Abstract
Background
Clostridium thermocellum is a promising candidate for production of cellulosic biofuels, however, its final product titer is too low for commercial application, and this may be due to thermodynamic limitations in glycolysis. Previous studies in this organism have revealed a metabolic bottleneck at the phosphofructokinase (PFK) reaction in glycolysis. In the wild-type organism, this reaction uses pyrophosphate (PPi) as an energy cofactor, which is thermodynamically less favorable compared to reactions that use ATP as a cofactor. Previously we showed that replacing the PPi-linked PFK reaction with an ATP-linked reaction increased the thermodynamic driving force of glycolysis, but only had a local effect on intracellular metabolite concentrations, and did not affect final ethanol titer.
Results
In this study, we substituted PPi-pfk with ATP-pfk, deleted the other PPi-requiring glycolytic gene pyruvate:phosphate dikinase (ppdk), and expressed a soluble pyrophosphatase (PPase) and pyruvate kinase (pyk) genes to engineer PPi-free glycolysis in C. thermocellum. We demonstrated a decrease in the reversibility of the PFK reaction, higher levels of lower glycolysis metabolites, and an increase in ethanol titer by an average of 38% (from 15.1 to 21.0 g/L) by using PPi-free glycolysis.
Conclusions
By engineering PPi-free glycolysis in C. thermocellum, we achieved an increase in ethanol production. These results demonstrate that optimizing the thermodynamic landscape through metabolic engineering can enhance product titers. While further increases in ethanol titers are necessary for commercial application, this work represents a significant step toward engineering glycolysis in C. thermocellum to increase ethanol titers.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13068-024-02591-5.
Keywords: Acetivibrio thermocellus, Biofuels, Clostridium thermocellum, Ethanol production, Metabolic bottleneck, Metabolic engineering, Phosphofructokinase, Thermodynamics
Background
Technologies are needed to decarbonize the transportation sector to help reduce the need for petroleum usage that is driving catastrophic climate change. While electrification may help replace the current usage of gasoline for light duty transportation, sustainable liquid fuel solutions are critically needed for aviation, rail, and marine sectors [1]. Conversion of lignocellulose into ethanol can be part of the solution because ethanol can be catalytically oligomerized to hydrocarbons that can be used for sustainable aviation fuel (SAF). However, better conversion technologies are needed to make SAF a reality.
Clostridium thermocellum (also known as Acetivibrio thermocellus, Hungateiclostridium thermocellum, and Ruminiclostridium thermocellum) is a thermophilic anaerobe that has been extensively studied and engineered for consolidated bioprocessing of lignocellulose to biofuels such as ethanol [2, 3], due to its strong native ability to solubilize and ferment lignocellulose [4, 5]. One of the unique properties of this organism is that it consumes crystalline cellulose (e.g., Avicel) at the same rate or faster than soluble substrates, such as cellobiose [6]. Previous metabolic engineering efforts aimed at transforming C. thermocellum into a commercial platform for lignocellulosic biofuel production have primarily concentrated on enhancing fuel yield. Notably, high ethanol yields exceeding 80% of the theoretical maximum have been achieved [7, 8]. Current research is now directed toward further engineering the bacterium to achieve higher ethanol titers. Existing engineered strains produce titers in the range of 25–30 g/L [7–9]. Concentrations in the 40–50 g/L range are thought to be necessary for commercial applications [1, 10].
The atypical Embden–Meyerhof–Parnas (EMP) glycolytic pathway found in wild-type C. thermocellum [11] was previously observed to function closer to thermodynamic equilibrium compared to organisms such as Thermoanaerobacterium saccharolyticum or Escherichia coli that utilize a canonical glycolytic pathway [12, 13], and may limit final product titer in C. thermocellum. For our purposes, we define a canonical glycolytic pathway as one that uses an ATP-consuming reaction to generate fructose 1,6-bisphosphate and an ATP-producing reaction to convert phosphoenolpyruvate to pyruvate, and both of these steps have a high thermodynamic driving force that allows for allosteric regulation.
Differences in cofactor usage for key glycolytic reactions were proposed as the underlying reason for the distinct pathway thermodynamics based on metabolic flux analysis [12]. Specifically, the pyrophosphate (PPi)-dependent phosphofructokinase (PPi-PFK) reaction is thought to significantly contribute to the lower thermodynamic driving force observed in C. thermocellum. During exponential growth, the driving force of this reaction has been calculated as ∆rGʹPPi-pfk = −1.45 kJ/mol [12]. In microbes like T. saccharolyticum, which utilize an ATP-dependent phosphofructokinase (ATP–PFK), the PFK reaction exhibits the largest drop in Gibbs free energy change of the entire pathway. During exponential growth, the driving force of this reaction has been calculated as ∆rGʹATP-pfk = −22.57 kJ/mol [12]. (Note: Thermodynamic annotation conventions are adapted from eQuilibrator [14].) Moreover, research findings have suggested that the ATP/ADP ratio tends to be significantly higher than the PPi to inorganic phosphate (PPi/Pi) ratio in many organisms [15], thus further widening the difference in thermodynamic driving force between the two reactions. Additionally, it has been previously noted that the two native pathways employed by C. thermocellum for the conversion of phosphoenolpyruvate (PEP) to pyruvate, namely, the pyruvate:phosphate dikinase (PPDK) that uses PPi to convert PEP and AMP to pyruvate and ATP [16] and the malate shunt that converts PEP to oxaloacetate to malate to pyruvate [16], also represent thermodynamic bottlenecks [17].
We recently engineered C. thermocellum to use ATP as the main cofactor for the PFK reaction [18]. Although this increased the thermodynamic driving force of the PFK reaction, the effects on intracellular metabolites were limited to a small region adjacent to this reaction, and there was no effect on ethanol titer. Furthermore, resequencing analyses found that the resulting strain (LL1660) had a partial genome duplication, complicating subsequent genetic engineering.
In this work, we make further progress toward engineering a PPi-free glycolysis in C. thermocellum glycolysis by eliminating the PPi-consuming reactions PFK and PPDK and providing an alternative pathway for PEP to pyruvate conversion via the pyruvate kinase (PYK) reaction. We define a “PPi-free glycolysis” strain as one that does not require pyrophosphate (PPi) for any of the reactions in glycolysis. A key enabling step of engineering this into C. thermocellum was the expression of a soluble pyrophosphatase (PPase). Finally, we show that PPi-free glycolysis increases the levels of downstream glycolysis metabolites, increasing ethanol titers. This demonstrates the potential of engineering pathway thermodynamics as a general strategy for improving product titers.
Material and methods
Strains used in this work
Plasmid and strain construction
Plasmids were constructed via isothermal assembly [20], using commercially available kits (New England Biolabs NEBuilder® HiFi DNA Assembly Master Mix, catalog number E2621). Plasmids were maintained and propagated from BL21 derivative E. coli strains (New England Biolabs T7 Express E. coli, catalog number C2566) to ensure proper methylation of the plasmid DNA [21]. Plasmid DNA was purified using commercial kits available from either New England Biolabs or Zymo Research.
Transformation of C. thermocellum was performed as previously described [22].
Media and growth conditions
All reagents used in this study were of molecular grade and obtained from Sigma Aldrich or Fisher Scientific, unless otherwise noted.
C. thermocellum strains were grown in either pH controlled bioreactor in MTC-7 media at 55 °C under anaerobic conditions in anaerobic chambers (Coy Laboratory Products, Grass Lakes, MI) with a gas phase of 85% N2, 10% CO2, and 5% H2, as previously described [25] or in MTC-5 media in sealed serum bottles purged with N2 gas as previously described [24].
Complex medium CTFUD was prepared as previously described [22] and used for culturing cells for use in transformations, or for preparing genomic DNA for strain resequencing. The CTFUD media contained 5 g/L cellobiose, 1.3 g/L (NH4)2SO4, 1.5 g/L KH2PO4, 0.13 g/L CaCl2·2H2O, 2.6 g/L MgCl2·6H2O, 0.001 g/L FeSO4·7H2O, 4.5 g/L yeast extract, 3 g/L sodium citrate tribasic dihydrate, 0.5 g/L L-cysteine–HCl monohydrate and 0.002 g/L resazurin. Defined MTC-5 medium was used for all other purposes [24].
The MTC-7 medium contained 100 g/L cellobiose, 9.3 g/L MOPS (morpholinepropanesulfonic acid) sodium salt, 2 g/L potassium citrate monohydrate, 1.3 g/L citric acid monohydrate, 1 g/L Na2SO4, 1 g/L KH2PO4, 2.5 g/L NaHCO3, 2 g/L urea, 1 g/L MgCl2·6H2O, 0.2 g/L CaCl2·2H2O, 0.1 g/L FeCl2·4H2O, 1 g/L L-cysteine HCl monohydrate, 0.02 g/L pyridoxamine HCl, 0.004 g/L p-aminobenzoic acid, 0.002 g/L D-biotin, 0.002 g/L Vitamin B12, 0.004 g/L thiamine, 0.0005 g/L MnCl2·4H2O, 0.0005 g/L CoCl2·6H2O, 0.0002 g/L ZnCl2, 0.0001 g/L CuCl2·2H2O, 0.0001 g/L H3BO3, 0.0001 g/L Na2MoO4·2H2O, 0.0001 g/L NiCl2·6H2O. The medium components were prepared as separate solutions, A–F. Solution A, concentrated 1.2-fold, contained cellobiose and MOPS sodium salt. Solution B, concentrated 25-fold, contained potassium citrate monohydrate, citric acid monohydrate, Na2SO4, KH2PO4 and NaHCO3. Solution C, concentrated 25-fold, contained urea. Solution D, concentrated 50-fold, contained MgCl2·6H2O, CaCl2·2H2O, FeCl2·4H2O, L-cysteine HCl monohydrate and trace minerals (solution F). Solutions E, concentrated 50-fold, contained pyridoxamine HCl, p-aminobenzoic acid, D-biotin, vitamin B12 and thiamine. Solution F, concentrated 1000×, contained MnCl2·4H2O, CoCl2·6H2O, ZnCl2, CuCl2·2H2O, H3BO3, Na2MoO4·2H2O and NiCl2·6H2O. MTC-5 medium is MTC-7 without thiamine.
For a 400-mL working volume with pH control in a bioreactor, 300 mL of solution A was prepared without MOPS sodium salt and autoclaved. The bioreactor was left inside the anaerobic chamber for 12 to 16 h for purging with the gas inlet/outlet tube open. Next, 16 mL of solution B, 8 mL of solution C, 8 mL of solution D, and 16 mL of solution E were added through a 0.22-μm filter (catalog no. 430517; Corning), and the final volume was adjusted to 400 mL using autoclaved distilled water stored in a nitrogen atmosphere. The pH was controlled at 7.0 ± 0.05 for using a Mettler–Toledo (Columbus, OH, USA) pH probe with the addition of 4 N KOH. To control for small random variations in media preparation and fermenter setup, biological replicate fermentations were performed on different days.
Enzyme assays
All chemicals were ordered from MilliporeSigma unless otherwise specified. All enzyme assays were performed anaerobically unless otherwise mentioned. For enzyme assays, cells were grown to mid-exponential phase (OD600 between 0.6 and 1.0), harvested and lysed to obtain cell extract as described earlier [24]. Protein concentrations were determined using Bradford assay (Coomassie Plus Reagent, Thermo Scientific), with bovine serum albumin used as a protein standard. All the enzyme assays were performed at 55 °C except for the phosphofructokinase assay, which was performed at 40 °C due to precipitation issues associated with the aldolase coupling enzyme. All enzyme assays were performed in (1.2 - 1.4) ml reaction volume in a diode-array spectrophotometer.
Phosphofructokinase (PFK) assay
PFK activity was assayed by measuring the formation of fructose 1,6-bisphosphate coupled with consumption of NADH as previously described [11, 18]. The assay reaction contained 100 mM Tris–HCl (pH 7.0), 5 mM MgCl2, 0.15 mM NADH, 1 mM fructose-6-phosphate, 4 U/mL fructose bisphosphate aldolase, 4 U/mL triosephosphate isomerase, 4 U/mL a-glycerophosphate dehydrogenase, cell extract, and 2 mM either PPi or ATP. The assay reaction was started by the addition of the phosphate donor (PPi or ATP).
Pyruvate phosphate dikinase (PPDK) assay
PPDK activity was assayed by measuring the consumption of NADH by coupled assay with lactate dehydrogenase (LDH) as previously described with slight modification [11]. The assay mixture contained 100 mM Tris–HCl (pH 7.0), 5 mM MgCl2, 2 mM AMP, 0.15 mM NADH, 20 mM NH4Cl, 2 mM PEP, 1 mM fructose-1,6-bisphosphate, 4 U/ml lactate dehydrogenase, cell extract, and 2 mM PPi. The assay reaction was started by the addition of PPi.
Pyruvate kinase (PYK) assay
PYK activity was measured by coupled assay with lactate dehydrogenase (LDH) as described earlier with slight modification [11, 16, 26]. The assay buffer contained 100 mM Tris–HCl, pH 7.5 (at 55 °C), 5 mM dithiothreitol (DTT), 10 mM KCl, 12 mM MgCl2, 10 mM ADP, 0.3 mM NADH, 0.1 mM 3-phosphoglyceric acid (3PG), 5 mM PEP, 12 U/ml LDH enzyme, and cell extract. This reaction could be started with either ADP or PEP. For this experiment, the assay reaction was started by the addition of ADP.
Soluble pyrophosphatase (PPase) assay
PPase activity was measured aerobically based on hydrolysis of pyrophosphate (PPi) to inorganic phosphate (Pi) using malachite green assay as defined earlier [27] with some modifications. The assay mixture contained 100 mM Tris–HCl at pH 8.0, 1 mM MgCl2, 1 mM sodium pyrophosphate and 2, 4 or 8 μl cell extract. This assay mixture was incubated at 55 °C for 2 min and the reaction was stopped by transferring the samples to ice. 2.5 μl of this sample was added to 40 μl of malachite green reagent (Malachite green:Ammonium molybdate at 3:1) in 96 wells plate. The wells with PPase activity turn green in color. 7.5 μl of 34% sodium citrate was added, mixed and incubated at room temperature for 30 min to allow full color development [28]. Phosphate concentration was calculated by measuring absorbance at 660 nm using a calibration curve with phosphate concentration ranging from 0 to 5 mM.
Aldehyde dehydrogenase (ALDH) assay
ALDH activity was measured anaerobically as described earlier [29]. The assay reaction contained 100 mM Tris–HCl (pH 7.0), 5 μM FeSO4, 0.25 mM NADH or NADPH, 1.25 mM acetyl-CoA, 1 mM DTT, and cell extract. The assay reaction was started by the addition of acetyl-CoA.
Alcohol dehydrogenase (ADH) assay
ADH activity was measured anaerobically as described earlier [29]. The assay reaction contained 100 mM Tris–HCl (pH 7.0), 5 μM FeSO4, 0.25 mM NADH or NADPH, 18 mM acetaldehyde, 1 mM DTT, and cell extract. The assay reaction was started by the addition of acetaldehyde.
13C labeling conditions
For 13C labeling experiments, C. thermocellum cells were grown anaerobically on MTC-5 (medium for thermophilic clostridia) medium consisting of 3 mM naturally labeled cellobiose (Sigma C7252) and 3 mM uniformly labeled 13C-cellobiose (Omicron Biomedicals, CEL-002) as mentioned earlier [18]. 10 mL of this media was inoculated with 10 µL freezer stock of LL1590, LL1592 and LL1711 and incubated at 55 °C. The freezer stocks were prepared by growing the cells to mid-exponential phase (OD600 ~ 0.6) on MTC-5 with 5 g/L naturally labeled cellobiose.
Metabolite samples were extracted at three different phases during growth, i.e., early log phase (OD600 ~ 0.4), late log phase (OD600 ~ 0.8) and stationary phase (12 h after maximum OD has reached). The growth was monitored continuously using a custom-built absorbance reader. Intracellular metabolites were collected using vacuum filtration as previously described [12, 25, 30, 31]. For each sample, 2.5 mL of culture medium was filtered through 3.0 µm hydrophilic nylon filter (SF14529, Tisch Scientific) to separate cells from medium components. The filter with cells was placed in 1.6 mL of cold extraction solvent (40% acetonitrile, 40% methanol, and 20% water) with the cell side facing down and kept on aluminum block from −80 °C to quench metabolism and extract metabolites. Cells were washed off the filter using extraction solvent and then centrifuged for 5 min to remove cell debris. The supernatant with metabolites was collected, dried using a sample concentrator (catalog no. EW-36620-40; Cole-Parmer) to remove the metabolite extraction buffer and resuspended in molecular-grade water. This metabolite extract was analyzed using LC–MS. Metabolites of interest were detected based on retention time and mass-to-charge ratio, compared with pure standards, using El-MAVEN software [32].
Quantification of PPi in cells
To quantify the concentration of PPi in cells, samples were resuspended in molecular-grade water as mentioned earlier (Sect. "13C labeling conditions"). Inorganic pyrophosphate (PPi) was quantified using a combination of two different kits; a PPiLight inorganic pyrophosphate assay kit (catalog no. LT07-610; Lonza) which measures the sum of PPi and ATP and an ADP/ATP ratio assay kit (catalog no. MAK135; Sigma-Aldrich) which measures only ATP without cross-reacting with PPi. Then, PPi in cells was calculated using the first kit, and corrected for the effects of ATP using the second kit using the formula below:
PPi (mM) per cell = (PPi (mM) in metabolite extract × Volume of metabolite extraction buffer)/Intracellular volume (ml) where,
PPi (mM) in metabolite extract was calculated by using the kits above as PPi (mM) = ((ATP + PPi)LT07-610 − ATPMAK135)/1000,
and Intracellular volume = cell volume × cell number [25].
pH-controlled bioreactor fermentation conditions and metabolites extraction
C. thermocellum cultures were grown anaerobically at 55 °C in a Coy (Ann Arbor, MI) anaerobic chamber with a gas phase of 85% N2, 10% CO2, and 5% H2. Fermentations were carried out in a 400 mL working volume in MTC-7 medium with 100 g/L cellobiose as substrate and pH controlled at 7.0, as mentioned earlier (sect. Media and Growth Conditions).
The extracellular metabolites/fermentation products (ethanol, lactate, formate, acetate, pyruvate) at different timepoints during the fermentation were quantified by HPLC (Waters, Milford, MA or LC-2030, Shimadzu) with refractive index (RI) and UV detection using an Aminex HPX-87H column (Bio-Rad, Hercules, CA) with a 5 mM sulfuric acid solution eluent at 0.6 mL/minute flow rate and column temperature of 60°C [33].
To quantify intracellular/glycolysis metabolites, samples were taken at the same timepoints as HPLC. Samples were prepared using a protocol we previously developed for metabolite quantification from high-substrate fermentations [25]. Briefly, samples were extracted using vacuum filtration followed by quenching. For each sample (0.5–8) mL of culture medium was filtered through 3.0 µm hydrophilic nylon filter based on OD600 of cells. The samples were then quenched, dried, resuspended in molecular grade water and analyzed using LC–MS.
Liquid chromatography–mass spectrometry (LC–MS) analysis
A Vanquish ultra-high-performance liquid chromatography (UHPLC) system (Thermo Scientific) coupled to a hybrid quadrupole-Orbitrap™ mass spectrometer (Q Exactive™; Thermo Scientific) equipped with electrospray ionization operating in negative-ion mode as defined earlier [12] was used for LCMS analysis. The chromatography was performed at 25 °C using a 2.1 × 100 mm reverse-phase C18 column with a 1.7 μm particle size (Water™; Acquity UHPLC BEH). Two chromatography gradients were used. The first used Solvent A (97:3 H2O: methanol + 10 mM tributylamine) and Solvent B (100% methanol) as follows: 0–2.5 min, 5% B; 2.5–17 min, linear gradient from 5% B to 95% B; 17–19.5 min, 95% B; 19.5–20 min, linear gradient from 95% B to 5% B; 20–25 min, 5% B. The second also used Solvent A and Solvent B (100% methanol) and was as follows: 0–2.5 min, 5% B; 2.5–7.5 min, linear gradient from 5% B to 20% B; 7.5–13 min, 20% B to 55% B; 13–18.5 min, 55% B to 95% B; 18.5–19 min linear gradient from 95% B to 5% B; 19–25 min, 5% B. The flow rate was held constant at 0.2 mL/min for both chromatograph methods. Metabolites of interest were identified by retention times (based on pure standards) and monoisotopic mass using MAVEN [34] and El-MAVEN [32] software.
Sequencing methods
Routine Sanger sequencing was outsourced to Azenta Life Sciences. Whole genome resequencing was performed by the Department of Energy Joint Genome Institute using the Illumina MiSeq sequencing platform, with a minimum of 100-fold coverage. Strains were analyzed with the software CLC Genomics Workbench (Qiagen) using strain DSM1313 as the reference genome (Genbank accession NC_017304.1); reads were filtered against strain LL1649 (accession number SRP222669) [18] to exclude inherited mutations. A summary of the identified mutations is provided in Additional File 4.
Statistical analysis
Statistical analyses were conducted using t tests to assess significant differences, with a threshold set at p = 0.05. A paired t-test was used to compare PPase enzyme activities between strains with and without PPase. A one-sided t-test was used to determine whether PPi metabolite levels were lower in strains expressing PPase. Specifically, PPi levels in the PPi-free glycolysis strain (LL1711) were compared to those in the wild-type glycolysis strains (LL1590 and LL1592), with significance set at p = 0.05.
Results
Strain construction to eliminate and replace PPi-dependent glycolytic reactions
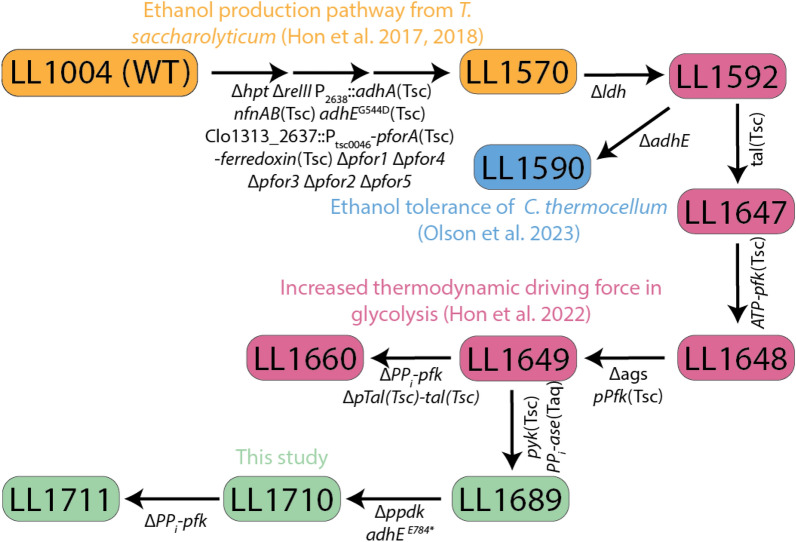
In our previous work, we created strain LL1660 (Fig. 1), where the PPi-pfk gene was replaced with an ATP-pfk gene [18], eliminating a major sink for PPi. We also observed the spontaneous occurrence of a large partial genome duplication. This partial genome duplication region encodes proteins that may serve as alternative sinks for PPi, including a membrane-bound pyrophosphatase gene (hppA–Clo1313_0823) and pyruvate:phosphate dikinase gene (ppdk–Clo1313_0949).
Fig. 1.
Strain lineage diagram showing the sequence of genetic modifications for strains presented in this work and relevant prior work. Strains shown in blue, orange and pink colors correspond to previous metabolic engineering work in our group. The green color represents the strains that were created in this study. Genetic modification between the strains are mentioned below specific arrows. Multiple arrows indicate multiple genetic modifications between different strains. The complete genotype of strains used in this work are presented in Table 1
We, therefore, hypothesized that the partial genome duplication event might have occurred to address an imbalance in the supply and demand of PPi created by the mutations in strain LL1660. To address this potential problem, we set out to construct a new strain where PPi-consuming reactions in glycolysis were eliminated, and where the excess PPi generated by metabolism was hydrolyzed using a soluble PPase [27]. Initially, we attempted to express soluble PPase in wild-type C. thermocellum, but we did not get any colonies in at least two transformations. This was not surprising, given our prior work showing the importance of PPi as a cofactor for the PFK reaction in glycolysis [11, 16].
Next, we tried to express the soluble PPase in a strain with ATP-linked PFK activity (strain LL1649), which we hypothesized would be more compatible with the metabolic effects of the PPase. In this strain, we occasionally obtained a small number of colonies with our PPase expression plasmid. However, in those colonies, the PPase gene was invariably inactivated either by a transposon insertion or inactivating nonsense mutation. In this phase, we tested PPases from Geobacillus stearothermophilus, Geobacillus thermoglucosidasius, Thermoplasma acidophilum, Thermus aquaticus, and Thermus thermophilus (plasmids pLL1501 to pLL1505, Table 2). Out of these, only PPase from Geobacillus thermoglucosidasius and Thermus aquaticus showed PPase activity (data not shown).
Table 2.
Plasmids used in this study
| Plasmid | Description | Accession no | References |
|---|---|---|---|
| pDGO143 | C. thermocellum expression vector | KX259110 | [23] |
| pDGO145 | Deletion/Integration vector backbone | KY852359 | [24] |
| pMU2051 | Plasmid for deletion of ppdk gene | KC146550 | [11] |
| pLL1392 | Deletion vector; deletes C. thermocellum PPi-pfk only in strain LL1647 and its derivatives | ON809513 | [18] |
| pLL1498 | pDGO143 with C. thermocellum Clo 1313_2638 promoter driving T. saccharolyticum ATP-pfk and T. aquaticus PPase | PP855251 | This work |
| pLL1499 | pDGO143 with C. thermocellum Clo 1313_2638 promoter driving T. aquaticus PPase | PP855252 | This work |
| pLL1500 | pDGO143 with C. thermocellum Clo 1313_2638 promoter driving Geobacillus stearothermophilus PPase | PP855253 | This work |
| pLL1501 | pDGO143 with T. saccharolyticum enolase promoter driving Geobacillus stearothermophilus PPase | PP855254 | This work |
| pLL1502 | pDGO143 with T. saccharolyticum enolase promoter driving Geobacillus thermoglucosidasius PPase | PP855255 | This work |
| pLL1503 | pDGO143 with T. saccharolyticum enolase promoter driving T. acidophilum PPase | PP855256 | This work |
| pLL1504 | pDGO143 with T. saccharolyticum enolase promoter driving T. aquaticus PPase | PP855257 | This work |
| pLL1505 | pDGO143 with T. saccharolyticum enolase promoter driving T. thermophilus PPase | PP855258 | This work |
| pLL1506 | pDGO145 with T. saccharolyticum pyk and G. thermoglucosidasius cytosolic PPase driven by T. saccharolyticum enolase promoter | PP855259 | This work |
| pLL1507 | pDGO145 with T. saccharolyticum pyk and T. aquaticus cytosolic PPase driven by T. saccharolyticum enolase promoter | PP855260 | This work |
We further hypothesized that in addition to eliminating the need for PPi in the PFK reaction, we also needed to eliminate the need for PPi in the PPDK reaction (which converts PEP to pyruvate) [16, 35]. To do this, we designed an integration construct to simultaneously introduce pyruvate kinase and a soluble PPase onto the chromosome, in this case using the PPases from G. thermoglucosidasius and T. aquaticus, since we had in some instances observed PPase activity from cell extracts of C thermocellum that expressed those PPases [27]. Successful strain construction was obtained only with the T. aquaticus PPase containing construct, pLL1507 (Table 2), which was validated by the gain of both pyruvate kinase and cytosolic PPase activity (Fig. 2). Our ability to observe functional PPase expression only in strains with both ATP–PFK and PYK activity provides further evidence of progress toward eliminating PPi as a key energy carrying cofactor in C. thermocellum.
Fig. 2.
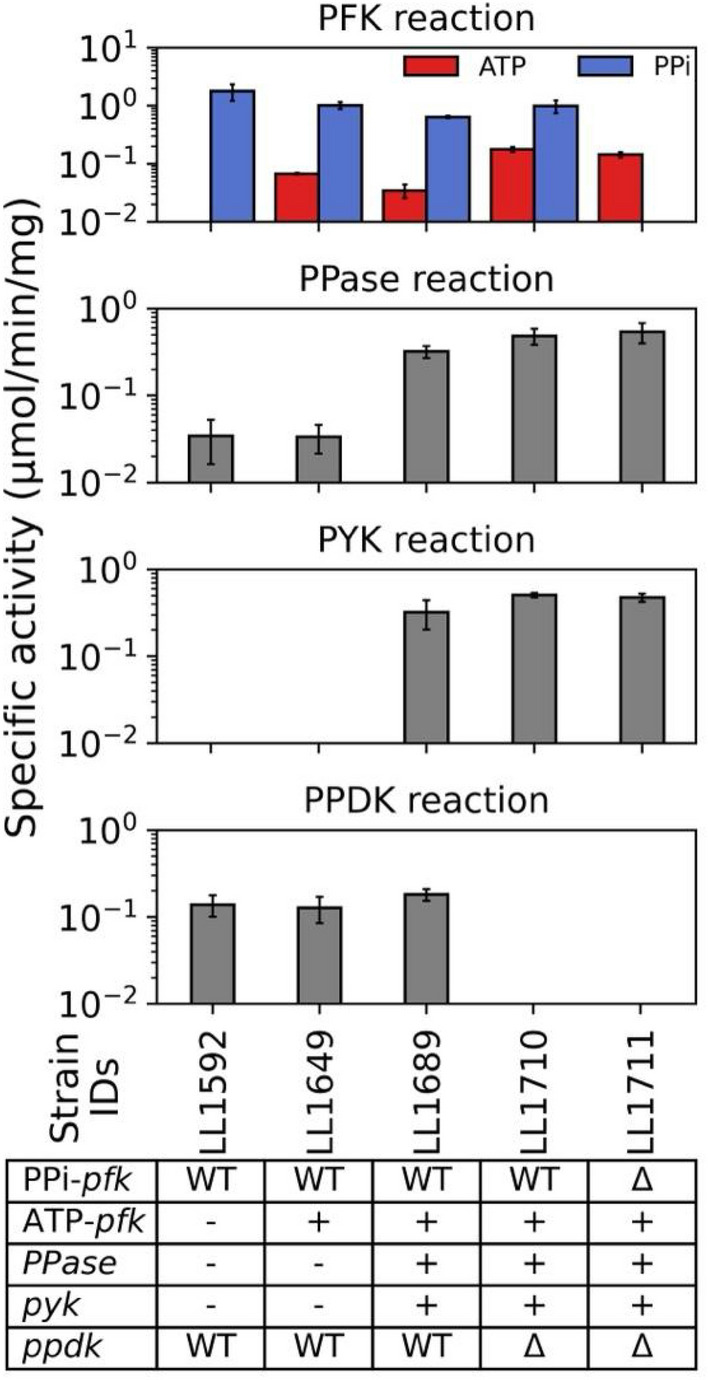
Enzyme activity of C. thermocellum strains for reactions related to PPi-linked reactions. These reactions include phosphofructokinase (PFK, EC 2.7.1.11 or 2.7.1.90), pyruvate phosphate dikinase (PPDK, EC 2.7.9.1), pyruvate kinase (PYK, EC 2.7.1.40) and thermostable soluble PPase (EC 3.6.1.1). In the PFK reaction panel, red bars and blue bars represent activity with either ATP or PPi as the cofactor. In the table at the bottom of the figure, the ‘+’ and ‘−’ signs indicate the presence or absence of heterologous genes at a given locus. The ‘WT’ and ‘Δ’ signs indicate the presence or absence (i.e., by deletion) of native genes. All the reactions were carried out using cell extracts. Missing bars indicate the absence of activity (i.e., below the limit of detection, <0.02 μmol/min/mg). Error bars represent 1 standard deviation (n ≥ 3 biological replicates). Raw enzyme assay data is available as Additional File 1
Strain LL1592, which uses the native C. thermocellum glycolytic pathway, with the T. saccharolyticum pyruvate-to-ethanol pathway [18] was selected as starting strain for further engineering. Strain LL1649 was derived from strain LL1592, by introduction of the T. saccharolyticum transaldolase, followed by simultaneous deletion of the ADP–glucose synthase operon and expression of T. saccharolyticum ATP-pfk. Strain LL1689 was derived from strain LL1649 by the simultaneous integration of the T. saccharolyticum pyruvate kinase and the cytosolic pyrophosphatase from Thermus aquaticus. The purpose of introducing a heterologous pyruvate kinase was to allow subsequent deletion of the ppdk gene. The purpose of introducing a soluble PPase was to serve as an alternative sink for PPi, once both the PPi-pfk and ppdk genes were deleted. With these modification in place, we were then able to successfully delete glycolytic sinks for PPi, including the ppdk gene (resulting in strain LL1710), and then, subsequently, the PPi-pfk gene (resulting in strain LL1711) (Fig. 1).
Biochemical assays were performed to confirm the effect of the genetic modifications of the strains used in this study. As expected, strain LL1592 has exclusively PPi-PFK activity.Strains LL1649, LL1689, and LL1710 have both ATP- and PPi-linked PFK activity, as expected (PFK reaction, Fig. 2). Finally, strain LL1711 has exclusively ATP-PFK activity as expected. Functional expression of the pyk gene was confirmed by the presence of PYK activity in strains LL1689, LL1710 and LL1711 (PYK reaction, Fig. 2). The effect of the ppdk gene deletion was confirmed in strains LL1710 and LL1711 based on elimination of PPDK activity (PPDK reaction, Fig. 2). Low levels of PPase activity were still detected in strains LL1592 and LL1649; this could be attributed to the native membrane bound PPase enzyme being present in the cell extracts. Previous studies have reported the presence of membrane-bound pyrophosphatase in C. thermocellum cells [27]. Nonetheless, strains LL1689, LL1710, and LL1711 exhibited PPase activities that were approximately 13-fold higher on average (paired t-test, p = 0.0003) than those of strains LL1592 and LL1649, which lack soluble PPase. We attribute this increase to the expression of a soluble PPase in the former strains. As such, it was determined that strain LL1711 had been engineered to not require PPi as a cofactor in glycolysis, and that it now possessed a more-canonical glycolytic pathway [12].
Use of ATP-dependent pathway alters thermodynamics of glycolysis
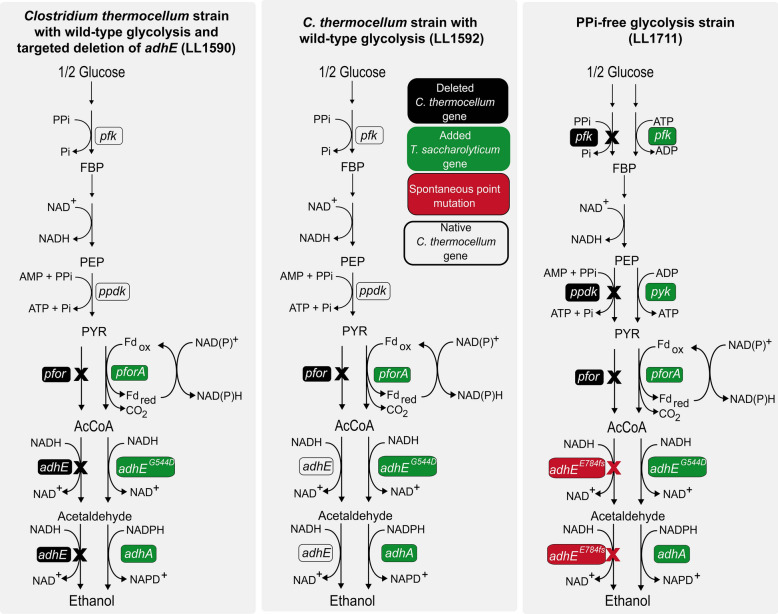
To determine the effect the genetic modifications had on the reversibility of the PFK reaction, we performed 13C labeling on C. thermocellum strains LL1590, LL1592 and LL1711. Initially, we were planning to compare only the parent strain (LL1592) and the PPi-free glycolysis strain (LL1711); however, whole genome sequencing of strain LL1711 revealed a frameshift mutation in the wild-type C. thermocellum adhE gene corresponding to amino acid position E784 that eliminated its function (Figure S1). We, therefore, included another control strain (LL1590) derived from the parent strain (LL1592) with a targeted deletion of the wild-type C. thermocellum adhE gene [19], to better understand the effects of our changes to the glycolytic pathway without the confounding effects of the modification to ethanol production pathway. Note that all three strains (LL1590, LL1592, and LL1711) still produce ethanol via the heterologous T. saccharolyticum adhEG544D (Figs. 1, 3).
Fig. 3.
Graphical representation of C. thermocellum strains used for fermentation and metabolite analysis. White boxes represent native C. thermocellum genes, black boxes represent knockouted genes, green boxes represent heterologous expressed T. saccharolyticum genes and red boxes indicate spontaneous mutation present in strain LL1711. X sign represents inactivated reactions. Strain LL1592 is the parent strain. Strain LL1590 is a control for loss of wild-type C. thermocellum AdhE activity (note that strain LL1590 is descended from strain LL1592 despite what might be assumed from the strain ID number, see Fig. 1). Strain LL1711 is the PPi-free glycolysis strain. Strain LL1590 (LL1592 with a targeted deletion of C. thermocellum adhE) is included to control for the effects of inactivation of the native adhE gene, to allow the effects of the changes to glycolysis to be seen more clearly. In strains LL1590 and LL1711, the only functional adhE gene is the one heterologously expressed from T. saccharolyticum. Key: fructose 1,6-bisphosphate (FBP); phosphoenolpyruvate (PEP); pyruvate (PYR); acetyl-CoA (AcCoA); phosphofructokinase (pfk); pyruvate phosphate dikinase (ppdk); pyruvate kinase (pyk); pyruvate ferredoxin/flavodoxin oxidoreductase (pfor); alcohol dehydrogenase (adh). Figure adapted from [36]
C. thermocellum does not readily consume glucose [37], and thus cellobiose is the most commonly used soluble sugar for laboratory growth experiments. Strains were cultured on a 1:1 ratio of naturally labeled to uniformly 13C-labeled cellobiose. We confirmed that 13C cellobiose and naturally labeled cellobiose are taken up equally by measuring the ratio of M+0 to M+6 isotopomers for glucose 6-phosphate, which ranged from 0.8 to 1.2, and is close to the expected value of 1.0 (Additional File 2). Samples were collected at three different phases (early log, late-log and stationary) during growth (Figure S2). As previously described [12], the forward glycolysis reaction should generate a 1:1 mixture of fully labeled (M+6) and fully unlabeled (M+0) glucose-6-phosphate (G6P), fructose-6-phosphate (F6P) and fructose-1,6-bisphosphate (FBP). The forward flux of fructose–bisphosphate aldolase reaction (FBA) will generate 50% unlabeled (M+0) and 50% fully labeled (M+3) dihydroxyacetone phosphate (DHAP) and glyceraldehyde-3-phosphate (G3P). However, the reverse FBA reaction flux generates a 1:2:1 mixture of M+0, M+3 and M+6 FBP. The reverse flux from phosphofructokinase (PFK) and phosphoglycerate isomerase (PGI) reactions transfer these M+3 species into the F6P and G6P pools. Appearance of the M+3 isotopomer in the F6P and G6P pools allows us to uniquely identify reverse flux through the PFK reaction. If the PFK reaction were completely irreversible, we would expect to see no M+3 species in either the F6P or G6P pools.
As expected, based on our prior work [18], we observed that the M+3 pool in the glycolytic intermediates G6P and F6P decreased substantially in the PPi-free glycolysis strain (LL1711) vs. the parent and control strains (LL1590 and LL1592), indicating that the genetic modifications introduced into LL1711 reduced the reversibility of the PFK reaction (G6P, F6P; Fig. 4). We saw an unexpected decrease in the M+3 fraction of FBP in mid-log phase in the PPi-free glycolysis strain (LL1711) (Figs. 4, S3). This suggests reduced reversibility of FBA reaction, although the reason for this change is not known.
Fig. 4.
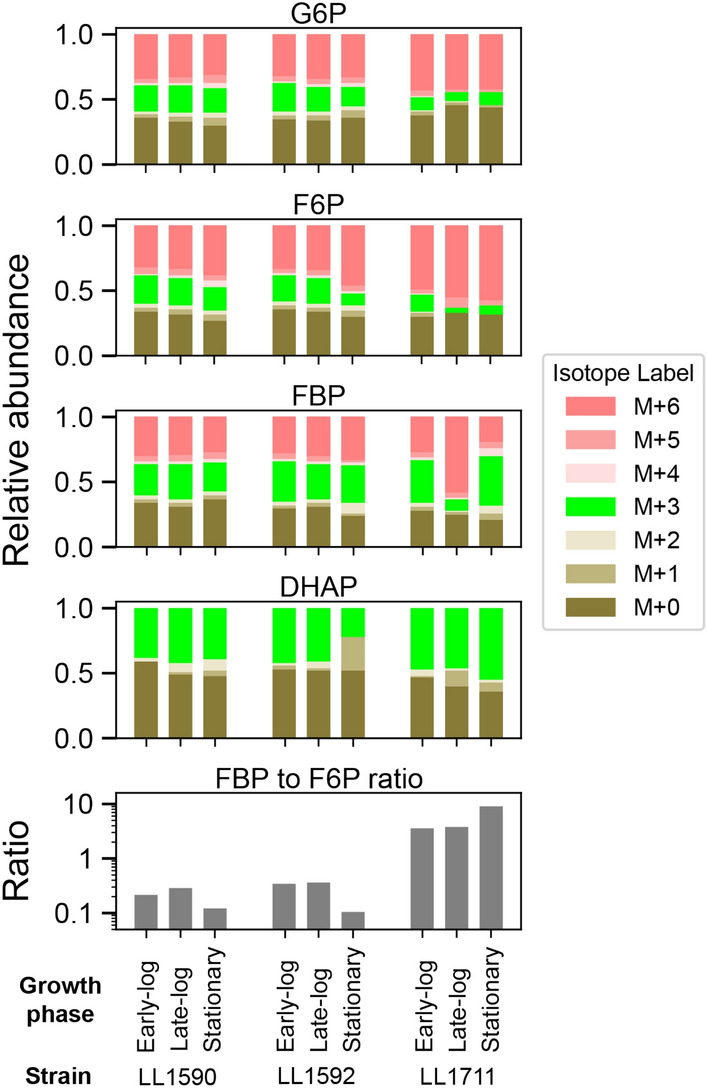
13C-labeling patterns of key glycolysis metabolites near the PFK reaction. The metabolites glucose-6-phosphate (G6P), fructose-6-phosphate (F6P), fructose 1,6-bisphosphate (FBP) and dihydroxyacetone phosphate (DHAP) were measured at different stages of growth for cells grown with a 50:50 mixture of uniformly labeled and naturally labeled cellobiose. ‘M+’ notation indicates the number of 13C-labeled carbon atoms. One set of representative data is shown for each strain (n = 2 biological replicates). Strain LL1592 is the parent strain, strain LL1590 is a control for loss of wild-type C. thermocellum AdhE activity, strain LL1711 is the PPi-free glycolysis strain. The ratio represents the ratio of absolute abundance of FBP to F6P. Figure S3 shows a comparison between biological duplicates of this experiment. Raw data used for the analysis is available as Additional File 2
In addition to comparing the isotope ratios, we also compared the ratio of the FBP and F6P metabolite pool sizes. This gives us an additional indication of the thermodynamic driving force of the reaction. A high thermodynamic driving force at the PFK reaction would be expected to give a high FBP/F6P ratio. Comparisons of FBP/F6P ratio showed a lower ratio for the parent and control strains that use the PPi-PFK reaction (LL1590 and LL1592) and a much higher ratio for the PPi-free glycolysis strain (LL1711), which uses the ATP–PFK reaction (Fig. 4). This ratio is 10-fold and 98-fold higher on average in ATP-pfk strain compared to PPi-pfk strains during log phases and stationary phases, respectively. The higher FBP/F6P ratio is similar to what is observed in organisms with canonical glycolytic pathways, such as E. coli and T. saccharolyticum [12, 38]. Taken together, these two lines of evidence (M+3 abundance changes and FBP/F6P ratio changes) support the conclusion that we have increased the thermodynamic driving force of the PFK reaction in our PPi-free glycolysis strain.
Expression of PPase results in lower PPi levels
To determine the effect of PPase expression in PPi levels of C. thermocellum strains with wild-type glycolysis (strains LL1590 and LL1592) and PPi-free glycolysis (strain LL1711), we quantified the concentration of PPi in cells.
We observed a significant decrease in PPi concentration in the PPi-free glycolysis strain (strain LL1711) (Fig. 5) during the early log and late-log growth phases. During the stationary phase, the difference between strain LL1592 and strain LL1711 was not as significant, which might be due to the depletion of the PPi pool during this phase of growth. We see consistently higher levels of PPi during the early log and late-log phases in strains LL1590 and LL1592, which then drop during the stationary phase in both strains.
Fig. 5.
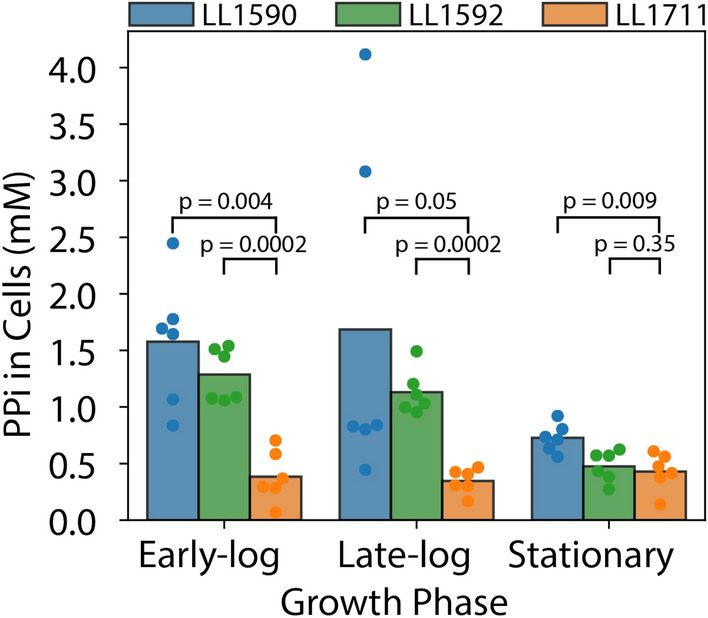
PPi concentration in C. thermocellum strains without PPase expression (strain LL1590 and strain LL1592) and with PPase expression (strain LL1711). The bar plot represents average values. The colored circles represent individual data points. PPi levels were measured in cell extracts and the intracellular concentration was calculated as described in the materials and methods section. p-values were calculated using one-sided paired t-test statistics. (n = 6; 3 biological replicates, 2 technical replicates)
Increased driving force leads to higher level of fermentation products in ATP-pfk strain
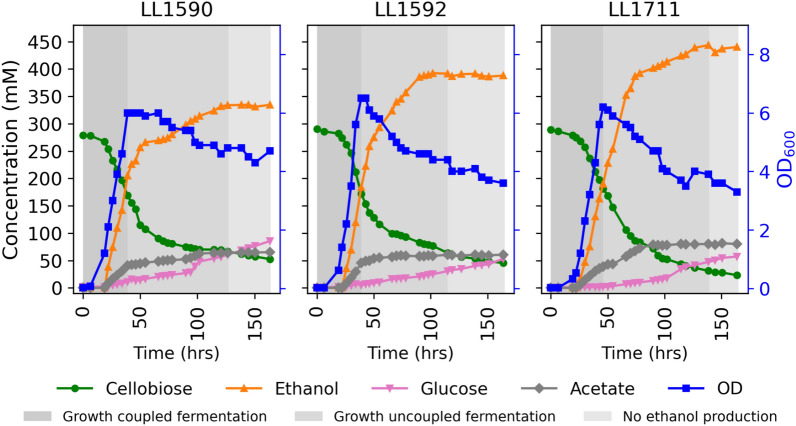
Having demonstrated increased thermodynamic driving force in glycolysis in the PPi-free glycolysis strain, we explored its impact on fermentation behavior and ethanol production using batch fermentations of all three strains with high substrate concentrations (100 g/L cellobiose). We optimized fermentation conditions to get repeatable results using strain LL1592 (Supplementary Figure S4), and used those conditions for all three strains. The fermentations had three distinct phases: (1) growth coupled fermentation, where both the cell growth and ethanol production are at the highest rate, (2) growth uncoupled fermentation, where cell growth stops and the rate of ethanol production decreases, and (3) cessation of both growth and ethanol production. During this final phase, cellobiose is hydrolyzed to glucose at a slow rate, but metabolism appears to be inactive.
The final ethanol titer was 329 ± 8 mM (15.1 ± 0.4 g/L) for strain LL1590, 403 ± 11 mM (18.6 ± 0.5 g/L) for strain LL1592, and 455 ± 12 mM (21.0 ± 0.6 g/L) for strain LL1711. Titer of another fermentation product, acetate, was 65 ± 0 mM (3.8 ± 0 g/L) for strain LL1590, 51 ± 9 mM (3.0 ± 0.5 g/L) for strain LL1592 and 88 ± 7 (5.2 ± 0.4 g/L) for strain LL1711. (Figs. 6, S5, Additional File 3). Comparison of the fermentation products between LL1592 and LL1590 suggests that deletion of C. thermocellum adhE impairs the production of ethanol. Taking the effect of the native adhE inactivation into account (i.e., comparing LL1711 to LL1590), allows us to observe the effect of PPi-free glycolysis on ethanol production without the confounding effects of changes in the ethanol production pathway. In this comparison (LL1711 vs. LL1590), we observe a 38% increase in ethanol titer, and a 35.4% increase in acetate titer. Together, these suggest that our PPi-free glycolysis strain exhibits increased glycolytic flux, resulting in an increased abundance of acetyl-CoA, which is then converted to more ethanol and acetate.
Fig. 6.
Batch fermentations of C. thermocellum strains with wild-type glycolysis (strains LL1590 and LL1592) and PPi-free glycolysis (strain LL1711). Fermentations were performed with 100 g/L cellobiose in MTC-7 chemically defined medium with pH maintained at 7.0 ± 0.05 by addition of 4 M potassium hydroxide. The shaded background represents different phases of fermentation: dark gray, growth coupled fermentation; medium gray, growth uncoupled fermentation; and light gray, no ethanol production phases. One representative fermentation profile is shown for each strain (n ≥ 2). Figure S5 shows a biological duplicate of this experiment. Additional File 3 (LL1590-1, LL1592-1, and LL1711-1) shows additional fermentation product data for this experiment.
Changes in glycolysis metabolite levels at elevated substrate concentrations in pH-controlled fermentations
To better understand the changes in central metabolism that allowed for increased ethanol titer in the PPi-free glycolysis strain, we measured intracellular glycolytic metabolites over the course of the fermentation using the same fermentation conditions from Fig. 6 (100 g/L cellobiose, pH-controlled batch fermentations). Since preliminary data showed similar pattern of glycolytic metabolites (Figure S6) and 13C labeling patterns were similar for both the PPi-pfk strains (Fig. 4, S3), and strain LL1590 produced lower levels of ethanol than strain LL1592 (Fig. 6, S5), we compared glycolytic metabolite levels in strains LL1592 and LL1711.
In C. thermocellum, cellobiose is transported into cells via an ATP-binding cassette (ABC) transport system [11, 39, 40]. Once inside, cellobiose is cleaved into glucose and glucose 1-phosphate (G1P) by the enzyme cellobiose phosphorylase (EC 2.4.1.20). G1P and glucose are then converted to glucose 6-phosphate (G6P) through the actions of phosphoglucomutase (EC 5.4.2.2) and GTP-linked glucokinase (EC 2.7.1.2), respectively. G6P subsequently enters the Embden–Meyerhof–Parnas (EMP) glycolysis pathway, as previously described [11].
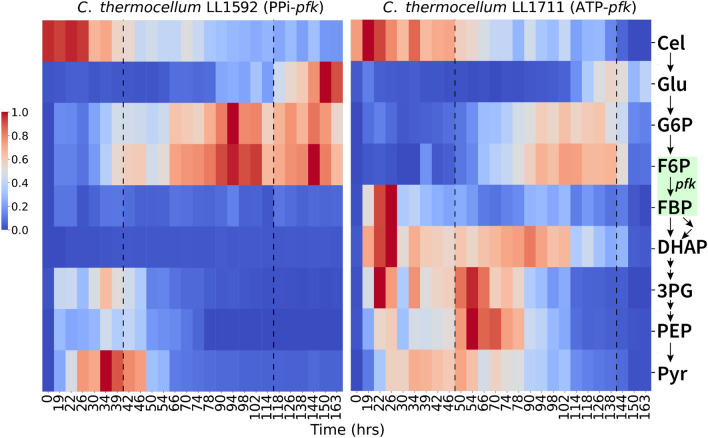
Three major changes were observed in the glycolytic metabolite profiles between these two strains. First, in the parent strain (LL1592, with wild-type glycolysis) we see gradual accumulation of hexose phosphates (G6P, F6P) as the fermentation progresses. In the PPi-free glycolysis strain (LL1711), hexose phosphates were still observed to accumulate, but to a lesser extent as the fermentation progressed. Second, during the growth-uncoupled fermentation phase, we observed a dramatic increase in the levels of lower glycolysis metabolites (DHAP, 3PG, and PEP) in the PPi-free glycolysis strain. Finally, in the phase where fermentation and ethanol production stop, the parent strain shows a large accumulation of upper glycolysis metabolites (glucose, G6P, and F6P), while the PPi-free glycolysis strain shows low levels of almost all metabolites (Figs. 7, S7).
Fig. 7.
Abundance of glycolytic metabolites in C. thermocellum strains LL1592 and LL1711 during the course of fermentation. The dotted lines differentiate different growth phases (from left to right, growth coupled fermentation, growth uncoupled fermentation and no ethanol production, respectively). Absolute metabolite concentrations were determined by using external standards. Metabolite concentrations were normalized (per metabolite) from 0 to 1, with 0 being the least and 1 being the highest concentration measured. The green box shows the location of the PFK reaction in glycolysis. Figure S7 shows a biological duplicate of this experiment. Additional File 6 shows absolute intracellular metabolite concentrations. Cel cellobiose, Glu glucose, G6P glucose 6-phosphate, F6P fructose 6-phosphate, FBP fructose 1,6-bisphosphate, DHAP dihydroxyacetone phosphate, 3PG 3-phosphoglycerate, PEP phosphoenolpyruvate, Pyr pyruvate
These intracellular metabolites data over the course of fermentation also underscores the importance of conducting metabolomics studies throughout the fermentation process. Most previous metabolomics studies in C. thermocellum and other microbes have focused on metabolite levels at a single-timepoint, typically during the mid-exponential growth phase [12, 38, 41]. However, analyzing metabolites at multiple timepoints during fermentation reveals important dynamic changes in the metabolite pools.
Discussion
We have successfully engineered a strain of C. thermocellum where PPi no longer plays a role in central metabolism, which increased the thermodynamic driving force in glycolysis. In our previous work, where we replaced the PPi-PFK reaction with an ATP-PFK reaction without engineering other aspects of PPi metabolism [18], we observed an increased thermodynamic driving force at the PFK reaction, and an increased FBP/F6P ratio, but this metabolic perturbation appeared to be confined to a localized region around the PFK reaction. For example, the FBP level increased, but the downstream metabolites (DHAP, 3PG, or PEP) did not. In this work, by contrast, the levels of these downstream metabolites do increase. There are several possible explanations for this difference. One is that changes in PPi levels resulting from expression of a soluble PPase affect glycolytic flux. It is known that PPi concentrations allosterically regulate enzymes in the malate shunt [42], and it is possible that PPi concentrations regulate other enzymes in central metabolism as well. Another possible explanation is that we previously measured intracellular metabolite concentrations in bottle fermentations where pH was only controlled by MOPS buffer, whereas in this work, the pH was actively controlled. Changes in pH may have limited the effects of the ATP–PFK reaction in those experiments. Finally, a third possible explanation is that the previously observed localization of the effect of ATP–PFK was a temporal effect, since our prior work only measured intracellular metabolites at a single timepoint in mid-log phase. Among these three possibilities, we have no evidence to distinguish between the first two. We can, however, tentatively reject the third one. In the PPi-free glycolysis strain (LL1711), the increased abundance of downstream metabolites (DHAP, 3PG, and PEP) is observed in both mid-log and early stationary phases (approximately 22–78 h). Comparing any of the points in this range with our prior results shows a clear difference with the much lower levels of these metabolites observed in mid-log phase with our previously developed ATP–PFK strain (LL1660) [18].
In our PPi-free glycolysis strain (strain LL1711), whole genome sequencing revealed a frameshift mutation (E784) in C. thermocellum adhE that eliminated its function. We have routinely observed mutations in adhE in strains that either have higher ethanol production or tolerance [19, 43, 44]. Previous studies have shown that inactivating mutations in adhE that eliminated NADH-linked alcohol dehydrogenase (ADH) activity increase ethanol tolerance in C. thermocellum [43, 45]. We believe that this is an adaptive response by the strain to avoid a thermodynamic limitation that can result from glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and ADH reactions using the same cofactor (i.e., NADH) [19, 44, 45].
Although the PPi-free glycolysis strain produced 455 mM (21.0 g/L) ethanol, it is likely that titers of 40–50 g/L are needed for commercial application [1]. Nonetheless this work represents another step along the path toward converting the atypical glycolysis of C. thermocellum to the canonical glycolysis [11, 12]. In our previous work, we replaced the PPi-linked PFK reaction with an ATP-linked PFK reaction [18]. In this work, we needed to recreate this modification due to the presence of a spontaneous partial genome duplication event in our prior strain, and additionally introduce a deletion of the ppdk gene and heterologous expression of a soluble PPase gene. The expression of a soluble PPase appears to be the enabling factor for engineering PPi-free glycolysis [27]. Subsequent steps along this path may include eliminating the malate shunt [16], and understanding the role of GTP vs. ATP in the glucokinase and phosphoglycerate kinase reactions [11].
Conclusions
In the field of Metabolic Engineering, strategies for improving product yield are relatively well developed compared to strategies for improving product titer. Engineering the thermodynamic landscape of a metabolic pathway provides an interesting approach to address product titer limitations. Although there is extensive prior literature on analysis of the thermodynamic landscape of metabolic pathways [12, 15, 17, 46], there have been far fewer attempts to apply this knowledge for targeted improvement of these pathways. Several prior studies have hypothesized that the low thermodynamic driving force of the atypical glycolysis of C. thermocellum, and/or a limited PPi supply for the PPi-consuming reactions might limit product titer [11, 27, 47]. Here we show that the improved thermodynamic driving force of our PPi-free glycolysis allowed a 38% increase in ethanol titer (Fig. 6), providing evidence of the utility of this approach.
Supplementary Information
Additional file 2. Quantification of different isotopes of Glucose 6-phosphate, Fructose 6-phosphate, Fructose 1,6-bisphosphate and Dihydroxyacetone phosphate.
Additional file 3. Fermentation data for the strains LL1590, LL1592 and LL1711
Additional file 4. Table of mutations identified by genome resequencing.
Additional file 5. Compiled supplementary information document with figures S1 through S7.
Additional file 6. Intracellular metabolite concentrations for the strains LL1590, LL1592 and LL1711.
Acknowledgements
Not applicable.
Author contributions
BDS: Conceptualization; Data curation; Formal analysis; Investigation; Methodology; Software; Validation; Visualization; Writing—original draft. SH: Conceptualization; Investigation; Strain development; Formal analysis; Visualization; Writing—Review and Editing. ET: Data curation; Investigation; Writing—Review and Editing. DMS: Data curation; Investigation. DAN: Writing—Review and Editing. AMG: Conceptualization, Writing—Review and Editing. LL: Funding Acquisition, Project Administration, Writing—Review and Editing. DGO: Conceptualization, Writing—Review and Editing, Supervision, Project Administration, Funding Acquisition.
Funding
This work was supported by the Center for Bioenergy Innovation (CBI), U.S. Department of Energy, Office of Science, Biological and Environmental Research Program under Award Number ERKP886. Whole genome resequencing was performed by the Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, and is supported by the Office of Science of the U.S. Department of Energy under contract number DE‑AC02–05CH11231
Availability of data and materials
Enzyme assay data, extracellular metabolite data, intracellular metabolite data, 13C labeling data and genome resequencing data are all provided in the supporting information. Strain resequencing data is available from the NCBI Sequence Read Archive https://www.ncbi.nlm.nih.gov/sra/ (Table 1). Plasmid DNA sequences and annotations are available from GenBank (Table 2). Materials available by request.
Table 1.
Strains used in this work are described below
| Organism | Strain ID | Description | Accession Number | References |
|---|---|---|---|---|
| C. thermocellum | LL1592 | DSM 1313 ∆hpt, ∆ldh, ΔreIII, P2638::adhA(Tsc), nfnAB(Tsc), adhEG544D(Tsc), Clo1313_2637::Ptsc0046-pforA(Tsc)-ferredoxin(Tsc), ∆pfor1, ∆pfor2, ∆pfor3, ∆pfor4, ∆pfor5 | SRX5290154 | [18] |
| C. thermocellum | LL1590 | LL1592 ∆adhE | SRX5290158 | [19] |
| C. thermocellum | LL1649 | LL1592 tal(Tsc), ATP-pfk(Tsc), ∆ags, pPfk(Tsc) | SRX6875980 | [18] |
| C. thermocellum | LL1689 | LL1649 pyk(Tsc), PPase(Taq) | SRX7724531 | This study |
| C. thermocellum | LL1710 | LL1689 ∆ppdk, adhE E784* | SRX9409012 | This study |
| C. thermocellum | LL1711 | LL1710 ∆PPi-pfk | SRX9409011 | This study |
Tsc and Taq refers to genes introduced from T. saccharolyticum and T. aquaticus, respectively.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
Lee R. Lynd is the co-founder and CTO of Terragia Biofuels, Inc (https://terragiabiofuel.com/). Shuen Hon is an employee of Terragia Biofuels. Terragia has a financial interest in commercialization of Clostridium thermocellum. There are no other competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Bishal Dev Sharma and Shuen Hon contributed equally to this work.
References
- 1.Lynd LR, Beckham GT, Guss AM, et al. Toward low-cost biological and hybrid biological/catalytic conversion of cellulosic biomass to fuels. Energy Environ Sci. 2022;15:938–90. [Google Scholar]
- 2.Lynd LR, Guss AM, Himmel ME, et al. Advances in consolidated bioprocessing using Clostridium thermocellum and Thermoanaerobacter saccharolyticum. In: Wittmann C, Liao JC, editors., et al., Industrial Biotechnology: Microorganisms. 1st ed. New York: Wiley; 2017. p. 365–94. [Google Scholar]
- 3.Mazzoli R, Olson DG. Clostridium thermocellum: a microbial platform for high-value chemical production from lignocellulose. Adv Appl Microbiol. 2020;113:111–61. [DOI] [PubMed] [Google Scholar]
- 4.Holwerda EK, Worthen RS, Kothari N, et al. Multiple levers for overcoming the recalcitrance of lignocellulosic biomass. Biotechnol Biofuels. 2019;12:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lynd LR, Liang X, Biddy MJ, Allee A, Cai H, Foust T, Himmel ME, Laser MS, Wang M, Wyman CE. Cellulosic ethanol: status and innovation. Curr Opin Biotechnol. 2017;45:202–11. [DOI] [PubMed] [Google Scholar]
- 6.Holwerda EK, Ellis LD, Lynd LR. Development and evaluation of methods to infer biosynthesis and substrate consumption in cultures of cellulolytic microorganisms. Biotechnol Bioeng. 2013;110:2380–8. [DOI] [PubMed] [Google Scholar]
- 7.Hon S, Holwerda EK, Worthen RS, Maloney MI, Tian L, Cui J, Lin PP, Lynd LR, Olson DG. Expressing the Thermoanaerobacterium saccharolyticum pforA in engineered Clostridium thermocellum improves ethanol production. Biotechnol Biofuels. 2018. 10.1186/s13068-018-1245-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tian L, Papanek B, Olson DG, et al. Simultaneous achievement of high ethanol yield and titer in Clostridium thermocellum. Biotechnol Biofuels. 2016;9:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holwerda EK, Olson DG, Ruppertsberger NM, Stevenson DM, Murphy SJL, Maloney MI, Lanahan AA, Amador-Noguez D, Lynd LR. Metabolic and evolutionary responses of Clostridium thermocellum to genetic interventions aimed at improving ethanol production. Biotechnol Biofuels. 2020;13:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dien BS, Cotta MA, Jeffries TW. Bacteria engineered for fuel ethanol production: current status. Appl Microbiol Biotechnol. 2003;63:258–66. [DOI] [PubMed] [Google Scholar]
- 11.Zhou J, Olson DG, Argyros DA, Deng Y, van Gulik WM, van Dijken JP, Lynd LR. Atypical glycolysis in Clostridium thermocellum. Appl Environ Microbiol. 2013;79:3000–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacobson TB, Korosh TK, Stevenson DM, Foster C, Maranas C, Olson DG, Lynd LR, Amador-Noguez D. In vivo thermodynamic analysis of glycolysis in Clostridium thermocellum and Thermoanaerobacterium saccharolyticum using 13 C and 2 H tracers. mSystems. 2020;5:e00736-e819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park JO, Rubin SA, Xu Y-F, Amador-Noguez D, Fan J, Shlomi T, Rabinowitz JD. Metabolite concentrations, fluxes and free energies imply efficient enzyme usage. Nat Chem Biol. 2016;12:482–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flamholz A, Noor E, Bar-Even A, Milo R. EQuilibrator - The biochemical thermodynamics calculator. Nucleic Acids Res. 2012;40:770–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noor E, Bar-Even A, Flamholz A, Reznik E, Liebermeister W, Milo R. Pathway thermodynamics highlights kinetic obstacles in central metabolism. PLoS Comput Biol. 2014;10: e1003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olson DG, Hörl M, Fuhrer T, Cui J, Zhou J, Maloney MI, Amador-Noguez D, Tian L, Sauer U, Lynd LR. Glycolysis without pyruvate kinase in Clostridium thermocellum. Metab Eng. 2017;39:169–80. [DOI] [PubMed] [Google Scholar]
- 17.Dash S, Olson DG, Hung S, Chan J, Amador-noguez D, Lynd LR, Maranas CD. Thermodynamic analysis of the pathway for ethanol production from cellobiose in Clostridium thermocellum. Metab Eng. 2019;55:161–9. [DOI] [PubMed] [Google Scholar]
- 18.Hon S, Jacobson T, Stevenson DM, Maloney MI, Giannone RJ, Hettich RL, Amador-Noguez D, Olson DG, Lynd LR. Increasing the thermodynamic driving force of the phosphofructokinase reaction in Clostridium thermocellum. Appl Environ Microbiol. 2022;88:e01258-e1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olson DG, Maloney MI, Lanahan AA, et al. Ethanol tolerance in engineered strains of Clostridium thermocellum. Biotechnol Biofuels Bioprod. 2023;16:137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gibson DG. Enzymatic assembly of overlapping DNA fragments. Methods Enzymol. 2011;498:349–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guss AM, Olson DG, Caiazza NC, Lynd LR. Dcm methylation is detrimental to plasmid transformation in Clostridium thermocellum. Biotechnol Biofuels. 2012;5:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olson DG, Lynd LR. Transformation of Clostridium thermocellum by electroporation. Methods Enzymol. 2012. 10.1016/B978-0-12-415931-0.00017-3. [DOI] [PubMed] [Google Scholar]
- 23.Hon S, Lanahan AA, Tian L, Giannone RJ, Hettich RL, Olson DG, Lynd LR. Development of a plasmid-based expression system in Clostridium thermocellum and its use to screen heterologous expression of bifunctional alcohol dehydrogenases (adhEs). Metab Eng Commun. 2016;3:120–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hon S, Olson DG, Holwerda EK, Lanahan AA, Murphy SJL, Maloney MI, Zheng T, Papanek B, Guss AM, Lynd LR. The ethanol pathway from Thermoanaerobacterium saccharolyticum improves ethanol production in Clostridium thermocellum. Metab Eng. 2017;42:175–84. [DOI] [PubMed] [Google Scholar]
- 25.Sharma BD, Olson DG, Giannone RJ, Hettich RL, Lynd LR. Characterization and amelioration of filtration difficulties encountered in metabolomic studies of Clostridium thermocellum at elevated sugar concentrations. Appl Environ Microbiol. 2023;89:e00406-e423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yayo J, Rydzak T, Kuil T, Karlsson A, Harding DJ, Guss AM, Van Maris AJA. The roles of nicotinamide adenine dinucleotide phosphate reoxidation and ammonium assimilation in the secretion of amino acids as byproducts of Clostridium thermocellum. Appl Environ Microbiol. 2023;89:e01753-e1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuil T, Hon S, Yayo J, Foster C, Ravagnan G, Maranas CD, Lynd LR, Olson DG, van Maris AJA. Functional analysis of H + -pumping membrane-bound pyrophosphatase, ADP-glucose synthase, and pyruvate phosphate dikinase as pyrophosphate sources in Clostridium thermocellum. Appl Environ Microbiol. 2022;88:e01857-e1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lanzetta PA, Alvarez LJ, Reinach PS, Candia OA. An improved assay for nanomole amounts of inorganic phosphate. Anal Biochem. 1979;100:95–7. [DOI] [PubMed] [Google Scholar]
- 29.Lo J, Zheng T, Hon S, Olson DG, Lynd LR. The bifunctional alcohol and aldehyde dehydrogenase gene, adhE, is necessary for ethanol production in Clostridium thermocellum and Thermoanaerobacterium saccharolyticum. J Bacteriol. 2015;197:1386–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bennett BD, Yuan J, Kimball EH, Rabinowitz JD. Absolute quantitation of intracellular metabolite concentrations by an isotope ratio-based approach. Nat Protoc. 2008;3:1299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Callaghan MM, Thusoo E, Sharma BD, Getahun F, Stevenson DM, Maranas C, Olson DG, Lynd LR, Amador-Noguez D. Deuterated water as a substrate-agnostic isotope tracer for investigating reversibility and thermodynamics of reactions in central carbon metabolism. Metab Eng. 2023;80:254–66. [DOI] [PubMed] [Google Scholar]
- 32.Agrawal S, Kumar S, Sehgal R, George S, Gupta R, Poddar S, Jha A, Pathak S. El-MAVEN: a fast, robust, and user-friendly mass spectrometry data processing engine for metabolomics. In: D’Alessandro A, editor. High-throughput metabolomics. New York, NY: Springer New York; 2019. p. 301–21. [DOI] [PubMed] [Google Scholar]
- 33.Holwerda EK, Thorne PG, Olson DG, Amador-Noguez D, Engle NL, Tschaplinski TJ, Van Dijken JP, Lynd LR. The exometabolome of Clostridium thermocellum reveals overflow metabolism at high cellulose loading. Biotechnol Biofuels. 2014;7:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clasquin MF, Melamud E, Rabinowitz JD. LC-MS data processing with MAVEN: a metabolomic analysis and visualization engine. Curr Protoc Bioinforma. 2012. 10.1002/0471250953.bi1411s37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deng Y, Olson DG, Zhou J, Herring CD, Joe Shaw A, Lynd LR. Redirecting carbon flux through exogenous pyruvate kinase to achieve high ethanol yields in Clostridium thermocellum. Metab Eng. 2013. 10.1016/j.ymben.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 36.Chiarelli DP, Sharma BD, Hon S, Bergamo LW, Lynd LR, Olson DG. Expression and characterization of monofunctional alcohol dehydrogenase enzymes in Clostridium thermocellum. Metab Eng Commun. 2024;19: e00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yayo J, Kuil T, Olson DG, Lynd LR, Holwerda EK, van Maris AJA. Laboratory evolution and reverse engineering of Clostridium thermocellum for growth on glucose and fructose. Appl Environ Microbiol. 2021;87:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bennett BD, Kimball EH, Gao M, Osterhout R, Van Dien SJ, Rabinowitz JD. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat Chem Biol. 2009;5:593–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lynd LR, Weimer PJ, van Zyl WH, Pretorius IS. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol Mol Biol Rev. 2002;66:506–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strobel HJ, Caldwell FC, Dawson KA. Carbohydrate transport by the anaerobic thermophile Clostridium thermocellum LQRI. Appl Environ Microbiol. 1995;61:4012–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Røst LM, Brekke Thorfinnsdottir L, Kumar K, Fuchino K, Eide Langørgen I, Bartosova Z, Kristiansen KA, Bruheim P. Absolute quantification of the central carbon metabolome in eight commonly applied prokaryotic and eukaryotic model systems. Metabolites. 2020;10:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taillefer M, Rydzak T, Levin DB, Oresnik IJ, Sparling R. Reassessment of the transhydrogenase/malate shunt pathway in Clostridium thermocellum ATCC 27405 through kinetic characterization of malic enzyme and malate dehydrogenase. Appl Environ Microbiol. 2015;81:2423–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Biswas R, Prabhu S, Lynd LR, Guss AM. Increase in ethanol yield via elimination of lactate production in an ethanol-tolerant mutant of Clostridium thermocellum. PLoS ONE. 2014;9: e86389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pech-Canul A, Hammer SK, Ziegler SJ, Richardson ID, Sharma BD, Maloney MI, Bomble YJ, Lynd LR, Olson DG. The role of AdhE on ethanol tolerance and production in Clostridium thermocellum. J Biol Chem. 2024;300:107559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tian L, Cervenka ND, Low AM, Olson DG, Lynd LR. A mutation in the AdhE alcohol dehydrogenase of Clostridium thermocellum increases tolerance to several primary alcohols, including isobutanol, n-butanol and ethanol. Sci Rep. 2019;9:1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khana DB, Callaghan MM, Amador-Noguez D. Novel computational and experimental approaches for investigating the thermodynamics of metabolic networks. Curr Opin Microbiol. 2022;66:21–31. [DOI] [PubMed] [Google Scholar]
- 47.Schroeder WL, Kuil T, Van Maris AJA, Olson DG, Lynd LR, Maranas CD. A detailed genome-scale metabolic model of Clostridium thermocellum investigates sources of pyrophosphate for driving glycolysis. Metab Eng. 2023;77:306–22. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 2. Quantification of different isotopes of Glucose 6-phosphate, Fructose 6-phosphate, Fructose 1,6-bisphosphate and Dihydroxyacetone phosphate.
Additional file 3. Fermentation data for the strains LL1590, LL1592 and LL1711
Additional file 4. Table of mutations identified by genome resequencing.
Additional file 5. Compiled supplementary information document with figures S1 through S7.
Additional file 6. Intracellular metabolite concentrations for the strains LL1590, LL1592 and LL1711.
Data Availability Statement
Enzyme assay data, extracellular metabolite data, intracellular metabolite data, 13C labeling data and genome resequencing data are all provided in the supporting information. Strain resequencing data is available from the NCBI Sequence Read Archive https://www.ncbi.nlm.nih.gov/sra/ (Table 1). Plasmid DNA sequences and annotations are available from GenBank (Table 2). Materials available by request.
Table 1.
Strains used in this work are described below
| Organism | Strain ID | Description | Accession Number | References |
|---|---|---|---|---|
| C. thermocellum | LL1592 | DSM 1313 ∆hpt, ∆ldh, ΔreIII, P2638::adhA(Tsc), nfnAB(Tsc), adhEG544D(Tsc), Clo1313_2637::Ptsc0046-pforA(Tsc)-ferredoxin(Tsc), ∆pfor1, ∆pfor2, ∆pfor3, ∆pfor4, ∆pfor5 | SRX5290154 | [18] |
| C. thermocellum | LL1590 | LL1592 ∆adhE | SRX5290158 | [19] |
| C. thermocellum | LL1649 | LL1592 tal(Tsc), ATP-pfk(Tsc), ∆ags, pPfk(Tsc) | SRX6875980 | [18] |
| C. thermocellum | LL1689 | LL1649 pyk(Tsc), PPase(Taq) | SRX7724531 | This study |
| C. thermocellum | LL1710 | LL1689 ∆ppdk, adhE E784* | SRX9409012 | This study |
| C. thermocellum | LL1711 | LL1710 ∆PPi-pfk | SRX9409011 | This study |
Tsc and Taq refers to genes introduced from T. saccharolyticum and T. aquaticus, respectively.