Abstract
Hippophae gyantsensis is a dioecious plant endemic to the Qinghai-Tibet Plateau and is significant for ecological restoration and sand stabilization. Its fruit is rich in bioactive compounds that offer economic potential. However, the inability to distinguish sexes before flowering and prolonged maturation hinder breeding and cultivation. We performed whole-genome resequencing on male and female plants, identified large insertion/deletion (InDel) variants, and developed two sex-specific primers (Higy_04 and Higy_06). These primers enable rapid, accurate PCR-based sex identification. All sex-specific sites were located on chromosome 2, suggesting its potential role as the sex chromosome. Additionally, we found a 1:1 sex ratio among offspring from the same mother plant, consistent with Mendelian inheritance, indicating that sex segregation is mainly genetically controlled. This work lays the foundation for developing molecular markers applicable across the entire genus Hippophae and contributes to understanding sex chromosome formation and adaptive evolution within the genus.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-024-05978-6.
Keywords: Hippophae gyantsensis, Sex-specific molecular markers, Whole-genome resequencing, Chromosome 2, Sex determination mechanisms
Introduction
Hippophae gyantsensis, belonging to the genus Hippophae of the family Elaeagnaceae, is an endemic species of the Qinghai-Tibet Plateau. It is mainly distributed in riverbed gravel lands and floodplains at elevations ranging from 3,500 to 5,000 m in the Yarlung Zangbo River Basin [1–3]. As a dioecious deciduous tree, H. gyantsensis exhibits remarkable environmental adaptability, capable of surviving extreme conditions, including cold, drought, and saline-alkali soils, and shows high adaptability to various soil types [4]. As a non-leguminous woody nitrogen-fixing plant, H. gyantsensis forms a symbiotic relationship with Frankia bacteria, fixing atmospheric nitrogen through root nodule formation, significantly improving soil fertility and enhancing ecosystem productivity [5]. Its rapid growth and strong asexual reproduction abilities make it play an important role in ecological restoration, vegetation reconstruction, and windbreak and sand fixation. In addition, the roots, stems, leaves, flowers, and fruits of H. gyantsensis are rich in flavonoids, vitamins, superoxide dismutase (SOD), and other bioactive substances. They have significant nutritional and medicinal value and can be used to treat diseases such as coronary heart disease and tumors, with broad applications in medicine and health products [6–8].
Although H. gyantsensis has outstanding ecological and economic value, research on its sex determination mechanisms is limited. Approximately 6% of angiosperm species are dioecious [9], and these species are widely distributed across different plant lineages [10, 11]. The evolution of dioecy and the mechanisms of sex determination in these plants have long been important topics in botanical research. Studies indicate that plants in the genus Hippophae are all diploid, utilizing an XY-type sex determination mechanisms [6, 12]. However, in natural communities, plants of the genus Hippophae often exhibit sex ratio bias, especially in populations of H. tibetana and H. rhamnoides subsp. turkestanica, where the proportion of male plants is significantly higher than that of females [12, 13]. This sex ratio bias may be related to differences in environmental adaptability between male and female plants. Male plants usually exhibit stronger adaptability when facing environmental stress, while female plants have relatively weaker adaptability due to higher energy and resource demands during the reproductive process [10, 11]. However, the specific causes of sex ratio bias remain unclear, necessitating further research to determine whether it is influenced by genetic factors in the seeds or environmental adaptability after germination.
Currently, research on the sex ratio bias and sex determination mechanisms of H. gyantsensis remains unclear, partly due to a lack of effective methods for sex identification. H. gyantsensis cannot be distinguished morphologically before flowering, typically requiring 3 to 4 years for seedlings to reach sexual maturity. Additionally, the small inflorescences and short flowering period make timely and accurate sex identification challenging, even during flowering [12, 14]. These factors hinder in-depth research into the adaptability differences and sex determination mechanisms between male and female H. gyantsensis. Thus, developing a rapid and accurate molecular marker technique for early sex identification in H. gyantsensis is crucial.
Recently, molecular marker technology has been widely applied for sex identification in dioecious plants, such as Actinidia chinensis [15], Spinacia oleracea [16], and Carica papaya [17]. By detecting genetic differences in sex-specific gene regions, molecular markers offer an effective means for early sex identification [12]. In plants of the genus Hippophae, some sex-specific molecular markers have been developed, but their applicability is often restricted to specific populations or geographical regions, lacking generality and reliability [12, 18–22]. Zeng et al. developed three pairs of sex-specific molecular markers via whole-genome resequencing of H. tibetana individuals, allowing for accurate male and female plant distinction through PCR. However, preliminary experimental results (Fig. S1) showed that these markers are only effective in H. tibetana and cannot be used universally in other species of the genus Hippophae, highlighting the complexity of sex evolution within the genus. H. gyantsensis is considered a homoploid hybrid of H. neurocarpa and H. rhamnoides subsp. yunnanensis [2], which provides a good basis for developing sex-specific molecular markers applicable to the entire genus Hippophae.
Advancements in high-throughput sequencing technology offer new opportunities for studying plant sex determination mechanisms [12]. By conducting whole-genome resequencing on male and female individuals and accurately detecting single nucleotide polymorphisms (SNPs) and insertion/deletion (InDel) variations, sex-specific molecular markers can be developed [12, 23]. For woody plants such as H. gyantsensis, which require many years to reach sexual maturity, molecular marker technology is crucial for achieving sex identification at the seedling stage, thereby optimizing planting strategies, enhancing breeding efficiency, and reducing costs.
This study aims to conduct whole-genome resequencing on H. gyantsensis individuals of known sex, utilizing the published female H. gyantsensis reference genome [4], to screen and develop molecular markers capable of accurately distinguishing the sex of H. gyantsensis. We will also validate the universality of these molecular markers in other species within the genus Hippophae, aiming to develop a sex identification tool applicable across the entire genus. In addition, we analyzed the sex ratio among progeny from the same mother plant of H. gyantsensis to explore the relationship between sex ratio and genetic characteristics. This study aims to elucidate the sex determination mechanisms of H. gyantsensis, promote breeding and propagation research of plants in the genus Hippophae, and provide a theoretical basis and technical support for ecological restoration and economic crop development.
Materials and methods
Sample collection and sequencing
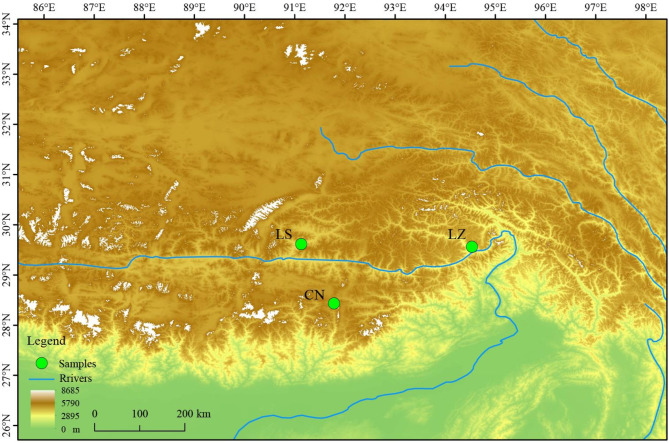
Field sampling was conducted across three distinct natural populations of H. gyantsensis in Tibet, China: Nyingchi (LZ; 29°27′31.10″ N, 94°34′39.28″ E), Lhasa (LS; 29°42′26.19″ N, 91°25′34.81″ E), and Qomolangma (CN; 28°18′11.27″ N, 91°48′49.88″ E) (sampling locations are detailed in Fig. 1). In each population, plants were first identified in the field by Dr. Wenju Zhan and Dr. La Qiong based on floral morphological characteristics. To prevent sampling genetically identical individuals due to clonal reproduction, a minimum linear distance of 50 m was maintained between sampled plants. Young leaves were then randomly collected from eight identified female and eight identified male plants in each population. The collected leaf samples were immediately dried in sealed containers with silica gel and stored at − 20 °C for future use. Voucher specimens of both female and male plants have been deposited at the Herbarium of the College of Ecology and Environmental Science, Tibet University (Voucher No.: Lq20230136).
Fig. 1.
Sampling locations of the three H. gyantsensis populations in Tibet. Generated using ggplot2 v. 3.3.5 and sf v. 1.0–3 packages in R v. 4.1.2. URL: https://ggplot2.tidyverse.org/
Genomic DNA was extracted using a modified CTAB method [24], and DNA quality and concentration were assessed with a NanoDrop 2000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). Qualified DNA samples were sent to Novogene Co., Ltd. (Beijing, China) for library construction and sequenced on the Illumina NovaSeq X Plus PE150 platform, with a target sequencing depth of 30×.
Whole-genome population variant detection
We utilized Fastp v0.21 [25] for quality control and cleaning of the raw sequencing data with default parameters. The quality of the filtered data was assessed using FastQC v0.11.9 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Next, we aligned the filtered reads to the H. gyantsensis reference genome using BWA v2.2.1 [26], converting alignment results to BAM format with Samtools v1.19 [27]. We then employed GATK4 v4.2 [28] for InDel identification and preliminary filtering, which included the removal of PCR duplicates, single-sample variant detection, and population variant detection. InDel and low-quality sites were marked and filtered based on the following criteria: “QD < 2.0 || FS > 200.0 || SOR > 10.0 || MQRankSum < − 12.5 || ReadPosRankSum < − 8.0.” Further filtering was done using Plink v1.9 [29], excluding sites with a minor allele frequency less than 0.01 and sites with missing genotype data exceeding 50%. Ultimately, from the initial 2,782,058 variant sites, 1,919,234 were selected for subsequent analysis.
Identification of sex-specific sites
Following the methodology of Zeng et al. [12], we identified sex-associated specific sites in H. gyantsensis. Given that H. gyantsensis is a diploid species with an XY sex determination mechanisms and that the reference genome is derived from a female plant, we focused on alignment discrepancies at identical genomic positions between male and female samples. Specifically, using the Python scripts provided by Zeng et al. [12], we screened for large InDel variants greater than 20 base pairs (bp) that were present in male samples but fully aligned in female samples. Subsequently, we performed manual validation of these candidate sites using the Integrative Genomics Viewer (IGV) [30]. During this process, we ensured that the selected insertions or deletions were consistently observed across all male individuals and entirely absent in all female individuals, thus minimizing the potential for false positives due to individual variability. The sex-specific sites confirmed through this rigorous validation were then used as candidate regions for subsequent primer design.
Design of sex-specific primers
We employed SeqKit v2.6.1 [31] to extract sequences of 1,000 bp upstream and downstream of the identified variant sites from the reference genome for primer design. Since the length differences between male and female individuals in the candidate variant regions were mostly 30–60 bp, the target PCR product length was set to 200–300 bp to facilitate the design of specific primers that could distinguish male and female plants. Following Zeng et al. [12], we evaluated the suitability of variant regions for primer development by checking the base distribution within approximately 200 bp on either side of the variant site, ensuring uniformity and avoiding excessive repetitive sequences that could impair primer design efficiency. We then used Primer3 v2.3.7 [32] for batch primer design, minimizing mismatches, dimers, and hairpin structures among primers during the design process. The designed primers were aligned to the reference genome using BLASTn v2.5.0 [33] to ensure specificity. Finally, primers with high specificity were selected and synthesized by Sangon Biotech (Shanghai) Co., Ltd.
Validation of sex-specific primer applicability
To verify primer specificity, we performed conventional PCR amplification using the synthesized primers on a small number of H. gyantsensis DNA samples (including two females and two males) to preliminarily screen for specific primers that can accurately distinguish male and female plants. The PCR reaction system had a total volume of 30 µL, comprising 15 µL of Taq Mix, 1 µL of DNA template, 1 µL each of forward and reverse primers, and 12 µL of deionized water. The PCR amplification program followed Zeng et al. [12], with the following settings: pre-denaturation at 95 °C for 3 min; 35 cycles of 95 °C for 15 s, 58 °C for 15 s, and 72 °C for 30 s; extension at 72 °C for 5 min; and final hold at 4 °C. PCR products were analyzed by 2% agarose gel electrophoresis. Ultimately, two pairs of specific primers that effectively distinguished male and female plants based on product length were selected. These primers were then applied to additional H. gyantsensis samples with identified sex to validate their universality among different populations.
Analysis of sex ratio in offspring of H. Gyantsensis
To explore the sex ratio among offspring from the same mother plant of H. gyantsensis, we collected seeds from a single mother plant in Lhasa and Nyingchi, Tibet. After returning to the laboratory, the pericarps were manually removed, and the seeds were air-dried to eliminate surface moisture, then stored at 4 °C. In the following spring, the seeds underwent moist sand stratification to promote germination before being sown in seedling trays for cultivation. When the seedlings reached a suitable size for DNA extraction, they were uprooted, surface impurities were cleaned, dried with silica gel, and stored at − 20 °C. Genomic DNA of the seedlings was extracted using the modified CTAB method [24], and DNA quality and concentration were assessed using a NanoDrop 2000. DNA samples of qualified quality were subjected to sex identification using the sex-specific molecular markers, and the sex ratio among the offspring was further analyzed.
Applicability of sex-specific primers in the Genus Hippophae
To assess the applicability of the primers designed in this study to other species within the genus Hippophae, we selected H. salicifolia, H. tibetana, H. rhamnoides, and H. rhamnoides subsp. turkestanica for testing. Six female and six male plants of each species were selected, and PCR amplification was performed using the previously screened sex-specific primers to determine their universality among different species in the genus Hippophae.
Results
Distribution of sex-specific sites in H. gyantsensis
Through the screening of large InDel variant sites in the whole-genome sequencing data of H. gyantsensis, we identified a total of 59 sex-specific sites that differ between male and female plants (Table S1). Notably, all these sites were located on chromosome 2. This finding aligns with the results of Wang et al. [34]. and Zeng et al. [12] in H. tibetana, suggesting that chromosome 2 may function as the sex chromosome in H. gyantsensis. This provides important insights for further research into its sex determination mechanisms.
Screening and preliminary validation of sex-specific markers
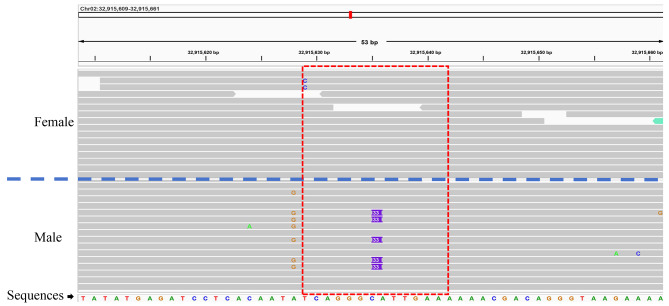
After further screening and validation of the sex-specific sites using IGV [30], we designed eight pairs of candidate molecular marker primers. We conducted preliminary screening of these primers through PCR amplification and gel electrophoresis analysis. The results showed that two pairs of primers (Higy_04 and Higy_06) were able to effectively distinguish between male and female individuals of H. gyantsensis (Table 1). The IGV [30] visualization results further confirmed the reliability of these variant sites: at the variant site corresponding to Higy_04, sequencing reads from female individuals aligned perfectly with the reference genome, whereas approximately 50% of the reads from male individuals exhibited a 33 bp insertion at this site (Fig. 2). Similarly, at the variant site corresponding to Higy_06, sequencing reads from female individuals also matched the reference genome completely, while about 50% of the reads from male individuals showed a 40 bp deletion at this site (Fig. S2). These findings further validated the effectiveness of these two primers as reliable sex-specific molecular markers.
Table 1.
Primer pairs for sex-linked markers of H. Gyantsensis. The presence of insertions and deletions at the variant sites corresponding to primers Higy_04 and Higy_06 resulted in two products being amplified in male samples, while female samples produced only one product; the actual product lengths are highlighted in bold
| Marker | Primer sequences (5′ to 3′) | Tm/℃ | Product length (bp) | Variant position |
|---|---|---|---|---|
| higy_04 | F: CCCCCTTAGTCACTACCAACAC | 55 | 218 | 251 | Chr02: 32,915,672 |
| R: ATCCCATCGATCATCACCATCC | ||||
| higy_06 | F: TAATCTGGTGCCTCTCGATCAATT | 55 | 310 | 270 | Chr02: 33,487,840 |
| R: TCTGTACAATTTATGCAGCACCAC |
Fig. 2.
Alignment results of next-generation sequencing sequences on chromosome 2 for one male and one female individual of H. gyantsensis. In the region marked by the red dashed box, approximately 50% of the sequences in the male individual show a 33 bp insertion in the Higy_04 primer amplification area
Accuracy test of sex-specific markers
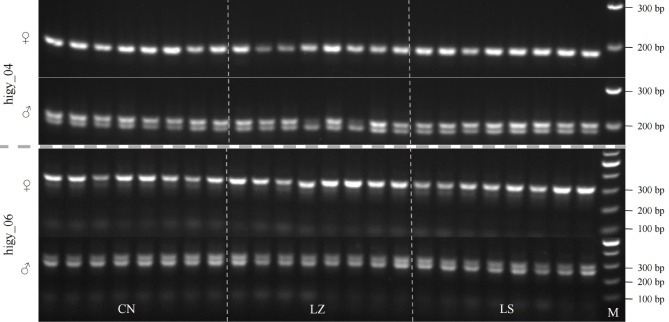
To assess the applicability and accuracy of primers Higy_04 and Higy_06 across different populations of H. gyantsensis, we selected 48 samples (24 females and 24 males) from three populations: Nyingchi, Lhasa, and Qomolangana in Tibet. PCR amplification was performed under the same conditions as the preliminary screening. The results confirmed that both primers could accurately distinguish male and female plants: PCR products from male samples displayed two bands, while those from female samples exhibited a single band (Fig. 3). Specifically, in female plants, primers Higy_04 and Higy_06 amplified single bands of 218 bp and 310 bp, respectively, aligning with design expectations; male plants presented an additional longer or shorter band, reflecting the male-specific insertion or deletion. These results indicate that primers Higy_04 and Higy_06 have good efficacy for sex identification across different populations of H. gyantsensis.
Fig. 3.
PCR amplification results of 48 H. gyantsensis samples from different regions using the two pairs of primers (Higy_04 and Higy_06). The symbols ♀ and ♂ represent female and male H. gyantsensis samples, respectively. Original gel images produced by primers Higy_04 and Higy_06 can be found in Figs. S3 and S4
Analysis of sex ratio in offspring of H. gyantsensis
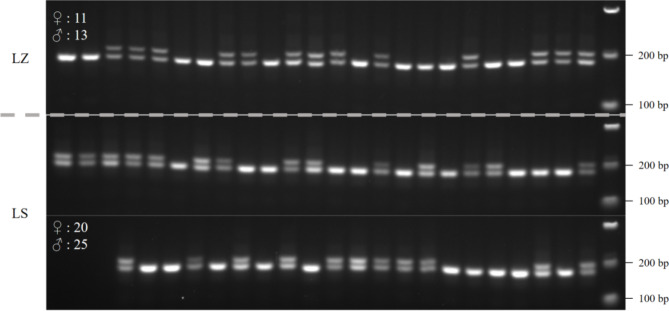
To investigate whether the sex ratio among offspring from the same mother plant of H. gyantsensis conforms to genetic expectations, we employed primer Higy_06 to perform sex identification on seedlings germinated from seeds collected from the same mother plant in Lhasa (LS) and Nyingchi (LZ). The results indicated that among 45 seedlings from LS, there were 20 females and 25 males; among 24 seedlings from LZ, there were 11 females and 13 males (Fig. 4; Table 2). Statistical analysis showed that the sex ratios among offspring in both locations were approximately 1:1, with no significant difference (χ² test, p > 0.05). This result aligns with Mendelian inheritance laws [35], suggesting that sex segregation in H. gyantsensis follows a random distribution pattern, with potential influences from environmental factors rather than genetic factors within the seeds.
Fig. 4.
Results of sex identification of H. gyantsensis seedlings using primer Higy_06. Female samples display one band, while male samples display two bands. Original gel images produced by primer Higy_06 can be found in Fig. S5 and S6
Table 2.
Sex ratios among progeny from the same mother plant of H. Gyantsensis from different provenances
| Sampling Location | Total Seedlings | Number of Female Plants | Number of Male Plants | Female-to-Male Ratio | Chi-Square Value | p Value |
|---|---|---|---|---|---|---|
| LS | 45 | 20 | 25 | 0.8 | 0.56 | > 0.05 |
| LZ | 24 | 11 | 13 | 0.85 | 0.17 | > 0.05 |
Applicability analysis of sex-specific primers in the Genus Hippophae
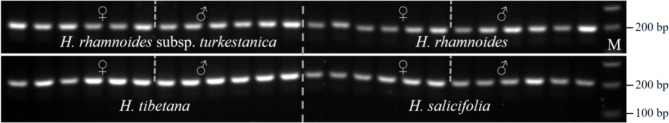
To evaluate the universality of primers Higy_04 and Higy_06 in other species of the genus Hippophae, we selected six female and six male plants from four species: H. salicifolia, H. tibetana, H. rhamnoides, and H. rhamnoides subsp. turkestanica, for PCR amplification tests. The results showed that primer Higy_04 amplified a single band of approximately 200 bp in all samples of the four species, failing to distinguish between male and female plants (Fig. 5). Primer Higy_06 did not produce the expected amplification products in these species, primarily resulting in primer dimers (Fig. S7). These findings indicate that the sex-specific primers developed based on H. gyantsensis are not applicable to other species within the genus Hippophae.
Fig. 5.
PCR amplification results of primer Higy_04 in other species of the genus Hippophae. In both female and male samples of all species, only a single band of approximately 200 bp appeared, preventing sex distinction
Discussion
H. gyantsensis is crucial for ecological conservation and economic development, particularly in fragile ecosystems like the Qinghai-Tibet Plateau. Its remarkable environmental adaptability and clonal reproduction make it a key species for ecological restoration, vegetation reconstruction, and sand fixation [5]. Additionally, its fruits are rich in bioactive compounds, such as flavonoids and vitamins, which have significant potential for pharmaceutical and health product development [6, 7]. However, as a dioecious woody plant, H. gyantsensis cannot be differentiated by morphological characteristics before flowering, which typically takes 3 to 4 years [12, 14]. This challenge in sex identification limits its applications in breeding and cultivation.
In this study, we successfully identified long InDel variant sites between male and female plants through high-throughput whole-genome resequencing and developed two pairs of sex-specific molecular markers (Higy_04 and Higy_06). These markers showed high accuracy across samples from various H. gyantsensis populations, allowing for rapid and precise identification of plant sex at the seedling stage. Compared to traditional methods like Random Amplified Polymorphic DNA (RAPD) and Amplified Fragment Length Polymorphism (AFLP), our approach, which leverages whole-genome resequencing data to screen variant sites on the reference genome, is an effective strategy for developing sex-specific markers in dioecious plants Notably, all sex-specific sites were located on chromosome 2, consistent with previous findings in H. tibetana [12], which suggests that chromosome 2 may play a role as the sex chromosome in H. gyantsensis. The origin and evolution of sex chromosomes in plants often involve the accumulation of sex-determining genes on specific chromosomes, leading to differentiation of sex chromosome regions [36]. A detailed examination of chromosome 2’s genomic structure and function is anticipated to elucidate the molecular mechanisms of sex determination in H. gyantsensis, contributing new insights into the evolution of plant sex chromosomes.
In our analysis of offspring from the same mother plant of H. gyantsensis, the sex ratio was approximately 1:1, consistent with Mendelian inheritance laws [35], indicating that sex segregation is primarily controlled by genetic factors. However, natural populations of the genus Hippophae often exhibit a male-biased sex ratio, with a higher proportion of male plants compared to female plants [12, 13]. This phenomenon may result from differential impacts of environmental factors on the survival and growth of male and female plants. Studies have shown that environmental stresses, such as cold, drought, and poor soil conditions, can affect the sex ratio distribution in plants [37, 38]. Male plants generally exhibit greater tolerance to environmental stresses, possibly because they invest fewer resources in reproduction, allowing them to allocate more energy to growth and maintenance [38]. In contrast, female plants require more energy and resources during reproduction, making them less adaptable to adverse environmental conditions, which may reduce their survival rates [38]. For example, in dioecious species like Myrica esculenta [39] and Pistacia lentiscus [40], research has found that unfavorable environmental conditions can lead to a decrease in the proportion of female plants, resulting in a male-biased sex ratio in the population. Therefore, although sex is genetically determined, environmental selection pressures may cause deviations in the sex ratio in natural populations. Future research should investigate how environmental factors influence the growth and reproductive success of male and female H. gyantsensis, exploring the relationship between environmental conditions and sex distribution to better understand the adaptation mechanisms of Hippophae species in extreme environments.
Although we successfully developed sex-specific molecular markers for H. gyantsensis, these markers were not universally applicable to other species within the genus Hippophae. Experimental results showed that these primers could not effectively distinguish between male and female individuals of H. salicifolia, H. tibetana, H. rhamnoides, and H. rhamnoides subsp. Turkestanica. This finding highlights the complexity of sex determination mechanisms within the genus Hippophae, suggesting that different species may possess unique sex-determining genes or regulatory elements. The reasons for this discrepancy may include genetic divergence between species and differences in the rate of sex chromosome evolution. On one hand, different species within the genus Hippophae may have undergone distinct genetic variations and genome rearrangements during evolution, leading to changes in the sequence and structure of sex-determining genes [41]. Such genetic divergence may cause sex-specific loci to no longer be conserved among different species, thereby affecting the universality of the markers. On the other hand, the evolution of sex chromosomes often exhibits rapid and dynamic characteristics, including duplication, loss, and rearrangement of sex-determining genes, which may result in inconsistencies of sex-specific markers among species [41, 42]. Additionally, different species may employ different sex determination mechanisms involving different genes and regulatory pathways [42, 43].
Similar phenomena have been reported in other dioecious plants. For example, in kiwifruit and persimmon [44, 45], sex-specific markers are effective in certain cultivars or populations but are not universally applicable. This may be due to independent evolution of sex determination mechanisms in these species, leading to diversification of sex-related genes. Therefore, developing universal sex-specific molecular markers applicable to the entire genus Hippophae remains a significant challenge. Future research should focus on utilizing more conserved sex-determining genes or sequences, or integrating whole-genome data from multiple species to identify common sex-related loci.
The sex-specific molecular markers developed in this study have considerable potential for applications in breeding and ecological restoration. By enabling rapid and accurate identification of H. gyantsensis sex at the seedling stage, breeders can optimize the male-to-female ratio to enhance fruit yield or improve population adaptability. For instance, increasing the proportion of female plants can boost yields in fruit production plantations, whereas in ecological restoration, enhancing the proportion of male plants may improve the population’s resilience to environmental stresses. Additionally, employing these markers can reduce planting costs and enhance breeding efficiency. Traditional methods for sex determination often require several years until flowering, making them time-consuming and costly. In contrast, molecular marker technology enables the selection of desired male and female plants at the seedling stage, thus saving time and resources and facilitating the development of industries related to H. gyantsensis.
While we successfully developed sex-specific molecular markers for early sex identification in H. gyantsensis, several limitations persist. First, the markers are only effective in H. gyantsensis and lack universality in other species of the genus Hippophae, limiting their broader application in breeding and research. This may stem from differences in sex-determining genes or regulatory regions among species. Second, the absence of a male H. gyantsensis reference genome assembly hampers our understanding of the precise locations and functions of the sex-determining genes. Although sex-specific sites were concentrated on chromosome 2, indicating its potential role in sex determination, comprehensive analysis requires high-quality male genome and transcriptome data. Future research should expand the sample scope to include more species within the genus Hippophae and utilize multi-omics data alongside molecular biology techniques and gene function analysis to further identify and characterize sex-determining genes, explore the structure and evolutionary mechanisms of sex chromosomes, and provide new perspectives for studying sex determination in both H. gyantsensis and the genus Hippophae.
Conclusion
In this study, we utilized high-throughput whole-genome resequencing technology to develop two pairs of sex-specific molecular markers capable of accurately distinguishing male and female individuals of H. gyantsensis. These markers demonstrated high accuracy across different populations and enabled rapid and precise sex identification at the seedling stage, providing an effective tool for the breeding, cultivation, and ecological restoration of H. gyantsensis. All sex-specific loci were located on chromosome 2, which supports the hypothesis that this chromosome may function as the sex chromosome in H. gyantsensis and suggests it may contain key sex-determining genes or regulatory regions. Additionally, the sex ratio among offspring was close to 1:1, indicating that sex segregation is primarily controlled by genetic factors. However, the developed sex-specific markers were not universally applicable to other species within the genus Hippophae, highlighting the complexity of sex determination mechanisms in these plants. Future research should focus on H. gyantsensis and other Hippophae species, integrating high-quality male genome and transcriptome data to precisely analyze the location and function of sex-determining genes, explore the structure and evolutionary mechanisms of sex chromosomes, and provide new insights into the study of sex biology in dioecious plants.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We would like to express our sincere gratitude to the editor and the anonymous reviewers for their valuable suggestions and insightful comments.
Author contributions
Z Z and J W: Conceptualization, Supervision, Methodology, Software, Resources, Investigation, Validation, and Writing—Original draft preparation. Z T, N N, Y C, and J C: Methodology, Software, Formal analysis, and Data curation. W Z and L Q: Conceptualization, Writing—Review and editing.
Funding
This research was funded by the Graduate High-level Talent Training Program of Tibet University (2022-GSP-B003), the National Natural Science Foundation of China (No. 31760127), the First-class Discipline Construction Project of Ecology (Zangcaijiaozhi [2023]01), and the Key R&D Project of Science and Technology Program of Tibet Autonomous Region (NO. XZ202301ZY0006G).
Data availability
In this study, the source data for the resequencing of H. gyantsensis has been submitted to the China National Center for Bioinformation and is publicly available as of the date of publication. The assigned accession number is CRA018242 and can be cited in your publication. Please access it via the following link: https://bigd.big.ac.cn/gsa/browse/CRA018242.
Declarations
Ethics approval and consent to participate
The research sample in this study is H. gyantsensis, which is not a protected plant species in China. All plant materials were collected and used with proper permissions and in compliance with relevant institutional, national, and international guidelines and legislation.
Consent for publication
Not applicable.
Clinical trial number
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Zhefei Zeng and Junwei Wang contributed equally to this work.
Contributor Information
Wenju Zhang, Email: wjzhang@fudan.edu.cn.
La Qiong, Email: lhagchong@163.com.
References
- 1.Jia DR, Bartish IV. Climatic changes and orogeneses in the late miocene of Eurasia: the main triggers of an expansion at a continental scale? Front Plant Sci. 2018;9:1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jia DR, Wang YJ, Liu TL, Wu GL, Kou YX, Cheng K, et al. Diploid hybrid origin of Hippophaë gyantsensis (Elaeagnaceae) in the western Qinghai–Tibet Plateau. Biol J Linn Soc. 2016;117(4):658–71. [Google Scholar]
- 3.Xu T, Wang R, La Q, Yonezawa T, Huang X, Sun K, et al. Climate heterogeneity shapes phylogeographic pattern of Hippophae gyantsensis (Elaeagnaceae) in the east Himalaya-Hengduan Mountains. Ecol Evol. 2023;13(6):e10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen M, Yang D, Yang S, Yang X, Chen Z, Yang T, et al. Chromosome-level genome assembly of Hippophae gyantsensis. Sci Data. 2024;11(1):126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao A, Xu W, Xu P, Zhang X, Wu Y, Xu A, et al. Establishment of tissue culture and regeneration system in Hippophae Gyantsensis Lian. Horticulturae. 2024;10(5):460. [Google Scholar]
- 6.Nybom H, Ruan C, Rumpunen K. The Systematics, Reproductive Biology, Biochemistry, and breeding of Sea Buckthorn—A Review. Genes. 2023;14(12):2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dhyani D, Maikhuri RK, Dhyani S. Seabuckthorn: an underutilized resource for the nutritional security and livelihood improvement of rural communities in Uttarakhand Himalaya. Ecol Food Nutr. 2011;50(2):168–80. [DOI] [PubMed] [Google Scholar]
- 8.Suryakumar G, Gupta A. Medicinal and therapeutic potential of Sea Buckthorn (Hippophae rhamnoides L). J Ethnopharmacol. 2011;138(2):268–78. [DOI] [PubMed] [Google Scholar]
- 9.He L, Jia KH, Zhang RG, Wang Y, Shi TL, Li ZC, et al. Chromosome-scale assembly of the genome of Salix Dunnii reveals a male‐heterogametic sex determination system on chromosome 7. Mol Ecol Resour. 2021;21(6):1966–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Renner SS, Müller NA. Plant sex chromosomes defy evolutionary models of expanding recombination suppression and genetic degeneration. Nat Plants. 2021;7(4):392–402. [DOI] [PubMed] [Google Scholar]
- 11.Razumova OV, Alexandrov OS, Bone KD, Karlov GI, Divashuk MG. Sex chromosomes and sex determination in dioecious agricultural plants. Agronomy. 2023;13(2):540. [Google Scholar]
- 12.Zeng Z, Wang R, Wang J, Chen Y, Wang Y, Song Z, et al. Development and validation of sex-linked molecular markers for rapid and accurate identification of male and female Hippophae tibetana plants. Sci Rep. 2024;14(1):19243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ali A, Kaul V. Seabuckthorn—a valuable resource of the cold desert (Ladakh). J Plant Dev Sci. 2012;4(2):151–5. [Google Scholar]
- 14.Jain A, Kumar A, Sharma PC. Repertoire of molecular markers and their applications in Seabuckthorn. InThe Seabuckthorn Genome 2022 (pp. 187–212). Cham: Springer International Publishing.
- 15.Guo D, Wang R, Fang J, Zhong Y, Qi X. Development of sex-linked markers for gender identification of Actinidia arguta. Sci Rep. 2023;13(1):12780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.She H, Xu Z, Zhang H, Li G, Wu J, Wang X, et al. Identification of a male-specific region (MSR) in Spinacia oleracea. Hortic. Plant J. 2021;7(4):341–6. [Google Scholar]
- 17.Bui TL, Truong NT, Do TK. The application of molecular marker in papaya sex determination: from the laboratory to the field. Sci Hortic. 2024;327:112872. [Google Scholar]
- 18.Sharma A, Zinta G, Rana S, Shirko P. Molecular identification of sex in Hippophae rhamnoides L. using isozyme and RAPD markers. Stud China. 2010;12:62–6. [Google Scholar]
- 19.Korekar G, Sharma RK, Kumar R, Meenu, Bisht NC, Srivastava RB, et al. Identification and validation of sex-linked SCAR markers in dioecious Hippophae rhamnoides L.(Elaeagnaceae). Biotechnol Lett. 2012;34:973–8. [DOI] [PubMed] [Google Scholar]
- 20.Chawla A, Kant A, Stobdan T, Srivastava RB, Chauhan RS. Cross-species application of sex linked markers in H. Salicifolia and H. Tibetana. Sci Hortic. 2014;170:281–3. [Google Scholar]
- 21.Das K, Ganie SH, Mangla Y, Dar TU, Chaudhary M, Thakur RK, et al. ISSR markers for gender identification and genetic diagnosis of Hippophae rhamnoides ssp. turkestanica growing at high altitudes in Ladakh region (Jammu and Kashmir). Protoplasma. 2017;254:1063–77. [DOI] [PubMed] [Google Scholar]
- 22.Zhou W, Wang Y, Zhang G, Luan G, Chen S, Meng J, et al. Molecular sex identification in dioecious Hippophae rhamnoides L. via RAPD and SCAR markers. Molecules. 2018;23(5):1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Varshney RK, Terauchi R, McCouch SR. Harvesting the promising fruits of genomics: applying genome sequencing technologies to crop breeding. Plos Biol. 2014;12(6):e1001883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aboul-Maaty NA, Oraby HA. Extraction of high-quality genomic DNA from different plant orders applying a modified CTAB-based method. Bull Natl Res Cent. 2019;43(1):1–0. [Google Scholar]
- 25.Chen S, Zhou Y, Chen Y, Gu J. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jung Y, Han D, BWA-MEME:. BWA-MEM emulated with a machine learning approach. Bioinformatics. 2022;38(9):2404–13. [DOI] [PubMed] [Google Scholar]
- 27.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The genome analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Briefings Bioinf. 2013;14(2):178–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen W, Le S, Li Y, Hu F. SeqKit: a cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLoS ONE. 2016;11(10):e0163962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. Primer3—new capabilities and interfaces. Nucleic Acids Res. 2012;40(15):e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Y, Ye W, Zhang Y, Xu Y. High speed BLASTN: an accelerated MegaBLAST search tool. Nucleic Acids Res. 2015;43(16):7762–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang R, Wu B, Jian J, Tang Y, Zhang T, Song Z, et al. How to survive in the world’s third poplar: insights from the genome of the highest altitude woody plant, Hippophae Tibetana (Elaeagnaceae). Front. Plant Sci. 2022;13:1051587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karl P. A Mendelian’s view of the law of ancestral inheritance. Biometrika. 1904;3(1):109–12.
- 36.Charlesworth D. Evolution of recombination rates between sex chromosomes. Philos Trans R Soc B. 2017;372(1736):20160456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hultine KR, Grady KC, Wood TE, Shuster SM, Stella JC, Whitham TG. Climate change perils for dioecious plant species. Nat Plants. 2016;2(8):1–8. [DOI] [PubMed] [Google Scholar]
- 38.Juvany M, Munné-Bosch S. Sex-related differences in stress tolerance in dioecious plants: a critical appraisal in a physiological context. J Exp Bot. 2015;66(20):6083–92. [DOI] [PubMed] [Google Scholar]
- 39.Khanduri VP, Sukumaran A, Sharma CM. Male-skewed sex ratio in Myrica esculenta: a dioecious tree species. Trees. 2019;33:1157–65. [Google Scholar]
- 40.Barradas MC, Correia O. Sexual dimorphism, sex ratio and spatial distribution of male and female shrubs in the dioecious species Pistacia lentiscus L. Folia Geobot. 1999;34:163–74. [Google Scholar]
- 41.Li J, Chen T, Gao K, Xue Y, Wu R, Guo B, et al. Unravelling the novel sex determination genotype with ‘ZY’and a distinctive 2.15–2.95 mb inversion among poplar species through haplotype-resolved genome assembly and comparative genomics analysis. Mol Ecol Resour. 2024;24(7):e14002. [DOI] [PubMed] [Google Scholar]
- 42.Charlesworth D. Plant sex chromosomes. Annu Rev Plant Biol. 2016;67(1):397–420. [DOI] [PubMed] [Google Scholar]
- 43.Leite Montalvão AP, Kersten B, Fladung M, Müller NA. The diversity and dynamics of sex determination in dioecious plants. Front Plant Sci. 2021;11:580488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo DD. Validation of kiwifruit sex molecular markers in Actinidia arguta. J Fruit Sci. 2019;36:549–56. [Google Scholar]
- 45.Zhang PX, Yang SC, Liu YF, Zhang QL, Xu LQ, Luo ZR. Validation of a male-linked gene locus (OGI) for sex identification in persimmon (Diospyros kaki Thunb.) And its application in F1 progeny. Plant Breeding. 2016;135(6):721–7. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
In this study, the source data for the resequencing of H. gyantsensis has been submitted to the China National Center for Bioinformation and is publicly available as of the date of publication. The assigned accession number is CRA018242 and can be cited in your publication. Please access it via the following link: https://bigd.big.ac.cn/gsa/browse/CRA018242.