Abstract
Background
Pancreatic Ductal Adenocarcinoma (PDAC) is among the most aggressive cancers, characterized by high mortality rates. Studies on various cancers across the globe indicate that regulatory miRNAs play a vital role in cellular signaling. However, the expression and interactions of these miRNAs in the Pakistani patients with PDAC is yet to be explored. Here, we aim to investigate a panel of four regulatory miRNAs (miRNA 34a, 30b, 142 and 137) in PDAC and their interaction with selected target proteins in the signaling pathway (KRAS, p53, BRCA1, APC).
Methods
We conducted a study on 109 PDAC patients to analyze the selected miRNAs and protein targets. Formalin Fixed Paraffin Embedded (FFPE) tumor samples were obtained from the hospital’s department of histopathology. After confirmation of diagnosis and appropriate tumor content, tissues were processed for RNA extraction. Based on the acceptable quality and quantity of RNA, 43 samples were proceeded for qRT-PCR. Relative expression of the miRNAs was determined through 2−[ΔΔCt] method. Further, FFPE tumor blocks were used to perform tissue sectioning followed by immunohistochemistry experiments. Stained slides were scored independently by two pathologists according to set criteria.
Results
Expression profiles revealed that miRNA 34a, 30b, and 142 showed high expression in approximately 69–70% of cases, while miRNA 137 had a lower high expression frequency (53.4%). Among protein biomarkers, KRAS, BRCA1, and APC were predominantly expressed, with high expression levels observed in 79.1%, 69.8%, and 51.2% of cases, respectively, whereas p53 showed positive expression in only 34.9% of cases. Statistical analysis showed that expression of miRNA 34a was significantly associated with the expression of BRCA1 (p = 0.034). No significant associations were observed for KRAS, p53, or APC with the selected miRNAs. Moreover, the expression of miRNA 34a independently showed significant association with miRNA 30b (p = 0.000) and miRNA 137 (p = 0.001). None of the miRNA showed an association with the overall survival, patient demographics or the clinicopathological characteristics.
Conclusion
Our study highlights a potential bi-directional regulatory relationship between BRCA1 and miRNA 34a, suggesting that miRNA 34a may both respond to and influence BRCA1 activity within cellular signaling pathways. This complex interaction points to a layered regulatory network that could play a crucial role in tumor suppression in PDAC, underscoring the therapeutic potential of targeting this miRNA-protein crosstalk.
Keywords: Pancreatic ductal adenocarcinoma, MiRNA 34a, MiRNA 30b, MiRNA 142, MiRNA 137, BRCA1
Introduction
Pancreatic cancer is among the most aggressive forms of cancer, responsible for the sixth-highest number of cancer-related deaths worldwide [1]. For patients with early-stage Pancreatic Ductal Adenocarcinoma (PDAC), surgery is the primary treatment option and can significantly improve survival rates. However, nearly 80% of patients are diagnosed at an advanced stage, where surgical intervention is no longer feasible, leaving chemotherapy as the only treatment option [2]. Despite advances in surgical techniques and chemotherapy, the 5-year survival rate remains alarmingly low at 12.5% [3]. Over the years, Pakistani population has been underrepresented in global pancreatic cancer research. According to the International Agency for Research on Cancer (IARC), the mortality rate for pancreatic cancer in Pakistan is notably high, reaching a staggering 97% [1]. Despite these concerning statistics, research focused on this population has been limited, both globally and locally [4].
To address this research gap, our group conducted a pilot study [5] to evaluate genetic alterations in Pakistani PDAC patients. Our findings identified pathogenic variants in key oncogenes and tumor suppressors, including KRAS, TP53, BRCA1, and APC, highlighting the unique genetic profile of PDAC in this underrepresented population. The identification of these key genes enabled an in-depth exploration of their associated regulatory pathways. Consequently, specific miRNAs (miRNA-34a, miRNA-30b, miRNA-142, and miRNA-137), known for their roles in regulating the expression of these genes, were highlighted. We hypothesize that transcriptomic alterations in these miRNAs may contribute to the pathology of PDAC.
It is well established that regulatory microRNAs (miRNAs) play crucial roles in key oncogenic signaling pathways. miRNAs are small, non-coding RNA molecules (~ 19–25 nucleotides in length) that regulate gene expression by binding to target mRNAs after transcription, leading to either inhibition or repression of the target gene expression. miRNAs are pivotal in regulating various biological processes, including cell proliferation, differentiation, apoptosis, the cell cycle, metabolism, and immune responses [6, 7]. Reduced expression of miRNA 34a, for instance, has been documented in multiple cancers, including leukemia and lung cancer. miRNA 30b, which is implicated in processes such as differentiation and inflammation, targets critical proteins like KRAS and p53, and its decreased expression is associated with poor prognosis in pancreatic cancer. Similarly, low levels of miRNA 142 and miRNA 137 have been linked to poor survival in gastric, colon, and pancreatic cancers, respectively [8–11]. Together, these miRNAs play significant roles in cancer biology by regulating key pathways and molecular targets. Understanding their functions and interactions in PDAC could open new avenues for developing targeted therapies and improving patient outcomes.
In the current study, we investigated these regulatory miRNAs (miRNA 34a, miRNA 30b, miRNA 142, and miRNA 137) and their involvement in the signaling pathways of KRAS, p53, BRCA1, and APC in PDAC. Our study further examines the associations of these miRNAs with clinicopathological characteristics and patient survival, aiming to deepen our understanding of PDAC pathogenesis in this unique patient population. This approach represents an essential step toward identifying potential therapeutic targets and advancing tailored treatment options for PDAC patients.
Methodology
Patient enrollment and sample collection
A retrospective cohort study was performed on patients with PDAC. Initially, a total of 109 Formalin-fixed paraffin-embedded (FFPE) tumor samples were obtained from the Department of Histopathology at Aga Khan University Hospital (AKUH), Karachi, Pakistan. The study was conducted in accordance with the Declaration of Helsinki and was approved by the Ethical Review Committee of AKUH (ERC No. 2023–6278-25,839). The inclusion criteria were PDAC patients aged 18 years and older who had undergone biopsy or surgery at AKUH between January 2014 and December 2022. Informed consent was obtained from all participants. Patients who did not meet these criteria were excluded from the study. There was no gender preference applied during enrollment. Clinical data were collected by reviewing patient medical records. Follow-up calls were made at six-month intervals to monitor their health status. The last follow-up call conducted in June 2024.
Total RNA extraction and quantification
RNA was extracted from the FFPE tumor samples and adjacent normal tissues using the miRNAeasy FFPE kit (Qiagen, USA, Cat No. 217504). Adjacent normal tissues, confirmed by the histopathologist to be free of malignant cells, served as controls. For both RNA extraction and quantification, the manufacturer’s recommended protocol was followed. RNA quantification was carried out using spectrophotometry DS-11vs (DeNovix, USA). The A260/280 ratio of 2.0 was considered optimal for all the samples. Based on the acceptable quality ratio and quantity of the RNA extracted from tumor and the adjacent normal tissues, a total of 43 samples were selected for downstream processing.
Synthesis of complementary DNA (cDNA)
cDNA synthesis was performed using TaqMan MicroRNA Reverse Transcription Kit miSCRIPT II RT kit (Qiagen, USA, Cat No. 218160). A total of 10 ng RNA sample was used to prepare 20 µl reaction mix. Each tube was placed in the thermal cycler for 60 min incubation at 37 °C followed by 5 min incubation at 95 °C. Further, the prepared cDNA samples were immediately placed on ice and diluted (1:20) in RNAse free-water before performing the miRNA quantification step.
Real-Time PCR (RT-PCR) quantification of miRNA expression
Real time PCR was performed using miRNA specific forward and reverse primers, cDNA and miRCURY LNA SYBR® Green PCR Kit (Qiagen, USA, Cat No. 339345). Reaction mixture was prepared and cycling conditions were set according to the manufacturer’s instructions. After the reaction completion, Ct values were obtained. For normalizing the expression of the target, U6 primers were used as internal control in order to calculate ΔCt for each sample. U6 primer sequence is as given: Forward: 5′-CTCGCTTCGGCAGCACA-3′, Reverse: 5′-AACGCTTCACGAATTTGCGT-3’ [12]. Relative expression of individual miRNA and U6 transcripts was calculated using 2−[ΔΔCt] method [13, 14]. miRNA primer sequences and cycling conditions are given in Table 1.
Table 1.
Cycling conditions and primer sequences for miRNAs
| miRNAs | Primers | Reference |
|---|---|---|
| miRNA 34a |
Forward: 5’—TGGCAGTGTCTTAGCTGGTTG—3’ Reverse: 5’—GGCAGTATACTTGCTGATTGCTT—3’ |
[15, 16] |
| miRNA 30b |
Forward: 5′- CGCGCTGTAAACATCCTACAC −3′ Reverse: 5′- GTGCAGGGTCCGAGGT −3′ |
[17] |
| miRNA 142 |
Forward: 5′-AACTCCAGCTGGTCCTTAG-3′ Reverse: 5′-TCTTGAACCCTCATCCTGT-3′ |
[12] |
| miRNA 137 |
Forward: 5′‐GCTCCTCAGGTCGAACCTATTG‐3′ Reverse: 5′‐CCGACGCTATTGCTTAAGAATACG‐3′ |
[18] |
Sectioning
A senior histopathologist in the research team conducted Hematoxylin and Eosin (H&E) staining on representative tissue blocks to confirm the tumor type diagnosis and ensure adequate tumor content. Tissues with over 20% tumor cells were selected for further processing. Following this, 4 µm sections were cut from each FFPE block using a semi-automatic microtome (pfm Rotary 3005 E, pfm medical, Germany). The sections were then transferred to a floating hot water bath to remove any wrinkles before being placed onto charged glass slides (FLEX IHC Microscope Slides, K8020, Dako, Denmark).
Immunohistochemistry
The optimal experimental conditions for each antibody were initially determined based on the manufacturer’s guidelines. Immunohistochemical analysis was performed on a specific panel of proteins (KRAS, p53, BRCA1, APC) using either the EnVision FLEX High pH (Link) system (K8000221, Dako, Denmark) or the Low pH (Link) system (K800521-2, Dako, Denmark). Tumor sections were deparaffinization and rehydrated using xylene followed by a graded series of ethanol solutions (100%, 90%, 70%, and 50%). Antigen retrieval was performed using a high pH method for p53, while the remaining antibodies were processed using the low pH method. The slides were then immersed in retrieval solution (K8004, Dako, Denmark) and incubated at 90 °C for 40 min. Cellular peroxidase activity was blocked using 0.03% hydrogen peroxide solution (S2023, Dako, Denmark).
Slides were incubated with primary antibodies at room temperature for the optimized durations as specified in Table 2, followed by incubation with Horse Radish Peroxidase (HRP)-conjugated EnVision secondary antibodies (labeled-polymer Rabbit/Mouse, Dako, Denmark) under the same conditions as the primary antibodies. Between each step, slides were washed using Tween 20 and Tris-buffered saline containing wash buffer (S3006, Dako, Denmark). Diaminobenzidine (DAB) chromogen (GV825, Dako, Denmark) was used to visualize the antibody-antigen reactions. All slides were counterstained with hematoxylin (CS70030, Dako, Denmark), then dehydrated through a reverse series of graded ethanol solutions (50%, 70%, 90%, and 100%) before coverslips were mounted using a toluene-free mounting medium (CS705, Dako, Denmark). Positive and negative controls were included in each batch to validate the results. Negative control slides were processed by incubating tissue with saline instead of the primary antibody. The evaluation and scoring of immunohistochemical results were carried out by two independent pathologists using a microscope at 20–40 × magnification. Scoring criteria were established for each antibody, and any discrepancies between the observers were resolved using a conference microscope. Details on positive controls, expression patterns and scoring criteria for each antibody are provided in Table 2.
Table 2.
Details for antibodies, immunohistochemistry optimization conditions and scoring criteria used in the study for detection of target protein expression
| SNo | Antibody | Company/ Clone/ RRID No |
Antigen Retrieval pH/ Duration | Ab Incubation Time | Positive Control | Dilution | Expression Pattern | Scoring Criteria | Reference |
|---|---|---|---|---|---|---|---|---|---|
| 1 | KRAS |
Abcam, UK Rabbit polyclonal anti human Cat# ab180772 RRID: AB_2884935 |
Low pH 40 min |
60 min | Known positive case of colorectal carcinoma | 1:100 | Membranous and Cytosolic |
0 mean no staining is observed or membrane staining is observed in < 10% of tumor cells; 1 + mean faint or partly membrane staining is found in > 10% of tumor cells; 2 + represented weak to moderate complete membrane staining is detected in > 10% of tumor cells; 3 + represented strong, complete membrane staining is observed in > 10% of tumor cells |
[19] |
| 2 | p53 |
Dako, Denmark Mouse monoclonal anti human Clone: DO-7 Cat# M7001 RRID: AB_2206626 |
High pH 40 min |
30 min | Known positive case of glioblastoma | Ready to use | Nuclear |
0 / negative (when no staining was observed), 1 (< 5% cells are stained), 2 (5%−50% cells stained), and 3 (> 50% of cells stained) |
[20] |
| 3 | BRCA1 |
Abcam, UK Mouse monoclonal anti human Clone: MS110 Cat# ab16780 RRID: AB_2259338 |
Low pH 20 min |
60 min | Known positive case of breast carcinoma | 1:100 | Membranous and Cytosolic |
(0) no staining and few scattered positive cells (< 5%), (1) 5%−25% of cells stained, (2) 26%−50% of cells stained, (3) 51%−75% of cells stained, and (4) 76%−100% of cells stained Staining intensity was categorized as: (0) no staining, (1) weak staining, (2) moderate staining, and (3) strong staining Positive staining results were divided into low– and high–staining intensity groups, where 1–2 were classified as a low-staining group and 3–4 as a high-staining group |
[21] |
| 4 | APC |
Abcam, UK Rabbit monoclonal anti human Clone: EP701Y Cat# ab40778 RRID: AB_2057497 |
Low pH 40 min |
60 min | Known positive case of colorectal carcinoma | 1:100 | Membranous and Cytosolic |
The intensity of staining was rated as either (0) negative, (1) low, (2) moderate, or (3) high Area of distribution was determined by percentage and it was scored from 0 to 100 The ultimate score was the outcome of multiplying the percentage of tumor cells stained (0 to 100) by staining intensity (0 to 3) Cutoff score of 100 was used to differentiate between negative (< 100) and positive (> 100) results |
[22] |
Statistical analysis
The statistical analysis was performed using SPSS® version 23. Paired t test was used to evaluate the difference in miRNA expression between the tumor and the normal tissues. Pearson’s Chi-square test or Fisher’s exact test (as appropriate) was employed to assess the association between categorical variables. Survival analysis was conducted using the Kaplan–Meier method and log-rank test. A value of p < 0.05 was considered significant for all analyses.
Results
Patient demographics and tumor clinicopathological characteristics
A total of 43 patients were analyzed in the validation cohort. The cohort comprised 21 males (49.8%) and 22 females (51.2%). The majority of patients were over 40 years old (90.6%), with only 4 patients (9.4%) aged 40 years or younger. The mean age of the cohort was 57 ± 12.668 years. Diabetes comorbidity was also assessed, revealing a positive status in 51.2% of patients. The primary tumor sites were distributed as follows: 28 patients (65%) had tumors in the pancreatic head, 3 (6.9%) had tumors in the body or tail, and the specific site in the pancreas was unknown in 12 patients (27.9%). In terms of T stage (tumor size), 6 patients (13.9%) were classified as T1, 24 (55.8%) as T2, and 13 (30.23%) as T3. Lymph node involvement, as indicated by the N stage, showed that 12 patients (27.9%) were classified as N0, 19 (44.1%) as N1, and 12 (27.9%) as N2. According to the American Joint Committee on Cancer (AJCC) staging, 6 patients (13.9%) were in Stage I, 28 (65.1%) in Stage II, and 9 (20.9%) in Stage III.
Histological differentiation revealed that 6 tumors (13.9%) were well differentiated, 30 (69.7%) were moderately differentiated, and 7 (16.2%) were poorly differentiated. Lymphovascular invasion was present in 7 patients (16.2%), while it was absent in 36 patients (83.7%). Perineural invasion was observed in 17 patients (39.5%), while 26 patients (60.4%) showed no evidence of perineural invasion. At the time of data collection, 39 patients (90.7%) were deceased, and 4 patients (9.3%) were alive. Detailed demographics and clinicopathological characteristics are described in Table 3.
Table 3.
Patient demographics and tumor clinicopathological characteristics of the validation cohort (N = 43, 100%)
| Variables | Frequency (%) |
|---|---|
| Gender | |
| Male | 21 (49.8) |
| Female | 22 (51.2) |
| Age Group | |
| ≤ 40 years | 4 (9.4) |
| > 40 years | 39 (90.6) |
| Diabetes Status | |
| Yes | 22 (51.2) |
| No | 21 (48.8) |
| Tumor site | |
| Head | 28 (65) |
| Body/tail | 3 (6.9) |
| Specific site in pancreas unknown | 12 (27.9) |
| T stage (tumor size) | |
| T1 | 6 (13.9) |
| T2 | 24 (55.8) |
| T3 | 13 (30.23) |
| N stage (lymph node involvement) | |
| N0 | 12 (27.9) |
| N1 | 19 (44.1) |
| N2 | 12 (27.9) |
| AJCC stage* | |
| I | 6 (13.9) |
| II | 28 (65.1) |
| III | 9 (20.9) |
| Histological differentiation | |
| Well differentiated | 6 (13.9) |
| Moderately differentiated | 30 (69.7) |
| Poorly differentiated | 7 (16.2) |
| Lymphovascular invasion | |
| Present | 7 (16.2) |
| Absent | 36 (83.7) |
| Perineural invasion | |
| Present | 17 (39.5) |
| Absent | 26 (60.4) |
| Health Status | |
| Dead | 39 (90.7) |
| Alive | 4 (9.3) |
*Staging criteria by American Joint Committee on Cancer
Expression profile of regulatory miRNAs and proteomic biomarkers
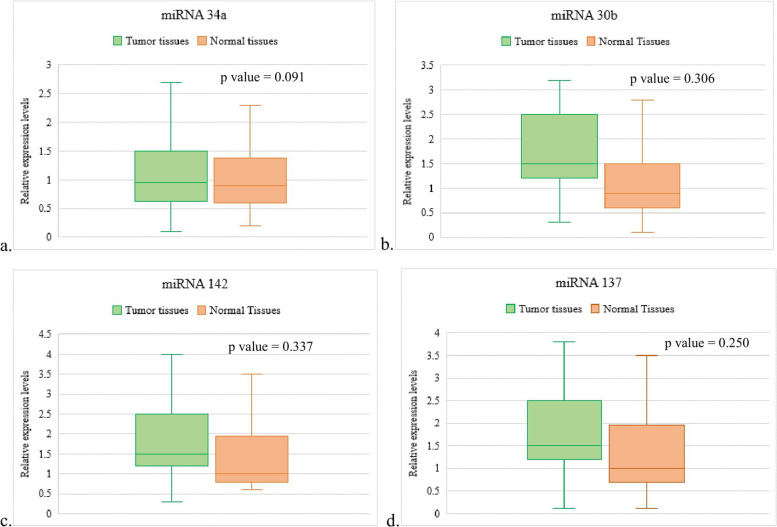
In our analysis of miRNA expression levels among the subset of 43 PDAC patients, the majority exhibited high expression levels across the selected miRNAs. Specifically, miRNA 34a showed high expression in 30 patients (69.7%) and low expression in 13 patients (30.2%). miRNA 30b was highly expressed in 29 patients (67.4%) and had low expression in 14 patients (32.5%). Similarly, miRNA 142 displayed high expression in 30 patients (69.7%) and low expression in 13 patients (30.2%). In contrast, miRNA 137 had a relatively lower prevalence of high expression, with 23 patients (53.4%) showing high expression and 20 patients (46.5%) showing low expression (Table 4). Moreover, no significant difference was observed between the expression of the selected miRNAs in the tumor tissues and normal tissues (Fig. 1).
Table 4.
Biomarkers (miRNAs and proteins) expression profile of the cohort (N = 43, 100%)
| Biomarker | Frequency (%) |
|---|---|
| miRNA 34a Expression | |
| High expression | 30 (69.7) |
| Low expression | 13 (30.2) |
| miRNA 30b Expression | |
| High expression | 29 (67.4) |
| Low expression | 14 (32.5) |
| miRNA 142 Expression | |
| High expression | 30 (69.7) |
| Low expression | 13 (30.2) |
| miRNA 137 Expression | |
| High expression | 23 (53.4) |
| Low expression | 20 (46.5) |
| KRAS Expression | |
| Positive expression | 34 (79.1) |
| - Mild | 10 (29.4)* |
| - Moderate | 7 (20.5)* |
| - Strong | 17 (50)* |
| Negative expression | 9 (20.9) |
| p53 Expression | |
| Positive expression | 15 (34.9) |
| - Mild | 6 (40)* |
| - Moderate | 6 (40)* |
| - Strong | 3 (20)* |
| Negative expression | 28 (65.1) |
| BRCA1 Expression | |
| Positive expression | 30 (69.8) |
| - High expression | 17 (56.6)* |
| - Low expression | 13 (43.3)* |
| Negative expression | 13 (30.2) |
| APC Expression | |
| High expression | 22 (51.2) |
| Low expression | 21 (48.8) |
Fig. 1.
Expression levels of the selected miRNas between tumor tissues and normal tissues – a miRNA 34a, b miRNA 30b, miRNA 142, d miRNA 137
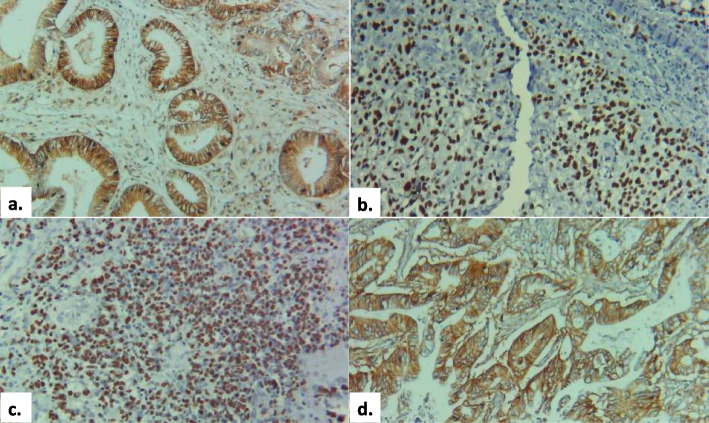
For the KRAS protein, 34 patients (79.1%) exhibited positive expression, stratified into mild (10 patients, 29.4%), moderate (7 patients, 20.5%), and strong expression (17 patients, 50%). Nine patients (20.9%) showed no detectable KRAS expression. p53 expression was positive in 15 patients (34.9%), with 6 patients (40%) each displaying mild and moderate expression, and 3 patients (20%) showing strong expression. Negative expression of p53 was observed in 28 patients (65.1%). BRCA1 protein expression was positive in 30 patients (69.8%), with high expression in 17 patients (56.6%) and low expression in 13 patients (43.3%). Thirteen patients (30.2%) were negative for BRCA1 expression. APC protein expression analysis showed that 22 patients (51.2%) had high expression, while 21 patients (48.8%) exhibited low expression (Table 4). Figure 2 show the expression of the selected proteomic biomarkers in PDAC tumor samples.
Fig. 2.
Positive expression of the selected proteins in the study cohort, a KRAS, b p53, c BRCA1, d APC (magnification – 10X, scale – 51 μm)
Association of regulatory miRNA with proteomic biomarkers, demographics and clinicopathological characteristics
In a cohort of 43 patients, the expression levels of miRNA biomarkers (miRNA 34a, miRNA 30b, miRNA 142, and miRNA 137) were analyzed in relation to patient demographics, tumor characteristics, and protein biomarker expression profiles. Among patients older than 40 years, high expression of miRNA 34a was observed in 86.7%, whereas no low expression was detected in those aged ≤ 40 years. Similar trends were noted for miRNA 30b and miRNA 137, which also showed predominant expression in patients over 40. Analysis of gender and diabetes status showed a relatively balanced distribution of miRNA expression, with no significant differences observed in the expression levels of any miRNA biomarkers based on gender or diabetes comorbidity status.
Tumor invasion characteristics revealed noteworthy trends. Patients exhibiting lymphovascular invasion tended to have higher miRNA 34a and miRNA 30b expression, though these associations were not statistically significant (p = 0.057 and p = 0.260, respectively). In contrast, a statistically significant association was identified between perineural invasion and elevated miRNA 34a (p = 0.000, OR = 21.667) as well as miRNA 137 expression (p = 0.001, OR = 12.833), suggesting these miRNAs may play a role in perineural invasion pathways. Analysis of protein biomarkers demonstrated that while miRNA expression levels did not show significant correlations with KRAS or p53 expression, a notable association was identified between BRCA1 expression and high miRNA 34a levels (p = 0.034, OR = 8.000), underscoring a potential interaction between miRNA 34a and BRCA1 in the tumor microenvironment. Details of association analysis are shown in Table 5.
Table 5.
Association of selected miRNA biomarkers with patient demographics, tumor clinicopathological characteristics and selected protein biomarkers – N = 43 (100%)
| miRNA 34a expression | miRNA 30b expression | miRNA 142 expression | miRNA 137 expression | |||||
|---|---|---|---|---|---|---|---|---|
| High^ | Low | High^ | Low | High^ | Low | High^ | Low | |
| Age group | ||||||||
| ≤ 40 years | 4 (13.3) | 0 (0) | 4 (13.8) | 0 (0) | 3 (10) | 1 (7.7) | 1 (4.3) | 3 (15) |
| > 40 years | 26 (86.7) | 13 (100) | 25 (86.2) | 14 (100) | 27 (90) | 12 (92.3) | 22 (95.7) | 17 (85) |
| p-value (odds ratio) | 0.167 (0.867) | 0.145 (0.862) | 0.811 (1.333) | 0.230 (0.258) | ||||
| Gender | ||||||||
| Female | 15 (50) | 7 (53.8) | 14 (48.3) | 8 (57.1) | 16 (53.3) | 7 (53.8) | 12 (52.2) | 10 (50) |
| Male | 15 (50) | 6 (46.2) | 15 (51.7) | 6 (42.9) | 14 (46.7) | 6 (46.2) | 11 (47.8) | 10 (50) |
| p-value (odds ratio) | 0.817 (1.167) | 0.586 (1.429) | 0.665 (0.750) | 0.887 (0.917) | ||||
| Diabetes status | ||||||||
| Yes | 14 (46.7) | 8 (61.5) | 16 (55.2) | 6 (42.9) | 18 (60) | 4 (30.8) | 12 (52.2) | 10 (50) |
| No | 16 (53.3) | 5 (38.5) | 13 (44.8) | 8 (57.1) | 12 (40) | 9 (69.2) | 11 (47.8) | 10 (50) |
| p-value (odds ratio) | 0.370 (1.829) | 0.449 (0.609) | 0.078 (0.296) | 0.887 (0.917) | ||||
| Lymphovascular invasion | ||||||||
| Present | 7 (23.3) | 0 (0) | 6 (20.7) | 1 (7.1) | 4 (13.3) | 3 (23.1) | 5 (21.7) | 2 (10) |
| Absent | 23 (76.7) | 13 (100) | 23 (79.3) | 13 (92.9) | 26 (86.7) | 10 (76.9) | 18 (78.3) | 18 (90) |
| p-value (odds ratio) | 0.057 (0.767) | 0.260 (3.391) | 0.427 (0.513) | 0.298 (2.500) | ||||
| Perineural invasion | ||||||||
| Present | 14 (11.9) | 0 (0) | 14 (48.3) | 3 (21.4) | 13 (43.3) | 4 (30.8) | 10 (43.5) | 7 (35) |
| Absent | 3 (23.1) | 10 (76.9) | 15 (51.7) | 11 (78.6) | 17 (56.7) | 9 (69.2) | 13 (56.5) | 13 (65) |
| miRNA 34a expression | ||||||||
| High | - | - | 26 (86.7) | 4 (13.3) | 21 (70) | 9 (30) | 21 (70) | 9 (30) |
| Low | - | - | 3 (23.1) | 10 (76.9) | 9 (69.2) | 4 (30.8) | 2 (15.4) | 11 (84.6) |
| p-value (odds ratio) | - | 0.000 (21.667)* | 0.960 (1.037) | 0.001 (12.833)* | ||||
| miRNA 30b expression | ||||||||
| High | 26 (89.7) | 3 (10.3) | - | - | 20 (69) | 9 (31) | 19 (65.5) | 10 (34.5) |
| Low | 4 (28.6) | 10 (71.4) | - | - | 10 (71.4) | 4 (28.6) | 4 (28.6) | 10 (71.4) |
| p-value (odds ratio) | 0.000 (21.667)* | - | 0.869 (0.889) | 0.023 (4.750)* | ||||
| miRNA 142 expression | ||||||||
| High | 21 (70) | 9 (30) | 20 (66.7) | 10 (33.3) | - | - | 18 (60) | 12 (40) |
| Low | 9 (69.2) | 4 (30.8) | 9 (69.2) | 4 (30.8) | - | - | 5 (38.5) | 8 (61.5) |
| p-value (odds ratio) | 0.960 (1.037) | 0.869 (0.889) | - | 0.193 (2.400) | ||||
| miRNA 137 expression | ||||||||
| High | 21 (91.3) | 2 (8.7) | 19 (82.6) | 4 (17.4) | 18 (78.3) | 5 (21.3) | - | - |
| Low | 9 (45) | 11 (55) | 10 (50) | 10 (50) | 12 (60) | 8 (40) | - | - |
| p-value (odds ratio) | 0.001 (12.833)* | 0.023 (4.750)* | 0.193 (2.400) | - | ||||
| KRAS expression | ||||||||
| Positive | 23 (76.7) | 11 (84.6) | 24 (82.8) | 10 (71.4) | 24 (80) | 10 (76.9) | 18 (78.3) | 16 (80) |
| Negative | 7 (23.3) | 2 (15.4) | 5 (17.2) | 4 (28.6) | 6 (20) | 3 (23.1) | 5 (21.7) | 4 (20) |
| p-value (odds ratio) | 0.556 (0.597) | 0.392 (1.920) | 0.820 (1.200) | 0.889 (0.900) | ||||
| p53 expression | ||||||||
| Positive | 9 (30) | 6 (46.2) | 12 (41.4) | 3 (21.4) | 11 (36.7) | 4 (30.8) | 9 (39.1) | 6 (30) |
| Negative | 21 (70) | 7 (53.8) | 17 (58.6) | 11 (78.6) | 19 (63.3) | 9 (69.2) | 14 (60.9) | 14 (70) |
| p-value (odds ratio) | 0.307 (0.500) | 0.198 (2.588) | 0.709 (1.303) | 0.531 (1.500) | ||||
| BRCA1 expression | ||||||||
| Positive | 18 (60) | 12 (92.3) | 19 (65.5) | 11 (78.6) | 23 (76.7) | 7 (53.8) | 16 (69.6) | 14 (70) |
| Negative | 12 (40) | 1 (7.7) | 10 (34.5) | 3 (21.4) | 7 (23.3) | 6 (46.2) | 7 (30.4) | 6 (30) |
| p-value (odds ratio) | 0.034 (8.000)* | 0.382 (0.518) | 0.135 (2.816) | 0.975 (0.980) | ||||
| APC expression | ||||||||
| Present | 16 (53.3) | 6 (46.2) | 14 (48.3) | 8 (57.1) | 13 (43.3) | 9 (69.2) | 13 (56.5) | 9 (45) |
| Absent | 14 (46.7) | 7 (53.8) | 15 (51.7) | 6 (42.9) | 17 (56.7) | 4 (30.8) | 10 (43.5) | 11 (55) |
| p-value (odds ratio) | 0.665 (1.333) | 0.586 (0.700) | 0.119 (0.340) | 0.451 (1.589) | ||||
*Significant association (p < 0.05), ^Reference for odds ratio
Association of regulatory miRNA, proteomic biomarkers, demographics and clinicopathological characteristics with overall survival
Mean survival of the cohort was 14.23 ± 16.308. Among the total of 43 patients, 39 (90.7%) died during the study period. We examined the association of demographics, clinicopathological characteristics, regulatory miRNAs, and proteomic biomarkers with overall survival. Age and gender were not significantly associated with survival outcomes, with median survival months of 11 for patients ≤ 40 years and 10 for those > 40 years (p = 0.578). Tumor location also showed no significant impact on survival (p = 0.416). However, significant associations were observed with tumor stage (T stage), nodal involvement (N stage), and AJCC stage. Patients with T1 tumors had a median survival of 21 months, significantly higher than those with T2 (11 months) or T3 tumors (2 months; p = 0.000). Similarly, patients with N0 nodal status had a median survival of 20 months, compared to 11 months for N1 and 1 month for N2 (p = 0.000). Higher AJCC stages were also associated with decreased survival, with stage I patients showing a median survival of 34 months, versus 11 months for stage II and 1 month for stage III patients (p = 0.000). Histological differentiation, lymphovascular invasion, and perineural invasion did not show significant associations with overall survival.
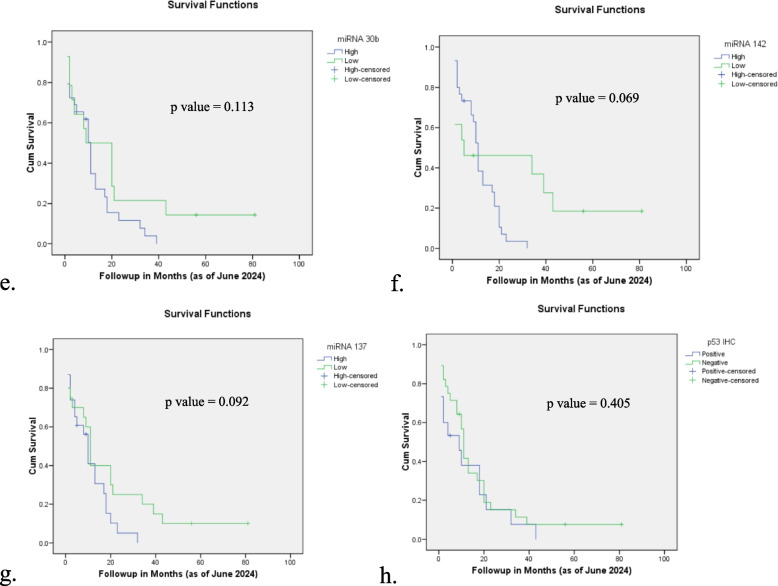
Among the miRNAs, higher expression levels of miRNA 34a, 30b, and 142 correlated with slightly better survival rates, though these associations did not reach statistical significance (p = 0.158, p = 0.113, and p = 0.069, respectively). miRNA 137 expression was also found to be non-significant, with patients exhibiting high miRNA 137 expression having a median survival of 10 months compared to 11 months in those with low expression (p = 0.092). Protein biomarkers including KRAS, p53, BRCA1, and APC were also evaluated. Survival duration of the patients with positive KRAS expression was skewed towards a higher median survival of 11 months compared to 4 months in KRAS-negative patients, though this trend was not statistically significant (p = 0.052). No significant associations with survival were found for p53, BRCA1, or APC expression. Details of association analysis are shown in Table 6 and Fig. 3.
Table 6.
Association of demographics, clinicopathological characteristics, regulatory miRNAs and proteomic biomarkers with the overall survival of the patients (N = 43, 100%)
| Overall Survival | |||
|---|---|---|---|
| Variables | Patients | Median Survival Months | p-value |
| Age Group | |||
| ≤ 40 years | 4 | 11 | 0.578 |
| > 40 years | 39 | 10 | |
| Gender | |||
| Male | 21 | 10 | 0.270 |
| Female | 22 | 11 | |
| Tumor site | |||
| Head | 28 | 11 | 0.416 |
| Body/tail | 3 | 13 | |
| Specific site in pancreas unknown | 12 | 9 | |
| T stage | |||
| T1 | 6 | 21 | 0.000* |
| T2 | 24 | 11 | |
| T3 | 13 | 2 | |
| N stage | |||
| N0 | 12 | 20 | 0.000* |
| N1 | 19 | 11 | |
| N2 | 12 | 1 | |
| AJCC stage | |||
| I | 6 | 34 | 0.000* |
| II | 28 | 11 | |
| III | 9 | 1 | |
| Histological differentiation | |||
| Well differentiated | 6 | 8 | 0.125 |
| Moderately differentiated | 30 | 11 | |
| Poorly differentiated | 7 | 2 | |
| Lymphovascular invasion | |||
| Present | 7 | 20 | 0.388 |
| Absent | 36 | 10 | |
| Perineural invasion | |||
| Present | 17 | 11 | 0.414 |
| Absent | 26 | 8 | |
| miRNA 34a expression | |||
| High | 30 | 11 | 0.158 |
| Low | 13 | 9 | |
| miRNA 30b expression | |||
| High | 29 | 11 | 0.113 |
| Low | 14 | 9 | |
| miRNA 142 expression | |||
| High | 30 | 11 | 0.069 |
| Low | 13 | 5 | |
| miRNA 137 expression | |||
| High | 23 | 10 | 0.092 |
| Low | 20 | 11 | |
| KRAS expression | |||
| Positive | 34 | 11 | 0.052 |
| Negative | 9 | 4 | |
| p53 expression | |||
| Positive | 15 | 9 | 0.405 |
| Negative | 28 | 11 | |
| BRCA1 expression | |||
| Positive | 30 | 11 | 0.185 |
| Negative | 13 | 8 | |
| APC expression | |||
| Positive | 22 | 9 | 0.452 |
| Negative | 21 | 11 | |
*Significant association (p < 0.05)
Fig. 3.
Association of clinicopathological characteristics and miRNA expression with overall survival of the patients – a T stage, b N stage, c AJCC stage, d miRNA 34a, e miRNA 30b, f miRNA 142, g miRNA 137, h KRAS expression, i p53 expression, j BRCA1 expression, k APC expression
Discussion
In this study, we investigated a panel of four regulatory miRNAs (miRNA 34a, 30b, 142, and 137) in PDAC and their interactions with specific target proteins within key signaling pathways (KRAS, p53, BRCA1, and APC). Precision medicine has become increasingly crucial in cancer treatment, providing tailored therapeutic strategies that enhance patient outcomes. Numerous studies have focused on identifying precision medicine targets, especially molecular biomarkers such as genetic mutations and specific protein expressions [5, 23]. Among these biomarkers, miRNAs stand out for their regulatory roles in gene expression and their potential as therapeutic targets. Our aim was to identify miRNA-based molecular biomarkers with potential for targeted therapies in cancer, advancing the field of personalized treatment [24, 25]. We initially examined miRNA expression in all 109 patient samples. However, RNA extraction issues in several cases (from both tumor and normal tissues) resulted in either inadequate RNA yields or A260/280 ratios outside the acceptable range. Consequently, we present miRNA expression analysis here as pilot data from a subset of 43 PDAC patients (Fig. 1). Most patients in this subset showed elevated expression levels across all examined miRNAs, with the highest frequency observed for miRNA 34a and miRNA 142 (69.7% each), followed by miRNA 30b (67.4%) and miRNA 137 (53.4%).
Our findings reveal a significant association between miRNA 34a expression and BRCA1 protein levels (p = 0.034). Prior studies have shown that BRCA1 upregulates both precursor and mature forms of several tumor-suppressive regulatory miRNAs, including miRNA 34a, miRNA 16, and miRNA 145 [26]. Moreover, significant associations between miRNA 34a and BRCA1, BRCA2, and p53 expression have been documented [27]. Further evidence suggests that BRCA1 overexpression can markedly enhance miRNA 34a maturation, accelerating primary transcript processing and boosting levels of both precursor and mature miRNA 34a forms [28]. Collectively, these studies demonstrate the regulatory impact of BRCA1 on miRNA 34a expression. However, it is plausible that miRNA 34a might also impact BRCA1 expression or activity. This bi-directional regulation could imply a complex regulatory interplay where miRNA 34a not only responds to BRCA1 but also potentially influences BRCA1’s function within the cellular signaling environment. Furthermore, a similar reciprocal relationship has been documented between miRNA 34a and the tumor suppressor protein p53, where miRNA 34a both responds to and regulates p53, establishing a feedback loop that can amplify tumor-suppressive responses [29–31]. This parallel raises the possibility of a broader regulatory network involving miRNA 34a, BRCA1, and p53, which may collectively influence key pathways in tumor suppression within PDAC. Further exploration of these bi-directional regulatory mechanisms is essential to elucidate novel therapeutic targets within the miRNA regulatory network, potentially contributing to advancements in personalized treatment strategies for cancer patients. Furthermore, the significant association between miRNA 34a and BRCA1 expression is reinforced by the presence of a significant association between miRNA 34a and two additional tumor-suppressor miRNAs, miRNA 30b (p = 0.000) and miRNA 137 (p = 0.001), within our panel. The interconnected expression patterns observed among these three miRNAs; each known for their regulatory functions in cell cycle control, apoptosis, and tumor suppression alongside BRCA1, point to potential cooperative interactions within a broader signaling network. This suggests that these miRNAs may not act in isolation; rather, they could interplay with BRCA1 and each other to modulate key oncogenic and tumor-suppressive pathways in PDAC. Such interactions highlight the need for further research to dissect the precise molecular mechanisms underlying these associations, which may uncover new insights into how miRNA-mediated regulation can impact cancer progression and response to therapy. Elucidating these relationships may enable the identification of novel combinatorial therapeutic targets that harness the synergistic tumor-suppressive roles of miRNA 34a, miRNA 30b, miRNA 137, and BRCA1. Further, studies have shown role of miRNA 34a in metabolic conditions such as diabetes [32, 33].
miRNA 30b plays a critical role in cellular processes such as differentiation and inflammation and is known to target KRAS and p53. Decreased expression of miRNA 30b is linked to poor prognosis in pancreatic cancer. Overexpression of miRNA 30b induces G1 cell cycle arrest and apoptosis by directly binding to the 3’UTR of KRAS mRNA, thereby downregulating KRAS. In contrast, KRAS promotes cell proliferation and apoptosis in colorectal cancer (CRC) cells, and increased miRNA 30b expression has been shown to significantly reduce cell invasion and migration in CRC [34]. Similarly, in hepatocellular carcinoma, miRNA 142 regulates the Wnt/PCP pathway by targeting Rac1, suppressing cancer cell migration [35, 36]. Likewise, miRNA 137 modulates the Wnt/β-catenin and TGF-β pathways, and its downregulation is associated with increased cell proliferation and growth, mediated by β-catenin nuclear translocation and inhibition of TGF-β signaling [10, 11]. This negative regulation of the Wnt pathway by miRNA 137 correlates with elevated APC expression [37]. However, in our study, we found no significant associations between the expressions of miRNAs (30b, 142, and 137) and any of the targeted proteins. Additionally, we observed no significant differences in miRNA expression between tumor and normal tissues, nor any associations between miRNA expression and overall survival. Although literature on the role of these miRNAs in PDAC remains limited, studies in other cancers offer some insights. For instance, miRNA 34a has been shown to be significantly downregulated in PDAC samples from Chinese and American populations [38, 39], with a significant association observed in a Chinese cohort (p < 0.001) [38]. In an Iranian population, miRNA 34a levels were also markedly lower in esophageal squamous cell carcinoma (ESCC) [40]. Similarly, studies on Chinese PDAC patients report decreased expression of miRNA 30b [41, 42], with further findings linking miRNA 30b expression to perineural invasion (p = 0.018), TNM stage (p < 0.001), tumor differentiation (p < 0.001), and overall survival (p = 0.0021). Lower expression of miRNA 30b has also been observed in non-small cell lung cancer (NSCLC) [43] and gallbladder tumors [44]. In addition, miRNA 142 is significantly downregulated in various cancers, including PDAC [45, 46] and oral squamous cell carcinoma (OSCC) [47], while miRNA 137 shows decreased expression in PDAC in Chinese populations [48] and in cholangiocarcinoma [49].
Although most studies report a significant downregulation of these miRNAs, the majority of investigations have focused on Chinese populations. Further research in diverse populations is essential to clarify the role of these miRNAs across different genetic backgrounds. There are Food and Drug Administration (FDA)-approved targeted drugs available for the protein biomarkers studied here, such as olaparib, niraparib, and rucaparib for treating BRCA-related cancers, and sotorasib and adagrasib for KRAS G12C mutated Non-Small Cell Lung Cancers (NSCLC) [50, 51]. Additionally, MRX34, a synthetic mimic of miRNA 34a encapsulated in a liposomal nanoparticle, was tested in phase 1 clinical trials in 2013 (NCT01829971), pioneering miRNA therapy in various solid tumors [52, 53]. Furthermore, studies have shown that decreased expression of miRNA 142 is associated with resistance to gemcitabine chemotherapy in pancreatic tumors [54]. However, before determining the clinical utility and implications of these targeted drugs in PDAC, it is crucial to evaluate the role of these regulatory miRNAs in the patient population. The insights gained from this study on protein profiles and their clinical implications in PDAC could pave the way for personalized treatment approaches and improved patient survival. This study has several limitations that should be acknowledged. Firstly, the sample size is relatively small, limiting the generalizability of our findings. Additionally, due to the retrospective nature of the study, there may be inherent biases associated with patient selection and data completeness. RNA extraction issues led to a reduced sample subset, impacting the scope of our analysis. Future studies with larger, multi-centric cohorts and prospective designs would help validate these findings and explore the role of these miRNAs in PDAC more comprehensively.
Conclusion
In conclusion, this study provides an insight on the expression of the selected regulatory miRNAs in the Pakistani PDAC population along with their association with the signaling pathway protein expression. To the best of our knowledge, this is the first study to investigate the combined panel of miRNA 34a, miRNA30b, miRNA 142 and miRNA 137 in PDAC, globally. The statistical analyses results indicated a significant association between the expression of miRNA 34a and BRCA1 protein. Moreover, the expression of miRNA 34a independently showed significant association with miRNA 30b and miRNA 137. This study underscores the regulatory effect of BRCA1 expression on miRNA 34a expression, with evidence suggesting a potential bi-directional relationship. While BRCA1 has been shown to enhance miRNA 34a maturation, our findings raise the possibility that miRNA 34a may also influence BRCA1 expression or activity. This complex interplay implies that miRNA 34a not only responds to BRCA1 but might actively modulate BRCA1’s function within cellular signaling pathways. Such bi-directional regulation suggests a layered regulatory network that could be central to tumor suppression mechanisms in PDAC. Future investigations are warranted to fully elucidate these interactions and their therapeutic implications, potentially advancing personalized treatment approaches that harness this miRNA-protein crosstalk for improved clinical outcomes in PDAC.
Acknowledgements
We acknowledge the funder as well as the patients who participated in the study.
Abbreviations
- PDAC
Pancreatic Ductal Adenocarcinoma
- US FDA
Food and Drug Administration of the United States
- IARC
International Agency for Research on Cancers
- NSCLC
Non-small cell lung cancers
- FFPE
Formalin-fixed paraffin-embedded
- AKUH
Aga Khan University Hospital
- miRNA
Micro RNA
- RT-PCR
Real-Time PCR
- cDNA
Complementary DNA
- H&E
Hematoxylin and eosin
- KRAS
Kirsten rat sarcoma viral oncogene homolog protein
- p53
Tumor protein 53
- BRCA1
BReast CAncer gene 1 protein
- APC
Adenomatous polyposis coli protein
- CRC
Colorectal cancer
- ESCC
Esophageal squamous cell carcinoma
Authors’ contributions
SMA: Data curation, designing methodology, performing experiments, data entry, data analysis, results interpretation, manuscript writing, manuscript editing and finalization YA: Research idea, designing methodology, project administration, data analysis, results interpretation, manuscript editing and finalization ZA: Data curation, designing methodology, performing experiments, manuscript editing and finalization TC: Data curation, designing methodology, project administration, manuscript editing and finalization HAF: Data curation, project administration, performing experiments SMAA: Research idea, funding acquisition, designing methodology, data analysis, bench work supervision, manuscript editing and finalization.
Funding
This work was supported by the University Research Council grant by Aga Khan University Hospital (183027SUR).
Data availability
The datasets generated during the current study are available in this manuscript.
Declarations
Ethical approval and consent to participate
The study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Ethical Review Committee of Aga Khan University Hospital, Karachi, Pakistan (2023–6278-25,839). Informed consent was obtained from all the participants.
Consent to publication
Not Applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ferlay J, Ervik M, Lam F, Laversanne M, Colombet M, Mery L, Piñeros M, Znaor A, Soerjomataram I, Bray F. Global Cancer Observatory: Cancer Today. Lyon, France: International Agency for Research on Cancer. Available from: https://gco.iarc.who.int/today. Accessed 5 Nov 2024.
- 2.Jiang Y, Sohal DP. Pancreatic adenocarcinoma management. JCO OP. 2023;19(1):19–32. [DOI] [PubMed] [Google Scholar]
- 3.SEER 2024, https://seer.cancer.gov/statfacts/html/pancreas.html. Accessed 5 Nov 2024.
- 4.Ali SM, Adnan Y, Ali SA. Pancreatic cancers: a review of studies from Pakistan and comparison with global trends. PJPH. 2021;11(2):120–7. [Google Scholar]
- 5.Ali SM, Adnan Y, Ahmad Z, Farooqui HA, Chawla T, Ali SA. Genetic landscape of pancreatic adenocarcinoma patients: a pilot study from Pakistan. Mol Biol Rep. 2022;49(2):1341–50. [DOI] [PubMed] [Google Scholar]
- 6.Bayraktar E, Bayraktar R, Oztatlici H, Lopez-Berestein G, Amero P, Rodriguez-Aguayo C. Targeting miRNAs and other non-coding RNAs as a therapeutic approach: an update. ncRNA. 2023;9(2):27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ali SM, Adnan Y, d SM AA. Role of mi-RNAs in Pancreatic Cancers. PJBMB. 2020;53(4):101–15. [Google Scholar]
- 8.Islam F, Gopalan V, Vider J, Lu CT, Lam AK. MiRNA 142–5p act as an oncogenic microRNA in colorectal cancer: Clinicopathological and functional insights. Exp Mol Pathol. 2018;104(1):98–107. [DOI] [PubMed] [Google Scholar]
- 9.Jelski W, Mroczko B. Potential Diagnostic Utility of microRNAs in Gastrointestinal Cancers. Cancer Manag Res. 2023;31:863–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jafari S, Motedayyen H, Javadi P, Jamali K, Moradi Hasan-Abad A, Atapour A, Sarab GA. The roles of lncRNAs and miRNAs in pancreatic cancer: a focus on cancer development and progression and their roles as potential biomarkers. Front Oncol. 2024;15(14):1355064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tesfaye AA, Azmi AS, Philip PA. miRNA and gene expression in pancreatic ductal adenocarcinoma. AJP. 2019;189(1):58–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wan J, Ling X, Peng B, Ding G. miRNA 142–5p regulates CD4+ T cells in human non-small cell lung cancer through PD-L1 expression via the PTEN pathway. Oncol Rep. 2018;40(1):272–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25(4):402–8. [DOI] [PubMed] [Google Scholar]
- 14.Masood KI, Rottenberg ME, Salahuddin N, Irfan M, Rao N, Carow B, Islam M, Hussain R, Hasan Z. Expression of M. tuberculosis-induced suppressor of cytokine signaling (SOCS) 1, SOCS3, FoxP3 and secretion of IL-6 associates with differing clinical severity of tuberculosis. BMC Infect Dis. 2013;13:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharma VK, Raimondi V, Ruggero K, Pise-Masison CA, Cavallari I, Silic-Benussi M, Ciminale V, D’Agostino DM. Expression of miRNA 34a in T-cells infected by human T-lymphotropic virus 1. Front Microbiol. 2018;4(9):832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei W, Tang H, Tang L. MicroRNA-34a inhibits metastasis in liver cancer cells. Oncol Lett. 2018;16(6):6960–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan W, Zheng H, Dong J, Liu C, Zuo Z, Liu X. MicroRNA-30b is involved in the pathological process of diabetes mellitus induced by pancreatic cancer by regulating plasminogen activator inhibitor-1. Biotechnol Biotechnol Equip. 2019;33(1):1741–9. [Google Scholar]
- 18.Huang Y, Zou Y, Zheng R, Ma X. MiR-137 inhibits cell proliferation in acute lymphoblastic leukemia by targeting JARID1B. Eur J Heamatol. 2019;103(3):215–24. [DOI] [PubMed] [Google Scholar]
- 19.Wu N, Huang Y, Liu F, Xu X, Liu B, Wei J. KRAS gene status in gastric signet-ring cell carcinoma patients and acts as biomarker of MEK inhibitor. J Gastrointest Oncol. 2021;12(3):1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ali SM, Shamim MS, Enam SA, Ahmad Z, Adnan Y, Farooqui HA. Immunohistochemical detection and prognostic significance of p53, epidermal growth factor receptor, murine double minute 2, and isocitrate dehydrogenase 1 in glioblastoma multiforme patients of pakistan. Clin Med Insights Oncol. 2022;16:11795549221119108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Somri-Gannam L, Meisel-Sharon S, Hantisteanu S, Groisman G, Limonad O, Hallak M, Bruchim I. IGF1R axis inhibition restores dendritic cell antitumor response in ovarian cancer. Trans Oncol. 2020;13(8):100790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mohammed KH, Siddiqui MT, Willis BC, Tsvetkova DZ, Mohamed A, Patel S, Sharma J, Weber C, Cohen C. Parafibromin, APC, and MIB-1 are useful markers for distinguishing parathyroid carcinomas from adenomas. Appl Immunohistochem Mol Morphol. 2017;25(10):731–5. [DOI] [PubMed] [Google Scholar]
- 23.Ali SMA, Adnan Y, Ali SM, Ahmad Z, Chawla T, Farooqui HA. Immunohistochemical analysis of a panel of cancer stem cell markers and potential therapeutic markers in pancreatic ductal adenocarcinoma. J Cancer Res Clin Oncol. 2023;149(6):2279–92. [DOI] [PubMed] [Google Scholar]
- 24.Diener C, Keller A, Meese E. The miRNA–target interactions: An underestimated intricacy. Nucleic Acids Res. 2024Feb 28;52(4):1544–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sindhu KJ, Venkatesan N, Karunagaran D. MicroRNA interactome multiomics characterization for cancer research and personalized medicine: an expert review. OMICS: Integr Biol. 2021;25(9):545–66. [DOI] [PubMed] [Google Scholar]
- 26.Farooqi AA, Tabassum S, Ahmad A. MicroRNA-34a: a versatile regulator of myriads of targets in different cancers. Int J Mol Sci. 2017;18(10):2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shaban NZ, Ibrahim NK, Saada HN, El-Rashidy FH, Shaaban HM, Farrag MA, ElDebaiky K, Kodous AS. miRNA 34a and miRNA 21 as biomarkers in evaluating the response of chemo-radiotherapy in Egyptian breast cancer patients. J Radiat Res Apl Sci. 2022;15(3):285–92. [Google Scholar]
- 28.Slabáková E, Culig Z, Remšík J, Souček K. Alternative mechanisms of miRNA 34a regulation in cancer. Cell Death Dis. 2017;8(10):e3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li W, Wang Y, Liu R, Kasinski AL, Shen H, Slack FJ, Tang DG. MicroRNA-34a: potent tumor suppressor, cancer stem cell inhibitor, and potential anticancer therapeutic. Front Cell Dev Biol. 2021;8(9):640587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chakravarty D, Johnson A, Sklar J, Lindeman NI, Moore K, Ganesan S, Lovly CM, Perlmutter J, Gray SW, Hwang J, Lieu C. Somatic genomic testing in patients with metastatic or advanced cancer: ASCO provisional clinical opinion. J Clin Oncol. 2022;40(11):1231–58. [DOI] [PubMed] [Google Scholar]
- 31.Fu J, Imani S, Wu MY, Wu RC. MicroRNA-34 family in cancers: role, mechanism, and therapeutic potential. Cancers. 2023;15(19):4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mone P, de Donato A, Varzideh F, Kansakar U, Jankauskas SS, Pansini A, Santulli G. Functional role of miR-34a in diabetes and frailty. Front Aging. 2022;3:949924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Macvanin MT, Gluvic Z, Bajic V, Isenovic ER. Novel insights regarding the role of noncoding RNAs in diabetes. World J Diabetes. 2023;14(7):958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ibrahim H, Lim YC. KRAS-associated microRNAs in colorectal cancer. Oncol Rev. 2020;14(2):454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Banerjee S, Mandal AK. Role of MicroRNA Modulated Wnt Pathway in Breast Cancer and Its Therapeutic Use. Cytol Genet. 2024;58(4):326–42. [Google Scholar]
- 36.Wu L, Cai C, Wang X, Liu M, Li X, Tang H. MicroRNA-142-3p, a new regulator of RAC1, suppresses the migration and invasion of hepatocellular carcinoma cells. FEBS Lett. 2011;585(9):1322–30. [DOI] [PubMed] [Google Scholar]
- 37.He Z, Guo X, Tian S, Zhu C, Chen S, Yu C, Jiang J, Sun C. MicroRNA-137 reduces stemness features of pancreatic cancer cells by targeting KLF12. J Exp Clin Cancer Res. 2019;38:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Long LM, Zhan JK, Wang HQ, Li S, Chen YY, Liu YS. The clinical significance of miRNA 34a in pancreatic ductal carcinoma and associated molecular and cellular mechanisms. Pathobiology. 2016;84(1):38–48. [DOI] [PubMed] [Google Scholar]
- 39.Akula SM, Ruvolo PP, McCubrey JA. TP53/miRNA 34a-associated signaling targets SERPINE1 expression in human pancreatic cancer. Aging (Albany NY). 2020;12(3):2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Asadi M, Shanehbandi D, Mohammadpour H, Hashemzadeh S, Sepehri B. Expression level of miRNA 34a in tumor tissue from patients with esophageal squamous cell carcinoma. J Gastrointest Cancer. 2019;15(50):304–7. [DOI] [PubMed] [Google Scholar]
- 41.Xiong Y, Wang Y, Wang L, Huang Y, Xu Y, Xu L, Guo Y, Lu J, Li X, Zhu M, Qian H. MicroRNA-30b targets Snail to impede epithelial-mesenchymal transition in pancreatic cancer stem cells. J Cancer. 2018;9(12):2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen K, Wang Q, Liu X, Wang F, Yang Y, Tian X. Hypoxic pancreatic cancer derived exosomal miRNA 30b–5p promotes tumor angiogenesis by inhibiting GJA1 expression. Int J Biol Sci. 2022;18(3):1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qi Z, Zhang B, Zhang J, Hu Q, Xu F, Chen B, Zhu C. MicroRNA-30b inhibits non-small cell lung cancer cell growth by targeting the epidermal growth factor receptor. Neoplasma. 2018;65(2):192–200. [DOI] [PubMed] [Google Scholar]
- 44.Cui K, Bian X. The microRNA cluster miRNA 30b/-30d prevents tumor cell switch from an epithelial to a mesenchymal-like phenotype in GBC. Mol Ther Methods Clin Dev. 2021;12(20):716–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu Y, Ji N, Wei W, Sun W, Gong X, Wang X. MiRNA 142 modulates human pancreatic cancer proliferation and invasion by targeting hypoxia-inducible factor 1 (HIF-1α) in the tumor microenvironments. Biol Open. 2017Feb 15;6(2):252–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu J, Zhou L, Wei B, Qian Z, Wang J, Hui H, Sun Y. miRNA 142–5p inhibits pancreatic cancer cell migration and invasion by targeting PIK3CA. Mol Med Rep. 2020;22(3):2085–92. [DOI] [PubMed] [Google Scholar]
- 47.Iizumi S, Uchida F, Nagai H, Takaoka S, Fukuzawa S, Kanno NI, Yamagata K, Tabuchi K, Yanagawa T, Bukawa H. MicroRNA 142–5p promotes tumor growth in oral squamous cell carcinoma via the PI3K/AKT pathway by regulating PTEN. Heliyon. 2021;7(10):e08086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiao J, Peng F, Yu C, Wang M, Li X, Li Z, Jiang J, Sun C. microRNA-137 modulates pancreatic cancer cells tumor growth, invasion and sensitivity to chemotherapy. IJCEP. 2014;7(11):7442. [PMC free article] [PubMed] [Google Scholar]
- 49.Chen T, Lei S, Zeng Z, Pan S, Zhang J, Xue Y, Sun Y, Lan J, Xu S, Mao D, Guo B. MicroRNA-137 suppresses the proliferation, migration and invasion of cholangiocarcinoma cells by targeting WNT2B. Int J Mol Med. 2020;45(3):886–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Food and Drugs Administration USA (2024) http://www.fda.gov. Accessed 5 Nov 2024.
- 51.Cascetta P, Marinello A, Lazzari C, Gregorc V, Planchard D, Bianco R, Normanno N, Morabito A. KRAS in NSCLC: state of the art and future perspectives. Cancers. 2022;14(21):5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hong DS, Kang YK, Borad M, Sachdev J, Ejadi S, Lim HY, Brenner AJ, Park K, Lee JL, Kim TY, Shin S. Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. Br J Cancer. 2020;122(11):1630–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim T, Croce CM. MicroRNA: trends in clinical trials of cancer diagnosis and therapy strategies. Exp Mol Med. 2023;55(7):1314–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rawat M, Kadian K, Gupta Y, Kumar A, Chain PS, Kovbasnjuk O, Kumar S, Parasher G. MicroRNA in pancreatic cancer: from biology to therapeutic potential. Genes. 2019;10(10):752. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during the current study are available in this manuscript.