Abstract
Purpose of review
With growing emphasis on data-driven research in pediatric oncology, particularly in the context of advances in molecular characterization and precision medicine, there is an urgent need for comprehensive data-sharing initiatives. This review explores how the Childhood Cancer Data Initiative (CCDI) addresses this critical need.
Recent findings
CCDI plays a key role in enhancing pediatric cancer research by improving data integration, sharing, and collaboration. Its Molecular Characterization Initiative advances the field by leveraging detailed molecular data to inform clinical trials and therapeutic strategies. For small patient populations, such as those with rhabdomyosarcoma, CCDI's efforts in integrating data across institutions are vital for advancing risk-based treatment strategies to achieve meaningful clinical outcomes.
Summary
CCDI's advancements in data sharing have profound implications for both clinical practice and research. By enabling precise diagnoses, tailoring treatments based on individual genetic profiles, and addressing the challenges associated with small patient populations, CCDI is driving transformative changes in pediatric oncology. Continued support and expansion of such initiatives are crucial to fully realizing their potential in improving outcomes for children with cancer.
Keywords: childhood cancer data initiative, data sharing, molecular characterization, pediatric oncology, precision medicine
INTRODUCTION
Cancers in children, adolescents, and young adults (AYAs) represent unique challenges in oncology. Despite the marked improvement in outcomes for children with cancer over the past five decades, cancer remains the leading cause of death from disease. Also, the quality of survival for more than 30% of successfully treated children is painfully diminished by debilitating and potentially life-threatening persistent and/or late effects of conventional treatments [1▪]. While certain cancers in children and AYAs (the patient group extending to age 39 who have been diagnosed with a pediatric cancer) are now curable, survival rates vary widely depending on the specific cancer diagnosis and extent of disease, with some cancers, including some brain tumors and metastatic sarcomas still having very poor prognoses [2,3]. Survivors of childhood and AYA cancers require long-term follow-up care due to the risk of late effects from cancer treatments, which can include secondary cancers, cardiovascular complications, and impaired growth and development [4].
Causes of childhood cancers are multifaceted, involving both genetic and environmental factors. Approximately 8–10% of childhood cancers are linked to inherited genetic mutations, such as those seen in retinoblastoma and other familial cancer syndromes [5]. Environmental exposures may also contribute, but identifying specific environmental causes is challenging due to the rarity and complexity of these cancers in contrast to the known association of lifestyle-linked exposures and association with many adult cancers [6▪,7]. Lack of full understanding of risk factors for children with cancer, other than known genetic predisposition syndromes, makes both screening and prevention strategies difficult to develop. Screening and prevention have had a substantial impact on reducing adult cancer incidence and mortality given the well documented, over many years, association between certain lifestyle-linked exposures and risk of many common adult cancers [8].
Despite the very collaborative nature of pediatric oncology research, data from clinical trials have not been easily or timely accessible for secondary research purposes. Several reports highlight the critical need for ‘big data’ and improved opportunities for broader access to data for new analyses, which could lead to significant discoveries in pediatric healthcare, particularly in cancer [9,10▪–12▪,13,14].
A specific call for the sharing of genomic data from children with cancer has been made to decrease the global burden of cancer [15]. The Childhood Cancer Data Initiative (CCDI) is a significant effort aimed at addressing this need [16▪,17▪]. By facilitating the sharing and harmonization of genomic data, CCDI seeks to accelerate research and improve clinical outcomes for pediatric cancer patients. This initiative is critical to advancing our understanding of pediatric cancers, identifying new therapeutic targets, and ultimately improving patient care. Through the CCDI Data Ecosystem, we can make substantial progress toward these goals, highlighting the need for continued support and innovation. This overview underscores the importance of ongoing investment in pediatric oncology research and clinical care to achieve better outcomes and enhance the quality of life for children and AYAs affected by cancer.
Box 1.
no caption available
NAVIGATING CHALLENGES: SMALL PATIENT POPULATIONS IN PEDIATRIC CANCER RESEARCH
A significant challenge in pediatric cancer research is the relatively small patient population, which profoundly impacts clinical trials and treatment, particularly new drug development. Rhabdomyosarcoma, affecting approximately 400 patients annually in the United States, exemplifies this issue [18,19▪]. Patients are subclassified now based on factors like stage, extent of disease, primary tumor site, and molecular characteristics, necessitating high participation rates in clinical trials for timely meaningful results [20]. This requirement mandates multiinstitutional collaborations, often extending across continents. The small patient population also means there is no free-market incentive for conducting or supporting the infrastructure for clinical trials designed to demonstrate the safety and effectiveness of new drugs, which could lead to regulatory approval [21].
The Research to Accelerate Cures and Equity (RACE) for Children Act addresses several challenges in pediatric cancer drug development. It mandates that new adult cancer drugs be evaluated in children earlier in development when the drug's molecular target may be relevant to one or more pediatric cancers. This legislation ensures that promising therapies developed for adults are also considered for children when appropriate, helping to bridge the gap between adult and pediatric cancer research. By requiring pharmaceutical companies to include preliminary pediatric assessments early in their drug development process, the RACE Act supports translational and clinical investigators in exploring new treatment strategies for pediatric cancers [22]. This complements the National Cancer Institute (NCI)'s efforts to advance research and improve patient outcomes, even in areas with limited market incentives.
GENETIC MUTATIONS AND ABNORMALITIES IN PEDIATRIC CANCERS
A critical advancement in modern oncology is the rise of precision medicine, made possible by molecular characterization of tumors at diagnosis and relapse to identify somatic aberrations when compared to germline, which has led to the development of numerous molecularly targeted agents [23,24]. The widespread application of molecular characterization as part of routine diagnostic evaluation of cancers in children has been delayed, causing subsequent delays in precision medicine approaches for children with cancer.
However, research teams globally have made significant strides over the past decade in uncovering the genomic landscapes of childhood cancers that may contribute to causation, influence disease progression, and impact treatment outcomes [25▪▪]. Initially hopeful that actionable oncogenes like activated tyrosine kinases would be prevalent, it is now evident that childhood cancer genomics are diverse and often distinct from adult cancers. Key genomic abnormalities, such as NPM-ALK fusions in anaplastic large cell lymphoma and BRAF alterations in pediatric low-grade gliomas, have provided immediate therapeutic insights [26].
Integrating molecular insights into clinical practice aims to tailor treatments based on individual genomic profiles, improving long-term outcomes for pediatric cancer patients [5,25▪▪,27]. Continued research is essential to further elucidate the molecular mechanisms underlying childhood cancers and develop innovative therapeutic strategies directed at improving efficacy while minimizing long-term adverse effects.
BUILDING A COLLABORATIVE COMMUNITY FOR ADVANCING CHILDHOOD CANCER RESEARCH
CCDI is an NCI initiative supported by a $50 million special Congressional appropriation passed in 2020, with an additional $50 million proposed each fiscal year for a total of 10 years. These funds allow NCI to build a community centered around childhood cancer care and research data. This funding supports a variety of projects to advance pediatric cancer research. It includes strategic investments in molecular and germline characterization of tumors, including rare cancers, as well as the development of preclinical models like patient-derived xenografts and organoids for use in translational medicine. The funding also facilitates data sharing and collaboration by integrating clinical, research, and registry data, employing federated data models, and providing visualization tools to enhance data accessibility and utility.
As part of CCDI's goals, comprehensive-omics characterization will offer a unique opportunity to understand the genomic landscapes of a diverse representation of cancers, creating a high-value resource for the cancer research community.
CHILDHOOD CANCER DATA INITIATIVE'S MOLECULAR CHARACTERIZATION INITIATIVE: A FLAGSHIP PROGRAM IN COLLABORATION WITH THE CHILDREN'S ONCOLOGY GROUP
Children and AYAs with cancer are typically treated at specialized children's cancer centers, where multidisciplinary teams provide comprehensive care tailored to young patients’ unique needs. These centers often participate in clinical trials through networks like the Children's Oncology Group (COG), facilitating access to cutting-edge treatments and improving outcomes through evidence-based research. COG, a component of NCI's National Clinical Trials Network (NCTN), operates across more than 200 sites in North America, Australia, and New Zealand. Annually, COG enrolls over 10 000 patients into research protocols, encompassing both first-line clinical trials and myriad nontherapeutic studies, including epidemiological studies. The NCTN Operations, Biopathology Center, and Statistical Data Management Core are collaborating to execute these protocols and several other clinical trials. Specimens from NCI-sponsored, phase 3 clinical trials are available through the NCTN Navigator online request system. Other NCI-supported specimens from earlier-phase COG clinical trials and other COG research protocols are available through a ‘direct-to-COG’ request process.
CCDI is sponsoring the Molecular Characterization Initiative (MCI), a program that currently characterizes molecular features of central nervous system (CNS) tumors, soft tissue sarcomas, rare cancers, and high-risk neuroblastoma (NBL) in pediatric and AYA patients. The goal of this initiative is to enhance understanding of genetic factors in pediatric cancers and to provide timely, clinically relevant findings at no cost to patients and families, aiding in treatment decisions. Nearly 66% of participants were diagnosed with CNS tumors, 20% with soft tissue sarcomas, 9% with rare tumors, 4% NBL, and 1% other tumor types. So far, nearly 4500 participants have been enrolled.
MCI currently uses COG's Project:EveryChild (APEC14B1) protocol for enrolling participants, collecting specimens, and annotating clinical information. APEC14B1 serves as a comprehensive study encompassing registry, eligibility screening, molecular characterization, biology, and outcome assessment for childhood cancer. This will also determine eligibility for COG clinical trials. The Childhood Cancer Survivorship, Treatment, Access, and Research (STAR) Act aims to advance research through support of biorepositories and research in cancer survivorship. STAR Act funding supports the critical infrastructure and resources necessary for MCI, including acquisition and management of biospecimens and ensuring timely processing for nucleic acid extraction, distribution of samples, and residual material banking.
MCI supports the tests which are conducted in a Clinical Laboratory Improvement Amendments (CLIA)-certified environment that allows for return of results to physicians, and patients, namely, enhanced paired tumor-normal exome sequencing, a targeted RNA solid tumor fusion assay (Archer FUSIONPlex Pan Solid Tumor panel), and a DNA-based methylation array assay for CNS tumor classification. Results from these tests are returned within 21 days of receipt of all required materials at Nationwide Children's Hospital's Steve and Cindy Rasmussen Institute for Genomic Medicine, and the Biopathology Center. Separately, molecular characterization data and deidentified clinical reports are submitted to the CCDI Data Ecosystem, along with additional data encompassing demographics, diagnosis, treatment, and follow-up directly from COG. See the instructions on how to access MCI data (accession number phs002790). For a preview of MCI data prior to going through the data authorization process, visit MCI's page in the CCDI Childhood Cancer Data Catalog.
FOSTERING DIVERSITY AND EQUITY IN PEDIATRIC, ADOLESCENT, AND YOUNG ADULT CANCER RESEARCH
CCDI is committed to enhancing diversity and equity by extending MCI to non-COG sites and will do so, in part, through the Coordinated Pediatric, Adolescent, and Young Adult Rare Cancer Initiative (CPAYARCI). This initiative focuses on pediatric and AYA patients with very rare cancers, excluding common adult cancers not typically found in children. This is an observational study aimed at collecting structured and real-world data, building a registry using standardized clinical and genomic data to identify genomic vulnerabilities that might inform drug discovery and development and future interventional trials, and potentially serving as a source for fit for purpose real-world data the construction of external controls.
By including non-COG sites, CCDI seeks to ensure a more diverse and representative sample of pediatric and AYA cancer patients. This expansion addresses disparities in cancer research and treatment, ultimately leading to better outcomes for all patients. NCI's National Community Oncology Research Program also helps expand MCI's reach by enrolling patients in rural areas with limited access to COG institutions and in areas with large minority populations. CPAYARCI will further expand MCI's reach to the young adult population (up to age 39).
CHILDHOOD CANCER DATA INITIATIVE ECOSYSTEM: ADVANCING PEDIATRIC CANCER RESEARCH THROUGH DATA INTEGRATION, VISUALIZATION, AND ACCESSIBILITY
The CCDI Data Ecosystem is a network of tools and resources designed to integrate and harmonize data from clinical, research, and registry sources. Its primary goal is to enhance data collection, sharing, and analysis by consolidating disparate data repositories, ultimately aiming to improve patient outcomes.
DATA INTEGRATION AND HARMONIZATION
Harmonization will enhance the utility of CCDI data by providing standardized outputs, eliminating the need for users to independently download and process large, raw genomic files. This will enable high-throughput and scalable analyses, driving cost savings and increasing research efficiency. Researchers will benefit from immediate access to well curated, annotated data, allowing for cohesive integration and data analysis from diverse sources without extensive preprocessing. Comprehensive data harmonization could foster a collaborative research environment that accelerates scientific breakthroughs in pediatric oncology.
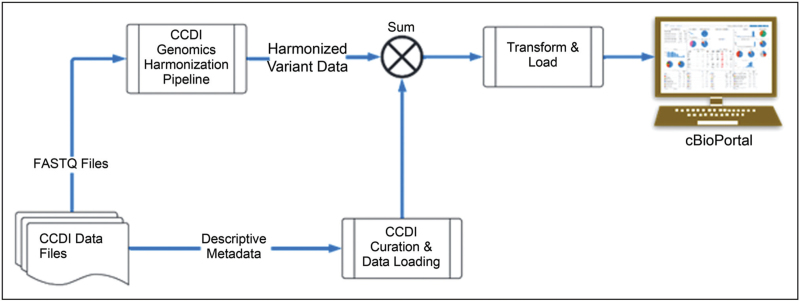
The CCDI Data Ecosystem includes data from multiple NCI pediatric programs, each potentially using different protocols and platforms. Genomic data collected from these projects are stored as static files within NCI's Cancer Research Data Commons. Due to the diverse origins and formats of these data, integrating these data sets presents challenges and requires postharmonization to ensure consistency, accuracy, and interoperability. Initial efforts have focused on collecting data and carefully curating and harmonizing descriptive metadata, including demographic and clinical phenotypic information. The harmonized clinical data are available through the Childhood Cancer Clinical Data Commons. CCDI will focus on harmonizing genomic data to align with common standards and making the unified data set accessible via visualization tools (Fig. 1). Aligning data processing pipelines and implementing quality control measures could significantly improve the reliability of genomic data analysis and interpretation.
FIGURE 1.
CCDI Data Ecosystem Data Integration, Harmonization, Visualization & Analysis. The figure is a high-level illustration of how CCDI data are harmonized and made available for analysis and visualization through tools like cBioPortal.
DATA VISUALIZATION AND ACCESSIBILITY
Genomic data visualization tools are essential for both basic scientists and clinicians. For basic science researchers, these tools facilitate the exploration of genetic variations, gene expression patterns, and genomic structures, aiding in the discovery of new genes and pathways relevant to specific diseases. For clinicians, visualizing genomic data in a clear and interactive manner enhances their understanding of underlying genetic factors contributing to a patient's condition, leading to more accurate diagnoses and tailored treatment plans.
To support these needs, CCDI is developing a data visualization capability using cBioPortal. This open-source platform is designed for the interactive exploration of multidimensional cancer genomics data sets. cBioPortal aims to simplify access to complex genomic data for cancer researchers by providing intuitive, easy access to molecular profiles and clinical attributes from large-scale cancer genomics projects. Users can explore specific genes or pathways across various cancer types, including querying mutations, copy number alterations, mRNA expression, and clinical data. CCDI plans to establish a bespoke instance of cBioPortal, ensuring seamless integration of harmonized CCDI data and compatibility with CCDI standards, particularly for clinical data. This tailored instance will further enhance the accessibility and utility of the data, supporting the broader goals of the initiative.
CHALLENGES AND FUTURE DIRECTIONS
Retrospective harmonization presents significant challenges compared to prospective efforts, due to the complexity of aligning data sets across different versions. For example, primary DNA sequence data from various projects exist in different forms, such as unaligned reads or reads aligned to different genome reference standards. Setting standards and providing clear guidelines for harmonizing CCDI data is a crucial milestone, as it establishes a robust foundation for future efforts.
CONCLUSION
CCDI's efforts aim to create unified, high-quality federated data sets that integrate clinical, genomic, transcriptomic, and epigenomic data from multiple pediatric cancer projects. Given the rarity of childhood cancer, the ability to analyze large, harmonized data sets is crucial for significant discoveries to drive advancements in the field and beyond. The long-term vision is to enhance data sharing, accessibility, and usability in a way that is most broadly beneficial. CCDI could serve as an example for how the larger adult research and participant communities can maximize their data value to benefit cancer research. By harmonizing diverse data types into a consistent and compatible format, the CCDI Data Ecosystem facilitates research that can uncover new disease mechanisms, identify genetic markers of cancer predisposition and molecular vulnerabilities to support targeted drug development, and predict patient outcomes.
Acknowledgements
The efforts described in this article are fully or partially funded by NCI's CCDI. The authors gratefully acknowledge Samantha Gonzalez and Rena Kingery for their assistance with editing and proofreading, and Mark Cunningham for his help in preparing the figure for this manuscript.
Financial support and sponsorship
None.
Conflicts of interest
None.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual period of review, have been highlighted as:
▪ of special interest
▪▪ of outstanding interest
REFERENCES
- 1▪.Siegle RL, Giaquinto AN, Jemal A. Cancer statistics, 2024. CA Cancer J Clin 2024; 74:12–49. [DOI] [PubMed] [Google Scholar]; These 2024 statistics from the American Cancer Society show that leukemia remains the most common childhood cancer, making up 28% of cases, followed by brain and other nervous system tumors. Despite reductions in mortality rates for childhood cancers, particularly leukemia, progress in adolescents has lagged due to differences in tumor biology, clinical trial enrollment, and treatment protocols, tolerance, and compliance.
- 2. American Cancer Society. Key statistics for childhood cancers. 2024. https://www.cancer.org/cancer/types/cancer-in-children/key-statistics.html. [Accessed 4 September 2024.] [Google Scholar]
- 3. National Cancer Institute Surveillance, Epidemiology, and End Results Program. Cancer Statistics Explorer Network. https://seer.cancer.gov/statistics-network/. [Accessed 4 September 2024]. in press. [Google Scholar]
- 4. Children's Oncology Group Long-Term Follow-Up Guidelines for Survivors of Childhood, Adolescent and Young Adult Cancers, Version 6.0. 2023. http://www.survivorshipguidelines.org/. [Accessed 4 September 2024.] [Google Scholar]
- 5.Zhang J, Walsh MF, Wu G, et al. Germline mutations in predisposition genes in pediatric cancer. N Engl J Med 2015; 373:2336–2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6▪.Siegel DA, King JB, Lupo PJ, et al. Counts, incidence rates, and trends of pediatric cancer in the United States, 2003–2019. J Natl Cancer Inst 2023; 115:1337–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study estimated recent trends in pediatric cancer using US Cancer Statistics, which analyzes registry data from all 50 states and the District of Columbia. The study found an overall increase in pediatric cancer incidence rates from 2003 to 2019, with specific increases in leukemia, lymphoma, hepatic tumors, bone tumors, and thyroid carcinomas, while melanoma rates decreased. The highest incidence rates were found in men, children aged 0–4 and 15–19 years, white children and adolescents, and those in the Northeast census region. These findings could guide future public health and research priorities for pediatric cancers.
- 7.Shakeel O, Lupo PJ, Strong S, et al. A brief review of the current knowledge on environmental toxicants and risk of pediatric cancers. Pediatr Hematol Oncol 2022; 39:193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robison LL, Hudson MM. Survivors of childhood and adolescent cancer: life-long risks and responsibilities. Nat Rev Cancer 2014; 14:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benedetti DJ, Marron JM. Ethical challenges in pediatric oncology care and clinical trials. Recent Results Cancer Res 2021; 218:149–173. [DOI] [PubMed] [Google Scholar]
- 10▪.Narang C, Ouvina M, Rees CA, Bourgeois FT. Data sharing for pediatric clinical trials funded by the U.S. National Institutes of Health. JAMA Network Open 2023; 6:e2325342. [DOI] [PMC free article] [PubMed] [Google Scholar]; This cross-sectional study highlights the importance of NIH's data management and sharing policy, effective January 25, 2023, which requires data management and sharing plans in grant applications for NIH funding. This study analyzed 213 pediatric clinical trials with results published prior to June 2022. Only 29.1% of studies declared that data were available, and the majority required researchers to request data directly from authors rather than providing direct access to readers. The study provides a benchmark for assessing changes in data sharing practices under the new policy.
- 11▪.Helms L, Guimera AE, Janeway KA, Bailey KM. Innovations in cancer treatment of children. Pediatrics 2023; 152:e2023061539. [DOI] [PMC free article] [PubMed] [Google Scholar]; This review of recent pediatric oncology literature discusses recent advances and areas for continued improvement in reducing toxicity of treatments, understanding cancer biology, developing novel therapies, detecting and monitoring childhood cancers, and improving access to care.
- 12▪.Vesoulis ZA, Hussain AN, Cole FS. Improving child health through Big Data and data science. Pediatr Res 2023; 93:342–349. [DOI] [PMC free article] [PubMed] [Google Scholar]; This review of recent literature suggests that applying Big Data and data science to child health questions could improve child health outcomes and might also help identify previously unrecognized patterns associated with childhood diseases. The review also provides examples of existing pediatric Big Data initiatives and identifies areas of future research.
- 13.Goulooze SC. Beyond the randomized clinical trial: innovative data science to close the pediatric evidence gap. Clin Pharm Ther 2020; 107:786–795. [DOI] [PubMed] [Google Scholar]
- 14.Major A, Cox SM, Volchenboum SL. Using big data in pediatric oncology applications and future directions. Semin Oncol 2020; 47:56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vaske OM, Hausler D. Data sharing for pediatric cancers. Science 2019; 363:1125. [DOI] [PubMed] [Google Scholar]
- 16▪.Flores-Toro JA, Jagu S, Armstrong GT, et al. The Childhood Cancer Data Initiative: using the power of data to learn from and improve outcomes for every child and young adult with pediatric cancer. J Clin Oncol 2023; 41:4045–4053. [DOI] [PMC free article] [PubMed] [Google Scholar]; The study discusses the CCDI, which aims to accelerate treatment development and improve outcomes for children with cancer through enhanced data sharing and analysis. The authors describe CCDI as a community-based initiative that will expand on existing projects and tools, while building a data ecosystem that will facilitate learning from every child and AYA with a pediatric cancer. They also introduce the CCDI MCI, a program that provides molecular testing for newly diagnosed patients, helping to generate data and guide treatment for children with cancer.
- 17▪.Jagu S, Mardis ER, Wedekind MF, et al. Childhood Cancer Data Initiative: status report. Pediatr Blood Cancer 2024; 71:e30745. [DOI] [PubMed] [Google Scholar]; This study provides an update on the CCDI. The authors describe key developments such as platforms and tools available through the CCDI Data Ecosystem, plans to connect data from multiple sources using a digital ID mapping and matching service, expansion of the MCI, and efforts to establish computable consent and a national initiative for studying rare cancers in children, adolescents, and young adults.
- 18.Ward ZJ, Yeh JM, Bhakta N, et al. Estimating the total incidence of global childhood cancer: a simulation-based analysis. Lancet Oncol 2020; 21:483–497. [DOI] [PubMed] [Google Scholar]
- 19▪.Zarrabi A, Perrin D, Kavoosi M, et al. Rhabdomyosarcoma: current therapy, challenges, and future approaches to treatment strategies. Cancers (Basel) 2023; 15:5269. [DOI] [PMC free article] [PubMed] [Google Scholar]; This review discusses the current treatment options for rhabdomyosarcoma (RMS) and the need for less toxic therapies. The authors highlight challenges in treating RMS, including drug resistance, tumor recurrence, and long-term side effects in survivors. The authors also discuss preclinical studies that investigate potential targeted therapies based on molecular characterization of tumors and tissue engineering techniques.
- 20.Weigel BJ, Lyden E, Anderson JR, et al. Intensive multiagent therapy, including dose-compressed cycles, with or without dose-extended maintenance therapy, for patients with high-risk embryonal rhabdomyosarcoma: a report from the Children's Oncology Group. J Clin Oncol 2016; 34:117–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laetsch TW, DuBois SG, Bender JG, et al. Opportunities and challenges in drug development for pediatric cancers. Cancer Discov 2021; 11:545–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reaman G, Karres D, Ligas F, et al. Accelerating the global development of pediatric cancer drugs: a call to coordinate the submissions of pediatric investigation plans and pediatric study plans to the European Medicines Agency and US Food and Drug Administration. J Clin Oncol 2020; 38:4227–4230. [DOI] [PubMed] [Google Scholar]
- 23.Mody RJ, Prensner JR, Everett J, et al. Precision medicine in pediatric oncology: lessons learned and next steps. Pediatr Blood Cancer 2017; 64:e26288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parsons DW, Janeway KA, Patton DR, et al. Actionable tumor alterations and treatment protocol enrollment of pediatric and young adult patients with refractory cancers in the National Cancer Institute-Children's Oncology Group Pediatric MATCH Trial. J Clin Oncol 2022; 40:2224–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25▪▪.Kolekar P, Balagopal V, Dong L, et al. SJPedPanel: a pan-cancer gene panel for childhood malignancies to enhance cancer monitoring and early detection. Clin Cancer Res 2024; 30:4100–4114. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study introduces and validates the SJPedPanel, a pan-cancer gene panel designed for sequencing and detecting clinically relevant genomic alterations in a wide range of childhood cancer samples. SJPedPanel shows superior coverage of genes relevant to pediatric cancers compared to existing panels. The authors validated the panel's effectiveness through in silico analysis of a real-time clinical genomics cohort, showing its potential in detecting rare variants and monitoring disease for relapse. This tool could significantly enhance diagnostic capabilities in pediatric oncology.
- 26.Krumbholz M, Woessmann W, Zierk J, et al. Characterization and diagnostic application of genomic NPM-ALK fusion sequences in anaplastic large-cell lymphoma. Oncotarget 2018; 9:26543–26555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Villani A, Davidson S, Kanwar N, et al. The clinical utility of integrative genomics in childhood cancer extends beyond targetable mutations. Nat Cancer 2023; 4:203–221. [DOI] [PMC free article] [PubMed] [Google Scholar]