Abstract
Background
The application of beneficial microbes in agriculture is gaining increasing attention as a means to reduce reliance on chemical fertilizers. This approach can potentially mitigate negative impacts on soil, animal, and human health, as well as decrease climate-changing factors. Among these microbes, yeast has been the least explored, particularly within the phyllosphere compartment. This study addresses this knowledge gap by investigating the potential of phyllosphere yeast to improve rice yield while reducing fertilizer dosage.
Results
From fifty-two rice yeast phyllosphere isolates, we identified three yeast strains—Rhodotorula paludigena Y1, Pseudozyma sp. Y71, and Cryptococcus sp. Y72—that could thrive at 36 °C and possessed significant multifarious plant growth-promoting traits, enhancing rice root and shoot length upon seed inoculation. These three strains demonstrated favorable compatibility, leading to the creation of a yeast consortium. We assessed the combined effect of foliar application of this yeast consortium and individual strains with two distinct recommended doses of chemical fertilizers (RDCFs) (75 and 100%), as well as RDCFs alone (75 and 100%), in rice maintained in pot-culture and field experiments. The pot-culture experiment investigated the leaf microbial community, plant biochemicals, root and shoot length during the stem elongation, flowering, and dough phases, and yield-related parameters at harvest. The field experiment determined the actual yield. Integrated results from both experiments revealed that the yeast consortium with 75% RDCFs was more effective than the yeast consortium with 100% RDCFs, single strain applications with RDCFs (75 and 100%), and RDCFs alone (75 and 100%). Additionally, this treatment improved leaf metabolite levels compared to control rice plants.
Conclusions
Overall, a 25% reduction in soil chemical fertilizers combined with yeast consortium foliar application improved rice growth, biochemicals, and yield. This study also advances the field of phyllosphere yeast research in agriculture.
Supplementary Information
The online version contains supplementary material available at 10.1186/s40793-024-00635-9.
Keywords: Yeast, Phyllosphere, Plant growth-promoting microbes, Rice, Crop improvement
Introduction
India, primarily an agricultural nation, ranks as the world's second-largest rice producer. The International Production Assessment Division's (IPAD) report from the United States Department of Agriculture (USDA) shows that in the 2022–23 season, rice production in India increased by 8.3 million tonnes from the previous year, totalling 135.7 million tonnes [1]. Annually, 30% of food is lost or wasted. Managing this loss and waste could potentially enhance food security and nutrition, although it's not a guaranteed outcome [2]. According to the report, a certain level of food loss and waste is needed as a buffer to ensure steady availability and access to food. Nonetheless, the output must be increased as agricultural land area diminishes and population size increases. Climate change, on the other hand, is known to have a negative effect on agricultural production and is expected to reduce the production of three cereal grains (rice, maize, and wheat) globally by 10 to 25%, with a 1 °C increase in mean surface temperature [3]. The use of agrochemicals has increased to improve crop yield and compensate for food loss. Annually, millions of tonnes of synthetic nutrients added to soils are not fully absorbed by plants. Excessive runoff from crop fields degrades soil fertility, harms human health, and contributes to climate change by emitting greenhouse gases, such as N2O from nitrogen fertilizers [4, 5]. Considering all these factors, there has been a global shift to a sustainable farming approach with the primary goal of improving yield by reducing the use of agrochemicals in the field. There are many suggested approaches; among these, biofertilizers are gaining popularity, especially plant growth-promoting microorganisms (PGPM).
The microbes that exert beneficial effects on plants are termed PGPM. These microbes have the potential to alter plant performance directly through the production of specific compounds that enhance plant growth and increase the availability and uptake of nutrients in the soil (i.e., mineral solubilization, phytohormone production, nitrogen fixation, siderophore production, and 1-aminocyclopropane-1-carboxylate (ACC) deaminase production) [6]. The indirect mechanisms of plant growth promotion involves HCN production, induced systemic resistance, and suppression of pests and pathogens through metabolite production [6]. PGPM are primarily isolated from soils, rhizospheres (soil influenced by plant roots) or phyllospheres (aerial surface of the plants), and endospheres (inner tissues of plants), and it was discovered that bacteria predominate in soil and each plant compartment [7]. The soil microbiome is the primary source of plant microbiomes, which are strongly and sequentially filtered by the rhizosphere, rhizoplane (root surface), endosphere, and phyllosphere. As a result, phyllosphere microbial diversity is lower than that of the rhizosphere, despite receiving microbial sources from air and insects [8, 9]. So, bacteria in the rhizosphere were the most extensively studied PGPM, known as plant growth-promoting rhizobacteria (PGPR). The second most widely studied microbes are from the phyllosphere compartment. The phyllosphere habitat includes all the above-ground plant parts (i.e., leaves, stems, flowers, and fruits). It is an unstable habitat where microbial life is shaped by multiple factors, like temperature, UV radiation, nutrient and water availability, and the chemical composition of the leaf surface. Bacteria dominate the leaf area, with an average of 106–107 bacteria per square cm of the leaf surface. However, the overall size of the phyllosphere's fungal population has not yet been estimated, but it is predicted to be smaller [10].
When it comes to the application of plant growth-promoting fungi (PGPF), the fungal origins were mostly from the soil and rhizosphere i.e., Trichoderma spp., Gliocladium virens, Penicillium digitatum, Aspergillus flavus, Actinomucor elegans, Podospora bulbillosa, and Arbuscular mycorrhizal fungi [11]. On the other hand, yeast is one of the least studied fungi, but it is gaining favor in crop improvement because it is generally recognized as safe (GRAS) for field application [12]. Since yeasts are unicellular, proliferate asexually and rapidly on simple carbohydrates, and typically utilize both fermentative and respiratory pathways, they are simpler to cultivate as a biofertilizer inoculant [13]. A diverse range of yeasts isolated from soil, rhizosphere, and, to a lesser extent, other plant parts demonstrate plant growth-promoting (PGP) characteristics, including the production of exopolysaccharide (EPS), Indole-3-acetic acid (IAA), gibberellic acid (GA), cytokinin, ACCD, siderophore [14–19], mineral solubilization [20–22], and biocontrol agents [23–25]. Given the beneficial effects of yeasts, it is critical to comprehend the role of yeast in compartments that have received little attention, such as the phyllosphere and endosphere. The significance of phyllosphere bacteria in improving rice yield and alleviating drought stress has previously been studied [26, 27], but no appropriate reports on the role of phyllosphere yeast in rice have been published. Since PGPMs alone do not achieve the same yield levels as chemical fertilizers, it is essential to explore their combined application for optimal results [28]. Nonetheless, more research is required to determine the role of yeast in crop plant improvement.
From the preceding discussion, it is clear that rice production in India must be enhanced using sustainable methods like biofertilizers. Nonetheless, to the best of the authors' knowledge, no previous publication has described the foliar application of rice phyllosphere yeast for improving rice yield. With the knowledge gathered above, the current study is about the isolation and characterization of yeast from the rice leaf surface; the characterization of PGP properties in yeast; and understanding the individual and consortium effects of best yeast strains as well as different fertilizer dose applications on rice growth, leaf biochemicals, microbial population, and yield. Finally, the microbial and plant data will be combined using suitable integration techniques to identify the most effective beneficial yeast strains for rice [29]. Thus, this study contributes to the knowledge of the role and significance of phyllosphere yeast in crop improvement, making it a valuable asset for sustainable approaches.
Materials and methods
Collection of rice leaf samples
During the months of January, February, and March of 2019, rice leaf samples from various stages (stem elongation, flowering, and dough stages) were collected from high yielding, short-duration rice cultivar ASD 16 grown in the Killikulam region (8°40′26.616''N, 77°51′55.3392''E), Thoothukudi district, Tamil Nadu, India. The samples were collected aseptically and transported to the laboratory in an icebox for microbial analysis. During the sample collection months, the lowest and highest atmospheric temperatures were 25 °C and 36 °C. The average atmospheric temperature in the three months was 30.5 °C.
Isolation and selection of yeast
The leaf samples were washed three times with sterile water and dried with sterile tissue paper for 30 s to isolate putative phyllosphere yeast colonized on the leaf surface. The leaf imprinting technique was used to isolate phylloplane yeast [30]. Adaxial and abaxial sides of leaves were imprinted on various yeast media such as yeast extract peptone dextrose (YEPD) agar media, supplemented with chloramphenicol (50 μl ml−1) and tetracycline (50 μl ml−1) at pH 7. The plates were incubated for 48 h at 30 ± 1 °C. Morphologically distinct yeast colonies were purified and maintained on a YEPD slant at 4 °C, and 60% glycerol stock was preserved at -80 °C for future use. Yeast isolates were screened for growth in different temperature levels. The cell density of the purified isolates was set to OD600nm 0.1, and 1 mL was added to 9 mL of YEPD broth test tubes at pH 7. Due to the variable temperature conditions observed in the phyllosphere, the test tubes were incubated at field-simulated temperatures of 25, 30, and 36 °C for five days. Yeast strains cultivated at these three temperature levels were subsequently selected for taxonomic identification.
Molecular characterization of yeast isolates and phylogenetic analysis
Genomic DNA from all the selected yeast isolates was extracted as per the described procedure [31]. The isolates were characterized by using ITS rRNA gene sequencing. Polymerase chain reaction (PCR) amplifications were performed using ITS1 as forward primer (5'-CTTGGTCATTTAGAGGAAGTAA -3') and ITS4 as reverse primer (5'-TCCTCCGCTTATTGATATGC -3'). The PCR conditions were as follows: initial denaturing at 94 °C for 5 min followed by 30 cycles of denaturation at 94 °C for 30 s. Annealing at 59 °C for 30 s and extension at 70 °C for 2 min, then final extension at 72 °C for 7 min. The PCR products were resolved by electrophoresis in 1% agarose gel and purified using FavorPrep GEL/ PCR purification kit. The products were sequenced using Sanger dideoxy sequencing. Based on the obtained sequence, species were identified in the NCBI database, and the sequences of closely related species were also retrieved. The phylogenetic tree was generated using MEGA 7.0 software with a maximum likelihood algorithm, and the bootstrap analysis was based on 100 resamplings [32]. The sequence files of the yeast strains were submitted to the NCBI database via BANKIT, and accession numbers were assigned to each strain.
Assessment of direct PGP activity in yeast strain
IAA production
Indolic compound production was quantitatively measured [33]. Yeast strains were inoculated in 10 mL of YEPD broth supplemented with 1% tryptophan, incubated for 48 h at 30 ± 1 °C on a rotary shaker, while uninoculated broth served as control. Cultures were centrifuged at 10,000 rpm for 20 min, and supernatants were collected. After that two drops of concentrated orthophosphoric acid and 2 mL of Salkowski's reagent (2% 0.5 M FeCl3 in 35% perchloric acid solution) were mixed with 1 mL of supernatant and incubated at room temperature for 25 min in the dark. The development of a reddish-pink color indicated IAA production. The absorbance was measured at 530 nm using a spectrophotometer (LAMBDA 365 UV–Vis spectrophotometer, PerkinElmer, Mumbai, India), and the quantitative estimation of IAA was performed by using a standard graph. The result is expressed as µg mL−1.
GA production
GA production by yeast strains was quantified by the spectrophotometric method [34]. The yeast strains were inoculated into 10 mL of YEPD broth and incubated at room temperature for seven days, and the uninoculated broth was used as a control. Cultures were centrifuged at 8,000 rpm for 10 min, and 2 mL of supernatant was transferred into a 15 mL reaction tube. Then, 2 mL of zinc acetate solution (21.9 g zinc acetate with 1 ml of glacial acetic acid and the volume was made up to 100 ml with distilled water) and 2 mL of 10.6% potassium ferrocyanide solution were added to the tubes, followed by centrifugation at 8,000 rpm for 10 min. A total of 5 mL of supernatant was added to 5 mL of 30% hydrochloric acid and incubated at 27 °C for 75 min. The absorbance was measured at 254 nm. The GA concentration in the samples was determined with reference to the standard GA curve. The result is expressed as µg mL−1.
Qualitative assay of phosphate and potassium solubilization
Solubilization of inorganic minerals such as phosphate and potassium by yeast strains was carried out using Pikovskaya medium and Aleksandrov medium [35]. In short, spot inoculation of each yeast strain was done on these media and incubated at 28 ± 2 °C for three days. The strains that exhibited a clear halo zone around the colony on the media were identified as P and K solubilizers. Based on the area of solubilization, the solubilization index (SI) was calculated using the following formula.
Assessment of indirect PGP traits in yeast strain
Siderophore production
Siderophore production was evaluated for all yeast strains using Chrome Azurol S (CAS) assay [36]. Briefly, the CAS medium was prepared by adding CAS solution to the melted King's B agar medium in a 1:15 ratio. Actively grown yeast strains were spot-inoculated onto the CAS medium, and the plates were incubated in the dark at 30 ± 1 °C for ten days. An uninoculated CAS blue agar plate served as a control. Colonies with a yellow-orange halo after incubation were considered positive for siderophore production. The yeast strains were grown in King's B broth and incubated for 48 h to quantify the siderophore. After incubation, the broth was centrifuged at 10,000 rpm for 10 min. The supernatant was mixed with CAS solution in a 1:1 ratio and incubated for 20 min, with sterile king's B broth containing CAS solution as a control reference. The absorbance of the solution was then measured using a spectrophotometer at 630 nm. The siderophore production was calculated using the following formula [37]:
whereas Ar- Absorbance of reference and As- Absorbance of the sample.
ACC deaminase activity
ACC deaminase activity of yeast strains was screened based on their ability to use ACC as a sole nitrogen source. Yeast strains were grown in Dworkin and Foster (DF) salts broth with ACC as a nitrogen source for 24 h. The inoculated broth was centrifuged at 10,000 rpm for 5 min at 4 °C, and the solution was washed using 1 mL of 0.1 M Tris HCl pH 7.6 and repeated several times, and the pH was maintained at 7.6 and stored the pellet at 20 °C for 30 min. The cells were suspended with 1 mL of 0.1 M Tris–HCl at pH 7.6 and centrifuged. The pellet was resuspended in 600 µL of 0.1 M Tris–HCl at pH 8.5. Toluene was added to the 30 µl and vortexed for 30 s. To this, 20 µL of 0.5 M ACC was added in one set, and blank was maintained without ACC addition. Both the tubes were vortexed and incubated at 30 °C for 15 min, and 1 mL of 0.56 M HCl was added, vortexed, and centrifuged for 5 min at 10,000 rpm at room temperature. Then 300 µL of 2,4-Dinitrophenylhydrazine (DNPH) was added, vortexed, and incubated for 30 min at 30 °C. Finally, 2 mL of 2 N NaOH was added, mixed well and absorbance was read at 540 nm [38]. The enzyme activity is expressed as nmol α-ketobutyrate mg−1protein h−1.
Biocontrol activity of phyllosphere yeast strains against rice fungal pathogens
The yeast strains were screened for direct antagonism against Helminthosporium oryzae, Pyricularia oryzae, and Saracladium oryzae (Standard cultures, Department of Soil Science and Agricultural Chemistry, AC & RI, Killikulam, India). In brief, a mycelial disk (5 mm diameter) of each rice pathogenic fungus was placed on the edge of potato dextrose agar (PDA) media (30 mm), and each yeast was streak inoculated close to the center of the plates. The plates were incubated for 5 to 6 days at 25 °C. Inhibition of mycelial growth was measured 5 to 6 days after the inoculation, and the percentages of inhibition by each yeast strain were calculated as follows [39]
Effect of yeast strains on the growth of rice seedlings
The effect of yeast strains on rice seedling growth was assessed in vitro using the standard roll paper towel method [40, 41]. Surface disinfection of rice seeds was performed for 1 min with 70% ethanol, followed by 20 min with a 1.65% sodium hypochlorite (NaOCl) solution. The seeds were rinsed five times with distilled water for one minute each. Yeast cells from the overnight-grown culture were pelleted after centrifugation and resuspended in the same volume of sterile water, with the final concentration adjusted to ~ 108 colony-forming units (CFU) mL−1. The seeds were soaked in yeast cell suspension for 2 h before being blotted at room temperature with sterile filter paper. As wet blotters, we used sterile filter papers with a diameter of 120 mm. Five seeds were placed in a straight line at equal distances on each filter paper. Each filter paper was rolled, placed vertically in a 350 mL plastic cup with water, and incubated at 28 °C for 8 days in the growth chamber. Adequate water was regularly poured into the cups 1 cm below the seed arrangement. As a control, seeds soaked in distilled water were used. The plant phenotypic traits, such as root and shoot lengths, were measured after 10 days.
Pot-culture experiment
Selected yeast strains were tested for compatibility in terms of growth by cross-streak assay in a YEPD medium [42]. A triangular streak of three yeast colonies was made on a media plate and incubated for three days. Growth without inhibition confirmed compatibility; otherwise, the strains were considered non-compatible.
Yeast inoculums were prepared by cultivating the yeast strains in YEPD broth for 24 h. After centrifuging the yeast cells for 10 min, the pellets from the three best strains were resuspended in sterile water, resulting in a final consortium concentration of approximately 108 CFU mL−1, with 0.2% Tween 80 as a sticking agent. The same protocol was followed for individual strain suspensions, each with a final concentration of 108 CFU mL−1.
A pot culture experiment using a completely randomized block design (CRBD) was carried out in rice variety ASD 16 at the unit of Agricultural Microbiology, Department of Soil Science and Agricultural Chemistry, Agricultural College and Research Institute, Killikulam, India.
Soils were sterilized in an autoclave at 121 °C for 15 min at 20 psi over three consecutive days. A 5 kg potting mixture (soil + farmyard manure in a 4:1 ratio) was placed in each mud pot. Rice seeds (ASD 16) were sown in trays, and seedlings were transplanted to the pots after 20 days, with each pot containing ten plants. Fertilizers such as urea, single superphosphate, and potassium chloride were applied at 100% and 75% recommended dose of chemical fertilizers (RDCFs) (nitrogen: phosphorus: potassium, 120:40:40 kg ha−1).
Treatment details
T1—Yeast 1 + 100% RDCFs.
T2—Yeast 2 + 100% RDCFs.
T3—Yeast 3 + 100% RDCFs.
T4—Yeast consortium (Yeast 1 + Yeast 2 + Yeast 3) + 100% RDCFs.
T5—100% RDCFs.
T6—Yeast 1 + 75% RDCFs.
T7—Yeast 2 + 75% RDCFs.
T8—Yeast 3 + 75% RDCFs.
T9—Yeast consortium (Yeast 1 + Yeast 2 + Yeast 3) + 75% RDCFs.
T10—75% RDCFs.
TC—Control.
The prepared yeast consortium and individual suspensions were foliar sprayed (five sprays per pot) on rice at the beginning of the stem elongation (55 DAS), flowering (85 DAS), and dough (100 DAS) stages. Three days after each foliar application, total chlorophyll, protein, proline, and antioxidant enzyme activity in the leaves were measured, along with the lengths of the shoots and roots. Additionally, leaf samples from each stage were stored at −4 °C for future use. Yield characteristics were recorded at the harvesting stage (115 DAS).
Estimation of the microbial population on the rice leaf surface
The microbial population on the rice leaf surface (bacteria, yeast, and actinomycetes) was assessed using the Most Probable Number (MPN) method in which the rice leaves were dipped into the sterile water for a minute, followed by serial dilution. Plating techniques involving microbial growth media viz. Luria–Bertani (LB), YEPD, and glycerol agar for bacteria, yeast, and actinomycetes, respectively. CFUs of bacteria, yeast, and actinomycetes were detected after incubating for 5–7 days. Microbial count and changes in microbe populations were studied at three different crop stages (stem elongation, flowering, and dough stage).
Estimation of total chlorophyll
To estimate the total chlorophyll content, 0.5 g of the leaf samples collected during all three stages were thoroughly ground in a pestle and mortar with 10 ml of 80% acetone. The sample mixture was then centrifuged at 10,000 rpm for 10 min in an ice-cold environment. After carefully transferring the supernatant into new tubes, 2 ml of the sample was aliquoted in a cuvette and tested for chlorophyll a, chlorophyll b, and carotenoid content by measuring multiple absorbances at 663, 646, and 470 nm, [43] and calculated using the formula as follows:
Estimation of total protein and proline content
The conventional method [44] was used to assess the total protein content (µg g−1 FW) at 650 nm against various bovine serum albumIn (BSA) concentrations. Proline content in rice leaves was measured at 520 nm using the acid ninhydrin and glacial acetic acid as described by Bates et al., (1973) [45]. The concentration of proline in the leaf samples was determined using the standard curve for free proline and is expressed as µg g−1 FW.
Assay of antioxidant enzymes
The sample extraction for enzyme assay was done according to the protocol described by Alici and Arabaci (2016) [46]. 1 g of fresh rice leaves were taken, cleaned and homogenized in an ice-cold mortar and pestle with 50 ml of a 100 mM sodium phosphate buffer (pH 7.0) for five minutes at 4 °C. Filter paper was used to filter the homogenate, and the filtrate was centrifuged at 5,000 rpm for 15 min. The supernatant was collected, and it was used for enzyme assay of catalase, peroxidase, and polyphenol oxidase.
The catalase activity (CAT) activity was determined by monitoring the disappearance of H2O2 at 240 nm using the method defined by Aebi [47]. The 5 mL reaction mixture consisted of 50 mM phosphate buffer (pH 7.0), 33 mM H2O2, and enzyme extract. The absorbance was recorded for 1 min once the reaction began. An extinction coefficient of 36 M cm−1 was used to calculate CAT activity.
The peroxidase (POD) activity was estimated [48] using guaiacol as a substrate. The 3 ml reaction mixture consisted of 100 mM potassium phosphate buffer with pH 7.0 (1.9 ml), 5 mM guaiacol (0.5 ml), 5 mM H2O2 (0.5 ml), and 100 μl of sample extract. The reaction mixture for blank was prepared in a similar way excluding the sample extract. The sample was added to the reaction mixture, and the absorbance was measured at 470 nm for 1 min. An extinction coefficient of 26.6 mM−1 cm−1 was used to calculate peroxidase activity.
The polyphenol oxidase (PPO) activity was assayed as per the method adopted [49]. The 3 ml reaction mixture contained 100 mM potassium phosphate buffer having pH 7.0 (2 ml), 5 mM catechol (0.5 ml), and 500 μl of sample extract (0.5 ml) in a test tube. And a blank without a sample extract. The sample was added to the reaction mixture, and the absorbance was measured at 420 nm for 1 min. The polyphenol oxidase activity was determined by measuring the increase in absorbance resulting from the oxidation of catechol (ɛ = 34.5 mM−1 cm−1) at 420 nm spectrophotometrically. All three enzyme activities are expressed as variations in mmol per time unit per mg total protein (mmol min−1 mg protein−1).
Measurement of plant phenotypic and yield parameters
Plant phenotypic traits such as shoot and root length were measured during stem elongation, flowering, and dough stages. Yield parameters such as the number of productive tillers, grains per panicle, and 1000-grain weight were recorded during the harvest stage.
Gas chromatography-mass spectrometry (GC–MS) analysis of leaf extracts
Leaf samples from the control and the most effective treatment during the stem elongation phase were chosen for metabolite comparison. Leaf extracts were prepared by grinding them in 10 mL of cold water. 1N HCl was used to modify the pH to 2.0, and an equal volume of chloroform was added. These mixtures were then shaken for 24 h. Following incubation, the chloroform layer was removed, and the metabolites were concentrated using a vacuum flask evaporator before being dissolved in methanol. GC–MS analysis of methanol extracts was performed on a GCMS-TQ8040 triple quadrupole fitted with a CrossbondTM silarylene phase column (1,4-bis (dimethylsiloxy) phenylene dimethyl polysiloxane) (30 m 250 m). The volume used for analysis was 10 µL, and the carrier gas used for GC–MS was helium at the flow rate of 1 mL min−1. The injector was operated under the maximum temperature of 470 °C, and the oven temperature was 350 °C constant till the entire analysis. The extracts from control and treatment samples were prepared in triplicate, with pure methanol serving as the control. The compounds were identified by comparing the obtained spectral configurations to those in the accessible mass spectral library (NIST and WILEY libraries).
Field experiment
A field experiment was carried out at the Agricultural Microbiology Unit, Department of Soil Science and Agricultural Chemistry, Agricultural College and Research Institute, Killikulam, India, from January to April 2020. Rice seeds (ASD 16) were planted in the nursery bed, and seedlings were transplanted to fields with plot sizes of 20 m2 for each treatment. The treatments are listed below.
FT1—Yeast 1 + 100% RDCFs.
FT2—Yeast 2 + 100% RDCFs.
FT3—Yeast 3 + 100% RDCFs.
FT4—Yeast consortium (Yeast 1 + Yeast 2 + Yeast 3) + 100% RDCFs.
FT5—100% RDCFs.
FT6—Yeast 1 + 75% RDCFs.
FT7—Yeast 2 + 75% RDCFs.
FT8—Yeast 3 + 75% RDCFs.
FT9—Yeast consortium (Yeast 1 + Yeast 2 + Yeast 3) + 75% RDCFs.
FT10—75% RDCFs.
FTC—Control.
The prepared yeast inoculum (~ 108 CFU mL−1) was foliar sprayed on rice during the stem elongation, flowering, and dough stages. Finally, the rice grains were harvested from each plot, and the yield was calculated and expressed as tons per hectare (t ha−1).
Statistical analysis
All experiments were conducted in triplicate, with results expressed as means with standard deviations. Statistical analyses were performed using R version 4.1.1. Tukey's post hoc test was applied using the stats package [50] to rank the yeast strains and treatments. A two-way ANOVA using the stats package was conducted to assess the effects of treatments, growth stages, and their interaction on various rice plant variables. A Venn diagram, created with the ggVennDiagram package, illustrated the percentage of unique and common rice leaf metabolites between control and yeast consortium-sprayed plants [51]. Changes in rice leaf metabolites in yeast consortium-treated plants were visualized using a volcano plot generated with ggplot2 [52]. For data integration approaches, the made4 package was used for coinertia analysis (CIA) [53], the FactoMineR package followed by the PCAtest package for Principal Component Analysis (PCA) [54, 55], the mixKernel package for Multiple Kernel Learning (MKL) [56], and the omicade4 package for Multiple Co-Inertia Analysis (MCIA) [57].
Data integration
We used unsupervised machine learning-based horizontal data integration methods, including MKL, MCIA, and PCA, to identify the optimal treatment for rice growth and yield. PCA reduces dimensionality linearly, highlighting principal components that explain the most variance. MKL combines multiple kernels to model non-linear relationships and interactions. MCIA uncovers common structures and correlations across multiple datasets, offering a comprehensive view of shared variations. These diverse techniques help select the best treatment and understand the underlying reasons for treatment groupings, as each method varies in data integration and dimension reduction approaches [29].
For the integration techniques, the parameters from the pot culture experiment were categorized into plant biochemical traits (total chlorophyll, protein, proline, and antioxidant enzyme activity: catalase, peroxidase, and polyphenol oxidase) and plant phenotypic traits (shoot and root length) and microbial population (CFUs of bacteria, actinomycetes, and yeast), separately for each growth stage. The yield-related parameters from pot culture and actual yield from the field experiment measured during harvest were grouped into one dataset. The dataset combinations used for the integration techniques are listed in Supplementary Table 1. Unlike MCIA, which can integrate multiple datasets, CIA can only integrate two. We used CIA to combine PGP traits of yeast strains with the shoot and root lengths of rice grown in roll paper towels.
Results
Isolation and characterization of yeast isolates
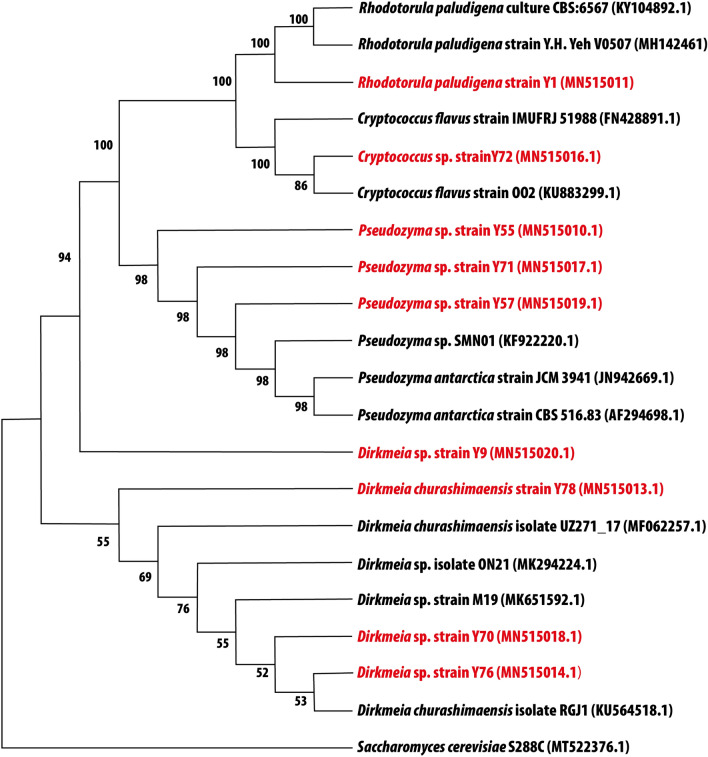
Fifty-two morphologically distinct isolates were obtained from the collected leaf samples using the leaf imprinting method on different media. Among 52 isolates, 11, 16, and 25 were isolated from the stem elongation, flowering, and dough phases, respectively. The isolates were subjected to growth at different temperature levels (25, 30, and 36 °C), and all 52 isolates were grown at 25 and 30 °C, but only nine isolates ( Y1, Y9, Y55, Y57, Y70, Y71, Y72, Y76, and Y78) were found to grow at 36 °C (Supplementary Table 2). DNA from the nine yeast isolates was extracted, and the ITS rRNA gene was sequenced. The sequences were compared in the NCBI database, and closely related species were obtained using BLAST. A phylogenetic tree was constructed for all the yeast isolates, and their close relatives were determined (Fig. 1). The isolated yeasts were identified as Rhodotorula paludigena Y1, Dirkmeia sp. Y9, Pseudozyma sp. Y55, Pseudozyma sp. Y57, Dirkmeia sp. Y70, Pseudozyma sp. Y71, Cryptococcus sp. Y72, Dirkmeia sp. Y76, Dirkmeia churashimaensis Y78. The DNA sequences of the identified yeast species were submitted to Genebank, and their accession numbers and plant host details are provided in Supplementary Table 3.
Fig. 1.
The maximum likelihood phylogenetic tree based on ITS1 and ITS4 gene sequences, showing the relationships between the yeast taxa identified in this study. The bootstrap values ≥ 50% (based on 100 replications) are shown at branching points. Strains in red are the yeast strains isolated from the rice phyllosphere
PGP of phyllosphere yeast strains
Direct PGP traits
Among the nine strains, R. paludigena Y1 exhibited the highest production of IAA, followed by, Pseudozyma sp. Y55, Dirkmeia sp. Y70, Pseudozyma sp. Y71, Cryptococcus sp. Y72, Dirkmeia sp. Y76, D. churashimaensis Y78 showed no significant difference in the production of IAA. The least IAA production was observed in Dirkmeia sp. Y9. In the case of GA production, R. paludigena Y1 recorded the highest GA production, followed by Pseudozyma sp. Y71 and Cryptococcus sp. Y72 had the maximum GA production with a less significant difference (Table 1). The minimum amount was recorded in yeast strains Cryptococcus sp. Y72, Dirkmeia sp. Y76 and D. churashimaensis Y78 with no significant difference in GA production. The potential of the yeast strains for the solubilization of phosphate and potassium was determined. All nine strains solubilized the tricalcium phosphate in the plate assay method. The highest phosphate solubilization index was recorded in the strain R. paludigena Y1 and Pseudozyma sp, with the least significant difference. Whereas low phosphate solubilizing index was recorded in Dirkmeia sp. Y76 (Table 1). The results of the potassium solubilization study showed that all strains had the inorganic potassium solubilization potential, but there was no considerable significant difference among them. The highest production was recorded in R. paludigena Y1, Pseudozyma sp. Y71, Cryptococcus sp. Y72, Pseudozyma sp. Y57, and Pseudozyma sp. Y55 and they are significantly different from the rest of the other strains, which were recorded with the least solubilization index (Table 1).
Table 1.
Mean and standard deviation values (n = 3) of direct plant growth-promoting traits such as Indole acetic acid (IAA) and gibberellic acid (GA) production and mineral solubilization potential of rice phyllosphere yeast strains
| Strains | IAA (µg mL−1) | GA (µg mL−1) | Solubilization index | |
|---|---|---|---|---|
| Phosphate (P) | Potassium (K) | |||
| Rhodotorula paludigena Y1 | 77.2 (9.13)a | 101 (6.60)a | 2.84 (0.12)a | 2.48 (0.12)a |
| Dirkmeia sp. Y9 | 5.01 (1.43) d | 59.4 (15.6)cd | 2.36 (0.17)bcd | 1.33 (0.28)b |
| Pseudozyma sp. Y55 | 34.2 (3.36)b | 6.11 (3.32)e | 2.06 (0.05)cde | 2.01 (0.01)a |
| Pseudozyma sp. Y57 | 12.6 (2.82)c | 54.7 (7.87)cd | 2.00 (0.01)de | 2.13 (0.15)a |
| Dirkmeia sp. Y70 | 30.2 (11.7)b | 44.4 (7.13)d | 2.32 (0.12)bcd | 1.21 (0.26)b |
| Pseudozyma sp. Y71 | 40.8 (2.61)b | 84.4 (7.84)ab | 2.61 (0.10)ab | 2.43 (0.13)a |
| Cryptococcus sp. Y72 | 40.6 (2.52)b | 74.2 (7.73)bc | 2.51 (0.10)abc | 2.35 (0.13)a |
| Dirkmeia sp. Y76 | 29.3 (2.80)b | 7.14 (2.26)e | 1.66 (0.28)e | 1.33 (0.28)b |
| Dirkmeia churashimaensis Y78 | 36.0 (2.02)b | 2.13 (1.81)e | 2.24 (0.10)bcd | 1.16 (0.28)b |
Parameter values with different letters are significantly different according to Tukey’s test. p < 0.05
Indirect PGP traits
All the yeast strains were tested for siderophore production and showed significant production (Table 2). The highest production was recorded in the strain R. paludigena Y1 followed by D. churashimaensis Y78 and Cryptococcus sp. Y72. While the least production was recorded in the strain Dirkmeia sp. Y9. The activity of ACC deaminase was assessed for all nine strains; among them, R. paludigena Y1 recorded the highest ACC deaminase activity, followed by Pseudozyma sp. Y71 and Cryptococcus sp. Y72. The strain Dirkmeia sp. Y70 exhibited the lowest ACC deaminase activity (Table 2). The results of the antagonistic activity of nine yeast strains against rice fungal pathogens showed that R. paludigena Y1 had the highest inhibition percentage against H. oryzae and P. oryzae, while R. paludigena Y1 and Pseudozyma sp. Y71 exhibited the highest inhibition percentage against S. oryzae (Table 2; Supplementary Fig. 1). Similarly, the lowest inhibition of all three fungal pathogens was recorded in D. churashimaensis Y78.
Table 2.
Mean and standard deviation values (n = 3) of indirect plant growth-promoting traits such as siderophore production, ACC deaminase activity, and growth inhibition of Helminthosporium oryzae, Pyricularia oryzae and Saracladium oryzae by rice phyllosphere yeast strains
| Strains | Siderophore (%) | ACC deaminase activity (nmol α-ketobutyrate·mg−1 protein·h−1) | Inhibition percentage (%) | ||
|---|---|---|---|---|---|
| Helminthosporium oryzae | Pyricularia oryzae | Saracladium oryzae | |||
| Rhodotorula paludigena Y1 | 26.8 (0.82)a | 10.3 (0.65)a | 46.6 (0.52)a | 50.8 (2.22)a | 40.2 (1.33)a |
| Dirkmeia sp. Y9 | 3.62 (0.51)g | 0.60 (0.16)ef | 15.2 (0.48)f | 15.6 (0.34)d | 15.5 (0.45)de |
| Pseudozyma sp. Y55 | 6.23 (0.40)f | 0.41 (0.06)ef | 28.6 (0.81)d | 15.5 (0.25)de | 20.0 (0.41)c |
| Pseudozyma sp. Y57 | 2.30 (0.65)gh | 1.33 (0.07)def | 18.1 (0.23)e | 15.5 (0.41)de | 17.1 (0.25)d |
| Dirkmeia sp. Y70 | 8.76 (0.76)e | 0.15 (0.06)f | 37.4 (1.75)b | 18.1 (0.22)cd | 15.9 (0.61)de |
| Pseudozyma sp. Y71 | 14.9 (0.83)d | 5.37 (1.11)b | 33.1 (0.21)c | 27.4 (1.35)b | 40.2 (1.32)a |
| Cryptococcus sp. Y72 | 17.2 (0.31)c | 2.14 (0.24)c | 38.6 (2.24)b | 20.4 (1.01)c | 38.0 (0.86)b |
| Dirkmeia sp. Y76 | 7.80 (0.71)ef | 0.32 (0.06)ef | 15.6 (0.32)ef | 16.1 (0.62)d | 13.8 (0.27)ef |
| Dirkmeia churashimaensis Y78 | 24.1 (0.31)b | 1.55 (0.11)de | 15.7 (0.33)ef | 12.9 (0.15)ef | 14.9 (0.44)ef |
Parameter values with different letters are significantly different according to Tukey’s test. p < 0.05
Relationship between yeast strains and rice growth parameters
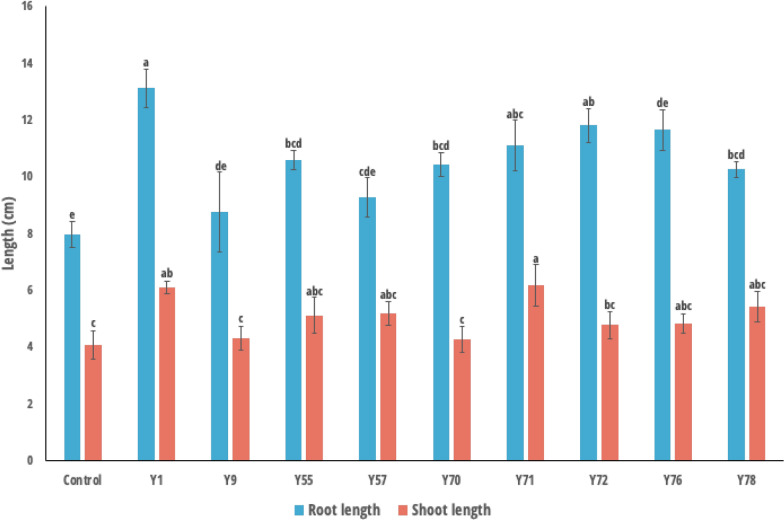
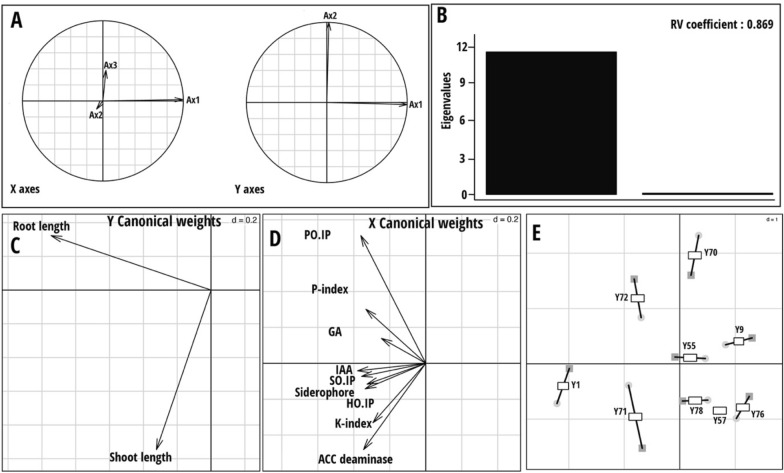
The root and shoot length of the rice plants treated with yeast strains grown on rolled paper were measured (Fig. 2). The root length was significantly higher in the rice plants treated with R. paludigena Y1, followed by those treated with Cryptococcus sp. Y72 and Pseudozyma sp. Y71 recorded the maximum root length. The shortestroot length was recorded in the control plants, and in the case of treated plants, the low length was noted in Dirkmeia sp. Y9, and Dirkmeia sp. Y76 treated plants. Interestingly, shoot length was observed to be higher in the rice treated with Pseudozyma sp. Y71. The shortest shoot length was observed in control plants as well as plants treated with Dirkmeia sp. Y9 and Dirkmeia sp. Y70 with no significant difference between them. Co-inertia analysis (CIA) was applied to assess the correspondence between two datasets (yeast PGP traits and rice phenotypic traits obtained from the roll paper towel method). The graphical output from this analysis is provided in Fig. 3. The CIA findings revealed a strong correlation (RV = 0.869, p < 0.001) between the two datasets with an eigenvalue of 11.8 in the first axis (Fig. 3B). The yeast PGP traits, specifically the P-solubilization index and P. oryzae inhibition percentage, were associated with root length, while the K-solubilization index and ACC deaminase activity were linked to shoot length (Fig. 3C, D). The first CIA axis captured the most variance (99%) in the two datasets and separates the R. paludigena Y1 strain from the rest of the yeast strains (Fig. 3E).
Fig. 2.
Results of root and shoot length of rice obtained from the roll paper towel method treated with yeast strains. The strain codes on the plot are as follows, Y1—Rhodotorula paludigena Y1, Y9—Dirkmeia sp. Y9, Y55—Pseudozyma sp. Y55, Y57—Pseudozyma sp. Y57, Y70—Dirkmeia sp. Y70, Y71—Pseudozyma sp. Y71, Y27—Cryptococcus sp. Y72, Y76—Dirkmeia sp. Y76, and Y78—Dirkmeia churashimaensis Y78. Bars with different letters are significantly different according to Tukey’s test. p < 0.05
Fig. 3.
Co-inertia analysis (CIA) results based on two datasets (yeast plant growth promoting traits and rice phenotypic traits). A Projections of the principal axes of the two datasets onto the axes of the co-inertia analysis. X axes: Yeast plant growth promoting traits; Y axes: rice phenotypic traits. B Scree plot of eigenvalues. C Correlation of plant phenotypic traits data with the first two axes of the co-inertia analysis. D Correlation of yeast plant growth promoting traits and plant phenotypic traits with the first two axes of the co-inertia analysis. E Plot of the first two components in the sample space. Each sample is represented by a square, where the two datasets for each sample are connected by lines to a center point (global score). The strain codes on the plot are as follows, Y1—Rhodotorula paludigena Y1, Y9—Dirkmeia sp. Y9, Y55—Pseudozyma sp. Y55, Y57—Pseudozyma sp. Y57, Y70—Dirkmeia sp. Y70, Y71—Pseudozyma sp. Y71, Y27—Cryptococcus sp. Y72, Y76—Dirkmeia sp. Y76, and Y78—Dirkmeia churashimaensis Y78
Impact of treatments on rice in pot-culture experiment
Treatments, rice growth stages, and their combined effects significantly impacted the microbial population, rice biochemical traits, and phenotypic traits (Supplementary Table 4).
Effect on leaf surface microbial population
In the case of the measured microbial population, the CFUs of bacteria, actinomycetes, and yeast in all the treatments were increased from the stem elongation to the dough stage. The highest bacterial, yeast, and actinomycetes population was recorded in plants treated with yeast consortium (R. paludigena Y1, Pseudozyma sp. Y71, and Cryptococcus sp. Y72) + 75% RDCFs (T9) in all stages and that was approximately double the population measured in control plants (TC) (Table 3). At all stages, the TC plants had the lowest microbial population.
Table 3.
Mean and standard deviation (n = 3) values of bacterial, yeast, and actinomycetes population on rice surface at the stem elongation, flowering, and dough stages grown in pots
| Treatments | Bacteria (107 CFU cm−1) | Yeast (105 CFU cm−1) | Actinomycetes (103 CFU cm−1) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Stem elongation stage | Flowering stage | Dough stage | Stem elongation stage | Flowering stage | Dough stage | Stem elongation stage | Flowering stage | Dough stage | |
| T1 | 7.40 (0.18)c | 14.4 (0.55)ab | 17.0 (0.58)ab | 11.6 (0.16)b | 17.9 (0.15)c | 21.7 (0.27)b | 1.61 (0.03)c | 1.80 (0.02)c | 1.98 (0.02)c |
| T2 | 7.10 (0.17)c | 13.9 (0.35)bc | 16.4 (0.55)abc | 10.5 (0.14)c | 17.5 (0.14)cd | 19.8 (0.25)d | 1.57 (0.03)c | 1.68 (0.02)d | 1.83 (0.02)d |
| T3 | 6.90 (0.17)c | 13.6 (0.52)bc | 16.8 (0.57)ab | 10.5 (0.14)c | 17.2 (0.14)d | 19.8 (0.25)d | 1.50 (0.02)cd | 1.55 (0.02)e | 1.68 (0.02)f |
| T4 | 9.40 (0.23)b | 14.6 (0.55)ab | 15.6 (0.53)bcd | 12.1 (0.16)b | 19.1 (0.16)b | 21.7 (0.27)b | 1.98 (0.03)b | 2.06 (0.02)b | 2.15 (0.02)b |
| T5 | 6.10 (0.18)d | 11.4 (0.46)d | 16.7 (0.63)ab | 9.77 (0.13)d | 16.1 (0.13)e | 19.6 (0.21)e | 1.46 (0.04)de | 1.54 (0.02)e | 1.73 (0.03)e |
| T6 | 7.20 (0.18)c | 14.1 (0.26)bc | 16.8 (0.57)ab | 10.8 (0.15)c | 17.5 (0.15)cd | 20.8 (0.26)c | 1.57 (0.03)c | 1.71 (0.02)d | 1.83 (0.02)d |
| T7 | 6.90 (0.17)c | 13.0 (0.50)c | 15.1 (0.51)cd | 9.85 (0.13)d | 17.2 (0.14)d | 19.7 (0.25)d | 1.43 (0.02)de | 1.51 (0.02)e | 1.64 (0.02)ef |
| T8 | 6.10 (0.15)d | 10.4 (0.40)d | 14.4 (0.49)d | 9.74 (0.13)d | 16.2 (0.13)e | 18.9 (0.24)f | 1.36 (0.02)f | 1.44 (0.01)f | 1.59 (0.02)gh |
| T9 | 10.1 (0.25)a | 15.4 (0.59)a | 17.6 (0.60)a | 13.4 (0.18)a | 20.2 (0.17)a | 23.6 (0.30)a | 2.23 (0.04)a | 2.89 (0.03)a | 3.15 (0.03)a |
| T10 | 5.40 (0.12)e | 11.2 (0.52)d | 14.3 (0.53)d | 9.56 (0.12)d | 16.9 (0.16)d | 18.8 (0.23)f | 1.36 (0.02)f | 1.39 (0.02)f | 1.52 (0.02)h |
| TC | 4.30 (0.10)f | 8.30 (0.32)e | 10.3 (0.35)e | 6.84 (0.09)e | 8.90 (0.07)f | 9.90 (0.12)g | 1.12 (0.02)g | 1.18 (0.01)g | 1.29 (0.01)i |
Parameter values with different letters are significantly different according to Tukey’s test. p < 0.05
T1, Rhodotorula paludigena Y1 + 100% recommended dose of chemical fertilizers (RDCFs); T2, Pseudozyma sp. Y71 + 100% RDCFs; T3, Cryptococcus sp. Y72 + 100% RDCFs; T4, Yeast consortium (Rhodotorula paludigena Y1, Pseudozyma sp. Y71 and Cryptococcus sp. Y72) + 100% RDCFs; T5, 100% RDCFs; T6, Rhodotorula paludigena Y1 + 75% RDCFs; T7; Pseudozyma sp. Y71 + 75% RDCFs; T8, Cryptococcus sp. Y72 + 75% RDCFs; T9, Yeast consortium (Rhodotorula paludigena Y1, Pseudozyma sp. Y71 and Cryptococcus sp. Y72) + 75% RDCFs; T10, 75% RDCFs; TC, Control
Effect on rice biochemical traits
In all treatments, the leaf chlorophyll content increased from the stem elongation stage to the flowering stage but declined slightly in the dough stage than in the stem elongation stage. The chlorophyll content was found to be higher in T9- treatment plants in stem elongation and flowering stages, and it was almost 3 and 2.5 times higher than the Tc plants (Table 4). R. paludigena Y1 + 75% RDCFs-treated plants (T6) had a higher chlorophyll level during the dough stage and TC, T3-, T7-, and T8- treatment plants contained comparatively lower chlorophyll content. Regarding protein and proline content in rice, T9- and Yeast consortium + 100% RDCFs-treatment plants (T4) had the highest levels with minimal significant variation between them (Table 4). Similarly, the TC plants had the least protein and proline content. Protein content was higher during the flowering and dough stages compared to the stem elongation stage, but there was no substantial difference between the flowering and dough stages. The activities of antioxidant enzymes such as catalase, polyphenol oxidase, and peroxidase revealed that T9- treatment plants had considerably higher activity than other treatment plants (Table 5). T4- treatment plants exhibited the next highest enzyme activity, with only minor differences from T9- treatment plants. The TC plants had the lowest levels of enzyme activity. Catalase activity increased in all treatment plants from the stem elongation to the flowering and dough stages. In contrast, polyphenol oxidase activity rose only from the stem elongation stage to the flowering stage but dropped below the stem elongation stage in the dough stage. Peroxidase activity was higher in the flowering stage, followed by the dough stage, and with the lowest activity recorded during the stem elongation stage in all treated plants.
Table 4.
Mean and standard deviation (n = 3) values of total chlorophyll, protein, and proline content in rice leaves at the stem elongation, flowering, and dough stages grown in pots
| Treatments | Total chlorophyll (mg g−1 FW) | Protein (µg g−1 FW) | Proline (µg g−1 FW) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Stem elongation stage | Flowering stage | Dough stage | Stem elongation stage | Flowering stage | Dough stage | Stem elongation stage | Flowering stage | Dough stage | |
| T1 | 10.0 (0.11)c | 13.5 (0.16)c | 7.2 (0.16)d | 35.4 (1.14)bc | 43.9 (1.46)bc | 43.0 (1.06)abc | 66.7 (1.06)ab | 66.8 (1.21)b | 69.0 (1.44)bc |
| T2 | 8.01 (0.13)e | 11.9 (0.29)d | 7.3 (0.20)d | 30.1 (2.00)de | 34.5 (1.09)e | 35.1 (1.08)de | 64.7 (1.27)bc | 66.4 (2.08)b | 65.8 (1.01)cd |
| T3 | 5.45 (0.10)h | 8.10 (0.06)f | 4.2 (0.16)e | 24.6 (1.23)f | 27.2 (0.98)f | 25.5 (1.17)gh | 55.9 (1.02)de | 60.3 (1.11)c | 58.9 (1.03)ef |
| T4 | 13.2 (0.10)b | 17.1 (0.38)b | 9.5 (0.34)b | 40.5 (2.00)ab | 51.0 (2.30)a | 44.4 (1.27)ab | 65.5 (1.00)abc | 76.2 (2.3)a | 73.7 (2.19)ab |
| T5 | 7.44 (0.12)f | 10.8 (0.23)e | 7.7 (0.23)cd | 33.8 (0.54)cd | 43.5 (1.34)bc | 39.7 (1.08)bcd | 61.6 (1.21)bc | 66.4 (1.45)b | 66.9 (1.17)cd |
| T6 | 10.2 (0.14)c | 13.4 (0.26)c | 10.5 (0.04)a | 37.0 (1.10)abc | 47.7 (1.17)ab | 41.5 (1.11)abc | 64.9 (1.40)bc | 67.0 (1.34)b | 72.6 (1.26)ab |
| T7 | 5.53 (0.08)g | 8.90 (0.18)f | 4.4 (0.08)e | 32.6 (0.78)cd | 35.1 (1.16)de | 33.8 (0.54)ef | 60.4 (0.80)cd | 65.9 (0.91)b | 63.2 (1.22)de |
| T8 | 5.61 (0.13)i | 8.80 (0.03)g | 4.6 (0.13)e | 26.4 (1.14)ef | 26.6 (0.86)f | 30.0 (1.08)fg | 55.7 (1.25)de | 58.9 (1.00)c | 55.6 (0.81)f |
| T9 | 17.4 (0.21)a | 20.5 (0.06)a | 9.5 (0.15)b | 41.5 (1.40)a | 48.0 (1.04)ab | 45.1 (2.14)a | 70.7 (1.97)a | 78.6 (1.27)a | 74.8 (1.00)a |
| T10 | 8.82 (0.15)d | 12.5 (0.31)d | 8.2 (0.12)c | 34.4 (0.67)cd | 40.6 (1.27)cd | 38.8 (1.13)cde | 63.7 (1.03)bc | 66.6 (1.18)b | 66.8 (1.32)cd |
| TC | 5.40 (0.10)j | 8.70 (0.05)h | 4.4 (0.15)e | 18.8 (1.14)g | 24.4 (0.90)f | 23.7 (0.84)h | 52.8 (0.75)e | 51.4 (1.02)d | 54.6 (0.94)f |
Parameter values with different letters are significantly different according to Tukey’s test. p < 0.05
T1, Rhodotorula paludigena Y1 + 100% recommended dose of chemical fertilizers (RDCFs); T2, Pseudozyma sp. Y71 + 100% RDCFs; T3, Cryptococcus sp. Y72 + 100% RDCFs; T4, Yeast consortium (Rhodotorula paludigena Y1, Pseudozyma sp. Y71 and Cryptococcus sp. Y72) + 100% RDCFs; T5, 100% RDCFs; T6, Rhodotorula paludigena Y1 + 75% RDCFs; T7; Pseudozyma sp. Y71 + 75% RDCFs; T8, Cryptococcus sp. Y72 + 75% RDCFs; T9, Yeast consortium (Rhodotorula paludigena Y1, Pseudozyma sp. Y71 and Cryptococcus sp. Y72) + 75% RDCFs; T10, 75% RDCFs; TC, Control
Table 5.
Mean and standard deviation (n = 3) values of antioxidant enzyme activity (catalase, polyphenol oxidase, and peroxidase) in rice leaves at the stem elongation, flowering, and dough stages grown in pots
| Treatments | Catalase (mmol min−1 mg protein−1) | Polyphenol oxidase (mmol min−1 mg protein−1) | Peroxidase (mmol min−1 mg protein−1) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Stem elongation stage | Flowering stage | Dough stage | Stem elongation stage | Flowering stage | Dough stage | Stem elongation stage | Flowering stage | Dough stage | |
| T1 | 0.83 (0.005)c | 0.99 (0.006)d | 1.34 (0.006)d | 1.62 (0.07)bc | 2.09 (0.02)cd | 1.37 (0.12)bcd | 1.55 (0.02)bc | 1.98 (0.07)bc | 1.60 (0.12)bcd |
| T2 | 0.46 (0.007)f | 0.60 (0.011)g | 0.80 (0.008)h | 1.42 (0.08)cd | 1.92 (0.02)cde | 1.30 (0.08)bcd | 1.43 (0.02)bcd | 1.75 (0.08)cde | 1.68 (0.09)bc |
| T3 | 0.26 (0.002)h | 0.47 (0.003)h | 0.74 (0.018)i | 1.35 (0.16)cde | 1.70 (0.04)e | 1.12 (0.01)de | 1.02 (0.04)f | 1.56 (0.16)e | 1.37 (0.01)d |
| T4 | 1.14 (0.005)a | 1.34 (0.002)b | 1.76 (0.001)b | 1.87 (0.04)ab | 2.41 (0.12)ab | 1.56 (0.03)bc | 1.72 (0.12)ab | 2.44 (0.04)a | 1.89 (0.03)b |
| T5 | 0.73 (0.003)d | 0.89 (0.003)e | 1.10 (0.002)f | 1.58 (0.05)bc | 2.02 (0.09)cd | 1.21 (0.05)d | 1.55 (0.07)bc | 1.83 (0.05)bcd | 1.48 (0.02)cd |
| T6 | 0.99 (0.009)b | 1.09 (0.013)c | 1.46 (0.013)c | 1.91 (0.10)a | 2.18 (0.01)bc | 1.58 (0.07)b | 1.65 (0.01)abc | 1.95 (0.11)b | 1.69 (0.07)bc |
| T7 | 0.55 (0.009)e | 0.81 (0.013)e | 1.19 (0.011)e | 1.34 (0.09)cd | 1.97 (0.03)cde | 1.32 (0.08)bcd | 1.26 (0.03)def | 2.06 (0.09)b | 1.63 (0.08)cd |
| T8 | 0.32 (0.001)g | 0.53 (0.001)f | 0.95 (0.004)f | 1.25 (0.05)de | 1.81 (0.08)de | 1.12 (0.16)d | 1.09 (0.08)ef | 1.96 (0.05)bcd | 1.35 (0.16)d |
| T9 | 1.16 (0.005)a | 1.60 (0.004)a | 1.98 (0.001)g | 1.93 (0.02)a | 2.68 (0.07)a | 1.98 (0.04)a | 1.91 (0.07)a | 2.61 (0.02)a | 2.21 (0.04)a |
| T10 | 0.74 (0.005)d | 1.00 (0.002)d | 1.22 (0.021)e | 1.46 (0.03)cd | 1.84 (0.07)de | 1.26 (0.03)cd | 1.36 (0.10)cde | 1.67 (0.04)de | 1.49 (0.02)cd |
| TC | 0.26 (0.007)h | 0.51 (0.009)hi | 0.67 (0.011)j | 1.11 (0.10)e | 1.27 (0.16)f | 0.81 (0.10)e | 0.98 (0.16)f | 1.11 (0.12)f | 0.71 (0.13)e |
Parameter values with different letters are significantly different according to Tukey’s test. p < 0.05
T1, Rhodotorula paludigena Y1 + 100% recommended dose of chemical fertilizers (RDCFs); T2, Pseudozyma sp. Y71 + 100% RDCFs; T3, Cryptococcus sp. Y72 + 100% RDCFs; T4, Yeast consortium (Rhodotorula paludigena Y1, Pseudozyma sp. Y71 and Cryptococcus sp. Y72) + 100% RDCFs; T5, 100% RDCFs; T6, Rhodotorula paludigena Y1 + 75% RDCFs; T7; Pseudozyma sp. Y71 + 75% RDCFs; T8, Cryptococcus sp. Y72 + 75% RDCFs; T9, Yeast consortium (Rhodotorula paludigena Y1, Pseudozyma sp. Y71 and Cryptococcus sp. Y72) + 75% RDCFs; T10, 75% RDCFs; TC, Control
Effect on rice phenotypic traits
The shoot length at the stem elongation stage varied significantly among the various treatments. The plants from T4 and T9 sets had the longest shoots in all three stages with no significant difference. The shoot length was approximately 38%, 15%, and 7.7% higher than TC plants in stem elongation, flowering, and dough stages (Table 6). While there was less significant variation in shoot length among the treatments during the flowering stage, no significant variation was observed at the dough stage, except in the TC plants. The root length measured at all three stages was found to be higher in the T9- treatment plants, and the shortest roots were recorded in the TC plants. In the case of other treatment plants in stem elongation and flowering stages, they do not have substantial differences in root length.
Table 6.
Mean and standard deviation (n = 3) values of shoot and root length of rice measured at the stem elongation, flowering, and dough stages grown in pots, as well as yield parameters such as the number of productive tillers, the number of grains per panicle, and the 1000-grain weight
| Treatments | Shoot length (cm) | Root length (cm) | Yield parameters | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Stem elongation stage | Flowering stage | Dough stage | Stem elongation stage | Flowering stage | Dough stage | No. productive tillers | No. grains per panicle | 1000-grain weight (g) | |
| T1 | 39.6 (1.7)abc | 59.7 (1.8)ab | 68.3 (1.1)ab | 22.7 (1.1)ab | 31.6 (1.4)abc | 43.8 (0.5)ab | 11.2 (0.2)b | 95.3 (0. 6)c | 23.9 (0.5)a |
| T2 | 35.7 (1.1)bcd | 58.2 (2.0)abc | 68.1 (1.5)ab | 21.8 (1.2)ab | 31.4 (1.1)abc | 43.1 (0.7)abc | 9.37 (0.6)cd | 91.3 (1.2)d | 23.8 (0.6)ab |
| T3 | 37.8 (0.9)bcd | 58.2 (0.5)abc | 67.8 (0.8)ab | 21.7 (0.1)ab | 30.7 (0.9)abc | 42.1 (1.8)bcd | 8.92 (0.3)de | 89.9 (1.1)d | 23.5 (0.6)ab |
| T4 | 44.6 (0.4)a | 61.9 (0.9)a | 70.1 (1.6)a | 23.3 (1.1)ab | 32.9 (1.0)ab | 44.3 (0.6)ab | 12.1 (0.3)ab | 97.3 (0.4)b | 24.1 (0.2)a |
| T5 | 41.2 (0.5)abc | 56.9 (0.8)bc | 67.2 (2.1)ab | 19.5 (0.9)bc | 30.0 (0.5)bc | 42.6 (0.7)bcd | 8.36 (0.4)ef | 89.1 (0.7)d | 23.4 (0.4)ab |
| T6 | 39.9 (0.5)abc | 59.2 (1.3)ab | 68.2 (0.8)ab | 21.9 (1.5)ab | 31.4 (1.1)abc | 43.6 (1.1)ab | 10.1 (0.2)c | 95.9 (0.3)bc | 23.8 (0.2)ab |
| T7 | 37.2 (0.4)bcd | 57.2 (1.2)bc | 67.1 (2.4)ab | 21.7 (1.0)ab | 31.3 (0.3)abc | 41.0 (0.8)bcd | 8.14 (0.2)efg | 83.1 (0.2)ef | 23.3 (0.5)ab |
| T8 | 33.2 (1.6)de | 57.1 (1.4)bc | 66.1 (2.2)ab | 21.1 (1.6)ab | 29.9 (1.0)bc | 41.9 (1.1)bcd | 7.61 (0.5)fg | 81.3 (0.4)f | 23.2 (0.3)ab |
| T9 | 43.9 (0.6)a | 62.1 (2.6)a | 70.0 (1.5)a | 24.6 (1.3)a | 33.5 (0.8)a | 45.3 (1.0)a | 12.8 (0.2)a | 112 (0.4)a | 24.2 (0.3)a |
| T10 | 36.5 (0.7)cde | 57.3 (1.3)bc | 66.2 (1.1)ab | 19.2 (1.1)bc | 29.9 (0.4)bc | 41.1 (0.9)cd | 7.19 (0.3)gh | 84.8 (0.5)e | 23.0 (0.5)ab |
| TC | 31.9 (1.6)e | 53.9 (0.4)c | 64.5 (0.2)b | 18.3 (1.1)c | 29.3 (1.2)c | 38.1 (1.1)d | 6.52 (0.4)h | 75.1 (0.4)g | 22.6 (0. 6)b |
Parameter values with different letters are significantly different according to Tukey’s test. p < 0.05
T1, Rhodotorula paludigena Y1 + 100% recommended dose of chemical fertilizers (RDCFs); T2, Pseudozyma sp. Y71 + 100% RDCFs; T3, Cryptococcus sp. Y72 + 100% RDCFs; T4, Yeast consortium (Rhodotorula paludigena Y1, Pseudozyma sp. Y71 and Cryptococcus sp. Y72) + 100% RDCFs; T5, 100% RDCFs; T6, Rhodotorula paludigena Y1 + 75% RDCFs; T7; Pseudozyma sp. Y71 + 75% RDCFs; T8, Cryptococcus sp. Y72 + 75% RDCFs; T9, Yeast consortium (Rhodotorula paludigena Y1, Pseudozyma sp. Y71 and Cryptococcus sp. Y72) + 75% RDCFs; T10, 75% RDCFs; TC, Control
Relationship between the measured variables in different rice growth stages
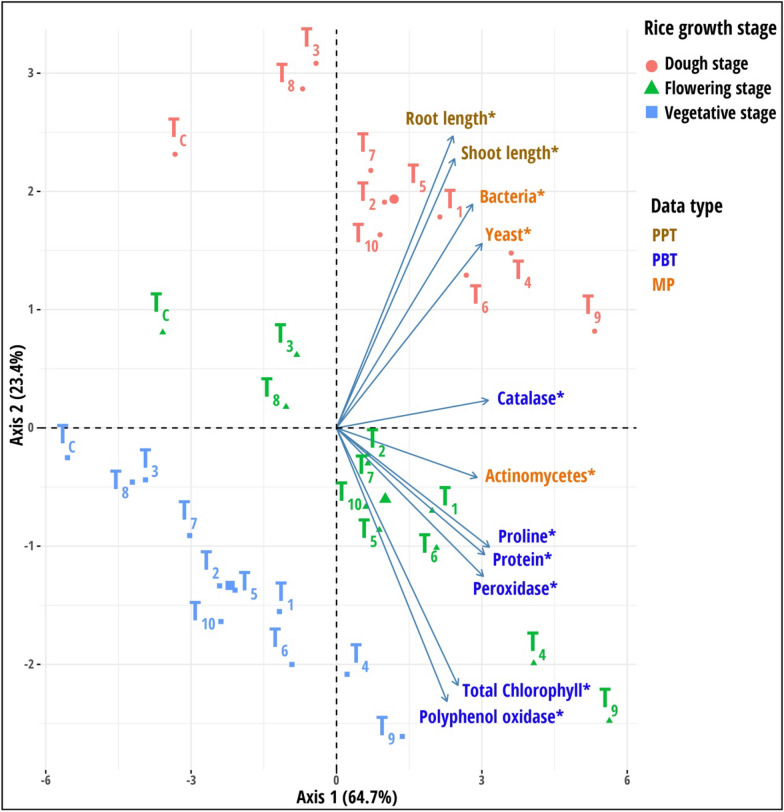
A PCA was applied to understand the variation among plants with different treatments across three rice growth stages using measured parameters (microbial population, plant biochemicals, and plant phenotypic traits) (Fig. 4). The total variance explained by the first two PC axes was 88.1%, indicating a strong treatment impact on plants at all stages of rice development. The PC1 axis was statistically significant (p < 0.05), accounting for 64.7% of the overall variance. Across all rice growing phases, the PC 1 axis separated plants treated with yeast consortium with 75% RDCFs (T9) and 100% RDCFs (T4) from other treatments. In the PC1 axis, all variables were determined to be statistically significant. The PC axis accounted for only 23.4% of the total variance and effectively separated plant samples based on rice development stages. At the dough stage, the bacterial and yeast population variables were dominant in plants treated with yeast consortium with 75% RDCFs (T9) and 100% RDCFs (T4). During the flowering period, peroxidase activity, proline, and protein content were prominent in the same treatment plants.
Fig. 4.
Results of principal component analysis (PCA) based on the integration of all dataset categories measured in pot culture experiment. Variables marked with an asterisk are significant along the first principal component axis obtained from the PCAtest analysis. The treatments are as follows: T1—Rhodotorula paludigena Y1 + 100% recommend dose of chemical fertilizers (RDCFs), T2—Pseudozyma sp. Y71 + 100% RDCFs, T3—Cryptococcus sp. Y72 + 100% RDCFs, T4—Yeast consortium (Rhodotorula paludigena Y1, Pseudozyma sp. Y71 and Cryptococcus sp. Y72) + 100% RDCFs, T5—100% RDCFs, T6—Rhodotorula paludigena Y1 + 75% RDCFs, T7—Pseudozyma sp. Y71 + 75% RDCFs, T8—Cryptococcus sp. Y72 + 75% RDCFs, T9—Yeast consortium (Rhodotorula paludigena Y1, Pseudozyma sp. Y71 and Cryptococcus sp. Y72) + 75% RDCFs, T10—75% RDCFs, and TC – Control. Data type abbreviations: PBT, plant biochemical traits; PPT, plant phenotypic traits; and MP, Microbial population
Comparison of metabolite profile
The PCA results based on rice leaf biochemicals, phenotypic traits, and microbial population showed that most variables were contributed by plants treated with the yeast consortium + 75% RDCFs (T9) in the flowering stage. Hence, the leaf metabolites were extracted and profiled in TC plants and T9 treatment plants collected during the flowering stage (Supplementary Table 5). A total of 51 and 84 metabolites were identified in the TC and T9- treatment plants, respectively, and 48 metabolites were discovered to be shared by the two treatment plants (Fig. 5A). In T9- treatment plants, 36 new metabolites were discovered, while 3 metabolites detected in TC plants were absent in T9- treatment plants (Fig. 5A). A volcano plot was created using the 48 metabolites identified to be common in TC and T9- treatment plants. T9- treatment plants had higher concentrations of 40 metabolites and lower concentrations of 6 metabolites than TC plants (Fig. 5B; Supplementary Table 6). Two metabolites (Hepta-2,4-dienoic acid and 3,7,11,15-Tetramethyl-2-hexadecen-1-ol) showed no significant changes (Fig. 5B).
Fig. 5.
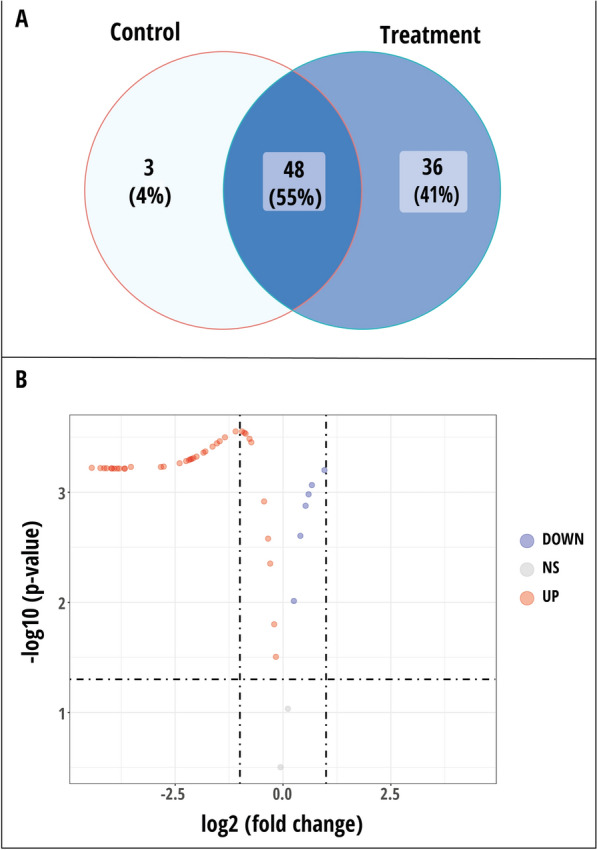
A Comparison of the leaf metabolite profiles between control plants and those treated with the yeast consortium plus 75% RDCFs B Volcano plot illustrating the changes in leaf metabolites of plants treated with the yeast consortium plus 75% RDCFs compared to control plants. NS—nonsignificant
Effect on rice yield-related parameters
The yield-related parameters were measured during the harvest stage. the number of productive tillers was significantly higher in T9- treatment plants, which was two times greater than in TC plants. Followed by T4- and T1 (R. paludigena Y1 + 100%)- treatment plants had maximum productive tillers (Table 6). T9- treatment plants produced the most seeds per panicle, followed by T4- treatment plants. The weight of 1000 grains was highest in plants that had T9, T4, and T1 treatments. Other treatment plants had similar values, indicating no significant differences. In all parameters, the TC plants recorded the least.
Treatment effect on rice yield under field condition
The yield results from the field experiment showed that FT9- treatment plants had the highest yield, which was 15.4% higher than the FTC plants (Table 7). Followed by, plants with FT4, FT6, and FT1 treatments had the maximum yield, which was 14.5%, 12.2%, and 11,3% higher than FTC plants, respectively (Table 7). Other treatment plants (FT2, FT3, FT5, FT7, and FT10) showed no significant difference in the rice yield and the lowest yield was recorded in FTC plants.
Table 7.
Mean and standard deviation (n = 3) values of yield from field experiment
| Treatments | Yield (t/ha) |
|---|---|
| FT1 | 3.68 (0.047)b |
| FT2 | 3.46 (0.045)c |
| FT3 | 3.42 (0.044)c |
| FT4 | 3.79 (0.048)ab |
| FT5 | 3.71 (0.047)ab |
| FT6 | 3.53 (0.045)c |
| FT7 | 3.45 (0.045)c |
| FT8 | 3.82 (0.048)a |
| FTC | 3.41 (0.044)c |
The yield unit is mentioned as tons per hectare (t/ha). Parameter values with different letters are significantly different according to Tukey’s test. p < 0.05
T1, Rhodotorula paludigena Y1 + 100% recommended dose of chemical fertilizers (RDCFs); T2, Pseudozyma sp. Y71 + 100% RDCFs; T3, Cryptococcus sp. Y72 + 100% RDCFs; T4, Yeast consortium (Rhodotorula paludigena Y1, Pseudozyma sp. Y71 and Cryptococcus sp. Y72) + 100% RDCFs; T5, 100% RDCFs; T6, Rhodotorula paludigena Y1 + 75% RDCFs; T7; Pseudozyma sp. Y71 + 75% RDCFs; T8, Cryptococcus sp. Y72 + 75% RDCFs; T9, Yeast consortium (Rhodotorula paludigena Y1, Pseudozyma sp. Y71 and Cryptococcus sp. Y72) + 75% RDCFs; T10, 75% RDCFs; TC, Control
Relationship between the various parameter categories
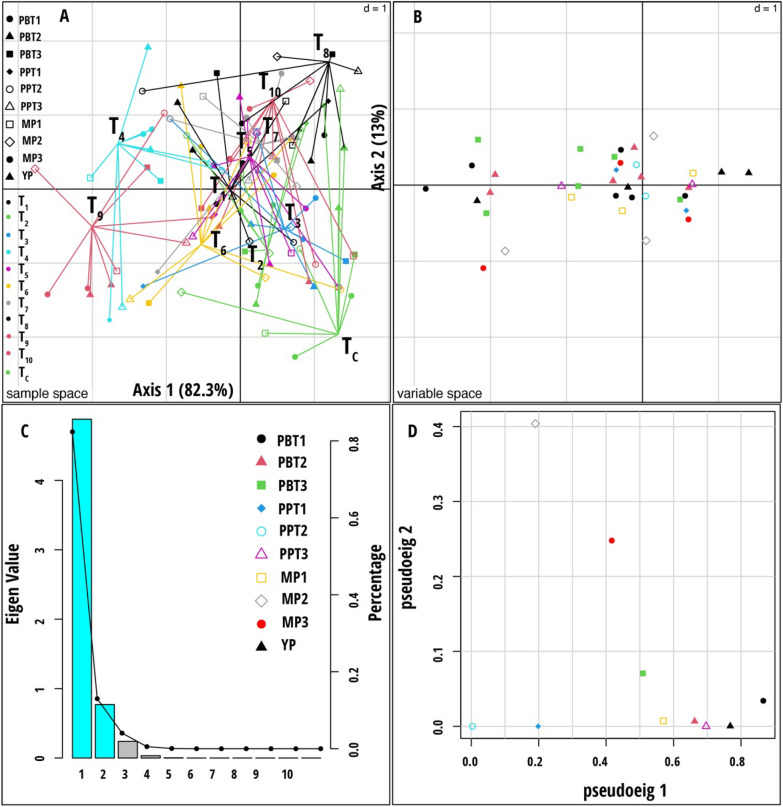
MCIA was applied to analyze the ten datasets jointly. The graphical outputs of this analysis are shown in Fig. 6. The total variance captured by the first two axes was 95.3%, with the first axis capturing the highest variance (82.3%) and separating the two treatment plants, yeast consortium with 75% RDCFs (T9) and 100% RDCFs (T4), from other treatments. The MCIA second axis, explaining 13.1% of the data variation, emphasized the separation of TC plants from other treatment plants (Fig. 6A). Total chlorophyll and catalase activity from all three datasets (PBT1, PBT2, and PBT3), shoot length from PPT1, root length from PPT3, bacteria from MP1, and the number of tillers from the YP dataset was well associated with T9- and T4- treatment plants (Supplementary Fig. 2). The RV coefficient matrix of the combined dataset showed a strong positive correlation within datasets of plant biochemical traits (PBT1, PBT2, and PBT3) and microbial population (MP2 and MP3) (Supplementary Fig. 3A). A significant positive correlation was noted between the yield parameter dataset (YP) and the plant biochemical traits dataset (PBT1, PBT2, and PBT3). However, no substantial correlation was found between other dataset categories. As a result, only all plant biochemical traits, microbial population, and yield parameters datasets were significant in separating the treatments in MCIA.
Fig. 6.
Multiple co-inertia analysis (MCIA) results based on nine data sets (Plant biochemical traits, plant phenotypic traits, and microbial population measured during stem elongation, flowering, and dough stages). A Plot of the first two components in the sample space. Each sample is represented by a colored shape, with lines connecting the nine datasets for each sample to a central point (MCIA global score). B Variable space for each data set. C A scree plot of absolute eigenvalues (bars) and the proportions of variance for the eigenvectors (line). D A plot of data weighting space that shows the pseudo-eigenvalues space of all data sets indicating how much variance of an eigenvalue is contributed by each data set. The treatment codes used are: T1—Rhodotorula paludigena Y1 + 100% recommend dose of chemical fertilizers (RDCFs), T2—Pseudozyma sp. Y71 + 100% RDCFs, T3—Cryptococcus sp. Y72 + 100% RDCFs, T4—Yeast consortium (Rhodotorula paludigena Y1, Pseudozyma sp. Y71 and Cryptococcus sp. Y72) + 100% RDCFs, T5—100% RDCFs, T6—Rhodotorula paludigena Y1 + 75% RDCFs, T7—Pseudozyma sp. Y71 + 75% RDCFs, T8—Cryptococcus sp. Y72 + 75% RDCFs, T9—Yeast consortium (Rhodotorula paludigena Y1, Pseudozyma sp. Y71 and Cryptococcus sp. Y72) + 75% RDCFs, T10—75% RDCFs, and TC – Control. The data set abbreviations PBT1, PBT2, and PBT3 are plant biochemical traits; PPT1, PPT2, and PPT3 are plant phenotypic traits; MP1, MP2, and MP3 are microbial population, and YP are yield parameters
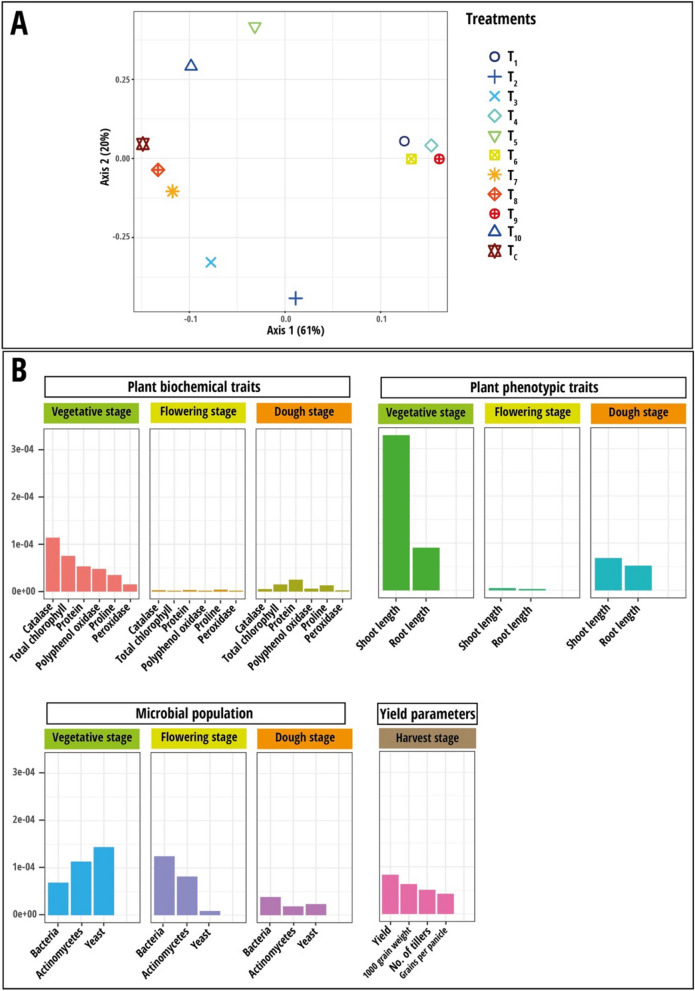
For MKL, the same datasets were used, and the combined kernel principal component analysis (KPCA) was applied in further exploratory analysis. The results revealed that the majority of the overall variance in the data was captured by the first axis of KPCA (61%), which clearly separated the four treatments, yeast consortium with 75% RDCFs (T9), 100% RDCFs (T4), R. paludigena Y1 + 75% RDCFs (T6) and R. paludigena Y1 + 100% RDCFs (T1) from other treatments but T9 and T4 had the highest variables impact than T6 and T1 (Fig. 7A). The second axis captured the least variance (20%), which separated the fertilizer-only treatments (100% RDCFs (T5) and 75% RDCFs (T10)) from other treatments. An important variable plot was computed for each dataset to determine the impact of the variables on KPCA (Fig. 7B). In terms of the microbial community, yeast was found to be a key factor during the stem elongation stage, whereas bacteria was the critical variable during the flowering and dough stages. Catalase activity and total chlorophyll content significantly influenced plant biochemical parameters during the stem elongation stage, while the other two stages did not show a significant impact on the variables. On the other hand, shoot length was found to be an important factor during the flowering and dough stages. In the case of yield parameters data, the actual yield measured after harvest emerged as the most important variable. Correlation using the RV coefficient matrix of the combined dataset revealed a strong positive correlation within the plant biochemical traits dataset (PBT1, PBT2, and PBT3) (Supplementary Fig. 3B). A substantial positive correlation was noted between PBTs and PPT1, PPT2, and YP datasets. Microbial population datasets (MPs) also showed significant positive correlations within themselves and with PPT2, PPT3, and YP datasets. As a result, all four parameter category datasets significantly separated the treatments in KPCA.
Fig. 7.
A Results of multiple kernel learning analysis (MKL). A Plot of kernel principal component analysis (KPCA) based on nine data sets (Plant biochemical traits, plant phenotypic traits and microbial population measured during stem elongation, flowering, and dough stages). B Plot for important variables in each dataset assessed using Crone-Crosby distance. The treatment codes used are: T1—Rhodotorula paludigena Y1 + 100% recommend dose of chemical fertilizers (RDCFs), T2—Pseudozyma sp. Y71 + 100% RDCFs, T3—Cryptococcus sp. Y72 + 100% RDCFs, T4—Yeast consortium (R. paludigena Y1, Pseudozyma sp. Y71 and Cryptococcus sp. Y72) + 100% RDCFs, T5—100% RDCFs, T6 – R. paludigena Y1 + 75% RDCFs, T7—Pseudozyma sp. Y71 + 75% RDCFs, T8—Cryptococcus sp. Y72 + 75% RDCFs, T9—Yeast consortium (Rhodotorula paludigena Y1, Pseudozyma sp. Y71 and Cryptococcus sp. Y72) + 75% RDCFs, T10—75% RDCFs, and TC – Control
MCIA determined that T9 was the most effective treatment and T4 was the next-most effective treatment. In the case of MKL, both T9 and T4 treatments were found to be the better treatments in KPCA, as they can separate the inseparable linear space between the samples. Additionally, PCA results support that the T9 treatment significantly improved rice biochemical and phenotypic traits (Fig. 4). Therefore, applying the yeast consortium to rice fields with 75% RDCFs better enhances rice yield. Overall, the foliar application of yeast consortium with 75% regular fertilizer application improved rice yield.
Discussion
Yeast, a key model organism, is extensively used in the food industry, biotechnology, and medicine due to its unicellular eukaryotic nature and ease of genetic manipulation [58]. Saccharomyces cerevisiae, the most studied yeast, is known for its efficient homologous recombination and was the first eukaryote to have a fully sequenced genome [59]. With advancements in genome editing, non-conventional yeasts such as Kluyveromyces lactis, Yarrowia lipolytica, Komagataella phaffii (formerly Pichia pastoris), and Schizosaccharomyces pombe have also become important in biotechnology and cellular biology research [60]. On the other hand, environmental yeasts have received little attention in terms of applications but have been extensively researched from an ecological standpoint. In contrast to conventional yeasts used in fermentation studies, which thrive in extremely rich culture conditions, environmental yeasts are constrained by a lack of readily available nutrients [61]. Notably, phyllosphere yeasts are subjected to environmental stresses such as high temperature, UV radiation, and dryness, which likely force them to use different metabolic routes and regulatory mechanisms for substrate utilization [10]. Thus, further efforts should be made to investigate their genetic and metabolic potential under natural conditions. They are also involved in many interactions with other microorganisms and plants, which include symbiosis, mutualism, parasitism, and competition. With these capabilities, recent reports suggest that yeast could be used for plant growth promotion and plant disease control. To fulfil the void in the search for new beneficial microbes to enhance crop production and protection, an attempt was made to isolate and develop potential yeast isolates with multiple beneficial traits for use in crop production and protection. The results obtained through various experiments have been discussed here under.
In the current study, 52 yeast isolates were obtained from the phyllosphere of rice at various phases of the rice crop. Nine of the isolates were discovered to grow at 36 °C. As previously stated, the phyllosphere is an unfavorable environment, so testing the phyllosphere microbes for specific stress levels is critical. Similarly, phyllosphere bacterial strains were tested for salinity, temperature, and osmotic stress before rice foliar treatments [26]. The ability of rice phylloplane yeast strains Rhodotorula taiwanensis, Cryptococcus aff. laurentii, and Cryptococcus flavescens to grow up to 35 °Cwas tested to ensure that they can withstand environmental temperatures [62]. Using ITS rRNA gene sequencing, the nine-temperature tolerant yeast genera were identified Rhodotorula, Dirkmeia, Pseudozyma, and Cryptococcus. Cryptococcus albidus and Rhodotorula mucilaginosa were primarily found on leaves, representing more than 50% of the community [63]. The isolation of Pseudozyma genera from plant leaves was also reported [64, 65]. Dirkmeia churashimaensis was the most common basidiomycetous yeast discovered on the sugarcane phylloplane, accounting for 12% of all yeast strains identified [66]. Oleaginous red yeasts, Rhodotorula species, are usually a promising source of carotenoids, lipids, and exopolysaccharides (EPS), which provide essential phyllosphere microbial characteristics such as UV protection, avoidance of desiccation, and increased adhesion to the plant leaf surface [67, 68].
IAA is the most common plant hormone of the auxin class, which regulates various processes in plants from development to senescence [69]. Similarly, GA is an essential plant hormone that aids stem elongation and germination. The most common form of GA is GA3, which is a fungal product, and plants can primarily utilize GA1, and other GA forms are used as precursors for GA1 synthesis [69]. The microbial origin of phytohormones and their application in plant growth and stress alleviation have been widely studied [70]. A diverse range of yeast strains isolated from plant phylloplane showed IAA production [71]. Rhodosporidium paludigenum isolated from rice phyllosphere was reported to produce IAA [72]. Root treatment with IAA-producing endophytic yeast has been shown to increase root and shoot length, and maximum IAA production was recorded by Williopsis saturnus (22.5 μg ml−1). In our study, maximum IAA was produced by R. paludigena Y1 (77.2 ± 9.1 μg ml−1), and treating rice seeds with yeast strains had minor and major improvements in shoot and root lengths, respectively [73]. The report on the production of GA by yeast is uncommon, and only production of GA in soil yeast through biochemical assay. Further confirmation of GA production needs to be studied through modern approaches. One of the most important needs of the plant is nutrients, which are provided by microbes via mechanisms such as mineralization (from organic to inorganic form), solubilization (from inorganic to soluble form), and mobilization (uptake) [74]. Plants require phosphorus and potassium as macronutrients. All nine yeast strains in this study demonstrated phosphate and potassium solubilization by forming a halo zone around the colony in the specific media. A similar study reported phosphorus solubilization by Drosera spatulata phyllosphere yeasts Rhodosporidium paludigenum, Cryptococcus laurentii, and Pseudozyma species [75]. Soil yeasts Pichia anomala and Rhodotorula glutini have been shown to solubilize potassium and enhance plant height, as well as root and stem dry weights [22]. According to our CIA results, K-index significantly increases the shoot length of plants whose seeds were treated with yeast strains.
Siderophore is a ferric iron-chelating compound with low molecular weight and strong affinity that is secreted by organisms. Siderophore-producing microbes promote plant growth by increasing iron availability and protecting plants from pathogens by increasing competition for iron sources [76]. Pseudozyma aphidis, a phyllosphere yeast, is known to produce the most siderophores of the 15 yeast species examined [77]. Similarly, in our study, among the nine strains, R. paludigena Y1 had the maximum siderophore production compared to other strains. Trichosporon ovoides and Saccharomyces cerevisiae are shown to exhibit a high production of siderophore and sidD gene expression under different levels of Cd2+ and Pb2+ toxicity stress [19]. Yeasts produce hydroxymate type of siderophores and are effective in controlling postharvest diseases of apples and pears [77]. So, the results indicated that phyllosphere yeast could increase the yield by enhancing the uptake of iron and also can control the disease by making the unavailability of iron for pathogens.
During plant stress, higher ethylene levels are produced than usual, which causes leaf senescence. ACC deaminase is an enzyme that breaks the ethylene precursor ACC, thereby reducing ethylene levels. Both plants and microbes produce ACC deaminase [78]. In our case, all the yeast strains showed positive ACC deaminase activity, and high activity was recorded in R. paludigena Y1. High production of ACC deaminase was reported in Pseudozyma sp. out of the eight isolates tested [75]. Two seed-borne yeasts, Pichia kudriavzevii and Issatchenkia terricola, were found to have ACC deaminase activity as well as many other PGP functions [79]. Similarly, Liu et al., [80] discovered that rhizosphere yeast, Cryptococcus sp., has ACC deaminase activity and inorganic phosphate solubilization and improves the shoot biomass of Sedum arboretum. In our study, CIA showed a significant relationship between shoot length and ACC deaminase activity. Hence, the isolated yeast species demonstrated their potential to decrease ACC levels in rice plants, ultimately reducing ethylene levels, enhancing plant growth, and protecting the host from various environmental stresses.
Fungal pathogens in plants cause serious crop losses with significant socioeconomic impacts. Increasingly, people recognize that fungal pathogens pose a global threat to agriculture [81]. Microbial tools such as yeast may protect plants from pathogens through various processes such as colonization, antifungal compound production, induced systemic resistance, and the mycoparasitism process [82]. Yeast isolates from the phyllosphere of rice were tested for antagonistic activity against rice pathogens. All the yeast isolates inhibited the growth of P. oryzae, H. oryzae, and S. oryzae. Among the nine strains R. paludigena Y1, Pseudozyma sp. Y71 and Cryptococcus sp. Y72 significantly controlled the rice pathogens. Pepper phyllosphere Pseudozyma churashimaensis conferred protection against Xanthomonas axonopodis and some pepper viral infections upon foliar application under field conditions [64]. Similarly, the antagonistic activity of phyllosphere yeast Rhodotorula glutinis against tomato Botrytis cinerea was reported [83]. Cryptococcus and some other yeast genera showed antagonistic activity against phytopathogenic fungi Penicillium, Aspergillus, and Botrytis of table grapes, wine grapes, and raisins [84]. The possible antagonistic mechanisms of Rhodotorula sp. against fungal pathogens could include siderophore production [85], chitinase production [86], and other inhibitory metabolites [87]. Hence our isolate can protect against rice foliar fungal pathogens.
The three yeast strains that were found to have prominent plant growth traits tested positive for compatibility. All three strains were found to be Basidiomycete. Basidiomycota yeasts, along with other endophytic bacteria and oomycetes, were prevalent in the Arabidopsis thaliana leaf microbial community [88]. Similarly, many Basidiomycota yeasts were reported in the rice phylloplane [62]. Hence it is possible to have good compatibility among the Basidomycota yeast. According to the review by [89], applying multiple microbial species of inoculum frequently enhances nutrient availability, promotes plant growth, and yields better than applying a single microbial species. In our study, we applied individual yeast strains and the consortium of three strains along with 75% and 100% RDCFs. The results showed that consortium application, along with 75% RDCFs was the best application in rice as it significantly improved plant biochemicals and yield. In a similar study, it was discovered that the application of Glomus mosseae (AMF), Bacillus subtilis (Phosphate solubilizing bacteria), and Nitrifying microorganisms (Nitrosomonas + Nitrobacter) along with 75% of the recommended urea + superphosphate improved the potato yield and quality more than individual microbial application and high dose fertilizers (100 and 125% RDCFs) [90]. Integrated biofertilizer consortium (Proteobacteria and some fungal species from Basdiomycota and Ascomycota) with 30% recommended chemical fertilizer better-improved oil palm growth and soil microbial diversity than zero and 100% dose chemical fertilizer [91]. As a result, the yeast consortium substituted 25% of the chemical fertilizers commonly used in rice cultivation while outperforming 100% chemical fertilizer applications.
Data integration approaches were invaluable in uncovering various aspects of this study. Within linear relationships, PCA and MCIA highlighted T9 and T4 as the best treatments, with a stronger emphasis on T9. MKL, which captures non-linear relationships, additionally identified T1 and T6 as strongly related to the measured parameters. PCA loadings revealed significant variables relevant to each growth stage. MCIA demonstrated that the significance of each measured variable at each rice growth stage was closely related to specific treatments. In contrast, MKL highlighted the order of importance of each variable across different rice growth stages, rather than focusing on relationships.
We also studied the impact of yeast consortium on rice leaf metabolites. Several compounds were identified in rice leaf extract at the critical stage of rice plants from both consortium-treated plants with fertilizer and control plants. The quantity of organic acids and volatile compounds was synthesized at higher levels in the treated plants compared to the control plants. Wheat seeds exposed to two rhizosphere bacteria were found to alter leaf and rhizosphere metabolites. The metabolite profile was found to have beneficial functions such as promoting plant growth, interacting with microbes, and chemotaxis. [92]. Deproteinized leaf extract has been used as a yeast growth medium, and the and the metabolites produced by yeast in that medium were reported to have plant-beneficial compounds [93]. Moreover, the yeast extract was reported to act as an elicitor and induced growth and secondary metabolite production in transgenic Red sage (Salvia miltiorrhiza) cells in 2012 suspension culture [94]. Hence in our study, the yeast strains foliar inoculated on the rice leaf likely utilized the leaf nutrient source and induced a wide range of beneficial metabolite production in rice.
Conclusion
The primary aim of this study was to reduce the dosage of chemical fertilizers in agriculture by integrating them with underexplored microorganisms that can enhance crop improvement. Consequently, we isolated rice phyllosphere yeast and selected nine strains capable of withstanding high temperatures (36 °C), enabling survival in the phyllosphere environment. Among these, three yeast strains, Rhodotorula paludigena Y1, Pseudozyma sp. Y71, and Cryptococcus sp. Y72, demonstrated better PGP traits and improved root and shoot lengths upon seed treatment. When these top three strains were combined and foliar-sprayed on rice in pot culture and field experiments with 75% of the RDCFs, the results showed significant improvements in plant biochemicals, growth, leaf microbial population, and yield compared to using 100% RDCFs alone. Thus, we found that the foliar application of the identified yeast consortium reduced fertilizer usage by 25% while achieving a better yield than regular fertilization. Therefore, this yeast consortium can be applied to rice fields with reduced fertilizer input to achieve a better yield. Reports on the role of phyllosphere yeast in crop development and biotic stress control are scarce. This study and its findings on rice phyllosphere yeast provide significant contributions to PGPM research. Future research could focus on comparing the rice yield improvement efficiency of phyllosphere yeast strains with that of bacterial strains.
Supplementary Information
Acknowledgements
The authors thank the Department of Biotechnology, Foldscope Project (E28ACN)—India.
Author contributions
GM and AKD designed the experiments. GM, SKG, and JM performed the experiments, and GM and AKD wrote the first draft of the manuscript. AKD did the data analyses. SKG, JM, KSS, RD, DJN, and PD did the proofreading of the first version. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the Department of Biotechnology, Ministry of Science and Technology, grant number BT/IN/Indo-US/Foldscope/39/2015.
Availability of data and materials
No datasets were generated or analysed during the current study.
Declarations
Ethics approval
This material is the authors' own original work, which has not been previously published elsewhere. The paper reflects the authors' own research and analysis in a truthful and complete manner.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Gomathy Muthukrishanan, Email: gomathymicro@gmail.com.
Sabarinathan Kuttalingam Gopalasubramaniam, Email: sabarinathan@tnau.ac.in, Email: sabarimicro@hotmail.com.
Arun Kumar Devarajan, Email: aruntechtnau@gmail.com.
References
- 1.IPAD. USDA. India Rice Area, Yield and Production. 2024. Retrive from https://ipad.fas.usda.gov/countrysummary/Default.aspx?id=IN&crop=Rice.
- 2.FAO. The state of food and agriculture. In: Moving forward on food loss and waste reduction. Rome: FAO; 2019. [Google Scholar]
- 3.Deutsch CA, Tewksbury JJ, Tigchelaar M, Battisti DS, Merrill SC, Huey RB, Naylor RL. Increase in crop losses to insect pests in a warming climate. Science. 2018;361(6405):916–9. 10.1126/science.aat3466. [DOI] [PubMed] [Google Scholar]
- 4.Koli P, Bhardwaj NR, Mahawer SK. Agrochemicals: harmful and beneficial effects of climate changing scenarios. In: Climate change and agricultural ecosystems. New Delhi: Woodhead Publishing; 2019. p. 65–94. [Google Scholar]
- 5.Malhi GS, Kaur M, Kaushik P. Impact of climate change on agriculture and its mitigation strategies: a review. Sustainability. 2021;13(3):1318. 10.3390/su13031318. [Google Scholar]
- 6.Naik K, Mishra S, Srichandan H, Singh PK, Sarangi PK. Plant growth promoting microbes: potential link to sustainable agriculture and environment. Biocatal Agric Biotechnol. 2019;21:101326. 10.1016/j.bcab.2019.101326. [Google Scholar]
- 7.Turner TR, James EK, Poole PS. The plant microbiome. Genome Boil. 2013;14(6):1–10. 10.1186/gb-2013-14-6-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hassani MA, Durán P, Hacquard S. Microbial interactions within the plant holobiont. Microbiome. 2018;6:1–17. 10.1186/s40168-018-0445-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh BK, Liu H, Trivedi P. Eco-holobiont: a new concept to identify drivers of host-associated microorganisms. Environ Microbiol. 2020;22(2):564–7. 10.1111/1462-2920.14900. [DOI] [PubMed] [Google Scholar]
- 10.Vorholt JA. Microbial life in the phyllosphere. Nat Rev Microbiol. 2012;10(12):828–40. 10.1038/nrmicro2910. [DOI] [PubMed] [Google Scholar]
- 11.Afeez AA, Babalola OO. Fungi that promote plant growth in the rhizosphere boost crop growth. J Fungus. 2023;9(2):239. 10.3390/jof9020239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nimsi KA, Manjusha K, Kathiresan K, Arya H. Plant growth-promoting yeasts (PGPY), the latest entrant for use in sustainable agriculture: a review. J Appl Microbiol. 2023. 10.1093/jambio/lxac088. [DOI] [PubMed] [Google Scholar]
- 13.Botha A. The importance and ecology of yeasts in soil. Soil Biol Biochem. 2011;43(1):1–8. 10.1016/j.soilbio.2010.10.001. [Google Scholar]
- 14.Nascimento FX, Rossi MJ, Soares CR, McConkey BJ, Glick BR. New insights into 1-aminocyclopropane-1-carboxylate (ACC) deaminase phylogeny, evolution and ecological significance. PLoS ONE. 2014;9(6):e99168. 10.1371/journal.pone.0099168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peng X, Wang Y, Tang LJ, et al. Yeasts from Nanfeng mandarin plants: occurrence, diversity and capability to produce indole-3-acetic acid. Biotechnol Biotechnol. 2018;32(6):1496–506. 10.1080/13102818.2018.1487337. [Google Scholar]
- 16.Silambarasan S, Logeswari P, Cornejo P, Kannan VR. Evaluation of the production of exopolysaccharide by plant growth promoting yeast Rhodotorula sp. strain CAH2 under abiotic stress conditions. Int J Biol Macromol. 2019;121:55–62. 10.1016/j.ijbiomac.2018.10.016. [DOI] [PubMed] [Google Scholar]
- 17.Ramya P, Velu G, Parimala D, Dananjeyan B. Isolation and screening of soil yeasts for plant growth promoting traits. Madras Agric J. 2019;106(7/9):439–43. [Google Scholar]
- 18.Streletskii RA, Kachalkin AV, Glushakova AM, Yurkov AM, Demin VV. Yeasts producing zeatin. PeerJ. 2019;7:e6474. 10.7717/peerj.6474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.El-Maraghy SS, Tohamy TA, Hussein KA. Expression of SidD gene and physiological characterization of the rhizosphere plant growth-promoting yeasts. Heliyon. 2020;6:e04384. 10.1016/j.heliyon.2020.e04384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Falih AM, Wainwright M. Nitrification, S-oxidation and P-solubilization by the soil yeast Williopsis californica and by Saccharomyces cerevisiae. Mycol Res. 1995;99(2):200–4. 10.1016/S0953-7562(09)80886-1. [Google Scholar]
- 21.Rosa-Magri MM, Avansini SH, Lopes-Assad ML, Tauk-Tornisielo SM, Ceccato-Antonini SR. Release of potassium from rock powder by the yeast Torulaspora globosa. Braz Arch Biol Technol. 2012;55:577–82. 10.1590/S1516-89132012000400013. [Google Scholar]
- 22.Mohamed HM, El-Homosy RF, Abd-Ellatef AEH, Salh FM, Hussein MY. Identification of yeast strains isolated from agricultural soils for releasing potassium-bearing minerals. Geomicrobiol J. 2017;34(3):261–6. 10.1080/01490451.2016.1186762. [Google Scholar]
- 23.Liu J, Sui Y, Wisniewski M, Droby S, Liu Y. Utilization of antagonistic yeasts to manage postharvest fungal diseases of fruit. Int J Food Microbiol. 2013;167(2):153–60. 10.1016/j.ijfoodmicro.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 24.Ignatova LV, Brazhnikova YV, Berzhanova RZ, Mukasheva TD. Plant growth-promoting and antifungal activity of yeasts from dark chestnut soil. Microbiol Res. 2015;175:78–83. 10.1016/j.micres.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 25.Mannazzu I, Domizio P, Carboni G, Zara S, Zara G, Comitini F, Budroni M, Ciani M. Yeast killer toxins: from ecological significance to application. Crit Rev Biotechnol. 2019;39(5):603–17. 10.1080/07388551.2019.1601679. [DOI] [PubMed] [Google Scholar]
- 26.Arun KD, Sabarinathan KG, Gomathy M, Kannan R, Balachandar D. Mitigation of drought stress in rice crop with plant growth-promoting abiotic stress-tolerant rice phyllosphere bacteria. J Basic Microbiol. 2020;60(9):768–86. 10.1002/jobm.202000011. [DOI] [PubMed] [Google Scholar]
- 27.Devarajan AK, Muthukrishanan G, Truu J, Truu M, Ostonen I, Panneerselvam P, Gopalasubramanian SK. The foliar application of rice phyllosphere bacteria induces drought-stress tolerance in Oryza sativa (L.). Plants. 2021;10(2):387. 10.3390/plants10020387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sedri MH, Niedbała G, Roohi E, Niazian M, Szulc P, Rahmani HA, Feiziasl V. Comparative analysis of plant growth-promoting rhizobacteria (PGPR) and chemical fertilizers on quantitative and qualitative characteristics of rainfed wheat. Agron. 2022;12(7):1524. 10.3390/agronomy12071524. [Google Scholar]
- 29.Devarajan AK, Truu M, Gopalasubramaniam SK, Muthukrishanan G, Truu J. Application of data integration for rice bacterial strain selection by combining their osmotic stress response and plant growth-promoting traits. Front Microbiol. 2022. 10.3389/fmicb.2022.1058772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yadav RKP, Kakamanoli K, Vokou D. Estimating bacterial population on the phyllosphere by serial dilution plating and leaf imprint methods. Ecoprint Int J Ecol. 2010;17:47–52. 10.3126/eco.v17i0.4105. [Google Scholar]
- 31.Harju S, Fedosyuk H, Peterson KR. Rapid isolation of yeast genomic DNA: bust n’Grab. BMC Biotechnol. 2004;4(1):1–6. 10.1186/1472-6750-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):547. 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bric JM, Bostock RM, Silverstone SE. Rapid in situ assay for indoleacetic acid production by bacteria immobilized on a nitrocellulose membrane. AEM. 1991;57(2):535–8. 10.1128/aem.57.2.535-538.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holbrook AA, Edge WJW, Bailey F. spectrophotometric method for determination of gibberellic acid. In: Gould RF, editor. Gibberellins. Washington: Journal of American Chemical Society; 1961. p. 159–67. [Google Scholar]
- 35.Hu X, Chen J, Guo J. Two phosphate-and potassium-solubilizing bacteria isolated from Tianmu Mountain, Zhejiang, China. World J Microbiol Biotechnol. 2006;22(9):983–90. 10.1007/s11274-006-9144-2. [Google Scholar]
- 36.Kesaulya H, Hasinu J, Tuhumury GN. Potential of Bacillus spp. produces siderophores insuppressing the wilt disease of banana plants. In: IOP conference series: earth and environmental science. IOP Pub-lishing, Bristol, England. 2018.
- 37.Ferreira CM, Vilas-Boas Â, Sousa CA, Soares HM, Soares EV. Comparison of five bacterial strains producing siderophores with ability to chelate iron under alkaline conditions. AMB Express. 2019;1:78. 10.1186/s13568-019-0796-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Penrose DM, Glick BR. Methods for isolating and characterizing ACC deaminase-containing plant growth-promoting rhizobacteria. Physiol Plant. 2003;118(1):10–5. 10.1034/j.1399-3054.2003.00086.x. [DOI] [PubMed] [Google Scholar]
- 39.Rabindran R, Vidhyasekaran P. Development of a formulation of Pseudomonas fluorescens PfALR2 for management of rice sheath blight. Crop Prot. 1996;15(8):715–21. 10.1016/S0261-2194(96)00045-2. [Google Scholar]
- 40.Kandasamy S, Loganathan K, Muthuraj R, et al. Understanding the molecular basis of plant growth promotional effect of Pseudomonas fluorescens on rice through protein profiling. Proteome Sci. 2009;7:1–8. 10.1186/1477-5956-7-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang W, Chen LN, Wu H, Zang H, Gao S, Yang Y, Xie S, Gao X. Comparative proteomic analysis of rice seedlings in response to inoculation with Bacillus cereus. Lett Appl Microbiol. 2013;56(3):208–15. 10.1111/lam.12035. [DOI] [PubMed] [Google Scholar]
- 42.Santiago CD, Yagi S, Ijima M, et al. Bacterial compatibility in combined inoculations enhances the growth of potato seedlings. Microbes Environ. 2017;32(1):14–23. 10.1264/jsme2.ME16127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lichtenthaler HK. Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. In: Methods in enzymology. Cambridge: Academic Press; 1987. p. 350–82. [Google Scholar]
- 44.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the folin phenol reagent. J Biol Chem. 1951;193:265–75. 10.1016/S0021-9258(19)52451-6. [PubMed] [Google Scholar]
- 45.Bates LS, Waldren RA, Teare ID. Rapid determination of free proline for water-stress studies. Plant Soil. 1973;39:205–7. 10.1007/BF00018060. [Google Scholar]
- 46.Alici EH, Arabaci G. Determination of SOD, POD, PPO and cat enzyme activities in Rumex obtusifolius L. Annu Res Rev Biol. 2016. 10.9734/ARRB/2016/29809. [Google Scholar]
- 47.Aebi H. Catalase. In: Bergmeyer HU, editor. Methods of enzymatic analysis. NewYork: Academic Press Inc.; 1974. p. 673–80. [Google Scholar]
- 48.Fielding JL, Hall JL. A biolchemical and cytochemical study of peroxidase activity in roots of Pisum sativum: I. a comparison of DAB-peroxidase and guaiacol-peroxidase with particular emphasis on the properties of cell wall activity. J Exp Bot. 1978;29(4):969–81. 10.1093/jxb/29.4.969. [Google Scholar]
- 49.Coseteng MY, Lee CY. Changes in apple polyphenoloxidase and polyphenol concentrations in relation to degree of browning. J Food Sci. 1987;52(4):985–9. [Google Scholar]
- 50.Team R. Core. R: a language and environment for statistical computing. r foundation for statistical computing, Vienna, Austria; 2013. Online: https://www.r-project.org
- 51.Gao CH, Yu G, Cai P. ggVennDiagram: an intuitive, easy-to-use, and highly customizable R package to generate Venn diagram. Front Genet. 2021;12:1598. 10.3389/fgene.2021.706907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wickham H. ggplot2. Wiley Interdiscip Rev Comput Stat. 2011;3(2):180–5. 10.1002/wics.147. [Google Scholar]
- 53.Culhane AC, Thioulouse J, Perrière G, Higgins DG. MADE4: an R package for multivariate analysis of gene expression data. Bioinformatics. 2005;21(11):2789–90. [DOI] [PubMed] [Google Scholar]
- 54.Lê S, Josse J, Husson F. FactoMineR: an R package for multivariate analysis. J Stat Softw. 2008;25(1):1–18. [Google Scholar]
- 55.Camargo A. PCAtest: testing the statistical significance of principal component analysis in R. PeerJ. 2022;10:e12967. 10.7717/peerj.12967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mariette J, Villa-Vialaneix N. Unsupervised multiple kernel learning for heterogeneous data integration. Bioinform. 2018;34(6):1009–15. 10.1093/bioinformatics/btx682. [DOI] [PubMed] [Google Scholar]
- 57.Meng C, Kuster B, Culhane AC, Gholami AM. A multivariate approach to the integration of multi-omics datasets. BMC Bioinform. 2014;15:1–13. 10.1186/1471-2105-15-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schindler D. Genetic engineering and synthetic genomics in yeast to understand life and boost biotechnology. Bioeng. 2020;4:137. 10.3390/bioengineering7040137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goffeau AEA, Aert R, Agostini-Carbone ML, et al. The yeast genome directory. Nature. 1997;387(6632):5–6. 10.1038/38784. [Google Scholar]
- 60.Stovicek V, Holkenbrink C, Borodina I. CRISPR/Cas system for yeast genome engineering: advances and applications. FEMS Yeast Res. 2017. 10.1093/femsyr/fox030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gouka L, Raaijmakers JM, Cordovez V. Ecology and functional potential of phyllosphere yeasts. Trends Plant Sci. 2022;22(11):1109–23. 10.1016/j.tplants.2022.06.007. [DOI] [PubMed] [Google Scholar]
- 62.Limtong S, Kaewwichian R. The diversity of culturable yeasts in the phylloplane of rice in Thailand. Ann Microbiol. 2015;65:667–75. 10.1007/s13213-014-0905-0. [DOI] [PubMed] [Google Scholar]
- 63.Glushakova AM, Chernov IY. Seasonal dynamics of the structure of epiphytic yeast communities. Microbiol. 2010;79:830–9. 10.1134/S0026261710060160. [PubMed] [Google Scholar]
- 64.Lee G, Lee SH, Kim KM, Ryu CM. Foliar application of the leaf-colonizing yeast Pseudozyma churashimaensis elicits systemic defense of pepper against bacterial and viral pathogens. Sci Rep. 2017;7(1):1–13. 10.1038/srep39432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Santhanam P, Labbé C, Fietto LG, Bélanger RR. A reassessment of flocculosinmediated biocontrol activity of Pseudozyma flocculosa through CRISPR/Cas9 gene editing. Fungal Genet Biol. 2021;153:03573. 10.1016/j.fgb.2021.103573. [DOI] [PubMed] [Google Scholar]
- 66.Srisuk N, Nutaratat P, Surussawadee J, Limtong S. Yeast communities in sugarcane phylloplane. Microbiology. 2019;88:353–69. 10.1134/S0026261719030135. [Google Scholar]
- 67.Mannazzu I, Landolfo S, Da Silva TL, Buzzini P. Red yeasts and carotenoid production: outlining a future for non-conventional yeasts of biotechnological interest. World J Microbiol Biotechnol. 2015;31:1665–73. 10.1007/s11274-015-1927-x. [DOI] [PubMed] [Google Scholar]
- 68.Beattie GA, Lindow SE. Bacterial colonization of leaves: a spectrum of strategies. Phytopathology. 1999;89(5):353–9. 10.1094/PHYTO.1999.89.5.353. [DOI] [PubMed] [Google Scholar]
- 69.Davies PJ. The plant hormones: their nature, occurrence, and functions. In: Davies PJ, editor. Plant hormones. Dordrecht: Springer; 2010. [Google Scholar]
- 70.Jha Y. Plant microbiomes with phytohormones: attribute for plant growth and adaptation under the stress conditions. Advances in Plant Microbiome and Sustainable Agriculture: Functional Annotation and Future Challenges. 2020;85–103.
- 71.Limtong S, Koowadjanakul N. Yeasts from phylloplane and their capability to produce indole-3-acetic acid. World J Microbiol Biotechnol. 2012;28:3323–35. 10.1007/s11274-012-1144-9. [DOI] [PubMed] [Google Scholar]
- 72.Nutaratat P, Amsri W, Srisuk N, Arunrattiyakorn P, Limtong S. Indole-3-acetic acid production by newly isolated red yeast Rhodosporidium paludigenum. J Gen Appl Microbiol. 2015;61(1):1–9. 10.2323/jgam.61.1. [DOI] [PubMed] [Google Scholar]
- 73.Nassar AH, El-Tarabily KA, Sivasithamparam K. Promotion of plant growth by an auxin-producing isolate of the yeast Williopsis saturnus endophytic in maize (Zea mays L.) roots. Biol Fertil Soils. 2005;42:97–108. 10.1007/s00374-005-0008-y. [Google Scholar]
- 74.Devi R, Kaur T, Kour D, Yadav A, Yadav AN, Suman A, Ahluwalia AS, Saxena AK. Minerals solubilizing and mobilizing microbiomes: a sustainable approach for managing minerals deficiency in agricultural soil. J Appl Microbiol. 2022;133(3):1245–72. 10.1111/jam.15627. [DOI] [PubMed] [Google Scholar]
- 75.Fu SF, Sun PF, Lu HY, Wei JY, Xiao HS, Fang WT, Cheng BY, Chou JY. Plant growth-promoting traits of yeasts isolated from the phyllosphere and rhizosphere of Drosera spatulata Lab. Fungal Biol. 2016;120(3):433–48. 10.1016/j.funbio.2015.12.006. [DOI] [PubMed] [Google Scholar]
- 76.Ahmed E, Holmström SJ. Siderophores in environmental research: roles and applications. Microb Biotechnol. 2014;7(3):196–208. 10.1111/1751-7915.12117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Calvente V, de Orellano ME, Sansone G, Benuzzi D, de Tosetti MI. A simple agar plate assay for screening siderophore producer yeasts. J Microbiol Meth. 2001;47(3):273–9. 10.1016/S0167-7012(01)00333-5. [DOI] [PubMed] [Google Scholar]
- 78.Glick BR, Todorovic B, Czarny J, Cheng Z, Duan J, McConkey B. Promotion of plant growth by bacterial ACC deaminase. Crit Rev Plant Sci. 2007;26(5–6):227–42. 10.1080/07352680701572966. [Google Scholar]
- 79.Bright JP, Karunanadham K, Maheshwari HS, et al. Seed-borne probiotic yeasts foster plant growth and elicit health protection in black gram (Vigna mungo L.). Sustainability. 2022;14(8):4618. 10.3390/su14084618. [Google Scholar]
- 80.Liu W, Wang B, Wang Q, Hou J, Wu L, Wood JL, Luo Y, Franks AE. Characteristics of metal-tolerant plant growth-promoting yeast (Cryptococcus sp. NSE1) and its influence on Cd hyperaccumulator Sedum plumbizincicola. Environ Sci Pollut Res. 2016;23:18621–9. 10.1007/s11356-016-7041-2. [DOI] [PubMed] [Google Scholar]
- 81.Almeida F, Rodrigues ML, Coelho C. The still underestimated problem of fungal diseases worldwide. Front Microbiol. 2019;10:214. 10.3389/fmicb.2019.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Morath SU, Hung R, Bennett JW. Fungal volatile organic compounds: a review with emphasis on their biotechnological potential. Fungal Biol Rev. 2012;26(2–3):73–83. 10.1016/j.fbr.2012.07.001. [Google Scholar]
- 83.Kalogiannis S, Tjamos SE, Stergiou A, Antoniou PP, Ziogas BN, Tjamos EC. Selection and evaluation of phyllosphere yeasts as biocontrol agents against grey mould of tomato. Eur J Plant Pathol. 2006;116:69–76. 10.1007/s10658-006-9040-5. [Google Scholar]
- 84.Di Canito A, Mateo-Vargas MA, Mazzieri M, Cantoral J, Foschino R, Cordero-Bueso G, Vigentini I. The role of yeasts as biocontrol agents for pathogenic fungi on postharvest grapes: a review. Foods. 2021;10(7):1650. 10.3390/foods10071650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Calvente V, Benuzzi D, de Tosetti MS. Antagonistic action of siderophores from Rhodotorula glutinis upon the postharvest pathogen Penicillium expansum. Int Biodeterior Biodegrad. 1999;43(4):167–72. 10.1016/S0964-8305(99)00046-3. [Google Scholar]
- 86.Ge L, Zhang H, Chen K, Ma L, Xu Z. Effect of chitin on the antagonistic activity of Rhodotorula glutinis against Botrytis cinerea in strawberries and the possible mechanisms involved. Food Chem. 2010;120(2):490–5. 10.1016/j.foodchem.2009.10.042. [Google Scholar]
- 87.Zhang X, Li B, Zhang Z, Chen Y, Tian S. Antagonistic yeasts: a promising alternative to chemical fungicides for controlling postharvest decay of fruit. J Fungi. 2020;6(3):158. 10.3390/jof6030158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Agler MT, Ruhe J, Kroll S, Morhenn C, Kim ST, Weigel D, Kemen EM. Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLoS Biol. 2016;14:e1002352. 10.1371/journal.pbio.1002352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tabacchioni S, Passato S, Ambrosino P, et al. Identification of beneficial microbial consortia and bioactive compounds with potential as plant biostimulants for a sustainable agriculture. Microorganisms. 2021;9(2):426. 10.3390/microorganisms9020426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Saini I, Kaushik P, Al-Huqail AA, Khan F, Siddiqui MH. Effect of the diverse combinations of useful microbes and chemical fertilizers on important traits of potato. Saudi J Biol Sci. 2021;28(5):2641–8. 10.1016/j.sjbs.2021.02.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zainuddin N, Keni MF, Ibrahim SAS, Masri MMM. Effect of integrated biofertilizers with chemical fertilizers on the oil palm growth and soil microbial diversity. Biocatal Agric Biotechnol. 2022;39:102237. 10.1016/j.bcab.2021.102237. [Google Scholar]
- 92.Mashabela MD, Tugizimana F, Steenkamp PA, Piater LA, Dubery IA, Mhlongo MI. Untargeted metabolite profiling to elucidate rhizosphere and leaf metabolome changes of wheat cultivars (Triticum aestivum L.) treated with the plant growth-promoting rhizobacteria Paenibacillus alvei (T22) and Bacillus subtilis. Front Microbiol. 2022. 10.3389/fmicb.2022.971836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jadhav RK. Yeast utilizing deproteinised leaf juice (DPJ) as a medium for growth and production of metabolites. Plant Arch. 2018;18(2):1716–20. [Google Scholar]
- 94.Chen H, Chen F. Effects of yeast elicitor on the growth and secondary metabolism of a high-tanshinone-producing line of the Ti transformed Salvia miltiorrhiza cells in suspension culture. Process Biochem. 2000;35(8):837–40. 10.1016/S0032-9592(99)00146-6. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed during the current study.