Abstract
Background
Sugar transporters (STs) play a critical role in the transportation of sugar, participating in plant growth and development, stress/defense responses, and signal transduction processes. Dendrobium catenatum (also known as Dendrobium officinale, hereinafter referred to as D. catenatum) is an important traditional Chinese medicinal herb with remarkable medicinal properties and possessing high economic value. Polysaccharides are the primary active components in D. catenatum, exhibiting diverse pharmacological activities. Sugar transporters function as material supplier and may play the essential roles in the polysaccharide biosynthesis, as well as the key reulators in the signaling and responses to abiotic stresses in Dendrobium plants. However, a comprehensive analysis of sugar transporters in D. catenatum remains elusive, thereby hindering our understanding of sugar partitioning within this species.
Results
In this work, the members belonging to MST, SUT, and SWEET gene families were identified in four Dendrobium plants. A comprehensive study of sugar transporters was conducted in D. catenatum, including phylogenetic relationship, structural arrangement, regulatory networks, expression profiles, and potential functions analysis. Seven sugar transporters were found to be involved in the process of polysaccharide biosynthesis in D. catenatum. Red-blue light is an effective way to enhance the accumulation of polysaccharides, and exerts an influence on the expression of polysaccharide biosynthesis related genes.
Conclusions
This study provide insight into the evolution and functional annotation of sugar transporters in Dendrobium, and establishing a foundation for future functional research of sugar transporters involved in polysaccharide biosynthesis and stress response.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-024-11121-4.
Keywords: MST, SUT, SWEET, Dendrobium catenatum, Polysaccharide biosynthesis
Introduction
Dendrobium is one of the largest genera in Orchidaceae, and naturally grows on the harsh environments as tree trunks and cliffs. Dendrobium plants possess enormous ornamental and medicinal values for their spectacular flowers and various types of secondary metabolites [1]. A typical characteristic of Dendrobium is its fleshy stems/leaves, which are rich of polysaccharide, a class of active macromolecules exhibiting antioxidant, anticancer, and immunomodulatory properties [2]. The transportation of photosynthetic carbohydrate products is critical to the biosynthesis and accumulation of polysaccharide in Dendrobium. The process of intercellular and intracellular sugar transport requires several essential membrane-crossing steps, in which sugar transporters play the pivotal roles [3]. Plants have three major types sugar transporters to coordinate the transport of monosaccharide and disaccharide: MST (monosaccharide transporter), SUT (sucrose transporter, also called SUC for sucrose carrier), and SWEET (Sugars Will Eventually be Exported Transporter) [4]. The genomes of several Dendrobium plants have been finished, including D. catenatum [5–7], D. huoshanense [8], D. nobile [9], and D. chrysotoxum [10]. In previous studies, 8 SUT and 25 SWEET genes were identified in D. catenatum [11–13]. DcSUTs are mainly expressing and functioning in the flower, while two members are highly expressed in the stem of D. catenatum [12]. Hao et al. reported that SWEET family undergo expansion in D. catenatum, mainly due to the tandem duplication events [11]. It also found that three DcSWEET genes are highly expressing in the stem, while two members are significantly regulated under cold, drought, and MeJA treatment [11]. However, there is still a lack of comprehensive analysis of sugar transporters in Dendrobium, and limits the understanding of sugar partitioning in Dendrobium.
MST belongs to the MFS (major facilitator superfamily) class of transporters, is the largest family of plant STs. There are 53 AtMSTs in Arabidopsis thaliana and 65 OsMSTs in Oryza sativa, respectively [14, 15]. MST family includes seven subfamilies which have one common origin [16]. Sugar transport proteins (STPs) are hexose/H+ symporters in the plasma membrane, and responsible for the uptake of hexoses of sink cells [17, 18]. Vacuolar glucose transporters (VGTs) are tonoplast or chloroplast localized glucose/H+ antiporter [17, 19]. Polyol/monosaccharide transporters (PLTs) are localized in the plasma membrane, and act as broad-spectrum H+-symporter for linear polyols and sugars, including sorbitol, mannitol, xylitol, ribose, myo-inositol, pentoses and hexoses [17, 20]. Inositol transporters (INTs) are localized in plasma membrane or tonoplast, and responsible for the transportation of myo-inositol which involved in the synthesis of raffinose [21, 22]. Tonoplast monosaccharide transporters (TMTs) are tonoplastic sucrose/H+ antiporters, and responsible for the influx of sugars into the vacuole [23]. Plastidic glucose translocators (pGLTs) are localized to chloroplast inner envelope membrane or Golgi stacks, act as hexose transporter [24, 25]. Early Response to Dehydration 6-like (ERD6-like or ESL) is the largest subfamily of MST family in Arabidopsis, some ERD6 proteins have been characterized as tonoplastic glucose exporters in Arabidopsis [26].
So far, SUT has been exclusively discovered in plants, functions as sucrose–H+ symporter and plays a pivotal role in the long-distance transportation of sucrose from the source to sink [27]. Most SUT proteins are responsible for transporting sucrose and maltose as their substrates, while some members also be capable of transporting esculin and biotin [28, 29]. SUT proteins belong to the of MFS superfamily, have 12 transmembrane (TM) domains and always be localized to plasma membrane, whereas some members localize to tonoplast and responsible for the efflux of sucrose from vacuole to cytosol [30]. There are nine AtSUC genes and five OsSUT genes in the genome of Arabidopsis and rice [18, 31]. In higher plants, SUT proteins could be divided into three distinct clades (Type I, II and III) which originated from two ancient groups (AG1 and AG2) in the ancestral bryophyte [32]. Type I SUTs are originated from AG1 and lost in the monocot lineages, the monocot-specific Type IIB SUTs are originated from Type II but exhibit highly variable gene structures [32]. Type I and Type IIB members are plasmalemmal localized and involved in apoplastic phloem loading [33, 34]. Type II members are plasmalemmal or cytoplasmic localized, but their function is still unclear [31, 32]. Type III members are tonoplastic-localized and responsible for sucrose efflux from vacuole [35].
SWEET is a recently identified sugar transport family, which mediate the translocation of sugars across cell membranes, especially the efflux of cell-to-cell transport and the secretion of sugars [36, 37]. In plants, SWEET is a class of low-affinity sugar transporter, and may functions as uniporters involved in the transportation of neutral sugars such as glucose, fructose, and sucrose [37]. SWEET proteins always have seven transmembrane helices (TMHs) that harbor two MtN3/saliva (PF03083) domains [36, 38], and are found in various cellular compartments including plasma membrane, tonoplast, and Golgi [39]. There are 17 AtSWEETs in Arabidopsis and 21 OsSWEET members in rice [38]. The evolutionary analysis showed that four major clades were perceived in plant SWEET proteins, from clade I to clade IV [40]. In Arabidopsis, SWEET members of clades I, II, and IV primarily transport hexose, clade III SWEETs mainly transport sucrose [39]. In addition to their roles in sugar loading and unloading, SWEETs are also implicated in plant development, resistance to environmental stresses, hormone regulation, plant-pathogen and symbiotic interactions [41].
In this study, the sugar transporters were identified in the genome of four Dendrobium plants and one Orchid plant Apostasia shenzhenica [42]. Then, the evolution history of sugar transporters was analyzed in Dendrobium plants. Furthermore, a comprehensive analysis of sugar transporters was conducted in D. catenatum, a well-known and widely cultivated Dendrobium plant, including the structural arrangement, cis-element analysis, regulatory networks, and expression profiles. Finally, the polysaccharides content and gene expression the leaf and stem of D. catenatum investigated under different light conditions. The results laid the foundation of functional studies of the sugar transporters, aiding in the understanding of polysaccharide biosynthesis and the germplasm improvement of Dendrobium plants.
Results
Genome‑wide identification and phylogenetic analysis of sugar transporter genes in Dendrobium plants
The members of the MST, SUT and SWEET sugar transporter gene families were identified using combined BLAST and HMM methods in the genome of A. shenzhenica and four Dendrobium plants (D. catenatum, D. chrysotoxum, D. huoshanense, and D. nobile). As shown in Table 1 and Table S1, a total of 341 sugar transporters were identified in these five Orchid plants, including 61 members in A. shenzhenica, 73 members in D. catenatum, 66 members in D. chrysotoxum, 60 members in D. huoshanense, and 81 members in D. nobile.
Table 1.
The number of different type sugar transporters in 7 plants
| Speices | MST | SUT | SWEET | Total | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| STP | PLT | VGT | TMT | pGlcT | INT | ERD6-like | Total | I | IIa | IIb | IIIa | IIIb | Total | I | IIa | IIb | III | IV | Total | ||
| A. thaliana | 14 | 6 | 3 | 3 | 4 | 4 | 19 | 53 | 1 | 1 | 7 | 0 | 0 | 9 | 3 | 5 | 0 | 7 | 2 | 17 | 79 |
| O. sativa | 29 | 15 | 2 | 6 | 4 | 3 | 6 | 65 | 1 | 1 | 0 | 3 | 0 | 5 | 6 | 8 | 1 | 5 | 1 | 21 | 91 |
| A. shenzhenica | 21 | 5 | 2 | 2 | 3 | 3 | 3 | 39 | 1 | 1 | 0 | 1 | 2 | 5 | 3 | 3 | 3 | 7 | 1 | 17 | 61 |
| D. catenatum | 15 | 6 | 2 | 2 | 3 | 3 | 3 | 34 | 1 | 1 | 0 | 2 | 3 | 7 | 5 | 5 | 11 | 10 | 1 | 32 | 73 |
| D. chrysotoxum | 14 | 4 | 2 | 1 | 1 | 3 | 0 | 25 | 1 | 1 | 0 | 2 | 3 | 7 | 3 | 3 | 18 | 8 | 2 | 34 | 66 |
| D. huoshanense | 12 | 9 | 1 | 2 | 0 | 3 | 1 | 28 | 1 | 0 | 0 | 3 | 5 | 9 | 5 | 4 | 5 | 7 | 2 | 23 | 60 |
| D. nobile | 18 | 5 | 2 | 2 | 3 | 3 | 3 | 36 | 1 | 1 | 0 | 2 | 4 | 8 | 5 | 6 | 14 | 10 | 2 | 37 | 81 |
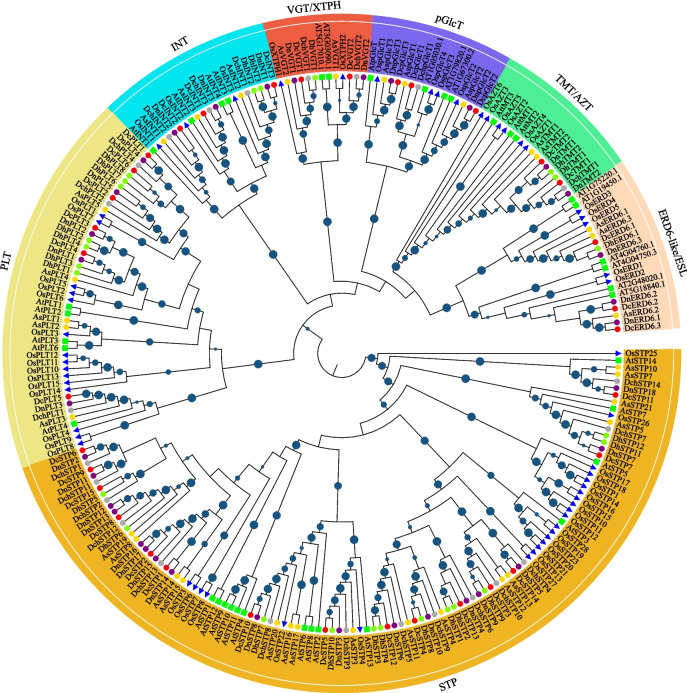
MST is the largest sugar transporter family in A. shenzhenica (39 members), D. catenatum (34 members), and D. huoshanense (28 members). The members of MST could be classified into seven subfamilies based their evolutionary relationship in most Dendrobium plants (Table 1 and Fig. 1), as well as that of Arabidopsis and rice. STP and ERD-6 like always be the largest subfamilies of MST gene family in plants [26, 43]. S In Dendrobium plants, STP is the largest subfamily; ERD-6 like subfamily undergoes contraction and loss events, especially in D. chrysotoxum and D. huoshanense; the subfamilies of PLT, VGT, TMT, INT, and pGlcT exhibit similar scales to other plants.
Fig. 1.
Phylogenetic tree and subgroup classification of MST proteins in A. thaliana, O. sativa, A. shenzhenica, D. catenatum, D. chrysotoxum, D. huoshanense, and D. nobile. The phylogenetic tree was constructed by the neighbor-joining (NJ) method with 1000 bootstrap replicates. The blue circles in the middle of branches represent the bootstrap value > 50. Green rectangle, blue triangles, yellow cycle, red cycle, grey cycle, yellowgreen cycle, purple cycle represent proteins from A. thaliana, O. sativa, A. shenzhenica, D. catenatum, D. chrysotoxum, D. huoshanense, and D. nobile
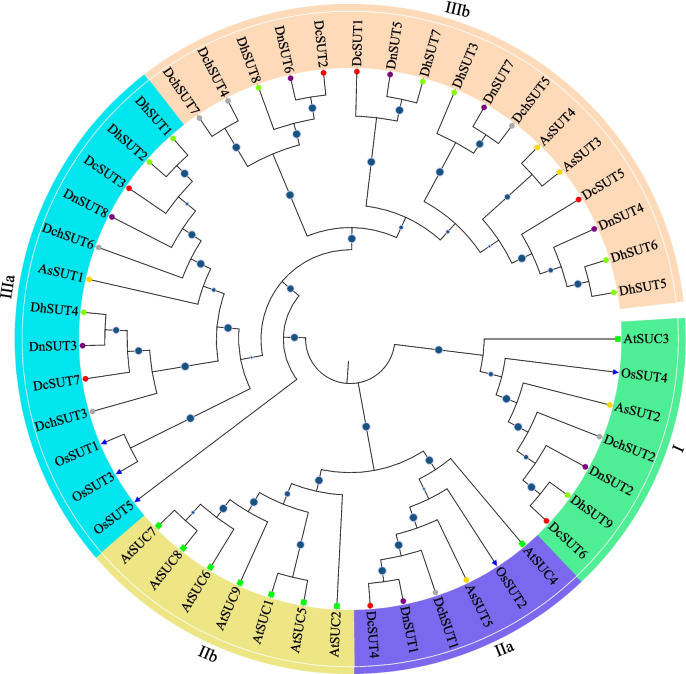
SUT contains five to nine members in Dendrobium plants and A. shenzhenica, and could be classified into four clades (Table 1 and Fig. 2), including clade I, clade IIa, clade IIIa/b. In most Dendrobium plants, clade I and clade IIa each contain a single member. Clade IIb is a dicotyledon-specific clade of SUT gene family, and is absent in rice, A. shenzhenica, and four Dendrobium plants. Clade III is a monocotyledon-specific clade of SUT gene family, and could be further subdivided into two subclades, including IIIa and the Orchid-specific IIIb. There are 17 IIIb members in five Orchid species: two members in A. shenzhenica (AsSUT3/4), three members in D. catenatum (DcSUT1/2/5) and D. chrysotoxum (DchSUT4/5/7), four members in D. nobile (DnSUT4/5/6/7), and five members in D. huoshanense (DhSUT3/5/6/7/8).
Fig. 2.
Phylogenetic trees and subgroup classification of SUT proteins in A. thaliana, O. sativa, A. shenzhenica, D. catenatum, D. chrysotoxum, D. huoshanense, and D. nobile. The phylogenetic tree was constructed by the neighbor-joining (NJ) method with 1000 bootstrap replicates. The blue circles in the middle of branches represent the bootstrap value > 50. The green rectangle, blue triangles, yellow cycle, red cycle, grey cycle, yellowgreen cycle, purple cycle represent proteins from A. thaliana, O. sativa, A. shenzhenica, D. catenatum, D. chrysotoxum, D. huoshanense, and D. nobile
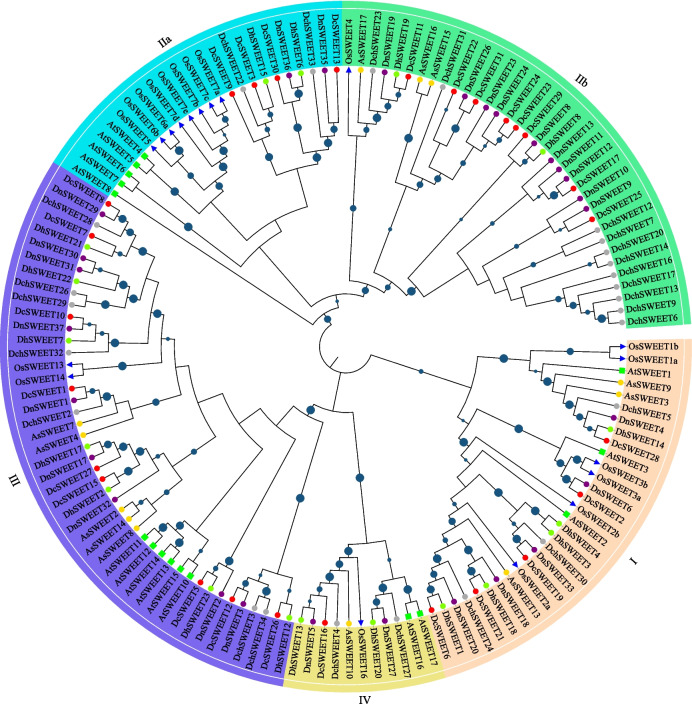
SWEET is the largest sugar transporter family in D. chrysotoxum (34 members) and D. nobile (37 members). As shown in Fig. 3, SWEET proteins could be classified into five clades: clade I, clade IIa, clade IIb, clade III, and clade IV. There are three to five members of clade I in A. shenzhenica and Dendrobium plants. In the clade IIa, the members of Arabidopsis, rice and Dendrobium plants cluster into the single clades, respectively. Clade IIb is a monocotyledon-specific clade of SWEET gene family, there are one member in rice (OsSWEET4), three members in A. shenzhenica (AsSWEET15/16/17), and expanded dramatically in Dendrobium plants. In clade III and IV, the members from different plants always mixed with each other.
Fig. 3.
Phylogenetic trees and subgroup classification of SWEET proteins in A. thaliana, O. sativa, A. shenzhenica, D. catenatum, D. chrysotoxum, D. huoshanense, and D. nobile. The phylogenetic tree was constructed by the neighbor-joining (NJ) method with 1000 bootstrap replicates. The blue circles in the middle of branches represent the bootstrap value > 50. The green rectangle, blue triangles, yellow cycle, red cycle, grey cycle, yellowgreen cycle, purple cycle represent proteins from A. thaliana, O. sativa, A. shenzhenica, D. catenatum, D. chrysotoxum, D. huoshanense, and D. nobile
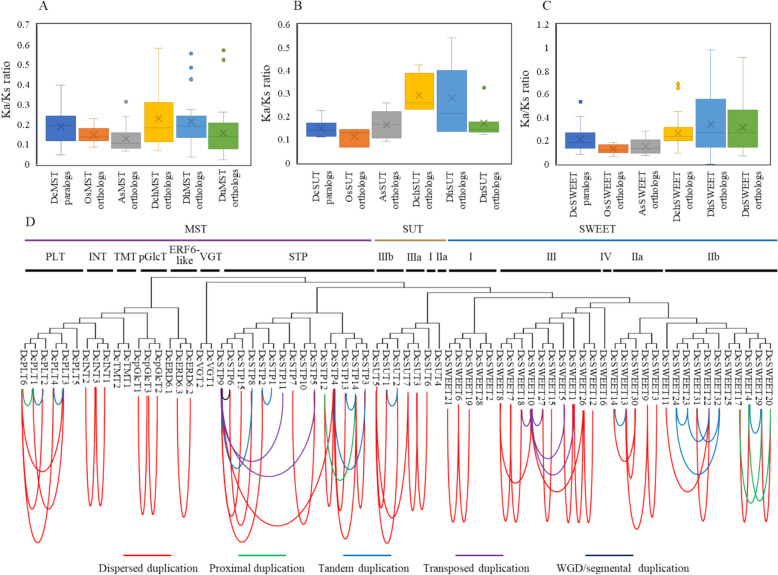
To investigate the evolution history of sugar transporter genes, the homologous gene pairs of DcSTs were identified and the Ka/Ks ratio were calculated. As shown in Table S2, there are 66 DcST paralogous gene pairs (including 31 DcMST gene pairs, 5 DcSUT gene pairs, and 30 DcSWEET gene pairs) in D. catenatum, 41 orthologous gene pairs (25 MST gene pairs, 3 SUT gene pairs, and 14 SWEET gene pairs) between D. catenatum and O. sativa, 74 orthologous gene pairs (38 MST gene pairs, 10 SUT gene pairs, and 26 SWEET gene pairs) between D. catenatum and A. shenzhenica, 77 orthologous gene pairs (23 MST gene pairs, 4 SUT gene pairs, and 50 SWEET gene pairs) between D. catenatum and D. chrysotoxum, 58 orthologous gene pairs (29 MST gene pairs, 7 SUT gene pairs, and 22 SWEET gene pairs) between D. catenatum and D. huoshanense, 74 orthologous gene pairs (35 MST gene pairs, 7 SUT gene pairs, and 32 SWEET gene pairs) between D. catenatum and D. nobile. The Ka/Ks ratio ranged from 0.05 to 0.40 (median 0.19) among the DcMST paralogs, 0.11 to 0.23 (median 0.15) among the DcSUT paralogs, 0.08 to 0.53 (median 0.21) among the DcSWEET paralogs (Table S2). As shown in Fig. 4A, the Ka/Ks ratio among the DcMST orthologous gene pairs is similar to that of DcMST paralogous gene pairs. However, the Ka/Ks ratio of DcSUT/DcSWEET orthologous gene pairs between Dendrobium plants is significantly higher than that of rice and A. shenzhenica (Fig. 4B and C). The results indicate that sugar transporter homologous genes in D. catenatum are under the purifying selection pressure, while SUT and SWEET genes among Dendrobium plants are under the relative positive selection.
Fig. 4.
Inference of DcST gene duplications. A Box and whisker plots representing the distribution of Ka/Ks ratio in MST Orthologues; B SUT Orthologues; C SWEET Orthologues; D Phylogenetic relationship and gene duplications pairs among DcSTs
The duplication events of DcST genes were analyzed using DupGen_finder [44]. As shown in Fig. 4D, a total of 66 duplicated gene pairs were identified among 61 DcST genes. Dispersed duplication (42 duplicated gene pairs) and tandem duplication (13 duplicated gene pairs) were the main duplicated types of sugar transporter genes in D. catenatum. In clade II type DcSWEETs, seven tandem duplication-derived DcSWEETs were identified, including a gene cluster that consisted of five DcSWEET genes (DcSWEET22/23/24/31/32) and a duplicated gene pair of DcSWEET4 and DcSWEET29. The results suggest that tandem duplication events have played a pivotal role in the expansion of clade II type DcSWEETs.
Characteristic features and structural arrangement of DcSTs in D. catenatum
A total of 73 DcST genes were identified in D. catenatum genome, including 34 DcMST, 7 DcSUT and 32 DcSWEET genes (Fig. 4D). The detailed information of these 73 ST genes were listed in Table 2, including the gene ID, genomic location, protein length, exon number, molecular weight (MW), and subcellular localization.
Table 2.
Characteristic features of sugar transporters in D. catenatum
| S. No | Protein ID | Protein length (aa) | Molecular weight | Exons | THMs | Subcellular location |
|---|---|---|---|---|---|---|
| DcERD6.1 | XP_020690096.1 | 497 | 53,619 | 17 | 12 | plasma membrane and vacuole |
| DcERD6.2 | XP_020701355.1 | 474 | 51,494.47 | 18 | 12 | plasma membrane and vacuole |
| DcERD6.3 | XP_020702150.1 | 481 | 51,494.84 | 17 | 12 | plasma membrane |
| DcINT1 | XP_020675104.1 | 581 | 63,279.1 | 6 | 12 | plasma membrane and vacuole |
| DcINT2 | XP_020676904.1 | 509 | 54,680.9 | 6 | 12 | plasma membrane |
| DcINT3 | XP_020696092.1 | 579 | 63,420.1 | 6 | 12 | plasma membrane and vacuole |
| DcpGlcT1 | XP_020699357.2 | 537 | 58,411.23 | 12 | 11 | plasma membrane and chloroplast |
| DcpGlcT2 | XP_020671882.1 | 491 | 52,786.05 | 13 | 10 | plasma membrane |
| DcpGlcT3 | XP_020672325.2 | 583 | 61,401.43 | 13 | 11 | plasma membrane and chloroplast |
| DcPLT1 | XP_020689528.1 | 496 | 53,417.9 | 2 | 10 | plasma membrane and vacuole |
| DcPLT2 | XP_020689572.2 | 505 | 54,039.13 | 2 | 10 | plasma membrane and vacuole |
| DcPLT3 | XP_020693533.2 | 525 | 56,358.75 | 3 | 12 | plasma membrane and vacuole |
| DcPLT4 | XP_020693540.1 | 522 | 55,692.14 | 3 | 12 | plasma membrane and vacuole |
| DcPLT5 | XP_020696863.1 | 549 | 58,933.98 | 2 | 11 | plasma membrane and vacuole |
| DcPLT6 | XP_028551540.1 | 461 | 49,985.54 | 1 | 9 | plasma membrane and vacuole |
| DcSTP1 | XP_020673769.1 | 504 | 55,621.81 | 1 | 11 | plasma membrane and vacuole |
| DcSTP2 | XP_020673820.1 | 508 | 55,929.32 | 1 | 12 | plasma membrane and vacuole |
| DcSTP3 | XP_020676035.2 | 511 | 57,155.97 | 3 | 8 | plasma membrane and vacuole |
| DcSTP4 | XP_020676038.1 | 512 | 56,568.53 | 3 | 11 | plasma membrane and vacuole |
| DcSTP5 | XP_020678679.1 | 522 | 57,723.61 | 4 | 12 | plasma membrane and vacuole |
| DcSTP6 | XP_020682728.1 | 479 | 52,510.74 | 2 | 10 | plasma membrane |
| DcSTP7 | XP_020690754.1 | 504 | 54,603.33 | 4 | 11 | plasma membrane and vacuole |
| DcSTP8 | XP_020692064.1 | 520 | 56,587.14 | 3 | 12 | plasma membrane and vacuole |
| DcSTP9 | XP_020692077.1 | 527 | 57,029.1 | 3 | 11 | plasma membrane and vacuole |
| DcSTP10 | XP_020695794.1 | 523 | 57,967.25 | 4 | 12 | plasma membrane and vacuole |
| DcSTP11 | XP_020700131.1 | 518 | 56,636.82 | 4 | 10 | plasma membrane and vacuole |
| DcSTP12 | XP_020704792.1 | 521 | 57,001.69 | 3 | 11 | plasma membrane and vacuole |
| DcSTP13 | XP_020704799.1 | 511 | 56,210.29 | 3 | 11 | plasma membrane and vacuole |
| DcSTP14 | XP_020704818.1 | 529 | 57,159.1 | 4 | 12 | plasma membrane and vacuole |
| DcSTP15 | XP_028548876.1 | 585 | 64,189.57 | 6 | 12 | plasma membrane |
| DcTMT1 | XP_020676096.1 | 733 | 79,105.15 | 5 | 10 | plasma membrane and vacuole |
| DcTMT2 | XP_020702404.1 | 754 | 81,183.79 | 5 | 10 | plasma membrane and vacuole |
| DcVGT1 | XP_020690641.1 | 555 | 59,351.64 | 13 | 12 | plasma membrane and chloroplast |
| DcVGT2 | XP_028551786.1 | 624 | 69,089.88 | 12 | 11 | plasma membrane and vacuole |
| DcSUT1 | XP_020695242.1 | 500 | 53,276.48 | 14 | 11 | plasma membrane and vacuole |
| DcSUT2 | XP_020695324.1 | 500 | 53,406.88 | 14 | 12 | plasma membrane |
| DcSUT3 | XP_020698795.1 | 518 | 55,639.42 | 14 | 10 | plasma membrane and vacuole |
| DcSUT4 | XP_020700410.1 | 538 | 57,434.5 | 5 | 12 | plasma membrane |
| DcSUT5 | XP_020702803.1 | 496 | 52,805.96 | 14 | 10 | plasma membrane and vacuole |
| DcSUT6 | XP_020703969.1 | 638 | 68,894.38 | 14 | 11 | plasma membrane |
| DcSUT7 | XP_028557242.1 | 491 | 53,217.55 | 14 | 9 | plasma membrane and vacuole |
| DcSWEET1 | XP_020671992.2 | 262 | 29,221.23 | 6 | 7 | plasma membrane and vacuole |
| DcSWEET2 | XP_020672052.1 | 277 | 30,999.59 | 5 | 6 | plasma membrane and vacuole |
| DcSWEET3 | XP_020674019.1 | 233 | 26,201.75 | 5 | 7 | plasma membrane |
| DcSWEET4 | XP_020674476.2 | 237 | 26,775.07 | 4 | 7 | plasma membrane and vacuole |
| DcSWEET5 | XP_020675146.2 | 296 | 33,103.11 | 7 | 7 | plasma membrane and vacuole |
| DcSWEET6 | XP_020675601.1 | 237 | 26,619.75 | 6 | 7 | plasma membrane, chloroplast, chloroplast |
| DcSWEET7 | XP_020681423.1 | 265 | 29,628.48 | 6 | 7 | plasma membrane and vacuole |
| DcSWEET8 | XP_020681431.1 | 265 | 29,548.47 | 6 | 7 | plasma membrane and vacuole |
| DcSWEET9 | XP_020682347.1 | 232 | 25,952.07 | 5 | 7 | plasma membrane |
| DcSWEET10 | XP_020686358.1 | 266 | 29,937.55 | 6 | 7 | plasma membrane and vacuole |
| DcSWEET11 | XP_020687552.1 | 248 | 27,452.87 | 5 | 7 | plasma membrane, chloroplast, chloroplast |
| DcSWEET12 | XP_020694055.1 | 204 | 23,383.16 | 4 | 5 | plasma membrane and vacuole |
| DcSWEET13 | XP_020694837.1 | 236 | 26,368.85 | 5 | 7 | plasma membrane and vacuole |
| DcSWEET14 | XP_020694842.2 | 235 | 26,270.7 | 5 | 7 | plasma membrane and vacuole |
| DcSWEET15 | XP_020695376.1 | 203 | 23,090.31 | 4 | 7 | plasma membrane and vacuole |
| DcSWEET16 | XP_020695524.1 | 296 | 33,105.05 | 6 | 6 | plasma membrane and vacuole |
| DcSWEET17 | XP_020698470.1 | 196 | 22,184.7 | 3 | 6 | plasma membrane and vacuole |
| DcSWEET18 | XP_020699529.1 | 271 | 30,501.38 | 6 | 7 | plasma membrane and vacuole |
| DcSWEET19 | XP_020700202.1 | 234 | 26,016.98 | 6 | 7 | plasma membrane and vacuole |
| DcSWEET20 | XP_020700756.1 | 237 | 26,594.95 | 4 | 7 | plasma membrane and vacuole |
| DcSWEET21 | XP_020701878.1 | 237 | 26,577.42 | 6 | 7 | plasma membrane, chloroplast, chloroplast |
| DcSWEET22 | XP_020706020.1 | 259 | 28,917.85 | 4 | 7 | plasma membrane and vacuole |
| DcSWEET23 | XP_020706032.1 | 266 | 30,208.04 | 4 | 6 | plasma membrane and vacuole |
| DcSWEET24 | XP_020706043.1 | 260 | 28,848.86 | 4 | 7 | plasma membrane |
| DcSWEET25 | XP_028548619.1 | 237 | 26,514.24 | 4 | 7 | plasma membrane and vacuole |
| DcSWEET26 | XP_028549267.1 | 258 | 29,397.21 | 6 | 7 | plasma membrane and vacuole |
| DcSWEET27 | XP_028551223.1 | 249 | 27,592.3 | 6 | 7 | plasma membrane and vacuole |
| DcSWEET28 | XP_028554265.1 | 211 | 23,202.44 | 4 | 5 | plasma membrane and vacuole |
| DcSWEET29 | XP_028556203.1 | 295 | 33,080.46 | 6 | 6 | plasma membrane and vacuole |
| DcSWEET30 | XP_028556661.1 | 201 | 22,443.16 | 4 | 6 | plasma membrane and vacuole |
| DcSWEET31 | XP_028557180.1 | 285 | 31,999.37 | 5 | 7 | plasma membrane |
| DcSWEET32 | XP_028557181.1 | 283 | 31,646.02 | 5 | 7 | plasma membrane |
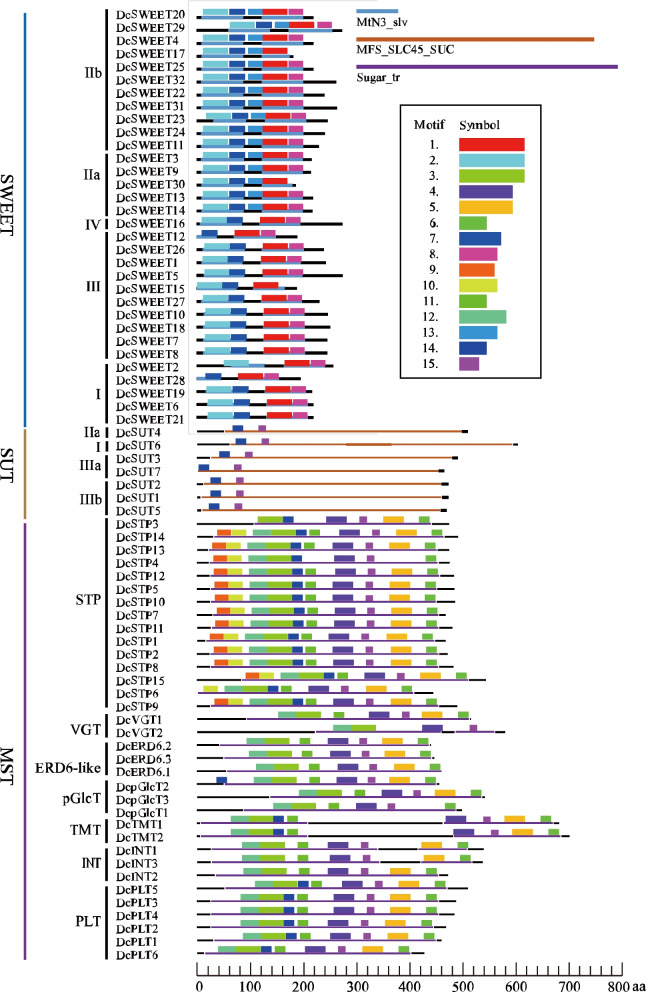
The protein length of DcMSTs ranges from 461 (DcPLT6) to 754 (DcTMT2) amino acids (aa). DcMST proteins always have 10–12 TMHs except DcSTP3 (8 exons) and DcPLT6 (9 TMHs) (Fig. S1 and Table 2). DcSUT proteins vary in length—from 491 (DcSUT7) to 638 aa (DcSUT6) and generally contain 10–12 TMHs. DcSWEETs are smaller in size, ranging from 196 (DcSWEET17 to 296 aa (DcSWEET6/16), and possess fewer TMHs (5–7). Most DcST proteins are localized in the plasma membrane, vacuole, and chloroplast.
As shown in Fig. 5A, most of the DcMST proteins possess a complete Sugar_tr (Sugar (and other) transporter, PF00083) domain except five DsMSTs (DcVGT2, DcTMT1/2, and DcINT1/3). The Sugar_tr domain consists of eight core conserved motifs (motif 3/4/5/6/11/12/14/15) in DcMSTs. DcSTPs always have two additional motifs (motif 9 and 10) in the N-terminal of their Sugar_tr domain. MFS_SLC45_SUC (Solute carrier family 45 and similar sugar transporters of the Major Facilitator Superfamily (MFS) of transporters, cd17313) domain is a conserved domain in plant sucrose transporters. Each of the seven DcSUTs have one complete MFS_SLC45_SUC domain and two conserved motifs (motif 14 and 15) in the N-terminal. All DcSWEET proteins harbor two MtN3_slv (Sugar efflux transporter for intercellular exchange, PF03083) domains, and each of MtN3_slv harbored two conserved motifs (motif 2 and 7 in N’-MtN3_slv, motif 1 and 8 in C’-MtN3_slv). Compared to other subfamilies of DcSWEET, clade II members have an additional conserved motif (motif 13) between the two MtN3_slv domains. There are some incomplete MtN3_slv domains in DcSWEETs, including the N-terminal MtN3_slv of DcSWEET2/12/28 and the C-terminal MtN3_slv of DcSWEET15/17/30.
Fig. 5.
Conserved domains and motifs (1–15) distribution of sugar transporter proteins in D. catenatum. Heterochromatic boxes represent different domains and motifs, indicated in the middle
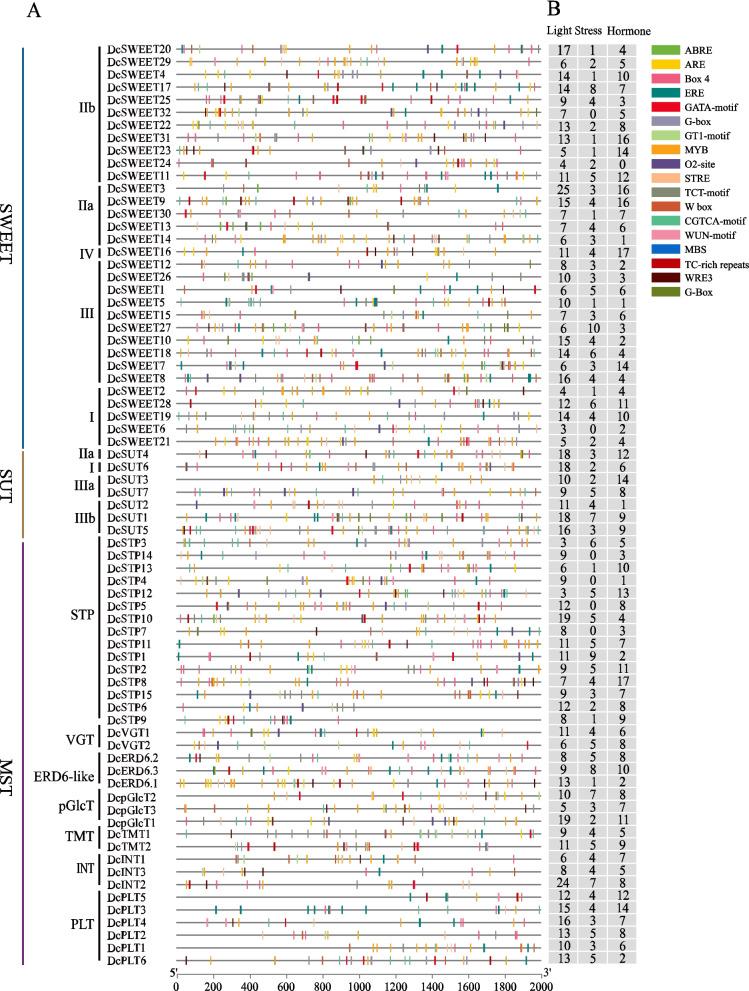
Cis‑regulatory element analysis of the sugar transporter genes
The upstream 2000 bp promoter regions of 73 DcSTs were retrieved for the analysis ofcis-acting elements. As shown in Fig. 6A, the top 20 most abundant cis-element encompass those involved in transcription factor (TF) recognition and binding (MYB, W box, MYB-like sequence, Myb-binding site), light responsiveness (Box 4, G-box, TCT-motif, GT1-motif, GATA-motif), stress responsiveness (STRE, ARE, WRE3, WUN-motif, MBS, TC-rich repeats), and hormones responsiveness (CGTCA-motif, TGACG-motif, ABRE, ERE). MYB (core cis-element for MYB TFs recognition and binding), Box 4 (conserved DNA module involved in light responsiveness), CGTCA-motif (cis-acting regulatory elements involved in the methyl jasmonate (MeJA) responsiveness) were the most abundant elements in DcSTs. Most DcSTs contain abundant light responsiveness and hormone responsiveness cis-elements (Fig. 6B), hinting at their potential roles in the growth and development of D. catenatum. DcSTP1/3, DcERD6.3 and DcSWEET27 contain amounts of stress responsiveness cis-elements, implying their involvement in the response to environmental stresses. Furthermore, the transcriptional regulatory network of sugar transporter genes and transcription factors were analyzed in D. catenatum. A total of 40 TF families are implicated in the transcriptional regulatory of DcSTs, with most DcST genes being regulated by multiple TFs (Fig. S2). In accordance with the results of cis‑regulatory element analysis (Fig. 6A), most DcST genes are targeted by MYB TFs. In addition, B3, C2H2, Dof, ERF, HD-ZIP, MIKC_MADS TFs also extensively participate in the regulation of DcSTs expression.
Fig. 6.
Cis-regulatory element analysis of sugar transporter proteins in D. catenatum. A) The distribution of 20 cis-acting elements in the 2000 bp upstream promoter; B) the frequency of the cis-element response to light, environmental stresses, and plant hormones
Post-transcription regulation of DcST genes
MicroRNAs (miRNAs) play the critical roles in the post-transcriptional regulatory networks. As shown in Fig. 7D, 27 out of 73 sugar transporters are regulated by 64 miRNAs belonging to 23 miRNA families. Most of these conserved miRNAs are involved in the plant development and responses to biotic/abiotic stresses, such as miR156, miR172, mR397, miR399, miR529, and miR535. Five DcSTs (DcINT3, DcSTP15, DcSUT5, and DcSWEET21/25) are regulated by miR397 members, which mainly target the laccase (LAC) genes that involved in lignin synthesis [45]. MiR156, miR529 and miR535 share the similarity sequence and all target SQUAMOSA promoter binding protein-like (SPL) genes to negatively regulate the immunity in plants. DcSWEET16 is mainly targeted by miR156 members while DcpGlcT2 is both targeted by miR156 and miR172, which are involved in reproductive growth and flower development. DcpGlcT3 andDcINT1 are mainly regulated by miR535, while DcSWEET8/15 are targeted by miR529. DcSWEET7/18 are targeted by miR399, which serves as a critical regulator of phosphate homeostasis in plants.
Fig. 7.
Interaction of miRNAs and sugar transporter genes. Blue cycles represent miRNAs, red triangles represent DcST genes
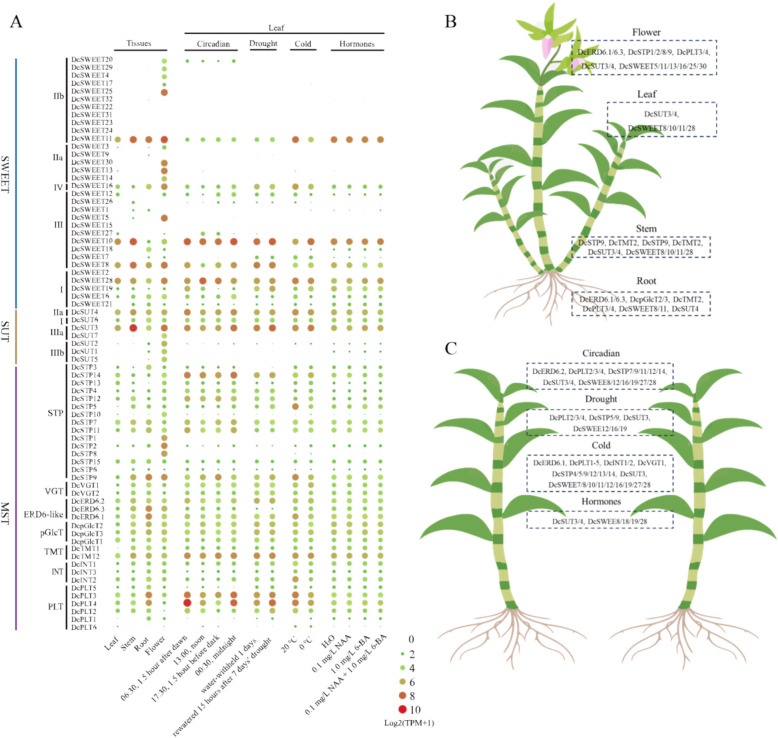
Transcriptional patterns of ST genes in D. catenatum
The RNA-seq data of D. catenatum was analyzed to investigate the expression patterns of sugar transporter genes (Fig. 8A). Compared to vegetative tissues (leaf, stem, and root), most ST genes are relatively high expressed in flowers. Several sugar transporter genes are flower-specific expression, including the members of clade II SWEET family (DcSWEET4/14/17/20/25/29/30), clade III SUT family (DcSUT5/7), and STP subfamily of MST (DcSTP1/8) (Fig. 8B). Most of MST genes, some members of SUT (DcSUT3/4/6) and SWEET (DcSWEET8/10/11/16/28) are constitutive expressing in D. catenatum. DcSUT3/4/6 show constitutive expression pattern in D. catenatum, and DcSUT3/4 are relatively high-expressed in stem. DcSWEET28 and DcSTP9 exhibit relatively high-expression levels in the daytime and downregulated during the night (Fig. 8C). Some DcST genes are down-regulated in the daytime, including DcSWEET8, DcSTP11, PLT2/3/4. DcSWEET16/19, DcSUT3, DcSTP5/9. DcPLT1/2/3/4 display differential expression patterns between the drought and rewatered treatments. in comparison. In comparison, a larger number of DcST genes are involved in the response to cold stress, including DcSWEET6/10/11/12/16/18/19/21, DcSTP4/5/9/12/13/14, DcVGT1/2, DcERD6.1, DcINT1/2, DcPLT1/2/3/4/5. DcSWEET8/19 are downregulated under exogenous hormone treatments, including NAA, 6-BA, combined NAA and 6-BA treatment. In contrast, DcSWEET28 is upregulated under these hormone treatments.
Fig. 8.
Expression profiles of DcST genes. The expression data were obtained from RNA-seq data and shown as log2 values based on TPM values (Transcripts Per Kilobase of exon model per Million mapped reads). A Heatmap of the expression pattern of DcST genes; B DcSTs with high expression level in different tissues of D. catenatum; C DcSTs involved in circadian, and the response to environmental stress and exogenous hormones
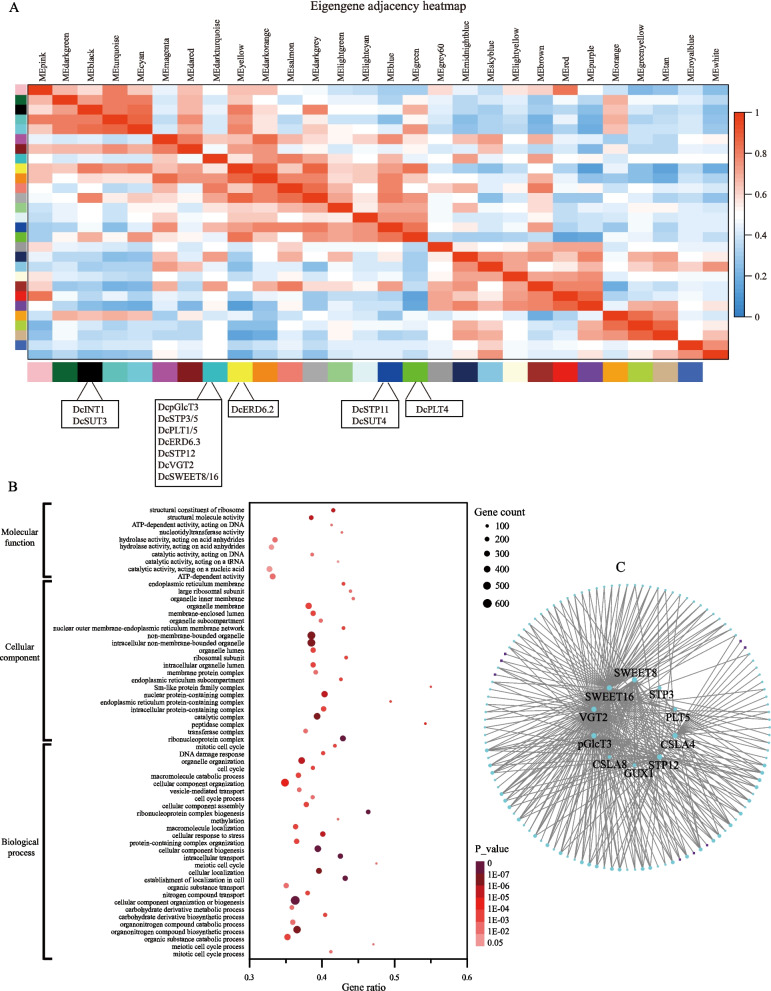
The co-expression network of sugar transporter genes in D. catenatum
To identify the potential cooperative genes of sugar transporters in D. catenatum. WGCNA (Weighted correlation network analysis) was performed using 26 RNA-seq data. As shown in Fig. 9A, 16 DcST genes are assigned to 5 modules: DcINT1 and DcSUT3 to MEblack module, 8 MST genes (DcpGlcT3, DcSTP3/5/12, DcPLT1/5, DcERD6.3, DcVGT2) and 2 SWEET genes (DcSWEET8 and DcSWEET16) to MEturquoise module, DcERD6.2 to MEyellow module, DcSTP11 and DcSUT4 to MEblue module, DcPLT4 to MEgreen module. The GO (Gene Ontology) enrichment analysis was performed in the genes sets of those five modules. Two polysaccharide biosynthesis-related GO terms are enriched in the MEturquoise module, including carbohydrate derivative binding (GO:0097367) and carbohydrate derivative metabolic process (GO:1,901,135) (Fig. 9B). GO:0097367 is also enriched in black module, whereas none of the polysaccharide biosynthesis-related GO terms were found in other three modules. The central genes involved in polysaccharide biosynthesis were identified in MEturquoise module (Fig. 9C), including seven sugar transporter genes (DcSWWT8/16, DcSTP3/12, DcVGT2, DcpGlcT4, and DcPLT5), two CSLA genes (CELLULOSE SYNTHASE-LIKE A genes, LOC110107854_DcCSLA4 and LOC110108926_DcCSLA8), and GUX1 (UDP-glucuronate:xylan alpha-glucuronosyltransferase 1, LOC110102381).
Fig. 9.
The key sugar transporter genes involved in polysaccharide biosynthesis. A Module eigengene (ME) adjacency heatmap of 28 co-expression modules; B GO enrichement analysis of the gene sets of MEturquoise module; C Co-expression network of DcST and polysaccharide biosynthesis related genes in the turquoise module, blue circle represents protein-coding genes, purple square represents noncoding genes
Polysaccharides content and expression profiles of central genes under different light conditions
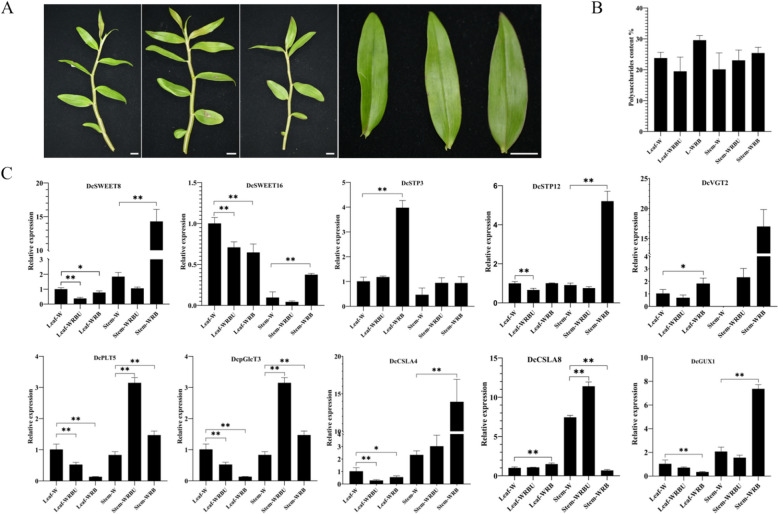
To further clarify the potential function of sugar transporter genes in the process of polysaccharide biosynthesis under different light conditions. D. catenatum were cultivated under white light (W, control, CK), white-red-blue-UV light (WRBU), and white-red-blue light (WRB) for seven days. Then, the leaves and stems were sampled for the analysis of polysaccharide content and expression patterns of central genes.
As shown in Fig. 10A, there were no significant morphological difference among three treatment groups, besides the weakly anthocyanin accumulation in the leaf of WRBU group, which may be induced by UV radiation. Compared CK group (23.79 ± 1.52%), the content of polysaccharides in the leaves was up-regulated slightly in the WRB group (29.57 ± 1.23%), while down-regulated weakly in the WRBU group (19.50 ± 3.74%), both without significant difference (Fig. 10B). In the stems, polysaccharides contents were upregulated under both WRBU and WRB treatments (CK 20.13 ± 4.34%, WRBU 23.05 ± 2.73%, WRB 25.41 ± 1.54%). It suggests that Red + Blue light could stimulate the biosynthesis and accumulation of polysaccharides in D. catenatum, whereas UV radiation tends to diminish this positive effect.
Fig. 10.
Morphological, polysaccharides content, and expression profiles of polysaccharide biosynthesis related central genes in D. catenatum under different light conditions. A Morphological of D. catenatum under different light conditions, from left to right: Stem-W, Stem-WRBU, Stem-WRB, Leaf-W, Leaf-WRBU, and Leaf-WRB; B the content of polysaccharides in leaf and stem; C the expression of ten central genes in leaf and stem. The Each data point represents mean value ± standard deviation (SD), n = 3 (Three biological replicates). Error bars indicate standard deviation. Asterisks indicate the significant degree of expression level compared to the value of the control (* < 0.05, ** < 0.01)
The expression profiles of seven sugar transporter genes (DcSWEET8/16, DcSTP3/12, DcVGT2, DcPLT5, DcpGlcT3) and three polysaccharide biosynthesis-related genes (DcCSLA4, DcCSLA8, DcGUX1) were verified using qRT-PCR. As shown in Fig. 10C, DcSWEET8/16 were down-regulated in the Leaf-WRBU and Leaf-WRB groups, while up-regulated in the Stem-WRB group. DcSTP3 was up-regulated in the Stem-WRB group. DcSTP12 was down-regulated in the Leaf-WRB group and up-regulated in the Stem-WRB group. DsVGT2 was up-regulated in Leaf-WRB group, although it did not be detected in the Stem-W group, the expression was increasing dramatically in Stem-WRBU and Stem-WRB groups. DcPLT5 and DcpGlcT3 both were down-regulated in Leaf-WRBU and Leaf-WRB groups, but up-regulated in Stem-WRBU and Stem-WRB groups. Three polysaccharide biosynthesis related genes both have the higher relative expression level in the stem than leaf. DcCSLA4 was down-regulated in Leaf-WRBU and Leaf-WRB groups, and up-regulated in Stem-WRB group. DcCSLA8 was up-regulated in Leaf-WRB group, Stem-WRBU and Stem-WRB groups. DcGUX1 was down-regulated in Leaf-WRB group and up-regulated in Stem-WRB group.
Discussion
Carbohydrates (sugars) serve as one of the most important carbon and energy resource in higher plants, also function as signal and functional molecules in many biological processes [46]. Plants synthesize sugars within the chloroplast of phototrophic cells during the daytime, and transport these sugars throughout the plant to fulfill consumption and storage requirements. Sucrose serves as the predominant sugar for long-distance transport through the phloem [35, 47]. It is mainly imported the sieve elements-companion cell complex (SE-CC) via symplasmic or apoplasmic pathways [35, 47], and then is directly transported into sink cells, or is hydrolyzed to monosaccharides (glucose and fructose) for sugar uptake of sink cells [17, 48]. Hexoses and sucrose are allocated to various subcellular compartments to maintain the carbon and energy supplies. Translocation of sugar plays an indispensable role in plant growth, development, stress/defense and signal transduction [37]. Sugar transports are mainly responsible for sugars translocation, and classified into MST, SUT, and SWEET gene families [37, 49]. The evolutional and functional research of sugar transporter have been well studied in some plants, such as Arabidopsis [15, 50, 51], rice [15, 52], sorghum [53], pineapple [54], and Lilies [55]. However, there is still a lack of the thorough comprehension of sugar transporters in Dendrobium plants.
In present study, the members belong to MST, SUT, and SWEET gene families were identified in four Dendrobium plants, including D. catenatum, D. chrysotoxum, D. huoshanense, and D. nobile. The members of sugar transporter gene families were identified in A. shenzhenica to study the evolution history of sugar transporters. The phylogenetic analysis showed that ERD6-like subfamily has experienced dramatically contraction and loss in Orchid plants. ERD6-like is the largest subfamily of MST gene family in Arabidopsis due to the segmental and tandem duplications events in the evolutionary histories [15, 56]. Previous reports showed that ERD6-like genes mainly participate in the transport of different hexoses, and the biological functions of most ERD6-like genes remain largely unknown [56]. The clade IIIb of SUT and clade IIb of SWEET have experienced expansion in Dendrobium plants. Clade IIIb of SUT is an Orchid-specific clade, which may be originated from the dispersed duplication event during the speciation of ancestor of Orchid and undergo expansion in Dendrobium plants (Figs. 2, 3, and 4D). Previous studies reported the expansion events of SWEET in D. catenatum and D. chrysotoxum, and speculated that these expansion events are related to the abundant medicinal compounds and fleshy stems of Dendrobium [10, 11]. Tandem duplication was found to be responsible for the expansion of the DcSWEET family, especially clade II type of DcSWEETs [10, 11].
The promoter region of DcSTs contained many cis-element involved in light/stress/hormones responsiveness and TF binding. The results indicate that most DsSTs may be influenced by light condition and involved in the partitioning of photosynthates in D. catenatum. The transcriptional regulatory network reveals that most DcST genes are targeted by MYB TFs (Fig. S2). MYB is one of the largest TF families in plants, play important roles in plant growth and development, and responses to environmental stimuli [57]. Previous study has demonstrated that MYB TFs are integral to the synthesis of various secondary metabolites, such as flavonoids, phenolic acids, terpenoids, alkaloids, and polysaccharides [58]. Some MYB TFs are essential for the polysaccharides biosynthesis, such as AtMYB5/46 targeting to AtCSLA9 in Arabidopsis [59, 60], and DoMYB75 in D. catenatum [61]. DcMYBs targeting DcSTs may also represent the crucial regulators of polysaccharides biosynthesis in D. catenatum. DcST genes are regulated by several conserved miRNA families at post-transcriptional level, it suggesting that these miRNAs also play important roles in the process of sugar partitioning.
Dendrobium plants are rich of polysaccharide in the stem and leaf. Previous studies reported that some members of DcSUT and DcSWEET are expressing at a relatively high level in the stem of D. catenatum [11, 12]. In this work, it is found that some DcSUT and DcSWEET genes are highly expressed in stem and leaf, while some DcMST genes are highly expressed in stem (Fig. 8A and B). The results indicate that the MST genes also play important roles in the biosynthesis of polysaccharide, as well as SUT and SWEET genes. In accordance with the reports of Petunia axillaris [62] and some Orchid plants [11–13], most DcSTs are relatively high expressed in flowers, while some members are flower-specific expression. Previous studies also reported that most of the DcSUT genes and three DcSWEET genes are relatively high expressed in flower, and the higher content of sucrose in flower [11, 12]. It suggests that DcSTs play pivotal roles in flower development, such as the production of nectar and sugar-rich secretion. The expression patterns of some DcSTs have obvious circadian rhythm, such as DcSWEET8/28 and DcSTP7/9/11, DcSUT3, DcPLT2/3/4 (Fig. 8A). There is a bidirectional relationship between the circadian oscillator and primary metabolism [63]. The expression of AtSUC and AtSWEET genes are rhythmic, such as AtSWEET11 expressed in leaves, AtSWEET14 and AtSUC1 expressed in roots [64, 65] (Covington et al., 2008; Durand et al., 2018). The results suggest that DcSTs are responsible for the sugar translocation to different sink organs and are implicated in various physiological process, such as photosynthesis, flowering, and the biosynthesis of polysaccharides. Sugar transporters modulate sugar partitioning to counterbalance the negative impacts of biotic stresses in plants [49]. In this study, some DcST genes were found to be responsive to the environment stimuli in D. catenatum, especially SWEET members. Mannan polysaccharides are promising bioactive polysaccharides and key ingredient in D. catenatum [2]. CSLA genes encode mannan synthases that are responsible for glucomannan biosynthesis [66, 67]. A total of 13 DcCSLAs have been identified, which play the important roles in the biosynthesis of mannan polysaccharides in D. catenatum [66, 68, 69]. Ten polysaccharide biosynthesis-related central genes were identified using WGCNA, including seven DcST genes, two DcCSLA genes, and DcGUX1.
Most DcST genes contains abundant cis-elements responsive to light in their promoter region (Fig. 6B). The results indicated that light conditions regulate the expression of DcSTs and play a crucial role in influencing polysaccharide biosynthesis in D. catenatum. Previous studies demonstrated that light condition, particularly light quality, are essential for controlling the growth and polysaccharide production in alga and plants [70–72]. Red-blue light has been shown to enhance the accumulation of dry matter and polysaccharides in Dendrobium plants [73, 74]. We further investigated the content of polysaccharides and the expression of central genes in D. catenatum under various light qualities. And found that red-blue light could promote the accumulation of polysaccharides in both leaf and stem, whereas UV light suppresses polysaccharide accumulation in leaf. The expression of ten central genes were influenced significantly by different light conditions, indicating their close association with light responsiveness and polysaccharide biosynthesis. The further work will focus on determining the functions of these DcSTs and elucidating their roles in the polysaccharide biosynthesis of D. catenatum.
Materials and methods
Identification of MST, SUT and SWEET proteins in D. catenatum
All the MST, SUT, and SWEET family members of A. thaliana and O. sativa were downloaded from The Arabidopsis Information Resource (http://www.arabidopsis.org/) and the Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/). The Hidden Markov Model (HMM) profiles of Sugar_tr (PF00083), MtN3/saliva (PF03083), and MFS_SLC45_SUC (cd17313) domains were downloaded from the Pfam database (http://pfam.xfam.org/) and Conserved Domain Database (https://www.ncbi.nlm.nih.gov/Structure/cdd). The genomes of D. catenatum (GCF_001605985.2), D. nobile (GCA_022539455.1), D. chrysotoxum (GCA_019925795.1), A. shenzhenica (GCA_002786265.1), and D. huoshanense (CNP0000830) was downloaded from NCBI (https://www.ncbi.nlm.nih.gov) and CNGBdb (https://db.cngb.org/).
The protein sequences of MST, SUT and SWEET in Arabidopsis and rice were used for seed to find the candidate proteins in the genomes of four Dendrobium plants and A. shenzhenica using BLASTP program with an E-value cutoff set as 1e–10. Then, HMMER program (http://hmmer.org/) was used to identify the candidate MST proteins harboring Sugar_tr domain, SUT proteins harboring MFS_SLC45_SUC domain, and SWEET proteins harboring MtN3/saliva domain with an E-value cutoff of 1e–5 [75].
Basic physicochemical properties and phylogenetic analysis of sugar transporters in D. catenatum
The molecular weight (MW), isoelectric point (pI) and grand average of hydropathicity (GRAVY) of sugar transporter proteins were calculated using ProtParam (https://web.expasy.org/protparam/) [76]. The subcellular localization information were predicted using WoLF PSORT (https://wolfpsort.hgc.jp/) [77], Cell-Ploc (http://www.csbio.sjtu.edu.cn/bioinf/Cell-PLoc-2/) [78] and Bologna Unified Subcellular Component Annotator (BUSCA, http://busca.biocomp.unibo.it) [79], and final decided based on consensus localization for two or more algorithms.
The information of transmembrane domain (TM) and conserved domain was obtained using InterPro (https://www.ebi.ac.uk/interpro/search/sequence/). The conserved motifs of MST, SUT, and SWEET proteins were analyzed by MEME (http://meme-suite.org) with the following parameters: maximum number of motifs, 15; motif length, 6 to 50 [80]. The structure of sugar transporter genes was visualized using the online tools Gene Structure Display Server 2.0 (GSDS 2.0, http://gsds.gao-lab.org/index.php) [81].
MST, SUT, and SWEET proteins were aligned respectively using MAFFT online service (https://mafft.cbrc.jp/alignment/server/) [82]. A neighbor-joining (NJ) phylogenetic tree was constructed in MEGAX (http://www.megasoftware.net/) based the multiple sequence alignment with 1000 bootstrap replicates, and displayed using Interactive Tree of Life (https://itol.embl.de/).
Gene duplication and Ka/Ks analysis
The duplication events of DcSTs were defined based on the collinearity analysis of candidate gene pairs using DupGen_finder [44]. The orthologous gene pairs were identified using OrthoFinder [83]. The non-synonymous substitution rate (Ka) and synonymous substitution rate (Ks) of the duplication and orthologous gene pairs were calculated using PAMLX [84]. Promoter analysis of DcST genes.
Cis-acting elements in the promoter region of DcST genes were predicted and analyzed using PLANTCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) [85]. The potential binding sites of TFs in the promoter region of DcST genes were predicted using PlantRegMAP (http://plantregmap.gao-lab.org/binding_site_prediction.php) and visualized using Cytoscape 3.7.0 [86, 87].
Interaction analysis of sugar transporter genes and miRNAs in D. catenatum
The miRNA data of D. catenatum were downloaded from the previous study [88]. The sugar transporter mRNAs targeted by Dca-miRNA were predicted using psRNATarget (http://plantgrn.noble.org/psRNATarget/) [89]. The mRNA-miRNA interaction network was visualized using Cytoscape 3.7.0 [87].
Transcript abundance analysis of sugar transporter genes in D. catenatum
To explore the expression patterens of DcST genes, a total of 49 RNA-seq of D. catenatum was downloaded from NCBI. All the Sequence Read Archive accession numbers were displayed in Table S3. The gene expression levels were calculated and normalized to the TPM (Transcripts Per Kilobase of exon model per Million mapped reads). The expression profile of DcST genes were extracted and visualized using RStudio.
WGCNA analysis
WGCNA was performed using WGCNA package 1.63 in R. The co-expression network modules were identified and the gene sets were extracted for GO enrichment analysis. The signed weighted correlation network was visualized using Cytoscape 3.7.0.
Plant materials and treatment
Two years old D. catenatum plants were sourced from Senhang Dendrobium catenatum plantation base (Zhejiang province, P.R. China), and cultivated in the greenhouse under the condition of 12/12 h photoperiod (white light, 180 μmol m−2 s−1) and temperature of 25 °C. D. catenatum were subjected to three light conditions: white light (control, CK), white-red-blue-UV light (60 white LED + 18 red LED (660 nm) + 16 blue LED (440 nm) + 2 UV LED (395 nm), WRBU), white-red-blue light (60 white LED + 18 red LED + 16 blue LED, WRB). The second to fifth adult leaves and stem nodes were sampled at 7th day after treatment, immediately frozen in liquid nitrogen and then then stored at -80 °C.
Measurement of polysaccharide content
Total polysaccharide was extracted by water extraction and alcohol precipitation method: 0.3 g dry powder, add 200 mL of H2O, heat and reflux for 2 h; transfer the extracting solution to a 250 mL volumetric flask, constant volume; take 2 mL of filtrate, add 10 mL ethanol, refrigerate for 1 h, centrifuge (20 min, 4000 r/min), wash the precipitate using 80% ethanol (8 mL) for twice; dissolve the precipitate using heat water, transfer to a 25 mL volumetric flask, constant volume. The content of total polysaccharide in leaf and stem were measured using the phenol–sulfuric acid method following the protocol provided in the Pharmacopeia of the People's Republic of China (2020). The absorbance at 488 nm was determined using Microplate Reader (FlexStation 3, Molecular Devices, USA). Three biological replicates were obtained for each treatment.
Quantitative real-time PCR
Total RNA was extracted using Fruit-mate™ for RNA Purification (TaKaRa, Dalian, China) and Ominiplant RNA Kit (CoWin Biosciences, Beijing, China). The cDNA was synthesized using the PrimeScript™ RT reagent Kit with gDNA Eraser (TaKaRa, Dalian, China). qRT-PCR was conducted on Bio-Rad CFX96 touch Real Time PCR System (Bio-Rad, Hercules, CA, USA) using the TB Green® Premix Ex Taq™ (TaKaRa, Dalian, China). The oligonucleotide primers were designed using primer-premier 6 (Table S4), actin (ACT) gene was used as an internal control [90]. Each reaction was analyzed in triplicate and the 2−△△CT method was used to analyze the data [91].
Conclusion
In this study, a total of 341 sugar transporters were identified in A. shenzhenica, D. catenatum, D. chrysotoxum, D. huoshanense, and D. nobile. The evolution history, expansion and contraction of those sugar transporter genes were analyzed. The phylogenetic analysis, structural arrangement, regulatory networks, expression profiles, and potential functions of 73 DcSTs were performed in D. catenatum. Seven DcSTs were found to play crucial roles in the biosynthesis of polysaccharides. Red-blue light is an effective way to improve the accumulation of polysaccharides in D. catenatum. This study provides insight into the evolution and functional annotation of MST, SUT, and SWEET genes in D. catenatum, and provide valuable information for further functional investigations of sugar transporter genes involved in stress responses and polysaccharides biosynthesis in Dendrobium plants.
Supplementary Information
Acknowledgements
Not applicable.
Authors’ contributions
Conceptualization, X.-Y.Z. and H.-W.H.; writing—original draft preparation, W.-W.F.; formal analysis, H.-S. G., L.Z and D.-D.M.; methodology, Y.-Y.L., L.-F.Z. and J.-T.L.; writing—review and editing, X.-Y.Z. and H.-W.H.; All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Natural Science Foundation of Henan (212300410217), Pingdingshan University PhD Startup Fund under Grant (PXY-BSQD-2014011), PhD Research Fund of Pingdingshan University (PXY-PYJJ2017006), the Key Scientific Research Project in Colleges and Universities of Henan Province of China (22B210008), Natural Science Foundation of Henan (222300420508), National Natural Science Foundation of China (32201285), Zhongke Technology Achievement Transfer and Transformation Center of Henan Province (2024128), Henan Province Science and Technology Research Projects (222102310237, 222102310479).
Data availability
All data used in this study are publicly available and included in supplementary information files. The genome of Dendrobium catenatum (GCF_001605985.2), Dendrobium nobile (GCA_022539455.1), Dendrobium chrysotoxum (GCA_019925795.1), Apostasia shenzhenica (GCA_002786265.1) was downloaded from NCBI (https://www.ncbi.nlm.nih.gov), and Dendrobium huoshanense (CNP0000830) was downloaded from CNGBdb (https://db.cngb.org/). The raw RNA-Seq data of Dendrobium catenatum were downloaded from NCBI BioProject PRJNA348403, PRJNA283237, PRJNA314400, PRJNA763165, PRJNA432825 (https://www.ncbi.nlm.nih.gov/bioproject/).
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interest
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xuyao Zhao, Email: zhaoxuyao@ihb.ac.cn.
Hongwei Hou, Email: houhw@ihb.ac.cn.
References
- 1.Ketsa S, Warrington IJ. The Dendrobium orchid: botany, horticulture, and utilization. Crop Sci. 2023;63(4):1829–88. [Google Scholar]
- 2.Ng TB, Liu JY, Wong JH, Ye XJ, Sze SCW, Tong Y, Zhang KY. Review of research on Dendrobium, a prized folk medicine. Appl Microbiol Biotechnol. 2012;93(5):1795–803. [DOI] [PubMed] [Google Scholar]
- 3.Williams LE, Lemoine R, Sauer N. Sugar transporters in higher plants - A diversity of roles and complex regulation. Trends Plant Sci. 2000;5(7):283–90. [DOI] [PubMed] [Google Scholar]
- 4.Doidy J, Grace E, Kühn C, Simon-Plas F, Casieri L, Wipf D. Sugar transporters in plants and in their interactions with fungi. Trends Plant Sci. 2012;17(7):413–22. [DOI] [PubMed] [Google Scholar]
- 5.Zhang GQ, Xu Q, Bian C, Tsai WC, Yeh CM, Liu KW, Yoshida K, Zhang LS, Chang SB, Chen F, et al. The Dendrobium catenatum Lindl. genome sequence provides insights into polysaccharide synthase, floral development and adaptive evolution. Sci Rep. 2016;6:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niu Z, Zhu F, Fan Y, Li C, Zhang B, Zhu S, et al. The chromosome-level reference genome assembly for Dendrobium officinale and its utility of functional genomics research and molecular breeding study. Acta Pharmaceutica Sinica B. 2021;11(7):2080–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan L, Wang X, Liu H, Tian Y, Lian J, Yang R, et al. The Genome of Dendrobium officinale Illuminates the Biology of the Important Traditional Chinese Orchid Herb. Mol Plant. 2015;8(6):922–34. [DOI] [PubMed] [Google Scholar]
- 8.Han BX, Jing Y, Dai J, Zheng T, Gu FL, Zhao Q, et al. A chromosome-level genome assembly of Dendrobium Huoshanense using long reads and Hi-C data. Genome Biol Evol. 2020;12(12):2486–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu Q, Niu SC, Li KL, Zheng PJ, Zhang XJ, Jia Y, Liu Y, Niu YX, Yu LH, Chen DF, et al. Chromosome-scale assembly of the Dendrobium nobile genome provides insights into the molecular mechanism of the biosynthesis of the medicinal active ingredient of Dendrobium. Front Genet. 2022;13:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang YX, Zhang GQ, Zhang DY, Liu XD, Xu XY, Sun WH, Yu X, Zhu XE, Wang ZW, Zhao X, et al. Chromosome-scale assembly of the Dendrobium chrysotoxum genome enhances the understanding of orchid evolution. Hortic Res-England. 2021;8(1):14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hao L, Shi X, Qin SW, Dong JH, Shi H, Wang YH, Zhang Y. Genome-wide identification, characterization and transcriptional profile of the SWEET gene family in Dendrobium officinale. BMC Genomics. 2023;24(1):15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang YZ, Chen Y, Wei QZ, Wan HJ, Sun CB. Phylogenetic relationships of sucrose transporters (SUTs) in plants and genome-wide characterization of SUT genes in Orchidaceae reveal roles in floral organ development. PeerJ. 2021;9:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang T, Song Z, Meng WL, Li LB. Identification, characterization, and expression of the SWEET gene family in Phalaenopsis equestris and Dendrobium officinale. Biol Plant. 2018;62(1):24–32. [Google Scholar]
- 14.Büttner M. The Arabidopsis sugar transporter (AtSTP) family: an update. Plant Biol. 2010;12:35–41. [DOI] [PubMed] [Google Scholar]
- 15.Johnson DA, Thomas MA. The monosaccharide transporter gene family in Arabidopsis and rice: a history of duplications, adaptive evolution, and functional divergence. Mol Biol Evol. 2007;24(11):2412–23. [DOI] [PubMed] [Google Scholar]
- 16.Wipf D, Pfister C, Mounier A, Leborgne-Castel N, Frommer WB, Courty PE. Identification of putative interactors of Arabidopsis sugar transporters. Trends Plant Sci. 2021;26(1):13–22. [DOI] [PubMed] [Google Scholar]
- 17.Büttner M. The monosaccharide transporter(-like) gene family in Arabidopsis. FEBS Lett. 2007;581(12):2318–24. [DOI] [PubMed] [Google Scholar]
- 18.Niño-González M, Novo-Uzal E, Richardson DN, Barros PM, Duque P. More transporters, more substrates: The Arabidopsis Major Facilitator Superfamily revisited. Mol Plant. 2019;12(9):1182–202. [DOI] [PubMed] [Google Scholar]
- 19.Aluri S, Büttner M. Identification and functional expression of the Arabidopsis thaliana vacuolar glucose transporter 1 and its role in seed germination and flowering. Proc Natl Acad Sci U S A. 2007;104(7):2537–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klepek YS, Geiger D, Stadler R, Klebl F, Landouar-Arsivaud L, Lemoine R, Hedrich R, Sauer N. Arabidopsis POLYOL TRANSPORTER5, a new member of the monosaccharide transporter-like superfamily, mediates H+-symport of numerous substrates, including myo-inositol, glycerol, and ribosele. Plant Cell. 2005;17(1):204–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schneider S, Schneidereit A, Konrad KR, Hajirezaei MR, Gramann M, Hedrich R, Sauer N. Arabidopsis INOSITOL TRANSPORTER4 mediates high-affinity H+ symport of myoinositol across the plasma membrane. Plant Physiol. 2006;141(2):565–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cridland C, Gillaspy G. Inositol pyrophosphate pathways and mechanisms: What can we learn from plants? Molecules. 2020;25(12):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schulz A, Beyhl D, Marten I, Wormit A, Neuhaus E, Poschet G, Büttner M, Schneider S, Sauer N, Hedrich R. Proton-driven sucrose symport and antiport are provided by the vacuolar transporters SUC4 and TMT1/2. Plant J. 2011;68(1):129–36. [DOI] [PubMed] [Google Scholar]
- 24.Wang HX, Weerasinghe RR, Perdue TD, Cakmakci NG, Taylor JP, Marzluff WF, Jones AM. A golgi-localized hexose transporter is involved in heterotrimeric G protein-mediated early development in Arabidopsis. Mol Biol Cell. 2006;17(10):4257–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weber A, Servaites JC, Geiger DR, Kofler H, Hille D, Gröner F, Hebbeker U, Flügge UI. Identification, purification, and molecular cloning of a putative plastidic glucose translocator. Plant Cell. 2000;12(5):787–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Endler A, Meyer S, Schelbert S, Schneider T, Weschke W, Peters SW, Keller F, Baginsky S, Martinoia E, Schmidt UG. Identification of a vacuolar sucrose transporter in barley and arabidopsis mesophyll cells by a tonoplast proteomic approach. Plant Physiol. 2006;141(1):196–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ayre BG. Membrane-transport systems for sucrose in relation to whole-plant carbon partitioning. Mol Plant. 2011;4(3):377–94. [DOI] [PubMed] [Google Scholar]
- 28.Rottmann TM, Fritz C, Lauter A, Schneider S, Fischer C, Danzberger N, Dietrich P, Sauer N, Stadler R. Protoplast-esculin assay as a new method to assay plant sucrose transporters: Characterization of AtSUC6 and AtSUC7 sucrose uptake activity in Arabidopsis Col-0 ecotype. Front Plant Sci. 2018;9:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ludwig A, Stolz J, Sauer N. Plant sucrose-H+ symporters mediate the transport of vitamin H. Plant J. 2000;24(4):503–9. [DOI] [PubMed] [Google Scholar]
- 30.Braun DM. Phloem loading and unloading of sucrose: What a long, strange trip from source to sink. Annu Rev Plant Biol. 2022;73:553–84. [DOI] [PubMed] [Google Scholar]
- 31.Aoki N, Hirose T, Scofield GN, Whitfeld PR, Furbank RT. The sucrose transporter gene family in rice. Plant Cell Physiol. 2003;44(3):223–32. [DOI] [PubMed] [Google Scholar]
- 32.Peng D, Gu X, Xue LJ, Leebens-Mack JH, Tsai CJ. Bayesian phylogeny of sucrose transporters: ancient origins, differential expansion and convergent evolution in monocots and dicots. Front Plant Sci. 2014;5:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sauer N. Molecular physiology of higher plant sucrose transporters. FEBS Lett. 2007;581(12):2309–17. [DOI] [PubMed] [Google Scholar]
- 34.Kuhn C, Grof CPL. Sucrose transporters of higher plants. Curr Opin Plant Biol. 2010;13(3):287–98. [DOI] [PubMed] [Google Scholar]
- 35.Braun DM, Wang L, Ruan Y-L. Understanding and manipulating sucrose phloem loading, unloading, metabolism, and signalling to enhance crop yield and food security. J Exp Bot. 2014;65(7):1713–35. [DOI] [PubMed] [Google Scholar]
- 36.Chen L-Q, Hou B-H, Lalonde S, Takanaga H, Hartung ML, Qu X-Q, Guo W-J, Kim J-G, Underwood W, Chaudhuri B, et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature. 2010;468(7323):527–U199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen LQ, Cheung LS, Feng L, Tanner W, Frommer WB. Transport of Sugars. In: Annual Review of Biochemistry, Vol 84. Edited by Kornberg RD, vol. 84. Palo Alto: Annual Reviews; 2015: 865–894. [DOI] [PubMed]
- 38.Yuan M, Wang SP. Rice MtN3/Saliva/SWEET family genes and their homologs in cellular organisms. Mol Plant. 2013;6(3):665–74. [DOI] [PubMed] [Google Scholar]
- 39.Feng L, Frommer WB. Structure and function of SemiSWEET and SWEET sugar transporters. Trends Biochem Sci. 2015;40(8):480–6. [DOI] [PubMed] [Google Scholar]
- 40.Patil G, Valliyodan B, Deshmukh R, Prince S, Nicander B, Zhao MZ, Sonah H, Song L, Lin L, Chaudhary J, et al. Soybean (Glycine max) SWEET gene family: insights through comparative genomics, transcriptome profiling and whole genome re-sequence analysis. BMC Genomics. 2015;16:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Breia R, Conde A, Badim H, Fortes AM, Gerós H, Granell A. Plant SWEETs: from sugar transport to plant-pathogen interaction and more unexpected physiological roles. Plant Physiol. 2021;186(2):836–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang GQ, Liu KW, Li Z, Lohaus R, Hsiao YY, Niu SC, Wang JY, Lin YC, Xu Q, Chen LJ, et al. The Apostasia genome and the evolution of orchids. Nature. 2017;549(7672):379-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Afoufa-Bastien D, Medici A, Jeauffre J, Coutos-Thévenot P, Lemoine R, Atanassova R, Laloi M. The Vitis vinifera sugar transporter gene family: phylogenetic overview and macroarray expression profiling. BMC Plant Biol. 2010;10:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qiao X, Li QH, Yin H, Qi KJ, Li LT, Wang RZ, Zhang SL, Paterson AH. Gene duplication and evolution in recurring polyploidization-diploidization cycles in plants. Genome Biol. 2019;20:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang SL, Zhou JJ, Gao L, Tang YL. Plant miR397 and its functions. Funct Plant Biol. 2021;48(4):361–70. [DOI] [PubMed] [Google Scholar]
- 46.Lastdrager J, Hanson J, Smeekens S. Sugar signals and the control of plant growth and development. J Exp Bot. 2014;65(3):799–807. [DOI] [PubMed] [Google Scholar]
- 47.Van Bel AJE. The phloem, a miracle of ingenuity. Plant, Cell Environ. 2003;26(1):125–49. [Google Scholar]
- 48.Sherson SM, Alford HL, Forbes SM, Wallace G, Smith SM. Roles of cell-wall invertases and monosaccharide transporters in the growth and development of Arabidopsis. J Exp Bot. 2003;54(382):525–31. [DOI] [PubMed] [Google Scholar]
- 49.Saddhe AA, Manuka R, Penna S. Plant sugars: Homeostasis and transport under abiotic stress in plants. Physiol Plant. 2021;171(4):739–55. [DOI] [PubMed] [Google Scholar]
- 50.Chen L-Q, Qu X-Q, Hou B-H, Sosso D, Osorio S, Fernie AR, Frommer WB. Sucrose efflux mediated by sweet proteins as a key step for phloem transport. Science. 2012;335(6065):207–11. [DOI] [PubMed] [Google Scholar]
- 51.Kühn C. A comparison of the sucrose transporter systems of different plant species. Plant Biol. 2003;5(3):215–32. [Google Scholar]
- 52.Hu Z, Tang ZJ, Zhang YM, Niu LP, Yang F, Zhang DC, Hu YB. Rice SUT and SWEET transporters. Int J Mol Sci. 2021;22(20):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiao QL, Li Z, Wang YY, Hou XB, Wei XM, Zhao X, Huang L, Guo YJ, Liu ZZ. Genome-wide identification, expression and functional analysis of sugar transporters in sorghum (Sorghum bicolor L.). J Integr Agr. 2022;21(10):2848–64. [Google Scholar]
- 54.Fakher B, Jakada BH, Greaves JG, Wang LL, Niu XP, Cheng Y, Zheng P, Aslam M, Qin Y, Wang XM. Identification and expression analysis of pineapple sugar transporters reveal their role in the development and environmental response. Front Plant Sci. 2022;13:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang ZY, Gao C, Xu YC, Liu J, Kang J, Ren ZM, Cui Q, Li DZ, Ma S, Xia YP, et al. Identification and expression analysis of putative sugar transporter gene family during bulb formation in Lilies. Int J Mol Sci. 2024;25(6):28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Slawinski L, Israel A, Paillot C, Thibault F, Cordaux R, Atanassova R, Dédaldéchamp F, Laloi M. Early Response to Dehydration Six-like transporter family: Early origin in streptophytes and evolution in land plants. Front Plant Sci. 2021;12:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pireyre M, Burow M. Regulation of MYB and bHLH transcription factors: a glance at the protein level. Mol Plant. 2015;8(3):378–88. [DOI] [PubMed] [Google Scholar]
- 58.Tong YQ, Xue JP, Li QZ, Zhang L. A generalist regulator: MYB transcription factors regulate the biosynthesis of active compounds in medicinal plants. J Exp Bot. 2024;75(16):4729–44. [DOI] [PubMed] [Google Scholar]
- 59.Li SF, Milliken ON, Pham H, Seyit R, Napoli R, Preston J, Koltunow AM, Parisha RW. The Arabidopsis MYB5 transcription factor regulates mucilage synthesis, seed coat development, and trichome morphogenesis. Plant Cell. 2009;21(1):72–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim WC, Reca IB, Kim Y, Park S, Thomashow M, Keegstra K, Han KH. Transcription factors that directly regulate the expression of CSLA9 encoding mannan synthase in Arabidopsis thaliana. Plant Mol Biol. 2014;84(4–5):577–87. [DOI] [PubMed] [Google Scholar]
- 61.He CM. Teixeira da Silva JA, Wang HB, Si C, Zhang MZ, Zhang XM, Li MZ, Tan JW, Duan J: Mining MYB transcription factors from the genomes of orchids (Phalaenopsis and Dendrobium) and characterization of an orchid R2R3-MYB gene involved in water-soluble polysaccharide biosynthesis. Sci Rep. 2019;9:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Iftikhar J, Lyu ML, Liu ZY, Mehmood N, Munir N, Ahmed MAA, Batool W, Aslam MM, Yuan Y, Wu BH. Sugar and hormone dynamics and the expression profiles of SUT/SUC and SWEET sugar transporters during flower development in Petunia axillaris. Plants-Basel. 2020;9(12):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dantas LLD, Eldridge BM, Dorling J, Dekeya R, Lynch DA, Dodd AN. Circadian regulation of metabolism across photosynthetic organisms. Plant J. 2023;116(3):650–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Covington MF, Maloof JN, Straume M, Kay SA, Harmer SL. Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 2008;9(8):R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Durand M, Mainson D, Porcheron B, Maurousset L, Lemoine R, Pourtau N. Carbon source-sink relationship in Arabidopsis thaliana: the role of sucrose transporters. Planta. 2018;247(3):587–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.He C, Wu K, Zhang J, Liu X, Zeng S, Yu Z, et al. Cytochemical localization of polysaccharides in Dendrobium officinale and the Involvement of DoCSLA6 in the synthesis of mannan polysaccharides. Front Plant Sci. 2017;8:173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu L, Shi D, Li J, Kong Y, Yu Y, Chai G, Hu R, Wang J, Hahn MG, Zhou G. CELLULOSE SYNTHASE-LIKE A2, a glucomannan synthase, is involved in maintaining adherent mucilage structure in Arabidopsis Seed. Plant Physiol. 2014;164(4):1842–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang YZ, Zhao KK, Chen Y, Wei QZ, Chen XY, Wan HJ, Sun CB. Species-specific gene expansion of the cellulose synthase gene superfamily in the orchidaceae family and functional divergence of mannan synthesis-related genes in Dendrobium officinale. Front Plant Sci. 2022;13:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gao Y-Q, Chen X-L, Chen D-H, Liu J-J, Si J-P. Genome-wide identification and expression analysis of CSLA gene family of Dendrobium catenatum. China J Chin Mater. 2020;45(13):3120–7. [DOI] [PubMed] [Google Scholar]
- 70.You T, Barnett SM. Effect of light quality on production of extracellular polysaccharides and growth rate of Porphyridium cruentum. Biochem Eng J. 2004;19(3):251–8. [Google Scholar]
- 71.Lin X, Lai Z. Effects of light quality on expression of PEPC and polysaccharide accumulation in Dendrobium officinale. Chin J Trop Crops. 2017;38(5):838–42. [Google Scholar]
- 72.Han P-p. Sun Y, Jia S-r, Zhong C, Tan Z-l: Effects of light wavelengths on extracellular and capsular polysaccharide production by Nostoc flagelliforme. Carbohydr Polym. 2014;105:145–51. [DOI] [PubMed] [Google Scholar]
- 73.Lin X, Lai Z. Effect of light quality on the proliferation of protocorm and active ingredient contents of Dendrobium officinale. Chin J Trop Crops. 2015;36(10):1796–801. [Google Scholar]
- 74.Gao T, Si J, Zhu Y, Huang H. Effects of light quality and germplasm on growth and effective ingredients of Dendrobium officinale germchit. China J Chin Mater. 2012;37(2):198–201. [PubMed] [Google Scholar]
- 75.Wheeler TJ, Eddy SR. nhmmer: DNA homology search with profile HMMs. Bioinformatics. 2013;29(19):2487–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31(13):3784–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Horton P, Park K-J, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35:W585–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chou KC, Shen HB. Cell-PLoc: a package of Web servers for predicting subcellular localization of proteins in various organisms. Nat Protoc. 2008;3(2):153–62. [DOI] [PubMed] [Google Scholar]
- 79.Savojardo C, Martelli PL, Fariselli P, Profiti G, Casadio R. BUSCA: an integrative web server to predict subcellular localization of proteins. Nucleic Acids Res. 2018;46(W1):W459–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hu B, Jin J, Guo A-Y, Zhang H, Luo J, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015;31(8):1296–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Katoh K, Rozewicki J, Yamada KD. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 2017;20(4):1160–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Emms DM, Kelly S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019;20(1):238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xu B, Yang ZH. pamlX: A Graphical User Interface for PAML. Mol Biol Evol. 2013;30(12):2723–4. [DOI] [PubMed] [Google Scholar]
- 85.Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouze P, Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30(1):325–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tian F, Yang DC, Meng YQ, Jin JP, Gao G. PlantRegMap: charting functional regulatory maps in plants. Nucleic Acids Res. 2020;48(D1):D1104–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Meng YJ, Yu DL, Xue J, Lu JJ, Feng SG, Shen CJ, Wang HZ. A transcriptome-wide, organ-specific regulatory map of Dendrobium officinale, an important traditional Chinese orchid herb. Sci Rep. 2016;6:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dai XB, Zhuang ZH, Zhao PXC. psRNATarget: a plant small RNA target analysis server (2017 release). Nucleic Acids Res. 2018;46(W1):W49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang TT, Cui Z, Li YX, Kang YQ, Song XQ, Wang J, Zhou Y. Genome-Wide Identification and Expression Analysis of MYB Transcription Factor Superfamily in Dendrobium catenatum. Front Genet. 2021;12:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used in this study are publicly available and included in supplementary information files. The genome of Dendrobium catenatum (GCF_001605985.2), Dendrobium nobile (GCA_022539455.1), Dendrobium chrysotoxum (GCA_019925795.1), Apostasia shenzhenica (GCA_002786265.1) was downloaded from NCBI (https://www.ncbi.nlm.nih.gov), and Dendrobium huoshanense (CNP0000830) was downloaded from CNGBdb (https://db.cngb.org/). The raw RNA-Seq data of Dendrobium catenatum were downloaded from NCBI BioProject PRJNA348403, PRJNA283237, PRJNA314400, PRJNA763165, PRJNA432825 (https://www.ncbi.nlm.nih.gov/bioproject/).