Lysitsas M, Triantafillou E, Chatzipanagiotidou I, Antoniou K, Spyrou V, Billinis C, et al. Phenotypic investigation and detection of biofilm-associated genes in Acinetobacter baumannii isolates, obtained from Companion animals. Trop Med Infect Dis. 2024;9(5). 10.3390/tropicalmed9050109. [DOI] [PMC free article] [PubMed]
- 12.Jalal D, Elzayat MG, Diab AA, El-Shqanqery HE, Samir O, Bakry U, et al. Deciphering Multidrug-Resistant Acinetobacter baumannii from a Pediatric Cancer Hospital in Egypt. mSphere. 2021;6(6):e0072521. 10.1128/mSphere.00725-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buchhorn de Freitas S, Clair Pinto Seixas Neto A, Aparecido Panagio L, Pereira Soares M, Drawanz Hartwig D. Hypothetical adhesin CAM87009.1 formulated in alum or biogenic silver nanoparticles protects mice from lethal infection by multidrug-resistant Acinetobacter baumannii. Vaccine. 2024;42(18):3802–10. 10.1016/j.vaccine.2024.04.094. [DOI] [PubMed] [Google Scholar]
- 14.Harding CM, Hennon SW, Feldman MF. Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat Rev Microbiol. 2018;16(2):91–102. 10.1038/nrmicro.2017.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Law SKK, Tan HS. The role of quorum sensing, biofilm formation, and iron acquisition as key virulence mechanisms in Acinetobacter baumannii and the corresponding anti-virulence strategies. Microbiol Res. 2022;260:127032. 10.1016/j.micres.2022.127032. [DOI] [PubMed] [Google Scholar]
- 16.Karampatakis T, Tsergouli K, Behzadi P. Pan-genome plasticity and virulence factors: a natural treasure trove for Acinetobacter baumannii. Antibiot (Basel). 2024;13(3). 10.3390/antibiotics13030257. [DOI] [PMC free article] [PubMed]
- 17.Ham SY, Chun JY, Song KH, Kang CK, Park JS, Jo HB, et al. Limited impact of bacterial virulence on early mortality risk factors in Acinetobacter baumannii bacteremia observed in a Galleria mellonella model. Sci Rep. 2024;14(1):14960. 10.1038/s41598-024-65940-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eijkelkamp BA, Stroeher UH, Hassan KA, Paulsen IT, Brown MH. Comparative analysis of surface-exposed virulence factors of Acinetobacter baumannii. BMC Genomics. 2014;15(1):1020. 10.1186/1471-2164-15-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu K, Wang M, Zhang Y, Fang C, Zhang R, Fang L, et al. Distribution of antibiotic resistance genes and their pathogen hosts in duck farm environments in south-east coastal China. Appl Microbiol Biotechnol. 2024;108(1):136. 10.1007/s00253-023-12842-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee CR, Lee JH, Park M, Park KS, Bae IK, Kim YB, et al. Biology of Acinetobacter baumannii: Pathogenesis, Antibiotic Resistance mechanisms, and prospective treatment options. Front Cell Infect Microbiol. 2017;7:55. 10.3389/fcimb.2017.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roy S, Chowdhury G, Mukhopadhyay AK, Dutta S, Basu S. Convergence of Biofilm formation and Antibiotic Resistance in Acinetobacter baumannii infection. Front Med (Lausanne). 2022;9:793615. 10.3389/fmed.2022.793615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bruzek S, Vestal G, Lasher A, Lima A, Silbert S. Bacterial whole genome sequencing on the Illumina iSeq 100 for clinical and Public Health Laboratories. J Mol Diagn. 2020;22(12):1419–29. 10.1016/j.jmoldx.2020.09.003. [DOI] [PubMed] [Google Scholar]
- 23.Ali A, Khatoon A, Mirza T, Ahmad F. Intensification in Genetic Information and Acquisition of Resistant Genes in genome of Acinetobacter baumannii: a pan-genomic analysis. Biomed Res Int. 2022;2022:3186343. 10.1155/2022/3186343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weinmaier T, Conzemius R, Bergman Y, Lewis S, Jacobs EB, Tamma PD, et al. Validation and application of Long-Read whole-genome sequencing for Antimicrobial Resistance Gene Detection and Antimicrobial susceptibility testing. Antimicrob Agents Chemother. 2023;67(1):e0107222. 10.1128/aac.01072-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bogaty C, Mataseje L, Gray A, Lefebvre B, Lévesque S, Mulvey M, et al. Investigation of a carbapenemase-producing Acinetobacter baumannii outbreak using whole genome sequencing versus a standard epidemiologic investigation. Antimicrob Resist Infect Control. 2018;7:140. 10.1186/s13756-018-0437-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu L, Wang H, Zhang X, Xue T. Oxidative stress response in avian pathogenic Escherichia coli. Res Vet Sci. 2024;180:105426. 10.1016/j.rvsc.2024.105426. [DOI] [PubMed] [Google Scholar]
- 27.Kho CJY, Lau MML, Chung HH, Chew IYY, Gan HM. Whole-genome sequencing of Pseudomonas koreensis isolated from diseased Tor tambroides. Curr Microbiol. 2023;80(8):255. 10.1007/s00284-023-03354-5. [DOI] [PubMed] [Google Scholar]
- 28.Tian C, Di L, Dong S, Tian X, Huang D, Zhao Y, et al. Whole genome sequencing and genomic characteristics analysis of carbapenem-resistant Acinetobacter baumannii clinical isolates in two hospitals in China. Infect Genet Evol. 2024;123:105642. 10.1016/j.meegid.2024.105642. [DOI] [PubMed] [Google Scholar]
- 29.Baleivanualala SC, Isaia L, Devi SV, Howden B, Gorrie CL, Matanitobua S, et al. Molecular and clinical epidemiology of carbapenem resistant Acinetobacter baumannii ST2 in Oceania: a multicountry cohort study. Lancet Reg Health West Pac. 2023;40:100896. 10.1016/j.lanwpc.2023.100896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang WJ, Lu Z, Schwarz S, Zhang RM, Wang XM, Si W, et al. Complete sequence of the bla(NDM-1)-carrying plasmid pNDM-AB from Acinetobacter baumannii of food animal origin. J Antimicrob Chemother. 2013;68(7):1681–2. 10.1093/jac/dkt066. [DOI] [PubMed] [Google Scholar]
- 31.Nasser F, Gaudreau A, Lubega S, Zaker A, Xia X, Mer AS, et al. Characterization of the diversity of type IV secretion system-encoding plasmids in Acinetobacter. Emerg Microbes Infect. 2024;13(1):2320929. 10.1080/22221751.2024.2320929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brovedan MA, Cameranesi MM, Limansky AS, Morán-Barrio J, Marchiaro P, Repizo GD. What do we know about plasmids carried by members of the Acinetobacter Genus? World J Microbiol Biotechnol. 2020;36(8):109. 10.1007/s11274-020-02890-7. [DOI] [PubMed] [Google Scholar]
- 33.Drobiazko AY, Kasimova AA, Evseev PV, Shneider MM, Klimuk EI, Shashkov AS, et al. Capsule-Targeting depolymerases Derived from Acinetobacter baumannii Prophage regions. Int J Mol Sci. 2022;23(9). 10.3390/ijms23094971. [DOI] [PMC free article] [PubMed]
- 34.de Sousa JAM, Buffet A, Haudiquet M, Rocha EPC, Rendueles O. Modular prophage interactions driven by capsule serotype select for capsule loss under phage predation. Isme j. 2020;14(12):2980–96. 10.1038/s41396-020-0726-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Artuso I, Lucidi M, Visaggio D, Capecchi G, Lugli GA, Ventura M, et al. Genome diversity of domesticated Acinetobacter baumannii ATCC 19606(T) strains. Microb Genom. 2022;8(1). 10.1099/mgen.0.000749. [DOI] [PMC free article] [PubMed]
- 36.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482(7385):331–8. 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 37.Yang Z, Li B, Bu R, Wang Z, Xin Z, Li Z, et al. A highly efficient method for genomic deletion across diverse lengths in thermophilic parageobacillus thermoglucosidasius. Synth Syst Biotechnol. 2024;9(4):658–66. 10.1016/j.synbio.2024.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu Y, Lu J, Zhao J, Zhang X, Yu HH, Velkov T, et al. Complete genome sequence and genome-scale metabolic modelling of Acinetobacter baumannii type strain ATCC 19606. Int J Med Microbiol. 2020;310(3):151412. 10.1016/j.ijmm.2020.151412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coyne S, Rosenfeld N, Lambert T, Courvalin P, Périchon B. Overexpression of resistance-nodulation-cell division pump AdeFGH confers multidrug resistance in Acinetobacter baumannii. Antimicrob Agents Chemother. 2010;54(10):4389–93. 10.1128/aac.00155-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tambat R, Kinthada RK, Saral Sariyer A, Leus IV, Sariyer E, D’Cunha N, et al. AdeIJK pump-specific inhibitors effective against Multidrug Resistant Acinetobacter baumannii. ACS Infect Dis. 2024;10(6):2239–49. 10.1021/acsinfecdis.4c00190. [DOI] [PubMed] [Google Scholar]
- 41.Wang J, Zhou Z, He F, Ruan Z, Jiang Y, Hua X, et al. The role of the type VI secretion system vgrG gene in the virulence and antimicrobial resistance of Acinetobacter baumannii ATCC 19606. PLoS ONE. 2018;13(2):e0192288. 10.1371/journal.pone.0192288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gaddy JA, Arivett BA, McConnell MJ, López-Rojas R, Pachón J, Actis LA. Role of acinetobactin-mediated iron acquisition functions in the interaction of Acinetobacter baumannii strain ATCC 19606T with human lung epithelial cells, Galleria mellonella caterpillars, and mice. Infect Immun. 2012;80(3):1015–24. 10.1128/iai.06279-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The complete genome sequences of MDR A. baumannii Y03 including 1 chromosome sequence and 4 plasmids sequences using whole-genome sequencing have been deposited into the NCBI Genbank nucleotide sequence database under accession numbers CP163382, CP163383, CP163384, CP163385, and CP163386. The raw sequencing data of MDR A. baumannii Y03 are available in the NCBI Sequence Read Archive (SRA) database with the SRA accession number SRR30247697.