Abstract
Background
Long-read technologies from Pacific Biosciences (PacBio) and Oxford Nanopore Technologies (ONT) have transformed genomics research by providing diverse data types like HiFi, Duplex, and ultra-long ONT. Despite recent strides in achieving haplotype-phased gapless genome assemblies using long-read technologies, concerns persist regarding the representation of genetic diversity, prompting the development of pangenome references. However, pangenome studies face challenges related to data types, volumes, and cost considerations for each assembled genome, while striving to maintain sensitivity. The absence of comprehensive guidance on optimal data selection exacerbates these challenges.
Results
Our study evaluates recommended data types and volumes required to establish a robust de novo genome assembly pipeline for population-level pangenome projects, extensively examining performance between ONT’s Duplex and PacBio HiFi datasets in the context of achieving high-quality phased genomes with enhanced contiguity and completeness. The results show that achieving chromosome-level haplotype-resolved assembly requires 20 × high-quality long reads such as PacBio HiFi or ONT Duplex, combined with 15–20 × of ultra-long ONT per haplotype and 10 × of long-range data such as Omni-C or Hi-C. High-quality long reads from both platforms yield assemblies with comparable contiguity, with HiFi excelling in phasing accuracies, while Duplex generates more T2T contigs.
Conclusion
Our study provides insights into optimal data types and volumes for robust de novo genome assembly in population-level pangenome projects. Reassessing the recommended data types and volumes in this study and aligning them with practical economic limitations are vital to the pangenome research community, contributing to their efforts and pushing genomic studies with broader impacts.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13059-024-03452-y.
Keywords: LRS special issue, Pangenome, De novo assembly, Sequencing platforms, Population-level studies
Background
A high-quality and complete human reference genome is the fundamental bedrock supporting genetic studies of human diseases and population structures. Over the past two decades, the human reference genome employed in genetic studies has been meticulously crafted from genomic segments sourced from thousands of individuals [1, 2]. Despite efforts to assemble high-quality, gapless genomes such as T2T-CHM13 [3], T2T-YAO [4], CN1 [5], I002C [6], or HG002 [7], such references raise concerns regarding their abilities to represent genetic variations across diverse human populations accurately. The prevailing consensus is that no singular reference sequence can adequately encapsulate the complex genomic diversity across global populations [8]. This understanding highlights the crucial need for high-quality reference genome panels that accurately resolve haplotypes, presenting the complex genetic variations observed within distinct populations [9–11]. In parallel, there is a growing trend to shift from a singular reference to a pangenomic approach, which supports a broader range of genomic diversity, acknowledging the complexities within and across diverse human populations [12–15]. This shift is supported by the rapid development of computational tools for pangenome construction and analysis [16–21].
Haplotype-resolved genome sequences are the building blocks for pangenome construction. However, despite the contradictory nature of cost and sensitivity, both of which play vital roles in pangenomic projects, most recent studies [14, 15] lack comprehensive evaluation and guidelines related to the optimal data types and volumes required, mostly relying on the propositions of assembly tool authors for volume and data type requirements.
At the forefront of long-read technology (LRT) innovation, Pacific Biosciences (PacBio) and Oxford Nanopore Technologies (ONT) stand out as the primary driving forces, spearheading advancements in this field through their groundbreaking contributions. PacBio’s long reads (LR) have excelled in read quality, while ONT has leveraged its competitive edge in providing substantial read lengths at a lower cost [22]. To address the disparity between read quality and length, ONT has recently introduced a novel technique termed “Duplex,” capable of achieving a quality level of Q30, thereby bridging the gap between read quality and read length (https://nanoporetech.com/about-us/news/oxford-nanopore-tech-update-new-duplex-method-q30-nanopore-single-molecule-reads-0). Recent comparisons suggest that the two platforms exhibit similar performance in structural variation (SV) analysis [23, 24]. Details on the current LR sequencing platforms, LR mapping, variant calling, and genome assembly approaches are discussed elsewhere [25]. However, HiFi vs Duplex performance in genome assembly has not been properly evaluated and compared.
In this study, we evaluated different data types and the minimum data volume required to establish a robust pipeline of genome assembly for population-level pangenome projects. Specifically, we conducted a performance comparison between ONT’s Duplex dataset and PacBio HiFi dataset. In this comparison, we extensively examined the performance of these datasets in the context of genome assembly, scrutinizing their effectiveness in achieving high-quality phased genomes with enhanced contiguity and completeness. Given the swift advancements in long-read technologies (LRT), it is prudent to reassess the recommended data types and volumes outlined in this study, aligning them with the practical economic limitations within the scope of your research endeavors.
Results
DNA sequencing
The I002C data used for this research are generated as a part of an ongoing effort to generate telomere-2-telomere diploid assembly of a male Singaporean of Indian ancestry I002C [6]. Through sequencing on various platforms, we obtained the following dataset for the child sample: 152.97 Gb (~ 50.99 ×) PacBio HiFi data, 193.66 Gb (~ 64.55 ×) ONT Duplex data, 441.07 Gb (~ 147 ×) ONT Ultralong data (ULONT) and 222.21 Gb (~ 74.07 ×) Omni-C data. For the paternal sample, 107.69 Gb (~ 35.90 ×), and maternal sample, 112.48 Gb (~ 37.49 ×), MGI paired-end data was sequenced (Table 1). A similar volume of publicly available HG002 dataset was utilized in this study (Table 1). On average, the Duplex reads were twice as long as the HiFi reads, yet they maintained a comparable level of read quality (Fig. 1).
Table 1.
Summary of sequencing data from different platforms
| Dataset | Sample | Platform | Data type | Reads | Total bases (bb) | Depth | Min Length | Max length | Average read length | N50 | %GC |
|---|---|---|---|---|---|---|---|---|---|---|---|
| I002C | Child | PacBio | HiFi | 8,986,857 | 152,977,790,522 | 50.99 | 8 | 63,921 | 17,022.40 | 17,132 | 40.78 |
| ONT | Duplex | 6,561,950 | 193,663,321,924 | 64.55 | 1 | 191,644 | 29,513.10 | 38,128 | 40.85 | ||
| ONT | Ultralong | 12,710,241 | 441,077,651,554 | 147.03 | 1 | 1,434,925 | 34,702.50 | 87,761 | 40.58 | ||
| MGI | Omni-C | 1,476,892,046 | 222,216,283,752 | 74.07 | 150 | 150 | 150 | 150 | 42.5 | ||
| I002A | Father | MGI | Short reads | 717,979,504 | 107,696,925,600 | 35.9 | 150 | 150 | 150 | 150 | 40.48 |
| I002B | Mother | MGI | Short reads | 749,860,990 | 112,479,148,500 | 37.49 | 150 | 150 | 150 | 150 | 40.28 |
| HG002 | Child | Pacbio | HiFi | 9,076,876 | 164,744,547,100 | 54.91 | 86 | 63,894 | 18,149.92 | 17,963 | 40.32 |
| ONT | Duplex | 6,110,824 | 147,724,801,341 | 49.24 | 1 | 187,925 | 24,174.29 | 34,205 | 40.89 | ||
| ONT | Ultralong | 1,347,597 | 204,361,988,910 | 68.12 | 100,000 | 2,486,048 | 151,649 | 147,890 | 40.54 | ||
| Illumina | Hi-C | 596,026,484 | 89,999,999,084 | 30 | 151 | 151 | 151 | 151 | 42.09 | ||
| HG003 | Father | Illumina | Short reads | 687,439,454 | 101,741,039,192 | 33.91 | 148 | 148 | 148 | 148 | 39.67 |
| HG004 | Mother | Illumina | Short reads | 684,524,092 | 101,309,565,616 | 33.77 | 148 | 148 | 148 | 148 | 39.95 |
The coverage depth calculation was based on a genome size estimation of 3 Gb. PacBio Pacific Biosciences, HiFi High-Fidelity reads, ONT Oxford Nanopore Technologies
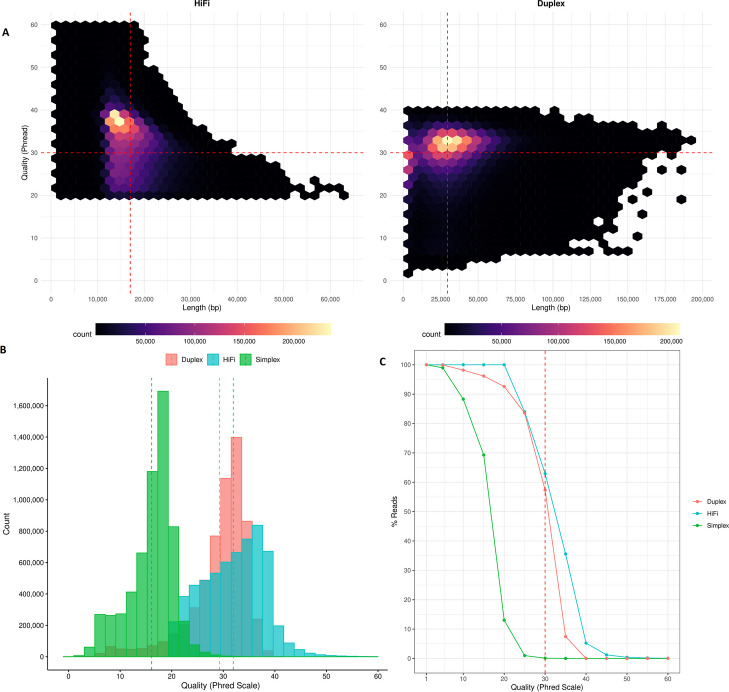
Fig. 1.
Comparison of read length and quality (Phred scale) between PacBio HiFi and ONT Duplex reads. A Distribution of read length vs quality of HiFi and Duplex reads with vertical dotted lines indicating the average lengths: 17 kbp for HiFi and 29.5 kbp for Duplex reads. On average, more than 50% of both Duplex and HiFi reads have quality scores ≥ Q30, a general cutoff for high-quality reads, indicated by the horizontal dotted line. B Comparison of read quality among ONT Simplex, ONT Duplex, and PacBio HiFi, with vertical dotted lines representing average quality scores: Q16 for Simplex, Q29 for Duplex, and Q32 for HiFi reads. C Percentage of reads with a quality score of Q30 and higher (dotted line). On average, 63% of HiFi reads and 57% of Duplex reads have a quality score of Q30 and higher
Coverage saturation analysis of population-scalede novoassembly
To leverage the potential of long reads, for genome assembly we utilized high-quality long reads (HQLR) such as HiFi and Duplex, which are 10 kb or longer and ULONT reads of at least 100 kb (Additional file 2: Table S1). We examined the importance of diverse data types and offered general observations on the sequencing depth or data volume required for genome assembly and its analyses at scale.
Data down-sampling
We generated varying coverage depths by randomly down-sampling different data types, considering a haploid genome size of 3 Gbp (Additional file 2: Table S2–S4).
-
i)
HiFi and Duplex reads: downsampled datasets at 20 × , 30 × , 35 × , 40 × , and 45 × coverage
-
ii)
ULONT: downsampled datasets at 10 × , 20 × , 30 × , 40 × , 50 × , and 60 × coverage
-
iii)
Omni-C/Hi-C: downsampled datasets at 10 × , 20 × , and 30 × coverage
Due to the longer length of Duplex reads, achieving the same sequencing depth requires, on average, twice as many HiFi reads as Duplex reads at any given coverage level, as demonstrated in Additional file 1: Fig. S1.
Evaluation of assembly results in terms of sequence saturation
To evaluate the impact of sequencing coverage on assembling performance and identify the coverage saturation point where assembly contiguity begins to plateau, we utilized hifiasm [26–28] to assemble HiFi/Duplex data independently (HQLR_Only) and in conjunction with ULONT data across varying coverage depths. The assembly results show a clear positive correlation between the augmentation of data coverage (HiFi/Duplex) and assembly performance for both primary assemblies (Additional file 1: Fig. S2), representing a mosaic of the two haplotypes and the two haplotypes derived from the phased assembly (Fig. 2).
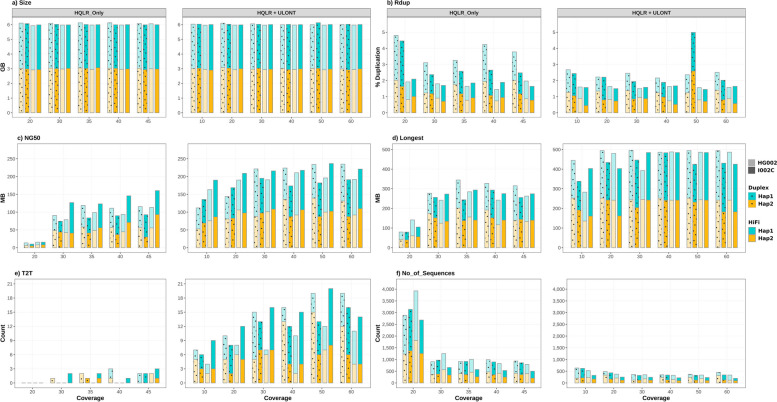
Fig. 2.
Comparison of assembly performance versus data coverage. “HQLR_Only” denotes assemblies generated solely with HiFi or Duplex data across various coverages. “HQLR + ULONT” signifies assemblies generated with a saturation coverage (35 ×) of HiFi or Duplex data combined with various ULONT coverages
At any given coverage, the assembled genome size aligns well with expected genome sizes (2.9 Gb paternal, 3 Gb maternal, and 3.1 Gb primary assembly). The inflated assembled genome size positively correlated with the duplication rate (Rdup). As the data coverage increases, key assembly contiguity features such as NG50, Longest contig length, and Telomere-2-Telomere [T2T] contigs exhibit an upward trend, while the “No_of_Sequences” demonstrate a downward trajectory. Assembly contiguity reaches plateaus when the HQLR-only (HiFi/Duplex) coverage exceeds 35 × (Fig. 2).
Furthermore, in combination with ULONT data, even as low as 10 × ULONT along with 35 × of HQLR plateau coverage significantly enhances assembly contiguity compared to that of 45 × HQLR-only assembly. The inclusion of ULONT data notably improves the assembly of telomere-to-telomere contigs. Assembly contiguity reaches a plateau with ULONT coverage exceeding 30 × . We observed a similar trend for primary assemblies (Additional file 1: Fig. S2). The detailed assembly statistics are provided in the Additional Materials (Additional file 2: Table S5–S10). Hereafter, in the figures and tables, “HQLR + ULONT” denotes HQLR (HiFi/Duplex) coverage of 35 × , representing the HQLR-only plateau coverage, combined with various ULONT coverages. Similarly, “HQLR + ULONT + Omni-C” signifies 35 × HQLR coverage combined with 30 × ULONT coverage, representing the ULONT plateau coverage, along with different levels of Omni-C (I002C)/Hi-C (HG002) coverage.
Improvement of phasing with Omni-C/Hi-C
The hifiasm tool is capable of producing pseudo-haplotypes or a dual assembly using HiFi/Duplex data alone (Fig. 3a) or in conjunction with ULONT (Fig. 3b). This process efficiently captures the heterozygous variances across the two haplotypes. HiFi-only assemblies demonstrate relatively fewer switch errors due to their higher quality compared to Duplex-only assemblies. Conversely, the longer read lengths of Duplex data contribute to achieving superior global phasing (hamming) compared to HiFi reads (Additional file 1: Fig. S3). Due to their lengths, ULONT reads additionally improve phasing [29]. However, even with ULONT reads, assemblers generate contigs with short phase blocks that often show increased phasing errors (Fig. 3b).
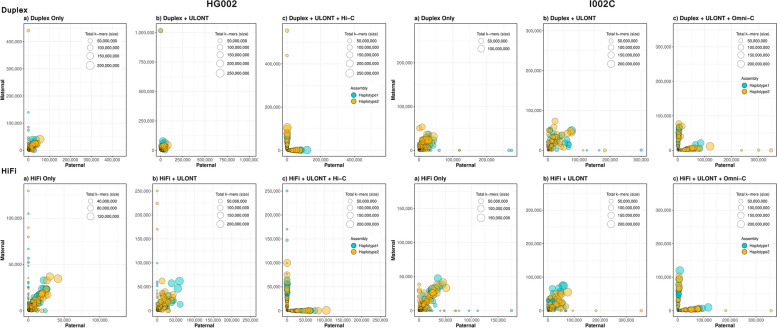
Fig. 3.
Comparison of phasing accuracies of different assemblies (Duplex assemblies—top row, HiFi assemblies—bottom row). a Phasing accuracy of dual assembly generated from HQLR-only (HiFi/Duplex). b Phasing accuracy of dual assembly in conjunction with ULONT. c Haplotype separated assemblies with Omni-C/Hi-C data. Each circle denotes a contig, size reflecting its length. Circle’s positions are determined by the number of maternal and paternal k-mers derived from high-quality short reads on respective contigs. Contigs positioned along the axis indicate higher phasing accuracy
Incorporating even low coverage of long-range chromatin interaction data like Omni-C/Hi-C, such as 10 × , results in a notable reduction in globally incorrectly phased variants (measured by hamming error), leveraging the long-range chromatin interaction information provided by Omni-C/Hi-C (Additional file 1: Fig. S3). Even though long-range interaction data can produce full-length phased contigs from different chromosomes, maternal and paternal origin contigs can be mixed in one haplotype (Fig. 3c). This intrinsic ambiguity in long-range interaction data phasing is attributed to the challenge of identifying markers that define paternal and maternal origin, a task not easily achievable with offspring data alone, except in the case of XY chromosomes. Despite this improvement, switch errors, which measure the local inaccuracies of heterozygous variants, remain largely unaffected due to the limitations in the information offered by long-range chromatin interaction data. Besides its phasing capability, Omni-C/Hi-C data can also be utilized for scaffolding. Omni-C coverage saturation concerning phasing can be observed at 10 × (Additional file 1: Fig. S4). Since hifiasm does not leverage long-range data for scaffolding to enhance contiguity (Additional file 1: Fig. S4), higher coverage may prove advantageous for scaffolding processes. Determining the optimal coverage for long-range chromatin interaction data (Omni-C/Hi-C) is beyond the scope of this study, as discussed elsewhere [30].
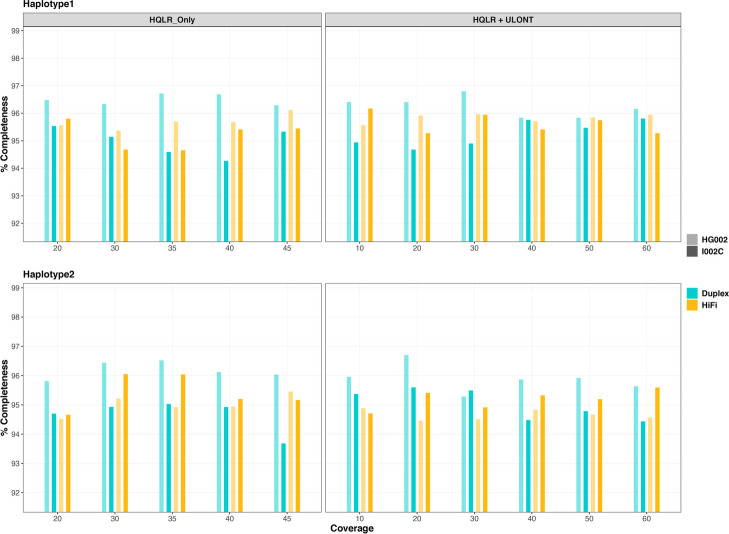
Genome completeness and quality
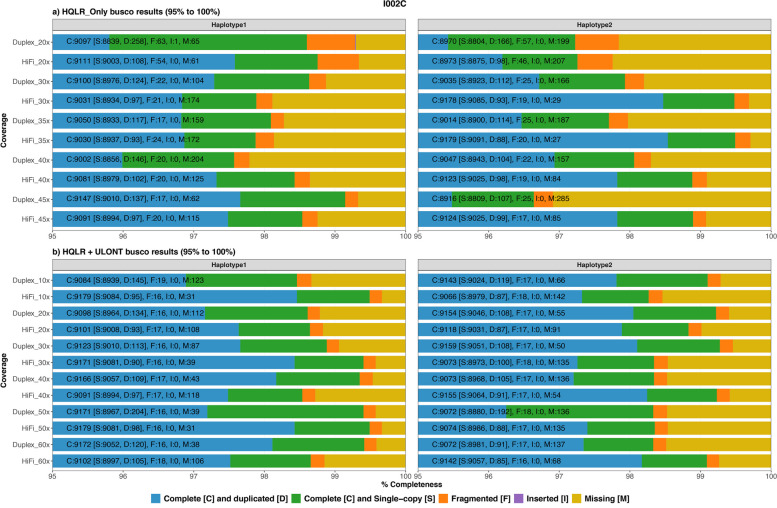
Genome completeness assessed through single-copy gene analysis revealed that assemblies from HQLR-only exhibited slightly lower performance with an average of 96.98% single copy, 1.40% duplicated, 0.30% fragmented, and 1.31% missing genes (Fig. 4a). Meanwhile, assemblies generated with HQLR + ULONT data showed higher completeness values with 97.53% single copy, 1.18% duplicated, 0.19% fragmented, and 1.10 missing genes (Fig. 4b). Combined haplotype results show increased coverage resulted in marginal improvements in gene completeness, with HQLR-only assemblies reaching saturation around 35 × coverage, and ULONT assemblies around 30–40 × (Fig. 4, Additional file 2: Table S11–12). Similar results were found for HG002 (Additional file 1: Fig. S5) and primary assemblies of both datasets (Additional file 1: Fig. S6–S7, Additional file 2: Table S13). Individual haplotypes from haplotype-resolved assemblies do not follow a clear trend of coverage saturation but show noticeable improvements by incorporating ULONT reads (Fig. 4, Additional file 1: Fig. S5).
Fig. 4.
Assessment of gene completeness analysis. a Output from the assemblies generated from HQLR-only. b Output from assemblies generated from HQLR + ULONT
The estimated k-mer completeness, which indicates the proportion of reliable k-mers from the reads found in the assembly, averaged 95.46% for haplotype-resolved assemblies (Fig. 5). In comparison, the primary assemblies averaged a slightly higher rate at 96.37 (Additional file 1: Fig. S8, Additional file 2: Table S14). This finding aligns with the gene completeness analysis results presented in (Fig. 4).
Fig. 5.
Assessment of k-mer-based genome completeness analysis
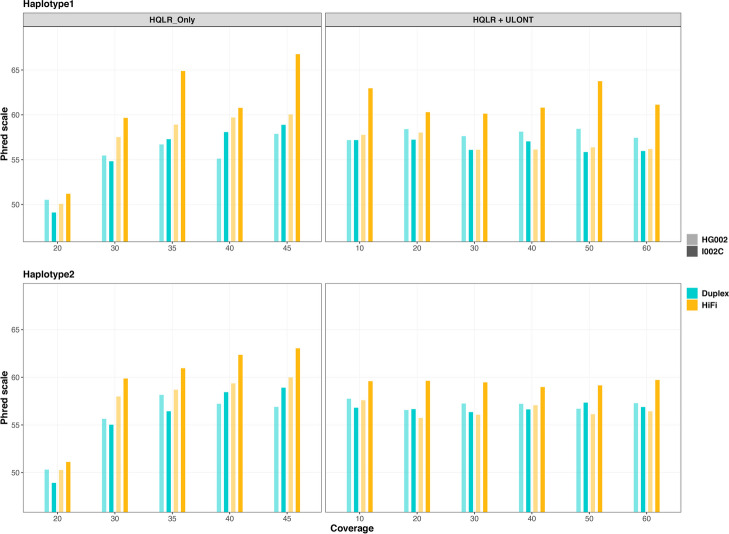
Assembly quality assessed from k-mers as measured by phred scale quality score (QV) generally showed improvement with increased coverage for both haplotype-resolved assemblies (Fig. 6, Additional file 2: Table S15) and primary assemblies (Additional file 1: Fig. S9, Additional file 2: Table S15).
Fig. 6.
K-mer-based genome quality scores
Computational requirements
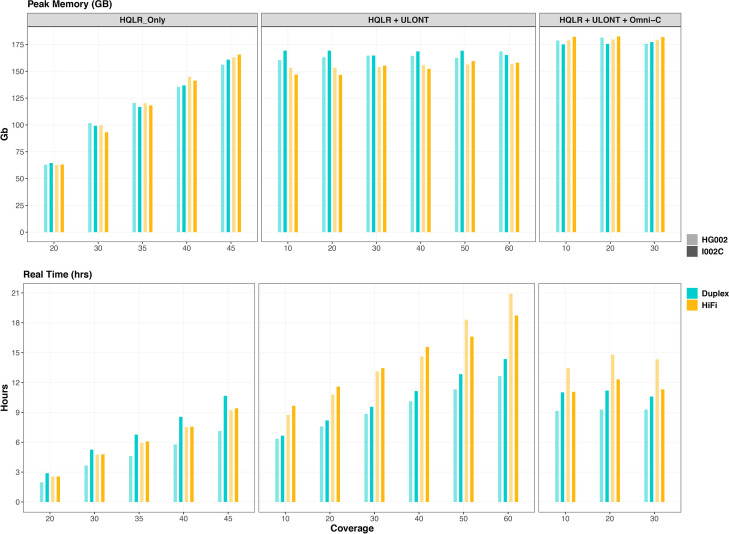
We conducted comparisons of both the runtime and peak memory consumption of assembling steps across various coverage levels for specific data types and assemblies resulting from the various combinations of different data types. The computational demands are of paramount importance, particularly in studies conducted at population scale and those utilizing cloud-based platforms for analysis.
The error correction process is a critical and most time-intensive step taking more than half of the total execution time, followed by the graph construction by long read assemblers. By default, hifiasm performs three rounds of error correction of input HiFi/Duplex reads. Consequently, the time and memory requirements exhibit an upward trajectory with increasing coverage when assemblies are derived solely from data (HQLR_only). In the case of “HQLR + ULONT,” where a fixed amount of HQLR data is employed, computational time shows an upward trend with the increased coverage of ULONT. At the same time, memory requirements remain stable across coverage levels. This stability in memory consumption is attributed to the implementation of ULONT data in their algorithm [28].
The incorporation of long-range chromatin interaction data (Omni-C) primarily utilized for phasing and resolving graph tangles reveals that both time and memory requirements remain more consistent across increased long-range data coverage (Fig. 7, Additional file 2: Table S16). However, compared to HQLR and “HQLR + ULONT,” the overall increase in memory requirements can be attributed to an additional step required to construct unique 31-mers from the initial assembly graph generated from HQLR reads for processing long-range data. This step depends on the coverage of HQLR, which remains fixed. In contrast, the variable coverage of long-range data has minimal influence on memory requirements, making it consistent across different levels of long-range data coverage. Computation time does not linearly increase with Hi-C coverage. Instead of using general-purpose read mappers to align Hi-C reads, hifiasm implements k-mer-based alignment to filter Hi-C reads that do not bridge heterozygous alleles or are mapped to homozygous unitigs reducing the computational burden as described in their algorithm [27].
Fig. 7.
Computation resources consumed by hifiasm across different data types and coverages
Comparison of HiFi and Duplex reads performance inde novoassembly
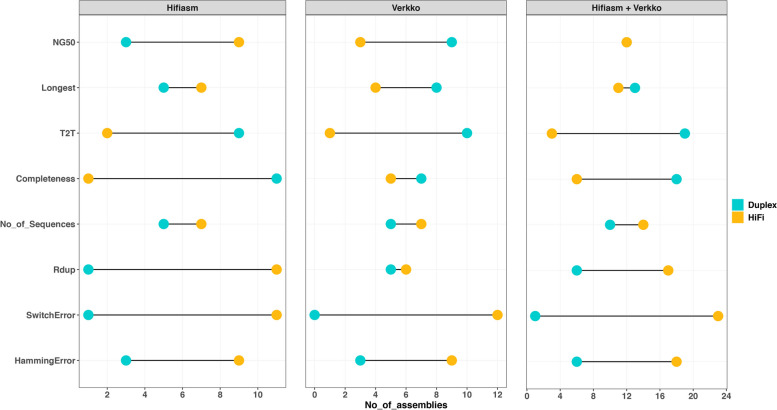
We conducted a comparison between the new data types regarding their performance in genome assembly using the current I002C and publicly available HG002 datasets. HG002 Duplex and Revio HiFi data were downloaded from HPRC. The assemblies were constructed with the same coverage (i.e., 35 × HiFi/Duplex + 30 × ULONT + 10 × Hi-C [HG002]/Omni-C [I002C]) data with default parameters of hifiasm and Verkko [31] across three independent replicates. We assigned a rank of 1 to the highest value and 0 otherwise for each assembly feature. The sum of these ranks was then computed for both HiFi and Duplex assemblies to evaluate their performance based on specific criteria, ranging from best to worst (Fig. 8). Overall HiFi assemblies demonstrated lower values for metrics such as Rduplication, Number of Sequences, Switch, and Hamming errors, indicating superior assembly quality. However, Duplex assemblies achieved a higher count of T2T contigs and k-mer completeness. The lower NG50 of hifiasm Duplex assemblies may be due to hifiasm’s use of a string graph-based method, which struggles with handling contained reads, with read length being the aggravating factor. Since Duplex reads are, on average, twice as long, this issue is exacerbated. RAFT algorithm [32], minimizes this problem. However, the RAFT-hifiasm workflow requires RAFT to be executed once and hifiasm three times, making it at least two times slower than a single run of hifiasm. This issue is absent in de Bruijn graph-based assemblers like Verkko. The quantitative values for assembly features across replicates are available in the additional materials (Additional file 1: Fig. S10, Additional file 2: Table S17).
Fig. 8.
The relative performance of HiFi vs Duplex assemblies from hifiasm and Verkko using I002C and HG002 dataset with plateau coverage, i.e., (35 × HiFi vs 35 × Duplex) + (30 × ULONT and 10 × Omni-C/Hi-C). For each assembly feature, we compared 24 assemblies obtained from 2 samples, 2 assemblers, 3 replicates, and 2 haplotypes, recording instances where the HiFi assembly outperformed the Duplex assembly and vice versa. “hifiasm + Verkko” shows the overall performance of HiFi vs Duplex across 24 assemblies. For example, 23 out of 24 HiFi assemblies had a lower switch error compared to Duplex assemblies, while 19 Duplex assemblies contained more T2T contigs than HiFi assemblies. Both platforms, however, produced comparable results in terms of NG50 and the length of the longest contigs
Discussion
The DNA sequencing landscape is continually evolving, with advancements in sequencing technologies offering unprecedented opportunities for genomic research. In this study, we conducted a comprehensive analysis of sequencing data obtained from PacBio HiFi, ONT Duplex, ONT Ultralong (ULONT), and Omni-C data in the context of genome assembly. We aimed to investigate coverage saturation for different data types and their implications for various aspects of genome assembly, including phasing, genome completeness, and assembly quality.
Our findings provide valuable insights into the optimal sequencing coverage depth required for genome assembly in large-scale analyses. Through coverage saturation analysis, we observed a positive correlation between sequencing coverage and assembly performance. Notably, assembly contiguity plateaued when the HQLR-only coverage exceeded 35 × . Furthermore, the integration of ULONT data significantly enhanced assembly contiguity, particularly for assembling telomere-to-telomere contigs, underscoring the importance of long-range data in improving assembly contiguity. The assembly contiguity plateaus with ULONT coverage exceeding 30 × .
We did not involve parental information in generating haplotype-resolved assemblies. Trio binning using parental data facilitates assembly and increases phasing accuracy compared to long-range data phasing [33]. However, it requires additional effort in the recruitment process and often parental information is not available. Even with high-quality long reads such as HiFi/Duplex and ULONT with substantial coverage, the assembled genome still can have higher switch and hamming errors. Our study demonstrates the efficacy of incorporating long-range chromatin interaction data like Omni-C/Hi-C to address this issue. By leveraging long-range contact information provided by long-range chromatin interaction data, we observed a notable reduction in globally incorrectly phased variants. However, challenges persist in accurately identifying the parental origin of phased contigs, highlighting the inherent ambiguity in long-range data phasing.
Genome completeness and quality assessments revealed marginal improvements with increased coverage, with assemblies incorporating ULONT data exhibiting higher completeness metrics compared to HQLR-only assemblies. Our analysis emphasizes the importance of considering both single-copy gene analysis and k-mer completeness for a comprehensive assessment of genome quality.
The computational demands associated with genomic analysis are substantial, particularly in population-scale studies. Our study highlights the time and memory requirements associated with the assembly process, emphasizing the need for efficient algorithms and computational resources to handle large datasets effectively.
As pioneers in long-read technology (LRT), PacBio and ONT continually refine their technologies and develop new advancements to deliver high-quality data at increasingly affordable prices. The recent launch of the PacBio Revio platform (https://www.pacb.com/revio/) stands as a testament to this commitment, elevating HiFi yield by 15 × while maintaining impeccable data quality compared to its predecessor, the PacBio Sequel II platform. The assembly contiguity achieved with HiFi data exhibits nearly identical performance on both the Sequel IIe and Revio platforms [24]. The substantial boost in data yield has effectively mitigated affordability concerns in comparison to competition. Similarly, ONT has unveiled the enhanced R10 flowcell and introduced the innovative “Duplex” method, which achieves read quality nearing Q30 by sequencing both the template and complement strands of a single molecule. The effectiveness of these cutting-edge data types has been demonstrated in variant calling [24] and methylation studies [34], showcasing their utility and performance across different genomic applications.
A comparative analysis between PacBio HiFi and ONT Duplex data for genome assembly shows that HiFi data consistently delivers superior assembly quality, particularly in reducing duplication rates, sequence count, switch errors, and Hamming errors. However, Duplex data outperformed in producing a higher number of T2T contigs and k-mer completeness. Despite these strengths, both platforms exhibit comparable performance in terms of NG50 and the length of the longest contigs, highlighting that each method offers unique benefits depending on the specific assembly objective. These findings are backed by a recent study by Koren et al. [35], who evaluated ONT Duplex data for non-human samples and HG002.
Conclusion
Recognizing the dynamic nature of genomic research and the evolution of sequencing technologies and analytical methodologies is essential. Through our exploration of various sequencing data types and algorithms, we offer several key insights and recommendations for population-level pangenome reference generation efforts. We highlight the pivotal role of integrating high-quality data sources such as Pacbio HiFi/ONT Duplex and ONT ULONT, alongside long-range contract data like Omni-C, to achieve phased telomere-to-telomere level assemblies. In general, HiFi/Duplex coverage of ≥ 20 × complemented with 15–20 × of ULONT per haplotype and 10 × long-range data are essential requisites for attaining high-quality contiguous and phased assembly. We offer our findings as practical guidelines to help users choose sequencing platforms and coverage effectively.
Methods
Sample selection and preparation for sequencing
One family (comprising a mother, father, and child) with an Indian ethnic background out of 15 families recruited from Singapore as a part of the human genome project was selected. The selection criteria for the family were (1) current generation (child sample) is a male and (2) no genetic diseases with normal phenotype. All the participants were provided with informed consent for sample collection and usage including making data publicly available via databases. Sample collection and usage were approved by SingHealth Centralised Institutional Review Board. Whole blood was collected from the family (I002).
Isolation of peripheral blood mononuclear cells (PBMCs)
Ten milliliters of human whole blood samples were collected, and PBMCs were isolated using density gradient centrifugation with Ficoll-Paque (GE Healthcare). Blood was diluted with 20 ml of phosphate-buffered saline (PBS) and carefully layered over with 15 ml of Ficoll-Paque solution before centrifugation at 225 g for 30 min at room temperature. The PBMC layer was harvested, washed twice with PBS, and resuspended in complete RPMI 1640 medium (Gibco) supplemented with 20% fetal bovine serum (FBS) and 1% penicillin–streptomycin.
Infection with B95-8 Epstein-Barr virus (EBV)
PBMCs were infected with the B95-8 strain of EBV by adding virus-containing supernatant derived from B95-8-infected marmoset B lymphocytes. The mixture was incubated at 37 °C with 5% CO₂ and left untouched for 8 days to facilitate virus entry into the B cells.
Cells were cryopreserved in a freezing medium containing 20% FBS and 10% dimethyl sulfoxide (DMSO), and stored in liquid nitrogen for long-term preservation.
Long-read sequencing (LRS) data generation
Pacbio data generation
The high molecular weight (HMW) DNA used for PacBio sequencing was extracted using the GentraPuregene kit (Qiagen; #158043) according to the manufacturer’s instructions. Briefly, 1 × 107 frozen cell pellets from the I002C cell line were used as input for extraction. All vortexing steps were replaced with gentle inversion throughout the process, and 300 µl of Qiagen EB buffer was used for elution. Eluted DNA was incubated at 12 °C with gentle shaking over a period of 7 to 10 days. To avoid shearing the high molecular weight DNA, wide bore tips with gentle pipetting were used during handling. DNA was stored at 4 °C to prevent freeze and thaw cycle. Quantity and purity of extracted HMW DNA were assessed using triplicate concentration measurements from the top, middle, and bottom sections of sample volume, using Qubit dsDNA BR (Broad-Range) assay (Thermofisher Scientific; Q32853) and NanoDrop 2000 spectrophotometer (ThermoFisher Scientific; ND-2000), according to manufacturer’s instructions.
After DNA extraction, DNA fragment lengths were then measured using TapeStation 4200 (Agilent). Sequencing libraries were created using the SMRTbell Express Template Prep Kit 2.0 (PacBio) per the manufacturer’s instructions. Libraries were sequenced on a Sequel IIe and Revio System (PacBio). After sequencing, CCS analyses were run using SMRTLink software v10 to produce HiFi reads.
ONT data generation
High nolecular weight (HMW) gDNA extraction (Duplex sequencing)
We obtained 12 × 106 frozen cell pellets from established lymphoblastoid cell line for the child sample and processed for HMW gDNA extraction using the Monarch HMW DNA Extraction Kit for Tissue (NEB; T3060). During the extraction, we excluded shaking during all incubation steps to preserve gDNA integrity. Quantity, purity, and integrity of extracted HMW gDNA were assessed using Qubit dsDNA BR assay (Thermofisher Scientific; Q32853), NanoDrop 2000 spectrophotometer (ThermoFisher Scientific; ND-2000), and 15-h pulsed-field gel electrophoresis runs with the Pippin Pulse system (Sage Science; PPI0200), respectively. Quality-assessed HMW gDNA was then used for ligation-based library preparation with the Ligation Sequencing Kit V14 (Oxford Nanopore Technologies; SQK-LSK114) to generate Duplex sequencing reads.
Ultra-high molecular weight (UHMW) gDNA extraction (ultra-long read sequencing)
We processed 15 × 106 frozen cell pellets from an established lymphoblastoid cell line for the child sample for UHMW gDNA extraction using the Monarch HMW DNA Extraction Kit for Tissue, following the extraction steps described in the Ultra-Long DNA Sequencing Kit V14 (Oxford Nanopore Technologies; SQK-ULK114) protocol. Quality assessment of UHMW gDNA was performed similarly to HMW gDNA extraction. Quality-assessed UHMW gDNA was then used for transposase-based library preparation and purification with the Ultra-Long DNA Sequencing Kit V14 for the generation of ultra-long sequencing reads.
Library preparation and PromethION sequencing (Duplex and high duplex)
We sheared 3 µg to 7.5 µg of extracted HMW gDNA to a target size of 55 kb to 60 kb and performed size-selective precipitation to remove DNA sizes < 25 kb. Repaired and end-prepped DNA was then used for library construction with SQK-LSK114 for both duplex and high duplex sequencing approaches. For standard duplex runs, libraries were loaded at 6 fmol to 7 fmol per load, while for high duplex runs, 7 fmol to 55 fmol of libraries were loaded and sequenced on PromethION 24 (Oxford Nanopore; PCA100024), R10.4.1 flowcells (Oxford Nanopore) FLO-PRO114M and FLO-PRO114HD respectively.
Library preparation and PromethION sequencing (ultra-long, UL)
We used 40 µg to 45 µg of UHMW gDNA for ultra-long read library preparation using SQK-ULK114. Final UL libraries were sequenced on PromethION 24 using FLO-PRO114M flowcells with nuclease flushes performed at 23-h intervals.
The detailed steps of the entire procedure are outlined in the Additional Materials.
Omni-C data generation
The Dovetail Omni-C library was prepared using the Dovetail Omni-C™ Proximity Ligation Assay kit (Dovetail Genomics, Scotts Valley, CA, USA), according to the manufacturer’s protocol (version 1.2). Briefly, after sample crosslinking with DSG and formaldehyde, chromatin was digested using a sequence-independent endonuclease and bound to chromatin capture beads. Proximity ligation was performed using a biotin-labeled bridge between the ends of the digested DNA. After reversal crosslinking, the DNA was purified and followed by library preparation. Finally, the biotinylated molecules were captured and amplified before sequencing on the Novaseq 6000 and HiSeq 4000 instruments (Illumina, San Diego, CA, USA) in paired-end mode.
Data analysis
All commands employed in the analysis are comprehensively listed in the Additional Materials file, providing readers with detailed procedures undertaken in this study.
Reads downsampling
To evaluate coverage saturation for both assembly contiguity and phasing efficiency, we downsampled the reads to various coverages. Reads were randomly subsampled to achieve the desired coverage utilizing Rasusa v0.7.1 [36], considering a genome size estimation of 3 gigabases (3 gb).
De novoassembly and assessment
The choice of assembler is critical for the evaluation process. We selected the assembler for coverage saturation analysis on the following criteria:
Ability to support different types of long reads
Native capability to generate haplotype-separated assemblies using a single data type and/or with additional data such as trio or long-range contact information
Computational demands
Active maintenance of the tool
Currently, the two most popular hybrid assemblers that support high-quality data, such as HiFi/Duplex, in addition to ultra-long (UL) reads, along with trio or long-range reads, to generate telomere-to-telomere haplotype-separated assemblies, are hifiasm [26–28] and Verkko [31]. However, when tested with the same dataset and computational configuration, Verkko’s runtime was more than twice that of hifiasm (Additional file 1: Fig. S11, Additional file 2: Table S18). Furthermore, like other ONT assemblers, Verkko cannot produce haplotype-resolved assemblies using only Duplex or HiFi data unless the reads are first binned by haplotype for individual assembly or a diploid assembly is recovered from a haploid assembly using tools like HapDup. The recently published ONT assembler PECAT [37] can generate haplotype-wise assemblies from Duplex data alone. Still, it does not support the integration of additional datasets like ULONT or long-range interaction data. Given these limitations and insights from previous benchmark studies [38, 39], we selected hifiasm as the assembler to evaluate coverage saturation.
Assembly statistics
Assembly contiguity metrics were computed utilizing minigraph v0.20 [18] and paftools v2.26-r1175 [40].
Phasing statistics
The phasing efficiency of an assembly was evaluated in terms of switch error and Hamming error rates with Yak v0.1-r69-dirty [41] using parental short reads. Switch error quantifies the frequency of adjacent phased variants incorrectly transitioning between maternal and paternal haplotypes. Meanwhile, the Hamming error rate denotes the total misphased variants within each assembled contig. Phasing statistics were generated for both the haplotypes separately.
Assembly completeness and quality
To evaluate the impact of coverage variations on the completeness, we employed compleasm v0.2.2 [42] to obtain the BUSCO assessment results. Concurrently, we applied a k-mer-based approach for assembly completeness evaluation, using the KMC tool v3.2.1 [43]. Identifying reliable k-mers within the reads followed a previously outlined methodology [44]. The assembly completeness was computed as the fraction of reliable k-mers in the read set that also appeared in the assembly. Assembly QV was estimated using Yak.
Supplementary Information
Additional file 1. Supplemental methods and supplemental Figures S1-S11.
Additional file 2. Supplemental tables S1-S18.
Acknowledgements
The authors thank Oxford Nanopore for providing duplex and ULONT flow cells, as well as PacBio for HiFi flow cells at no cost. We would like to thank Mr Low Hwee Meng and Ms See Ting Leong from the Genome Institute of Singapore (GIS) Integrated Genomics Platform for their help with generating ONT data. Moreover, we would like to thank Mr Li Zhihui also from GIS Integrated Genomics Platform, for base calling the data as part of ONT-T2T collaboration. Finally, we would like to thank Ms Yao Fei for generating Omni-C libraries for the I002C T2T project and Ms Sia YY for maintaining the cell line, both from the GIS Laboratory of Human Genomics. This work was supported by the A*STAR Computational Resource Centre through the use of its high-performance computing facilities.
Peer review information
Editorial Board Member Shilpa Garg and Andrew Cosgrove were the primary editors of this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Review history
The review history is available as Additional file 3.
Authors' contributions
JJ.L. and M.Š. conceived the project. P.S. designed the pipeline. P.S. and J.L. prepared datasets and conducted the data analysis with the help of F.T. for genome completeness analysis. P.S. wrote the manuscript, and J.L. and F.T. helped with the organization of it. M.Š. and JJ.L. supervised the project and provided mentorship.
Funding
This study was supported by the Agency for Science, Technology and Research (A*STAR) and the Singapore National Precision Medicine (NPM) Program (MOH-000588–01). Additional funding was provided by the European Union through the European Regional Development Fund under grant KK.01.1.1.01.0009 (DATACROSS), as well as by the Croatian Science Foundation under grant IP-2018–01-5886 (SIGMA).
Data availability
The I002C data utilized in this research are generated as part of an ongoing initiative to develop a telomere2-telomere diploid assembly of I002C [6]. The reads are submitted to NCBI under project ID PRJNA1150503. Yak files for parental data, used for assembly phasing analysis are added to zenodo (https://doi.org/10.5281/zenodo.14242314 [45]). Additionally, the HG002 dataset used in this study is accessible via AWS from the Human Pangenome Reference Consortium (HPRC).
Pacbio Revio HiFi data downloaded from: https://human-pangenomics.s3.amazonaws.com/index.html?prefix=submissions/80d00e88-7a92-46d8-88c7-48f1486e11ed--HG002_PACBIO_REVIO/.
ONT Duplex data downloaded from: https://human-pangenomics.s3.amazonaws.com/index.html?prefix=submissions/0CB931D5-AE0C-4187-8BD8-B3A9C9BFDADE--UCSC_HG002_R1041_Duplex_Dorado/Dorado_v0.1.1/.
ONT ultra long data downloaded from: https://s3-us-west-2.amazonaws.com/human-pangenomics/index.html?prefix=NHGRI_UCSC_panel/HG002/nanopore/ultra-long/.
Hi-C data downloaded from: https://s3-us-west-2.amazonaws.com/human-pangenomics/index.html?prefix=NHGRI_UCSC_panel/HG002/hic/.
Declarations
Ethics approval and consent to participate
All the participants were provided with informed consent for sample collection, and usage including making data publicly available via databases. Sample collection and usage were approved by SingHealth Centralised Institutional Review Board (IRB Reference: 2024–069). All experimental methods comply with the Helsinki Declaration.
Consent for publication
Not applicable.
Competing interests
M.Š. has been jointly funded by Oxford Nanopore Technologies and AI Singapore for the project AI-driven De Novo Diploid Assembler. The remaining authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mile Šikić, Email: miles@gis.a-star.edu.sg.
Jianjun Liu, Email: liuj3@gis.a-star.edu.sg.
References
- 1.International Human Genome Sequencing Consortium, Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Schneider VA, Graves-Lindsay T, Howe K, Bouk N, Chen H-C, Kitts PA, et al. Evaluation of GRCh38 and de novo haploid genome assemblies demonstrates the enduring quality of the reference assembly. Genome Res. 2017;27:849–64. 10.1101/gr.213611.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nurk S, Koren S, Rhie A, Rautiainen M, Bzikadze AV, Mikheenko A, et al. The complete sequence of a human genome. Science. 2022;376:44–53. 10.1126/science.abj6987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He Y, Chu Y, Guo S, Hu J, Li R, Zheng Y, et al. T2T-YAO: a telomere-to-telomere assembled diploid reference genome for Han Chinese. Genomics Proteomics Bioinformatics. 2023. 10.1016/j.gpb.2023.08.001. [DOI] [PMC free article] [PubMed]
- 5.Yang C, Zhou Y, Song Y, Wu D, Zeng Y, Nie L, et al. The complete and fully-phased diploid genome of a male Han Chinese. Cell Res. 2023;33:745–61. 10.1038/s41422-023-00849-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lab of Human Genomics. I002C: Telomere-to-Telomere diploid Indian Genome. Available from: https://github.com/LHG-GG/I002C or https://github.com/lbcb-sci/I002C. Accessed 24 Mar 2024.
- 7.Telomere-to-Telomere (T2T) consortium. HG002: A complete diploid human genome. Available from: https://github.com/marbl/HG002. Accessed 24 Mar 2024.
- 8.Yang X, Lee W-P, Ye K, Lee C. One reference genome is not enough. Genome Biol. 2019;20:104. 10.1186/s13059-019-1717-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lou H, Gao Y, Xie B, Wang Y, Zhang H, Shi M, et al. Haplotype-resolved de novo assembly of a Tujia genome suggests the necessity for high-quality population-specific genome references. Cell Syst. 2022;13:321-333.e6. 10.1016/j.cels.2022.01.006. [DOI] [PubMed] [Google Scholar]
- 10.Deng L, Xie B, Wang Y, Zhang X, Xu S. A protocol for applying a population-specific reference genome assembly to population genetics and medical studies. STAR Protoc. 2022;3:101440. 10.1016/j.xpro.2022.101440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ballouz S, Dobin A, Gillis JA. Is it time to change the reference genome? Genome Biol. 2019;20:20. 10.1186/s13059-019-1774-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sherman RM, Salzberg SL. Pan-genomics in the human genome era. Nat Rev Genet. 2020;21:243–54. 10.1038/s41576-020-0210-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang T, Antonacci-Fulton L, Howe K, Lawson HA, Lucas JK, Phillippy AM, et al. The human pangenome project: a global resource to map genomic diversity. Nature. 2022;604:437–46. 10.1038/s41586-022-04601-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liao W-W, Asri M, Ebler J, Doerr D, Haukness M, Hickey G, et al. A draft human pangenome reference. Nature. 2023;617:312–24. 10.1038/s41586-023-05896-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao Y, Yang X, Chen H, Tan X, Yang Z, Deng L, et al. A pangenome reference of 36 Chinese populations. Nature. 2023;619:112–21. 10.1038/s41586-023-06173-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garrison E, Sirén J, Novak AM, Hickey G, Eizenga JM, Dawson ET, et al. Variation graph toolkit improves read mapping by representing genetic variation in the reference. Nat Biotechnol. 2018;36:875–9. 10.1038/nbt.4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eizenga JM, Novak AM, Sibbesen JA, Heumos S, Ghaffaari A, Hickey G, et al. Pangenome graphs. Annu Rev Genomics Hum Genet. 2020;21:139–62. 10.1146/annurev-genom-120219-080406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H, Feng X, Chu C. The design and construction of reference pangenome graphs with minigraph. Genome Biol. 2020;21:21. 10.1186/s13059-020-02168-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vernikos GS. A review of pangenome tools and recent studies. The Pangenome. Cham: Springer International Publishing; 2020. p. 89–112. [PubMed] [Google Scholar]
- 20.Ebler J, Ebert P, Clarke WE, Rausch T, Audano PA, Houwaart T, et al. Pangenome-based genome inference allows efficient and accurate genotyping across a wide spectrum of variant classes. Nat Genet. 2022;54:518–25. 10.1038/s41588-022-01043-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hickey G, Monlong J, Ebler J, Novak AM, Eizenga JM, Gao Y, et al. Pangenome graph construction from genome alignments with minigraph-cactus. Nat Biotechnol. 2023;42:663. 10.1038/s41587-023-01793-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Logsdon GA, Vollger MR, Eichler EE. Long-read human genome sequencing and its applications. Nat Rev Genet. 2020;21:597–614. 10.1038/s41576-020-0236-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du X, Li L, Liang F, Liu S, Zhang W, Sun S, et al. Robust benchmark structural variant calls of an Asian using state-of-the-art long-read sequencing technologies. Genomics Proteomics Bioinformatics. 2022;20:192–204. 10.1016/j.gpb.2020.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harvey WT, Ebert P, Ebler J, Audano PA, Munson KM, Hoekzema K, et al. Whole-genome long-read sequencing downsampling and its effect on variant calling precision and recall. bioRxiv. 2023. 10.1101/2023.05.04.539448. [DOI] [PMC free article] [PubMed]
- 25.De Coster W, Weissensteiner MH, Sedlazeck FJ. Towards population-scale long-read sequencing. Nat Rev Genet. 2021;22:572–87. 10.1038/s41576-021-00367-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng H, Concepcion GT, Feng X, Zhang H, Li H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat Methods. 2021;18:170–5. 10.1038/s41592-020-01056-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng H, Jarvis ED, Fedrigo O, Koepfli K-P, Urban L, Gemmell NJ, et al. Haplotype-resolved assembly of diploid genomes without parental data. Nat Biotechnol. 2022;40:1332–5. 10.1038/s41587-022-01261-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng H, Asri M, Lucas J, Koren S, Li H. Scalable telomere-to-telomere assembly for diploid and polyploid genomes with double graph. Nat Methods. 2024;21:967–70. 10.1038/s41592-024-02269-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jain M, Koren S, Miga KH, Quick J, Rand AC, Sasani TA, et al. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat Biotechnol. 2018;36:338–45. 10.1038/nbt.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sur A, Noble WS, Myler PJ. A benchmark of Hi-C scaffolders using reference genomes and de novo assemblies. bioRxiv. 2022. 10.1101/2022.04.20.488415.
- 31.Rautiainen M, Nurk S, Walenz BP, Logsdon GA, Porubsky D, Rhie A, et al. Telomere-to-telomere assembly of diploid chromosomes with Verkko. Nat Biotechnol. 2023;41:1474–82. 10.1038/s41587-023-01662-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kamath SS, Bindra M, Pal D, Jain C. Telomere-to-telomere assembly by preserving contained reads. bioRxiv. 2023. 10.1101/2023.11.07.565066. [DOI] [PMC free article] [PubMed]
- 33.Koren S, Rhie A, Walenz BP, Dilthey AT, Bickhart DM, Kingan SB, et al. De novo assembly of haplotype-resolved genomes with trio binning. Nat Biotechnol. 2018;36:1174–82. 10.1038/nbt.4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ni Y, Liu X, Simeneh ZM, Yang M, Li R. Benchmarking of nanopore R10.4 and R9.4.1 flow cells in single-cell whole-genome amplification and whole-genome shotgun sequencing. Comput Struct Biotechnol J. 2023;21:2352–64. 10.1016/j.csbj.2023.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koren S, Bao Z, Guarracino A, Ou S, Goodwin S, Jenike KM, et al. Gapless assembly of complete human and plant chromosomes using only nanopore sequencing. bioRxiv. 2024. 10.1101/2024.03.15.585294. [DOI] [PMC free article] [PubMed]
- 36.Hall M. Rasusa: Randomly subsample sequencing reads to a specified coverage. J Open Source Softw. 2022;7:3941. 10.21105/joss.03941. [Google Scholar]
- 37.Nie F, Ni P, Huang N, Zhang J, Wang Z, Xiao C, et al. De novo diploid genome assembly using long noisy reads. Nat Commun. 2024;15:1–15. 10.1101/2022.09.25.509436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jarvis ED, Formenti G, Rhie A, Guarracino A, Yang C, Wood J, et al. Semi-automated assembly of high-quality diploid human reference genomes. Nature. 2022;611:519–31. 10.1038/s41586-022-05325-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H, Durbin R. Genome assembly in the telomere-to-telomere era. arXiv [q-bio.GN]. 2023. 10.48550/arXiv.2308.07877.
- 40.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–100. 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H. yak: Yet another k-mer analyzer. Available from: https://github.com/lh3/yak. Accessed 24 Mar 2024.
- 42.Huang N, Li H. compleasm: a faster and more accurate reimplementation of BUSCO. Bioinformatics. 2023;39:39. 10.1093/bioinformatics/btad595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kokot M, Długosz M, Deorowicz S. KMC 3: counting and manipulating k-mer statistics. Bioinformatics. 2017;33:2759–61. 10.1093/bioinformatics/btx304. [DOI] [PubMed] [Google Scholar]
- 44.Rhie A, Walenz BP, Koren S, Phillippy AM. Merqury: reference-free quality, completeness, and phasing assessment for genome assemblies. Genome Biol. 2020;21:21. 10.1186/s13059-020-02134-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sarashetti P. parental yak files. 10.5281/zenodo.14242314.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Supplemental methods and supplemental Figures S1-S11.
Additional file 2. Supplemental tables S1-S18.
Data Availability Statement
The I002C data utilized in this research are generated as part of an ongoing initiative to develop a telomere2-telomere diploid assembly of I002C [6]. The reads are submitted to NCBI under project ID PRJNA1150503. Yak files for parental data, used for assembly phasing analysis are added to zenodo (https://doi.org/10.5281/zenodo.14242314 [45]). Additionally, the HG002 dataset used in this study is accessible via AWS from the Human Pangenome Reference Consortium (HPRC).
Pacbio Revio HiFi data downloaded from: https://human-pangenomics.s3.amazonaws.com/index.html?prefix=submissions/80d00e88-7a92-46d8-88c7-48f1486e11ed--HG002_PACBIO_REVIO/.
ONT Duplex data downloaded from: https://human-pangenomics.s3.amazonaws.com/index.html?prefix=submissions/0CB931D5-AE0C-4187-8BD8-B3A9C9BFDADE--UCSC_HG002_R1041_Duplex_Dorado/Dorado_v0.1.1/.
ONT ultra long data downloaded from: https://s3-us-west-2.amazonaws.com/human-pangenomics/index.html?prefix=NHGRI_UCSC_panel/HG002/nanopore/ultra-long/.
Hi-C data downloaded from: https://s3-us-west-2.amazonaws.com/human-pangenomics/index.html?prefix=NHGRI_UCSC_panel/HG002/hic/.