Abstract
Radiation-induced lung injury (RILI) is a common adverse effect of radiation therapy that negatively affects treatment progression and the quality of life of patients. Identifying biomarkers for RILI can provide reference for the prevention and treatment of RILI in clinical practice. In this study, to explore key proteins related to RILI, we constructed a rat model of RILI and analyzed RILI tissues and normal lung tissues using tandem mass spectrometry labeling and quantitative proteomics technology. We used Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment, Gene Ontology (GO) enrichment and protein–protein interaction (PPI) networks for bioinformatics analysis of Differentially expressed proteins (DEPs). The results identified 185 differentially expressed proteins in lung tissue from the RILI group compared with the controls, including 110 up-regulated proteins and 75 down-regulated proteins. GO analysis showed that the differentially expressed proteins were involved oxidation-reduction process, cellular biosynthetic processes and extracellular matrix. KEGG results demonstrated that the differentially expressed proteins were mainly involved in the PI3K-Akt, ECM receptor interactions, arachidonic acid metabolism, glutathione metabolism and other pathways. These results on the functions and signaling pathways of the differentially expressed proteins provide a theoretical basis for further study of the mechanism of RILI.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12014-024-09518-0.
Keywords: Radiation-induced lung injury, Proteomic, TMT
Background
Radiation therapy plays an important role in both curative and palliative tumor treatment [1]. However, during radiotherapy, normal tissues inevitably are exposed to irradiation. Radiation-induced lung injury (RILI) is a common complication of chest radiotherapy and includes early-stage radiation pneumonitis and late-stage radiation pulmonary fibrosis [2]. Two main mechanisms of RILI have been proposed: direct DNA damage and indirect DNA damage [3]. The common symptoms of RILI include cough, dyspnea, and low-grade fever. Imaging by CT scan of the chest also shows typical changes of poorly defined hyperdense shadows [4]. In lung cancer, while toxic side effects after radiotherapy have become less frequent through the improvement of modern radiotherapy equipment, the development of more advanced stereotactic radiotherapy techniques and the wide application of adjuvant drugs, the probability of symptomatic pneumonia still ranges from 9.4–28% [5, 6]. In patients with severe radiation pneumonitis, treatment progression and the quality of life are seriously affected [7]. Therefore, the search for biomarkers of RILI is essential to prevent the onset of the disease and improve the quality of life of patients.
Quantitative proteomics is a mass spectrometry–based high-throughput screening technique commonly used to quantify proteins and metabolites in complex samples. It is widely used to identify biological pathways and targets for disease treatment and to elucidate changes in target proteins involved in disease pathogenesis and drug intervention [8–11]. High-throughput proteomics combined with bioinformatics for data mining an effective approach for large-scale screening and the identification of disease-related proteins, the prediction of drug targets, and mechanisms of action [12]. Tandem mass tag (TMT) is a nanoscale LC-MS/MS-based labeling technique that allows accurate comparison of multiple samples simultaneously for quantitative analysis of proteins and peptides.
In this study, we established a model of RILI in rats and screened differential proteins in lung tissues using proteomic methods. To explore the potential mechanism of action of the differential proteins, we performed biological function and signaling pathway analyses. These findings may provide promising markers for RILI.
Materials and methods
Experimental animals
A total of 50 healthy male rats weighing 220 ± 20 g were purchased from Liaoning Changsheng Biotechnology Co. (China). The animal experiment protocol was approved by the Animal Experimentation Ethics Committee of the General Hospital of the Northern Theater of Operations of the Chinese People’s Liberation Army.
Establishment of the RILI rat model
The rats were randomly divided into an experimental group (R) and a control group (CG). The rats in the experimental group were anesthetized with isoflurane using a Reward R500 small animal respiratory anesthesia machine and fixed in the supine position. The irradiation field was positioned under a CT simulator; the right lung tissue was outlined in the Elekta Monaco planning system, and the right lung was vertically irradiated with a single 30 Gy using 6MV-X-rays with a source skin distance of 100 cm and a dose rate of 300 cGy/min. The rats in the control group were positioned and mock irradiated (no treatment) after anesthesia.
The model rats were treated as described above. On days 1, 3, 7, 14, and 28 (n = 5/group), the rats were weighed, anesthetized, and fixed on the dissection board in the supine position. After execution of the rats, the thoracic cavity was dissected immediately. The lung tissues were separated from the heart, and both lungs were stripped out of the thoracic cavity. The right lungs were stored at -80 °C in a freezer tube. The reagents and instruments are listed in Table 1 of the Supplementary Material. Lung tissues were fixed, dehydrated and paraffin-embedded. The specimen blocks were cut into 5-µm-thick sections. The sections were stained with hematoxylin-eosin (H&E) to observe inflammatory infiltration and the structural integrity of the alveoli.
Table 1.
The top 10 up-regulated proteins in lung tissue from RILI model rats
| Gene | Description | FC | Pvalue |
|---|---|---|---|
| TNC | tenascin precursor | 2.121856 | 0.000716 |
| BPIFB1 | BPI fold-containing family B member 1 isoform X1 | 1.854901 | 0.003632 |
| COL14A1 | collagen alpha-1(XIV) chain isoform X1 | 1.796428 | 0.008273 |
| AGR2 | anterior gradient protein 2 homolog isoform X1 | 1.726592 | 0.028186 |
| COL3A1 | collagen alpha-1(III) chain precursor | 1.63969 | 0.000204 |
| TOP2A | DNA topoisomerase 2-alpha | 1.594113 | 0.01909 |
| SCN3B | sodium channel subunit beta-3 isoform X1 | 1.580324 | 0.004611 |
| COL1A1 | collagen alpha-1(I) chain precursor | 1.578288 | 0.002988 |
| ACSM1 | acyl-coenzyme A synthetase ACSM1 | 1.548452 | 0.000884 |
| SCGB3A2 | secretoglobin family 3 A member 2 | 1.54808 | 0.001611 |
Sample preparation
Samples were ground individually in liquid nitrogen and lysed with PASP lysis buffer (100 mM NH4HCO3, 8 M Urea, pH 8), followed by 5 min of ultrasonication on ice. The lysate was centrifuged at 12,000 g for 15 min at 4 °C; the supernatant was reduced with 10 mM DTT for 1 h at 56 °C and subsequently alkylated with IAM for 1 h at room temperature in the dark. Then samples were mixed with 4 times the volume of precooled acetone by vortexing and incubated at -20 °C for at least 2 h. Samples were then centrifuged at 12,000 g for 15 min at 4 °C, and the precipitant was collected. The pellet was washed in 1 mL cold acetone and dissolved in dissolution buffer (8 M Urea, 100 mM TEAB, pH 8.5). A Bradford protein quantification kit was used for protein quality inspection.
TMT labeling of peptides
DB dissolution buffer (8 M Urea, 100 mM TEAB, pH 8.5) was added to protein samples to a final volume of 100 µL. Trypsin and 100 mM TEAB buffer were added; the sample was mixed and digested at 37 °C for 4 h. Trypsin and CaCl2 were added, and the sample was digested overnight. Formic acid was added and the pH was adjusted to under 3; the sample was centrifuged at 12,000 g for 5 min at room temperature. The supernatant was slowly loaded to a C18 desalting column, washed with washing buffer (0.1% formic acid, 3% acetonitrile) three times, and eluted by elution buffer (0.1% formic acid, 70% acetonitrile). The eluents were collected and lyophilized. Next, 100 µL of 0.1 M TEAB buffer was added to reconstitute the sample, and 41 µL of acetonitrile-dissolved TMT labeling reagent was added. The sample was mixed with shaking for 2 h at room temperature. The reaction was stopped by adding 8% ammonia. All labeling samples were mixed with equal volume, desalted and lyophilized.
Separation of fractions
Mobile phase A (2% acetonitrile, adjusted pH to 10.0 using ammonium hydroxide) and B (98% acetonitrile) were used for gradient elution. The lyophilized powder was dissolved in solution A and centrifuged at 12,000 g for 10 min at room temperature. The sample was fractionated using a C18 column (Waters BEH C18, 4.6 × 250 mm, 5 μm) on a Rigol L3000 HPLC system; the column oven was set as 45 °C. The eluates were monitored at UV 214 nm, collected for a tube per minute and combined into 10 fractions. All fractions were dried under vacuum and reconstituted in 0.1% (v/v) formic acid (FA) in water.
LC-MS/MS analysis
Shotgun proteomics analyses were performed using an EASY-nLCTM 1200 UHPLC system (Thermo Fisher) coupled with a Q Exactive™ HF-X mass spectrometer (Thermo Fisher) operating in the data-dependent acquisition (DDA) mode. The sample (1 µg) was injected into a homemade C18 Nano-Trap column (4.5 cm×75 μm, 3 μm). Peptides were separated on a homemade analytical column (15 cm×150 μm, 1.9 μm) using a linear gradient elution as listed in Table 2. The separated peptides were analyzed by Q Exactive™ HF-X mass spectrometer (Thermo Fisher), with an ion source of Nanospray Flex™ (ESI), spray voltage of 2.3 kV and ion transport capillary temperature of 320 °C. The full scan ranged from m/z 350 to 1500 with a resolution of 60,000 (at m/z 200); the automatic gain control (AGC) target value was 3 × 106 and maximum ion injection time was 20 ms. The top 40 precursors of the highest abundance in the full scan were selected and fragmented by higher energy collisional dissociation (HCD) and analyzed in MS/MS. The resolution was 45,000 (at m/z 200) for 10 plex, the AGC target value was 5 × 104, the maximum ion injection time was 86 ms, the normalized collision energy was set as 32%, intensity threshold was 1.2 × 105, and the dynamic exclusion parameter was 20 s.
Table 2.
The top 10 down-regulated proteins in lung tissue from RILI model rats
| Gene | Description | FC | Pvalue |
|---|---|---|---|
| PRG3 | proteoglycan 3 isoform X1 | 0.38806 | 0.023746 |
| MCPT1L1 | mast cell protease 1 preproprotein | 0.389339 | 0.003543 |
| PRG2 | bone marrow proteoglycan precursor | 0.392083 | 0.025202 |
| MCPT2 | mast cell protease 2 precursor | 0.392472 | 0.024099 |
| TLN1 | talin-1 isoform X2 | 0.429919 | 0.013958 |
| INMT | indolethylamine N-methyltransferase | 0.463709 | 0.004766 |
| LOC100361814 | eosinophil cationic protein | 0.471696 | 0.031394 |
| EPX | eosinophil peroxidase precursor | 0.487341 | 0.03147 |
| EAR1 | eosinophil cationic protein | 0.632912 | 0.019473 |
| VIPR1 | vasoactive intestinal polypeptide receptor 1 isoform X1 | 0.640019 | 0.000847 |
Data analysis
Proteome Discoverer 2.4 software was used for identification and quantitative analysis of raw data, with the following parameter settings: a mass tolerance of 10 ppm for precursor ions and 0.02 Da for fragment ions. Fixed modification was alkylation modification of cysteine, variable modification was oxidation modification of side chain methionine, peptide side chain TMT tag modification, N-terminal was TMT tag modification, acetylation modification, methionine loss, and methionine loss + acetylation, allowing up to two missing sites. In order to improve the quality of analysis results, the software PD 2.4 further filtered the retrieval results: Peptide Spectrum Matches (PSMs) with a credibility of more than 99% was identified PSMs. The identified protein contains at least 1 unique peptide. The identified PSMs and protein were retained and performed with FDR no more than 1.0%. The samples were also subjected to Principal ComponentAnalysis (PCA) and Repeatability Analysis. In the analysis of differentially expressed proteins between the experimental group and the control group, up-regulated proteins were defined as those with FC ≥ 1.2 and P-value ≤ 0.05 and down-regulated proteins were defined as those with FC ≤ 0.83 and P-value ≤ 0.05.
Bioinformatics analysis
To study the biological function of differential proteins, we applied R software to perform Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. We enriched the structural domains of proteins by InterProScan. CELLO software was used for subcellular localization analysis.The potential protein–protein interactions (PPI) were analyzed using the STRING database (https://string-db.org) and CytoScape 3.9.0 software.
Western blotting
Protein concentration was determined using the BCA kit (coolaber, China). Protein samples were separated using sodium dodecyl sulfate poly acrylamide gel electrophoresis and transferred onto a polyvinylidene fluoride membraneand blocked with 5% non-fat milk for 2 h.The membrane was incubated with the primary antibody overnight at 4 ◦C. Then the membrane was washed in Tris-buffered saline and Tween (TBST) and incubated with the corresponding secondary antibody for 2 h at room temperature. The membrane was washed with TBST four times and then developed using the ECL kit (Beyotime, China) to visualize the target protein band. The primary antibodies used were as follows: TNC (1:1000, abcam, UK), COL1A1 (1:1000, Santa, USA), COL3A1 (1:1000, Servicebio, China),α-Tubulin(1:1000, Santa, USA).
Statistical analysis
Data analysis and visualization were conducted using GraphPad Prism 9.0 software. Comparison of means between two groups was performed utilizing Student’s t-test, and statistical significance was considered at P < 0.05. (*P < 0.05, ** P < 0.01 and *** P < 0.001).
Results
Confirmation of the RILI rat model
We established the RILI model in rats as described in the Methods and monitored animals after irradiation. After 1 day of irradiation the model group rats showed a decrease in activity and reduced eating compared with controls. After 3 days of irradiation, the rats occasionally coughed and exhibited dry and dull fur; the irradiation field area began to shed hair. After 7 days of irradiation, the activity of the rats was average, with coughing; some rats had oral and nasal secretions. After 14 days of irradiation, the exudation and ulceration symptoms in the irradiation field area of the model rats gradually improved. After 28 days of irradiation, the skin in the irradiation field area of the rats was repaired, and the hair increased compared compared to before. No animal deaths were observed during the observation period.
Pathological characteristics of irradiated rats
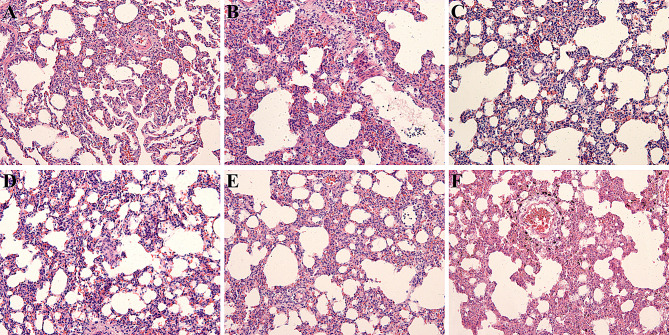
We next examined lung injury pathology on samples obtained on days 1, 3, 7, 14, and 28. The alveolar structure of the normal lung tissue of the control group rats was relatively clear, with uniform alveolar size and intact mucosal epithelial structure. In the RILI model group, some alveolar walls collapsed and thickened, and interstitial cells became congested, with varying degrees of inflammatory cell infiltration (Fig. 1). Pathology assessment of lung tissue showed the highest inflammatory reaction in lung tissue on day 7 after irradiation in rats. We thus used the right lung specimen of rats on day 7 after irradiation in proteomic analysis.
Fig. 1.
Pathologic changes in lung tissue. A. Pathologic changes in lung tissue samples in the control group. B–F. Pathologic changes in lung tissue samples in the irradiated groups on days 1, 3, 7, 14, and 28
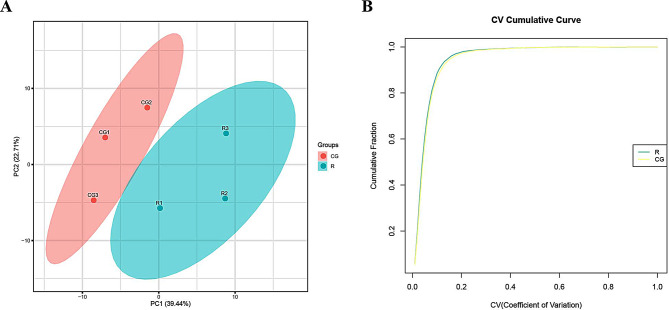
Principal component analysis (PCA) and repeatability analysis
We performed principal component analysis (PCA) and repeatability analysis on both sets of samples. Principal component analysis reflects the differences in samples between and within groups. Figure 2A shows the distribution of lung tissue samples in the two groups; a higher degree of aggregation of the samples represents less inter-sample variability and better reproducibility. The Coefficient of Variance (CV), which reflects the degree of dispersion of the data and determines the reproducibility, indicates that the smaller the CV value, the better the reproducibility. Figure 2B shows the cumulative graph of the CV values of the two groups of samples, the faster the curve rises, the better the repeatability of the sample as a whole.
Fig. 2.
Principal component analysis (PCA) and repeatability analysis. A. PCA Analysis. B. Repeatability CV analysis
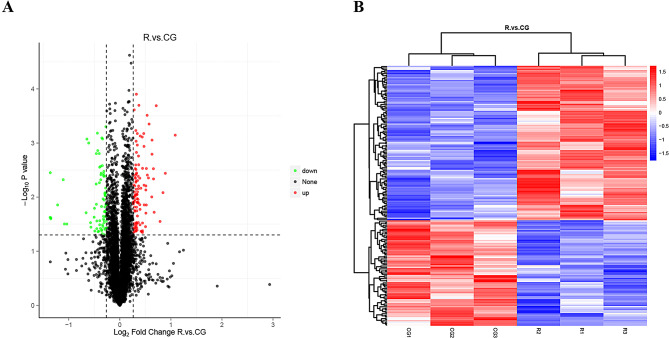
Identification of differentially expressed proteins
We next analyzed lung tissues in RILI model rats after 7 days of irradiation and controls by proteomic screening. A total of 185 differential proteins were identified, including 110 up-regulated proteins and 75 down-regulated proteins. The volcano and heat maps of the differential proteins are shown in Fig. 3. The top 10 up- and down-regulated proteins are listed in Tables 1 and 2. These include mainly Tenascin C (TNC)、(collagen alpha-1(III) chain precursor) COL3A1、(collagen alpha-1(I) chain precursor) COL1A1、BPI fold-containing family B member 1 (BPIFB1) and topoisomerase IIalpha(TOP2A).
Fig. 3.
Volcano plot and heat map of DEPs between RILI tissue and normal lung tissue. A. Volcano plot of DEPs. B. Heat map of DEPs
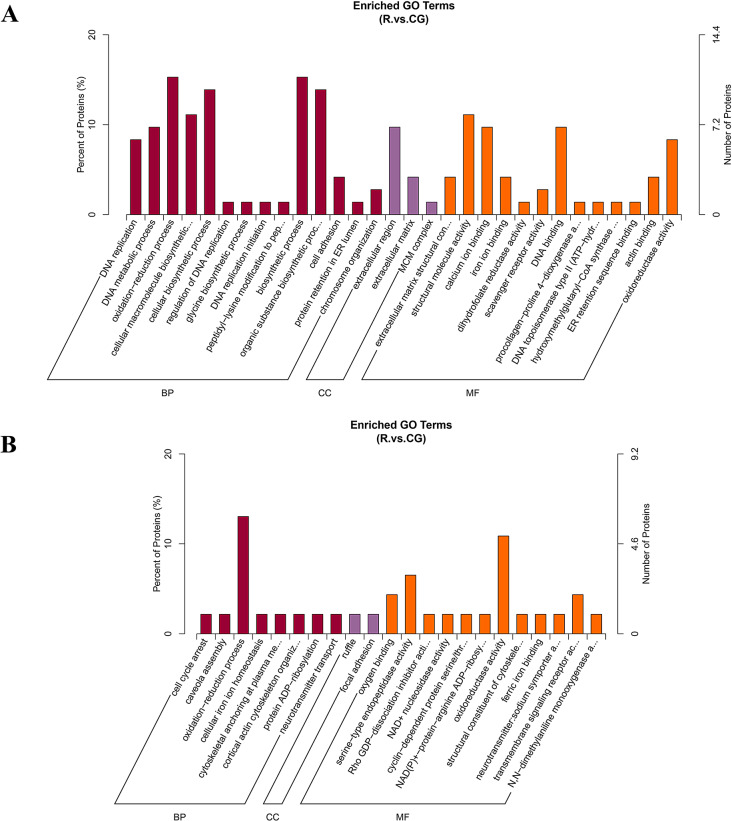
GO and KEGG annotation analyses of the differentially expressed proteins
To explore the function of differential proteins and the related signaling pathways of the differentially expressed proteins, we performed GO and KEGG analyses. The GO analysis results are shown in Fig. 4. The up-regulated proteins were mainly involved in oxidation-reduction process, biosynthetic processes, and cellular biosynthetic processes in the biological process (BP) category. In the cellular component (CC) category, the proteins were mainly involved in the extracellular region and extracellular matrix. In the molecular function (MF) category, they mainly regulate structural molecule activity and DNA binding. The down-regulated proteins were mainly involved in oxidation-reduction process, focal adhesion, oxidoreductase activity, and serine-type endopeptidase activity.
Fig. 4.
GO enrichment analysis. A. GO enrichment analysis of up-regulated differentially expressed proteins. B. GO enrichment analysis of down-regulated differentially expressed proteins
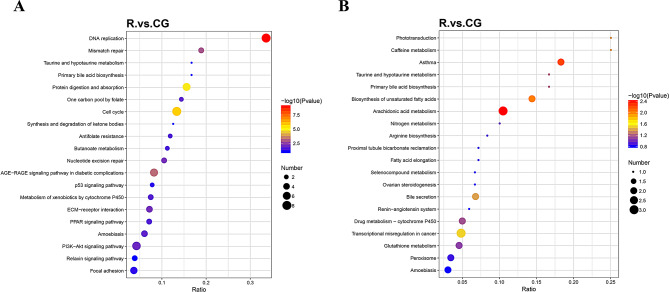
KEGG pathway enrichment analysis revealed that the up-regulated proteins were mainly enriched in cell cycle, DNA replication, PI3K-Akt signaling pathway, and ECM receptor interaction. The down-regulated proteins were mainly enriched in arachidonic acid metabolism, transcriptional misregulation in cancer, and glutathione metabolism. KEGG analysis results are shown in Fig. 5.
Fig. 5.
KEGG enrichment analysis. A. KEGG enrichment analysis of up-regulated differentially expressed proteins. B. KEGG enrichment analysis of down-regulated differentially expressed proteins
Subcellular localization and domain analysis of the differentially expressed proteins
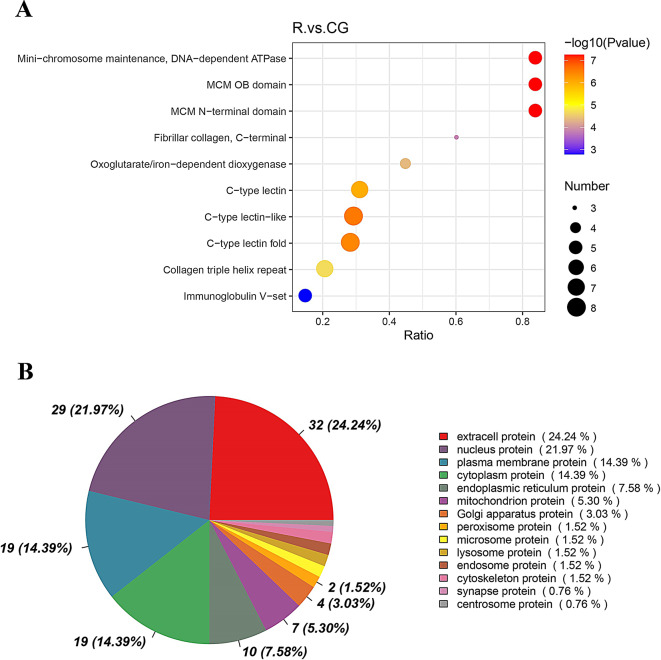
Structural domain analysis revealed a high number of differential proteins containing C-type lectin-like, C-type lectin folded, and collagen triple helix repeat domains. Identifying the subcellular localization of proteins can help provide clues into the cellular function of proteins. The differential proteins were mainly distributed in extracellular protein, nuclear proteins, and plasma membrane proteins (Fig. 6).
Fig. 6.
Classification of DEPs based on domain and subcellular localization. A. Number of proteins containing the indicated structural domains. B. Distribution of the differential proteins in each subcellular organelle
Protein–protein interaction analysis
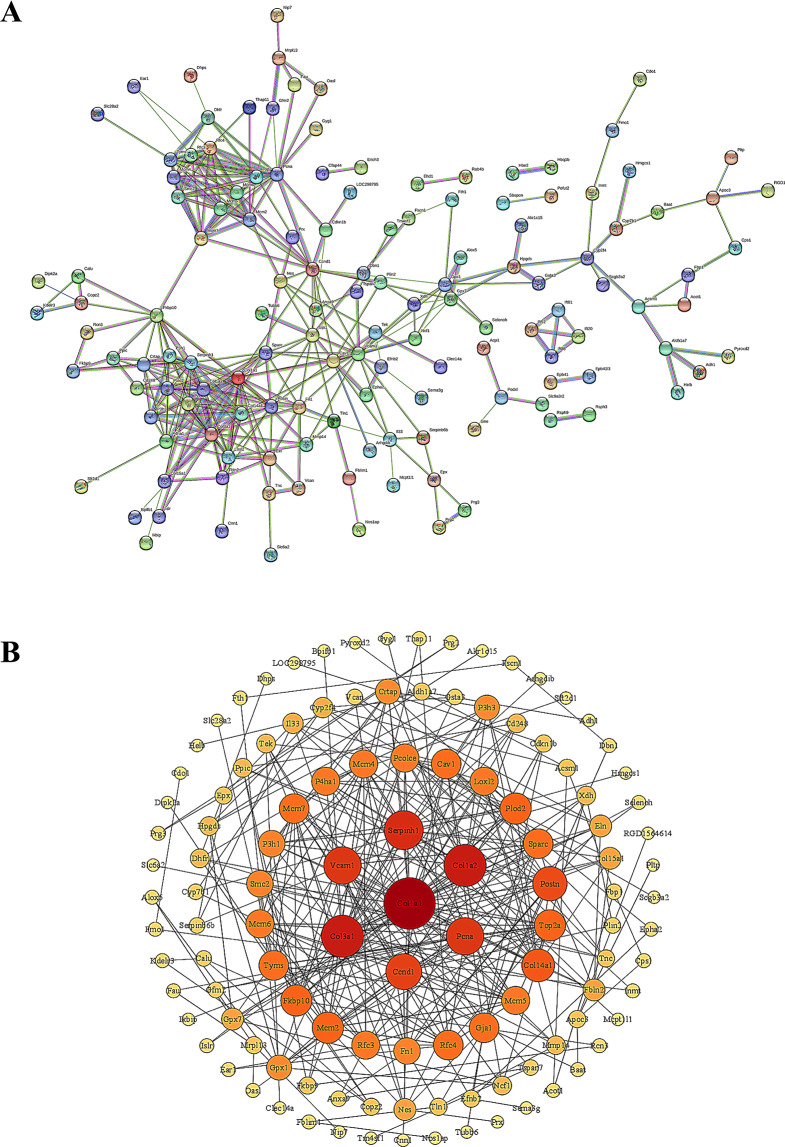
To explore the relationship between the differential proteins, we used the STRING database to construct PPI networks. CytoScape software was applied to visualize the protein–protein interaction network. We selected protein interactions with confidence score > 0.4. The results showed that Col1a1 was the core of the PPI network and serve as the key proteins that interact with other differential proteins, suggesting that they may have an important role in the pathogenesis of RILI (Fig. 7).
Fig. 7.
PPI analysis and CytoScape visualization. A. PPI analysis of DEPs. B. CytoScape visualization of DEPs
Western blotting
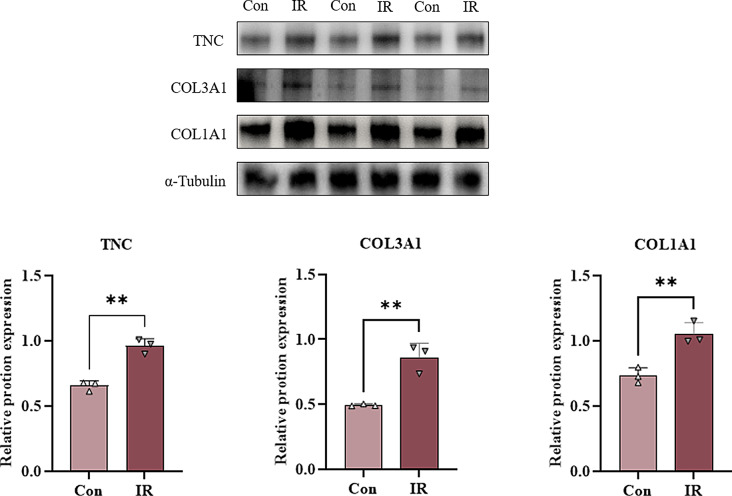
We performed western blotting experiments on the screened differentially expressed proteins TNC, COL3A1, and COL1A1, and the results showed that the expression of TNC, COL3A1, and COL1A1 was up-regulated in the irradiation group compared with the control group, and the difference was statistically significant (Fig. 8).
Fig. 8.
Western blotting for TNC, COL3A1, COL1A1 protein expression in normal and radiotherapy groups of rats. (Con: control group IR: irradiation group; *P < 0.05, **P < 0.01 and ***P < 0.001)
Discussion
RILI is a dynamic response process involving multiple cells and factors and is characterized by an inflammatory phenotype and impaired cellular repair [13]. Inflammation is a key stage in RILI, and the characteristic pathological changes of RILI are the accumulation of alveolar and interstitial macrophages [14]. The lung is one of the tissues most sensitive to ionizing radiation, and DNA and organelle damage cause the release of transforming growth factor beta, platelet-derived growth factor, and interleukin 1 within minutes of exposure [15]. In clinical practice, RILI not only affects the treatment of patients but also affects their quality of life. Therefore, the search for reliable biomarkers can help to screen sensitive populations for RILI and target these individuals for prevention and treatment.
High-throughput proteomics technology has been widely used to find biomarkers and drug targets for diseases. TMT-based proteomics is an in vitro peptide-labeled quantitative technique that involves amino labeling to simultaneously achieve qualitative and quantitative proteomic characterization of multiple samples by high-precision mass spectrometry analysis with high reproducibility and sensitivity [16].
In this study, we screened differentially expressed proteins in RILI by establishing a rat model of RILI and performed TMT proteomics in RILI lung tissues. The results identified 185 differentially expressed proteins, including 110 up-regulated proteins and 75 down-regulated proteins. These include mainly Tenascin C (TNC)、COL3A1 (collagen alpha-1(III) chain precursor)、COL1A1 (collagen alpha-1(I) chain precursor)、BPI fold-containing family B member 1 (BPIFB1) and topoisomerase IIalpha (TOP2A).
Tenascin C (TNC) is a multifunctional extracellular matrix (ECM) protein involved in many cellular processes in embryonic development. In a study analyzing primary fibroblast cultures from fibrotic and healthy patients, TNC was barely detected in normal lungs and significantly up-regulated in fibrotic lungs, the results indicate that differences in ECM glycoprotein content in interstitial lung diseases may be related to the development of pulmonary fibrosis [17]. TNC also plays an important role in lung cancer, and novel therapeutic agents targeting TNC or TNC-regulated pathways may offer new avenues of treatment for lung disease [18].
Collagen alpha-1(III) chain, also known as the alpha 1 chain of type III collagen, is a protein that in humans is encoded by the COL3A1 gene, Accumulation of type III collagen is a hallmark of several chronic human diseases that involve fibrotic processes including systemic sclerosis, cardiac fibrosis, lung fibrosis, liver cirrhosis and renal fibrosis [19]. It was found that in a mouse model to study the mechanisms of chronic allergic airway inflammation, proteins of nerve growth factor induced the expression of type III collagen, which exerts pro-fibrotic activity in the airways [20]. It has also been shown that radiotherapy significantly upregulates the expression of COL1A1 and COL3A1 genes in breast cancer tumour tissues, and their upregulation may be a risk factor for radiation-associated secondary non-breast diseases in breast cancer patients [21].Collagen type I alpha 1 (COL1A1) is a member of group I collagen which include COL1A1 and COL1A2. Collagen is the major protein of bone, tendon and teeth and is involved in tumor cell adhesion, gap junctions and extracellular matrix (ECM) [22]. Studies have shown that radiation-induced tissue fibrosis occurs as a result of collagen over-synthesis [23]. In a mouse model receiving hepatocyte growth factor (HGF) gene therapy with mesenchymal stem cells (MSCs), it was found that MSCs-HGF reduced the expression levels of transforming growth factor-β, fibrotic factors of Col1a1 and Col3a1, and inhibited the fibrotic process [24]. One study found that Col1a1 and Col1a2 may be important markers of obesity-induced cardiac fibrosis by single-cell transcriptomics, with fibroblasts playing a key role [25]. We demonstrated that the above proteins were significantly up-regulated in the radiotherapy group of rats by westren blot results of TNC, COL3A1, COL1A1 proteins, which was consistent with our high-throughput TMT proteomics results and proved the reliability of the assay results.
BPI fold-containing family B member 1 (BPIFB1) is member of the PI-fold-containing family [26]. This protein is specifically expressed in the respiratory system. Previous studies have shown that BPIFB1 is abnormally expressed in cancerous tissues such as nasopharyngeal and gastric cancers and regulates chronic infection and inflammation, suggesting that it may play an important role in the development of tumors and inflammation [27]. Topoisomerase 2-α (TOP2A) is a key nuclear enzyme involved in metabolic processes such as replication, transcription, and chromosome segregation during mitosis, and several studies have demonstrated that TPO2A is highly expressed in tumors and correlates with prognosis [28–31]. Studies indicated that TOP2A may be a potential diagnostic biomarker for severe COVID-19 [32], COVID-19 is also an inflammatory response in which viral invasion causes diffuse lung injury, edema, and hyaline membrane formation. Thus, they may also be involved in the biological processes of radiation lung injury and play a role in the development of the disease.
The above results suggest that the differential proteins such as TNC that were screened in this study may play an important role in the inflammatory response induced by RILI. These proteins represent promising candidates as biomarkers for RILI.
GO enrichment analysis demonstrated that DEPs were significantly associated with redox processes, DNA metabolic processes, and regulation of oxidoreductase activity. KEGG analysis results indicated that differential proteins were mainly involved in DNA replication, cell cycle, arachidonic acid metabolism, PI3K-Akt signaling pathway, and ECM receptor interactions. Previous studies showed that cell cycle alterations and cell loss in late G2/M blockade after radiotherapy promotes the manifestation of RILI [33]. At the cellular level, radiation activates the production of free radicals, leading to DNA damage, apoptosis, cell cycle alterations, and reduced cell viability [34]. Most cancers exhibit hyperactivation of the PI3K/AKT/mTOR signaling pathway [35]. The PI3K/AKT pathway protects or promotes regeneration after radiation injury [36]. SOX9-expressing cells promote regeneration after RILI through the PI3K/AKT pathway [37]. A previous study found that serum TGF-β levels were predictive of RILI in patients treated with radiation for advanced lung cancer [38]. The PI3K/AKT pathway is an important downstream pathway of TGFβ; it is involved in the pathogenesis of inflammatory and fibrotic diseases and regulates several key events in the inflammatory response to injury [39]. A prospective study indicated that genetic variation in the PI3K/AKT pathway was significantly associated with grade ≥ 3 radiation pneumonitis and may serve as a predictor of radiation pneumonitis [40]. Our KEGG analysis indicated that the differential proteins identified in RILI model rats were significantly enriched in the PI3K/AKT pathway. Therefore, this pathway may be involved in the mechanism underlying the development of RILI.
The extracellular matrix (ECM) is an important structure that provides structure and support for all organs, including the lungs, and is critical for the regulation of organ development, homeostasis, and damage repair. ECM-based therapeutic and bioengineering research has become a new hotspot for lung regeneration and treatment of chronic lung diseases [41]. Choi et al. [42] showed that TNC contributes to the integrity of the ECM. Of all the lung ECM proteins, type I collagen is the most abundant; it has tensile strength and in many fibrotic lung diseases, it increases collagen expression to influence pericellular stiffness. One study found that TNC upregulates type I collagen expression by activating the TGF-β signaling pathway and increasing TGF-β1 mRNA levels [43]. Velázquez et al. [44] analyzed TNC, fibronectin 1 (FBN1), and α-2 collagen chain (α-2) proteins by LC-MS/MS quantitative proteomics analysis in fibroblast cell lines isolated from idiopathic pulmonary fibrosis (IPF) donors compared with extracellular vesicles (EVs) derived from fibroblast cell lines isolated from healthy donors. MS label-free quantitative proteomics analysis revealed that differential proteins such as TNC, fibrillogenin 1 (FBN1), α-2 collagen chain (I) (COL1A2), and α-1 collagen chain (I) (COL1A1), which are involved in the fibrotic process, were detected in EVs isolated from IPF cell lines. Our PPI protein network analysis results also indicated that COL1A2 and COL1A1 occupy important positions in the interaction network and may be the key proteins that interact with other differential proteins. Therefore, the differential proteins identified in this study may influence the occurrence of RILI through ECM interactions.
Arachidonic acid (AA) is a 20-carbon chain polyunsaturated fatty acid with four double bonds. The four double bonds of AA make it susceptible to oxidation, which leads to excess metabolites important for the normal function of the immune system, the promotion of allergies and inflammation, and anti-inflammatory properties [45]. Modification of the protein lysine by the arachidonic acid–derived γ-ketoaldehyde isomer was found in a mouse model of radiation-induced pulmonary fibrosis, suggesting that this modification may contribute to the development of radiological lung injury [46]. Several other studies have found that some herbal medicines exert anti-inflammatory effects through the arachidonic acid metabolic pathway and attenuate lung injury in mouse models [47, 48]. These studies suggest that arachidonic acid metabolism plays an important role in lung injury and repair.
In summary, the differential proteins such as TNC screened in this study are closely related to the pathophysiological conditions of radiological lung injury and may regulate the occurrence and development of radiological lung injury through a variety of pathways such as the PI3K/AKT pathway and the ECM. These findings provide a basis for subsequent research and potential clinical application. However, there are some limitations of this study, such as the biological functions of the differentially expressed proteins were not thoroughly investigated. Further exploration such as some in vitro experiments such as cellular experiments are needed in the future.
Conclusion
In this study, TMT-tagged proteomics technology was used to identify a variety of differentially expressed proteins to explore the pathogenesis of radiological lung injury in model rats. Our results suggest that the differentially expressed proteins, such as TNC, and pathways, such as PI3KA-Akt and ECM receptor interactions, may play important roles in RILI.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Author contributions
JL and KG conduct experiments and write manuscripts, XR, DY and HW organize and analyze data, HZ and TW are responsible for image processing, YX and YY are responsible for organizing ideas and editing manuscripts, all the authors listed here read and approved the publication of the manuscript.
Funding
None to declare.
Data availability
No datasets were generated or analysed during the current study.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jing Liu and Kuanke Gao contributed equally to this work.
Contributor Information
Ying Xu, Email: 514705167@qq.com.
Ying Yan, Email: yanyingdoctor@sina.com.
References
- 1.Hanania AN, Mainwaring W, Ghebre YT, Hanania NA, Ludwig M. Radiation-Induced Lung Injury. Chest. 2019;156(1):150–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bledsoe TJ, Nath SK, Decker RH. Radiation Pneumonitis. Clin chest med. 2017;38(2):201–8. [DOI] [PubMed] [Google Scholar]
- 3.Azzam EI, Jay-Gerin JP, Pain D. Ionizing radiation-induced metabolic oxidative stress and prolonged cell injury. Cancer lett. 2012;327(1–2):48–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang X, Shin YK, Zheng Z, Zhu L, Lee IJ. Risk of radiation-induced pneumonitis after helical and static-port tomotherapy in lung cancer patients and experimental rats. Radiat Oncol. 2015;10(null):195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palma DA, Senan S, Tsujino K, Barriger RB, Rengan R, Moreno M, Bradley JD, Kim TH, Ramella S, Marks LB, et al. Predicting radiation pneumonitis after chemoradiation therapy for lung cancer: an international individual patient data meta-analysis. Int j radiat oncol. 2013;85(2):444–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ueki N, Matsuo Y, Togashi Y, Kubo T, Shibuya K, Iizuka Y, Mizowaki T, Togashi K, Mishima M, Hiraoka M. Impact of pretreatment interstitial lung disease on radiation pneumonitis and survival after stereotactic body radiation therapy for lung cancer. J thorac oncol. 2015;10(1):116–25. [DOI] [PubMed] [Google Scholar]
- 7.Bernchou U, Christiansen RL, Asmussen JT, Schytte T, Hansen O, Brink C. Extent and computed tomography appearance of early radiation induced lung injury for non-small cell lung cancer. Radiother oncol. 2017;123(1):93–8. [DOI] [PubMed] [Google Scholar]
- 8.Eckhard U, Marino G, Butler GS, Overall CM. Positional proteomics in the era of the human proteome project on the doorstep of precision medicine. Biochimie. 2016;122(null):110–8. [DOI] [PubMed] [Google Scholar]
- 9.Pan S, Ding A, Li Y, Sun Y, Zhan Y, Ye Z, Song N, Peng B, Li L, Huang W, et al. Small-molecule probes from bench to bedside: advancing molecular analysis of drug-target interactions toward precision medicine. Chem soc rev. 2023;52(16):5706–43. [DOI] [PubMed] [Google Scholar]
- 10.Pinto AFM, Diedrich JK, Moresco JJ, Yates JR. Differential Precipitation of Proteins: A Simple Protein Fractionation Strategy to Gain Biological Insights with Proteomics. J am soc mass spectr. 2023;34(9):2025–33. [DOI] [PubMed] [Google Scholar]
- 11.Sahu I, Zhu H, Buhrlage SJ, Marto JA. Proteomic approaches to study ubiquitinomics. Bba-gene regul mech. 2023;1866(2):194940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan ZH, Yu X, Duan YD, Liu P, Jiang YY, Zhang W, Liu B. [Method and application of proteomics in study of targets of traditional Chinese medicines]. Zhongguo Zhong Yao Za Zhi. 2020;45(5):1034–8. [DOI] [PubMed] [Google Scholar]
- 13.Lu L, Sun C, Su Q, Wang Y, Li J, Guo Z, Chen L, Zhang H. Radiation-induced lung injury: latest molecular developments, therapeutic approaches, and clinical guidance. Clin exp med. 2019;19(4):417–26. [DOI] [PubMed] [Google Scholar]
- 14.Roy S, Salerno KE, Citrin DE. Biology of Radiation-Induced Lung Injury. Semin Radiat Oncol. 2021;31(2):155–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khan MA, Van Dyk J, Yeung IW, Hill RP. Partial volume rat lung irradiation; assessment of early DNA damage in different lung regions and effect of radical scavengers. Radiother oncol. 2003;66(1):95–102. [DOI] [PubMed] [Google Scholar]
- 16.Lan Y, Zeng X, Xiao J, Hu L, Tan L, Liang M, Wang X, Lu S, Long F, Peng T. New advances in quantitative proteomics research and current applications in asthma. Expert rev proteomic. 2021;18(12):1045–57. [DOI] [PubMed] [Google Scholar]
- 17.Estany S, Vicens-Zygmunt V, Llatjós R, Montes A, Penín R, Escobar I, Xaubet A, Santos S, Manresa F, Dorca J, et al. Lung fibrotic tenascin-C upregulation is associated with other extracellular matrix proteins and induced by TGFβ1. BMC Pulm Med. 2014;14(null):120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donovan C, Bai X, Chan YL, Feng M, Ho KF, Guo H, Chen H, Oliver BG. Tenascin C in Lung Diseases. Biology (Basel). 2023;12(2):null. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuivaniemi H, Tromp G. Type III collagen (COL3A1): Gene and protein structure, tissue distribution, and associated diseases. Gene. 2019;707(null):151–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kılıç A, Sonar SS, Yildirim AO, Fehrenbach H, Nockher WA, Renz H. Nerve growth factor induces type III collagen production in chronic allergic airway inflammation. J Allergy Clin Immunol. 2011;128(5):1058–e10661051. [DOI] [PubMed] [Google Scholar]
- 21.Yao G, Zhao K, Bao K, Li J. Radiation increases COL1A1, COL3A1, and COL1A2 expression in breast cancer. Open Med (Wars). 2022;17(1):329–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu S, Liao G, Li G. Regulatory effects of COL1A1 on apoptosis induced by radiation in cervical cancer cells. Cancer Cell Int. 2017;17(null):73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang J, Wang Q, Hu Y, Qi Z, Lin Z, Ying W, Zhou M. Proteomic Profiling for Serum Biomarkers in Mice Exposed to Ionizing Radiation. Dose Response. 2019;17(4):1559325819894794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang H, Yang YF, Zhao L, Xiao FJ, Zhang QW, Wen ML, Wu CT, Peng RY, Wang LS. Hepatocyte growth factor gene-modified mesenchymal stem cells reduce radiation-induced lung injury. Hum gene ther. 2013;24(3):343–53. [DOI] [PubMed] [Google Scholar]
- 25.Pan X, Chen X, Ren Q, Yue L, Niu S, Li Z, Zhu R, Chen X, Jia Z, Zhen R, et al. Single-cell transcriptomics identifies Col1a1 and Col1a2 as hub genes in obesity-induced cardiac fibrosis. Biochem Biophys Res Commun. 2022;618:30–7. [DOI] [PubMed] [Google Scholar]
- 26.Wei F, Tang L, He Y, Wu Y, Shi L, Xiong F, Gong Z, Guo C, Li X, Liao Q, et al. BPIFB1 (LPLUNC1) inhibits radioresistance in nasopharyngeal carcinoma by inhibiting VTN expression. Cell Death Dis. 2018;9(4):432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J, Xu P, Wang L, Feng M, Chen D, Yu X, Lu Y. Molecular biology of BPIFB1 and its advances in disease. Ann transl med. 2020;8(10):651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cai H, Zhu X, Qian F, Shao B, Zhou Y, Zhang Y, Chen Z. High expression of TOP2A gene predicted poor prognosis of hepatocellular carcinoma after radical hepatectomy. Transl cancer res. 2020;9(2):983–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gong MC, Chen WQ, Jin ZQ, Lyu J, Meng LH, Wu HY, Chen FH. Prognostic Value and Significant Pathway Exploration Associated with TOP2A Involved in Papillary Thyroid Cancer. Int J Gen Med. 2021;14(null):3485–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ong KH, Lai HY, Sun DP, Chen TJ, Huang SK, Tian YF, Chou CL, Shiue YL, Chan TC, Li CF, et al. Prognostic Significance of DNA Topoisomerase II Alpha (TOP2A) in Cholangiocarcinoma. Front biosci-landmrk. 2023;28(4):75. [DOI] [PubMed] [Google Scholar]
- 31.Zeng S, Liu A, Dai L, Yu X, Zhang Z, Xiong Q, Yang J, Liu F, Xu J, Xue Y, et al. Prognostic value of TOP2A in bladder urothelial carcinoma and potential molecular mechanisms. BMC Cancer. 2019;19(1):604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu S, Long J, Liang T, Lv M, Huang X, Liang X, Su L, Zhou L. Bioinformatics analysis based on high-throughput sequencing data to identify hub genes related to different clinical types of COVID-19. Funct integr genomic. 2023;23(1):71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Willner J, Vordermark D, Schmidt M, Gassel A, Flentje M, Wirtz H. Secretory activity and cell cycle alteration of alveolar type II cells in the early and late phase after irradiation. Int j radiat oncol. 2003;55(3):617–25. [DOI] [PubMed] [Google Scholar]
- 34.Rajan Radha R, Chandrasekharan G. Pulmonary injury associated with radiation therapy – Assessment, complications and therapeutic targets. Biomed Pharmacother. 2017;89:1092–104. [DOI] [PubMed] [Google Scholar]
- 35.Thorpe LM, Yuzugullu H, Zhao JJ. PI3K in cancer: divergent roles of isoforms, modes of activation and therapeutic targeting. Nat rev cancer. 2015;15(1):7–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang L, Wang R, Gao Y, Xu X, Fu K, Wang S, Li Y, Peng R. The protective role of interleukin-11 against neutron radiation injury in mouse intestines via MEK/ERK and PI3K/Akt dependent pathways. Digest dis sci. 2014;59(7):1406–14. [DOI] [PubMed] [Google Scholar]
- 37.Chen S, Li K, Zhong X, Wang G, Wang X, Cheng M, Chen J, Chen Z, Chen J, Zhang C et al. Sox9-expressing cells promote regeneration after radiation-induced lung injury via the PI3K/AKT pathway. Stem Cell Res Ther 2021, 12(1). [DOI] [PMC free article] [PubMed]
- 38.Seto Y, Kaneko Y, Mouri T, Shimizu D, Morimoto Y, Tokuda S, Iwasaku M, Yamada T, Takayama K. Changes in serum transforming growth factor-beta concentration as a predictive factor for radiation-induced lung injury onset in radiotherapy-treated patients with locally advanced lung cancer. Transl lung cancer r. 2022;11(9):1823–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hawkins PT, Stephens LR. PI3K signalling in inflammation. Biochim biophys acta. 2015;1851(6):882–97. [DOI] [PubMed] [Google Scholar]
- 40.Tang Y, Liu B, Li J, Wu H, Yang J, Zhou X, Yi M, Li Q, Yu S, Yuan X. Genetic variants in PI3K/AKT pathway are associated with severe radiation pneumonitis in lung cancer patients treated with radiation therapy. Cancer Med. 2016;5(1):24–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou Y, Horowitz JC, Naba A, Ambalavanan N, Atabai K, Balestrini J, Bitterman PB, Corley RA, Ding BS, Engler AJ, et al. Extracellular matrix in lung development, homeostasis and disease. Matrix biol. 2018;73(null):77–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choi YE, Song MJ, Hara M, Imanaka-Yoshida K, Lee DH, Chung JH, Lee ST. Effects of Tenascin C on the Integrity of Extracellular Matrix and Skin Aging. Int J Mol Sci. 2020;21(22):null. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hackett TL, Vriesde N, Al-Fouadi M, Mostaco-Guidolin L, Maftoun D, Hsieh A, Coxson N, Usman K, Sin DD, Booth S, et al. The Role of the Dynamic Lung Extracellular Matrix Environment on Fibroblast Morphology and Inflammation. Cells. 2022;11(2):null. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Velázquez-Enríquez JM, Santos-Álvarez JC, Ramírez-Hernández AA, Reyes-Jiménez E, López-Martínez A, Pina-Canseco S, Aguilar-Ruiz SR, Romero-Tlalolini M, Castro-Sánchez L, Arellanes-Robledo J, et al. Proteomic Analysis Reveals Key Proteins in Extracellular Vesicles Cargo Associated with Idiopathic Pulmonary Fibrosis In Vitro. Biomedicines. 2021;9(8):null. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hanna VS, Hafez EAA. Synopsis of arachidonic acid metabolism: A review. J adv res. 2018;11(null):23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mont S, Davies SS, Roberts Second LJ, Mernaugh RL, McDonald WH, Segal BH, Zackert W, Kropski JA, Blackwell TS, Sekhar KR et al. Accumulation of isolevuglandin-modified protein in normal and fibrotic lung. Sci Rep 2016, 6(null):24919. [DOI] [PMC free article] [PubMed]
- 47.Ruan Y, Fan Y, Xie Y, Ma C, Mo B, Lai Y, Li G, Liu X, Kuang W. Modified Xiaoqinglong decoction alleviates lipopolysaccharide-induced acute lung injury in mice by regulating arachidonic acid metabolism and exerting anti-apoptotic and anti-inflammatory effects. Anat rec. 2022;305(7):1672–81. [DOI] [PubMed] [Google Scholar]
- 48.Tang C, Tang Y, Wang Q, Chu D, Zhou J, Zhou Y. Yangyinqingfei decoction attenuates PM2.5-induced lung injury by enhancing arachidonic acid metabolism. Front Pharmacol. 2022;13(null):1056078. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed during the current study.