Abstract
Polycystic ovary syndrome (PCOS) is among the most prevalent endocrine and metabolic disorders affecting women of reproductive age. Multiple factors, including genetic predisposition, environmental influences, and lifestyle choices, are considered significant contributors to the development of PCOS. A kind of long noncoding RNA—C-Terminal binding protein 1 antisense (lncRNA CTBP1-AS) has been proven to be a new androgen receptor regulator. Previous studies showed that the lncRNA CTBP1-AS gene was highly expressed in a small sample of PCOS patients and was associated with the risk of PCOS, but its specific function and mechanism have not been clearly reported. In this study, the expression of lncRNA CTBP1-AS was detected by real-time quantitative PCR (RT-qPCR) in PCOS patients. In addition, lncRNA CTBP1-AS was overexpressed in KGN cells to explore its effect on granulocyte function. The results showed that the expression levels of lncRNA CTBP1-AS were increased in peripheral blood mononuclear cells and follicular fluid granulosa cells of PCOS patients compared with controls, which correlated with androgen levels and sinus follicle number; overexpression of lncRNA CTBP1-AS increased apoptosis and decreased cell migration ability, thus promoting the progression of PCOS. This study explores new biomarkers and therapeutic targets for the clinical individualized diagnosis and treatment of PCOS.
Keywords: Polycystic ovary syndrome, lncRNA, CTBP1-AS, Expression regulation, Granulosa cells
Introduction
Polycystic ovary syndrome (PCOS), a common heterogeneous endocrine and metabolic disorder, is one of the most common reproductive endocrine disorders in women of childbearing age, affecting up to 20% of women worldwide [1, 2]. Nevertheless, the etiology and pathogenesis of PCOS are complex and influenced by environmental, psychological, and genetic factors, but the exact etiology has not yet been identified [3]. At the same time, the clinical manifestations of PCOS are highly heterogeneous, and there is no specific treatment in the clinic. For women of childbearing age, the main treatment is ovulation promotion and assisted reproduction techniques, and for women who have no requirement for childbearing, the prevention and treatment of long-term complications and metabolic syndrome should also be carried out, and such metabolic and endocrine disorders may affect the whole life of a woman [4, 5]. Therefore, the diagnosis and treatment of PCOS still need to be further researched into its pathogenesis-related factors and progression mechanisms, to identify new diagnostic and therapeutic targets, provide early diagnosis of PCOS patients, and provide new and better individualized diagnostic and therapeutic methods.
Long noncoding RNA of C-Terminal binding protein 1 antisense (lncRNA CTBP1-AS) is a newly discovered long noncoding RNA [6]. It has been demonstrated that lncRNA CTBP1-AS, a potential androgen receptor regulator, functions in the androgen receptor (AR) signaling pathway [7, 8]. Notably, aberrant expression of AR can lead to enhanced activation of otherwise normal levels of androgens (testosterone and dihydrotestosterone) in the body, resulting in hyperandrogenemia [9]. The available evidence suggests that metabolic disturbances due to hyperandrogenemia are one of the important features of the pathological changes in PCOS [10]. Recently, lncRNA CTBP1-AS as a new androgen-regulated long noncoding RNA has gradually attracted the attention of many scholars [11]. Previous studies have found that lncRNA CTBP1-AS is associated with the pathogenesis of PCOS, but their samples are small and limited to the detection of a single type of cells in patients with PCOS. The influence of lncRNA CTBP1-AS on the occurrence and development of PCOS as well as a clear mechanism of action still requires further study [6, 12]. Therefore, the effect of lncRNA CTBP1-AS in the development of PCOS and its definitive mechanism remains to be further explored.
Our study aimed to further explore the correlation between lncRNA CTBP1-AS and the pathogenesis of PCOS by expanding the sample size and investigate the functional effects of lncRNA CTBP1-AS on ovarian GCs by cellular assays. This study confirmed the functional role of lncRNA CTBP1-AS in the occurrence and development of PCOS, and explored new targets for molecular diagnosis and gene therapy of PCOS.
Materials and methods
Study design
This study employed a case-control design and was conducted at the Reproductive Medicine Center of the Affiliated Hospital of Youjiang Medical University for Nationalities in Guangxi from January 2018 to March 2019.
Study population
Patients diagnosed with PCOS constituted the case group, while age-matched patients undergoing assisted reproductive technology for infertility due to tubal or male factors served as the control group.
Peripheral blood specimens
125 peripheral blood specimens were collected, comprising 85 cases from the PCOS group and 40 controls. The mean age of participants was 27.38 ± 4.35 years.
Inclusion criteria of peripheral blood specimens
Inclusion criteria were as follows: ①Case group: The diagnostic criteria for PCOS were based on the Rotterdam criteria recommended by ESHRE/ASRM in 2003: (1) oligo-ovulation and/or anovulation (OA); (2) increased serum androgen levels and/or hyperandrogenism (HA); (3) polycystic ovarian changes. The presence of two of the above three items at the same time needs to rule out other diseases, such as Cushing syndrome, congenital adrenal hyperplasia, and other diseases causing high androgen, as well as hyperprolactinemia and other diseases causing abnormal ovulation. ②Control group: patients with infertility due to tubal or male factors, regular menstruation (menstrual cycle of 28 ± 7 days), no clinical signs of hyperandrogenism, normal ovulation by ultrasound monitoring, and normal morphology of both ovaries during the same period.
Clinical indicators of peripheral blood specimens
Table 1 presents the clinical indicators for peripheral blood specimens from patients with PCOS and controls.
Table 1.
Clinical indicators of peripheral blood samples
| Clinical indicators | PCOS (N = 85) | Control(N = 40) | t / Z | p value |
|---|---|---|---|---|
| Age(years) | 26(24,29) | 28(24, 31) | -1.397 | 0.163 |
| Duration of infertility (years) | 2(1, 4) | 2(1, 4) | -0.51 | 0.61 |
| Menstrual cycles(days) | 68(60, 90) | 31(30, 40) | -7.907 | < 0.001* |
| Antral follicle count | 31(28, 40) | 18(15, 20) | -8.358 | < 0.001* |
| Basic sex hormone | ||||
| FSH (IU/L) | 5.78 ± 1.84 | 5.81 ± 1.78 | -0.095 | 0.925 |
| LH (IU/L) | 8.67(5.53, 12.60) | 4.16(3.64, 5.39) | -5.547 | < 0.001* |
| E2 (pg/mL) | 56.0(45.0, 68.5) | 47.0(38.0, 59.7) | -2.844 | 0.004* |
| P (ng/mL) | 0.46(0.30, 0.70) | 0.49(0.22, 0.60) | -0.506 | 0.613 |
| T (ug/L) | 0.76 ± 0.27 | 0.47 ± 0.19 | 6.253 | < 0.001* |
| PRL (ug/L) | 12.58 ± 5.74 | 10.80 ± 4.84 | 1.7 | 0.092 |
| AMH (ng/mL) | 7.55(5.00,10.62) | 4.48(2.37, 6.48) | -4.701 | < 0.001* |
| FPG (mmol/L) | 4.87(4.60, 5.23) | 4.75(4.52, 5.15) | -1.5 | 0.134 |
| FINS (pmol/L) | 75.0(58.1, 89.1) | 52.0(40.5, 68.5) | -4.01 | < 0.001* |
| HOMAIR | 2.44(1.77, 2.91) | 1.57(1.26, 2.08) | -4.319 | < 0.001* |
| BMI (kg/m2) | 22.7(20.1, 24.9) | 21.3(19.1, 24.9) | -1.527 | 0.127 |
Data are presented as the mean ± standard deviation, or median (interquartile range). FSH, follicle-stimulating hormone; LH, luteinizing hormone; E2, estradiol; P, progestogens; T, testosterone; PRL, prolactin; AMH, anti-mullerian hormone; FPG, Fasting Plasma Glucose; FINS, Fasting Insulin; HOMAIR, homeostatic model assessment of insulin resistance; BMI, body mass index; P-value from Mann-Whitney U test or independent t-test for continuous variables. *P < 0.05 indicates a statistically significant association between the variables
Follicular fluid specimens
A total of 30 follicular fluid specimens were collected, 13 in the case group and 17 in the control group; the mean age was 32.87 ± 4.15 years.
Inclusion criteria of follicular fluid specimens
The inclusion criteria were as follows: ① Case group: patients with PCOS who underwent in vitro fertilization and embryo transfer (IVF-ET) for the first time due to female factor (diagnostic criteria as before). ②Control group: patients who underwent IVF-ET for the first time due to tubal factor infertility, with regular menstruation, no clinical manifestations of hyperandrogenism, and no abnormality in the morphology of bilateral ovaries under ultrasound.
Clinical indicators of follicular fluid specimens
Table 2 presents the clinical indicators for follicular fluid specimens from patients with PCOS and controls. At the same time, Table 3 presents the clinical indicators of IVF-ET for follicular fluid specimens from patients with PCOS and controls.
Table 2.
Clinical indicators of follicular fluid samples
| Clinical indicators | PCOS (N = 13) | Control(N = 17) | t / Z | p value |
|---|---|---|---|---|
| Age(years) | 31.92 ± 4.59 | 33.59 ± 3.76 | 1.093 | 0.284 |
| Duration of infertility (years) | 3(2, 5) | 3(2,4) | -0.301 | 0.764 |
| Menstrual cycles(days) | 60(37, 90) | 30(30, 33) | -3.656 | < 0.001* |
| Antral follicle count | 24(19,30) | 14(11, 16) | -3.558 | < 0.001* |
| Basic sex hormone | ||||
| FSH (IU/L) | 5.70(5.1, 6.35) | 6.68(5.09, 8.64) | -1.612 | 0.107 |
| LH (IU/L) | 5.43(2.75, 11.48) | 3.60(2.71, 4.73) | -1.78 | 0.075 |
| E2 (pg/mL) | 43.31 ± 12.61 | 54.18 ± 22.97 | 1.535 | 0.136 |
| P (ng/mL) | 0.36 ± 0.19 | 0.45 ± 0.18 | 1.352 | 0.187 |
| T (ug/L) | 0.50 ± 0.17 | 0.43 ± 0.16 | -1.192 | 0.243 |
| PRL (ug/L) | 11.37(8.63,13.59) | 12.3(9.99,17.45) | -1.089 | 0.276 |
| AMH (ng/mL) | 7.12 ± 3.13 | 3.42 ± 1.77 | -3.829 | 0.001* |
| FPG (mmol/L) | 5.05(4.58, 5.36) | 4.80(4.60, 5.15) | -1.219 | 0.223 |
| FINS (pmol/L) | 58.92 ± 10.78 | 51.01 ± 15.15 | -1.596 | 0.122 |
| HOMAIR | 1.88 ± 0.32 | 1.59 ± 0.53 | -1.749 | 0.091 |
| BMI (kg/m2) | 21.44 ± 1.73 | 21.95 ± 2.36 | 0.66 | 0.515 |
P-value from Mann-Whitney U test or independent t-test for continuous variables. *P < 0.05 indicates a statistically significant association between the variables
Table 3.
Clinical indicators of IVF-ET of follicular fluid samples
| Clinical indicators | PCOS(N = 13) | Control(N = 17) | t / Z / χ2 | p value |
|---|---|---|---|---|
| Gn Days of use(d) | 9(9, 10) | 10(10,12) | 2.329 | 0.022* |
| Total Gn(IU) | 1550(1200, 1562) | 2125(1775,2575) | 3.440 | < 0.001* |
| HCG days | ||||
| Endometrial thickness(mm) | 11.0(11.0, 12.5) | 11.0(10.0, 12.5) | 0.021 | 0.983 |
| E2(pg/mL) | 4082.46 ± 1997.17 | 3379.39 ± 1194.54 | 1.201 | 0.240 |
| No. of oocytes retrieved(n) | 20.15 ± 4.93 | 12.18 ± 4.49 | 4.623 | < 0.001* |
| No. of MII oocytes(n) | 15.08 ± 5.01 | 9.35 ± 3.84 | 3.548 | 0.001* |
| Number of fertilized(n) | 11(7, 19) | 9(6, 11) | 1.643 | 0.100 |
| No. of cleavage embryos(n) | 10(7, 18) | 9(5, 11) | 1.368 | 0.171 |
| No. of good-quality embryos(n) | 7(5, 8) | 4(3, 5) | 2.147 | 0.035* |
| OHSS occurrence rate (%) | 46.2(6/13) | 11.8(2/17) | 4.455 | 0.045* |
| Transplant cancellation rate (%) | 46.2(6/13) | 29.4(5/17) | 0.889 | 0.287 |
| Pregnancy rate (%) | 42.9(3/7) | 41.7(5/12) | 0.003 | 0.663 |
P-value from Mann-Whitney U test or independent t-test for continuous variables, and Chi-square test or Fisher’s exact test for categorical variables. *P < 0.05 indicates a statistically significant association between the variables
Cell culture and cell transfection
Human ovarian granulosa cells KGN were purchased from the Cell Bank of the Chinese Academy of Sciences. Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (Ausgenex), penicillin (100 U/mL, Solarbio), and puromycin (100 µg/mL, Thermofisher).
293 A and 293T cells were purchased from the Cell Bank of the Chinese Academy of Sciences, and the lncRNA CTBP1-AS overexpression plasmid was purchased from Suzhou Ltd. Adenovirus after successful packaging were transfected into the cultured KGN cells at Polybrene 4 µg/ml, MOI = 5 conditions, and the fluorescence expression was observed 48–72 h after infection.
CCK-8 assay
CCK-8 assay was used to measure cell proliferation. In the presence of 1-Methoxy PMS, WST-8 in the reagent was reduced by dehydrogenases in the cells to a highly water-soluble yellow formazan product (Formazan dye). The number of produced formazan products was proportional to the number of living cells. The OD value of the optical density at 450 nm, measured using an enzyme marker, reflected the change in the number of living cells, thus detecting differences in proliferation produced by different cells over time.
Flow cytometry
The cells were digested and centrifuged at 200 g. The supernatant was discarded. The cells were resuspended with 50 µL 1× Binding Buffer, and the dyes were added according to the following groups: without any dye (negative control), 5 µL Annexin V-APC (single positive control), 10µL 7-AAD (single positive control), 5 µL Annexin V-FITC and 10 µL PI (sample tube). The mixture was incubated for 15 minutes in the dark. The apoptosis rate was analyzed by flow cytometer (BD Biosciences).
Cell scratching assay
Cells were seeded in transwell chambers, and the chambers were immersed in 2% serum medium outside the chambers. 24 h later, the chambers were removed, and the cells flanking the outside of the chambers were stained and photographed with a light microscope.
Primer information
For the RT-qPCR analysis of lncRNA CTBP1-AS expression, specific primers were designed as follows: lncRNA CTBP1-AS Forward Primer: 5’-ACAACACAAAGCCCCGGAA-3’, lncRNA CTBP1-AS Reverse Primer: 5’-AGTGAAGAATGGTCTCGCCC-3’, GAPDH Forward Primer (Housekeeping Gene): 5’-GGGAAACTGTGGCGTGAT-3’, GAPDH Reverse Primer: 5’- GAGTGGGTGTCGCTGTTGA-3’. These primers were synthesized by Sangon Biotech (Shanghai) Co., Ltd.
Reverse transcription quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted by using AxyPrep Total RNA Small Volume Preparation Kit, and the extracted RNA was detected by applying a nucleic acid quantifier, including the content and purity. The RNA with qualified purity and concentration was subjected to reverse transcription using the Reverse Transcription Reaction Kit (Fermentas Inc., USA). Then, lncRNA CTBP1-AS and the internal reference GAPDH were amplified by PCR, and a sample from the same normal group was added as a reference sample for each RT-qPCR. The reaction system for RT-qPCR was 20 µl, consisting of 3 µl F, 3 µl R, 3 µl DEPC, 1 µl cDNA and 10 µl SYBR. The reaction conditions were: 95 ℃ for 10 min pre-denaturation followed by 95 ℃ for 15 s and 60 ℃ for 1 min for 40 cycles. Three replicate measurements were taken for each sample to ensure accurate results. The Ct mean values of each sample lncRNA CTBP1-AS and GAPDH were first calculated separately, and then the data were used to calculate the relative quantity (RQ) of expression using the 2-ΔΔCT method.
Statistical analysis
SPSS 27.0 software was used for statistical processing. Measurement data conforming to normal distribution were expressed as x̅ ± s, and comparison of data between groups should be made by t-test or ANOVA according to the nature of the data; if they did not conform to normal distribution they were expressed as median (M) and quartiles (P25, P75), and comparison between groups was changed to rank sum test. Count data were expressed as rates (%), and comparisons between groups were made using the four-cell χ2 test or Fischer’s exact test. Correlations between variables were analyzed by Spearman’s rank correlation analysis. A value of P < 0.05 was considered statistically significant.
Results
General clinical information
In the clinical data of peripheral blood samples, age, duration of infertility, basal FSH, P, PRL, FPG, and BMI levels of two study groups were not statistically differences. The menstrual cycle, antral follicle count, basal LH, E2, T, AMH, FINS, and HOMAIR levels were higher in the PCOS group than in the control group(p < 0.05)(Table 1). In the clinical data of granulocyte samples, The menstrual cycle, antral follicle count, and AMH were higher in the PCOS group than in the control group. After pretreatment of the study subjects who entered the IVF-ET cycle, the differences in basal FSH, LH, E2, PRL, P, and T were not statistically significant when comparing the pre-superovulation sex hormone levels between the two groups. It suggested the two groups were more balanced in terms of sex hormones and other indicators at the time of entry into the treatment cycle, and the follow-up studies were comparable (Table 2). When comparing the clinical indicators of IVF-ET between the PCOS group and the control group, Gn days of use (d) and total Gn (IU) in the PCOS group were lower than those in the control group (P < 0.05), whereas the number of oocytes retrieved, the number of MII oocytes, the number of good-quality embryos, and OHSS occurrence rate in the PCOS group were higher (P < 0.05) (Table 3).
The expression of lncRNA CTBP1-AS in peripheral blood mononuclear cells and follicular fluid GCs
In our study, we found that patients with PCOS had a higher lncRNA CTBP1-AS level in peripheral blood mononuclear cells, compared with controls(0.86 ± 0.43 vs. 0.71 ± 0.29, P = 0.034) (Fig. 1A). To better study the clinical heterogeneity of PCOS, we further divided the PCOS group into several subgroups according to the Rotterdam criteria and the main clinical characteristics (hyperandrogenic and insulin resistance), respectively, and analyzed them statistically using the control group as a reference. They were categorized into 4 subtypes according to the Rotterdam criteria: complete OA + HA + PCO (n = 33), ovulatory HA + PCO (n = 4), non-hyperandrogenic OA + PCO (n = 39), and classic OA + HA (n = 9); Four subgroups were categorized according to the two main clinical features of hyperandrogenic and insulin resistance: the hyperandrogenic group HA (n = 36), the insulin-resistant group IR (n = 14), the hyperandrogenic + insulin-resistant group HA + IR (n = 10), and the no hyperandrogenic and insulin-resistant group NONE (n = 25).
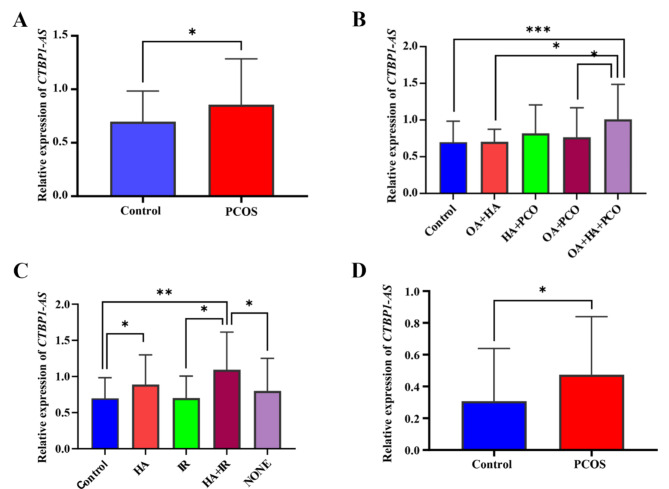
Fig. 1.
The expression level of lncRNA CTBP1-AS significantly increased in patients with PCOS. (A) Relative expression levels of lncRNA CTBP1-AS in peripheral blood mononuclear cells of the control and PCOS groups. (B) Relative expression of lncRNA CTBP1-AS in peripheral blood mononuclear cells across PCOS subtypes. (C) Relative expression of lncRNA CTBP1-AS in peripheral blood mononuclear cells based on subgroup clinical characteristics. (D) Relative expression of lncRNA CTBP1-AS in granulosa cells from the control and PCOS groups
Statistical analysis of the four subtypes of the Rotterdam criteria for PCOS (Fig. 1B) revealed a statistically significant difference between the elevated expression of the lncRNA CTBP1-AS in OA + HA + PCO only compared with control group (1.01 ± 0.47 vs. 0.71 ± 0.29, P = 0.001). In contrast, the expression of lncRNA CTBP1-AS in HA + PCO (0.81 ± 0.38) and OA + PCO (0.76 ± 0.40) was elevated compared with the control group, but the difference was not statistically significant; the expression of lncRNA CTBP1-AS in OA + HA (0.70 ± 0.16) was not significantly different from the control group. Further two-by-two comparisons revealed that lncRNA CTBP1-AS expression in OA + HA + PCO also differed from OA + HA, OA + PCO and not from HA + PCO.
Statistical analysis of the four subgroups with the main clinical features of PCOS (hyperandrogenemia and insulin resistance) versus the control group (Fig. 1C) revealed that the expression of lncRNA CTBP1-AS was elevated in HA, and in HA + IR, and the difference was statistically significant (0.88 ± 0.41 vs. 0.71 ± 0.29, P = 0.031; 1.09 ± 0.51 vs. 0.71 ± 0.29, P = 0.004); the mean expression level of lncRNA CTBP1-AS was also elevated in NONE (0.80 ± 0.44) compared to the control group, but none of the differences were statistically significant (P > 0.05); in IR (0.70 ± 0.31) the mean expression level of lncRNA CTBP1-AS was not significantly different from the control group. Further two-by-two comparisons revealed that the lncRNA CTBP1-AS expression in HA + IR also differed from IR, the NONE group (P < 0.05), and did not differ from the HA group. The mean expression level in GCs was higher in PCOS patients than in controls [0.39 (0.18, 0.66) vs. 0.17 (0.10, 0.34), P = 0.043] (Fig. 1D).
The results in this part illustrate that the expression level of lncRNA CTBP1-AS is up-regulated in PCOS, especially elevated most significantly in OA + HA + PCO subtype and HA + IR subgroup.
The correlation between lncRNA CTBP1-AS transcript levels and clinical parameters
Spearman’s rank correlation analysis was applied to analyze the correlation between lncRNA CTBP1-AS transcript levels and clinical parameters. The results showed that lncRNA CTBP1-AS in peripheral blood mononuclear cells was correlated with sinus follicle number and androgen levels (Table 4); lncRNA CTBP1-AS transcript levels in GCs were negatively correlated with the age of the subjects and positively correlated with menstrual cycle; lncRNA CTBP1-AS levels in GCs were negatively correlated with the total amount of Gn in the subjects (Table 5). The results of the correlation analysis between granulosa cells lncRNA CTBP1-AS transcript levels and IVF parameters showed a negative correlation between lncRNA CTBP1-AS levels and the total amount of Gn in the subjects (Table 6). Therefore, we hypothesized that lncRNA CTBP1-AS might be involved in the central part of PCOS pathogenesis through certain pathways.
Table 4.
Correlation between lncRNA CTBP1-AS expression levels in peripheral blood mononuclear cells and clinical parameters
| Clinical indicators | Expression of lncRNA CTBP1-AS(N = 125) | |
|---|---|---|
| r | p | |
| Age(years) | 0.173 | 0.053 |
| Duration of infertility (years) | 0.088 | 0.328 |
| Menstrual cycles(days) | 0.134 | 0.136 |
| Antral follicle count | 0.194 | 0.030* |
| Basic sex hormone | ||
| FSH (IU/L) | 0.054 | 0.549 |
| LH (IU/L) | 0.083 | 0.357 |
| E2 (pg/mL) | -0.056 | 0.537 |
| P (ng/mL) | -0.032 | 0.726 |
| T (ug/L) | 0.202 | 0.024* |
| PRL (ug/L) | 0.048 | 0.593 |
| AMH (ng/mL) | 0.149 | 0.098 |
| FPG (mmol/L) | -0.019 | 0.835 |
| FINS (pmol/L) | 0.087 | 0.332 |
| HOMAIR | 0.065 | 0.473 |
| BMI (kg/m2) | 0.056 | 0.536 |
Statistical analysis of the data was performed using Spearman’s test. *P < 0.05 indicates a statistically significant association between the variables
Table 5.
Correlation between lncRNA CTBP1-AS expression levels in GCs and clinical parameters
| Clinical indicators | Expression of lncRNA CTBP1-AS(N = 30) | |
|---|---|---|
| r | p | |
| Age(years) | -0.369 | 0.045* |
| Duration of infertility (years) | 0.262 | 0.161 |
| Menstrual cycles(days) | 0.455 | 0.012* |
| Antral follicle count | 0.312 | 0.093 |
| Basic sex hormone | ||
| FSH (IU/L) | -0.157 | 0.408 |
| LH (IU/L) | 0.005 | 0.98 |
| E2 (pg/mL) | -0.09 | 0.636 |
| P (ng/mL) | 0.098 | 0.608 |
| T (ug/L) | -0.056 | 0.771 |
| PRL (ug/L) | 0.101 | 0.595 |
| AMH (ng/mL) | 0.212 | 0.261 |
| FPG (mmol/L) | 0.305 | 0.102 |
| FINS (pmol/L) | 0.044 | 0.818 |
| HOMAIR | 0.122 | 0.521 |
| BMI (kg/m2) | -0.294 | 0.114 |
Statistical analysis of the data was performed using Spearman’s test. *P < 0.05 indicates a statistically significant association between the variables
Table 6.
Correlation between lncRNA CTBP1-AS expression levels in GCs and IVF-ET indicators
| Clinical indicators | Expression of lncRNA CTBP1-AS(N = 30) | |
|---|---|---|
| r | p | |
| Gn Days of use (d) | -0.233 | 0.215 |
| Total Gn (IU) | -0.399 | 0.029* |
| HCG day | ||
| Endometrium(mm) | -0.211 | 0.262 |
| E2 (pg/mL) | 0.334 | 0.071 |
| No. of oocytes retrieved | 0.026 | 0.891 |
| No. of MII oocytes | 0.018 | 0.927 |
| No.of fertilized oocytes | -0.065 | 0.734 |
| No. of cleavage embryos | -0.118 | 0.534 |
| No. of good-quality embryos | 0.15 | 0.429 |
Statistical analysis of the data was performed using Spearman’s test. *P < 0.05 indicates a statistically significant association between the variables
Adenovirus of lncRNA CTBP1-AS overexpression infects ovarian granulosa cells
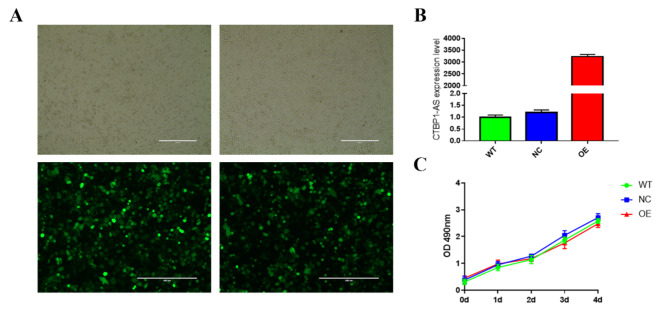
We hypothesized that lncRNA CTBP1-AS may be involved in the pathologic process of PCOS development by affecting the function of GCs and normal follicular development. To confirm the role of lncRNA CTBP1-AS in the pathogenesis of PCOS, lncRNA CTBP1-AS overexpression adenovirus was applied to infect KGN cells. According to the results of the pre-experiment, the optimal viral transfection conditions were obtained as Polybrene 4 µg/ml, MOI = 5, and the infection efficiency reached more than 90% as seen under the inverted fluorescence microscope after 48 h of transfection (Fig. 2A), which can be used for the subsequent cell function experiments. Total RNA was extracted from KGN cells after 48 h of adenovirus infection, and the overexpression and silencing efficiencies were detected by RT- qPCR using empty virus (Negative Control, NC) and wild type KGN cells without virus infection (Wild Type, WT) as controls, and the results showed that the expression of lncRNA CTBP1-AS was upregulated more than 3,000-fold in KGN cells overexpressing lncRNA CTBP1-AS (Over Expression, OE), whereas the difference in lncRNA CTBP1-AS expression between NC and WT was not statistically significant (Fig. 2B)
Fig. 2.
The effect of overexpression of lncRNA CTBP1-AS on the proliferative function of KGN cells. (A) KGN cells were infected by adenovirus for 48 h. (B) The expression level of lncRNA CTBP1-AS in KGN cells were detected by RT-qPCR. (C) Cell viability was detected by CCK-8 assay in the Negative Control (NC), Wild Type (WT) without virus, and Overexpression of lncRNA CTBP1-AS (Over Expression (OE)) groups
lncRNA CTBP1-AS overexpression affects follicular development by disturbing with the function of GCs
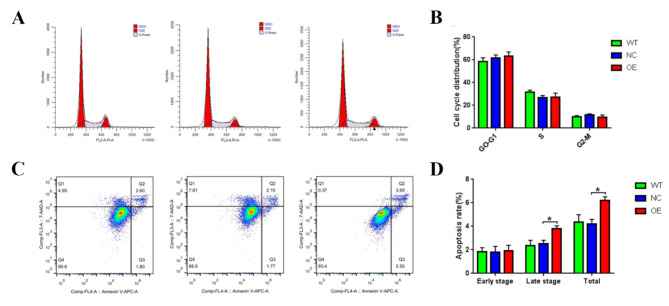
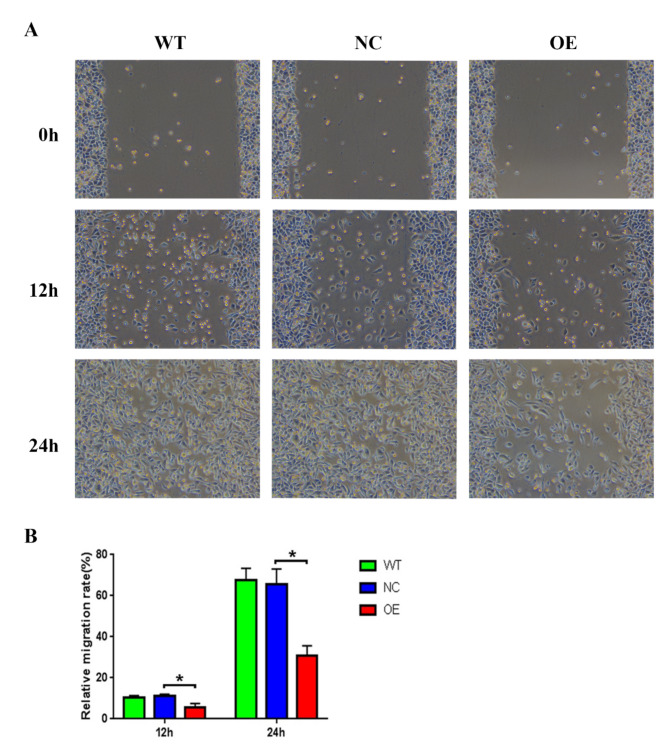
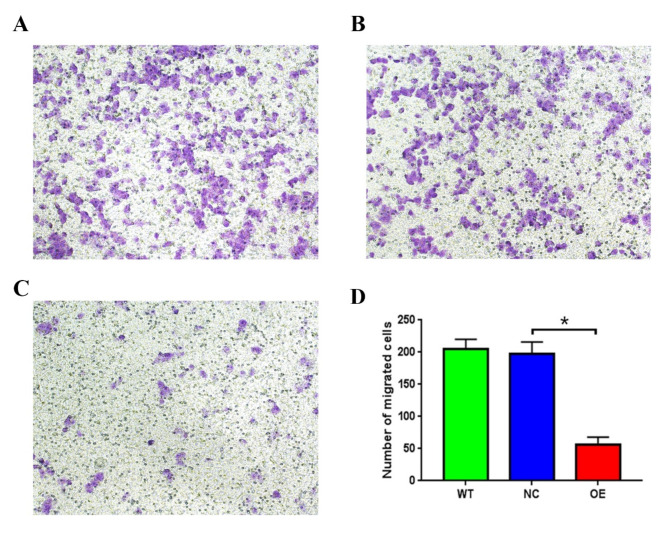
To investigate the effect of lncRNA CTBP1-AS on granulosa cell function, we overexpressed lncRNA CTBP1-AS in KGN cells and detected changes in cell growth, apoptosis, cell cycle, cell migration, and other functions. The proliferation of KGN cells was detected by CCK-8, and it was found that compared with WT and NC, the cell proliferation ability of KGN cells overexpressed lncRNA CTBP1-AS was reduced, but the difference was not yet statistically significant (Fig. 2C). The flow cytometry results indicated that there was no statistically significant difference in the proportion of cells in the G0-G1, S, and G2-M phases of KGN cells overexpressed lncRNA CTBP1-AS (Table 7; Fig. 3A-B), suggesting that overexpression of lncRNA CTBP1-AS did not affect the cell cycle of KGN cells. In addition, flow cytometry was used to examine the effect of lncRNA CTBP1-AS overexpression on KGN cell apoptosis, and the results showed that the proportion of late apoptotic cells was increased in KGN cells overexpressed lncRNA CTBP1-AS as compared to WT and NC (Fig. 3C-D), which suggests that overexpression of lncRNA CTBP1-AS promotes apoptosis in KGN cells. Cell scratch wound assay and Transwell assay were used to investigate the effect of lncRNA CTBP1-AS on the cell migration profile. The results of cell scratch wound assay revealed that the migration ability of KGN cells was significantly decreased after overexpression of lncRNA CTBP1-AS (Fig. 4A-B); Transwell assay illustrated that overexpression of lncRNA CTBP1- AS was detrimental to cell migration (Fig. 5A-D). Both experimental results suggested that overexpression of lncRNA CTBP1-AS was unfavorable to cell migration. It can be seen that overexpression of lncRNA CTBP1-AS did cause corresponding functional changes in GCs, and the results of this part confirmed that lncRNA CTBP1-AS was involved in the pathological process of PCOS development by affecting the function of ovarian granulosa cells.
Table 7.
The results of cell cycle
| Cell Cycle(%) | WT | NC | OE |
|---|---|---|---|
| G0-G1 | 58.47 ± 3.25 | 61.65 ± 2.58 | 63.14 ± 3.84 |
| S | 31.64 ± 1.65 | 26.81 ± 1.84 | 27.25 ± 3.54 |
| G2-M | 9.89 ± 1.10 | 11.54 ± 0.98 | 9.62 ± 1.93 |
Data are presented as the mean ± standard deviation
Fig. 3.
The effect of overexpression of lncRNA CTBP1-AS on cell cycle and apoptosis in KGN cells. (A) Cell cycle of KGN cells in NC, WT and OE groups was detected by flow cytometry; (B) The results were visualised using bar graphs. (C) The apoptosis of KGN cells in NC, WT and OE groups was detected by flow cytometry; (D) The results were visualised using bar graphs
Fig. 4.
The effect of overexpression of lncRNA CTBP1-AS on cell migration in KGN cells. (A) The cell migration of KGN cells in NC, WT and OE groups was detected by cell scratch wound assay. (B) The results were visualised using bar graphs
Fig. 5.
The effect of overexpression of lncRNA CTBP1-AS on cell migration in KGN cells. Migratory capacity of WT (A), NC (B), OE (C) group was detected with Transwell cell migration; (D) Number of technically migrated cells in 100x field of view, the difference between groups was analysed by ANOVA, *P < 0.05
Discussion
In this randomized clinical study, we analyzed the expression levels of lncRNA CTBP1-AS in peripheral blood mononuclear cells and ovarian granulosa cells of PCOS patients and controls who met the inclusion criteria, and found that the expression levels of lncRNA CTBP1-AS in peripheral blood mononuclear cells and ovarian granulosa cells were significantly higher in the PCOS group compared with that in the normal control group. This suggests that the high expression of lncRNA CTBP1-AS may be related to the pathogenesis of PCOS, and is a potential diagnostic marker and drug treatment target. Studies have been conducted to support this view, Liu [13] first reported that lncRNACTBP1-AS expressed at elevated levels in peripheral blood mononuclear cells of PCOS patients and that abnormal expression of lncRNA CTBP1-AS was a risk factor for PCOS. In addition, a case-control study involving Kashmiri women also found that subjects with higher levels of lncRNA CTBP1-AS expression had a significantly higher risk of PCOS (OR = 11.36, 95% CI = 5.59–23.08, P < 0.001), further confirming the association between lncRNA CTBP1-AS and PCOS [6].
Hyperandrogenemia is one of the significant features of PCOS, which is often accompanied by elevated levels of androgens, including testosterone and androstenedione [14, 15]. At the same time, lncRNA CTBP1-AS was initially identified by researchers as a novel androgen receptor modulator [16, 17], suggesting that lncRNA CTBP1-AS may play an important role in the pathogenesis of PCOS by regulating androgen levels. lncRNA CTBP1-AS expression was commonly upregulated in prostate cancer, and lncRNA CTBP1-AS promoted hormone-dependent and desmoplasia-resistant tumor growth. Mechanistically, lncRNA CTBP1-AS directly inhibited lncRNA CTBP1 expression by recruiting RNA-binding transcriptional repressors (PSFs) and histone deacetylases; lncRNA CTBP1-AS also suppressed tumor suppressor genes and promoted cell cycle progression through PSF-dependent mechanisms, thus exhibiting overall androgen-dependent functions [11, 18]. In our study, we found a close correlation between the expression level of lncRNA CTBP1-AS in peripheral blood mononuclear cells of PCOS patients and androgens, which indicates that lncRNA CTBP1-AS may be involved in the central part of the pathogenesis of PCOS through certain pathways.
Jin [19] applied high-density microarray technology to study differentially expressed lncRNAs in ovarian granulosa cells from PCOS patients (n = 4), and the screening revealed that lncRNA CTBP1-AS expression was up-regulated in the PCOS-T group, by 1.3-fold and 1.5-fold, as respectively compared with the control and PCOS-N groups. A total of 30 follicular fluid specimens were collected in our study, and the average expression level of granulosa cells was higher in PCOS patients than in the control group, and the granulosa cell lncRNA CTBP1-AS level was positively correlated with the menstrual cycle. The length of the menstrual cycle mainly depends on the time frame of the follicular phase. Ovarian granulosa cells (GCs) exist in a proliferative manner in developing follicles and undergo multiple biochemical processes during folliculogenesis. In developing follicles, GCs communicate bi-directionally with oocytes through gap junctions [20]. Numerous studies have reported that the survival and physiological status of granulosa cells can have an impact on follicular development [21, 22]. High levels of lncRNA CTBP1-AS may have affected the function of ovarian granulosa cells, leading to abnormal follicular development and prolonged follicular phase, which in turn caused menstrual abnormalities in PCOS.
The primary feature of PCOS is anovulatory infertility, the critical feature of which is impaired follicular development or ovulation, which is manifested by the presence of many growing follicles at all stages of follicular development but ultimately the failure to form a mature dominant follicle [23, 24]. Numerous studies have revealed that the survival and physiological status of granulosa cells can affect follicular development and oocyte quality, and that rapid apoptosis of granulosa cells may lead to follicular dysfunction and follicular atresia, as well as a significant reduction in the maturation rate of the oocyte and oocyte developmental potential. On the other hand, enhancing granulosa cell activity and intracellular expression of related genes may improve oocyte quality to a certain extent and promote embryonic development potential [25–27]. Overexpression of the lncRNA CTBP1-AS may also affect ovarian granulosa cell function involved in the development of the pathological process of ovulatory disorders associated with PCOS. A study found that CTBP1-AS interacts with Polycomb histone (enhancer of zeste homolog 2 and embryonic (eZH2) and ectoderm development protein (eed)) in ovarian granulosa cells to regulate PCOS, and cryptotanshinone can reduce the expression level of CTBP1-AS in granulosa cells, which may be a new direction for PCOS treatment [12]. lncRNA CTBP1-AS overexpression may be involved in the pathogenesis of PCOS by affecting the function of ovarian granulosa cells. In our study, we found that the proportion of apoptotic cells in granulosa cells was increased after overexpression of lncRNA CTBP1-AS, especially the proportion of apoptotic cells in the late stage was significantly increased, which is consistent with the findings of increased apoptosis of granulosa cells in PCOS patients [28].
Furthermore, we found that the migratory ability of granulosa cells was significantly decreased after overexpression of the lncRNA CTBP1-AS, and hypothesized that it might be involved in the mechanism related to ovulation disorder in PCOS. Normally, ovulation is a process in which the oocyte is expelled together with its surrounding granulosa cells that form the oval corpuscle complex. Before ovulation, ovarian mound expansion occurs after mucoidization of the granulosa cells of the ovarian mound. If the expansion of the mound is impaired, it will lead to abnormal ovulation [29]. Consequently, the reduced migratory capacity of granulosa cells may cause abnormal mound expansion and abnormal follicular migration to the ovarian surface, which in turn leads to impaired ovulation in PCOS and increases the incidence of its unruptured follicular luteinization syndrome. In the microenvironment of the ovary, there are other complex mechanisms for the regulation of ovulation, such as the regulatory role between steroid hormones, prostaglandins, proteolytic enzymes, and other cytokines. We still need to conduct more exploratory studies to elucidate and validate this hypothesis. In conclusion, granulosa cells overexpressing lncRNA CTBP1-AS exhibited an increased proportion of apoptotic cells and a significant decrease in migration ability, and the corresponding functional alterations in ovarian granulosa cells provide a positive experimental basis for elucidating the mechanism of action of PCOS development.
Conclusion
lncRNA CTBP1-AS may affect the function of human ovarian granulosa cells, resulting in an increase in apoptosis and a decrease in migration capacity, which causes impaired follicular development, ovulation disorders, and hyperandrogenemia in PCOS. However, the exact molecular genetic mechanism still needs to be further investigated.
Acknowledgements
Not applicable.
Author contributions
L Q, and C T contributed to the study design and data acquisition, drafted the manuscript, and were the co-first authors. L H, X Q, S L, B H, J W, and L L contributed to contributed experimental data. X L is considered a correspondence author. All authors have read and approved the final manuscript.
Funding
This study was financially supported by the 2019 Natural Science Foundation of Guangxi, China (No. 2019GXNSFBA245020), the High-Level Talent Scientific Research Project of the Affiliated Hospital of Youjiang Medical University for Nationalities, China (No. Y20196316), and Self-financed scientific research projects of Guangxi Autonomous Region Health and Wellness Commission (NO.Z-L20230901).
Data availability
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
The present study was approved by the ethics committee of The Affiliated Hospital of Youjiang Medical University for Nationalities (Guangxi, China; approval no. YYFY-LL-2022-61); all subjects provided signed consent forms prior to recruitment to the study.
Patient consent for publication
All the patients in this project signed the consent forms.
Conflict of interest
The authors declare that the research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Li Qin and Chun Tian share first authorship.
References
- 1.Herman R, Sikonja J, Jensterle M, et al. Insulin metabolism in polycystic ovary syndrome: secretion, signaling, and clearance[J]. Int J Mol Sci. 2023;24(4):3140. 10.3390/ijms24043140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sadeghi HM, Adeli I, Calina D, et al. Polycystic ovary syndrome: a comprehensive review of pathogenesis, management, and drug repurposing[J]. Int J Mol Sci. 2022;23(2):583. 10.3390/ijms23020583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Szukiewicz D, Trojanowski S, Kociszewska A, et al. Modulation of the inflammatory response in polycystic ovary syndrome (pcos)—searching for epigenetic factors[J]. Int J Mol Sci. 2022;23(23):14663. 10.3390/ijms232314663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Helvaci N, Yildiz BO. Current and emerging drug treatment strategies for polycystic ovary syndrome[J]. Expert Opin Pharmacother. 2023;24(1):105–20. 10.1080/14656566.2022.2108702. [DOI] [PubMed] [Google Scholar]
- 5.Millán-De-Meer M, Luque-Ramírez M, Nattero-Chávez L, et al. PCOS during the menopausal transition and after menopause: a systematic review and meta-analysis[J]. Hum Reprod Update. 2023;29(6):741–72. 10.1093/humupd/dmad015. [DOI] [PubMed] [Google Scholar]
- 6.Nabi M, Andrabi SM, Rasool SUA, et al. Androgen receptor coregulator long noncoding rna ctbp1-as is associated with polycystic ovary syndrome in kashmiri women[J]. Endocrine. 2022;75(2):614–22. 10.1007/s12020-021-02894-9. [DOI] [PubMed] [Google Scholar]
- 7.Takayama K, Tsutsumi S, Katayama S, et al. Integration of cap analysis of gene expression and chromatin immunoprecipitation analysis on array reveals genome-wide androgen receptor signaling in prostate cancer cells[J]. Oncogene. 2011;30(5):619–30. 10.1038/onc.2010.436. [DOI] [PubMed] [Google Scholar]
- 8.Sung YY, Cheung E. Antisense now makes sense: dual modulation of androgen-dependent transcription by ctbp1-as[J]. EMBO J. 2013;32(12):1653–4. 10.1038/emboj.2013.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walters KA. Polycystic ovary syndrome: is it androgen or estrogen receptor?[J]. Curr Opin Endocr Metabolic Res. 2020;12:1–7. 10.1016/j.coemr.2020.01.003. [Google Scholar]
- 10.Liao B, Qi X, Yun C, et al. Effects of androgen excess-related metabolic disturbances on granulosa cell function and follicular development[J]. Front Endocrinol. 2022;13:815968. 10.3389/fendo.2022.815968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu G, Dong Z, Dong Y, et al. LncRNA ctbp1-as inhibits tp63‐mediated activation of s100a14 during prostate cancer progression[J]. Cancer Sci. 2024;115(5):1492–504. 10.1111/cas.16138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wen M, Dou X, Zhang S, et al. CTBP1–AS upregulation is associated with polycystic ovary syndrome and can be effectively downregulated by cryptotanshinone[J]. Mol Med Rep. 2022;26(1):245. 10.3892/mmr.2022.12761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Z, Hao C, Song D et al. Androgen receptor coregulator ctbp1-as is associated with polycystic ovary syndrome in Chinese women: a preliminary study[J]. Reproductive Sciences (Thousand Oaks, Calif.), 2015, 22(7): 829–37. 10.1177/1933719114565037 [DOI] [PMC free article] [PubMed]
- 14.Grassi G, Polledri E, Fustinoni S, et al. Hyperandrogenism by liquid chromatography tandem mass spectrometry in pcos: focus on testosterone and androstenedione[J]. J Clin Med. 2020;10(1):119. 10.3390/jcm10010119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carmina E, Longo RA. Increased prevalence of elevated dheas in pcos women with non-classic (b or c) phenotypes: a retrospective analysis in patients aged 20 to 29 years[J]. Cells. 2022;11(20):3255. 10.3390/cells11203255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang D, Li M, Li J et al. Comprehensive characterization of androgen-responsive lncrnas mediated regulatory network in hormone-related cancers[J]. Disease Markers, 2020, 2020: 1–18. 10.1155/2020/8884450 [DOI] [PMC free article] [PubMed]
- 17.Zhang Y, Pitchiaya S, Cieślik M, et al. Analysis of the androgen receptor–regulated lncrna landscape identifies a role for arlnc1 in prostate cancer progression[J]. Nat Genet. 2018;50(6):814–24. 10.1038/s41588-018-0120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takayama K, Horie-Inoue K, Katayama S, et al. Androgen-responsive long noncoding rna ctbp1-as promotes prostate cancer[J]. EMBO J. 2013;32(12):1665–80. 10.1038/emboj.2013.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin L, Yang Q, Zhou C, et al. Profiles for long non-coding rnas in ovarian granulosa cells from women with pcos with or without hyperandrogenism[J]. Reprod Biomed Online. 2018;37(5):613–23. 10.1016/j.rbmo.2018.08.005. [DOI] [PubMed] [Google Scholar]
- 20.Jozkowiak M, Hutchings G, Jankowski M, et al. The stemness of human ovarian granulosa cells and the role of resveratrol in the differentiation of mscs-a review based on cellular and molecular knowledge[J]. Cells. 2020;9(6):1418. 10.3390/cells9061418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brązert M, Iżycki D, Kranc W, et al. Genes involved in hormone metabolism and cellular response in human ovarian granulosa cells[J]. J Biol Regul Homeost Agents. 2019;33(2):461–8. [PubMed] [Google Scholar]
- 22.Zhu G, Fang C, Li J, et al. Transcriptomic diversification of granulosa cells during follicular development in chicken[J]. Sci Rep. 2019;9(1):5462. 10.1038/s41598-019-41132-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao Y, Zou Y, Wu G, et al. Oxidative stress and mitochondrial dysfunction of granulosa cells in polycystic ovarian syndrome[J]. Front Med. 2023;10:1193749. 10.3389/fmed.2023.1193749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumariya S, Ubba V, Jha RK, et al. Autophagy in ovary and polycystic ovary syndrome: role, dispute and future perspective[J]. Autophagy. 2021;17(10):2706–33. 10.1080/15548627.2021.1938914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen Q, Liu Y, Li H, et al. Effect of mitophagy in oocytes and granulosa cells on oocyte quality†[J]. Biol Reprod. 2021;104(2):294–304. 10.1093/biolre/ioaa194. [DOI] [PubMed] [Google Scholar]
- 26.Xing J, Qiao G, Luo X et al. Ferredoxin 1 regulates granulosa cell apoptosis and autophagy in polycystic ovary syndrome[J]. Clinical Science (London, England: 1979), 2023, 137(6): 453–468. 10.1042/CS20220408 [DOI] [PubMed]
- 27.Liu S, Jia Y, Meng S, et al. Mechanisms of and potential medications for oxidative stress in ovarian granulosa cells: a review[J]. Int J Mol Sci. 2023;24(11):9205. 10.3390/ijms24119205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gong Y, Luo S, Fan P, et al. Growth hormone activates pi3k/akt signaling and inhibits ros accumulation and apoptosis in granulosa cells of patients with polycystic ovary syndrome[J]. Volume 18. Reproductive biology and endocrinology: RB&E; 2020. p. 121. 110.1186/s12958-020-00677-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y, Wang H, Zhou D, et al. Up-regulation of long noncoding rna sra promotes cell growth, inhibits cell apoptosis, and induces secretion of estradiol and progesterone in ovarian granular cells of mice[J]. Med Sci Monitor: Int Med J Experimental Clin Res. 2018;24:2384–90. 10.12659/msm.907138. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No datasets were generated or analysed during the current study.