Abstract
Background
In recent years, human microbiome research has flourished and has drawn attention from both healthcare professionals and general consumers as the human microbiome is now recognized as having a significant influence on human health. This has led to the emergence of companies offering microbiome testing services. Some of these services are sold directly to the consumer via companies’ websites or via medical laboratory websites.
Methodology
In order to provide an overview of the consumer experience proposed by these microbiome testing services, one single faecal sample was sent to six different companies (five based in Europe and one based in the USA). Two out of the six testing kits were commercialized by medical laboratories, but without any requirement for a medical prescription. The analyses and reports received were discussed with a panel of experts (21 experts from 8 countries) during an online workshop.
Results
This workshop led to the identification of several limitations and challenges related to these kits, including over-promising messages from the companies, a lack of transparency in the methodology used for the analysis and a lack of reliability of the results. The experts considered the interpretations and recommendations provided in the different reports to be premature due to the lack of robust scientific evidence and the analyses associated with the reports to be of limited clinical utility. The experts also discussed the grey areas surrounding the regulatory status of these test kits, including their positioning in the European market. The experts recommended a distinction between regulatory requirements based on the intended use or purpose of the kit: on the one hand, test kits developed to satisfy consumer curiosity, with a clear mention of this objective, and no mention of any disease or risk of disease, and on the other hand, in vitro diagnostic (IVD) CE-marked test kits, which could go deeper into the analysis and interpretation of samples, as such a report would be intended for trained healthcare professionals.
Conclusions
Recommendations or actions, specific to the context of use of microbiome testing kits, are listed to improve the quality and the robustness of these test kits to meet expectations of end users (consumers, patients and healthcare professionals). The need for standardization, robust scientific evidence, qualification of microbiome-based biomarkers and a clear regulatory status in Europe are the main issues that will require attention in the near future to align laboratory development with societal needs and thus foster translation into daily health practice.
Supplementary Information
The online version contains supplementary material available at 10.1186/s40168-024-01991-x.
Keywords: Microbiome testing, Direct-to-consumer tests, In vitro diagnostic, Regulatory status
Introduction
The human gut microbiome is the most extensively studied microbiome, and its central role in human health has been extensively documented by multiple research teams. Numerous studies have reported associations between gut microbiome alterations (compositional and/or functional changes) and the onset or progression of many pathologies (including metabolic disorders, allergies, inflammatory bowel diseases, cognitive disorders, cancer) [1, 2]. As microbiome research expands, so does the public’s interest in being aware of their own microbiome. In 2023, approximately 6500 participants from 7 countries took part in an international survey conducted by the Biocodex Microbiota Institute and IPSOS which aimed to better understand the knowledge and behaviour of the European public regarding their microbiota [3]. Of those who had received comprehensive information from their healthcare professional, 95% had adopted behaviours to maintain a balanced microbiome, and almost two-thirds thought it would be useful to test their microbiome. In addition, an international initiative is underway with the “Million Microbiome of Humans Project” (MMHP), a collaboration initiated by scientists from China, Sweden, Denmark, France and Latvia, which aims to analyze one million microbiome samples (from different body sites) to build the largest database of the human microbiome [4]. In parallel, governments across Europe begin to promote prevention within public health measures, and a high interest in health self-management by both healthcare professionals and consumers is observed, and the microbiome follows this trend. Indeed, while consumer curiosity has led to a growing interest in microbiome testing kits, analysis of microbial signatures also represents an interesting option for healthcare professionals and patients to enable improved diagnostics or prognosis, to assess safety, to predict susceptibility and to develop a disease or response to treatment [5]. This dual utility is quite encouraging for the field and demonstrates (i) the importance of supporting healthcare professionals to integrate microbiome considerations into their daily clinical practice and (ii) the need for appropriate information to general consumers, who also express high interest in microbiome research and their wish to integrate this new component into their wellness practices. However, as microbiome research is still in its early stages, one can question whether some of the products that are already available on the market are actually useful for monitoring one’s health based on the characteristics of their gut microbiome.
Microbiome testing presents some similarities with genetic testing (which is well-known to citizens), but specific challenges distinguish the two [6]. Microbiome testing is much more complex than genetic testing due to the high number of genes present and expressed by each constituent of the microbiome, as compared to human genes [7], and a higher interpersonal variability. In addition, microbiome samples must often be associated with personal data (referred to as “metadata”) to help to interpret the results, as the sample alone is not sufficient. Indeed, the microbiome is constantly remodeling in response to multiple environmental factors (diet, drugs, transit time…), making its analysis and interpretation highly complex [1].
Currently, the majority of microbiome testing services available are dedicated to the analysis of faecal samples, as a well-established proxy for the gut microbiome, while some are emerging for vaginal, oral or skin microbiome analysis. With the availability of a growing number of microbiome testing products, it is now possible for citizens to integrate microbiome analysis as part of their wellness practices; some citizens may even feel that some of their health problems are not fully understood or addressed by physicians, particularly digestive disorders, which are common. Increased public awareness could extend this questioning to other clinical situations where a link with the microbiome has been described (neurological disorders, auto-immune diseases or degenerative diseases).
Currently in Europe, microbiome testing kits can be commercialized in two different ways, either sold directly by companies or commercialized through medical laboratories. Those sold directly by commercial companies are aimed at general consumers, whereas those commercialized through medical laboratories are generally intended for patients, although the distinction between “consumers” and “patients” is not strictly defined. In both cases, the purpose of these kits is to provide an analysis of the microbiome composition based on the examination of a specimen collected at home. However, these novel test kits deserve a more rigorous consideration from a scientific, ethical and regulatory point of view, in the interest of managing the expectations of consumers, patients and healthcare professionals who would use these services. “Promissory” or over-promising communication of sequencing results runs the risk of being exaggerated and can lead to reputational damage and loss of credibility for science in general and microbiome research in particular [8].
Despite the current intense interest in these new devices, two recent publications warn of their lack of analytical and clinical validity, the urgent need for validation of all the steps of the testing pathway and the lack of federal oversight (in the USA) to prevent any potential harm to the consumer [9,10]. Previous studies have also evaluated the data protection policies and questioned the scientific validity and clinical relevance of these devices, based on the existing literature or promotional messages mentioned on the companies’ websites [6,11].
As part of the larger International Human Microbiome Coordination and Support Action (IHMCSA) consortium (EU-funded project [12]), the European context of currently marketed faecal microbiome testing kits has been assessed.
Experimental approach
Non-exhaustive benchmarking analysis of microbiome testing kits available in Europe
To recreate the experience of consumers, a non-exhaustive benchmarking analysis of six microbiome testing kits available in Europe (5 EU-based kits and 1 kit sold by a company based in the USA) was conducted. Four out of the six kits were bought directly from the companies’ websites, while the other two were bought from medical laboratories websites, without any requirement for a medical prescription. There were no strict selection criteria for the choice of kits. However, particular attention was paid to include at least one in vitro diagnostic (IVD) CE-marked kit and commercialized by a medical laboratory. Many products require CE (Conformité Européenne) marking before they can be sold in the European Union (EU). CE marking indicates that the conformity of the product has been assessed by the manufacturer (and in certain cases a notified body) and deemed to meet safety, performance, health and environmental protection EU requirements. It is required for products manufactured anywhere in the world that are then marketed in the EU. Thus, in the case of microbiome testing kits, the CE mark indicates that the test kit complies with the requirements of the Directive 98/79/EC [13] on in vitro diagnostic medical devices or the new and stricter Regulation on In Vitro Diagnostic Medical Devices [14] (IVDR), as the transition period is still ongoing. It should be specified that the CE mark applied on the product included in this analysis was granted under previous Directive 98/79/EC and its associated requirements. The transition from Directive 98/79/EC to Regulation 2017/746 brought stricter harmonization, higher safety requirements and increased transparency to improve patient protection. Key changes include the extension of the scope to more devices, a new risk classification system, increased involvement of notified bodies and clear post-market surveillance obligations to ensure long-term safety and performance. It is now clear that such tests would require assessment by a notified body, a conformity assessment body designated in accordance with the IVDR (Supplementary Table 1). For the other kits, devices sold in different European countries were randomly selected, and one kit outside of Europe (USA) was purchased. The overview of the six kits, their components and their description on the respective companies’ websites is described in Table 1.
Table 1.
Benchmark analysis: general overview of the six microbiome testing kits
| 1 | 2 | 3 | 4 | 5 | 6 | ||
|---|---|---|---|---|---|---|---|
| Location | EU | EU | EU | EU | EU | USA | |
| Price | 167 € | 150 € | 250 € | 205 € | 300 € | 165 € | |
| Commercialization channel | Medical laboratory | Company website | Company website | Medical laboratory | Company website | Company website | |
| CE mark | No | CE mark on the fæcal swab only | CE mark on the fæcal swab only | Yes, IVD CE marked (granted under Directive 98/79/EC) | No | Not applicable | |
| Consumer-informed consent | No | No | No | Yes | No | No | |
| Time for analysis | < 3 weeks | 3 weeks | > 11 weeks | 6 weeks | > 12 weeks | 4 weeks | |
| Pre-analytical and analytical methodology | Sampling tube | Tube with integrated spoon | Tube with mention “for research only, not for use in diagnostic procedures” | Tube expired (expiry date) | Tube with CE mark “for in vitro diagnostic” | Tube with mention “for research only, not for use in diagnostic procedures” | Tube, original tag was removed |
| Tool for fæcal sample collection | Spoon included | Fæcal swab | Fæcal swab | Fæcal swab | Spoon included | No particular tool | |
| The presence of stabilizer | No | Yes | Yes | Yes | Yes | Yes | |
| Sample homogenization | No | No | No | No | No | Yes | |
| Sample shipping condition | Ice packed | Room temperature | Room temperature | Room temperature | Room temperature | Room temperature | |
| Sequencing method | 16S rRNA gene sequencing | 16S rRNA gene sequencing | Shotgun sequencing | Not reported | Not reported | Not reported | |
| Reference cohort | Not described | Not described | Healthy individuals without chronic diseases | Healthy cohort from Western Europe | Not described | “Population from the company” | |
| Metadata requested | Questionnaire | No | On the application | On the website | Medical questionnaire provided with the kit (paper version) | On the website | On the application |
| Information collected | Not applicable |
General Anthropometric & clinical Gastrointestinal symptoms Nutrition Activity — sedentarity Medicines and addiction |
General Anthropometric & clinical Gastrointestinal symptoms Nutrition |
General Anthropometric & clinical Medical history |
General Gastrointestinal symptoms Nutrition Activity — sedentarity Medicines and addiction |
General Anthropometric & clinical Gastrointestinal symptoms Nutrition Activity — sedentarity Medicines and addiction |
|
| Traceability | ID for identifier number | No ID | Activation key/ID for the kit | Kit ID + sample ID | Sample ID | Sample ID | Kit ID |
| Recommendations | Nutritional advice | Basic and general lifestyle recommendations | Nutritional advice | No recommendations | Nutritional advice + advice for use of food supplements |
Nutritional advice “Superfood” |
|
| Results provided | |||||||
| Microbiota composition | Bacterial diversity | Yes | Yes●☑ | Yes | Yes● | Yes● | Yes● |
| Enterotype | Yes●☑ | Yes | Yes● | Yes● | |||
| Dysbiosis index | Yes | Yes | |||||
| Taxonomy level | Phyla | Phyla● | Phyla● | Phyla | Phyla | ||
| Gut microbes precision | Bacteria associated with SCFA production, H2S production, inflammation, gut barrier, mucus degradation, fermentation | Probiotic bacteria and bacteria associated with inflammation, intestinal mucosa, irritable bowel syndrome, gut-brain/gut-skin/gut-heart or gut-liver axis, insulin balance and joint health | Bacteria of interest (10 species, 4 genera) | Probiotic bacteria and bacteria associated with the control of body weight control, the SCFA production, allergies | 43 bacteria (species or strain level) | Probiotic microbes, “active” gut microbes | |
| Health | Gut health index | Health index | Gut health score ● | ||||
| Functionality | Physical, neuropsychological performances, digestion, SCFA production, immunity & intestinal inflammation | Scores for many functions (inflammation, production of sulfure-butyrate-uric acid-ammoniac-putrescine-vitamins-TMA, bile acid metabolism, LPS biosynthesis) | Digestion efficiency protein fermentation, gas/putrescine/uric acid/butyrate/LPS production, bile acid metabolism, inflammation… |
Description of the different kits included in the benchmark analysis, including their cost, commercialization channel, the presence of a CE mark, consent, time for analysis, methodology used for sample collection and sample analysis, details of their reference cohorts, personal metadata required for the analysis, traceability and services offered (recommendations and different results provided after analysis). ●Indicates that a definition (or at least an explanation) is given by the company in the report. ☑Indicates that the report refers to a scientific publication
One faecal sample from a single healthy (absence of known disease) adult donor was collected at home. After manual homogenization, an aliquot of this sample was sent on the same day to the different companies following the instructions provided with each kit. It was performed with the intention of having a representative overview of a consumer experience with these test kits. For this reason, one sample was sufficient to receive the different reports and compare the results, interpretations and recommendations provided by the different companies or medical laboratories.
To summarize, six different analyses were obtained:
One from a European IVD CE-marked kit (Directive 98/79/EC) commercialized by a medical laboratory
One from a European kit commercialized by a medical laboratory but without being an IVD CE-marked kit
Three European kits commercialized directly via the companies
One from a kit from the USA commercialized directly via the company
Workshop with microbiome experts
Based on the reports provided by the six companies, several critical points were listed (Table 2). These points were discussed with an expert panel during an online workshop organized on 7 March 2023 to assess the current state of these new innovative test kits. These observations were shared and discussed with an interdisciplinary panel of 21 experts from 8 different countries with various expertise in the microbiome field (38% from academic research, 9% from food industry, 19% from pharma and biotechnology industries, 5% from reference laboratories, 10% from regulatory science, 5% dieticians and 5% consultants). This approach (including the overall consumer experience) gave us the opportunity to envisage avenues for the improvement of these test kits and to better meet end-user expectations, both from the consumers’, the patients’ and the healthcare professionals’ points of view. There is, therefore, no intention referring to a particular kit or supplier, nor to influence the choice of one kit over another. For all these reasons, the names of the companies that market the kits shall remain anonymous. This methodology has allowed the expert panel to identify several current gaps and limitations and to propose a series of recommendations to improve the reliability and robustness of these test kits but also to clarify the differences between consumer-directed products and diagnostic devices and the implications this could have for the management of expectations, regulatory policies and consumer communication.
Table 2.
Examples of results and interpretations provided in the different reports
| 1 | 2 | 3 | 4 | 5 | 6 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Health index | Poor gut health — strongly altered microbiome | Improvable health | Average for gut health | |||||||||
| Bacterial diversity index | Good (based on Shannon index) |
Values Diversity — Shannon index: 6.08 Species richness: 277 Species evenness: 0.75 Average for the three parameters but interpreted as “Excellent” |
Value = 250 Average |
Value = 5.82 Average (based on Shannon index) |
Unfavourable | Good | ||||||
| Gut dysbiosis index | High dysbiosis | Small dysbiosis | ||||||||||
| Enterotypes | Ruminococcus | Bacteroides | Bacteroides? | Bacteroides | ||||||||
| Inflammation |
Anti-inflammatory (Lactobacillus ↑) Pro-inflammatory (Proteobacteria ↑) |
Excellent | Average | High = pro-inflammatory | Average | |||||||
| (Value in % of relative abundance) | Value | Interpretation | Value | Interpretation | Value | Interpretation | Value | Interpretation | Value | Interpretation | Value | Interpretation |
| Bacillota | 43.88 | 47.63 | 47.2 | 26.46 | 19 | No value: the results are qualitative and only mention the presence or absence of identified bacteria | ||||||
| Bacteroidota | 46.36 | 42.83 | 46.5 | 63.93 | 77 | |||||||
| Actinobacteriota | 2.27 | 1.77 | 2.3 | 3.72 | 2 | |||||||
| Pseudomonata | 2.39 | 0.93 | 2 | 3.05 | 1 | |||||||
| Verrucomicrobiota | 3.08 | 1.45 | 2 | 0.36 | 1 | |||||||
| Examples of bacteria generally presented in the reports (value in % of relative abundance) | ||||||||||||
| Akkermansia | 0.53 | Normal | 0.34 | High | ||||||||
| Akkermansia muciniphila | 2.44 | High | Normal | 2.04 | Optimal | High | Favourable | |||||
| Bacteroides | 29.16 | Average | 32.21 | High | 36.3 | High | ||||||
| Bifidobacterium | 1.19 | High | 0.53 | Normal | Optimal | 1.68 | High | |||||
| Faecalibacterium | 14.16 | High | 8.41 | Low | ||||||||
| Faecalibacterium prausnitzii | 6.26 | High | High | 5.7 and 0.22* | Optimal | Low | Favourable | |||||
Presentation of some results obtained in the different reports (non-exhaustive list of examples). Conflicting results are obtained for the different indices evaluated (health index/diversity index/gut dysbiosis index), for the determination of enterotypes, for the estimation of inflammation and for the relative abundance of specific taxa (at the phyla or at the genera level)
*The company 3 reported 2 different relative abundances regarding Faecalibacterium prausnitzii for an unknown reason
Results
The lack of standardized and validated methods hinders confidence in microbiome testing services
The analysis of one sample by six microbiome tests showed conflicting results and interpretations
Considering the microbiome analyses performed by the six companies, all the experts expressed concern about the conflicting results received in the different reports. Although this benchmark was not designed to compare results in a statistically significant way, the level of heterogeneity is such that at least some of the test results must be inaccurate. The experts considered the lack of validation of analytical methods as the most plausible explanation for these conflicting results. To illustrate this point, some examples of conflicting results and interpretations are presented in Table 2.
The first example is the conflicting conclusions on the bacterial diversity measurements between the different companies. Whereas three kits concluded an “excellent” or “good” bacterial diversity in the samples (companies 1, 2 and 6), one estimated that the diversity was “unfavourable” (company 5), and two estimated the diversity as “average” (companies 3 and 4). Looking deeper at the values reported for the bacterial diversity, the differences observed in the range of the values were quite important (Table 2), suggesting that the different companies do not refer to the same indices for evaluating diversity. Diversity indices are generally not comparable, even when using the common Shannon diversity; this index is still discordant as it depends upon species abundance estimation. This means that while the specific numeric values might differ, the relative ranking or categorization (e.g. high vs. low diversity) should remain consistent between the different reports, here again raising the question of the representativity of the reference cohort.
The second example relates to the enterotype determination, which also differed among the four companies providing this information in their report. The faecal sample was considered as either Bacteroides-dominant (companies 3 and 5) or Ruminococcus-dominant (company 2), and one of the reports even showed the data without giving a clear interpretation (company 4). Enterotypes determination algorithm makes consensus and is likely the same for all companies. However, this algorithm is dependent upon species abundance estimation and sensitive to the reference cohort. The lack of methodology description prevents to make any conclusion on whether the “enterotype” determination was consistent with the original definition [15].
The third example refers to the relative abundance of different bacterial taxa which varied greatly from one report to another, which despite the uniqueness of the benchmarking sample is hardly compatible with the expected accuracy for such measurements. For instance, the relative abundance of Faecalibacterium was reported as “high” with a relative abundance of 14.16% in one report (company 2, based on 16S rRNA gene amplicon sequencing), whereas it was considered as “low” with a relative abundance of 8.41% in another report (company 4, sequencing approach not reported). A third company (company 1, based on 16S rRNA gene amplicon sequencing) reports the relative abundance of Faecalibacterium prausnitzii, despite the low accuracy of short regions of the 16S rRNA gene that are usually sequenced (e.g. variable regions V3, V4) to resolve taxonomy at low taxonomic ranks, and thus, species-level assumptions should be taken with caution. The absence of transparency in the bioinformatic methodology conducted did not allow us to discern whether the companies assessed differences in gene copy number for 16S rRNA surveys, which might affect relative abundance values (median of 3 and 6 16S rRNA gene copy numbers for A. muciniphila and F. prausnitzii, respectively; search performed in the rrnDB database on 30 July 2024 [16]). While companies 1, 2 and 3 showed similar relative abundance values at the phyla level, companies 4 and 5 widely differed (Table 2). However, these two companies did not provide any information on the sequencing approach and methodology applied, so it was impossible to deduce where the differences might come from. Obviously, the variability in the reference cohorts can induce different interpretations of the levels of some bacteria (considered as low, average, normal or high depending on the report) even though their respective relative abundance did not seem so numerically different (see for instance Bifidobacterium in Table 2).
These interlaboratory variations were already reported in a recent publication evaluating the analytical performance of microbiome testing service in the USA [17]. This variability in next-generation sequencing (NGS) methodologies was also captured during a large, international multicentre evaluation of reference materials developed by NIBCS and recently endorsed by the World Health Organization (WHO) [18, 19]. These studies also proposed a set of Minimum Quality Criteria (MQC) to be reported along with the results of the study, to allow for transparency and comparability between studies.
All steps in analytical pipelines need validation to provide reliable results
The analytical performance of the different kits was not evaluated in the current study. However, this benchmarking comparison enabled us to observe a considerable variability in the sample collection and processing procedures as well as in the analytical methods between the different kits, which could explain the contradictory results (Table 1). The need for validation of all the steps and tools required in the microbiome analysis pipeline was called for during the workshop. Again, the main recommendation is to use, develop and provide standards for each step of microbiome sample collection, processing, sequencing and analysis [20]: sample collection and storage, DNA extraction, primers (if applicable), sequencing technology and platform, bioinformatic software/pipeline and databases, and reference cohorts and/or reference data for results interpretation. These standards are susceptible to evolve following technological improvements.
In addition, a large variability in the extent of personal data to be provided with the faecal sample was observed. Personal data are essential for the interpretation of microbiome data, and such discrepancies in the extent of personal data collected could also explain the differences in the interpretations and recommendations received. Another point which is key in the interpretation of the results is the reference cohort used. In fact, the results of the analysis are compared to a “reference cohort” to allow for interpretation. Some values were considered as “high”, “low”, “favourable” or “average”. However, these interpretations are critically dependent on the characteristics of the reference cohort, and none of the companies or laboratories described their reference cohorts in the report or instructions. It is unclear whether they used publicly available cohorts or an internal database. For the first point, it raises the difficult question of the comparability of the collection, DNA extraction and sequencing procedures of the kit and the corresponding procedures of these public cohorts. For the latter, the reference cohort is sometimes based on the data of the previous consumers without any clear mention to that effect. This raises three important potential issues regarding the reference cohort: firstly, this would question the representativity of the reference cohort due to the interindividual variability of the microbiome; secondly, the consumer should be clearly aware of the potential reuse of their data to constitute this “reference cohort”; and thirdly, the reliability of data, e.g. on users’ health status, is questionable as they do not undergo medical examination.
On the contrary, if the companies do not use their own clients/users for building their databases, but instead use cohorts described in the scientific literature, the references should be clearly stated in the reports, as is required for CE-marked IVD tests. The experts’ recommendations mainly support the need for more transparency in each step of the analysis and call for the development of standards and guidelines for each of these steps.
Some interpretations and recommendations formulated in the different reports are premature and not supported by scientific data
The hype and hope of microbiome testing
Some analyses were performed almost systematically by the companies, such as the assessment of bacterial diversity, the determination of bacterial enterotypes [15], the microbiome composition (at different taxonomic levels) or the relative abundance (or just the presence/absence) of some specific bacteria. Some companies evaluated indices that they named “dysbiosis index” or “gut health index”. There were no scientific references associated with the measures of these indices (“dysbiosis index” and “gut health index”), which raises the question of how these indices are calculated, validated and interpreted. Some of these assessments raised the question of scientific and clinical relevance: is it particularly useful to analyse and interpret microbiome composition at the phylum level? What is the meaning of “high dysbiosis”, “small dysbiosis” or “strongly altered microbiome”? All these measurements highlight an important need to have a consensus on certain specific concepts or definitions that are essential for microbiome analysis and to not use them carelessly. To date, without a scientific consensus on thresholds, it is impossible to say whether a microbiome is healthy [21], has a “good” or “unfavourable” diversity or contains some specific bacteria at “high”/ “low” or even “optimal” levels — this being the latest and most conflictual point as it requires the placement of a certain bacterial abundance between thresholds (“high” or “low” values only refer to the relative abundance overall) and being highly influenced by the methodology conducted. In addition, some reports presented over-interpretations. For example, some reports provided interpretations regarding short-chain fatty acids (SCFA), whereas the SCFA production was not directly assessed but only the relative abundance of a few bacteria known to produce SCFA. Most kits were based on 16S rRNA gene amplicon sequencing; thus, the real capacity of a community to produce SCFA cannot be evaluated. Company 3 claimed to perform shotgun sequencing, but, as the analytical methods were not specified, it is impossible to know if they assessed the metabolic potential of a community based on the evaluation of the presence/absence of genes involved in a certain metabolic pathway or if they operate on the assumption that some metabolites are produced simply because certain taxa were reported in previous studies to produce such metabolites (a biased assumption, as closely related organisms might harbour different genetic pools). Transparency should be a priority in the reporting. Although it might not be necessary to detail the methods in the report intended for a consumer, they are still entitled to an informative feedback, and it is essential to inform the healthcare professional about the technologies used and their possible applications (16S rRNA gene sequencing which only allows for taxonomic evaluation; whole genome shotgun sequencing which allows for more resolutive taxonomic evaluation and, in some cases, for functional profiling and must be easily reachable for any user/practitioner looking for this information, e.g. a link in the report linking to the web page). It is impossible to interpret microbiome data without having the information of the methodology followed [22].
A recommendation was made to distinguish between analysis and reporting for a consumer on the one hand and for a healthcare professional on the other. The analysis and report for the consumer could be mostly descriptive, with straightforward and simple interpretations, whereas the analysis for an IVD purpose would be based on a few validated biomarkers, possibly accompanied by a more detailed report, as these different end users have different levels of knowledge and understanding of the results. In the case of IVD purpose, the trained healthcare professional is responsible for the information and recommendations they provide to their patients.
The lack of clinical relevance for certain interpretations
Several reports mentioned associations between some specific bacteria and various pathological or physiological situations, referring to some of the following examples: “association between increased level of Dorea and irritable bowel syndrome”; “association between increased level of Alistipes and irritable bowel syndrome, colon cancer and depression”; “Desulfovibrio piger participates to intestinal inflammation”; “Collinsella, Bilophila and Sutterella are associated with intestinal mucosa, whereas Lactobacillus, Oscillibacter, F. prausnitzii, Ruminococcus and A. muciniphila are protective bacteria for intestinal mucosa”; or “Christensenella has been associated with the gut microbiome from lean people”. Indeed, a myriad of data are available in the scientific literature about associations between pathologies and modification of microbiome composition and/or function, including emerging models predicting response to cancer treatment [2,23]. However, there remain key scientific questions to answer, such as the cause/consequences relationship of composition or function changes and the causality link between microbiome modification and the onset or progression of disease. Furthermore, these correlations between microbiome modification and disease may not be sufficient to discriminate between healthy and diseased subjects. To begin with, the experts agree that it is premature to mention an association between some specific bacteria and various pathologies in this kind of report and secondly, and such interpretations and conclusions should only be permitted within the framework of IVD. However, in the context of IVD, the current lack of qualified microbiome-based biomarkers makes these interpretations (and their clinical use) impossible at this time [5]. Therefore, the experts considered the interpretations to be premature due to the lack of robust scientific evidence, and the analyses associated with the reports to be of limited clinical utility. In addition, the experts were clear that microbiome tests performed in the context of “curiosity” and “wellbeing” should not mention disease or risk of disease.
The interpretations must be based on peer-reviewed scientific literature (with their corresponding references clearly cited in the report), and no results in the reports should be over-interpreted. No health claims or health associations, direct or implied, should be made in a report intended for a consumer.
The premature or unfounded recommendations based on microbiome testing
In most of the reports, and based on the microbiome analysis, recommendations and especially nutritional advice were provided (Table 1). One company (company 2) provided only well-known and very general nutritional recommendations such as “eat at least 5 fruits or vegetables a day”. These recommendations are aligned with nutritional recommendations made for the general population and are not directly related to microbiome analysis itself. Other reports made inconsistent nutritional recommendations. For example, one report recommended reducing garlic consumption (associating this recommendation to the relative abundance of Desulfovibrio) but further on recommended consuming garlic (now linking this opposite recommendation to the relative abundance of Methanobrevibacter). This type of irreconcilable recommendation may induce the consumer to adopt inappropriate changes in their dietary habits and may even lead to food exclusion behaviours which could be detrimental to health [24]. The experts also pointed out that no known peer-reviewed method exists which can accurately predict the effect of specific dietary recommendation on the microbiome and questioned the ability of foods eaten in small amounts, such as garlic or dill, to have a significant impact on the composition of the microbiome. In addition, changing eating habits should be accompanied by nutrition specialists to ensure that a balanced diet is maintained. It is important to note that one report did not make any recommendations (company 4), and the experts agreed that this was fair as it was too early to make the kind of recommendations that the other reports made. They also agreed that complex and expensive microbiome analyses are not needed to make general dietary recommendations such as “eat 5 fruits or vegetables a day” and could lead to a certain consumer frustration as they are expecting actionable recommendations.
The regulatory framework of microbiome testing is unclear in Europe
There are two different avenues of commercialization for microbiome testing kits in Europe
Whereas some of the microbiome testing kits used in this analysis were directly commercialized by companies, others were made available by medical laboratories. For all the kits included in this analysis, a medical prescription was not required, and the results of the analyses (reports) were sent directly to the consumer whatever the commercialization model. This situation can create confusion regarding the intended use of these test kits, as one could expect that test kits provided by medical laboratories would be proposed as “in vitro diagnostic medical device”. However, in our benchmark, only one medical laboratory offered a kit with a CE mark (granted under Directive 98/79/EC) with the intended use specified on the kit as “diagnosis of the loss of bacterial diversity in the gut microbiota”.
In Europe, the “in vitro diagnostic medical device” is governed by the regulation 2017/746 of the European Parliament and the Council of 5 April 2017 [14]. This new regulation applies for new devices intended for diagnostic use since May 2022, but a transition period is still ongoing for devices CE-IVD marked under the Directive 98/79/EC [13] before this date. This transition period has been extended several times as the medical device competent authorities, and all the stakeholders encountered challenges in the implementation of the new regulation. Medical device competent authorities recently issued a statement recognizing that there is a need to improve the regulatory system and calling for medical devices to be made a priority of the European Commission’s next mandate [25].
The regulation 2017/746 defines the IVD as “any medical device which is a reagent, reagent product, calibrator, control material, kit, instrument, apparatus, piece of equipment, software or system, whether used alone or in combination, intended by the manufacturer to be used in vitro for the examination of specimens, including blood and tissue donations, derived from the human body, solely or principally for the purpose of providing information on one or more of the following: (a) concerning a physiological or pathological process or state; (b) concerning congenital physical or mental impairments; (c) concerning the predisposition to a medical condition or a disease; (d) to determine the safety and compatibility with potential recipients; (e) to predict treatment response or reactions; (f) to define or monitor therapeutic measures.”
The regulation 2017/746 also defines “device for self-testing” as “any device intended by the manufacturer to be used by lay persons, including devices used for testing services offered to lay persons by means of information society services”. This definition is very broad, but the guidance on classification rules for IVD published by the Medical Device Coordination Group (MDCG) clarifies that devices “are considered devices for self-testing when the lay persons carry out at least a part of the testing procedure, such as adding a reagent or placing the specimen on a test cassette. Such actions do not include those needed to collect the specimen or to ensure specimen integrity and stability” [26].
According to these definitions, microbiome testing kits referring to a medical condition or a disease would fall under the scope of the IVDR. However, the question of whether they are considered “self-testing devices” or not could benefit from further clarification from the competent authorities.
The Regulation 2017/746 proposes a classification of IVD into different classes A, B, C and D, taking into account the intended purpose of the devices and inherent risks, from lower to higher risk (Supplementary Table 1). According to the classification rules, IVD microbiome tests would fall at minimum under Class B (following the rule 6 of the IVDR), and self-testing would fall at minimum under Class C (following the rule 4 of the IVDR). To reach the European market, a Class B and higher IVD must undergo conformity assessment through evaluation of their quality management system (QMS, under ISO13485:2016) [27] and/or technical documentation for compliance towards appropriate General Safety and performance Requirements by a notified body.
Another important aspect to consider for the processing of IVD tests is the regulatory framework of medical laboratories and the International Standard ISO15189:2022 [28]. This standard defines the medical laboratory as an “entity for the examination of materials derived from the human body for the purpose of providing information for the diagnosis, monitoring, management, prevention and treatment of disease, or assessment of health” and specifies the requirements for quality and competence in medical laboratories. No mention to the compliance for ISO15189 has been made in the different reports received from the six companies or medical laboratories.
In conclusion, despite the increasing number of microbiome self-testing kits available on the European market, only a few developers have decided to position their microbiome tests as in vitro diagnostic tests and undergone the associated conformity assessment. Among the five European kits purchased, only one affixed the CE mark granted under Directive 98/79/EC (Table 1). To our knowledge, none has been granted to date under the new Regulation 2017/746. A second confusing situation was found where only one component (the faecal swab in our benchmarking) in the kit had the CE mark, while the entire kit itself was not CE marked. This could create further confusion for the consumer. Globally, except for the IVD CE-marked kit, the positioning and regulatory status of these self-management test kits are unclear in Europe and could be confused by some with medical tests.
The USA situation
In the USA, microbiome testing kits such as those investigated in this analysis fall under the “Direct-to-consumer” tests (also referred to as DTC tests) according to FDA categorization [29]. DTC tests are in vitro diagnostics (IVDs) that are marketed directly to consumers without the involvement of a healthcare provider. These tests generally ask the consumer to collect a specimen and send it to the company for testing and analysis. Interestingly, some DTC tests are reviewed by the FDA, while others are not. Indeed, DTC tests for non-medical, general wellness or low-risk medical purposes are not reviewed by the FDA before being marketed [30]. DTC tests for moderate- to high-risk medical purposes, which may have a higher impact on medical care, are generally reviewed by the FDA to determine the validity of test claims. Microbiome DTC tests are marketed without being reviewed by the FDA, as they are considered “low-risk general wellness products” [30]. A general wellness product has “(1) an intended use that relates to maintaining or encouraging a general state of health or a healthy activity, or (2) an intended use that relates to the role of healthy lifestyle with helping to reduce the risk or impact of certain chronic diseases or conditions and where it is well understood and accepted that healthy lifestyle choices may play an important role in health outcomes for the disease or condition” [30]. Two categories of claims can be associated with a general wellness product. The first are claims about sustaining or offering a general improvement to functions associated with a general state of health without making any reference to diseases or conditions. The second are claims about sustaining or offering a general improvement to functions associated with a general state of health while referring to diseases or conditions: for (a) promoting, tracking and/or encouraging choice(s), which, as part of a healthy lifestyle, may help to reduce the risk of certain chronic diseases or conditions and (b) promoting, tracking and/or encouraging choice(s) which, as part of a healthy lifestyle, may help living well with certain chronic diseases or conditions (this should be generally accepted, supported by peer-reviewed publications or official statements by healthcare professional organizations). Two recent publications claim that microbiome DTC tests available in the USA lack analytical and clinical validity, and experts recommend more federal oversight to protect the consumer [9,10]. However, for the tests that do not fall under general wellness due to their claims, one can expect an increased FDA oversight as the FDA has recently adopted a final rule for laboratory developed in vitro diagnostic (IVD) tests [31]. For example, if a commercial lab develops a new microbiome test that does not fall under general wellness due to its claims, such a test would be required to have New York State CLEP (Clinical Laboratory Evaluation Program) Test Approval or have a premarket authorization submission in review with FDA by either 2027 or 2028. Previously, this test could claim to be a “Laboratory Developed Test” exempt from FDA authorization.
The kit purchased in USA for our benchmarking clearly mentioned (on both the website and the report) that the kit is intended for educational and informational use only, and not for a diagnostic purpose. No mention of any disease or condition was found in the report. The company also informed the customer in full transparency that the kit had not been cleared or approved by the FDA.
Different regulatory statuses for different intended uses of microbiome testing should be proposed in the EU
In the European Union, the absence of a clear intended purpose and clear regulatory status of microbiome testing surveys has led to a confusing situation. The experts recommend a need to clarify the intended purpose of these microbiome testing services. Indeed, it appears that some microbiome tests could fulfil a consumer demand to satisfy their curiosity only and in the context of lifestyle/well-being monitoring. However, it is important to specify that based on the IVD regulation 2017/746, a device with the purpose of providing information concerning a physiological process or state qualifies as an IVD. In parallel, healthcare professionals are also particularly interested in these microbiome testing offers for an in vitro diagnostic use. Based on these different intended purposes, it can be envisioned to apply distinct regulatory frameworks of microbiome testing services in the EU, according to the specific nature of both categories (non-IVD versus IVD) (Fig. 1). The first category could be a “wellness”-like category, intended solely for personal information/curiosity with no reference to any risk of diseases or medical conditions but potentially providing nutritional advice, lifestyle and dietary recommendations when the state of the art will fully support these recommendations. The second category would be the IVD CE-marked test, intended for diagnostic, prognostic or prediction of treatment response, in addition to the potential recommendations allowed by the first category. Based on the IVD Regulation 2017/746, microbiome test could be considered:
As minimum Class B if the test is not a self-test, considering dysbiosis is not a disease itself but a risk factor to develop a disease or a marker of a disease.
As minimum Class C if the test is a self-test and/or based on the risk profile of the test. In the case of self-testing devices, this classification could be an issue as the conformity assessment could be burdensome for developers and not proportionate to risk. What is critical for the market is that the framework in place establishes controls that are proportionate to the level of risk of the product.
As Class D, if the test is designed for specific purposes, such as detection of the presence of a transmissible agent before transplantation.
Fig. 1.
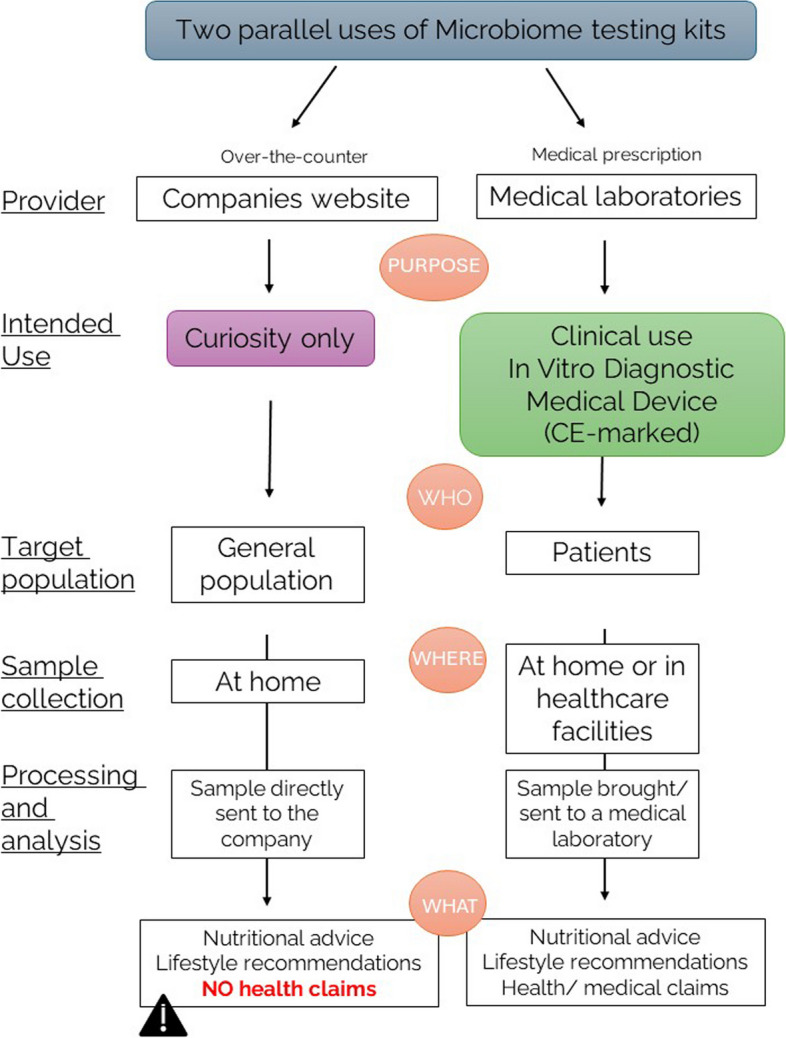
Two different regulatory statuses, depending on the intended use, need to be proposed for the microbiome testing placed on the European market. In terms of intended use, two different microbiome testing kits can be proposed, but currently, the distinction between these two different purposes is still unclear and undefined. In conclusion of a multistakeholder workshop, two different frameworks to separate the microbiome testing kits intended for consumer curiosity only and the microbiome testing kits intended for clinical use and diagnostic were proposed. Microbiome testing kits, designed to satisfy consumer curiosity, can be purchased over the counter (as is currently the case) and are being proposed to the general population. The sample to be analysed is collected at home and sent directly to the company offering the analysis. Based on the results and analysis, general recommendations (dietary or lifestyle recommendations) can be made directly to the consumer, without the assistance of a healthcare professional. However, no health claims or health recommendations can be made in the report. On the other hand, microbiome testing kits intended for clinical use should be regulated as in vitro diagnostic medical devices that comply with the currently applicable regulation on the medical devices (Regulation (EU) 2017/746 of the European Parliament and of the Council of 5 April 2017 on in vitro diagnostic medical devices and repealing Directive 98/79/EC and Commission Decision 2010/227/EU). These devices are intended for a diagnostic purpose and should therefore require a medical prescription. The sample can be collected at home and brought directly or sent to a medical laboratory. In this context, medical claims can be included in the report, which should be sent to a doctor or health professional
The regulatory status, especially for the non-IVD kits, and the classification of IVD kits, will need to be clarified as soon as possible. Reinforcement of the oversight by the competent authorities is also recommended to prevent any misconduct or fraud [6].
The accessibility to the legal notice and privacy policies
In general, access to the privacy policies was not easy, which is consistent with previous observations shared in 2021 by Knoppers and collaborators [6]. The privacy policy was only provided in the IVD CE-marked kit. Data governance is mandatory for protection, duration and retention practices or potential secondary uses such as an inclusion of those data in the reference cohort used by the companies for analysis. Indeed, companies may reuse consumers’ microbiome and personal data to create their own database and internal reference cohort. However, this consent was not provided with the kit, and consumers would not be fully informed and aware of this potential practice if they do not read the policies on the website. Information and policies regarding the samples’ retention time, transfer or reuse should also be mandatory. One of the reports (from a European company) indicated that the analysis was done “in collaboration” with another company (based outside of Europe). This element had not been shared prior to the analysis, raising questions on how and where these samples are really processed, analysed and stored.
Appropriate measures should be implemented to ensure the security of data. Indeed, beyond microbiome data, the companies also process personal data such as identification data (name, address…) and sensitive data from health questionnaires that should be handled appropriately. Considering the IVD marked test, medical information regulatory frameworks should apply. Indeed, the European Commission published a proposal in May 2022 for a Regulation on the European Health Data Space (EHDS) [32]. This Regulation is expected to come into force in autumn 2024. Given the extent of metadata required for test interpretation, such requirements could legitimately apply to non-IVD tests. Some tests may even include artificial intelligence (AI) algorithms, which could require the very new European AI Act (Regulation 2024/1689) [33] to apply in the near future.
It is highly recommended for those companies to adopt the best practices covering transparency related to data and sample use, retention and sharing with different entities. This information should appear on the company website upon purchase and should also be included in the kit with an information notice and a consent form to sign and return to the company with the sample.
Are microbiome data considered as personal information?
The delineation between personal and non-personal data but also between sensitive and non-sensitive data is of paramount importance in determining how to handle these data following the General Data Protection Regulation (GDPR) [34]. Whereas genetic and health-related data are considered as personal sensitive data, it is more difficult to know whether microbiome data fall under the scope of personal sensitive data. On the basis of the criteria listed by the European Commission defining personal data [35], the experts were asked whether they considered microbiome sequencing data deprived of host DNA as sensitive personal data, and this question was hotly debated. No clear answer was obtained, which confirmed the complex nature of the question. Indeed, as certain scientists think that such data cannot be considered as personal sensitive data, the potential presence of human cells and human genetic material in the sample creates confusion. When shotgun sequencing is performed, a good practice to promote could be the systematic deletion of human data from the sequencing dataset prior to any microbiome investigation and/or storage and reuse of the data. The time variable was also discussed, since, based on the current state of the art, it seems impractical to identify the donor; however, some experts thought it could be possible in the near future.
Ethical concerns surrounding microbiome testing
While microbiome testing can potentially promote self-care and microbiome stewardship practices, the relationship between microbiota, lifestyle and health is fluid and dynamical [36]. As already mentioned, the expert panel agreed that the association between microbiome characteristics and the risks of developing certain diseases should never appear in any report intended to satisfy consumer curiosity. Indeed, a consumer could suffer from psychological consequences (e.g. anxiety induced by unwarranted health concerns) and adopt inappropriate dietary or lifestyle habits, upon reading alarming messages without the assistance of a healthcare professional. Even if sufficient scientific evidence becomes available in the future, this type of information should only be included in the report of an IVD test and addressed to a healthcare professional until the technology has reached the required level of maturity. Secondly, some experts expressed the need for worldwide protection of metadata sets. Thirdly, the business model of these companies, which requires the collection of information from consumers to interpret the results, also raised ethical questions among the experts. However, the experts also believe that the creation of large data sets (including microbiome data and personal metadata) could help the research field and lead to the identification of clinical outcomes while respecting privacy policies by complete anonymization. Fourthly, the recommendations of some companies to perform additional and repeated tests over time are also questionable in view of the possible issues related to the comparability of the sequencing data over time due to the current lack of analytical method validation. Additionally, none of the companies mentioned the possibility of making the raw data available, nor provided the data; as such, the question arises as to whether the data still belongs to the donor. The consumer should have the right to access these raw data to carry out other analyses elsewhere. The high price of some kits is another ethical concern. Finally, the experts questioned the fact that some companies also proposed products such as food supplements (such as probiotics, prebiotics and vitamins), including those produced and marketed by these companies.
Discussion and conclusion
The interest in microbiome testing is growing, and the clinical application of microbiome testing kits is seen as useful for the future. However, this investigation highlighted that some microbiome tests are available on the market without authorities’ oversight, without a clear regulatory status, and are making nutritional and health recommendations based on analysis using non-validated analytical methods, leading to a problematic situation.
There are currently two commercialization channels for microbiome testing kits in the European market: (1) those commercialized directly by the companies and (2) those commercialized by medical laboratories. The lack of a clear distinction regarding the regulatory status and commercialization channels of these kits creates great confusion for all end users (consumers, patients and healthcare professionals). While the kits commercialized directly by companies can be offered to satisfy the curiosity of consumers wishing to know the composition of their microbiome, the kits commercialized by medical laboratories could be associated with the “medical expectations” of consumers, patients and healthcare professionals. As with the current classification in the USA, the kits sold in Europe could thus be proposed according to two different regulatory frameworks: on the one hand, the test kits developed to satisfy consumers’ curiosity, with a clear mention of this objective and no association with any medical information, and on the other hand, the IVD CE-marked test kits, which could go further in the analysis and interpretation of the samples, as the report would be intended for trained healthcare professionals. Based on this report, it would remain the responsibility of the healthcare professional to make any recommendation to the patient. However, the increasing availability of microbiome testing kits has led to more patients presenting their results to healthcare providers, many of whom may feel unprepared to interpret or utilize this data. This disconnect can result in patient frustration and, potentially, a loss of trust in the medical system, particularly if patients perceive their microbiome data as being dismissed without adequate explanation. A key question is how training in microbiome science can be integrated or enhanced within medical education to better equip clinicians to interpret these results effectively. The current “complexity” of microbiome testing reports also reflects the already mentioned immaturity of these tests. Thus, when learned societies will accept the clinical demonstration of some medical devices and microbiome-based biomarkers in medical practice, medical schools will undoubtedly follow.
Thanks to our benchmarking and to the multi-stakeholder workshop, a number of avenues for improvement that could be beneficial for both purposes (curiosity and clinical) was identified, and a number of recommendations and actions for improving the reliability of these test kits (Table 3) was proposed. It is important to insist on the fact that the objective of this investigation was not to evaluate or rank the performance of the different microbiome tests, and the first and only goal was to have an understanding of the overall consumer experience and encourage the different companies to improve the quality of their microbiome self-management kits by aligning their development with the end users’ expectations (lay users or healthcare professionals).
Table 3.
List of recommendations suggested by microbiome experts
| Non-IVD testing kits | IVD testing kits | ||
|---|---|---|---|
| Intended use | Curiosity only | Clinical use, diagnostic use | |
| Commercialization channel | Commercialized directly by the companies, publicly available | Commercialized by medical laboratories, medical prescription should be required | |
| Recommendations related to the scientific validity and reliability of the results | Avoid bias in sample collection |
•The companies should provide standard operating procedures (SOP) or instructions to the consumer for the sample collection, based on available international standards in order to improve the reproducibility and reliability of their analysis •The kits components for sample collection must be adapted to the intended analytical method •Use of validated sampling devices as well as compliance with international standards (such as CEN/TS 17626:2021) should be highly recommended |
•The medical laboratories must provide standard operating procedures (SOP) to the patients for the sample collection, based on available international standards in order to ensure the reproducibility and reliability of their analysis •The kit components for sample collection must be validated and suitable for the intended analytical method •Use of validated sampling devices as well as compliance with international standards (such as CEN/TS 17626:2021) is mandatory |
| Harmonize shipping and storage conditions |
•Guidelines/consensus should be developed by the scientific community in order to ensure appropriate storage and shipping conditions (stabilizing buffer, temperature, time…) by the different companies •In the case where the consumer lives in another country and it is not possible to ensure good conditions and a reasonable delivery time for the preservation of the sample, the company should not offer its service |
•Medical laboratories should provide a standardized sampling and clear instructions regarding the delay when shipping the sample. They then should ensure appropriate shipping and storage conditions for intended analysis, even if the sample is processed and analysed in another location | |
| Report methodology and its validation |
•The company should provide at least a general description of the analytical method used and their potential validation. This should be adapted to the level of understanding of the consumer •The company must avoid over-interpretation of the data. The consumer must not be misled •As the reproducibility and comparability of the results can be limited, the company should not advise the consumer for repeated testing over time. In case of repeated testing, the company should propose to the consumer to re-analyse the previous sample(s) with a new sample in exchange for additional costs. This will ensure the comparison between different samples |
•Medical laboratories must provide clear and complete information on the methods used and their validation (“more information leads to more confidence”). This should be adapted to the level of understanding of the end user (different information should be provided to the patient versus healthcare professional) •Medical laboratories must be clear about all of the limitations of the current analytical methods and not provide over-interpretation of the results (for example mentioning SCFA production based only on taxonomic data) •Interpretation of the results and medical consequences should be restricted to healthcare professionals |
|
| Describe the reference cohort |
•Transparency is mandatory for reference cohorts, and the main participants characteristics must be provided prior to analysis as well as in the final report •Reference cohorts should be appropriate to the claims •If a proprietary reference cohort is used, the update of the reference cohort (addition of new user/participants in the cohort) must be mentioned as well as the date of the latest update. If a publicly available reference cohort is used, the reference number of the registered cohort must be indicated prior to the analysis and in the report •The authorities should provide a list of main criteria for the cohort constitution, based on scientific evidence |
•In order to support the development of IVD, collaborative efforts should be engaged to establish consensus on some important microbiome-related concepts such as “healthy microbiome”, “dysbiosis”,…. A regular update/revision of these concepts is greatly needed •In the future, if microbiome-based biomarkers were to be qualified, this would foster the development of IVD. All stakeholders should engage in collaborative efforts to support the qualification of microbiome-based biomarkers in order to support the development of useful tests for clinical routine |
|
| Harmonize and clarify the status of metadata |
•Metadata are necessary for the interpretation of the results. The authorities should provide and/or engage collaborative efforts to establish a validating questionnaire as a “gold metadata standard” to support company and medical laboratories in their activity and to ensure the application of the General Data Protection Regulation •A worldwide coverage concerning the metadata sets is claimed by the experts |
||
| Ensure reliability of analytical methods and interpretation | •Collaborative efforts must be engaged and supported to lead to the standardization of all the steps required for microbiome testing (from sample collection up to downstream analysis of sequencing data) | ||
| Recommendations related to the interpretation of the results | Avoid over-promising interpretation of data |
•Species claims should be made with caution, when analysis was performed using 16S rRNA gene sequencing. Additional evidence concerning species or strain level claims need to be provided regarding the identification (e.g. coverage, average nucleotide identity to type strain) •The different measures, scores and indices must be explained and supported by scientific validation •The results must not be over-interpreted since it dramatically affects the reliability of the results and the credibility of the field •The report for the consumer must be simple, easy to read and to understand without any mention of health and medical claims or any association to particular diseases or risk of disease •Nutritional and lifestyle advice based on microbiome characteristics are too premature, the scientific evidence does not support such recommendations at this time and the messages are often confusing. Specific nutritional recommendations linked to microbiome profiling should be removed from the reports |
•Species claims should be made with caution, when analysis was performed using 16S rRNA gene sequencing. Additional evidence concerning species or strain level claims need to be provided regarding the identification (e.g. coverage, average nucleotide identity to type strain) •An indication of whether the species level is sufficient to assign clinical relevance to any organism mentioned in the report should be clearly stated •The different measures, scores and indices must be explained and supported by scientific validation (gut health index, dysbiosis index, …) •Only a trained healthcare professional should interpret the results of this test and make medical recommendations or a diagnostic •The report sent to the healthcare professional should include details of the analysis and analytical methods used. Another, simpler report could also be made available to the patient |
| Recommendations related to the regulatory status in Europe | Clarify the regulatory status |
•A reinforced oversight from the European authorities is requested •Clarification of the regulatory status and framework are highly needed in Europe |
|
| Avoid over-promising messages |
•Health claims must be removed from commercial marketing messages •It is critical to be clear and honest on the goals achieved by this analysis, so as to avoid over-promising expectations •There is a clear need for educational information for the consumer |
•No promotional messages, the analysis must be done under a medical prescription for a specific health condition •Health/medical claims are supported with scientific evidence •Regular training for the healthcare professionals to ensure an appropriate interpretation of microbiome data |
|
| Recommendations related to the legal notice, privacy policies and ethical aspects | Protect the consumer/patient data (microbiome data and personal data) |
•The adoption of the best practices covering transparency related to data and samples use, retention and sharing with different entities should be mandatory •The privacy policy should be easily accessible on the company’s website (upon purchase) and should be included in the kit with a consent form to sign and to return to the company with the sample •Data policy for the samples’ retention time must be clear •For ensuring that consumer rights are upheld, data protections must extend to scenarios involving the sale or liquidation of a company |
•For ensuring that consumer rights are upheld, data protections must extend to scenarios involving the closure or transfer of a medical laboratory |
| Clarify the status of microbiome data (sensitive personal data or not) | •The authorities should help to define whether microbiome sequencing data are personal sensitive data or not (based on the GDPR) | ||
| Respect ethical principles |
•No reference to pathologies/diseases should appear on the report as the consumer can suffer from psychological consequences of receiving alarming messages •The price of these kits is a key concern, especially regarding the limited interpretation and recommendation that can be done today based on microbiome data. Repeated testing should not be recommended due to the price and limited added value of repeated testing (see above, lack of comparison of the data obtained during consecutive tests) •The business model of these companies is not clearly described, and the consumer has to be fully aware of the use of personal data to create private databases • “Name-dropping” should be avoided in the reports provided by the companies |
•The sequencing raw data should be sent to the healthcare professional | |
Based on the analysis of the different services offered and the reports received after the analysis, and on the discussion with experts in the microbiome field during a workshop held on 7 March 2023, a list of recommendations has been established. Common and specific recommendations to support and encourage the development of microbiome testing services have been listed for non-IVD kits (intended for curiosity only) and for the in vitro diagnostic medical device (IVD, intended for diagnostic or clinical use)
The lack of consensus on the extent of analytical methods validation required for these tests has raised concerns from different stakeholders in the microbiome field. The current investigation unfortunately indicates a lack of consistency of the results among different companies or medical laboratories. This observation is aligned with a recent publication in which the authors concluded that regulators should develop requirements for the industry to document and demonstrate the consistency and validity of methods and claims [10]. One worry from some microbiome researchers is that validation of analytical methods could impair innovation in the field. However, the purpose of a validation process is not to force the companies to share their analytical pipelines, or to impose a single analytical pipeline that would be incompatible with innovation and anti-trust policy, but on the contrary to support them by increasing the quality of their service by providing robust, reproducible, reliable and consistent results. Collaborative efforts must be engaged to foster the validation of all steps required for microbiome testing (from sample collection all the way to downstream processing of sequencing data). Promoting awareness of the test kits already available for the standardization of microbiome analytical methods (international standards, standard operating procedures (SOPs) or reference materials…) is also essential to increase their adoption [5,18,37–39]. The dialogue between standardization bodies/developers and academic or clinical end users also needs to be engaged to improve the development of the different tools necessary for the objective of validation. As is the case for other medical analyses, external quality assessments (EQA) should also be proposed for the IVD test kits dedicated to a diagnostic or other clinical purpose. These EQA will include external audits as well as proficiency testing with samples sent by reference nodal laboratories to be analysed and described [40]. Regular proficiency testing will improve the reliability of these tools and will be essential to allow the translation of microbiome research into a more standardized and routine diagnostic procedure. Moreover, different analytical methods and pipelines could be envisioned according to the intended use of microbiome testing, meaning that the methodology used for a diagnostic purpose could be different from those adopted for the purpose of satisfying one’s curiosity. In this context, IVD test kits (with a clinical purpose) will require standardized and validated technologies to ensure the reliability, reproducibility and comparability of the results.
To avoid over-promising messages, an effective, efficient and proportionate governance approach must be implemented, one which combines scientific insights with user experience and priorities. The lack of reliability, reproducibility, standardization and authorities’ oversight has led to a situation that is neither beneficial for the consumer nor for the companies and that will in turn affect the entire microbiome research sector: (1) consumers spending a large sum of money to obtain an unreliable analysis and premature recommendations, (2) companies having poor feedback about their service and seeing a loss of confidence from end users (consumers, patients and healthcare professionals) in their device, and (3) credibility of the entire microbiome field being affected by the lack of reproducible results, inconsistent interpretations and conflicting results apparent in the scientific literature. However, some tools, which involve components sometimes similar to the tests described here, help the research sector since the analysis of large cohorts can help to identify new avenues and support the development of microbiome-based biomarkers [5,21]. Nevertheless, their pertinence still depends on the availability of the raw sequencing data, the transparency of the analytical methods and the appropriate quality of collected personal metadata. In return, the qualification of microbiome-based biomarkers will foster the development of IVD applicable in clinical routine and will strongly support the integration of microbiome data in clinical practice.
The key message from this multi-stakeholder investigation remains that collaborative efforts and an understanding the needs of scientists, consumers, patients and healthcare professionals are essential to enhance the quality and usefulness of microbiome testing devices, which are both sought after by consumers and eagerly expected in clinical practice.
Supplementary Information
Additional file 1: Supplementary Table 1. List of definitions
Acknowledgements
At the time of workshop, Austin Udocor was affiliated with DNA Genotek but is now at GeneDx.
Authors are very grateful to Joseph Simmons for English proofreading.
Human Microbiome Action Consortium
Alessio Fasano, Federica Carraturo, Jonel Trebicka, Yolanda Godoy, Robert Schierwagen, Peer Bork, Anandhi Iyappan, Typas Nassos, Hazenbrink Dienty Hendrina Maria Johanna, Hub Zwart, Laurence Zitvogel, Lisa Derosa, Carolina Alves Costa Silva, Joel Doré, Hervé Blottière, Aicha Kriaa, Emmanuelle Maguin, Moez Rhimi, Patrick Veiga, Nicolas Pons, Zahra Hassani, Pierre-Louis Prost, Fay Betsou, Celine Druart, Magali Cordaillat-Simmons, Julie Rodriguez, Alexander Jarde, Isabelle Boutron, Philippe Ravaud, Aleksander Krag, Mads Israelsen, Ida Falk Villesen, Dirk Haller, Amira Metwaly, Paul Ross, Paul O’Toole, Aonghus Lavelle, Marcus Claesson, Raphaela Joos, Colin Hill, Andrey Shkoporov, Saba Loftus, Katy Boucher, Manimozhayan Arumugam, Arjun Sarathi, Vitalina Morozova, Nicola Segata, Francesco Asnicar, and Federica Pinto.
Authors’ contributions
JR, MCS and CD designed the study, performed the non-exhaustive benchmarking analysis and organised the discussions and workshop. All authors participated in the discussions and workshop. All authors discussed the recommendations in the manuscript. JR, JD, CD made the synthesis of all the discussions. JR and CD drafted the manuscript. All authors read, revised and approved the final manuscript.
Funding
This paper is the result of a collaborative workshop performed in the context of a project entitled International Human Microbiome Coordination and Support Action (IHMCSA) which receives funding from the European Union’s Horizon 2020 Research and Innovation Program under grant agreement no. 964590.
Contribution of J. D. was supported in part by the European Commission in the context of ERC-2017-AdG No. 788191 and in the context of MetaGenoPolis grant ANR-11-DPBS-0001.
N. M. Q. is currently funded by the European Union’s Horizon 2020 Research and Innovation Program under the Marie Skłodowska-Curie grant agreement no. 101034371.
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Donor has consent for publication.
Competing interests
JS is co-founder and CSO of MyMicroZoo. RdL is co-founder and CTO of GMT Science.
The other authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Julie Rodriguez, Email: julie@pharmabiotic.org.
Human Microbiome Action Consortium:
Julie Rodriguez, Magali Cordaillat-Simmons, Aonghus Lavelle, Amira Metwaly, Hub Zwart, Alessio Fasano, Federica Carraturo, Jonel Trebicka, Yolanda Godoy, Robert Schierwagen, Peer Bork, Anandhi Iyappan, Typas Nassos, Hazenbrink Dienty Hendrina Maria Johanna, Laurence Zitvogel, Lisa Derosa, Carolina Alves Costa Silva, Joel Doré, Hervé Blottière, Aicha Kriaa, Moez Rhimi, Patrick Veiga, Nicolas Pons, Zahra Hassani, Pierre-Louis Prost, Fay Betsou, Celine Druart, Alexander Jarde, Isabelle Boutron, Philippe Ravaud, Aleksander Krag, Mads Israelsen, Ida Falk Villesen, Dirk Haller, Paul Ross, Paul O’Toole, Marcus Claesson, Raphaela Joos, Colin Hill, Andrey Shkoporov, Saba Loftus, Katy Boucher, Manimozhayan Arumugam, Arjun Sarathi, Vitalina Morozova, Nicola Segata, Francesco Asnicar, and Federica Pinto
References
- 1.Schmidt TSB, Raes J, Bork P. The human gut microbiome: from association to modulation. Cell. 2018;172:1198–215. [DOI] [PubMed] [Google Scholar]
- 2.Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. N Engl J Med. 2016;375:2369–79. [DOI] [PubMed] [Google Scholar]
- 3.The International Microbiota Observatory. The International Microbiota Observatory2023 results. https://www.biocodexmicrobiotainstitute.com/en/international-microbiota-observatory.
- 4.Million Microbiomes from Humans Project - MMHP. https://db.cngb.org/mmhp/ (accessed Aug 21, 2024).
- 5.Rodriguez J, Hassani Z, Alves Costa Silva C, et al. State of the art and the future of microbiome-based biomarkers: a multidisciplinary Delphi consensus. Lancet Microbe 2024; : 100948. [DOI] [PubMed]
- 6.Knoppers T, Beauchamp E, Dewar K, et al. The omics of our lives: practices and policies of direct-to-consumer epigenetic and microbiomic testing companies. New Genetics and Society. 2021;40:541–69. [Google Scholar]
- 7.Gilbert J, Blaser MJ, Caporaso JG, Jansson J, Lynch SV, Knight R. Current understanding of the human microbiome. Nat Med. 2018;24:392–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chadwick R, Zwart H. From ELSA to responsible research and promisomics. Life Sciences, Society and Policy. 2013;9:3. [Google Scholar]
- 9.Hepatology TLG. Direct-to-consumer microbiome testing needs regulation. Lancet Gastroenterol Hepatol. 2024;9:583. [DOI] [PubMed] [Google Scholar]
- 10.Hoffmann DE, von Rosenvinge EC, Roghmann M-C, Palumbo FB, McDonald D, Ravel J. The DTC microbiome testing industry needs more regulation. Science. 2024;383:1176–9. [DOI] [PubMed] [Google Scholar]
- 11.Gimbert C, Lapointe F-J. Self-tracking the microbiome: where do we go from here? Microbiome. 2015;3:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Human Microbiome Action Project. https://humanmicrobiomeaction.eu/. Accessed 21 Aug 2024.
- 13.Directive 98/79/EC of the European Parliament and of the Council of 27 October 1998 on in vitro diagnostic medical devices. 1998 http://data.europa.eu/eli/dir/1998/79/oj (accessed Aug 21, 2024).
- 14.The European Parliament and Council of the European Union. Regulation (EU) 2017/746 of the European Parliament and of the Council of 5 April 2017 on in vitro diagnostic medical devices and repealing Directive 98/79/EC and Commission Decision 2010/227/EU. 2017 https://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:32017R0746.
- 15.Arumugam M, Raes J, Pelletier E, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stoddard SF, Smith BJ, Hein R, Roller BRK, Schmidt TM. rrnDB: improved tools for interpreting rRNA gene abundance in bacteria and archaea and a new foundation for future development. Nucleic Acids Res. 2015;43:D593-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Servetas SL, Hoffmann D, Ravel J, Jackson SA. Evaluating the analytical performance of direct-to-consumer gut microbiome testing services. 2024; : 2024.06.05.596628.
- 18.Sergaki C, Anwar S, Hassall J, et al. WHO/BS/2022.2416: a WHO collaborative study to evaluate the candidate 1st WHO International Reference Reagents for Gut Microbiome analysis by Next-Generation Sequencing. https://www.who.int/publications/m/item/who-bs-2022.2416 (accessed March 29, 2024).
- 19.WHO/BS/2023.2455 WHO 1st Reference Reagent for DNA extraction of gut microbiome. https://www.who.int/publications/m/item/who-bs-2023.2455 (accessed Aug 22, 2024).
- 20.Barcenilla C, Cobo-Díaz JF, De Filippis F, et al. Improved sampling and DNA extraction procedures for microbiome analysis in food-processing environments. Nat Protoc. 2024;19:1291–310. [DOI] [PubMed] [Google Scholar]
- 21.Joos R, Boucher K, Lavelle A, et al. Examining the healthy human microbiome concept. Nat Rev Microbiol In Press. [DOI] [PubMed]
- 22.Sharon I, Quijada NM, Pasolli E, et al. The core human microbiome: does it exist and how can we find it? A critical review of the concept. Nutrients. 2022;14:2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Derosa L, Iebba V, Silva CAC, et al. Custom scoring based on ecological topology of gut microbiota associated with cancer immunotherapy outcome. Cell. 2024;187:3373-3389.e16. [DOI] [PubMed] [Google Scholar]
- 24.Sergeant P, Morisset M, Beaudoin É, Renaudin J-M, Kanny G. Les conséquences nutritionnelles des régimes d’éviction pour allergies alimentaires : le point de vue de la diététicienne. Revue Française d’Allergologie. 2009;49:143–6. [Google Scholar]
- 25.Competent authorities for medical devices. Medical device competent authority statement on the status of the EU regulatory. 2024; published online July 12. https://www.camd-europe.eu/regulatory/publication-of-a-consensus-statement/.
- 26.MDCG 2020–16 Rev.3 - Guidance on Classification Rules for in vitro Diagnostic Medical Devices under Regulation (EU) 2017/746 - July 2024 - European Commission. https://health.ec.europa.eu/latest-updates/mdcg-2020-16-rev3-guidance-classification-rules-vitro-diagnostic-medical-devices-under-regulation-eu-2024-07-08_en (accessed Sept 26, 2024).
- 27.ISO: the International Organization for Standardization. ISO 13485:2016. ISO. https://www.iso.org/standard/59752.html (accessed Aug 28, 2024).
- 28.ISO: the International Organization for Standardization. ISO 15189:2022. ISO. https://www.iso.org/standard/76677.html (accessed Aug 21, 2024).
- 29.Direct-to-Consumer Tests. U.S. Food and Drug Administration. https://www.fda.gov/medical-devices/in-vitro-diagnostics/direct-consumer-tests#list.
- 30.General Wellness: Policy for Low Risk Devices Guidance for Industry and Food and Drug Administration Staff. U.S. Food and Drug Administration. 2019. https://www.fda.gov/regulatory-information/search-fda-guidance-documents/general-wellness-policy-low-risk-devices.
- 31.Health C for D and R. Laboratory developed tests. FDA. 2024; published online Aug 14. https://www.fda.gov/medical-devices/in-vitro-diagnostics/laboratory-developed-tests (accessed Aug 28, 2024).
- 32.Proposal for a Regulation of the European Parliament and of the Council on the European Health Data Space. 2022 https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX%3A52022PC0197 (accessed Aug 22, 2024).
- 33.Regulation (EU) 2024/1689 of the European Parliament and of the Council of 13 June 2024 laying down harmonised rules on artificial intelligence and amending Regulations (EC) No 300/2008, (EU) No 167/2013, (EU) No 168/2013, (EU) 2018/858, (EU) 2018/1139 and (EU) 2019/2144 and Directives 2014/90/EU, (EU) 2016/797 and (EU) 2020/1828 (Artificial Intelligence Act) (Text with EEA relevance). 2024 http://data.europa.eu/eli/reg/2024/1689/oj/eng (accessed Aug 22, 2024).
- 34.Finck M, Pallas F. They who must not be identified—distinguishing personal from non-personal data under the GDPR. International Data Privacy Law 2020; 10.
- 35.What personal data is considered sensitive? European Commission. https://commission.europa.eu/law/law-topic/data-protection/reform/rules-business-and-organisations/legal-grounds-processing-data/sensitive-data/what-personal-data-considered-sensitive_en.
- 36.Rook O, Zwart H. Awareness of the human microbiome may promote healthier lifestyle and more positive environmental attitudes. Communications Medicine Under review.
- 37.Amos GCA, Logan A, Anwar S, et al. Developing standards for the microbiome field. Microbiome. 2020;8:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sergaki C, Anwar S, Fritzsche M, et al. Developing whole cell standards for the microbiome field. Microbiome. 2022;10:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.© IHMS Consortium © Dore, J., Ehrlich, S.D., Levenez, F., Pelletier, E., Alberti, A., Bertrand, L., Bork, P., Costea, P.I., Sunagawa, S., Guarner, F., Manichanh, C., Santiago, A., Zhao, L., Shen, J., Zhang, C., Versalovic, J., Luna, R.A., Petrosino, J., Yang, H., Li, S., Wang, J., Allen‐Vercoe, E., Gloor, G., Singh, B.and IHMS Consortium (2015). International Human Microbiome Standards. International Human Microbiome Standards. https://human-microbiome.org/index.php?id=sop_all.
- 40.Scherz V, Greub G, Bertelli C. Building up a clinical microbiota profiling: a quality framework proposal. Crit Rev Microbiol. 2022;48:356–75. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary Table 1. List of definitions
Data Availability Statement
No datasets were generated or analysed during the current study.


